Abstract
The discovery that androgens play an important role in the progression of prostate cancer (PCa) led to the development of androgen deprivation therapy (ADT) as a first line of treatment. However, paradoxical growth inhibition has been observed in a subset of PCa upon administration of supraphysiological levels of testosterone (SupraT), both experimentally and clinically. Here we report that SupraT activates cytoplasmic nucleic acid sensors and induces growth inhibition of SupraT-sensitive PCa cells. This was initiated by the induction of two parallel autophagy-mediated processes, namely, ferritinophagy and nucleophagy. Consequently, autophagosomal DNA activated nucleic acid sensors that converge on NF-κB to drive immune signaling pathways. Chemokines and cytokines secreted by the tumor cells in response to SupraT resulted in increased migration of cytotoxic immune cells to tumor beds in xenograft models and patient tumors. Collectively, these findings indicate that SupraT may inhibit a subset of PCa by activating nucleic acid sensors and downstream immune signaling.
Keywords: Prostate cancer, NF-kappaB, Ferroptosis, Autophagy
Introduction
In 1941, Charles Huggins discovered the benefits of androgen deprivation therapy, which has become the mainstay for advanced prostate cancer (PCa) treatment (1). However, from the outset, it was recognized that all men eventually develop castration-resistant prostate cancer (CRPC) (2). Evaluation of clinical specimens demonstrates that CRPC cells remain highly reliant on Androgen receptor (AR) signaling (3, 4). These studies suggested that the adaptive reliance on AR signaling by CRPC cells becomes a therapeutic liability that can be exploited through the administration of SupraT, a concept we have termed bipolar androgen therapy (BAT) (5, 6). In this regard, we and others have demonstrated that the growth of some AR-positive human PCa cells can be inhibited by exposure to SupraT (6). The mechanisms underlying this paradoxical effect of SupraT on PCa cells are likely multifactorial as the androgen is the key mediator of prostate cancer cell metabolism, proliferation and death. Haffner et al. and others showed that androgens generate double-strand DNA breaks (DSBs) in PCa cells through the recruitment of AR and topoisomerase II beta to androgen response elements (7, 8). Hypothetically, in prostate tumors with DNA repair mutations, SupraT-induced DSBs would trigger DNA repair stress and lead to either growth inhibition or cell death (9). Intuitively, these tumors would be acutely susceptible to BAT. In agreement with this postulate, we have recently discovered an association between germline and/or somatic DNA repair gene mutations and favorable response to BAT (10–12). A link between DNA repair gene mutation or transcriptome repression and response to BAT has also been reported by others (13, 14). In this study, we report SupraT induces ferroptosis and nucleophagy-mediated immune activation resulting in growth inhibition of PCa.
Material & Methods
Cell culture
LNCaP, LAPC4, HEK293T, and NK92 cells were purchased from the American Type Culture Collection (ATCC). LNCaP, LAPC4, and 22Rv1 cells were cultured in phenol red-free RPMI (Thermo Fisher), and HEK293T cells were cultured in DMEM-high glucose (Sigma) supplemented with 10% FBS (Gemini Bio). VCaP cells were cultured in DMEM media (ATCC) containing 1.5 gram/L sodium-bi-carbonate. NK92 cells were cultured in CTS™ AIM V® SFM (Thermo Fisher) with 200 U/mL recombinant IL-2 (Peprotech), 12.5% horse serum (Thermo Fisher), and 12.5% FBS according to manufacturer’s instructions. All cell lines were tested for mycoplasma using the PCR based mycoplasma detection kit (Agilent).
Knockout/knockdown cell line generation
To generate the gene knockouts, tetracycline induced cas9 vector (Addgene # 50661) was stably expressed in prostate cancer lines. sgRNAs against target genes given in were cloned in pLXsgRNA vector (Addgene# 50662), and lentiviral particles for sgRNA were produced in HEK293T cells by co-transfecting pLXsgRNA plasmid with pMD2.G (Addgene# 12259) and psPAX2 (Addgene #12260). (Supplementary Table S1A) Viruses were harvested after 48h, and cells were infected with lentiviral particles. Transduced cells were treated with 1 μg/mL doxycycline to induce cas9 before selection with 10 μg/mL blasticidin. Following selection, cells were transferred in 96-well plates to select individual clones. Knockouts were verified by western blots, and confirmed clones were expanded and cryopreserved for future experiments.
For NCOA4 knockdown in LNCaP cells, MISSION shRNA constructs (TRCN0000236184, TRCN0000236186, TRCN0000236187, TRCN0000236188 and TRCN000019724) were purchased from Sigma, and lentiviral particles were generated in HEK293T cells by co-transfecting shRNA plasmid construct with pMD2.G (Addgene#12259) and psPAX2 (Addgene#12260). LNCaP cells were infected with NCOA4 lentivirus and were selected with 1.0 μg/mL puromycin. The level of NCOA4 knockdown was measured by western blots, and confirmed cells were expanded, cryopreserved, and used for the experiments.
Quantitative real-time PCR
Total RNA was extracted from cells using Trizol reagent (Thermo Fisher) as per manufacturer’s instructions. cDNA was synthesized from RNA by reverse transcription using SuperScript IV reverse (Thermo Fisher). Quantitative PCR was performed on samples mixed with SYBR master mix (BioRad) and gene-specific primers using a real-time thermocycler (BioRad). Data analysis was performed using the ΔΔCt method, and fold change (2^−ΔΔCt) was calculated after double normalization with the housekeeper gene and respective untreated control. Each PCR reaction was run in triplicates. Details of primer sequences used in qPCR are provided in Supplementary Table S1B.
Immunoprecipitation
NCOA4-FLAG-HA (CTAP) plasmid was received as a gift from Prof. J Wade Harper (Harvard Medical School, Boston). LNCaP cells were transfected with NCOA4-FLAG plasmid for immunoprecipitation studies or kept as untransfected control (C). NCOA4-FLAG transfected cells were either treated with 10 nM R1881 (T) or kept as untreated control (C*). Following treatment, cells were washed with 3 mL PBS, harvested in ice-cold PBS, and pelleted using a refrigerated centrifuge. Cells were lysed with 750 μL 1X lysis buffer (Promega) containing protease and phosphatase inhibitor for 20 minutes on ice. After lysis, the lysates were centrifuged at 13200 × g at 4 °C, and 10 percent of supernatant was stored as input. The remaining supernatant was used for immunoprecipitation using anti-FLAG M2 agarose beads (Thermo Fisher) as per manufacturer’s instructions. Affinity separated FLAG-tagged protein was eluted using 35 μL IgG elution buffer (Thermo Fisher), denatured with an equal volume of Laemmli buffer, and stored at −80 °C before western blot analysis. Each experiment was repeated at least three times.
Western blotting
Cell lysates were prepared either in 1X denaturing lysis buffer (Cell Signaling #9803) or non-denaturing lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Nonidet P-40, and supplemented with phosphatase and protease inhibitors). For denaturing gels, lysates were mixed with Laemmli buffer containing β-mercaptoethanol, boiled for 5mins and stored at −80 °C until use. Native lysates were mixed with native loading buffer (BioRad), and semi-native lysates were mixed with Laemmli buffer without beta-mercaptoethanol and stored at −80 °C without boiling. Native gels were resolved as described (15); denaturing and semi-native gels were resolved on 4–15% precast polyacrylamide gels (BioRad) followed by transfer to PVDF membrane. Membranes were blocked in 5% nonfat milk in wash buffer (Tris-buffered saline+0.1% Tween-20). After blocking, immunoblotting was performed with primary antibodies overnight at 4 °C, followed by incubation with HRP-conjugated secondary antibodies for 1h at room temperature. Blots were analyzed using the chemiluminescence method. Blots were developed using ECL Western blot detection reagent (GE Healthcare) for highly expressed proteins and Super Signal West Femto (Thermo Fisher) reagent for low expression proteins. Each experiment was repeated at least two to three times. The antibodies used are listed in Supplementary Table S2.
Immunofluorescence
Cells were grown on sterile coverslips in low density and were treated with R1881 (T) or vehicle control (C). After treatment, media was removed, and cells were fixed and permeabilized using chilled methanol for 10 min in deep-freezer followed by 10 min fixation in 10% natural buffered formalin at room temperature. Post-fixation, cells were blocked overnight in sterile-filtered 5% BSA in PBS at 4°C. Cells were incubated with a primary antibody, followed by a suitable fluorochrome tagged secondary antibody at 4 °C overnight. Both primary and secondary antibodies were diluted with 5% BSA in PBS. Cells were postfixed with 10 % NBF for 2–3 min before mounting on a clean glass slide using Vectashield antifade mounting media (Vector Laboratories). Image acquisition was performed on the LSM 700 laser confocal microscope (Zeiss). Experiment was repeated at least three times and images from at least five random fields from each experiment were used for analysis. Images were analyzed using the image processing package Fiji.
Measurement of cell death
Cells were stained with 0.4% trypan blue (Sigma) and incubated for 2–3 min at room temperature to allow dead cells to take up the blue stain. Cell viability was measured using an automated cell counter, Cellometer, (Nexcelom). Each experiment was repeated at least three times. Dead cells were calculated from subtracting the percentage of viable cells from 100. Mean percentage of dead cells was plotted on histograms.
Measurement of the labile iron pool (Fe2+)
An iron assay kit (Sigma) was used to measure the labile iron pool in control and R1881 treated cells. LNCaP and LAPC4 cells were culture in a 100 mm tissue culture dish and were treated for 72 before lysis in the buffer provided with the kit by snap freezing and thawing method. The lysate was cleared by centrifuging at 13000x g, and iron was measured by iron detection reagents supplied in the kit. Iron levels were calculated and normalized by the total protein present in the cell lysate. Levels of Fe2+ were analyzed in SupraT treated cells and were normalized to vehicle control cells.
