Summary
Here we describe a protocol for identifying metabolites in respiratory specimens of patients that are SARS-CoV-2 positive, SARS-CoV-2 negative, or H1N1 positive. This protocol provides step-by-step instructions on sample collection from patients, followed by metabolite extraction. We use ultra-high-pressure liquid chromatography (UHPLC) coupled with high-resolution mass spectrometry (HRMS) for data acquisition and describe the steps for data analysis. The protocol was standardized with specific customization for SARS-CoV-2-containing respiratory specimens.
For complete details on the use and execution of this protocol, please refer to Maras et al. (2021).
Subject areas: Health Sciences, Clinical Protocol, Immunology, Metabolomics, Mass Spectrometry
Graphical abstract

Highlights
-
•
High-resolution mass spectrometry (HRMS)-based metabolomics of respiratory specimens
-
•
Heat inactivation, homogenization, and extraction of metabolites from patient samples
-
•
Peak picking and workflow for untargeted metabolomics data analysis
Here we describe a protocol for identifying metabolites in respiratory specimens of patients that are SARS-CoV-2 positive, SARS-CoV-2 negative, or H1N1 positive. This protocol provides step-by-step instructions on sample collection from patients, followed by metabolite extraction. We use ultra-high-pressure liquid chromatography (UHPLC) coupled with high-rResolution rass spectrometry (HRMS) for data acquisition and describe the steps for data analysis. The protocol was standardized with specific customization for SARS-CoV-2 containing respiratory specimens.
Before you begin
Note: This protocol has been used by our group (Maras et al., 2018, 2020, 2021; Bhat et al., 2020), and other groups (Weiss et al., 2016; Moreau et al., 2020) and (Castelli et al., 2021) for metabolome analysis.
Respiratory specimens were collected under the guidelines provided by the Centers for Disease Control and Prevention (Control and Prevention, 2020). The Viral Transport Media utilized is made of Hanks Balanced Salt Solution and contains a protective protein, antibiotics to control microbial and fungal contamination, and buffers to control the pH. Phenol red is used as a pH indicator. The medium also contains a cryoprotectant which helps in preserving the viruses if specimens are frozen for prolonged storage.
Nasopharyngeal specimen (NP) collection
Tilt the head of the patient 70° to the back. Gently insert and roll the swab with a flexible shaft (wire or plastic) through the nostril parallel to the palate (not upwards) until resistance is encountered or the distance is equivalent to that from the ear to the nostril of the patient, indicating contact with the nasopharynx. Leave the swab in place for a few seconds to absorb secretions. Slowly remove the swab while rotating it. Place swab, tip first, into the viral transport media containing tube provided.
Note: Specimens can be collected from both sides using the same swab, but it is not necessary to collect specimens from both sides if the swab is saturated with fluid from the first collection.
Oropharyngeal (OP) (throat) specimen collection
Insert swab into the posterior pharynx and tonsillar areas. Rub swab over both tonsillar pillars and posterior oropharynx and avoid touching the tongue, teeth, and gums. Place swab, tip first, into the viral transport media to make the respiratory specimen. Mix the NP and the OP specimen to form a respiratory specimen which can be used for RT-PCR based detection of viral presence and also could be used as a starting material for multi-omics analysis.
