Significance
The Epstein–Barr virus (EBV) is an oncogenic virus that encodes multiple transforming proteins. EBV transforms resting normal B cells and is associated with multiple types of B cell lymphomas. How EBV transforms these cells is incompletely understood, in particular in tumor cells such as Burkitt’s lymphomas that do not express EBV’s oncogenes. Here, we show that the EBV noncoding RNA EBER2 accelerates B cell division through overexpression of cyclin B1 and of Aurora kinases. Mechanistically, we found that EBER2 induces the expression of the UCHL1 deubiquitinase that itself modulates expression of the cell-cycle proteins. Interestingly, this effect is modulated by the lineage of the infected cell and by polymorphisms within the viral genome.
Keywords: Epstein–Barr virus, B cell lymphomas, UCHL1, noncoding RNA EBER2
Abstract
The Epstein–Barr virus (EBV) transforms resting B cells and is involved in the development of B cell lymphomas. We report here that the viral noncoding RNA EBER2 accelerates B cell growth by potentiating expression of the UCHL1 deubiquitinase that itself increased expression of the Aurora kinases and of cyclin B1. Importantly, this effect was also visible in Burkitt’s lymphoma cells that express none of the virus’s known oncogenes. Mechanistically, EBER2 bound the UCHL1 messenger RNA (mRNA), thereby bringing a protein complex that includes PU.1, a UCHL1 transactivator, to the vicinity of its promoter. Although the EBV oncogene LMP1 has been suggested to induce UCHL1, we show here that EBER2 plays a much more important role to reach significant levels of the deubiquitinase in infected cells. However, some viruses that carried a polymorphic LMP1 had an increased ability to achieve full UCHL1 expression. This work identifies a direct cellular target of a viral noncoding RNA that is likely to be central to EBV’s oncogenic properties.
The Epstein–Barr virus (EBV) infects the large majority of the human population in which it induces a usually silent chronic infection (1). It also causes in excess of 200,000 cancer cases each year, mainly lymphomas such as Burkitt’s lymphomas (BL), and carcinomas (1). EBV efficiently infects B cells that initiate cell growth under the combined action of a set of viral proteins called the latent proteins and of viral noncoding RNAs. EBV-mediated B cell proliferation is visible in the blood and lymph nodes of infected individuals who undergo an infectious mononucleosis syndrome following primary infection (1). Infected B cells are eventually cleared by the immune response, but the virus can establish long-term persistence in his host’s B cells to form the virus reservoir (2). Thus, the ability of the virus to induce B cell proliferation can be seen as a strategy to expand its reservoir and increase the chances of persistence (2). Multiple latent proteins such as EBNA2 or EBNA3A are known to be indispensable to EBV-mediated B cell proliferation. In their absence, infected B cells die after a few days or even do not initiate cell growth at all (3). Noncoding RNAs play a subtler role in this process. Viruses that lack the BHRF1 microRNAs are still able to induce B cell growth, but the infected cells grow more slowly than those infected with their wild-type counterparts (4–6). The EBV encoded RNAs (EBER) are noncoding RNAs that bind multiple cellular proteins and serve a multitude of functions in infected cells, including a modulation of the interferon alpha pathway and of cytokine secretion (7–11). The EBERs have been shown to modulate lytic replication, the process that leads to new virus progeny, and to stimulate lytic replication through activation of the TLR7 pathway, resulting in increased CXCL8 expression (12). Whether EBER contributes to EBV’s ability to induce cell growth is a matter of debate. Some groups reported that EBER potentiates B cell transformation, but others could not confirm the result (13–16). Although EBERs have been shown to modulate the transcription of the viral latent membrane protein LMP2, they do not modify its translation (12, 17, 18). Finding out whether EBERs can modulate B cell growth is also essential to understand EBV’s contribution to tumor development. The viral expression pattern in EBV-positive BLs is restricted to the EBNA1 latent protein, the EBERs, and the EBV BART microRNAs (3). However, the BART microRNAs are dispensable for B cell transformation, and the oncogenic potential of EBNA1 has been controversially debated (19–22). We now report that EBER2 accelerates cell growth by enhancing the expression of crucial modulators of the cell cycle. Interestingly, this function was controlled by the type of infected cell, but also by the viral strain that infected it.
Results
The M81 EBERs Control Growth Transformation in Infected Cells In Vitro and In Vivo.
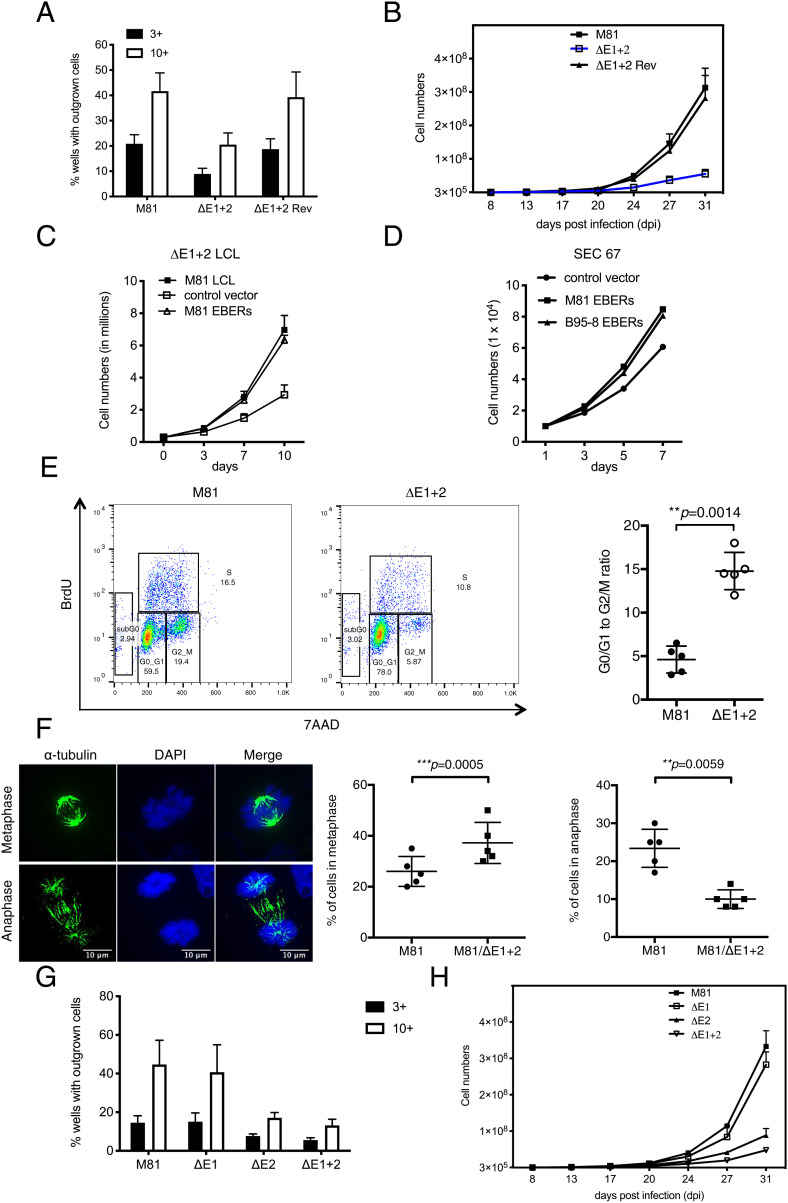
While one group previously reported that the noncoding RNAs EBERs enhance B cell growth, others could not confirm this effect (13–16). To clarify this issue, we performed infection assays with a virus triplet including the wild-type M81 virus, an EBER1 and 2 M81 double knockout (M81/ΔE1 + 2), and a revertant thereof (M81/ΔE1 + 2 Rev), in which primary B cells were infected at the same multiplicity of infection. In parallel, we infected B cells with wild-type B95-8 virus and with a B95-8 EBER1 and 2 double knockout (B95-8/ΔE1 + 2). Three days later, the cells were stained for EBNA2, a latent protein expressed shortly after infection. The number of EBNA2-positive cells was very similar after infection with deletion mutants or their wild-type controls. Then, 96-well cluster plates were seeded at a concentration of three or 10 EBNA2-positive cells per well. Six weeks after infection, the M81 EBER double mutant generated on average only half as many colonies as the two wild-type controls (Fig. 1A). In contrast, deletion of the B95-8 EBERs did not significantly influence cell growth (SI Appendix, Fig. 1A). We extended the characterization of EBER’s functions by performing bulk infections followed by the drawing of growth curves with the M81 triplet. This assay confirmed that the deletion of the EBERs significantly slows down the growth of B cells infected with the M81 virus, with six times less cells produced after infection with the mutant after 31 d in culture (Fig. 1B). Similar experiments with the B95-8 EBER deletion mutants confirmed previous results (13) showing that EBER plays no role in the growth of B cells infected with B95-8 EBV (SI Appendix, Fig. 1B), in line with earlier reports with the closely related P3HR1 strain (16). The effect of EBERs on M81-mediated cell growth was even more visible when the EBV-infected cells were grown under low-serum conditions (SI Appendix, Fig. 1C). At a medium concentration of 2.5% fetal calf serum, B cells transformed by M81/ΔE1 + 2 hardly grew at all. Although the M81 revertant behaved exactly as the wild-type virus, we wished to confirm that the phenotype observed after infection with M81/ΔE1 + 2 was exclusively due to the EBERs by performing complementation assays. To this end, we introduced multiple copies of the EBERs cloned in tandem (23, 24) under