Mismatch repair (MMR) systems are evolutionarily conserved and play a primary role in mutation avoidance by removing base-base and small insertion-deletion mismatches that arise during DNA replication (31). In addition, MMR factors are required for the repair of mismatches in heteroduplex DNA (hDNA) that form as a result of sequence heterologies between recombining sequences (6, 41, 43). MMR also acts to inhibit recombination between moderately divergent (homeologous) sequences (11, 42). The roles of MMR during recombination are believed to reflect the interaction of MMR factors with mismatches that arise in hDNA or possibly with other structures such as Holliday junctions (2, 33). The full range of effects that MMR can exert on mitotic and meiotic recombination have been discussed elsewhere (11) and will only be summarized briefly here. The purpose of this review is to highlight recent results that have furthered our understanding of interactions between MMR factors and mitotic recombination intermediates.
BRIEF OVERVIEW OF MMR IN PROKARYOTES AND EUKARYOTES
The best-understood MMR pathway is the Escherichia coli methyl-directed MutHLS system that has been reconstituted in vitro from purified components (35). To initiate MMR, MutS forms a homodimeric complex that binds base-base mismatches and loop insertions-deletions that result primarily from polymerase misincorporation and slippage errors, respectively. The MutS-mispair complex then recruits a MutL homodimer to activate MutH endonuclease activity on newly synthesized DNA. ATP binding and hydrolysis by MutS and MutL are hypothesized to induce conformational changes in these factors that regulate mismatch binding and interactions with downstream factors such as MutH. Following activation, MutH endonuclease incises the newly replicated (unmethylated) DNA strand at hemimethylated sites formed after the passage of the replication fork. The nicked strand is then unwound by helicase II and degraded back past the mismatch by the action of 5′-to-3′ or 3′-to-5′ exonucleases (35; M. Viswanathan, V. Burdett, C. Baitinger, P. Modrich, and S. T. Lovett, submitted for publication), and repair synthesis fills in the resulting gap.
In eukaryotes, mismatch recognition is accomplished by Msh2 (MutS homolog 2) protein forming a heterodimer with either Msh3p or Msh6p to bind to a distinct but overlapping spectra of mismatches (reviewed in reference 31). In both the yeast Saccharomyces cerevisiae and humans, the repair of base-base mismatches appears to be solely dependent on Msh2p-Msh6p, while both Msh2p-Msh6p and Msh2p-Msh3p can participate in the repair of small (1 to 12 nucleotide nt) loop insertions. In yeast, analysis of the mutation spectra in msh mutant strains and in vitro studies of the mismatch binding and ATPase activities of wild-type and mutant heterodimers have led to a clearer picture of the early steps in eukaryotic MMR. Currently, it is thought that Msh heterodimer binding to a mismatch triggers ATP-dependent steps that allow interactions with Mlh (MutL homolog) heterodimers composed of Mlh1p-Pms1p or Mlh1p-Mlh3p (19, 24, 62; reviewed in reference 37). No MutH homolog has been identified in eukaryotes, and the exact details of strand discrimination and error removal are not known, although in both yeast and humans PCNA and the 5′-to-3′ exonuclease Exo1p have been implicated in steps following mismatch recognition (9, 29, 49, 52, 58, 60).
Genetic and biochemical analyses of E. coli MutS and human Msh2p-Msh6p suggest that MutS homolog proteins can bind to DNA in at least two different modes (4, 22). In the case of MutS, the first mode allows mismatch recognition, and the second mode allows MutS to translocate along DNA with MutL so that it can activate MutH at GATC sites (4, 23). Support for the presence of a second binding mode was obtained from DNA binding assays showing that the addition of ATP resulted in the loss of MutS mismatch binding specificity (23). Further support was obtained by Allen et al. (4), who showed in electron microscopic analysis that MutS can form ATP-dependent loop structures on mismatched DNA substrates. They hypothesized that MutS can bind to a mismatch substrate in an ATP-independent step. After recognition, a second binding mode is activated through a conformational change in MutS that allows translocation along DNA away from a mismatch site via ATP hydrolysis-dependent mechanisms (4, 7). An alternative model has been proposed by Gradia and coworkers in which ATP binding acts as a molecular switch analogous to G protein switches (21, 22). In this model, MutS family proteins are competent to bind to a mispair when in the ADP-bound form. Mispair binding then provokes the exchange of ADP for ATP that allows the MutS family proteins to form a hydrolysis-independent sliding clamp that can slide along DNA to interact with downstream MMR components. In this model, MutL family proteins could act as regulators of MutS family proteins either by stimulating MSH2-MSH6 ATPase activity or by promoting the exchange of ADP for ATP (21).