Note: The mixing of NP or OP is not mandatory and can be analyzed individually. An aliquot of the respiratory specimens was sent for SARS-CoV-2 detection, and the other was utilized for metabolomics analysis. Further, respiratory specimens can be stored in −80°C for a year. For better results, it should be used within 2 months for analysis
Key resources table
| REREAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Acetonitrile for LCMS | J.T. Baker | Cat#14650359 |
| Methanol for LCMS | Sigma-Aldrich | Cat#34860 |
| Pierce™ Positive Ion Calibration Solution | Thermo Scientific | Cat#88323 |
| Pierce™ Negative Ion Calibration Solution | Thermo Scientific | Cat#88324 |
| LC-MS grade Formic acid | Merck | CAS#64-18-6 |
| Milli-Q water (18 MU) | Thermo Scientific | Cat#W6-4 |
| Metformin | Sigma-Aldrich | CAS#1115-70-4 |
| Ethylmalonic acid | Sigma-Aldrich | CAS#601-75-2 |
| Dihydrostreptomycin | Sigma-Aldrich | CAS#5490-27-7 |
| Colchicine | Sigma-Aldrich | CAS#64-86-8 |
| Imipramine | Sigma-Aldrich | CAS#113-52-0 |
| Roxithromycin | Sigma-Aldrich | CAS#80214-83-1 |
| Amiloride | Sigma-Aldrich | CAS#2016-88-8 |
| Atropine | Sigma-Aldrich | CAS#51-55-8 |
| 2-aminoanthracene | Sigma-Aldrich | CAS#613-13-8 |
| Prednisolone | Sigma-Aldrich | CAS#50-24-8 |
| Dinoseb | Sigma-Aldrich | CAS#88-85-7 |
| MCPA 2-methyl-4-chlorophenoxyacetic acid | Sigma-Aldrich | CAS#2436-73-9 |
| Dimetridazole | Sigma-Aldrich | CAS#551-92-8 |
| AMPA 2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl) propanoic acid | Sigma-Aldrich | Cat#A6816 |
| Viral Transport Media | Himedia | Cat#MS2760A |
| Software and algorithms | ||
| Compound Discoverer Software 3.1 | Thermo Fisher Scientific | Compound Discoverer™ Software |
| MetaboAnalyst 4.0 | Web server | https://www.metaboanalyst.ca/ |
| Other | ||
| Q Exactive Orbitrap Mass Spectrometer | Thermo Scientific | Cat#IQLAAEGAAPFALGMBDK |
| Hypersil GOLD™ C18 Selectivity HPLC Columns | Thermo Scientific | Cat#25003102130 |
| 200μL pipette tips, Sterile grade | Eppendorf® Combitips advanced | Z762997 |
| 9mm Target DP 300μL HPLC Vial (Polypropylene) | Thermo Scientific | C4000-11 |
| 9mm Autosampler Vial Screw Thread Caps (Blue), PTFE/White Silicone Septum | Thermo Scientific | C5000-54B |
| Benchtop centrifuge | Eppendorf | Cat#5409000012 |
| Sonicator | Helix Biosciences | I.T NO.#HBSNII-92 |
| Vortex | Sigma-Aldrich | Z258423 |
| Vacuum evaporator | Genevac | Cat#75871-454 |
Materials and equipment
Internal standards for untargeted analysis
| Reagent | Concentration | Amount |
|---|---|---|
| Dinoseb | 5 μg/mL | 25 μg |
| MCPA 2-methyl-4-chlorophenoxyacetic acid | 5 μg/mL | 25 μg |
| Dimetridazole | 5 μg/mL | 25 μg |
| AMPA 2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl) propanoic acid | 5 μg/mL | 25 μg |
| Milli-Q water | n/a | Make-up volume |
| Total | 5 mL |
External standard for untargeted analysis
| Reagent | Concentration | Amount |
|---|---|---|
| Dihydrostreptomycin | 20 μg/mL | 100 μg |
| Colchicine | 0.5 μg/mL | 2.5 μg |
| Imipramine | 0.5 μg/mL | 2.5 μg |
| Roxithromycin | 20 μg/mL | 100 μg |
| Amiloride | 10 μg/mL | 50 μg |
| Atropine | 1 μg/mL | 5 μg |
| 2-aminoanthracene | 1 μg/mL | 5 μg |
| Prednisolone | 1 μg/mL | 5 μg |
| Metformin | 1 μg/mL | 5 μg |
| Ethylmalonic acid | 3 μg/mL | 15 μg |
| Milli-Q water | Make-up volume | |
| Total | 5 mL |
Solvent A for untargeted analysis
| Reagent | Concentration | Amount (500 mL) |
|---|---|---|
| Formic Acid | 0.1% | 1 mL |
| Milli-Q water | 499 mL |
Note: Always prepare fresh Solvent A. To prevent contamination of any particulate matter filter the solvent with 0.45 μm filter. Then place the bottle in an ultrasonic bath for 10 min to degas the solvent. To avoid shift in the retention time, the same sample set should be run within 2–3 days with the fresh mobile phase solvent.
