Abstract
A PCR method to amplify DNA segments of the glycoprotein G gene of monkey B virus (BV) was achieved by adding betaine to the PCR mixture, in spite of the high G+C content of this gene. No product was obtained when DNA of human herpes simplex viruses (HSVs) was used as the template under the same conditions. Thus, this PCR method is useful in discriminating BV from HSVs.
Herpesvirus simiae (monkey B virus [BV]) is a member of the alphaherpesvirus subfamily containing the human herpes simplex viruses (HSV types 1 and 2 [HSV-1 and -2]) (3). This virus is a common pathogen in macaque monkeys (7). BV infection is usually asymptomatic in monkeys, but its transmission to humans leads to high mortality unless proper medical care is taken immediately after infection (4). Accurate and rapid diagnosis of BV infection is critical for the safety of research or animal care staffs who happen to have contact with BV-positive monkeys or tissues.
Swab culture is a definitive method to detect BV, but it takes a few days and requires a level 4 facility (4). PCR has been applied to in vitro diagnosis to detect viral DNA with rapidity and safety, and several PCR methods to identify BV have been proposed (2, 9, 10, 11). However, one of these methods is not applicable to rhesus monkey-derived strains (11), which have caused most of the fatalities to date (7). The other methods require digestion with restriction enzymes or Southern hybridization with radioactive probes in order to determine whether the PCR product is derived from BV or from one of the two HSVs (2, 9, 10).
There is considerable difference between the gG gene of BV and that of HSVs (12). BVgG is, however, high in G+C content, which makes it difficult for this gene to be amplified by PCR. Dimethyl sulfoxide (DMSO) is effective in the amplification of some G+C-rich sequences (8). Recently, some investigators demonstrated that the addition of betaine (1-carboxy-N,N,N-trimethylmethanammonium inner salt) further improved the amplification of G+C-rich sequences (1, 6, 13). To discriminate BV from HSVs in a single step, we sought to apply betaine to the amplification of BVgG.
Amplification of segments of BVgG.
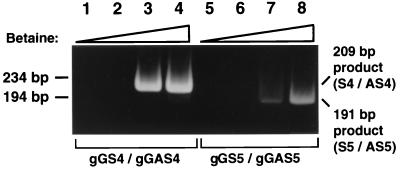
E-2490, one of the BV strains from rhesus monkeys, was grown in a confluent monolayer of Vero cells in serum-free medium at Kalter's Laboratory (Virus Reference Laboratory, South Texas Medical Center, San Antonio). After UV irradiation in the presence of psoralen, the virions were transported to Japan. Viral DNA was purified by the sodium iodide method (5). Briefly, viral protein was solubilized in 4.5 M sodium iodide, and then the viral DNA was coprecipitated with glycogen in 50% isopropanol. Two primer sets were synthesized to amplify two segments of the BVgG gene, based on the published sequence (12). The acronyms and sequences of the primers are as follow: gGS4 (forward, 5′-CCGCGTACGACTACGAGATCC-3′), gGAS4 (reverse, 5′-GTTCGCGGCCACGATCCA-3′), gGS5 (forward, 5′-CCCAGGACATGGCCTACGTG-3′), and gGAS5 (reverse, 5′-CGTCCCCTCCGTCGTTAC-3′). Using an Advantage-GC Kit (Clontech), we prepared a PCR mixture (50 μl) containing 5% DMSO, 1 μl of BV DNA solution, and a 0.4 μM concentration of each of the forward and reverse primers. This 1-μl solution was estimated to contain BV DNA extracted from 800 50% tissue culture infective dose (TCID50) virions (described below). After the initial denaturation (94°C, 5 min), PCR was conducted for 35 cycles (94°C, 1 min; 55°C, 1 min; 72°, 2 min), followed by a final extension (72°C, 7 min). The PCR products were subjected to electrophoresis on a 5% polyacrylamide gel, and the DNA bands were visualized under illumination at 234 nm after staining with SYBR Green I (Molecular Probes). In the presence of DMSO alone, no product was observed with primers gGS4 and gGAS4 or with gGS5 and gGAS5 (Fig. 1, lanes 1 and 5). By the addition of 1.0 M betaine, however, 209- and 191-bp products were amplified with primer pairs gGS4-gGAS4 and gGS5-gGAS5, respectively (Fig. 1, lanes 3 and 7). We confirmed by cycle sequencing that the two products corresponded to nucleotides 1073 to 1281 and 1340 to 1530, respectively, in the published sequence of BVgG (accession no. AB032191 and AB032192 in the DNA Database of Japan [DDBJ]). We used the reaction mixture containing 1.5 M betaine in the experiments described below (Fig. 1, lanes 4 and 8).
