LETTER
Correct identification of a (genome-sequenced) strain is an essential step in evolutionary and comparative genomic studies. It came to our attention that the number of publicly available misidentified genome-sequenced strains is increasing. By using the order Eurotiales (Aspergillus, Penicillium, Talaromyces, and related genera) as an example, in this letter we want to increase awareness among readers of Microbiology Resource Announcements of this ongoing problem and give recommendations to ensure availability and correct strain identification in the future.
Species identification is an important step in biological research. A correct name is vital for optimal communication and is the link between studies in various fields. Currently, the identification of fungi relies mainly on (single) gene sequencing, and this approach has largely replaced identification methods using phenotypic and physiological characteristics. The internal transcribed spacer (ITS) region was accepted as the primary fungal barcode (1) and is recommended for the identification of uncharacterized fungal strains (without any a priori knowledge). Although the ITS is the most commonly used region, with a good track record for identifying species, there are many genera for which it does not provide enough resolution. Various studies showed that this locus cannot be used for identification of species in well-known genera such as Aspergillus, Cladosporium, Fusarium, Penicillium, and Talaromyces (2–6). In these genera, protein-coding genes are commonly used for identification and generally have higher interspecies variability than the ITS region. Unfortunately, there is no standard choice of a protein-coding gene for the identification of fungal isolates across different groups. Efforts have been made to assess potential candidate gene regions (and corresponding universal primer pairs) as secondary DNA barcodes (7). Translation elongation factor 1-α (tef1-α) is widely used as a phylogenetic marker in mycology and is used as a secondary identification barcode for various genera; however, standardization is lacking. While it has sufficient resolution in many genera (e.g., Cladosporium and Fusarium), tef1-α has never been extensively studied in Aspergillus, Penicillium, and related genera (order Eurotiales); therefore, databases (e.g., GenBank) lack reference sequences of this locus for these genera. With the exception of Aspergillus, partial β-tubulin (BenA) gene sequencing is recommended for Penicillium, Talaromyces, Paecilomyces, and related genera (8–10). Partial calmodulin (CaM) gene sequencing is recommended as an identification barcode for Aspergillus; however, BenA sequencing generally also works well. Both species markers perform better than ITS (8–11). Two examples are given in Fig. 1. The (ex-)type cultures of Aspergillus aflatoxiformans, Aspergillus austwickii, Aspergillus cerealis, Aspergillus flavus, Aspergillus minisclerotigenes, Aspergillus oryzae, and Aspergillus pipericola (9) have the same ITS sequence, while the majority have unique BenA and CaM sequences (with the exception of A. flavus and its domesticated form A. oryzae). Similarly, Penicillium cavernicola, Penicillium discolor, Penicillium echinulatum, Penicillium solitum, and Penicillium speluncae share the same ITS sequence but differ in their BenA and CaM gene sequences. In summary, ITS is the primary barcode but might lack resolution in some genera at the species level; in those cases, an additional marker is needed. There is no consensus regarding a secondary marker, and this needs to be determined for each genus. If needed, contact a taxonomist who can advise on the barcode(s) to use.
FIG 1.