the control of a tetracycline-inducible promoter that also drives the expression of a truncated rat CD2 protein. After transfection, the CD2-positive cells were purified to obtain a homogeneous cell population that expresses EBERs at 60 to 75% of the levels seen in cells infected with wild-type virus (12). Comparison with the nontransfected cells and with wild-type infected cells showed that the EBER reintroduction led to cell doubling time close to wild-type levels (Fig. 1C). Complementation of lymphoblastoid cell lines generated with B95-8/ΔE1 + 2 with B95-8 EBERs had no effect on cell growth (SI Appendix, Fig. 1 D and E). We extended these complementation experiments to primary epithelial cells and could confirm that the M81 EBERs stimulate cell growth in these cells (Fig. 1D). We then assessed the mitotic rate of B cells infected with M81 wild-type or M81/ΔE1 + 2 virus by staining them for phospho-histone H3 expression. This assay revealed that the mitotic rate dropped 50% after infection with the EBER deletion mutant relative to wild-type infections (SI Appendix, Fig. 1F). Therefore, we performed BrdU incorporation assays coupled to 7AAD staining at day 45 postinfection that revealed a different cell-cycle distribution in B cells transformed with the M81/ΔE1 + 2 mutant in comparison to M81 wild-type. Cells infected with the EBER deletion mutant showed a decrease in the percentage of cells present in the S and particularly in the M phase, coupled to an increase in the percentage of cells present in the G0/G1 phase of the cell cycle, relative to wild-type controls (Fig. 1E). As a result, the ratio between cells in the G0/G1 phase and those in the G2/M phase was three times higher in cells infected with M81/ΔE1 + 2 compared to wild-type controls (Fig. 1E). Similarly, the G0/G1 to S ratio was twice as high in B cells infected with the deletion mutant (SI Appendix, Fig. 2A). Similar results were obtained after propidium iodide (PI) staining (SI Appendix, Fig. 2 B–E). We then performed a morphological analysis of the mitotic figures displayed by cells transformed by M81/ΔE1 + 2 or by wild-type controls using an antibody specific to alpha-tubulin. This analysis revealed that cells infected with M81/ΔE1 + 2 have a lower percentage of cells undergoing anaphase than the controls (Fig. 1F). Reciprocally, the percentage of cells in metaphase was higher, suggesting that the absence of EBERs impedes the metaphase–anaphase transition. We did not observe differences in the frequency of prometaphases in the two cell populations (SI Appendix, Fig. 2F). We then determined which of the EBERs was responsible for the decreased transformation abilities. To this end, we repeated the aforementioned transformation experiments with viruses that lack EBER1 or EBER2. These experiments showed that EBER2 but not EBER1 potentiates EBV-mediated B cell transformation (Fig. 1 G and H).
Fig. 1.
The M81 EBERs enhance cell growth. (A) We compared B cell growth after infection with M81, M81/ΔE1 + 2, and M81/ΔE1 + 2 Rev by counting the number of outgrowing wells in 96-well plates seeded with infected primary B cells containing either three or 10 EBNA2-positive cells after 5 wk (n = 5). The bar chart shows the arithmetic mean of five independent experiments and their SD. (B) Growth of M81-infected B cells during the first weeks after infection (n = 5). Mean values of five independent B cell infection experiments are shown. (C) An LCL generated with M81/ΔE1 + 2 was stably transfected with a plasmid that encodes a truncated form of NGFR and the M81 EBERs or with a plasmid that encodes NGFR only (control vector) (n = 5). After 21 d, the NGFR-positive cells were purified with a specific antibody, and we monitored cell growth for 10 d. Mean values of five independent B cell infection experiments are shown. (D) Primary epithelial cells were infected with a lentivirus that expresses the M81 or B95-8 EBERs. We monitored cell growth in these cells and in cells infected by a control lentivirus for 7 d. We analyzed three independent primary samples and show here one sample (SEC67). (E) BrdU incorporation assays were performed with independent B cell samples transformed with wild-type M81 virus or M81/ΔE1 + 2 mutant at day 45 postinfection (n = 5). The ratio of cells in the G0/G1 and in the G2/M phase for each sample is given in the scatter plot. Central horizontal lines represent means, and error bars indicate SD. All P values were obtained from two-tailed paired t tests performed with the two types of LCLs. (F) B cells infected by M81 or M81/ΔE1 + 2 were cytospinned and stained for α-tubulin to visualize the mitotic spindle and with DAPI (n = 5). The figures show cells in metaphase and anaphase. (Scale bar, 10 μm.) For each sample, at least 100 mitotic cells were examined. The graphs give the percentage of cells undergoing metaphase or anaphase. Central horizontal lines represent means, and error bars indicate SD. All P values were obtained from two-tailed paired t tests performed with the two types of LCLs. (G) We compared B cell growth after infection with M81, M81/ΔE1, M81/ΔE2, and M81/ΔE1 + 2 by counting the number of outgrowing wells in 96-well plates seeded with infected primary B cells containing either 3 or 10 EBNA2-positive cells after 5 wk (n = 5). The bar chart shows the arithmetic mean of five independent experiments and their SD. (H) Growth of M81- and M81 deletion mutant-infected B cells (see G) during the first weeks after infection (n = 5). Mean values of five independent B cell infection experiments are given.
The M81 EBERs Control M81-Driven B Cell Growth in an Animal Model.
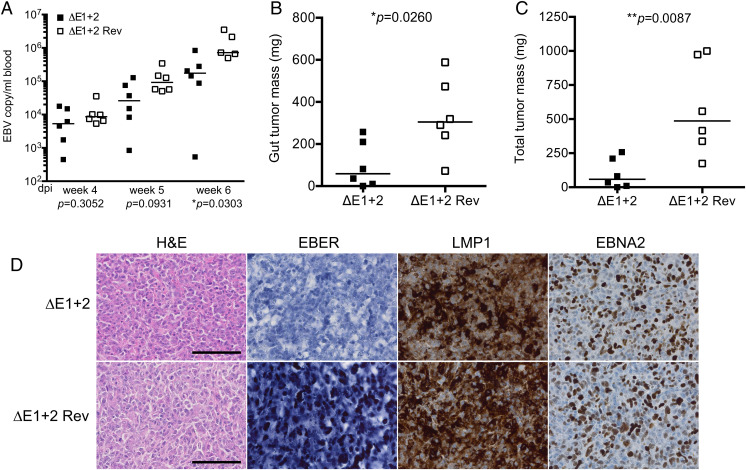
We wished to confirm the data gathered in vitro in an in vivo model of EBV infection and injected resting B cells exposed to M81/ΔE1 + 2 or M81/ΔE1 + 2 Rev into immunodeficient mice (19). Sequential recording of the virus load in the blood by qPCR starting 4 wk after infection showed that all mice carried infected cells but that the mice infected with the EBER null mutant had on average slightly lower blood titers than wild-type counterparts (Fig. 2A). Autopsy of the animals 6 wk postinfection indeed revealed that all except one mouse infected with M81/ΔE1 + 2 had developed tumors. However, total tumor mass and tumor mass in the gastrointestinal tract of mice that were infected with the M81/ΔE1 + 2 virus were significantly lower relative to mice infected with the virus revertant (5.8 and 3.3 times, respectively) (Fig. 2 B and C). These data fit with the results of the in vitro experiments and confirm that B cells infected with M81/ΔE1 + 2 grew at lower rate that their wild-type counterparts. We analyzed the tumor tissues for EBER, LMP1, and EBNA2 expression. As expected, the mice infected with the mutant did not express the EBERs as evidenced by both in situ hybridization and qPCR (Fig. 2D). Cells infected with either mutant or controls showed a similar pattern of LMP1 and EBNA2 expression, and we did not find any evidence of mutations within these genes during passaging of the tumor cells in the animal (Fig. 2D). We conclude that M81 EBERs boost B cell growth in vitro and in vivo.
Fig. 2.