MMR AND THE REJECTION OF MISMATCHED RECOMBINATION INTERMEDIATES
In bacteria, yeast, and mammalian cells, recombination beween homeologous DNA substrates containing a few mismatches (<1%) occurs much less efficiently than between identical sequences. The frequency of recombination (gene conversions and/or crossovers) between homeologous sequences, however, is often dramatically elevated in MMR-defective cell lines (3, 8, 13, 15, 26, 44, 50, 51). The antirecombinogenic activity of MMR has been proposed to play a role in preventing interspecific gene transfer, which could be important in establishing a genetic barrier between closely related organisms (42, 53, 61). Furthermore, recombination between diverged repeats present in the genome could lead to chromosomal translocations, deletions, or inversions which in higher eukaryotes are thought to contribute to tumor formation. msh2-deficient mice, for example, display hematological malignancies that are hypothesized to arise through chromosomal rearrangements (1, 45).
How are mismatch repair factors thought to prevent homeologous recombination? Current models of genetic recombination suggest that recombination is accomplished through a double-strand break (DSB) repair (DSBR) mechanism. In such a model, heteroduplex DNA is initially formed during the invasion of single-stranded (ss) DNA from a recipient chromosome into a homologous region in a donor chromosome. In DSBR, DSBs are first processed by 5′- 3′ exonucleases to yield 3′-ended ss tails that invade a homologous duplex. DNA synthesis is then primed from the 3′ end of the invading strand which results in the copying of donor information (gene conversion). Recombination may be completed through branch migration and resolution of a Holliday junction intermediate to form noncrossover or crossover products (57), or by pairing of the extended ss end with its original partner followed by ligation (synthesis-dependent strand-annealing model (41).
Genetic studies of homeologous recombination during mitosis in yeast (10, 12, 13, 50) suggest that if recipient and donor sequences are too divergent (>10%), recombination is severely repressed, presumably due to an inability to form a sufficiently stable base-paired intermediate. At lower levels of divergence, MMR imposes an additional barrier to recombination so that the formation of hDNA occurs with a probability that declines exponentially with increasing sequence divergence. Several investigators (10, 12, 13, 51) have proposed that a minimal region of completely homologous sequence is required to initiate heteroduplex formation (∼20 bp in yeast) and that a minimal heterology-free region is necessary to escape rejection by MMR (∼610 bp in yeast). A random walk model has also been applied to these data to explain the rapid and nonlinear drop-off of recombination triggered by a small number of mismatches (20).
The Msh2p-Msh6p and Msh2p-Msh3p heterodimers show substrate specificity for the type of mismatch that leads to the repression of genetic recombination. Nicholson et al. (39) examined the requirement for different Msh complexes in repression of homeologous recombination. They employed an indirect repeat assay system that selects for reorientation of an intron DNA segment shared between homologous or homeologous recombination substrates. Divergent cassettes were designed to generate defined mismatch types within the recombination intermediate in order to test the specificity of msh2, msh3, and msh6 deletions on the rate of recombination. Compared to the wild type, an msh2 deletion mutation caused the greatest increase (loss of rejection) in recombination rate for all homeologous substrates. An msh6Δ strain displayed an increase in recombination between homeologous cassettes bearing various arrays of base-base and 1-nt loop insertions but did not affect substrates containing 4-nt (and presumably larger) loops, consistent with the mismatch binding and repair specificity of the Msh2p-Msh6p heterodimer. Surprisingly, an msh3Δ mutation led to an increase in recombination between substrates predicted to exclusively form base-base mismatches in the recombination intermediate, even though the Msh2p–Msh3p complex is considered to recognize only extrahelical loops during MMR. This suggested an unexpected role for Msh3p in recognition of base-base mismatches in recombination intermediates. msh3Δ msh6Δ and msh2Δ strains displayed similar results, providing further evidence for overlap between MSH3 and MSH6 gene functions.
