Abstract
Hepatitis C Virus (HCV) infection is a major risk factor that can leads to chronic liver disease including fibrosis, cirrhosis, and hepatocellular carcinoma. Progression of chronic liver disease by HCV infection is caused by a complex intercellular reaction. Specially, exosomes and microRNAs (miRNAs) from HCV-infected hepatocytes play a role in the pathogenesis of liver disease by facilitating cellular communication between parenchymal and non-parenchymal cells. However, the underlying mechanism of secretions of exosome and miRNAs during HCV infection is still unknown. In this study, we demonstrated a novel pathway for the release of exosome and exosomal miRNAs via caspase-3/Panx1/P2X4 activation during HCV infection in hepatocytes. We found that HCV infection induced the stimulation of exosome release and activation of caspase-3/Panx1/P2X4 pathway in Huh7.5.1 cells. In addition, miR-122 and miR-146a levels in extracellular exosomes from HCV-infected cells were dramatically increased while intracellular miR122 and miR-146a expression had no large changes. Notably, the secretions of exosomes and exosomal miRNAs were decreased by inhibition of caspase 3, Panx1 and P2X4 while inhibition of ROCK-1 cleavage did not affect that during HCV infection in Huh7.5.1 cells. Conclusion: These results suggested that HCV infection caused secretions of exosomes and exosomal miRNAs dependent on caspase 3/Panx1/P2X4 pathway. Our study provides the possible therapeutic intervention using Panx1 suppression for liver disease development mediated by exosome from HCV-infected hepatocytes.
Keywords: HCV, exosome, miRNAs, Pannexin, purinergic receptor
Introduction
Cells exchange information through the release of soluble factors or by direct interaction. Recently, it has been suggested that cells can also communicate by circular membrane fragments called extracellular vesicles (EVs) (1). Normal or diseased cells release different types of extracellular vesicles, including microvesicles (MVs) and exosomes, depending on their cellular origin. Exosomes (40–100 nm) are formed by the fusion between multivesicular bodies (MVBs) and the plasma membrane, while MVs (100–2000 nm) bud directly from the plasma membrane (1, 2). Exosomes have been shown to provide a means of intercellular communication as contributing factors in the development of several diseases by the spread of proteins, mRNAs, and microRNAs (miRNA) (3). During virus infection, exosomes released from virus-infected cells contain viral proteins, viral RNAs, and certain specific miRNAs that are able to spread the infection and alter the cellular response in uninfected target cells during the immune response and pathogenesis (4, 5).
Hepatitis C virus (HCV) is a positive-sense single-stranded RNA virus of the family Flaviviridae; HCV infection is a global health problem and a major cause of chronic hepatitis, which leads to fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (6). HCV infection induced-development of liver disease is caused by complex intercellular reactions between cells (6, 7). Exosome plays a critical role in mediating this cellular communication (8). Syntenin has been reported to be involved on the secretion of E2 via exosomes (9). Recently, accumulating evidence demonstrated that exosomes and exosomal miRNAs from HCV-infected hepatocytes lead to polarization and differentiation of macrophages and mediate pro-fibrotic responses in stellate cells and T follicular regulatory cells expansion (8, 10, 11). This suggested that the development of liver disease involves intercellular communication during HCV infection. Interestingly, some studies have shown increased release of specific miRNAs, such as miR-122, miR-146a and miR-155 in HCV infection (12, 13). However, the underlying mechanism of HCV-mediated secretions of exosome and exosomal miRNAs is still unknown.
The release of MVs or exosomes can be stimulated by stress signals, including DNA damage, intracellular calcium, and extracellular adenosine triphosphate (ATP) (2, 14, 15). Savina et al. have shown that a calcium influx inducer caused formation of MVBs and induced exosome release from K562 cells (14). In addition, it has been demonstrated that exosome release is induced by extracellular ATP which is associated with purinergic receptor activation (15, 16). In this regard, we speculated that the Pannexin 1 (Panx1) pathway is associated with the secretions of exosomes and miRNAs. Panx1 is a transmembrane channel that mediates ATP release when it is activated by the stretch of the plasma membrane during changes in osmolality or mechanical injuries or by proteolysis via caspase-3 and −7 during early apoptosis (17). The ATP released by Panx1 activation can bind to the purinergic receptor, leading to a calcium influx (18). Some studies have shown increased Panx-1 and purinergic receptor expression in HCV-infected hepatocytes and patients’ liver (19, 20). However, the participation of Panx1 pathway-mediated exosome release in viral infection has not been examined yet. In this study, we demonstrated that secretions of exosomes and specific miRNAs are associated with the Panx-1/purinergic receptor pathway in HCV-infected hepatocytes.
Materials and Methods
Cell culture and HCV and dengue virus infection
The human hepatoma cell line, Huh7.5.1, was obtained from the American Type Culture Collection (ATCC). Huh7.5.1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100 mg/L penicillin-streptomycin, and 2 mmol/L glutamine. Cells were maintained at 37°C under a humidified atmosphere of 5% CO2. Huh7.5.1 cells were infected with HCV (JFH-1 strain, genotype 2a) and dengue virus (type 2) at a multiplicity of infection (MOI) of 0.1. After 3 days, the culture medium was changed to exosome-depleted FBS medium. After 24 h, we used the cells or supernatants for assays. Cryopreserved primary human hepatocytes (PHHs) were obtained from Thermo Fisher Scientific and cultured with Williams’ Medium E and Hepatocyte Maintenance Supplement Pack on Collagen I-coated plates, according to the manufacturer’s protocols (Thermo Fisher Scientific), and were then inoculated with normal (n=3) sera or HCV-infected patient (n=3) sera for 12 days.
Exosome isolation and quantification
Culture media from Huh7.5.1 or PHHs cultured with exosome-depleted serum were collected on day 4 or day 12 post-infection, respectively, and centrifuged at 1500 rpm for 5 minutes. Supernatants were centrifuged at 2500 rpm for 15 minutes, and then passed through 0.4-μm and 0.2-μm filters. ExoQuick-TC (System Biosciences) was added and the supernatants were incubated overnight. The supernatants were centrifuged at 1500 × g for 30 minutes and the exosome pellet was resuspended in phosphate-buffered saline (PBS). Quantification of exosomes was performed using a NanoSight LM10 system and the amount of protein in the exosomes was measured by a bicinchoninic acid (BCA) assay.
Transfection and inhibitor treatment
Huh 7.5.1 cells were infected with HCV at 0.1 MOI and 20 μM carbenoxolone (CBX) (Panx-1 inhibitor, Sigma) dissolved in medium or 5 μM BX430 (P2X4 inhibitor, Tocris Bioscience) dissolved in DMSO were added every day. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with CBX or BX430. After 24 h, exosomes were isolated from cell supernatants using ExoQuick-TC solution, and miRNAs were extracted from cells and exosomes. Huh 7.5.1 cells were transfected with AllStars Negative Control siRNA (Qiagen), PANX1 Silencer (Ambion) or P2RX4 Silencer (Ambion) using Lipofectamine™ 2000 Transfection Reagent (Thermo Fisher Scientific) at 1 day post infection. The culture medium was replaced with exosome-depleted FBS medium at 3 days post infection. After 24 h, exosomes were isolated from cell supernatants using ExoQuick-TC solution, and miRNAs were extracted from cells and exosomes.
Huh 7.5.1 cells were infected with HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with 20 μM Pan Caspase Inhibitor z-VAD-fmk (R&D Systems), 20 μM z-DQMD-fmk (Biovision), 50 μM Probenocid (Sigma), 50 μM Scrambled 10Panx (Tocris Bioscience), 50 μM 10Panx (Tocris Bioscience) or 50 μM Y-27632 dihydrochloride (Abcam). After 24 h, exosomes were isolated from cell supernatants using ExoQuick-TC solution, and miRNAs were extracted from cells and exosomes.
Western blotting
Cells were harvested, lysed in a Radioimmunoprecipitation assay (RIPA) Lysis Buffer System (Santa Cruz Biotechnology), and centrifuged at 12,000 × g for 20 min at 4°C. The protein content of the clear lysates was estimated using a Pierce BCA Protein Assay Kit according to the manufacturer’s protocols (Thermo Fisher Scientific). Equal amounts of total protein were dissolved in NuPAGE® LDS sample buffer (4×; Life Technologies). Protein samples were separated on a 10% SDS-polyacrylamide gel and then transferred to a nitrocellulose membrane (Bio-Rad Laboratories). Membranes were incubated for 1 h in a blocking solution containing 5% nonfat milk in Tris-buffered saline, and then incubated for 12 h at 4°C with antibodies recognizing Pannexin-1 (D9M1C), P2X4 (purinergic receptor P2X4) (D9R1H), Caspase-3, CD63, ROCK1 (Rho associated coiled-coil containing protein kinase 1), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Cell Signaling Technology). After incubation with the primary antibody, the membranes were incubated with a secondary antibody (anti-rabbit IgG HRP-linked antibody, 1:5,000, Cell Signaling Technology, Inc.,) for 1 h at room temperature. The immunoreactive protein bands were then developed.
