Abstract
The interplay between signaling molecules and transcription factors during retinal development is key to controlling the correct number of retinal cell types. Zeb2 (Sip1) is a zinc-finger multidomain transcription factor that plays multiple roles in central and peripheral nervous system development. Haploinsufficiency of ZEB2 causes Mowat-Wilson syndrome, a congenital disease characterized by intellectual disability, epilepsy and Hirschsprung disease. In the developing retina, Zeb2 is required for generation of horizontal cells and the correct number of interneurons; however, its potential function in controlling gliogenic versus neurogenic decisions remains unresolved. Here we present cellular and molecular evidence of the inhibition of Muller glia cell fate by Zeb2 in late stages of retinogenesis. Unbiased transcriptomic profiling of control and Zeb2-deficient early-postnatal retina revealed that Zeb2 functions in inhibiting Id1/2/4 and Hes1 gene expression. These neural progenitor factors normally inhibit neural differentiation and promote Muller glia cell fate. Chromatin immunoprecipitation (ChIP) supported direct regulation of Id1 by Zeb2 in the postnatal retina. Reporter assays and ChIP analyses in differentiating neural progenitors provided further evidence that Zeb2 inhibits Id1 through inhibition of Smad-mediated activation of Id1 transcription. Together, the results suggest that Zeb2 promotes the timely differentiation of retinal interneurons at least in part by repressing BMP–Smad/Notch target genes that inhibit neurogenesis. These findings show that Zeb2 integrates extrinsic cues to regulate the balance between neuronal and glial cell types in the developing murine retina.
Keywords: Zeb2, retinogenesis, BMP, retinal progenitor cell, Muller glia
Introduction
The vertebrate neural retina is a complex, highly specialized neural network comprised of seven major cell types organized in three nuclear layers. Rod and cone photoreceptors are found in the outer nuclear layer (ONL), and three types of retinal interneurons—horizontal, amacrine and bipolar (BPL) cells—are located in the inner nuclear layer (INL), together with Muller glia (MG), which are the major glial cell type of the retina. In addition, retinal ganglion cells, which extend axons to the brain, are located in the ganglion cell layer (GCL) together with some “displaced” amacrine cells. These cell types are generated from a population of multipotent retinal progenitor cells (RPCs) located in the inner part of the optic cup. Their differentiation follows an evolutionarily conserved, partially overlapping temporal order, in which retinal ganglion cells are the first to exit the cell cycle and initiate differentiation, followed by cone photoreceptors, horizontal cells, amacrine cells, rod photoreceptors and finally, BPL and MG cells (Cepko, 2014; Morrow et al., 2008; Young, 1985).
The generation of diverse retinal cell types, in appropriate numbers and at the right times, is regulated by many different transcription factors (TFs) which act at different developmental stages. Some TFs promote specification and differentiation to specific cell lineages. Ptf1a, for example, is essential for the generation of the early-born amacrine and horizontal cells (Fujitani et al., 2006; Nakhai et al., 2007). Bhlhe22 (Bhlhb5), in turn, is required to generate the correct number of subtypes of amacrine and BPL interneurons (Huang et al., 2014). Extrinsic signals also affect retinal differentiation. For instance, active Notch signaling inhibits cell-cycle exit and photoreceptor specification, whereas it promotes MG differentiation (Jadhav et al., 2006a; Jadhav et al., 2006b; Mizeracka et al., 2013; Yaron et al., 2006). BMP signaling also inhibits photoreceptor differentiation, but promotes the differentiation of MG and BPL cells (Kuribayashi et al., 2014; Ueki et al., 2015).
Because of the temporally dynamic nature of retinal histogenesis, changes in the timing and dynamics of differentiation processes may result in changes in cell-type composition of the retina. For example, early genetic inactivation of the TF Lhx2 or its cofactors Ldb1 and Ldb2 results in premature cell-cycle exit, which leads to overproduction of retinal ganglion cells, as they are the earliest retinal cell type to be generated (Gordon et al., 2013; Gueta et al., 2016). Inactivation of Lhx2 at later stages of retinal differentiation inhibits gliogenesis (de Melo et al. 2016a; de Melo et al., 2016b; de Melo et al. 2018). Similarly, Notch signaling promotes MG generation by inhibiting the differentiation of other cell types and in doing so, increases the number of retinal progenitors, which in turn differentiate to MG cells in late stages of retinal development (Jadhav et al., 2009).
We have previously demonstrated that the TF Zeb2 (Sip1, Zfhx1b) regulates interneuron specification in the murine neural retina (Menuchin-Lasowski et al., 2016). The mouse Zeb2 gene contains 9 translated exons and 8 introns (Nelles et al., 2003), and encodes a 1,215-amino acid protein, which binds DNA via specific zinc fingers in two separate zinc finger-rich domains (Remacle et al., 1999; Verschueren et al., 1999b). Zeb2 binds to BMP receptor-activated Smad proteins (Conidi et al., 2013; Verschueren et al., 1999), and also interacts with the chromatin-remodeling complex NuRD (Verstappen et al., 2008) and the corepressors Ctbp1 and Ctbp2 (van Grunsven et al., 2003; van Grunsven et al., 2007).
Zeb2 plays many different roles in the development of the central nervous system. During the early stages of mouse embryonic development, Zeb2 is essential for neural induction and neural tube closure (Van de Putte et al., 2003), whereas at later stages, it directs differentiation and tangential migration of cortical interneurons from the ganglionic eminences (McKinsey et al., 2013; van den Berghe et al., 2013), regulates the timing of cortical neurogenesis and gliogenesis within the cortex (Seuntjens et al., 2009), and by generating anti-BMP (and anti-Wnt) activity, promotes oligodendrocyte differentiation in the central nervous system (Weng et al., 2012).
In humans, mutation in one allele of ZEB2 is the only known cause of Mowat-Wilson Syndrome (MOWS, OMIM #235730), which is characterized by intellectual disability and multiple congenital defects (Cacheux et al., 2001; Mowat et al., 1998; Wakamatsu et al., 2001). Ocular abnormalities, including microphthalmia, coloboma, and retinal aplasia, are also found in some MOWS patients (Ariss et al., 2012; Garavelli et al., 2009; Gregory-Evans et al., 2004; Ivanovski et al., 2018; Zweier et al., 2005). A genome-wide association study also identified ZEB2 as a susceptibility locus for severe myopia (Khor et al., 2013), emphasizing its important role in ocular tissues.
Conditionally removing Zeb2 from peripheral RPCs using the Cre/loxP system resulted in complete loss of horizontal interneurons and a reduction in the number of amacrine and BPL interneurons, while the number of MGs increased. A thorough analysis of the development of such Zeb2-deficient retinas indicated that Zeb2 has an important role in controlling differentiation of the inner retinal neurons, and a more recent has study shown its role in inhibiting the differentiation/specification of photoreceptor cells (Menuchin-Lasowski et al., 2016; Wei et al., 2018). The functions of Zeb2 during early retinogenesis were attributed to regulation of the downstream TF Ptf1a, an important factor for the differentiation of amacrine and horizontal cells (Menuchin-Lasowski et al., 2016). In the current study, we employed unbiased transcriptomic analyses, chromatin immunoprecipitation (ChIP) and reporter assays to further delve into the roles of Zeb2 during late stages of retinogenesis, and to uncover its role in regulating the balance between retinal neurons and MG cells.
Materials and Methods
Animals
Mice were kept at the Tel Aviv University Animal House Facility. Animal use was approved by the Tel Aviv University Animal Care and Use Committee (M-08–092). Zeb2loxp/loxp;αCre and Zeb2loxp/loxp were generated from crossing between Zeb2 loxp and αCre (Higashi et al., 2002; Marquardt et al., 2001) maintained on C57BL6/J genetic background. The primers used for genotyping are listed in Supplemental Table S1.
Immunofluorescence and quantification
Paraffin retinal sections (10 μm) were used for conventional indirect immunofluorescence analyses. Briefly, the sections were treated with Unmasking Solution (Vector Laboratories, H-3300) for antigen retrieval. To decrease nonspecific binding, sections were incubated for 2 h in blocker PBSTG (PBS, 0.2% gelatin, 0.2% Tween 20). Sections were then washed in PBS, incubated overnight with a primary antibody (Supplemental Table S2) at 4°C, washed with PBSTG, incubated with a secondary fluorescent antibody for 2 h, washed three times in PBS and then with PBSTG, and sealed with fluorescent mounting medium containing DAPI (GBI Labs, E19–18). Slides were viewed with an Olympus BX61 fluorescence microscope or Nikon C2+ laser-scanning confocal microscope. The number of marker-positive cells was counted using Imaris image analysis software (Bitplane AG) or ImageJ. For cell counting, at least three eyes were analyzed from each genotype in three different peripheral sections of each eye (in a fixed area or length as indicated in the figure legends). The number of cells in the two or three sections of each eye was averaged, and this average was considered to be one biological replicate.
In-situ hybridization
Retinas were dissected in cold PBS, washed, fixed in 4% paraformaldehyde at 4°C overnight, transferred for 1 h to 15% sucrose in PBS and later to 30% sucrose in PBS overnight at 4°C. The tissue was then immersed in Tissue-plus O.C.T. compound (Fisher Healthcare, 23–730-571) for 1 h at room temperature (RT) and snap-frozen on dry ice. Sections (16-μm thick) were placed on slides and air-dried for 40 min; hybridization was then conducted overnight at 65°C with digoxigenin-labeled probes (1 μg/ml). The slides were treated with RNaseA, washed, blocked with 20% normal goat serum and incubated with sheep anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (1:250, Roche, 10713023001) in blocking solution overnight at 4°C. Non-specific antibody was removed and slides were incubated in BM Purple (Roche, 11442074001). The probes used in this study were Zeb2 (Maruhashi et al., 2005) and Hes1 (Gueta et al., 2016).
For hybridization chain reaction (HCR), fluorescence in-situ hybridization probe sets of 20 V3.0 split-inhibitor probe pairs that target Id1 transcripts were from Molecular Technologies (moleculartechnologies.org), and the in-situ hybridization protocol was performed as described by Choi et al. (Choi et al., 2018). The signal intensity was quantified using ImageJ. The mean signal of a retinal area (50 × 50 μm2) within the neuroblastic and differentiating INL, where the signal was stronger, was measured and compared to background measurements of smaller retinal areas in the outer nuclear layer and ganglion cell layer, where almost no signal was detected. For each biological replicate, three different peripheral sections were analyzed.
RNA extraction
Two whole retinas from the same mouse were taken for each biological replicate. The samples were homogenized by QIAshredder kit (Qiagen, 79654), and the RNA was extracted using the RNeasy mini kit (Qiagen, 74104). DNase was used to remove genomic DNA from the samples.
Quantitative real-time PCR (qPCR)
Retinal cDNA was amplified with Power SYBR Green Mix (Applied Biosystems, 4309155) in a 384-well optical reaction plate using a Via7 real-time PCR machine (Applied Biosystems). The PCR mixture consisted of 20 ng cDNA, 0.5 μM of each primer, and 10 μl of 2X SYBR green fluorescent dye master mix in a final volume of 20 μl. Dimer formation was assessed by analyzing the dissociation curve at the end of each amplification reaction. Results were calibrated in relation to the housekeeping gene Tbp. Raw data were processed using the comparative Ct method by the formula 2-ΔΔCT. Primers used for the qPCR analysis are listed in Supplemental Table S1.
