ABSTRACT
This work reports on the whole-genome sequencing of Leishmania infantum chagasi from Honduras (Central America) and Brazil (South America).
ANNOUNCEMENT
Until the end of the past century, Leishmania chagasi was regarded as the causal agent of American visceral leishmaniasis (1) but, after recent genomic evidence showing that L. chagasi and Leishmania infantum are similar, L. chagasi has come to be considered synonymous with L. infantum (2–4). However, in light of evidence about the original, enzootic cycle of Leishmania infantum chagasi in the New World (5–7), we describe here the whole-genome sequence of L. infantum chagasi from Honduras (Central America) and compare it with the sequence of the same parasite from Brazil (South America) and that of L. infantum from Europe available in GenBank.
The two Leishmania species isolates used in this work were (i) L. infantum chagasi MHOM/HD/2017/M32502/Amapala District/Honduras, which was isolated from a human case of nonulcerated cutaneous leishmaniasis according to procedures approved by the Research Ethics Committee of the Medical School of São Paulo University (CAAE protocol number 64223917.1.0000.006) (8), and (ii) L. infantum chagasi MCER/BR/1981/M6445/Salvaterra/Pará State/Brazil, which was isolated from a crab-eating fox (Cerdocyon thous), the wild reservoir (6). Both L. infantum chagasi isolates were grown at 25°C for 7 days in Schneider medium supplemented with 10% fetal bovine serum, 10 μg/ml 1% l-glutamine, and 100 IU/mL ampicillin. A 3-mL aliquot was collected and used for DNA extraction with the ReliaPrep genomic DNA (gDNA) miniprep system (Promega, Madison, WI, USA).
The total DNA quality was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The genomic library was prepared using the Nextera XT DNA sample preparation kit (Illumina, USA). The quality of the library was verified using a Bioanalyzer 2010 system (Agilent Technologies), and the library was sequenced on a HiSeq 2500 instrument (Illumina) with a 2 × 100-bp paired-end format sequencing kit v.4. The DNA concentration used for sequencing was 1 ng/μL in the Illumina HiSeq system. The reads generated were trimmed with Trimmomatic v.0.39 (9) and assembled using a de novo strategy with SPAdes v.3.12 (10). Genomes were manually curated, and the final genomes were compared with that of the L. infantum reference strain using Geneious v.8.1.9 (11). A molecular clock analysis was also performed to compare the origin and ancestry of these parasites, using BEAST v.1.10.4 with three independent runs and the strict Yule-coalescent model of epidemiological dispersion, with 100 million generations and with the DNA polymerase alpha subunit gene (a highly conserved genomic region related to the evolutionary process of Leishmania parasites). All tools were used with default parameters unless otherwise specified.
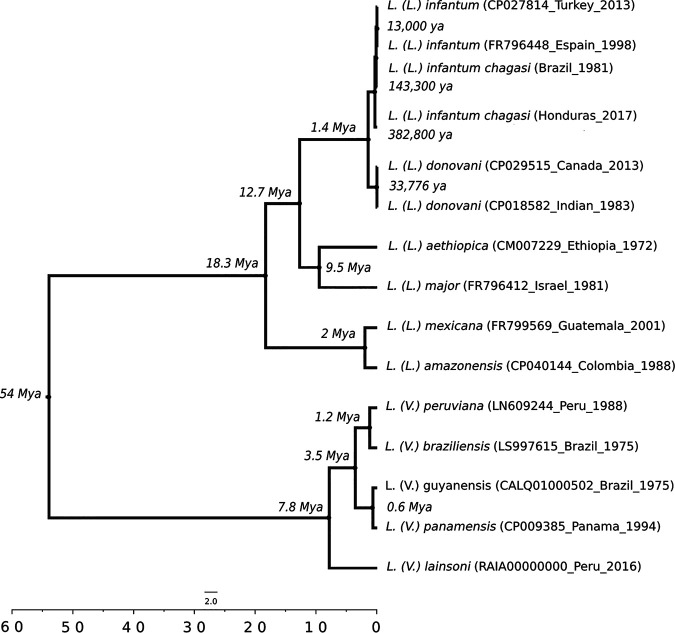
The genomes of the L. infantum chagasi isolates from Honduras and Brazil showed ∼99.9% similarity to that of L. infantum when all genome chromosomes were compared (Table 1); however, the molecular clock comparisons revealed that L. infantum chagasi from Honduras proved to be considerably more ancestral (∼382,800 years ago) than L. infantum chagasi from Brazil (∼143,300 years ago) and L. infantum (∼13,000 years ago). In addition, it should also be emphasized that the DNA polymerase alpha subunit gene was able to reveal significant differences in ancestry between some Leishmania parasites belonging to the subgenera Leishmania and Viannia (Fig. 1).
TABLE 1.
