Summary
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019. Few studies have compared replication dynamics and host responses to SARS-CoV-2 in cell lines from different tissues and species. Therefore, we investigated the role of tissue type and antiviral genes during SARS-CoV-2 infection in nonhuman primate (kidney) and human (liver, respiratory epithelial, gastric) cell lines. We report different viral growth kinetics and release among the cell lines despite comparable ACE2 expression. Transcriptomics revealed that absence of STAT1 in nonhuman primate cells appeared to enhance inflammatory responses without effecting infectious viral titer. Deletion of RL-6 in respiratory epithelial cells increased viral replication. Impaired infectious virus release was detected in Huh7 but not Huh7.5 cells, suggesting a role for RIG1. Gastric cells MKN45 exhibited robust antiviral gene expression and supported viral replication. Data here provide insight into molecular pathogenesis of and alternative cell lines for studying SARS-CoV-2 infection.
Subject areas: Immunology, Microbiology, Virology
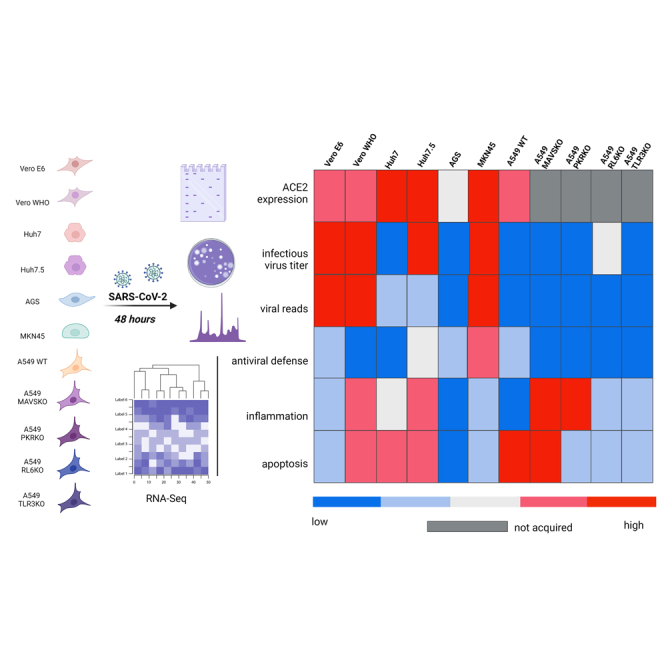
Graphical abstract
Highlights
-
•
Tissue- and species-specific response to SARS-CoV-2 were profiled in vitro
-
•
Cell lines expressing similar levels of ACE2 exhibit distinct antiviral responses
-
•
Alteration of RIG1, STAT1, or RIG1 differentially effect infectious virus production
-
•
Human gastric cells exhibit robust antiviral responses and SARS-CoV-2 replication
Immunology; Microbiology; Virology
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a positive-sense, single-stranded RNA virus belonging to Coronaviridae family and Betacoronavirus genus (Lu et al., 2020; Zhou et al., 2020; Zhu et al., 2020). Similar to other betacoronaviruses, SARS-CoV-2 causes an upper respiratory tract infection. This infection, called coronavirus disease 2019 (COVID-19), ranges in severity from asymptomatic to severe disease including death. A majority (∼80%) of patients exhibit self-limiting disease (Guan et al., 2020; Zheng et al., 2020). COVID-19 symptoms include involvement of the respiratory tract (e.g., cough, rhinitis), the gastrointestinal tract (e.g., diarrhea), and the nervous system (e.g., anosmia as a sign of peripheral nervous system involvement and ischemic stroke central nervous involvement) (Harrison et al., 2020; Hornuss et al., 2020; Lopez-Leon et al., 2021; Merkler et al., 2020). Recent approval/emergency use authorization of several vaccines, remdesivir, and monoclonal antibody cocktails by the Food and Drug administration has contributed to pandemic control (Creech et al., 2021; Kyriakidis et al., 2021). However, our understanding of cellular mechanisms modulating SARS-CoV-2 pathogenesis remains incomplete.
SARS-CoV-2 and other coronaviruses are primarily spread through virus-laden respiratory droplets (Harrison et al., 2020). Infection of respiratory cells with SARS-CoV-2 largely depends on the interaction between the viral spike protein and host ACE2 receptor (Hoffmann et al., 2020, 2021; Murgolo et al., 2021; Shang et al., 2020). The interaction between ACE2 and spike, as well as host proteases (e.g., TMPRSS2), leads to cytosolic replication followed by assembly and release of viral progeny through the lysosomal trafficking pathway (Ghosh et al., 2020). In humans and non-human primates (NHPs), ACE2 is highly expressed on epithelial cells throughout the oropharyngeal and respiratory tracts, in addition to within the liver and along the gastro-intestinal tract (Hou et al., 2020; Zhang et al., 2020; Ziegler et al., 2020). The ubiquitous expression of ACE2 throughout the human body has raised questions concerning the susceptibility and permissivity of various cells and tissues to SARS-CoV-2 infection, which may explain extra-pulmonary symptoms.
Various proteins encoded by SARS-CoV-2, such as nsp6, nsp13, orf3b, and orf6, antagonize the host antiviral response to permit robust viral replication following cell entry (Xia et al., 2020; Shemesh et al., 2021; Yuen et al., 2020; Lei et al., 2020). The initial detection of viral double-stranded RNA in the cytosol activates pattern recognition receptors (PRRs), leading to the production of type I interferons (IFNs) via IRF3 activation (Jensen and Thomsen, 2012). Type I IFN then induces the synthesis of interferon stimulated genes (ISGs) in infected and neighboring cells through activation of STAT1 and STAT2 transcription factors. Both SARS-CoV-2 nonstructural proteins (NSPs) and open reading frames (ORFs) have been shown to prevent the activation and downstream signaling of the type I IFN signaling pathway, including nsp6 and nsp13, which inhibit IRF3 phosphorylation; orf3b, which is proposed to inhibit IRF3 nuclear translocation; and orf6, which blocks nuclear import of STAT (Lee et al., 2021; Lei et al., 2020; Miorin et al., 2020; Xia et al., 2020; Yuen et al., 2020; Shemesh et al., 2021).
In vitro systems have been used to evaluate the function of antiviral drugs and elucidate mechanisms of viral pathogenesis. Almost all systems have demonstrated that defects in IFN signaling machinery are essential for efficient replication of SARS-CoV-2 (Hoffmann et al., 2020; Chu et al., 2020; Ogando et al., 2020). For instance, Vero E6 and Vero CCL81 (WHO) cells, both derived from African Green monkey kidney cells, are the widely used cell lines to grow SARS-CoV-2 stocks (Li et al., 2021; Hoffmann et al., 2020; Ogando et al., 2020; Pohl et al., 2021). Vero WHO cells were derived from Vero E6 cells by the World Health Organization for human vaccine production. Both cell lines express ACE2, lack type I interferon, and encode an inefficient IRF3, while Vero WHO cell line also lacks the expression of STAT1 (Chew et al., 2009; Desmyter et al., 1968; Emeny and Morgan, 1979; Osada et al., 2014; Ren et al., 2006; Young et al., 2003). Human liver cells Huh7 and Huh7.5 are also used to grow virus (Chen et al., 2021; Hoffmann et al., 2020; Ramirez et al., 2021; Kawamoto et al., 2020; Sumpter et al., 2005). Huh7 cells harbor mutations in IFNA10 and TRIM56, which are key components of type I interferon signaling, while Huh7.5 cells additionally possess a missense mutation in RIG-I that interferes with double-stranded RNA recognition (Chen et al., 2021; Hoffmann et al., 2020; Ramirez et al., 2021; Kawamoto et al., 2020; Sumpter et al., 2005). Although various studies have investigated the role of specific IFN signaling components in SARS-CoV-2 infection, a limited number of studies have determined tissue-specific responses (Murgolo et al., 2021; Gagliardi et al., 2021; Lamers et al., 2020; Liu et al., 2021; Zang et al., 2020). The ultimate impact of these differences on viral replication and molecular responses remains unclear but is critical to further elucidate mechanisms of SARS-CoV-2 pathogenesis.
In this study, we determined responses to and replication kinetics of SARS-CoV-2 in various canonical and noncanonical cell lines. Multi-step viral growth assays demonstrated increased replication levels in human and NHP cell lines possessing defective type I IFN signaling machinery. In contrast, human respiratory epithelial cells lines (A549) and liver (Huh7) and gastric (AGS) cells poorly supported infectious virus production. Interesting, ACE2 expression was not associated with cell permissivity and the magnitude of viral replication. Our data suggest that RL6 may be a restriction factor that interferes with the production of infectious virus in human epithelial cell lines. Furthermore, baseline levels of STAT1 modulated inflammatory responses following SARS-CoV-2 infection in Vero cells. We report for the first time that the human gastric adenocarcinoma cell line MKN45 supports SARS-CoV-2 replication while generating a robust antiviral response akin to what has been recently described for alveolar epithelial cell line A459 transfected with ACE2, suggesting that this cell line can function as an alternative to NHP cell lines lacking antiviral machinery for virus stock production (Li et al., 2021). Collectively, data presented here provide novel insight into molecular mechanisms of SARS-CoV-2 pathogenesis, as well as insight into alternative cell lines for SARS-CoV-2 propagation and in vitro studies.