FIG. 1.
PCR in the presence of various concentrations of betaine. Lanes 1 and 5, no betaine (0 M); lanes 2 and 6, 0.5 M betaine; lanes 3 and 7, 1 M betaine; lanes 4 and 8, 1.5 M betaine. The two triangles indicate that the concentration of betaine increased from lanes 1 to 4 and from lanes 5 to 8. Lanes 1 to 4, PCR with primers gGS4 and gGAS4; lanes 5 to 8, PCR with primers gGS5 and gGAS5. The positions of the 194- and 234-bp fragments of HaeIII-digested φX174 replicative-form DNA are indicated to the left of the panel. The product obtained with gGS4-gGAS4 was 209 bp; that obtained with gGS5-gGAS5 was 191 bp.
Specificity of the PCR assays.
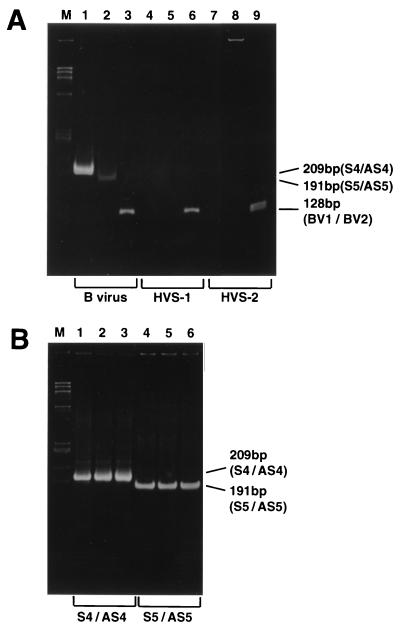
HSV-1 (strain HF) and HSV-2 (strain UW268) were grown, and the viral DNA was purified by the sodium iodide method. We prepared a PCR mixture containing HSV DNA extracted from virions whose titer was 800 TCID50. No product was obtained when either HSV-1 or HSV-2 was subjected to PCR with gGS4-gGAS4 (Fig. 2A, lanes 4 and 7, respectively) or with gGS5-gGAS5 (Fig. 2A, lanes 5 and 8, respectively) in the presence of 1.5 M betaine. To confirm that the viral DNA was extracted from HSV virions, control experiments were done with primers BV1 and BV2 in accordance with the PCR method of Scinicariello et al. (9, 10), but the cycle number was increased to 35 in order to match that of our assays. As reported previously, 128-bp fragments of the ICP 18.5 genes of HSV-1 and HSV-2 were amplified with primers BV1 and BV2 (Fig. 2A, lanes 6 and 9). These results show the recovery of HSV DNA, excluding the possibility of false negativity caused by insufficiency of the template (Fig. 2A, lanes 4, 5, 7, and 8). Thus, we have developed a PCR method to specifically amplify segments of the BVgG gene.
FIG. 2.