The M81 EBERs potentiate cell growth in vivo. (A) Primary B cells exposed to M81/ΔE 1+ 2 or M81/ΔE1 + 2 Rev were injected into NSG mice (n = 6 per group). Viral titers in the peripheral blood of infected mice were determined by qPCR at different weeks postinfection. One mouse from the M81/ΔE1 + 2 rev group developed a tumor and died between week 5 and week 6. Central horizontal lines represent the median. The results were analyzed with a Mann–Whitney U test. (B) The dot plot shows the gut tumor mass in animals that developed a tumor. Central horizontal lines represent median. The results were analyzed with a Mann–Whitney U test. (C) same as in B for the analysis of total tumor mass. (D) These pictures show immunohistochemistry stains of tumors that developed in the gut. (Scale bar, 100 µm.) Continuous tissue sections were stained with hematoxylin and eosin (H&E), immunostained with antibodies specific for LMP1, EBNA2, or subjected to an in situ hybridization with an EBER-specific probe.
B95-8 EBERs Can Potentiate Cell Growth in M81/ΔE1 + 2 Infected B cells.
The different results obtained with either the M81 or the B95-8 EBER deletion mutants could result from interstrain polymorphisms within the EBER1 and 2 genes. Alternatively, it could indicate that the EBERs are dispensable after infection with B95-8, irrespective of their sequence. We tested these hypotheses by introducing the B95-8 EBERs in an LCL generated with M81/ΔE1 + 2. This experiment showed that the B95-8 EBERs can perfectly complement the phenotype observed in cells infected with M81/ΔE1 + 2 (SI Appendix, Fig. 3A) (12). Furthermore, we used previously constructed hybrid viral genomes in which the EBER genes are swapped between the viral strains to transform B cells (12). The B cell growth potential of a M81 virus that carries B95-8 EBERs was not modified relative to M81 wild-type, demonstrating that the polymorphisms between M81 and B95-8 have no influence on M81-mediated transformation (SI Appendix, Fig. 3 B and C). Reciprocal experiments performed with a B95-8/ΔE1 + 2 virus that carries M81 EBERs confirmed that EBERs, independently of their origin, have no influence on B cell growth in this case (SI Appendix, Fig. 4 A–C). In conclusion, these experiments show that the EBERs, whatever their origin, are dispensable for B95-8’s but are important for M81-mediated B cell growth, regardless of their polymorphisms.
EBER2 Enhances the Expression of Essential Cell-Cycle Proteins.
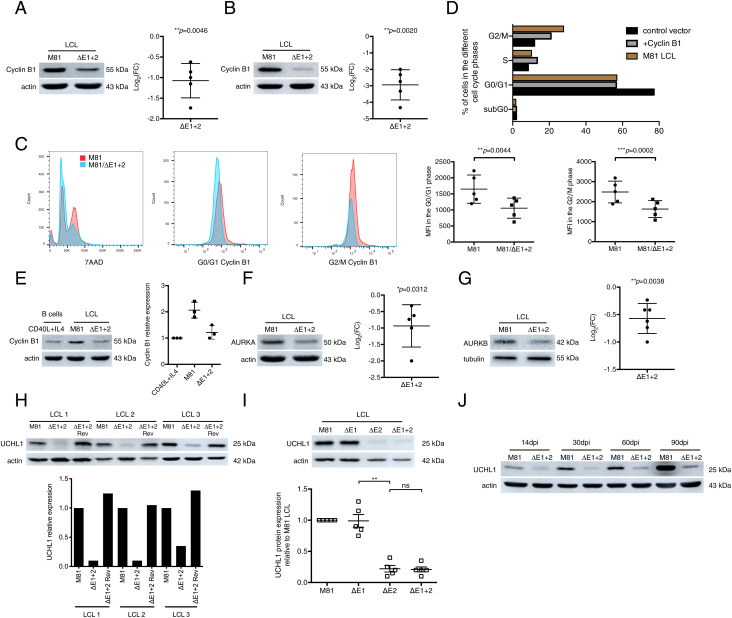
We performed a proteome and a phosphoproteome analysis with B cells infected with M81/ΔE1 + 2 or with wild-type controls to identify differential expression of proteins and phosphorylation events involved in the regulation of the cell cycle. While we could not detect any significant alteration of the phosphoproteome between these two sets of cells, we identified cyclin B1, Aurora kinase A (AURKA), and Aurora kinase B (AURKB) as potential EBER targets in this screen (Datasets S1 and S2). Western blot analyses showed that EBER deletion on average halved cyclin B1 expression, thereby confirming the proteome results (Fig. 3A; all blot results are available in SI Appendix, Fig. 8). The same assay performed with synchronized LCLs showed that mitotic cells after infection with M81/ΔE1 + 2 expressed six times less protein than wild-type counterparts (Fig. 3B). Similarly, intracellular flow cytometry staining showed that LCLs generated by M81/ΔE1 + 2 produced even less cyclin B1 protein in the G2/M phase than wild-type controls do in the G0/G1 phase when its expression is minimal (Fig. 3C). We then expressed cyclin B1 from a plasmid equipped with a tetracyclin-inducible promoter in B cells transformed by M81/ΔE1 + 2. This complementation led to cyclin B1 expression levels close to those seen in wild-type controls and improved both the percentage of cells present in the G2/M phase and the growth rate of the transfected cells to reach ∼70% of wild-type levels (Fig. 3D and SI Appendix, Fig. 5A). Altogether, these results show that EBERs potentiate cyclin B1 expression and that this effect largely if not entirely explains the phenotype displayed by B cells transformed with M81/ΔE1 + 2. However, cyclin B1 expression in B cells infected with M81/ΔE1 + 2 was very close to the expression level seen in CD40L-stimulated B cells, suggesting that EBERs enhance cyclin B1 levels above those required in normal proliferating B cells (Fig. 3E). We then generated growth curves under optimal cell culture conditions with these different cell populations and found that B cells infected with the EBER deletion mutant grew to levels closer to those seen in CD40L-stimulated B cells, in comparison with wild-type EBV controls (SI Appendix, Fig. 5B). To refine this analysis, we performed a comparative cell-cycle analysis of B cells stimulated by CD40L and IL4 together with B cells transformed by wild-type M81 or its EBER deletion mutant (SI Appendix, Fig. 5C). This assay revealed that CD40L-stimulated B cells and B cells infected by M81/ΔE1 + 2 display a very similar cell-cycle profile with a decreased proportion of cells in the S and M phase relative to wild-type M81-infected cells. This suggests that wild-type EBV infection accelerates transit through the cell cycle in comparison to normal proliferating B cells. Western blot analyses confirmed a decrease in AURKA and AURKB expression in cells infected with the null mutant relative to wild-type controls (Fig. 3 F and G). Importantly, a previously performed expression array performed with B cells infected with the same panel of viruses showed that EBERs do not affect AURKA, AURKB, and cyclin B1 transcription, suggesting a posttranslational impact of these noncoding RNAs (12). Our proteome assay indeed revealed an EBER-mediated upregulation of UCHL1, a deubiquitinase that was previously identified as an EBV target (SI Appendix, Table S1) and was previously found to stimulate cyclin B1 expression in ovarian tumors (25–30). A Western blot with LCLs infected with ΔE1 + 2 or ΔE2 and their respective wild-type controls confirmed that EBER2 enhances UCHL1 protein expression (Fig. 3 H and I). UCHL1 transcription was down-regulated in expression arrays interrogating B cells infected with an EBER deletion mutant, suggesting that EBER2 stimulates its transcription (12). Noninfected primary B cells or CD40L+IL4-stimulated B cells did not express UCHL1 at all, confirming the role of the virus in its induction (SI Appendix, Fig. 5D). However, there was a residual UCHL1 expression in B cells infected with M81/ΔE1 + 2, suggesting that EBV genetic elements other than EBER2, presumably latent proteins, are also involved in UCHL1 expression (Fig. 3H). Monitoring UCHL1 expression over time revealed a steady increase in B cells infected with wild-type viruses as described before but only a marginal one in those infected with M81/ΔE1 + 2, underscoring the importance of EBER for its expression and suggesting that the effect of the EBERs is potentiated with time (27) (Fig. 3J). We assessed the growth characteristics of the same LCLs at an early and late time point postinfection when the UCHL1 expression levels are respectively low and high and found indeed that late-passage LCLs grow quicker than their early passage counterparts (SI Appendix, Fig. 5E). Transfection of an expression plasmid encoding UCHL1 into LCLs generated with the M81/ΔE1 + 2 genome enhanced both cyclin B1 and Aurora kinases expression to levels seen in LCLs generated by wild-type M81 and restored cell growth to almost wild-type levels in B cells infected with M81/ΔE1 + 2 (Fig. 4A). Thus, UCHL1 appears to be a crucial mediator of EBER2’s effects on the cell cycle.
Fig. 3.