In addition to identifying the substrate specificities of the MutS homologs that act to repress homeologous recombination, Nicholson et al. (39) found that strains containing null mutations in the mutL homologs MLH1 and PMS1 (individually or in combination) elevated homeologous recombination levels to only a fraction of the levels observed in msh2Δ or msh3Δ msh6Δ strains, suggesting that the mechanism of homeologous rejection may be distinct from MMR where the Msh and Mlh heterodimers are equally required (8, 10, 26, 39). A caveat in this interpretation is that the contribution of the mutL homologs MLH2 and MLH3 in repressing homeologous recombination has yet to be assessed (37). Nicholson et al. (39) also found that strains lacking the Exo1p or Rad1p–Rad10p nucleases showed increased recombination between homeologous substrates, suggesting a role for these proteins in the repression of homeologous recombination which may reflect a physical and/or functional association with Msh2p (39). The effects of rad1, exo1, and pms1 deletion mutations were not epistatic, indicating that distinct pathways or complexes regulate homeologous recombination. Further analysis revealed that while msh6 and pms1 mutants displayed an epistatic effect on homeologous recombination, msh3 and pms1 mutants did not, raising the possibility that Msh3p may play a separate role in preventing homeologous recombination when complexed with other factors such as Rad1p-Rad10p or Exo1p (39).
How can only a subset of MMR factors participate in the repression of homeologous recombination? One possibility is that similar signals may initiate recognition and DNA translocation steps by the MutS homolog proteins in both the MMR and homeologous rejection pathways. For example, binding of MutS homolog proteins to mispaired or perturbed DNA structures could result in the activation of translocating complexes that then encounter downstream factors specific to MMR or recombinational repair pathways (see below).
MMR FACTORS ACT TO REMOVE NONHOMOLOGOUS DNA DURING GENETIC RECOMBINATION
In addition to their roles in MMR and homeologous recombination, MMR proteins play an important role in removing nonhomologous DNA during gene conversion and single-strand annealing (SSA) events (30, 48, 56). During gene conversion, nonhomologous ends of DSBs must be removed to enable the invading or annealed 3′ ss end to prime new DNA synthesis from its template. Genetic studies suggest that nonhomologous ends are cleaved by the Rad1p-Rad10p endonuclease and their removal is facilitated by Msh2p-Msh3p (18, 28, 40, 48, 56). In addition, the Msh2p-Msh3p complex participates with Rad1p-Rad10p to remove nonhomologous ends during repair by SSA and also to repair extrahelical loops of intermediate size (30). Importantly, downstream factors in MMR (MLH1 and PMS1) are not required for the removal of nonhomologous ends, suggesting that Msh2p-Msh3p and Rad1p-Rad10p are part of a distinct complex that excises heterologies larger than a few nucleotides (56).
Sugawara et al. (56) proposed a model for tail removal during DSBR (Fig. 1 and 2) in which Msh2p-Msh3p stabilizes annealed intermediates by binding to the unpaired single-stranded DNA (ssDNA) at the ends of the annealed region. This allows Rad1p-Rad10p to locate and cleave the 3′-ended tail, possibly facilitated by physical interactions between these complexes (5). This model is based on the fact that Msh2p-Msh3p is not required for SSA when paired homology blocks are long (>1 kb) but is required during Rad1p-Rad10p-dependent gene conversion regardless of the length of homology available for pairing. Furthermore, Msh2p-Msh3p is not needed for gene conversion if nonhomologous tails are very short (<30 nt). Sugawara et al. (56) proposed that in gene conversion, the invading strand can only form a side-by-side paranemic joint between homologous sequences in a three-stranded intermediate prior to tail removal. Paranemic joints are unstable in vitro and require protein binding, in addition to base pairing, for stability (46). During SSA, the authors propose that interwound, plectonemic molecules can form in a two-stranded intermediate, and when the homologous segments are long enough, the inherent stability of this structure is sufficient to recruit Rad1p-Rad10p independently of Msh2p-Msh3p.
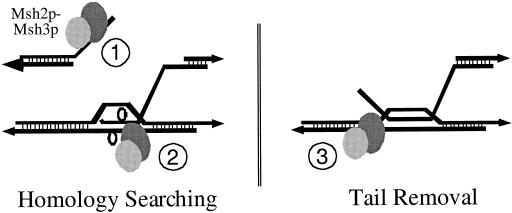
FIG. 1.