Immunofluorescence microscopy
Cells were fixed using 4% paraformaldehyde in PBS for 15 min, and then washed. They were blocked and permeabilized using 3% bovine serum albumin (BSA), 10% FBS, and 0.3% triton X-100 in PBS overnight. The fixed cells were probed with a CD63 Monoclonal Antibody (MEM-259) (Alexa Fluor 488, Invitrogen) at a final dilution of 1:100 (2 μg/mL) overnight. The cells were then washed using PBS and overlaid with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma). Images were captured using a LSM700 (Carl Zeiss, Oberkochen, Germany) confocal microscope.
RNA extraction and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis
Cells were lysed in the presence of Trizol. Synthetic Caenorhabditis elegans miR-39 (Thermo Fisher Scientific) was spiked into the lysates when assessing the miRNAs in exosomes. Total RNA, including miRNAs, was extracted from the cell lysate or exosomes using a miRNeasy mini kit, according to the manufacturer’s protocols (Qiagen). Complementary DNA (cDNA) was then synthesized using High-Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific) for mRNA assay or TaqMan MicroRNA Reverse Transcription Kit and TaqMan MicroRNA Assay (Thermo Fisher Scientific) for miRNA assay. Real-time PCR (Applied Biosystems) was performed on triplicate samples using 1 μL of cDNA with Fast SYBR Green Master Mix (Thermo Fisher Scientific) for mRNA assay or 1.33 μL with the TaqMan® Universal PCR Master Mix, no AmpErase® UNG (Thermo Fisher Scientific). The cDNA for mRNA assay was amplified for 40 cycles of denaturation (95°C for 3 s) and annealing and extension (60°C for 30 s) using the primers shown in supplemental Table S2. The cDNA for miRNA assay was amplified for 40 cycles of denaturation (95°C for 15 s) and annealing and extension (60°C for 60 s) using a specific TaqMan MicroRNA Assay (Thermo Fisher Scientific). The reaction was performed using StepOne Real-Time PCR Systems (Applied Biosystems). HPRT or U18 were used as an endogenous control for normalization in cells, and C. elegans miR-39 was used as an exogenous control for normalization in exosomes.
Flow cytometry analysis
Cells were detached by treatment with trypsin, washed, and centrifuged with PBS three times, and then stained with Annexin V-fluorescein isothiocyanate (FITC)/TO-PRO-3 (BioLegend) for TO-PRO-3 uptake and apoptosis assays. For CD63 staining, cells were fixed and stained with CD63-FITC (BioLegend), and then washed and measured using a flow cytometer (CytoFLEX Flow Cytometer, Beckman Coulter, Inc.).
ATP measurement
Huh7.5.1 cells were infected with HCV or dengue virus at an MOI of 0.1. After 3 days, the culture medium was changed to exosome-depleted FBS medium. After 24 h, extracellular ATP levels in the culture medium from Huh7.5.1 were measured by the CellTiter-Glo® luminescent cell viability assay, according to the manufacturer’s protocols (Promega Corporation).
Calcium level measurement
Huh7.5.1 cells were infected with HCV or dengue virus at an MOI of 0.1. After 3 days, the culture medium was changed to exosome-depleted FBS medium. After 24 h, intracellular calcium levels were measured using a fluorometric Fluo-4 NW calcium assay kit according to the manufacturer’s protocols (Thermo Fisher Scientific).
Neutral sphingomyelinase (nSMase) activity measurement
Huh7.5.1 cells were infected with HCV or dengue virus at an MOI of 0.1. After 3 days, the culture medium was changed to exosome-depleted FBS medium. After 24 h, the cells were harvested, lysed in the RIPA Lysis Buffer System (Santa Cruz Biotechnology), and centrifuged at 12,000 × g for 20 min at 4°C. The protein content of the clear lysates was estimated using the Pierce BCA Protein Assay Kit, according to the manufacturer’s protocols (Thermo Fisher Scientific). Equal amounts of total protein were used for nSMase activity measurement using Sphingomyelinase Assay Kit according, to the manufacturer’s protocols (Abcam).
Statistical analysis
All experimental data were expressed as mean ± standard deviation (SD). The significance of the treatment effects was analyzed using an unpaired student t-test in the SPSS statistical package (SPSS PASW Statistic 20.0, SPSS Inc.,). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant.
Results
HCV infection stimulates the secretion of exosomes, but dengue virus infection does not
Recently, a study by Lambrecht et al. showed that plasma from HCV fibrotic patients was significantly enriched with circulating vesicles compared with that of healthy controls (21). Hence in this study, we investigated whether HCV infection could affect the release of exosome from hepatocytes in vitro. We compared dengue virus-infected cells with HCV-infected cells. Dengue virus is also a positive-sense single-stranded RNA virus of the family Flaviviridae that can replicate in hepatocytes (22). We isolated the accumulated exosomes between day 3 and 4 post infection, the time when HCV replication is at its highest (S1A Fig), from HCV or dengue virus-infected Huh 7.5.1 cells. The number of exosomes and their protein content were significantly increased in HCV-infected cells compared with those in uninfected Huh 7.5.1 cells, while dengue virus infection did not induce a change in exosome release (Fig 1A and 1B) even though the virus replicated well (S1B Fig). We also observed significantly increased protein content in exosome from primary human hepatocytes (PHHs) exposed to HCV patient sera compared with normal sera-exposed PHHs (Fig 1B). Next, we measured CD63, an exosome protein marker and dependent on exosome particle number. We found that intracellular CD63 was decreased in HCV-infected cells but not in dengue virus infected cells using immunofluorescence microscopy (Fig 1C) and western blotting (Fig 1D). However, HCV infection increased the levels of CD63 and GAPDH in extracellular exosomes from HCV infected Huh 7.5.1 cells (Fig 1D). These data indicated that HCV infection stimulates exosome release from hepatocytes.