EBER2 upregulates UCHL1, cyclin B1, and Aurora kinase expression in infected cells. (A) We show representative immunoblot analyses performed on LCLs generated with M81 or M81/ΔE1 + 2 and stained with antibodies specific for cyclin B1 and actin (n = 5). We determined the relative intensity of the signals quantified by the ImageJ software over the loading control (actin). For each sample, the log2-transformed fold change (FC) is given based on the relative signals displayed by M81/ΔE1 + 2 and M81 wild-type (n = 5). Central horizontal lines represent means, and error bars indicate SD. The statistical significance of the assay was evaluated with a one-sample t test. (B) Same as A with antibodies specific for cyclin B1 and actin using LCLs generated with M81 or M81/ΔE1 + 2 and synchronized by double thymidine block (n = 5). (C) LCLs generated with M81 or M81/ΔE1 + 2 at day 60 postinfection were intracellularly stained with 7AAD and cyclin B1. We show one representative example out of five independent experiments that showed similar results. The MFI (Mean Fluorescence Intensity) of cells in the G0/G1 and in the G2/M phase for each sample is given in the scatter plot. Central horizontal lines represent means and error bars indicate SD. The statistical significance of the assay was evaluated with two-tailed paired t tests. (D) An LCL generated by M81/ΔE1 + 2 was stably transfected with a plasmid that encodes a truncated form of NGFR and the cyclin B1 gene or with a plasmid that encodes NGFR only (control vector). The NGFR-positive cells were purified with a specific antibody and synchronized by double thymidine block. PI staining was performed with NGFR-positive cells. M81 LCL was used as a positive control. The graph of bars gives the percentage of cells in the various phases of the cell cycle for a representative example (n = 2). (E) We determined cyclin B1 expression levels in primary B cells stimulated with CD40L and IL4 for 10 d relative to B cells infected with M81 or M81/ΔE1 + 2 using Western blot (n = 3). We show one representative example out of three independent experiments that showed similar results. The graph of bars shows the relative intensity of the signals quantified by the ImageJ software from three independent samples. The data are given relative to values obtained in primary B cells stimulated with CD40L and IL4. Central horizontal lines represent means and error bars indicate SD. (F) Same as A with antibodies specific for AURKA and actin (n = 5). (G) Same as A with antibodies specific for AURKB and tubulin (n = 6). (H) Same as A with antibodies specific for UCHL1 and actin (n = 10). The graph of bars shows the relative intensity of the signals quantified by the ImageJ software. We show three representative samples. (I) Immunoblot analysis performed on LCLs infected with M81, M81/ΔE1, M81/ΔE2, and M81/ΔE1 + 2 with antibodies specific for UCHL1 and actin (n = 5). The scatter plot shows the relative intensity of the signals quantified by the ImageJ software from five independent samples. The data are given relative to values obtained in LCLs generated with M81. Central horizontal lines represent means, and error bars indicate SD. All P values were obtained from two-tailed paired t tests performed with the two types of LCLs. ns, not significant. **P = 0.0012. (J) We monitored UCHL1 expression levels in LCLs exposed to M81 or M81/ΔE1 + 2 over time using Western blot (n = 2). The graph shows one representative example of UCHL1 expression at multiple days postinfection (dpi). We show one representative example out of two independent experiments that showed similar results.
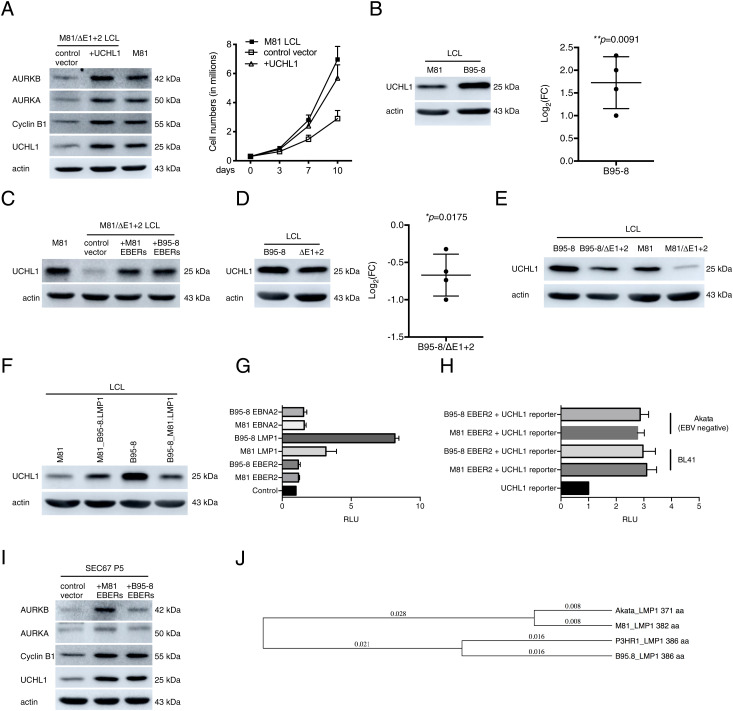
Fig. 4.
UCHL1 enhances cyclin B1 and Aurora kinase expression in infected cells. (A) An LCL generated with M81/ΔE1 + 2 was stably transfected with a plasmid that encodes a truncated form of NGFR and the UCHL1 gene or with a plasmid that encodes NGFR only (control vector). The NGFR-positive cells were purified with a specific antibody and immunoblotted with antibodies specific to UCHL1, cyclin B1, AURKA, and AURKB. The data are representative of three independent experiments (n = 3). The same cells were kept in culture and growth was monitored for 10 d. Mean values of three independent B cell infection experiments are shown (n = 3). (B) We show representative immunoblot analyses on LCLs generated with M81 or B95-8 using antibodies specific for UCHL1 and actin (n = 4). We determined the relative intensity of the signals quantified by the ImageJ software over the loading control (actin). For each sample, the log2-transformed FC is given based on the relative signals displayed by M81/ΔE1 + 2 and M81 wild-type. Central horizontal lines represent means and error bars indicate SD. The statistical significance of the assay was evaluated with a one-sample t test. (C) An LCL transformed by M81/ΔE1 + 2 was stably transfected with a plasmid that encodes a truncated form of NGFR and the EBERs, from either M81 or from B95-8, or with a plasmid that encodes NGFR only (control). The NGFR-positive cells were purified with a specific antibody and subjected to immunoblotting using a UCHL1-specific antibody. M81 wild-type LCLs were used as a positive control. The graph shows one representative experiment (n = 5). (D) We show representative immunoblot analyses on LCLs transformed with B95-8 and B95-8/ΔE1 + 2 with antibodies specific for UCHL1 and actin (n = 4). For each sample, the log2-transformed FC is given based on the relative signals displayed by M81/ΔE1 + 2 and M81 wild-type. Central horizontal lines represent means, and error bars indicate SD. The statistical significance of the assay was evaluated with a one-sample t test. (E) We show representative immunoblot analyses on LCLs infected with B95-8, B95-8/ΔE1 + 2, M81, and M81/ΔE1 + 2 with antibodies specific for UCHL1 and actin (n = 3). (F) Western blot performed on LCLs transformed with M81, M81_B95-8.LMP1, B95-8, and B95-8_M81.LMP1 using antibodies specific for UCHL1 and actin. The data are representative of two independent experiments. (G) NIH 3T3 cells were transfected with plasmids encoding LMP1 or EBNA2 or EBER2 from either M81 or B95-8 along with a UCHL1p-Luc reporter plasmid and 200 ng of the pRL Renilla expression vector. The total amount of DNA in all transfections was kept constant by adding control vectors. Luciferase assays were performed 48 h posttransfection. Renilla and Firefly luciferase signals were measured using the Dual-Luciferase Assay System Kit. Renilla luciferase signals were used for normalization of the Firefly luciferase signal. The relative light units (RLU) between the luminescence levels generated by expression of the expression constructs and the control constructs are given in a graph of bars that represent the mean of five independent experiments with error bars representing the SD (n = 5). (H) BL41 (EBV-negative) and Akata (EBV-negative) were transfected with each of the given constructs along with UCHL1p-Luc reporter plasmid and the pRL Renilla expression vector. Luciferase assays were performed as described in G (n = 3). (I) Primary epithelial cells were infected with a lentivirus that expresses the M81 or B95-8 EBERs. We determined the UCHL1, cyclin B1, AURKA, and AURKB expression in these cells relative to controls using Western blot. The data are representative of three independent experiments (n = 3). (J) Distance trees based on available EBV LMP1 protein sequences.
EBER2 Is Dependent on LMP1 to Induce UCHL1 Expression in B Cells but Not in Epithelial Cells or in BL Cells.