Two steps in recombination in which the Msh2p-Msh3p complex may interact with recombination intermediates. (Left) Msh2p-Msh3p loads onto DSB sites at recessed ends (1) and/or plays an active role in scanning hDNA and interacts with loops formed during pairing of homeologous sequences (2), leading to their rejection from the heteroduplex. (Right) Msh2p-Msh3p binds at the junction of homologous and nonhomologous DNA allowing for cleavage of unpaired tails by Rad1p-Rad10p (3) (adapted from reference 17).
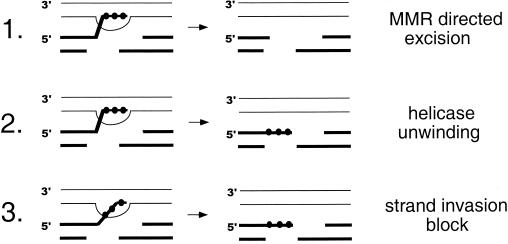
FIG. 2.
Models to explain rejection of heteroduplex intermediates containing mispairs via MMR proteins. In this figure, base pair differences between the recipient and donor chromosomes are indicated by the solid circles. (1) The mismatch correction process itself could lead to resection of nicked strands and the creation of a single-stranded gap that destroys the recombination intermediate. (2) hDNA rejection results in the unwinding of the annealed strands by a helicase that takes its cue from interactions with bound Msh factors. (3) Binding of MMR factors blocks attempted hDNA formation (Sugawara et al., unpublished).
How do mismatch repair proteins participate in the removal of unpaired ssDNA tails? Analogous to events proposed for postreplication mismatch repair, the removal of nonhomologous DNA could be mediated by the loading of Msh2p-Msh3p heterodimers onto the branched DNA structures at or near the double-strand DNA–ssDNA junction. In such a model, Msh2p-Msh3p loading at or between junctions would result in the formation of ATP hydrolysis-dependent (7) or -independent (21, 22) sliding DNA clamps that translocate along DNA and interact with downstream excision factors. Alternatively, Msh2p-Msh3p loading at branched DNA structures could serve as a target for the Rad1p-Rad10p endonuclease or mismatch repair nucleases. Few studies have been done to examine these possible mechanisms. An initial analysis of msh2 and msh3 ATP binding domain mutants suggested that ATP binding and/or hydrolysis was critical for the removal of nonhomologous DNA during gene conversion (54); however, this analysis did not identify which steps were compromised in the recombination pathway. In vitro binding studies involving Msh2p-Msh3p, Rad1p-Rad10p, model branch DNA structures, and ATP have yet to be performed; such an approach could provide evidence to support or refute the models presented above.
INTERACTION OF MMR FACTORS AND RECOMBINATION INTERMEDIATES: A ROLE FOR MSH PROTEINS IN THE SEARCH FOR HOMOLOGY?
The first biochemical demonstration of an impact of MMR proteins on the recombination machinery was observed in strand transfer reactions performed in vitro. In these reactions, the E. coli RecA protein catalyzes heteroduplex DNA formation between a duplex linear plasmid and a homologous ss circular substrate. Worth et al. (65) showed that the addition of purified MutS blocked full-length heteroduplex formation between 3% divergent ss circular and linear duplex DNAs. Analysis of MutS mutants defective in ATPase activity suggested that mismatch binding itself blocked further strand exchange (66). Addition of MutL enhanced this effect, possibly by stabilizing MutS on the mismatch. Although the effect of MMR on in vitro strand exchange in eukaryotes has not been measured, yeast Msh2p can to bind model substrates that resemble Holliday junction recombination intermediates in vitro (2, 33) and may well interact with recombination factors in vivo.
Interaction of MMR factors with recombination intermediates in vivo was recently examined by Evans et al. (17) who employed cross-linking, immunoprecipitation, and PCR (by a chromatin immunoprecipitation assay) to examine the specific association of S. cerevisiae Msh2p with plasmids undergoing DSB repair. In the repair substrates used, one copy of the lacZ gene (the recipient for repair) contains an internal HO endonuclease cleavage site, and induced expression of the HO endonuclease leads to formation of a unique DSB within the recipient sequence that can be repaired by different pathways, depending on the nature of the available lacZ donor sequence (40, 55, 56). Following the induction of a DSB in the recipient copy of lacZ, Msh2p was found to specifically localize to recipient sequences in a time frame consistent with repair events as measured in physical assays (56). Msh2p localization was strongest in cases where nonhomologous DNA was present at the DNA ends, and little localization was observed when fully homologous lacZ substrates were tested. Msh2p localization to donor sequences was observed in plasmid substrates that contain nonhomologous DNA within the HO endonuclease DSB site. During recombination, these plasmids are predicted to form recombination intermediates bearing ss tails that are removed through the action of Msh2p-Msh3p and Rad1p-Rad10p (56) (Fig. 1).