NK cell migration assay
This assay was performed in a trans-well co-culture set up, using tumor cells in the bottom chamber (culture plate) and NK-92 cells in the upper chamber (insert) (Corning). LNCaP and LAPC4 cells were first seeded in a 24-well plate for 24h (day 1). The following day, tumor cells were treated with vehicle (C) or 10 nM R1881 (T) for 72h. After 72h, NK-92 cells, stained with 5 μM CFSE (Biolegend) for 10 min at 37 °C, were then added to a 5-micron pore size insert placed in each culture well already containing the vehicle/R1881 treated tumor cells. The culture plate was incubated for 6h, and microscopy was performed to image the CFSE labeled NK-92 cells that migrated to the lower chamber of the plate. Images were analyzed using the Image J software to determine the cell count. At least five fields were counted per experiment.
Cell cycle analysis
Harvested cell pellets were resuspended in ice-cold 70% ethanol and incubated at −20 °C overnight. Cells were then washed with 1X PBS, resuspended in 50 μg/ml propidium iodide (Sigma) in PBS with 50 μg/ml RNase A (Sigma), and incubated for 30 min at room temperature. Cell cycle analysis was performed using BD FACSCelesta cytometer equipped with BD FACS DIVA. Data were analyzed using the FlowJo software version 10 (FlowJo). The distribution of cells in the cell cycle phases was calculated using the Watson-Pragmatic algorithm. Each experiment was repeated at least three times.
NanoString immune profiling assay
Total RNA was isolated from vehicle/R1881 treated cells using Trizol (Thermo Fisher), and RNA quality was determined using the Bioanalyzer (Agilent). Gene signature was determined by using the NanoString PanCancer Immune Profiling Panel according to the manufacturer’s instructions (NanoString Technologies). House-keeping genes and built-in positive controls were used for data normalization and quality control assessments. Data analysis and gene expression analysis was performed using the nSolver 4.0 software (NanoString Technologies).
Luciferase reporter assay
Dual-Luciferase® reporter assay (Promega) was used to measure NF-kB, STING, and AIM-2 promoter activity. LNCaP and LAPC4 cells were transfected with Ig-IFN-Luc and Renila-Luc plasmids in 12-well plates and were incubated with vehicle or 10 nM R1881 treatment for 72h. Post-treatment, cells were washed with PBS, followed by lysis with 250 μL 1X Passive lysis buffer (Promega) at room temperature for 15 min. The lysate was cleared by isolating cell debris by centrifugation and stored at −80 °C unless used immediately for luminescence measurement. For luminometric measurement, 100 μL LARII buffer containing D-luciferin was dispensed in each well of the luminometer plate followed by 20 μL of cell lysate to measure Firefly luminescence. After measuring firefly luminescence, 100 μL Stop & Glo® reagent containing quencher and substrate for Renilla luciferase was added to measure respective responsive luminescence. Data were normalized using individual well’s Renilla luminescence. Each experiment was performed in triplicates and repeated three times.
Bioplex assay for chemokine and cytokine measurement
The concentration of 17 chemokines, cytokines, and growth factors in culture supernatants of control or R1881 treated LNCaP cells was determined using the Bioplex Pro Human Cytokine 17-plex assay system (Bio-Rad) according to manufacturer’s protocol. Culture supernatant from vehicle and R1881 treated LNCaP, and LAPC4 cells were collected at 3 and 6-day time points, and stored at −80 °C until further analysis. Data was collected on the Bio-Rad BioPlex 200 instrument and analyzed using the Bio-Plex Manager (Bio-Rad Laboratory).
Autophagosome Flux measurement
Cells were plated on 60 mm dishes to obtain 80% confluence on the next day. Cells were transfected with tandem mRFP-GFP fluorescent-tagged LC3 (ptfLC3) (Addgene #21074). 24h post-transfection, cells were trypsinized and plated on sterile glass-bottom dishes at a 40–50% confluency and treated with androgens. Autophagosomes are labelled as green and red dual positive punctas and autolysosomes appear as red puncta as the green puncta is pH sensitive and quenched by the acidic lysosomal pH. Cells were then imaged using a live cell Zeiss LSM780-FCS Single-point, laser scanning confocal microscope. Images were processed using the ImageJ software. Cells with predominantly yellow RFP/GFP (autophagosome) or red RFP (autolysosome) punctae were counted and analyzed from at least twenty fields. Each experiment had images from at least five random fields and repeated three times.
Mouse xenografts and treatment
Using a Johns Hopkins Animal Care and Use Committee approved protocol, adult athymic nude mice were inoculated subcutaneously in the flank with the LNCaP human prostate cancer cell lines in 200 μL of Matrigel. Mice were divided into two groups, and the treatment group was implanted with 2 one cm long silastic implants filled with testosterone as described previously (6). Tumors were harvested 2- and 4-days post-treatment and fixed in 10% buffered formalin and processed for IHC and H&E staining. NK cells were identified using antiCD57-PE (Santa Cruz Biotechnology) and antiCD49b-FITC (Santa Cruz Biotechnology) antibodies. F4/80-Alexafluor 488 and Ly-6G- Alexafluor 488 antibodies (Biolegend) were used to stain the macrophages and neutrophils cells respectively. Stained sections were imaged using Zeiss LSM 700 laser confocal microscope. Images were analyzed using the image processing package Fiji. NK cells, macrophages and neutrophils were counted per field from at least five fields and plotted as mean values.
Patient Materials
The study was conducted in accordance with ethical guidelines as outlined by the Declaration of Helsinki. The Institutional Review Board approved this study at Johns Hopkins, and all accrued patients provided written informed consent. Biopsies from metastatic lesions were obtained from patients with castration-resistant prostate cancer, under a prospective protocol (clinicaltrials.gov: NCT03554317) examining the use of SupraT as a treatment for metastatic castration-resistant prostate cancer (mCRPC). These patients had previously been treated with standard androgen deprivation therapy as well as next-generation anti-androgen therapy using abiraterone and/or enzalutamide. Biopsies were typically obtained from soft-tissue metastatic sites (lymph nodes, liver, and lung) using an 18-gauge core biopsy needle and were collected before and 12 weeks after starting the SupraT treatment. For each metastatic biopsy, at least two fresh cores were collected for immediate flash-freezing in liquid nitrogen (Frozen tissue method), and at least two separate cores for formalin-fixed and paraffin-embedded sections (FFPE method). A dedicated tissue technician was called at the biopsy suite to initiate processing steps, including FFPE preparation within 30 minutes of biopsy collection. All samples were evaluated for adequacy and assessment of tumor-to-normal ratio by an expert urological pathologist. FFPE specimens were processed according to the standard procedures at the Johns Hopkins pathology department.
Immunohistochemical Staining and CD8 density measurement by image analysis
Chromogenic IHC for CD8 was performed as follows. Sections of 4-micron thickness were cut and placed on super frost plus slides. Paraffin sections were baked on a hot plate at 60 oC for 10 minutes, dewaxed using xylene, rehydrated in a series of graded alcohols to distilled water, and finally rinsed in distilled water with 0.1% Tween 20. Slides were transferred to a glass jar filled with a suitable antigen retrieval solution. The glass jar was irradiated in a microwave oven at full power for 1 minute, followed by 15 minutes at power level 20. Slides were cooled for 5 minutes at room temperature and washed 2X in tris-buffered saline with 0.1% Tween 20 (TBST). Tissues were subjected to endogenous peroxidase blocking using hydrogen peroxide for 5 minutes. Slides were then incubated with CD8 (DAKO, Clone: C8/144B) antibody for 45 minutes at room temperature, rinsed with TBST, and incubated with the secondary antibody (PowerVision Poly-HRP Anti-Mouse IgG, Leica, PV6119) for 30 minutes. Following incubation with the chromogen, 3-Amino-9-Ethylcarbazole (AEC) for 20 minutes, slides were counterstained with hematoxylin. After counterstaining, slides were washed with tap water for 2 minutes and distilled water for 1 minute and mounted using VectaMount AQ, Vector H-5501.
Whole biopsy slides stained for CD8 were scanned on a Roche-Ventana DP200 whole slide scanner and analyzed using the HALO 3.0 (Indica Labs) software. Regions of interest (ROI) consisted of tumor tissue that was delineated manually by a pathologist with expertise in prostate pathology. In cases with clear lymph node tissue from lymph node biopsies, regions were chosen to avoid encompassing the lymphoid tissue apart from the tumor. T cells were delineated using the cytonuclear IHC module in HALO. CD8 density was calculated as the number of T cells per mm2 of ROI. Cell density measured using HALO were verified by manual counting in a subset of cases.
Statistical analysis
Suitable central tendency values were calculated for all the quantitatively measurable variables and were used for the analysis of statistical significance. Parametric analysis was performed to compare mean values after estimating the normal distribution unless otherwise specified. These analyses were performed using Prism version 6.0 (GraphPad Software), and a value of p≤0.05 was considered significant in all the statistical analyses.
Results
Our previous work demonstrated that sensitivity to SupraT differed among PCa cell lines (6). Cell cycle analysis revealed that SupraT causes a growth inhibitory accumulation of LNCaP cells in the G0/G1 phase, while LAPC4 cells show the opposite response and continue to proliferate (Supplementary Figure. S1A and B). Furthermore, the clonogenic analysis revealed that SupraT significantly decreases the clonogenic potential of SupraT-sensitive LNCaP cells (Supplementary Figure S1C and D). To understand whether sensitivity to SupraT reflected DNA repair mutations in these cancer lines, we searched the COSMIC cell line database for mutations in DNA repair genes in PCa lines (Supplementary Table S3). Strikingly, our analysis revealed that sensitivity to hormone exposure correlated directly with the number of DNA repair mutations harbored by the cell types (LNCaP > VCaP > LAPC4), mirroring our clinical data (6, 11). 22R-v1, another AR positive line that is castration resistant (16) was not inhibited by SupraT (Supplementary Figure S1E). We hypothesized that due to the prevalence of DNA repair mutations, LNCaP and VCaP cells (the intermediate sensitivity line) treated with SupraT might not repair DSBs induced by androgens and instead undergo a DNA repair crisis leading to apoptosis. However, contrary to our hypothesis, none of the prostate cancer cell lines treated with synthetic androgen R1881 had any substantial enhancement of PARP cleavage (Supplementary Figure S2A). We also measured Annexin-V positivity, an early marker of apoptosis, in the highly sensitive LNCaP cells and found no significant increase in Annexin-V after treatment (Supplementary Figure S2B). This suggested that the decrease in cell number upon testosterone treatment in the sensitive cell lines might likely also involve a nonapoptotic cell death mechanism. We, therefore, probed whether necrotic markers were induced upon SupraT treatment (Supplementary Figure S2C), and found them to be markedly reduced or unchanged, ruling out necrotic induction by SupraT. Recently, ferroptosis, a nonapoptotic cell death mechanism, has been linked to autophagy (17, 18). We first determined whether SupraT can induce autophagy in PCa lines. Treatment of PCa cell lines demonstrated that the synthetic androgen, R1881, notably induces autophagy in the SupraT sensitive LNCaP and VCaP cells, while basal autophagy in the SupraT-insensitive LAPC4 and 22Rv1 cells remained largely unaffected upon treatment (Supplementary Figure S3A). We did not find global upregulation of key autophagy proteins such as Beclin or ATG12 (Supplementary Figure S3B). However, we did find both the number of autophagosomes and autophagy flux (evaluated using an autophagy flux sensor) increased only in LNCaP cells as compared to LAPC4 cells upon treatment with androgens (Supplementary Figure S3C). These results prompted us to determine whether SupraT is able to differentially induce ferroptosis in PCa cells that are sensitive to androgen.
