Abstract
Many models of motor control emphasize the role of sensorimotor cortex in movement, principally through the projections corticospinal neurons (CSNs) make to the spinal cord. Additionally, CSNs possess expansive supraspinal axon collaterals, the functional organization of which is largely unknown. Using anatomical and electrophysiological circuit mapping techniques in the mouse, we reveal dorsolateral striatum as the preeminent target of CSN collateral innervation. We found this innervation is biased so that CSNs targeting different striatal pathways show biased targeting of spinal cord circuits. Contrary to more conventional perspectives, CSNs encode not only individual movements, but information related to the onset and offset of motor sequences. Furthermore, similar activity patterns are broadcast by CSN populations targeting different striatal circuits. Our results reveal a logic of coordinated connectivity between forebrain and spinal circuits, where separate CSN modules broadcast similarly complex information to downstream circuits, suggesting that differences in postsynaptic connectivity dictate motor specificity.
Corticospinal neurons (CSNs), the principle output of sensorimotor cortex, relay motor command signals to the spinal cord, where their main axons synapse on several classes of interneurons involved in patterning motor output and shaping sensory feedback1–4. Corticospinal output is topographically organized: CSNs within regions that control specific body parts preferentially innervate spinal segments containing the circuits that control the muscles for those body parts2,3,5–7. CSNs arising from motor regions of isocortex form synapses spread across intermediate and ventral laminae of the spinal grey, home to premotor spinal interneurons; while sensory cortical CSNs have terminal fields mostly restricted to superficial regions of spinal cord, home to the dorsal horn interneurons that regulate peripheral sensory feedback8,9. Partly because of this anatomy, motor cortical CSNs have been hypothesized to control the activity of individual muscles or muscle synergies through their direct projections to the spinal cord10–12. Concordantly, many CSNs are active at discrete phases of movement or during the recruitment of single muscles. However, CSNs often display complex activity patterns that do not closely correspond to muscle output, suggesting their role in movement may be nuanced and task-dependent13–15. For instance, some pyramidal tract-projecting neurons in non-human primates act as mirror neurons, displaying activity that is modulated during both execution of a forelimb movement and the passive observation of an experimenter’s forelimb movement16. Complex activity in corticospinal populations has led to hypotheses that CSNs subserve diverse modulatory control over spinal cord, including motor inhibition, plasticity of spinal circuits, reafference suppression, and state-dependent gain control9,17,18. This complexity of corticospinal function is further reflected in peculiarities of corticospinal organization. Foremost, CSNs give rise to axon collaterals that project to a broad range of brain structures, affording them remarkable – yet largely uncharted – influence over nearly all levels of the motor control neuraxis19–21. Despite the widespread supraspinal collateralizations of CSNs, a detailed characterization of their anatomical and functional organization has been elusive. Therefore, we sought to 1) comprehensively map the structural organization of CSNs and their brainwide axon collaterals, 2) uncover synaptic and circuit principles that mediate coordinated spinal and supraspinal connectivity, and 3) characterize the behaviorally-relevant activity of CSNs with identified cell type-specific supraspinal targets.
Uncertainty surrounding the anatomical and electrophysiological properties of corticospinal output is partly result of difficulties in capturing, mapping, and monitoring large populations of CSNs, particularly during complex skilled behaviors. In this study we overcame these challenges by first using a suite of intersectional anatomical methods to map the input and output organization of corticospinal neurons. While CSNs formed widespread synapses in many supraspinal structures, we discovered that striatum is the preeminent target of collateral innervation. The striatum is composed of two distinct populations of spiny projection neurons (SPNs) defined in part by the expression of dopamine receptor type 1 (D1) or type 2 (D2)22–25. Using anatomical and electrophysiological circuit mapping techniques, we showed that separate populations of CSNs synapse on D1 and D2 SPNs, that more synapses are formed on D1 SPNs than D2 SPNs, and that this biased connectivity translates to distinct innervation of the cervical spinal cord. D1 and D2 SPNs are both necessary for the learned sequencing of body movements, and encode sequence-related information, but to different degrees26–29. Therefore, we leveraged calcium imaging during a striatal-dependent lever press sequence behavior, identifying neurons that linearly encoded muscle activity as well as ones that encoded higher order features of behavior, including sequence onset and offset. Combining these experiments with a novel use of in vivo transsynaptic tracing revealed that both muscle-related and higher-order activity are transmitted to both D1 and D2 striatal SPNs. Our results support a model wherein CSNs broadcast copies of muscle and non-muscle correlated information throughout the nervous system, and differences in postsynaptic connectivity determine motor specificity.
Corticospinal collaterals project prominently to striatum
Corticospinal neurons collateralize throughout the brain, but the degree to which CSNs innervate each target structure was unclear. We combined intersectional viral expression of fluorescent makers with unbiased anatomical reconstruction to quantify collateral innervation of brain regions by CSNs. First, we labeled inputs to the spinal cord by injecting a retrogradely-transported adeno-associated virus encoding Cre recombinase fused to RFP (AAV-retro-Cre.RFP) into right forelimb cervical spinal segments C3-C730. In the same animals, we injected a Cre-dependent AAV encoding GFP (AAV-FLEX-GFP) into forelimb-control regions6 of left sensorimotor cortex, resulting in expression of GFP exclusively in CSNs (Figure 1A–I). We then imaged antibody-enhanced GFP and RFP labeling, and used the imaging analysis pipeline BrainJ, which incorporates machine learning based methods to distinguish cell bodies and processes and maps their positions to a common brain atlas (Figure 1J–L, see Methods for details)31. We first noted the widespread brain regions that project to cervical spinal cord, spanning all levels of the motor neuraxis (Extended Data Figure 1A–C). Isocortical structures dominated, comprising 47 percent of the total input (Extended Data Figure 1C inset, 47±0.03%, N=3). Imaging GFP+ labeled (i.e. CSN) axons in the spinal cord revealed widespread varicosities around cervical spinal injection sites, but also collateralization in distant thoracic, and to a lesser degree, lumbar segments (Extended Data Figure 1D–F). Quantification of GFP+ cellular labeling revealed these axons arose from neurons in deep layers of sensorimotor cortex (Extended Data Figure 1I); this labeling was consistent across animals (Figure 1M inset, correlation coefficient: 0.98±0.01). CSN labeling in the brain revealed axonal processes in many forebrain, midbrain, and hindbrain regions, several of which are themselves implicated in motor control, including brainstem nuclei that mediate skilled forelimb actions (Figure 1N, Figure 1I, Extended Data Figure 1J–K, correlation coefficient: 0.93±0.003)32,33. Notably, CSNs project most prominently to the dorsolateral striatum (DLS; Figure 1N–P, 9.63±0.69% of all neurites), and form the most abundant fraction of synapses in this region as confirmed using a synaptophysin-fused reporter (Extended Data Figure 1L–P). Because direct cortical injections of AAV-FLEX-GFP capture GFP-labeled CSNs only around the injection area, we confirmed our results using an intersectional approach to label only CSNs that project to DLS (CSNsDLS). We injected AAV-retro-Cre.RFP into right cervical spinal cord, and AAV-retro-FLEX-GFP into left DLS, resulting in Cre-mediated recombination in cortical inputs to striatum that also project to spinal cord, regardless of their cortical origin (Extended Data Figure 2A). Inspecting the distribution of CSNDLS cell bodies revealed widespread labeling in sensorimotor cortex, with sparse labeling in other isocortical structures (Extended Data Figure 2B–E). Quantifying all axonal projections from these CSNsDLS revealed this population projects throughout the brain, including brainstem regions implicated in motor control (Extended Data Figure F–I). Consistently, of all regions targeted by these neurons, the largest fraction of axons was found in DLS (Extended Data Figure 2F, N=3).
