Purpose of review
Translation of genetic information encoded within mRNA molecules by ribosomes into proteins is a key part of the central dogma of molecular biology. Despite the central position of the ribosome in the translation of proteins, and considering the major proteomic changes that occur in the joint during osteoarthritis development and progression, the ribosome has received very limited attention as driver of osteoarthritis pathogenesis.
Recent findings
We provide an overview of the limited literature regarding this developing topic for the osteoarthritis field. Recent key findings that connect ribosome biogenesis and activity with osteoarthritis include: ribosomal RNA transcription, processing and maturation, ribosomal protein expression, protein translation capacity and preferential translation.
Summary
The ribosome as the central cellular protein synthesis hub is largely neglected in osteoarthritis research. Findings included in this review reveal that in osteoarthritis, ribosome aberrations have been found from early-stage ribosome biogenesis, through ribosome build-up and maturation, up to preferential translation. Classically, osteoarthritis has been explained as an imbalance between joint tissue anabolism and catabolism. We postulate that osteoarthritis can be interpreted as an acquired ribosomopathy. This hypothesis fine-tunes the dogmatic anabolism/katabolism point-of-view, and may provide novel molecular opportunities for the development of osteoarthritis disease-modifying treatments.
Keywords: osteoarthritis, protein translation, ribosome, rRNA, snoRNA
INTRODUCTION
The dogmatic long-standing view on the molecular pathobiology of osteoarthritis is that of an imbalance between anabolism and catabolism of tissues and cells of the joint [1]. From a plethora of studies reported over the past decades, it has become overwhelmingly clear that the low turn-over homeostatic balance of a healthy joint becomes compromised in osteoarthritis and a net loss of tissue occurs. This is caused by a compromised balance between anabolic reparative capacity and catabolic degenerative activity, resulting in the destruction of anatomical joint function. A major part of the extracellular matrix of joint tissues and many of the molecules involved in anabolic and catabolic cellular events are proteins. Osteoarthritis-related aberrations in gene and protein expression are widely reported in the literature. However, the central cellular hub that critically catalyses the biosynthesis of these proteins from dedicated gene transcripts; the ribosome, has been largely neglected in osteoarthritis research. In addition to a variety of genetic and epigenetic regulatory mechanisms of protein expression, it has now become clear that regulation of protein expression also takes place at the level of the ribosome itself [2]. Environmental, developmental and pathological conditions are all able to influence the protein translation characteristics of the cellular ribosome pool [3], including ribosome heterogeneity as a mechanism for preferential translation [4]. This has major consequences for total protein translation capacity but also for the synthesis of specific proteins involved in development, homeostasis and pathology.
Box 1.
no caption available
THE RIBOSOME IN OSTEOARTHRITIS
Disrupted ribosome biogenesis in osteoarthritis
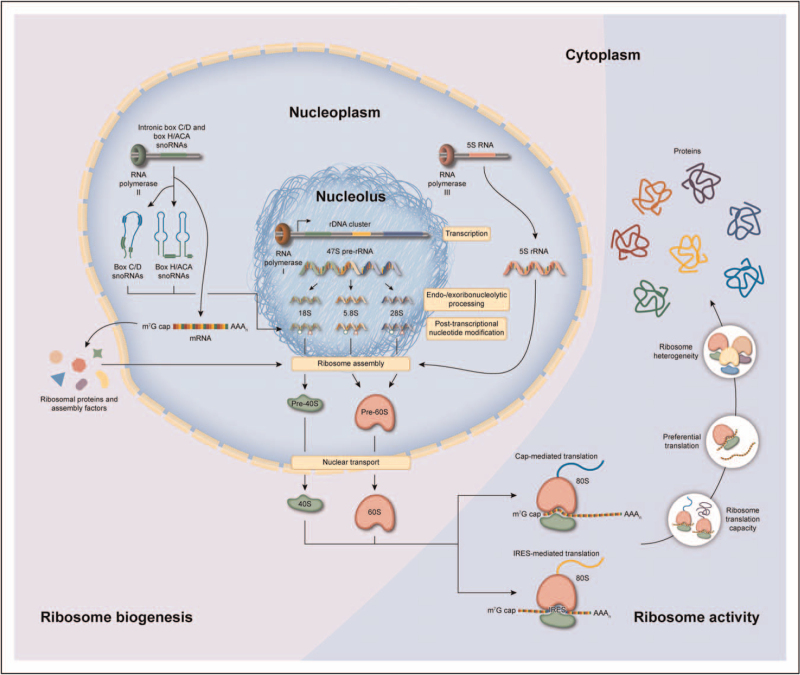
In order to build a ribosome, the cell is equipped with sophisticated mechanisms that support the biogenesis of a ribosome in a highly orchestrated and regulated manner (Fig. 1). Although the pathway of ribosome biogenesis is tightly integrated in the cell, the high complexity of this process makes it relatively vulnerable to aberrations that may result in pathological consequences. In the field of oncology, a wide variety of alterations in ribosome biogenesis are known to promote carcinogenesis [5–7], and a number of anticancer drugs target ribosome biogenesis pathways [8,9]. In osteoarthritis, however, the role of ribosome biogenesis is only beginning to be studied. Human ribosome biogenesis is initiated in the nucleolus and starts with the transcription of the 47S preribosomal RNA (rRNA) precursor by the dedicated RNA polymerase I transcription complex [10] (Fig. 1). There are ±200 