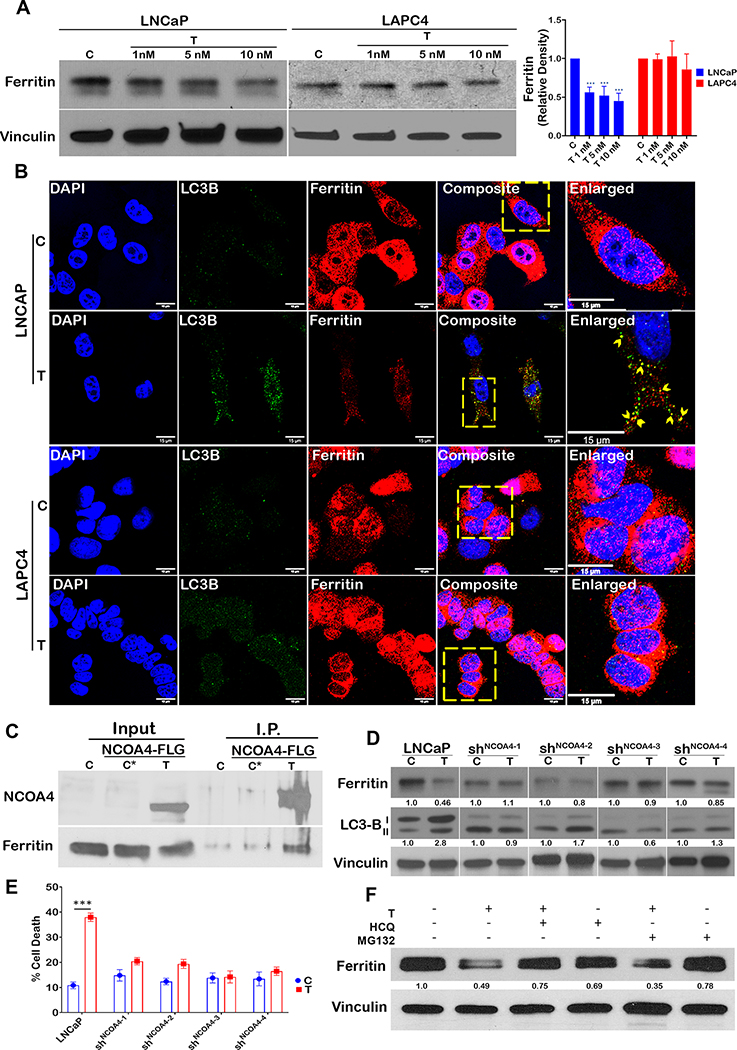
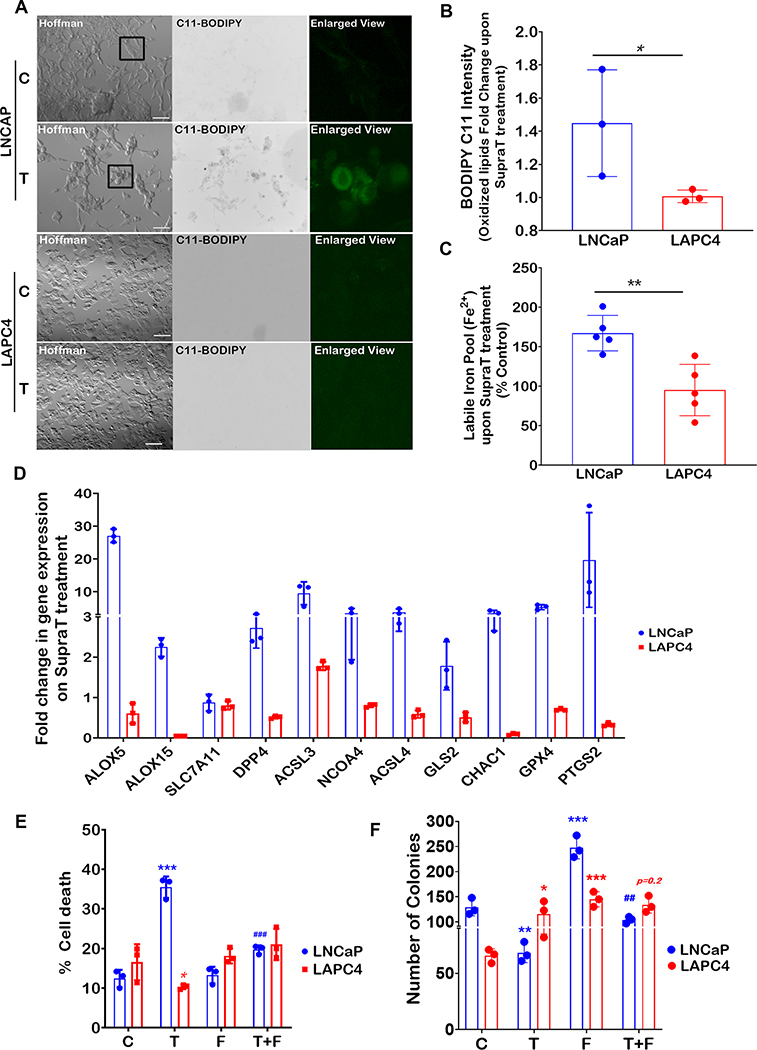
Ferroptosis involves iron-dependent accumulation of toxic lipid peroxides that leads to cell death (19). Degradation of the iron storage protein ferritin through a specialized form of autophagy, termed ferritinophagy, increases the labile pool of iron, leading to an increase in lipid peroxides (20). We sought to determine whether ferritinophagy is induced by SupraT. As depicted in Figure 1A and Supplementary Figure S4A, SupraT causes a dose-dependent decrease in ferritin levels in LNCaP and VCaP cells compared to LAPC4 and 22Rv1 cells, where ferritin levels remain unchanged. We confirmed whether decrease in ferritin level is indeed through testosterone by treating cells with Dihydrotestosterone (DHT), a high affinity testosterone metabolite. As shown in Supplementary Figure S4B, supraphysiological levels of DHT decreased ferritin levels in LNCaP cells but not in LAPC4 cells. Confocal images of SupraT treated LNCaP cells revealed that ferritin colocalized with LC3B positive autophagosomes (Figure 1B). Since NCOA4 interacts with ferritin and mediates its autophagic degradation (20–22), we probed for NCOA4 and found that NCOA4 is induced by SupraT (Supplementary Figure S4C), and interacts with ferritin in a SupraT dependent manner (Figure 1C). Further, knockdown of NCOA4 in LNCaP cells (Supplementary Figure S4D) inhibited R1881 induced ferritin degradation and decreased cell death. (Figure 1D, and E). Knockdown of NCOA4 also decreased SupraT induced autophagy (Figure 1D). To determine whether ferritin is indeed degraded through autophagy, we treated cells with hydroxychloroquine, an autophagy inhibitor, and MG-132, a proteasome inhibitor, and evaluated its effect on ferritin degradation upon SupraT treatment. As seen in Figure 1F, hydroxychloroquine prevented the degradation of ferritin by SupraT, suggesting that ferritin is degraded through autophagy. We evaluated the functional consequence of decreased ferritin on lipid peroxide formation using the lipid peroxide sensor, C11-BODIPY (19, 23), which fluoresces green upon oxidation. As shown in Figure 2A and B, R1881 treated LNCaP cells but not LAPC4 had increased lipid peroxides and labile iron pool (Figure 2C) compared to vehicle treated controls. Next, we investigated whether SupraT induces pro-ferroptotic gene expression (17, 24). Pro-ferroptotic genes such as ALOX5, PTGS2, and NCOA4 were increased many folds over vehicle controls (Figure 2D). We further confirmed that SupraT induced cell death in LNCaP is indeed through ferroptosis, as treatment with a combination of R1881 and a ferroptosis inhibitor, ferrostatin-1, abrogated cell death (Figure 2E and F). A similar abrogation of SupraT induced cell death was observed in VCaP cells that demonstrates intermediate sensitivity to SupraT (Supplementary Figure S4E). Moreover, treatment of LNCaP and LAPC4 cells with DHT or a combination of DHT and ferrostatin-1 had the same effect (Supplementary Figure S4F).
Figure 1: SupraT induces ferritinophagy in PCa cells.