Transcriptomic analysis
Retinal cDNA libraries were prepared from the RNA using an in-house protocol from G-INCPM (Weizmann Institute of Science, Israel). Samples were sequenced on an Illumina NextSeq machine, using the Single-Read 60 protocol. The output was ~36 million reads per sample. Reads were trimmed using cutadapt (Martin, 2011) and mapped to the mouse genome (GRCm38) using STAR(Dobin et al., 2013)) v2.4.2a (with EndToEnd option and outFilterMismatchNoverLmax set to 0.04). Counting proceeded over genes annotated in Ensembl release 88, using htseq-count (Anders et al., 2015) (intersection-strict mode). Differential expression analysis was performed using DESeq2 (Love et al., 2014) with the betaPrior, cooksCutoff and independent filtering parameters set to False. Raw P-values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. A pipeline was constructed using Snakemake (Köster and Rahmann, 2012).
Luciferase reporter assay and western blot analysis
Neuroblastoma line N2a cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C and 5% CO2. Cells were transfected using jetPEI DNA transfection reagent (Polyplus-transfection, 101–10N). Each well was cotransfected with vectors in a total amount of 1 μg DNA: 500 ng of Id1-luciferase reporter (Id1-luc, Korchynskyi and ten Dijke, 2002) and 20 ng of Renilla pRL-TK, with 500 ng of either wild-type pCAG-Zeb2 or mutant pCAGZeb2(AxAx)2 (as described in Conidi et al., 2013), or an empty vector control (pCAG). After 24 h in complete medium, cells were starved in medium with 0.5% fetal calf serum (6 h) and incubated with or without 10 nM BMP2 in low serum for an additional 24 h. Cells were harvested and luminescence was evaluated using the Dual-Luciferase Reporter Assay System (Promega, E1910), and normalized relative to the luminescence of the pRL-TK normalizing vector. For regulation of Ptf1a-luciferase, the reporter activity was tested 48 h after cotransfection with the expression vectors into HeLa cells as previously described (((Menuchin-Lasowski et al., 2016; Raviv et al., 2014). To examine protein-expression levels of each expression vector, whole-cell lysates were subjected to western blot analyses using antibodies against Zeb2 and β-tubulin or GAPDH (Supplemental Fig. S2B) and the Crescendo ECL kit (Millipore).
Chromatin immunoprecipitation (ChIP)
E14 murine embryonic stem cells (ESCs) were subjected to neural differentiation up to day 8 (D8) according to a previously described protocol (Stryjewska et al., 2017a). Samples were collected on D0 (undifferentiated), D6 and D8 (early and late neural commitment, respectively). The cells (100 × 106) were fixed in 1% formaldehyde by gently shaking for 10 min at RT, and the reaction was quenched by incubating with 0.125 M glycine at RT for 5 min. Cells were then washed twice with PBS and lysed with sonication buffer (10 mM TrisHCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA) supplemented with protease and phosphatase inhibitors (Roche). Cells were sonicated using a BioRuptor system (20 min, maximum amplitude, 15 s on, 15 s off). The sonicated material (50 μl) was stored at −80°C as control input. Sonicated material was brought to a final volume of 2 ml with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 17 mM Tris-HCl pH 8, 170 mM NaCl) and incubated with 15 μg anti-Zeb2 antibody (H260, Santa Cruz) or 10 μg anti-Smad1/5 antibody (D4G2, Cell Signaling Technologies) overnight at 4°C on a rotating wheel. The next day, protein A/G agarose beads (Santa Cruz) were added and incubated for 1 h. Beads were spun down at 1,000 gfor 1 min and washed as follows: once with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 150 mM NaCl) and transferred to nonstick low-binding 1.5-ml tubes, washed once with high-salt buffer (low-salt buffer with 500 mM NaCl), once with LiCl buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 10 mM Tris-HCl pH 8.0, 1 mM EDTA), and twice with 10 mM Tris-HCl pH 8.0, 1 mM EDTA. Incubation with the different buffers was performed for 5 min each on a rotating wheel at 4°C.
Protein–chromatin complexes were eluted from the beads by incubation at RT for 15 min with elution buffer (1% SDS, 0.1 M NaHCO3). Samples were de-crosslinked in 5 M NaCl overnight at 65°C with shaking at 950 rpm. Proteinase K was then added, then immunoprecipitated chromatin was purified by phenol:chloroform:isoamyl alcohol, and regions of interest were amplified by qPCR. ChIP was performed twice on postnatal day 2 (P2) murine retinas. For each repeat, 20 retinas were dissociated with papain and 20 × 106 cells were used for the ChIP using Zeb2 antibody (H260) as previously described(Sailaja et al., 2012). Graphs present the means and standard deviations of three or two independent biological experiments in cells or P2 retinas respectively, calculated using GraphPad’s PRISM software.
Statistical analysis
Statistical analysis of qPCR data, and cell number and fluorescence intensity quantification was performed using Student’s t-test. In cases of multiple testing, the P-value was adjusted by the Benjamini and Hochberg procedure. Statistical analysis of the luciferase assay results was performed using one-way repeated measures ANOVA with post-hoc test to identify differences among the groups as indicated in figure legends. The analysis was performed using Microsoft Excel and Graphpad’s PRISM software.
Results
Postnatal retinal phenotype of Zeb2loxp/loxp;αCre supports a role for Zeb2 in inhibition of MG cell fate
In the retina, a common pool of multipotent progenitors gives rise to neurons and MGs. As in other regions of the central nervous system, neurogenesis precedes gliogenesis and thus a change in the temporal dynamics of cell differentiation and survival will impact the number of MGs. In a previous study, we documented an increase in the number of MG cells in the retinas of Zeb2loxp/loxp;αCre mice on P14, based on indirect immunofluorescence of the MG markers Cdkn1b (P27/Kip1) and glutamine synthetase (Glul) (Menuchin-Lasowski et al., 2016). In contrast to these findings, Wei et al., (2018) reported a reduction of all of the INL cell types, including MGs, in Zeb2loxp/loxp;Six3-Cre mice based on analyses of Sox9 expression on P21 ((Wei et al., 2018)). The different outcome led to different views on Zeb2 functions in retinal development; Wei et al. concluded a role for Zeb2 in the differentiation of all postmitotic INL precursors.
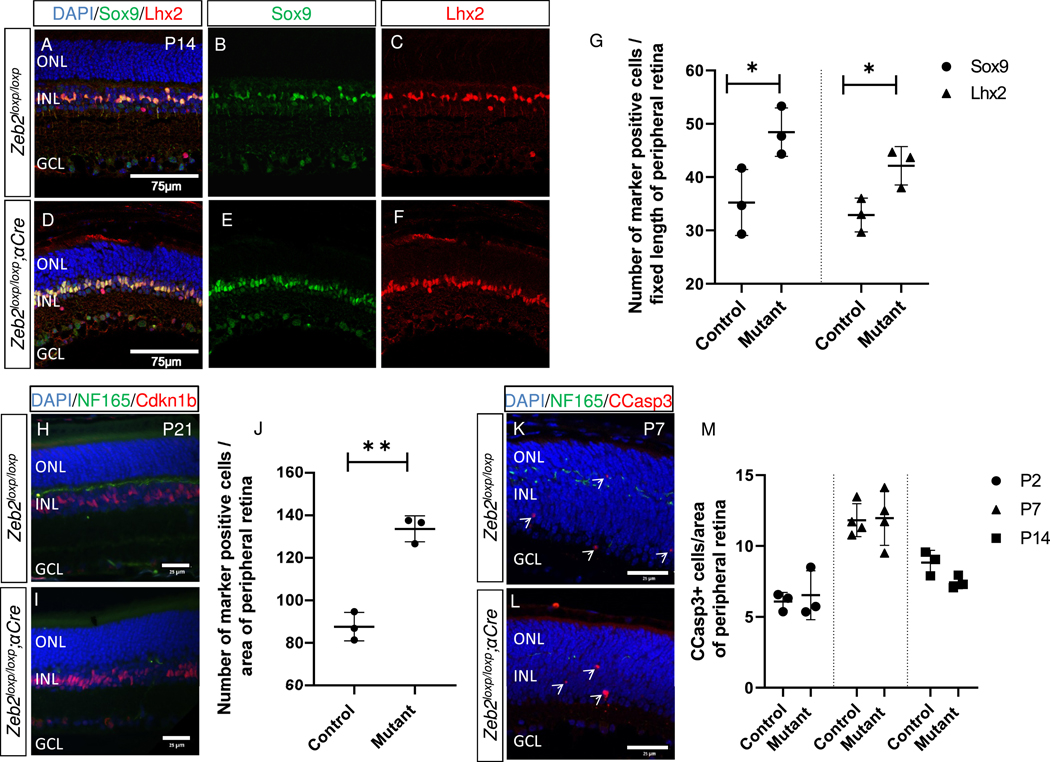
To further examine the possible involvement of Zeb2 in the neural-to-MG cell fate decision, and whether the discrepancy between the two studies is due to the different markers or stages analyzed, we extended our quantitative analyses by immunolabeling control and Zeb2loxp/loxp;αCre peripheral retinas using antibodies against Lhx2 and Sox9 (P14, Fig. 1A–G) and Cdkn1b (P21, Fig. 1H–J). Consistent with our previous report, this analysis showed an increase in the three MG markers in Zeb2loxp/loxp;αCre compared to control peripheral retinas (Fig. 1G, J; 37% increase in Sox9+ cells, SD = 12.9%, and 28% increase in Lhx2+ cells, SD = 10.9%, P14, P < 0.05; 50% increase in Cdkn1b, SD = 7.5%, P21, P < 0.01). These new results confirmed that loss of function of Zeb2 in RPCs, using αCre, leads to an increase in the generation of MG cells in the mouse retina.
Fig. 1.
Increased number of Muller glia in the Zeb2-deficient retina. Antibody labeling for detection of Sox9 and Lhx2 in control (A–C) and Zeb2loxp/loxp;αCre mutant (D–F) P14 peripheral retinas. (G) Quantification of the number of Sox9+ and Lhx2+ cells in each of the groups (P = 0.0406 for Sox9 and P = 0.029 for Lhx2). The number of cells detected in three different peripheral sections from each eye, in a peripheral retinal area of fixed length (225 μm), was averaged and represents one biological replicate (N = 3). (H, I) Antibody labeling for the detection of Nf-165 (green) and Cdkn1b (red) in control (H) and Zeb2loxp/loxp;αCre (I) P21 retinas. (J) Quantification of the number of Cdkn1b-expressing cells in the Zeb2loxp/loxp;αCre and control eyes (P = 0.0009, N = 3). The mutant regions were defined by loss of Nf-165+ horizontal precursors in the Zeb2loxp/loxp;αCre but not control retina. (K, L) Detection of cleaved caspase 3 (CCasp3+, red) and Nf-165 (green) by antibody labeling in the peripheral retina; representative image shown for P7, mutant regions were defined by loss of Nf-165+ horizontal precursors in the Zeb2loxp/loxp;αCre but not control retina. (M) Quantification of the number of CCasp3+ cells in the Zeb2loxp/loxp;αCre and control eyes on P2 (P = 0.7, N = 3), P7 (P = 0.9, N = 4), and P14 (P = 0.07, N = 3). Each value in J and M is an average of the number of cells detected in two adjacent regions of 50,000 μm2 of DAPI+ peripheral retina. Significance for differences between groups was determined by two-tailed Student’s t-test. *P < 0.05, **P < 0.01. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
Together with a reduction in MG cells, Wei et al. observed a significant increase in apoptosis in the Zeb2loxp/loxp;Six3-Cre postnatal retina(Wei et al., 2018). To determine whether the two genetic models also differ with respect to cell death, we quantified the number of cleaved-caspase-3+ (CCasp3+) cells during postnatal stages P2, P7 and P14 in the peripheral retina of Zeb2loxp/loxp;αCre compared to controls. In contrast to the outcome of deleting Zeb2 using Six3-Cre, we did not detect any change in the number of CCasp3+ cells compared to the control peripheral retina at the three examined stages (at P2 we detected 6.5 cells, SD = 1.7 in Zeb2loxp/loxp;αCre versus 6.1 cells, SD = 0.64 in controls; at P7, the respective values were 12 cells, SD = 1.9 versus 11.8, SD = 1.6; at P14, they were 7.4 cells, SD = 0.5 versus 8.8 cells, SD = 0.86, respectively).
