Abstract
To investigate the checkpoint response to aberrant initiation, we analyzed the cell cycle checkpoint response induced by mutations of Schizosaccharomyces pombe DNA primase. DNA primase has two subunits, Spp1 and Spp2 (S. pombe primases 1 and 2). Spp1 is the catalytic subunit that synthesizes the RNA primer, which is then extended by DNA polymerase α (Polα) to synthesize an initiation DNA structure, and this catalytic function of Polα is a prerequisite for generating the S-M phase checkpoint. Here we show that Spp2 is required for coupling the function of Spp1 to Polα. Thermosensitive mutations of spp2+ destabilize the Polα-primase complex, resulting in an allele-specific S phase checkpoint defect. The mutant exhibiting a more severe checkpoint defect also has a higher extent of Polα-primase complex instability and deficiency in the hydroxyurea-induced Cds1-mediated intra-S phase checkpoint response. However, this mutant is able to activate the Cds1 response to S phase arrest induced by temperature. These findings suggest that the Cds1 response to the S-phase arrest signal(s) induced by a initiation mutant is different from that induced by hydroxyurea. Interestingly, a polαts mutant with a defective S-M phase checkpoint and an spp2 mutant with an intact checkpoint have a similar Polα-primase complex stability, and the Cds1 response induced by hydroxyurea or by the mutant arrests at the restrictive temperature. Thus, the Cds1-mediated intra-S phase checkpoint response induced by hydroxyurea can also be distinguished from the S-M phase checkpoint response that requires the initiation DNA synthesis by Polα.
To maintain genomic integrity, eukaryotic cells have the checkpoint mechanisms to delay progression of the cell cycle when cells encounter perturbation of DNA replication or DNA damage (18). In fission yeast, a group of proteins, Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1, known as checkpoint Rad proteins, function early in the surveillance of both the replication perturbation and DNA damage (1, 13). These checkpoint Rad proteins are thought to sense and transduce signals of aberrant replication and DNA damage to activate two downstream protein kinases, Cds1 and Chk1, to arrest the cell cycle (5–8, 40). In response to S phase arrest by hydroxyurea, cdc mutant arrest, or DNA damage induced during S phase, Cds1 is phosphorylated and activated (19). Cds1 activation delays the progression of S phase (termed intra-S phase checkpoint) and contributes to preventing mitosis (3, 19, 26). The Cds1 structural counterpart of budding yeast, RAD53, has been shown to be an essential factor for maintaining the intra-S phase checkpoint to prevent the firing of late replication origins when the progression of replication forks from early-firing origins is blocked by hydroxyurea (12, 33, 34).
Another downstream kinase, Chk1, is required to arrest mitosis when DNA is damaged in late S phase and G2 phase and is also required to prevent mitosis when cdc replication mutants are used to perturb S phase (7). Chk1 is not required to prevent mitosis in response to hydroxyurea block. Following DNA damage, Chk1 protein is phosphorylated in a cell-cycle-specific manner (23), and Chk1 phosphorylation is correlated to cell cycle arrest (4). Chk1 phosphorylation allows binding of 14-3-3 proteins with Chk1 that is thought to direct Chk1 for specific substrate (9). Although Chk1 is not phosphorylated in hydroxyurea block or during early S phase perturbation (23), Chk1 is phosphorylated when S phase is blocked by hydroxyurea in a cds1Δ background (4, 19). Chk1 kinase has been shown to phosphorylate in vitro two Cdc2 kinase regulators, Wee1 kinase and Cdc25 phosphatase (15, 30, 32). Phosphorylation of Cdc25 by Chk1 allows Cdc25 to associate with 14-3-3 proteins, leading to nuclear exclusion of Cdc25 (21, 31, 42).
These findings strongly suggest that checkpoint signals generated from early-S-phase perturbation are different from those generated during ongoing or late S phase. Early-S-phase perturbation activates Cds1 kinase to maintain an intra-S phase checkpoint, while ongoing or late-S-phase perturbation results in Chk1 phosphorylation to prevent mitosis. Thus, Cds1 and Chk1 function in two distinct but mutually reinforced ways in the cell cycle surveillance mechanisms.
We are interested in defining the requirements for generating the checkpoint response to aberrant S phase initiation. To achieve this goal, we investigated the effect of mutations of DNA polymerase α (Polα)-primase on the cell cycle events. DNA Polα-primase, a four-subunit enzyme complex, is the principal enzyme that initiates DNA replication on both leading and lagging strands. DNA primase synthesizes an RNA primer which is then extended by Polα to synthesize an initiation DNA structure (39, 41). DNA primase is a heterodimeric enzyme complex, consisting of a catalytic subunit that synthesizes the RNA primer, named p49 in mammalian cells and PRI1 in budding yeast, and a second subunit that has no detectable enzymatic activity, named p58 in mammalian cells and PRIII in budding yeast (41).
We and others have previously demonstrated that deletion or mutation of DNA Polα results in the cells entering inappropriate mitosis (2, 11). We have previously shown that germinating spores derived from a Schizosaccharomyces pombe heterozygous diploid containing one copy of the polα+ gene with a mutation at a critical metal activator binding residue (Asp984 in region I) display an abnormal mitotic phenotype (see Fig. 2 of reference 2). In vitro studies have shown that Polα-primase complex containing this specific Polα mutation abolishes only the catalytic function and not the physical association of Polα and primase, and the mutant complex is able to synthesize the RNA primer in vitro but is unable to extend the RNA primer (10). These experiments strongly suggest that the catalytic function of Polα is required for generating the checkpoint to prevent inappropriate mitotic entry.
We isolated the fission yeast genes and cDNAs of both primase subunits, named spp1+ and spp2+ for S. pombe primases 1 and 2, respectively, and generated conditional mutants of spp1+ and spp2+. In this report, we investigate the effect of spp2 mutations on the cell cycle checkpoint response. The results of our studies indicate that Spp2 is the subunit that couples the function of Spp1 with Polα. Mutations of spp2 cause instability of Polα-primase complex. Analyses of the checkpoint effector kinase response to mutations of spp2 indicate that the requirement for Cds1 checkpoint response to an S phase initiation mutant arrest is different from that for Cds1 response to replication stall induced by the DNA synthesis inhibitor hydroxyurea. The requirements for intra-S phase Cds1 response can also be distinguished from the Polα activity-dependent S-M phase checkpoint response.
MATERIALS AND METHODS
Yeast strains, media, and genetic, cell biological, and molecular methods.
The strains used in this study were listed in Table 1. Cells were maintained either in rich medium (YE5S) or Edinburgh minimal medium with appropriate supplements as described elsewhere (25). All genetic methods were performed as described previously (17). Molecular biology techniques were performed as described by Maniatis et al. (22). Transformation of fission yeast was performed as described previously (16). Growth and viability analysis of cells were performed as described previously (2, 16). Cell extracts were prepared by glass bead disruption as described by Al-Khodairy et al. (2).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| KG2 | h− ade6-M216 leu1-32 ura4-D18 his3-D1 | K. Gould |
| KG23 | h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 | K. Gould |
| KG3 | h+ ade6-M210 leu1-32 ura4-D18 his3-D1 | K. Gould |
| NW222 | h− chk1:ep ade6-M216 leu1-32 | N. Walworth |
| DBts13 | h+ polαΔ::polαts13 leu+ his+ ade6-M210 leu1-32 ura4-D18 his3-D1 | D. Bhaumik |
| ST108 | h− spp2-8 ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| ST109 | h− spp2-9 ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| ST118 | h− spp2-8 ura4+ | This study |
| ST119 | h− spp2-9 ura4+ | This study |
| ST101 | h−/h+ spp2+/spp2Δ::ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 | This study |
| ST128 | h− spp2-8 ura4+ chk1:ep ade6-M216 leu1-32 his3-D1 | This study |
| ST129 | h+ spp2-9 ura4+ chk1:ep ade6-M216 leu1-32 his3-D1 | This study |
| ST138 | h−/h+ spp2+/spp2-8 ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 | This study |
| ST23 | h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4+ his3-D1/his3-D1 | This study |
| spp2-8 cds1Δ | h− spp2-8 cds1::ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spp2-8 chk1Δ | h− spp2-8 chk1::ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spp2-8 rad3Δ | h+ spp2-8 rad3::ura4+ ade6-M210 leu1-32 ura4-D18 his3-D1 | This study |
| spp2-9 cds1Δ | h− spp2-9 cds1::ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spp2-9 chk1Δ | h− spp2-9 chk1::ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| spp2-9 rad3Δ | h+ spp2-9 rad3::ura4+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
Plasmids.