Figure 1. Anatomical characterization of corticospinal neurons.
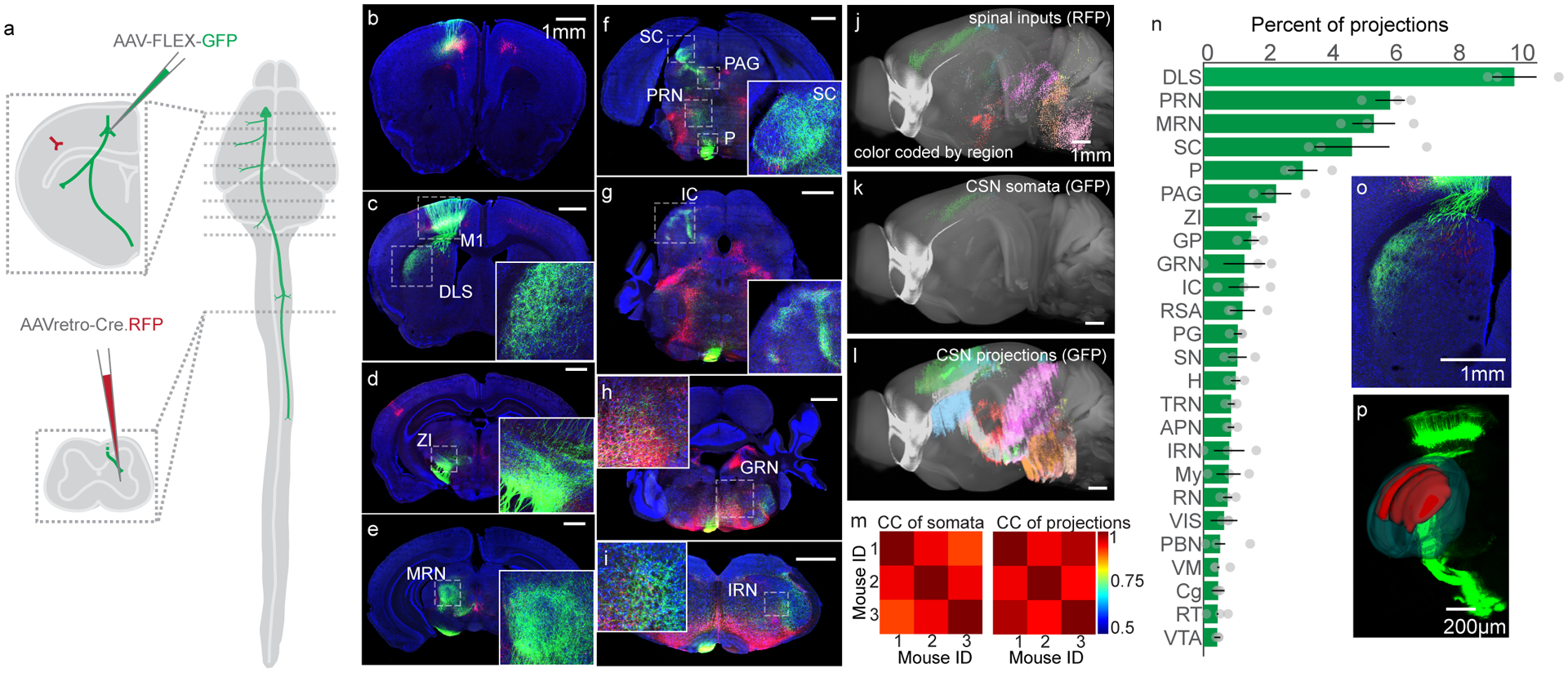
(A) Schematic illustrating the viral injection sites in motor cortex and the spinal cord, and their relative positions in the nervous system. Dashed lines indicate the position of representative images to follow. (B-I) Confocal micrographs of transverse sections throughout the brain, illustrating GFP+ CSNs and their axonal projections (green), along with all spinal inputs made to express Cre.RFP (red). Some regions of interest are boxed by dashed lines and include: primary motor cortex (M1), dorsolateral striatum (DLS), zona incerta (ZI), midbrain reticular nucleus (MRN), superior colliculus (SC), pons (P), periaqueductal grey (PAG), pontine reticular nucleus (PRN), inferior colliculus (IC), gigantocellular reticular nucleus (GRN), and intermediate reticular nucleus (IRN). DAPI is in blue. Representative of N=3. (J-L) Three dimensional reconstructions of spinal inputs (J), CSN somata (K), and CSN neurites (L) throughout the brain. Colors correspond to major brain divisions in which they reside. Note that the most caudal portion of brainstem is not included in these analyses. (M) Correlation analyses of mesoscopic animal-to-animal CSN somata locations (left) and CSN neurites locations (right). (N) Top brain regions to which CSNs project, measured as what fraction of all bins of neurites are found within those brain structures, excluding sensorimotor cortex and fiber tracts (additional structures not defined above: GP: globus pallidus, RSA: retrosplenial area, PG: pontine grey, SN: substantia nigra, H: hypothalamus, TRN: tegmental reticular nucleus, APN: anterior pretectal nucleus, IRN: intermediate reticular nucleus, My: medulla, RN: red nucleus, VIS: visual areas, PBN: parabrachial nucleus, VM: ventromedial nucleus of the thalamus, Cg: cingulate, RT: reticular nucleus of the thalamus, VTA: ventral tegmental area. N=3 mice (O) A high-magnification micrograph of DLS. Representative of N=3. (P) An Imaris 3D reconstructions of DLS (dark green) and CSN axonal labeling in DLS (red) superimposed over a 3D projection of GFP labeling. Representative of N=3. Error bars are SEM.
CSNs synapse on a range of interneurons with distinct roles in motor control and sensory processing2,34. We wondered whether CSNsDLS also innervate interneuron subtypes with well-defined functions, such as the GAD2-expressing neurons responsible for presynaptic inhibition of sensory afferents35, as well as Chx10-expressing propriospinal excitatory neurons36, and dorsal somatostatin (SST)-expressing mechanoreceptive neurons37. We used an intersectional approach to drive expression of two viral constructs in GAD2-, Chx10-, or SST-expressing spinal interneurons: one encoding the avian receptor for EnVA glycoprotein (AAV-FLEX-TVA), and the other encoding the rabies glycoprotein necessary for transsynaptic spread (AAV-FLEX-N2cG; Extended Data Figure 2J). We then injected AAV-FRT-GFP into forelimb motor cortex. Two weeks later, we injected spinal cord with the improved pseudotyped, G-deficient CVS-N2c rabies construct expressing FlpO recombinase fused to mCherry (EnVa-N2cΔG-FlpO.mCherry). This construct infects neurons expressing TVA, and from a subset of those also expressing N2cG, infects and expresses FlpO in presynaptic input neurons, in turn driving expression of GFP through Flp-mediated recombination (see Extended Data Figure 3 for validation of transsynaptic rabies constructs)38. We then mapped the position of GFP-labeled neuronal processes throughout the brain. First, we note from the sizable population of transynaptically labeled neurons that CSNs synapse onto all of the spinal interneuron subtypes tested (GAD2: 585±316, N=3, Chx10: 592±425, N=4, SST: 869±187, N=2). Importantly, CSNs that synapse on each interneuron subtype of interest all formed consistently widespread axonal arborizations in DLS (Extended Data Figure 2K–M).
