Abstract
mRNA-1273 vaccine efficacy against SARS-CoV-2 Delta wanes over time; however, there are limited data on the impact of durability of immune responses on protection. Here, we immunized rhesus macaques and assessed immune responses over 1 year in blood and upper and lower airways. Serum neutralizing titers to Delta were 280 and 34 reciprocal ID50 at weeks 6 (peak) and 48 (challenge), respectively. Antibody-binding titers also decreased in bronchoalveolar lavage (BAL). Four days after Delta challenge, the virus was unculturable in BAL, and subgenomic RNA declined by ∼3-log10 compared with control animals. In nasal swabs, sgRNA was reduced by 1-log10, and the virus remained culturable. Anamnestic antibodies (590-fold increased titer) but not T cell responses were detected in BAL by day 4 post-challenge. mRNA-1273-mediated protection in the lungs is durable but delayed and potentially dependent on anamnestic antibody responses. Rapid and sustained protection in upper and lower airways may eventually require a boost.
Keywords: SARS-CoV-2, Delta, COVID-19, mRNA vaccine, anamnestic, antibody, B cells, T cells, mucosal immunity, nonhuman primates
Graphical abstract
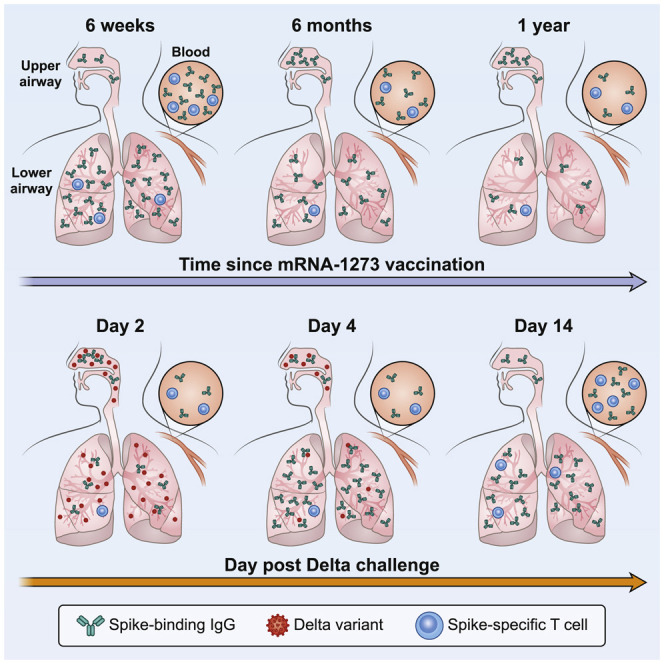
A study looking at immunity against the SARS-CoV-2 Delta variant one year after mRNA vaccination found durable but delayed anamnestic antibody responses in the lung but limited protection in the upper airway and low neutralizing responses, thus advocating for booster shots for sustained upper and lower airway protection.
Introduction
COVID-19 vaccines designed to express the spike (S) protein of SARS-CoV-2, including the mRNA-based vaccines mRNA-1273 (Baden et al., 2021b) and BNT162b2 (Polack et al., 2020) and the adenovirus-vectored vaccines Ad26.COV2.S (Sadoff et al., 2021) and AZD1222 (Ramasamy et al., 2021) have shown remarkable protection against vaccine-matched virus strains. mRNA-1273 had an efficacy of 94% in a phase III clinical trial (Baden et al., 2021b) and 96.3% among United States healthcare workers (Pilishvili et al., 2021). mRNA-1273-elicited neutralizing antibodies were still detectable 6 months after immunization (Doria-Rose et al., 2021). However, new SARS-CoV-2 variants have mutations that decrease the sensitivity of vaccine-elicited neutralization and increase viral replication, raising concerns about the durability of protection provided by mRNA-based and other COVID-19 vaccines (Baden et al., 2021a; Bruxvoort et al., 2021; Puranik et al., 2021).
Delta (henceforth referred to by its Pango lineage, B.1.617.2) is a WHO-designated variant of concern (VOC) and is currently the dominant circulating strain of SARS-CoV-2 worldwide, although it may soon be outcompeted in prevalence by Omicron (B.1.1.529). B.1.617.2 was first identified in India in October 2020 amidst substantial levels of community transmission (Cherian et al., 2021; Mishra et al., 2021; Mlcochova et al., 2021). This strain contains the mutations L452R, T478K, D614G, and P681R in the receptor-binding domain (RBD) and C terminus of the S1-binding subdomain. These substitutions contribute to both increased receptor binding and reduced neutralization by vaccine-elicited and monoclonal antibodies (mAbs) (Ozono et al., 2021; Planas et al., 2021; Tada et al., 2021; Wang et al., 2021b). In addition, B.1.617.2 has acquired several unique mutations in the N-terminal domain (NTD) including T19R and G142D and a deletion at positions 156–158 accompanied by a G insertion that substantially decrease antibody binding and neutralization sensitivity (Liu et al., 2021; Planas et al., 2021). Neutralizing antibody titers from mRNA-1273 and BNT162b2 vaccinee sera to B.1.617.2 are reduced 3-fold compared with the vaccine-matched strain USA-WA1/2020 (WA1) shortly after immunization (Edara et al., 2021b). A 7-fold reduction in neutralizing titers to B.1.617.2 for Ad26.CoV2.S sera in comparison with WA1 or WA1 with a D614G substitution (henceforth referred to as D614G) has also been reported (Barouch et al., 2021; Tada et al., 2021).
Recent studies in the United Kingdom, United States, and Qatar have shown a reduced efficacy of mRNA-based vaccines against asymptomatic and symptomatic, but not severe, B.1.617.2 infection (Bruxvoort et al., 2021; Chemaitelly et al., 2021; Lopez Bernal et al., 2021; Puranik et al., 2021; Tang et al., 2021). Antibody titers significantly decrease over a 6-month interval after the initial immunization series (Canaday et al., 2021; Corbett et al., 2021a; Levin et al., 2021), raising the concern that protection may wane. We and others reported that binding and neutralizing antibody titers in nonhuman primates (NHPs) and humans are key correlates of protection for mRNA and adenovirus-vectored COVID-19 vaccines (Corbett et al., 2021b; Gilbert et al., 2021; Khoury et al., 2021; Roozendaal et al., 2021). Retrospective analysis in Israel found that breakthrough cases in BNT162b2 vaccinees during a period of substantial B.1.617.2 transmission were statistically correlated with the length of time elapsed since vaccination, suggesting a role for waning antibody titers in vaccine efficacy reduction (Goldberg et al., 2021). Similarly, participants in the mRNA-1273 efficacy trial (COVE) who initially received a placebo prior to vaccination had reduced rates of B.1.617.2 breakthrough infections and severe disease compared with study participants who received mRNA-1273 at an earlier time (Baden et al., 2021a). These observations were consistent with previous findings showing that breakthrough infections with Alpha (B.1.1.7) were associated with lower BNT162b2-elicited binding and neutralizing antibody titers immediately prior to infection (Bergwerk et al., 2021). While BNT162b2 efficacy against breakthrough infections with B.1.617.2 has been estimated as 42% in Minnesota, USA and 51.9% in Qatar, those same studies reported mRNA-1273 efficacy as 76% and 73.1%, respectively (Puranik et al., 2021; Tang et al., 2021). Likewise, additional analysis in California, USA indicates a mRNA-1273 vaccine efficacy against B.1.617.2 of 87% (Bruxvoort et al., 2021). While any differences between mRNA-1273 and BNT162b2 may diminish with increased time since vaccination, this observation warrants continued investigation. Furthermore, there is no analysis of mRNA-1273-elicited immunity out to 1 year in the context of protection against mild and severe disease in the upper and lower airways.
The NHP model has been used extensively to assess vaccine candidates against SARS-CoV-2 (Corbett et al., 2020; Francica et al., 2021; He et al., 2021; Mercado et al., 2020; van Doremalen et al., 2020) and has been reliable for predicting protective efficacy with mRNA-1273 in humans (Corbett et al., 2021b; Gilbert et al., 2021). Thus, this model is an ideal tool for examining the effect of waning antibody titers on long-term protection in the context of a challenge with B.1.617.2.
Here, we immunized rhesus macaques with 100 μg of a pre-clinical formulation of mRNA-1273 at weeks 0 and 4 and then challenged them with B.1.617.2 approximately 1 year later. To provide insights into potential mechanisms of protection, we assessed B.1.617.2-binding antibody titers from the blood and both the upper and lower airways after vaccination and challenge. Serum neutralizing titers and the longevity of virus-specific memory B and T cell responses were also analyzed.
Results
mRNA-1273 immunization elicits binding and neutralizing antibodies to B.1.617.2
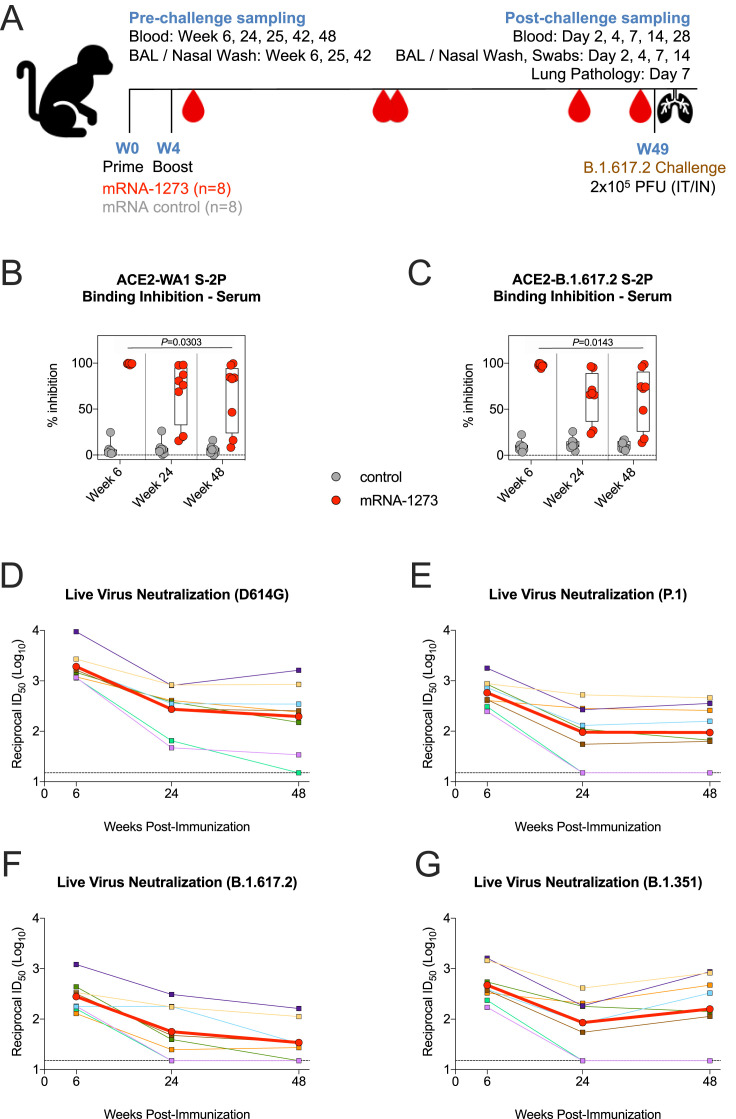
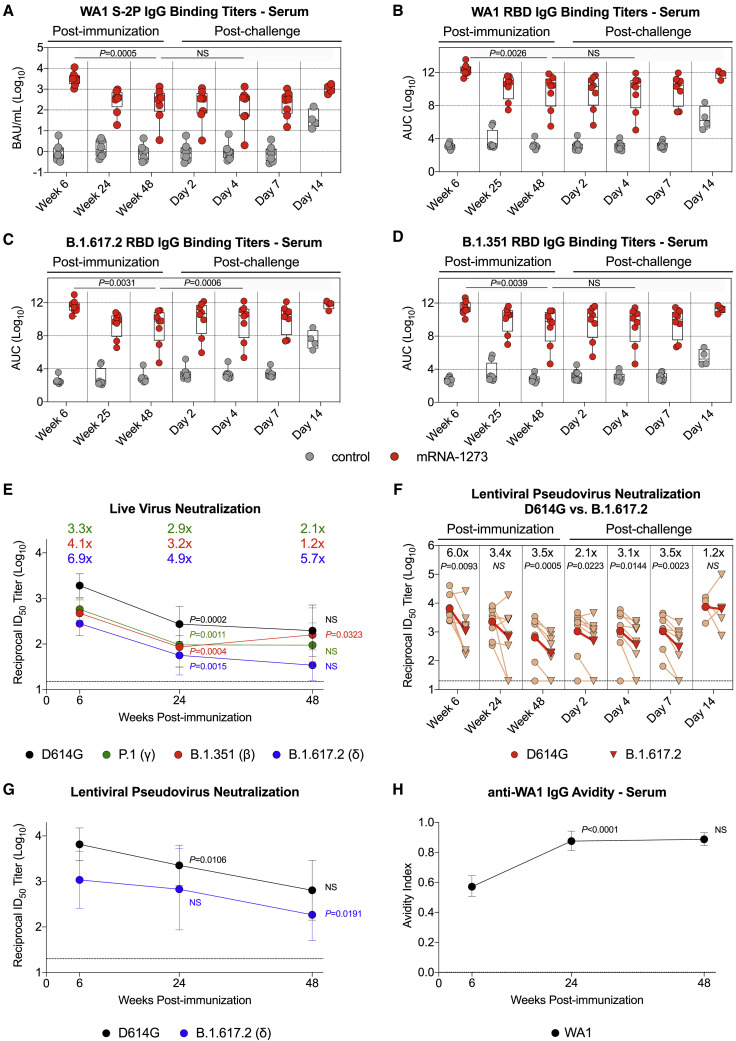
Indian-origin rhesus macaques (n = 8/group) were immunized with 100 μg mRNA-1273 or an mRNA control formulated in lipid nanoparticles at weeks 0 and 4 (Figure S1 A). Serum immunoglobulin G (IgG) binding titers to prefusion stabilized S protein (S-2P) of the vaccine-matched WA1 strain were assessed at weeks 6 (peak), 24 (memory), and 48 (memory and time of challenge) after vaccination. Geometric mean titers (GMTs) significantly decreased from ∼3,000 WHO International Standard binding antibody units (BAU)/mL at week 6 to ∼260 BAU/mL at week 24. However, the rate of decline was less over the next 6 months, reaching ∼188 BAU/mL at week 48 (Figure 1 A). Seven of the 8 animals in the mRNA-1273 group had higher binding titers than all control NHPs did at week 48. The kinetics observed for WA1 RBD-binding titers showed a 130-fold reduction in geometric mean area under curve (AUC) between weeks 6 and 25 from ∼2 × 1012 AUC to ∼1.5 × 1010 AUC but only an additional 3-fold reduction by week 48 (Figure 1B). GMTs to B.1.617.2 RBD at week 6 were ∼3.7 × 1011 AUC, a reduction of 5.4-fold compared to WA1 (Figure 1C). Despite the lower binding titers to B.1.617.2, antibodies retained potent function, as sera from vaccinated NHPs inhibited almost 100% of binding between the SARS-CoV-2 receptor, angiotensin-converting enzyme 2 (ACE2), and S-2P of both variants at week 6 (Figures S1B–S1C).
Figure S1.
Serum antibody kinetics after mRNA-1273 immunization, related to Figure 1
(A) Experimental timeline showing immunization schedule and B.1.617.2 challenge. 8 NHPs vaccinated with mRNA-1273, and 8 NHPs given mRNA control.
(B-C) Sera were collected at weeks 6, 24, and 48 post-immunization and diluted 1:40. SARS-CoV-2 WA1 (B) and B.1.617.2 (C) S-2P binding to ACE2 measured both alone and in the presence of sera to calculate % inhibition. Circles denote individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines set to 0% inhibition. 8 NHP per group. Statistical analysis shown for mRNA-1273 cohort only.
(D-G) Sera were collected at weeks 6, 24, and 48 post-immunization. Reciprocal 50% inhibitory dilution (ID50) titers calculated for neutralization of live virus D614G (D), P.1 (E), B.1.617.2 (F) and B.1.351 (G). Color-coded squares and lines indicate individual NHPs. Red circles and thick lines indicate geometric means. Dotted lines indicate assay limit of detection. 8 NHPs per group.
Figure 1.
mRNA-1273 elicits SARS-CoV-2 binding and neutralizing antibodies
(A–D) Sera were collected at weeks 6, 24 or 25, and 48 post-immunization and days 2, 4, 7, and 14 post-challenge. IgG binding titers to (A) WA1 S-2P, (B) WA1 RBD, (C) B.1.617.2 RBD, and (D) B.1.351 RBD. IgG-binding titers in (A) are expressed in WHO units. Circles in (A)–(D) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. 4–8 NHPs per group. Dotted lines in (A)–(D) are for visualization purposes and denote 1-log10 (A) or 4-log10 (B–D) increases in binding titers. Statistical analysis shown for mRNA-1273 cohort only.