Fig 1. HCV infection stimulates the secretion of exosomes, but dengue virus infection does not.
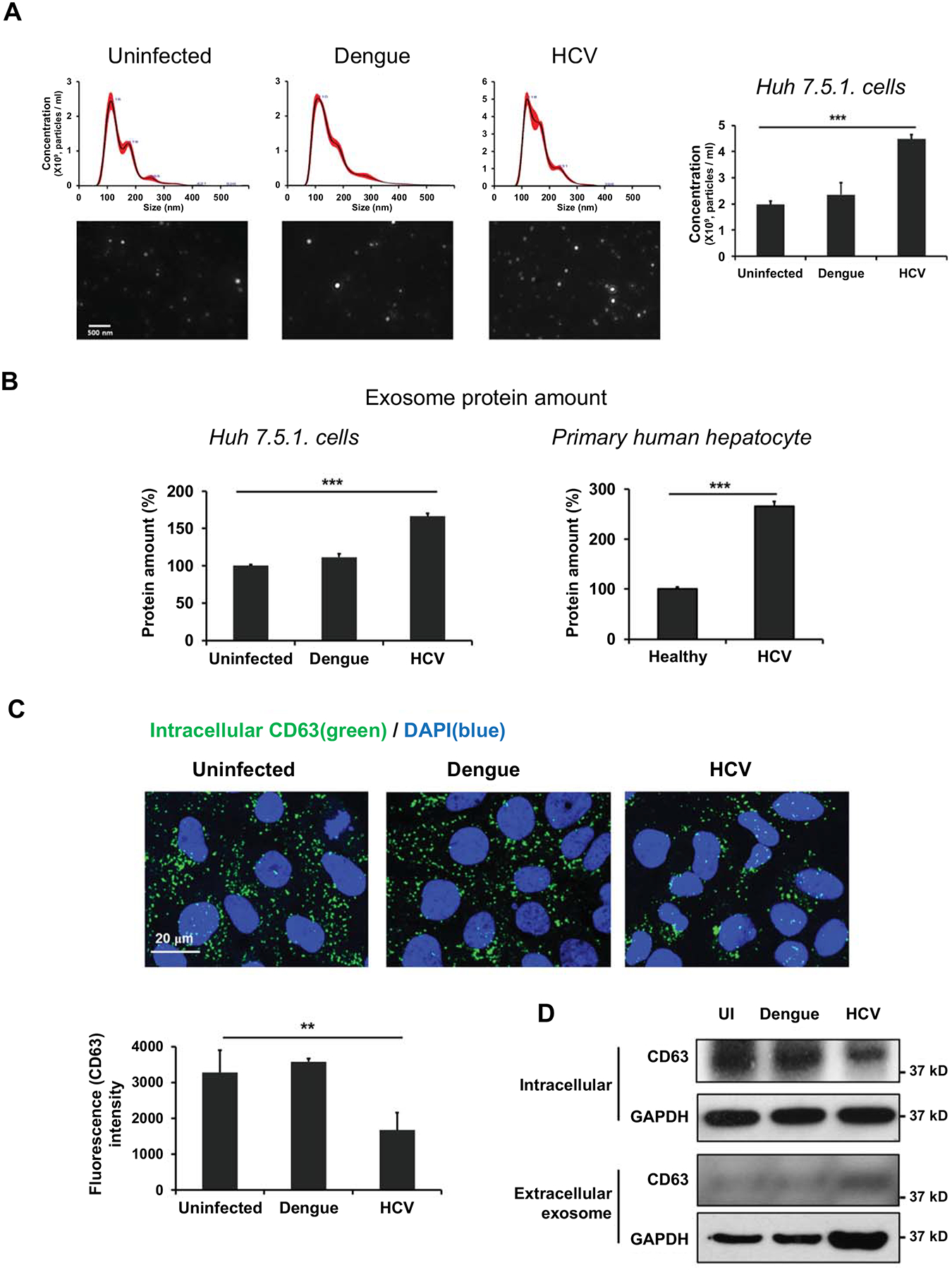
Huh 7.5.1 cells were infected HCV or dengue virus at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS media. After 24 h, exosomes were isolated from the cell supernatant. Primary human hepatocytes were exposed to healthy or HCV-infected patient sera and exosomes were isolated from the cell supernatants after 12 days. (A) The exosome number was measured using Nano-sight and (B) exosome protein amount was measured using a BCA assay. (C) Intracellular CD63 (green) localization and density in Huh 7.5.1 cells were measured by fluorescence microscopy, and (D) the protein expression of CD63 and GAPDH in exosomes and cell lysates from Huh 7.5.1 cells were measured by western blotting. Data are expressed as mean ± SD (n=3). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
HCV infection stimulates the secretion of specific miRNAs in exosome from hepatocytes
miRNAs are small non-coding RNA molecules that regulate gene expression post transcriptionally by repressing specific target mRNAs via degradation and/or translational repression. Circulating miRNAs are packaged as exosomes, allowing their stable delivery into target cells from body fluids (23, 24). Serum levels of certain specific miRNAs, including miR122, miR-146a, and miR-155, are increased in patients with chronic HCV infection (12). We expected that the miRNAs are secreted using exosome from hepatocytes. Therefore, we compared the levels of intracellular miRNAs and extracellular exosomal miRNAs during HCV infection in Huh 7.5.1 cells. We found that intracellular miR122 and miR-146a expression showed no significant changes at 4 post-infection (Fig 2A) while miR-122 and miR-146a levels in extracellular exosomes from HCV-infected Huh 7.5.1 cells between day 3 and 4 post-infection were dramatically increased (Fig 2B). However, the exosomal miR155 level was similar to the intracellular miR-155 level from HCV-infected Huh 7.5.1 cells at 4 post-infection (Fig 2A and 2B). In addition, HCV sera-exposed PHHs showed significantly increased exosomal miR-122 and miR-146a levels compared with those in PHHs exposed to normal sera (Fig 2D); meanwhile, there were no difference in intracellular miRNA expression (Fig 2C). Thus, we suggest that HCV infection induces the secretion of specific miRNA, such as miR-122 and miR-146a, in exosome from hepatocytes.
Fig 2. HCV infection stimulates the secretion of specific miRNAs in exosome from hepatocytes.
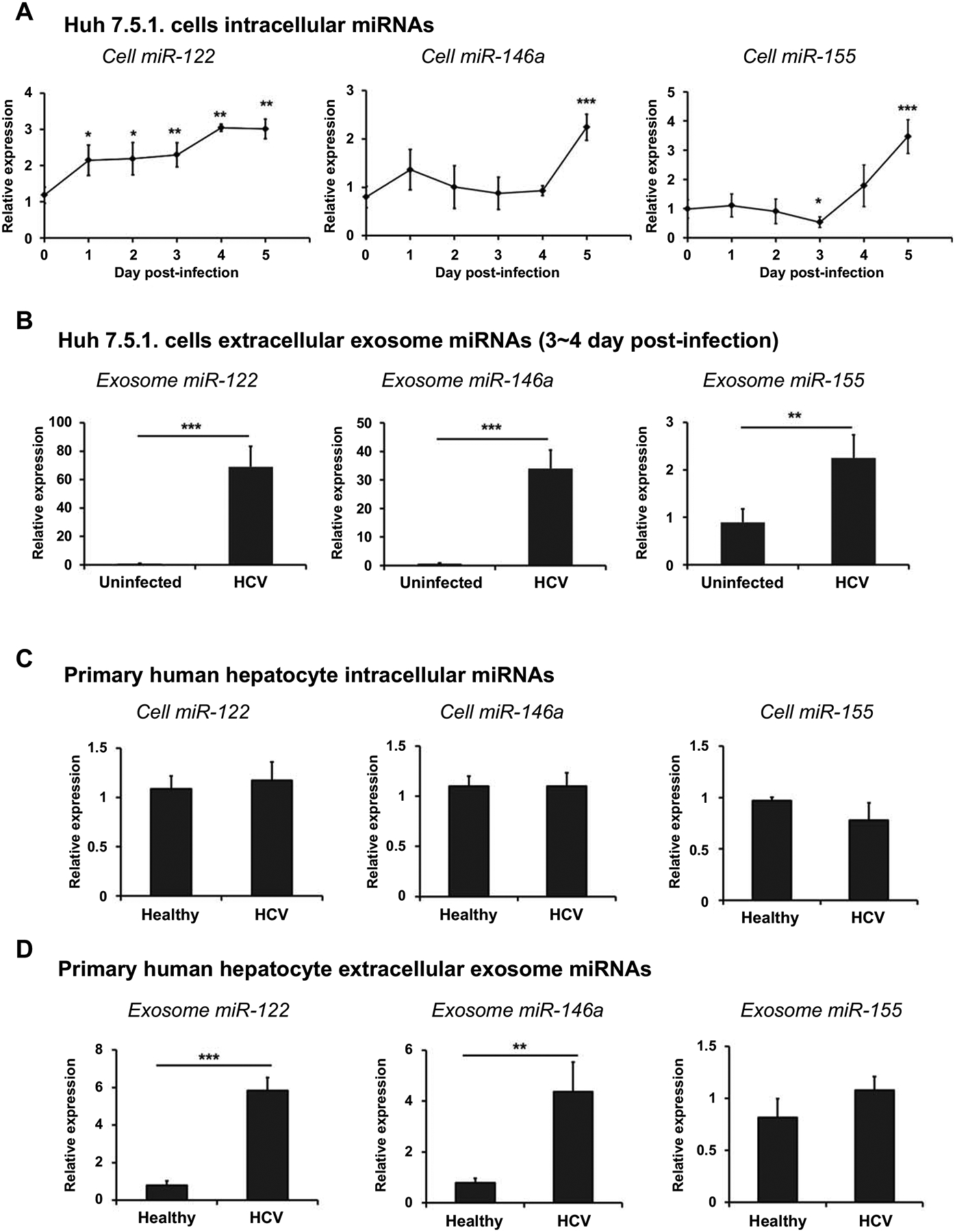
(A) Intracellular miRNAs were extracted after Huh 7.5.1 cells infected HCV with 0.1 MOI 4 days. (B) Huh 7.5.1 cells were infected with HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium. After 24 h, exosomes were isolated from cell supernatants and used for miRNA extraction. (C) Primary human hepatocytes were exposed healthy or HCV-infected patient sera. After 12 days, intracellular miRNAs were extracted, and (D) exosomes were isolated from cell supernatants and used for miRNA extraction. Intracellular miRNAs and extracellular exosomal miRNAs were extracted using a miRNeasy mini kit, and miRNAs expression was analyzed by real-time PCR. U18 was used as an endogenous control in cells for normalization, and Caenorhabditis elegans miR-39 was used as an exogenous control for normalization in the exosomes. Data are expressed as mean ± SD (n=3). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
HCV infection induces the activation of caspase-3, Panx1 and P2X4, but dengue virus infection does not
Exosomes are derived from the intraluminal vesicles (ILV) contained within the MVBs, which fuse with the cell membrane in an exocytic manner. The MVB formation mechanism is dependent on the endosomal sorting complex required for transport (ESCRT) machinery or is independent of the ESCRT machinery. Although the best studied factor in MVB formation is known to be depend on the ESCRT machinery, ESCRT-independent MVB formation or the exosome release pathway has also been reported in several studies. ESCRT-independent pathway involves the incorporation of ceramide into the endosomal membrane by nSMase activation (25, 26). Additionally, Rab GTPases play important roles in exosome production, being involved in the transport of MVBs to the plasma membrane and their release from the cell (27). Therefore, we first investigated whether HCV infection affects the ESCRT machinery or the Rab protein pathway. We found that HCV infection did not affect the Alix (encoding apoptosis-linked gene 2-interacting protein X, an ESCRT protein) and Rab27a (a member the RAS oncogene family) mRNA expression levels up to 5 days post-infection of HCV in Huh 7.5.1 cells (S2A and S2B Fig). These results indicated that the pathway of exosome release induced by HCV infection is not mediated by ESCRT and Rab27a-mediated pathways. Thus, we focused on the ESCRT-independent pathway of exosome release in HCV-infected hepatocytes.
