Abstract
Infection with group A streptococci (GAS) can lead to the development of severe postinfectious sequelae such as rheumatic fever (RF). In Thailand, RF and rheumatic heart disease (RHD) remain important health problems. More than 80% of GAS circulating in this population are non-M antigen typeable by conventional M serotyping methods. In this study, we determine the M protein sequence types of GAS isolates found in northern Thailand. The emm genes from 53 GAS isolates, collected between 1985 and 1995 from individuals with pharyngitis, impetigo, acute RF (ARF), RHD, or meningitis as well as from individuals without infections, were amplified by PCR and sequenced. Thirteen new sequence types that did not show homology to previously published sequences were characterized. Six of these sequence types could be isolated from both skin and throat sites of impetigo and pharyngitis/ARF patients, respectively. In many cases we could not specifically differentiate skin strains or throat strains that could be associated with ARF or acute glomerulonephritis. Antigenic variations in the emm gene of the isolates investigated, compared to published M protein sequences, were predominantly due to point mutations, small deletions, and insertions in the hypervariable region. One group of isolates with homology to M44 exhibited corrected frameshift mutations. A new M type isolated from an RHD patient exhibited nucleotide sequence corresponding to the N terminus of M58 and the C terminus of M25, suggesting that recombination between the two types may have occurred. This study provided epidemiological data relating to GAS endemic to northern Thailand which could be useful for identification of vaccine candidates in a specific region of endemicity.
Infection with group A streptococci (GAS) can lead to diseases ranging from impetigo and pharyngitis to the postinfectious sequelae rheumatic fever (RF), rheumatic heart disease (RHD), and acute glomerulonephritis. The incidence of RF has declined in developed countries since World War II, but in the last decade RF outbreaks have been described in several United States cities and new M antigen types of GAS, previously not associated with RF, have been isolated (7). In Thailand, RF remains an important health problem in children aged 5 to 15 years (17); its prevalence of 0.38 per 1,000 in Thailand (12, 22) is comparable to that in other developing countries in the western Pacific, Africa, and the Americas (22).
The M protein, a cell surface protein, may play an important role in the pathogenesis of disease. More than 80 GAS M types have been identified by serological M typing. However, most GAS isolated from patients and carriers in developing countries such as Thailand (8, 14, 17), aboriginal communities in Australia (5), and Kuwait (9) cannot be classified into M types by conventional M serotyping. DNA sequencing of the M protein gene permits the typing of strains which cannot be serologically classified (2, 10, 20, 21).
In vaccine development, many studies have defined protective epitopes from the N-terminal and C-terminal regions of the M protein (3, 4, 15, 16, 18). However, the vast number of isolates from specific regions of endemicity remain largely uncharacterized, with over 80% of isolates being classified as non-M typeable (5, 8, 17). The non-M-typeable strains in Thailand have not yet been characterized. Identification of predominant M types in this area would facilitate the development of a vaccine targeted to this population. This study examines non-M-typeable GAS isolates from patients and carriers in northern Thailand. The sequence or sequence types of the M protein genes were identified and their relatedness to published M protein sequences was determined. These data provide useful information for epidemiological studies of GAS in Thailand.
Isolation of GAS.
Throat swab and skin lesion swab specimens were obtained from persons living in Chiang Mai, Thailand, with sore throat, acute RF (ARF), RHD, meningitis, or impetigo as well as from individuals without disease. The swab specimens were then cultured on blood agar plates, with incubation at 37°C in an atmosphere with 5% CO2 for 24 to 48 h. The organisms that produced beta-hemolytic colonies were identified as GAS by susceptibility to 0.04 U of bacitracin and agglutinated with group-specific antiserum by the latex agglutination test (bioMerieux, Marcy Létoile, France). M typing was performed by a slide precipitation test with type-specific antiserum (19). All GAS isolates were stored in glycerol storage medium (6) at −40°C until required. Fifty-three non-M-typeable isolates were included in the study.
DNA isolation.