(E) Sera were collected at weeks 6, 24, and 48 post-immunization. Neutralizing antibody titers assessed from a live virus assay with D614G, P.1, B.1.351, and B.1.617.2. Circles indicate GMTs, and error bars represent 95% confidence interval (CI). Values above circles represent fold reduction in GMTs for each variant in comparison to D614G at each time point. Dotted line indicates assay limit of detection. 8 NHPs per group. Statistical analysis shown for variant-specific responses at weeks 24 and 48 in comparison to corresponding variant-specific responses at preceding time point.
(F and G) Sera were collected at weeks 6, 24, and 48 post-immunization and days 2, 4, 7, and 14 post-challenge. Neutralizing antibody titers assessed from a lentiviral pseudovirus assay with either D614G or B.1.617.2 S. 4–8 NHPs per group.
(F) Light red lines and symbols indicate individual NHPs, while dark red lines and symbols indicate GMTs. Values above symbols represent fold reduction in neutralizing titers to B.1.617.2 compared with D614G at each time point. Dotted line indicates assay limit of detection. Statistical analysis shown for neutralizing titers to B.1.617.2 in comparison with D614G at each time point.
(G) Circles indicate GMTs, and error bars represent 95% CI. Dotted line indicates assay limit of detection. Statistical analysis shown for variant-specific responses at weeks 24 and 48 in comparison to corresponding variant-specific responses at preceding time point.
(H) Sera were collected at weeks 6, 24, and 48 post-immunization to determine anti-WA1 S-2P IgG avidity index. Circles indicate GMTs, and error bars represent 95% CI. Statistical analysis shown for avidity at weeks 24 and 48 in comparison to avidity at preceding time point. 8 NHPs per group.
We next measured binding titers to Beta (B.1.351), as this variant has the greatest impact on reducing neutralization by vaccinee and convalescent sera (Geers et al., 2021; Tada et al., 2021; Wang et al., 2021c) and was associated with reduced vaccine efficacy (Madhi et al., 2021; Shinde et al., 2021). At week 6, there was an 8-fold reduction in binding titers to B.1.351 RBD compared to WA1 RBD (Figure 1D). At week 48, GMTs to B.1.351 (∼1.2 × 109 AUC) were similar to B.1.617.2 (∼1.2 × 109 AUC) and were 4-fold less than GMTs to WA1 (∼4.6 × 109 AUC).
Serum neutralization of the D614G prototype virus and a panel of variants were then measured using a live virus assay. mRNA-1273-vaccinated NHPs had GMTs to D614G of 1,900 at week 6, which declined to 275 at week 24 and 200 at week 48 (Figure 1E). The kinetics of a rapid decline in neutralizing antibody titers during the first 6 months (week 6 versus week 24: p = 0.0002) followed by a slower decline at approximately 1 year (week 24 versus week 48: p > 0.05) were consistent with the analysis of binding titers (Figures 1A–1D). Compared to D614G, neutralizing GMTs to Gamma (P.1), B.1.351, and B.1.617.2 were reduced 3.3-fold, 4.1-fold, and 6.9-fold, respectively, at week 6. Neutralizing titers to B.1.617.2 declined over the following year, with 3 of the 8 NHPs having undetectable responses against B.1.617.2 at week 48 (Figure S1F).
Decreased serum neutralization capacity over time was substantiated with a lentiviral pseudovirus assay. At week 6, pseudovirus-neutralizing GMTs to B.1.617.2 were detectable in all NHPs but reduced 6-fold as compared to D614G (p = 0.0093). By week 24, the reduction in GMTs for B.1.617.2 compared to D614G declined to 3.5-fold, and 2 of the 8 vaccinated NHPs had undetectable neutralizing titers to B.1.617.2 at both weeks 24 and 48 (Figures 1F and 1G).
In contrast to the data with B.1.617.2, live virus neutralizing titers to B.1.351 showed a modest increase from week 24 to 48 (p = 0.0323). This observation, together with a difference in the kinetics of neutralizing titers to B.1.617.2 and B.1.351, suggests a change in serum epitope dominance and/or increased antibody affinity maturation, consistent with data showing a continued evolution of antibody responses induced by mRNA and adenovirus-vectored vaccines in NHPs and humans (Barouch et al., 2021; Corbett et al., 2021a; Gonzalez Lopez Ledesma et al., 2021).
Last, antibody avidity was measured from sera over 48 weeks (Figure 1H). The geometric mean avidity index of WA1 S-2P-binding IgG antibodies increased from 0.6 at week 6 to 0.9 at week 24 (p < 0.0001) and was maintained through week 48 (p > 0.05), suggesting either affinity maturation within the original antibody pool or immuno-focusing toward antibodies targeting more potently neutralizing epitopes.
Serum epitope analysis reveals immuno-focusing on epitopes associated with neutralization of B.1.617.2
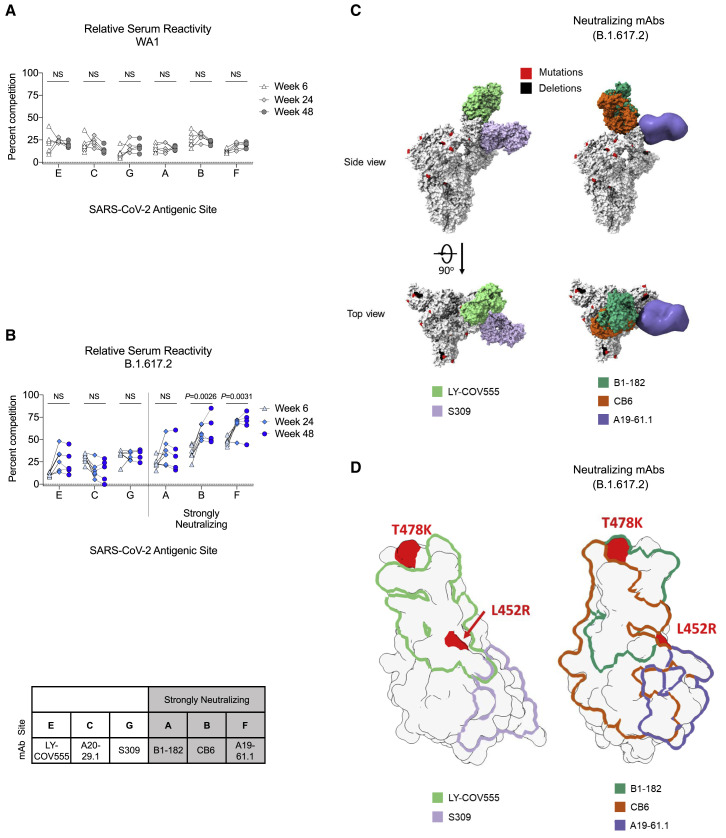
To explore the mechanistic basis for differences in serum neutralization and identify antibodies that could be contributing to the increased avidity, we performed serum antibody epitope mapping at weeks 6, 24, and 48 after immunization. Using a surface plasmon resonance (SPR)-based competition assay, we determined the relative proportion (percentage of competition) of serum antibodies targeting cross-reactive epitopes on both WA1 and B.1.617.2 SARS-CoV-2 S-2P (Table S2). In this analysis, serum binding to the respective S-2P proteins is assessed in the presence of mAbs with defined binding to specific epitopes. These mAbs can compete sterically or allosterically with serum antibodies recognizing partially overlapping or nearby epitopes on S-2P.
Cross-reactive antigenic sites A, B, C, E, F, and G were defined by mAbs B1-182, CB6, A20-29.1, LY-COV555, A19-61.1, and S309, respectively, all targeting the RBD of SARS-CoV-2 S, with sites A, B, and F being associated with strong neutralization of SARS-CoV-2 B.1.617.2 (Table S2) (Corbett et al., 2021a; Wang et al., 2021b).
Longitudinal analysis of the epitope-specific serum antibody responses to WA1 S-2P showed no significant differences in the relative serum reactivity over time (Figure 2 A). This indicates that while the quantity of serum antibodies targeting WA1 S-2P decreased over time (Figure 1A), the composition of serum antibodies targeting cross-reactive epitopes remained unchanged.
Figure 2.
B.1.617.2 S-2P-binding serum antibodies recognize epitopes associated with neutralization
(A and B) Longitudinal analysis of relative serum reactivity to cross-reactive RBD epitopes on both WA1 (A) and B.1.617.2 S-2P (B) was evaluated at 6, 24, and 48 weeks post-immunization. Relative serum reactivity was measured as percentage of competition of total measured serum antibody S-2P binding competed by single monoclonal antibody (mAb) targeting cross-reactive RBD epitopes on both WA1 and B.1.617.2 S-2P. Antigenic sites are defined by mAbs LY-COV555 (site E), A20-29.1 (site C), S309 (site G), B1-182 (site A), CB6 (site B), and A19-61.1 (site F). 5–8 NHPs per group. Statistical analysis shown for percentage of competition of binding to indicated epitopes at week 48 in comparison to week 6.
(C) SARS-CoV-2 S models with B.1.617.2 mutations indicated in red and deletions in black shown in complex with neutralizing (B1-182, CB6, A19-61.1) and other (LY-COV555, S309) mAbs.
(D) Footprints of both neutralizing (B1-182, CB6, A19-61.1) and other (LY-COV555, S309) mAbs indicate areas of binding on B.1.617.2 RBD with mutations highlighted in red.
See also Table S2.
In contrast, longitudinal analysis of serum reactivity to cross-reactive epitopes on B.1.617.2 S-2P showed significantly increased relative reactivity to antigenic sites B and F (represented by neutralizing mAbs CB6 and A19-61.1) from week 6 to 48 (B: p = 0.0026; F: p = 0.0031) (Figure 2B). Structural models show angles of approach for strongly neutralizing (B1-182, CB6, A19-61.1) and other mAbs (LY-COV555, S309) evaluated in complex with B.1.617.2 S-2P (Figure 2C). Modeling of binding footprints on B.1.617.2 RBD shows that immuno-focusing occurs on strongly neutralizing epitopes with areas of binding outside mutations T478K and L452R (CB6 and A19-61.1) (Figure 2D). These data indicate that although serum antibodies targeting B.1.617.2 S-2P decreased over time (Figure 1C), the proportion of serum antibodies targeting epitopes associated with neutralization increased, suggesting affinity maturation and immuno-focusing on conserved epitopes associated with neutralization against B.1.617.2.
Kinetics of B.1.617.2 RBD-binding IgG and IgA antibodies in the upper and lower airway
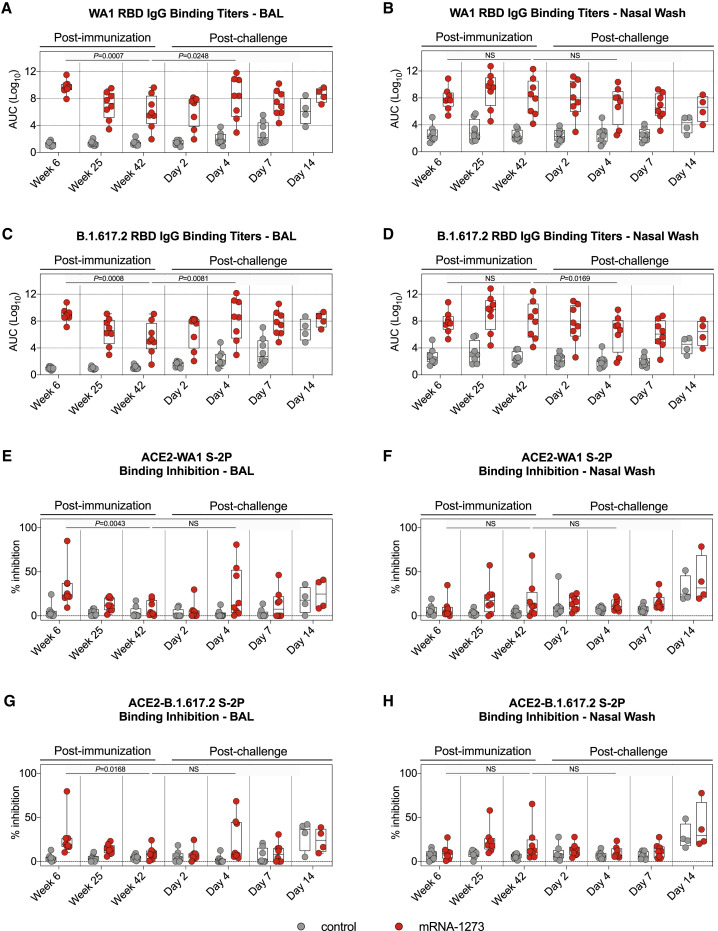
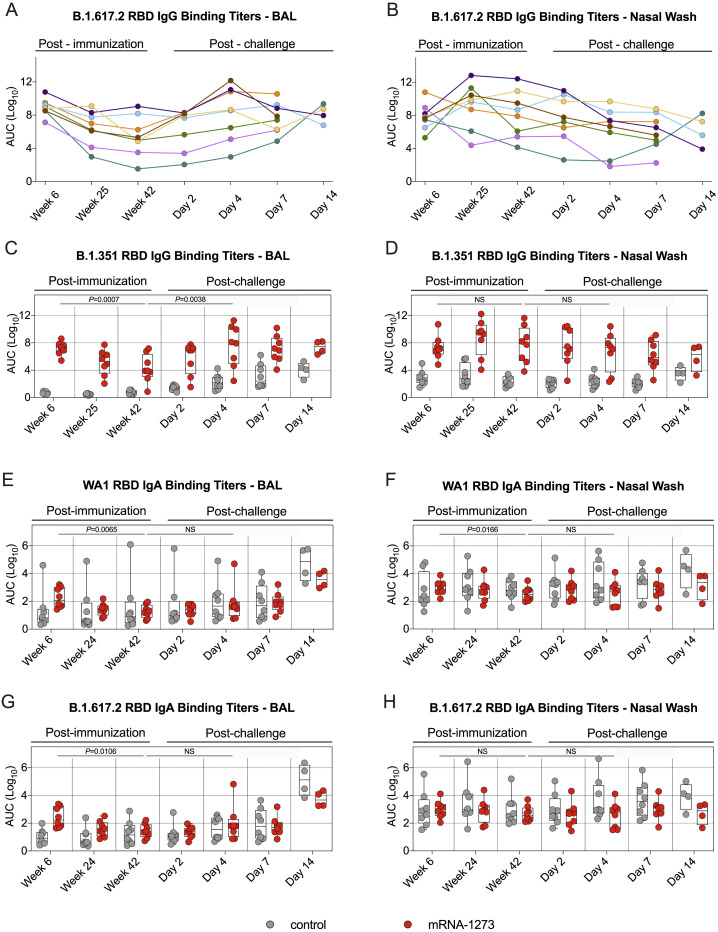
Antibodies are likely to provide the initial immune mechanism to control viral replication in the upper and lower airways (Corbett et al., 2021b; Francica et al., 2021; Fröberg et al., 2021; Gilbert et al., 2021; Khoury et al., 2021; Roozendaal et al., 2021). To determine the persistence of antibody titers in these relevant anatomical sites, we collected bronchoalveolar lavage (BAL) fluid and nasal washes at weeks 6, 25, and 42. WA1 RBD IgG binding titers in the BAL were highest at week 6, with GMTs of ∼5.2 × 109 AUC, and declined almost 5,000-fold to ∼1.1 × 106 AUC by week 42 (p = 0.0007) (Figure 3 A). B.1.617.2 RBD-binding IgG titers were lower in the BAL, with GMTs of ∼8.7 × 108 AUC at week 6 and ∼2.9 × 105 AUC at week 42, reductions of 6-fold and 4-fold compared to WA1 at the same time points (Figures 3C and S2 A). B.1.351 RBD-binding titers were lowest, with GMTs of ∼1.5 × 107 AUC at week 6 and ∼1.9 × 104 AUC at week 42 (Figure S2C). In addition, B.1.617.2 and B.1.351 RBD-binding titers followed a similar trend as WA1 RBD-binding titers with all antibody levels decreasing by week 42 (B.1.617.2: p = 0.0008; B.1.351: p = 0.0007). In contrast to the decreased antibody responses in BAL over time, WA1, B.1.351, and B.1.617.2 RBD-binding titers in the nasal washes were highest at week 25 and remained stable by week 42 (week 6 versus week 42: p > 0.05) (Figures 3B, 3D, S2B, and S2D).
Figure 3.
RBD-binding mucosal antibodies observe distinct kinetic patterns in the upper and lower airways
(A–D) BAL and nasal washes were collected at weeks 6, 25, and 42 post-immunization and days 2, 4, 7, and 14 post-challenge. (A and B) WA1 and (C and D) B.1.617.2 RBD-binding IgG titers in the lower (A and C) or upper (B and D) airways. Circles in (A)–(D) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines are for visualization purposes and denote 4-log10 increases in binding titers. 4–8 NHPs per group.