A, Western blot analysis showing ferritin and loading control vinculin in LNCaP and LAPC4 cells treated with indicated concentrations (nM) of R1881 (T) and vehicle (C) for 72h. A representative of three independent experiments. Ferritin ~ 21 kDa; loading control Vinculin 124 kDa. Bar graph shows mean relative density from three experiments. B, A representative single confocal section from three independent experiments is shown. Each experiment had at least 5 random fields LNCaP and LAPC4 cells were stained for LC3B (green) and ferritin (red). Right most panels show enlarged inset images of a region of interest, and the arrow-head indicates co-localization of ferritin in autophagosomes (LC3B). C, Western blot for immunoprecipitated NCOA4-FLAG for interaction with ferritin in LNCaP cells transfected with human NCOA4α-pHAGE-C-FLAG-HA plasmid. Non-transfected (C) and NCOA4α-pHAGE-C-FLAG-HA transfected vehicle treated cells (C*) were used as controls. A representative of three independent experiments. NCOA4 70 kDa and Ferritin ~ 21 kDa. D, Ferritin and LC3B levels probed by western blot in vehicle (C) or R1881 (T) treated LNCaP or NCOA4 shRNA lentiviral particle transduced LNCaP shNCOA4(1−4) cells. A representative of three independent experiments. Ferritin ~ 21 kDa; LC3-B 14,16 kDa; loading control Vinculin 124 kDa. E, Cell death in LNCaP and LNCaPshNCOA4 (1−4) measured using trypan blue exclusion assay, Control (C), 10 nM R1881 (T). Bar graph shows mean % cell death from three independent experiments and error bar shows standard deviation. F, Western blot analysis for ferritin LNCaP cells treated with vehicle (C), 10 nM R1881 (T), 10 μM hydroxychloroquine (HCQ), 10 μM MG132 (MG132) alone or combination of T with either of HCQ or MG132. A representative of three independent experiments. Ferritin ~ 21 kDa; loading control Vinculin 124 kDa. Scale bars: 15 μm (B).
Figure 2: SupraT induces ferroptosis in PCa cells.
A, A representative fluorescence microscopy images of control (C) and 10 nM R1881 treated (T) LNCaP and LAPC4 cells from three independent experiments showing C11-BODIPY staining for oxidized membrane lipid (green fluorescence). Each experiment had at least 5 random field images. Enlarged inset images in the rightmost panels show a region of interest. B, Bar graph showing fold change in levels of control normalized oxidized lipids (Bodipy C11) in LNCaP and LAPC4 cells, measured by image analysis using ImageJ. C, Measurement of labile iron pool (Fe2+) in LNCaP and LAPC4 cells. Bar graph shows fold change in labile iron pool calculated by normalizing to untreated control cells. D, Quantitative RT-PCR analysis for a panel of pro-ferroptotic genes in 10 nM R1881 treated LNCaP, and LAPC4 cells post 72h. Vehicle control was used for normalization to calculate the fold change. A representative of three independent experiments. Bar graph indicates fold change upon SupraT treatment. E, Cell death measured using trypan blue exclusion assay. Control (C), 10 nM R1881 (T), 10 μM Ferrostatin (F), or combination (T+F) shown as mean from three measurements with error bar showing standard deviation. F, Bar graph showing the mean of the number of colonies from three independent measurements counted using Fiji software for treatment conditions stated in (E) with error bars showing standard deviation. Asterisk * compares T or F vs. C and # indicates comparison between T and T+F. Both * and # indicates statistically significant differences (p<0.05). Scale bars: 100 μm (A).
To investigate whether there is a direct link between response to SupraT, autophagy, and DNA repair, we treated SupraT sensitive LNCaP cells, which harbor several DNA repair gene mutations (Supplementary Table S3), with R1881 and stained cells for autophagosomes and DSBs. We postulated that cells harboring damaged DNA would have increased autophagy in response to DNA damage stress. Interestingly, LNCaP cells with increased autophagosomes displayed a smaller number of γ-H2AX puncta (Pearson’s correlation = −0.78) (Figure 3A and B). This suggested that cells undergoing autophagy in response to SupraT might clear their damaged DNA more efficiently. Autophagy, being a dynamic event, renders visualization of fractions of autophagosomes carrying damaged DNA a challenge. Hydroxychloroquine prevents the fusion of autophagosomes to lysosomes, leading to accumulation of autophagosomes (25). Treatment of PCa cell lines with a combination of hydroxychloroquine and R1881 revealed marked localization of cytoplasmic DNA in LNCaP cells compared to the fewer cytoplasmic DNA puncta displayed in VCaP cells, which have intermediate DNA repair gene mutations (Supplementary Table S3). The SupraT-insensitive LAPC4 cells, however, did not show any cytoplasmic DNA (Figure 3C). Intriguingly, although we did find induction of DSBs in SupraT treated LNCaP NCOA4 knockdown cells, similar to wild type cells (Supplementary Figure S5A), we did not find any cytoplasmic DNA (Supplementary Figure S5B) in these cells after SupraT treatment. This could be as a result of abrogation of autophagy induction as evident from the number of autophagosomes between control and SupraT treated cells (Supplementary Figure S5C). To ensure that the observed cytoplasmic DNA was not mitochondrial DNA, we stained cells with antibodies against mitochondrial complex IV subunit I and LC3B. We did not find any notable co-localization of mitochondria with autophagosomes (Figure. 3D). However, we did find marked co-localization of cytoplasmic DAPI staining with LC3B in SupraT treated cells, indicating that the cytoplasmic DNA is present in autophagosomes (Figure 3E). DAPI intensity peaked in the lumen of autophagosomes indicating that the DNA was indeed present within the autophagosomes (Figure 3F). To ascertain whether the DNA within the autophagosomes harbored damaged DNA, we stained cells treated with a combination of hydroxychloroquine and SupraT with γ-H2AX and LC3B. As seen in Figure 3G, LC3B and γ-H2AX colocalize with DAPI signals in the cytoplasm. This suggests that SupraT induced damaged DNA can be shuttled to the cytoplasm for autophagosome-mediated degradation. This implies that SupraT might induce two parallel autophagy-mediated phenomena: ferritinophagy and nucleophagy, both of which may be responsible for the growth inhibitory effects of SupraT.
Figure 3: SupraT induces nucleophagic degradation of unrepaired damaged DNA.
A, A representative single confocal section is shown from three independent experiments. LNCaP cells stained for γ-H2AX (red) and LC3B (green) after treatment with 1 and 5 nM R1881. Arrow-heads indicate cells harboring γ-H2AX foci with lower LC3B puncta. Each experiment had at least 5 random field images. B, Scatter plot showing a correlation between the number of LC3B puncta and the number of γ-H2AX foci counted (n=19 measurements). Counting of puncta was performed using the image analysis software Fiji. C, Confocal microscopy images for PCa cell lines treated with 10 nM R1881 (T) and hydroxychloroquine (HCQ). Each experiment had at least 5 random field images. The lower panel is an inverted and magnified image of a single cell in the view-field for better visualization. Arrows indicate the localization of cytoplasmic DNA. D, Confocal images showing mitochondrial staining in LNCaP cells treated with vehicle (C), 10 nM R1881 (T), 10 μM HCQ, or combination of T+HCQ for 72h. Lower panel shows a magnified view for enhanced visualization. Each experiment had at least 5 random field images. E, Photomicrographs showing colocalization of LC3B (green) and DAPI (Gray) in LNCaP cells treated with control (C), 10 nM R1881 (T), HCQ (10 μM) or T+HCQ treated LNCaP cells (72h). Rightmost panel shows colocalized pixels. F, Graphical representation of fluorescence intensities of LC3B and DAPI on individual autophagosomes (n=16 measurements). G, Photomicrographs showing LC3B (green) and γ-H2AX (red) in LNCaP cells after 72h of treatment with T+HCQ. Lower panels show the enlarged insets with a region of interest. Arrow-heads in the lower-left image indicate the presence of DNA (DAPI) in autophagosomes (LC3B), and lower-right image show γ-H2AX positivity in DNA present in those autophagosomes. Each experiment had at least 5 random field images. Scale bars: 5μm (A), 10μm (C), 10μm (D), 10μm (E), and 10μm (G).
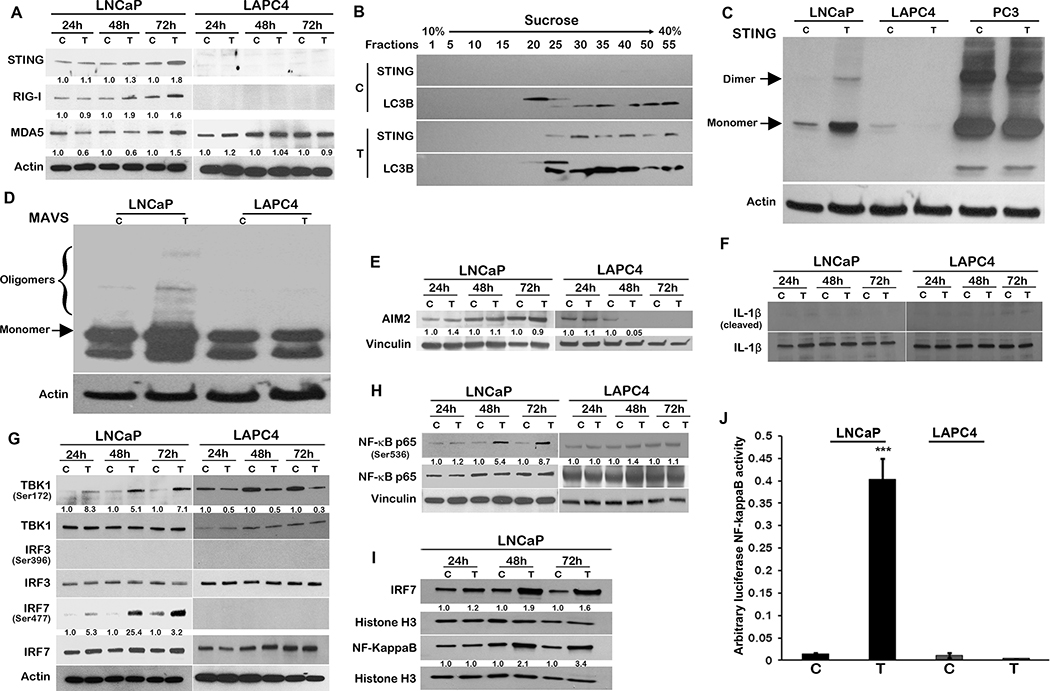
Cytosolic DNA is seen as a stimulant by the innate immune system as it is detected by DNA sensors in the cytoplasm that activate the adaptor protein STING and downstream innate immune signaling (26, 27). The STING promoter harbors an AR binding motif (Supplementary Figure S6A); however, we did not find any induction of the STING transcript or STING promoter by treatment with SupraT (Supplementary Figure S6B and C). Intriguingly, treatment with SupraT induced the STING protein as well as the RNA sensors RIG-I and MDA5 proteins in the SupraT sensitive LNCaP cells (Figure 4A). Upon activation, monomeric STING dimerizes and translocates from the ER to autophagosome-like vesicles (28, 29). Separation of cellular homogenates on sucrose gradients revealed STING cofractionates with LC3B positive autophagosomes in SupraT treated cells (Figure 4B). To confirm that STING is indeed activated upon SupraT treatment, we performed a STING dimerization assay, a gold standard for STING activation (27). As shown in Figure 4C, treatment with SupraT activates STING in LNCaP compared to LAPC4 cells. Interestingly, SupraT also activated MAVS oligomerization, indicating that the RNA sensing pathway was activated as well (Figure 4D). Mislocalized and damaged DNA can also be detected by AIM2 (30). The AIM2 promoter harbors an AR binding motif (Supplementary Figure S6D); hence we first measured AIM2 transcript and protein levels. We did not find any induction of AIM2 promoter activity, AIM2 transcript levels or protein by SupraT (Supplementary Figure S6E and F and Figure 4E). Neither did we find SupraT mediated activation of downstream inflammasomes, as evaluated by IL-1β cleavage, ruling out the involvement of AIM2 mediated inflammasome signaling by SupraT (Figure 4F). We found a similar activation of STING, albeit less robustly, in VCaP cells (Supplementary Figure S7A). Interestingly in VCaP cells, STING appeared as a tetramer as reported for STING activation in some studies (31). We did not find any activation of STING in the SupraT insensitive 22Rv1 cells (Supplementary Figure S7A), neither did we find any activation of MAVS in both VCaP and 22Rv1 cells (Supplementary Figure S7C). We next investigated whether the observed effect is indeed due to testosterone by treating cells with DHT. In agreement with our findings using R1881, we found DHT robustly increased autophagy in LNCaP cells but not in LAPC4 cells (Supplementary Figure S4B). DHT also activated STING and MAVS preferentially in LNCaP cells (Supplementary Figure S7B and 7D), however, the activation was not as robust as R1881. This may be due to the fact that persistent H2AX foci are not observed in DHT treated cells as compared to R1881(9), as DHT is rapidly metabolized (32, 33).