Results
SARS-CoV-2 exhibits species- and tissue-specific viral replication kinetics
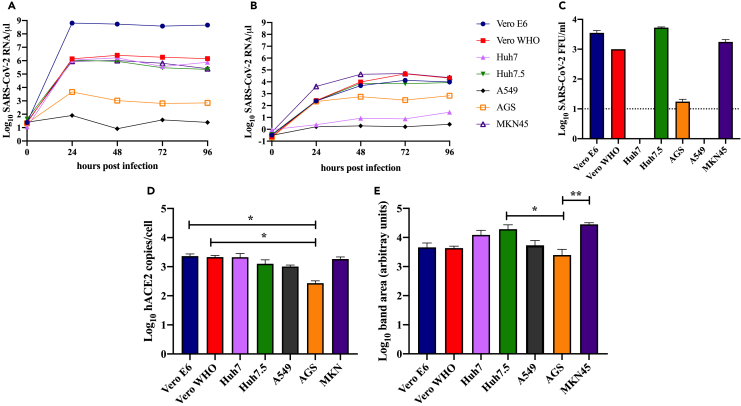
To determine differences in SARS-CoV-2 replication kinetics between and among human (A549, AGS, Huh7, Huh7.5, MKN45) and NHPs (Vero WHO and Vero E6), we performed multi-step growth assays with these cell lines in vitro (Figures 1A–1D, Table 1). Cells were infected with SARS-CoV-2 (isolate USA-WA1/2020) and monitored every 24 h over the course of 4 days. Analysis of cellular viral genome copies revealed peak levels at approximately 24 h post infection (HPI) in all cell lines (Figure 1A). Vero E6 achieved the highest cellular genome copy levels at 6.44 × 108 copies/μL, whereas levels from A549 cells remained near baseline (80 copies/μL) throughout the experiment. Vero WHO, Huh7, and Huh7.5 cells achieved similar peak levels at approximately 1.1–5 x 106 copies/μL at 24–48 HPI. Consistent with delayed release of viral progeny following viral entry, peak viral genome copies were detected at 48–72 HPI in most supernatants with peak titers ranging from 1.3 to 4.7 x 104 copies/μL, mirroring levels of cellular viral RNA (Figure 1B). The magnitude and kinetics of SARS-CoV-2 replication in MKN45 cells was comparable with that observed in Vero cells, whereas those in AGS cells were slightly lower (Figures 1A and 1B).
Figure 1.
Viral replication and growth kinetics, and ACE2 expression in immortalized nonhuman primate and human cell lines
SARS-CoV-2 genome copy number per microliter in cells (A) or supernatants (B) as determined by qRT-PCR for human (Huh7/7.5, A549, AGS, MKN45) and nonhuman primate cell lines (Vero E6/WHO) infected at an MOI of 0.5 with SARS-CoV-2 and sampled at 24-h intervals. Total cellular RNA was extracted and normalized to an RNaseP and GADPH control as well as an in-house virus copy control. (C) Infectious virus in supernatant was quantified at 48 h by focus-forming assay (LOD = 10 FFU/ml). Quantification of ACE2 (D) mRNA (p = 0.048 AS versus Vero E6; p = 0.048 AGS versus Vero WHO) and (E) protein expression (p = 0.048 AGS versus Huh7.5; p = 0.004 AGS versus MKN45) by qRT-PCR and western blot, respectively, in infected cell lines.
Table 1.
Cell line characteristics and SARS-CoV-2 viral reads, infectious virions, and amino acid mutations
| Cell line | Organism | Tissue | Genes impacted | % SARS-CoV-2 viral reads (RNA-Seq) replicate 1; replicate 2 | cellular SARS-CoV-2 RNA/μL |
log10 supernatant SARS-CoV-2 RNA/μL |
Average FFU/mL | Viral amino acid changes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 HPI | 24 HPI | 48 HPI | 72 HPI | 96 HPI | 0 HPI | 24 HPI | 48 HPI | 72 HPI | 96 HPI | |||||||
| A549 | Human | Alveolar basal epithelial adenocarcinoma | Wild type | 0.022; 0.008 | 25.64 | 80.59 | 8.34 | 37.64 | 24.49 | 0.31 | 1.62 | 1.95 | 1.64 | 2.62 | 0 | |
| MAVS | 0.011; 0.013 | 1.00 | 32.62 | 13.91 | 26.52 | 12.07 | 0.05 | 0.16 | 2.77 | 2.09 | 8.06 | 17.76 | 3′ UTR T152C | |||
| PKR | 0.008; 0.012 | 1.00 | 30.84 | 16.89 | 19.41 | 17.83 | 0.01 | 0.17 | 2.19 | 11.32 | 13.32 | 27.15 | ||||
| RL6 | 0.014; 0.023 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 249.19 | ||||
| TLR3 | 0.005; 0.007 | 1.00 | 26.34 | 13.86 | 12.25 | 15.60 | 0.07 | 0.22 | 7.78 | 10.05 | 6.50 | 23.78 | ||||
| Huh7 | Human | Epithelial-like hepatoma | IFNA10, TRIM56 | 2.560; 3.041 | 11.28 | 1,104,317 | 1,633,247 | 361,097 | 770,922 | 0.93 | 2.46 | 8.46 | 7.52 | 27.30 | 0 | |
| Huh7.5 | Human | Epithelial-like hepatoma | IFNA10, TRIM56, RIG1 | 7.352; 57.657 | 43.50 | 1,089,127 | 898,185 | 293,625 | 226,267 | 0.37 | 254 | 7,368 | 6,890 | 8,625 | 5,250 | |
| Vero E6 | African green monkey | Kidney epithelial | IRF3, type I IFN | 47.622; 50.618 | 24.12 | 644,000,000 | 541,000,000 | 383,000,000 | 452,000,000 | 0.32 | 245 | 4,777 | 13,532 | 9,750 | 3,500 | |
| Vero WHO | African green monkey | Kidney epithelial | IRF3, STAT1, type I IFN | 30.429; 54.789 | 23.48 | 1,368,663 | 2,533,407 | 1,810,978 | 1,431,811 | 0.24 | 265 | 9,696 | 47,295 | 20,685 | 1,000 | |
| AGS | Human | Gastric (stomach) adenocarcinoma | Unknown | 0.007; 0.008 | 21.02 | 4,582 | 1,049 | 639 | 705 | 0.21 | 222 | 554 | 288 | 671 | 17.5 | |
| MKN45 | Human | Gastric adenocarcinoma | STAT1 | 43.456; 40.915 | 27.00 | 902,338 | 1,025,334 | 643,455 | 254,758 | 0.58 | 4,069 | 43,231 | 49,837 | 22,492 | 1,750 | |
ND – not determined
We additionally measured infectious viral particles in the supernatant at 48 HPI (Figure 1C). Consistent with our measurements of viral genome copy number, we detected large quantities of infectious virus in Vero WHO, Vero E6, Huh 7.5, and MKN45 cell supernatant (1–5 x103 FFU/mL). No infectious virus was detected in supernatants from infected A549 and Huh7 cells, whereas low levels were detected in AGS cells. Furthermore, sequencing of viral genomes isolated in the supernatant revealed no mutations in cell lines that produced infectious virus (Table 1).
To uncover potential reasons for differences in viral production between these various cell lines, we compared RNA and protein expression of ACE2 post infection (Figures 1D and 1E, S1A). Levels of ACE2 mRNA were comparable in all cell lines, except for AGS, which expressed the lowest levels of ACE2 (Figure 1D). At the protein level, MKN45 and Huh7.5 cells expressed the highest level of ACE2, whereas AGS cells expressed the lowest (Figure 1E).
SARS-CoV-2-infected Vero E6 and Vero WHO cells maintain both distinct and shared expression signatures associated with cellular stress and death
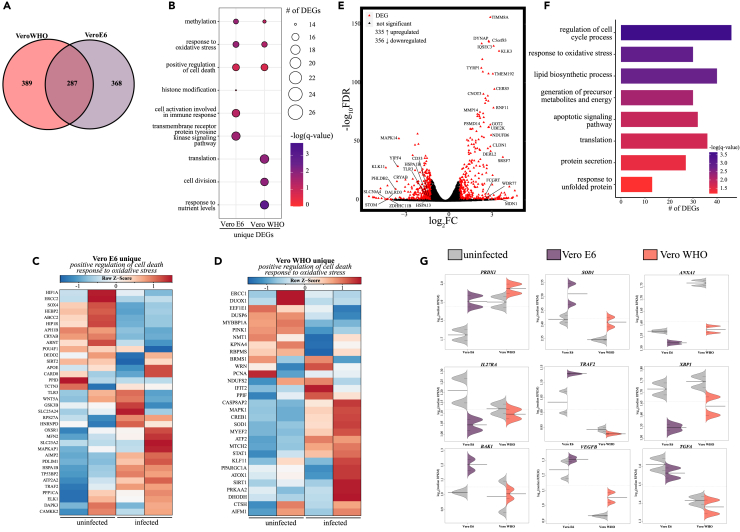
Given that Vero cells are commonly used to grow and titer SARS-CoV-2 viral stocks as well as a screening platform for potential therapeutics and antivirals, we profiled transcriptional responses 48 h after SARS-CoV-2 infection (MOI = 0.5, Figure 2) (Hoffmann et al., 2020; Chu et al., 2020; Ogando et al., 2020; Li et al., 2021; Pohl et al., 2021). Although Vero cells express low levels of the furin enzyme, SARS-CoV-2 furin site is also cleaved by TMPRSS2, TMPRSS4, TMPRSS13, and cathepsins, all of which are expressed by Vero cells (Bollavaram et al., 2021; Hoffmann et al., 2020; Kishimoto et al., 2021; Zang et al., 2020; Shapiro et al., 1997). Over 40% of the aligned transcripts originated from SARS-CoV-2 for both Vero E6 and Vero WHO cell lines, with reads originating from all open reading frames (Figure S1B, Table 1). Principle component analysis (PCA) of the transcriptional profiles revealed a clear separation between Vero E6 and Vero WHO cells accounting for 72% of the transcriptional differences, whereas viral infection accounted for approximately 23% of the transcriptional differences (Figure S2A). We detected a similar number (∼650) of differentially expressed genes (DEGs) following SARS-CoV-2 infection in each of the cell lines with approximately 40% overlap (Figure 2A). DEGs unique to Vero E6 enriched to Gene Ontology (GO) terms associated with immune activation (e.g., “cell activation involved in immune response” CD46, DOCK11) and regulation of gene expression (e.g., “methylation”; METTL8, METTL16) (Figure 2B). In contrast, DEGs unique to Vero WHO played a role primarily in cell division (e.g., “cell division”; CDC23, CCNY) and metabolism (e.g., “response to nutrient levels”; SIRT1, TYMS) (Figure 2B). Moreover, DEGs unique to either Vero E6 or Vero WHO enriched to GO terms associated with cellular stress (e.g., response to oxidative stress” and “positive regulation of cell death”) (Figure 2B). DEGs mapping to these two GO terms from Vero E6 were mainly downregulated (e.g., HIF1A, HIP1R, POU4F1, CARD8) (Figure 2C), whereas those detected in Vero WHO were upregulated (e.g., CASP8AP2, KLF11, SIRT1, SOD1, ATOX1) (Figure 2D).