(E–H) BAL and nasal washes were collected at weeks 6, 25, and 42 post-immunization and days 2, 4, 7, and 14 post-challenge. All samples diluted 1:5. SARS-CoV-2 WA1 (E and F) and B.1.617.2 (G and H) S-2P binding to ACE2 measured both alone and in the presence of BAL (E and G) or nasal wash (F and H) in order to calculate the percentage of inhibition. Circles denote individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines set to 0% inhibition. 4–8 NHPs per group.
Statistical analysis in (A)–(H) shown for mRNA-1273 cohort only.
Figure S2.
Kinetics of RBD-binding IgG and IgA responses in the upper and lower airways, related to Figure 3
(A-D) BAL and nasal washes were collected at weeks 6, 25, and 42 post-immunization, and days 2, 4, 7, and 14 post-challenge. B.1.617.2 (A-B) and B.1.351 (C-D) RBD-binding IgG titers in the lower (A, C) or upper airway (B, D). Circles indicate individual NHPs. 4-8 NHPs per group. (A-B) Color-coded lines indicate temporal changes in B.1.617.2 RBD-binding titers for individual NHPs. All animals received mRNA-1273. (C-D) Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines are for visualization purposes and denote 4-log10 increases in binding titers.
(E-H) BAL and nasal washes were collected at weeks 6, 24, and 42 post-immunization, and days 2, 4, 7, and 14 post-challenge. (E-F) WA1 and (G-H) B.1.617.2 RBD-binding IgA titers in the lower (E, G) or upper airway (F, H). Circles in (E-H) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines are for visualization purposes and denote 2-log10 increases in binding titers. 4-8 NHPs per group.
Statistical analysis in (C-H) shown for mRNA-1273 cohort only.
SARS-CoV-2 RBD-binding IgA titers in nasal washes and BAL were also assessed (Figures S2E–S2H). At week 6, GMTs in the BAL of vaccinated NHPs to WA1 RBD and B.1.617.2 RBD were ∼176 and 259 AUC, respectively. At the same time, GMTs in controls were <20 AUC. IgA titers were indistinguishable from controls by week 24 in the BAL. IgA titers in the nasal washes of vaccinated NHPs were similar to those from controls at all time points.
In our previous NHP studies, we have been unable to detect neutralizing antibodies in the upper or lower airway following mRNA-1273 immunization using either pseudovirus or live virus assays. Here, we used the S-2P-ACE2 binding inhibition assay which provides a highly sensitive assessment of antibody function to extend the analysis of binding titers in the airways. Consistent with the findings from the sera (Figures S1B and S1C), the highest S-2P-ACE2 binding in BAL was detected at week 6 and declined by week 42 (WA1: p = 0.0043; B.1.617.2: p = 0.0168) (Figures 3E and 3G). By contrast, S-2P-ACE2 binding inhibition in the upper airway was not statistically different between weeks 6 and 42 (Figures 3F and 3H). Together, these results suggest that the kinetics and durability of antibody responses in the upper respiratory tract (nasal washes) are distinct from those in the blood or lungs (BAL).
Kinetics of S-specific memory B cells responses
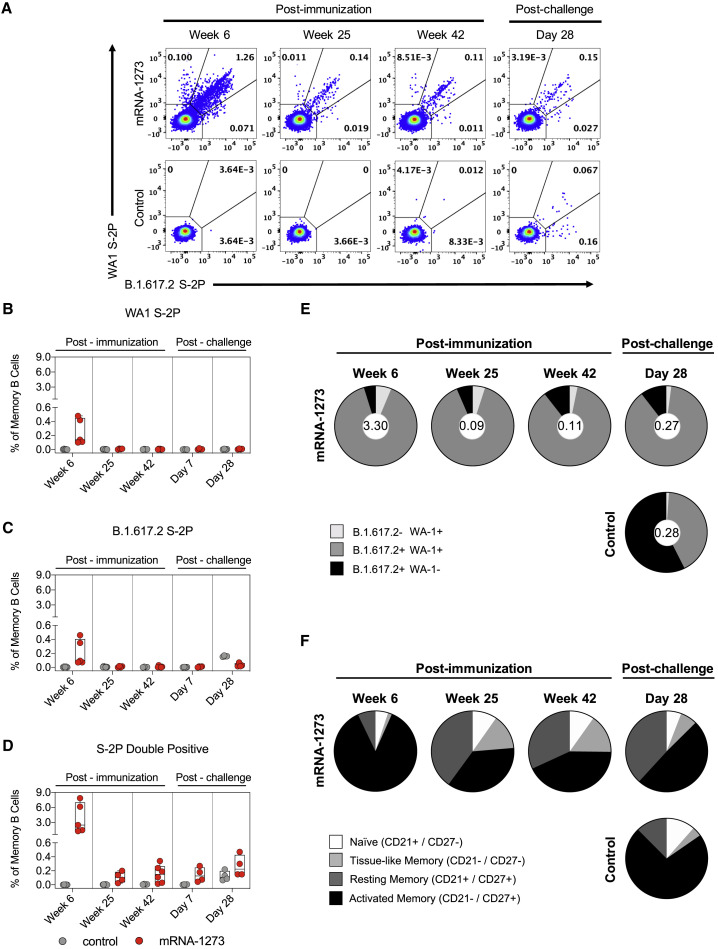
Sustained antibody production and increased secondary responses following a vaccine boost or a viral challenge are dependent on antigen-specific memory B cell responses (Gaebler et al., 2021; Goel et al., 2021a). A number of human studies have shown that mRNA S-2P vaccines and/or SARS-CoV-2 infection induce S-specific memory B cells that persist over time (Dan et al., 2021; Goel et al., 2021b; Rodda et al., 2021; Turner et al., 2021; Wang et al., 2021d, 2021e). Here, the frequency of WA1 S- and B.1.617.2 S-specific memory B cells following mRNA-1273 vaccination in NHPs was assessed by flow cytometry using fluorescent probes (Figure S3 A). At week 6, a median value of 2.5% of all memory B cells were dual-specific for WA-1 and B.1.617.2 in comparison to 0.14% for WA-1 and 0.09% for B.1.617.2 S-specific alone (Figures 4 A–4D). Although the total percentage of probe-binding memory B cells decreased through week 42, the high frequency of dual-binding to single-binding cells remained constant, with a geometric mean proportion of dual-binding cells greater than 85% at all time points (Figure 4E). These data are consistent with recent reports that mRNA-1273 and BNT162b2 elicit S-specific memory B cells that are capable of binding both the vaccine-matched strain and VOC (Corbett et al., 2021a; Goel et al., 2021b). The memory phenotype of these cells was also determined over time after vaccination (Figure S3B). Over the course of 1 year, S-2P-binding B cells shifted from 87% having an activated memory phenotype to a more balanced distribution of 43% activated memory, 32% resting memory, and 15% tissue-like memory cells (Figure 4F). Together, these data suggest that mRNA-1273 induces durable, broad cross-strain B cell memory to SARS-CoV-2.
Figure S3.
B and T cell gating strategy, related to Figures 4 and 5
(A) Representative flow cytometry plots showing gating strategy for B cells in Figure 4. Cells were gated as singlets and live cells on forward and side scatter and a live/dead aqua blue stain. CD3- cells were then gated on absence of CD14 and CD16 expression and positive CD20 expression. Memory B cells were selected based on lack of IgD or IgM. Finally, WA1 S-2P and B.1.617.2 S-2P probes were used to determine binding specificity.
(B) Representative flow cytometry plots showing gating strategy for B cell phenotypes in Figure 4F. CD20+ B cells were gated as described above. WA1 S-2P and B.1.617.2 S-2P probes were used to determine binding specificity. Probe-binding cells were further characterized as having a phenotype consistent with naive (N), tissue-like memory (TL), activated memory (AM), or resting memory (RM) cells according to expression of CD27 and CD21.
(C) Representative flow cytometry plots showing gating strategy for T cells in Figures 5 and S4. Cells were gated as singlets and live cells on forward and side scatter and a live/dead aqua blue stain. CD3+ events were gated as CD4+ or CD8+ T cells. Total memory CD8+ T cells were selected based on expression of CCR7 and CD45RA. Finally, SARS-CoV-2 S-specific memory CD8+ T cells were gated according to co-expression of CD69 and IL-2, TNF, or IFNγ. The CD4+ events were defined as naive, total memory, or central memory according to expression of CCR7 and CD45RA. CD4+ cells with a TH1 phenotype were defined as memory cells that co-expressed CD69 and IL-2, TNF, or IFNγ. CD4+ cells with a TH2 phenotype were defined as memory cells that co-expressed CD69 and IL-4 or IL-13. TFH cells were defined as central memory CD4+ T cells that expressed CXCR5, ICOS, and PD-1. TFH cells were further characterized as IL-21+, CD69+ or CD40L+, CD69+.
Figure 4.
mRNA-1273 elicits memory B cells that bind both WA1 and B.1.617.2
(A) Representative flow cytometry plots showing WA1 S-2P-specific, B.1.617.2 S-2P-specific, and dual-binding memory B cells at weeks 6, 25, and 42 post-immunization and day 28 post-challenge. Top row, mRNA-1273 group. Bottom row, mRNA control group.
(B–D) Percentage of all memory B cells that are specific for WA1 S-2P (B), B.1.617.2 S-2P (C), or both (D) at weeks 6, 25, and 42 post-immunization and days 7 and 28 post-challenge. Circles in (B)–(D) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. 4–7 NHPs per group. Break in y axis indicates a change in scale without a break in the range depicted.
(E) Pie charts indicating the proportion of SARS-CoV-2 S-specific memory B cells that bind to WA1 S-2P alone (light gray), B.1.617.2 S-2P alone (black), or both (dark gray) at weeks 6, 25, and 42 post-immunization and day 28 post-challenge. Values inside pie charts represent the percentage of all memory B cells that bind any SARS-CoV-2 S-2P probe at each time point. 4–7 NHPs per group. Top row, mRNA-1273 group. Bottom row, mRNA control group.
(F) Pie charts indicating the proportion of SARS-CoV-2 S-specific B cells with a phenotype consistent with naive (white), tissue-like memory (light gray), resting memory (dark gray), or activated memory (black) cells at weeks 6, 25, and 42 post-immunization and day 28 post-challenge. 4–7 NHPs per group. Top row, mRNA-1273 group. Bottom row, mRNA control group.
mRNA-1273 induces TH1 and TFH responses
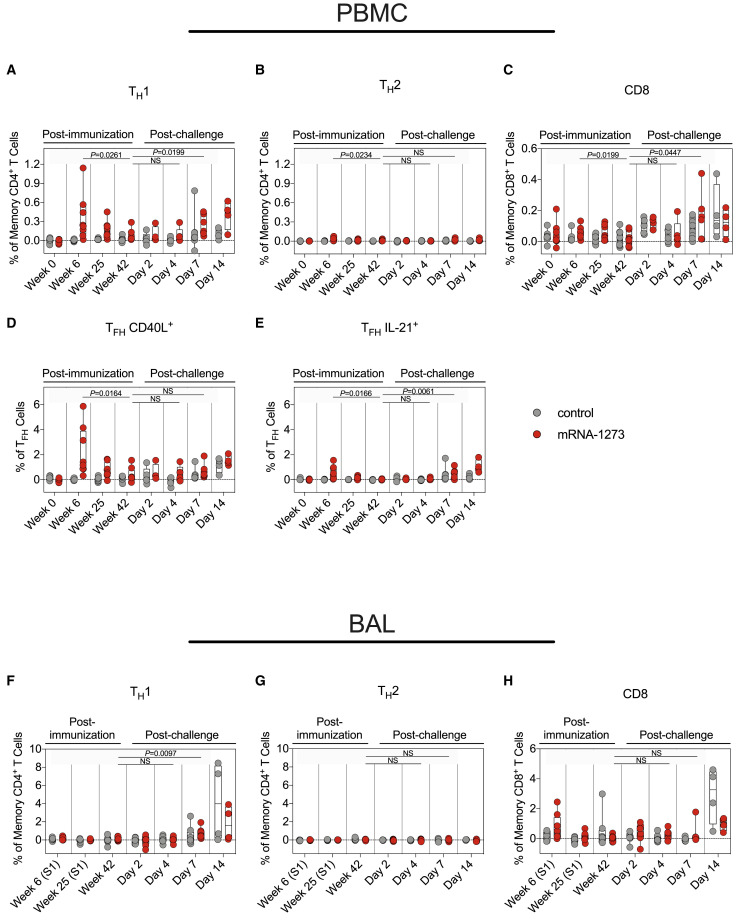
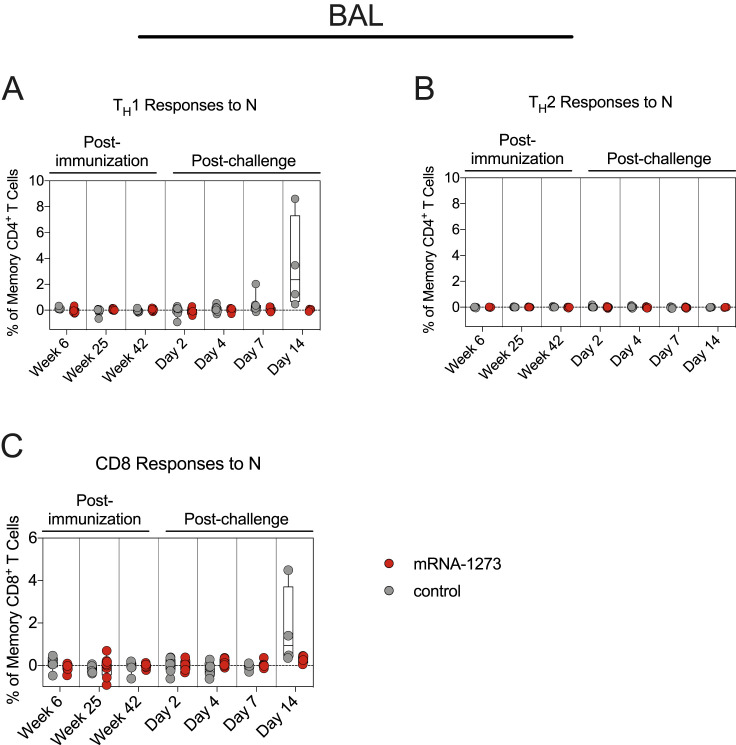
Our previous reports showed that mRNA-1273 immunization elicits SARS-CoV-2 S-specific T helper type 1 (TH1), CD40L+ T follicular helper (TFH), and IL-21+ TFH cells, which decrease over a 6-month period in NHPs (Corbett et al., 2021a). Here, S-specific T cell responses were assessed through week 42 in blood and BAL (Figure S3C). Consistent with our prior studies, TH1 but not T helper type 2 (TH2) responses were detected in the blood at week 6, decreasing by week 25. TH1 levels were low to undetectable by week 42 (week 6 versus week 42: p = 0.0261) (Figures 5A and 5B). mRNA-1273 induced both CD40L+ and IL-21+ TFH cells at week 6. By week 42, both populations had declined (CD40L+: p = 0.0164; IL-21+: p = 0.0166) (Figures 5D and 5E). Median CD8+ T cell responses in the blood were low (Figure 5C), although we did detect a low frequency of CD8+ T cells in the BAL at week 6 (Figure 5H).
Figure 5.
B.1.617.2 challenge induces an anamnestic T cell response in the periphery of vaccinated NHPs
(A–E) Peripheral blood mononuclear cells (PBMCs) collected at weeks 0, 6, 25, and 42 post-immunization and days 2, 4, 7, and 14 post-challenge. Cells were stimulated with SARS-CoV-2 S1 and S2 peptide pools and then measured by intracellular cytokine staining (ICS).
(A and B) Percentage of memory CD4+ T cells with (A) TH1 markers (interleukin [IL]-2, tumor necrosis factor [TNF], or interferon [IFN]γ) or (B) TH2 markers (IL-4 or IL-13) following stimulation.
(C) Percentage of CD8+ T cells expressing IL-2, TNF, or IFNγ.
(D and E) Percentage of TFH cells that express (D) CD40L or (E) IL-21.
(F–H) BAL fluid was collected at weeks 6, 25, and 42 post-immunization and days 2, 4, 7, and 14 post-challenge. Lymphocytes in the BAL were stimulated with S1 and S2 peptide pools and responses measured by ICS using TH1 (F), TH2 (G), and CD8 (H) markers. Samples from weeks 6 and 25 were only stimulated with S1 peptides.
Circles in (A)–(H) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines set at 0%. Reported percentages may be negative due to background subtraction. 4–8 NHPs per group. Statistical analysis shown for mRNA-1273 cohort only.
See also Figures S3 and S4.
