Significance
Forward genetic screens in Drosophila have played an integral role in elucidating cellular and molecular pathways that govern almost every facet of biology. However, current screening methods in Drosophila are either fast, but limited in their specificity, or rely on imaging, requiring substantial expertise, time, and cost. We developed a rapid GFP-based ELISA that, when paired with the wealth of genetic tools available in Drosophila, can be used to screen for regulators of many subpopulations of cells, transcriptional programs, and proteins. Using this assay, we identified genes required for astrocytic synapse elimination. This technique provides a screening platform that is fast, accessible, and broadly applicable to many pathways and processes, making Drosophila an even more powerful screening platform.
Keywords: screening tools, transcriptional reporters, GFP-tagged proteins, synaptic development, astrocytes
Abstract
Drosophila is a powerful model in which to perform genetic screens, but screening assays that are both rapid and can be used to examine a wide variety of cellular and molecular pathways are limited. Drosophila offer an extensive toolbox of GFP-based transcriptional reporters, GFP-tagged proteins, and driver lines, which can be used to express GFP in numerous subpopulations of cells. Thus, a tool that can rapidly and quantitatively evaluate GFP levels in Drosophila tissue would provide a broadly applicable screening platform. We developed a GFP-based enzyme-linked immunosorbent assay (ELISA) that can detect GFP in Drosophila lysates collected from whole animals and dissected tissues across all stages of Drosophila development. We demonstrate that this assay can detect membrane-localized GFP in a variety of neuronal and glial populations and validate that it can identify genes that change the morphology of these cells, as well as changes in STAT and JNK transcriptional activity. We found that this assay can detect endogenously GFP-tagged proteins, including Draper, Cryptochrome, and the synaptic marker Brp. This approach is able to detect changes in Brp-GFP signal during developmental synaptic remodeling, and known genetic regulators of glial synaptic engulfment could be identified using this ELISA method. Finally, we used the assay to perform a small-scale screen, which identified Syntaxins as potential regulators of astrocyte-mediated synapse elimination. Together, these studies establish an ELISA as a rapid, easy, and quantitative in vivo screening method that can be used to assay a wide breadth of fundamental biological questions.
Drosophila are an excellent model system for forward genetic screens. Their fast generation time, balance of relative genetic simplicity and cellular and organismal complexity, and the high degree of conservation in genes and pathways between Drosophila and vertebrates, make Drosophila a powerful system for gene discovery.
Historically, there has been a tradeoff between designing screens with efficient readouts and designing screens to identify genes specifically involved in a desired phenotype of interest (1). In the first category, many classic and extremely productive screens have been performed using easy to identify phenotypes, such as lethality (2) or sterility (3). Others devised behavioral assays that could be performed with many flies in parallel (4–6) or studied processes that affect easily recognizable external features, such as using patterning of the fly’s cuticle to screen for genes involved in embryonic development (7) or studying genes that are involved in gross development of the eye (8). These types of screens can be performed rapidly, but are limited in the specificity of the cellular and molecular pathways of the genes they identify.
In order to interrogate more specific questions, screens in Drosophila have traditionally relied on imaging-based assays. These can be performed in a variety of different tissues and cell types and assess a wide variety of molecular pathways (9, 10). In these screens, the tissue of interest is often dissected, labeled, and then samples are imaged and typically qualitatively scored by the investigator. Even in systems designed to dramatically reduce the time required for these screens (11), they still take multiple person-years to complete. These screens are also costly and require specialized skills.
To address the limitations of these current approaches, we adapted an enzyme-linked immunosorbent assay (ELISA) to work with Drosophila lysates. ELISAs provide a quantitative readout of the concentration of a protein of interest, and thus can be used to answer a wide variety of specific molecular questions. ELISAs are fast and high throughput, require little technical expertise, and provide quantitative results. In addition, compared to imaging-based screens, they are relatively inexpensive and require no specialized equipment, making this screening method accessible to a wide variety of laboratory environments.
ELISAs have been used extensively for rapid screening in vitro. However, many questions simply cannot be addressed in cultured cells. Cells in vitro often lack the complex morphology observed in vivo, and thus cannot be used to investigate how diverse cell types establish and maintain these morphological features. Many cells also undergo large-scale transcriptional changes when cultured, making it difficult to study regulation of their transcriptional programs. Even in coculture systems, it is challenging to assess questions about how different cell types interact in complex tissues in vitro. Finally, it is difficult to accurately recapitulate complex organismal events, like development, injury, or disease in a dish. Drosophila provide a highly genetically tractable system in which these questions can be studied in vivo, and so adapting an ELISA for use with Drosophila lysates extends the types of questions that can be addressed using this rapid screening method.
While ELISAs can be used to identify many protein targets, we chose to develop an assay to quantify levels of GFP to maximize its versatility. Paired with the large array of GFP-based tools already available in Drosophila, this assay can be used to address a wide variety of cellular and molecular questions with minimal or no troubleshooting of new assay conditions. GFP can be driven in most cellular compartments and in almost any desired cell type using the vast array of Gal4 and other driver lines that have been developed (12–16), allowing a GFP-based ELISA to potentially assess the abundance of virtually any subpopulation of cells and their subcellular components. There are GFP-based transcriptional reporters that can be used to assess activity of a variety of different transcription factors, as well as reporters of signaling pathways and cellular metabolism and physiology (17–20). In addition, multiple approaches have been used in ongoing, large-scale efforts to insert endogenous GFP tags into all conserved Drosophila proteins (21–25). Thus, a GFP-based ELISA could be used to screen for regulators of almost any protein in any tissue type.
Here, we provide six independent examples of how this assay can be used to identify known regulators of cellular morphology, transcription, and protein levels in the Drosophila nervous system. We provide additional data demonstrating that it can be used across developmental stages, in additional tissue types, and in lysates prepared from whole animals. Further, we apply this assay to perform a small-scale screen that reveals Syntaxins are key mediators of astrocyte-neuron interactions during the process of developmental synapse elimination.
Results
Validation of a GFP-Based ELISA Assay for Use with Drosophila Tissue.
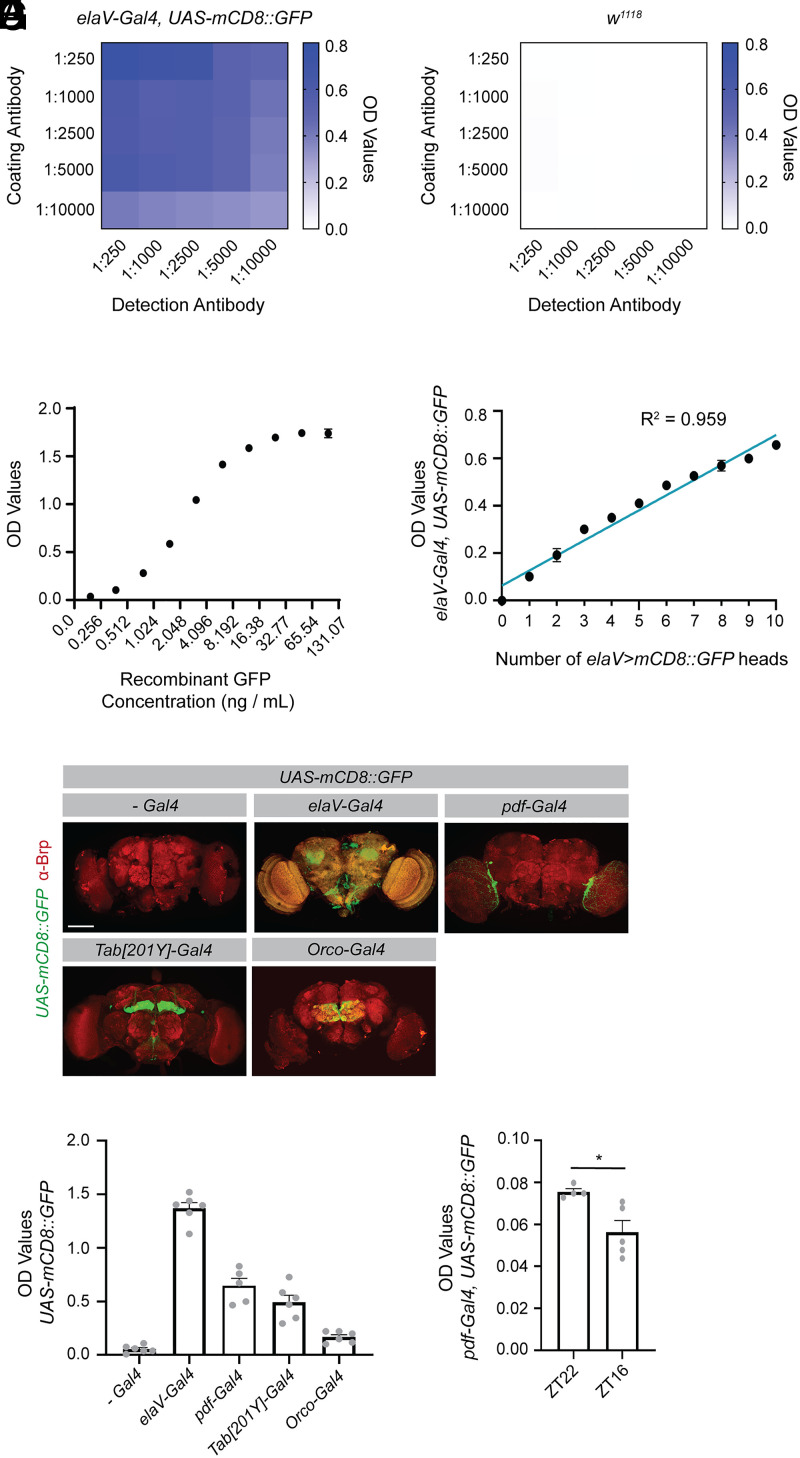
In order to use an ELISA as a screening method, we first needed to optimize and validate its use with Drosophila lysates. After optimizing tissue preparation and assay conditions, we were able to achieve substantial enrichment of signal in lysates prepared from homogenized heads of adult flies expressing GFP in all neurons (Fig. 1A), relative to lysates prepared from wild-type fly heads (Fig. 1B and SI Appendix, Fig. S1A), even with relatively low antibody concentrations. To evaluate the detection limits of the method, we used recombinant GFP to assay its dynamic range (Fig. 1C) and established that the assay was sufficiently sensitive to detect concentrations as low as 25 pg/mL GFP (SI Appendix, Fig. S1B). Using Drosophila tissue, we found that the assay could detect signal ranging from 1 to 10 heads of adult flies expressing GFP in all neurons within the assay’s linear dynamic range (Fig. 1D). We developed this in-house ELISA using commercially available antibodies and solutions that are commonly available in most laboratories to ensure it was sufficiently cost effective to perform large-scale screens. However, we found that a rapid commercial GFP ELISA kit can also perform well at detecting GFP in Drosophila adult head lysates (SI Appendix, Fig. S1C). While the cost of this commercial assay is ∼25 times higher than our assay, it can be performed in under 2 h, compared to our assay, which uses a multiday protocol. Together, these studies demonstrate that an ELISA can robustly detect GFP signal in Drosophila tissue, with high resolution between GFP-expressing flies and non-GFP–expressing controls, even in single fly heads. This assay also has a linear dynamic range that allows signal detection across a sufficient scale to detect physiologically meaningful differences in GFP levels.