Figure 2.
Transcriptional analysis of SARS-CoV-2-infected VeroE6 and Vero WHO cell lines
(A) Venn diagram of differentially expressed genes (DEGs) for SARS-CoV-2-infected Vero E6 and Vero WHO cell lines relative to uninfected samples.
(B) Bubbleplot representing functional enrichment of DEGs unique to Vero E6 and Vero WHO. Color intensity of each bubble represents the –log(q-value), and the relative size of each bubble represents the number of DEGs belonging to the specified Gene Ontology (GO) term.
(C and D) Heatmaps representing DEGs enriching to GO terms in (B) “positive regulation of cell death” and “response to oxidative stress” for (C) Vero E6 and (D) Vero WHO. Columns of all heatmaps represent the reads per kilobase per million mapped reads (rpkm) of an individual uninfected or infected sample. Range of colors for each heatmap is based on scaled and centered rpkm values of the represented DEGs.
(E) Volcano plot of global gene expression changes shared between Vero E6 and Vero WHO. DEGs (average rpkm ≥5) are denoted in red. Exemplar DEGs are labeled.
(F) Functional enrichment of DEGs shared by both Vero E6 and Vero WHO. Horizontal bars represent the number of genes enriching to each GO term with color intensity representing the negative log of the FDR-adjusted p value (-log(q-value)). Range of colors based on the GO terms with the lowest and highest –log(q-value) values.
(G) Beanplots representing the expression (rpkm) of the indicated gene in uninfected and infected Vero E6 or Vero WHO cells. Median and standard deviation plotted.
We next identified DEGs that were common to both infected cell lines following correction for cell-specific transcriptional differences (Figures 2E–2G). The 691 DEGs identified in this manner enriched to GO terms reflecting cellular stress (e.g., “response to oxidative stress,” “apoptotic signaling pathway”; BCL2L1, MMP14), metabolism (e.g., “generation of precursor metabolites and energy”; GOT2, CERS5), and protein unfolding (e.g., “response to unfolded protein”; HSPA1B, HSPA13), a process seen in viral infection (Figures 2E–2G). Inhibitors of oxidative stress (PRDX1, SOD1) were upregulated while mediators of inflammation (e.g., ANXA1, TRAF2, IL27RA) were downregulated in both cell lines but to different extents (Figure 2G). XBP1, which plays a role in the unfolded protein response, was downregulated to a greater extent in Vero E6 compared with Vero WHO (Figures 2E–2G). In addition, BAK1 and VEGFB were upregulated to a greater extent in Vero E6 cells while TGFA was downregulated to a comparable extent in both cell lines (Figures 2E–2G).
Infection of Huh7.5 cells versus Huh7 cells leads to more diverse transcriptional response
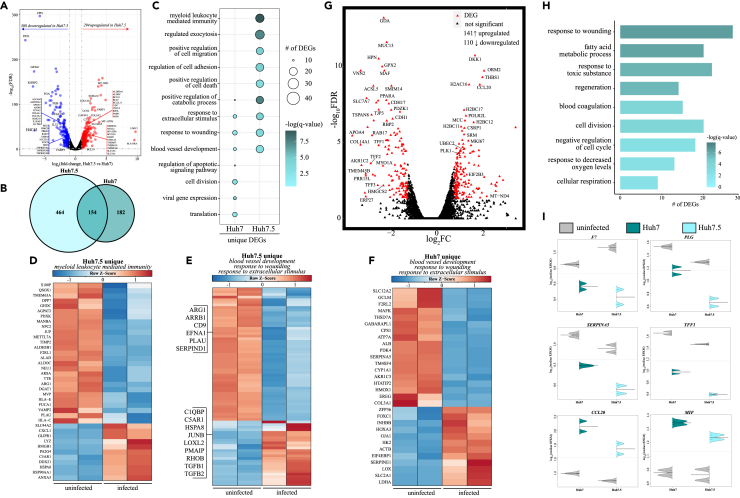
We further investigated the importance of type I IFN signaling in the restriction SARS-CoV-2 growth by profiling transcriptional responses in human liver carcinoma cell lines, Huh7 and Huh7.5, at 0 and 48 HPI (Figure 3). Previous studies with hepatitis C, dengue, and yellow fever viruses have shown that enhanced viral permissivity in Huh7.5 cells compared with Huh7 cells is due to defective RIG-1-mediated signaling (both cell lines lack IFNA10 and TRIM56) (Beauclair et al., 2020; Chatel-Chaix et al., 2016; Chen et al., 2021; Sumpter et al., 2005). In line with these previous studies, Huh7.5 cells supported greater viral growth and release compared with Huh7 cells (Figures 1A–1C). Moreover, in one of the replicates, over half of the aligned transcripts in Huh7.5 cells originated from SARS-CoV-2, whereas less than 3% was observed in both duplicate Huh7 cell samples (Figure S1B, Table 1). Of interest, levels of cellular SARS-CoV-2 mRNA were comparable between Huh7 and Huh7.5 cell lines (Figure 1A), suggesting additional defects outside of viral nucleic acid sensing may be important. Likewise, cell line type accounted for 62% of the transcriptional variance while infection accounted for approximately 24% of the variance (Figure S2B). Direct comparison of uninfected Huh7.5 cells with uninfected Huh7 cells revealed increased expression of genes important for cell survival (e.g., BCL9, BCL2L11), fatty acid metabolism (e.g., ALDH9A1, FASN), and lipid transport (e.g., TMEM97) in Huh7.5 cells (Figure 3A). On the other hand, expression of several cytokine receptors (e.g., IL1RAP, IL17RB) and genes involved in innate immune signaling and defense (e.g., ISG15, PRKCD, STAT6), as well as genes promoting apoptosis (e.g., TNFRSF10A), was higher in the Huh7 cells (Figure 3A).
Figure 3.
Transcriptional analysis of SARS-CoV-2-infected Huh7 and Huh7.5 cell lines
(A) Volcano plot of global gene expression changes in uninfected Huh7.5 relative to uninfected Huh7. DEGs (average rpkm ≥5) are denoted in red. Exemplar DEGs are labeled.
(B) Venn diagram of differentially expressed genes (DEGs) for SARS-CoV-2-infected Huh7 and Huh7.5 cell lines relative to uninfected samples.
(C) Bubbleplot representing functional enrichment of DEGs unique to Huh7 and Huh7.5. Color intensity of each bubble represents the –log(q-value) and the relative size of each bubble represents the number of DEGs belonging to the specified Gene Ontology (GO) term.
(D–F) Heatmap in (D) represents GO term “myeloid leukocyte mediated immunity” unique to Huh7.5. Heatmaps representing DEGs from GO terms in (C) blood vessel development,” “response to wounding,” and “response to extracellular stimulus” for (E) Huh7 and (F) Huh7.5. Columns of all heatmaps represent the reads per kilobase per million mapped reads (rpkm) of an individual uninfected or infected sample. Range of colors per heatmap is based on scaled and centered rpkm values of the represented DEGs.
(G) Volcano plot of global gene expression changes shared between infected Huh7 and Huh7.5. DEGs (average rpkm ≥5) are denoted in red. Exemplar DEGs are labeled.
(H) Functional enrichment of DEGs shared by both Huh7 and Huh7.5. Horizontal bars represent the number of genes enriching to each GO term with color intensity representing the negative log of the FDR-adjusted p value (-log(q-value)). Range of colors based on the GO terms with the lowest and highest –log(q-value) values.
(I) Beanplots representing the rpkm of the indicated gene in uninfected and infected Huh7 or Huh7.5 cells. Median values for each condition are indicated.
Next, we compared DEGs detected during infection in these two cell lines (Figures 3B–3F). We identified a smaller number of DEGs in Huh7 (336) than in Huh7.5 (618) cells, with 154 DEGs shared between the two cell lines (Figure 3B). DEGs unique to either Huh7 or Huh7.5 both enriched to GO terms “response to wounding,” “blood vessel development,” and “response to extracellular stimulus” (Figure 3C). DEGs detected in Huh7 cells that enriched to these GO terms included genes involved in angiogenesis, coagulation, and energy production (e.g., upregulated COL3A, EREG, PDK4; downregulated HK2, LOX, SERPINE1) (Figure 3D). Similarly, DEGs unique to Huh7.5 also played a role in angiogenesis and coagulation (e.g., downregulated, PLAU, EFNA1; upregulated, LOXL2, TGFB2), as well as cell growth and stress in response to external stimuli (e.g., downregulated, ARRB1, HSPA8; upregulated, RHOB, TGFB1) (Figure 3D). Only DEGs expressed by Huh7.5 enriched to GO term “myeloid leukocyte mediated immunity” (Figure 3D). This included many downregulated DEGs important for lipid metabolism (e.g., AGAPT2, NPC2). Several DEGs also played roles in regulating membrane activity, such as phagocytosis and vesicular trafficking (e.g., JUP, MVP, RAB11FIP1, VAMP2). Other DEGs upregulated only in Huh7.5 cells regulate cell differentiation (e.g., GLIPR1) and complement-mediated immunity (e.g., C5AR1) (Figure 3E). Most DEGs unique to Huh7 that enriched to GO term “translation” were upregulated and encoded ribosomal proteins and translation initiation/elongation factors (e.g., EIF4EBP1, RPL10/11/12/36) (Figure 3F).