Protection against B.1.617.2 1 year after mRNA-1273 vaccination
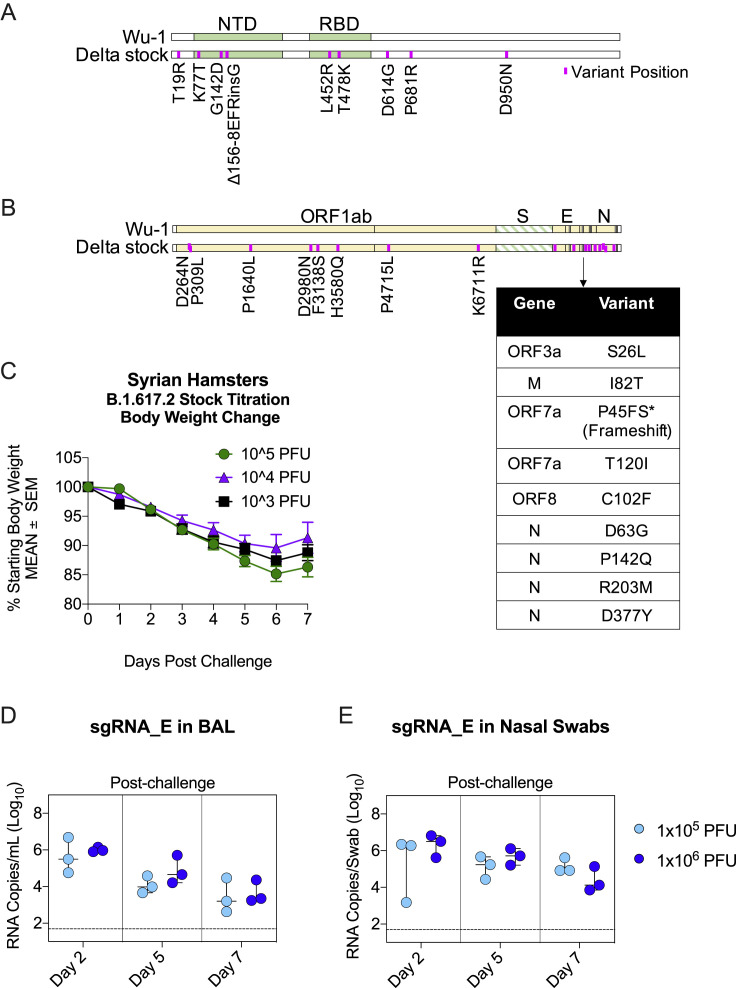
We evaluated the durability of vaccine protection against B.1.617.2 containing the canonical S mutations T19R, L452R, T478K, D614G, and P681R, as well as the NTD deletion (Figure S5 A), which were verified by sequencing. Pathogenicity of this viral stock was confirmed in hamsters (Figure S5C), and titration in NHPs (Figures S5D and S5E) was performed to define the experimental challenge dose.
Figure S5.
B.1.617.2 challenge stock characterization, related to Figure 6
(A-B) Alignment of B.1.617.2 stock to Wuhan-Hu-1 reference sequence. (A) S gene only. (B) Whole genome. Variant positions depicted in purple. Multiple mutations at the 3′ end of genome shown in table for visualization purposes. ∗Non-canonical frameshift in ORF7a produced by 172 nucleotide deletion. Deletion matched original isolate.
(C) B.1.617.2 stock titration in golden Syrian hamsters at 3 doses spanning 2-logs PFU. Arithmetic mean body weight across indicated days in comparison to starting weight shown. Error bars indicate standard error of the mean. 4 hamsters per group.
(D-E) B.1.617.2 stock titration in rhesus macaques at dose of 1x105 or 1x106 PFU. Copies sgRNA_E per mL BAL (D) or per NS (E). Circles indicate individual NHPs. Median value denoted by horizontal line. Dotted lines indicate assay limit of detection. 3 NHPs per group.
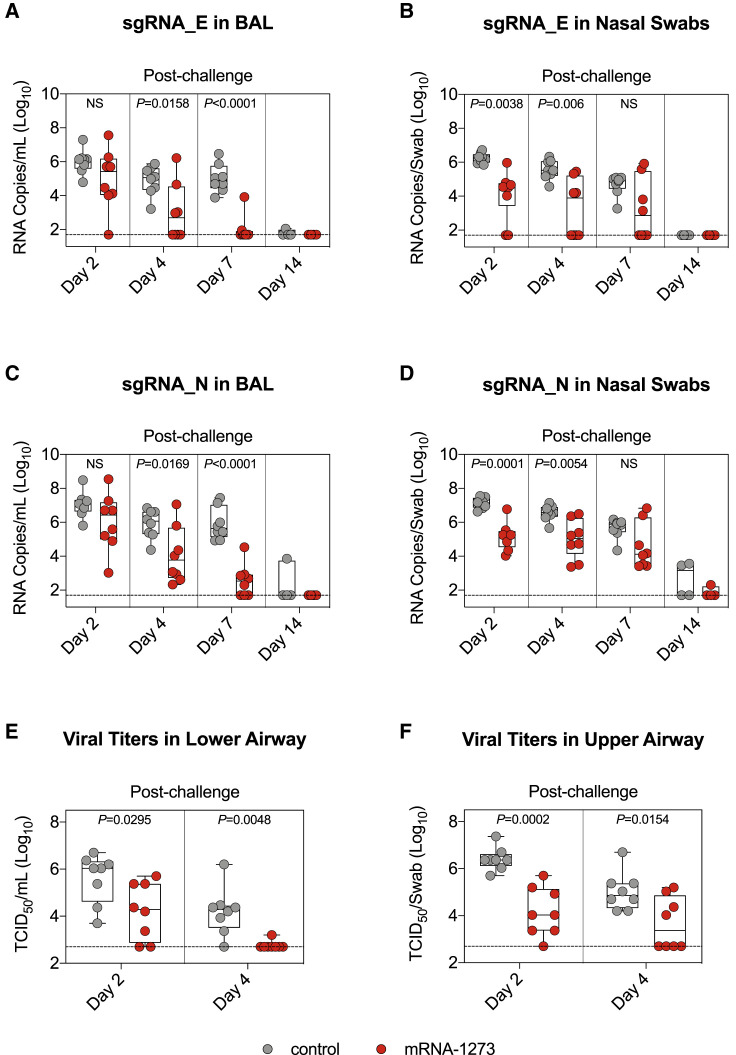
mRNA-1273 and mRNA control NHPs were challenged 49 weeks after the initial immunization with 2 × 105 plaque forming units (PFU) of virus via intratracheal (IT) and intranasal (IN) routes (Figure S1A). BAL and nasal swabs (NS) were collected on days 2, 4, 7, and 14 following challenge, and qRT-PCR was used to measure viral replication by assessing subgenomic RNA (sgRNA) copies encoding for the SARS-CoV-2 E and N genes. On day 2, geometric mean sgRNA_E copies in the lower airway of controls and vaccinated NHPs were 1 × 106 and 9 × 104 per mL, respectively (p > 0.05) (Figure 6 A). sgRNA_E copies in the lower airway of vaccinated NHPs declined rapidly over the following days, with geometric mean copies of 9 × 102 on day 4 and 1 × 102 on day 7. sgRNA_E copies in the lungs of unvaccinated NHPs remained significantly elevated compared to vaccinated NHPs, with a geometric mean of 1 × 105 on day 7 (mRNA-1273 versus control: p < 0.0001). These results differed from our earlier findings that NHPs vaccinated with 100 μg mRNA-1273 and then challenged only 4–8 weeks later largely controlled WA1 or B.1.351 replication in the lower airway by day 2 (Corbett et al., 2020, 2021c).
Figure 6.
B.1.617.2 replication is reduced in the upper and lower airways of mRNA-1273-immunized NHPs
(A–D) BAL and nasal swabs (NS) were collected 2, 4, 7, and 14 days after challenge with 2 × 105 PFU B.1.617.2. Copy numbers of sgRNA_E (A and B) and sgRNA_N (C and D) in the BAL (A and C) or nose (B and D). 4–8 NHPs per group.
(E and F) Viral titers per mL of BAL (E) or per NS (F) collected 2 and 4 days after challenge. 8 NHPs per group.
Circles in (A)–(F) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines indicate assay limit of detection. Statistical analysis shown for mRNA-1273 cohort in comparison to control NHPs at indicated time points.
See also Figures S5, S6, and S7 and Table S3.
In contrast to the lower airway, there was a more modest reduction in viral replication in the upper airway following mRNA-1273 immunization (Figure 6B). On day 2, geometric mean copies of sgRNA_E were 2 × 106 and 6 × 104 per NS of controls and vaccinated NHPs, respectively (p = 0.0038). On days 4 and 7, copies declined to 2 × 103 and 1 × 103 for the mRNA-1273 group and 4 × 105 and 5 × 104 for the control group (day 4: p = 0.006; day 7: p > 0.05). By day 7, 6 of the 8 and 4 of the 8 vaccinated NHPs had undetectable sgRNA_E levels in the lower airway and upper airway, respectively. All control NHPs still had detectable sgRNA_E copies at that time point. We observed similar trends with sgRNA_N (Figures 6C and 6D) but with higher geometric mean copy numbers, in agreement with our prior publications (Corbett et al., 2021b, 2021c). We euthanized 4 of the 8 NHPs in each cohort on day 7 for an assessment of lung pathology but continued to monitor the remaining animals for an additional week. On day 14, 1 of the 4 and 2 of the 4 control NHPs had detectable sgRNA_N in the lower and upper airways, respectively. In contrast, 0 of the 4 and 1 of the 4 vaccinated NHPs had detectable sgRNA_N in the same compartments. It is noteworthy that the only animal in the mRNA-1273 cohort with detectable sgRNA_N at day 14 had undetectable pseudovirus-neutralizing antibody titers to both D614G and B.1.617.2 at week 48 (Figure 1F).
To provide an additional assessment of protection, we measured the tissue culture infectious dose (TCID50) in samples taken from the lungs and nose on days 2 and 4 (Figures 6E and 6F). By day 4, only 1 of the 8 animals in the mRNA-1273 group had detectable virus in the lower airway compared to 7 of the 8 in the control mRNA group (p = 0.0048). In the upper airway, 4 of the 8 vaccinated animals had detectable virus, while all control animals had detectable virus (p = 0.0154).
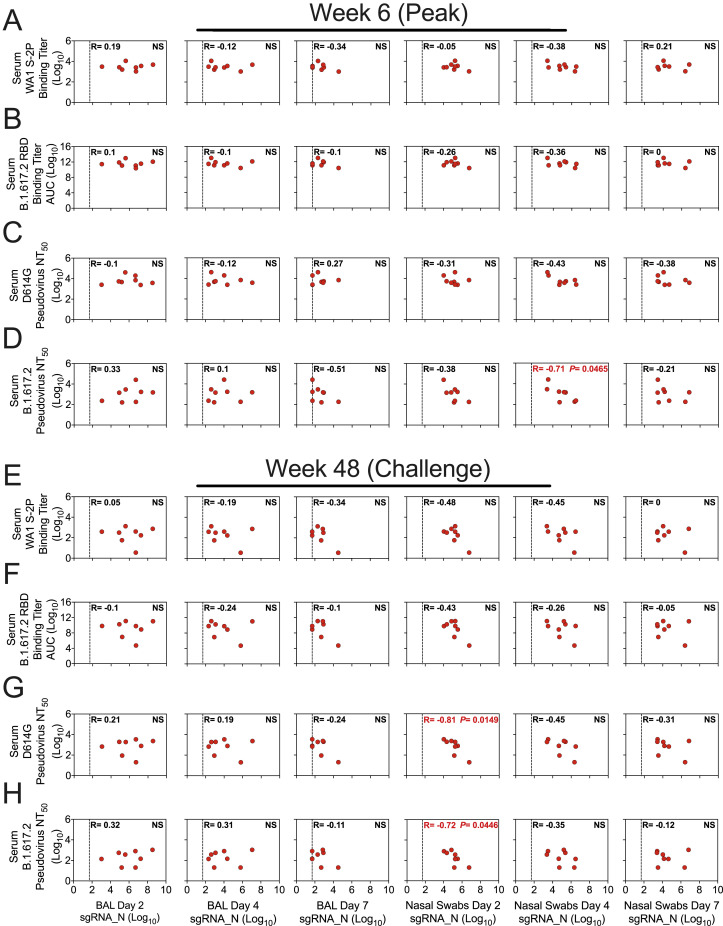
We previously established binding and neutralizing antibody titers in the sera as immune correlates of protection from WA1 and B.1.351 replication in NHPs, when both challenges were given 4–8 weeks after boost (Corbett et al., 2021b, 2021c). Here, when B.1.617.2 challenge was approximately 1 year after mRNA-1273 vaccination, we looked to see whether these associations held between viral replication and serum WA1 S-2P-binding titers (WHO units), B.1.617.2 RBD-binding IgG titers, D614G lentiviral pseudovirus-neutralizing titers, and B.1.617.2 lentiviral pseudovirus-neutralizing titers at peak (week 6) and immediately prior to challenge (week 48) (Figure S6 ; Table S3). Unlike in the challenges that were conducted soon after vaccination, none of these measurements correlated with viral replication in the lower airway. However, neutralizing titers at week 48 were significantly inversely correlated with viral replication in the upper airway at day 2 after challenge (D614G: p = 0.0149; B.1.617.2: p = 0.0446) but not on any subsequent days (Figures S6G and S6H).
Figure S6.
Serum antibody titers are not a correlate for protection 1 year after vaccination, related to Figure 6
(A-H) Correlations between sgRNA_N copies per mL of BAL or per NS at days 2, 4, and 7 post-challenge and indicated binding or neutralizing titers. Antibody responses include serum binding titers to WA1 S-2P in WHO units (A, E), serum binding titers to B.1.617.2 RBD (B, F), serum lentiviral pseudovirus-neutralizing titers to D614G (C, G), or serum lentiviral pseudovirus-neutralizing titers to B.1.617.2 (D, H) at week 6 (A-D) or week 48 (E-H) post-immunization. R denotes Spearman correlation coefficient. Dotted line indicates qRT-PCR limit of detection. 8 NHPs per group. Correlates analysis limited to vaccinated NHPs.
B.1.617.2 challenge elicits an anamnestic antibody response in the lower airway
Analysis of immune responses after challenge provides data on the kinetics of anamnestic responses, which may reveal potential mechanisms of protection. First, IgG binding titers and neutralizing antibody titers were assessed in sera at days 2, 4, 7, and 14 following challenge. We observed a clear primary response by day 14 in control NHPs, with increased binding to WA1, B.1.351, and B.1.617.2 RBDs (Figures 1B–1D). By day 14 after challenge, B.1.617.2 GMTs had increased 37,000-fold relative to week 48 after immunization, from ∼870 to ∼3.2 × 107 AUC, and were higher than WA1 GMTs. In contrast, B.1.617.2 GMTs in the mRNA-1273 group had increased only 475-fold during that time span, from ∼1.2 × 109 AUC to ∼5.7 × 1011 AUC, and were equivalent to WA1 GMTs at day 14. In addition, WA1 S-2P-binding GMTs in control NHPs increased 39-fold from ∼0.9 BAU/mL to ∼35 BAU/mL between week 48 after immunization and day 14 after challenge, while GMTs in vaccinated NHPs rose 5-fold from ∼189 to ∼1,030 BAU/mL (Figure 1A).
Pseudovirus-neutralizing responses were also measured to D614G and B.1.617.2 at days 2, 4, 7, and 14. The hierarchy of neutralizing antibody titers elicited by the vaccine remained stable until day 14, when titers to B.1.617.2 approached those of D614G (p > 0.05) (Figure 1F). These findings show that while serum antibody responses did not increase at a faster rate in vaccinated NHPs than in controls, they were boosted by infection.
We next measured mucosal antibody levels in the upper and lower airways. RBD-binding IgG titers in the upper airways of vaccinated animals showed a modest decline (Figures 3D and S2B) despite virus persistence in the nose (Figures 6B, 6D, and 6F), which may reflect clearance of viral protein-bound antibodies. We detected a strikingly different trend in the lower airway, however. B.1.617.2-binding GMTs in the control group increased slowly over the post-challenge observational period, rising 18-fold to ∼230 AUC and 213-fold to ∼2,750 AUC on days 4 and 7, respectively, from pre-challenge GMTs of 12.9 AUC at week 42. The antibody binding response to B.1.617.2 in vaccinated NHPs was more rapid, rising 590-fold to ∼1.7 × 108 AUC on day 4 before falling to 155-fold (∼4.5 × 107 AUC) on day 7 compared with week 42 after immunization (∼2.9 × 105 AUC) (Figure 3C). Antibody titers to all 3 variants were significantly greater 4 days after challenge than 42 weeks after immunization in the mRNA-1273 vaccinated group (WA1: p = 0.0248; B.1.617.2: p = 0.0081; B.1.351: p = 0.0038) (Figures 3A, 3C, and S2). Last, we did not detect differential IgA responses or ACE2 binding inhibition between vaccinated animals or controls in either the lower or upper airway (Figures 3E–3H and S2). These data show that the mRNA-1273-vaccinated NHPs made an anamnestic IgG response in the lower airways.