Figure 4: SupraT activates cytoplasmic nucleic acid sensors and subsequent NF-kappaB signaling.
A, Western blot analysis showing DNA (STING) and RNA (RIG-I and MDA5) specific sensors along with actin as a loading control in LNCaP and LAPC4 cells treated with vehicle control (C) or 10 nM R1881 (T) in a time-dependent manner. A representative of three independent experiments. STING 33–35 kDa; RIG-I 102 kDa; MDA5 135 kDa; loading control Actin 43 kDa. B, Immunoblotting for LC3B and STING on fractions isolated using sucrose gradient centrifugation from LNCaP cell homogenates after 72h of 10 nM R1881 treatment with 10 nM R1881 (T) or vehicle control (C). A representative of at least three independent experiments. STING 33–35 kDa; LC3B 14, 16 kDa. C-D, Native western blot probing for STING (C), and MAVS (D) in PCa cell lines treated vehicle (C) or 10 nM R1881 (T). A representative of three independent experiments; Loading control Actin 43 kDa. PC3 cells with constitutive expression of active STING were used as a positive control (C). E-F, Western blot analysis for AIM2 induction (AIM2 (E)) and signaling (IL-1β (F)) in PCa cell lines treated with vehicle (C) or 10 nM R1881 (T) in a time-dependent manner. A representative of three independent experiments. AIM2 40 kDa; IL-1β 17,31 kDa; Cleaved IL-1β 17 kDa; loading control Vinculin 124 kDa. G-H Immunoblotting for interferon regulatory genes (IRF7, IRF3, TBK1 (G), and NF-kappaB (H) in PCa cell lines treated with vehicle (C) or10 nM R1881 (T) at 24h, 48h, and 72h. Activated forms of all interferon regulatory genes were probed by respective phosphorylation specific antibodies. A representative of at least two independent experiments. Phospho-TBK1 84 kDa; TBK1 84 kDa; Phospho IRF3 45–55 kDa; IRF3 50–55 kDa, Phospho-IRF7 65 kDa; IRF7 65 kDa; loading control Actin 43 kDa. I, Nuclear localization of IRF-7 and NF-kappaB in LNCaP cells after treatment with vehicle (C) or 10 nM R1881 (T) at indicated time points. A representative of at least two independent experiments. IRF7 65 kDa; NF-KappaB 65 kDa, loading control Histone H3 17 kDa. J, Bar graphs showing mean of luciferase NF-kappaB activity in LNCaP cells after treatment with vehicle (C) or 10 nM R1881 (T) using luciferase-based NF-kappaB reporter assay from three independent measurements and error bars show standard deviation. An asterisk indicates statistically significant differences (p<0.05).
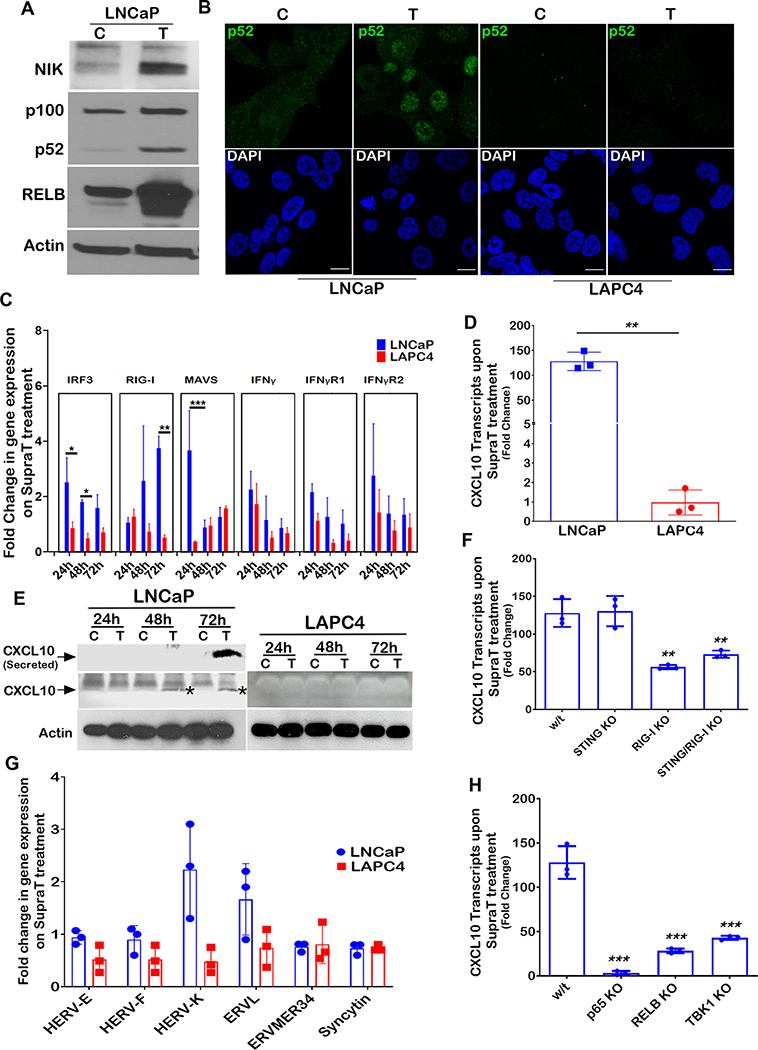
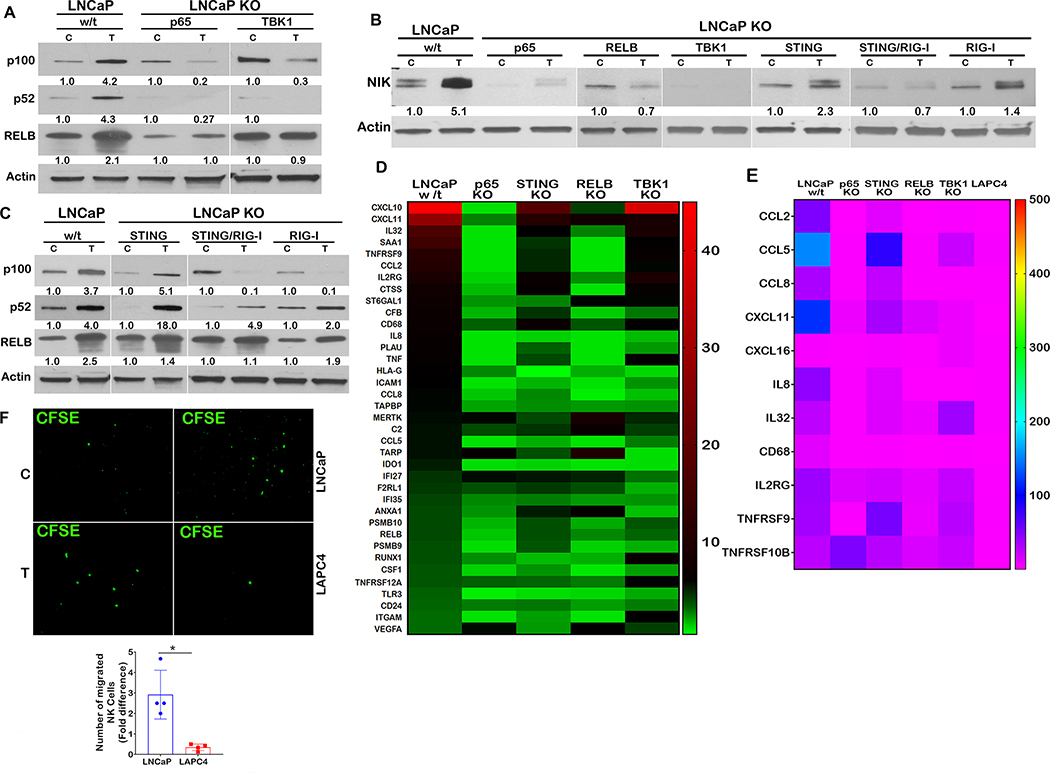
Following its own activation, STING activates the Tank binding kinase 1 (TBK1), which in turn phosphorylates and activates interferon regulatory factors (IRFs), including IRF3, IRF7, and NF-kappaB, leading to the induction of immune response genes (26). Although we did not find any activation of IRF3 by SupraT treatment (Figure 4G and Supplementary Figure S7E), our data indicates that TBK1, IRF7, and NF-kappaB canonical p65 subunit were activated upon SupraT treatment in LNCaP and VCaP cells (Figure 4G and H and Supplementary Figure S7F). Activated p65 and IRF7 increased in the nucleus (Figure 4I). Similar to LAPC4 cells, 22Rv1 cells did not show an induction of NF-kappaB or IRF7 signaling (Supplementary Figure S7F). We performed an NF-kappaB promoter-reporter assay, which confirmed the functional activation of NF-kappaB in SupraT sensitive cells (Figure 4J). Intriguingly, we found SupraT stabilized NF-kappaB inducing kinase (NIK), a kinase central to the non-canonical NF-kappaB pathway (Figure 5A). NIK phosphorylates the non-canonical p100 subunit of NF-kappaB and marks it for processing by a proteasome into the smaller p52 subunit, which subsequently dimerizes with RELB to activate transcription of target genes. Treatment with SupraT led to the formation of p52 and induction of RELB in the SupraT sensitive LNCaP cells but not in VCaP cells (Figure 5A and Supplementary Figure S7F); probably reflecting differences in SupraT sensitivity of these cells. Immunofluorescence confirmed the nuclear translocation of p52 upon SupraT treatment (Figure 5B). Further, in line with nucleic acid sensor activation, treatment with DHT led to stabilization of RelB and NIK and phosphorylation of the canonical and non-canonical NF-kappaB subunits downstream of nucleic acid sensors (Supplementary Figure S7G). This data indicates that SupraT induced DNA damage activates nucleic acid sensors and downstream NF-kappaB signaling.