Taken together, although the number of interneurons is reduced in both conditional mutants, we identified important differences between the two genetic models. Zeb2 conditional mutants using Six3-Cre resulted in elevated cell death and reduced MG cells, whereas the Zeb2 conditional mutation using αCre resulted in an increase in MGs with no change in cell survival. The different phenotypic outcomes are probably due to differences in the onset, distribution and possibly the level of Cre (see discussion and (Lam et al., 2019)). Considering that we did not detect compromised cell survival in ourZeb2loxp/loxp;αCre mice, we concluded that this genetic model is suitable for further study of the roles of Zeb2 during late stages of retinogenesis.
Transcriptomic analysis of the effects of Zeb2-mutant retina supports its roles in interneuron differentiation and inhibition of photoreceptor gene expression
Most photoreceptors, BPL and MG cells are generated postnatally in mice (Rapaport et al., 2004; Young, 1985). To further determine the molecular changes in the Zeb2-mutant retina, we analyzed transcriptomic changes at P2 using bulk RNA-Seq.
The αCre transgene is only active in the peripheral retina (Marquardt et al., 2001). As a result, approximately 50% of the cells of the Zeb2loxp/loxp;αCre retina remain wild type, and changes in gene expression may be masked by the Zeb2+ cells in the central retina. Indeed, the number of reads of the floxed critical exon 7 (Higashi et al., 2002) was reduced in the P2 Zeb2loxp/loxp;αCre retina to 58 ± 16% of the control level (Supplemental Fig. S1A). Despite this limitation, principal component analysis of the RNA-Seq data showed that the control and Zeb2-deficient samples do group together according to their genotype (Supplemental Fig. S1B), confirming that the RNA-Seq analysis includes effects of Zeb2 genetic inactivation on the transcriptome of P2 retinas.
This analysis identified 787 genes that were significantly altered in the Zeb2loxp/loxp;αCre retina compared to the control (P-value adjusted by FDR ≤ 0.05, max count ≥ 30, N = 3); 489 of these differentially expressed genes were upregulated in the Zeb2deficient retinas, and 298 were downregulated (Supplemental Table S3). The list of downregulated genes in the mutant mice included many that are known to be expressed in amacrine cells and reduced on P2 in Zeb2loxp/loxp;αCre retinas (Menuchin-Lasowski et al., 2016). These included Pax6 (1.2-fold change), Barhl2 (1.3-fold), Tfap2a (AP-2α, 1.4-fold) and Tfap2c (AP-2γ, 1.5-fold) (Bassett et al., 2012; Ding et al., 2009; Shaham et al., 2012). Tfap2b (AP-2β) was downregulated (1.4-fold), but did not pass the FDR cutoff. Ptf1a, previously shown to be a direct target of Zeb2 (Menuchin-Lasowski et al., 2016), was also significantly downregulated, by 1.5-fold, as was Bhlhe22 (1.3-fold). Similarly, many genes which are highly expressed in horizontal cells were downregulated in the Zeb2loxp/loxp;αCre retina. One of the most severely downregulated genes found in the analysis was Lhx1 (2.2-fold change), which is specific for horizontal cells (Poche et al., 2007). This downregulation nicely reflects the complete loss of horizontal cells from the Zeb2-deficient retina (MenuchinLasowski et al., 2016).
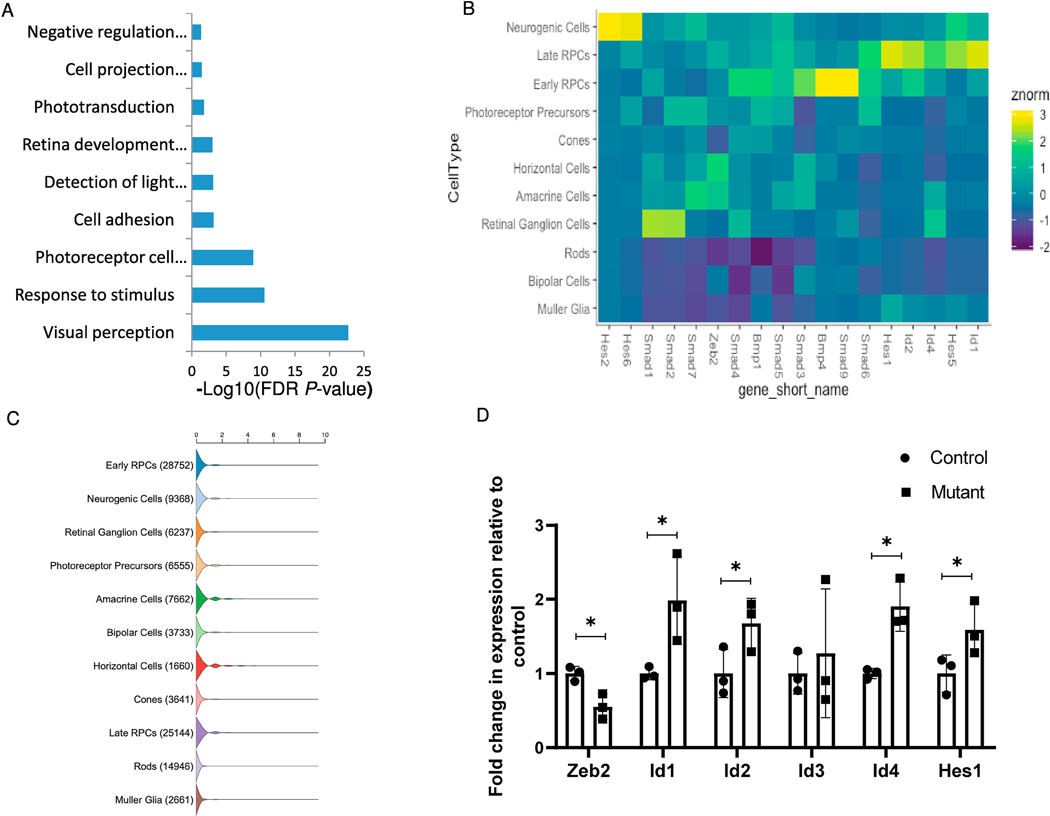
Gene-ontology enrichment analysis using the DAVID functional annotation tool (Huang et al., 2008; Huang et al., 2009) identified several enriched annotation categories in the list of genes that were upregulated in the Zeb2loxp/loxp;αCre retina (Fig. 2A). Among these are functional categories related to photoreceptor cells, including visual perception, response to stimulus, photoreceptor maintenance, detection of light stimulus involved with visual perception, and phototransduction. Upregulated photoreceptor genes in the mutant retinas included both genes that are specific to rod photoreceptors such as rhodopsin (Rho, 2.1-fold change) and Gnat1 (2.3-fold), and cone photoreceptor-specific genes such as s-opsin (Opn1sw, 1.5-fold change) and cone arrestin (Arr3, 2.3-fold).
Fig. 2.
Transcriptomic analysis of control and Zeb2loxp/loxp;αCre retinas during early postnatal development reveals upregulation of BMP and Notch target genes. (A) Significantly enriched functional categories in the list of genes upregulated in Zeb2loxp/loxp;αCre retinas according to DAVID gene-ontology analysis. (B) Heatmap representing normalized enrichment of select genes across individual retinal cell types as annotated through single-cell RNA-Seq. Individual genes shown represent upregulated transcripts in the Zeb2loxp/loxp;αCre retinas from the RNA-Seq data. (C) Violin plot representation of the number of transcripts of Zeb2 in developing and mature retinal cell types based on single-cell analyses (Clark et al., 2019).(D) Relative quantification of the transcript levels of Zeb2, Id1, Id2, Id3, Id4 and Hes1 by realtime PCR analysis (P = 0.008 for Zeb2, 0.022 for Id1, 0.033 for Id2, 0.31 for Id3, 0.004 for Id4, 0.039 for Hes1, N = 3, one-tailed Student’s t-test). *P < 0.05.
Taken together, the results of the bulk RNA-Seq and validation of gene expression using antibody labeling were consistent, overall, with previous reports on the role of Zeb2 in the generation of retinal interneurons and inhibition of photoreceptor differentiation. These functions could be mediated by Zeb2 regulation of lineage-specific TFs, such as Ptf1a, regulation of the balance between progenitor/glial and differentiating neurons, or both.
Zeb2 inhibits the expression of inhibitors of neural differentiation
Another interesting enriched functional category in the list of upregulated genes in the Zeb2-knockout retina was “negative regulation of oligodendrocyte differentiation” (Fig. 2A), which altogether corresponds with the known role of Zeb2 in embryonic myelinogenesis in the central nervous system and in Schwann cells of the postnatal peripheral nervous system (Quintes et al., 2016; Weng et al., 2012; Wu et al., 2016). Among the genes in this category were three that are known to be directly inhibited by Zeb2 in oligodendrocyte precursors: Hes1 (1.3-fold change), Id2 (1.3-fold) and Id4 (2-fold). Interestingly, Hes1 has a documented role as an inhibitor of neural differentiation and promoter of MG cell differentiation in the retina (Furukawa et al., 2000; Takatsuka et al., 2004; Tomita et al., 1996). Id2 and Id4 are members of the Id gene family, which consists of four well-documented BMP-induced members that are known to primarily act as inhibitors of differentiation (Ling et al., 2014; Ruzinova and Benezra, 2003; Tzeng, 2003). The two other members of the Id family, Id1 (1.3-fold change) and Id3 (1.2-fold), were also found to be upregulated in the Zeb2loxp/loxp;αCre retinas and, like Hes1, are known to inhibit neural differentiation and promote MG cell differentiation (Du and Yip, 2011; Mizeracka et al., 2013). In line with these known functions of Hes1 and Id genes, single-cell RNA-Seq analysis demonstrated that Id1–4 and Hes1 are strongly expressed in late-stage RPCs and their transcripts are also detected in MG cells (Clark et al., 2019; Fig. 2B, C). Zeb2 transcripts are detected in late-stage RPCs but their level is low or undetectable in MGs. Combined, this further suggests a role for Zeb2 in controlling cell-fate of RPCs or of post mitotic precursors, rather than in the differentiation of MG cells.