Fig. 1.
An ELISA can be used to quantify GFP levels in Drosophila in diverse cell populations. An ELISA was optimized to detect GFP in Drosophila lysates. (A) For each sample, lysates were prepared from 5 heads collected from flies expressing a membrane-localized GFP under the control of the pan-neuronal elaV driver. Optical density (OD) values were recorded with different concentrations of coating antibody (chicken αGFP) and detection antibody (mouse αGFP). Depicted OD values represent the mean of two biological replicates. (B) A parallel analysis was performed as in A but in lysates containing 5 heads from w1118 flies. (C) Indicated concentrations of recombinant GFP were evaluated by ELISA and the resulting OD values are shown. Six replicates were performed for each concentration. (D) Ten fly heads were prepared for each sample, with the number on the x axis representing the number of heads from elaV > mCD8::GFP-expressing flies. The remaining heads in each sample were from w1118 flies. OD values were recorded from three replicates. A correlation analysis was performed using a simple linear regression. (E) Flies expressing the indicated drivers were crossed with UAS-mCD8::GFP flies, their brains dissected at 10 to 12 d posteclosion (dpe) and stained for Brp. The GFP depicted is from the endogenous GFP signal. Images were acquired at 20x magnification and represent maximum projections of z slices taken every 3 μm through the brain. (F) Five fly heads from each of the indicated genotypes were prepared for each sample and the OD values assessed by ELISA (no Gal4 (-Gal4) n = 6, elaV-Gal4 n = 6, pdf-Gal4 n = 5, Tab[201Y]-Gal4 n = 6, Orco-Gal4 n = 6). (G) Five fly heads were collected for each sample from flies expressing a membrane-localized GFP under the control of the pdf driver. Flies were collected at ZT22, shortly before the onset of light, and at ZT16 shortly after the onset of the dark phase. OD values for these samples were assessed by ELISA (unpaired, two-sided t test; ZT22 n = 4, ZT16 n = 5). Graphs represent the mean ± SEM *P < 0.05. (Scale bar, 100 μm.)
Detecting GFP Expression in Small Subpopulations of Cells.
While we had established that the ELISA could be used to detect GFP when expressed in all neurons, we next wanted to assess whether the assay was also sensitive enough to detect GFP when expressed in smaller subpopulations of cells. We homogenized heads from adult flies expressing a membrane-localized GFP under the control of several Gal4 drivers that express in subpopulations of neurons. GFP expression was readily detectable in all populations tested, including mushroom body neurons, olfactory receptor neurons, and even the ∼150 pdf expressing neurons (Fig. 1 E and F). We next wanted to test whether this assay could be used to detect relevant changes in these neuronal subpopulations. It has previously been shown that pdf neurons undergo circadian remodeling, with elimination of a subset of their processes in the early dark phase (26). To determine whether we could detect these circadian differences using the ELISA, we collected heads from flies at zeitgeber time 22 (ZT22), just preceding the onset of the light phase and ZT16, early in the dark phase. We were able to detect a significant decrease in GFP expression between these two time points (Fig. 1G), in line with the imaging studies that established a reduction in processes across circadian time. The sensitivity of signal detection in the additional driver lines used here (SI Appendix, Table S1) also suggests that the assay is likely to be compatible with many of the thousands of cell-specific driver lines available in Drosophila, which would allow one to explore a diverse array of questions in a variety of cell populations using quantification of GFP levels by ELISA as an initial readout.
Identifying Molecules Involved in Establishing Complex Cellular Morphology.
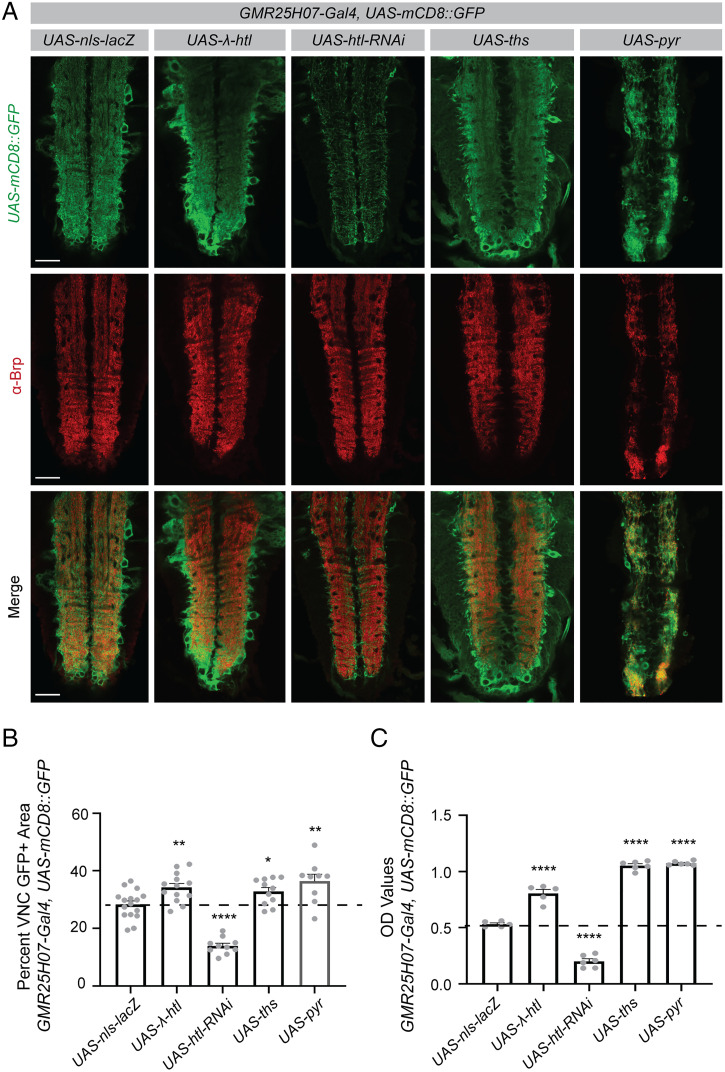
It has long been a challenge to screen for molecules involved in establishing the complex morphology of cells like astrocytes, which fail to recapitulate these morphologies in vitro. We wanted to test whether we could use our ELISA to identify regulators of astrocyte morphology in vivo. Using a mosiac analysis with a repressible cell marker (MARCM)-based clonal expression system, it has previously been established that signaling by ligands Thisbe (Ths) and Pyramus (Pyr) through the FGF receptor Heartless (Htl) promotes astrocyte growth (27). Indeed, when we expressed a membrane-localized GFP in astrocytes, we were able to detect by imaging an increased astrocyte area within the neuropil of fly larvae in which astrocytes overexpress ths and pyr and in flies that express a constitutively active form of the Htl receptor (Fig. 2 A and B). Conversely, knockdown of htl resulted in a reduced astrocyte area (Fig. 2 A and B). These findings were consistent with quantification of images using antibody staining for GAT (SI Appendix, Fig. S2 A and B), a membrane-localized astrocyte marker (27), suggesting that the GFP signal reliably correlates with the astrocyte membrane area. We were able to recapitulate all of the differences observed in these imaging studies by measuring GFP signal in dissected larval central nervous system (CNS) tissue using the ELISA (Fig. 2C). Not only did this assay require fewer animals and less time, but the variability in these results was much lower than when using image quantification (Fig. 2 B and C). We did not detect differences in GFP signal by ELISA with different numbers of UAS sequences in the background (SI Appendix, Fig. S2C), indicating that UAS dosage effects were not responsible for the differences we observed in the experimental genotypes used in this study. These results establish that the ELISA can be used to identify genes that have previously been shown to alter astrocyte morphology and could be used as an initial screening platform to identify novel regulators of astrocyte morphogenesis and maintenance. It could also be applied to screens aimed at identifying genes required to establish the morphology of other cell types in an in vivo system.