A disruption vector pUS-Ura-DS, which contains an ura4+ gene placed between upstream and downstream of spp2+ coding sequence, was constructed as follows. First, the ura4+ gene was cloned into SalI/BamHI sites of pBluescript SK(−) (Stratagene) to generate pBS-Ura. Then, the ∼600-bp downstream region of spp2+ coding sequence was amplified from genomic DNA by PCR using Spp2-5′-DS/Pst and Spp2-3′-DS/Kpn as primers and subcloned into the PstI/KpnI sites of pBS-Ura to create pUra-DS. Finally, the ∼500-bp upstream region of spp2+ coding sequence was amplified from genomic DNA by PCR using Spp2-5′-US/Xba and Spp2-3′-US/Bam as primers and subcloned into the XbaI/BamHI sites of pUra-DS to create pUS-Ura-DS. The coding region of spp2+ cDNA was amplified from the spp2+-cDNA clone (as described below) by PCR using Spp2-5′-Xho and Spp2-3′-Bam as primers and subcloned into pET expression vector to create pET-Spp2. The oligonucleotides used for PCR were as follows: Spp2-5′-US/Xba, AGATCTAGAAGTCTTTACGATGCATTATCTAATG; Spp2-3′-US/Bam, CGCGGATCCGGTGGTTAGGGAAGAGTCTATTTG; Spp2-5′-DS/Pst, AAACTGCAGTAAGCTAAGATACTTTTAGTTCACG; SPP2-3′-DS/Kpn, AAAGGTACCGGTGGTGTTCTTATGCTTATCG; Spp2-5′-Xho, CCGCTCGAGATGTTCAGAACGACCAAAAGTCGAG; and Spp2-3′-Bam, AAGGGATCCTTATGATTCTAAACTAAGTTGAAAATATTG.
Identification and isolation of spp2+ gene and cDNA.
Spp2 protein was initially identified as a 54-kDa polypeptide tightly associated with DNA Polα purified from S. pombe cell extract (R. Davis and T. Wang, unpublished). Mass spectrometric analysis of the 54-kDa protein identified several peptide sequences overlapping with an open reading frame in chromosome II (cosmid SPBC17D11) from the Sanger fission yeast genome database. The spp2+ cDNA was isolated by screening a λ-ZAP cDNA library from S. pombe (S. Tan and T. Wang, unpublished results). A full-length spp2+ cDNA was assembled from overlapping clones and was subcloned into pET expression vector. Homology searching through GenBank found that Spp2 shares 37, 37, and 40% sequence identity to the primase subunits p58 of humans (35) and mice (24) and of PRIII of S. cerevisiae (14), respectively.
Construction of spp2Δ strain.
Heterozygous diploid strain ST101 (spp2+/spp2Δ) carrying a deletion of one copy of spp2+ was constructed by one-step gene replacement. The DNA fragment, which contains a ∼500-bp spp2+ upstream sequence, a functional ura4+ gene, and a ∼600-bp spp2+ downstream region, was released from pUS-Ura-DS by digesting it with XbaI and KpnI and transformed into the diploid strain KG23. The transformants were selected on minimal medium lacking uracil and screened for several rounds for the stability of the ura4+ marker. Successful replacement of one copy of spp2+ coding region by ura4+ gene was confirmed by genomic Southern analysis. Tetrad dissection of spores derived from ST101 (spp2+/spp2Δ) resulted in two viable uracil auxotrophic spores, confirming that Spp2 is essential for cell growth. To analyze the phenotype of spp2Δ germinating spores, the spores isolated from ST101 (spp2+/spp2Δ) were selectively germinated in minimal medium lacking uracil. Samples were taken at indicated times to determine the DNA contents by fluorescence-activated cell sorter (FACS) analysis. A heterozygous ST23 containing only one copy of ura4+ was constructed and used as a wild-type control.
Isolation of spp2 temperature-sensitive mutants.
Mutations were introduced into the spp2+ gene by mutagenic PCR as described earlier (37). Briefly, spp2+ cDNA was amplified from pET-Spp2 by Taq DNA polymerase in a buffer containing 20 mM Tris (pH 8.4), 50 mM KCl, 7 mM MgCl2, 0.5 mM MnCl2, 0.1% gelatin, 200 μM dATP, 200 μM dGTP, 1 mM dCTP, and 1 mM dTTP. The primers for PCR amplification were Spp2-3′-BamH and T7 primer (AATACGACTCACTATAG). The PCR reaction was carried out in a 100-μl reaction and performed with 25 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min. An spp2 mutation library was constructed by cloning the mutagenized spp2 cDNA into XbaI/BamHI sites of pUra-DS. The DNA fragments, which contain the mutagenized spp2 cDNA, a ura4+ gene, and ∼600 bp of downstream of the spp2+ coding region, were isolated by digesting the library with XbaI and KpnI and transformed into the haploid strain KG2. Transformants were screened by replica plating onto minimal medium lacking uracil and then tested for temperature sensitivity. Stable temperature-sensitive ura+ transformants were selected and backcrossed with wild-type strain three times. Correct chromosomal integration of the spp2 mutant was verified by genomic Southern blot analysis. All of the temperature-sensitive spp2 mutants isolated can be rescued by pREP81 plasmid carrying spp2+.
Generation and purification of Spp2 antibody.
Recombinant Spp2 protein was expressed in bacteria by using pET expression system (Novagen). The entire coding region of spp2+ was cloned into pET vector. The resulting plasmid, pET-Spp2, which contains the translation initiation codon ATG, a six-histidine tag, and the spp2+ coding region, was transformed into strain BL21(DE3). Spp2 protein expressed from BL21(DE3) harboring pET-Spp2 was purified on Ni2+-nitrilotriacetic acid column chromatography as described elsewhere (36). The recombinant Spp2 protein eluted from the Ni2+-nitrilotriacetic acid column was further purified to near homogeneity by Mono S chromatography via the SMART system (Pharmacia Biotech). Briefly, an Spp2 protein fraction isolated from the Ni2+-nitrilotriacetic acid column was first dialyzed against buffer containing 50 mM HEPES (pH 7.9), 0.5 mM EDTA, and 50 mM KCl and then applied to a 0.12-ml Mono S column (Pharmacia Biotech). Spp2 protein was eluted with a 1-ml linear gradient from 50 to 500 mM KCl in the same buffer. The purified recombinant Spp2 protein was used as antigen to raise antibodies against Spp2 in rabbit. The antisera were affinity purified on an Spp2 protein column prepared by immobilizing the recombinant Spp2 on N-hydroxysuccinimide-activated Sepharose (Pharmacia Biotech).
Cds1 kinase assay.
In vitro Cds1 kinase assay was performed as described elsewhere (2, 19) with the following modifications. One milligram of total protein from cell extract was mixed with 2 μl of affinity-purified rabbit anti-Cds1 antibody at 4°C for 2 h. Twenty microliters of protein A-agarose (as 50% slurry) was added to the mixture and incubated at 4°C for an additional hour. The immunocomplexes were precipitated and washed two times in a lysis buffer containing 150 mM HEPES (pH 7.9), 250 mM KCl, 1 mM EDTA, 6 μM leupeptin, 2 μM pepstatin A, 2 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride, followed by washing three times with the kinase buffer containing 10 mM HEPES (pH 7.5), 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, and 1 mM dithiothreitol. As a control for equal amounts of Cds1 used for the kinase assay, 3% of each sample was removed prior to immunoprecipitation and quantitated by Western blotting with anti-Cds1 antibodies. A kinase reaction was performed at 30°C for 15 min in the kinase buffer containing the Cds1 immunoprecipitate, 5 μg of myelin basic protein (MBP), 5 μCi of [α-32P]ATP, and 100 μM ATP. Reactions were terminated by the addition of sodium dodecyl sulfate (SDS) sample buffer. After being boiled for 3 min, the samples were fractionated on an SDS–15% polyacrylamide gel. Gels were fixed in 40% methanol and 10% acetic acid and dried. The kinase activity was quantitated using an IS-1000 digital imaging system (Alpha Innotech, San Leandro, Calif.).
Immunoprecipitation of Polα-primase complex.
The Polα-primase complex was immunoprecipitated from cell lysates by using anti-Polα antibody (27) immobilized on protein A-agarose. One milligram of total cell extract protein was mixed with 15 μl of the anti-Polα antibody beads in the cell lysis buffer described above. After incubation at 4°C for 4 h, the immunocomplexes were collected and washed five times with the lysis buffer. The pellets were resuspended in 30 μl of SDS sample loading buffer and analyzed by Western blotting using antibodies against Polα, Spp1, and Spp2 as probes.
Flow cytometry analysis.
Cells were harvested, washed in water, and fixed in 70% ethanol prior to staining with propidium iodide as described earlier (28). DNA contents were measured by using a Coulter Electronics FACS.
Cytology analysis.
Cells were fixed in 70% ethanol and stained by DAPI (4′,6′-diamidino-2-phenylindole) followed by calcofluor as described previously (38).
RESULTS
Characterization of cells with spp2 mutations.