Amplification of the 209- and 191-bp fragments. Lane M, HaeIII-digested φX174 replicative-form DNA. (A) PCR with BV, HSV-1, or HSV-2 DNA as the template. Lanes 1 to 3, BV (strain E-2490) DNA as the template; lanes 4 to 6, HSV-1 (strain HF); lanes 7 to 9, HSV-2 (strain UW268). Lanes 1, 4, and 7, PCR with primers gGS4 and gGAS4; lanes 2, 5, and 8, PCR with primers gGS5 and gGAS5; lanes 3, 6, and 9, PCR with primers BV1 and BV2 according to the modified method of Scinicariello et al. (9, 10). Note that 128-bp products were obtained with BV and HSVs as the DNA templates. (B) PCR in the presence of contaminating viral DNA. Lanes 1 and 4, BV DNA alone as the template; lanes 2 and 5, BV DNA plus HSV-1 and HSV-2 DNA (800 TCID50 each); lanes 3 and 6, BV DNA plus 1,000 copies of hepatitis B virus genomic DNA. Lanes 1 to 3, PCR with primers gGS4 and gGAS4; lanes 4 to 6, PCR with primers gGS5 and gGAS5.
We found that the band intensity of the 128-bp product obtained after PCR using 1 μl of BV DNA solution was equal to that of HSV-1 or HSV-2 DNA extracted from 800 TCID50 virions. An example is shown in Fig. 2A, lanes 3, 6, and 9. Thus, assuming the yield of viral DNA is constant in the sodium iodide method and that the template BV DNA works as efficiently as the HSV DNA, we estimated that the amount of BV DNA in the 1-μl solution corresponded to the titer of 800 TCID50.
To further confirm the specificity of our PCR assays, we amplified segments of the BVgG gene in the presence of both HSV-1 and HSV-2 DNA or hepatitis B virus genomic DNA (1,000 copies). As shown in Fig. 2B, contamination either by HSV-1 and HSV-2 DNA or by hepatitis B virus DNA caused no interference with amplification of the 209- and 191-bp fragments of the BVgG gene.
Sensitivity of the PCR assays.
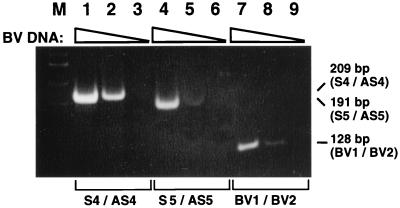
Serially diluted BV DNA was used to estimate the detection limit of our PCR method. At a 10-fold dilution, the 209-bp product was clearly observed after amplification with primer pair gGS4-gGAS4, and the 191-bp product was slightly detected with primer pair gGS5-gGAS5 (Fig. 3, lanes 2 and 5). The 100-fold-diluted BV DNA yielded no product with either gGS4-gGAS4 or gGS5-gGAS5 (Fig. 3, lanes 3 and 6). The 128-bp fragment amplified with primers BV1 and BV2 was seen at dilutions up to 10-fold (Fig. 3, lanes 7 and 8). Thus, we could detect 80 TCID50 of BV at a 10-fold dilution but could not detect 8 TCID50 of BV at a 100-fold dilution, by either our method or that of Scinicariello et al. (9, 10). The sensitivity of these two methods appeared comparable on the template BV DNA used in this study.
FIG. 3.
PCR with serially diluted BV DNA as the template. Lanes 1, 4, and 7, nondiluted BV DNA; lanes 2, 5, and 8, 10-fold dilution; lanes 3, 6, and 9, 100-fold dilution. The three triangles indicate that the concentration of BV DNA decreased from lanes 1 to 3, from lanes 4 to 6, and from lanes 7 to 9. Lane M, HaeIII-digested φX174 replicative-form DNA. Lanes 1 to 3, PCR product obtained with primers gGS4 and gGAS4; lanes 4 to 6, PCR product obtained with primers gGS5 and gGAS5; lanes 7 to 9, PCR product obtained with primers BV1 and BV2 according to the modified method of Scinicariello et al. (9, 10).
To our knowledge, this study is the first one to distinguish BV from HSVs by PCR alone, when BV DNA of a rhesus monkey-derived strain is used. Furthermore, our PCR method utilizing betaine may be applicable to the identification of viruses, other than BV, whose genomes are also high in G+C content.