It has been reported that ATP or calcium influx inducer treatment caused the MVBs formation and exosome release (14, 15). P2X receptors mediate calcium influx when activated by the binding of ATP (15). Qu et al. (16) and Turola et al. (28) reported that activation of purinergic receptor P2X triggered EVs shedding and release. Furthermore, a recent study showed that virus infection induced the release of ATP from cells in Panx1 dependent manner (29). In the context of these findings, we hypothesized that P2X activation by Panx1-induced ATP release would be the pathway responsible for exosome release during HCV infection. We measured P2X1, P2X4, and P2X7 expression in Huh 7.5.1 cells and found P2X4 expression was the highest (S2C Fig). Therefore, to elucidate the mechanism of exosome release, we further investigated whether HCV infection activates Panx1 and P2X4 channels. We found that HCV infection stimulated the release of ATP (Fig 3A) and calcium influx (Fig 3B), but dengue virus infection did not. In addition, HCV-infected Huh 7.5.1 cells showed significantly increased nSMase activation compared with uninfected cells, while dengue virus infection had no effect on such activation (Fig 3C). While nSMase activation was increased in HCV-infected cells (Fig 3C), intracellular CD63 expression decreased (Fig 1C and 1D). These data indicated that HCV infection stimulates both exosome formation and release.
Fig 3. HCV infection induces the activation of caspase-3, Panx1 and P2X4, but dengue virus infection does not.
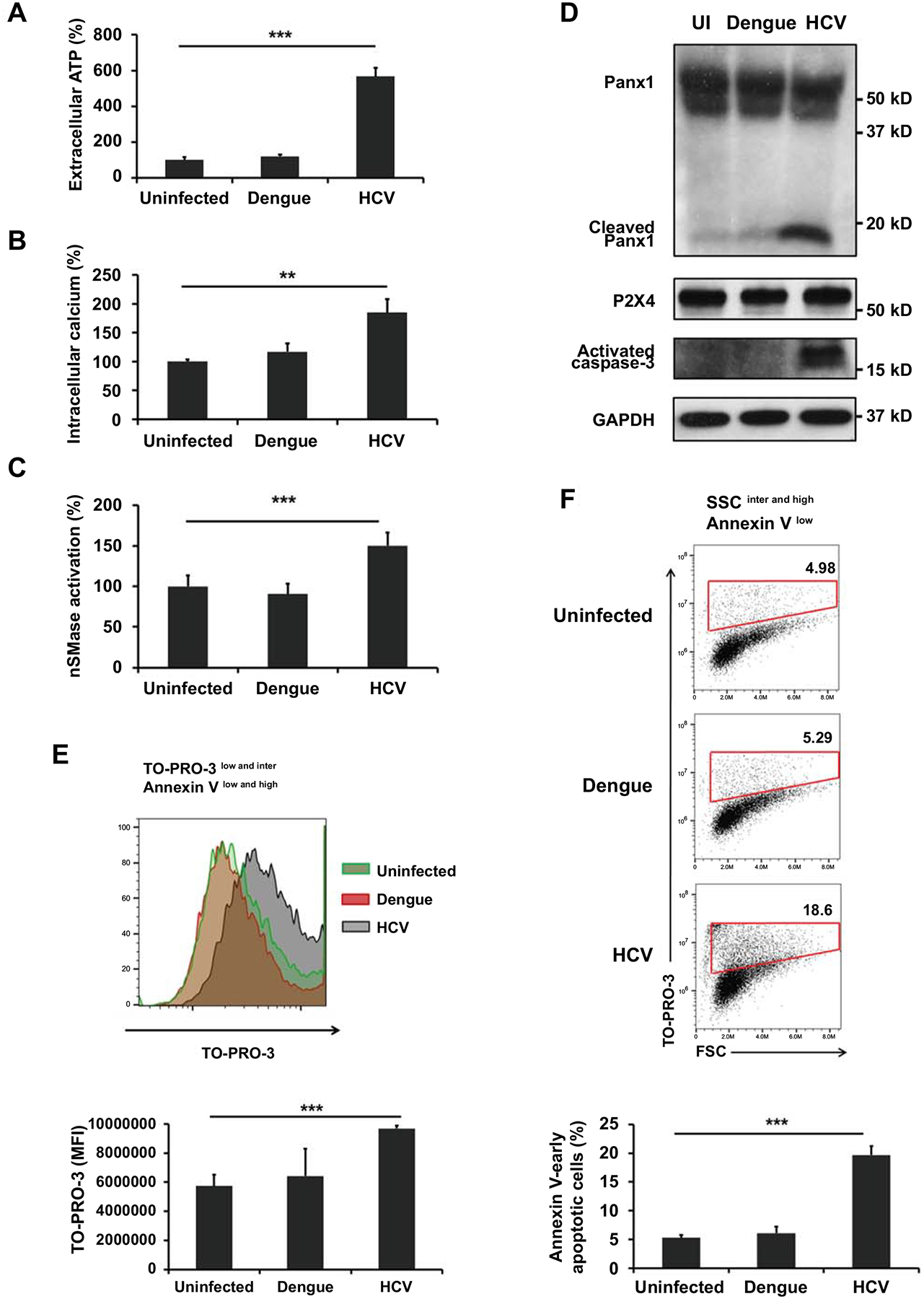
Huh 7.5.1 cells were infected with HCV or dengue virus at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium. After 24 h, (A) extracellular ATP levels were measured using a luminescent cell viability assay and (B) intracellular calcium levels were measured using a fluorometric calcium assay. (C) Neutral sphingomyelinase (nSMase) activity and (D) protein levels of Panx1, P2X4, activated caspase-3, and GAPDH were measured from cell lysates. Cells were stained with TO-PRO-3 and annexin V and determined by flow cytometry. (E) TO-PRO-3 uptake was measured in TO-PRO-3 low and intermediate/Annexin V low and high cells. (F) Annexin− early apoptotic cells were identified by forward scatter (FSC) intermediate and high/TO-PRO-3 high in side scatter (SSC) intermediates and high/annexin V low cells. Data are expressed as mean ± SD (n=3). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
Next, we measured the levels of cleaved Panx1 and caspase-3 using western blotting, because Panx1 can allow efflux of ATP by proteolysis via caspase-3 during the early apoptotic stage (30). As shown in Fig 3D, only HCV-infected Huh 7.5.1 cells contained both cleaved Panx1 and caspase-3. However, P2X4 protein levels did not change, even when calcium influx increased in HCV-infected cells. These results indicated that HCV infection affected the activation of P2X4 and induced the cleavage of Panx1 and caspase-3, but did not stimulate P2X4 protein synthesis.
A recent study proposed that Panx1 activated by caspase-mediated cleavage allows the entry of TO-PRO-3 (a very small size nucleic acid–binding dye) during the early apoptotic stage (31); therefore, we examined a TO-PRO-3 uptake and apoptosis status to confirm the activation Panx1 and the apoptosis stage. HCV infection induced an increase in TO-PRO-3 uptake significantly compared to uninfected cells and dengue virus infected cells (Fig 3E) in sorted viable and apoptotic cells (TO-PRO-3low and intermediate/Annexin Vlow and high) (S3 Fig). Additionally, Annexin V− early apoptotic cells contained high levels of TO-PRO-3 was significantly higher numbers in the HCV-infected cells compared with uninfected cells and dengue virus-infected cells (Fig 3F). The results indicated that caspase-3 activation during HCV infection (Fig 3D) is caused by not only late apoptosis but also early apoptosis. We suggest that HCV infection can leads to caspase-3-mediated Panx1 cleavage and P2X4 activation during early stages of apoptosis.
The secretions of exosomes and exosomal miRNAs are dependent on the activation of caspase-3 in HCV-infected hepatocytes
In a study by Böing et al., MCF-7 cells transfected with caspase-3 showed an approximately 5-fold increase in EVs release compared with that from untransfected cells (32). Thus, we treated HCV-infected Huh 7.5.1 cells with caspase-3 inhibitors, z-VAD-fmk and z-DQMD-fmk, to investigate whether the secretions of exosomes and exosomal miRNAs are dependent on the activation of caspase-3 during HCV infection. We found that these inhibitors did not affect HCV replication (Fig 4A). However, the caspase-3 inhibitors suppressed extracellular ATP (Fig 4B) and intracellular calcium levels (Fig 4C) during HCV infection. Western blotting result shows that cleaved Panx1 was also decreased when activated caspase-3 was suppressed by inhibitor treatment during HCV infection (Fig 4D). In addition, we found that caspase-3 inhibitor treatment caused an increase in intracellular CD63 expression (Fig 4D) and a decrease in the extracellular exosome protein content (Fig 4E) in HCV-infected Huh 7.5.1 cells. These results suggested that caspase-3 activation stimulates Panx1 cleavage and exosome release during HCV infection.