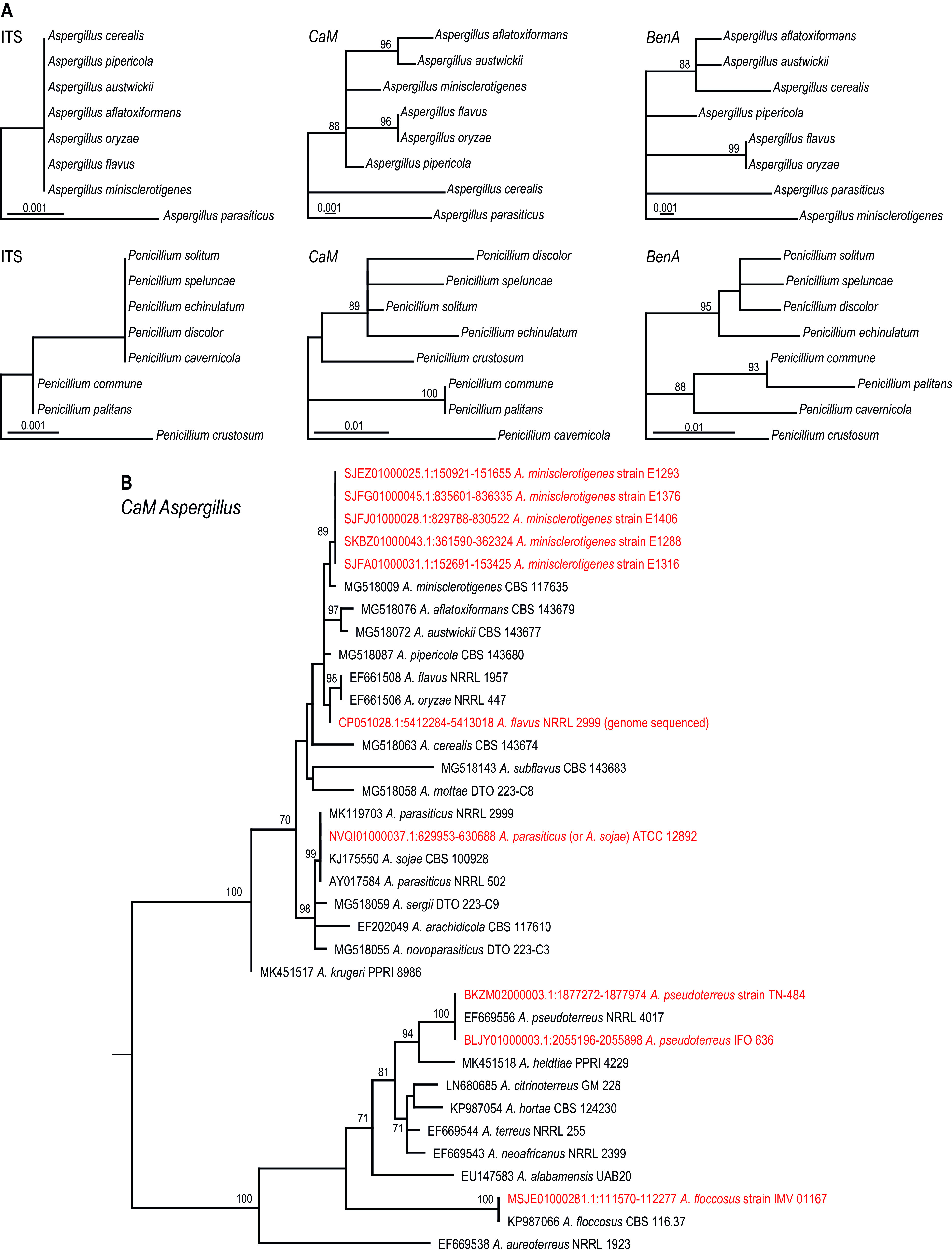
(A) Phylogenetic analysis showing the relationships of Aspergillus flavus and related species (top row) and Penicillium solitum and related species (bottom row). The phylogram based on the ITS barcode has low resolution, and greater variability is present in the BenA and CaM trees. (B) Phylogram based on CaM gene sequences of Aspergillus section Flavi and Terrei reference strains (9) and incorrectly identified genome-sequenced strains (indicated in red). The GenBank accession numbers are listed before the species name and strain numbers are listed afterward.
Correct identification also depends on the quality of the database. GenBank is generally used for strain identification, and users should be aware that sequences of incorrectly identified strains occur in GenBank (12, 13), leading to incorrect identifications. To date, there are no cutoff scores for species identification, and the variability differs according to marker and species (14). In the future, when more fungal genome sequences become available, average nucleotide identity (ANI) analyses could be applied to determine species boundaries and to confirm identifications, a method that is already used in bacteriology, where more genome sequences are available (15). In cases in which BLAST analysis results are not conclusive, it is recommended to construct a phylogenetic tree to determine the closest relatives of the strain. Lists of accepted Eurotialean species that include reference partial β-tubulin and calmodulin gene sequence data have been constructed and are a helpful aid for researchers to create phylogenetic trees based on reference sequence data and to obtain a correct identification (9). Similar lists have been prepared for other genera (16–18).