We followed our output mapping experiments by determining the sources of input to CSNsDLS. We first used an intersectional approach to drive expression of TVA and N2cG in CSNsDLS (Figure 2A). Two weeks later, we injected motor cortex with EnVa-N2cΔG-tdTomato, resulting in expression of tdTomato in synaptic inputs to CSNsDLS (Figure 2B–J)38. Using anatomical reconstructions, we found isocortical regions like S1 and M2 provide the main source of input to CSNsDLS (Figure 2K–M, N=3). Interestingly, thalamic regions predominated non-cortical input to CSNsDLS, including regions that are thought to serve as relays for the output of the basal ganglia (Fig 2M, inset)39. These anatomical experiments reveal the structural complexity of CSNs, and highlight their unique capacity to influence diverse brain regions involved in motor control, most notably the main input to the basal ganglia, the striatum.
Figure 2. Identifying brainwide inputs to CSNsDLS.
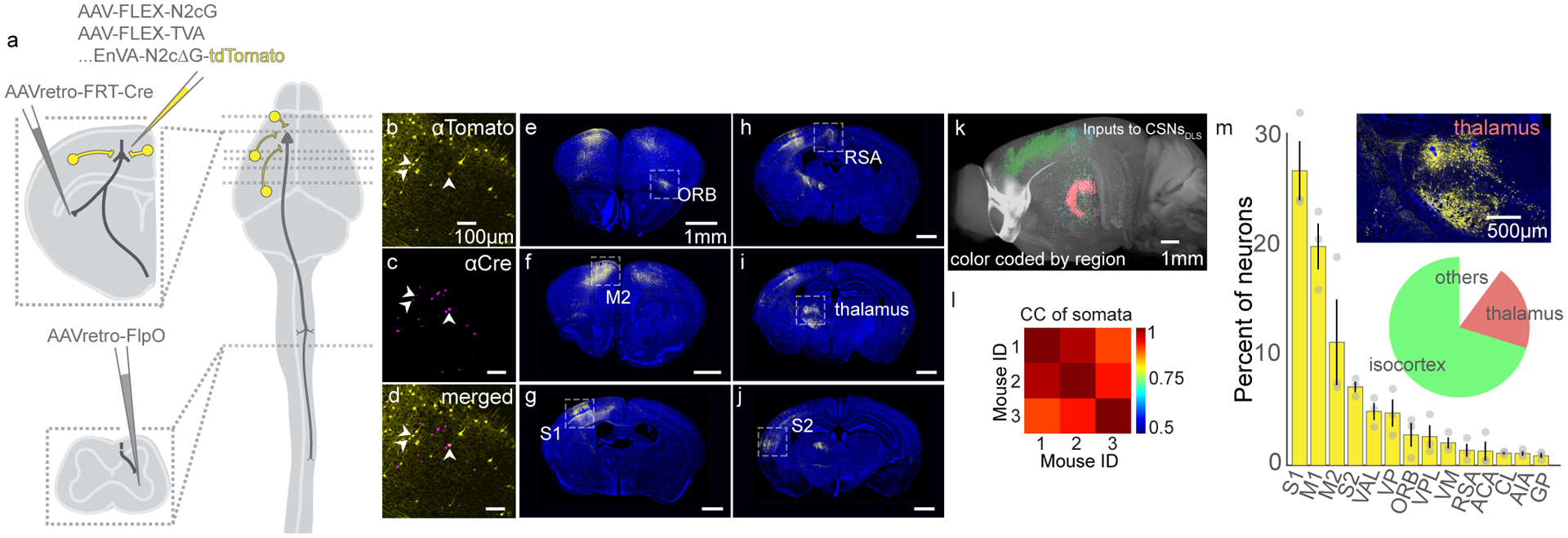
(A) Strategy to use intersectional transsynaptic tracing to label inputs to CSNsDLS. (B-D) Identification of starter cells (arrowheads) through coexpression of tdTomato (B) and Cre (C). Overlay in (D). Representative of N=3. (E-J) Example confocal micrographs of tdTomato labeling throughout the brain. DAPI is blue. Representative of N=3. (K) 3D reconstruction of tdTomato+ neurons, color coded by brain group. (L) Correlation analysis of somata population positions across animals. (M) Quantification of the top brain regions giving rise to neurons that form synapses on CSNsDLS. The pie chart indicates the major brain groups providing input to CSNsDLS. The inset image displays neuronal labeling in subdivisions of the thalamus. N=3 mice. Error bars are SEM.
We next sought to determine if CSNs are unique in their capacity to directly modulate both spinal cord and striatum. We injected AAV-retro-GFP into cervical spinal cord and AAV-retro-tdTomato into contralateral DLS (Figure 3A). Using anatomical reconstruction, we then identified the brain-wide sources of input to DLS and spinal cord. The only structures that contained appreciable numbers of both DLS inputs and spinal inputs were in isocortex (Figure 3B–D, N=4). Within isocortex, DLS- and spinal cord-projecting neurons may be interspersed, or there may be neurons that simultaneously project to both structures. To disambiguate these possibilities, we marked neurons that were positive for both GFP and tdTomato, and confirmed that nearly all cells that project to both DLS and cervical spinal cord were located in sensorimotor cortex (Figure 3E–H, N=4).
Figure 3. CSNs are likely the only neurons that project to both DLS and spinal cord.
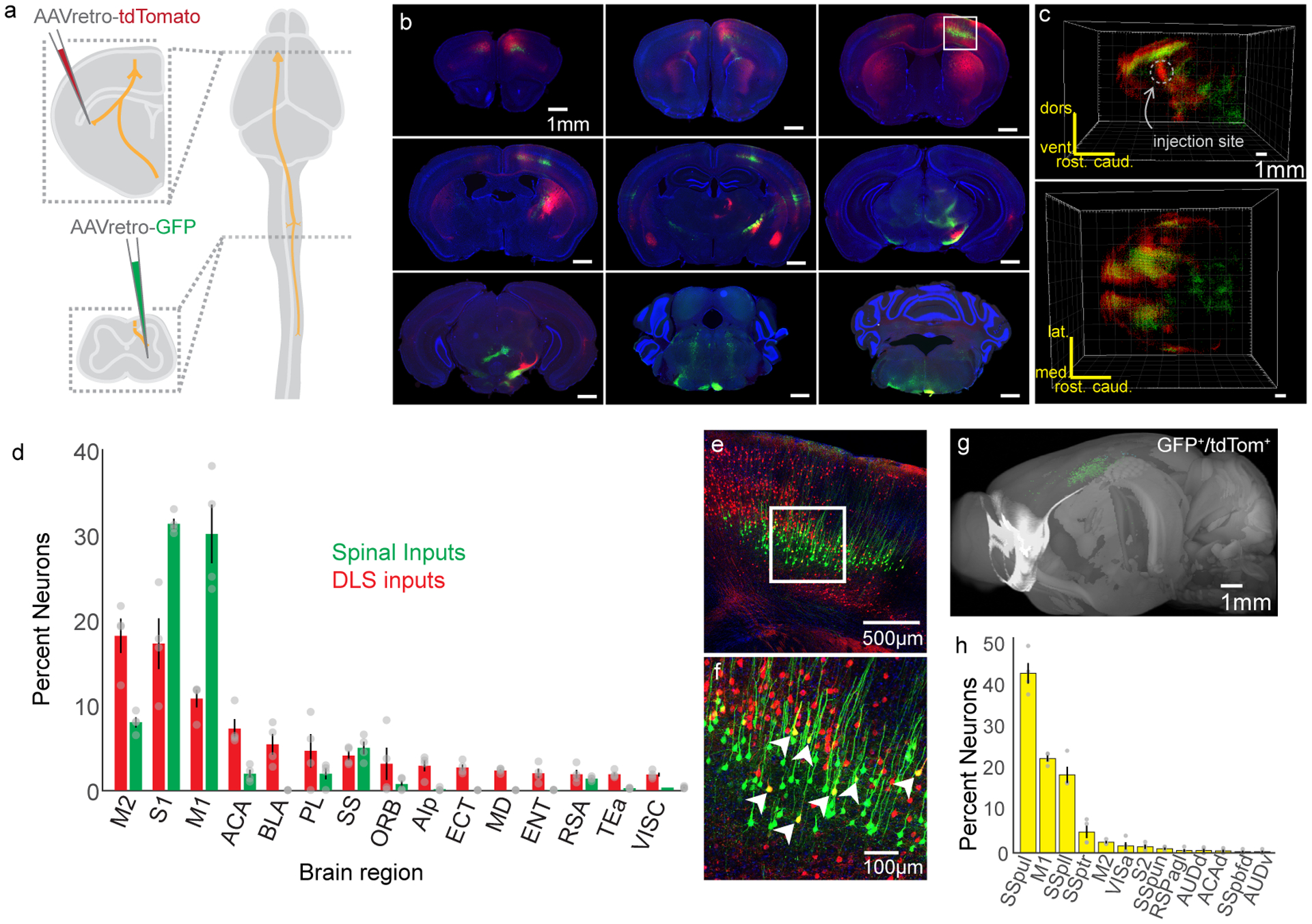
(A) Experimental strategy to label inputs to DLS in red and inputs to spinal cord in green. (B) Photomicrographs of labeling throughout the brain. The boxed region indicates sensorimotor cortex, which is highlighted in (E). (C) 3D reconstructions of cell body positions from two vantages. The DLS injection site is indicated, which corresponds to a bolus of red cell bodies. (D) Quantification of brain regions with inputs to DLS (red bars) and spinal cord (green bars). (E) Confocal photomicrograph from sensorimotor cortex showing dual labeling. (F) High magnification view from the inset in (E). Arrowheads indicate double-positive cells. (G) 3D view of double positive neurons. (H) The brain regions with double positive neurons. (structures not defined before: Primary Somatosensory Areas: SSpul (upper limb), SSpll (lower limb), SSptr (trunk), SSpun (unassigned), SSpbfd (barrel field), VISa: Anterior Area, RSPagl: Retrosplenial Area, Agranular Part, AUDd: Dorsal Auditory Area, ACAd: Anterior Cingulate Area, dorsal part, AUDv: Ventral Auditory Area). N=4. Error bars are SEM.