CRITICAL: Formic acid is highly caustic and flammable liquid, with a strong pungent odor. It can cause damage to skin, eyes, and mucosal surfaces. If contact occurs, the person should flush the affected areas immediately with plenty of water, followed by washing with soap and water. Wear protective gloves, laboratory coat, eye and face protection while working with this chemical
Solvent B for untargeted analysis
| Reagent | Concentration | Amount (500 mL) |
|---|---|---|
| Acetonitrile for LC-MS | 100% | 500 mL |
Note: Always prepare fresh Solvent B. To prevent contamination of any particulate matter filter the solvent with 0.45 μm filter. Mix the solvent gently. Then place the bottle in an ultrasonic bath for 10 min to degas the solvent. To avoid shift in the retention time, the same sample set should be run within 2–3 days with the fresh mobile phase solvent.
CRITICAL: Acetonitrile is a colorless volatile liquid with an aromatic odor, highly flammable and hazardous if inhaled or absorbed by skin. One should use a fume hood to minimize exposure to this substance. Wear protective clothing to avoid skin or eye contact, inhalation or ingestion. A long-sleeved laboratory coat or gown, rubber gloves, safety goggles and a face mask as a minimum standard.
-
•
LC-MS setup for untargeted analysis:
-
•
Metabolomic analysis is performed on the Thermo ScientificTM UHPLC system combined with Q Exactive Orbitrap Mass Spectrometers.
-
•
Samples are analyzed in single runs for positive and negative modes using the parameters shown in Table 1.
-
•
Hypersil GOLD™ C18 Selectivity HPLC Column is used as a stationary phase to separate respiratory metabolites. Column Specifications are mentioned in Table 2.
-
•
The mobile phases consist of (A) Acetonitrile with 1% formic acid and (B) Milli-Q-water with 0.1% formic acid, used for gradient elution (Table 3).
Note: Overall run cycle time per sample is 25 min.
Table 1.
LC-MS parameter settings
| Ionization | Electrospray ionization |
|---|---|
| MS1 and MS2 mass ranges | m/z 150–m/z 2,000 |
| Sheath gas flow rate | 54 |
| Auxiliary gas flow rate | 13 |
| Resolution | 70,000 m/Δ m |
| Collision gas | Nitrogen |
| Collision energy (positive mode/negative mode) | +40/-42 eV |
| Collision energy spread | 15 eV |
| Ion source gas 1 (air; positive mode/negative mode) | 40/50 psi |
| Ion source gas 2 (air; positive mode/negative mode) | 80/50 psi |
| Curtain gas (nitrogen) | 30 psi |
| Ion source temperature (positive mode/negative mode) | 250°C/300°C |
| Ion spray voltage floating (positive mode/negative mode) | 5500–4500 V |
| Declustering potential (positive mode /negative mode) | 80/−80 eV |
Table 2.
Column formats
| Column format | Analytical column |
|---|---|
| For Use With | LC/MS |
| Max. Pressure | 5800 psi (400 bar) |
| pH | 1 to 11 |
| Surface Area | 220 m2/g |
| USP Type | L1 |
| Product Line | Hypersil GOLD |
| Carbon Load | 11% |
| Endcapped | Yes |
| Diameter (Metric) | 2.1 mm |
| Length (Metric) | 100 mm |
| Particle Size | 3 μm |
| Pore Size | 175 Å |
| Temperature | 60°C |
| Stationary Phase | C18 Selectivity |
| Packing Material | Spherical, Fully Porous, Ultrapure Silica |
| Column Type | Reversed Phase |
Table 3.
LC-MS gradient for untargeted metabolomics
| S. No. | Time (min) | Flow (mL/min) | % B | Curve |
|---|---|---|---|---|
| 1 | 0 | 0.500 | 5.0 | 5 |
| 2 | 1 | 0.500 | 5.0 | 5 |
| 3 | 17 | 0.500 | 99.0 | 5 |
| 4 | 21 | 0.500 | 99.0 | 5 |
| 5 | 22 | 0.500 | 5.0 | 5 |
| 6 | 25 | Stop Run | ||
Step-by-step method details
An outline of the procedures is described in Figure 1. A total of 500 μL respiratory specimens was heat-inactivated using heat block at 95°C for 15 min in a BSL3 lab. Heat inactivated respiratory specimens were subjected to homogenization by probe sonicator (steps 1–6). Respiratory metabolites were extracted by organic phase extraction method (steps 7–18) for untargeted metabolomics using LCMS (steps 19–20). The obtained data are analyzed using the Compound discoverer version 3.1 (steps 21–31).