The number of genome sequencing projects has increased tremendously in recent years. It came to our attention that there is a continuing problem regarding incorrect identification and the unavailability of genome-sequenced strains. An overview of the genome-sequenced strains published in Microbiology Resource Announcements (including Genome Announcements) that belong to the order Eurotiales (Aspergillus, Penicillium, and related genera) was generated (19). Fifty-eight articles (from February 2013 to 31 March 2021), covering 141 Eurotialean strains, were published in Microbiology Resource Announcements and Genome Announcements (19). Of those strains, 18% (n = 26) were inadequately or wrongly identified (see Table 1 and the example of Aspergillus sections Flavi and Terrei in Fig. 1B), and 75% (n = 106) were not deposited in a public culture collection. These misidentifications can lead to incorrect conclusions. For example, the genome sequence of ATCC 48735, an environmental strain of Penicillium capsulatum (20), is actually that of Penicillium canescens. The genome data were later used in a comparative genomic analysis with a clinical P. capsulatum strain (21). Another example is the genome sequences of two P. solitum strains (22, 23) that are reported to be used for deepening the understanding of the genetic differences in, for example, mycotoxin production. Reidentification based on the available genome data showed that those P. solitum strains were actually Penicillium polonicum (RS1) and Penicillium crustosum (NJ1). More recently, the genomes of a set of 16 Aspergillus flavus and Aspergillus parasiticus strains were sequenced (24). These strains were selected based on the genetic fingerprints of 25 insertion/deletion markers within the aflatoxin biosynthesis pathway. Analyses of these markers will give insight into the potential of these strains to produce aflatoxin but are not recommended for species identification (11). Reidentification of the strains using the calmodulin barcode gene extracted from the genome sequence showed that 5 strains are actually A. minisclerotigenes (listed as A. flavus S-type, referring to the small-sized sclerotia the strain produces) (25). In 2008, Pildain et al. (26) showed that production of small-sized sclerotia is not a characteristic that can be attributed to one species but multiple A. flavus-like species (Aspergillus series Flavi), including A. minisclerotigenes, can produce these. This example illustrates that, besides a rigid sequence comparison, it is also important to use the most up-to-date taxonomic schemes and insights. The focus of our letter was the order Eurotiales, but similar issues may exist for other groups of fungi as well. For example, Cladosporium sp. strain TM138 (27) can be identified as Cladosporium halotolerans (based on partial tef1 and actin gene sequences) and Aureobasidium pullulans var. aubasidani (28) as Aureobasidium pullulans (based on ITS and partial RNA polymerase II second largest subunit sequence data).
TABLE 1.
Overview of inaccurately and inadequately genome-sequenced Eurotiales strains published in Genome Announcements and Microbiology Resource Announcements between February 2013 and 31 March 2021
| Strain | Reported identity | Correct identity | Remarks | Reference |
|---|---|---|---|---|
| IFM 58123 | Aspergillus awamori | Aspergillus welwitschiae | Incorrect identification | 31 |
| Strain E1288 | Aspergillus flavus | Aspergillus minisclerotigenes | Incorrect identification | 24 |
| Strain E1293 | Aspergillus flavus | Aspergillus minisclerotigenes | Incorrect identification | 24 |
| Strain E1316 | Aspergillus flavus | Aspergillus minisclerotigenes | Incorrect identification | 24 |
| Strain E1376 | Aspergillus flavus | Aspergillus minisclerotigenes | Incorrect identification | 24 |
| Strain E1406 | Aspergillus flavus | Aspergillus minisclerotigenes | Incorrect identification | 24 |
| NRRL 5109 | Aspergillus neoellipticus | Aspergillus fumigatus | Incorrect identification | 32 |
| Strain An76 | Aspergillus niger | Aspergillus tubingensis | Incorrect identification | 33 |
| ATCC 12892 | Aspergillus oryzae | Aspergillus parasiticus (or Aspergillus sojae) | Incorrect identification | 34 |
| NRRL 2999 | Aspergillus parasiticus | Aspergillus flavus | Original strain differs from genome-sequenced strain | 35 |
| Strain TN-484 | Aspergillus terreus | Aspergillus pseudoterreus | Incorrect identification | 36 |
| Strain IMV 01167 | Aspergillus terreus | Aspergillus floccosus | Incorrect identification | 37 |