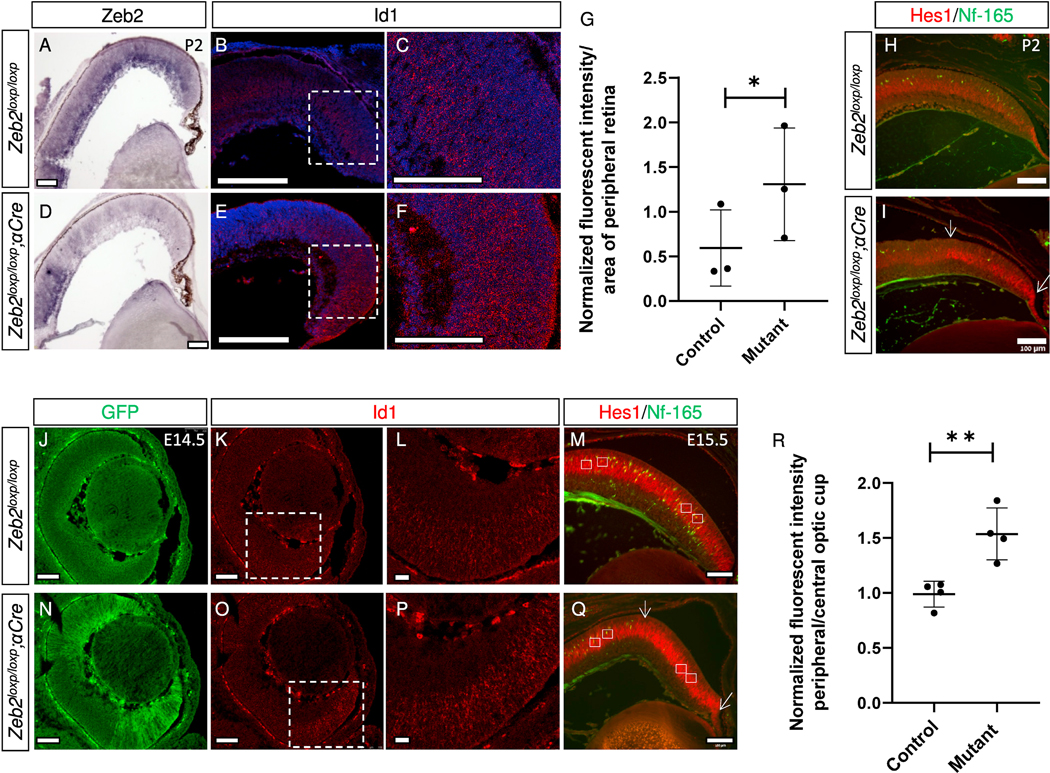
The increased expression of Id1, Id2, Id4, and Hes1 was validated using real-time PCR (Fig. 2D). We next examined, by HCR-based fluorescence in-situ hybridization (Choi et al., 2018), the expression patterns and levels of Id1 in the P2 control and Zeb2-mutant retinas (Fig. 3A–G; Choi et al., 2018). Quantification of the fluorescent signal derived from Id1 mRNA revealed an ~1.8-fold increase in intensity in the periphery of the mutant retinas (Fig. 3G), further validating the increase in Id1 expression. Similarly, Hes1 protein, detected by antibody labeling, was higher in the Zeb2-mutant region on P2 (the mutant region was defined by loss of Nf-165+ horizontal precursors in the mutant but not control retina, Fig. 3H and I, green) compared to the control (Fig. 3H and I, red). These results demonstrated that during the postnatal stages of retinal development, Zeb2 has a role in inhibiting the expression of genes that promote MG differentiation and inhibit neural differentiation. This role is probably at least partially responsible for the delayed generation of bipolar cells and overproduction of MG cells observed in the Zeb2 mutants.
Fig. 3.
Validation of increased levels of Id1 and Hes1 mRNA transcripts in Zeb2loxp/loxp;αCre retinas compared to controls. In-situ hybridization (ISH) was used to detect the expression pattern of Zeb2 in control (A) and Zeb2loxp/loxp;αCre (D) P2 retinas, while hybridization chain reaction fluorescent in-situ hybridization (HCR–FISH) was used to detect the expression pattern of Id1 in control (B, C) and Zeb2loxp/loxp;αCre (E, F) P2 retinas (magnification of the marked areas in B and E are shown in C and F, respectively). (G) Quantification of the average mean intensity of the HCR–FISH signal in control and Zeb2loxp/loxp;αCre P2 peripheral retinas in an area of 50 × 50 μm2 (N = 3, P = 0.02). Antibody labeling for detection of horizontal precursors (Nf-165, green) and Hes1 (red) in P2 control (H) and Zeb2loxp/loxp;αCre (I) retinas. Visualization of the EGFP signal generated by the Z/EG transgene in E14.5 Zeb2loxp/loxp Z/EG (J) and Zeb2loxp/loxp;αCre Z/EG (N) retinas. HCR–FISH was used to detect the expression pattern of Id1 (K, L, O, P) in control (K, L) and Zeb2loxp/loxp;αCre (O, P) E14.5 retinas (magnification of the marked areas in K and O are presented in L and P, respectively). This experiment was repeated in two control and Zeb2loxp/loxp;αCre retinas. Antibody labeling for detection of horizontal precursors (Nf-165, green) and Hes1 (red) in E15.5 control (M) and Zeb2loxp/loxp;αCre (Q) retinas. Fluorescence intensity was quantified by measuring the intensity of the fluorescence in a peripheral area versus a central area in each eye (average intensity measured in an area of 1,170 μm2, white rectangles) following normalization to the background (R). Significance for differences between groups was determined by two-tailed Student’s t-test (P = 0.0060). *P < 0.05, **P < 0.01. Scale bars = 100 μm.
The differentiation of earlier born amacrine interneurons is delayed in the Zeb2loxp/loxp;αCre retina, much like the delay in bipolar cell differentiation seen at later stages of retinal development (Menuchin-Lasowski et al., 2016). Therefore, it is possible that upregulation of differentiation inhibitors may also underlie this earlier phenotype. To test this possibility, HCR in-situ hybridization was used to document Id1 expression, while antibody labeling was used to detect Hes1 in the retinas of control and Zeb2loxp/loxp;αCre embryos. These analyses revealed an increase in Id1 transcript (Fig. 3J–L, N–P), as well as a significant elevation in Hes1 protein in the Zeb2-mutant retinas (Fig. 3M and Q, and quantification of fluorescence intensity in Fig. 3R (mutant region was defined by loss of Nf-165+ horizontal precursors in the mutant but not control retina). Altogether, these results show that Zeb2 inhibits Id1 and Hes1 expression in the embryonic retina.
Zeb2 is required for the generation of most of the BPL precursors
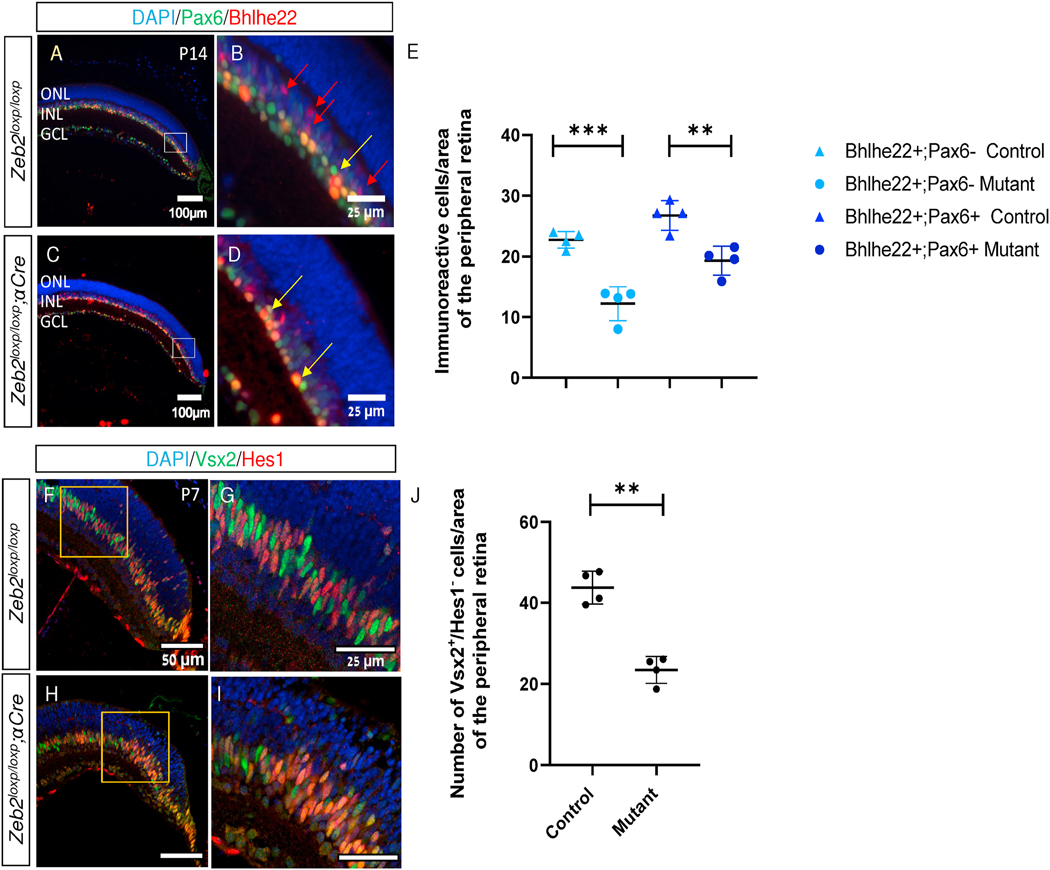
Zeb2 ablation from the developing retina results in a reduction in the numbers of BPL cell markers, including rod and OFF cone bipolar cell TFs Isl1 and Bhlhe22, respectively (Huang et al., 2014; Menuchin-Lasowski et al., 2016; Wei et al., 2018)). We also detected a significant reduction in Bhlhe22-expressing amacrine (Bhlhe22+;Pax6+) and BPL cells (Bhlhe22+;Pax6-) in the Zeb2loxp/loxp;αCre P14 retina (Fig. 4A–E) The reduction in BPL TFs (Isl1, Bhlbe22) could be due to delayed onset of expression of the two genes due to activity of Zeb2 in BPL differentiation, or it could represent failure to generate the early BPL precursors. To discern between these two possibilities, we co-immunostained control and Zeb2loxp/loxp;αCre retinas using antibodies against Hes1, which labels RPCs and MG cells, and Vsx2, which marks RPCs but is also maintained in the BPL precursors (P7, Fig. 4F–I). We then quantified the number of Vsx2+/Hes1- cells, representing early BPL precursors. This quantification revealed a significant (2-fold) reduction in the number of BPL precursors generated in the Zeb2loxp/loxp;αCre retinas compared to controls (Fig. 4J, mean of 23.4 cells, SD = 3.3 in Zeb2loxp/loxp;αCre versus 43.3 cells, SD = 4 in controls, P = 0.0099, two-tailed Student’s t-test). This result supports a role for Zeb2 in promoting the generation of postmitotic BPL precursors in late RPCs.
Fig. 4.
Reduced bipolar precursors in Zeb2-mutant retinas. Antibody labeling for the detection of amacrine cells (Pax6, green) and subtypes of amacrines and the OFF cone bipolar cells (Bhlhe22, red) in control (A, B) and Zeb2loxp/loxp;αCre (C, D) P14 retinas, respectively (B, D are higher magnifications of the marked insets in A and C, respectively). (E) Quantification of cells expressing Bhlhe22 but not Pax6 (OFF cone bipolar cells, P = 0.0005) or Bhlhe22 and Pax6 (subtype of amacrine cells, P = 0.0048) by two-tailed Student’s t-test P14 control and mutant peripheral retinas. Each value is an average of the number of cells detected in two adjacent regions of 20,000 μm2 of DAPI+ peripheral retina. (F–I) Antibody labeling for detection of Hes1 (red) and Vsx2 (green) in control (F, G) and Zeb2loxp/loxp;αCre (H, I) P7 peripheral retinas. (J) Quantification of the number of Vsx2+/Hes1- cells in the two genotypes (P = 0.0099). Each value is an average of the number of cells detected in two adjacent regions of 6,000 μm2 of DAPI+ peripheral retina. Significance determined by two-tailed Student’s t-test, N = 4, **P < 0.01, ***P < 0.001.