copies of the 47S gene spread over five chromosomes, forming the nucleolus organizer regions. Following transcription, the 47S prerRNA transcript is endoribonucleolytically and exoribonucleolytically processed into the 18S, 5.8S and 28S rRNAs, and a number of these processing steps take place co-transcriptionally. The 5S rRNA is transcribed separately in the nucleus by RNA polymerase III and imported in the nucleolus to be integrated into the ribosome biogenesis pathway. In ageing (a major risk factor for osteoarthritis), mouse bone marrow cells have been shown to have an increased rDNA copy number [11]. However, these rDNA copies have increased CpG methylation levels [11,12], probably leading to the observed reduction of rRNA expression [11]. Recent work demonstrated regulation of rRNA expression by a Chromobox 4 (CBX4)-dependent mechanism. This mechanism of nucleolar homeostasis protects against mesenchymal stem cell senescence and against murine osteoarthritis development [13]. In addition, cartilage ageing led to lower expression of the RNA component of mitochondrial RNA processing endoribonuclease (RMRP) in equine chondrocytes [14]. This small nucleolar RNA (snoRNA) is a key factor in the endoribonucleolytic processing of the 47S prerRNA [15] and provides an indication of age-related impairment of chondrocyte rRNA processing. This was confirmed in human chondrocytes, when RMRP snoRNA expression was found to be enriched in hypertrophic chondrocytes in a single-cell sequencing analysis of osteoarthritis cartilage [16▪,17]. Another key factor in the endoribonucleolytic processing of the 47S prerRNA is U3 snoRNA [18]. The expression of U3 snoRNA was reduced in human osteoarthritis cartilage and chondrocytes, as well as in murine joints in which experimental osteoarthritis [destabilization of the medial meniscus (DMM)] was induced [19▪▪]. Osteoarthritis-dependent inhibition of U3 snoRNA transcription was identified as one of the causes of reduced U3 snoRNA expression in osteoarthritis chondrocytes and resulted in a decrease of chondrocyte rRNA levels [19▪▪].
FIGURE 1.
An overview of ribosome biogenesis and activity. 47S prerRNA is transcribed in the nucleolus from rDNA clusters by RNA polymerase I. The 47S precursor is simultaneously processed by endoribonucleases and exoribonucleases and posttranscriptionally modified by snoRNPs. These snoRNPs consist of an enzyme, accessory proteins and box C/D or box H/ACA snoRNAs that guide site-specific 2’-O ribose methylation or pseudouridylation of rRNA nucleotides. These canonical snoRNAs originate from intronic regions of RNA polymerase II-transcribed mRNAs and are liberated by the splicing machinery and subsequent processing. Together with mature rRNA, ribosomal proteins assemble into the small 40S and large 60S ribosomal subunits. This highly coordinated process requires additional assembly factors. After nuclear export, the 40S subunit can form a 43S preinitiation complex together with eukaryotic translation initiation factors that recognize the m7G cap of mRNAs and initiate protein translation after recruitment of the 60S large subunit, resulting in the formation of an active 80S ribosome. In addition to cap-mediated translation, Internal Ribosome Entry Sites (IRESs) can mediate direct recruitment of the ribosome to a translation start site. This process is of paramount importance under cellular stress conditions, where cap-mediated translation is generally inhibited. Ribosome activity is tightly regulated by a multitude of signalling pathways (e.g. AKT/mTOR, TGF/BMP, FGF), and other important factors are energy status and amino acid availability for aminoacyl-tRNA formation. The most rate-limiting step of ribosome activity is translation initiation by eIF4E, which is counteracted by 4E-BP1. In addition, ribosomes can preferentially translate a certain mRNA because of specific (IRES) trans-acting factors. To add to the complexity, the ribosome can generate multiple protein variants from a single mRNA, when more than one or alternative translation initiation sites are present (e.g. in VEGF, MYC and FGF2 mRNAs). Ribosome core protein composition and rRNA posttranscriptional modification levels can vary [4,10], which leads to heterogeneous ribosomes with distinct translational characteristics.