| IFO 6365 | Aspergillus terreus | Aspergillus pseudoterreus | Incorrect identification | 38 |
| Strain BYSS01 | Byssochlamys sp. | Monascus floridanus | Incorrect identification | 39 |
| Strain AF001 | Byssochlamys sp. | Paecilomyces dactylethromorphus | Incorrect identification | 40 |
| Strain no. 5 (= NBRC 109023) | Byssochlamys spectabilis/Paecilomyces variotii | Paecilomyces formosus | Incorrect identification | 41 |
| Strain FENG | Paecilomyces hepiali | Samsoniella sp. (Cordycipitaceae) | Incorrect identification | 42 |
| ATCC 48735 | Penicillium capsulatum | Penicillium canescens | Incorrect identification | 20 |
| Strain P2niaD18 | Penicillium chrysogenum | Penicillium rubens | Incorrect identification | 43 |
| ATCC 18224 | Penicillium marneffei | Talaromyces marneffei | Incorrect identification | 44 |
| Strain 113 | Penicillium sclerotiorum | Penicillium maximae | Incorrect identification | 45 |
| Strain NJ1 | Penicillium solitum | Penicillium crustosum | Incorrect identification | 22 |
| Strain RS1 | Penicillium solitum | Penicillium polonicum | Incorrect identification | 23 |
| Strain SPG-F1 | Penicillium sp. | Penicillium solitum | Inadequate identification | 46 |
| Strain SPG-F15 | Penicillium sp. | Penicillium commune (or Penicillium camemberti, depending on colony morphology) | Inadequate identification | 46 |
| Strain Y-94 (= CBS 136886) | Talaromyces cellulolyticus | Talaromyces pinophilus | Incorrect identification | 47 |
Finally, we would like to highlight that species and genus names can change due to new taxonomic insights. However, old names remain in the literature and, for scientists who are unaware of these taxonomic changes, literature with old taxonomic names might be overlooked or misinterpreted. For example, the genome-sequenced strain Trichoderma harzianum T6776 was correctly identified in 2015, but this strain is identified as Trichoderma afroharzianum using the current taxonomic classification (18). Similarly, Talaromyces marneffei was originally described in Penicillium (as Penicillium marneffei), and both names can occur in the literature (29).
Here, we want to increase awareness among scientists to use up-to-date taxonomic schemes in order to avoid incorrect identification and to ensure that a strain is available for the scientific community. We recommend the following steps before publication of genome sequences in the public domain. (i) Perform an identification using the latest taxonomic insights. If needed, contact a taxonomist who can advise regarding the current identity of the strain. (ii) Compare the identity of the original strain with the genome-sequenced strain. For example, NRRL 2999 was originally an Aspergillus parasiticus strain but is A. flavus based on the genome sequence (19, 30). In addition to the strain identification before genome sequencing, it is recommended to extract relevant gene regions from the genome obtained to confirm a correct identification. (iii) The strains should be deposited in at least one, but preferably two or more, recognized, public culture collections (from two countries). This would guarantee that the strain is (easily) accessible for other researchers and for future research purposes. (iv) If the project involves sequencing a representative of a species, make sure that the selected strain is typical of the species. In this case, it is important to study the phenotype of the strains. Type strains (and other [old] strains in culture collections) are not always the best choice, because these strains might have been preserved over a long time and could be deteriorated.
Data availability.
The data that support the findings of this study are openly available in Figshare (https://doi.org/10.6084/m9.figshare.c.5360423.v1) (19).
Footnotes
For the author replies, see https://doi.org/10.1128/mra.00458-21, https://doi.org/10.1128/mra.00460-21, https://doi.org/10.1128/mra.00464-21, https://doi.org/10.1128/mra.00466-21, https://doi.org/10.1128/mra.00469-21, https://doi.org/10.1128/mra.00472-21, https://doi.org/10.1128/mra.00473-21, https://doi.org/10.1128/mra.00475-21, and https://doi.org/10.1128/mra.00784-21. The authors of genomeA01162-13, genomeA01559-14, genomeA00014-15, genomeA00307-15, genomeA01700-15, genomeA00363-16, genomeA00606-16, genomeA00251-18, MRA01453-18, and MRA01613-18 elected not to submit a formal reply.
Contributor Information
Jos Houbraken, Email: j.houbraken@wi.knaw.nl.
Antonis Rokas, Vanderbilt University.