Fig 4. The secretions of exosomes and exosomal miRNAs are dependent on the activation of caspase-3 in HCV-infected hepatocytes.
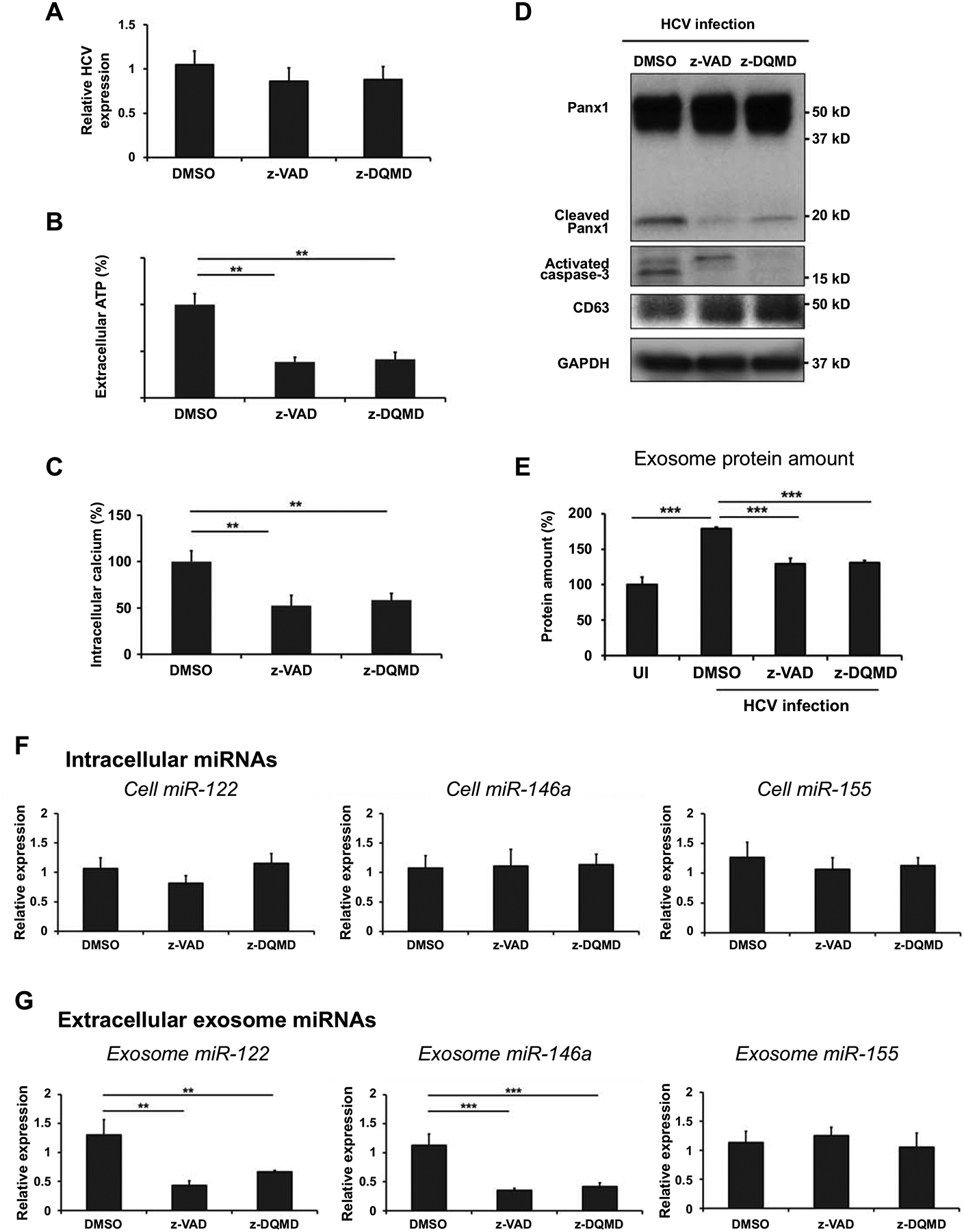
Huh 7.5.1 cells were infected with HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium, and 20 μM z-VAD-fmk or z-DQMD-fmk dissolved in DMSO was added. After 24 h, exosomes were isolated from cell supernatants using ExoQuick-TC solution, and miRNAs were extracted from cells and exosomes. (A) HCV RNA level in the cells was analyzed by qRT-PCR and HPRT was used as an endogenous control for normalization. (B) Extracellular ATP levels in cell supernatant were measured using a luminescent cell viability assay and (C) intracellular calcium levels were measured using a fluorometric calcium assay. (D) Protein levels of Panx1, activated caspase-3, CD63, and GAPDH were measured in the cell lysates. (E) Exosome protein contents were measured using the BCA assay. (F) Intracellular miRNAs and (G) extracellular exosomal miRNAs were extracted using a miRNeasy mini kit and miRNA expression was analyzed by qRT-PCR. U18 was used as an endogenous control in cells for normalization, and Caenorhabditis elegans miR-39 was used as an exogenous control for normalization in exosomes. Data are expressed as mean ± SD (n=3). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
Next, we measured miRNAs expression in released exosomes and intracellular levels to determine whether caspase-3 activation affects exosomal miRNAs release in HCV-infected hepatocytes. Fig 4F and 4G show that the caspase-3 inhibitors suppressed exosomal miR-122 and miR-146a levels (Fig 4G) but did not affect intracellular miR-122 and miR-146a expression (Fig 4F) in HCV-infected Huh 7.5.1 cells. However, exosomal and intracellular miR-155 levels were affected by caspase-3 inhibitors in HCV-infected Huh 7.5.1 cells (Fig 4F and 4G). Thus, these observations indicate that caspase-3 activation causes the secretion of exosomal specific miRNAs during HCV infection.
The secretions of exosomes and exosomal miRNAs are dependent on the activation of Panx1 and P2X4 in HCV-infected hepatocytes
In order to demonstrate the role of Panx1 and P2X4, we treated the cells with pharmacological blockers and transfected Panx1 short interfering RNAs (siRNAs; siPanx1) or P2X4 siRNAs (siP2X4) into HCV-infected Huh 7.5.1 cells. We first tested the effects of pharmacological Panx1 inhibitors, including CBX, probenecid, and 10Panx-1 (a Panx-1 mimetic inhibitor) on Panx1 cleavage during HCV infection (S4A Fig). We found that only CBX inhibited Panx1 cleavage in HCV-infected Huh 7.5.1 cells; therefore, we used CBX as the Panx1 inhibitor in subsequent experiments. Treatment with pharmacological inhibitors for Panx1 (CBX) and P2X4 (BX430) and siRNA transfection did not affect HCV replication in HCV-infected Huh 7.5.1 cells (Fig 5A). CBX treatment and siPanx1 transfection inhibited ATP release (Fig 5B), calcium influx (Fig 5C), TO-PRO-3 uptake (Fig 5E), and the level of cleaved Panx1 (Fig 5F) in HCV-infected Huh 7.5.1 cells. Unexpectedly, we found that CBX treatment also induced a decrease in the protein level of activated caspase-3 (Fig 5F). CBX treatment also induced the suppression of caspase activation (33); therefore, we could not determine whether the suppression of Panx1 cleavage was caused directly by CBX treatment or indirectly by inhibition of caspase-3 activation. nSMase activation (Fig 5D) and the exosome protein content (Fig 5G) were suppressed and the intracellular CD63 level (Fig 5F) was increased by CBX treatment and siPanx1 transfection in HCV-infected Huh 7.5.1 cells, which suggested that Panx1 activation affects both exosome formation and release. Both P2X4 pharmacological inhibition and siP2X4 transfection were associated with significant decreases in calcium influx (Fig 5C), nSMase activation (Fig 5D), and exosome protein content (Fig 5G), and an increase in intracellular CD63 expression (Fig 5F) in HCV-infected Huh 7.5.1 cells. BX430 treatment did not reduce the level of P2X4 (Fig 5F), but did suppress calcium influx, which showed that BX430 treatment affects P2X4 activation but not its protein expression. ATP release (Fig 5B), TO-PRO-3 uptake (Fig 5E), and caspase-3 activation (Fig 5F) were not altered by P2X4 treatment and siP2X4 transfection, suggesting that activation of Panx1 and caspase-3 is upstream of P2X4 activation in the signaling pathway during HCV infection.
Fig 5. The secretions of exosomes and exosomal miRNAs are dependent on the activation of Panx1 and P2X4 in HCV-infected hepatocytes.