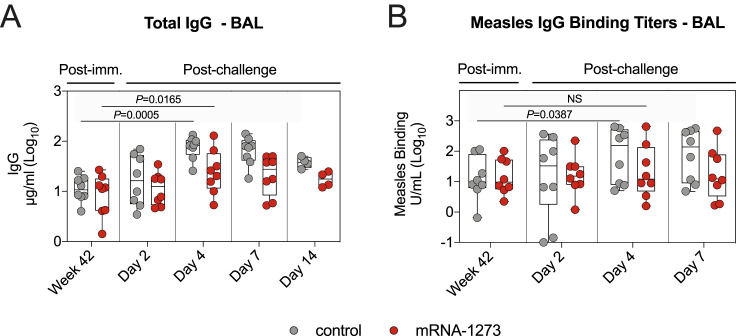
Although the total IgG concentration in the lungs of both vaccinated and unvaccinated NHPs increased following challenge, this increase was greater in the control cohort. Geometric mean IgG concentrations in the vaccinated animals rose from 8.3 μg/mL at week 42 after immunization to 11.2 and 24.2 μg/mL on days 2 and 4 after challenge, respectively. Simultaneously, IgG concentrations in the controls rose from a baseline of 11 μg/mL to 16.2 and 73.9 μg/mL on days 2 and 4, respectively (Figure S7 A). This rapid response after challenge is consistent with our prior experience with a protein SARS-CoV-2 vaccine in which we observed a rapid increase in both total IgG and measles morbillivirus (MeV) titers in primates that had also been vaccinated against MeV (Francica et al., 2021). In the present study, although binding titers were similar between both cohorts prior to challenge, responses to MeV increased more rapidly in the control group. GMTs to MeV in the lower airway of vaccinated NHPs increased from 14 units/mL (U/mL) at week 42 after immunization to 20 U/mL on day 4 after challenge, while GMTs in control NHPs increased from 12 to 67 U/mL at the same time points (Figure S7B).
Figure S7.
B.1.617.2 challenge induces a broad antiviral immune response, related to Figure 6
(A-B) BAL fluid was collected at week 42 post-immunization, and days 2, 4, 7, and 14 post-challenge. (A) Total IgG. (B) Measles morbillivirus-binding titers. Circles in (A-B) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. 4-8 NHPs per group. Statistical analysis shown for values at week 42 in comparison to day 4 post-challenge for both control NHPs (lower line) and mRNA-1273 cohort (upper line).
An anamnestic S-specific B cell response in the lower airways would likely be the underlying mechanism for an increase in local antibody titers. While we were unable to collect a sufficient number of B cells in BAL to analyze after B.1.617.2 challenge, we were nevertheless able to interrogate these cells in blood. Memory B cells, which bound both WA1 S-2P and B.1.617.2 S-2P, dominated the S-specific immune response of vaccinated NHPs. While the total frequency of all S-binding memory B cells increased, the proportion of these dual-specific cells remained between 85%–90% (Figure 4E). In contrast, B.1.617.2 challenge in control NHPs elicited a higher frequency of memory B cells specific only for B.1.617.2 S, with a geometric mean frequency of 57% at day 28 (Figure 4E). At that time, 72% of S-binding B cells in control NHPs had a phenotype consistent with activated memory B cells, in contrast to the vaccinated cohort in which 49% of cells had an activated memory phenotype and 38% of cells had a resting memory phenotype (Figure 4F).
T cell responses in blood and BAL following vaccination and B.1.617.2 challenge
After showing a potential role for memory B cell responses in controlling virus replication in the lower airways, we also measured T cell responses to SARS-CoV-2 S in the blood and BAL following challenge. By day 7, TH1 responses were elevated as compared with week 42 in both compartments (serum: p = 0.0199; BAL: p = 0.0097). However, the range of S-specific TH1 CD4+ T cell frequencies in the BAL of control and vaccinated NHPs overlapped (Figures 5A and 5F). While we observed increased IL-21+ TFH responses in vaccinated NHPs by day 7 (p = 0.0061) (Figure 5E), we did not detect any TH2 responses following challenge in vaccinated or control animals (p > 0.05) (Figures 5B and 5G). In contrast to the minimal vaccine-elicited CD8+ T cells prior to challenge, both control and vaccinated NHPs mounted CD8+ T cell responses following challenge (Figures 5C and 5H). Finally, TH1 and CD8+ responses in the lungs to SARS-CoV-2 N, which was not encoded by mRNA-1273, were only present in control NHPs (Figure S4 ). Together, these data demonstrate that vaccination with mRNA-1273 not only elicited B and T cellular responses to SARS-CoV-2 S following challenge but also protected NHPs from encountering sufficient levels of SARS-CoV-2 N to mount a primary response to that antigen.
Figure S4.
Vaccinated NHPs do not mount a primary T cell response to SARS-CoV-2 N peptides following challenge, related to Figure 5
(A-C) BAL fluid was collected at weeks 6, 25, and 42 post-immunization, and days 2, 4, 7, and 14 post-challenge. Lymphocytes in the BAL were stimulated with N peptide pools and responses measured by ICS. (A-B) Percentage of memory CD4+ T cells with (A) TH1 markers (IL-2, TNF, or IFNγ) or (B) TH2 markers (IL-4 or IL-13) following stimulation. (C) Percentage of CD8+ T cells expressing IL-2, TNF, or IFNγ. Circles in (A-C) indicate individual NHPs. Boxes represent interquartile range with the median denoted by a horizontal line. Dotted lines set at 0%. Reported percentages may be negative due to background subtraction. 4-8 NHPs per group. See also Figure S3 for gating strategy.
mRNA-1273 protects the lower airway from severe inflammation
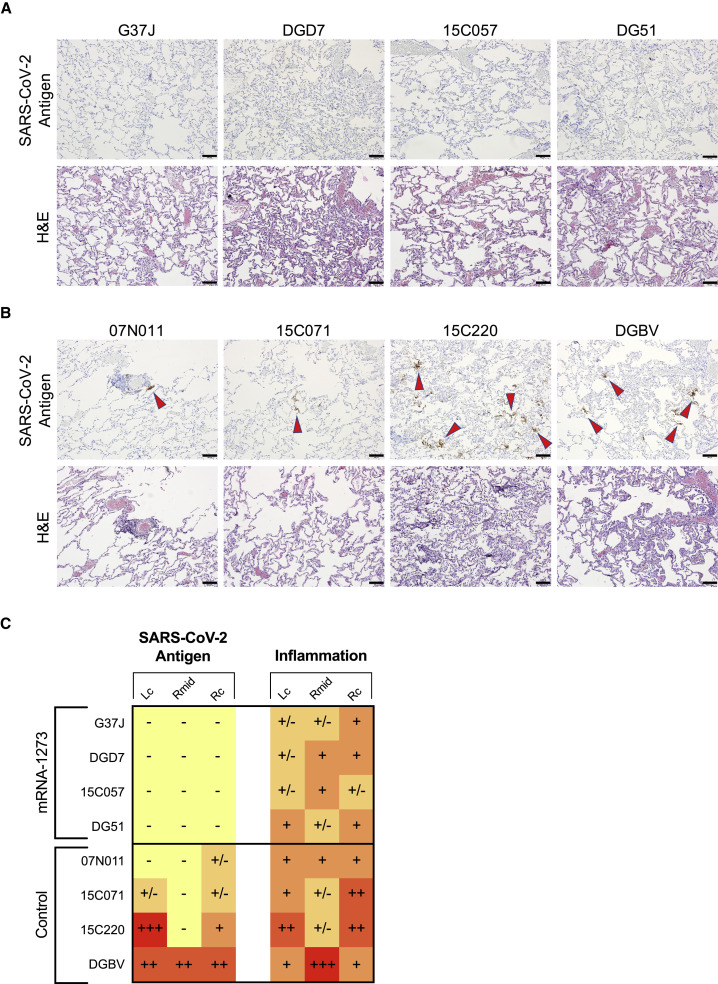
Because sufficient virus was present in the lower airways of vaccinated NHPs to elicit both a local anamnestic antibody response and increased S-specific B and T cell populations, we assessed if mRNA-1273 vaccination protected the lungs from gross pathologic changes. Lung samples from 4 of the 8 NHPs in each group were evaluated for pathology and detection of viral antigen (VAg) 7 days after B.1.617.2 challenge. SARS-CoV-2 VAg (red arrowheads) was detected in the lungs of 4 of the 4 control animals (Figures 7B and 7C) and was not detected in any of the animals in the vaccinated cohort (Figures 7A and 7C). Inflammation ranged from minimal to moderate across lung samples from animals that received mRNA-1273 and from minimal to severe in control NHPs (Figure 7C). The inflammatory changes in the lungs of vaccinated NHPs were characterized by a mixture of macrophages and polymorphonuclear cells present within some alveolar spaces and mild to moderate expansion of alveolar capillaries with mild type II pneumocyte hyperplasia (Figure 7A). Changes in control NHPs were more consistent with lymphocytes, histiocytes, and fewer polymorphonuclear cells associated with more prominent and expanded alveolar capillaries, collections of cells with alveolar air spaces, occasional areas of perivascular and peribronchiolar inflammation, and type II pneumocyte hyperplasia (Figure 7B). The protection observed in the lungs suggests that although binding and neutralizing antibody titers had declined 1 year after vaccination (Figure 1), leaving the lower airway susceptible to virus replication in the first 2 days after challenge (Figure 6), a local anamnestic response to antigen previously encountered in the vaccine proved sufficient to prevent severe disease.
Figure 7.
mRNA-1273 provides durable protection in the lower airway from B.1.617.2
(A and B) Representative images of lung samples 7 days after B.1.617.2 challenge from 4 NHPs that received mRNA-1273 (A) or mRNA control (B). Top row, detection of SARS-CoV-2 antigen by immunohistochemistry with a polyclonal anti-N antibody. Antigen-positive foci are marked by red arrows. Bottom row, hematoxylin and eosin (H&E) staining illustrating the extent of inflammation and cellular infiltrates. Images at 10× magnification with black bars for scale (100 μm).
(C) SARS-CoV-2 antigen and inflammation scores in the left cranial (Lc) lobe, right middle (Rmid) lobe, and right caudal (Rc) lobe of the lungs 7 days after B.1.617.2 challenge. Antigen scoring legend: –, no antigen detected; +/−, rare to occasional foci; +, occasional to multiple foci; ++, multiple to numerous foci; +++, numerous foci. Inflammation scoring legend: –, minimal to absent inflammation; +/−, minimal to mild inflammation; +, mild to moderate inflammation; ++, moderate to severe inflammation; +++, severe inflammation. Horizontal rows correspond to individual NHPs depicted above (A and B).
Discussion
mRNA-1273 vaccine efficacy against both symptomatic and asymptomatic infections with B.1.617.2 is reduced compared to ancestral strains, a result of both variant-specific mutations that confer diminished sensitivity to neutralization and waning antibody titers over time following vaccination (Baden et al., 2021a; Bruxvoort et al., 2021; Choi et al., 2021; Puranik et al., 2021; Tada et al., 2021). Here, we studied protection against B.1.617.2 infection by mRNA-1273 almost 1 year after vaccination in NHPs. Our principal findings were: (1) protection in the lower airway was complete by day 4 but delayed compared to prior NHP studies in which viral challenges were done much closer to the time of vaccination; (2) anamnestic antibody responses were detected in BAL at day 4 in vaccinated animals; and (3) binding and neutralizing antibody titers in blood and BAL decreased over 48 weeks.
Prior studies by us and others in NHPs (Corbett et al., 2021a) and humans (Doria-Rose et al., 2021; Goel et al., 2021b) show a reduction of neutralizing antibodies in sera over 6 months following mRNA vaccination. Here, antibody binding and neutralizing titers also decline in the sera over 1 year, and we extend these data by showing similar findings in BAL, suggesting that antibody measurements in serum may be a surrogate for such responses in the lung. By contrast, antibodies remain relatively stable from week 24–48 in the upper airway. The IgG stability in the nasal tract is notable considering the source of this immunoglobulin class is primarily transudation from the blood (Renegar et al., 2004). These data are consistent with a prior study showing that subcutaneous vaccines elicit antibodies that are detectable in the nose for at least 6 months (Clements and Murphy, 1986). The differential kinetics for the stability of IgG responses in the upper respiratory and lower respiratory tracts suggest unique mechanisms of turnover within these different anatomical sites.
The limited reduction in viral replication in the lower airway 2 days after challenge in this present study stands in contrast to prior studies in which mRNA-1273-vaccinated NHPs were challenged 4–8 weeks after vaccination and lower airway viral replication was largely controlled by day 2 (Corbett et al., 2020, 2021c). The decline of antibody titers in the lungs over 1 year, which are replete with epithelial target cells, may explain the different kinetics in virus control. Importantly, vaccinated NHPs had a striking anamnestic antibody response in the lower airways, with a 590-fold increase in B.1.617.2 GMTs by day 4 after challenge compared with the pre-challenge time point, and there was no detectable sgRNA_E in 6 of the 8 vaccinated NHPs by day 7. This suggests that protection in the lower airway is durable but somewhat delayed and may be dependent on a recall antibody response. It is also notable that while we were able to detect binding and some functional antibodies in the blood and mucosa through the S-2P-ACE2 inhibition assay at the time of challenge, neutralizing titers to B.1.617.2 were low to undetectable. Thus, vaccine-elicited protection in the lower airway could be mediated by neutralizing antibodies below our limit of detection or through fragment crystallizable (Fc)-mediated functions, which have been shown to be important for protection with both neutralizing and non-neutralizing mAbs (Li et al., 2021; Winkler et al., 2021). It is also noteworthy that although the quantity of antibodies had decreased 1 year after vaccination, the quality was improved, as evidenced by the increase in avidity and the shift in epitope dominance of B.1.617.2 S-binding antibodies toward sites associated with neutralization. In the nose, the 1-log10 reduction in sgRNA_N copy numbers on days 4 and 7 was consistent with what we have observed after our prior B.1.351 challenge with the same dose and regimen of mRNA-1273 (Corbett et al., 2021c) and shows that there is a higher threshold of antibodies required for protection in the upper airway compared to the lower airway (Corbett et al., 2021b). These data are also consistent with studies in human vaccinees showing that there can be a significantly higher level of protection against severe disease than mild or asymptomatic infections (Baden et al., 2021a; Bruxvoort et al., 2021; Puranik et al., 2021; Tang et al., 2021).
Rapid control of virus replication in both upper and lower airways has potential implications for transmission. Smaller aerosols (<20 μm) enriched with virus are generated mainly in the lower airways, and their inhalation may lead to transmission (Wang et al., 2021a). Therefore, in the context of limited virus control in the upper airway and slower kinetics of control in the lower airway, a boost to increase antibody titers may eventually be warranted.
Antibodies elicited by mRNA-1273 are capable of binding multiple VOC in NHPs and humans (Corbett et al., 2021a; Goel et al., 2021b; Wang et al., 2021e). We extend these findings here to show that the majority of antigen-specific B cells bind both WA1 and B.1.617.2 proteins. It is noteworthy that in NHPs, there is a more substantial contraction of the S-specific memory B cell compartment following vaccination than reported for humans (Goel et al., 2021b). Species-specific differences and the high percentage of memory B cells that bound SARS-CoV-2 S in our NHPs immediately after vaccination may account for these observations. Memory B cell expansion after challenge likely provides a mechanism for the anamnestic antibody response detected in BAL. Although we detected this expansion in blood, it probably reflects a similar local response in lung lymphoid tissue (Poon et al., 2021). Importantly, the extent of anamnestic B cell activation may be dependent on the viral challenge dose and consequent virus replication.
The increase in total IgG and measles-binding titers following challenge provides further evidence of a rapid polyclonal B cell response in the lung, likely driven by Toll-like receptor 7 (TLR7) activation by SARS-CoV-2, a single-stranded RNA virus (Bortolotti et al., 2021; Hornung et al., 2002). Indeed, transient innate stimulation of B cells in the lung may be critical for the initial adaptive immune response to respiratory pathogens and appears to precede T cell expansion.
There are multiple potential roles for T cells following vaccination by mRNA. This vaccine platform has been shown by us and others in NHPs and humans to induce TH1 and TFH and a low frequency of CD8+ T cells. TFH cells are important for inducing and sustaining antibody responses (Corbett et al., 2021b; Lederer et al., 2020; Painter et al., 2021; Pardi et al., 2018). Here, we show that S-specific T cells decreased over the course of 1 year but were still detectable in a subset of vaccinated NHPs at the time of challenge. A second potential role for T cells would be mediating the control of viral replication in the lower airway to limit severe disease. Notably, we did not see increased TH1 or CD8+ T cell responses in the BAL of vaccinated NHPs compared with controls following challenge. Interestingly, we detected CD8+ T cells in the blood before their detection in the lungs post-challenge, which requires further investigation. A formal demonstration for whether T cells have a role in protection in the lungs would require depletion at the time of challenge.