Figure 5: SupraT induced NF-kappaB signaling drives innate immune response.
A, Immunoblotting for NIK, p100, p52, and RELB proteins in vehicle control (C) or 10 nM R1881 treated (T) LNCaP cells. A representative of at least three independent experiments. NIK 125 kDa; NF-kappaB 2 p100 and p52 120 and 52 kDa respectively; RELB 70 kDa; loading control Actin 43 kDa. B, Immunofluorescence images for the LNCaP and LAPC4 cells treated with vehicle (C) or 10 nM R1881 (T) and stained with p52 (green) and DAPI (blue) for visualization of nuclear p52 protein. A representative of at least two independent experiments. Each experiment had at least 5 random field images. C, Quantitative RT-PCR for the interferon related genes in 10 nM R1881 treated LNCaP and LAPC4 cells for 24h, 48h, and 72h time points. Mean fold change data between LNCaP and LAPC4 cells were compared statistically, and an asterisk indicates a statistically significant difference (n=3). Bar graph indicates fold change upon SupraT treatment. D, CXCL10 transcript analysis using quantitative RT-PCR from total RNA isolated from vehicle or 10 nM R1881 treated LNCaP and LAPC4 cells (n=3). Bar graph indicates fold change upon SupraT treatment. E, Western blot for CXCL10 protein (lysates for cellular and culture media for secreted) for vehicle (C) and 10nM R1881 (T) treated LNCaP and LAPC4 cells made after 24h, 48h, or 72h post-treatment (band marked with asterisk*). A representative of at least two independent experiments. CXCL10 10 kDa; loading control Actin 43 kDa. F, RT-PCR based measurement for CXCL10 transcripts in LNCaP or knockouts of RIG-I, STING, or both after treatment with 10 nM R1881 for 72h. Bar graph indicates mean fold change upon SupraT treatment (n=3) and an asterisk indicates a statistically significant difference (p<0.05). G, Histogram showing endogenous retroviral transcripts in 10 nM R1881 treated LNCaP or LAPC4 cells. Bar indicates a mean fold change upon SupraT treatment (n=3), and the error bar indicates the standard deviation from three replicate values. H, Expression of CXCL10 transcripts in wild type, p65 KO, RELB KO, and TBK1 KO LNCaP cells treated with 10 nM R1881. Statistical significance is calculated between the wild type and knockout variants of LNCaP cells. Bar graph indicates fold change upon SupraT treatment (n=3) and an asterisk indicates a statistically significant difference (p<0.05). Scale bars: 15 μm (B).
In order to find out whether NF-kappaB responsive innate immune genes were activated by the nucleic acid sensors, we measured transcript levels of innate immune genes and found many genes were significantly upregulated in the SupraT-sensitive LNCaP and VCaP cell lines (Figure 5C and Supplementary Figure S7H). CXCL10, a chemokine, was increased (34) several folds in treated LNCaP cells both at transcript and protein level (Figure 5D and E). CXCL10 was also induced (less robustly) in VCaP cells but not in SupraT insensitive 22Rv1 cells (Supplementary Figure S7I). Furthermore, DHT was able to induce CXCL10 (less robustly than R1881) preferentially in LNCaP cells as compared to LAPC4 cells (Supplementary Figure S7J). Bioplex assays confirmed the induction and secretion of CXCL10 and other chemokines upon SupraT treatment (Supplementary Table S4). To determine whether STING or RIG-I sensors play a role in the activation of downstream innate immune signaling, we created knockouts for STING, RIG-I, and STING/RIG-I double knockouts (Supplementary Figure S8A and B) and tested whether they abrogate CXCL10 induction. Knockout of STING did not abrogate CXCL10 induction, but double knockouts and knockout of RIG-I alone decreased CXCL10 expression (Figure 5F). This suggested that both the nucleic acid sensors are activated in SupraT-sensitive cells and that RIG-I may play an essential role in amplifying the signal. RIG-I can also be activated by the expression of endogenous retroviruses (35). Our analysis revealed that some of the endogenous retroviruses harbor androgen response elements (Supplementary Figure S8C). However, none of the endogenous retroviral transcripts investigated were upregulated by SupraT (Figure 5G), suggesting activation of RIG-I may be primarily through cytoplasmic DNA, as indicated by others (36).
To tease out whether the canonical or the non-canonical NF-kappaB pathway is important for innate immune gene signaling by SupraT, we made knockouts of TBK1 and other key components of the NF-kappaB pathway (Supplementary Figure S8D-F). Knockouts of p65, TBK1, and RELB each abrogated downstream CXCL10 induction by SupraT without altering the growth inhibitory effect of SupraT on of these cells (Figure 5H and Supplementary Fig S8G). Induction or proteolytic processing of non-canonical subunits RELB and p100, respectively, by SupraT was totally abrogated in p65 and TBK1 knockouts (Figure 6A), indicating that the canonical pathway is important for the activation of the non-canonical pathway and TBK1 may play a critical role in linking the two. Furthermore, knockouts of p65, RELB, and TBK1 each abrogated NIK stabilization compared to parental cells, knockouts of STING and RIG-I decreased NIK stabilization, and STING/RIG-I double knockouts totally diminished the NIK stabilization, mirroring that of the p65 and TBK1 knockouts (Figure 6B). Intriguingly, knockouts of STING did not revoke induction or processing of p100, but knockouts of RIG-I alone decreased p100 induction. Further, STING/RIG-I double knockouts had lower induction of RELB and p100 processing as compared to STING knockouts (Figure 6C). While this data corroborates the involvement of both the nucleic acid sensors, STING and RIG-I may have opposite effects on the non-canonical NF-kappaB signaling induced by SupraT - STING being suppressive and RIG-I being supportive.
Figure 6: SupraT activates both canonical and non-canonical NF-kappaB signaling.
A, Immunoblotting for p100, p52, and RELB protein to show non-canonical NF-kappaB signaling in wild type and knockout LNCaP cells treated with vehicle (C) or 10 nM R1881 (T). A representative of three independent experiments. p100 and p52 120 and 52 kDa respectively; RELB 70 kDa; loading control Actin 43 kDa. B, Blot depicting stabilization of NIK in wild type and all knockout cells treated with vehicle (C) or 10 nM R1881 (T). A representative of three independent experiments. NIK 125 kDa; loading control Actin 43 kDa. C, Levels of p100 and p52 protein in DNA and RNA sensors (RIG-I and STING) single and double knockouts in LNCaP cells treated with vehicle (C) or 10 nM R1881 (T). A representative of three independent experiments. NF-kappaB2 p100 and p52 120 and 52 kDa respectively; RELB 70 kDa; loading control Actin 43 kDa. D, Heat map depicting fold change for expression of a number of genes selected from pan-cancer immune profiling panel. Total RNA extracted from indicated samples was analyzed for 770 immune-related human genes using nCounter human PanCancer Immune Profiling Panel (Nanostring). E, Heatmap showing relative expression levels of genes selected from the list of 57 genes that showed higher expression in Nanostring analysis. Total RNA was analyzed using qRT-PCR, and data are shown as heatmap from a representative experiment of three independent experiments. F, Representative photomicrographs depict CFSE labeled NK-92 cells migrated towards either vehicle (C) or 10 nM R1881 (T) treated LNCaP or LAPC4 cells through 5 μM Boyden chamber. Vehicle control normalized count of NK-92 cells plotted as a mean of four independent experiments on histograms with error bars showing standard deviation. Each experiment had at least 5 random field images. An asterisk indicates a statistically significant difference (p<0.05) Scale bars: 100 μm (F).