Huh 7.5.1 cells were infected with HCV at 0.1 MOI and medium (NT, non-treatment), 20 μM carbenoxolone (CBX) (Panx-1 inhibitor) dissolved in medium, DMSO or 5 μM BX430 (P2X4 inhibitor) dissolved in DMSO were added every day. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with CBX or BX430. After 24 h, exosomes were isolated from cell supernatants using ExoQuick-TC solution, and miRNAs were extracted from cells and exosomes. Huh 7.5.1 cells were transfected with a negative control (NC), Panx1 short interfering RNA (siRNA) (siPnax1) or a P2X4 siRNA (siP2X4) at 1 day post infection. The culture medium was replaced with exosome-depleted FBS medium at 3 days post infection. After 24 h, exosomes were isolated from cell supernatants using ExoQuick-TC solution, and miRNAs were extracted from cells and exosomes. (A) HCV RNA level was analyzed by qRT-PCR and HPRT was used as an endogenous control for normalization. (B) Extracellular ATP levels in cell supernatants were measured using a luminescent cell viability assay and (C) intracellular calcium levels were measured using a fluorometric calcium assay. (D) Neutral sphingomyelinase (nSMase) activity and (F) protein levels of Panx1, P2X4, activated caspase-3, CD63, and GAPDH were measured in the cell lysates. (E) TO-PRO-3 uptake was measured by flow cytometry. (G) Exosome protein contents were measured by the BCA assay. (H) Extracellular exosomal miRNAs were extracted using a miRNeasy mini kit and miRNA expression was analyzed by qRT-PCR. Caenorhabditis elegans miR-39 was used as an exogenous control for normalization in exosomes. Data are expressed as mean ± SD (n=3). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
In addition, inhibition of Panx1 and P2X4 by pharmacological inhibitors and siRNAs transfection suppressed the levels of miR-122 and miR-146a in exosomes (Fig 5H) while they did not change the intracellular expression of miR-122 and miR-146a in HCV-infected Huh 7.5.1 cells (S4B Fig). These results identify a novel pathway for the release of exosome and exosomal miRNAs via Panx1/P2X4 activation during HCV infection.
ATP treatment induces the secretions of exosomes and exosomal miRNAs.
To assess whether ATP induces exosome release via P2X4 activation, we treated Huh 7.5.1 cells with different concentrations of ATP. ATP induced calcium influx in a dose dependent manner (Fig 6A). In addition, ATP treatment increased nSMase activation (Fig 6B) and the release of exosomes (Fig 6E), and decreased the intracellular CD63 level (Fig 6C and 6D) in Huh 7.5.1 cells. However, ATP treatment did not affect Panx1 cleavage and caspase activation (Fig 6C) in Huh 7.5.1 cells, which suggested that Panx1 cleavage and caspase-3 activation are not dependent on the extracellular ATP level. ATP treatment also increased the levels of miR-122, miR-146a, and miR-155 in exosomes from Huh 7.5.1 cells (Fig 6G), while not altering intracellular miRNA expression (Fig 6F). These results demonstrated that extracellular ATP induces the secretions of exosomes and exosomal miRNAs in normal cells.
Fig 6. ATP treatment induces the secretions of exosomes and exosomal miRNAs.

Huh 7.5.1 cells were treated ATP at 1 mM and 5 mM for 2 h. Thereafter, (A) intracellular calcium levels were measured using a fluorometric calcium assay. (B) Neutral sphingomyelinase (nSMase) activity and (C) protein levels of CD63, Panx1, P2X4, activated caspase-3, and GAPDH were measured in cell lysates. (D) Intracellular CD63 levels were measured by flow cytometry. Huh 7.5.1 cells were treated with ATP 5 mM, and then exosomes were isolated from cell supernatant using ExoQuick-TC solution after 2 h. (E) Protein contents of the isolated exosomes was measured using a BCA assay. (F) Intracellular miRNAs and (G) extracellular exosomal miRNAs were extracted using a miRNeasy mini kit and miRNA expression was analyzed by qRT-PCR. U18 was used as an endogenous control in cells for normalization, and Caenorhabditis elegans miR-39 was used as an exogenous control for normalization in exosomes. Data are expressed as mean ± SD (n=3). **P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
The secretion of exosomes induced by HCV infection is not dependent on ROCK1.
Caspase-3 cleaves and activates ROCK1, which leads to plasma membrane blebbing and release of EVs in the pro-apoptotic stage (34, 35). Morelli et al. reported that extracellular ATP mediated P2X7 activation caused membrane blebbing in a ROCK1 activation-dependent manner (35). Thus, we investigated whether ROCK1 cleavage is necessary to stimulate exosome release during HCV infection. We determined whether HCV infection induces ROCK1 cleavage. As shown in Fig 7A, HCV infection resulted in ROCK1 cleavage in Huh 7.5.1 cells, as expected. However, Panx1 cleavage, caspase-3 activation, intracellular CD63 expression (Fig 7B), and exosome release (Fig 7C) did not change when HCV-infected Huh 7.5.1 cells were treated with Y-27632, a pharmacological blocker of ROCK1 activity. These results indicated that HCV infection-mediated exosome release is not dependent on ROCK1 activation.
Fig 7. The secretion of exosomes induced by HCV infection is not dependent on ROCK1.

Huh 7.5.1 cells were infected with HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium and 50 μM Y27632 was added as a ROCK1 blocker. After 24 h, (A,B) the protein levels of ROCK1, Panx1, activated caspase-3, CD63, and GAPDH in cell lysates were measured using western blotting. (C) Exosomes were isolated from cell supernatants using ExoQuick-TC solution, and the exosome protein content was measured by a BCA assay. Data are expressed as mean ± SD (n=3). *P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
Discussion
Exosomes are released from almost every cell, not only normal cells, but also from diseased cells. Several reports have demonstrated that exosome composition, including proteins and miRNAs, is affected by the origin and physiological state of the cells, and can alter the physiological processes in the recipient cells after uptake (36). These characteristics have resulted in an explosion of interest in exosomes research, which has focused on potential biomarkers for the diagnosis and prognosis of diseases (37). In addition, recent studies have identified factors that caused the release exosome and that altered the exosome composition including miRNAs (38, 39). Although the MVBs formation for exosomes release is known to depend on the ESCRT machinery (25), we found Alix expression was unchanged up to 5 days post-infection of HCV in Huh 7.5.1 cells. Therefore, in the present study, we determined the effect of HCV infection on the ESCRT-independent pathway of exosome release in hepatocytes.
Numerous recent studies have demonstrated that miRNAs can affect physiological processes in cells, thus, the altered levels of circulating miRNAs packaged as exosomes can be used as candidate biomarkers for many diseases. Importantly, it has demonstrated that exosomal miRNAs levels do not always reflect intracellular miRNAs expression, and cells and exosomes have different enriched miRNAs repertoires (39, 40). The mechanisms that control miRNAs loading into exosomes are unknown; however, miRNAs are not randomly loaded into exosomes, and certain cellular responses to stress signals can lead to the release of exosomes loaded with a specific repertoire of miRNAs (37, 39, 40). In the present study, we found that HCV infection dramatically altered the secretion of miR-122 and miR-146a into exosomes from hepatocytes. miR-122 is the most abundant miRNA in the liver and contributes to stability, translation, and replication of HCV via its direct binding to the 5′ UTR of HCV RNA (13). In addition, Bandiera et al. observed an increased miR-146a level in HCV-infected hepatocytes and its function on HCV replication and pathogenesis of liver disease and HCC development (41). In this study, we found that inhibition of caspase-3, Panx1 and P2X4 suppressed the secretions of exosomes and exosomal miR-122 and miR-146a in HCV-infected hepatocytes. Thus, we hypothesized that one of the possible mechanisms of exosome release and controlling miRNAs loading into exosomes in HCV-infected hepatocytes is associated with caspase-3-mediated Panx1 and P2X4 activation.
The function of Panx1 has been studied in relation to the release of interleukin-1β in innate immunity (42), find-me signals during the early stage of apoptosis (30), HIV infection (43), cellular migration through ATP release, and interaction with purinergic receptors (42). In this study, we provided evidence of Panx1 function in the secretions of exosomes and exosomal miRNAs during HCV infection. We showed that the Panx1/P2X4 pathway and nSMase activation were stimulated, while Alix and Rab27a expression were unchanged during HCV infection.
EVs contain microvesicles, apoptotic bodies, exosomes, and other intracellular constituents (44), and are secreted from inside cell into the extracellular space. Rab27a is shown to be involved in EVs secretion by regulating membrane fusion and exocytosis (45). Although Rab27a has been reported to play a role in releasing of HCV EVs (46), it is not clear that Rab27a is involved in secretion of HCV exosome, a part of EVs. Moreover, Chen et al. reported that Rab27a affects HCV replication independent from its role in exosome secretion (47). Our studies indicate that secretion of HCV exosome occurs in part by panx1/P2X4 pathway.