The organisms were streaked out on blood agar plates, and a single colony was used to inoculate 50 ml of Todd-Hewitt broth. After incubation at 37°C overnight, the culture was spun down and the pellet was washed three times with phosphate-buffered saline (pH 7.0), resuspended in 0.5 ml of a lysozyme solution (100 mg/ml), and incubated at 37°C for 1 h. Sodium dodecyl sulfate (200 μl of a 20% solution) and proteinase K (100 μl of a 10-mg/ml solution) were added, and the suspension was incubated at 55°C overnight. One-third volume of a saturated NaCl solution was added, and the mixture was incubated at 4°C for 20 min. The mixture was then centrifuged to sediment the protein, the supernatant was transferred to a new tube, and 95% ethanol (3 volumes) was added to precipitate the DNA. The tube was rocked gently until the DNA flocculated. The DNA was then washed once in 70% ethanol and retrieved with a bent-tip pipette, allowed to air dry for 1 min, resuspended in 0.5 ml of Tris-EDTA buffer (pH 7.8), and stored at 4°C until used.
Primers, PCR, and sequencing analysis.
The forward primer, 5′ CAGTATTCGCTTAGAAAATTAAAA 3′, was derived from leader sequence of the M protein gene (10). The antisense primer, 5′ CCCTTACGGCTTGCTTCTGA 3′, was derived from the C repeat region of the M protein gene, which is conserved in several of the GAS isolates. These primers were also used for cycle sequencing.
The PCR conditions were as follows: denaturation at 94°C for 30 s, annealing at 45°C for 30 s, and extension at 72°C for 2 min for 35 cycles. The PCR products were purified by 0.8% low-melting-point agarose gel extraction, using a PrepAGen DNA purification kit (Bio-Rad Laboratories); they were quantitated and then kept at −20°C until used.
The DNA was sequenced by using an ABI Dye Terminator Cycle Sequencing Ready Reaction Kit in accordance with the manufacturer's instructions and an ABI 310 automated sequencer (both from The Perkin-Elmer Corporation). Each reaction product sequence was confirmed twice.
DNA sequences were transferred to the DNASIS program for sequence comparisons between isolates. Pairwise nucleotide sequence identity comparisons were included. The percentages of homology were used for the arrangement of Table 1. The BLAST 2 program (National Center for Biotechnology Information) was used to determine levels of homology with published sequences in the GenBank (1).
TABLE 1.
Sources and M antigen types of GAS isolated from noninfected subjects and patientsa
| No. | Strain | Patient condition | Source of isolation | Yr of collection | Endemic area | Homology to M typea | Accession no. |
|---|---|---|---|---|---|---|---|
| 1 | cmu104 | Impetigo | Skin | 1985 | Chiang Mai Hospital | ST1 | AF091805 |
| 2 | cmu328 | Impetigo | Skin | 1985 | Chiang Mai Hospital | ST1 | |
| 3 | Cmuh7-6 | Normal | Throat | 1985 | Chiang Rai School | ST1 | |
| 4 | cmu68 | Impetigo | Skin | 1985 | Chiang Mai Hospital | M44 | |
| 5 | Cmud14-5 | Normal | Throat | 1985 | Chiang Mai School | M44 | |
| 6 | cmuj63 | Impetigo | Skin | 1990 | Chiang Mai Hospital | M44 | |
| 7 | cmus665 | Sore throat | Throat | 1985 | Chiang Mai Hospital | M44 | |
| 8 | cmu42 | Impetigo | Skin | 1985 | Chiang Mai Hospital | M25 | |
| 9 | cmuj59 | Impetigo | Skin | 1990 | Chiang Mai Hospital | M25 | |
| 10 | cmuak19 | Sore throat | Throat | 1995 | Chiang Mai Hospital | M25 | |
| 11 | cmus14-6 | Normal | Throat | 1985 | Chiang Mai School | M27 | |
| 12 | cmuh92 | RHD | Throat | 1985 | Chiang Mai Hospital | ST2 | AF093817 |
| 13 | cmuarf19 | ARF | Throat | 1985 | Chiang Mai Hospital | M22 | |