REFERENCES
- 1.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houbraken J, Frisvad JC, Seifert KA, Overy DP, Tuthill DM, Valdez JG, Samson RA. 2012. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 29:78–100. doi: 10.3767/003158512X660571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen AJ, Hubka V, Frisvad JC, Visagie CM, Houbraken J, Meijer M, Varga J, Demirel R, Jurjević Ž, Kubátová A, Sklenář F, Zhou YG, Samson RA. 2017. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium) and its occurrence in indoor environments and food. Stud Mycol 88:37–135. doi: 10.1016/j.simyco.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seifert KA, Samson RA, Dewaard JR, Houbraken J, Levesque CA, Moncalvo JM, Louis-Seize G, Hebert PD. 2007. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci USA 104:3901–3906. doi: 10.1073/pnas.0611691104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell K, Ward TJ, Robert VARG, Crous PW, Geiser DM, Kang S. 2015. DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43:583–595. doi: 10.1007/s12600-015-0484-z. [DOI] [Google Scholar]
- 6.Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, Zalar P, de Hoog GS, Crous PW. 2007. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol 58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stielow JB, Lévesque CA, Seifert KA, Meyer W, Iriny L, Smits D, Renfurm R, Verkley GJM, Groenewald M, Chaduli D, Lomascolo A, Welti S, Lesage-Meessen L, Favel A, Al-Hatmi AMS, Damm U, Yilmaz N, Houbraken J, Lombard L, Quaedvlieg W, Binder M, Vaas LAI, Vu D, Yurkov A, Begerow D, Roehl O, Guerreiro M, Fonseca A, Samerpitak K, van Diepeningen AD, Dolatabadi S, Moreno LF, Casaregola S, Mallet S, Jacques N, Roscini L, Egidi E, Bizet C, Garcia-Hermoso D, Martín MP, Deng S, Groenewald JZ, Boekhout T, de Beer ZW, Barnes I, Duong TA, Wingfield MJ, de Hoog GS, Crous PW, Lewis CT, Hambleton S, Moussa TA, Al-Zahrani HS, Almaghrabi OA, Louis-Seize G, Assabgui R, McCormick W, Omer G, Dukik K, Cardinali G, Eberhardt U, de Vries M, Robert V. 2015. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35:242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CH, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. 2014. Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houbraken J, Kocsubé S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Bensch K, Samson RA, Frisvad JC. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol 95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yilmaz N, Visagie CM, Houbraken J, Frisvad JC, Samson RA. 2014. Polyphasic taxonomy of the genus Talaromyces. Stud Mycol 78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsube S, Szigeti G, Yaguchi T, Frisvad JC. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH, Koljalg U. 2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One 1:e59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofstetter V, Buyck B, Eyssartier G, Schnee S, Gindro K. 2019. The unbearable lightness of sequenced-based identification. Fungal Divers 96:243–284. doi: 10.1007/s13225-019-00428-3. [DOI] [Google Scholar]
- 14.Hubka V, Barrs V, Dudová Z, Sklenář F, Kubátová A, Matsuzawa T, Yaguchi T, Horie Y, Nováková A, Frisvad JC, Talbot JJ, Kolařík M. 2018. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 41:142–174. doi: 10.3767/persoonia.2018.41.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M. 2018. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392. doi: 10.1099/ijsem.0.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marin-Felix Y, Groenewald JZ, Cai L, Chen Q, Marincowitz S, Barnes I, Bensch K, Braun U, Camporesi E, Damm U, de Beer ZW, Dissanayake A, Edwards J, Giraldo A, Hernandez-Restrepo M, Hyde KD, Jayawardena RS, Lombard L, Luangsa-Ard J, McTaggart AR, Rossman AY, Sandoval-Denis M, Shen M, Shivas RG, Tan YP, van der Linde EJ, Wingfield MJ, Wood AR, Zhang JQ, Zhang Y, Crous PW. 2017. Genera of phytopathogenic fungi: GOPHY 1. Stud Mycol 86:99–216. doi: 10.1016/j.simyco.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin-Felix Y, Hernández-Restrepo M, Iturrieta-González I, García D, Gené J, Groenewald JZ, Cai L, Chen Q, Quaedvlieg W, Schumacher RK, Taylor PWJ, Ambers C, Bonthond G, Edwards J, Krueger-Hadfield SA, Luangsa-Ard JJ, Morton L, Moslemi A, Sandoval-Denis M, Tan YP, Thangavel R, Vaghefi N, Cheewangkoon R, Crous PW. 2019. Genera of phytopathogenic fungi: GOPHY 3. Stud Mycol 94:1–124. doi: 10.1016/j.simyco.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai F, Druzhinina IS. 2021. In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Divers 107:1–69. doi: 10.1007/s13225-020-00464-4. [DOI] [Google Scholar]