Figure 1.
Showing an outline of the respiratory specimen collection, preparation, and data analysis
Heat inactivation of virus
Timing: 15 min/sample
This step is crucial to inactivate the virus as a safety measure to mitigate the risk of infection during sample preparation.
-
1.
The collected respiratory specimens are placed on a heating block at 95°C for 15 min (Batejat et al., 2021) to inactivate the virus.
Pause point: Respiratory specimen can be stored in −80°C upto 4 weeks
Homogenization of respiratory specimen
Timing: 5–7 min/sample
Respiratory specimens may contain host cells, mucus, bacterial and viral components. Thus, it is essential to homogenize the samples.
CRITICAL: The high frequency sound emitted by the sonicator can damage hearing. Therefore place the sample in a noise isolating chamber and always close the door while operating. Do not grasp an activated horn or touch the tip of a vibrating probe. It can cause severe burns and tissue damage.
-
2.
Take 100 μL of respiratory specimen in a new microcentrifuge tube (MCT) and keep it on ice during homogenization.
-
3.
Place the sample in a noise isolating chamber and submerge the sonicator probe into the sample.
Note: The probe should not touch the walls of MCT as it will break the tube and destroy the sample.
-
4.
Close the door of the chamber after correctly placing the sample tube.
-
5.
Run the program at Power 20%, Run time 5 min (Cycle - 10 s ON, 10 s OFF) and temperature 22°C.
-
6.
Remove and clean the probe with ethanol.
Note: Keep the sample on ice during all the homogenization steps step 2–6.
Pause point: Respiratory specimen can be stored in −80°C upto 4 weeks
Organic phase metabolite extraction
Timing: ∼15 h/sample
To reduce the complexity of the sample, proteins must be removed. Hence, organic phase extraction will remove the proteins through precipitation and dissolve the metabolites in solution.
-
7.
Add 100 uL of homogenized respiratory specimen to 400 uL of 100% chilled methanol.
-
8.
Vortex it for 10 s.
-
9.
Keep it in −20°C for 10–12 h.
-
10.
Centrifuge the sample at 18,000g for 10 min.
-
11.
Take the supernatant into a new tube and discard the pellet.
-
12.
Freeze-dry the sample completely using a vacuum concentrator.
-
13.
Dissolve the sample in 105 uL of solvent (5% ACN (Acetonitrile), 5% Internal Standard, 95% water).
-
14.
Vortex the sample for 10 s.
-
15.
Pipette 80 uL of sample solution into HPLC tube.
-
16.
Add the remaining 25 μL in a different tube to prepare a pooled quality control sample (In this tube all the samples prepared for run are pooled together).
Note: QC samples are required to correct the small levels of variation within samples from the same group, to quantitatively measure technical reproducibility and to integrate data from different analytical experiments. The peak features exceeding 30% coefficient of variation (CV) values and less than 0.7 r2 for Coefficient of correlation in QC samples are excluded.
-
17.
Make a dilution of the Pool solution.
| Sample – solvent ratio | Sample% | Solvent% |
|---|---|---|
| 1:1 | 100% sample | |
| 1:2 | 50% sample | 50% solvent |
| 1:4 | 25% sample | 75% solvent |
| 1:8 | 12.5% sample | 87.5% solvent |
-
18.
Make a blank with 100% ACN.
Sample run and analysis
Timing: 30–40 min/sample
This step provides you MS.raw file for each sample for further analysis.
-
19.
Lay down the sample sequence as exemplified in Figure 2.
-
20.
Set the run with LC-MS parameters described in Table 1 (Boudah et al., 2014) and run gradient is described in Table 3. Further details are provided in Table S1.
Figure 2.
Showing an exemplary sample sequence list
Peak picking and software analysis
Timing: 6–8 h/sample
MS.raw file is mapped on different metabolome databases for metabolite annotation that is exported in excel sheet for statistical analysis in order to interpret the data.