| 14 | cmucsf3 | Meningitis | CSF | 1995 | Chiang Mai Hospital | ST3 | AF140798 |
| 15 | cmuk16 | Sore throat | Throat | 1995 | Chiang Mai Hospital | ST4 | AF091806 |
| 16 | cmuk3 | Sore throat | Throat | 1995 | Chiang Mai Hospital | ST4 | |
| 17 | cmuk8 | Sore throat | Throat | 1995 | Chiang Mai Hospital | ST4 | |
| 18 | cmuk9 | Sore throat | Throat | 1995 | Chiang Mai Hospital | ST4 | |
| 19 | cmuh338 | RHD | Throat | 1985 | Chiang Mai Hospital | ST5 | AF091807 |
| 20 | cmuj27 | Impetigo | Skin | 1990 | Chiang Mai Hospital | ST6 | |
| 21 | cmuj71 | Impetigo | Skin | 1990 | Chiang Mai Hospital | ST6 | |
| 22 | cmuj82 | Impetigo | Skin | 1990 | Chiang Mai Hospital | ST6 | AF140227 |
| 23 | cmuk17 | Sore throat | Throat | 1995 | Chiang Mai Hospital | ST6 | |
| 24 | cmu20 | Impetigo | Skin | 1985 | Chiang Mai Hospital | M12 | |
| 25 | cmuarf2 | ARF | Throat | 1985 | Chiang Mai Hospital | ST7 | AF091804 |
| 26 | cmu52T | Normal | Throat | 1985 | Lab personal | M3 | |
| 27 | Cmu64 | Impetigo | Skin | 1985 | Chiang Mai Hospital | M70 | |
| 28 | cmus546 | Sore throat | Throat | 1985 | Chiang Mai Hospital | M70 | |
| 29 | cmuo426 | Normal | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 30 | cmus122 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 31 | cmus14 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 32 | cmus142 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 33 | cmus148 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 34 | cmus182 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 35 | cmus19 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 36 | cmus219 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 37 | cmus330 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 38 | cmus431 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 39 | cmus578 | Sore throat | Throat | 1985 | Chiang Mai Hospital | Potter41 | |
| 40 | cmus744 | Sore throat | Throat | 1985 | Chiang Mai Hospital | M1 | |
| 41 | cmu38 | Impetigo | Skin | 1985 | Chiang Mai Hospital | M76 | |
| 42 | cmuh9 | RHD | Throat | 1985 | Chiang Mai Hospital | M81 | |
| 43 | cmut006 | Normal | Throat | 1985 | Chiang Rai School | M11 | |
| 44 | cmuarf15 | ARF | Throat | 1985 | Chiang Mai Hospital | M63 | |
| 45 | cmuarf3 | ARF | Throat | 1985 | Chiang Mai Hospital | M63 | |
| 46 | cmuj65 | Impetigo | Skin | 1995 | Chiang Mai Hospital | ST8 | AF089870 |
| 47 | cmuarf1 | ARF | Throat | 1985 | Chiang Mai Hospital | ST8 | |
| 48 | cmuarf39 | ARF | Throat | 1995 | Chiang Mai Hospital | ST9 | AF140797 |
| 49 | cmuh140 | RHD | Throat | 1985 | Chiang Mai Hospital | ST10 | AF104406 |
| 50 | cmuak1 | Impetigo | Throat | 1995 | Chiang Mai Hospital | ST10 | |
| 51 | cmuj76 | Impetigo | Skin | 1990 | Chiang Mai Hospital | ST11 | AF104407 |
| 52 | cmuk2 | Sore throat | Throat | 1990 | Chiang Mai Hospital | ST12 | AF104408 |
| 53 | cmu417 | Impetigo | Skin | 1985 | Chiang Mai Hospital | ST13 | AF104409 |
DNA sequences from GAS isolated from carriers and patients were aligned with DNA sequences of reference strains in the literature. CSF, cerebrospinal fluid.
Fifty-three non-M-typeable GAS isolates from northern Thailand were sequence typed by PCR amplification of the M protein genes of their DNAs. M protein genes were amplified from all 53 isolates. Thirty (59%) of the 53 isolates had DNA sequences with more than 98% homology to published M protein gene sequences. The remaining 23 isolates had novel M protein gene sequences designated ST1 to ST13 (Table 1). The level of homology between isolates of a given sequence type is 98 to 100%.