CSNsDLS form biased synapses onto distinct striatal pathways
To understand how CSN axon collaterals influence the brain, it is critical to understand the cell types that these axons innervate. Within the striatum, CSNsDLS could synapse on two interspersed populations of spiny projection neurons, defined in part by expression of either dopamine receptor 1 or 2 (D1 or D2 SPNs). Previous research revealed that corticopontine neurons (i.e. PT neurons) drive larger currents in D1 SPNs than D2 SPNs, despite the fact that D1 SPNs, on average, are larger than D2 SPNs and have lower input resistances24,40. Yet, PT neurons are a diverse population that encompasses both corticobulbar neurons and functionally heterogeneous corticospinal neurons41,42. Additionally, the synaptic mechanisms underlying differential connectivity between PT neurons and striatal SPNs is unclear. Therefore, we combined an intersectional optogenetic expression strategy with whole-cell voltage clamp recordings to characterize synapses arising exclusively from cervical spinal cord-projecting CSNs. First, we expressed channelrhodopsin-2 (ChR2) in CSNs by injecting AAV-retro-ChR2.tdTomato into cervical spinal cord of adult D1-tdTomato or D2-GFP reporter mice (Figure 4A). Weeks later, we made targeted whole-cell recordings from D1 and D2 SPNs in brain slices, identified in part by the presence or absence of reporter gene expression in cell bodies visualized under DIC optics (Figure 4B–H). Recordings were made from neighboring (within 50um) D1 and D2 SPNs in sequence (N=3, n=12 pairs), or simultaneously (N=3, n=8 pairs), and were pooled for further analysis (N=6, n=20). Brief (10ms) photostimulation of ChR2-expressing CSN axons drove excitatory postsynaptic currents (EPSCs) in both D1 and D2 SPNs when measured at membrane holding potentials of −70mV (Figure 4I). Comparing ChR2-evoked current and charge revealed that CSN collaterals drive larger responses in D1 SPNs when compared to D2 SPNs (Figure 4J–L, 33.44±5.69 pA for D1, 17.79±3.67 pA for D2, d.f.=19, t=3.3067, p=0.0037, d=0.7314; 7.17±1.12 nC for D1, 3.90±0.871 nC for D2, d.f.=19, t=3.021 p=0.006, d=0.7641). Repeating these experiments using stimulation of intratelencephalic axon collaterals resulted in equivalently-sized EPSCs in D1 and D2 neurons, suggesting biased innervation might be unique to CSNs (Extended Data Figure 4). Our results comport with previous studies of PT versus IT innervation of SPNs, and build on those results by finding cervical CSNs in particular are biased in their innervation of D1 SPNs over D2 SPNs43.
Figure 4. Optogenetics-assisted mapping of corticospinal collateral synapses in the striatum.
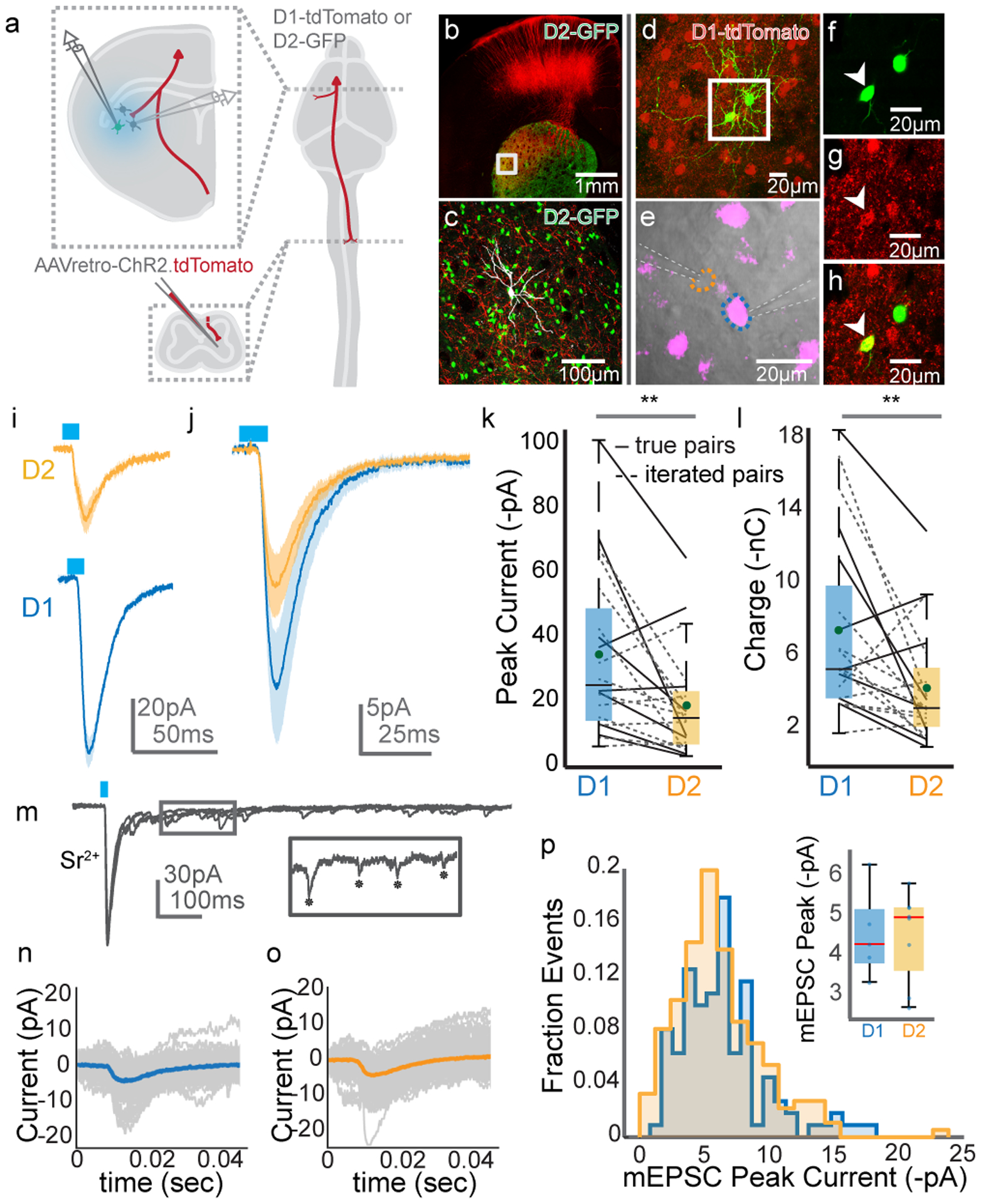
(A) Schematic of the experimental strategy. (B) Confocal micrograph of a brain slice, showing ChR2.tdTomato labeling (red) and D2 SPNs (green). (C) High magnification view of the boxed region from (B). An SPN targeted for recording and filled with neurobiotin is in grey. (D) Micrograph of two SPNs targeted for simultaneous recordings (green). D1 SPNs are in red. (E) DIC image of one D1 SPN (magenta, blue outline) and one D2 SPN (orange outline) targeted for simultaneous recording. Recording electrodes indicated with dashed white lines. (F-H) High magnification micrographs from (D) showing GFP (F), tdTomato (G), and overlay (H). The arrowhead indicates a D1 SPN. Micrographs from (B-H) are representative from N=6 experiments. (I) Recordings from a D1 SPN (blue) and D2 SPN (orange) in response to stimulation (blue bar) of ChR2-expressing axons. Holding potential is −70mV; shaded region indicates SEM. (J) Grand average response of all D1 (blue) and D2 (orange) SPNs to stimulation. Shaded area is SEM. (K-L) Pairwise comparison of ChR2-evoked amplitude (K) and charge (L) in D1 versus