-
21.
Standard determination at both positive and negative mode as shown in Figure 3.
Figure 3.
Showing mass spectral peak of standard metabolites
Data analysis using Compound discoverer ver. 3.1:
Compound Discoverer is a unique small molecule structure recognition software. It uses accurate mass data, isotope pattern matching, fragment matching, and mass spectral library search to identify the structure of small molecules. It is a qualitative data processing application that can process accurate mass spectra of the entire Thermo Scientific high-resolution mass spectrometer product line. Compound Discoverer provides a set of processing tools called Nodes, which can be combined into a processing workflow in many different ways according to the type of experiment you perform and the questions you must answer. These workflows can be saved and reused as templates. Additionally, users can even design and write their own nodes for use in Compound Discoverer.
-
22.
Open the Compound discoverer ver. 3.1 application. Click ‘‘New Study and Analysis” (in red borders; Figure 4).
-
23.
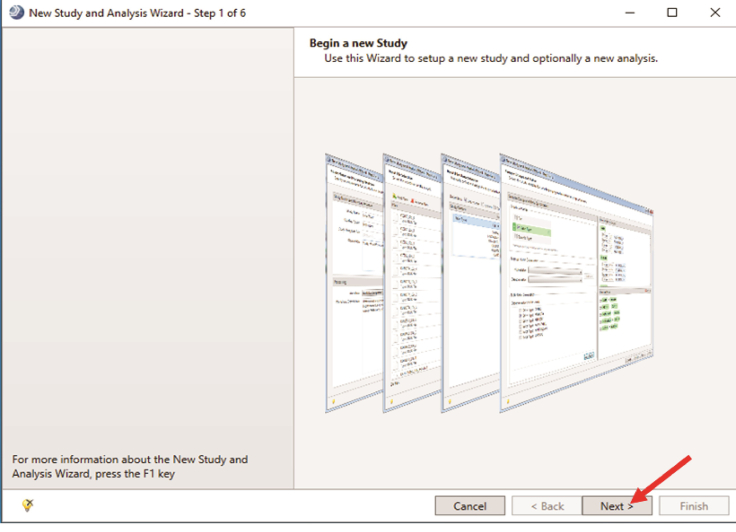
The New Study and Analysis Wizard opens. Click “Next” (indicated by the red arrow in Figure 5).
-
24.
Define the study type, name of study and storage location (underlined in red). Study template file and workflow can also be optionally selected. Then click “Next” (red arrow; Figure 6).
-
25.
Click “Add Files” on the Input File Selection page of the wizard to select the sample data files. The file names of the selected files appear in the Files box. And then click “Next” (indicated by the red arrow in Figure 7).
-
26.
In the Input File Characterization dialog box, define the Sample Type for blanks, samples, and pools and click on “Finish” (Figure 8).
-
27.
Select the metabolomics workflow in the Workflows tab appear in the set of tabbed pages.
Note: The workflow is utilized by Maras et al. (2021) shown below in Figure 9: each node is explained in user manual provided at https://assets.thermofisher.com › CMD › manuals
-
28.
Name the result file in the Analysis pane and click Run to submit the analysis to the job queue (Figure 10).
-
29.
When the job is done, a result layout file will appear (Figure 11), export the result file in “.xlsx” format.
Note: Peak picking based on “mass + retention time, mass + retention time + spectral match and mass alone.”
-
30.
In the excel file obtained, remove the duplicates (red borders) based on average area intensity of the samples (Figure 12).
-
31.
Create a comma delimited file of annotated metabolites along with their sample wise area intensity and do statistical analysis using software such as Metaboanalyst webserver (Chong et al., 2019) (Figure 13).
Figure 4.
Graphical representation of step 22
Compound discoverer 3.1 start page for new study and analysis.
Figure 5.
Graphical representation of step 23
Showing New Study and Analysis Wizard.
Figure 6.
Graphical representation of step 24
Name the study and describe the path of study.
Figure 7.
Graphical representation of step 25, showing Input File Selection Dialogue Box
Figure 8.
Graphical representation of step 26
In the Input File Characterization dialog box define the sample type.
Figure 9.