Except for 5S rRNA, all rRNAs are targets of snoRNA-mediated site-specific posttranscriptional modification (PTM) by 2’O-ribose methylation and pseudouridylation by fibrillarin and dyskerin, respectively. A total of 226 of these PTMs have been identified on human rRNAs [20]. A large family of canonical snoRNAs [active as small nucleolar ribonucleoprotein particles (snoRNPs)] ensures the site-directionality of these PTMs [21]. Differential expression of canonical snoRNAs was demonstrated in human ageing and osteoarthritis cartilage [22▪▪], in murine DMM joints [23], and in equine ageing cartilage [14]. Mechanistic analysis of the role of a number of these snoRNAs in chondrocyte biology demonstrated that SNORD26 and SNORD96A are involved in determining the chondrocyte phenotype [22▪▪] and SNORD32A, SNORD33 and SNORD35A in oxidative stress responses [24]. A great number of differentially expressed snoRNAs await further mechanistic studies and their consequences for rRNA PTM and ribosome function in cells from joints tissues need to be dissected. In this respect, our group mapped the rRNA PTM landscape in an in-vitro model for osteoarthritis chondrocytes and identified osteoarthritis-dependent changes in rRNA PTMs with consequences for the modus of ribosome translation initiation [25]. Differential expression of canonical snoRNAs was also detected in synovial fluid of early equine osteoarthritis [26], in the serum of DMM mice and horses [26] and in the serum of an anterior cruciate ligament injury cohort [27]. These studies may provide snoRNA-based biomarkers for musculoskeletal ageing and osteoarthritis development.
In addition to the rRNAs, the ribosome consists of 79 proteins [33 RPS (ribosomal protein small subunit) proteins in the 40S subunit and 46 RPL (ribosomal protein large subunit) proteins in the 60S subunit]. These proteins are imported into the nucleus and depending on the protein species and its position in the ribosome's biomolecular architecture, are assembled in a highly orchestrated sequence, which requires over 100 ancillary proteins to achieve this task [10,28]. This number excludes snoRNP-related proteins. The 40S and 60S ribosomal subunits are then transported to the cytoplasm and undergo several final maturation steps before they are ready to engage in the translation of mRNA into protein. Emerging evidence indicates that in osteoarthritis, the proteinaceous part of the ribosome's architecture may change. In recent single-cell sequencing work, it was demonstrated that increased RPS29 (ribosomal protein S29) expression in chondrocytes is associated with osteoarthritis progression [16▪]. The involvement of ribosomal protein expression in osteoarthritis was also highlighted in a recent single-cell sequencing study that investigated cross-talk between the synovium and cartilage in osteoarthritis [29]. A great number of ribosomal proteins were found to be differentially expressed in cell subpopulations of osteoarthritis synovium and osteoarthritis cartilage from damaged and nondamaged areas [29]. Although their association with functional ribosomes needs to be investigated, these observations at least highlight an interaction between the osteoarthritis disease stage, the chondrocyte phenotype, and the regulation of the expression of core ribosomal proteins. Indeed, recent work from our group demonstrated that in chondrogenesis, the expression of the chondrogenic transcription factor Sox9 regulates the expression of ribosomal proteins as well as proteins involved in the ribosome biogenesis pathway [30▪▪]. The osteoarthritis disease-dependent regulation of expression of core ribosomal subunits hints towards ribosome heterogeneity, which is a level of protein translational control that was recently discovered [31].