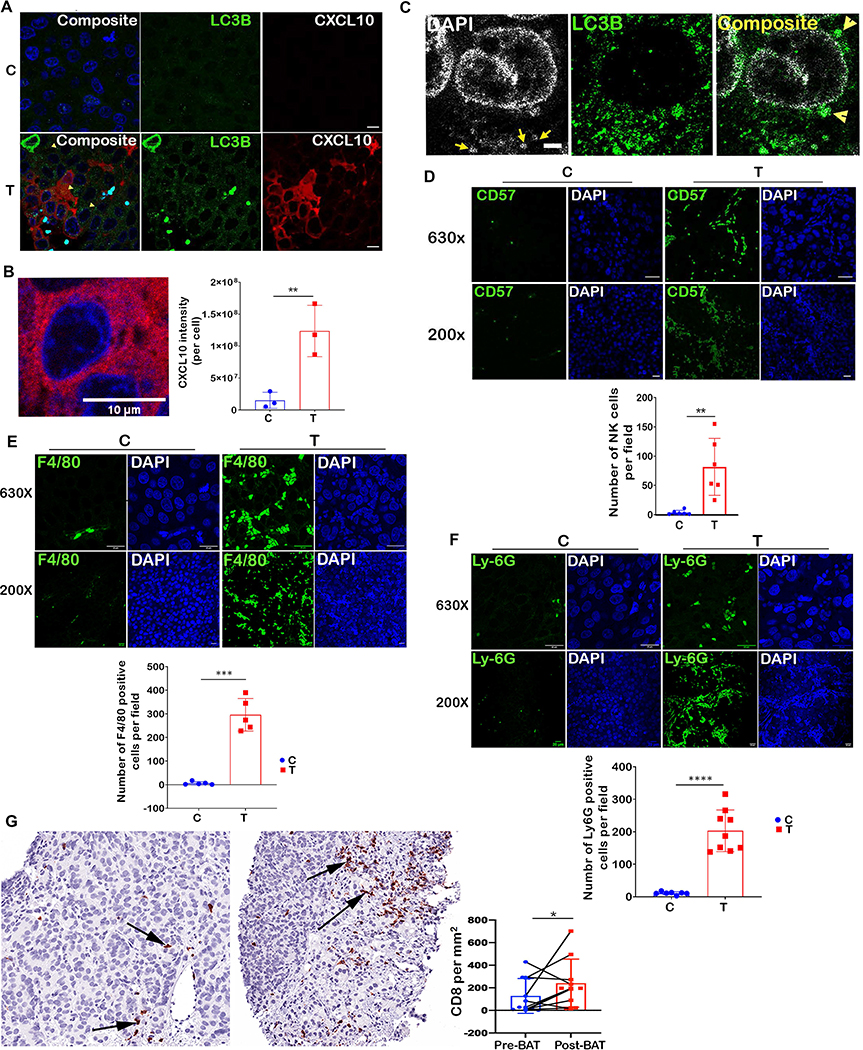
To identify immune genes activated by SupraT and discern those regulated by the STING-TBK1-NFkappaB axis, we performed a Nanostring PanCancer IO 360 Gene Expression analysis using a panel of 770 unique immune-gene signatures. Out of 57, differentially (>2.5 fold) expressed genes, 38 percent of immune genes were induced in a STING-TBK1-NFkappaB dependent manner (Figure 6D). We validated a panel of chemokines and cytokines that play a role in attracting immune cells using quantitative PCR, and in concurrence with our Nanostring data, these genes were altered in a STING-TBK1-NFkappaB dependent manner (Figure 6E). The gene expression data further indicates that both innate and adaptive immune cells might home to and get activated by SupraT induced cytokines and chemokines. To investigate whether SupraT is able to induce migration of NK cells, we conducted a transwell migration assay with human NK-92 cells and found the SupraT-sensitive cell line significantly increased migration of NK-92 cells upon treatment (Figure 6F). We next established LNCaP xenografts in athymic nude mice, which exhibit robust NK cell activation and harbor neutrophils and macrophages (37). In concordance with in vitro findings, SupraT led to an increase in autophagosomes, the presence of cytoplasmic DNA, and an increase in CXCL10 expression by tumor cells (Figure 7A and B). The presence of autophagosomal DNA without any autophagy inhibition indicated that the dynamics of autophagosomal degradation differed in vitro and in vivo (Figure 7C). Staining for NK cell markers CD57 and CD49b revealed a 17-fold induction in NK cell migration to the tumor bed (Figure 7D and Supplementary Figure S9). A similar result was obtained when tumors were stained for macrophages and neutrophils (Figure 7E and 7F). Prostate tumors are considered immunologically cold tumors with limited cytotoxic T cell infiltration (38). The clinical significance of our data was investigated using biopsy from patients undergoing BAT therapy. As seen in Figure 7G, BAT administration significantly increased the infiltration of CD8 T cells. In summary, these results indicate that SupraT may be able to activate immune cells through the NF-kappaB pathway by activating nucleic acid sensors, especially in cells with defects in DNA repair pathways. Our data also indicates that SupraT may induce ferroptosis, a potentially immunogenic cell death mechanism.
Figure 7: SupraT induces tumor infiltration of immune cells.
A, Immunofluorescence staining of tumor xenograft specimens for LC3B (green) and CXCL10 (red) from tumor-bearing mice treated with testosterone cypionate (T) (n=4) in the lower panel as compared to tumor isolated from vehicle (C) treated animals (n=4) shown in the top panel. Each sample had at least 5 random field images. B, Magnified view of a tumor cell showing cytoplasmic localization of CXCL10 (red). Bar graph shows CXCL10 measured by analyzing images (at least five fields) on ImageJ in control and testosterone treated groups. C, Tumor xenografts specimens from testosterone cypionate treated mice stained with DAPI (grey) and LC3B (green) to show the presence of cytoplasmic autophagosomal DNA. At least 5 random images were collected for each sample. D, E, and F, Representative immunofluorescence images for tumor sections from the vehicle (C) (n=4) or testosterone cypionate treated (T) mice (n=4) stained with Alexafluor488-antiCD57 (D), Alexafluor488-antiF4/80 (E), and Alexafluor488-antiLy-6G (F). The number of stained NK cells, Macrophages and Neutrophils were counted using Fiji image analysis software and plotted as mean on histogram. Each sample had at least 5 random field images. G, Immunohistochemical staining for CD8 in matched biopsy before and during BAT treatment (n=10). The left photomicrograph shows a medium power image of tumor with sparse CD8 cell infiltrates involving tumor, most of the cells seen represent tumor cells. Note an increase in the extent of infiltrate in the treated sample in image on the right. Arrows indicate CD8 positive T cells. The histogram shows area normalized number of CD8 T cells in biopsies collected from PCa patients before and after BAT regimen (n=10). An asterisk indicates a statistically significant difference (p<0.05). Scale bars: 10μm (A), 2μm (B) 25μm (C) 20 μm (D and E).
Discussion
Strategies to overcome resistance to ADT can make a significant impact on the current outcomes of therapy. Several complementary mechanisms for the paradoxical effect of SupraT on PCa have been described, including cellular senescence and cell death (39–42). Understanding how BAT works at the molecular and cellular levels might help in rationally combining BAT with other agents to achieve increased efficacy and tumor response. Our findings that the administration of SupraT may lead to ferroptosis mediated by lipid peroxides is intriguing. It is speculated that PCa cells rely on lipid metabolism for their growth, and targeting lipid metabolism to overcome prostate cancer growth is viewed as a possible therapeutic strategy (43). SupraT may also influence both lipid uptake and synthesis, as androgens have been linked to regulating lipid metabolism (44–46). The increased cellular lipids may serve as substrates for the generation of lipid peroxides, leading to ferroptosis. Future work will provide more insights into the role of SupraT induced lipid metabolism in ferroptosis.
Based on our previous experimental and clinical reports that SupraT induces DSBs and extreme responders to BAT therapy harbor DNA repair mutations (6, 11, 47), we speculated that PCa harboring mutations in the DSB repair pathway would be sensitive to SupraT. We were surprised to note that the damaged DNA was shuttled for autophagosomal degradation. Such nucleophagic degradation of damaged DNA has been noted for radiation- and chemo-therapy (48–50). A key finding of our study is that SupraT induces STING and downstream NF-kappaB-driven immune genes to activate immune infiltration in vitro, in animal xenografts, and in patient resected tumors. Our results further provide insights into cross-talk between the canonical and non-canonical NF-kappaB pathways induced by SupraT. Previous studies have shown that the canonical pathway is rapidly induced upon stimulation, while chronic stimulation is required for the non-canonical pathway to become activated (51). Our results indicate that the canonical pathway is induced forty-eight hours after SupraT administration, which synchronizes with the induction of autophagy. This early induction suggests that the activation of the canonical pathway might not be a direct response to SupraT, but maybe an indirect response to SupraT-mediated nucleophagy. A key question that remains to be addressed is whether autophagy induction occurs in response to the presence of damaged DNA in the cytoplasm or is induced by SupraT irrespective of DNA repair competency. We did not detect a global increase in autophagic proteins like Beclin or ATG12. Moreover, autophagy was not induced in AR-positive SupraT-insensitive LAPC4 or 22Rv1 cells, indicating that autophagy induction may not solely depend on AR status or SupraT treatment. A decrease in autophagy induction upon NCOA4 knockdown in SupraT treated cells indicated that NCOA4 may play an important role SupraT mediated autophagy-a feature that requires further investigation. Another noteworthy observation was the induction of both the STING and RIG-I pathways. While STING activation is mediated by non-self and damaged-self cytoplasmic DNA, RIG-I is stimulated by 5′-triphosphorylated short double-stranded RNA (52). Through mechanisms not yet fully understood, RIG-I is also activated by double-stranded DNA (36). We found MDA5, another RNA sensor, was also induced by SupraT. While the contributions of MDA5 to SupraT mediated innate immune signaling remain to be elucidated, it is likely to play a minor role as double knockouts of STING/RIG-I were sufficient to abrogate NIK stabilization and NF-kappaB signaling.
STING signaling is considered a double-edged sword as chronic STING signaling is considered pro-tumorigenic, while acute STING signaling is considered anti-tumorigenic (53). Our data indicates that STING is acutely induced by SupraT, a feature that may contribute to its anti-tumor effect. Infiltration of immune cells in tumor xenografts and patient biopsies further indicates that the immune system is engaged upon SupraT administration. This observation has clinical significance as the presence of immune infiltrates is a key parameter and correlates with therapeutic response to immunotherapy (54). Recently, immune checkpoint therapy has been shown to induce ferroptosis in tumor cells (55). Ferroptosis itself may likely potentiate the immune clearance of tumors through the release of damage-associated molecular patterns (24). Understanding the role of ferroptosis in SupraT induced immune signaling, may provide additional mechanistic insights into cellular immune response. Further, cytokines and chemokine genes induced by the STING-TBK1-NF-kappaB pathway may serve as predictors of therapeutic response in the future. Impending clinical investigations involving the combination of SupraT with immune checkpoint inhibitors may be further informative.