As described above for Vero cell lines, we also identified DEGs in Huh7 and Huh7.5 after infection when corrected for cell-specific differences (Figures 3G–3I). Overall, ∼250 DEGs were uncovered that enriched to GO terms associated with metabolism (e.g., “fatty acid metabolic process,” “cellular respiration”; GDA, PPARA), coagulation (e.g., “response to wounding” and “blood coagulation”; F7, PLG, SERPINA5), and cell proliferation (e.g., “cell division” and “cell cycle”; MIK167, PLK) (Figures 3G–3I). DEGs were expressed at different magnitudes in Huh7 and Huh7.5. For instance, F7, PLG and SERPINA5, and TFF1, which are involved in coagulation and barrier function, were downregulated in both cell lines, albeit to a greater extent in Huh7.5 (Figure 3I). In contrast, genes involved in inflammation (e.g., MIF) and immune cell recruitment (e.g., CCL20) were more upregulated in Huh7 (Figure 3I).
A549 cells lacking antiviral defense genes exhibit distinct transcriptomes following SARS-CoV-2 exposure
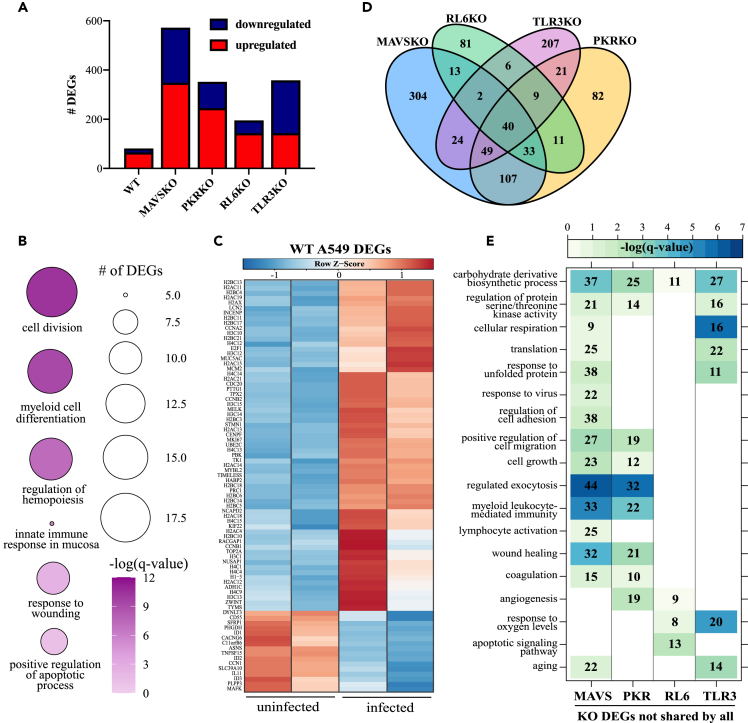
We noted that cell lines with intact antiviral sensing and type I interferon (IFN) signaling pathways tended to be less permissive to viral replication than those with defective pathways regardless of ACE2 expression (Figure 1). For instance, A549 cells poorly support SARS-CoV-2 replication unless ACE2 is over-expressed (Li et al., 2021). To further interrogate the role of viral RNA sensors, we profiled the transcriptional response of A549 cells deficient in MAVS, PKR, RL6, orTLR3 genes following SARS-CoV-2 infection (Figure S3) (Li et al., 2016, 2021). TLR3, RL6, and PKR bind dsRNA, whereas MAVS is downstream of RIG-I-like receptors (Lester and Li, 2014; Wu and Hur, 2015). Activation of MAVS leads to the transcription of type I interferon and interferon-stimulated genes (ISGs), such as RL6, which degrades viral RNA (Silverman, 2007). PKR is also activated by dsRNA and halts protein synthesis (Dauber and Wolff, 2009). Multi-step growth curves showed that little to no viral RNA was detected in WT and knock-out (KO) A549 cells and corresponding supernatants except for RL6KO (Figure S3C).
We then performed transcriptome analysis 48 h post infection with SARS-CoV-2 as described above (Figure 4). In line with the viral growth data, few viral reads were detected in WT and KO cells with one mutation detected in the noncoding 3’ UTR in A549 MAVSKO (Figure S1B, Table 1). PCA shows that WT A459 cells were transcriptionally distinct from the various knockout cell lines (Figure S2C). Separation between uninfected and infected cells was minimal for WT and RL6-KO cells and more evident for MAVSKO, PKRKO, and TLR3KO (Figure S2C). The ∼80 DEGs expressed in WT infection were mainly upregulated and enriched to GO terms associated with innate immunity (e.g., “myeloid cell differentiation,” “innate response in mucosa”; CD55, LCN2, MUC5AC), apoptosis (e.g., “positive regulation of apoptotic process”; MELK, TNFSF15), and cell division (e.g., “cell division”; CDC20, CCNB1, TIMELESS, TPX2) (Figures 4A and 4B). Numerous DEGs encoding histone proteins (e.g., H2AC11, H4C4) were also upregulated (Figure 4C). The few downregulated DEGs played a role in inflammation (e.g., TNFSF15, IL11) (Figure 4C).
Figure 4.
Transcriptional analysis of SARS-CoV-2-infected A549 wild-type and knockout cell lines
(A) Bar graph of DEGs detected in SARS-CoV-2-infected wild-type (WT) and knockout (KO) A549 cell lines relative to uninfected counterparts.
(B) Bubbleplot representing functional enrichment of WT infection DEGs. Color intensity of each bubble represents the –log(q-value), and the relative size of each bubble represents the number of DEGs belonging to the specified Gene Ontology (GO) term.
(C) Heatmap representing all WT DEGs. Columns of heatmap represent the reads per kilobase per million mapped reads (rpkm) of an individual uninfected or infected sample. Range of colors per heatmap is based on scaled and centered rpkm values of the represented DEGs.
(D) Venn diagram of DEGs in (A) for KO only.
(E) GO network depicting functional enrichment of the 40 DEGs expressed by all KO using Metascape. Clustered nodes of identical color correspond to one GO term. Node size represents the number of DEGs associated with the GO term. Gray lines represent shared interactions between GO terms, with density and number indicating the strengths of connections between closely related GO terms.
In contrast to WT A459 infection, ∼200 DEGs were detected in RL6KO infection and over 300 DEGs were detected in MAVSKO, PKRKO, and TLR3KO infections (Figure 4A). The 40 DEGs shared by all A549KO cell lines related primarily to DNA metabolism (e.g., H3C7, METTL7B) and innate immunity (e.g., FYB, TFPI2, TNFRSF10D) (Figure 4D). We next performed functional enrichment of DEGs detected in each KO excluding the 40 shared among all KOs (Figures 4D and 4E). DEGs unique to MAVSKO and PKRKO related to similar GO terms including “regulation of protein serine/threonine kinase activity,” “myeloid leukocyte mediated immunity,” “wound healing,” and “coagulation.” Interesting, DEGs found only in MAVSKO following infection enriched to “lymphocyte activation” (e.g., BCL3, ARG2) and “response to virus” (Figure 4E). The majority of antiviral DEGs encoded nucleic acid sensors (e.g., RSAD2, DDX60) and ISGs (e.g., HERC5, IFIT2, ISG15, STAT1) and were downregulated with SARS-CoV-2 infection. DEGs unique to RL6KO primarily enriched to GO terms indicative of hypoxia (e.g., “angiogenesis,” downregulated VEGFC) and cell death (e.g., “apoptotic signaling pathway,” upregulated BLCAF1) (Figure 4E), whereas DEGs unique to TLR3KO enriched to metabolic GO terms (e.g., “cellular respiration hypoxia,” “response to oxygen levels,” downregulated CCN2) and cellular senescence (e.g., “aging,” downregulated MARCHF5).
To further understand the molecular effects of deficient antiviral sensing and defense in SARS-CoV-2 infection, we compared the transcriptional profiles of all infected KO cells with infected WT A549 cell line after adjusting for cell-specific differences (Figure S4). These analyses uncovered 783 DEGs in KO relative to WT, most of which were upregulated (Figure S4A). These DEGs enriched to GO terms associated with cell morphogenesis and adhesion (e.g., “regulation of cell adhesion,” “extracellular structure organization”), tissue repair (e.g., “response to wounding”), and metabolism (e.g., “cellular amino acid metabolic process”) (Figure S4B). Expression analysis of key DEGs revealed different magnitudes of gene expression across the KO cell lines (Figure S4A and S4C). For instance, genes important for angiogenesis (TNFRSF12A and VEGFA) as well as master transcription factor NFKB2 were expressed at higher levels in RL6KO (Figure S4C). Inflammatory genes ANXA1, STAT3, and IFNGR1 were downregulated to a greater extent in TLR3KO. Expression of PAK1 was upregulated the least in infected TLR3KO. HSPA2 and TGFBR2, which play a role in the stress response, were downregulated most in RL6KO and TLR3KO cells, whereas expression of XBP1 was increased in all cells except RL6KO (Figure S4C).