Altered ribosome activity in osteoarthritis
The primary function of the ribosome is the translation of mRNAs into proteins (Fig. 1). Multiple studies have gathered compelling evidence of major proteomic changes in fluids, cells and tissues of the osteoarthritis joint [32–39]. For a long time, it was assumed that these changes originate from major mRNA transcriptomic changes only. An additional important level of epitranscriptomic regulation was later introduced by the identification of microRNA networks that control the translation of mRNAs involved in osteoarthritis development [40]. However, it was only recently demonstrated that protein synthesis in osteoarthritis is also regulated at the level of ribosome activity (Fig. 1). It was shown that ribosome protein translation activity was increased in human osteoarthritis chondrocytes, in a rat model for traumatic osteoarthritis, and in an IL-1β-dependent in-vitro chondrocyte model for osteoarthritis [41]. The identified mechanism behind this observation was a mammalian target of rapamycin complex 1 (mTORC1)-mediated inhibition of eIF4E-binding protein 1 (4E-BP1). As the activity of 4E-BP1 is rate-limiting for the activity of eIF4E, the cap-binding protein responsible for cap-dependent protein translation, this indicates that this mechanism primarily involves cap-mediated translation initiation. In concert with involvement of mTORC1 in cartilage homeostasis, mTOR activity was shown to be increased in osteoarthritis but was then linked to cartilage autophagy [42]. The finding that the mTORC1-mediated inhibition of 4E-BP1 precedes cartilage degeneration in rat osteoarthritis knees [43] strongly suggests its involvement in early osteoarthritis. In contrast to an overall increased level of chondrocyte protein translation activity, work from our group demonstrated that protein translation activity was reduced in chondrocytes isolated from end-stage knee osteoarthritis cartilage [19▪▪]. This was accompanied by lower levels of rRNA in osteoarthritis chondrocytes and chondrocytes treated with osteoarthritis synovial fluid. In addition, in a study comparing end-stage osteoarthritis cartilage with normal cartilage, it was found that expression of 4E-BP1 was higher in osteoarthritis cartilage, which is indicative of a reduction of translational activity [44▪]. Fibroblast growth factor (FGF) signalling [45] was described as another mechanism underlying a reduction in chondrocyte protein translation [46]. However, this mechanism was mTOR-independent. In addition, the recent single-cell sequencing work in chondrocytes demonstrated differential expression of translation initiation factors 4E-BP1 [in proliferative chondrocytes (ProCs)] and EIF4A1, EIF4A2, EIF4A3, EIF1 and EIF5 [in homeostatic chondrocytes (HomCs)] [16▪,29]. Interestingly, in an osteoarthritis serum biomarker study, patients with knee osteoarthritis had significantly lower serum levels of 4E-BP1, which was found be positively correlated with osteoarthritis pain intensity [34]. Together, this suggests that the mechanisms and involvement of deregulated ribosome protein translation activity in osteoarthritis chondrocytes are far more complex and depend on the stage of osteoarthritis progression and the chondrocyte phenotype, warranting further studies to dissect its complexity.
Rather than performing protein translation in a textbook manner, it has become clear that there is a large level of translational regulation that drives preferential translation of specific mRNAs [47]. Protein translation initiation can occur via multiple mechanisms and two well described mechanisms are cap-dependent translation and IRES-dependent translation (Fig. 1). Although cap-dependent translation is considered to constitute the majority of translation events, the cell preferentially uses IRES-dependent translation for the synthesis of many of its stress-related proteins. Indeed, ongoing work by our group demonstrates that in chondrocytes TNFα induces protein translation from the FGF1 IRES [48]. In addition, treatment of chondrocytes with TGFβ induced their protein translational activity [44▪,49] but skewed their preferential mode of translation toward cap-dependent translation [49]. The balance between cap-dependent and IRES-dependent translation is amongst others determined by specific snoRNA-mediated PTMs on the rRNA [18] and by expression of IRES-transacting factors (ITAFs). With the mapping of differential expression of snoRNAs [14,22▪▪,23,26,50] and rRNA PTMs [25], as well as high-resolution proteomics [30▪▪,39] in cells from joint tissues as a function of ageing [51,52] and osteoarthritis, it is expected that insight into this level of ribosome translation regulation will further unfold. The existence of other mechanisms of preferential translation in cell types from the joint is only starting to emerge. Recently ribosome profiling coupled to protein mass-spectrometry demonstrated that treatment of chondrocytic cells with IL-1β induced the preferential translation of proteins associated with inflammatory responses and oxidative stress [53▪▪]. IL-1β -induced preferential translation of osteoarthritis-related proteins in chondrocytes was suggested to be mediated by their 5′ untranslated regions [54].