In an effort to understand the molecular events underlying localization of Msh2p on nonhomologous DNA ends, Evans et al. (17) tested a series of repair mutants in the chromatin immunoprecipitation assay. Msh2p localization to DNA sequences adjacent to a DSB required Msh3p but not Msh6p, supporting the interpretation that the Msh2p–Msh3p complex is the primary species being detected. Localization was greatly reduced in the absence of Rad50p. The Rad50p-Mre11p-Xrs2p complex binds to ssDNA formed at DSB sites in vivo (25, 34, 38). This complex is believed to regulate the processing of DSB ends, as well as their participation in recombination and nonhomologous end-joining pathways (41). Importantly, rad50Δ and mre11Δ mutants display a slower rate of exonucleolytic processing of DSBs (28, 32, 36, 59). The reduction of Msh2p localization in a rad50 deletion strain suggested that localization depends on the formation and/or activities of the ss tails that engage in the search for homology.
Rad52p is essential for most types of recombination in yeast and has been implicated in early strand exchange steps (41). Studies of mitotic recombination at the MAT locus in S. cerevisiae indicated that the processing of DSBs proceeds in the absence of Rad52p but that primer extension and completion of recombination do not occur (64). Evans et al. (17) found that localization of Msh2p to DNA substrates that contained nonhomologous ends proceeded normally in a rad52Δ strain, providing further evidence that Msh2p localization relates to early events in recombination, prior to strand-annealing steps that involve Rad52p. This is consistent with the interpretation of Westmoreland et al. (63), who found that MMR is not effective in rejecting preformed heteroduplexes, suggesting that MMR proteins act to block the extension of intermediates that are tested during the homology search steps of recombination. Based on a chromosome spread analysis of Schizosaccharomyces pombe msh2 mutants in meiotic prophase, Rudolph et al. (47) argued that Msh2p acted to reject homeologous pairings during interactions with unstable and/or mismatched hDNA intermediates. This hypothesis is based on their observation that in meiosis msh2 mutants accumulate nuclei containing aggregated linear elements; such structures are thought to result from the failure to reject ectopic chromosomal interactions. Interestingly, Evans et al. (17) found that although Msh2p did not normally localize to ends undergoing fully homologous gene conversion, increased association was observed when homologous repair was blocked in a rad52Δ strain, suggesting that stalled repair intermediates either retained the binding of Msh2p from an earlier step or that Msh2p association to homologous tails was triggered in the absence of Rad52p, possibly by initiating new rounds of homology searching. Although the precise role of MMR factors located in DSBR mechanisms in vivo remains speculative, interactions can be further tested by examining their localization to DSBR substrates bearing specific mismatches.
Figure 2 presents a model where Msh2p-Msh3p associates with intermediates early in DSBR to participate in the rejection of homeologous pairing during heteroduplex DNA formation and can further act, where needed, to bind unstable intermediates, facilitating cleavage by Rad1p-Rad10p nuclease. Presumably, Msh heterodimers can recognize mismatches that form due to homeologous pairings, which may trigger loading of the heterodimer onto DNA as a sliding clamp that can translocate away from the mismatch. How might this trigger heteroduplex rejection? Mismatch repair-directed excision could lead to resection of nicked strands and the creation of a single-stranded gap that could destroy the recombination intermediate (Fig. 2, model 1). However, the data outlined above argues that rejection and repair are genetically distinct. Sugawara et al. (N. Sugawara, B. Studamire, E. Alani, and J. E. Haber, unpublished material) hypothesized a role for an endonuclease and/or exonuclease that excises hDNA in concert with Msh factors. Their analysis of exo1 mutants, however, did not support a role for Exo1p in heteroduplex rejection but did not exclude the possibility that exonucleases play a redundant role. hDNA rejection could involve unwinding of the annealed strands by a helicase that takes its cue from interactions with bound Msh factors: at present, candidate helicases have not been tested (Fig. 2, model 2).