Fig. 2.
The ELISA can be used to screen for changes in complex cellular morphology. The driver line, GMR25H07-Gal4 in which Gal4 is expressed under the control of an enhancer associated with the astrocytic alrm gene, was used to express a membrane-localized GFP and to manipulate expression of genes previously implicated in regulating astrocyte morphogenesis. (A) Images of the larval ventral nerve cord (VNC) were acquired at 20x magnification and single z planes are shown for each genotype. Immunohistochemistry was performed for GFP to detect the membrane-localized reporter. Brp was used to identify the boundaries of the neuropil in the VNC. (B) Brp was used to define a region of interest encompassing the neuropil of the VNC and GFP+ area within that region of interest was quantified using single z planes, as shown above (UAS-nls-lacZ n = 16, UAS-λ-htl n = 13, UAS-htl-RNAi n = 10, UAS-ths n = 11, UAS-pyr n = 9). (C) The CNS was dissected from three larvae for each sample and the GFP signal within these samples quantified by ELISA (UAS-nls-lacZ n = 5, UAS-λ-htl n = 5, UAS-htl-RNAi n = 6, UAS-ths n = 6, UAS-pyr n = 6). Graphs represent mean ± SEM and each experimental genotype was statistically compared using an unpaired, two-sided t test to the UAS-nls-lacZ control sample (mean depicted as a dotted line). *P < 0.05, **P < 0.01, ****P < 0.0001. (Scale bar, 50 μm.)
Quantifying Signal from GFP-Based Transcriptional Reporters.
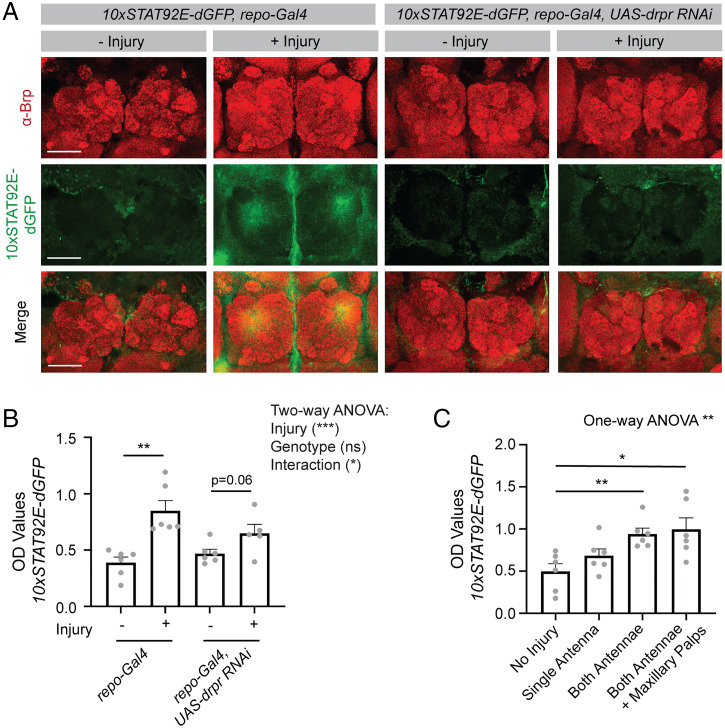
There is a large collection of GFP-based transcriptional reporters available in Drosophila, and we sought to determine whether we could use the ELISA to measure GFP signal from these transcriptional reporter tools. As an example, we chose to use a STAT92E-based GFP reporter, in which a destabilized GFP is expressed under the control of STAT92E enhancer sequences (19). We found that using this destabilized GFP was necessary in this context because the long half-life of GFP resulted in high levels of signal at baseline. This line has previously been used to evaluate up-regulation in STAT signaling following injury in Drosophila (28). Using imaging, we were able to recapitulate the expected increase in GFP signal within glial cells surrounding severed axons in the antennal lobe of adult flies following antennal ablation (Fig. 3A). While we were able to see this injury-induced increase by imaging, we did need to amplify the signal using a GFP antibody, indicating that this reporter is relatively weakly activated in this context. Despite this, we were able to detect robust increases in GFP signal by ELISA after injury in lysates prepared from adult fly heads (Fig. 3B), likely due to the high sensitivity of the ELISA (SI Appendix, Table S1). This response was attenuated by expression of an RNAi directed against draper (drpr) in glia (Fig. 3 A and B), consistent with Drpr’s known role in mediating glial injury responses (28). We were able to detect changes in signal from the transcriptional reporter by ELISA in response to a range of severities of injury (Fig. 3C). These results demonstrate that the ELISA can be used to detect glial transcriptional changes in response to injury in Drosophila and could be used to screen for genes involved in neuronal signaling that an injury has occurred or glial responses to those signals. More broadly, these results demonstrate that our ELISA can be used to measure signal from GFP-based transcriptional reporters, even those that are activated at relatively low levels.
Fig. 3.
The ELISA can be used to screen for transcriptional regulators using GFP transcriptional reporters. The STAT92E-dGFP reporter was expressed ubiquitously and gene expression manipulated in all glia using the repo-Gal4 driver. (A) Both antennae were removed from flies at 9 to 11 dpe and their brains were dissected after 24 ± 2 h. Brains were stained for Brp to label the neuropil and GFP to identify reporter activity. Images were acquired at 20x and a maximum projection of z sections, taken 1 μm apart through the antennal lobes, is shown. (B) Flies were injured as described in A and three fly heads were collected for each sample 24 h after injury. Lysates were prepared, and GFP OD values assessed by ELISA. Statistical comparisons were performed between uninjured (−) and injured (+) flies for each genotype using a two-way ANOVA, and unpaired two-sided t tests were performed between injury conditions within each genotype (repo-Gal4 – injury n = 6, repo-Gal4 + injury n = 6, repo-Gal4 UAS-drpr RNAi – injury n = 6, repo-Gal4 UAS-drpr RNAi + injury n = 6). (C) Graded severities of injury, as indicated on the x axis, were performed in flies expressing the STAT92E-GFP reporter and GFP OD values were measured by ELISA. A one-way ANOVA was performed across all injury conditions and unpaired, two-sided t tests were performed between the no injury condition and each of the injury conditions (no injury n = 6, single antenna n = 6, both antennae n = 6, antennae + maxillary palps n = 6). Graphs represent mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001, or as indicated. (Scale bar, 50 μm.)
Detecting Proteins with Endogenously Expressed GFP Tags.
Drosophila have a wealth of endogenously GFP-tagged proteins available, with the eventual goal of tagging every conserved protein with GFP (22–25, 29). Other strategies have also created comprehensive libraries of GFP-tagged proteins inserted as bacterial artifical chromosomes (BACs) (30, 31) or fosmids (32). Thus, if the ELISA could detect these endogenously tagged proteins, it could in principle be used to screen for regulators of virtually any protein of interest. We tested whether it could be used to detect expression of two example endogenously GFP-tagged proteins, Drpr, which was tagged within the endogenous drpr locus, and Cryptochome (Cry), which was inserted with a GFP tag using a BAC.
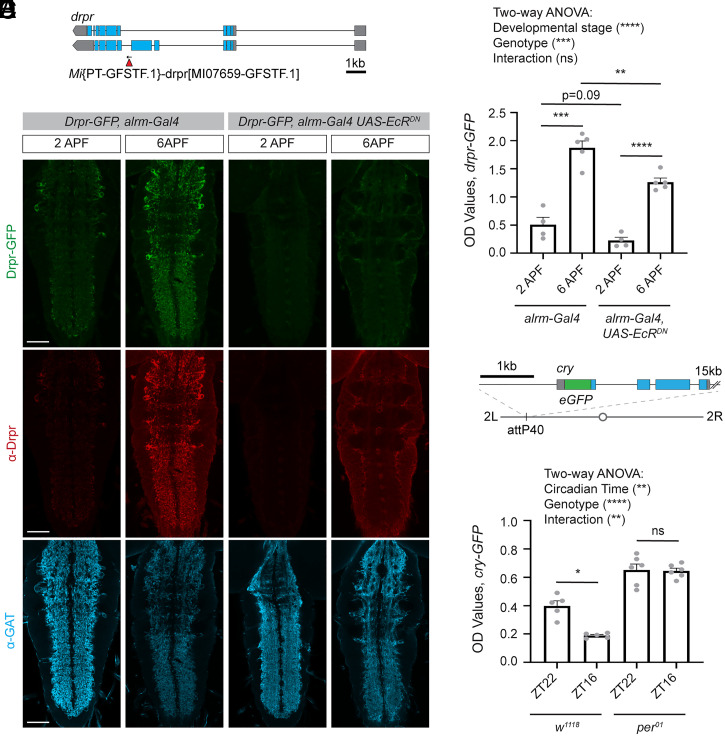
It had previously been shown that the phagocytic protein Drpr is up-regulated during metamorphosis (33), a developmental period during which the nervous system undergoes substantial remodeling, thereby placing high phagocytic demands on glia to clear extensive cellular debris. Indeed, we found using imaging that flies exhibited a substantial increase in Drpr-GFP expression during the period of metamorphosis from 2 to 6 h after puparium formation (APF) (Fig. 4A), similar to what we detected using an antibody against Drpr (Fig. 4B). This up-regulation was attenuated when astrocytes expressed a dominant negative form of the Ecdysone Receptor (EcRDN), which largely prevents these cells from responding to the cue that drives metamorphosis. Using the ELISA, we were similarly able to detect a robust increase in GFP signal in dissected CNS lysates from flies expressing Drpr-GFP from 2 to 6 h APF, and this response was partially attenuated in flies expressing EcRDN in astrocytes (Fig. 4C), similar to what we observed by imaging and by Western blot (SI Appendix, Fig. S3 A and B). This validates that the assay can detect GFP signal from flies expressing endogenously GFP-tagged Drpr, and that it is able to report robust increases in protein expression over the course of development. This could be used to screen for genes involved in establishing appropriate regulation of Drpr protein levels, or more broadly for genes that are required for glia to respond to developmental cues to change their function in appropriate contexts.