Comparison of UCHL1 expression levels in B cells infected with M81 or B95-8 showed that the latter cells express, on average, 3.5 times more UCHL1 than their M81 counterparts, although transfection of either B95-8 or M81 EBERs into a LCL infected with M81/ΔE1 + 2 led to a similar increase in UCHL1 production, suggesting a difference between M81 and B95-8 independent of the EBERs (Fig. 4 B and C). However, deletion of EBERs in B95-8 also led to a decrease in UCHL1 expression (60% of wild-type levels), suggesting that the link between EBERs and UCHL1 is also valid in the B95-8 context (Fig. 4D). Nevertheless, UCHL1 levels in LCLs generated with B95-8/ΔE1 + 2 were close to those observed after infection of the same cells with wild-type M81 and therefore much higher than in cells transformed with M81/ΔE1 + 2 (Fig. 4E). The observation that B cells infected with B95-8/ΔE1 + 2 expressed higher UCHL1 levels than their M81 counterparts suggested the existence of a polymorphism within the latent genes encoded by these viruses. Therefore, we generated a series of mutant viruses in which the EBV latent genes EBNA2, LMP1, or LMP2 from one viral strain were exchanged with its homolog from the other strain. The first two genes were chosen on the basis of their previously published ability to induce UCHL1 expression and on their degree of polymorphism across EBV strains (28, 29). Comparison between these exchange viruses and their wild-type counterparts showed that the exchange of LMP1 but not EBNA2 or LMP2 had a substantial impact on UCHL1 expression. Indeed, B cells infected with the B95-8 virus that carries M81 LMP1 expressed lower levels of UCHL1 than B cells infected with wild-type B95-8 (Fig. 4F and SI Appendix, Fig. 5 F and G). Reciprocally, B cells infected with the M81 virus that carries B95-8 LMP1 expressed higher levels of UCHL1 than B cells infected with wild-type M81. To confirm that LMP1 polymorphisms explain the variable UCHL1 levels after infection with M81 or B95-8, we cloned the UCHL1 promoter in front of the luciferase gene as previously reported (Fig. 4G) (28). This reporter construct was transiently transfected in NIH 3T3 cells that are negative for UCHL1 with EBER2, LMP1, or EBNA2 either from M81 or from B95-8 (SI Appendix, Fig. 5H). While transfection of EBER2 from either strain in these cells did not affect UCHL1 transcription at all, EBNA2 from both M81 and B95-8 only slightly increased UCHL1 expression (×1.5). M81 LMP1 more potently transactivated the UCHL1 promoter (×3.5), but B95-8 LMP1 induced UCHL1’s expression twice as much as M81 LMP1. LMP1 is known to be one of the most polymorphic EBV proteins, and these polymorphisms have been found to influence NF-κB induction (31, 32). Therefore, we performed a reporter assay to compare B95-8’s and M81’s abilities to induce NF-κB. However, we could not identify any difference, suggesting that the effect on UCHL1 is independent of NF-κB (SI Appendix, Fig. 5I). We then infected primary B cells with M81 and B95-8 viruses that lack LMP1 and assessed UCHL1 expression. B cells infected by the knockout viruses only showed background levels expression, confirming the role of this protein in UCHL1 induction and showing that in the absence of LMP1, EBER2 cannot substantially increase UCHL1 expression in peripheral blood B cells (SI Appendix, Fig. 5J). Similarly, transfection of EBER in HeLa cells that are UCHL1 negative did not lead to an expression of this protein (SI Appendix, Fig. 5H). We then turned our attention to the function of EBER2 in BLs that are derived from germinal center cells and express UCHL1 (25–27). To this end, we used two pairs of cell lines that are EBV negative or EBV positive. While BL41 is an EBV-negative BL that has been superinfected with B95-8, Akata is an EBV-positive BL for which a clone that has lost the virus is available (33). In both cases, the EBV-positive cell line expressed more UCHL1 than their negative counterparts (SI Appendix, Fig. 5K). We then went on to cotransfect EBER2 from either M81 or B95-8 and the UCHL1 reporter plasmid into the EBV-negative BL41 and Akata cell clones. This assay led to a clear activation of the UCHL1 promoter (3× in all cases, Fig. 4H). Finally, we introduced EBER2 into primary epithelial cells derived from a respiratory epithelium using a lentivirus (Fig. 4I). These cells naturally express low levels of UCHL1 but introduction of EBER2-enhanced expression of UCHL1 (2.5×), cyclin B1 (3×), and of the Aurora kinases (1.5× and 5×). We conclude that EBER2 can strongly potentiate UCHL1 expression but only in cells that already express this gene to some extent. This is the case in LCLs that express LMP1 in primary epithelial cells and in BL cells that derive from the germinal center.
EBER2 Enhances UCHL1 Expression through Interaction with the UCHL1 RNA.
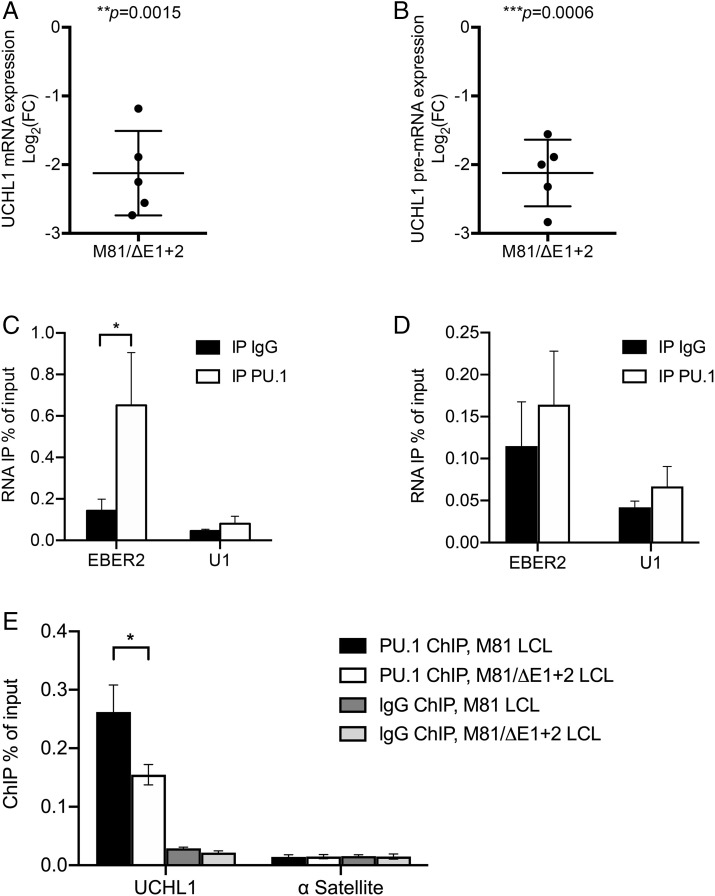
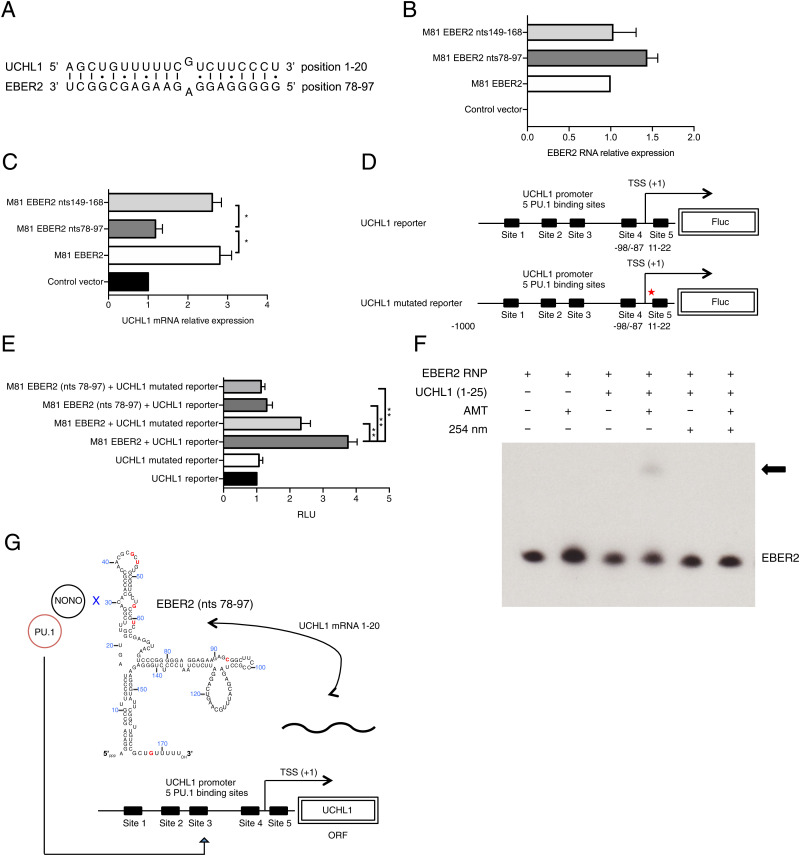
qRT-PCR studies performed on LCLs transformed with M81/ΔE1 + 2 and its wild-type control showed that the absence of EBERs reduced both UCHL1 pre-messenger RNA (mRNA) and mature mRNA expression, both confirming the results of the expression microarray assays and suggesting that the EBERs regulate UCHL1 transcription but not its splicing or maturation (Fig. 5 A and B). Moreover, the half-life of the UCHL1 transcript was independent of the presence or absence of EBERs, suggesting that EBER does not influence its stability (SI Appendix, Fig. 5L). Previous work has shown that the PU.1 transcription factor is recruited to the UCHL1 promoter and activates UCHL1 transcription (28). Furthermore, the NONO protein was previously shown to interact with PU.1 and to bind to EBER2 (34, 35). Therefore, we tested whether EBER2 could be recruited to the UCHL1 promoter via an interaction between PU.1 and NONO. To this end, we performed an RNA immunoprecipitation (RIP) assay with or without formaldehyde treatment using an anti-PU.1 antibody and an EBER-specific qRT-PCR on M81-transformed LCLs. This assay showed that EBER2 but not EBER1 was enriched in the precipitate but only after formaldehyde treatment (Fig. 5 C and D and SI Appendix, Fig. 6 A and B). This suggests that EBER2 and PU.1 interact indirectly. We then quantified PU.1 recruitment at the UCHL1 promoter region in B cells infected with M81 or M81/ΔE1 + 2 using a Chromatin immunoprecipitation (ChIP) assay with a PU.1 specific antibody coupled to qPCR to capture protein-bound DNA. This assay showed that the deletion of the EBERs reduced PU.1 recruitment to the UCHL1 promoter by 40% (Fig. 5E). Controls with a histone H3-specific ChIP showed no difference between these cell lines (SI Appendix, Fig. 6C). The data gathered so far suggested a possible physical association between EBER2 and the UCHL1 gene. We found that UCHL1 mRNA and EBER2 sequences could potentially hybridize at multiple sites, but an in silico analysis revealed that the interaction was potentially strongest between the beginning of the UCHL1 transcript (nucleotides 1 through 20) and the EBER2 nucleotides 78 through 97 (Fig. 6A). The free energy included in this potential interaction (ΔG = −27.35 kcal/mol) was similar to those previously recorded for the proven interaction between EBER2 and the terminal repeat RNA (ΔG = −28.10 kcal/mol) (17). Interestingly, the 1 through 20 UCHL1 region contains a previously identified PU.1 binding site, suggesting that an interaction with EBER2 could influence recruitment of PU.1 to this binding site (28). To determine whether this potential interaction is important for UCHL1 expression, we