Alternatively, the binding of Msh factors might block or reverse attempted hDNA formation (Fig. 2, model 3), possibly through an interaction between the bound Msh proteins and the recombination machinery, similar to the mechanism proposed for E. coli (65). In such a model, homeologous pairings would be prevented from entering recombination intermediates, and the presynaptic filament would disengage so that the homology search would continue elsewhere (27; Sugawara et al. unpublished). Similar models to explain the role of mismatch repair in heteroduplex rejection have been developed to explain the role of MutS and MutL proteins in preventing interspecies recombination in bacteria (53, 61). Studies by Stambuk et al. (53) have suggested that the prevention of homeologous recombination could occur through two distinct mechanisms. The first mechanism is thought to occur by UvrD helicase acting to abort initiation steps in recombination. The second is likely to involve an incomplete long-patch mechanism that requires excision functions directed by MutH endonuclease activity.
MMR factors have been implicated in cell cycle checkpoints and the p53-dependent apoptotic response to DNA damage (14, 16, 67), suggesting there may be additional roles for the MMR factors in signaling the presence of DSBs or other aberrant structures. Given the complex interplay of recombination and repair factors, it is worth considering models for the interaction of these factors on DNA which can be tested by combining genetic, biochemical, and physical approaches in vitro and in vivo.
ACKNOWLEDGMENTS
We thank James Haber, Neal Sugawara, and Barbara Studamire for engaging us in many stimulating discussions and members of the Alani Laboratory for critical reading and discussion of the manuscript. We are also grateful for the contributions of N. Sugawara and J. Haber that led to the development of Fig. 2.
This work was supported by NIH grant GM53085 and Hatch grant NYC-165-6424.
REFERENCES
- 1.Abuin A, Zhang H, Bradley A. Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol Cell Biol. 2000;20:149–157. doi: 10.1128/mcb.20.1.149-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 3.Alani E, Reenan R A G, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen D J, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith J D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand P, Tishkoff D X, Filosi N, Dasgupta R, Kolodner R D. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc Natl Acad Sci USA. 1998;95:14278–14283. doi: 10.1073/pnas.95.24.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop D K, Williamson M S, Fogel S, Kolodner R D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987;328:362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell LJ, Martik D, Bjornson K P, Bjornson E S, Modrich P. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J Biol Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 8.Chambers S R, Hunter N, Louis E J, Borts R H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Merrill B J, Lau P J, Holm C, Kolodner R D. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol Cell Biol. 1999;19:7801–7815. doi: 10.1128/mcb.19.11.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crouse G F. Mismatch repair systems in Saccharomyces cerevisiae, In: Nickoloff J A, Hoekstra M, editors. DNA damage and repair. Vol. 1. Totowa, N.J: Humana Press; 1998. pp. 411–448. [Google Scholar]
- 12.Datta A, Adjiri A, New L, Crouse G F, Jinks-Robertson S. Crossovers between diverged sequences are regulated by mismatch repair proteins in yeast. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis T W, Wilson-Van Patten C, Meyers M, Kunugi K A, Cuthill S, Reznikoff C, Garces C, Boland C R, Kinsella T J, Fishel R, Boothman D A. Defective expression of the DNA mismatch repair protein, MLH1, alters G2-M cell cycle checkpoint arrest following ionizing radiation. Cancer Res. 1998;58:767–778. [PubMed] [Google Scholar]
- 15.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse MSH2 gene results in postreplicational mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to tumorigenesis. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 16.Duckett D R, Bronstein S M, Taya Y, Modrich P. hMutsα and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Pro Natl Acad Sci USA. 1999;96:12384–12388. doi: 10.1073/pnas.96.22.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans E, Sugawara N, Haber J E, Alani E. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Mol Cell. 2000;5:789–799. doi: 10.1016/s1097-2765(00)80319-6. [DOI] [PubMed] [Google Scholar]