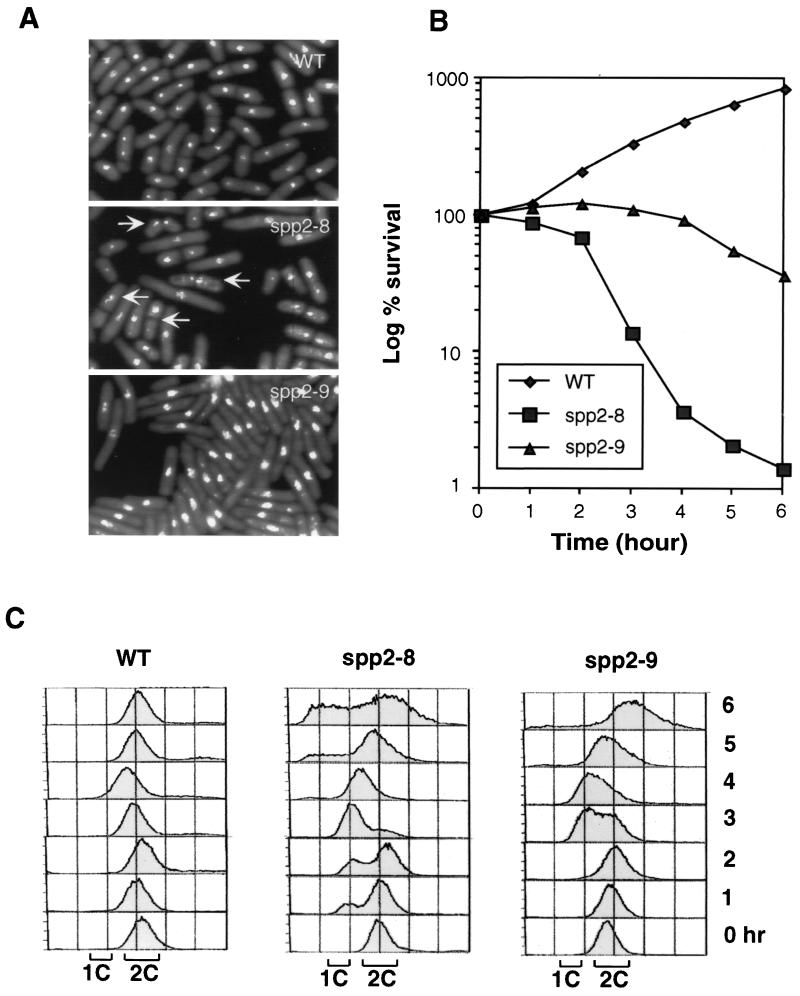
To investigate the effect of spp2+ mutation on the cell cycle checkpoint response, we isolated a panel of thermosensitive mutants of spp2+. At the restrictive temperature of 36°C, the spp2 mutants exhibited two distinct phenotypes. Two representative mutants of each phenotype, spp2-8 and spp2-9, were characterized in this study. At 6 h after the shift to 36°C, spp2-8 exhibited heterogeneous cell size, with ∼60% of the cells displaying aberrant mitotic nuclear phenotype. In contrast, spp2-9 cells had elongated cell morphology with nominal levels of cells displaying abnormal mitotic nuclear phenotype (Fig. 1A). Consistent with their phenotypes, spp2-8 cells died more rapidly than spp2-9 cells when mid-log-phase cultures were shifted to 36°C (Fig. 1B). At 6 h after the shift to 36°C, <2% of the spp2-8 cells remained viable, while >30% of the spp2-9 cells were viable (Fig. 1B). FACS profiles of these two mutants correlated with their phenotypes and their kinetics of viability loss (Fig. 1C). Wild-type and mutant cells initially had 2C DNA contents, since asynchronous S. pombe cultures are predominantly in the G2 phase. The DNA content of wild-type cells remained at 2C, the peak of the FACS profile of spp2-8 and spp2-9 cells shifted to about 1.5C after 3 h at 36°C and then shifted toward 2C. After 6 h, the FACS profile of spp2-8 showed that a portion of the cells had a less than 1C DNA content and a portion of the spp2-8 cells had a greater than 2C DNA content, which reflected the observed heterogeneous cell size and aberrant mitotic nuclear phenotype (Fig. 1A). At 6 h after the shift to 36°C, the majority of the spp2-9 cells exhibited a greater-than-2C profile, which correlated with the observed elongated phenotype of spp2-9 (Fig. 1A). None of the spp2 mutants was substantially sensitive to UV or hydroxyurea at 30°C.
FIG. 1.
Characterization of temperature-sensitive mutants of spp2+. Early-log-phase wild-type 972h−, ST118 (spp2-8), and ST119 (spp2-9) cells grown at 25°C were shifted to 36°C. Samples were removed at the indicated times and processed for morphological, survival, and FACS analysis. (A) Photomicrographs of wild-type and spp2 mutant cells 6 h after the shift to 36°C. Arrows indicate spp2-8 cells that entered inappropriate mitosis. (B) Viability of wild-type and spp2 mutant cells. (C) FACS analysis of wild-type and spp2 mutant cells.
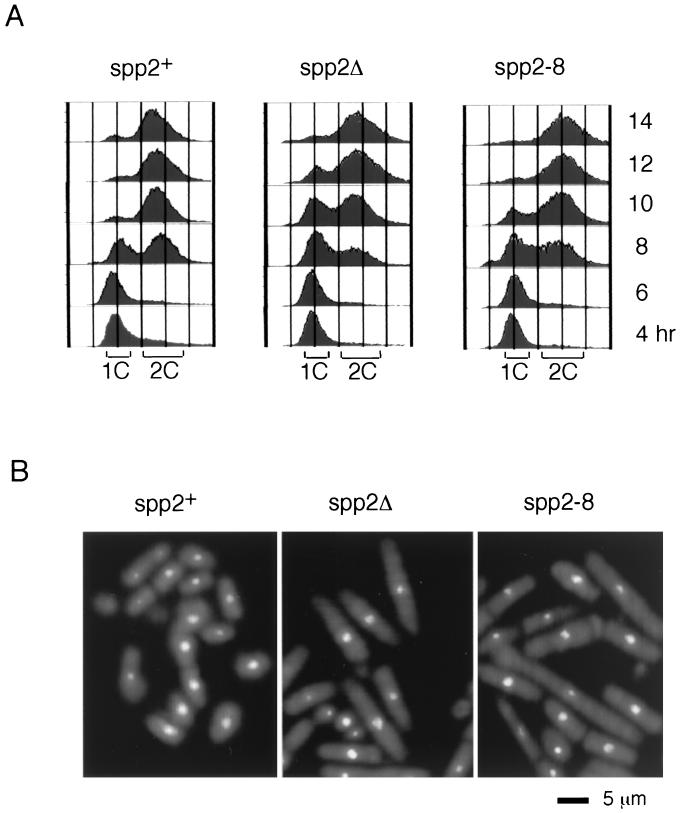
To determine the phenotype of absence of spp2+, a heterozygous diploid of spp2Δ and spp2+ was constructed as described in Materials and Methods. Tetrad analysis of the diploid yielded two viable ura4− spores, indicating that spp2+ is essential for cell viability. We have previously shown that germinating spores carrying polαΔ enter mitosis with a 1C DNA content (2). Since Spp2 is a component of the Polα-primase complex, we expect that the germinating spores carrying spp2Δ would display a phenotype similar to that of polαΔ. Spores carrying spp2Δ derived from the spp2+/spp2Δ diploid were selected for germination in medium lacking uracil. As a control, spores carrying spp2+ derived from a ura4-D18/ura4+ diploid strain were analyzed in parallel. Spores carrying spp2+ entered S phase 8 h after inoculation, and most cells completed S phase with 2C DNA content after 10 h (Fig. 2A, 8 to 10 h). In contrast, 10 h after inoculation, spores carrying spp2Δ showed a delay in entering S phase, with a fraction of the cells having a 2C DNA profile similar to the wild-type spores and a fraction of the cells having 1C DNA content (Fig. 2A, 8 to 10 h). After 14 h, spp2Δ cells completed S phase and had a DNA content identical to that of the spp2+ cells (Fig. 2A, 14 h). The germinating spp2Δ spores exhibited elongated cell morphology with <3% of the cells exhibiting aberrant mitotic nuclear morphology (Fig. 2B, middle panel). This is in striking contrast to polαΔ, where ∼60% of polαΔ germinating spores display aberrant mitotic phenotype (2).
FIG. 2.
Germinating spores harboring spp2Δ and spp2-8 arrested with a cdc phenotype. (A) FACS profiles of spp2+ (left panel), spp2Δ (middle panel), and spp2-8 (right panel) germinating spores at 36°C. (B) Phenotype of germinating spores carrying spp2+ (left panel), spp2Δ (middle panel), and spp2-8 14 h after inoculation into selective medium. The diploid strains used for spores harboring spp2+, spp2Δ, and spp2-8 were ST23, ST101, and ST 138, respectively.
Since spp2-8 mutant exhibits aberrant mitotic phenotype at 36°C (Fig. 1A), it is surprising that germinating spp2Δ spores display an elongated phenotype. It is possible that Spp2 is a stable protein and that the residual Spp2 carried over from the original diploid has allowed the spp2Δ cells to initiate S phase in the absence of Spp2 transcription and translation. To test this possibility, we constructed a heterozygous diploid of spp2+ and spp2-8 and selectively germinated the spores harboring spp2-8 at 36°C. The spp2-8 germinating spores had a similar FACS profile to that of the spores harboring spp2Δ (Fig. 2A). At 14 h following inoculation, the spp2-8 germinating spores exhibited an elongated cell morphology with normal nuclear morphology (Fig. 2B, right panel) and not the aberrant mitotic phenotype of spp2-8 shown in Fig. 1A. It is worth mentioning that spp2+/spp2-8 and spp2+/ssp2-9 diploids grow normally at 36°C, indicating that both mutant alleles are recessive.