Future perspectives
It is becoming clear that alterations in ribosome biogenesis and ribosome function find their place in the molecular pathobiology of osteoarthritis. However, considering the incredible complexity of ribosome biogenesis and the many mechanisms by which ribosome activity and modus can be influenced, this field of osteoarthritis research is only just emerging. In discovery-driven approaches, we need to further chart the levels and identify the molecules by which ribosome biogenesis and function are being disturbed in osteoarthritis. Cellular stress signalling provoked by environmental factors like growth factors, cytokines, chemokines, damage-associated molecular patterns, senescence, metabolites, mechanosensing mechanisms and the extracellular matrix are all, in one way or the other, involved in osteoarthritis development and its progression. The osteoarthritis-related proteomic changes in the tissues of the joint induced by these pathological stress signalling events can all be candidates for regulation at the translational level by the ribosome. This can either be caused by changes in ribosome biogenesis, total protein translation capacity, or by mechanisms of preferential translation of mRNAs. An interesting connection between ribosome biogenesis and cellular stress is p53. P53 expression is upregulated in senescent and osteoarthritis chondrocytes [55,56]. In addition, p53 is a major regulator of ribosome biogenesis stress and p53 activation shuts down ribosome biogenesis at multiple levels [57]. This can lead to impairment of the total protein translation capacity with consequences for cartilage proteostasis [52]. Another ribosome-related stress factor relevant to chondrocytes is endoplasmic reticulum (ER) stress [58]. ER stress triggers the unfolded protein response (UPR). UPR activation has been demonstrated in osteoarthritis chondrocytes [59,60] and recently it was shown that ER stress and the UPR are specifically involved in the onset of experimental osteoarthritis but not in its progression [61]. Whether alterations in ribosome biogenesis are involved in osteoarthritis chondrocyte ER stress is currently unclear. However, ribosome translation activity can be connected to ER stress and the UPR via inactivation of eukaryotic translation initiation factor (eIF) 2α and inhibition of 80S ribosome assembly [58,62]. Recent evidence demonstrates that translation of catabolic proteins in chondrocytic cells is preferred in osteoarthritis-mimicking environments [53▪▪,54]. The finding that mechanisms of such preferential translation can also include heterogeneity of the cellular ribosome pool is an exciting development that may further unveil how cells in joints tissues can translationally respond to changes in their environment at the protein level (Fig. 1). However, the potential ribosome biogenesis-related and ribosome activity-related consequences for the proteomes of different tissue types from the joint still need to be determined.
CONCLUSION
As outlined above, a slowly increasing amount of experimental evidence highlights that molecular mechanisms involved in ribosome biogenesis and ribosome activity are deregulated in osteoarthritis. The fact that we can find these aberrations from early-stage ribosome biogenesis, through ribosome build-up and maturation, up to preferential translation by the ribosome is fascinating. Underlying genetic factors do not seem to be the cause of these translational deficits. In contrast, ageing and many environmental factors are clearly connected to disturbances in ribosome biogenesis and ribosome activity in general, and also represent main risk factors for the development and progression of osteoarthritis. Classically, osteoarthritis has been explained as a disbalance between joint tissue anabolism and catabolism. Considering the current evidence collected in this article on osteoarthritis-related aberrations in ribosome biogenesis and ribosome function, we therefore hypothesize that osteoarthritis can be molecularly interpreted as an acquired ribosomopathy [63▪]. This hypothesis further fine-tunes the dogmatic anabolism/catabolism point-of-view by adding aberrations in total protein translation capacity and preferential translation to the molecular pathogenesis of osteoarthritis. This may provide novel molecular opportunities for the development of osteoarthritis disease-modifying treatments.
Acknowledgements
The authors thank Bobby Li (Sketchy Pipette) for graphical design support for the figure.
Financial support and sponsorship
T.J.M.W. is funded through a grant from Stichting de Weijerhorst (Bewegen zonder Pijn) and grants from the Dutch Arthritis Foundation (LLP14 and 17-2-401). M.J.P. is funded through a Wellcome Trust Clinical Intermediate Fellowship (grant 107471/Z/15/Z).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Guus G.H. van den Akker and Marjolein M.J. Caron are shared first authors.