MKN and AGS infections are characterized by transcriptional indicators of antiviral defense
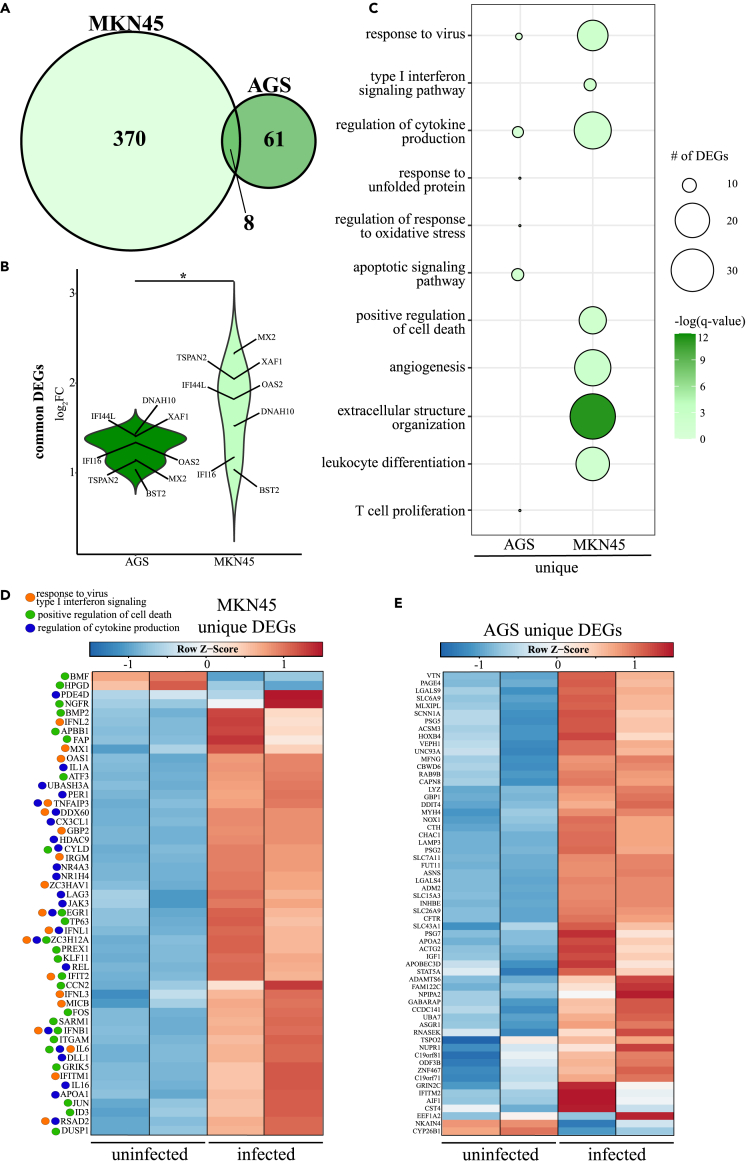
Owing to the high expression of ACE2 in the gastrointestinal tract and COVID-19-related gastrointestinal symptoms, we next characterized the transcriptional response of AGS and MKN45 cell lines following SARS-CoV-2 infection (Figure 5). Both cell lines are derived from human gastric adenocarcinomas, but AGS is deficient in STAT1 synthesis owing to a persistent parainfluenza virus 5 infection (Barranco et al., 1983; Wignall-Fleming et al., 2016; Young et al., 2003). As noted in WT A549 cell lines, but unlike Vero and Huh cell lines, these cells possess intact interferon signaling pathways. Surprisingly, SARS-CoV-2 viral reads made up 42.19% of the aligned transcripts detected in MKN45 cells but not AGS cells (Figure S1, Table 1). PCA revealed that ∼90% of transcriptional differences can be explained by infection (Figure S2D, E). We detected 378 DEGs in MKN45 cells, whereas 69 DEGs were detected in AGS cells in response to SARS-CoV-2 infection (Figure 5A). The eight DEGs upregulated in both MKN45 and AGS cells were ISGs (e.g., BST2, IFI16, IFI44L, MX2, OAS1) and inflammatory genes (e.g., DNAH10, TSPAN2, XAF1) (Figure 5B). With the exception of IFI16 and BST2, shared DEGs were upregulated to a greater degree in MKN45.
Figure 5.
Transcriptional analysis of SARS-CoV-2-infected AGS and MKN45 cell lines
(A) Venn diagram of differentially expressed genes (DEGs) for SARS-CoV-2-infected AGS and MKN45 cell lines relative to uninfected samples.
(B) Violin plot representing the log2(fold-change) of the eight DEGs shared by both cell lines.
(C) Bubbleplot representing functional enrichment of DEGs unique to AGS and MKN45. Color intensity of each bubble represents the –log(q-value), and the relative size of each bubble represents the number of DEGs belonging to the specified Gene Ontology (GO) term.
(D and E) Heatmaps of (D) DEGs unique to MKN45 and enriching to GO terms “response to virus” (orange), “positive regulation of cell death” (green), and “regulation of cytokine production” (blue), and (E) all DEGs unique to AGS. Columns of heatmaps represent the reads per kilobase per million mapped reads (rpkm) of an individual uninfected or infected sample. Range of colors per heatmap is based on scaled and centered rpkm values of the represented DEGs.
The 370 DEGs unique to MKN45 enriched to GO terms associated with antiviral defense (e.g., “response to virus,” “type I interferon signaling pathway”), leukocyte immunity (e.g., “leukocyte differentiation”), cytokine production (e.g., “regulation of cytokine production”), and apoptosis (e.g., “positive regulation of cell death”) (Figure 5C). Expression analysis revealed that DEGs enriching to GO terms related to antiviral processes, cell death, and cytokine production were primarily upregulated (Figure 5D). This included genes encoding type I and III interferon (e.g., IFNB, IFNL1/2/3), key components of T cell-mediated immunity (e.g., CX3CL1, JAK3, LAG3), and pro-apoptotic mediators (e.g., APBB1, KLF11) (Figure 5D). Similar to MKN45, DEGs unique to AGS also enriched to GO terms with roles in antiviral immunity (e.g., “response to virus”; APOBEC3D, IFITM2), cell stress and protein unfolding (e.g., “response to unfolded protein”; ASNS, NOX1), and leukocyte-mediated immunity (e.g., “T cell proliferation”; LGALS9) (Figures 4C and 4E). Most unique DEGs detected in AGS were upregulated (Figure 5E).
Discussion
Cell susceptibility and permissivity to SARS-CoV-2 infection is regulated by species, tissue type, receptor expression, and mobilization of host antiviral defense. In particular, the expression of ACE2 and genes involved in antiviral type I IFN signaling dictate the ability and kinetics of SARS-CoV-2 replication in vitro and in vivo. There is an urgent need for more data to inform the development of novel antiviral therapies against SARS-CoV-2 and other oncornaviruses using in vitro models. In this study, we examined the replication kinetics of SARS-CoV-2 in nonhuman and human cell lines lacking or possessing intact type I IFN signaling machinery. We further characterized transcriptional responses to understand molecular basis for differences in cell permissiveness and replicative capacity of SARS-CoV-2.
We noted peaks in intracellular viral replication at 24 HPI, whereas peaks in cell-free virus occurred later (48–96 HPI) for most cell lines, suggesting a delay in viral egress. Despite similar levels of ACE2 protein expression and peak levels of intracellular viral RNA, levels of infectious virus particles were lower in the supernatant of Huh7 cells compared with Vero WHO and Huh7.5 cells. Transcriptional analysis revealed that these two cell lines are divergent in several pathways that could explain the differences in infectious virus production. Chief among these differences are genes that play a critical role in innate immune responses to microbes (e.g., decreased expression of ISG-15, IL-32, and STAT-6) as well as lipid transport/breakdown (APOM, APO5A, ALDH3A2, and ALCAM). In addition, Huh7.5 encodes a mutation in RIG-I that results in a non-functional protein (Beauclair et al., 2020; Chatel-Chaix et al., 2016; Chen et al., 2021; Sumpter et al., 2005; Yamada et al., 2021). Several additional genes were differentially expressed between uninfected Huh7.5 and Huh7 that could contribute to increased virus release in Huh7.5 cells. For instance, expression of CAV1 was increased in Huh7.5. Caveolin 1 plays a critical role for virus trafficking, assembly, and egress (Xing et al., 2020). Similarly, levels of CXCL5 were increased in Huh 7.5, and levels of this chemokine correlated with increased SARS-CoV-2 replication in a hACE-2 transgenic mouse model (Liang et al., 2020). Increased expression of anti-apoptotic genes (e.g., BCL10) could also facilitate viral production (Kvansakul, 2017; Liang et al., 2015). The role of these various changes in modulating virus production still remains to be determined empirically using knockin and knockdown experiments. Finally, as described in these previous studies, we did not detect significant induction of ISGs or antiviral defense genes, as expected given the absence of functional IFNA10, TRIM56 (both Huh7 and Huh7.5), and RIG-I (Huh7.5 only).
Several genes encoding translation initiation/elongation factors and ribosomal subunits (e.g., EIF4EBP, RPL10/11/12) were uniquely upregulated in Huh7 cells, suggesting that, despite adequate protein synthesis machinery, Huh7 cells are incapable of supporting infectious virion production. This was accompanied by the upregulation of inflammatory and chemotactic genes, suggesting a more robust antiviral response to SARS-CoV-2 in Huh7 cells than in Huh7.5 cells. Genes related to membrane morphology/dynamics and cell adhesion (e.g., NPC2, RAB11FIP1) were unique to Huh7.5 and downregulated, suggesting viral egress potential may be altered in Huh 7.5. For instance, NPC2 has been shown to interact with orf8 and its suppression maybe required for virion morphogenesis, in line with greater infectious virus titers in Huh7.5 (Hoffmann et al., 2021).
Vero E6 and Vero WHO cells, which are used to propagate virus stocks and carry out large pan-coronavirus compound screening, displayed similar growth kinetics, but the latter achieved higher peak viral loads (Li et al., 2021; Hoffmann et al., 2020; Ogando et al., 2020; Pohl et al., 2021; Chu et al., 2020). Similar levels of infectious virus were found in the supernatant of these two cell lines, suggesting a lower rate of defective virion production in Vero WHO cells. Differences in type I interferon signaling between Vero E6 (IFN−/-, IRF3 deficient) and Vero WHO (IFN−/-, IRF3 deficient, STAT1 deficient) could drive differences in viral growth. Our transcriptional analysis did not identify any canonical antiviral defense or interferon stimulated genes, such as dsRNA detector RIG-I or ISG-inducing transcription factor STAT2, in either cell line, confirming inefficient type I interferon signaling mechanisms. However, expression of genes associated with innate defense such as those involved in inflammation (e.g., ANXA1), oxidative stress (e.g., SOD1), and apoptosis (e.g., BAK1) was altered in both cell lines. The upregulation of pro-inflammatory and pro-apoptotic genes in Vero WHO, but not Vero E6, suggests greater sensitivity of these cells to SARS-CoV-2 infection and NFκB-mediated inflammatory responses. Both Vero E6 and Vero WHO also downregulated genes related to protein folding, a process commonly seen in viral infection (Paladino et al., 2020). These results collectively support the greater viral replication in Vero WHO, whereas the higher levels of infectious SARS-CoV-2 in Vero E6 may be associated with the greater number of translation- and cell cycle-associated genes.