Acknowledgments
We thank Denka Seiken Co., Ltd., for the HSV-1 sample. We are grateful to Shuya Shirahama (Gene Analysis Section, Gene and Chromosome Analysis Center, SRL Co., Ltd.) for his technical assistance in DNA sequencing.
This work was supported by a grant (H10-Genome-016 to S.N.) from the Health Science Research Grants for Research on the Human Genome and Gene Therapy from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Baskaran N, Kandpal R P, Bhargava A K, Glynn M W, Bale A, Weissmann S M. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 1996;6:633–638. doi: 10.1101/gr.6.7.633. [DOI] [PubMed] [Google Scholar]
- 2.Black D H, Eberle R. Detection and differentiation of primate α-herpesviruses by PCR. J Vet Diagn Invest. 1997;9:225–231. doi: 10.1177/104063879700900301. [DOI] [PubMed] [Google Scholar]
- 3.Boulter E A. The isolation of monkey B virus (Herpesvirus simiae) from the trigeminal ganglia of a healthy seropositive rhesus monkey. J Biol Stand. 1975;3:279–280. doi: 10.1016/0092-1157(75)90031-1. [DOI] [PubMed] [Google Scholar]
- 4.Holmes G P, Chapman L E, Stewart J A, Straus S E, Hilliard J K, Davenport D S the B Virus Working Group. Guidelines for the prevention and treatment of B-virus infections in exposed persons. Clin Infect Dis. 1995;20:421–439. doi: 10.1093/clinids/20.2.421. [DOI] [PubMed] [Google Scholar]
- 5.Ishizawa M, Kobayashi Y, Miyamura T, Matsuura S. Simple procedure of DNA isolation from human serum. Nucleic Acids Res. 1991;19:5792. doi: 10.1093/nar/19.20.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mytelka D S, Chamberlin M J. Analysis and suppression of DNA-polymerase pauses associated with a trinucleotide repeat. Nucleic Acids Res. 1996;24:2774–2781. doi: 10.1093/nar/24.14.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer A E. B virus, herpesvirus simiae: historical perspective. J Med Primatol. 1987;16:99–130. [PubMed] [Google Scholar]
- 8.Pomp D, Medrano J F. Organic solvents as facilitators of polymerase chain reaction. Biotechniques. 1991;10:58–59. [PubMed] [Google Scholar]
- 9.Scinicariello F, Eberle R, Hilliard J K. Rapid detection of B virus (herpesvirus simiae) DNA by polymerase chain reaction. J Infect Dis. 1993;168:747–750. doi: 10.1093/infdis/168.3.747. [DOI] [PubMed] [Google Scholar]
- 10.Scinicariello F, English W J, Hilliard J K. Identification by PCR of meningitis caused by herpes B virus. Lancet. 1993;341:1660–1661. doi: 10.1016/0140-6736(93)90791-e. [DOI] [PubMed] [Google Scholar]
- 11.Slomka M J, Brown D W G, Clewley J P, Bennett A M, Harrington L, Kelly D C. Polymerase chain reaction for detection of herpesvirus simiae (B virus) in clinical specimens. Arch Virol. 1993;131:89–99. doi: 10.1007/BF01379082. [DOI] [PubMed] [Google Scholar]
- 12.Slomka M J, Harrington L, Arnold C, Norcott J P N, Brown D W G. Complete nucleotide sequence of the herpesvirus simiae glycoprotein G gene and its expression as an immunogenic fusion protein in bacteria. J Gen Virol. 1995;76:2161–2168. doi: 10.1099/0022-1317-76-9-2161. [DOI] [PubMed] [Google Scholar]
- 13.Weissensteiner T, Lanchbury J S. Strategy for controlling preferential amplification and avoiding false negatives in PCR typing reactions. Biotechniques. 1996;21:1102–1108. doi: 10.2144/96216rr03. [DOI] [PubMed] [Google Scholar]