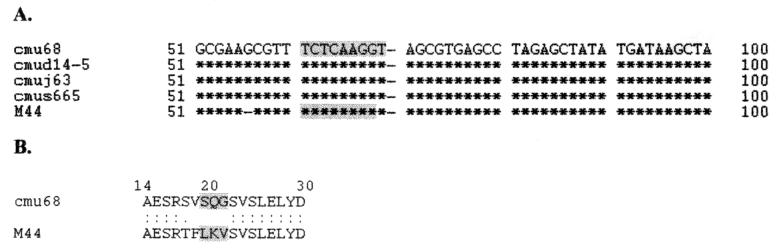
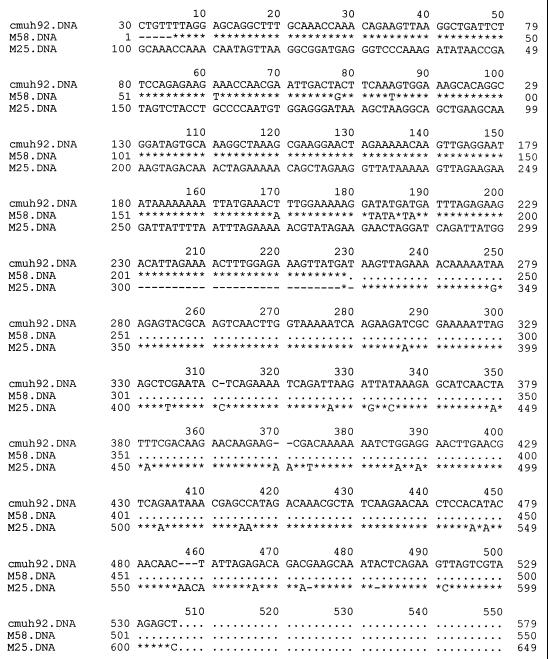
Of the M types with homology to published sequences, Potter41 predominated, representing up to 21% of the non-M-typeable isolates investigated, all of which were collected from patients with pharyngitis in 1985. The other M sequences, homologous to M44, M25, M27, M22, M12, M3, M70, M76, M81, M1, M11, and M63, represented up to 36% of the isolates sequenced. The translated sequences of the M proteins from isolates corresponding to M1, M3, M11, M22, Potter41, M70, and M80 showed complete homology in the hypervariable region (data not shown). Isolates that were homologous to M12, M25, M63, and M75 differed in sequence by point mutations which resulted in no more than three amino acid substitutions in the hypervariable region. The sequences of isolates cmu68, cmud14-5, cmuj63, and cmus665 differed from that of M44 by only a compensatory frameshift mutation spanning 5 amino acids (Fig. 1). Interestingly, the isolate represented by ST2 (cmuh92), which was obtained from an RHD patient, showed N-terminal sequence homology to M58 and C-terminal sequence homology to M25 (Fig. 2). GAS isolates with homology to M25 were found within the Thai population investigated in this study (cmu42-1985, cmuj59-1990, and cmuak19-1995) (Table 1).
FIG. 1.
(A) Alignment of nucleotide sequences of GAS isolates cmu68, cmud14-5, cmuj63, and cmus665 with that of M44 in the region of the frameshift mutation. Asterisks represent identity to the corresponding nucleotides; dashes represent missing nucleotides. The numbers attached to the sequence represent positions in the M44 nucleotide sequence published in GenBank. The region of the frameshift mutation is shaded. (B) Translated sequence of cmu68 and M44 in the region of the frameshift mutation. The region of the frameshift mutation is shaded.
FIG. 2.
Alignment of nucleotide sequence of GAS isolate cmuh92 with M58 and M25 DNA sequences (derived from GenBank), showing 96 and 89% homology, respectively. Asterisks represent identity to the corresponding nucleotides, dashes represent missing nucleotides, and dots indicate that no nucleotide sequence was given.