Mandy J. Peffers and Tim J.M. Welting are shared last authors.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Mueller MB, Tuan RS. Anabolic/catabolic balance in pathogenesis of osteoarthritis: identifying molecular targets. PM R 2011; 3: (6 Suppl 1): S3–11. [DOI] [PubMed] [Google Scholar]
- 2.Dalla Venezia N, Vincent A, Marcel V, et al. Emerging role of eukaryote ribosomes in translational control. Int J Mol Sci 2019; 20:1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Z, Barna M. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu Rev Cell Dev Biol 2015; 31:31–54. [DOI] [PubMed] [Google Scholar]
- 4.Shi Z, Fujii K, Kovary KM, et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 2017; 67:71.e7–83.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Wang J. Ribosome heterogeneity in stem cells and development. J Cell Biol 2020; 219:e202001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piskol R, de Sousa EMF. Colon cancer heterogeneity: welcome to the RiboZone. Cell Stem Cell 2020; 26:797–799. [DOI] [PubMed] [Google Scholar]
- 7.Turi Z, Lacey M, Mistrik M, Moudry P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany, NY) 2019; 11:2512–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catez F, Dalla Venezia N, Marcel V, et al. Ribosome biogenesis: an emerging druggable pathway for cancer therapeutics. Biochem Pharmacol 2019; 159:74–81. [DOI] [PubMed] [Google Scholar]
- 9.Ford D. Ribosomal heterogeneity - a new inroad for pharmacological innovation. Biochem Pharmacol 2020; 175:113874. [DOI] [PubMed] [Google Scholar]
- 10.Bohnsack KE, Bohnsack MT. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J 2019; 38:e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watada E, Li S, Hori Y, et al. Age-dependent ribosomal DNA variations in mice. Mol Cell Biol 2020; 40:e00368-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Lemos B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res 2019; 29:325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren X, Hu B, Song M, et al. Maintenance of Nucleolar Homeostasis by CBX4 Alleviates Senescence and Osteoarthritis. Cell Rep 2019; 26:3643.e7–3656.e7. [DOI] [PubMed] [Google Scholar]
- 14.Peffers M, Liu X, Clegg P. Transcriptomic signatures in cartilage ageing. Arthritis Res Ther 2013; 15:R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfarb KC, Cech TR. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev 2017; 31:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16▪.Ji Q, Zheng Y, Zhang G, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis 2019; 78:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study represents the first single cell sequencing analysis of osteoarthritic human cartilage. Seven chondrocyte sub-populations were defined by clustering analyses. RPS29 protein expression could distinguish osteoarthritis from normal cartilage.
- 17.Steinbusch MMF, Caron MMJ, Surtel DAM, et al. Expression of RMRP RNA is regulated in chondrocyte hypertrophy and determines chondrogenic differentiation. Sci Rep 2017; 7:6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 2015; 22:11–19. [DOI] [PubMed] [Google Scholar]
- 19▪▪.Ripmeester EGJ, Caron MMJ, van den Akker GGH, et al. Impaired chondrocyte U3 snoRNA expression in osteoarthritis impacts the chondrocyte protein translation apparatus. Sci Rep 2020; 10:13426. [DOI] [PMC free article] [PubMed] [Google Scholar]; U3 snoRNA, a crucial mediator of rRNA processing, is decreased in end-stage osteoarthritis and be rescued with BMP7 stimulation. Gain and loss-of-function analysis revealed that U3 snoRNA modulates the translational activity of chondrocytes.
- 20.Motorin Y, Quinternet M, Rhalloussi W, Marchand V. Constitutive and variable 2’-O-methylation (Nm) in human ribosomal RNA. RNA Biol 2021; 1–10. doi: 10.1080/15476286.2021.1974750. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bratkovic T, Bozic J, Rogelj B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res 2020; 48:1627–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22▪▪.Peffers MJ, Chabronova A, Balaskas P, et al. SnoRNA signatures in cartilage ageing and osteoarthritis. Sci Rep 2020; 10:10641. [DOI] [PMC free article] [PubMed] [Google Scholar]; Human healthy and osteoarthritis cartilage was profiled for snoRNA expression. Functional follow-up of SNORD26/96 by gain and loss-of-function experiments revealed that pertrubation of these snoRNAs induces osteoarthritis-like changes.