To further analyze the phenotype of spp2Δ germinating spores in the absence of DNA replication, we treated the germinating spp2Δ spores with hydroxyurea, an inhibitor of DNA synthesis, at 0 and 6 h following inoculation in selective medium lacking uracil. Under these conditions, germinating spores carrying spp2+ remained arrested with 1C DNA for up to 12 to 14 h postinoculation with elongated cell morphology. Similarly, germinating spores carrying spp2Δ arrested with 1C DNA content with normal nuclear morphology (data not shown). These experiments indicate that the observed elongated phenotype of spp2Δ is indeed due to residual wild-type Spp2 carried over from the original diploid.
Analysis of the stability of Polα-primase complex in spp2 and polαts mutants.
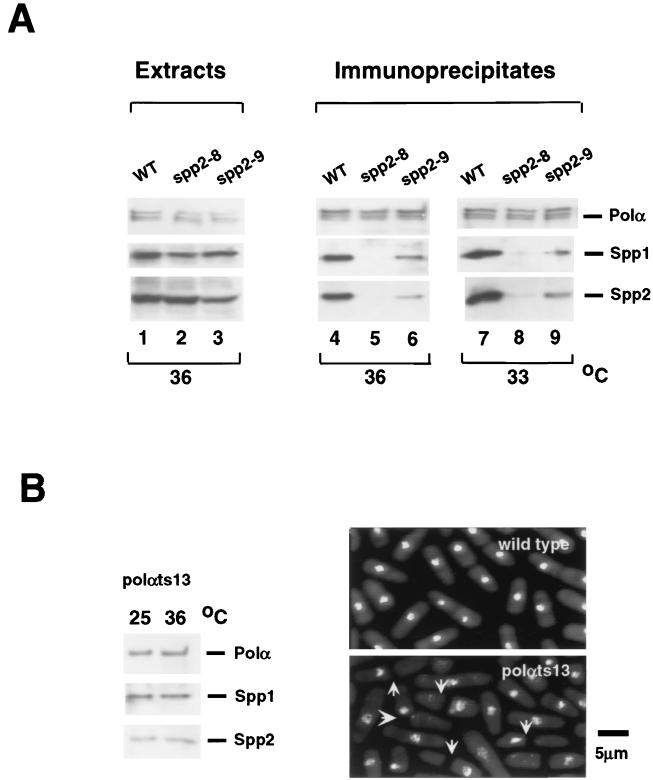
Since the two spp2 mutants have a difference in their phenotype, we tested the possible protein structural differences in the mutant's Polα-primase complex. To this end, we immunoprecipitated Polα by anti-Polα antibody from cell lysates of wild type and of polαts13, spp2-8 and spp2-9 mutants and tested for coimmunoprecipitation of Spp1 and Spp2 proteins by Western blot using antibodies against Polα, Spp1, and Spp2 as probes. To ensure that mutations of spp2+ did not affect the expression of Polα, Spp1, and Spp2 in cells, lysates from each strain grown for 4.5 h at 36°C were probed with appropriate antibodies. Wild-type and mutant cells had comparable levels of Polα, Spp1, and Spp2 (Fig. 3A, lanes 1 to 3). We then tested the stability of the Polα-primase complex in these strains after incubation at semipermissive and restrictive temperatures for 4.5 h. Spp1 and Spp2 coimmunoprecipitated with Polα from wild-type-cell extracts, indicating that the Polα-primase complex of wild-type cells was stable at either 33 or 36°C (Fig. 3B, lanes 4 and 7). In contrast, the amounts of Spp1 and Spp2 coimmunoprecipitated with Polα from spp2-8 and spp2-9 cell extracts varied substantially. Cell lysates prepared from spp2-9 at both 33 and 36°C had reduced levels of Spp1 and Spp2 coimmunoprecipitated with Polα (Fig. 3A, lanes 6 and 9). This indicated that at either a semipermissive or a restrictive temperature, the Polα-primase complex in spp2-9 was slightly compromised. Mutant spp2-8 grown at 33°C had barely detectable Spp1 and Spp2 coimmunoprecipitated with Polα (Fig. 3B, lane 8). At 36°C, no detectable Spp1 and Spp2 coimmunoprecipitated with Polα from spp2-8 cell lysates when the blot was exposed for the same length of time as the immunoblots from wild-type and spp2-9 cells (Fig. 3A, lane 5). However, with a longer exposure of the spp2-8 blot, coimmunoprecipitation of Spp1 and Spp2 with Polα was detectable (data not shown). These results indicated that the Polα-primase complex in spp2-8 cells was severely compromised.
FIG. 3.
Stability of Polα-primase complex in wild-type and mutant cells. (A) Expression of Polα, Spp1, and Spp2 in wild-type and spp2 mutant cells. Wild-type 972h−, ST118 (spp2-8), and ST119 (spp2-9) cells grown at 25°C were incubated at 36°C for 4.5 h. Equal amounts of protein from cell extracts were analyzed by Western blot using antibodies against Polα, Spp1, and Spp2 as probes (lanes 1 to 3). To test the coimmunoprecipitation of primase and Polα, cells were grown at the indicated temperatures for 4.5 h. Polα-primase complex was immunoprecipitated from the cell extracts by using anti-Polα antibody and probed with appropriate antibodies as described in Materials and Methods (lanes 4 to 9). (B) Checkpoint-defective polαts13 mutant has a stable Polα-primase complex. (Left panel) Spp1 and Spp2 coimmunoprecipitated with Polα in DBts13 (polαts13) cells. DBts13 (polαts13) cells were grown at 25 or 36°C for 4.5 h. Polα-primase complex was immunoprecipitated from the cell extracts by using anti-Polα antibody and probed with antibodies against Polα, Spp1, and Spp2. (Right panel) Phenotype of wild type and DBts13 (polαts13) grown at 36°C for 6 h. The arrows indicate polαts13 cells that displayed an abnormal mitotic phenotype.
We observed equal proportions of Spp1 and Spp2 reproducibly being coimmunoprecipitated with Polα from either wild-type cells or spp2 mutants. This suggests that neither spp2 mutant alleles affect the affinity between Spp1 and Spp2. In our separate mutational studies of spp1+, under the condition when Spp1 protein failed to coimmunoprecipitate with Polα, Spp2 protein was still able to coimmunoprecipitate with Polα in all spp1 mutants (D. Griffiths, V. Liu, P. Nurse, and T. Wang, manuscript submitted). The spp1 mutation results indicate that Spp1 is not require for the Polα-Spp2 complex formation and strongly suggest that Spp2 physically couples Spp1 with Polα. The spp1 results and the concomitant decrease of Spp1 and Spp2 coimmunoprecipitated with Polα in spp2 mutants shown in Fig. 3 indicate that Spp2 is the bridge protein between Spp1 and Polα. These immunoprecipitation experiments thus indicated that thermosensitive mutations of spp2+ affected the stability of Polα-primase complex. The Polα-primase complex in spp2-8 cells was severely compromised, while the complex in spp2-9 was mildly compromised.
Finding that mutations of spp2+ affect the stability of Polα-primase complex and spp2-8 has a severe instability of its Polα-primase complex, as well as an abnormal mitotic phenotype, led us to analyze the stability of the complex in the checkpoint-defective polαts13 at the restrictive temperature. Polα was able to coimmunoprecipitate comparable amounts of Spp1 and Spp2 from the polαts13 mutant when cells were grown at either 36 or 25°C for 4.5 h (Fig. 3B, left panel). The amounts of Spp1 and Spp2 coimmunoprecipitated with Polα in the polαts13 mutant at either 25 or 36°C, however, were lower than that of the wild type, indicating that the complex, although intact, was slightly compromised (compare Fig. 3B to 3A, lanes 4 and 6). Although the integrity of the four-subunit Polα-primase complex was not substantially affected by mutations in polαts13, the mutant had large fractions of the cells that entered aberrant mitosis exhibiting anucleated phenotype (Fig. 3B, right panel [2]). It is important to point out that the Polα-primase complex of the S-M phase checkpoint-defective polαts13 had a stability similar to that of the complex of the S-M phase checkpoint-intact spp2-9. These indicate that the checkpoint defect of polαts13 mutant is not due to the mutant-Polα protein's inability to associate with the primase proteins.
Mutations of spp2+ affect S phase entry and progression.