We and others have previously shown that antibodies are a correlate of protection (Corbett et al., 2021b; Gilbert et al., 2021). These studies are based on a short interval between vaccination and virus exposure or challenge. Here, we did not find a clear immune correlate of protection between binding or neutralizing antibody responses in the blood at the peak of the response or the time of challenge 1 year later. Additional analysis from human clinical trials with much larger numbers of vaccinated individuals than the 8 NHPs in this study will be important for determining whether serum antibody titers remain as a correlate of protection in the long term. Indeed, the data presented here raise the possibility that the correlate of protection may be due to the ability of tissue-resident memory B cells to expand after infection.
In summary, mRNA-1273 provided durable protection against B.1.617.2 challenge in the lower airway likely through anamnestic induction of antibody responses in the lung. An important consideration is that control of the virus was briefly delayed in the lungs and limited in the nose, which together may provide the virus with a greater opportunity for transmission, particularly if variants emerge that may be more transmissible, virulent, or resistant to neutralization such as B.1.1.529 (Basile et al., 2021, Hoffmann et al., 2021). These concerns highlight the importance of boosting for sustaining high-level protection against severe disease and limiting mild infections and transmission.
Limitations of the study
A potential limitation relates to the extent of viral replication following B.1.617.2 challenge. The challenge stock used here was passaged once and fully matched the canonical B.1.617.2 S sequences obtained from humans. However, we did not observe greater sgRNA copy numbers in the upper airway than in previous challenges with B.1.351 or WA1 (Corbett et al., 2021b, 2021c). Clinical reports have described substantially higher viral titers in the upper respiratory tract from B.1.617.2 infections compared with ancestral strains (Ong et al., 2021; Williams et al., 2021). It is possible that the NHP model does not precisely recapitulate human infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti human antibody (goat) sulfo-tag labeled | Meso Scale Diagnostics | Cat#R32AJ-1 |
| Human/NHP IgA detection antibody product | Meso Scale Diagnostics | Cat#D20JJ-6 |
| SARS-CoV-2 spike antibody (biotin, CR3022) | Novus Biologicals | Cat#CR3022; RRID:AB_2848080 |
| Goat anti-monkey IgG (H+L) secondary antibody, HRP (polyclonal) | Invitrogen | Cat#PA1-84631; RRID:AB_933605 |
| B1-182 | Vaccine Research Center, NIH (Wang et al., 2021b) | N/A |
| CB6 | Shi et al., 2020 | N/A |
| A20-29.1 | Vaccine Research Center, NIH (Corbett et al., 2021a) | N/A |
| LY-COV555 | Jones et al., 2021 | N/A |
| A19-61.1 | Vaccine Research Center, NIH (Wang et al., 2021b) | N/A |
| S309 | Pinto et al., 2020 | N/A |
| Goat anti-human IgD-FITC (polyclonal) | Southern Biotech | Cat#2030-02; RRID:AB_2795624 |
| PerCP-Cy5.5 mouse anti-human IgM (clone G20-127) | BD Biosciences | Cat#561285; RRID:AB_10611998 |
| DyLight 405 AffiniPure goat anti-human serum IgA, α chain specific (polyclonal) | Jackson ImmunoResearch | Cat#109-475-011; RRID:AB_2337789 |
| Brilliant Violet 570 anti-human CD20 antibody (clone 2H7) | Biolegend | Cat#302332; RRID:AB_2563805 |
| Brilliant Violet 650 anti-human CD27 antibody (clone O323) | Biolegend | Cat#302828; RRID:AB_2562096 |
| Brilliant Violet 785 anti-human CD14 antibody (clone M5E2) | Biolegend | Cat#301840; RRID:AB_2563425 |
| BUV496 mouse anti-human CD16 (clone 3G8) | BD Biosciences | Cat#612944; RRID:AB_2870224 |
| Alexa Fluor 700 mouse anti-human IgG (clone G18-145) | BD Biosciences | Cat#561296; RRID:AB_10612406 |
| APC-Cy7 mouse anti-human CD3 (clone SP34-2) | BD Biosciences | Cat#557757; RRID:AB_396863 |
| Anti-human CD38 PE (clone OKT10) | Caprico Biotechnologies | Cat#100826 |
| PE-Cy5 mouse anti-human CD21 (clone B-ly4) | BD Biosciences | Cat#551064; RRID:AB_394028 |
| Mouse anti-human CD185 (CXCR5) monoclonal antibody, PE-Cyanine7, eBioscience (clone MU5UBEE) | ThermoFisher Scientific | Cat#25-9185-42; RRID:AB_2573540 |
| Mouse anti-human CD4 monoclonal antibody, PE-Cyanine5.5 (clone S3.5) | ThermoFisher Scientific | Cat#MHCD0418; RRID:AB_10376013 |
| Brilliant Violet 570 anti-human CD8a antibody (clone RPA-T8) | Biolegend | Cat#301038; RRID:AB_2563213 |
| PE-Cy5 mouse anti-human CD45RA (clone 5H9) | BD Biosciences | Cat#552888; RRID:AB_394517 |
| Brilliant Violet 650 anti-human CD197 (CCR7) antibody (clone G043H7) | Biolegend | Cat#353234; RRID:AB_2563867 |
| Mouse anti-human CD185 (CXCR5) monoclonal antibody, PE, eBioscience (clone MU5UBEE) | ThermoFisher Scientific | Cat#12-9185-42; RRID:AB_11219877 |
| BV711 mouse anti-human CD183 (clone 1C6/CXCR3) | BD Biosciences | Cat#563156; RRID:AB_2738034 |
| BUV737 mouse anti-human CD279 (PD-1) (clone EH12.1) | BD Biosciences | Cat#565299; RRID:AB_2739167 |
| PE/Cyanine7 anti-human/mouse/rat CD278 (ICOS) antibody (clone C398.4A) | Biolegend | Cat#313520; RRID:AB_10643411 |
| Mouse anti-human CD69-ECD, RUO (clone TP1.55.3) | Beckman Coulter | Cat#6607110; RRID:AB_1575978 |
| Alexa Fluor 700 anti-human IFN-γ antibody (clone B27) | Biolegend | Cat#506516; RRID:AB_961351 |
| BV750 rat anti-human IL-2 (clone MQ1-17H12) | BD Biosciences | Cat#566361; RRID:AB_2739710 |
| High parameter custom BB700 conjugate (rat anti-human IL-4) (clone MP4-25D2) | BD Biosciences | Cat#624381 |
| FITC mouse anti-human TNF (clone MAb11) | BD Biosciences | Cat#554512; RRID:AB_395443 |
| BV421 rat anti-human IL-13 (clone JES10-5A2) | BD Biosciences | Cat#563580; RRID:AB_2738290 |
| Brilliant Violet 605 anti-human IL-17A antibody (clone BL168) | Biolegend | Cat#512326; RRID:AB_2563887 |
| Alexa Fluor 647 mouse anti-human IL-21 (clone 3A3-N2.1) | BD Biosciences | Cat#560493; RRID:AB_1645421 |
| Brilliant Violet 785 anti-human CD154 antibody (clone 24-31) | Biolegend | Cat#310842; RRID:AB_2572187 |
| SARS-CoV-2 (COVID-19) nucleocapsid antibody (polyclonal) | GeneTex | Cat#GTX135357; RRID:AB_2868464 |
| Bacterial and virus strains | ||
| SARS-CoV-2 B.1.617.2 (challenge stock) | This paper | N/A |
| SARS-CoV-2, isolate hCoV-19/Japan/TY7-503/2021 (Brazil P.1) | BEI Resources | Cat#NR-54982 |
| SARS-CoV-2 D614G (EHC-083E) | Mehul Suthar, Emory (Edara et al., 2021a) | N/A |
| SARS-CoV-2 B.1.351 | Mehul Suthar, Emory (Vanderheiden et al., 2021) | N/A |
| SARS-CoV-2 B.1.617.2 (neutralization assay) | Mehul Suthar, Emory (Edara et al., 2021b) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| MSD read buffer T (4X) | Meso Scale Diagnostics | Cat#R92TC |
| MSD blocker A kit | Meso Scale Diagnostics | Cat#R93AA-1 |
| WA1 RBD | Vaccine Research Center, NIH (Corbett et al., 2020) | N/A |
| B.1.617.2 RBD | This paper | N/A |
| B.1.351 RBD | Vaccine Research Center, NIH (Corbett et al., 2021c) | N/A |
| FuGENE 6 transfection reagent | Promega | Cat#E2692 |
| Sodium thiocyanate solution | Millipore-Sigma | Cat#80518-500ML-F |
| SARS-CoV-2 S-2P (Epitope Mapping) | Vaccine Research Center, NIH (Corbett et al., 2021b; Francica et al., 2021) | N/A |
| SARS-CoV-2 B.1.617.2 S-2P (Epitope Mapping) | Vaccine Research Center, NIH | N/A |
| SARS-CoV2-WT-S2P-bio-AVI (B cell assays) | Vaccine Research Center, NIH (Francica et al., 2021) | N/A |
| SARS-CoV2-B.1.617.2-S2P-bio-AVI (B cell assays) | This paper | N/A |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | ThermoFisher Scientific | Cat#L34966 |
| PepMix SARS-CoV-2 (S1+S2) (custom p.K986P and p.V987P) | JPT Peptide Techologies | N/A |
| PepMix SARS-CoV-2 (NCAP) | JPT Peptide Techologies | Cat#PM-WCPV-NCAP |
| Critical commercial assays | ||
| NEBNext ultra II RNA library prep kit for Illumina | New England Biolabs | Cat#E7770 |
| NEBNext multiplex oligos | New England Biolabs | Cat#E6440 |
| Monkey anti measles IgG | Alpha Diagnostic International | Cat#530-170-MMG |
| Isotyping panel 1 human/NHP kit | Meso Scale Diagnostics | Cat#K15203D-2 |
| V-PLEX SARS-CoV-2 panel 13 kit | Meso Scale Diagnostics | Cat#K15466U (ACE 2) |
| V-PLEX SARS-CoV-2 384 panel 2 (IgG) kit | Meso Scale Diagnostics | Cat#K25420U |
| His capture kit type 2 | Cytiva | Cat#29234602 |
| Amine coupling kit | Cytiva | Cat#BR100633 |
| Series S sensor chip CM5 | Cytiva | Cat#29149603 |
| RNAzol BD Column Kit | Molecular Research Center | Cat#RC 292 |
| TaqMan Fast Virus 1-Step Master Mix | ThermoFisher Scientific | Cat#4444436 |
| Experimental models: Cell lines | ||
| Vero (clone E6) | ATCC | Cat#CRL-1586; RRID:CVCL_0574 |
| VeroE6-TMPRSS2 | Vaccine Research Center, NIH (Corbett et al., 2021c) | N/A |
| HEK293T/17 | ATCC | Cat#CRL-11268; RRID:CVCL_1926 |
| HEK293T-ACE2 | Michael Farzan and Huihui Mu, Scripps Research | N/A |
| Experimental models: Organisms/strains | ||
| Indian-origin rhesus macaques | Vaccine Research Center, NIH | N/A |
| Syrian hamsters: HsdHan: AURA | Envigo | Cat#089 |
| Oligonucleotides | ||
| Primer: sgLeadSARSCoV2_F: 5′-CGATCTCTTGTAGATCTG TTCTC-3′ |
Integrated DNA Technologies (Francica et al., 2021; Wölfel et al., 2020) | N/A |
| Probe: E_Sarbeco_P: 5′-FAM-ACACTAGCCATCCTTACTGC GCTTCG-BHQ1-3′ |
Integrated DNA Technologies (Francica et al., 2021; Wölfel et al., 2020) | N/A |
| Primer: E_Sarbeco_R: 5′-ATATTGCAGCAGTACGCACACA-3′ | Integrated DNA Technologies (Francica et al., 2021; Wölfel et al., 2020) | N/A |
| Probe: wtN_P: 5′-FAM-TAACCAGAATGGAGAACGCAGTG GG-BHQ1-3′ |
Integrated DNA Technologies (Corbett et al., 2021b; Saunders et al., 2021) | N/A |
| Primer: wtN_R: 5′-GGTGAACCAAGACGCAGTAT-3′ | Integrated DNA Technologies (Corbett et al., 2021b; Saunders et al., 2021) | N/A |
| Recombinant DNA | ||
| VRC5601: pHR’CMV Luc | Naldini et al., 1996 | N/A |
| VRC5602: pCMV ΔR8.2 | Naldini et al., 1996 | N/A |
| VRC9260: TMPRSS2 | Vaccine Research Center, NIH (DiPiazza et al., 2021) | N/A |
| Spike_SARS-CoV-2 D614G | Vaccine Research Center, NIH (Corbett et al., 2021a) | N/A |
| Spike_SARS-CoV-2 B.1.351 | Vaccine Research Center, NIH (Corbett et al., 2021a) | N/A |
| Spike_SARS-CoV-2 B.1.617.2 | Vaccine Research Center, NIH (Corbett et al., 2021a) | N/A |
| Software and algorithms | ||
| CLC Genomics Workbench v.21.0.3 | QIAGEN | https://digitalinsights.qiagen.com/downloads/product-downloads/ |
| Viridot program | Katzelnick et al., 2018 | https://github.com/leahkatzelnick/Viridot |
| GraphPad Prism v8.2.0, v8.4.3, v9.0.2, v9.2.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Biacore Insight Evaluation Software | Cytiva | Cat#29310602 |
| FlowJo v10.7.2, v10.8.0 | Becton Dickinson | https://www.flowjo.com |
| R v4.0.2 | The R Foundation | https://www.r-project.org |
| Other | ||
| Amicon Ultra-15 centrifugal filter unit (50,000 MWCO) | Millipore Sigma | Cat#UFC905096 |
| Streptavidin multi array 384 well plate | Meso Scale Diagnostics | Cat#L21SA-1 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Robert A. Seder (rseder@mail.nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Rhesus macaque model and immunizations
All experiments conducted according to National Institutes of Health (NIH) regulations and standards on the humane care and use of laboratory animals as well as the Animal Care and Use Committees of the NIH Vaccine Research Center and Bioqual, Inc. (Rockville, Maryland). 4- to 14-year-old Indian-origin rhesus macaques were housed at Bioqual, Inc. NHPs were stratified based on age, weight, and gender into 2 cohorts (8 NHPs per group). One group received 100 μg of a pre-clinical formulation of mRNA-1273 at weeks 0 and 4, while the other group received 100 μg of an mRNA control at weeks 0 and 4, as described previously (Corbett et al., 2021a). Both products were formulated in lipid nanoparticles and prepared in 1 mL PBS. Vaccinations were given intramuscularly (IM) in the right quadriceps.
Titration of Delta stock in Syrian hamsters
All experiments conducted according to NIH regulations and standards on the humane care and use of laboratory animals as well as the Animal Care and Use Committees of the NIH Vaccine Research Center and Bioqual, Inc. (Rockville, Maryland). 6-week-old male Syrian hamsters (Envigo) were housed at Bioqual, Inc. Hamsters were stratified based on weight into 3 cohorts of 4 hamsters each. Hamsters were infected with B.1.617.2 challenge doses of 105, 104, and 103 PFU diluted in 100 μL PBS and split between both nostrils. Weight changes and clinical observations were collected daily.
Titration of Delta stock in nonhuman primates
All experiments conducted according to NIH regulations and standards on the humane care and use of laboratory animals as well as the Animal Care and Use Committees of the NIH Vaccine Research Center and Bioqual, Inc. (Rockville, Maryland). 4-year-old male Indian-origin rhesus macaques were housed at Bioqual, Inc. NHPs were stratified based on weight into 2 cohorts (3 NHPs per group). Groups received 1x105 or 1x106 PFU of SARS-CoV-2 B.1.617.2. 75% of the challenge dose was resuspended in 3mL PBS and administered intratracheally, while the remaining virus was resuspended in 1mL PBS and administered intranasally, with half of the volume in each nostril. BAL fluid and nasal swabs were collected on days 2, 5, and 7 following challenge for sgRNA quantification.
Method details
Cells and viruses
VeroE6 cells were obtained from ATCC (clone E6, ATCC, #CRL-1586) and cultured in complete DMEM medium consisting of 1x DMEM (VWR, #45000-304), 10% FBS, 25mM HEPES Buffer (Corning Cellgro), 2mM L-glutamine, 1mM sodium pyruvate, 1x non-essential Amino Acids, and 1x antibiotics. VeroE6-TMPRSS2 cells were generated at Vaccine Research Center, NIH, Bethesda, MD. The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/Japan/TY7-503/2021 (Brazil P.1), NR-54982, contributed by National Institute of Infectious Diseases. Isolation and sequencing of EHC-083E (D614G SARS-CoV-2), B.1.351, and B.1.617.2 for live virus neutralization assays were previously described (Edara et al., 2021a, 2021b; Vanderheiden et al., 2021). Viruses were propagated in Vero-TMPRSS2 cells to generate viral stocks. Viral titers were determined by focus-forming assay on VeroE6-TMPRSS2 cells. Viral stocks were stored at −80°C until use.