D2 SPNs, recorded simultaneously (solid lines) or in sequence (dashed lines). N=6, n=20, two-sided paired t-test, p=0.0037 for (K), p=0.006 for (L). (M) Voltage clamp recordings from an SPN following ChR2 stimulation with strontium replacing calcium. The inset shows single mEPSCs, indicated with asterisks. (N-O) Trial average of mEPSC evoked from an example D1 (N) and D2 (O) SPN. Individual trials are grey. (P) Distribution of all mEPSCs ordered by mEPSC peak current, recorded in D1 (blue) or D2 (orange) SPNs. The inset box-and-whisker plot compares average mEPSC amplitude in individual D1 versus D2 SPNs. N=5, n=5 for D1, n=8 for D2. The horizontal dashes in the box plots in (K), (L), and the inset for (P) represent the median. The greens dots in (K) and (L) indicate the means. The blue dots in the inset for (P) indicate peak mESPC amplitudes for individual neurons. The bottom and top edges of boxes indicate the 25th and 75th percentiles, while the whiskers indicate minima and maxima.
Results from the above experiments could be explained by CSNs forming either larger synapses onto D1 SPNs than D2 SPNs, or more numerous similarly sized synapses. To disambiguate between these possibilities, we replaced extracellular calcium with the divalent cation strontium, which desynchronizes neurotransmitter release from the pre-synapse (Figure 4M)44. We reasoned that measuring the amplitude of isolated miniature EPSCs (mEPSCs) evoked by photostimulation would allow us to infer the size of single synapses made by CSN axons on SPNs45. To this end, the averages of mEPSCs recorded from D1 or D2 SPNs were indistinguishable, implying that CSNs form similarly-sized synapses on both populations (Figure 4N–P, 4.47±0.51 pA for D1, n=5; 4.45±0.40 pA for D2, n=8, d.f.=11 t=0.0366, p=0.97, N=5). Stimulation of intratelencephalic axons also resulted in equivalently-sized mEPSCs in D1 and D2 SPNs (Extended Data Figure 4I–L). By extension, we concluded that CSNs, on average, form more synapses on D1 SPNs than on D2 SPNs. Together, these electrophysiological experiments reveal a synaptic and circuit basis by which CSNs interact with two distinct pathways of the basal ganglia.
CSNsD1 and CSNsD2 synapse in distinct spinal compartments
The CSNs that synapse on D1 or D2 SPNs could belong to the same population, or could belong to at least partially distinct populations. Furthermore, if they represent at least partially distinct populations, CSNs that innervate D1 or D2 SPNs could project into different regions of the spinal cord and synapse onto different spinal interneurons. To address these possibilities, we used rabies tracing to map the spinal projections of CSNsD1 and CSNsD2. Into DLS of D1-Cre or A2a-Cre mice, we injected a cocktail of AAV-FLEX-TVA and AAV-FLEX-N2cG. We later injected EnVa-N2cΔG-tdTomato into the same site, labeling inputs to D1 or D2 SPNs with tdTomato (Figure 5A). Because the input to DLS that projects to spinal cord is sensorimotor cortex (Figure 3), we concluded that axons found in spinal cord arose from CSNs. We then took high resolution confocal images throughout cervical spinal cord, visualizing antibody-enhanced tdTomato labeling, along with co-expression of vGlut1 in order to identify presynaptic boutons (Figure 5B). We first noted the expansive terminal fields formed by all CSNDLS from C2 to C7 (Figure 5C, N=5 for D1-Cre; N=5 for A2a-Cre). CSNDLS synapses were found throughout the spinal grey, with densest innervation in intermediate and superficial laminae. Still, we identified vGlut1+ boutons deep within the ventral horn, including those that were closely apposed to putative Renshaw neurons, highlighting their capacity to intimately influence motor output (Extended Data Figure 5A–G)46,47.
Figure 5. Mapping the distribution spinal synapses from CSNsDLS.