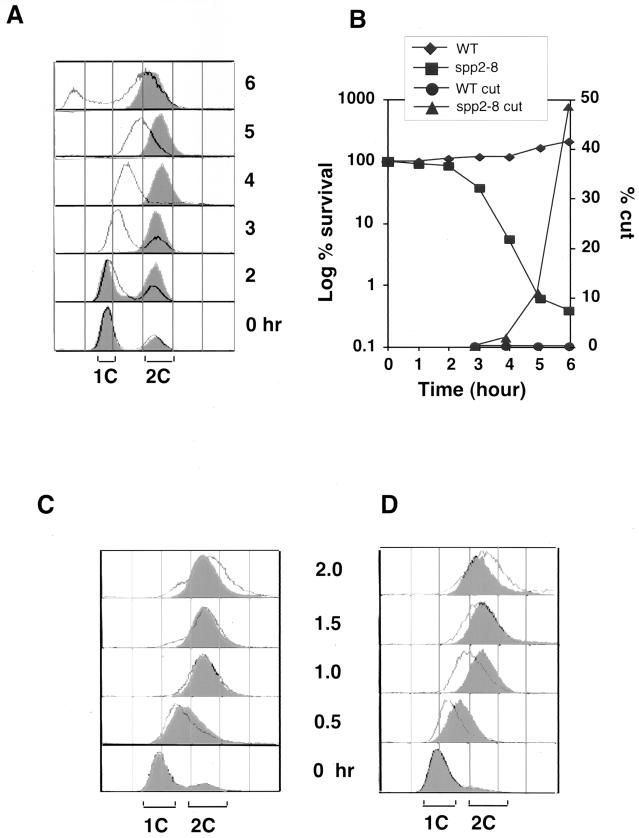
Since Spp2 couples the Spp1 and Polα, we analyzed whether mutations of spp2+ could affect the cells' S phase entry. Wild-type and spp2-8 mutant cells were synchronized in G1 phase by nitrogen starvation for 18 h at 25°C. Cells were then released into rich medium at 36°C. FACS analysis showed that wild-type cells completed S phase within 2 to 3 h after release, whereas spp2-8 cells remained with 1C DNA content for 2 to 3 h (Fig. 4A) and required about 6 h to reach a near-2C DNA content. Thus, when spp2 cells were released from nitrogen starvation, mutations of spp2 caused an approximately 3-h delay in entering S phase. As shown in Fig. 4B, this defect resulted in spp2-8 cells losing viability after 2 h. After 6 h, <1% of spp2-8 cells remained viable and >40% of spp2-8 cells displayed an abnormal mitotic phenotype.
FIG. 4.
Spp2 is essential for S phase entry and progression. Wild-type 972h− and mutant ST118 (spp2-8) cells were synchronized in G1 phase by maintaining them in minimal medium lacking a nitrogen source for 18 h at 25°C. The cultures were then released into YES medium at 36°C and collected at the indicated times for analysis. (A) FACS analysis of wild-type and spp2-8 cells. The wild type is shown in the overlaid histograms, and the spp2-8 mutant is shown in the shaded histograms. (B) Viability and percentage of cells displaying an abnormal mitotic “cut” phenotype quantified by microscopic examination of DAPI-stained cells. (C) Wild-type 972h− and ST118 (spp2-8) mutant cells were synchronized in 12 mM hydroxyurea for 4 h at 25°C and then released into YES medium at 36°C. Samples were removed after release from the hydroxyurea block at the indicated times for FACS analysis. The wild type is shown in the shaded histograms, and the spp2-8 mutant is shown in the overlaid histograms. (D) Wild-type 972h− and ST118 (spp2-8) mutant cells were synchronized in 12 mM hydroxyurea for 4 h at 25°C, followed by 1 h of incubation at 36°C, and then released into YES medium at 36°C. Samples were removed for FACS analysis at the indicated times after release. The wild type is shown in the shaded histograms, and the spp2-8 mutant is shown in the overlaid histograms.
We then tested whether mutations of spp2+ could affect the progression of S phase. Wild-type and spp2-8 cells were arrested postinitiation in early S phase by using hydroxyurea for 4 h at 25°C. Cells were then released into fresh medium at 36°C, and samples were taken every 30 min for FACS analysis. Both wild-type and spp2-8 cells were arrested by hydroxyurea at 25°C with a 1C DNA content. After release from the hydroxyurea block, wild-type and spp2-8 cells had nearly identical FACS profiles (Fig. 4C). We believe this observation may be due to the S. pombe having a short S phase. After the shift to 36°C, it requires a period of time for the temperature to have an effect on the spp2 mutant. It is possible that the time required to remove the hydroxyurea and shift the temperature is sufficient to allow the cells to progress through S phase before the 36°C temperature exerts an effect on the Spp2 protein. To circumvent this possibility, we arrested the cells for 4 h in hydroxyurea at 25°C, shifted them to 36°C for 1 h before release from the hydroxyurea block, and continued incubating them at 36°C. With this approach, wild-type cells completed S phase within 1 h after release from hydroxyurea, whereas spp2-8 cells showed a 0.5- to 1-h delay of progression through S phase (Fig. 4D). Thus, the spp2 mutant delayed S phase entry and progression at both before and after the hydroxyurea arrest point. Similar results were observed with the spp2-9 mutant (data not shown).
spp2 mutants require checkpoint Rad and Cds1, but not Chk1, for growth at the semipermissive temperature.
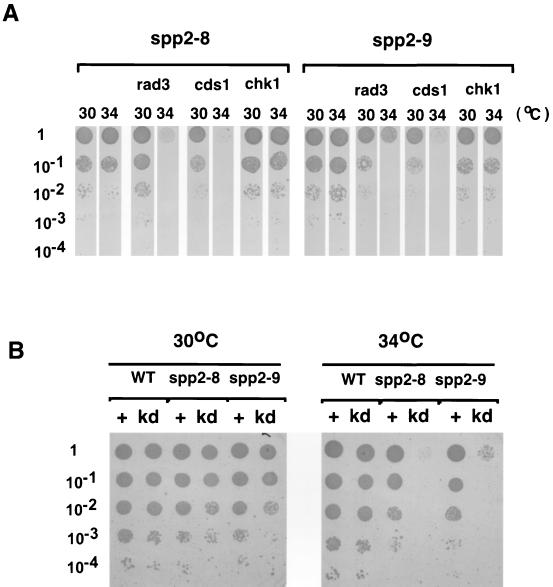
The finding that at the restrictive temperature spp2-8 displayed aberrant mitotic phenotype and that spp2-9 exhibited cdc phenotype led us to investigate the cell cycle checkpoint response induced by these two mutant alleles. We found that in the rad1Δ, rad3Δ, rad9Δ, rad17Δ, rad26Δ, or hus1Δ background, spp2-8 and spp2-9 were viable in liquid culture at up to 32°C and on plates at up to 33°C. This is different from the polαts mutants, which are synthetic lethal in these checkpoint rad deletion backgrounds at 25°C (2). At the semipermissive temperature of 34°C on solid media, both spp2-8 and spp2-9 required all six checkpoint Rad proteins and Cds1 for viability but not Chk1 (Fig. 5A). To test whether the kinase activity of Cds1 is required for the viability of spp2 mutants, cds1+ or a cds1 kinase dead mutant, cds1-kd, was ectopically expressed from the pREP1 vector in the presence of thiamine in the spp2-8 cds1Δ or spp2-9 cds1Δ double mutant. Moderate expression of cds1+ rescued the viability of the double mutants at 34°C. In contrast, the expression of kinase dead mutant of cds1 (cds1-kd) was unable to rescue the double mutants at 34°C (Fig. 5B), suggesting that Cds1 kinase activity is required for the viability of spp2 mutants at the semipermissive temperature. Thus, at the semipermissive temperature, cells with S phase perturbation due to mutations of spp2+ require the checkpoint Rad proteins and Cds1 kinase, but not Chk1 kinase, for maintaining cell viability.
FIG. 5.
spp2 mutants require checkpoint Rads and Cds1, but not Chk1, for growth at the semipermissive temperature. (A) Serial dilutions of spp2-8 and spp2-9 mutants and the spp2-8 and spp2-9 alleles in rad3, cds1, and chk1 deletion background were spotted on YES plates in duplicate. One set of plates was incubated at 30°C, and the other set was incubated at 34°C. (B) Ectopic expression of Cds1, but not a kinase-dead Cds1 mutant, rescues the growth of spp2 cds1Δ double mutants at 34°C. pREP1 plasmid, carrying either cds1+ or kinase-dead cds1 mutant, was introduced into spp2 cds1Δ double mutants in minimal medium lacking leucine. Thiamine was included in the medium to reduce the expression of Cds1 since the overexpression of Cds1 is toxic to the cells. Serial dilutions of transformants were spotted on the selective medium in duplicates and incubated independently at 30 and 34°C. The transformants of cds1+ and kinase-dead cds1 mutant are indicated as “+” and “kd,” respectively. Cell density, 104 cells per spot.
Chk1 response in spp2 mutants.
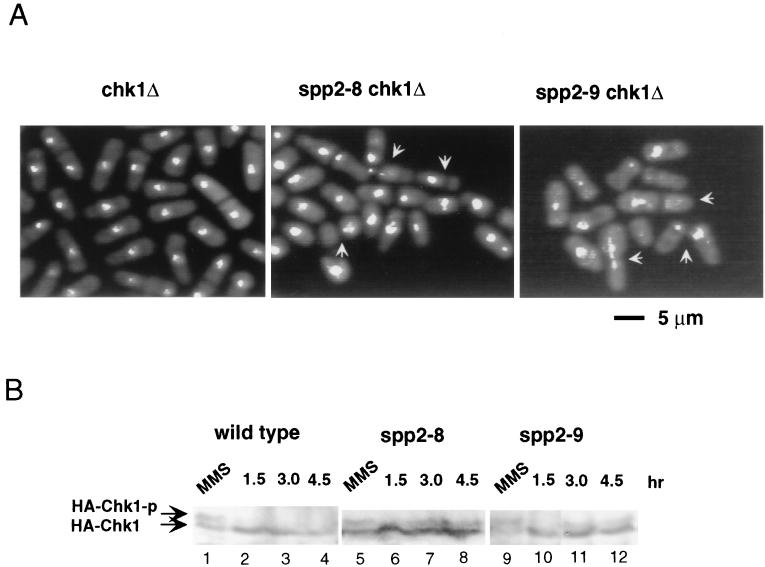
Since Chk1 was not required for maintaining growth of spp2 mutants at the semipermissive temperature, we tested how Chk1 responds to mutations of spp2+ at the restrictive temperature. As shown above, majority of the spp2-9 cells and a fraction of the spp2-8 cells displayed an elongated phenotype with normal nuclear morphology at the restrictive temperature (Fig. 1A). In the chk1Δ deletion background, both spp2-8 and spp2-9 cells died rapidly and exhibited a small cell size with aberrant mitotic nuclear morphology at 36°C (Fig. 6A). In contrast, chk1Δ cells grew normally and exhibited normal nuclear morphology. Thus, the elongated phenotype seen in spp2-8 and spp2-9 cells requires the function of Chk1.