- 23.Steinbusch MM, Fang Y, Milner PI, et al. Serum snoRNAs as biomarkers for joint ageing and post traumatic osteoarthritis. Sci Rep 2017; 7:43558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimlott Z, Hontoir F, Ashraf Kharaz Y, et al. Small nucleolar RNAs as mediators of oxidative stress in cross species cartilage and osteoarthritis. Osteoarthritis Cartilage 2020; 28: (Suppl 1): S342. [Google Scholar]
- 25.Chabronova A, Akker G, Ripmeester E, et al. Evidence of specialized ribosomes in osteoarthritic chondrocytes. Osteoarthritis Cartilage 2021; 29: (Suppl 1): 2. [Google Scholar]
- 26.Castanheira C, Balaskas P, Falls C, et al. Equine synovial fluid small noncoding RNA signatures in early osteoarthritis. BMC Vet Res 2021; 17:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Yang M, Marks P, et al. Serum noncoding RNAs as biomarkers for osteoarthritis progression after ACL injury. Osteoarthritis Cartilage 2012; 20:1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peculis BA. Ribosome biogenesis: ribosomal RNA synthesis as a package deal. Curr Biol 2002; 12:R623–R624. [DOI] [PubMed] [Google Scholar]
- 29.Chou CH, Jain V, Gibson J, et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep 2020; 10:10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30▪▪.Caron MMJ, Eveque M, Cillero-Pastor B, et al. Sox9 determines translational capacity during early chondrogenic differentiation of ATDC5 cells by regulating expression of ribosome biogenesis factors and ribosomal proteins. Front Cell Dev Biol 2021; 9:686096. [DOI] [PMC free article] [PubMed] [Google Scholar]; An important functional link between the master regulator of chondrogenesis Sox9 and ribosome modus and activity was established in ATDC5 murine progenitor cells.
- 31.Genuth NR, Barna M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol Cell 2018; 71:364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Folkesson E, Turkiewicz A, Ali N, et al. Proteomic comparison of osteoarthritic and reference human menisci using data-independent acquisition mass spectrometry. Osteoarthritis Cartilage 2020; 28:1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folkesson E, Turkiewicz A, Englund M, Onnerfjord P. Differential protein expression in human knee articular cartilage and medial meniscus using two different proteomic methods: a pilot analysis. BMC Musculoskelet Disord 2018; 19:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giordano R, Petersen KK, Andersen HH, et al. Serum inflammatory markers in patients with knee osteoarthritis: a proteomic approach. Clin J Pain 2020; 36:229–237. [DOI] [PubMed] [Google Scholar]
- 35.Hsueh MF, Khabut A, Kjellstrom S, et al. Elucidating the molecular composition of cartilage by proteomics. J Proteome Res 2016; 15:374–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lourido L, Balboa-Barreiro V, Ruiz-Romero C, et al. A clinical model including protein biomarkers predicts radiographic knee osteoarthritis: a prospective study using data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2021; 29:1147–1154. [DOI] [PubMed] [Google Scholar]
- 37.Mobasheri A. Applications of proteomics to osteoarthritis, a musculoskeletal disease characterized by aging. Front Physiol 2011; 2:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shabestari M, Shabestari YR, Landin MA, et al. Altered protein levels in bone marrow lesions of hip osteoarthritis: analysis by proteomics and multiplex immunoassays. Int J Rheum Dis 2020; 23:788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timur UT, Jahr H, Anderson J, et al. Identification of tissue-dependent proteins in knee OA synovial fluid. Osteoarthritis Cartilage 2021; 29:124–133. [DOI] [PubMed] [Google Scholar]
- 40.Swingler TE, Niu L, Smith P, et al. The function of microRNAs in cartilage and osteoarthritis. Clin Exp Rheumatol 2019; 37 Suppl 120:40–47. [PubMed] [Google Scholar]
- 41.Katsara O, Attur M, Ruoff R, et al. Increased activity of the chondrocyte translational apparatus accompanies osteoarthritic changes in human and rodent knee cartilage. Arthritis Rheumatol 2017; 69:586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Vasheghani F, Li YH, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis 2015; 74:1432–1440. [DOI] [PubMed] [Google Scholar]
- 43.Katsara O, Kolupaeva V. mTOR-mediated inactivation of 4E-BP1, an inhibitor of translation, precedes cartilage degeneration in rat osteoarthritic knees. J Orthop Res 2018; 36:2728–2735. [DOI] [PubMed] [Google Scholar]
- 44▪.Hwang HS, Lee MH, Kim HA. TGF-beta1-induced expression of collagen type II and ACAN is regulated by 4E-BP1, a repressor of translation. FASEB J 2020; 34:9531–9546. [DOI] [PubMed] [Google Scholar]; 4E-BP1 is the foremost inhibitor of protein translation, its expression is increased in osteoarthritis cartilage and TGF-beta1 can reverse this. Notably, 4E-BP1 is associated with preferential translation of SMADs and inhibitory SMADs.