Fig. 4.
The ELISA can be used to detect endogenously GFP-tagged proteins. The ELISA was used to detect levels of two endogenously GFP-tagged proteins. (A) A line expressing Draper-GFP (Drpr-GFP) was previously generated using a Minos-mediated integration cassette (MiMIC) insertion of GFP into the endogenous drpr locus. (B) Drpr levels were assessed at 2 h after puparium formation (2APF) and at 6APF. This was performed in a control background, in which the astrocyte driver alrm-Gal4 was expressed alone, and in flies expressing a dominant negative ecdysone receptor (EcRDN) under the control of the alrm-Gal4 driver. Pupae were dissected and immunohistochemistry was performed for Drpr and the astrocytic marker GAT. The GFP signal represents the endogenous signal from Drpr-GFP. Images were acquired at 20x magnification and represent a single z plane of the VNC. (C) The CNS from five pupae were dissected for each sample and GFP OD levels assessed by ELISA. A two-way ANOVA was performed and unpaired, two-sided t tests were performed between relevant genotypes (alrm-Gal4 2APF n = 4, alrm-Gal4 6APF n = 5, alrm-Gal4 UAS EcRDN 2APF n = 4, alrm-Gal4 UAS EcRDN 6APF n = 5). (D) A line expressing Cryptochrome-GFP (Cry-GFP) was previously generated by inserting a GFP sequence into the N terminus of cry in a plasmid containing a 20-kb genomic region, which includes the cry gene. This sequence was then inserted at the attP40 site on the second chromosome. (E) Cry-GFP levels were assessed by ELISA from three heads per sample collected at ZT22 just preceding the onset of light and at ZT16, shortly after the beginning of the dark phase. This was also performed in a per01 mutant background. A two-way ANOVA was performed and unpaired two-sided t tests were performed between time points for each genotype (w1118 ZT22 n = 5, w1118 ZT16 n = 5, per01 ZT22 n = 6, per01 ZT16 n = 6). Graphs represent mean ± SEM; ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, or as indicated. (Scale bar, 50 μm.)
We next attempted to use the ELISA to measure a GFP-tagged Cry (Fig. 4D), which was inserted in a BAC and was previously shown to act as a functional Cry protein (34) and to maintain its endogenous pattern of circadian regulation (35). We were able to detect this endogenously tagged Cry-GFP in adult Drosophila heads using the ELISA, and similar to what had been previously described, found that its levels were higher at circadian times shortly before the onset of light (ZT22) and lower early in the dark period (ZT16) (Fig. 4E). These results were similar to what we observed by Western blot (SI Appendix, Fig. S3 C and D) and imaging (SI Appendix, Fig. S3E), though Cry-GFP levels were only detected at low levels in these contexts, highlighting the sensitivity of the ELISA (SI Appendix, Table S1). Mutants in the gene period (per) have previously been shown to abrogate circadian rhythms (6), and we show that per01 mutants no longer show circadian changes in Cry protein expression by ELISA (Fig. 4E). Our ELISA results also indicate that Cry protein levels are elevated in per01 mutants, consistent with the known role for Cry and Per complexes in inhibiting cry transcription (35). These results demonstrate that the ELISA can detect endogenously GFP-tagged Cry protein and report expected circadian changes in its expression. Thus, this assay could be used to screen for modifiers of circadian regulation of Cry protein. Taken together, these results provide a proof of principle that the ELISA can be used to sensitively detect endogenously GFP-tagged proteins, which promises to provide a platform to screen for regulators of many of the thousands of proteins for which GFP-tagged versions are available.
Identifying Glial Genes Involved in Regulation of Synapse Elimination.
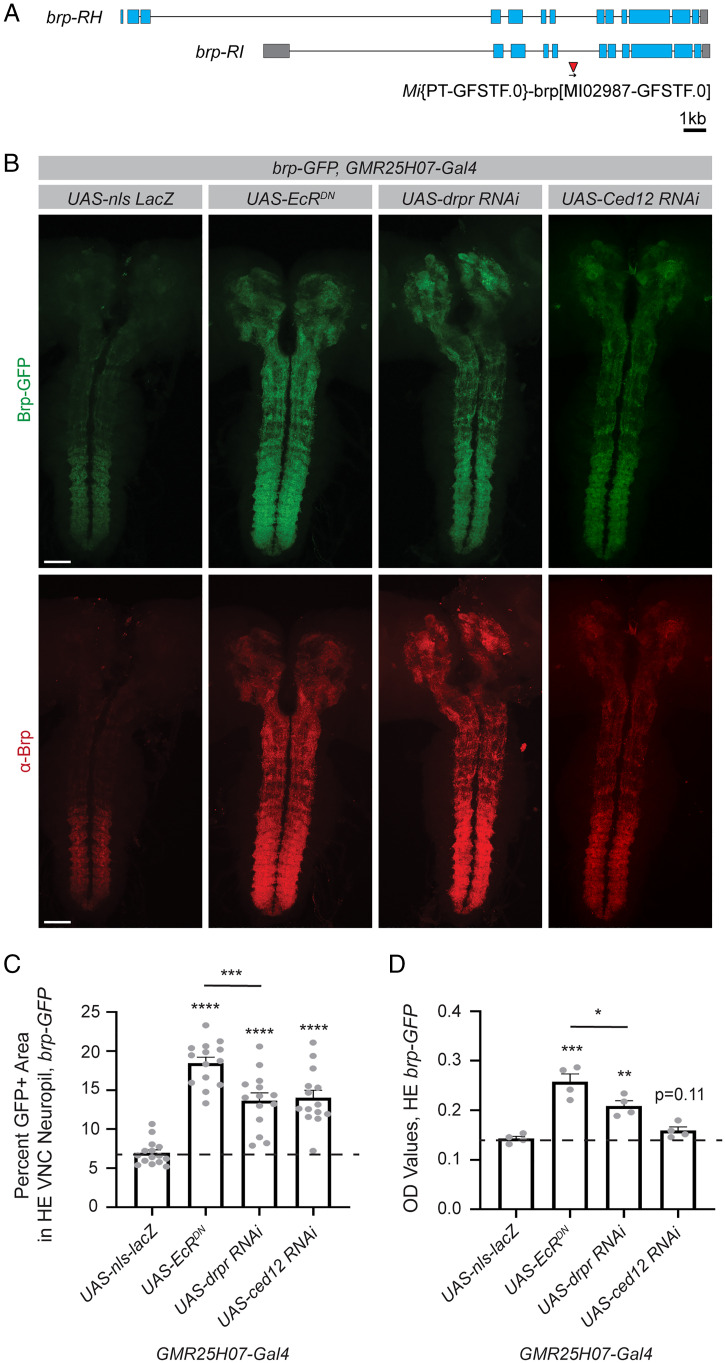
We took advantage of the ability of the ELISA to detect endogenously GFP-tagged proteins to measure a GFP-tagged Bruchpilot (Brp) (Fig. 5A), a protein integral to presynaptic structure (36) which is commonly used to evaluate synapses by immunohistochemistry (37). We specifically wanted to evaluate glial genes involved in the regulation of synaptic engulfment. To study this process in Drosophila, we evaluated Brp-GFP levels at a period of glial-mediated synapse elimination during metamorphosis (33). We evaluated Brp-GFP expression at head eversion (HE), ∼12 h into metamorphosis, when glia have already cleared a substantial number of existing synapses in wild-type flies (Fig. 5 A and B). For these experiments, we modified the ELISA to only detect full-length Brp-GFP by substituting the GFP detection antibody for a Brp-directed antibody. This was done because of concern that during synaptic engulfment by glia, Brp-GFP might initially be only partially broken down and GFP signal may persist even after synapses were engulfed. However, when this was tested empirically, we found that the results using either approach were indistinguishable (SI Appendix, Fig. S4 A–D). This protocol modification can, in principle, be tailored to any protein of interest with antibodies available to evaluate full-length tagged proteins or even used to compare full-length to cleaved GFP-tagged proteins to identify regulators of protein processing events.
Fig. 5.
The ELISA can be used to screen for molecules involved in glial-mediated regulation of synaptic protein levels during developmental synapse elimination. The ELISA was used to assess synapse elimination during development using the endogenously tagged synaptic protein, Bruchpilot-GFP (Brp-GFP). Gene expression was manipulated in astrocytes using the GMR25H07 driver. (A) An endogenously tagged Brp protein was previously generated using a MiMIC insertion of GFP into the endogenous brp locus. (B) The CNS was dissected from pupae at HE, ∼12 h into metamorphosis. Immunohistochemistry was performed for Brp, and the endogenous Brp-GFP signal was used to evaluate synapses. Images were acquired across the entire CNS and images shown are maximum z projections from two stitched 20x images. (C) GFP+ area within the VNC neuropil was quantified from single z planes (UAS-nls-lacZ n = 15, UAS-EcRDN n = 14, UAS-drpr RNAi n = 14, UAS-ced12 RNAi n = 14). (D) Five dissected CNSs were collected for each sample and GFP levels were assessed by ELISA (UAS-nls-lacZ n = 4, UAS-EcRDN n = 4, UAS-drpr RNAi n = 4, UAS-ced12 RNAi n = 4). Graphs represent mean ± SEM and the dotted line indicates the mean of the control UAS-nls-lacZ genotype. Statistical comparisons were performed using unpaired two-sided t tests between each experimental genotype and the control. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, or as indicated. (Scale bar, 50 μm.)