We characterized SARS-CoV-2 replication and host responses in human gastric cell lines (AGS, MKN45) given the involvement of the gastrointestinal tract in COVID-19 pathology (Fu et al., 2020; Liang et al., 2015; Zhang et al., 2020). Both AGS and MKN45 cell lines possess functional antiviral sensing machinery, whereas AGS produces low levels of STAT1 due to persistent PIV5 antagonism (Young et al., 2007). Consistent with higher levels of cellular and supernatant viral RNA, and titers of infectious virus, we detected robust changes in gene expression reflecting antiviral immunity in MKN45 cells. Canonical antiviral genes including RSAD2 and IFNL1 were uniquely expressed in MKN45, although several (e.g., OAS2) were also shared between AGS and MKN45, suggesting a minimal degree of viral infection in AGS. However, significant infectious virus titers were only detected in MKN45 cells-propagated infectious virus. This is consistent with recent studies identifying robust type III IFN release in primary human enterocytes following SARS-CoV-2 infection (Lamers et al., 2020; Vanderheiden et al., 2020; Zang et al., 2020). This is also unexpected given that persistent infection of AGS with PIV5 is associated with defective type I interferon signaling (Young et al., 2007). In contrast, absence of STAT1 in NHP kidney cells (Vero WHO) exacerbated transcriptional indicators of inflammation while suppressing viral replication, suggesting species-specific regulation of infection. Cellular factors, as well as the expression of co-receptors like TMPRSS2 and TMPRSS4, restricting production of infectious virions in AGS remain to be investigated (Lamers et al., 2020). MKN45 may serve as an ideal in vitro human model for evaluating SARS-CoV-2 pathogenesis and therapeutics given its relevance to COVID19 pathology and full expression of antiviral defense genes. Future studies will also investigate the role of key interferon signaling components (e.g., MAVS, PKR, RL6, TLR3) in MKN45 cells, which were investigated using A549 cells.
SARS-CoV-2 initially infects the respiratory epithelium (Hui et al., 2020; Vanderheiden et al., 2020). Here, we examined viral replication in human lung adenocarcinomas cells A549. Notably, low levels of viral RNA were detected in the cells and supernatants of A549 cells as reported elsewhere (Chu et al., 2020; Salgado-Benvindo et al., 2020; Felgenhauer et al., 2020). Previous studies have also reported low levels of A549 infectivity in the absence of ACE2 overexpression (Chu et al., 2020; Mossel et al., 2005; Sheahan et al., 2020). Whether this is due to restriction on viral entry or other factors is not clear. As we were able to detect hACE2 expression in the A549 cells, we hypothesized that removal of components of the viral sensing pathway would enhance the susceptibility of the A549 cells. We evaluated responses in cells lacking TLR3, a double-stranded (ds)RNA sensor, as well as lacking MAVS, which functions downstream of viral dsRNA sensor RIG-I (Lester and Li, 2014; Wu and Hur, 2015; Alexopoulou et al., 2001). Outcomes in RL6- and PKR-deficient A549 cells were also evaluated to elucidate the influence of interferon-stimulated genes that destroy viral RNA and halt protein synthesis, respectively (Dauber and Wolff, 2009; Silverman, 2007). With the exception of the RL6 knockout (KO), we were unable to detect viral RNA in the other wild-type and KO A459 cells through either qRT-PCR or RNA-seq. This may be enhanced with ectopic ACE2 expression (Li et al., 2021). However, the small number of genes detected in infection of wild-type A549 cells, including those related to innate immunity and apoptosis, suggests low levels of productive viral infection.
A recent study by Li et al. also evaluated the role of MAVS, PKR, and RL6 in SARS-CoV-2-infected A549 cells (Li et al., 2021). Unlike the study here, ACE2 was ectopically expressed in WT and KO cell lines (A549ACE2), which permitted robust viral replication at 6–48 hours post infection regardless of genotype. Similar to our findings, RL6 was shown to restrict viral replication, supporting a role for RL6 in SARS-CoV-2 pathogenesis.
There has been concern that in vitro passaging can induce mutations in SARS-CoV-2 (Davidson et al., 2020; Lau et al., 2020; Liu et al., 2020; Sasaki et al., 2021). Interesting, only a single mutation in the 3′ UTR (T152C) of MAVSKO was detected. Although the impact of this mutation is likely limited, viral evolution over several passages should be investigated to understand selective pressures in cell culture. In summary, we have identified key roles for viral sensors (e.g., MAVS) and ISGs (STAT1, RL6) in SARS-CoV-2 pathogenesis in various NHP and human cell types. However, various ISGs, like STAT1, may have species-specific effects in vitro. Furthermore, we report a lack of association between ACE2 expression and the permissivity and susceptibility of cells to SARS-CoV-2 infection. Future profiling of coreceptor expression profiling (e.g., TMPRSS2, TMPRSS4) will be needed to confirm this hypothesis. Findings here are critical for future in vitro studies aiming to replicate both human pathobiology for the evaluation for SARS-CoV-2 therapeutics and pathogenesis.
Limitations of the study
In this study we assessed SARS-CoV-2 infection, replication, and cellular transcriptional response using the variant isolated in Washington, USA early in 2020. Findings here may not extend to recently emerged variants of concern. In addition, infectious virus was not measured longitudinally. Transcriptional studies are limited by replicate size (n = 2), potential for lack of translatability between nonhuman primate and human species, absence of longitudinal sampling, and lack of confirmatory in vivo studies. Future longitudinal experiments will utilize greater number of replicates and investigate host responses in organoid and small animal models.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal Anti-SARS Coronavirus (antiserum, Guinea Pig) | BEI | NR-10361 |
| Monoclonal anti-ACE2 antibody produced in mouse | Millipore Sigma | AMAB91262 |
| IgG (H+L) Cross-Adsorbed Goat anti-mouse Alexa Fluor™ 647 | Invitrogen | A21235 |
| Bacterial and virus strains | ||
| SARS-CoV-2 Isolate USA-WA1/2020 | BEI | Catalog #NR-52281 |
| Chemicals, peptides, and recombinant proteins | ||
| PageRuler™ Prestained Protein Ladder, 10 to 180 kDa | Thermo Fischer Scientific | 26616 |
| Critical commercial assays | ||
| Pierce™ BCA Protein Assay | Thermo Fischer Scientific | 23227 |
| Next Ultra II Directional RNA Library Prep Kit | New England Biolabs | E7765 |
| Deposited data | ||
| Transcriptomic and viral genome data | NCBI BioProject | PRJNA745219 |
| Experimental models: Cell lines | ||
| Vero E6 CRL-1586™ | ATCC | |
| Vero CCL-81™ | ATCC | |
| AGS | R.J. DiPaolo | |
| MKN45 | R.J. DiPaolo | |
| A549 wild-type and knock-out (MAVS, PKR, RL6, TLR3) | S. Weiss | |
| Huh7 | M. Buller | |
| Huh7.5 | M. Buller | |
| Oligonucleotides | ||
| SARS-CoV-2 Forward 5’ GAC CCC AAA ATC AGC GAA AT 3’ | Integrated DNA Technologies | |
| SARS-CoV-2 Reverse 5’ TCT GGT TAC TGC CAG TTG AAT CTG 3’ | Integrated DNA Technologies | |
| SARS-CoV-2 5’ ACC CCG CAT TAC GTT TGG TGG ACC 3’ | Integrated DNA Technologies | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be dreicted to and will be fulfilled by the lead contact, Ilhem Messaoudi (imessaou@uci.edu).
Materials availability
Sequencing data generated in this study have been despite to NCBI BioProject PRJNA745219. Code for RNA-Seq can be accessed at https://github.com/MessaoudiLab/RNA-Seq-Analysis/tree/master/SystemPipeR.
Experimental model and subject details
Viruses and cells
SARS-CoV-2 viral stocks were derived from Isolate USA-WA1/2020 from BEI (Catalog #NR-52281). Virus was passaged two times in Vero E6 cells (ATCC® CRL-1586™, female) before clarification by centrifugation (3000 rpm for 30 min) and storage at −80°C until further use. To determine viral titers by focus forming assay, we used the Vero WHO cell line (ATCC® CCL-81™, female). Unless otherwise specified, all cells were cultured in Dulbecco's Modified Eagle Medium (Sigma- D5796-500ML) containing 1% HEPES (Sigma- H3537-100ML) and 5% FBS (Sigma- F0926) at 37°C, 5% CO2 (5% DMEM). AGS (female) and MKN45 (female) cells were gifted by R. J. DiPaolo. A549 (male) wild-type and knockout cells (MAVS, PKR, RL6, TLR3) were obtained from Dr. Susan Weiss. Huh7 and Huh7.5 (male) cells were gifted by Mark Buller. For virus growth curves, cells were plated at a density of 1x105 cells/well in 6 well plates and infected at MOI of 0.5.
Method details
qRT-PCR
hACE2 expression was measured by qRT-PCR using Taqman primer and probe sets from IDT (assay ID Hs.PT.58.27645939). SARS-CoV-2 viral burden was measured by qRT-PCR using Taqman primer and probe sets from IDT with the following sequences: Forward 5’ GAC CCC AAA ATC AGC GAA AT 3’, Reverse 5’ TCT GGT TAC TGC CAG TTG AAT CTG 3’, Probe 5’ ACC CCG CAT TAC GTT TGG TGG ACC 3’. Synthesized hACE2 RNA was used as a copy control to quantify the number of hACE2 molecules present in each sample. Similarly, a SARS- CoV-2 copy number control (available from BEI) was used to quantify SARS-CoV-2 genomes.