Six isolates were collected from ARF patients. Their sequences exhibited homology to M22 (n = 1) and M63 (n = 2), and three were new sequence types, ST7, ST8, and ST9. One isolate from an RHD patient exhibited nucleotide sequence homology to M81, and three were new sequence types, ST2, ST5, and ST10. Several isolates were collected from both throat and skin sites of patients with impetigo or sore throat or from noninfected individuals (ST1, ST6, ST7, ST8, M25, M44, and M70).
Sequence analysis of the 53 isolates used in this study revealed 13 novel-sequence M types which were not identifiable with previously published emm sequences. This finding shows the diversity of GAS strains found in northern Thailand. The majority of strains with more than one isolate (e.g., ST1, ST6, ST8, M25, M44, and M70) were isolated from both throat and skin sites. These strains, as with strains of other M types (10), cannot be exclusively categorized as rheumatogenic or nephritogenic.
Antigenic variation in the M proteins of the isolates investigated, compared to published M protein sequences, was predominantly due to point mutations, small deletions, and insertions in the hypervariable region. One group of isolates with homology to M44 exhibited corrected frameshift mutations (Fig. 1). Studies of isolates from the Northern Territory of Australia had previously revealed a number of M family groups that showed compensatory frameshift mutations, including M52/M53/M80, M5, emm49, emm13, emm33, and emm70 (5). Interestingly, one new sequence M type, ST2, shows N-terminal homology to M58 (96%) and C-terminal homology to M25 (89%) (Fig. 2). Our data show that M25 is endemic to this area (isolate cmuj59 and cmuak19); therefore, this sequence type may have been the result of intergenomic recombination between two isolates of M25 and M58, possibly while the host harbored two GAS strains at the same time. Recent studies suggest that this may be a mechanism for transfer of DNA between strains, resulting in emm-like genes and vir regulons with mosaic structures (5, 13). These recombination events would alter the amino acid sequence of M and M-like genes, which may contribute to pathogen virulence, thereby effecting host immune responses.
GAS are endemic in Thailand, and ARF is a severe health problem in that area (12). M sequence typing is a useful tool for conducting epidemiological studies of streptococcal infections, particularly in an area where nearly all GAS isolates are non-M typeable by conventional M serotyping methods. It allows not only monitoring of streptococcal carriage within regions of endemicity but also identification of types of circulating streptococci. This information provides a useful guideline for developing a vaccine for RF in a specific area of endemicity.
Nucleotide sequence accession numbers.
DNA sequences with no homology to any published emm gene sequence were submitted to GenBank (accession numbers are given in Table 1).
Acknowledgments
This study was supported by The Thailand Research Fund, grant no. BR/06/2539.
We are very grateful to Diana Martin and Teiko Murai for performing M serotyping.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandt E R, Hayman W A, Currie B, Pruksakorn S, Good M F. Human antibodies to the conserved region of the M protein: opsonization of the heterologous strains of group A streptococci. Vaccine. 1997;15:1805–1812. doi: 10.1016/s0264-410x(97)00178-3. [DOI] [PubMed] [Google Scholar]
- 4.Fischetti V A, Hodges W M, Hruby D E. Protection against streptococcal pharyngeal colonization with a vaccinia:M protein recombinant. Science. 1989;244:1487–1490. doi: 10.1126/science.2660266. [DOI] [PubMed] [Google Scholar]
- 5.Gardiner D L, Sriprakash K S. Molecular epidemiology of impetiginous group A streptococcal infections in aboriginal communities of northern Australia. J Clin Microbiol. 1996;34:1448–1452. doi: 10.1128/jcm.34.6.1448-1452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gherna R L. Preservation. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 208–217. [Google Scholar]