Zeb2 interaction with phospho-Smads can inhibit BMP-mediated activation of Id1 transcription
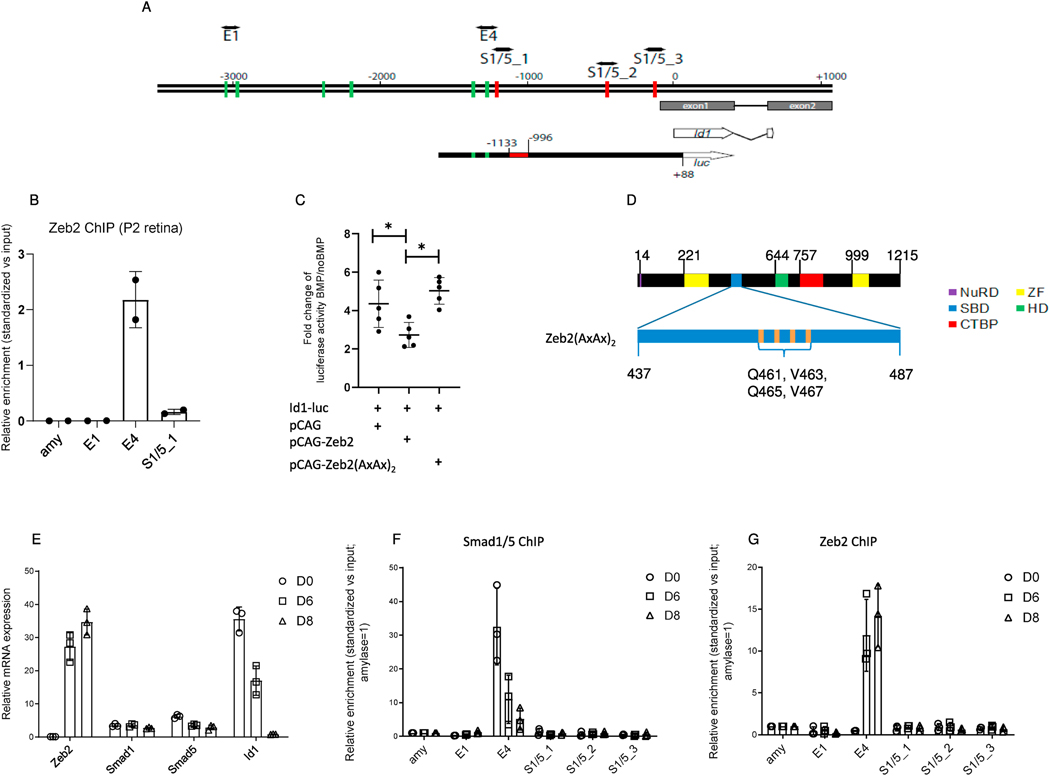
BMP promotes MG cell differentiation and activates Id1 in the murine retina (Ueki et al., 2015). We therefore examined whether Zeb2 can directly repress BMP-dependent activation of Id1, similar to its inhibitory activity on Id2 and Id4 in oligodendrocyte differentiation (Weng et al., 2012). We scanned 3.5 kb upstream of the transcriptional start site (TSS) of the mouse Id1 gene for Zeb2-binding E-boxes (Remacle et al., 1999) using the JASPAR database and 90% stringency. In the same region, we also identified potential BMP– Smad::Smad4-binding sites (Fig. 5A) as described (Morikawa et al., 2011). This region contains a BMP-responsive regulatory region (named E4) located near the Smad1/5-binding motif (S1/5_1, Fig. 5A). To further examine whether Zeb2 is associated with Id1 regulatory regions in vivo, we performed a quantitative ChIP–qPCR assay on P2 mouse retinas. We detected striking enrichment for this E4 region and lower enrichment for the S1/5_1 motif in the Zeb2 immunoprecipitated chromatin, but no evidence for binding to neither the E-box sequences located ~3 kb upstream of the TSS nor the amylase promoter (Fig. 5B). This result supports direct binding of Zeb2 to the BMP-Smad responsive Id1 regulatory region in the developing murine retina. To test whether Zeb2 can repress BMP-Smad dependent activation of the Id1 regulatory region, we used a luciferase reporter assay in line N2a; these cells have neuronal characteristics and are amenable to transfection and reporter assays (Klebe and Ruddle, 1969). The reporter used for this analysis included the region between 1,575 bp upstream and 89 bp downstream of the Id1 TSS and contained the BMP-responsive regulatory region (Fig. 5A, Id1-luc, putative Zeb2- and Smad-binding sites are marked by green and red lines, respectively (Korchynskyi and ten Dijke, 2002; Lopez-Rovira et al., 2002).
Fig. 5.
BMP-mediated activation of the Id1 promoter. (A) Schematic representation of the mouse Id1 promoter (−3500/+1000, ATG = 0) and comparison with the sequence cloned upstream of the luciferase (luc) gene used for the luciferase assay (Id1-luc). Green lines: Eboxes representing putative Zeb2-binding sites; red lines: putative BMP–Smad-binding elements (indicated as S1/5_1 to S1/5_3). The red box in Id1-luc is the BMP-responsive regulatory region (−1133/−996) in the Id1 promoter. (B) Zeb2 chromatin immunoprecipitation (ChIP) in P2 retinas (N = 2). The enrichment relative to input of the putative Zeb2- (E4, E1) or Smad-binding region (S1/5_1) quantified by PCR is indicated. amy, amylase (C) Scatter plot indicating mean and median fold-change (N = 5) of luciferase activity units of Id1 reporter described in BMP-treated N2a cells expressing either empty vector, wild-type Zeb2 or Zeb2(AxAx)2, compared to non-BMP-treated cells. Significance for differences between the groups was determined by one-way repeated measures ANOVA (F = 11.52, P = 0.0044). Tukey post-hoc test revealed significant differences in BMP-mediated activation between the empty and wild-type Zeb2 groups (P = 0.026), and the wild-type Zeb2 and Zeb2(AxAx)2 groups (P = 0.0041). *P < 0.05. (D) Protein location of the four point mutations in the Smadbinding domain of Zeb2(AxAx)2. (E) mRNA levels of Zeb2, Smad1, Smad5 and Id1 in differentiating murine embryonic stem cells (mESCs). (F) Smad1/5 ChIP and (G) Zeb2 ChIP in neural differentiated mESCs. Putative BMP–Smads::Smad4-binding regions are indicated as S1/5_1 to S1/5_3, while E-boxes are indicated as E1 and E4. BMP–Smads and Zeb2 are recruited in region E4 (−1287/−1147). (F, G) ChIP values are normalized against the input and using amylase (amy) as a negative control (set to 1).
Activity of the Id1-luc reporter was about 4-fold higher when treated with BMP2 and transfected with a control plasmid (pCAG), relative to untreated transfected cells, confirming previous report on BMP activation of the Id1 regulatory region (Korchynskyi and ten Dijke, 2002). Co-expression of wild-type Zeb2 (from vector-encoded cDNA) with Id1-luc significantly decreased this BMP2-dependent reporter activation (Fig. 5C), suggesting that Zeb2 can inhibit BMP-dependent activation of Id1 transcription. Since Zeb2 directly binds receptor-activated Smads (Verschueren et al., 1999b)), we next examined whether its inhibition of BMP-dependent transcription of Id1 is dependent on its ability to bind the activated phospho-Smads. To this end, we overexpressed a mutant form of Zeb2 in which four amino acids in its Smad-binding domain (Q461, V463, Q465 and V467) were converted to alanine making this Zeb2 mutant unable to bind activated Smads (Zeb2(AxAx)2) (Conidi et al., 2013, Fig. 5D). This Zeb2(AxAx)2 mutant was previously shown to no longer bind phospho-Smad. Consistent with a model in which Zeb2 directly inhibits Smad-dependent transcription of Id1, Zeb2(AxAx)2 was not able to inhibit the BMP-mediated activation of the Id1 promoter-containing construct, and levels of Id1-luc transcription remained similar to those seen with the empty control plasmid (Fig. 5C). Western blot analysis was conducted to exclude the possibility that loss of inhibition resulted from lower expression levels of the mutant form, rather than loss of Smad binding (Supplemental Fig. S2B).
We further tested whether the Zeb2(AxAx)2 mutation alters transcriptional activation mediated by Zeb2 at cis-regulatory regions of Ptf1a, which have been previously shown to be directly targeted by Zeb2 (Menuchin-Lasowski et al., 2016). This was examined by comparing the activation of the Ptf1a enhancer by pCAG–Zeb2 versus the pCAG–Zeb2(AxAx)2 mutation. Both expression constructs significantly induced expression of the Ptf1a-based reporter compared with a control plasmid (pCAG). Thus, the Zeb2(AxAx)2 mutation does not seem to interfere with transcriptional activation by Zeb2 (Supplemental Fig. S2B). Taken together, these findings suggest that Zeb2 inhibits the BMP-dependent transcription of Id1 at least in part via interaction with activated phospho-Smad proteins.
Mutual exclusive binding of Zeb2 and BMP-Smads on the Id1 promoter during neural differentiation
Next we wanted to functionally assess whether the antagonism between BMP–Smads and Zeb2 might occur through differential recruitment at the Id1 promoter in the course of neural differentiation. We generated neural precursors on day 6 and 8 D6 and D8 using an established model of neural differentiation of mouse ESCs (Stryjewska et al., 2017). The expression levels of transcripts for Smad1, Smad5, Zeb2 and Id1 were characterized by qPCR during this neural differentiation protocol (Fig. 5E). Smad1 and Smad5 seemed to be expressed at constant levels at the three stages (D0, D6 and D8), whereas Zeb2 and Id1 showed opposite expression levels: Zeb2 mRNA was barely detected at D0 (undifferentiated state) and increased on D6/D8, whereas the Id1 mRNA level was high on D0, had decreased by D6, and was almost undetectable on D8.
We next performed a quantitative ChIP–PCR assay on the Id1 promoter using antibodies against Zeb2 or Smad1/5 (Fig. 5F, G). On D0, when Zeb2 mRNA levels were barely detectable, BMP–Smads were bound to region E4, which is near a BMP–Smad-binding element and contains two E-boxes (Fig. 5F). As neural differentiation of the ESCs proceeded, Zeb2 was recruited to the same E4 region (Fig. 5G). Interestingly, the mRNA levels of Smad1 and Smad5 did not change dramatically during differentiation, suggesting, that Zeb2 provides a functional brake that inhibits the transcriptional activity of BMP–Smads, leading to downregulation of Id1 during neural differentiation. This is also in accordance with the requirement for Smad interaction during BMP inhibition suggested by the luciferase assay.