FIG. 6.
Chk1 response to spp2 mutation at 36°C. (A) DAPI staining of chk1Δ, spp2-8 chk1Δ, and spp2-9 chk1Δ cells after incubation at 36°C for 6 h. (B) Chk1 phosphorylation induced by mutation of spp2 at 36°C. Mid-log-phase cells grown at 25°C were shifted to 36°C. At the indicated times, cells were collected and lysed. A portion (30 μg) of cell lysates was fractionated on an SDS-polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and probed with mouse anti-HA monoclonal antibody (12CA5) as described previously (2). As a control, each strain was treated with 0.2% MMS at 25°C for 1 h. The HA-tagged chk1+ strains used for wild-type, spp2-8, and spp2-9 cells were NW222, ST128, and ST129, respectively. The phosphorylated and unphosphorylated HA-Chk1 proteins are indicated as HA-Chk1-p and HA-Chk1, respectively.
We then analyzed the phosphorylation status of Chk1 protein in spp2 mutant cells. Mutant spp2 cells containing three hemagglutinin (HA) epitopes tagged chk1+ (40) were constructed for the detection of Chk1 phosphorylation by 12CA5 monoclonal antibody. As a control, wild-type cells containing the HA tagged chk1+ were treated with the DNA-damaging agent methyl methane sulfonate (MMS), which had been shown to induce Chk1 phosphorylation (40). As expected, Chk1 became readily phosphorylated in response to MMS treatment (Fig. 6B). In both spp2-8 and spp2-9 cells, only after 3 to 4.5 h of incubation at 36°C was a weak, slower-migrating Chk1 protein detectable. This is consistent with the finding that phosphorylation of Chk1 primarily responds to late S phase or G2 phase perturbation and not early S phase perturbation (23). These results indicated that Chk1 was not required for spp2 mutants to maintain viability at the semipermissive temperature and that mutation of spp2+ also did not substantially induce Chk1 phosphorylation at 36°C. However, at the restrictive temperature, Chk1 did play a critical role in preventing aberrant mitotic entry of spp2-9 cells and of a fraction of the spp2-8 cells.
Cds1 response in spp2 and polα mutants.
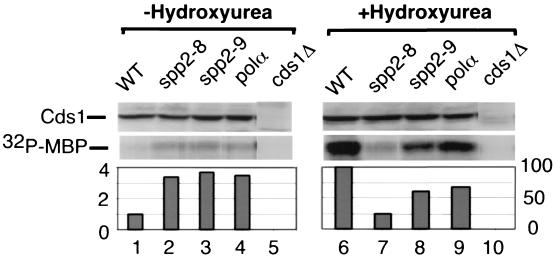
Since Cds1 is required for both spp2-8 and spp2-9 cells to maintain viability at the semipermissive temperature (Fig. 5) and since Cds1 kinase is activated in thermosensitive polα mutants (2, 19), we analyzed the Cds1 response to S phase arrest by spp2 mutants and compared it to that of the polαts13 mutant. Cds1 protein immunoprecipitated from cell extracts was assayed for kinase activity in vitro using MBP as the substrate (Fig. 7). The Cds1 kinase in spp2-8, spp2-9, and polαts13 was activated three- to fourfold over the wild-type level after incubation of the cells at 36°C for 4.5 h (Fig. 7, lanes 1 to 4). The kinase activity measured under these conditions was Cds1 specific because the phosphorylation of MBP was not detected in cds1Δ cells (Fig. 7, lane 5). It is important to mention that the Polα-primase complex is extremely unstable in spp2-8 cells, whereas the complex is only mildly compromised in spp2-9 and polαts13 cells (Fig. 3). Nonetheless, the Cds1 kinase was activated to a similar extent when all three mutants were arrested in early S phase at the restrictive temperature. These results suggest that early S phase arrest by these initiation mutants, regardless of the extent of their Polα-primase complex instability, generates a signal to moderately activate Cds1 kinase.
FIG. 7.
Cds1 response to spp2 mutations. Cds1 kinase activity of wild-type 972h−, ST118 (spp2-8), and ST119 (spp2-9) in the absence or presence of 12 mM hydroxyurea at 36°C. Mid-log-phase cells grown at 25°C were shift to 36°C for 1 h, and incubation continued at 36°C for 3.5 h in the absence (−Hydroxyurea) or presence (+Hydroxyurea) of 12 mM hydroxyurea. Cds1 kinase was purified from cell extracts and assayed for kinase activity as described in Materials and Methods. The relative amounts of Cds1 proteins used in the kinase assay were estimated by Western blot (upper panel, Cds1). The kinase activity was measured by phosphorylation of MBP (lower panel, 32P-MBP) and quantified by PhosphorImager analysis.
Cds1 kinase is highly activated when S phase is arrested by hydroxyurea (19). We therefore examined the hydroxyurea-induced Cds1 response in spp2 and polα mutants. Mid-log-phase cells grown at 25°C were shifted to 36°C for 1 h and then incubated for an additional 3.5 h at 36°C in 12 mM hydroxyurea. As expected, Cds1 kinase was activated >20-fold by hydroxyurea in wild-type cells (Fig. 7, compare lane 6 to lane 1). The hydroxyurea-induced Cds1 kinase activity in spp2-9 cells was about 65 to 70% of the wild-type level (Fig. 7, lane 8), whereas the hydroxyurea-induced Cds1 kinase activity in spp2-8 cells was only about 25% of the wild-type level (Fig. 7, lane 7). Therefore, although the Cds1 kinase can be activated to a similar extent in spp2-8 and spp2-9 mutants when they are arrested by temperature, there is a striking difference in their hydroxyurea-induced Cds1 kinase activation at 36°C. Thus, at the restrictive temperature, the Cds1 response to signals generated from S phase arrest induced by an initiation mutant is different from that induced by hydroxyurea arrest.
As shown in this study and elsewhere (2), at 36°C polαts13 cells exhibit heterogeneous cell morphology with >40% of the cells displaying abnormal nuclear phenotype; however, the Polα-primase complex in polαts13 was only slightly compromised (Fig. 3B). We therefore tested the hydroxyurea-induced Cds1 response in the polαts13 mutant. Surprisingly, the Cds1 kinase was activated to approximately 75% of the wild-type level (Fig. 7, lane 9), which was comparable to the levels of the checkpoint-proficient mutant spp2-9. Thus, the abnormal mitotic phenotype of polαts13 is due to a checkpoint defect that can be distinguished from the hydroxyurea-induced Cds1-mediated checkpoint pathway.
Maintenance of the hydroxyurea-induced Cds1 kinase in spp2 and polα mutants.
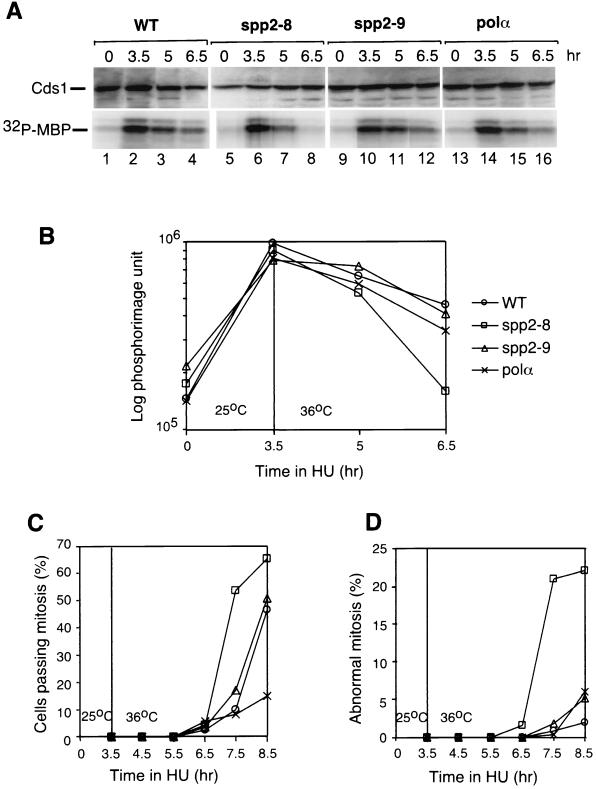
We then tested the ability of the mutants to maintain the hydroxyurea-induced Cds1 kinase activation. Wild-type and mutant cells were first incubated in hydroxyurea at 25°C for 3.5 h to activate the Cds1 kinase. Cells were then shifted to 36°C with fresh hydroxyurea added and further incubated at 36°C. At the indicated times, cell samples were taken for analysis of the Cds1 kinase activity (Fig. 8A), the percentage of cells passing mitosis (Fig. 8B), and the percentage of cells entering inappropriate mitosis (Fig. 8C). After incubation of the cells for 3.5 h in hydroxyurea at 25°C, Cds1 kinase was activated in spp2-8, spp2-9, and polαts13 cells to 80 to 90% of the wild-type cells levels (Fig. 8A, 3.5 h, compare lanes 6, 10, and 14 to lane 2; quantified in the lower graph). After 1.5 h at 36°C (Fig. 8A, 5-h time point), the Cds1 kinase activity in wild-type cells decreased to approximately 75 to 80% of the original level (Fig. 8A, 5-h time point, lane 3 [quantified in the lower graph]). After 3 h at 36°C (Fig. 8A, 6.5-h time point), the hydroxyurea-induced Cds1 kinase activity established at the permissive temperature in wild-type cells decreased to approximately 60% of the original level (Fig. 8A, 6.5-h time point, lane 4 [quantified in the lower graph]). The level of Cds1 kinase activity in spp2-8 after 1.5 h at 36°C decreased to 50% of its original levels that had been established at 25°C (Fig. 8A, 5-h time point, compare lane 7 to lane 6 [quantified in the lower graph]), whereas the Cds1 kinase activity in spp2-9 was maintained at its original level established at 25°C (Fig. 8A, compare lanes 10 and 11). The Cds1 kinase activity in polαts13 after 1.5 h at 36°C decreased to approximately 75% of the levels established at 25°C (Fig. 8A, compare lanes 14 and 15 [quantified in the lower graph]). Interestingly, after 3 h at 36°C (Fig. 8A, 6.5-h time points), the Cds1 kinase activity in spp2-8 was maintained at 15% of its original levels established at 25°C, while both spp2-9 and polαts13 decreased to approximately 40% of the levels established at 25°C (Fig. 8A, lanes 8, 12, and 16 [lower quantification graph]).