De novo genomic assembly of L. infantum chagasi isolates from Brazil and Honduras and their identity comparisons with L. infantum (from Europe)
| Isolate | No. of contigs | Minimum size (bp) | Maximum size (bp) | N50 (bp) | Coverage (×) | GC content (%) | Identity (%) |
|---|---|---|---|---|---|---|---|
| MCER/BR/1981/M6445a | 15,315 | 200 | 60,666 | 8,354 | 130 | 56.9 | 99.99 |
| MHOM/HD/2017/M32502b | 7,193 | 200 | 57,312 | 8,724 | 53.47 | 59.3 | 99.98 |
L. infantum chagasi isolate from Brazil.
L. infantum chagasi isolate from Honduras.
FIG 1.
Bayesian divergence-time analysis under the relaxed molecular clock model for Leishmania species from the Leishmania and Viannia subgenera, using the concatenated data set for the DNA polymerase alpha subunit gene. The x axis shows absolute time in millions of years, and nodes are located at the mean divergence. The molecular clock analysis shows that New World Leishmania infantum chagasi isolates (Brazil_1981 [SRA accession number SRR8842312] and Honduras_2017 [SRA accession number SRR8608748]) experienced divergence ∼143,300 years ago (ya) and ∼382,800 years ago, respectively, and thus are more ancestral than L. infantum isolates from the Old World (Turkey_2013 and Espain_1998), with divergence ∼13,000 years ago. All nodes on the tree are supported with a posterior probability of 1.
Data availability.
The de novo whole-genome assemblies and raw data for Leishmania infantum chagasi from Brazil and Honduras have been deposited in the GenBank and SRA databases. For Leishmania infantum chagasi from Brazil, the GenBank accession number is JAGRQE000000000 and the SRA accession number is SRR8842312. For Leishmania infantum chagasi from Honduras, the GenBank accession number is JAGRQD000000000 and the SRA accession number is SRR8608748. Both are under BioProject number PRJNA722301.
Contributor Information
Fernando Tobias Silveira, Email: fernandotobias@iec.gov.br.
Jason E. Stajich, University of California, Riverside
REFERENCES
- 1.Lainson R, Shaw JJ. 1987. Evaluation, classification and geographical distribution, p 1–120. In Peters W, Killick- Kendrick R (ed), The leishmaniases in biology and medicine. Academic Press, London, England. [Google Scholar]
- 2.Lukes J, Mauricio IL, Schönian G, Dujardin J-C, Soteriadou K, Dedet J-P, Kuhls K, Tintaya KWQ, Jirků M, Chocholová E, Haralambous C, Pratlong F, Oborník M, Horák A, Ayala FJ, Miles MA. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci USA 104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhls K, Alam MZ, Cupolillo E, Ferreira GEM, Mauricio IL, Oddone R, Feliciangeli MD, Wirth T, Miles MA, Schönian G. 2011. Comparative microsatellite typing of New World Leishmania infantum reveals low heterogeneity among populations and its recent Old World origin. PLoS Negl Trop Dis 5:e1155. doi: 10.1371/journal.pntd.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, Sereno D. 2016. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis 10:e0004349. doi: 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lainson R, Rangel EF. 2005. Lutzomyia longipalpis and the eco-epidemiology of American visceral leishmaniasis, with particular reference to Brazil: a review. Mem Inst Oswaldo Cruz 100:811–827. doi: 10.1590/s0074-02762005000800001. [DOI] [PubMed] [Google Scholar]
- 6.Silveira FT, Lainson R, Shaw JJ, Póvoa MM. 1982. Leishmaniasis in Brazil: XVIII. Further evidence incriminating the fox Cerdocyon thous (L.) as a reservoir of Amazonian visceral leishmaniasis. Trans R Soc Trop Med Hyg 76:830–832. doi: 10.1016/0035-9203(82)90119-5. [DOI] [PubMed] [Google Scholar]
- 7.Moreno ES, Sabioni LA, de Seixas MMM, de Souza Filho JA, Marcelino AP, Shimabukuro PHF. 2019. Evidence of a sylvatic enzootic cycle of Leishmania infantum in the State of Amapá, Brazil. Rev Soc Bras Med Trop 53:e20190169. doi: 10.1590/0037-8682-0169-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosa-Ochoa W, Zúniga C, Chaves LF, Flores GVA, Pacheco CMS, da Matta VLR, Corbett CEP, Silveira FT, Laurenti MD. 2020. Clinical and immunological features of human Leishmania (L.) infantum-infection, novel insights Honduras, Central America. Pathogens 9:554. doi: 10.3390/pathogens9070554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger AM, Marc L, Bjoern U. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The de novo whole-genome assemblies and raw data for Leishmania infantum chagasi from Brazil and Honduras have been deposited in the GenBank and SRA databases. For Leishmania infantum chagasi from Brazil, the GenBank accession number is JAGRQE000000000 and the SRA accession number is SRR8842312. For Leishmania infantum chagasi from Honduras, the GenBank accession number is JAGRQD000000000 and the SRA accession number is SRR8608748. Both are under BioProject number PRJNA722301.