- 18.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 19.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujitani Y, Kobayashi I. Effect of DNA sequence divergence on homologous recombination as analyzed by a random-walk model. Genetics. 1999;153:1973–1988. doi: 10.1093/genetics/153.4.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 22.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 23.Grilley M, Welsh K M, Su S S, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 24.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 25.Hopfner K P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 26.Hunter N, Chambers S R, Louis E J, Borts R H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 27.Inbar O, Kupiec M. Homology search and choice of homologous partner during mitotic recombination. Mol Cell Biol. 1999;19:4134–4142. doi: 10.1128/mcb.19.6.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov E L, Sugawara N, White C I, Fabre F, Haber J E. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson R E, Kovvali G K, Guzder S N, Amin N S, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 30.Kirkpatrick D T, Petes T D. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 31.Kolodner R D, Marsischky G T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Moore J, Holmes A, Umezu K, Kolodner R D, Haber J E. Saccharomyces Ku70, mre11/rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 33.Marsischky G T, Lee S, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2/6 complex interacts with Holliday junctions and facilitates their cleavage by phage resolution enzymes J. Biol Chem. 1999;274:7200–7206. doi: 10.1074/jbc.274.11.7200. [DOI] [PubMed] [Google Scholar]
- 34.Maser R S, Monsen K J, Nelms B E, Petrini J H. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 36.Moreau S, Ferguson J R, Symington L S. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa T, Datta A, Kolodner R D. Multiple functions of MutS- and MutL-related heterocomplexes. Proc Natl Acad Sci USA. 1999;96:14186–14188. doi: 10.1073/pnas.96.25.14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson A, Hendrix M, Jinks-Robertson S, Crouse G F. Regulation of mitotic homeologous recombination in yeast. Functions of mismatch repair and nucleotide excision repair genes. Genetics. 2000;154:133–146. doi: 10.1093/genetics/154.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pâques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pâques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radman M. Mismatch repair and the fidelity of genetic recombination. Genome. 1989;31:68–73. doi: 10.1139/g89-014. [DOI] [PubMed] [Google Scholar]
- 43.Ray B L, White C I, Haber J E. Heteroduplex formation and mismatch repair of the “stuck” mutation during mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5372–5380. doi: 10.1128/mcb.11.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 45.Reitmair A H, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker H W, Wakeham A, Liu B, et al. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 46.Riddles P, Lehman I. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1985;260:165–169. [PubMed] [Google Scholar]
- 47.Rudolph C, Kunz C, Parisi S, Lehmann E, Hartsuiker E, Fartmann B, Dramer W, Kohli J, Fleck O. The msh2 gene of Schizosaccharomyces pombe is involved in mismatch repair, mating-type switching, and meiotic chromosome organization. Mol Cell Biol. 1999;19:241–250. doi: 10.1128/mcb.19.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmutte C, Marinescu R C, Sadoff M M, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- 50.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen P, Huang H. Effect of base pair mismatches on recombination via the RecBCD pathway. Mol Gen Genet. 1989;18:358–360. doi: 10.1007/BF00331291. [DOI] [PubMed] [Google Scholar]
- 52.Sokolsky T, Alani E. EXOI and MSH6 are high copy suppressors of conditional mutations in the MSH2 mismatch repair gene of S. cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stambuk S, Radman M. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics. 1998;150:533–542. doi: 10.1093/genetics/150.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studamire B, Price G, Sugawara N, Haber J E, Alani E. Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination. Mol Cell Biol. 1999;19:7558–7567. doi: 10.1128/mcb.19.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugawara N, Pâques F, Colaiácovo M P, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break repair-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szostak J W, Orr-Weaver T L, Rothstein R, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 58.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, et al. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 61.Vulic M, Lenski R E, Radman M. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proc Natl Acad Sci USA. 1999;96:7348–7351. doi: 10.1073/pnas.96.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang T F, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA. 1999;96:13914–13909. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westmoreland J, Porter G, Radman M, Resnick M A. Highly mismatched molecules resembling recombination intermediates efficiently transform mismatch repair proficient Escherichia coli. Genetics. 1997;145:29–38. doi: 10.1093/genetics/145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Worth L, Jr, Bader T, Yang J, Clark S. Role of MutS ATPase activity in MutS,L-dependent block of in vitro strand transfer. J Biol Chem. 1998;273:23176–23182. doi: 10.1074/jbc.273.36.23176. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H, Richards B, Wilson T, Lloyd M, Cranston A, Thorburn A, Fishel R, Meuth M. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]