Focus forming assay for detection of infectious virus
Vero WHO cells were plated in a 96-well flat bottom plate to a confluency of 90-95% on the day prior to the assay. Tenfold serial dilutions of supernatant samples collected from infected wells were then made in a 96-well round bottom plate containing 5% DMEM media before being added to the Vero WHO cell monolayer and allowed to adsorb for one hour in an incubator with 37°C, 5% CO2. Following virus adsorption, a solution of 2% methylcellulose (Sigma-M0512-250G) was diluted 1:1 in 5% DMEM and warmed to room temperature. The methylcellulose-media mixture overlay was added to the plate by adding 125 μL of overlay media to each well and returned to an incubator with 37°C, 5% CO2 for 24 hours. Plates were then fixed in a solution of 5% paraformaldehyde (PFA) diluted in tissue culture grade 1X PBS, washed, and subjected to another fix in 5% formalin buffered phosphate (Fisher SF100-4) in 1X PBS for 15 minutes before washing and removal from BSL-3 containment. Foci were visualized by an immunostaining protocol using polyclonal anti-SARS-CoV-2 guinea pig sera (BEI NR-10361) diluted 1:15,000 with FFA staining buffer (1X PBS, 1mg/ml saponin, Sigma 47036) as a primary detection antibody overnight at 4°C. The secondary antibody consisted of goat anti-mouse conjugated horseradish peroxidase (Sigma A-7289) diluted 1:5,000 in FFA staining buffer and allowed to incubate for 2 hours at room temperature. Foci were visualized using KPL TrueBlue HRP substrate and allowed to develop for 10-15 minutes, or until blue foci are visible. The reaction was then quenched by washing with Millipore water and plates were imaged immediately thereafter with a CTL machine to quantify foci.
Western blot analysis
To generate cell lysates for western blot analysis of hACE2 expression, cells were seeded at 1x105 cells/well in 6 well plates and infected at an MOI of 0.5. 48 hours post infection, supernatants were removed, and cells were washed 2x with 1X PBS. 100 μl RIPA buffer (ThermoFisher 89900) was added to each well and plates sat on ice for 30 minutes. Cells were then scraped and collected into Eppendorf tubes for centrifugal clarification. A BCA assay (ThermoFisher 23227) was used to determine the protein concentration of each sample, and 30 μg of protein was used for each western blot. Protein samples were mixed 1:1 with Laemmli buffer (Sigma S3401) and heated to 95°C for 5 minutes. PageRuler prestained protein ladder (ThermoFisher 26616) and reduced samples were then loaded into NuPAGE 4-12% Bis-Tris pre-cast gels (ThermoFisher NP0322BOX) and run for 50 minutes at 200V in 1X MOPS SDS running buffer (ThermoFisher NP0001). Gels were then wet transferred overnight in transfer buffer (25 mM Tris, 192 mM glycine, 20% v/v methanol pH 8.3, mqH2O to 1L) at 4°C, 30V to nitrocellulose membranes (BioRad BR-9267005). Membranes were then blocked for 1 hour at room temperature while rocking in blocking buffer (15 ml 10X TBS, 135 ml mqH2O, 7.5 g BSA, 150 μl TWEEN20). The primary antibody for hACE2 detection (Sigma AMAB91262) was diluted 1:10000 in blocking buffer and membranes were stained for 2 hours at room temperature while rocking. Blots were washed 3x with 1X TBST (5% w/v BSA, 10X TBS, mqH2O, 0.1%TWEEN20) and the secondary antibody (goat α-mouse A647, ThermoFisher A-21235) was diluted 1:5000 in blocking buffer and incubated on the membrane for 1 hour at room temperature while rocking. Blots were then washed 3x with 1X TBST and imaged on a Typhoon FLA 9500.
cDNA library construction and sequencing
Quality and quantity of RNA from cells or supernatant were determined using an Agilent 2100 Bioanalyzer. cDNA libraries were constructed using the NEB Next Ultra II Directional RNA Library Prep Kit (New England Biolabs, E7765). RNA was treated with RNase H and DNase I following depletion of ribosomal RNA (rRNA). Adapters were ligated to cDNA products and the subsequent ∼300 base pair (bp) amplicons were PCR-amplified and selected by size exclusion. cDNA libraries were assessed for quality and quantity prior to 150 bp single-end sequencing using the Illumina NovaSeq platform.
Bioinformatic analysis
Preliminary data analysis was performed with RNA-Seq workflow module of systemPipeR, developed by Backman et al.32. RNA-Seq reads were demultiplexed, quality-filtered and trimmed using Trim Galore (average Phred score cut-off of 30, minimum length of 50 bp). FastQC was used to generate quality reports. Hisat2 was used to align reads to the reference genome. For A549, Huh7 and Huh7.5 cell lines, reads were aligned to and the Homo_sapiens.GRCh38.dna.primary assembly.fa reference genome and Homo_sapiens.GRCh38.85.gtf was used for annotation. For Vero E6 and Vero WHO cell lines, reads were first aligned to reference genome Macaca mulatta (Macaca_mulatta.Mmul_8.0.1.dna.toplevel.fa) and the Macaca_mulatta.Mmul_8.0.1.97.gtf was used for annotation. Unaligned reads were aligned to the SARS-CoV-2 (NC_045512.2) genome and the GCF_009858895.2_ASM985889v3_genomic.gff annotation file was used. Aligned reads were combined before annotation with input virus MT246667.1 used for comparison. Raw expression values (gene-level read counts) were generated using the summarize Overlaps function and normalized (read per kilobase of transcript per million mapped reads, rpkm) using the edgeR package. Differentially expressed genes (DEGs) were identified using edgeR was used with the following criteria: genes with median rpkm of ≥5, a false discovery rate (FDR) corrected p-value ≤ 0.05 and a log2fold change ≥ 1. To identify commonly regulated genes with adjustments for cell-specific differences, edgeR was performed with GLM capabilities. Batch-effects were adjusted for using the Limma R package. Functional enrichment of DEGs was performed using Metascape to identify relevant Gene Ontology (GO) biological process terms78. Heatmaps, bubbleplots, Venn diagrams and violin plots were generated using R packages ggplot2 and VennDiagrams. GO network plots were generated in Cytoscape (Version 3.5.1). Graphs were generated using GraphPad Prism software (version 8).
Quantification and statistical analysis
Kruskal-Wallis test with multiple comparisons was used to compare ACE2 mRNA and protein expression among cell lines. Standard deviation displayed on all bar graphs. p-value ≤ 0.05∗, p-value ≤ 0.01∗∗.
Acknowledgments
We thank the Brien, Messaoudi, and Pinto lab members for healthy discussions about the development of in vitro SARS-CoV-2 models. Support was received from Saint Louis University School of Medicine COVID-19 research seed funding to A.K.P. and J.D.B. NCI Seronet (U01CA260541) awarded to A.K.P. and J.D.B. supported this work. The National Center for Advancing Translation Sciences, NIH grant UL1 TR001414 and NIH grant R21 AI143301-02 supported I.M., A.N.P., and M.Z.Z. We thank Dr. Yize Li and Dr. Susan R. Weiss (University of Pennsylvania, Department of Microbiology) for graciously providing us with A549 wild-type and knockout cell lines.
Author contributions
A.K.P., I.M., J.D.B., E.G., and T.E.S. conceived and designed the experiments. E.G., A.N.P., I.M., and A.K.P. developed the methodology. E.G., M.Z.Z., K.J.M., and A.N.P. performed the experiments. A.K.P., J.D.B., T.E.S., E.G., I.M., and A.N.P. analyzed the data and wrote the manuscript. I.M. and A.K.P. supervised the study. All authors reviewed and approved the final version of the manuscript.
Declaration of interests
The authors declare no conflicts of interest.
Inclusion and diversity
We worked to ensure diversity in experimental samples through the selection of the cell lines. We worked to ensure diversity in experimental samples through the selection of the genomic datasets. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103553.
Contributor Information
Ilhem Messaoudi, Email: imessaou@uci.edu.
Amelia K. Pinto, Email: pintoak@slu.edu.
Supplemental information
Data and code availability
All sequencing data are available at BioProject PRJNA745219. Code for RNA-Seq can be accessed at https://github.com/MessaoudiLab/RNA-Seq-Analysis/tree/master/SystemPipeR.