We validated that the ELISA could detect expected differences in Brp-GFP levels in genotypes known to affect glial-mediated synapse elimination. As expected based on previous work (33), we found that astrocyte expression of EcRDN, an RNAi directed against drpr, or an RNAi directed against a downstream phagocytic pathway component, Ced12, all resulted in increased retention of Brp-GFP signal into metamorphosis by imaging (Fig. 5 B and C). There were no significant differences evident at larval stages across genotypes (SI Appendix, Fig. S4 E and F), indicating that these genetic manipulations to astrocytes specifically affected Brp-GFP levels during this period of synapse elimination, rather than causing aberrant synaptic development. By ELISA, we were able to detect differences in Brp-GFP expression at HE with astrocyte expression of EcRDN, and drpr RNAi, but not with knockdown of Ced12 (Fig. 5D). This indicates that the ELISA can detect physiologically relevant changes in Brp-GFP levels that correlate with expected differences in synapse number during development, though it is less sensitive than imaging and quantification to subtle changes in protein levels in this context. Similar to what we observed by imaging, no differences in Brp-GFP levels were detected in larval samples by ELISA (SI Appendix, Fig. S4G). We also validated that quantification of Brp signal by immunohistochemistry showed similar results to those obtained with the endogenously expressed Brp-GFP (SI Appendix, Fig. S4 H and I). Together, these results demonstrate that the ELISA can detect expected differences in Brp-GFP levels during glial-mediated synapse elimination in Drosophila.
Validating the ELISA in Diverse Tissue Types and in Whole Animal Lysates across All Stages of Drosophila Development.
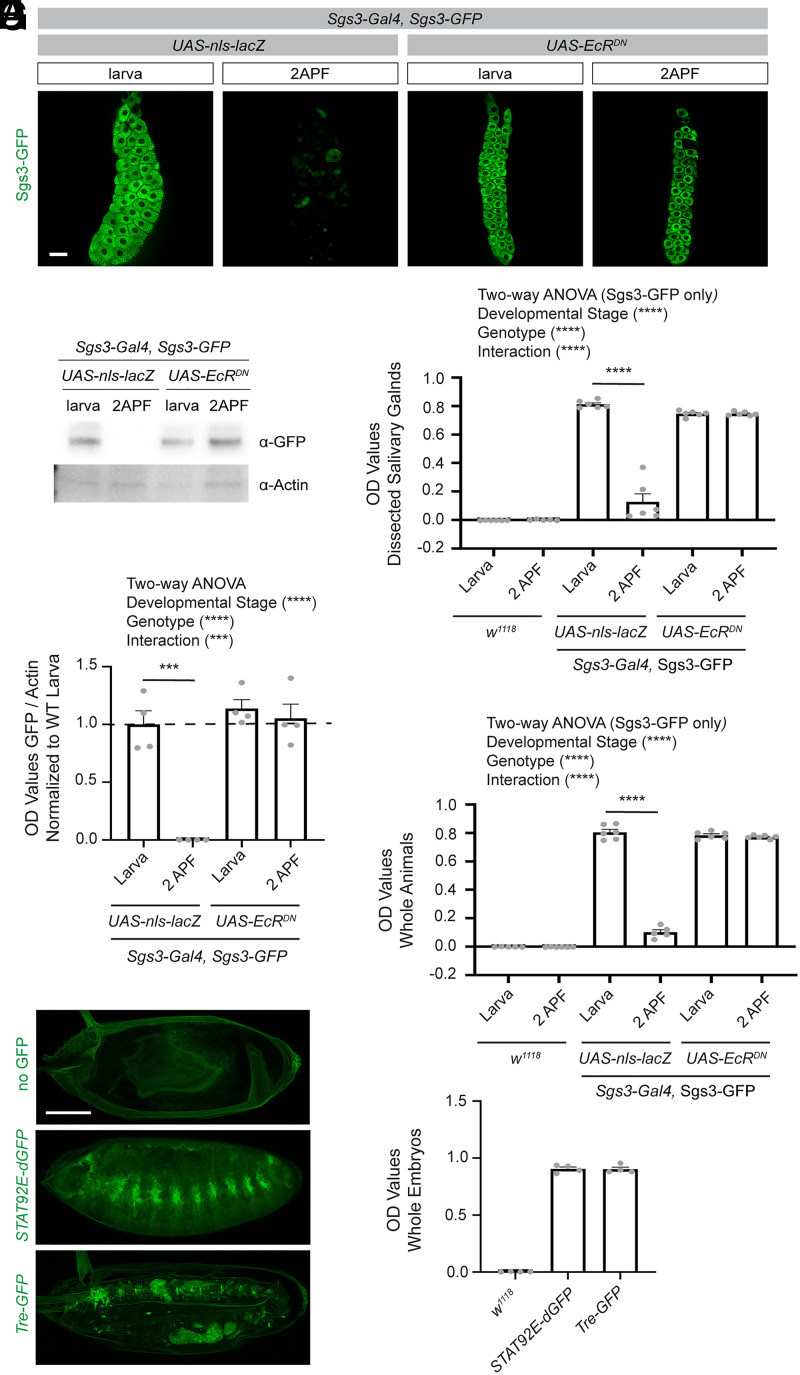
While the examples have so far focused on the utility of an ELISA-based assay in performing screens focused on the nervous system, we wanted to assess whether it might be more broadly applicable as a screening tool in other tissues and determine whether it would be suitable for screens across all stages of Drosophila development. To address this, first we evaluated whether the ELISA could detect relevant changes in levels of endogenously tagged salivary gland secretion 3 (Sgs3)-GFP, a protein expressed in salivary glands in late larval stages, which is later secreted at the onset of metamorphosis. This is a commonly used system to assay regulated exocytosis and tissue hormone responses (38). Similar to what we observed by imaging (Fig. 6A) and Western blot (Fig. 6 B and C), the ELISA was able to report a dramatic reduction in GFP signal from larval stages to 2 h APF, which did not occur in flies expressing EcRDN. ELISA results were comparable when using lysates prepared from dissected salivary glands (Fig. 6D) and whole larvae and pupae (Fig. 6E), indicating that, in this context, dissections would not be required to perform a screen to identify regulators of exocyotosis.
Fig. 6.
The ELISA can be used to measure GFP in tissues outside of the nervous system and in whole animals across all stages of Drosophila development. (A) Salivary glands were dissected from flies expressing a GFP-tagged Sgs3, a glue protein, which is produced at late larval stages and secreted at the onset of metamorphosis. Dissected glands were imaged at larval stages and 2 h APF in flies of the indicated genotypes. (B) Western blots were performed on dilutions of lysates prepared from single larva or pupa at 2 h APF and blotted with antibodies against GFP and Actin. (C) OD values from GFP were normalized to Actin for each sample, and these values normalized to the average of the larval control genotype. A two-way ANOVA was performed on all groups and unpaired two-sided t tests were performed to compare between time points within each genotype (UAS-nls-lacZ larvae n = 4, UAS-nls-lacZ 2APF n = 4, UAS-EcRDN larvae n = 4, UAS-EcRDN 2APF n = 4). (D) ELISAs were performed on lysates prepared from dissected salivary glands at a 1:20 dilution, the same protein amount as run in the Western blot assay for the indicated genotypes above, and OD values were recorded. A two-way ANOVA was performed on the Sgs3-GFP expressing groups (excluding the w1118 samples since these were run solely as technical controls [larvae n = 6, 2APF n = 6]). Unpaired two-sided t tests were performed between each time point within samples of the same genotype (UAS-nls-lacZ larvae n = 6, UAS-nls-lacZ 2APF n = 5, UAS-EcRDN larva n = 6, UAS-EcRDN 2APF n = 6). (E) Lysates were prepared from a whole larva for each sample, diluted 1:20 as described above, and OD values determined by ELISA. A two-way ANOVA was performed on the Sgs3-GFP expressing groups as in D. Unpaired two-sided t tests were performed between each time point within samples of the same genotype (UAS-nls-lacZ larvae n = 6, UAS-nls-lacZ 2APF n = 6, UAS-EcRDN larva n = 6, UAS-EcRDN 2APF n = 6). (F) Embryos were collected across all stages of embryonic development from w1118, STAT92E-GFP, and Tre-GFP flies. After fixation, GFP was imaged using confocal slices 3 μm apart through the whole embryo. Images shown are maximum z projections of these images. (G) Lysates were prepared from whole embryos across all stages of embryonic development for the indicated genotypes (n = 4 samples were prepared from independent collections for each group). Protein concentration for each sample was determined using a BCA assay and the concentration normalized across groups. These lysates were then analyzed by ELISA and OD values were recorded. ***P < 0.001, ****P < 0.0001. (Scale bars, 20 μm.)