FIG. 8.
Maintenance of hydroxyurea-induced Cds1 kinase activity. Cds1 activation was induced in wild-type and mutant cells by 12 mM hydroxyurea at 25°C for 3.5 h. The cells were then shifted to 36°C with 12 mM hydroxyurea added. At the indicated times, samples were removed and analyzed for Cds1 kinase activity (A and B), the percentage of cells passing mitosis (C), and the percentage of cells entering into aberrant mitosis described as the cut phenotype (D). The relative amounts of Cds1 proteins used in the kinase assay were estimated by Western blot (A, upper panel, Cds1). The kinase activity was measured by phosphorylation of MBP (A, lower panel, 32P-MBP) and was quantified by PhosphorImager analysis (B).
The observed hydroxyurea-induced Cds1 response in these mutants was also consistent with their kinetics of passing mitosis (Fig. 8B) and the appearance of abnormal mitotic phenotype (Fig. 8C). At 4 h after the shift to 36°C (7.5-h time point of Fig. 8B and C), >50% of the spp2-8 cells passed mitosis and >20% of the spp2-8 cells displayed an abnormal mitotic phenotype. At the same time point, <20% of the wild-type, spp2-9, and polαts13 cells passed mitosis and ∼2% of the cells exhibited an abnormal mitotic phenotype. Interestingly, after the shift to 36°C for 5 h (8.5-h time point of Fig. 8B and C), <15% of the polαts13 cells entered mitosis, while >45% of the wild-type and spp2-9 cells entered mitosis, suggesting that polαts13 and hydroxyurea arrest the cell cycle at close proximity.
Together, these results indicated that both the induction and the maintenance of the hydroxyurea-induced Cds1 response were severely compromised in spp2-8 cells and moderately compromised in spp2-9 and polαts13 cells. It is important to note that spp2-8 which has the most unstable Polα-primase complex, also has the lowest ability to induce and maintain the hydroxyurea-induced Cds1 response.
DISCUSSION
The goal of this study was to determine the checkpoint response(s) to aberrant initiation of S phase. Here, we analyzed the mutational effects of the DNA primase gene spp2+ encoding the primase subunit, Spp2, on the cell cycle checkpoint response and compared to that of polα mutation. Four main points emerged from our studies. First, Spp2 is required for coupling RNA primer synthesis by Spp1 to initiation DNA synthesis by Polα, suggesting that Spp2 is the bridge protein between Spp1 and Polα. Second, mutations of spp2+ destabilize the mutant's Polα-primase complex. The spp2 mutant that has a severely compromised Polα-primase complex also exhibits the aberrant mitotic phenotype at the restrictive temperature, suggesting a correlation between the stability of the complex and the aberrant mitotic phenotype. Third, the spp2 mutant that has the most severely compromised Polα-primase complex also has the lowest ability to activate and maintain the hydroxyurea-induced Cds1 response; however, this mutant is able to activate the Cds1 response induced by its temperature arrest. Fourth, a checkpoint defective polαts mutant has an intact Cds1 response in either the presence or the absence of hydroxyurea and a similar Polα-primase complex stability as a checkpoint intact spp2 mutant.
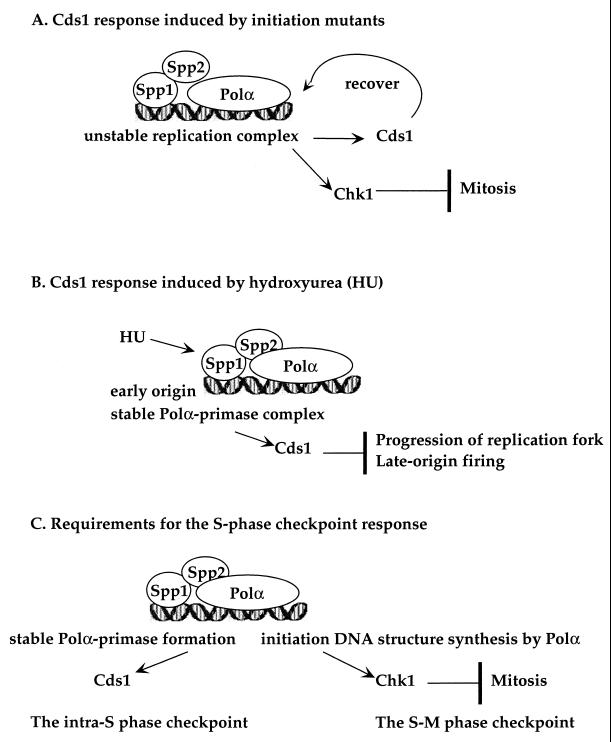
Based on these findings, we propose a model of how different types of perturbation of S phase initiation could induce different checkpoint responses: an unstable initiation complex or a stalled initiation generates a signal to moderately activate the Cds1 kinase to stabilize and/or recover the initiation complex to prevent replication fork collapse and accumulation of mutations (Fig. 9A). Following an early S phase stall caused by hydroxyurea, a stable and intact Polα-primase complex is required to signal high levels of Cds1 kinase activation to prevent progression of the early replication fork and premature initiation of the late-firing replicons (Fig. 9B). Once DNA replication is initiated, the synthesis of an initiation DNA structure by Polα is required for generating the S phase checkpoint to prevent inappropriate mitotic entry (Fig. 9C).
FIG. 9.
Model illustrating the proposed Cds1 responses to the aberrant initiation of S phase. The proposed Cds1 response to unstable replication complex (A), a stalled replication fork (B), and the requirement for cells to generate a signal (C) for the intra-S phase checkpoint and the S-M phase checkpoint as described in the Discussion.
A moderately activated Cds1 kinase is required to maintain an unstable Polα-primase complex.
Cds1 was originally identified as a high-copy suppressor of a temperature-sensitive polα mutant, swi7-H4 (26). The Cds1 structural counterpart in budding yeast, RAD53, has been reported to modulate phosphorylation of the B subunit of Polα-primase complex (29). We showed in this study that mutations of spp2 destabilize the Polα-primase complex (Fig. 3) and that Cds1 kinase is moderately activated in the spp2 and polα mutants at the restrictive temperature (Fig. 7). We have previously shown that deletion of Cds1 decreases the semipermissive temperature of several thermosensitive mutant alleles of polα and exacerbates the mutator phenotypes of these mutants (20). Thus, mutations of either spp2 or polα require the function of Cds1 kinase at both the semipermissive and the restrictive temperatures. It is not yet clear whether Cds1 directly or indirectly interacts with the Polα-primase complex and what the substrates of Cds1 kinase are. The results of this study and in combination with our previous studies (20) suggest that an unstable or stalled Polα-primase complex of polα or spp2 mutants requires a moderate levels of Cds1 kinase activation to stabilize and/or to prevent the formation of unrecoverable replication complex, replication fork collapse, and/or the accumulation of mutations (Fig. 9A).
It is important to mention that both spp2-8 and polαts13 display a mixed phenotype. The elongated cells seen in the spp2 population (Fig. 1A) and polαts13 mutants (Fig. 3B and reference 2) can be attributed to the Chk1 response to prevent aberrant mitotic entry of these initiation mutants (Fig. 6A). Chk1 is not significantly phosphorylated during early-S-phase arrest by spp2 mutants (Fig. 6B). It is possible that the unstable initiation complex in spp2-8 and/or the stalled initiation complex in polαts13 are not sufficient to activate Chk1 to a level that can fully prevent inappropriate mitotic entry, thus resulting in a mixed phenotype.