References
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of Nf-Kappab by toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Barranco S.C., Townsend C.M., Jr., Casartelli C., Macik B.G., Burger N.L., Boerwinkle W.R., Gourley W.K. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res. 1983;43:1703–1709. [PubMed] [Google Scholar]
- Beauclair G., Streicher F., Chazal M., Bruni D., Lesage S., Gracias S., Bourgeau S., Sinigaglia L., Fujita T., Meurs E.F., et al. Retinoic acid inducible gene I and protein kinase R, but not stress granules, mediate the proinflammatory response to yellow fever virus. J. Virol. 2020;94 doi: 10.1128/JVI.00403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollavaram K., Leeman T.H., Lee M.W., Kulkarni A., Upshaw S.G., Yang J., Song H., Platt M.O. Multiple sites on Sars-Cov-2 spike protein are susceptible to proteolysis by cathepsins B, K, L, S, and V. Protein Sci. 2021;30:1131–1143. doi: 10.1002/pro.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Cortese M., Romero-Brey I., Bender S., Neufeldt C.J., Fischl W., Scaturro P., Schieber N., Schwab Y., Fischer B., et al. Dengue virus perturbs mitochondrial morphodynamics to Dampen innate immune responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Saccon E., Appelberg K.S., Mikaeloff F., Rodriguez J.E., Vinhas B.S., Frisan T., Vegvari A., Mirazimi A., Neogi U., Gupta S. Type-I interferon signatures in Sars-Cov-2 infected Huh7 cells. Cell Death Discov. 2021;7:114. doi: 10.1038/s41420-021-00487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew T., Noyce R., Collins S.E., Hancock M.H., Mossman K.L. Characterization of the interferon regulatory factor 3-mediated antiviral response in A cell line deficient for Ifn production. Mol. Immunol. 2009;46:393–399. doi: 10.1016/j.molimm.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of Sars-Cov-2 and Sars-Cov with implications for clinical manifestations, transmissibility, and laboratory studies of Covid-19: an observational study. Lancet Microbe. 2020;1:E14–E23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech C.B., Walker S.C., Samuels R.J. Sars-Cov-2 vaccines. JAMA. 2021;325:1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- Dauber B., Wolff T. Activation of the antiviral kinase Pkr and viral Countermeasures. Viruses. 2009;1:523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of Sars-Cov-2 reveals A cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmyter J., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in A line of African green monkey kidney cells (Vero) J. Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny J.M., Morgan M.J. Regulation of the interferon system: evidence that Vero cells have A genetic defect in interferon production. J. Gen. Virol. 1979;43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- Felgenhauer U., Schoen A., Gad H.H., Hartmann R., Schaubmar A.R., Failing K., Drosten C., Weber F. Inhibition of Sars-Cov-2 By type I and type Iii interferons. J. Biol. Chem. 2020;295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Wang B., Yuan T., Chen X., Ao Y., Fitzpatrick T., Li P., Zhou Y., Lin Y.F., Duan Q., et al. Clinical characteristics of coronavirus disease 2019 (Covid-19) in China: a systematic review and meta-analysis. J. Infect. 2020:656–665. doi: 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi S., Poloni E.T., Pandini C., Garofalo M., Dragoni F., Medici V., Davin A., Visona S.D., Moretti M., Sproviero D., et al. Detection of Sars-Cov-2 genome and whole transcriptome sequencing in frontal Cortex of Covid-19 patients. Brain Behav. Immun. 2021;97:13–21. doi: 10.1016/j.bbi.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., Takvorian P.M., Bleck C., Hsu V.W., Fehr A.R., et al. Beta-coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535 E14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of Sars-Cov-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Sanchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of A Sars-Cov-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280 E5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. Sars-Cov-2 cell entry depends on Ace2 and Tmprss2 and is blocked by A clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornuss D., Lange B., Schröter N., Rieg S., Kern W.V., Wagner D. Anosmia in Covid-19 patients. Clin. Microbiol. Infect. 2020;26:1426–1427. doi: 10.1016/j.cmi.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. Sars-Cov-2 Reverse genetics reveals A variable infection gradient in the respiratory tract. Cell. 2020;182:429–446 e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Cheung M.C., Perera R., Ng K.C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.W., et al. Tropism, replication competence, and innate immune responses of the coronavirus Sars-Cov-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Thomsen A.R. Sensing of Rna viruses: a review of innate immune receptors involved in recognizing rna virus invasion. J. Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto M., Yamaji T., Saito K., Shirasago Y., Satomura K., Endo T., Fukasawa M., Hanada K., Osada N. Identification of characteristic genomic markers in human hepatoma Huh-7 and Huh7.5.1-8 cell lines. Front Genet. 2020;11:546106. doi: 10.3389/fgene.2020.546106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto M., Uemura K., Sanaki T., Sato A., Hall W.W., Kariwa H., Orba Y., Sawa H., Sasaki M. Tmprss11d and Tmprss13 activate the Sars-Cov-2 spike protein. Viruses. 2021;13:384. doi: 10.3390/v13030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvansakul M. Viral infection and apoptosis. Viruses. 2017;9:356. doi: 10.3390/v9120356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis N.C., Lopez-Cortes A., Gonzalez E.V., Grimaldos A.B., Prado E.O. Sars-Cov-2 vaccines strategies: a comprehensive review of phase 3 candidates. Npj Vaccin. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., Van Der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul Van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. Sars-Cov-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.Y., Wang P., Mok B.W., Zhang A.J., Chu H., Lee A.C., Deng S., Chen P., Chan K.H., Song W., et al. Attenuated Sars-Cov-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.G., Huang W., Lee H., Van De Leemput J., Kane M.A., Han Z. Characterization of Sars-Cov-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by selinexor. Cell Biosci. 2021;11:58. doi: 10.1186/s13578-021-00568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by Sars-Cov-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester S.N., Li K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Banerjee S., Wang Y., Goldstein S.A., Dong B., Gaughan C., Silverman R.H., Weiss S.R. Activation of Rnase L is dependent on Oas3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U S A. 2016;113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Renner D.M., Comar C.E., Whelan J.N., Reyes H.M., Cardenas-Diaz F.L., Truitt R., Tan L.H., Dong B., Alysandrato K.D., et al. Sars-Cov-2 induces double-stranded Rna-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2022643118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Oh B.H., Jung J.U. Novel functions of viral anti-apoptotic factors. Nat. Rev. Microbiol. 2015;13:7–12. doi: 10.1038/nrmicro3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Li H., Li J., Yang Z.N., Li J.L., Zheng H.W., Chen Y.L., Shi H.J., Guo L., Liu L.D. Role of neutrophil chemoattractant Cxcl5 in Sars-Cov-2 infection-induced lung inflammatory innate immune response in an in vivo Hace2 transfection mouse model. Zool. Res. 2020;41:621–631. doi: 10.24272/j.issn.2095-8137.2020.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., et al. Sars-Cov-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J., et al. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Leon S., Wegman-Ostrosky T., Perelman C., Sepulveda R., Rebolledo P., Cuapio A., Villapol S. More than 50 long-term effects of Covid-19: a systematic review and meta-analysis. Res. Square. 2021;11:16144. doi: 10.1038/s41598-021-95565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkler A.E., Parikh N.S., Mir S., Gupta A., Kamel H., Lin E., Lantos J., Schenck E.J., Goyal P., Bruce S.S., et al. Risk of ischemic stroke in patients with coronavirus disease 2019 (Covid-19) vs patients with influenza. JAMA Neurol. 2020;77:1–7. doi: 10.1001/jamaneurol.2020.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. Sars-Cov-2 Orf6 hijacks Nup98 to block stat nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U S A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel E.C., Huang C., Narayanan K., Makino S., Tesh R.B., Peters C.J. Exogenous Ace2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J. Virol. 2005;79:3846–3850. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgolo N., Therien A.G., Howell B., Klein D., Koeplinger K., Lieberman L.A., Adam G.C., Flynn J., Mckenna P., Swaminathan G., et al. Sars-Cov-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog. 2021;17:e1009225. doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R., Van Der Meer Y., Caly L., Druce J., De Vries J.J.C., Kikkert M., Barcena M., et al. Sars-Coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T., Kuroda M., Hanada K. The genome landscape of the African green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino L., Vitale A.M., Caruso Bavisotto C., Conway De Macario E., Cappello F., Macario A.J.L., Gammazza A.M. The role of molecular chaperones in virus infection and implications for understanding and treating Covid-19. J. Clin. Med. 2020;9:3518. doi: 10.3390/jcm9113518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl M.O., Busnadiego I., Kufner V., Glas I., Karakus U., Schmutz S., Zaheri M., Abela I., Trkola A., Huber M., et al. Sars-Cov-2 variants reveal features critical for replication in primary human cells. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001006. e3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S., Fernandez-Antunez C., Galli A., Underwood A., Pham L.V., Ryberg L.A., Feng S., Pedersen M.S., Mikkelsen L.S., Belouzard S., et al. Overcoming culture restriction for Sars-Cov-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.00097-21. e0009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Glende J., Al-Falah M., De Vries V., Schwegmann-Wessels C., Qu X., Tan L., Tschernig T., Deng H., Naim H.Y., Herrler G. Analysis of Ace2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J. Gen. Virol. 2006;87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- Salgado-Benvindo C., Thaler M., Tas A., Ogando N.S., Bredenbeek P.J., Ninaber D.K., Wang Y., Hiemstra P.S., Snijder E.J., Van Hemert M.J. Suramin inhibits Sars-Cov-2 infection in cell culture by interfering with early steps of the replication cycle. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00900-20. e00900-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., Hall W.W., Orba Y., Sawa H. Sars-Cov-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in Tmprss2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. e1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of Sars-Cov-2. Proc. Natl. Acad. Sci. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J., Sciaky N., Lee J., Bosshart H., Angeletti R.H., Bonifacino J.S. Localization of endogenous furin in cultured cell lines. J. Histochem. Cytochem. 1997;45:3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits sars-Cov-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M., Aktepe T.E., Deerain J.M., Mcauley J.L., Audsley M.D., David C.T., Purcell D.F.J., Urin V., Hartmann R., Moseley G.W., et al. Sars-Cov-2 suppresses Ifnbeta production mediated by Nsp1, 5, 6, 15, Orf6 and Orf7b but does not suppress the effects of added interferon. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009800. e1009800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R.H. Viral encounters with 2',5'-oligoadenylate synthetase and Rnase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R., Jr., Loo Y.M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S.M., Gale M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus Rna replication through A cellular Rna helicase, Rig-I. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., et al. Type I and type Iii interferons restrict Sars-Cov-2 infection of human airway epithelial cultures. J. Virol. 2020;94 doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall-Fleming E., Young D.F., Goodbourn S., Davison A.J., Randall R.E. Genome sequence of the parainfluenza virus 5 strain that persistently infects ags cells. Genome Announc. 2016;4 doi: 10.1128/genomeA.00653-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Hur S. How Rig-I like receptors activate mavs. Curr. Opin. Virol. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by sars-Cov-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Wen Z., Gao W., Lin Z., Zhong J., Jiu Y. Multifaceted functions of host cell caveolae/caveolin-1 in virus infections. Viruses. 2020;12 doi: 10.3390/v12050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. Rig-I triggers A signaling-abortive anti-Sars-Cov-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- Young D.F., Andrejeva L., Livingstone A., Goodbourn S., Lamb R.A., Collins P.L., Elliott R.M., Randall R.E. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 2003;77:2174–2181. doi: 10.1128/JVI.77.3.2174-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.F., Carlos T.S., Hagmaier K., Fan L., Randall R.E. Ags and other tissue culture cells can unknowingly Be persistently infected with Piv5; A virus that blocks interferon signalling by degrading Stat1. Virology. 2007;365:238–240. doi: 10.1016/j.virol.2007.03.061. [DOI] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K., Chan J.F., et al. Sars-Cov-2 Nsp13, Nsp14, Nsp15 and Orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., Mccune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. Tmprss2 and Tmprss4 promote Sars-Cov-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li H.B., Lyu J.R., Lei X.M., Li W., Wu G., Lyu J., Dai Z.M. Specific Ace2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-Ncov infection. Int. J. Infect Dis. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Liao C., Fan Q.H., Chen H.B., Zhao X.G., Xie Z.G., Li X.L., Chen C.X., Lu X.X., Liu Z.S., et al. Clinical characteristics of children with coronavirus disease 2019 in Hubei, China. Curr. Med. Sci. 2020;40:275–280. doi: 10.1007/s11596-020-2172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with A new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. Sars-Cov-2 receptor Ace2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035 e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available at BioProject PRJNA745219. Code for RNA-Seq can be accessed at https://github.com/MessaoudiLab/RNA-Seq-Analysis/tree/master/SystemPipeR.