Discussion
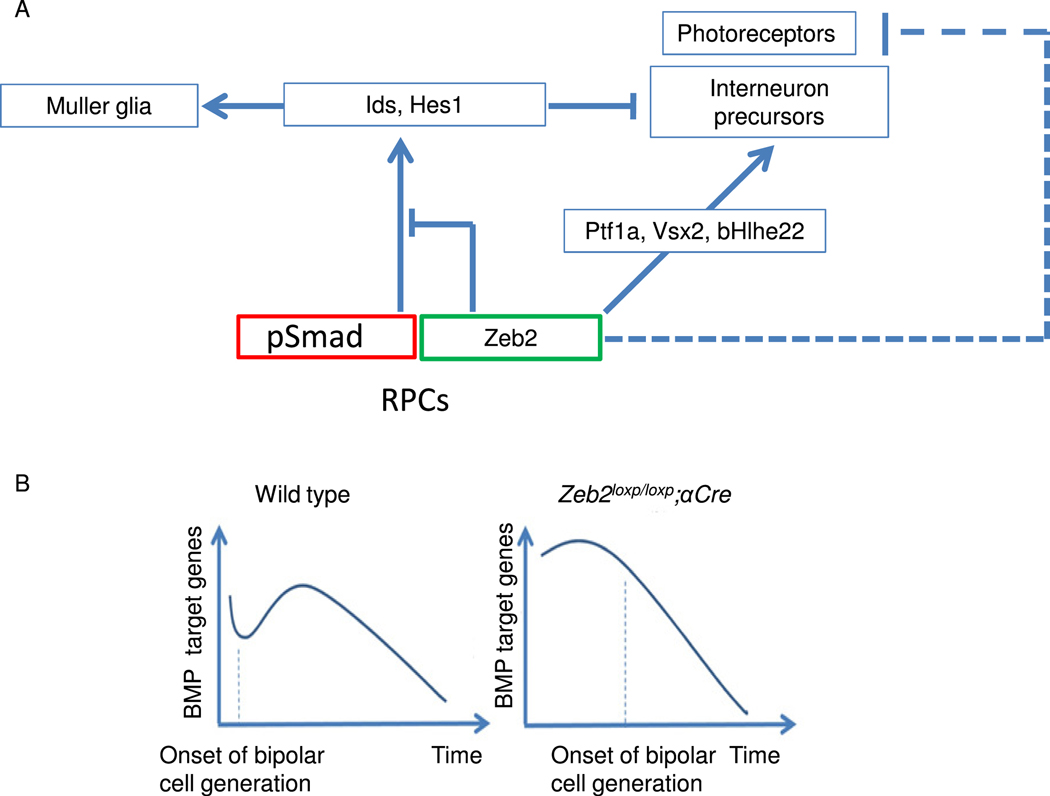
The neural-promoting activity of Zeb2 is well documented in different stages of nervous system development. At as early as neural plate formation, Zeb2 promotes the expression of neural factors, while inhibiting pluripotency genes and blocking signaling pathways that promote the formation of other germ layers (Chng et al., 2010; Tang et al., 2015). At later stages, in oligodendrocyte precursors for example, Zeb2 both activates oligodendrocyte differentiation-promoting genes and inhibits the BMP-mediated activation of several inhibitors of differentiation (Id2, Id4 and Hes1 (Weng et al., 2012)). Here we show that Zeb2 coordinates the normal balance between progenitors, MG cells and neurons in the retina. We document a role for Zeb2 in inhibiting MG cell specification, and our findings suggest that this is mediated by inhibition of Id and Hes1 expression. The results of cell-based reporter assays and ChIP analyses in vivo further document Zeb2’s direct interaction with the Id1 promoter and its likely repression of BMP-activated Smads. These findings point to a mechanism by which Zeb2 modulates extrinsic cues triggered by key signaling pathways to control the timing of cell specification and differentiation during retinogenesis (Fig. 6).
Fig. 6.
Schematic of the role of Zeb2 in postnatal retinogenesis. (A) BMP- and Notchsignaling act as activators of Id genes and Hes1. Ids and Hes1 selectively inhibit photoreceptor specification, and at high levels globally inhibit neural differentiation and promote Muller glial differentiation. Low or medium levels of Id genes and Hes1 are needed to enable bipolar cell differentiation at the expense of photoreceptors. Zeb2 promotes interneuron differentiation by balancing the level of Id genes and Hes1, directly activating bipolar cell-differentiation genes and inhibiting photoreceptor differentiation. (B) BMP signaling is downregulated in the developing retina after P6. Downregulation of BMP might be needed for the onset of bipolar cell generation. Deletion of Zeb2 increases the expression of BMP target genes, inhibiting bipolar cell differentiation. Later, as BMP activity decreases with time, the levels of BMP target genes become low enough to allow bipolar cell generation. Bipolar cell differentiation is thus delayed, but not abolished, in the Zeb2loxp/loxp;αCre retina.
Overproduction of MG cells in the αCre but not Six3-Cre Zeb2-mutant retina
Although Zeb2 has been documented in previous studies to be required for the generation of retinal interneurons (Menuchin-Lasowski et al., 2016)(Wei et al., 2018), its requirement for retinal cell survival and MG differentiation remains controversial: Menuchin-Lasowski et al. reported a reduction in cell-cycle exit and an increase in MGs, whereas Wei et al. reported reductions in both cell survival and MG number in Zeb2-mutant retinas.
These seemingly contradictory findings are most likely due to the differences in the temporal and spatial patterns of activity of the different Cre transgenes used by each team. Activity of αCre, used by Menuchin-Lasowski et al., is initiated around embryonic day 10.5 (E10.5) and is restricted to the peripheral retina from the early optic cup stage, but is not active in the central retina (Marquardt et al., 2001). In contrast, Six3-driven Cre employed by Wei et al. is first active at least 1 day earlier in the ventral forebrain, as well as the optic vesicle and optic nerve head, and targets both the central and peripheral retina (Furuta et al., 2000). Thus, the differences between the results of the two studies may emanate from differences in the timing of Zeb2 loss of function. Earlier deletion using Six3-Cre may potentially having a stronger effect on early-stage RPCs, resulting in extensive cell death and eventually, reduction in MG cell number. It is also possible that cells in the genotypically normal and central retina in mice carrying the αCre transgene rescue, in a non-cell-autonomous manner, some aspects of the phenotype seen in Zeb2-deficient cells at the retinal periphery. Non-cell autonomous actions of Zeb2 in vivo have been demonstrated, including in brain cortex development (Seuntjens et al., 2009).
Surprisingly, Zeb2 overexpression was shown to promote the generation of all nonphotoreceptor cell types, including Sox9+ MG cells (Wei et al., 2018). This might result from inhibition of photoreceptor specification, which in turn might increase the pool of undifferentiated cells in a manner that compensates for the promotion of neural differentiation by Zeb2. In addition, in our bulk RNA-Seq data, Sox9 mRNA is downregulated, suggesting that while promoting neuronal fate, Zeb2 might also promote the expression of Sox9, albeit to a lesser degree, and its resultant overexpression may thus result in an increase in Sox9+ cells. Additional MG-specific markers need to be tested to fully evaluate the effects of Zeb2 misexpression on MG cell formation and differentiation.
As developmental processes are dynamic and occur over short periods of time, even seemingly small differences between Cre lines in onset and distribution may result in different phenotypic outcomes. This has been previously noted in studies on the retina. For example, a Dicer1 conditional mutation generated using the Chx10-Cre transgene identified a requirement for microRNAs in photoreceptor survival, but observed no effects on retinal cell specification (Damiani et al., 2008). In contrast, conditional deletion of Dicer1 with αCre revealed its essential activity for maintaining the retinal progenitor pool and inhibiting ganglion cell generation (Davis et al., 2011; Georgi and Reh, 2010). Overall, comprehensive analyses of conditional mutagenesis using different Cre lines are important for broadening our understanding of the complex and dynamically TF-regulated processes controlling cell fate, survival and differentiation.
Zeb2 inhibits inhibitors of neural differentiation during retinogenesis
Interestingly, both Id1 and Hes1 have been documented to inhibit neural differentiation and promote MG cell generation (Du and Yip, 2011; Furukawa et al., 2000; Mizeracka et al., 2013; Tomita et al., 1996), and they are known to be regulated by both activated Notch and BMP receptor/Smad signaling in different systems (Mizeracka et al., 2013; Quintes et al., 2016; Ueki et al., 2015; Weng et al., 2012; Wu et al., 2016). Both Notch and BMP signaling also promote MG cell differentiation (Jadhav et al., 2006b; Kuribayashi et al., 2014; Mizeracka et al., 2013; Ueki et al., 2015).
The results presented in this study suggest that, in the retina, Zeb2 inhibits the targets of the BMP–Smad-mediated activation of neural differentiation inhibitors. While our luciferase reporter experiment suggested that an interaction with Smad proteins is necessary for Zeb2’s ability to inhibit Id1, the ChIP results indicated that during neural differentiation the binding of Zeb2 to Id1’s promoter is accompanied by a decrease in Smad binding in the same area. These results suggest a complex relationship between Zeb2 and the Smad proteins in the context of Id1 inhibition. One possibility is that interaction with the Smad proteins might be necessary for Zeb2 to remove them from their direct binding sites. Interaction with Smad proteins might be also necessary for Zeb2 to locate and bind in the E4 area in Id1, before preventing any further binding of the Smads to their target sequence. Notably, in Schwann cell differentiation, Zeb2 was demonstrated to function by recruiting HDAC1/2-NuRD corepressor complexes to antagonize Notch effector genes that inhibit Schwann cell differentiation (Wu et al., 2016). With this in mind, the interaction between Zeb2, Smads, Notch signaling as well as chromatin organization of the target loci should be comprehensively investigated during retinal development using unbiased transcriptomic and epigenomic approaches.
A possible triple role for Zeb2 in the differentiation of retinal interneurons
Following Zeb2 loss in RPCs, there is a marked reduction in different classes of bipolar cells, based on the reduction in cells expressing PKCa, Isl1 and Bhlhe22 proteins (Menuchin-Lasowski et al., 2016; Wei et al., 2018,Fig. 5). Wei and colleagues concluded that Zeb2 disrupts the differentiation of postmitotic BPL cell precursors, because Zeb2 expression was detected in these bipolar cell subtypes. Our study shows that Zeb2 also controls the relative balance between BPL and MG cells that are generated postnatally. We conclude this based on the elevated levels of Hes1 protein and Id1 transcript in the neuroblastic layer of Zeb2 mutants (Figs. 2, 3), the significant reduction in the number of BPL precursor (Vsx2+/Hes1-) cells detected in mutant P7 retina (Fig. 4), and the increase in the number of MGs in the mature mutant retina (Fig. 1). Furthermore, our observations (Fig 5) suggest that Zeb2 inhibits MG specification by blocking Smad-mediated activation of Id1 transcription. Notably, Wei et al. reported a 44% reduction in the number of rod bipolar cells based on PKCa staining (Fig. 5 in Wei et al., 2018), yet they detected Zeb2 in only 14% of the rod bipolar cells (Fig. 3 in Wei et al. 2018). Thus, Zeb2 most likely acts before bipolar cell differentiation to inhibit the expression of progenitor- and MG-specific genes, although it may also have a separate function in a subset of differentiating Bhlhe22-positive bipolar cell precursors. Future studies should directly examine the role of Zeb2 in bipolar cell differentiation using cell type-specific Cre lines (Lu et al., 2013).