A stable Polα-primase complex is required for activation of the Cds1-mediated intra-S phase checkpoint.
We propose that following an early S phase stall caused by hydroxyurea, a stable and intact Polα-primase complex is required to signal the activation of high levels of Cds1 kinase to prevent the premature initiation of the late-firing replicons. In budding yeast, it has been shown that, following hydroxyurea-induced arrest, the initiation complexes of the late-firing replicons remain in an initiation-competent state for a long period of time (12, 33). In fission yeast, after hydroxyurea-induced arrest, the Cds1-mediated checkpoint response is also thought to prevent the initiation of late-firing replicons, and/or stabilize existing replicons (19). This process is termed intra-S phase checkpoint.
We found that at the restrictive temperature, the hydroxyurea-induced Cds1 kinase of spp2-8 was only activated at 25% of the wild-type cell's level (Fig. 7). After 3 h at 36°C, spp2-8 was unable to maintain the hydroxyurea-induced Cds1 kinase established at 25°C (Fig. 8A). As shown in Fig. 8B and in our previous mutational studies of polα (2), hydroxyurea arrests the S phase at point close to the arrest point of polαts mutant, causing a stalled replication. It is not yet clear at the molecular level how Cds1 kinase inhibits the progression of the replication fork of the early-firing replicons and prevents initiation of the late-firing replicons following hydroxyurea-induced arrest. It is possible that a stable Polα-primase complex in the replication complex is required to generate the signal for high levels of Cds1 kinase activation to prevent progression of the early-firing replication fork and initiation of late-firing replicons. The rationale for this hypothesis is our finding that spp2-8 with a severely compromised Polα-primase complex also has the lowest ability to activate and maintain the hydroxyurea-induced Cds1 kinase (Fig. 3, 7, and 8). The inability of spp2-8 to activate and to maintain the hydroxyurea-induced Cds1 kinase activity may cause the cell to be unable to prevent the progression of early-firing replication fork and/or premature firing of the late replicons. Thus, spp2-8 has a higher percentage of cells entering abnormal mitosis than does spp2-9 (Fig. 8B and C). spp2-9 and polαts13 both have mildly compromised Polα-primase complex. The mildly compromised complex might be sufficient to generate a signal to activate sufficient levels of Cds1 to prevent the firing of late replicons. The activated Cds1 kinase in spp2-9 and polαts13, however, is insufficient to fully prevent inappropriate mitotic entry as the wild-type cells after 3.5 h at 36°C (Fig. 8A). Thus, spp2-9 and polαts13, despite having the wild-type-like percentage of cells passing mitosis and mitotic phenotype after 5 h in 36°C (Fig. 8B and C, 8.5-h time point), these mutants eventually die. These results suggest that, upon hydroxyurea-induced arrest, a functional Spp2 coupling Spp1 and Polα to establish a stable Polα-primase complex is required to generate a signal for high levels of Cds1 activation for the intra-S phase checkpoint (Fig. 9B).
As shown in Fig. 7, in all of the mutants, the levels of Cds1 kinase activity induced by hydroxyurea are much higher than the levels induced by spp2 or polαts mutant arrest (Fig. 7). It is not clear whether, following hydroxyurea-induced arrest or early S-phase arrest by spp2 and polαts, Cds1 proteins are being phosphorylated at the same or different sites, thus being activated at different levels. However, our results suggest that mutations of spp2+ cause an unstable initiation complex, resulting in an aberrant initiation that induces a moderate level of Cds1 response to recover from the perturbation. Upon hydroxyurea inhibition, a stable Polα-primase complex is required to induce higher levels of Cds1 kinase for the intra-S phase checkpoint response. Thus, Cds1 response(s) to the early-S-phase arrest signals induced by a replication initiation mutant is different from that induced by hydroxyurea.
The requirement for the S-M phase checkpoint can be distinguished from the requirement for the intra-S phase checkpoint.
We showed in this study that at the restrictive temperature, polαts mutant exhibits an aberrant mitotic phenotype but has an intact but mildly compromised Polα-primase complex (Fig. 3B). Importantly, the checkpoint-defective polαts13 and the checkpoint-intact spp2-9 at 36°C have similar Polα-primase complex stabilities and similar abilities to activate and maintain the hydroxyurea-induced Cds1 kinase. These suggest that the S-M phase checkpoint defect of polαts mutant most likely is not due to an unstable Polα-primase complex or to inability to activate and maintain Cds1 kinase activity. This also suggests that Cds1 activation does not play a major role in preventing the inappropriate mitotic entry of polαts13. We have previously shown that the catalytic activity of Polα to synthesize an initiation DNA structure is required for generating the replication checkpoint to prevent inappropriate mitosis during S phase (2). Thus, we propose that, upon S phase initiation, the requirement for generating the replication checkpoint to prevent inappropriate mitotic entry is the initiation DNA synthesis of Polα (Fig. 9C) and that this requirement is distinguished from the requirements for generating the hydroxyurea-induced Cds1-mediated intra-S phase checkpoint response.
ACKNOWLEDGMENTS
We thank A. M. Carr for providing us the checkpoint rad deletion strains, D. Bhaumik for help in isolation of the spp2+ gene, and T. Enoch for pREP1-cds1+ and pREP-cds1-kd (kinase dead mutant) plasmids. We especially thank members of our laboratory for helpful discussion during the course of this work.
S. Tan is sponsored by a postdoctoral training grant CA09151 awarded by National Cancer Institute. This study was supported by a grant CA54415 from The National Cancer Institute of The National Institutes of Health.
REFERENCES
- 1.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik D, Wang T S-F. Mutational effect of fission yeast Polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 4.Brondello J-M, Broddy M N, Furnari B, Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr A M. Analysis of fission yeast DNA structure checkpoints. Microbiology. 1998;144:5–11. doi: 10.1099/00221287-144-1-5. [DOI] [PubMed] [Google Scholar]
- 6.Carr A M. Checkpoints take the next step. Science. 1996;271:314–315. doi: 10.1126/science.271.5247.314. [DOI] [PubMed] [Google Scholar]
- 7.Carr A M. DNA-structure checkpoint in fission yeast. Semin Cell Biol. 1995;6:65–72. doi: 10.1016/1043-4682(95)90002-0. [DOI] [PubMed] [Google Scholar]
- 8.Carr A M, Hoekstra M F. The cellular responses to DNA damage. Trends Cell Biol. 1995;5:32–40. doi: 10.1016/s0962-8924(00)88934-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Liu T-H, Walworth N C. Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev. 1999;13:675–685. doi: 10.1101/gad.13.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland W C, Wang T S-F. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993c;268:26179–26189. [PubMed] [Google Scholar]
- 11.D'Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- 12.Desany B A, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enoch T, Carr A M, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 14.Foiani M, Santocanale C, Plevani P, Lucchini G. A single essential gene, PRI2, encodes the large subunit of DNA primase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3081–3087. doi: 10.1128/mcb.9.7.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnari B, Rhind N, Russell P. cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths D J F, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics 1. I. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 18.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 19.Lindsay H D, Griffiths D J F, Edwards R, Murray J M, Christensen P U, Walworth N, Carr A M. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in S. pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu V F, Bhaumik D, Wang T S-F. Mutator phenotype induced by aberrant replication. Mol Cell Biol. 1999;19:1126–1135. doi: 10.1128/mcb.19.2.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, carr A M, Bentley N J. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa H, Izumi M, Tada S, Takada R, Masutani M, Ui M, Hanaoka F. Molecular cloning of the cDNAs for the four subunits of mouse DNA polymerase alpha-primase complex and their gene expression during cell proliferation and the cell cycle. J Biol Chem. 1993;268:8111–8122. [PubMed] [Google Scholar]
- 25.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 26.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 27.Park H, Francesconi S, Wang T S-F. Cell cycle expression of two replicative DNA polymerases α and δ from Schizosaccharomyces pombe. Mol Biol Cell. 1993;4:145–157. doi: 10.1091/mbc.4.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 29.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Fiore P P D, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng C-Y, Graves P R, Thoma R S, Wu Z, Shaw S A, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 31.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Conservation of the chk1 checkpoint pathway in mammals: linkage of DNA damage to cdk regulation through cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 33.Santocanale S, Diffley J F X. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 34.Shirahige K, Hori Y, Shirishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 35.Stadlbauer F, Brueckner A, Rehfuess C, Eckerskorn C, Lottspeich F, Forster V, Tseng B Y, Nasheuer H P. DNA replication in vitro by recombinant DNA-polymerase-alpha-primase. Eur J Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- 36.Tan S, Conaway R C, Conaway J W. A bacteriophage vector suitable for site-directed mutagenesis and high level expression of multisubunit proteins in E. coli. BioTechniques. 1994;16:824–828. [PubMed] [Google Scholar]
- 37.Tatebayashi K, Katoa J, Ikedaa H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–58. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchiyama M, Galli I, Griffiths D J F, Wang T S-F. A novel mutant allele of Schizosaccharomyces pombe rad26 defective in monitoring S phase progression to prevent premature mitosis. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 40.Walworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 41.Wang T S-F. Cellular DNA polymerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- 42.Zeng Y, Piwnica-Worms H. DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14-3-3 binding. Mol Cell Biol. 1999;19:7410–7419. doi: 10.1128/mcb.19.11.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]