In summary, our findings suggests that SupraT induces two autophagy-mediated pathways, namely ferritinophagy and nucleophagy. While ferritinophagy may induce ferroptosis, consequently, leading to cell death; nucleophagy induces innate immune signaling (through nucleic acid sensing-NF kappaB signaling) and infiltration of immune cells.
Supplementary Material
Significance.
This study demonstrates that supraphysiological testosterone induces two parallel autophagy-mediated processes, ferritinophagy and nucleophagy, which then activate nucleic acid sensors to drive immune signaling pathways in prostate cancer.
Acknowledgement
We thank Dr. Sabatini and Dr. Lander for pCW-Cas9 (Addgene#50661) and pLXsgRNA (Addgene#50662), Dr. Trono for pMD2.G (Addgene 12259) and psPAX2 (Addgene#12260), and Dr. Wade Harper for the NCOA4α-pHAGE-C-FLAG-HA plasmid. This work is supported by the DOD grants W81XWH1910724 to SK and W81XWH-14–2-0189 to SD; Allegheny Health Network-Johns Hopkins Cancer Research Fund, Shared Instrumentation grant S10OD016374, and the NCI CORE Grant P30CA006973. ESA and SK are partially supported by a PCF 2018 Challenge Award. M. Coffey was supported by the CUPID program at Hopkins. Research reported in this publication was also supported by the Office of the Director of the National Institutes of Health under award number S10OD016374 and S10OD023548.
Footnotes
Conflict of interest
E.S.A. is a paid consultant/advisor to Janssen, Astellas, Sanofi, Dendreon, Pfizer, Amgen, Eli Lilly, Bayer, AstraZeneca, Bristol-Myers Squibb, Clovis, and Merck; and has received research funding from Janssen, Johnson & Johnson, Sanofi, Dendreon, Genentech, Novartis, Tokai, Bristol Myers-Squibb, AstraZeneca, Clovis, and Merck.
R.K., J.M., O.O., K.B., N.T., S.Kn., M.C., D.T., C.G., B.O., T.J., M.R., L.D., S.W., W.N.B., J.T.I., A.M.D. M., M.C.M., D.Z.Q., K.J.P., D.M.P. M.A.C., S.R.D., S.K.K. declare no potential conflicts of interest.
References
- 1.Huggins CHC. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Research. 1941;1(293–7. [DOI] [PubMed] [Google Scholar]
- 2.Harris WP, Mostaghel EA, Nelson PS, and Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nature clinical practice Urology. 2009;6(2):76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, and Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61(9):3550–5. [PubMed] [Google Scholar]
- 4.Taplin ME, and Balk SP. Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence. J Cell Biochem. 2004;91(3):483–90. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs JT, D’Antonio JM, Chen S, Antony L, Dalrymple SP, Ndikuyeze GH, Luo J, and Denmeade SR. Adaptive auto-regulation of androgen receptor provides a paradigm shifting rationale for bipolar androgen therapy (BAT) for castrate resistant human prostate cancer. Prostate. 2012;72(14):1491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweizer MT, Antonarakis ES, Wang H, Ajiboye AS, Spitz A, Cao H, Luo J, Haffner MC, Yegnasubramanian S, Carducci MA, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med. 2015;7(269):269ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42(8):668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, and Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326(5957):1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee P, Schweizer MT, Lucas JM, Coleman I, Nyquist MD, Frank SB, Tharakan R, Mostaghel E, Luo J, and Pritchard CC. Supraphysiological androgens suppress prostate cancer growth through androgen receptor–mediated DNA damage. The Journal of clinical investigation. 2019;129(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teply BA, and Antonarakis ES. Treatment strategies for DNA repair-deficient prostate cancer. Expert Rev Clin Pharmacol. 2017;10(8):889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teply BA, Kachhap S, Eisenberger MA, and Denmeade SR. Extreme Response to High-dose Testosterone in BRCA2- and ATM-mutated Prostate Cancer. Eur Urol. 2017;71(3):499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mark C Markowski SK, De Marzo Angelo M, Samuel R. Denmeade, Emmanuel S. Antonarakis. Molecular and clinical characterization of metastatic castration-resistant prostate cancer (mCRPC) patients achieving deep PSA responses to bipolar androgen therapy (BAT). J Clin Oncol. 2021;39( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee P, Schweizer MT, Lucas JM, Coleman I, Nyquist MD, Frank SB, Tharakan R, Mostaghel E, Luo J, Pritchard CC, et al. Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage. J Clin Invest. 2019;129(10):4245–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam HM, Nguyen HM, Labrecque MP, Brown LG, Coleman IM, Gulati R, Lakely B, Sondheim D, Chatterjee P, Marck BT, et al. Durable Response of Enzalutamide-resistant Prostate Cancer to Supraphysiological Testosterone Is Associated with a Multifaceted Growth Suppression and Impaired DNA Damage Response Transcriptomic Program in Patient-derived Xenografts. Eur Urol. 2020;77(2):144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robitaille AC, Mariani MK, Fortin A, and Grandvaux N. A High Resolution Method to Monitor Phosphorylation-dependent Activation of IRF3. J Vis Exp. 2016. 107):e53723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sramkoski RM, Pretlow TG 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, and Jacobberger JW. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35(7):403–9. [DOI] [PubMed] [Google Scholar]
- 17.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171(2):273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon SJ, and Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10(1):9–17. [DOI] [PubMed] [Google Scholar]
- 20.Doll S, and Conrad M. Iron and ferroptosis: A still ill-defined liaison. IUBMB Life. 2017;69(6):423–34. [DOI] [PubMed] [Google Scholar]
- 21.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16(11):1069–79. [DOI] [PubMed] [Google Scholar]
- 22.Mancias JD, Wang X, Gygi SP, Harper JW, and Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naguib YM. A fluorometric method for measurement of peroxyl radical scavenging activities of lipophilic antioxidants. Anal Biochem. 1998;265(2):290–8. [DOI] [PubMed] [Google Scholar]
- 24.Friedmann Angeli JP, Krysko DV, and Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19(7):405–14. [DOI] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burdette DL, and Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14(1):19–26. [DOI] [PubMed] [Google Scholar]
- 27.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, and Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46(6):735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, and Yan N. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe. 2015;18(2):157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106(49):20842–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroder K, Muruve DA, and Tschopp J. Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19(6):R262–5. [DOI] [PubMed] [Google Scholar]
- 31.Shang G, Zhang C, Chen ZJ, Bai XC, and Zhang X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature. 2019;567(7748):389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown TR, Rothwell SW, and Migeon CJ. Comparison of methyltrienolone and dihydrotestosterone binding and metabolism in human genital skin fibroblasts. J Steroid Biochem. 1981;14(10):1013–22. [DOI] [PubMed] [Google Scholar]
- 33.Kemppainen JA, Lane MV, Sar M, and Wilson EM. Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. J Biol Chem. 1992;267(2):968–74. [PubMed] [Google Scholar]
- 34.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, and Lenz H-J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation–a target for novel cancer therapy. Cancer treatment reviews. 2018;63(40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162(5):974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, and Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10(10):1065–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheil JM, Gallimore PH, Zimmer SG, and Sopori ML. Susceptibility of Adenovirus 2-transformed rat cell lines to natural killer (NK) cells: direct correlation between NK resistance and in vivo tumorigenesis. J Immunol. 1984;132(3):1578–82. [PubMed] [Google Scholar]
- 38.Fong L, Carroll P, Weinberg V, Chan S, Lewis J, Corman J, Amling CL, Stephenson RA, Simko J, Sheikh NA, et al. Activated lymphocyte recruitment into the tumor microenvironment following preoperative sipuleucel-T for localized prostate cancer. J Natl Cancer Inst. 2014;106(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vander Griend DJ, Litvinov IV, and Isaacs JT. Stabilizing androgen receptor in mitosis inhibits prostate cancer proliferation. Cell Cycle. 2007;6(6):647–51. [DOI] [PubMed] [Google Scholar]
- 40.Roediger J, Hessenkemper W, Bartsch S, Manvelyan M, Huettner SS, Liehr T, Esmaeili M, Foller S, Petersen I, Grimm MO, et al. Supraphysiological androgen levels induce cellular senescence in human prostate cancer cells through the Src-Akt pathway. Mol Cancer. 2014;13(214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen S, Niu Y, Lee SO, and Chang C. Androgen receptor (AR) positive vs negative roles in prostate cancer cell deaths including apoptosis, anoikis, entosis, necrosis and autophagic cell death. Cancer Treat Rev. 2014;40(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isaacs JT. Antagonistic effect of androgen on prostatic cell death. Prostate. 1984;5(5):545–57. [DOI] [PubMed] [Google Scholar]
- 43.Schlaepfer IR, Rider L, Rodrigues LU, Gijon MA, Pac CT, Romero L, Cimic A, Sirintrapun SJ, Glode LM, Eckel RH, et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol Cancer Ther. 2014;13(10):2361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swinnen JV, Brusselmans K, and Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9(4):358–65. [DOI] [PubMed] [Google Scholar]
- 45.Suburu J, and Chen YQ. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat. 2012;98(1–2):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler LM, Centenera MM, and Swinnen JV. Androgen control of lipid metabolism in prostate cancer: novel insights and future applications. Endocr Relat Cancer. 2016;23(5):R219–27. [DOI] [PubMed] [Google Scholar]
- 47.Teply BA, Wang H, Luber B, Sullivan R, Rifkind I, Bruns A, Spitz A, DeCarli M, Sinibaldi V, Pratz CF, et al. Bipolar androgen therapy in men with metastatic castration-resistant prostate cancer after progression on enzalutamide: an open-label, phase 2, multicohort study. Lancet Oncol. 2018;19(1):76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, and Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5(6):795–804. [DOI] [PubMed] [Google Scholar]
- 49.Eapen VV, Waterman DP, Bernard A, Schiffmann N, Sayas E, Kamber R, Lemos B, Memisoglu G, Ang J, Mazella A, et al. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc Natl Acad Sci U S A. 2017;114(7):E1158–E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41(5):843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun SC. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17(9):545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehwinkel J, and Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20(9):537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon J, and Bakhoum SF. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020;10(1):26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou W, Wolchok JD, and Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, Green M, Choi JE, Gijon M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.