We showed that the inhibition of caspase-3/Panx1/P2X4 pathway suppressed the secretions of exosomes and exosomal miRNAs during HCV infection. Moreover, HCV infection increased TO-PRO-3 uptake in not only Annexin V+ cells but also Annexin V- cells, suggesting that caspase-3 activation-mediated Panx1 cleavage was not caused by apoptosis during HCV infection. However, we found that dengue virus infection did not affect exosome release or caspase-3/Panx1/P2X4 pathway activation even HCV and dengue virus are both members of the family Flaviviridae and share many features in their viral life cycles. Corrêa et al. shown that dengue virus infection caused an increase in P2X7 mRNA expression in human monocytes (48), but our experiment system did not find the changes of P2X4 protein level and calcium influx in dengue virus-infected Huh 7.5.1 cells. HCV and dengue virus have different functional properties of replication and assembly sites and the molecular strategies underlying RNA replication and virus production in the cells (22). We hypothesized that one of the possible reasons of different effect in exosome release is the different molecular mechanism of RNA replication and virus production which could affect the Panx1-mediated exosome biogenesis pathway.
We showed that the increased exosome release was not dependent on ROCK1 cleavage in HCV-infected hepatocytes. Hirsova et al. reported that lipotoxicity increased the release of EVs from hepatocytes depending on ROCK1, which can be cleaved by caspase-3 like Panx1. However, they found that inhibitor of exosome release did not affect the EVs release amount in lipotoxicity-induced hepatocytes (34). We did not confirm the change in MVs amount during HCV infection, but could assume that ROCK1 cleavage can be associated with MVs release rather than exosome release during caspase-3 activation while Panx1 cleavage causes P2X4 activation-mediated exosome release (Fig 8).
Fig 8.

A schematic diagram showing the possible mechanism of regulation of exosome release associated with the caspase-3/Panx-1/P2X4 pathway during HCV infection in hepatocytes.
In summary, we investigated the exosome biogenesis pathway during HCV infection in hepatocytes and found that HCV infection stimulated exosome release and miR-122 and miR-146a loading into exosomes, dependent on the caspase-3/Panx1/P2X4 pathway. This result contributes to understanding of the mechanism underlying exosome release during HCV infection. Future studies will investigate the effect of Panx1 suppression in HCV-infected hepatocytes in the development of liver disease via exosome-mediated intercellular communication which can identify the mechanism of exosome-mediated pathogenesis of liver disease.
Supplementary Material
S1 Fig. HCV and dengue virus RNA expression in hepatocytes. (A) Huh 7.5.1 cells were infected HCV at 0.1 MOI and RNAs were extracted every day for 5 days. (B) Huh 7.5.1 cells were infected with HCV or dengue virus at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium. After 24 h, RNAs were extracted. Primary human hepatocytes were exposed to healthy or HCV-infected patient sera and RNAs were extracted after 12 days. HCV or dengue virus RNA levels in the cells were analyzed by qRT-PCR and HPRT was used as an endogenous control for normalization.
S2 Fig. Huh 7.5.1 cells were infected HCV at 0.1 MOI and (A) Alix and (B) Rab27a mRNA were extracted and analyzed every day for 5 days. (C) P2X1, P2X4 and P2X7 expressions in normal Huh 7.5.1 cells were analyzed by qRT-PCR and HPRT was used as an endogenous control for normalization.
S3 Fig. Flow cytometry gating strategy for separation of different population of cells undergoing apoptosis. Huh 7.5.1 cells were infected HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium. After 24 h, cells were stained with TO-PRO-3 and annexin V and determined by flow cytometry.
S4 Fig. The effects of pharmacological blockers of Panx1 on the Panx1 cleavage during HCV infection. (A) Huh 7.5.1 cells were infected with HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with CBX (carbenoxolone), Probenocid (Pro), 10Panx (Panx-1 mimetic inhibitor) or Scrambled 10Panx. After 24 h, protein levels of Panx1 and GAPDH were measured in the cell lysates. (B) Huh 7.5.1 cells were infected with HCV at 0.1 MOI and medium (NT, non-treatment), 20 μM CBX dissolved in medium, DMSO or 5 μM BX430 (P2X4 inhibitor) dissolved in DMSO were added every day. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with CBX or BX430. After 24 h, miRNAs were extracted from cells. Huh 7.5.1 cells were transfected with a negative control (NC), Panx1 short interfering RNA (siRNA) (siPnax1) or a P2X4 siRNA (siP2X4) at 1 day post infection. The culture medium was replaced with exosome-depleted FBS medium at 3 days post infection. After 24 h, miRNAs were extracted from cells. Intracellular miRNAs were extracted using a miRNeasy mini kit and miRNA expression was analyzed by qRT-PCR. U18 was used as an endogenous control in cells for normalization. Data are expressed as mean ± SD (n=3). **P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
S1 Table. List of antibody used in this study.
S2 Table. Origin and primer sequences used for qPCR.
Acknowledgments
The authors thank the members of the Hahn laboratory for providing advice and constructive criticism on this work.
Financial Support
This work was supported by U.S. National Institutes of Health (NIH) Grant DK122737 (to Y.S.H.).
Abbreviations
- ATP
adenosine triphosphate
- BCA
bicinchoninic acid
- CBX
carbenoxolone
- DMEM
Dulbecco’s modified Eagle’s medium
- ESCRT
endosomal sorting complex required for transport
- EVs
extracellular vesicles
- FBS
fetal bovine serum
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- ILV
intraluminal vesicles
- miRNAs
microRNAs
- MVBs
multivesicular bodies
- MVs
microvesicles
- nSMase
neutral sphingomyelinase
- Panx1
pannexin 1
- PHHs
primary human hepatocytes
- qRT-PCR
quantitative real-time reverse transcription polymerase chain reaction
- siRNA
short interfering RNA
References
- 1.Ratajczak MZ, Ratajczak D, Pedziwiatr D. Extracellular Microvesicles (ExMVs) in Cell to Cell Communication: A Role of Telocytes. Adv Exp Med Biol. 2016;913:41–9. [DOI] [PubMed] [Google Scholar]
- 2.Sluijter JPG, Verhage V, Deddens JC, van den Akker F, Doevendans PA. Microvesicles and exosomes for intracardiac communication. Cardiovasc Res. 2014;102(2):302–11. [DOI] [PubMed] [Google Scholar]
- 3.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78(9):838–48. [DOI] [PubMed] [Google Scholar]
- 4.Fu YX, Zhang L, Zhang F, Tang T, Zhou Q, Feng CH, et al. Exosome-mediated miR-146a transfer suppresses type I interferon response and facilitates EV71 infection. Plos Pathog. 2017;13(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schorey JS, Cheng Y, Singh PP, Smith VL. Exosomes and other extracellular vesicles in host-pathogen interactions. Embo Rep. 2015;16(1):24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandiera S, Bian CB, Hoshida Y, Baumert TF, Zeisel MB. Chronic hepatitis C virus infection and pathogenesis of hepatocellular carcinoma. Curr Opin Virol. 2016;20:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30(17):1969–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. Exosome Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells (vol 91, e02225–16, 2017). J Virol. 2017;91(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng LB, Jiang W, Wang XN, Merz A, Hiet MS, Chen YJ, et al. Syntenin regulates hepatitis C virus sensitivity to neutralizing antibody by promoting E2 secretion through exosomes. J Hepatol. 2019;71(1):52–61. [DOI] [PubMed] [Google Scholar]
- 10.Saha B, Kodys K, Adejumo A, Szabo G. Circulating and Exosome-Packaged Hepatitis C Single-Stranded RNA Induce Monocyte Differentiation via TLR7/8 to Polarized Macrophages and Fibrocytes. J Immunol. 2017;198(5):1974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobb DA, Kim OK, Golden-Mason L, Rosen HR, Hahn YS. Hepatocyte-derived exosomes promote T follicular regulatory cell expansion during hepatitis C virus infection. Hepatology. 2018;67(1):71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, et al. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90. Plos Pathog. 2014;10(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278(22):20083–90. [DOI] [PubMed] [Google Scholar]
- 15.Takenouchi T, Tsukimoto M, Iwamaru Y, Sugama S, Sekiyama K, Sato M, et al. Extracellular ATP induces unconventional release of glyceraldehyde-3-phosphate dehydrogenase from microglial cells. Immunol Lett. 2015;167(2):116–24. [DOI] [PubMed] [Google Scholar]