transfected wild-type or mutated versions of EBER2 into an LCL transformed by M81/ΔE1 + 2. This assay revealed that the mutation of the EBER2 nucleotide sequences 78 through 97 reduced UCHL1 expression by a threefold factor while moderately enhancing EBER2 expression levels (Fig. 6 B and C). Mutation of another EBER2 region (149 through 168) had no influence on UCHL1 transcription. We then assessed the consequences of mutating the putative EBER2–UCHL1 mRNA interaction sites on UCHL1 promoter–driven transcription and luciferase expression. While mutation of the UCHL1 nucleotides 1 through 20 on the UCHL1 luciferase reporter, which includes this sequence, halved luciferase expression in M81/ΔE1 + 2 LCL cells, simultaneous mutation of the 78 through 97 EBER2 sequences had an even more pronounced effect with a threefold reduction in expression (Fig. 6 D and E). Mutation of both putative interacting sites on EBER2 and UCHL1 mRNA led to a nearly fourfold reduction in UCHL1 reporter expression. This suggests that the complemented sequences on EBER2 and UCHL1 are important for UCHL1 expression and that both RNAs directly interact (SI Appendix, Fig. 7). Therefore, we performed hybridization studies with EBER2 and the 1 through 25 region of the UCHL1 transcript using the recently described psoralen cross-linking assay (Fig. 6F) (17). This experiment confirmed that both transcripts directly interact.
Fig. 5.
EBER2 mediates an indirect and potent interaction between PU.1 and the UCHL1 transcript. (A) We determined UCHL1 mRNA expression in B cells infected with M81 or M81/ΔE1 + 2 by RT-qPCR (n = 5). For each sample, the log2-transformed fold change (FC) is given based on the relative signals displayed by M81/ΔE1 + 2 and M81 wild-type. Central horizontal lines represent means, and error bars indicate SD. The statistical significance of the assay was evaluated with a one-sample t test. (B) Same as in A for UCHL1 pre-mRNA expression. (C) We determined EBER2 level by qRT-PCR following IP with IgG (negative antibody control) or with anti-PU.1 antibody in samples subjected to prior formaldehyde cross-linking. U1 was utilized as negative control located in the nucleus. Values are given as the average of three independent experiments ± SD (n = 3). Statistical analysis used a two-tailed paired Student’s t test. *P = 0.0472. (D) same as in C but without formaldehyde cross-linking. (E) PU.1 localization at the UCHL1 promoter region in B cells infected with M81 or M81/ΔE1 + 2 was measured by ChIP-qPCR. IgG was used as a negative antibody control in immunoprecipitations. The human α Satellite repeat region not bound by PU.1 served as a negative control. All data represent the mean of three independent experiments ± SD (n = 3). Statistical analysis used a two-tailed paired Student’s t test. *P = 0.02.
Fig. 6.
Base pairing between EBER2 and the UCHL1 mRNA potentiates UCHL1 expression. (A) Predicted RNA–RNA interaction between EBER2 and the UCHL1 transcript. (B) B cells infected with M81/ΔE1 + 2 were transiently cotransfected with multiple copies of EBER2 and a rat CD2 expression plasmid. We assessed EBER2 expression in purified CD2 populations using qRT-PCR. The pEGFP-C1 vector that encodes the enhanced GFP protein served as a negative control. M81 EBER2 is the pEGFP-C1 plasmid on which M81 EBER2 is cloned, together with its native promoter. M81 EBER2 nts 78 through 97 encodes EBER2 with mutations in the (78 through 97) region that is predicted to bind to the UCHL1 transcript. M81 EBER2 nts 149 through 168 encodes EBER2 with mutations in the (149 through 168) region that is not predicted to bind to the UCHL1 transcript. The data were given relative to values obtained in LCLs transfected with M81 EBER2. The data represents the mean of three independent experiments. (C) We assessed UCHL1 expression in the CD2 positive populations described in B using qRT-PCR. The data were normalized by the expression of EBER2 and are given relative to values obtained in LCLs transfected with the pEGFP-C1 plasmid. The data represent the mean of three independent experiments ± SD (n = 3). Statistical analysis used a two-tailed paired Student’s t test. *P < 0.05. (D) A schematic overview of a wild-type or mutated UCHL1 reporter plasmid. Five putative PU.1 binding sites are shown. The UCHL1 mutated reporter carries mutations in the region (nts 1 through 10) that is predicted to bind to the EBER2. TSS: transcription start site. (E) M81/ΔE1 + 2 LCLs were transfected with each of the given constructs along with UCHL1p-Luc reporter plasmid and the pRL Renilla expression vector. The total amount of DNA in all transfections was kept constant by adding control vector. Luciferase assays were performed 48 h posttransfection. Renilla and Firefly luciferase signals were measured using the Dual-Luciferase Assay System Kit. Renilla luciferase signals were used for normalization of the Firefly luciferase signal. The relative light units (RLU) between the luminescence levels generated by the expression constructs and the control constructs are given in a graph of bars that represent the mean of three independent experiments with error bars representing the SD (n = 3). The result of a two-tailed paired Student’s t test is given. **P < 0.01. (F) EBER2-RNP–containing cell lysate was incubated with a 25-nt UCHL1 fragment from the UCHL1 region (nts 1 through 25) predicted to anneal with EBER2 as shown in A. AMT was added where indicated, and the reaction was exposed to long-wave ultraviolet (UV) light (365 nm). Some samples were then treated with short-wave UV light irradiation (254 nm) to reverse cross-links. RNA was isolated, and Northern blotting was carried out on a denaturing urea-polyacrylamide gel using EBER2 as a RNA probe. The arrow indicates the position of EBER2 cross-linked to the UCHL1 RNA fragment. (G) Model depicting how the viral noncoding RNA EBER2 enables recruitment of the transcription factor PU.1 to the UCHL1 promoter.
Discussion
The ability to induce primary B cell growth quickly and efficiently is unique to the EBV, and how the virus drives this process is a central aspect of the host–virus interactions in both healthy and diseased hosts. We have now used a genetic approach to show that a noncoding RNA, EBER2, potentiates growth in primary B cells infected with the M81 strain. Under low-serum concentrations, the EBERs become even nearly indispensable for cell growth. Cells infected with EBV have an increased entry in the S phase and in the M phase with a relative excess of cells in anaphase, in comparison to primary B cells stimulated with CD40L and IL-4 or to B cells infected with the EBER deletion mutant. At the molecular level, the only alterations identified by the proteome and phosphoproteome assays in B cells infected with an EBER deletion mutant were lower levels of cyclin B1 and AURKA and AURKB. Complementation assays with cyclin B1 in LCLs generated with M81/ΔE1 + 2 show that the impact of EBER2 on cell growth can be largely ascribed to its ability to enhance the expression of this crucial cell-cycle mediator. Interestingly, B cells infected with the EBER null mutant and CD40L/IL-4-stimulated B cell expressed comparable levels of cyclin B1. This suggests that wild-type viral infection leads to a higher division rate though increased stimulation of cell-cycle regulators than seen in proliferating normal peripheral B cells. However, different B cell populations might divide at variable speeds in vivo. For example, centroblasts located in the dark zone of germinal centers proliferate rapidly (36).