Loss of function of Zeb2 resulted in increased expression levels of photoreceptorspecific genes. This matches the results of Wei at al., who recently showed that Zeb2 inhibits the expression of photoreceptor-specific genes in murine retinas (Wei et al., 2018). Wei et al. attributed the elevation in photoreceptor-specific gene expression to increased generation of photoreceptor precursors at the expense of INL cell types. While this is plausible, it is worth noting that the level of Crx, which is highly and selectively expressed in photoreceptor precursors, was not significantly upregulated (fold-change 1.1) in our bulk RNA-Seq analysis of Zeb2-deficient peripheral retina, and the increase in Crx expression reported by Wei et al. based on qRT-PCR may simply reflect altered cellular composition in the mutant retina. This suggests that Zeb2 primarily inhibits photoreceptor maturation rather than specification similar to its reported function in specific cell types of the immune system (van Helden et al., 2015). Furthermore, in the retina this inhibition could also include non-cell autonomous actions (see also Seuntjens et al., 2009), as the change in cellular composition in the Zeb2deficient retina may result in an altered response to temporally dynamic extrinsic signals that regulate photoreceptor differentiation. The inhibition of photoreceptor gene expression could also occur in an intrinsic manner, where Zeb2 may function to repress the expression of both photoreceptor genes, such as Rho and Opn1sw (Wei et al., 2018; this study), and genes that inhibit photoreceptor differentiation, such as Hes1, Id1 and Ptn (3.1-fold change, Roger et al., 2006). This might be an important aspect of Zeb2’s role as a TF that promotes the differentiation of retinal interneurons. To allow bipolar, amacrine and horizontal interneuron differentiation, Zeb2 may repress the expression of genes that broadly inhibit interneuron generation, while also inhibiting genes controlling photoreceptor-fate specification to prevent the generation of excess numbers of photoreceptors.
Given that Zeb2 is important for the timely activation of differentiation genes such as Ptf1a and Prox1 in horizontal and amacrine cells, and possibly Isl1 and Bhlhe22 in bipolar precursors, it seems that Zeb2 may promote the generation of retinal interneurons through three different mechanisms (Fig. 6A): by activating the expression of genes controlling the differentiation of retinal interneurons such as Ptf1a in amacrine and horizontal cells and Isl1 and Bhlhe22 in bipolar cells, by inhibiting genes encoding inhibitors of neural differentiation (Id1–4 and Hes1) to promote the generation of interneurons, and finally by inhibiting photoreceptor differentiation.
The levels of BMP signaling in the postnatal developing mouse retina have been reported to be highly dynamic (Ueki et al., 2015). The amount of phospho-Smad1/5/9 decreases between P2 and P4 and then increases again to a peak at around P6, only to sharply decrease after P8 (Ueki et al., 2015). If Zeb2 indeed promotes the generation of bipolar cells by inhibiting the expression of genes that are activated by BMP signaling, it is possible that these wave-like changes in BMP signaling contribute to the partial recovery of bipolar cell generation seen in Zeb2-loss-of-function mutants (Menuchin-Lasowski et al., 2016). Similar (in that case cyclic) waves that involve a functional interaction between Hes and Smad have been seen in BMP and Notch-dependent dynamic choice options between stalk- and tip-cell phenotypes in growing blood vessels, as analyzed in Smad1/5-deficient mouse models (Moya et al., 2012). When BMP–Smad-signaling levels are high, Zeb2 may be needed to balance the expression of BMP target genes and allow some of the cells to continue differentiating into bipolar cells (Fig. 6B). Without Zeb2, expression levels of BMP target genes will remain high, and will prevent neuronal differentiation. The partial recovery of bipolar cell differentiation observed at later ages in the Zeb2-deficient retina may result from reduced BMP signaling after P8, allowing differentiation of a subset of precursors to a bipolar fate.
Supplementary Material
Fig. S1. Additional data derived from the transcriptomic analysis of control and Zeb2loxp/loxp;αCre P2 retinas. (A) Normalized number of reads of the floxed seventh translated exon of Zeb2 was extracted from the transcriptomic analysis. (B) Principal component analysis of the transcriptomes of the different samples in the RNA-Seq analysis.
Fig. S2. Zeb2(AxAx)2 mutation does not alter the transcriptional activation by Zeb2 of the Ptf1a enhancer. (A) Luciferase reporter assay demonstrates activation of the Ptf1a enhancer by pCAG-Zeb2 and pCAG-Zeb2(AxAx)2 expression vectors relative to pCAG vector controls. Significance for differences between the groups was determined by one-way ANOVA (F = 12.67, P = 0.0159). Dunnett's multiple comparisons test show significant differences in reporter activation when comparing means between the pCAG empty vector to the Zeb2 and Zeb2(AxAx)2 groups (P = 0.0188, P = 0.046 respectively, N = 5). There is no significance difference was detected between Zeb2 and Zeb2(AxAx)2 groups (P = 0.796, two-tailed Student’s t-test). *P < 0.05. (B) Protein lysates of N2a cells transiently expressing constructs of pCAG empty vector, pCAG-Zeb2 and pCAG-Zeb2(AxAx)2, used for the luciferase reporter assay, were further analyzed by western blotting using Zeb2 antibody (upper panel) and β-tubulin antibody (lower panel) as loading controls, to verify transfection efficiencies.
Table S1 – List of primers used in the study
Table S2 - List of antibodies and their working dilutions
Table S3 – List of differentially expressed genes in the Zeb2loxp/loxp;αCre versus control P2 retinas
Highlights (EACH POINT: MAXIMUM 85 CHARACTERS AND SPACES)
Zeb2 mutation in developing retina reduces bipolar precursors and increases Muller glia
Zeb2 mutation in developing retina increases expression of inhibitors of neural differentiation
Zeb2 binds to Id1 promoter region in developing mouse retina and differentiating neural progenitors
Zeb2 inhibits Id1 expression through inhibition of BMP/Smad pathway
Zeb2 integrates extrinsic cues to regulate balance between neuronal and Muller cell types in developing mouse retina
Acknowledgments
We thank Benjamin Amram, Assaf Biran and Neta Hert for technical help and Victor Tarabykin for reagents. Research in the RA-P laboratory is supported by grants from the Israel Science Foundation (228/14), INCPN-ISF (2246/16), and Claire and Amedee Maratier Institute for the Study of Blindness and Visual Disorders, Sackler Faculty of Medicine, TelAviv University, Israel. RA-P and SB are supported by the Binational Science Foundation (2013016). SB and BSC are supported by grants from the NIH (R01EY020560 and U01EY027267 to SB, F32EY024201 and R00EY027844 to BSC) and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness to BSC. DH by the FWO-V (G.0A31.16).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT and Huber W. (2015). HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariss M, Natan K, Friedman N. and Traboulsi EI (2012). Ophthalmologic Abnormalities in Mowat-Wilson Syndrome and a Mutation in ZEB2. Ophthalmic Genet. 33, 159–160. [DOI] [PubMed] [Google Scholar]
- Bassett EA, Korol A, Deschamps PA, Buettner R, Wallace VA, Williams T. and West-Mays JA (2012). Overlapping expression patterns and redundant roles for AP-2 transcription factors in the developing mammalian retina. Dev. Dyn. 241, 814–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacheux V, Dastot-Le Moal F, Kääriäinen H, Bondurand N, Rintala R, Boissier B, Wilson M, Mowat D. and Goossens M. (2001). Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum. Mol. Genet. 10, 1503–10. [DOI] [PubMed] [Google Scholar]
- Cepko C. (2014). Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 15, 615–627. [DOI] [PubMed] [Google Scholar]
- Chng Z, Teo A, Pedersen R. a and Vallier L. (2010). SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell 6, 59–70. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A. and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, et al. (2019). Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102, 1111–1126.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conidi A, van den Berghe V, Leslie K, Stryjewska A, Xue H, Chen YG, Seuntjens E. and Huylebroeck D. (2013). Four Amino Acids within a Tandem QxVx Repeat in a Predicted Extended ??-Helix of the Smad-Binding Domain of Sip1 Are Necessary for Binding to Activated Smad Proteins. PLoS One 8, e76733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O’Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD and Strettoi E. (2008). Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J. Neurosci. 28, 4878–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N, Mor E. and Ashery-Padan R. (2011). Roles for Dicer1 in the patterning and differentiation of the optic cup neuroepithelium. Development 138, 127–138. [DOI] [PubMed] [Google Scholar]
- Ding Q, Chen H, Xie X, Libby RT, Tian N. and Gan L. (2009). BARHL2 Differentially Regulates the Development of Retinal Amacrine and Ganglion Neurons. J. Neurosci. 29, 3992–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M. and Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y. and Yip HK (2011). The expression and roles of inhibitor of DNA binding helixloop-helix proteins in the developing and adult mouse retina. Neuroscience 175, 367–79. [DOI] [PubMed] [Google Scholar]
- EM M, CM C. and CL C. (2008). Temporal order of bipolar cell genesis in the neural retina. Neural Dev. 3, 2–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, et al. (2006). Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133, 4439–50. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM and Cepko CL (2000). rax, Hes1, and notch1 promote the formation of Müller glia by postnatal retinal progenitor cells. Neuron 26, 383–94. [DOI] [PubMed] [Google Scholar]
- Garavelli L, Zollino M, Mainardi PC, Gurrieri F, Rivieri F, Soli F, Verri R, Albertini E, Favaron E, Zignani M, et al. (2009). Mowat-Wilson syndrome: facial phenotype changing with age: study of 19 Italian patients and review of the literature. Am. J. Med. Genet. A 149A, 417–26. [DOI] [PubMed] [Google Scholar]
- Georgi SA and Reh TA (2010). Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J. Neurosci. 30, 4048–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PJ, Yun S, Clark AM, Monuki ES, Murtaugh LC and Levine EM (2013). Lhx2 balances progenitor maintenance with neurogenic output and promotes competence state progression in the developing retina. J. Neurosci. 33, 12197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory-Evans CY, Vieira H, Dalton R, Adams GGW, Salt A. and Gregory-Evans K. (2004). Ocular coloboma and high myopia with Hirschsprung disease associated with a novel ZFHX1B missense mutation and trisomy 21. Am. J. Med. Genet. A 131, 86–90. [DOI] [PubMed] [Google Scholar]
- Gueta K, David A, Cohen T, Menuchin-Lasowski Y, Nobel H, Narkis G, Li L, Love P, de Melo J, Blackshaw S, et al. (2016). The stage-dependent roles of Ldb1 and functional redundancy with Ldb2 in mammalian retinogenesis. Development 143,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Maruhashi M, Nelles L, Van de Putte T, Verschueren K, Miyoshi T, Yoshimoto A, Kondoh H. and Huylebroeck D. (2002). Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 32, 82–4. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT and Lempicki RA (2008). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT and Lempicki RA (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Hu F, Feng L, Luo X-J, Liang G, Zeng X-Y, Yi J-L and Gan L. (2014). Bhlhb5 is required for the subtype development of retinal amacrine and bipolar cells in mice. Dev. Dyn. 243, 279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovski I, Djuric O, Caraffi SG, Santodirocco D, Pollazzon M, Rosato S, Cordelli DM, Abdalla E, Accorsi P, Adam MP, et al. (2018). Phenotype and genotype of 87 patients with Mowat–Wilson syndrome and recommendations for care. Genet. Med. 20, 965–975. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA and Cepko CL (2006a). Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913–23. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Cho S-H and Cepko CL (2006b). Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc. Natl. Acad. Sci. U. S. A. 103, 18998–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Roesch K. and Cepko CL (2009). Development and neurogenic potential of Muller glial cells in the vertebrate retina. Prog. Retin. Eye Res. 28, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor CC, Miyake M, Chen LJ, Shi Y, Barathi VA, Qiao F, Nakata I, Yamashiro K, Zhou X, Tam POS, et al. (2013). Genome-wide association study identifies ZFHX1B as a susceptibility locus for severe myopia. Hum. Mol. Genet. 22, 5288–94. [DOI] [PubMed] [Google Scholar]