- 16.Qu Y, Dubyak GR. P2X7 receptors regulate multiple types of membrane trafficking responses and non-classical secretion pathways. Purinerg Signal. 2009;5(2):163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandilos JK, Chiu YH, Chekeni FB, Armstrong AJ, Walk SF, Ravichandran KS, et al. Pannexin 1, an ATP Release Channel, Is Activated by Caspase Cleavage of Its Pore-associated C-terminal Autoinhibitory Region. J Biol Chem. 2012;287(14):11303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woehrle T, Yip L, Elkhal A, Sumi Y, Chen Y, Yao YL, et al. Pannexin-1 hemichannel-mediated ATP release together with P2X1 and P2X4 receptors regulate T-cell activation at the immune synapse. Blood. 2010;116(18):3475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manzoor S, Idrees M, Ashraf J, Mehmood A, Butt S, Fatima K, et al. Identification of ionotrophic purinergic receptors in Huh-7 cells and their response towards structural proteins of HCV genotype 3a. Virol J. 2011;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty Acid and Endotoxin Activate Inflammasomes in Mouse Hepatocytes that Release Danger Signals to Stimulate Immune Cells. Hepatology. 2011;54(1):133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambrecht J, Poortmans PJ, Verhulst S, Reynaert H, Mannaerts I, van Grunsven LA. Circulating ECV-Associated miRNAs as Potential Clinical Biomarkers in Early Stage HBV and HCV Induced Liver Fibrosis. Front Pharmacol. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatel-Chaix L, Bartenschlager R. Dengue Virus- and Hepatitis C Virus- Induced Replication and Assembly Compartments: the Enemy Inside-Caught in the Web. J Virol. 2014;88(11):5907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation (vol 5, pg 522 2004). Nat Rev Genet. 2004;5(8):522–+. [DOI] [PubMed] [Google Scholar]
- 24.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, Polchi A, et al. Signaling Pathways in Exosomes Biogenesis, Secretion and Fate. Genes-Basel. 2013;4(2):152–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo BB, Bellingham SA, Hill AF. The Neutral Sphingomyelinase Pathway Regulates Packaging of the Prion Protein into Exosomes. J Biol Chem. 2015;290(6):3455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Campbell EC, Lucocq J, Riches A, Powis SJ. Monitoring the Rab27 associated exosome pathway using nanoparticle tracking analysis. Exp Cell Res. 2013;319(12):1706–13. [DOI] [PubMed] [Google Scholar]
- 28.Turola E, Furlan R, Bianco F, Matteoli M, Verderio C. Microglial microvesicle secretion and intercellular signaling. Front Physiol. 2012;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang CF, He HW, Wang L, Zhang N, Huang HJ, Xiong QQ, et al. Virus-Triggered ATP Release Limits Viral Replication through Facilitating IFN-beta Production in a P2X7-Dependent Manner. J Immunol. 2017;199(4):1372–81. [DOI] [PubMed] [Google Scholar]
- 30.Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467(7317):863–U136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang LZ, Tixeira R, Caruso S, Atkin-Smith GK, Baxter AA, Paone S, et al. Monitoring the progression of cell death and the disassembly of dying cells by flow cytometry. Nat Protoc. 2016;11(4):655–63. [DOI] [PubMed] [Google Scholar]
- 32.Boing AN, Stap J, Hau CM, Afink GB, Ris-Stalpers C, Reits EA, et al. Active caspase-3 is removed from cells by release of caspase-3-enriched vesicles. Bba-Mol Cell Res. 2013;1833(8):1844–52. [DOI] [PubMed] [Google Scholar]
- 33.de Pina-Benabou MH, Szostak V, Kyrozis A, Rempe D, Uziel D, Urban-Maldonado M, et al. Blockade of gap junctions in vivo provides neuroprotection after perinatal global ischemia. Stroke. 2005;36(10):2232–7. [DOI] [PubMed] [Google Scholar]
- 34.Hirsova P, Ibrahim SH, Krishnan A, Verma VK, Bronk SF, Werneburg NW, et al. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology. 2016;150(4):956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morelli A, Chiozzi P, Chiesa A, Ferrari D, Sanz JM, Falzoni S, et al. Extracellular ATP causes ROCK I-dependent bleb formation in P2X7-transfected HEK293 cells. Mol Biol Cell. 2003;14(7):2655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael A, Bajracharya SD, Yuen PST, Zhou H, Star RA, Illei GG, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16(1):34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corrado C, Raimondo S, Chiesi A, Ciccia F, De Leo G, Alessandro R. Exosomes as Intercellular Signaling Organelles Involved in Health and Disease: Basic Science and Clinical Applications. Int J Mol Sci. 2013;14(3):5338–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Chen RJ, Kemper S, Brigstock DR. Pathways of production and delivery of hepatocyte exosomes. J Cell Commun Signal. 2018;12(1):343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, et al. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016;17(3):799–808. [DOI] [PubMed] [Google Scholar]
- 40.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandiera S, Pernot S, El Saghire H, Durand SC, Thumann C, Crouchet E, et al. Hepatitis C Virus-Induced Upregulation of MicroRNA miR-146a-5p in Hepatocytes Promotes Viral Infection and Deregulates Metabolic Pathways Associated with Liver Disease Pathogenesis. J Virol. 2016;90(14):6387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanjanamekanant K, Luckprom P, Pavasant P. P2X7 receptor-Pannexin1 interaction mediates stress-induced interleukin-1 beta expression in human periodontal ligament cells. J Periodontal Res. 2014;49(5):595–602. [DOI] [PubMed] [Google Scholar]
- 43.Orellana JA, Velasquez S, Williams DW, Saez JC, Berman JW, Eugenin EA. Pannexin1 hemichannels are critical for HIV infection of human primary CD4(+) T lymphocytes. J Leukocyte Biol. 2013;94(3):399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanez-Mo M, Siljander PRM, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–U61. [DOI] [PubMed] [Google Scholar]
- 46.Grunvogel O, Colasanti O, Lee JY, Kloss V, Belouzard S, Reustle A, Esser-Nobis K, et al. Secretion of Hepatitis C Virus Replication Intermediates Reduces Activation of Toll-Like Receptor 3 in Hepatocytes. Gastroenterology. 2018;154:2237–+. [DOI] [PubMed] [Google Scholar]
- 47.Chen TC, Hsieh CH, Sarnow P. Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect. Plos Pathog. 2015;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Correa G, Lindenberg CD, Fernandes-Santos C, Gandini M, Paiva FP, Coutinho-Silva R, Kubelka CF. The purinergic receptor P2X7 role in control of Dengue virus-2 infection and cytokine/chemokine production in infected human monocytes. Immunobiology 2016;221:794–802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 Fig. HCV and dengue virus RNA expression in hepatocytes. (A) Huh 7.5.1 cells were infected HCV at 0.1 MOI and RNAs were extracted every day for 5 days. (B) Huh 7.5.1 cells were infected with HCV or dengue virus at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium. After 24 h, RNAs were extracted. Primary human hepatocytes were exposed to healthy or HCV-infected patient sera and RNAs were extracted after 12 days. HCV or dengue virus RNA levels in the cells were analyzed by qRT-PCR and HPRT was used as an endogenous control for normalization.
S2 Fig. Huh 7.5.1 cells were infected HCV at 0.1 MOI and (A) Alix and (B) Rab27a mRNA were extracted and analyzed every day for 5 days. (C) P2X1, P2X4 and P2X7 expressions in normal Huh 7.5.1 cells were analyzed by qRT-PCR and HPRT was used as an endogenous control for normalization.
S3 Fig. Flow cytometry gating strategy for separation of different population of cells undergoing apoptosis. Huh 7.5.1 cells were infected HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium. After 24 h, cells were stained with TO-PRO-3 and annexin V and determined by flow cytometry.
S4 Fig. The effects of pharmacological blockers of Panx1 on the Panx1 cleavage during HCV infection. (A) Huh 7.5.1 cells were infected with HCV at 0.1 MOI. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with CBX (carbenoxolone), Probenocid (Pro), 10Panx (Panx-1 mimetic inhibitor) or Scrambled 10Panx. After 24 h, protein levels of Panx1 and GAPDH were measured in the cell lysates. (B) Huh 7.5.1 cells were infected with HCV at 0.1 MOI and medium (NT, non-treatment), 20 μM CBX dissolved in medium, DMSO or 5 μM BX430 (P2X4 inhibitor) dissolved in DMSO were added every day. After 3 days, the culture medium was replaced with exosome-depleted FBS medium with CBX or BX430. After 24 h, miRNAs were extracted from cells. Huh 7.5.1 cells were transfected with a negative control (NC), Panx1 short interfering RNA (siRNA) (siPnax1) or a P2X4 siRNA (siP2X4) at 1 day post infection. The culture medium was replaced with exosome-depleted FBS medium at 3 days post infection. After 24 h, miRNAs were extracted from cells. Intracellular miRNAs were extracted using a miRNeasy mini kit and miRNA expression was analyzed by qRT-PCR. U18 was used as an endogenous control in cells for normalization. Data are expressed as mean ± SD (n=3). **P < 0.05, ** P < 0.01 and *** P < 0.001 were considered statistically significant, as assessed using an unpaired student t-test.
S1 Table. List of antibody used in this study.
S2 Table. Origin and primer sequences used for qPCR.