Using a proteome-based approach, we identified the deubiquitinase UCHL1 as an EBER2 target. Western blot analysis showed that this noncoding RNA potentiates UCHL1 protein expression by a 10-fold factor on average. EBER2 is a protein-binding RNA that recruits the La antigen but also a large complex of interacting proteins that includes NONO, SFPQ, RBM14, PAX5, and PU.1 (SI Appendix, Fig. 7) (17, 34, 35, 37). NONO binds to EBER2 and recruits the other members of the complex though protein–protein interactions (34, 35). The UCHL1 promoter has five PU.1 binding sites, and this transactivator has previously been found to be important for UCHL1 expression (28). Thus, physical association between EBER2 and the UCHL1 transcript would facilitate recruitment of the transactivator to the UCHL1 promoter. We found that this indeed is the case and could identify an EBER2 20-ribonucleotide region as important for the interaction with the UCHL1 mRNA. Crucially, EBER2 could potentiate but hardly initiate UCHL1 protein expression as shown by transfection experiments in UCHL1-negative cells such as NIH 3T3 or HeLa or after infection of B cells with a LMP1 knockout. This fits with a model in which EBER2 interacts with a nascent mRNA and not with the promoter regions directly. Thus, EBER2 can only act if the UCHL1 promoter is firing. EBER2 has previously been shown to modulate transcription of LMP2 using a similar mechanism, although it did not influence its protein expression (12, 17). We now show that this property extends to cellular genes with, in this case, a major impact on protein expression.
The effect of EBER2 on the M phase regulators was mediated by UCHL1, as transfection of this deubiquitinase activated cyclin B1 and AURKA expression. However, EBER2 does not modulate their transcription, suggesting that it induces posttranslational modifications. Indeed, UCHL1 was previously shown to target cyclin B1 in an epithelial cellular model (30).
The increase in cyclin B1 expression induced by EBER2 though UCHL1 activation expression thus broadens the spectrum of cell-cycle regulators modified by the viral infection (reviewed in ref. 38). While the EBV latent proteins EBNA3A and EBNA3C induce the epigenetic repression of p14ARF, p15INK4b, p16INK4a, and p21WAF1 or their degradation (39–45), LMP2A induces p27KIP1 degradation (46).
A high UCHL1 expression has been observed in multiple types of carcinomas including colorectal and lung cancer (47, 48). UCHL1 is also expressed in B cell malignancies but only those derived from the germinal center, in particular BLs and diffuse large B cell lymphomas of the germinal center type (25, 27, 49). UCHL1 is likely to contribute to the acquisition of the malignant phenotype, as it increases the cell division rate, but it also facilitates tumor invasion through integrin upregulation (50). Reciprocally, downregulation of UCHL1 reduces cell division in B cell lymphoma cell lines (50). This suggests that UCHL1 acts as a pro-oncogenic signal, and its identification in proliferating EBV-infected B cells reinforces this view. However, UCHL1 is also physiologically expressed in reactive germinal center B cells and could contribute to the rapid proliferation of the centroblasts (25). Altogether, we suggest that EBER2-mediated induction of UCHL1 endows EBV-infected B cells with the ability to divide rapidly, thereby allowing an expansion of the B cell reservoir before the initiation of the immune response but at the same time also boosting the oncogenic potential of the virus.
Comparison of the EBER deletion mutants generated on the basis of M81 and B95-8 revealed that only M81-infected B cells require EBER2 for optimal growth. This can be explained by the much-higher levels of UCHL1 protein expression in B cells infected with the B95-8 EBER knockout, rendering EBER2 dispensable for this function. However, it is important to note that EBER2 also potentiates UCHL1 expression in B95-8-infected cells as B cells infected with the EBER deletion mutant express lower levels of this deubiquitinase, relative to its wild-type counterpart. Nevertheless, the UCHL1 expression levels in B cells infected with B95-8/ΔEBERs remained higher than in the same cells infected with wild-type M81. Thus, B95-8-infected cells do not need EBER2 to activate UCHL1 at levels required for optimal B cell transformation. A genetic tree of the LMP1 proteins expressed by P3HR1, B95-8, Akata, and M81 showed that the first two viral proteins are closely related but are more distant from the latter two that are themselves closely related (Fig. 4J). Thus, it is likely that the phenotypes displayed by the Akata and P3HR1 EBER knockouts can be explained by the features of their M81 and B95-8 counterparts. This likely explains previous contradictory reports that EBERs are not required for B cell transformation using B95-8 but are important for transformation mediated by the Akata virus (13–15).
The differences between M81 and B95-8 were likely to be due to differences in proteins or noncoding RNAs expressed during latency, and indeed, we could identify LMP1, which was previously known to induce UCHL1, as the reason for the different behaviors of B95-8 and M81. Indeed, B95-8 LMP1 led to a much-more-potent activation of the UCHL1 promoter than its M81 counterpart. Given the role of UCHL1 on the cell cycle and cell growth, this suggests that the various ability of LMP1 variants to activate this deubiquitinase is the first functionally relevant consequence of these polymorphisms. While LMP1 variants have previously been reported to activate NF-κB at variable levels, they did not lead to different phenotypes of the infected cells (31, 32).
The observation that both EBER2 and LMP1 activate UCHL1 expression shows the importance of this deubiquitinase for optimal B cell growth. LMP1 and EBER2 have been proposed to be functionally redundant because they both activate AKT in a model of BL EBV infection (24, 51). Our data indeed show that EBER2 is functionally redundant but only in the context of B95-8 infection.
The recognition that EBER2 upregulates UCHL1 expression and positively regulates the cell cycle has consequences for the pathogenesis of EBV-associated tumors. Well-characterized EBV transforming proteins such as LMP1 are not expressed in a large number of EBV-associated tumors, in particular BLs. We found that EBER2 can activate UCHL1 in BL cells and in primary epithelial cells without the help of LMP1, thereby directly contributing to the pathogenesis of these tumors, independently of other EBV products. Our work provides a molecular mechanism for the increased tumorigenicity of EBV-negative BLs carrying EBERs (23). Similarly, the ability of EBER2 to activate UCHL1, cyclin B1, and the Aurora kinases in primary epithelial cells might contribute to the development of EBV-associated carcinomas.
Materials and Methods
Detailed descriptions of the materials and methods are provided in SI Appendix and include the following: ethics statement; construction of recombinant viruses and virus production; cell lines and primary cells; oligonucleotides and probes; transfections; stable transfection of EBV-bacterial artificial chromosome and plasmid rescue into Escherichia coli; virus induction; quantification of viral titers; B cell infections; Western blot analysis; complementation experiments; antibodies; immunofluorescence staining; proteomics; Lentivirus infection in epithelial cells; analysis of mitosis and the cell-cycle profile; real-time RT-PCR; luciferase reporter assays; UCHL1 mRNA decay rate; infection experiments in NSG mice; immunohistochemistry; construction of mutated EBER2 expression plasmids; cell-cycle synchronization; EBV sequencing from mice tumors; psoralen cross-linking of RNAs.
ChIP.
The assay was carried out using a SimpleChIP Plus Sonication ChIP Kit (Cat#57976, Cell Signaling Technology) following the manufacturer’s instructions. For each reaction, 4 × 106 cells were used for immunoprecipitation. Briefly, LCLs were fixed with 37% formaldehyde (1% final concentration) for 10 min at room temperature; the reaction was stopped in a glycine solution for 5 min at room temperature and rinsed twice with cold phosphate-buffered saline (PBS). The cells were lysed in ChIP Sonication Lysis Buffer containing protease inhibitor mixture and RNase inhibitor. The nuclei were collected by centrifugation and lysed in ChIP Sonication Nuclear Lysis Buffer containing a protease inhibitor mixture and RNase inhibitor. Chromatin was sonicated briefly to obtain an average of 200- to 1,000-bp fragments. Sheared Chromatin was incubated with PU.1 antibody (Cat#2266) or Normal Rabbit lgG antibody (#2729) overnight at 4 °C with rotation. Protein G magnetic beads were added to each IP reaction and incubated for 2 h at 4 °C with rotation. Cross-linking was reversed by incubating immunoprecipitated complexes with NaCl (final concentration 0.2 M) and Proteinase-K (final concentration 0.25 mg/mL) for 2 h at 65 °C. DNA purification was performed after reversal of cross-links following the instructions. Real-time qPCR was performed with 2 μL precipitated DNA with primer pairs flanking consensus PU.1 binding sites. PCR conditions were: one cycle at 95 °C for 3 min and 40 cycles at 95 °C for 15 s and 60 °C for 60 s. The primers used for reaction are listed in SI Appendix, Table S2.
RIP Assay.