- Korchynskyi O. and ten Dijke P. (2002). Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277, 4883–91. [DOI] [PubMed] [Google Scholar]
- Köster J. and Rahmann S. (2012). Snakemake-a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Kuribayashi H, Baba Y. and Watanabe S. (2014). BMP signaling participates in late phase differentiation of the retina, partly via upregulation of Hey2. Dev. Neurobiol. 1–12. [DOI] [PubMed] [Google Scholar]
- Lopez-Rovira T, Chalaux E, Massaguo J, Rosa JL and Ventura F. (2002). Direct Binding of Smad1 and Smad4 to Two Distinct Motifs Mediates Bone Morphogenetic Protein-specific Transcriptional Activation of Id1 Gene. J. Biol. Chem. 277, 3176–3185. [DOI] [PubMed] [Google Scholar]
- Lam PT, Padula SL, Hoang TV, Poth JE, Liu L, Liang C, LeFever AS, Wallace LM, Ashery-Padan R, Riggs PK, et al. (2019). Considerations for the use of Cre recombinase for conditional gene deletion in the mouse lens. Hum. Genomics 13, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Kang B. and Sun X-H (2014). Id proteins: small molecules, mighty regulators. Curr. Top. Dev. Biol. 110, 189–216. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F. and Gruss P. (2001). Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43–55. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10. [Google Scholar]
- Maruhashi M, Van De Putte T, Huylebroeck D, Kondoh H. and Higashi Y. (2005). Involvement of SIP1 in positioning of somite boundaries in the mouse embryo. Dev. Dyn. 234, 332–8. [DOI] [PubMed] [Google Scholar]
- McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio L. a, Huylebroeck D, Higashi Y. and Rubenstein JLR (2013). Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron 77, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuchin-Lasowski Y, Oren-Giladi P, Xie Q, Ezra-Elia R, Ofri R, Peled-Hajaj S, Farhy C, Higashi Y, Van de Putte T, Kondoh H, et al. (2016). Sip1 regulates the generation of the inner nuclear layer retinal cell lineages in mammals. Development 143, 2829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka K, DeMaso CR and Cepko CL (2013). Notch1 is required in newly postmitotic cells to inhibit the rod photoreceptor fate. Development 140, 3188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin C-H, Aburatani H. and Miyazono K. (2011). ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 39, 8712–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat DR, Croaker GD, Cass DT, Kerr BA, Chaitow J, Adès LC, Chia NL and Wilson MJ (1998). Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J. Med. Genet. 35, 617–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya IM, Umans L, Maas E, Pereira PNG, Beets K, Francis A, Sents W, Robertson EJ, Mummery CL, Huylebroeck D, et al. (2012). Stalk cell phenotype depends on integration of Notch and Smad1/5 signaling cascades. Dev. Cell 22, 501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GIW and Schmid RM (2007). Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development 134, 1151–60. [DOI] [PubMed] [Google Scholar]
- Nelles L, Van de Putte T, van Grunsven L, Huylebroeck D. and Verschueren K. (2003). Organization of the mouse Zfhx1b gene encoding the two-handed zinc finger repressor Smad-interacting protein-1☆. Genomics 82, 460–469. [DOI] [PubMed] [Google Scholar]
- Poche RA, Kwan KM, Raven MA, Furuta Y, Reese BE and Behringer RR (2007). Lim1 Is Essential for the Correct Laminar Positioning of Retinal Horizontal Cells. J. Neurosci. 27, 14099–14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintes S, Brinkmann BG, Ebert M, Fröb F, Kungl T, Arlt FA, Tarabykin V, Huylebroeck D, Meijer D, Suter U, et al. (2016). Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nat. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D. and LaVail MM (2004). Timing and topography of cell genesis in the rat retina. J. Comp. Neurol. 474, 304–24. [DOI] [PubMed] [Google Scholar]
- Raviv S, Bharti K, Rencus-Lazar S, Cohen-Tayar Y, Schyr R, Evantal N, Meshorer E, Zilberberg A, Idelson M, Reubinoff B, et al. (2014). PAX6 regulates melanogenesis in the retinal pigmented epithelium through feed-forward regulatory interactions with MITF. PLoS Genet. 10, e1004360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC and Huylebroeck D. (1999). New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 18, 5073–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger J, Brajeul V, Thomasseau S, Hienola A, Sahel J-A, Guillonneau X. and Goureau O. (2006). Involvement of Pleiotrophin in CNTF-mediated differentiation of the late retinal progenitor cells. Dev. Biol. 298, 527–539. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB and Benezra R. (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13, 410–418. [DOI] [PubMed] [Google Scholar]
- Sailaja BS, Takizawa T. and Meshorer E. (2012). Chromatin immunoprecipitation in mouse hippocampal cells and tissues. Methods Mol. Biol. 809, 353–64. [DOI] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave K-A, Huylebroeck D. and Tarabykin V. (2009). Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat. Neurosci. 12, 1373–80. [DOI] [PubMed] [Google Scholar]
- Shaham O, Menuchin Y, Farhy C. and Ashery-Padan R. (2012). Pax6: A multi-level regulator of ocular development. Prog. Retin. Eye Res. 31, 351–376. [DOI] [PubMed] [Google Scholar]
- Stryjewska A, Dries R, Pieters T, Verstappen G, Conidi A, Coddens K, Francis A, Umans L, van IJcken WFJ, Berx G, et al. (2017a). Zeb2 Regulates Cell Fate at the Exit from Epiblast State in Mouse Embryonic Stem Cells. Stem Cells 35, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryjewska A, Dries R, Pieters T, Verstappen G, Conidi A, Coddens K, Francis A, Umans L, van IJcken WFJ, Berx G, et al. (2017b). Zeb2 Regulates Cell Fate at the Exit from Epiblast State in Mouse Embryonic Stem Cells. Stem Cells 35, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka K, Hatakeyama J, Bessho Y. and Kageyama R. (2004). Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 1004, 148–155. [DOI] [PubMed] [Google Scholar]
- Tang K, Peng G, Qiao Y, Song L. and Jing N. (2015). Intrinsic regulations in neural fate commitment. Dev. Growth Differ. 57, 109–120. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F. and Kageyama R. (1996). Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 16, 723–34. [DOI] [PubMed] [Google Scholar]
- Tzeng S-F (2003). Inhibitors of DNA binding in neural cell proliferation and differentiation. Neurochem. Res. 28, 45–52. [DOI] [PubMed] [Google Scholar]
- Ueki Y, Wilken MS, Cox KE, Chipman LB, Bermingham-McDonogh O. and Reh T. a (2015). A transient wave of BMP signaling in the retina is necessary for Müller glial differentiation. Development 142, 533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D. and Higashi Y. (2003). Mice lacking ZFHX1B, the gene that codes for Smadinteracting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am. J. Hum. Genet. 72, 465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berghe V, Stappers E, Vandesande B, Dimidschstein J, Kroes R, Francis A, Conidi A, Lesage F, Dries R, Cazzola S, et al. (2013). Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron 77, 70–82. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Michiels C, Van de Putte T, Nelles L, Wuytens G, Verschueren K. and Huylebroeck D. (2003). Interaction between Smad-interacting protein-1 and the corepressor C-terminal binding protein is dispensable for transcriptional repression of E-cadherin. J. Biol. Chem. 278, 26135–45. [DOI] [PubMed] [Google Scholar]
- van Grunsven L. a, Taelman V, Michiels C, Verstappen G, Souopgui J, Nichane M, Moens E, Opdecamp K, Vanhomwegen J, Kricha S, et al. (2007). XSip1 neuralizing activity involves the co-repressor CtBP and occurs through BMP dependent and independent mechanisms. Dev. Biol. 306, 34–49. [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su M, Bodmer R, et al. (1999a). SIP1, a Novel Zinc Finger / Homeodomain Repressor, Interacts with Smad Proteins and Binds to 5 -CACCT Sequences in Candidate Target Genes *. J. Biol. Chem. 274, 20489–20498. [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su M-T, Bodmer R, et al. (1999b). SIP1, a Novel Zinc Finger/Homeodomain Repressor, Interacts with Smad Proteins and Binds to 5’-CACCT Sequences in Candidate Target Genes. J. Biol. Chem. 274, 20489–20498. [DOI] [PubMed] [Google Scholar]
- Verstappen G, van Grunsven LA, Michiels C, Van de Putte T, Souopgui J, Van Damme J, Bellefroid E, Vandekerckhove J. and Huylebroeck D. (2008). Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum. Mol. Genet. 17, 1175–1183. [DOI] [PubMed] [Google Scholar]
- Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, et al. (2001). Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat. Genet. 27, 369–70. [DOI] [PubMed] [Google Scholar]
- Wei W, Liu B, Jiang H, Jin K. and Xiang M. (2018). Requirement of the MowatWilson Syndrome Gene Zeb2 in the Differentiation and Maintenance of Nonphotoreceptor Cell Types During Retinal Development. [DOI] [PubMed] [Google Scholar]
- Weng Q, Chen Y, Wang H, Xu X, Yang B, He Q, Shou W, Chen Y, Higashi Y, van den Berghe V, et al. (2012). Dual-mode modulation of Smad signaling by Smadinteracting protein Sip1 is required for myelination in the central nervous system. Neuron 73, 713–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LMN, Wang J, Conidi A, Zhao C, Wang H, Ford Z, Zhang L, Zweier C, Ayee BG, Maurel P, et al. (2016). Zeb2 recruits HDAC-NuRD to inhibit Notch and controls Schwann cell differentiation and remyelination. Nat. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M. and Ashery-Padan R. (2006). Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133, 1367–78. [DOI] [PubMed] [Google Scholar]
- Young RW (1985). Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199–205. [DOI] [PubMed] [Google Scholar]
- Zweier C, Thiel CT, Dufke A, Crow YJ, Meinecke P, Suri M, Ala-Mello S, Beemer F, Bernasconi S, Bianchi P, et al. (2005). Clinical and mutational spectrum of Mowat-Wilson syndrome. Eur. J. Med. Genet. 48, 97–111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Additional data derived from the transcriptomic analysis of control and Zeb2loxp/loxp;αCre P2 retinas. (A) Normalized number of reads of the floxed seventh translated exon of Zeb2 was extracted from the transcriptomic analysis. (B) Principal component analysis of the transcriptomes of the different samples in the RNA-Seq analysis.
Fig. S2. Zeb2(AxAx)2 mutation does not alter the transcriptional activation by Zeb2 of the Ptf1a enhancer. (A) Luciferase reporter assay demonstrates activation of the Ptf1a enhancer by pCAG-Zeb2 and pCAG-Zeb2(AxAx)2 expression vectors relative to pCAG vector controls. Significance for differences between the groups was determined by one-way ANOVA (F = 12.67, P = 0.0159). Dunnett's multiple comparisons test show significant differences in reporter activation when comparing means between the pCAG empty vector to the Zeb2 and Zeb2(AxAx)2 groups (P = 0.0188, P = 0.046 respectively, N = 5). There is no significance difference was detected between Zeb2 and Zeb2(AxAx)2 groups (P = 0.796, two-tailed Student’s t-test). *P < 0.05. (B) Protein lysates of N2a cells transiently expressing constructs of pCAG empty vector, pCAG-Zeb2 and pCAG-Zeb2(AxAx)2, used for the luciferase reporter assay, were further analyzed by western blotting using Zeb2 antibody (upper panel) and β-tubulin antibody (lower panel) as loading controls, to verify transfection efficiencies.
Table S1 – List of primers used in the study
Table S2 - List of antibodies and their working dilutions
Table S3 – List of differentially expressed genes in the Zeb2loxp/loxp;αCre versus control P2 retinas