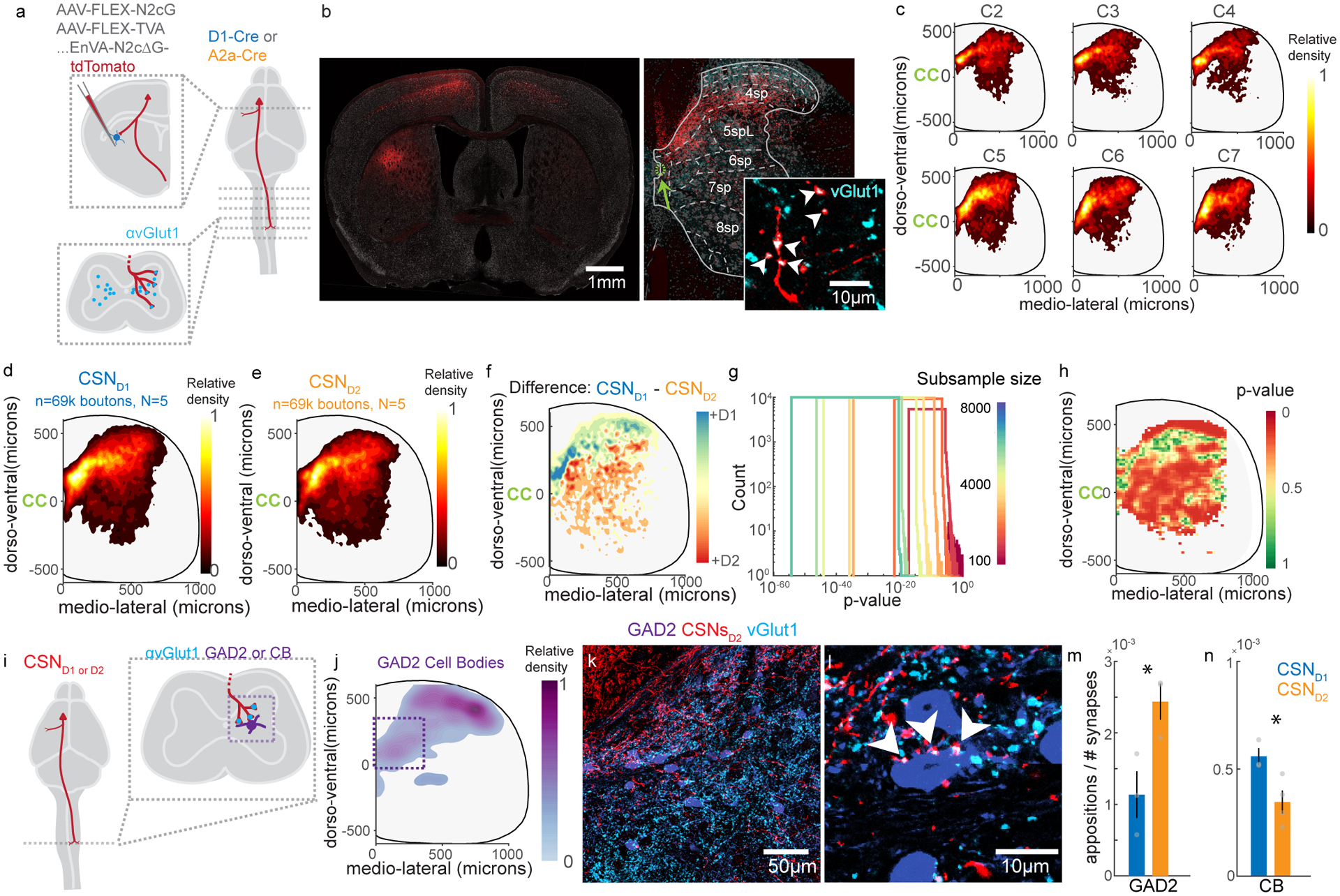
(A) Experimental strategy to transynaptically label CSNsD1 and CSNsD2. (B) Photomicrograph of tdTomato-labeled CSNs with identified synapses on striatal SPNs (left), and the synapses formed by these neurons in spinal cord (right). tdTomato+ synapses are identified by coincident expression of vGlut1 (cyan, inset). Arrowheads indicate vGlut1+ boutons. The green arrow indicates the central canal. Fluorescent Nissl stain is grey. Representative of N=10. (C) Contour plots illustrating the relative distribution of synapses arising from all CSNsDLS, ordered by cervical spinal segment. The central canal is indicated by ‘CC’. N=10, C2: n=22664, C3: n=30418, C4: n=26091, C5: n=30479, C6: n=23963, C7: n= 23571. (D–E) Contour plots illustrating the relative distribution of synapses arising from CSNsD1 (D) and CSNsD2 (E). N=5 for each genotype, n=69,636 each. (F) The difference between contour plots in (D) and (E). (G) Random resampling analysis. The dataset was resampled with different sample sizes (color, n=1100:1000:8000), and statistical analysis was repeated many times. (H) Statistical differences between the spatial distribution of CSNsD1 and CSNsD2 synapses. N=5 for each genotype, See Methods for details. (I) Experimental strategy to transynaptically label CSNsD1 and CSNsD2 and measure close appositions on spinal cell types. (J) Contour plot illustrating the relative distribution of GAD2 cell bodies for one mouse. n=531. (K) Photomicrograph showing GAD2 neurons (purple), CSND2 axons (red) and vGlut1 immunolabeling (cyan) in cervical spinal cord, C6 segment. The corticospinal tract is visible in the upper left corner. Representative of N=3. (L) High magnification view showing vGlut1+ CSND2 varicosities closely apposed to a GAD2+ neuron, indicated with arrowheads. Representative of N=3. (M-N) The number of close appositions on GAD2 neurons (M) or CB neurons (N) divided by the total number of vGlut1+ CSND1 or CSND2 varicosities in the field of view. N=3 for each genotype for GAD2, N=3 for CSND1 and N=4 for CSND2 for CB. Significance is measured with an unpaired two-sided t-test. p=0.0345 for (M). p=0.029 for (N). Error bars are SEM.
We next separately analyzed the distribution of synapses arising from CSNsD1 and CSNsD2. While both populations of neurons formed synapses throughout cervical spinal cord, CSND1 synapses were biased to more rostral coordinates, home to interneurons that modulate sensory feedback (e.g. SST interneurons in Extended Data Figure 2J), while CSND2 synapses were skewed to the ventral regions of spinal cord, where there is an enrichment of premotor interneuron populations (e.g. Chx10 interneurons) (Figure 5D–F, Extended Data Figure 5H, 147.41±0.79μm for D2 versus 188.17±0.78 μm for D1, dorso-ventral relative to the central canal, d.f. = 139266, t=36.72, p=1.08×10−293, d=0.20)34. Random subsampling revealed these results were statistically robust (Figure 5G, i.e. 1100 out of 69,636 resampled coordinates, median of repeated t-tests, dorsoventral: p=2.37×10−6). Next, we performed regional comparative statistics, revealing highly distributed regions of spinal grey with statistically significant innervation differences (Figure 5H, Extended Data Figure 5I–J). These data reveal that CSNsD1 and CSNsD2 are at least partially distinct populations, with biased spinal projection patterns.
These results raise the question of whether CSNsD1 and CSNsD2 are biased in their innervation of spinal interneuron subtypes. We reasoned that a stringent test for this possibility is to measure relative innervation of an interneuron species found in a region with equivalent innervation density by CSNsD1 and CSNsD2. Our experiments using transsynaptic tracing from spinal interneurons showed that GAD2-expresing putative GABApre neurons are innervated by CSNsDLS (Extended Data Figure 2J–M), making this population a prime candidate for study, since a group of these neurons are enriched in intermediate laminae of cervical spinal cord where we found equivalent innervation by CSNsD1 and CSNsD2 (compare Figure 5F with Figure 5J). We therefore generated double transgenic mice that 1) express Cre recombinase in D1 or D2 striatal SPNs and 2) express GFP in GAD2-expressing spinal interneurons48. We then used rabies virus to express tdTomato in CSNs that innervate either D1 or D2 SPNs, and quantified the number of vGlut1+ CSND1 or CSND2 appositions on the somata and proximal dendrites of GAD2 interneurons, normalized by the total number of vGlut1+ corticospinal synapses in that region of interest (Figure 5I–L). These experiments revealed that GAD2 neurons receive more appositions from CSNsD2 compared to CSNsD1 (Figure 5M, 0.0024±0.000218 appositions/total CSN boutons for CSND2, 0.0011±0.000283 for CSND1, d.f.=4, t=3.15, p=0.0345, d=2.57). We questioned whether increased innervation of interneurons by CSNsD2 is a general feature, or if other interneuron types receive different patterns of innervation. To test this, we analyzed innervation of calbindin 28k (CB)-immunolabeled interneurons, many of which are found in the same intermediate regions as GAD2-expressing neurons. In contrast to GAD2 neurons, CB neurons received more synapses from CSNsD1 compared to CSNsD2 (Figure 5N, 0.000556±0.0000336 appositions/total CSN boutons for CSNsD1, 0.000340±0.0000535 for CSNsD2, d.f.=5, t=3.034, p=0.029, d=2.42). Altogether, these results presented reveal coordinated connectivity between CSNs and cell types in striatum and spinal cord.