- 7.Kaplan E L, Johnson D J, Cleary P P. Group A streptococcal serotypes isolated from patients and sibling contacts during the resurgence of rheumatic fever in the United States in the mid-1980s. J Infect Dis. 1989;159:101–103. doi: 10.1093/infdis/159.1.101. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan E L, Johnson D R, Nanthapisud P, Sirilertpanrana S, Chumdermpadetsuk S. A comparison of group A streptococcal serotypes isolated from the upper respiratory tract in the USA and Thailand: implications. Bull W H O. 1992;70:433–437. [PMC free article] [PubMed] [Google Scholar]
- 9.Majeed H A, Khuffash F A, Yousof A M, Farwana S S, Chugh T D, Moussa M A, Rotta J, Havlickova H. The concurrent associations of group A streptococcal serotypes in children with acute rheumatic fever or pharyngitis-associated glomerulonephritis and their families in Kuwait. Zentbl Bakteriol Mikrobiol Hyg Ser A. 1986;262:346–356. doi: 10.1016/s0176-6724(86)80007-4. [DOI] [PubMed] [Google Scholar]
- 10.Manjula B N, Khandke K M, Fairwell T, Relf W A, Sriprakash K S. Heptad motifs within the distal subdomain of the coiled-coil rod region of M protein from rheumatic fever and nephritis associated serotypes of group A streptococci are distinct from each other: nucleotide sequence of the M57 gene and relation of the deduced amino acid sequence to other M proteins. J Protein Chem. 1991;10:369–383. doi: 10.1007/BF01025251. [DOI] [PubMed] [Google Scholar]
- 11.Martin D R. Streptococcal infection: rheumatogenic streptococci reconsidered. N Z Med J. 1988;101:394–396. [PubMed] [Google Scholar]
- 12.Phornphutkul C, Markowitz M. Secondary prophylaxis in patients with rheumatic fever: use of outlying health centers. Chiang Mai Med Bull. 1981;23:275–279. [Google Scholar]
- 13.Podbielski A, Krebs B, Kaufhold A. Genetic variability of the emm-related genes of the large vir regulon of group A streptococci: potential intra- and intergenomic recombination events. Mol Gen Genet. 1994;243:691–698. doi: 10.1007/BF00279579. [DOI] [PubMed] [Google Scholar]
- 14.Pruksachatkunakorn C, Vaniyapongs T, Pruksakorn S. Impetigo: an assessment of etiology and appropriate therapy in infants and children. J Med Assoc Thail. 1993;76:222–229. [PubMed] [Google Scholar]
- 15.Pruksakorn S, Galbraith A, Houghten R A, Good M F. Conserved T and B cell epitopes on the M protein of group A streptococci: induction of bactericidal antibodies. J Immunol. 1992;149:2729–2735. [PubMed] [Google Scholar]
- 16.Pruksakorn S, Currie B, Brandt E, Phornphutkul C, Hunsakunachai S, Manmontri A, Robinson J H, Kehoe M A, Galbraith A, Good M F. Toward a vaccine for rheumatic fever: identification of a conserved target epitope on the M protein of group A streptococci. Lancet. 1994;344:639–642. doi: 10.1016/s0140-6736(94)92083-4. [DOI] [PubMed] [Google Scholar]
- 17.Pruksakorn S, Phornphutkul C, Boonchoo C, et al. Prevalence of group A streptococci from school children and patients in Chiang Mai, Thailand. Chiang Mai Med Bull. 1990;29:15–26. [Google Scholar]
- 18.Relf W A, Cooper J, Brandt E R, Hayman W A, Ander R F, Pruksakorn S, Currie B, Saul A, Good M F. Mapping a conserved conformational epitope from the M protein of group A streptococci. Pept Res. 1996;9:12–20. [PubMed] [Google Scholar]
- 19.Rotta J, Facklam R R. Manual of microbiological diagnostic methods for streptococcal infections and their sequelae. Geneva, Switzerland: World Health Organization; 1980. [Google Scholar]
- 20.Saunders N A, Hallas G, Gaworzewska E T, Metherell L, Efstratiou A, Hookey J V, George R C. PCR–enzyme-linked immunosorbent assay and sequencing as an alternative to serology for M-antigen typing of Streptococcus pyogenes. J Clin Microbiol. 1997;35:2689–2691. doi: 10.1128/jcm.35.10.2689-2691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization. WHO programme for the prevention of rheumatic fever/rheumatic heart disease in 16 developing countries: report from phase I (1986–90) Bull W H O. 1992;70:213–221. [PMC free article] [PubMed] [Google Scholar]