Graphical representation of step 27, showing untargeted metabolome workflow
Figure 10.
Graphical representation of step 28, name the result file and click ‘Run’
Figure 11.
Showing result layout file (step 29)
Figure 12.
Showing excel sheet composed of metabolite name, formula, molecular weight, area(max), retention time, sample wise area intensity, etc
Figure 13.
Showing expected outcomes: of VTM metabolome
(A) Volcano plot differentially regulated metabolites in COVID-19 +ve compared to COVID-19 −ve samples (p < 0.05, FC > 1.5).
(B) Pathway and metabolite set enrichment analysis (KEGG) for the upregulated and downregulated metabolites (FC > 1.5, p < 0.05) in COVID19-positive respiratory specimen.
(C) Mean decrease in accuracy of the metabolites (Red = upregulated and Green = downregulated and yellow = unchanged) in COVID-19 +ve as compared to COVID-19 −ve. Also, AUROC analysis of N-acetylserotonin (C00978) and azelaic acid (C08261) with AUC = 0.987 CI (0.98–1) p < 0.05 along with prediction class probability score plot showing segregation of COVID-19 positive and negative.
Expected outcomes
Untargeted metabolomics analysis of the respiratory specimen could yield close to 20 thousand to 40 thousand ions depending on the machine sensitivity and specificity. Those ions which qualify analytical quality determination are subjected to annotation using Spectral Database and database of pure compound which document RT and intact mass for the same in the compound discoverer software. The compound discoverer software returns annotation for the probable ions which need to be checked individually and processed for statistical analysis for the study group. Statistical analysis of the metabolites within the VTM showed 106 metabolites that were significantly dysregulated in COVID-19 positive patients. Metabolites like N-acetylserotonin (C00978) and azelaic acid (C08261) had the highest mean decrease in accuracy and showed a combined diagnostic efficiency of 0.987 (0.98–1) for SARS-CoV-2 positive segregation from negatives. Pathway analysis revealed significant increase in pathways linked to biosynthesis of unsaturated fatty acids, glycerophospholipid metabolism, ubiquinone/terpenoid-quinone biosynthesis, aminoacyl-tRNA biosynthesis and amino acid metabolism including phenylalanine, tyrosine and tryptophan biosynthesis whereas, pathways linked to thiamine metabolism, one carbon pool by folate, vitamin B6 metabolism, riboflavin metabolism and steroid biosynthesis were decreased (Figure 13). Therefore, SARS-CoV-2 infection tends to change the metabolic phenotype of respiratory specimens.
Limitations
The precise determination of the actual quantitate of the metabolites could not be determined by this method. In turn this method returns quantity in arbitrary units. Further this method cannot distinguish between host and virus or microbe linked metabolites nor could assess the flux of these metabolites.
Troubleshooting
Problem 1
Leaks in the system (step 20).
Potential solution
Inspect all the fittings for leaks. Tighten any loose fittings, without over tightening them as this may cause damage to the fitting’s threads and cause leaks. Replace the fitting and ferrule if they may be damaged.
Problem 2
No peaks/very small peaks (step 20).
Potential solution
First of all, check the lamp is on and cables are well connected. Next make sure if the flow is normal and automatic sampler vials have sufficient liquid and no air bubbles in the sample. If the problem still persists, then evaluate the system performance with fresh standards to confirm if sample is the source of problem.
Problem 3
If the pressure is higher than normal (step 20).
Potential solution
Remove the guard and analytical column. Check the pressure, then isolate the cause by systematically eliminating system components, starting with detector, then in-line filter, and working back to pump. Replace filter in pump if present. Replace guard column if necessary. If the analytical column is obstructed, reverse and flush the column while disconnected from the detector. If the problem persists, the column may be clogged with strongly retained contaminants. Therefore, change the column.
Problem 4
If there is a shift in Retention Times (step 20).
Potential solution
Check the system for loose fittings. Check pump for leaks, salt buildup, unusual noises. Change pump seals if necessary. Also check make-up of mobile phase and be sure mobile phase is degassed. Purge air from pump head or check valves. Higher column temperatures increase column efficiency. For optimum results, heat eluent before introducing it into the column. Inject smaller volume (e.g., 10 μL vs. 100 μL) or inject the same volume after 1:10 or 1:100 dilutions of sample
Problem 5
If the Resolution is less (step 21).