Sequencing of B.1.617.2 virus stock
We used NEBNext Ultra II RNA Prep reagents and multiplex oligos (New England Biolabs) to prepare Illumina-ready libraries, which were sequenced on a NextSeq 2000 (Illumina) as described previously (Corbett et al., 2021c). Demultiplexed sequence reads were analyzed in the CLC Genomics Workbench v.21.0.3 by (1) trimming for quality, length, and adaptor sequence, (2) mapping to the Wuhan-Hu-1 SARS-CoV-2 reference (GenBank: MN908947.3), (3) improving the mapping by local realignment in areas containing insertions and deletions (indels), and (4) generating both a sample consensus sequence and a list of variants. Default settings were used for all tools.
Delta challenge
NHPs were challenged 49 weeks after prime with 2x105 PFU of SARS-CoV-2 B.1.617.2. 1.5x105 PFU was resuspended in 3 mL PBS and administered intratracheally, while 0.5x105 PFU was resuspended in 1 mL PBS and administered intranasally, with half of the volume in each nostril.
Serum and mucosal antibody titers
Determination of antibody responses in the blood and mucosa were performed as previously described for measurement of mucosal antibody responses (Corbett et al., 2020). Briefly, heat inactivated plasma was serially diluted 1:4. BAL fluid and nasal washes were concentrated 10-fold with Amicon Ultra centrifugal filter devices (Millipore Sigma) and then serially diluted 1:5. Total IgG and IgA antigen-specific antibodies to variant SARS-CoV-2 RBD-derived antigens were determined by enzyme-linked immunosorbent assay (ELISA) using MULTI-ARRAY 384-well streptavidin-coated plates (Meso Scale Discovery, MSD). Plates were blocked with Blocker A kit (MSD) for 30 minutes at room temperature (RT) and washed. Plates were then coated with variant-specific biotinylated antigens (see Table S1) at a concentration of 0.18 μg/mL in 1% BSA/PBS for 1 hour at RT. After washing plates, serial dilutions of BAL fluid and nasal washes in 1% BSA and 0.05% Tween-20 (1x PBS) were added for 1 hour at RT. Plates were then washed 5 times and either 1 μg/mL anti-human IgG sulfo-tag or 2 μg/mL anti-human/NHP IgA sulfo-tag (MSD) in 1% BSA and 0.05% Tween-20 (1x PBS) were added for 1 hour. We then washed plates 5 times, added 1x Read Buffer T (MSD), and measured chemiluminescence using MSD plate reader (Sector Imager 600). WHO international units were calculated using the MSD Reference Standard 1 against the WHO International Standard (NIBSC code: 20/136) from the V-Plex SARS-CoV-2 384 Panel 2 (IgG) kit (MSD).
Total IgG antibody titers in the BAL were quantitated by using the Human/NHP IgG Kit (MSD), and antibody titers to measles were quantitated by using the Monkey Anti-Measles IgG ELISA Kit (Alpha Diagnostic International) according to the manufacturer’s instructions.
S-2P-ACE2 binding inhibition
Heat inactivated plasma was diluted 1:40. BAL fluid and nasal washes were concentrated 10-fold with Amicon Ultra centrifugal filter devices (Millipore Sigma) and then diluted 1:5. ACE2 binding inhibition assay was performed with V-Plex SARS-CoV-2 Panel 13 (ACE2) Kit (MSD) per manufacturer’s instructions.
Focus reduction neutralization assay
Focus reduction neutralization test (FRNT) assays were performed as previously described (Vanderheiden et al., 2020). Briefly, samples were diluted 3-fold in 8 serial dilutions using DMEM (VWR, #45000-304) in duplicates with an initial dilution of 1:10 in a total volume of 60 μl. Serially diluted samples were incubated with an equal volume of SARS-CoV-2 (100-200 foci per well) at 37°C for 1 hour in a round-bottomed 96-well culture plate. The antibody-virus mixture was then added to Vero cells and incubated at 37°C for 1 hour. Post-incubation, the antibody-virus mixture was removed and 100 μl of prewarmed 0.85% methylcellulose (Sigma-Aldrich, #M0512-250G) overlay was added to each well. Plates were incubated at 37°C for 24 hours. After 24 hours, methylcellulose overlay was removed, and cells were washed three times with PBS. Cells were then fixed with 2% paraformaldehyde in PBS (Electron Microscopy Sciences) for 30 minutes. Following fixation, plates were washed twice with PBS and 100 μl of permeabilization buffer (0.1% BSA [VWR, #0332], Saponin [Sigma, 47036-250G-F] in PBS) was added to the fixed Vero cells for 20 minutes. Cells were incubated with an anti-SARS-CoV-2 S primary antibody directly conjugated to biotin (CR3022-biotin) for 1 hour at room temperature. Next, the cells were washed three times in PBS and avidin - horseradish peroxidase (HRP) was added for 1 hour at room temperature followed by three washes in PBS. Foci were visualized using TrueBlue HRP substrate (KPL, # 5510-0050) and imaged on an ELISPOT reader (CTL).
Antibody neutralization was quantified by counting the number of foci for each sample using the Viridot program (Katzelnick et al., 2018). The neutralization titers were calculated as follows: 1 - (ratio of the mean number of foci in the presence of sera and foci at the highest dilution of respective sera sample). Each specimen was tested in duplicate. The FRNT50 titers were interpolated using a 4-parameter nonlinear regression in GraphPad Prism 8.4.3. Samples that did not neutralize at the limit of detection at 50% were plotted at 15, which was used for geometric mean calculations.
Lentiviral pseudovirus neutralization
Pseudotyped lentiviral reporter viruses were produced as previously described (Wu et al., 2021). Briefly, HEK293T/17 cells (ATCC CRL-11268) were transfected using FuGENE 6 transfection reagent (Promega) with the following: plasmids encoding S proteins from Wuhan-Hu-1 strain (GenBank: MN908947.3) with a p.Asp614Gly mutation, a luciferase reporter, lentivirus backbone, and the human transmembrane protease serine 2 (TMPRSS2) genes. For pseudoviruses encoding the S from B.1.351 and B.1.617.2, the plasmid was altered via site-directed mutagenesis to match the S sequence to the corresponding variant sequence as previously described (Corbett et al., 2021a). Plasma and/or sera, in duplicate, were tested for neutralizing activity against the pseudoviruses via mixing serial dilutions of sample with previously titrated amount of pseudovirus. After incubating for 1 hour at 37°C, 5% CO2, we added 7.5x103 HEK293T-ACE2 cells (Michael Farzan and Huihui Mu, Scripps Research) to each well. After 72-hour incubation, cell were lysed, and luciferase signal was measured. Neutralizing activity was calculated by quantification of luciferase activity in relative light units (RLU). The percentage of neutralization was normalized, with luciferase activity in uninfected cells defined as 100% neutralization and luciferase activity in cells infected with pseudovirus alone as 0% neutralization. ID50 titers were calculated using a log(agonist) versus normalized-response (variable slope) nonlinear regression model in Prism v9.0.2 (GraphPad). For samples that did not neutralize at the limit of detection at 50%, a value of 20 was plotted and used for geometric mean calculations.
Serum antibody avidity
Avidity was measured as described previously (Francica et al., 2021) in an adapted ELISA assay. Briefly, ELISA against S-2P was performed in the absence or presence of sodium thiocyanate (NaSCN) and developed with HRP-conjugated goat anti-monkey IgG (H+L) secondary antibody (Invitrogen) and SureBlue 3,3′,5,5′-tetramethylbenzidine (TMB) microwell peroxidase substrate (1-Component; SeraCare) and quenched with 1 N H2SO4. The avidity index (AI) was calculated by determining the ratio of IgG binding to S-2P in the absence or presence of NaSCN. The reported AI is the average of two independent experiments, each containing duplicate samples.
Epitope mapping
Serum epitope mapping competition assays were performed using the Biacore 8K+ surface plasmon resonance system (Cytiva) as previously described (Corbett et al., 2021a). Briefly, anti-histidine antibody was immobilized on Series S Sensor Chip CM5 (Cytiva) through primary amine coupling using Amine Coupling and His Capture kits (Cytiva), allowing His-tagged SARS-CoV-2 S protein containing 2 proline stabilization mutations (S-2P) to be captured on active sensor surface.
Competitor human IgG mAbs used for these analyses include: RBD-specific mAbs B1-182, CB6, A20-29.1, LY-COV555, A19-61.1, and S309. Negative control antibody or competitor monoclonal antibody (mAb) was injected over both active and reference surfaces. Then, NHP sera (diluted 1:100) was flowed over both active and reference sensor surfaces. Active and reference sensor surfaces were regenerated between each analysis cycle.
Beginning at the serum association phase, sensorgrams were aligned to Y (Response Units) = 0, using Biacore 8K Insights Evaluation Software (Cytiva). Relative “analyte binding late” report points (RU) were collected and used to calculate relative percent competition (% C) using the following formula: % C = [1 – (100 ∗ ((RU in presence of competitor mAb) / (RU in presence of negative control mAb)))]. Results are reported as percent competition. Assays were performed in duplicate, with average data point represented on corresponding graphs.
B cell probe binding
Previously isolated and cryopreserved PBMCs were centrifuged into warm RPMI/10% FBS and resuspended in wash buffer (4% heat inactivated newborn calf serum/0.02% NaN3/ phenol-free RPMI). PBMCs were transferred to 96-wells, washed twice in 1X PBS, and incubated with Aqua Blue live/dead cell stain (Thermo Fisher Scientific) at room temperature for 20 minutes. Following washing, cells were incubated with primary antibodies for 20 minutes at room temperature. The following antibodies were used (monoclonal unless indicated): IgD FITC (goat polyclonal, Southern Biotech), IgM PerCP-Cy5.5 (clone G20-127, BD Biosciences), IgA Dylight 405 (goat polyclonal, Jackson Immunoresearch Inc), CD20 BV570 (clone 2H7, Biolegend), CD27 BV650 (clone O323, Biolegend), CD14 BV785 (clone M5E2, Biolegend), CD16 BUV496 (clone 3G8, BD Biosciences), IgG Alexa 700 (clone G18-145, BD Biosciences), CD3 APC-Cy7 (clone SP34-2, BD Biosciences), CD38 PE (clone OKT10, Caprico Biotechnologies), CD21 PE-Cy5 (clone B-ly4, BD Biosciences), and CXCR5 PE-Cy7 (clone MU5UBEE, Thermo Fisher Scientific). Cells were washed twice in wash buffer and then incubated with streptavidin-BV605 (BD Biosciences) labeled B.1.617.2 S-2P and streptavidin-BUV661 (BD Biosciences) labeled WA1 S-2P for 30 minutes at 4°C (protected from light). Cells were washed twice in wash buffer and residual red blood cells were lysed using BD FACS Lysing Solution (BD Biosciences) at room temperature for 10 minutes. Following two final washes, cells were fixed in 0.5% formaldehyde (Tousimis Research Corp). All antibodies titrated to determine the optimal concentration. Samples were acquired on an BD FACSymphony cytometer and analyzed using FlowJo version 10.7.2 (BD, Ashland, OR).
Intracellular cytokine staining
Cryopreserved PBMCs and BAL cells were thawed and rested overnight in a 37°C/5% CO2 incubator. The next morning, cells were stimulated with SARS-CoV-2 S protein (S1 and S2, matched to vaccine insert) and N peptide pools (JPT Peptides) at a final concentration of 2 μg/ml in the presence of 3 mM monensin for 6 hours. The S1, S2, and N peptide pools are comprised of 158, 157, and 102 individual peptides, respectively, as 15mers overlapping by 11 aa in 100% DMSO. Negative controls received an equal concentration of DMSO instead of peptides (final concentration of 0.5%). ICS was performed as described (Donaldson et al., 2019). The following monoclonal antibodies were used: CD3 APC-Cy7 (clone SP34-2, BD Biosciences), CD4 PE-Cy5.5 (clone S3.5, Invitrogen), CD8 BV570 (clone RPA-T8, BioLegend), CD45RA PE-Cy5 (clone 5H9, BD Biosciences), CCR7 BV650 (clone G043H7, BioLegend), CXCR5 PE (clone MU5UBEE, Thermo Fisher), CXCR3 BV711 (clone 1C6/CXCR3, BD Biosciences), PD-1 BUV737 (clone EH12.1, BD Biosciences), ICOS Pe-Cy7 (clone C398.4A, BioLegend), CD69 ECD (cloneTP1.55.3, Beckman Coulter), IFN-g Ax700 (clone B27, BioLegend), IL-2 BV750 (clone MQ1-17H12, BD Biosciences), IL-4 BB700 (clone MP4-25D2, BD Biosciences), TNF-FITC (clone Mab11, BD Biosciences), IL-13 BV421 (clone JES10-5A2, BD Biosciences), IL-17 BV605 (clone BL168, BioLegend), IL-21 Ax647 (clone 3A3-N2.1, BD Biosciences), and CD154 BV785 (clone 24-31, BioLegend). Aqua live/dead fixable dead cell stain kit (Thermo Fisher Scientific) was used to exclude dead cells. All antibodies were previously titrated to determine the optimal concentration. Samples were acquired on a BD FACSymphony flow cytometer and analyzed using FlowJo version 10.8.0 (BD, Ashland, OR).
BAL samples from weeks 6 and 25 were stimulated with S1 peptide pools only. One BAL sample from a vaccinated NHP on Day 2 post-challenge had extensive background staining for CD8 markers and was excluded from analysis of both S and N responses.
Subgenomic RNA quantification
sgRNA was isolated and quantified as previously described (Corbett et al., 2021c). Briefly, total RNA was extracted from BAL fluid and nasal swabs using RNAzol BD column kit (Molecular Research Center). PCR reactions were conducted with TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems), forward primer in the 5′ leader region, and gene-specific probes and reverse primers as follows:
sgLeadSARSCoV2_F: 5′-CGATCTCTTGTAGATCTGTTCTC-3′
E gene
E_Sarbeco_P: 5′-FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1-3′
E_Sarbeco_R: 5′-ATATTGCAGCAGTACGCACACA-3′
N gene
wtN_P: 5′-FAM-TAACCAGAATGGAGAACGCAGTGGG-BHQ1-3′
wtN_R: 5′-GGTGAACCAAGACGCAGTAT-3′
Amplifications were performed with a QuantStudio 6 Pro Real-Time PCR System (Applied Biosystems). The assay lower limit of detection was 50 copies per reaction.
TCID50 assay
Vero-TMPRSS2 cells (obtained from Adrian Creanga, Vaccine Research Center-NIAID) were plated at 25,000 cells/well in DMEM with 10% FBS and gentamicin, and the cultures were incubated at 37°C, 5.0% CO2. Media was aspirated and replaced with 180 μL of DMEM with 2% FBS and gentamicin. 10-fold serial dilutions of samples starting from 20 μL of material were added to the cells in quadruplicate and incubated at 37°C for 4 days. Positive (virus stock of known infectious titer) and negative (medium only) controls were included in each assay. The plates were incubated at 37°C, 5.0% CO2 for 4 days. Cell monolayers were visually inspected for cytopathic effect (CPE). The TCID50 was calculated using the Read-Muench formula.
Histopathology and immunohistochemistry
Histopathology and immunohistochemistry (IHC) were performed as described previously (Corbett et al., 2020). Briefly, H&E staining and IHC were conducted and analyzed by a board-certified veterinary pathologist with an Olympus BX51 light microscope. Pathologist was blinded to vaccination status of NHP. A rabbit polyclonal anti-SARS-CoV-2 antibody (GeneTex, GTX135357) at a dilution of 1:2000 was used for IHC. Photomicrographs were taken on an Olympus DP73 camera.
Quantification and statistical analysis
Comparisons between groups, or between time points within a group, are based on unpaired and paired t-tests, respectively. All analysis for serum epitope mapping was performed using unpaired, two-tailed t-test. Binding, neutralizing, and viral assays are log-transformed as appropriate and reported with geometric means and corresponding CIs where indicated. Correlations are based on Spearman’s nonparametric rho, and the associated asymptotic p values. There are no adjustments for multiple comparisons, so all p values and significance testing should be interpreted as suggestive rather than conclusive. All analyses are conducted using R version 4.0.2 and GraphPad Prism version 8.2.0 unless otherwise specified.
P values are shown within figures, and the sample n is listed in corresponding figure legends. NS denotes that the indicated comparison was not significant, with p > 0.05. We did not perform statistical analysis at any time point in which we had samples from fewer than 5 NHPs.