- 45.Xie Y, Zinkle A, Chen L, Mohammadi M. Fibroblast growth factor signalling in osteoarthritis and cartilage repair. Nat Rev Rheumatol 2020; 16:547–564. [DOI] [PubMed] [Google Scholar]
- 46.Ruoff R, Katsara O, Kolupaeva V. Cell type-specific control of protein synthesis and proliferation by FGF-dependent signaling to the translation repressor 4E-BP. Proc Natl Acad Sci U S A 2016; 113:7545–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James CC, Smyth JW. Alternative mechanisms of translation initiation: an emerging dynamic regulator of the proteome in health and disease. Life Sci 2018; 212:138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Akker G, Chabronova A, Housmans B, et al. TNF-alpha induces FGF1 ires mediated messenger RNA translation in chondrocytes. Osteoarthritis Cartilage 2021; 29: (Suppl 1): 2. [Google Scholar]
- 49.van den Akker G, Surtel D, Chabronova A, et al. TGF-beta induces ribosome activity, alters ribosome composition and inhibits ires-mediated translation in chondrocytes. Osteoarthritis Cartilage 2020; 28: (Suppl 1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balaskas P, Green JA, Haqqi TM, et al. Small non-coding RNAome of ageing chondrocytes. Int J Mol Sci 2020; 21:5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonskikh Y, Polacek N. Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev 2017; 168:30–36. [DOI] [PubMed] [Google Scholar]
- 52.Steffen KK, Dillin A. A ribosomal perspective on proteostasis and aging. Cell Metab 2016; 23:1004–1012. [DOI] [PubMed] [Google Scholar]
- 53▪▪.McDermott BT, Peffers MJ, McDonagh B, Tew SR. Translational regulation contributes to the secretory response of chondrocytic cells following exposure to interleukin-1beta. J Biol Chem 2019; 294:13027–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]; A unique combination of ribo-sequencing and proteome analyses revealed translational control of the chondrocyte's cytokine response in vitro.
- 54.Kolupaeva V, Katsara O, Attur M. The translational landscape in articular chondrocytes treated with interleukin-1 reveals novel potential players in osteoarthritis. Osteoarthritis Cartilage 2018; 26: (Suppl): 1.29042267 [Google Scholar]
- 55.Ashraf S, Cha BH, Kim JS, et al. Regulation of senescence associated signaling mechanisms in chondrocytes for cartilage tissue regeneration. Osteoarthritis Cartilage 2016; 24:196–205. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto S, Nishiyama T, Hayashi S, et al. Role of p53 in human chondrocyte apoptosis in response to shear strain. Arthritis Rheum 2009; 60:2340–2349. [DOI] [PubMed] [Google Scholar]
- 57.Golomb L, Volarevic S. Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett 2014; 588:2571–2579. [DOI] [PubMed] [Google Scholar]
- 58.Boot-Handford RP, Briggs MD. The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res 2010; 339:197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruiz-Romero C, Carreira V, Rego I, et al. Proteomic analysis of human osteoarthritic chondrocytes reveals protein changes in stress and glycolysis. Proteomics 2008; 8:495–507. [DOI] [PubMed] [Google Scholar]
- 60.Uehara Y, Hirose J, Yamabe S, et al. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthritis Cartilage 2014; 22:1007–1017. [DOI] [PubMed] [Google Scholar]
- 61.Kung LHW, Mullan L, Soul J, et al. Cartilage endoplasmic reticulum stress may influence the onset but not the progression of experimental osteoarthritis. Arthritis Res Ther 2019; 21:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal 2007; 9:2357–2371. [DOI] [PubMed] [Google Scholar]
- 63▪.Venturi G, Montanaro L. How altered ribosome production can cause or contribute to human disease: the spectrum of ribosomopathies. Cells 2020; 9:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review article describes pure and mixed ribosomopathies and proposes the definition of acquired ribosomopathies with a more complex pathobiology.