RIP was carried out following Millipore’s Nuclear RIP (Cross-Linked) Assay kit following the manufacturer’s instructions with some modifications. Briefly, 107 LCLs were washed with PBS and cross-linked in PBS containing 0.3% formaldehyde for 10 min at RT, quenched with glycine, and rinsed twice with cold PBS. Nuclear pellets were isolated and lysed, and IPs was performed by incubating antibody overnight followed by stringent washing of protein A/G bead pellets with final resuspension in TRIzol. RNA was isolated and quantified for EBER2 and U1 RNA by qRT-PCR analysis.
Statistical Analysis.
All results obtained in in vitro studies with LCLs generated by EBV wild-type or mutants with B cells from the same blood donors were paired and analyzed by paired Student’s t test. Mann–Whitney U test was applied to analyze NSG mice infected by either M81/ΔE1 + 2 or M81/ΔE1 + 2 Rev virus. All P values were analyzed as two-tailed, and values equal to 0.05 or less were considered significant unless indicated. The statistical analyses were performed with the GraphPad Prism 5 software.
Acknowledgments
We thank the staff of the animal facility at German Cancer Research Centre (DKFZ) for their excellent assistance with the animal experiments. The mass spectrometry/proteomics analysis was performed at the Zentrum für Molekulare Biologie der Universität Heidelberg Core facility for mass spectrometry and proteomics. This study was supported by the DKFZ (F100, and the Institut National de la Santé et de la Recherche Médicale (U1074). M.-H.T. is supported by the Ministry of Science and Technology (MOST) of Taiwan under grant no. MOST 109-2636-B-010-007.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115508118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Münz C., Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 17, 691–700 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Thorley-Lawson D. A., EBV persistence--Introducing the virus. Curr. Top. Microbiol. Immunol. 390, 151–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longnecker R. M., Kieff E., Cohen J. I., Epstein-Barr virus. Wolters Kluwer Health Adis 2, 1898–1959 (2013). [Google Scholar]
- 4.Feederle R., et al., A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 7, e1001294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhardt K., et al., A viral microRNA cluster regulates the expression of PTEN, p27 and of a bcl-2 homolog. PLoS Pathog. 12, e1005405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seto E., et al., Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 6, e1001063 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L., Aozasa K., Oshimi K., Takada K., Epstein-Barr virus (EBV)-encoded RNA promotes growth of EBV-infected T cells through interleukin-9 induction. Cancer Res. 64, 5332–5337 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Iwakiri D., Sheen T. S., Chen J. Y., Huang D. P., Takada K., Epstein-Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene 24, 1767–1773 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Samanta M., Iwakiri D., Takada K., Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 27, 4150–4160 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Nanbo A., Inoue K., Adachi-Takasawa K., Takada K., Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 21, 954–965 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwakiri D., et al., Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 206, 2091–2099 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., et al., Epstein-Barr virus ncRNA from a nasopharyngeal carcinoma induces an inflammatory response that promotes virus production. Nat. Microbiol. 4, 2475–2486 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Gregorovic G., et al., Cellular gene expression that correlates with EBER expression in Epstein-Barr Virus-infected lymphoblastoid cell lines. J. Virol. 85, 3535–3545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yajima M., Kanda T., Takada K., Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 79, 4298–4307 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y., Maruo S., Yajima M., Kanda T., Takada K., Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 81, 11236–11245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaminathan S., Tomkinson B., Kieff E., Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc. Natl. Acad. Sci. U.S.A. 88, 1546–1550 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee N., Moss W. N., Yario T. A., Steitz J. A., EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 160, 607–618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregorovic G., et al., Epstein-Barr viruses (EBVs) deficient in EBV-encoded RNAs have higher levels of latent membrane protein 2 RNA expression in lymphoblastoid cell lines and efficiently establish persistent infections in humanized mice. J. Virol. 89, 11711–11714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin X., et al., The Epstein-Barr virus BART miRNA cluster of the M81 strain modulates multiple functions in primary B cells. PLoS Pathog. 11, e1005344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang M. S., et al., Epstein-Barr virus nuclear antigen 1 does not induce lymphoma in transgenic FVB mice. Proc. Natl. Acad. Sci. U.S.A. 102, 820–825 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson J. B., Bell J. L., Levine A. J., Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 15, 3117–3126 (1996). [PMC free article] [PubMed] [Google Scholar]
- 22.Gnanasundram S. V., et al., PI3Kδ activates E2F1 synthesis in response to mRNA translation stress. Nat. Commun. 8, 2103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komano J., Maruo S., Kurozumi K., Oda T., Takada K., Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt’s lymphoma cell line Akata. J. Virol. 73, 9827–9831 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimienta G., et al., Proteomics and transcriptomics of BJAB cells expressing the Epstein-Barr Virus noncoding RNAs EBER1 and EBER2. PLoS One 10, e0124638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedekovics T., Hussain S., Feldman A. L., Galardy P. J., UCH-L1 is induced in germinal center B cells and identifies patients with aggressive germinal center diffuse large B-cell lymphoma. Blood 127, 1564–1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain S., et al., The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia 24, 1641–1655 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovaa H., et al., Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. U.S.A. 101, 2253–2258 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bheda A., Yue W., Gullapalli A., Shackelford J., Pagano J. S., PU.1-dependent regulation of UCH L1 expression in B-lymphoma cells. Leuk. Lymphoma 52, 1336–1347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentz G. L., et al., KSHV LANA and EBV LMP1 induce the expression of UCH-L1 following viral transformation. Virology 448, 293–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwan S. Y., et al., Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) promotes uterine serous cancer cell proliferation and cell cycle progression. Cancers (Basel) 12, E118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fielding C. A., et al., Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 75, 9129–9141 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mainou B. A., Raab-Traub N., LMP1 strain variants: Biological and molecular properties. J. Virol. 80, 6458–6468 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu N., Tanabe-Tochikura A., Kuroiwa Y., Takada K., Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: Malignant phenotypes of BL cells are dependent on EBV. J. Virol. 68, 6069–6073 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee N., Yario T. A., Gao J. S., Steitz J. A., EBV noncoding RNA EBER2 interacts with host RNA-binding proteins to regulate viral gene expression. Proc. Natl. Acad. Sci. U.S.A. 113, 3221–3226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallier M., Tavitian A., Moreau-Gachelin F., The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb. J. Biol. Chem. 271, 11177–11181 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Allen C. D., Okada T., Cyster J. G., Germinal-center organization and cellular dynamics. Immunity 27, 190–202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lerner M. R., Andrews N. C., Miller G., Steitz J. A., Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. U.S.A. 78, 805–809 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allday M. J., EBV finds a polycomb-mediated, epigenetic solution to the problem of oncogenic stress responses triggered by infection. Front. Genet. 4, 212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Nions J., Turner A., Craig R., Allday M. J., Epstein-Barr virus selectively deregulates DNA damage responses in normal B cells but has no detectable effect on regulation of the tumor suppressor p53. J. Virol. 80, 12408–12413 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruo S., et al., Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc. Natl. Acad. Sci. U.S.A. 103, 19500–19505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruo S., et al., Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc. Natl. Acad. Sci. U.S.A. 108, 1919–1924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tursiella M. L., et al., Epstein-Barr virus nuclear antigen 3A promotes cellular proliferation by repression of the cyclin-dependent kinase inhibitor p21WAF1/CIP1. PLoS Pathog. 10, e1004415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee S., et al., EBNA3C augments Pim-1 mediated phosphorylation and degradation of p21 to promote B-cell proliferation. PLoS Pathog. 10, e1004304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skalska L., White R. E., Franz M., Ruhmann M., Allday M. J., Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 6, e1000951 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skalska L., et al., Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog. 9, e1003187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fish K., Sora R. P., Schaller S. J., Longnecker R., Ikeda M., EBV latent membrane protein 2A orchestrates p27kip1 degradation via Cks1 to accelerate MYC-driven lymphoma in mice. Blood 130, 2516–2526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamazaki T., et al., PGP9.5 as a marker for invasive colorectal cancer. Clin. Cancer Res. 8, 192–195 (2002). [PubMed] [Google Scholar]
- 48.Hibi K., et al., Serial analysis of gene expression in non-small cell lung cancer. Cancer Res. 58, 5690–5694 (1998). [PubMed] [Google Scholar]
- 49.Bedekovics T., Hussain S., Galardy P. J., Walking the tightrope: UCH-L1 as an mTOR inhibitor and B-cell oncogene. Oncotarget 10, 5124–5125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolén U., et al., The ubiquitin C-terminal hydrolase UCH-L1 regulates B-cell proliferation and integrin activation. J. Cell. Mol. Med. 13, 1666–1678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herbert K. M., Pimienta G., Consideration of Epstein-Barr virus-encoded noncoding RNAs EBER1 and EBER2 as a functional backup of viral oncoprotein latent membrane protein 1. MBio 7, e01926–e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or supporting information.