CSNs signal both single movements and sequences
The anatomical complexities of CSNs – particularly their prominent projections to the striatum – inspired us to characterize their activity during a behavioral task relevant to basal ganglia. Previous research showed that striatum is necessary for sequential lever press behaviors, and that many striatal SPNs are selectively active around the onset or offset of sequences, while other neurons fire throughout execution28,29. We sought to determine whether inputs to striatum from CSNs display similar encoding properties during sequence performance, or if these striatal activity patterns arise downstream of cortex. Water-restricted mice were trained to depress a narrow lever positioned in front of their right forepaw four times in succession to receive a water reward (Figure 6A). Mice were acclimated to handling and head fixation, and rewarded for each lever press they made. After 7 days of this training, mice were then rewarded only after every fourth lever press. Next, the maximum sequence length that issued reward was lowered first to 3, then 2 seconds. Mice learn this task successfully, as evidenced by the rapid execution of grouped lever presses, and the increased performance of four press sequences and decrease of two press sequences (Figure 6B–D, two-way ANOVA, F=84.77, d.f.=9, p=0, post hoc t-test, p=0.0284 and p=0.026, respectively, N=8). Across training, both the inter-press interval (speed) and coefficient of variation (variability) of the inter-press interval for lever press sequences decreased and stabilized (Figure 6E, Extended Data Figure 6A–B). To analyze muscle activity with high resolution, we implanted electrodes made for recording electromyographic (EMG) signals into four forelimb muscles. To monitor the activity of CSNs, we injected AAV-retro-GCaMP6f into right cervical spinal cord of D1-Cre or A2a-Cre mice (Extended Data Figure 6C), and implanted a cranial window over left forelimb motor cortex. Two-photon (2p) imaging was used to record activity in dendritic trunks of CSNs approximately 300μm below the pial surface (Figure 6F–G). These dendritic signals are highly correlated with and faster than somatic calcium activity15,49,50. Calcium signals were extracted using CNMF and highly correlated (rho > 0.8) processes were treated as belonging to the same neuron to minimize overrepresentation by branching dendrites (Figure 6H)15,49,51. To overcome the temporal limitations of calcium transients, we used deconvolution developed with CNMF to convert calcium signals to an event signal, which we aligned to lever press sequences51. Event-triggered averaging of both raw fluorescence and CNMF-extracted signal revealed deconvolved activity corresponds well to underlying fluorescence signals (Extended Figure 6D–H). Z scored deconvolved activity from one mouse (Extended Data Figure 7A–B) and across all mice (Figure 6I) was visibly faster than calcium activity, and there was a strong trend for neuronal activity to be enhanced around lever press (Extended Data Figure 7C). Heatmaps of Z scored activity aligned to single lever press revealed a temporal distribution of peak responses (Extended Data Figure 7D). Binning neurons by the time of their peak responses revealed most neurons were active immediately around lever press (Extended Data Figure 7E, median time to peak response ~93ms), and the responses of these neurons were larger than those active further in time relative to lever press (Extended Data Figure 7F).
Figure 6. Two-photon calcium imaging of corticospinal neurons during a sequential forelimb behavior.
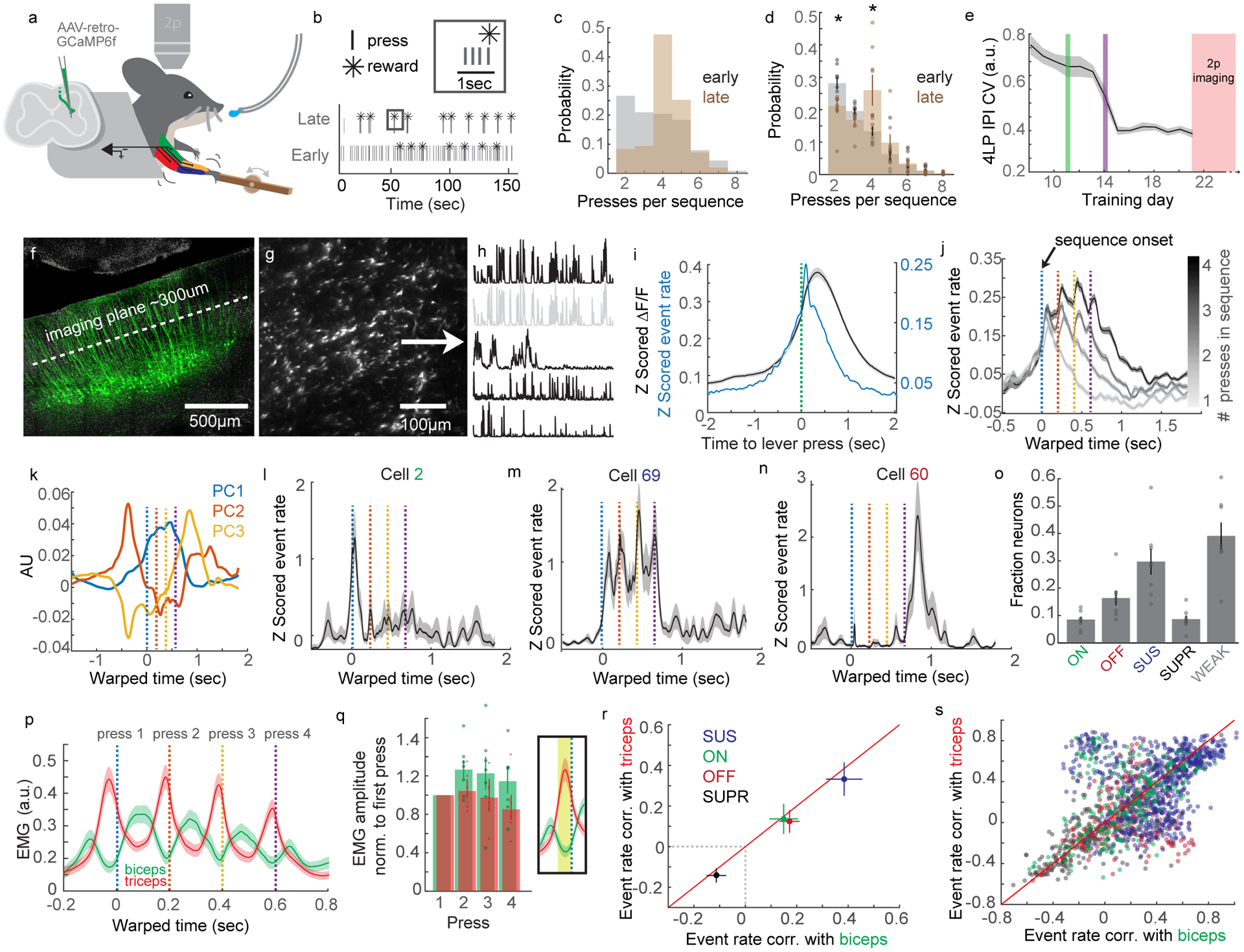
(A) Cartoon of behavior, 2p imaging, GCaMP6f labeling strategy, and EMG. (B) Example of lever press sequencing at training day 1 and day 7. (C) Probability of sequences containing different numbers of lever presses early (grey) and late in training (brown) from a single mouse. (D) Same as C, for all mice. Two-way ANOVA, F=84.77, d.f.=9, p=0, post hoc t-test, p=0.0284 and p=0.026, respectively, N=8. (E) Coefficient of variation of the inter-press interval (IPI) of four press sequences across training. The green vertical line indicates the day at which the maximum rewarded IPI is limited to 3 seconds. Before this, reward was issued every four presses, regardless of IPI. The purple line indicates when the IPI is limited to 2 seconds. (F) Micrograph of GCaMP6f-labeled CSNs, with the imaging plane indicated. DAPI is grey. (G) 2p image of GCaMP6f expression in dendrites of CSNs. (H) Calcium events derived using CNMF. The second trace is greyed out to indicate it is highly correlated to the top trace. (I) Average Z scored calcium (grey) and deconvolved events (blue) for all neurons and all mice aligned to single lever presses. n=2,374. (J) Average Z scored events aligned to the onset of lever press sequences, segregated by sequence length. IPIs are standardized using time warping; press times are indicated with colored dashed lines. n=2,374. (K) The top three PCs of time-warped activity. (L–N) Examples of neurons with activity coincident with the onset (L), individual presses (M), or offset (N) of lever press sequences. (O) The fraction of onset (ON), offset (OFF), sustained (SUS), suppressed (SUPR), or weak (WEAK) neurons, across all mice. (P) Time warped biceps and triceps EMG aligned to lever press sequences, for all mice. (Q) Quantification of the mean biceps and triceps activity preceding each press in a sequence. The inset indicates the time window for averaging activity. (R) Mean correlation of activity with biceps versus triceps EMG for ON, OFF, SUS, or SUPR neurons. (S) Same as (R), but showing the individual neuron correlations with biceps versus triceps. N=8 for all analyses. Error bars are SEM. Shaded areas are SEM.