Next, we tested whether our ELISA method could be used to measure GFP in embryos. We selected two transcriptional reporters that are known to be active during embryonic development (Fig. 6F), STAT92E-dGFP, which we had previously validated could be measured by ELISA as a readout of STAT activation in the context of adult nervous system injury, and Tre-GFP, in which GFP is expressed downstream of AP1 binding sites as a reporter of JNK activation (20). In both cases, we found that we were able to detect GFP from these reporter lines by ELISA, with signal substantially higher than what was observed in non–GFP-expressing controls (Fig. 6G). Because we collected embryos across multiple developmental stages, rather than normalizing to the number of animals, we normalized the protein concentration of the lysates before running the ELISA in this case. Loading 0.2 μg/μL protein concentration, or roughly 25 embryos for each sample produced quite robust signal by ELISA (SI Appendix, Table S1), at least for these transcriptional reporters, and the number of animals could thus be reduced for similarly highly expressed reporters or proteins in the context of a screen. Together, these examples illustrate that the ELISA can be used across every developmental stage of the fly, and that, at least in the contexts tested here, no dissections are required, greatly accelerating the speed at which large-scale screens could be performed using this system. In addition, these data demonstrate that the ELISA can be used in tissues beyond just the nervous system, expanding the possible applications of this technique to an even broader array of biological questions.
A Small-Scale ELISA-Based Screen to Identify Novel Glial Regulators of Developmental Synapse Elimination.
We have so far shown eight examples, which have validated that a GFP-based ELISA can reproduce expected results from a wide variety of different GFP-based tools, across diverse tissue types and developmental stages. We next wanted to use this ELISA method to perform a small-scale screen to uncover new biology, and chose to measure Brp-GFP signal to identify novel genes potentially involved in the process of glial-mediated synapse elimination. Work in mammalian systems has established critical roles for glial synaptic engulfment in nervous system development and in modulating neurological diseases (39–41). However, it is difficult in mammalian systems to simultaneously manipulate neuronal and glial gene expression in vivo, and it is extremely time and cost intensive to perform forward genetic screens in mammals. Thus, our knowledge about how neurons and glia communicate during this process and the full array of genes required remains incomplete.
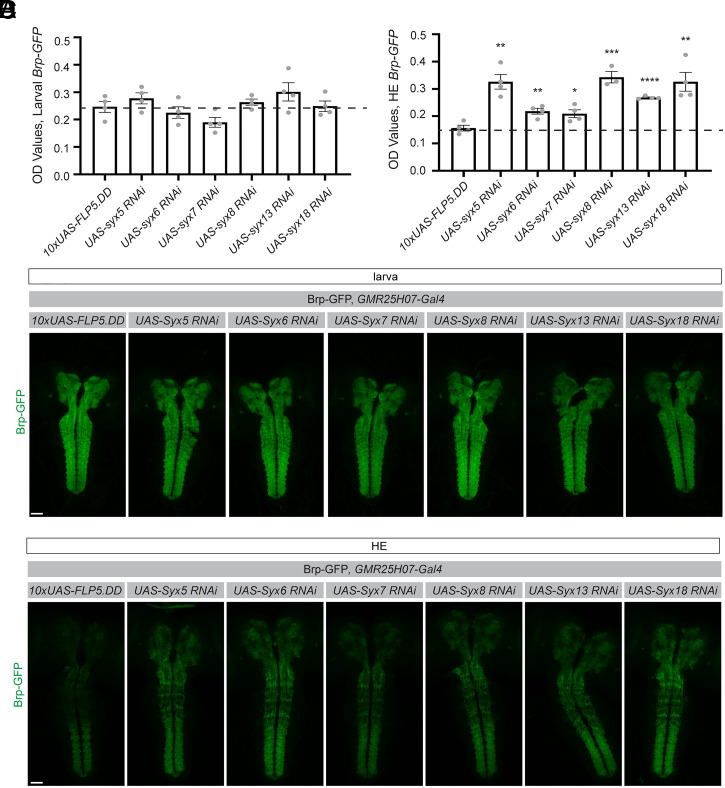
Using our ELISA, we identified a class of syntaxin genes that did not affect baseline levels of Brp-GFP signal at larval stages (Fig. 7A), but all increased retention of Brp-GFP levels into metamorphosis to varying degrees (Fig. 7B). Syntaxins are known to play roles in vesicular fusion and transport (42) and have been extensively studied in the context of neuronal transmission (43). However, their roles in glia are not well understood. We validated these results by imaging (Fig. 7 C and D), which showed similar trends as the ELISA results. Notably, it was difficult to assess the relative differences in Brp signal among different genotypes by imaging, while they were clearly distinguishable by ELISA.
Fig. 7.
A small-scale ELISA-based screen was performed to identify genes involved in astrocyte-mediated synapse elimination during metamorphosis. (A) The CNS was dissected from larvae and lysates were prepared from five CNSs for each sample. The ELISA was used to assess endogenously tagged Brp-GFP levels. Genes were knocked down in astrocytes by expressing UAS-driven RNAi lines using the GMR25H07-Gal4 driver. Statistical comparisons were performed using unpaired, two-sided t tests between each experimental genotype and the control 10xUAS-FLP5.DD line, the mean of which is shown as a dotted line (10xUAS-FLP5.DD n = 4, UAS-syntaxin5 RNAi n = 4, UAS-syntaxin6 RNAi n = 4, UAS-syntaxin7 RNAi n = 4, UAS-syntaxin8 RNAi n = 4, UAS-syntaxin13 RNAi n = 4, UAS-syntaxin18 RNAi n = 4). (B) ELISAs were similarly performed on five dissected CNSs per sample from flies collected at HE. Unpaired two-sided t tests were performed between each experimental group and the control genotype, as described in A above (10xUAS-FLP5.DD n = 4, UAS-syntaxin5 RNAi n = 4, UAS-syntaxin6 RNAi n = 4, UAS-syntaxin7 RNAi n = 4, UAS-syntaxin8 RNAi n = 3, UAS-syntaxin13 RNAi n = 4, UAS-syntaxin18 RNAi n = 4). (C) Results from the ELISA were confirmed by evaluating Brp-GFP in larvae and (D) pupae by imaging. Images were acquired at 20x and two images were stitched to show maximum intensity projections through the CNS. Graphs represent mean ± SEM, **P < 0.01, ***P < 0.001, ****P < 0.0001. (Scale bar, 50 μm.)
To investigate how these Syntaxins might regulate glial-mediated alterations in synaptic protein levels, we examined the morphology of astrocytes during this developmental process. At larval stages, astrocytes infiltrate the neuropil and closely associate with synapses (27). Just 6 h into metamorphosis (6 h APF), astrocytes transform into highly active phagocytes that engulf synaptic material and other cellular debris (33), accompanied by a loss of their highly branched morphology and accumulation of intracellular vesicular structures (SI Appendix, Fig. S5 A and B). With the exception of syntaxin 5, knockdown of most of the syntaxin genes did not significantly disrupt astrocyte morphology at larval stages (SI Appendix, Fig. S5 C and D). However, at 6 h APF, manipulation of all except syntaxin 13 resulted in alterations in astrocyte area relative to controls (SI Appendix, Fig. S5 E–G), with a range of phenotypic impacts on astrocyte morphology. These data provide a proof of principle that the ELISA can identify novel molecules involved in glial regulation of synaptic protein markers, and revealed surprising roles for syntaxins in mediating the changes in astrocyte morphology that are likely required for successful elimination of synapses during Drosophila development. Furthermore, this successful screen demonstrates that an ELISA-based screening method can uncover new biological insights in an in vivo system.
Discussion
Our work demonstrates that ELISAs can be used to rapidly perform quantitative screens in an in vivo system. In conjunction with the array of tools available in Drosophila to express GFP in numerous different subsets of cells, GFP-based transcriptional reporters, and thousands of endogenously GFP-tagged proteins, this assay can be used to screen for regulators of an extensive array of cellular and molecular pathways. In this study, we demonstrated the ease of adapting this assay to address a wide variety of questions related to fundamental topics in neuroscience and beyond and were able to validate eight completely independent sets of screening paradigms that could be performed with this ELISA platform.
First, we showed that the ELISA was capable of detecting GFP expression in a variety of different cell populations and demonstrated that it could detect changes in GFP signal, which correlated with morphological remodeling of pdf neurons. The example subpopulations evaluated here represent only a small handful of the subsets of neurons that can be labeled in Drosophila using the extensive array of driver lines that have been developed (12–14). This ELISA could be used with any drivers that induce high enough levels of expression. Detection of GFP signal within the small number of pdf neurons, ∼150 per brain, suggests that this assay could be used even with drivers that express in small subsets of cells.
We further demonstrate that our ELISA can be used to identify genes known to be involved in the morphogenesis of astrocytes. While the complex morphology of many cells in the nervous system has long been a topic of interest, the genes that establish these unique morphologies have been difficult to screen for in a systematic manner. It is challenging to quantitatively distinguish different patterns of infiltration of these densely organized cell types in vivo, and even if this were possible, the imaging and analysis would be prohibitively time consuming to perform a large-scale screen. Thus, the ELISA could provide a platform for a large-scale screen of genes that are involved in establishing the morphological features of glia or other cell types with interesting morphological characteristics, which have previously not been possible to study. Here, we used a membrane-localized GFP as a proxy to reflect the membrane content of these cells. However, Drosophila lines are available with GFP localized to virtually every cellular compartment, and thus the same principle could be used to screen for regulators of mitochondrial density, endoplasmic reticulum development, lysosomal content, or other subcellular features, in any desired subset of cells.
In addition to evaluating features of cells themselves, we also demonstrate that this assay can be used to assess changes in signaling pathways within desired cellular subsets, specifically, using a GFP-based transcriptional reporter for activation of the STAT pathway following injury. A wide variety of GFP-based transcriptional reporters have been developed in Drosophila to assess activity of specific transcription factors (20) and could be paired with this ELISA to screen to regulators of these diverse transcriptional programs. In addition, transcriptional reporters have been developed that respond to cytoplasmic Ca2+ levels, which can be used, among other things, to provide a readout of neuronal activity (17, 18). Thus, ELISAs could be used to screen for modulators of neuronal activity in any desired subset of neurons in response to environmental or genetic perturbations.