Potential solution
Prepare a fresh mobile phase and check problem 3 for an obstructed guard or analytical column.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Jaswinder Singh Maras (jassi2param@gmail.com).
Materials availability
The study did not generate any materials.
Acknowledgments
We would like to acknowledge the Science and Engineering Research Board Department of Science and Technology government of INDIA, for providing funds for the building of the manuscript. The work was supported by project DST (DST-SERB) (EMR/2016/004829).
Author contributions
J.S.M. conceptualized the work. The manuscript was written by N.S. and J.S.M., with help from S.H.B., M.Y., G.T., B.M., V.B., S.S., E.G., and S.K.S. The manuscript was read and approved by all authors.
Declaration of interests
There is no conflict of interest from any of the authors included in the manuscript.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.101051.
Contributor Information
Jaswinder Singh Maras, Email: jassi2param@gmail.com.
Shiv Kumar Sarin, Email: shivsarin@gmail.com.
Supplemental information
Data and code availability
The processed data is provided as supplemental information, and the raw data for the manuscript is available on request to the lead contact Dr. Jaswinder Singh Maras (jassi2param@gmail.com).
References
- Batejat C., Grassin Q., Manuguerra J.C., Leclercq I. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J. Biosaf. Biosecur. 2021;3:1–3. doi: 10.1016/j.jobb.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat A.H., Chaudhary S., Yadav G., Prasar A., Bihari C., Maras J.S., Sarin S.K. Multi-omics analysis of aspirin treatment response in mice provide molecular insights and targets linked to liver fibrosis regression. bioRxiv. 2020 doi: 10.1101/2020.07.03.186015. [DOI] [Google Scholar]
- Boudah S., Olivier M.F., Aros-Calt S., Oliveira L., Fenaille F., Tabet J.C., Junot C. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2014;966:34–47. doi: 10.1016/j.jchromb.2014.04.025. [DOI] [PubMed] [Google Scholar]
- Castelli F.A., Rosati G., Moguet C., Fuentes C., Marrugo-Ramirez J., Lefebvre T., Volland H., Merkoci A., Simon S., Fenaille F., Junot C. Metabolomics for personalized medicine: the input of analytical chemistry from biomarker discovery to point-of-care tests. Anal. Bioanal. Chem. 2021 doi: 10.1007/s00216-021-03586-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics. 2019;68:e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Control and Prevention C.F.D. 2020. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. [Google Scholar]
- Maras J., Bhat A., Yadav G., Thakur A., Sarin S.K. Temporal change in the plasma metabolome profile is indicative of outcome in severe alcoholic hepatitis. J. Hepatol. 2020;73:S180. [Google Scholar]
- Maras J.S., Das S., Sharma S., Shasthry S.M., Colsch B., Junot C., Moreau R., Sarin S.K. Baseline urine metabolic phenotype in patients with severe alcoholic hepatitis and its association with outcome. Hepatol. Commun. 2018;2:628–643. doi: 10.1002/hep4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maras J.S., Sharma S., bhat A., Rooge S., Aggrawal R., Gupta E., Sarin S.K. Multi-omics analysis of respiratory specimen characterizes baseline molecular determinants associated with SARS-CoV-2 outcome. iScience. 2021;24:102823. doi: 10.1016/j.isci.2021.102823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau R., Claria J., Aguilar F., Fenaille F., Lozano J.J., Junot C., Colsch B., Caraceni P., Trebicka J., Pavesi M., et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J. Hepatol. 2020;72:688–701. doi: 10.1016/j.jhep.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Weiss N., Barbier Saint Hilaire P., Colsch B., Isnard F., Attala S., Schaefer A., Amador M.D., Rudler M., Lamari F., Sedel F., et al. Cerebrospinal fluid metabolomics highlights dysregulation of energy metabolism in overt hepatic encephalopathy. J. Hepatol. 2016;65:1120–1130. doi: 10.1016/j.jhep.2016.07.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed data is provided as supplemental information, and the raw data for the manuscript is available on request to the lead contact Dr. Jaswinder Singh Maras (jassi2param@gmail.com).