Acknowledgments
Ethan Tyler designed the graphical abstract. We would like to thank G. Alvarado for experimental organization and administrative support. The VRC Production Program (VPP) provided the WA1 protein for the avidity assay. We would also like to especially thank A. Olia and C. Liu for providing the B.1.617.2 protein and J. Misasi, C. Stringham, C. Schramm, Y. Zhang, M. Choe, A. Henry, and B. Zhang for providing critical reagents, sequencing analysis, or expert technical assistance for serum antibody epitope mapping assays. D. Flebbe, S. Provost, E. Lamb, J. Marquez, A. Mychalowych, and M. Donaldson processed samples and prepared reagents for both ICS and B cell probe binding assays. A. Van Ry, B. Narvaez, Z. Flinchbaugh, and D. Valentin provided technical assistance for the TCID50 assays. We would like to thank M. Boursiquot and K. Steingrebe for assistance with NHP experiments. C. Case coordinated hamster experiments. M. Porto, C. Kitajewski, R. Stone, N. Daham, M. Hamilton, J. Wear, A. Faudree, B. Chang, J. Vazquez, and T. Wheatley provided technical assistance on hamster experiments. VPP contributors include C. Anderson, V. Bhagat, J. Burd, J. Cai, K. Carlton, W. Chuenchor, N. Cibelli, G. Dobrescu, M. Figur, J. Gall, H. Geng, D. Gowetski, K. Gulla, L. Hogan, V. Ivleva, S. Khayat, P. Lei, Y. Li, I. Loukinov, M. Mai, S. Nugent, M. Pratt, E. Reilly, E. Rosales-Zavala, E. Scheideman, A. Shaddeau, A. Thomas, S. Upadhyay, K. Vickery, A. Vinitsky, C. Wang, C. Webber, and Y. Yang.
This project has been funded in part by both the Intramural Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services and under HHSN272201400004C (NIAID Centers of Excellence for Influenza Research and Surveillance, CEIRS) and NIH P51 OD011132 awarded to Emory University. This work was also supported in part by the Emory Executive Vice President for Health Affairs Synergy Fund award, COVID-Catalyst-I3 Funds from the Woodruff Health Sciences Center and Emory School of Medicine, the Pediatric Research Alliance Center for Childhood Infections and Vaccines and Children’s Healthcare of Atlanta, and Woodruff Health Sciences Center 2020 COVID-19 CURE Award.
Author contributions
M.R., M.C.N., N.J.S., D.C.D., and R.A.S. designed experiments. M.G., K.S.C., B.J.F., K.E.F., D.A.W., S.F.A., J.-P.M.T., C.C.H., L.M., S.T.N., M.E.D.-G., L.P., K.W.B., B.M.N., M.M., A.P.W., J.I.M., C.T., C.G.L., B.Z., E.M., A. Cook, A.D., P.M., J.R.-T., F.L., L.W., A.G., S.K., A.P., N.D.-R., A. Carfi, J.R.M., E.A.B., D.K.E., H.A., M.G.L., M.S.S., B.S.G., M.R., I.N.M., M.C.N., N.J.S., D.C.D., and R.A.S. performed, analyzed, and/or supervised experiments. M.G., K.E.F., D.A.W., S.F.A., B.Z., E.A.B., and I.N.M. designed figures. I.-T.T., L.W., S.B.-B., E.S.Y., and W.S. provided critical reagents. M.G., D.C.D., and R.A.S. wrote manuscript. All authors edited the manuscript and provided feedback on research.
Declaration of interests
K.S.C. and B.S.G. are inventors on US patent no. 10,960,070 B2 and International patent application no. WO/2018/081318 entitled “Prefusion Coronavirus Spike Proteins and Their Use”. K.S.C. and B.S.G. are inventors on US patent application no. 62/972,886 entitled “2019-nCoV Vaccine”. L.W., E.S.Y., W.S., J.R.M., M.R., N.J.S. and D.C.D are inventors on US patent application no. 63/147,419 entitled “Antibodies Targeting the Spike Protein of Coronaviruses”. L.P., A. Cook, A.D., A.G., S.K., H.A., and M.G.L. are employees of Bioqual. K.S.C., L.W., W.S., and B.S.G are inventors on multiple US patent applications entitled “Anti-Coronavirus Antibodies and Methods of Use”. A. Carfi and D.K.E. are employees of Moderna. M.S.S. serves on the scientific board of advisors for Moderna. The other authors declare no competing interests.
Published: December 3, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.12.002.
Supporting citations
The following references appear in the supplemental information: Barnes et al. (2020); Hastie et al., 2021.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Baden L.R., El Sahly H.M., Essink B., Follmann D., Neuzil K.M., August A., Clouting H., Fortier G., Deng W., Han S., et al. Phase 3 Trial of mRNA-1273 during the Delta-Variant Surge. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2115597. Published online November 3, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., Stephenson K.E., Sadoff J., Yu J., Chang A., Gebre M., McMahan K., Liu J., Chandrashekar A., Patel S., et al. Durable Humoral and Cellular Immune Responses 8 Months after Ad26.COV2.S Vaccination. N. Engl. J. Med. 2021;385:951–953. doi: 10.1056/NEJMc2108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile K., Rockett R.J., McPhie K., Fennell M., Johnson-Mackinnon J., Agius J., Fong W., Rahman H., Ko D., Donavan L., et al. Improved neutralization of the SARS-CoV-2 Omicron variant after Pfizer-BioNTech BNT162b2 COVID-19 vaccine boosting. bioRxiv. 2021;2021.2012.2012.472252 doi: 10.3390/v14092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Gal Levin E., Rubin C., Indenbaum V., et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti D., Gentili V., Rizzo S., Schiuma G., Beltrami S., Strazzabosco G., Fernandez M., Caccuri F., Caruso A., Rizzo R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms. 2021;9:1820. doi: 10.3390/microorganisms9091820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruxvoort K.J., Sy L.S., Qian L., Ackerson B.K., Luo Y., Lee G.S., Tian Y., Florea A., Aragones M., Tubert J.E., et al. Effectiveness of mRNA-1273 against Delta, Mu, and other emerging variants. medRxiv. 2021 doi: 10.1136/bmj-2021-068848. 2021.2009.2029.21264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaday D.H., Oyebanji O.A., Keresztesy D., Payne M., Wilk D., Carias L., Aung H., Denis K.S., Lam E.C., Rowley C.F., et al. Significant reduction in humoral immunity among healthcare workers and nursing home residents 6 months after COVID-19 BNT162b2 mRNA vaccination. medRxiv. 2021 2021.2008.2015.21262067. [Google Scholar]
- Chemaitelly H., Tang P., Hasan M.R., AlMukdad S., Yassine H.M., Benslimane F.M., Al Khatib H.A., Coyle P., Ayoub H.H., Al Kanaani Z., et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2114114. Published online October 6, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., Team N. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A., Koch M., Wu K., Dixon G., Oestreicher J., Legault H., Stewart-Jones G.B.E., Colpitts T., Pajon R., Bennett H., et al. Serum Neutralizing Activity of mRNA-1273 against SARS-CoV-2 Variants. J. Virol. 2021;95:e0131321. doi: 10.1128/JVI.01313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M.L., Murphy B.R. Development and persistence of local and systemic antibody responses in adults given live attenuated or inactivated influenza A virus vaccine. J. Clin. Microbiol. 1986;23:66–72. doi: 10.1128/jcm.23.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Gagne M., Wagner D.A., O’ Connell S., Narpala S.R., Flebbe D.R., Andrew S.F., Davis R.L., Flynn B., Johnston T.S., et al. Protection against SARS-CoV-2 beta variant in mRNA-1273 vaccine-boosted nonhuman primates. Science. 2021 doi: 10.1126/science.abl8912. Published online October 21, 2021. [DOI] [PubMed] [Google Scholar]
- Corbett K.S., Nason M.C., Flach B., Gagne M., O’Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Werner A.P., Connell S.O., Gagne M., Lai L., Moliva J.I., Flynn B., Choi A., Koch M., Foulds K.E., et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat. Immunol. 2021;22:1306–1315. doi: 10.1038/s41590-021-01021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPiazza A.T., Leist S.R., Abiona O.M., Moliva J.I., Werner A., Minai M., Nagata B.M., Bock K.W., Phung E., Schafer A., et al. COVID-19 vaccine mRNA-1273 elicits a protective immune profile in mice that is not associated with vaccine-enhanced disease upon SARS-CoV-2 challenge. Immunity. 2021;54:1869–1882.e6. doi: 10.1016/j.immuni.2021.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson M.M., Kao S.F., Foulds K.E. OMIP-052: An 18-Color Panel for Measuring Th1, Th2, Th17, and Tfh Responses in Rhesus Macaques. Cytometry A. 2019;95:261–263. doi: 10.1002/cyto.a.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N., Suthar M.S., Makowski M., O’Connell S., McDermott A.B., Flach B., Ledgerwood J.E., Mascola J.R., Graham B.S., Lin B.C., et al. mRNA-1273 Study Group Antibody Persistence through 6 Months after the Second Dose of mRNA-1273 Vaccine for Covid-19. N. Engl. J. Med. 2021;384:2259–2261. doi: 10.1056/NEJMc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.V., Norwood C., Floyd K., Lai L., Davis-Gardner M.E., Hudson W.H., Mantus G., Nyhoff L.E., Adelman M.W., Fineman R., et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 2021;29:516–521.e3. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.V., Pinsky B.A., Suthar M.S., Lai L., Davis-Gardner M.E., Floyd K., Flowers M.W., Wrammert J., Hussaini L., Ciric C.R., et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francica J.R., Flynn B.J., Foulds K.E., Noe A.T., Werner A.P., Moore I.N., Gagne M., Johnston T.S., Tucker C., Davis R.L., et al. Protective antibodies elicited by SARS-CoV-2 spike protein vaccination are boosted in the lung after challenge in nonhuman primates. Sci. Transl. Med. 2021;13:eabi4547. doi: 10.1126/scitranslmed.abi4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröberg J., Gillard J., Philipsen R., Lanke K., Rust J., van Tuijl D., Teelen K., Bousema T., Simonetti E., van der Gaast-de Jongh C.E., et al. SARS-CoV-2 mucosal antibody development and persistence and their relation to viral load and COVID-19 symptoms. Nat. Commun. 2021;12:5621. doi: 10.1038/s41467-021-25949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6:eabj1750. doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune Assays Team§. Moderna, Inc. Team§. Coronavirus Vaccine Prevention Network (CoVPN)/Coronavirus Efficacy (COVE) Team§. United States Government (USG)/CoVPN Biostatistics Team§ Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2021 doi: 10.1126/science.abm3425. Published online November 23, 2021. [DOI] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. UPenn COVID Processing Unit‡ mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Y., Mandel M., Bar-On Y.M., Bodenheimer O., Freedman L., Haas E.J., Milo R., Alroy-Preis S., Ash N., Huppert A. Waning Immunity after the BNT162b2 Vaccine in Israel. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2114228. Published online October 27, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Lopez Ledesma M.M., Sanchez L., Ojeda D.S., Rouco S.O., Rossi A.H., Varese A., Mazzitelli I., Pascuale C.A., Miglietta E.A., Rodríguez P.E., et al. Longitudinal Study after Sputnik V Vaccination Shows Durable SARS-CoV-2 Neutralizing Antibodies and Reduced Viral Variant Escape over Time. medRxiv. 2021 doi: 10.1128/mbio.03442-21. 2021.2008.2022.21262186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. CoVIC-DB team1 Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.-t., Yuan M., Callaghan S., Musharrafieh R., Song G., Silva M., Beutler N., Lee W., Yong P., Torres J., et al. Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques. bioRxiv. 2021 doi: 10.1126/scitranslmed.abl9605. 2021.2007.2005.451222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization - implications for control of the COVID-19 pandemic. bioRxiv. 2021;2021.2012.2012.472286 doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdörfer B., Giese T., Endres S., Hartmann G. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick L.C., Coello Escoto A., McElvany B.D., Chávez C., Salje H., Luo W., Rodriguez-Barraquer I., Jarman R., Durbin A.P., Diehl S.A., et al. Viridot: An automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl. Trop. Dis. 2018;12:e0006862. doi: 10.1371/journal.pntd.0006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Lederer K., Castaño D., Gómez Atria D., Oguin T.H., 3rd, Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P., et al. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2114583. Published online October 6, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Arase N., Kishikawa J.-i., Hirose M., Li S., Tada A., Matsuoka S., Arakawa A., Akamatsu K., Ono C., et al. The SARS-CoV-2 Delta variant is poised to acquire complete resistance to wild-type spike vaccines. bioRxiv. 2021 2021.2008.2022.457114. [Google Scholar]
- Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. NGS-SA Group. Wits-VIDA COVID Group Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Mindermann S., Sharma M., Whittaker C., Mellan T.A., Wilton T., Klapsa D., Mate R., Fritzsche M., Zambon M., et al. COVID-19 Genomics UK (COG-UK) Consortium Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England. EClinicalMedicine. 2021;39:101064. doi: 10.1016/j.eclinm.2021.101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gage F.H., Trono D., Verma I.M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Chiew C.J., Ang L.W., Mak T.M., Cui L., Toh M., Lim Y.D., Lee P.H., Lee T.H., Chia P.Y., et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta) Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab721. Published online August 23, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilishvili T., Gierke R., Fleming-Dutra K.E., Farrar J.L., Mohr N.M., Talan D.A., Krishnadasan A., Harland K.K., Smithline H.A., Hou P.C., et al. Effectiveness of mRNA Covid-19 Vaccine among U.S. Health Care Personnel. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2106599. Published online September 22, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon M.M.L., Rybkina K., Kato Y., Kubota M., Matsumoto R., Bloom N.I., Zhang Z., Hastie K.M., Grifoni A., Weiskopf D., et al. SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci. Immunol. 2021;6:eabl9105. doi: 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik A., Lenehan P.J., Silvert E., Niesen M.J.M., Corchado-Garcia J., O’Horo J.C., Virk A., Swift M.D., Halamka J., Badley A.D., et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. medRxiv. 2021 2021.2008.2006.21261707. [Google Scholar]
- Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., et al. Oxford COVID Vaccine Trial Group Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renegar K.B., Small P.A., Jr., Boykins L.G., Wright P.F. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J. Immunol. 2004;173:1978–1986. doi: 10.4049/jimmunol.173.3.1978. [DOI] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal R., Solforosi L., Stieh D.J., Serroyen J., Straetemans R., Dari A., Boulton M., Wegmann F., Rosendahl Huber S.K., van der Lubbe J.E.M., et al. SARS-CoV-2 binding and neutralizing antibody levels after Ad26.COV2.S vaccination predict durable protection in rhesus macaques. Nat. Commun. 2021;12:5877. doi: 10.1038/s41467-021-26117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. ENSEMBLE Study Group Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S., et al. 2019nCoV-501 Study Group Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Samanovic M.I., Dcosta B.M., Cornelius A., Mulligan M.J., Landau N.R. Comparison of Neutralizing Antibody Titers Elicited by mRNA and Adenoviral Vector Vaccine against SARS-CoV-2 Variants. bioRxiv. 2021 2021.2007.2019.452771. [Google Scholar]
- Tang P., Hasan M.R., Chemaitelly H., Yassine H.M., Benslimane F.M., Al Khatib H.A., AlMukdad S., Coyle P., Ayoub H.H., Al Kanaani Z., et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 2021 doi: 10.1038/s41591-021-01583-4. Published online November 2, 2021. [DOI] [PubMed] [Google Scholar]
- Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Edara V.V., Floyd K., Kauffman R.C., Mantus G., Anderson E., Rouphael N., Edupuganti S., Shi P.Y., Menachery V.D., et al. Development of a Rapid Focus Reduction Neutralization Test Assay for Measuring SARS-CoV-2 Neutralizing Antibodies. Curr. Protoc. Immunol. 2020;131:e116. doi: 10.1002/cpim.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Thomas J., Soung A.L., Davis-Gardner M.E., Floyd K., Jin F., Cowan D.A., Pellegrini K., Shi P.Y., Grakoui A., et al. CCR2 Signaling Restricts SARS-CoV-2 Infection. MBio. 2021;12:e0274921. doi: 10.1128/mBio.02749-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.C., Prather K.A., Sznitman J., Jimenez J.L., Lakdawala S.S., Tufekci Z., Marr L.C. Airborne transmission of respiratory viruses. Science. 2021;373:eabd9149. doi: 10.1126/science.abd9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., Pegu A., Oloniniyi O.K., Henry A.R., Darko S., et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373:eabh1766. doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.H., Llewelyn A., Brandao R., Chowdhary K., Hardisty K.M., Loddo M. SARS-CoV-2 testing and sequencing for international arrivals reveals significant cross border transmission of high risk variants into the United Kingdom. EClinicalMedicine. 2021;38:101021. doi: 10.1016/j.eclinm.2021.101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.