Perhaps the largest array of GFP-based tools available in Drosophila is endogenously GFP-tagged proteins. Thanks to large scale efforts and multiple approaches toward the goal of tagging every conserved protein in Drosophila (22–25), there are already thousands of these lines available. We showed that this assay could detect GFP expression in two such lines, expressing Drpr-GFP and Cry-GFP. In both cases we found that the ELISA could detect expected changes in the expression of these proteins, even over short time scales, during the course of development and across circadian time. In addition, when comparing these results to Western blots, the ELISA was more sensitive, both in its ability to detect lower levels of GFP and in the dynamic range of protein levels it was able to capture (also see SI Appendix, Table S1). These two examples serve as a proof of principle that this assay could be used to evaluate expression of a variety of proteins, and thus could be used as a highly sensitive screening method for regulators of expression of virtually any protein of interest.
We took advantage of our assay’s capability to detect endogenously tagged proteins to evaluate Brp-GFP levels as a correlative proxy to assess synapse number during the process of metamorphosis. We first validated that the assay could detect aberrant retention of synapses with knockdown of genes within astrocytes, which had previously been shown to regulate this process (33). We then used this platform to perform a small-scale screen to identify novel potential regulators of glial-mediated synaptic engulfment and identified a previously unknown role for glial expression of syntaxins in regulating Brp-GFP labeled synapses and Brp-GFP protein levels. While this screen was designed to identify novel regulators of synapse elimination, we cannot exclude that these results are due to alterations in glial-mediated regulation of neuronal gene transcription or other cellular processes. Indeed, as in all screening methods, genes identified in ELISA-based screens will require thoughtful secondary methods of validation. In this case, we found that many syntaxins were required to regulate the changes in astrocytic phenotypes required for these cells to engage in synapse elimination during metamorphosis. Syntaxins 7 and 13 have previously been shown to be associated with vesicles in cultured phagocytes (44), but broader roles for this class of proteins in phagocytic function and their role in the nervous system was unknown. We recognize that these data are incomplete, and future work will be required to follow up on the results of this small-scale screen to definitively identify the functional roles of Syntaxins within astrocytes that are responsible for shaping nervous system development.
For our analyses using Brp-GFP levels to reflect possible changes in synapses, we used a modified ELISA to identify full-length Brp-tagged GFP by pairing GFP coating and Brp detection antibodies. This protocol modification could be used in other studies aimed at assessing relative levels of full-length and cleaved versions of different protein products or even to study regulators of posttranslational protein modifications by pairing GFP with antibodies against phosphate, ubiquitin, glycosides, or other desired functional groups. Together, these examples demonstrate that this GFP-based ELISA method can be used to screen for regulators of a wide variety of specific cellular and molecular pathways in an in vivo system.
Our screening method offers several advantages over the current methods that are used to perform screens aimed at identifying genes involved in specific cellular and molecular pathways, which most frequently rely on imaging. First, samples are simply frozen at the time of collection, and thus can be collected over time, and the assay itself later run on desired batches of samples together at the investigator’s convenience. In several contexts we tested, no tissue dissections were required, greatly increasing the speed of sample preparation and requiring little technical skill. We did find that larval and pupal nervous systems needed to be dissected in the contexts of evaluating membrane-localized GFP in astrocytes and to assess Brp-GFP using this assay, however, so it is an important consideration that dissections may be required for some desired screening applications. Regardless, after samples are collected, the assay itself requires little active time, making it substantially faster than mounting, imaging, and manually evaluating samples in imaging-based screens. While the exact time savings will vary based on the setup of the screen, we anticipate that running an ELISA-based assay for 1,500 lines would take weeks, while most imaging-based screens require years to complete. In addition to being faster, this ELISA-based screening method also requires no equipment beyond a colorimetric plate reader and basic supplies commonly available in the laboratory. We estimate that material costs of performing the ELISA would not exceed $1,500 for 1,500 lines. Finally, this screening method produces quantitative results. Thus, one does not have to rely on multiple investigators to employ consistent qualitative and subjective scoring criteria to establish hits from a screen, and one can go back at a later time to assess the precise relative strength of the effects of additional genes or conditions within the original screening dataset. Together, this ELISA-based screening assay offers a rapid, low-cost, accessible, and quantitative method that can be easily adapted to address a wide variety of cellular and molecular questions.
It is important to note that this method also has limitations. In imaging-based screens, one can see patterns of multiple different phenotypes. The ELISA simply provides a readout of the concentration of GFP molecules, changes in which may indicate a range of phenotypes and be caused by multiple underlying mechanisms depending upon how the screen is designed. For example, when we assayed astrocyte morphology in this study, the ELISA produced nearly identical results in conditions in which ths and pyr were overexpressed. However, by imaging, it was clear that while ths overexpression seemed to promote relatively uniform increases in astrocyte density within the neuropil, pyr overexpression resulted in highly concentrated clumps of astrocyte processes and disruption of the neuropil structure. Thus, possible hits identified in ELISA-based screens will need to be validated using imaging or other modalities to extract more information about how genes identified as hits might act. This assay would also not be compatible with most MARCM-based screening methods due to the stochastic nature of recombination in this system, and thus could not address questions that require use of a clonal system in which the number of labeled cells varies from animal to animal. In addition, we found that the assay could not always be used to reliably compare differences between different developmental stages of the fly. For example, Brp levels in pupal samples should be strongly reduced relative to larvae, but we detected only modest changes in signal between these developmental stages. So, while results within each stage of the life cycle were internally consistent, we could not compare between these groups in this context. The same may be true for comparisons across different tissue types within the fly as well, as we have not tested this directly. Finally, while the ELISA is sufficiently sensitive to detect relatively low levels of GFP, this screening method is not an ideal platform to study things with very low levels of expression in very small populations of cells. Thus, not all cells and proteins will be amenable to screening using this technique.
We have primarily focused on demonstrating the utility of a GFP-based ELISA for screening in the nervous system of Drosophila, but have provided proof-of-principle examples that this method could also be extended to use in other tissues and across all developmental stages. While we have used Drosophila to demonstrate its utility, this assay could also be adapted to work in other organisms that are amenable to forward genetic screens, such as Caenorhabditis elegans. In this paper, we developed the ELISA to work with GFP, since numerous GFP-based tools are already available, such that this assay could be used with virtually no troubleshooting of the assay itself for a wide variety of applications. However, ELISAs can work with many antibodies, and in cases where two antibodies are available against a given protein that recognize different epitopes, an ELISA may be able to be optimized to detect that native protein. In this case, there would be no need to use any lines that express GFP tags, and the protein itself could be measured directly. This approach and the GFP-based ELISA developed in this work could also be used for purposes beyond screening. These assays produce highly quantitative readouts of protein concentrations, and we demonstrate that it is a faster and more sensitive tool to quantify relative changes in protein levels compared to Western blots. Thus, an ELISA could be used to assess efficiency of knockdowns at the protein level if a GFP-tagged version of that protein is available, determine the relative strength of different driver lines by expressing GFP under their control, or simply to assess the effect of targeted manipulations on the levels of a given protein of interest.
In this study, we have demonstrated that a GFP-based ELISA can be used to rapidly screen for regulators of a wide variety of cellular and molecular pathways in Drosophila, demonstrating its utility as a tool to perform in vivo forward genetic screens. This assay makes Drosophila an even more powerful system to perform forward genetic screens that aim to understand a wide variety of targeted phenotypes and processes. This method promises to make these screens faster to validate and perform, accelerating the discovery of novel genes involved in diverse biological processes.
Materials and Methods
ELISA Assay.
ELISA sample preparation.
For larval and pupal CNS samples, the nervous system was dissected in phosphate buffered saline (PBS) containing 0.1% Tween-20 (0.1% PBST) and kept in 1.5-mL Ependorf tubes on ice until all samples were collected. Samples were frozen and stored at −80 °C until use. For whole animals, larvae and pupae were collected at the appropriate stage of development, washed, and placed in a 1.5-mL Eppendorf tube before being stored at −80 °C until use. For larval and pupal dissected salivary glands, larvae were staged to ensure they were late third instars prior to dissection. Salivary glands were removed, placed in 30 μL 0.1% PBST in 1.5-mL Eppendorf tubes, and placed at −80 °C until use. After homogenization (see below), samples using Sgs3-GFP were diluted 1:20 before being loaded into the ELISA plate.
Embryos were collected at 0 to 22 h after egg laying from grape agar plates using a paint brush and transferred to a 40-μm cell strainer in a Petri dish containing ddH2O. Cell strainers were then transferred to a 50% bleach solution for 2 min to dechorionate the embryos. Embryos were then washed briefly in ddH2O and then transferred from cell strainers into a 1.5-mL Eppendorf tube containing ddH2O using a paint brush. These tubes were then spun down at 13,000 rpm for 10 s. Excess ddH2O was removed and samples were stored at −80 °C. After homogenization (see below), a bicinchoninic acid (BCA) assay was performed using a BCA protein assay kit (Pierce) according to instructions in order to assess the protein concentration of each sample. Samples were then diluted such that they all contained the same concentration of protein (0.2 μg/μL) before being loaded into the ELISA plate.
For adult samples, flies were anesthetized using CO2 and collected in 1.5-mL Eppendorf tubes. Flies were then frozen on dry ice and stored at −80 °C. To remove heads, tubes were vortexed at high speed for 15 to 20 s. Tubes were then inverted onto a white index card and the desired number of heads for each sample were collected in fresh tubes. These tubes were then stored at −80 °C until use. We detected no substantial changes in signal in any of the applications used in this study when heads were thawed at this collection step. If large numbers of heads are required, flies can be collected in 15-mL conical tubes, frozen, vortexed, and passed through a series of two sieves (size no. 25 and no. 40, respectively). Heads will settle into the second sieve and can be gently removed using a paintbrush.