- 19.Houbraken J. 2020. Phylograms and datasets used for the identification of genome sequenced strains published in the journals Genome Announcements and Microbiology Resource Announcement. doi: 10.6084/m9.figshare.c.5360423.v1. [DOI]
- 20.Yang Y, Chen M, Li Z, Al-Hatmi AMS, Ye Q, Chen H, Wang S, Liao W, Wang J. 2015. Genome sequence of Penicillium capsulatum strain ATCC 48735, a rare Penicillium species used in paper manufactories but that recently caused invasive Infection. Genome Announc 3:e00307-15. doi: 10.1128/genomeA.00307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Chen M, Li Z, Al-Hatmi AMS, de Hoog S, Pan W, Ye Q, Bo X, Li Z, Wang S, Wang J, Chen H, Liao W. 2016. Genome sequencing and comparative genomics analysis revealed pathogenic potential in Penicillium capsulatum as a novel fungal pathogen belonging to Eurotiales. Front Microbiol 7:1541. doi: 10.3389/fmicb.2016.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin G, Zhang Y, Pennerman KK, Hua SST, Yu J, Guo A, Liu Z, Bennett JW. 2016. Draft genome sequence of the fungus Penicillium solitum NJ1. Genome Announc 4:e01176-16. doi: 10.1128/genomeA.01176-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Wu G, Jurick WM, Gaskins VL, Yin Y, Yin G, Bennett JW, Shelton DR. 2016. Genome sequence of Penicillium solitum RS1, which causes postharvest apple decay. Genome Announc 4:e00363-16. doi: 10.1128/genomeA.00363-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arias RS, Mohammed A, Orner VA, Faustinelli PC, Lamb MC, Sobolev VS. 2020. Sixteen draft genome sequences representing the genetic diversity of Aspergillus flavus and Aspergillus parasiticus colonizing peanut seeds in Ethiopia. Microbiol Resour Announc 9:e00591-20. doi: 10.1128/MRA.00591-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frisvad JC, Hubka V, Ezekiel CN, Hong S-B, Nováková A, Chen AJ, Arzanlou M, Larsen TO, Sklenář F, Mahakarnchanakul W, Samson RA, Houbraken J. 2019. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol 93:1–63. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pildain MB, Frisvad JC, Vaamonde G, Cabral D, Varga J, Samson RA. 2008. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int J Syst Evol Microbiol 58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- 27.Gioti A, Siaperas R, Nikolaivits E, Le Goff G, Ouazzani J, Kotoulas G, Topakas E. 2020. Draft genome sequence of a Cladosporium species isolated from the mesophotic ascidian Didemnum maculosum. Microbiol Resour Announc 9:e00311-20. doi: 10.1128/MRA.00311-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vignolle GA, Mach RL, Mach-Aigner AR, Derntl C. 2021. Genome sequence of the black yeast-like strain Aureobasidium pullulans var. aubasidani CBS 100524. Microbiol Resour Announc 10:e01293-20. doi: 10.1128/MRA.01293-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol 70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang P-K. 2021. Authentication of Aspergillus parasiticus strains in the genome database of the National Center for Biotechnology Information. BMC Res Notes 14:111. doi: 10.1186/s13104-021-05527-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu M, Kusuya Y, Alimu Y, Bian C, Takahashi H, Yaguchi T. 2019. Draft genome sequence of Aspergillus awamori IFM 58123NT. Microbiol Resour Announc 8:e01453-18. doi: 10.1128/MRA.01453-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Hsiang T, Li Q, Wang L, Yu Z. 2018. Draft genome sequence of NRRL 5109, an ex-type isolate of Aspergillus neoellipticus. Microbiol Resour Announc 7:e01262-18. doi: 10.1128/MRA.01262-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong W, Cheng Z, Zhang H, Liu L, Gao P, Wang L. 2016. Draft genome sequence of Aspergillus niger strain An76. Genome Announc 4:e01700-15. doi: 10.1128/genomeA.01700-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng S, Pomraning KR, Bohutskyi P, Magnuson JK. 2018. Draft genome sequence of Aspergillus oryzae ATCC 12892. Genome Announc 6:e00251-18. doi: 10.1128/genomeA.00251-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fountain JC, Clevenger JP, Nadon B, Wang H, Abbas HK, Kemerait RC, Scully BT, Vaughn JN, Guo B. 2020. Draft genome sequences of one Aspergillus parasiticus isolate and nine Aspergillus flavus isolates with varying stress tolerance and aflatoxin production. Microbiol Resour Announc 9:e00478-20. doi: 10.1128/MRA.00478-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanamasa S, Minami T, Okabe M, Park EY, Fujimoto T, Takahashi A, Murase M, Fukuyoshi S, Oda A, Satou K, Takahashi H. 2019. Draft genome sequence of Aspergillus terreus high-itaconic-acid-productivity mutant TN-484. Microbiol Resour Announc 8:e01170-19. doi: 10.1128/MRA.01170-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh NK, Blachowicz A, Romsdahl J, Wang C, Torok T, Venkateswaran K. 2017. Draft genome sequences of several fungal strains selected for exposure to microgravity at the International Space Station. Genome Announc 5:e01602-16. doi: 10.1128/genomeA.01602-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi H, Minami T, Okabe M, Park EY, Fujimoto T, Takahashi A, Murase M, Fukuyoshi S, Satou K, Kanamasa S. 2020. Draft genome sequence of the Aspergillus terreus high-itaconic-acid-productivity strain IFO6365. Microbiol Resour Announc 9:e00080-20. doi: 10.1128/MRA.00080-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radwan O, Gunasekera TS, Ruiz ON. 2018. Draft genome sequence of Byssochlamys sp. isolate BYSS01, a filamentous fungus adapted to the fuel environment. Genome Announc 6:e00164-18. doi: 10.1128/genomeA.00164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamps BW, Andrade OC, Lyon WJ, Floyd JG, Nunn HS, Bojanowski CL, Crookes-Goodson WJ, Stevenson BS. 2018. Genome sequence of a Byssochlamys sp. strain isolated from fouled B20 biodiesel. Genome Announc 6:e00085-18. doi: 10.1128/genomeA.00085-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oka T, Ekino K, Fukuda K, Nomura Y. 2014. Draft genome sequence of the formaldehyde-resistant fungus Byssochlamys spectabilis No. 5 (anamorph Paecilomyces variotii no. 5) (NBRC109023). Genome Announc 2:e01162-13. doi: 10.1128/genomeA.01162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y, Wang W, Wang L, Pang F, Guo L, Song L, Liu G, Feng C. 2016. Draft genome sequence of Paecilomyces hepiali isolated from Cordyceps sinensis. Genome Announc 4:e00606-16. doi: 10.1128/genomeA.00606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Specht T, Dahlmann TA, Zadra I, Kürnsteiner H, Kück U. 2014. Complete sequencing and chromosome-scale genome assembly of the industrial progenitor strain P2niaD18 from the penicillin producer Penicillium chrysogenum. Genome Announc 2:e00577-14. doi: 10.1128/genomeA.00577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nierman WC, Fedorova-Abrams ND, Andrianopoulos A. 2015. Genome sequence of the AIDS-associated pathogen Penicillium marneffei (ATCC18224) and its near taxonomic relative Talaromyces stipitatus (ATCC10500). Genome Announc 3:e01559-14. doi: 10.1128/genomeA.01559-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin G, Zhang Y, Pennerman KK, Hua SST, Huang Q, Guo A, Liu Z, Bennett JW. 2016. Genome sequencing and analysis of the filamentous fungus Penicillium sclerotiorum 113, isolated after Hurricane Sandy. Genome Announc 4:e01153-16. doi: 10.1128/genomeA.01153-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobol MS, Hoshino T, Futagami T, Inagaki F, Kiel Reese B. 2019. Draft genome sequences of Penicillium spp. from deeply buried oligotrophic marine sediments. Microbiol Resour Announc 8:e01613-18. doi: 10.1128/MRA.01613-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujii T, Koike H, Sawayama S, Yano S, Inoue H. 2015. Draft genome sequence of Talaromyces cellulolyticus strain Y-94, a source of lignocellulosic biomass-degrading enzymes. Genome Announc 3:e00014-15. doi: 10.1128/genomeA.00014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in Figshare (https://doi.org/10.6084/m9.figshare.c.5360423.v1) (19).