At the time of the assay, all samples were removed from −80 °C and placed on ice. The 0.1% PBST (adult head and dissected samples 120 μL, whole larvae and pupae 150 μL, and embryos 130 μL) was added to each sample and samples were homogenized using a pestle with a hand-held homogenizer in 1.5-mL Eppendorf tubes for 5 s. Longer homogenization times, addition of protease inhibitors, and sonication were tested, but had no impact on signal in the applications used in this study. Samples were placed on ice until all samples had been processed. Samples were centrifuged at 13,000 rpm for 10 min at 4 °C and placed on ice until loading into the plate. See summary in SI Appendix, Fig. S6A.
ELISA assay.
Plates (Immunolon 2HB flat bottom, Thermo no. 3455) were coated in 100 μL buffer (30 mM sodium carbonate, 70 mM sodium bicarbonate in ddH2O, pH 9.6) containing a 1:1,000 dilution of chicken αGFP antibody (Abcam no. ab13970). Our results in Fig. 1 A and B indicate that lower concentrations, down to 1:5,000 could also be used, but because of the wide range of applications and signal intensities anticipated throughout our studies, we used an excess of antibody to ensure we captured even samples with relatively high signal within the assay’s linear dynamic range. We also tested alternative antibodies and found that a chicken αGFP antibody (Aves no.1010) produced slightly lower signal with a 1:5,000 antibody dilution, but is a lower cost alternative that would also be suitable for most applications. The plate was sealed using Glad Press-n-Seal and kept on an orbital shaker at 4 °C overnight. At all steps, we found that the assay was tolerant of a variety of shaking speeds, but typically used a setting of ∼40 rpm. The plate cover was removed and the plate washed with 0.1% PBST. For washes, the plate was inverted over a sink and tapped on a paper towel to remove most of the liquid from the wells. The wash solution was applied using a squirt bottle aimed at the sides of the wells. The first and last wash were performed for 4 min on an orbital shaker at room temperature (RT) and two additional quick washes were performed in between in which wash solution was added and then removed from the plate immediately. We did find that substantially extending wash times resulted in reduced signal. After these washes, 350 μL of 2% nonfat dry milk (Carnation) in 0.1% PBST was added to each well. The plate was sealed and placed on an orbital shaker at 4 °C overnight. The seal and blocking solution were removed and the plate was washed as described above. After the last wash solution was removed from the plate, samples were added. In our validation experiments, we found that 0.1% PBST alone produced a signal indistinguishable from that of lysates from flies that did not express GFP, and thus 0.1% PBST was used as our negative control sample. Recombinant GFP (Abcam no. ab84191) was diluted in 0.1% PBST. Samples should be loaded quickly to prevent the plate from drying out, but we did perform optimization experiments in which samples were not loaded for up to 45 min and detected no substantial differences in signal. The plate was then sealed and placed in an incubator at 37 °C for 90 min. The plate was then inverted to remove the sample lysates and washed as described above. A total of 100 μL of 2% milk prepared in 0.1% PBST containing 1:1,000 concentration of mouse αGFP antibody (Life Technologies no. A-11120, reconstituted as instructed, then diluted 1:1 in glycerol to a final concentration of 0.1 mg/mL and stored at −20 °C) was added, and the plate was sealed and placed on an orbital shaker at 4 °C overnight. This solution was then removed and the plate washed as described above. A 1:2,000 solution of donkey α-mouse horseradish peroxidase (HRP) (Jackson ImmunoResearch no. 715-035-150) was then added in 2% milk in 0.1% PBST, and the plate was sealed and placed on an orbital shaker at 4 °C overnight. This solution was then discarded, the plate washed, and 100 μL of a 1:1 solution of tetramethylbenzidine (TMB) solution:peroxide solution (TMB substrate kit, Thermo no. 34021) added. The assay was allowed to develop for 30 s to 10 min, depending on the intensity of the signal for each given application. After this time, 100 μL of 1N HCl was added to each well to stop the reaction and the plate briefly tapped to mix. The plate was then read using a colorimetric plate reader (ClarioStar Plus, BMG LabTech) at 450 and 630 nm. See summary in SI Appendix, Fig. S6B. In Figs. 5 and 6, a similar protocol was used, except that 5% milk was used in place of 2% milk throughout the protocol and the mouse αGFP detection antibody was substituted with a 1:250 concentration of mouse αBrp (nc82, Developmental Studies Hybridoma Bank).
ELISA analysis.
The signal for each well was determined by subtracting the 630-nm reading from the 405-nm reading. The average of the blank wells in each plate was then determined and subtracted from all the samples. These numbers represent the optical density (OD) values reported for each sample throughout the manuscript. In cases where experiments from multiple plates were combined (Figs. 1G and 4C), samples from the second experiment were scaled, either by comparing to a GFP standard curve run on the same plate (Fig. 1G) or by determining the scale using a set of common internal control samples (Fig. 4C) to scale to OD values so they could be compared across experiments.
ELISA kit.
In SI Appendix, Fig. S1C, a GFP ELISA kit was used (Abcam no. ab171581). This assay was performed following the manufacturer’s instructions, with the exception of sample preparation. Samples were prepared by homogenizing in PBS with 0.1% Tween-20 as described above.
Methodological details regarding fly strains and maintenance, immunohistochemistry, imaging, image analyses, Western blots, and statistical analysis are available as SI Appendix.
Acknowledgments
We would like to thank members of the M.R.F. laboratory for their valuable feedback on this work. This work was supported by NIH Grant R37-NS053538 to M.R.F., NIH Grant RO1-NS112215 to M.R.F., NIH Grant F32-NS117647 to T.R.J., and by Damon Runyon Fellowship 2329-18 to Y.K.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107427118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.St Johnston D., The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3, 176–188 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Ligoxygakis P., Pelte N., Hoffmann J. A., Reichhart J.-M., Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297, 114–116 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Gans M., Audit C., Masson M., Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics 81, 683–704 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benzer S., Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. U.S.A. 58, 1112–1119 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudai Y., Jan Y.-N., Byers D., Quinn W. G., Benzer S., Dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. U.S.A. 73, 1684–1688 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konopka R. J., Benzer S., Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nüsslein-Volhard C., Wieschaus E., Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). [DOI] [PubMed] [Google Scholar]
- 8.Simon M. A., Bowtell D. D. L., Dodson G. S., Laverty T. R., Rubin G. M., Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67, 701–716 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Gao F. B., Brenman J. E., Jan L. Y., Jan Y. N., Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 13, 2549–2561 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeger M., Tear G., Ferres-Marco D., Goodman C. S., Mutations affecting growth cone guidance in Drosophila: Genes necessary for guidance toward or away from the midline. Neuron 10, 409–426 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Neukomm L. J., Burdett T. C., Gonzalez M. A., Züchner S., Freeman M. R., Rapid in vivo forward genetic approach for identifying axon death genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, 9965–9970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenett A., et al., A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeiffer B. D., et al., Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 9715–9720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeiffer B. D., et al., Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee P. T., et al., A gene-specific T2A-GAL4 library for Drosophila. eLife 7, e35574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellen H. J., et al., The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics 188, 731–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuyama K., Zhang Y., Rao Y., Wang J. W., Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J. Neurogenet. 26, 89–102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X. J., et al., A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat. Neurosci. 18, 917–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach E. A., et al., GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323–331 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee N., Bohmann D., A versatile ΦC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One 7, e34063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spradling A. C., et al., The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin X., Daneman R., Zavortink M., Chia W., A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 15050–15055 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellen H. J., et al., The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buszczak M., et al., The carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venken K. J. T., et al., MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández M. P., Berni J., Ceriani M. F., Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 6, e69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stork T., Sheehan A., Tasdemir-Yilmaz O. E., Freeman M. R., Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83, 388–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doherty J., et al., PI3K signaling and Stat92E converge to modulate glial responsiveness to axonal injury. PLoS Biol. 12, e1001985 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelso R. J., et al., Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32, D418–D420 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venken K. J., et al., Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venken K. J. T., He Y., Hoskins R. A., Bellen H. J., P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Sarov M., et al., A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, e12068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tasdemir-Yilmaz O. E., Freeman M. R., Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 28, 20–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agrawal P., et al., Drosophila CRY entrains clocks in body tissues to light and maintains passive membrane properties in a non-clock body tissue independent of light. Curr. Biol. 27, 2431–2441.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Emery P., So W. V., Kaneko M., Hall J. C., Rosbash M., CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Kittel R. J., et al., Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Rein K., Zöckler M., Mader M. T., Grübel C., Heisenberg M., The Drosophila standard brain. Curr. Biol. 12, 227–231 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Biyasheva A., Do T. V., Lu Y., Vaskova M., Andres A. J., Glue secretion in the Drosophila salivary gland: A model for steroid-regulated exocytosis. Dev. Biol. 231, 234–251 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Chung W. S., et al., Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong S., et al., Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer D. P., et al., Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng F. Y., Wang Y., Tang B. L., The syntaxins. Genome Biol. 2, Review s3012 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett M. K., Calakos N., Scheller R. H., Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 257, 255–259 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Collins R. F., Schreiber A. D., Grinstein S., Trimble W. S., Syntaxins 13 and 7 function at distinct steps during phagocytosis. J. Immunol. 169, 3250–3256 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.