Is CSN activity linearly related to motor output, or can CSNs display sequence-level activity similar to what is seen in striatal SPNs? To answer this question, we identified and grouped sequences of lever presses of one, two, three, or four presses. We used time warping to standardize the inter-press interval within sequences to 200ms, allowing us to preserve temporal resolution when averaging across trials (Extended Data Figure 7G–I, see Methods for details). Aligning activity from the total population of neurons (n=2,374, N=8) to lever press sequences revealed that, on average, CSN activity scales in duration to lever press sequences of increasing length (Figure 6J, see Extended Data Figure 7J–K for calcium signal). Yet, when plotted individually, the top three PCs of Z scored CSN activity each displayed unique activity signatures. The component accounting for the most variance was elevated in activity throughout sequence execution, while the next two PCs were active most strongly at the onset or offset of sequence (Figure 6K). This result motivated us to characterize the activity patterns of single neurons. We aligned time warped Z-scored deconvolved events of single neurons to four-lever press sequences. Doing so, we found a heterogenous population of neurons, including those with activity around the first or final press in a sequence, and neurons that were active around each press in a sequence (Figure 6L–N), all of which were intermingled in the same fields of view. Aligning Z scored event rates to each press in a sequence recapitulated these results (Extended Data Figure 8A).
Motivated by these results, we next aligned binned Z-scored events to time warped lever press sequences, and identified neurons with significant modulation at different windows of the lever press sequence. We identified 8.26% and 16.04% of CSNs responding at the onset (ON) and offset (OFF) of sequence, respectively, as well as a larger population of neurons with activity sustained (SUS) throughout sequence execution (Figure 6O, 29.18%). We further identified a population of neurons with activity significantly suppressed (SUPR) relative to baseline (8.45%), as well as population of neurons that did not meet criteria for significant modulation (38.41%). Averaging the Z scored activity of categorized neurons revealed the relative stereotypy of these responses, and this was recapitulated by inspecting the timing of peak responses of categorized neurons (Extended Data Figure 8B–C).
CSNs encode muscle- and non-muscle-correlated activity
Muscle activity may change from lever press to press, raising the possibility that the variability we observe in neuronal activity could be due to differential recruitment of musculature at the onset or offset of sequences, or preferential correlation with individual muscles. To directly address these possibilities, we analyzed the EMG activity of biceps and triceps during behavior and in relation to neuronal activity (Figure 6P–S, Extended Data Figure 9). First, aligning all biceps activity to local peaks in triceps activity revealed a robust alternation of the activity between these two antagonist muscles (Extended Data Figure 9A). Biceps and triceps activity alternated preceding lever press, consistent with their flexor and extensor identity (Extended Data Figure 9B). Importantly, biceps and triceps activity strongly alternated during sequences, and the amplitude of EMG activity preceding lever press events was similar throughout the sequence (Figure 6P–Q, biceps: one-way ANOVA, d.f.=3, F=0.98, p=0.4199; triceps: one-way ANOVA, d.f.=3, F=1.1, p=0.3684, N=8, Extended Data Figure 9C). We leveraged our EMG dataset by correlating CSN event rate to biceps and triceps activity during concatenated periods of behavioral quiescence or concatenated lever press sequences. On average, CSNs were more correlated with triceps activity than biceps during random periods of activity and quiescence, but this preference was lost when correlating neural activity with only concatenated lever press sequences, controlling for the number of samples in each condition (Extended Data Figure 9D). We next measured the correlation between average time warped biceps and triceps EMG activity and average time warped events of ON, OFF, SUS, and SUPR neurons during lever press sequences. SUS neurons were more correlated with biceps and triceps activity than ON and OFF CSNs, and SUPR neurons were anticorrelated with muscle activity (Figure 6R). While some individual neurons were more strongly correlated with biceps or triceps EMG, on average no group of neurons was consistently more correlated with one muscle over the other, suggesting encoding of muscle identity cannot explain sequence encoding properties of CSNs (Figure 6R–S, Extended Data Figure 9E–F). Finally, we addressed the possibility that ON neurons are encoding gross body movements that precede the onset of lever press sequences. We measured the variance of regions of interest encompassing various body parts in videos of trained animals performing the task, and showed that before arm movement onset, there were no overt body, mouth, or nose movements (Extended Data Figure 9G–L). These data indicate that, in general, muscle output is not the exclusive determinant of CSN activity during skilled motor sequences.
Diverse CSN activity is broadcast to both striatal pathways
Our results thus far show CSNs can encode sequence-level activity similar to what is observed in striatal SPNs. Although similar proportions of D1 and D2 SPNs display onset and offset signals, more D1 than D2 SPNs show activity that is sustained through sequence execution, while more D2 than D1 SPNs are suppressed during sequences28. Therefore, it is plausible to hypothesize that CSNs that synapse onto D1 SPNs are more likely to be SUS neurons, while CSNs that synapse on D2 SPNs are more likely to be SUPR neurons. To address this possibility, we combined our 2p calcium imaging experiments with transsynaptic rabies tracing from D1 or D2 SPNs. In the same mice as above (i.e. D1-Cre, N=4 or A2a-Cre, N=4), we injected AAV-FLEX-N2cG and AAV-FLEX-TVA into DLS before cranial window implantation (Figure 7A). After all functional calcium imaging data was acquired, EnVA-N2cΔG-tdTomato was injected into the same location of DLS (Figure 7B–D). Ten days following rabies injection, we took structural images of GCaMP labeling and Z stacks of tdTomato labeling (Figure 7E, Extended Data Figure 10). We then used 3D reconstruction to improve detection of tdTomato+ dendrites at the functional imaging plane, and generated binary masks from this dataset. We then used the GCaMP structural reference images to align masks of rabies labeling to the functional imaging dataset. This approach allowed us to identify CSNsD1 and CSNsD2 post hoc, avoiding any deleterious effect rabies expression has on response properties. Because our anatomical experiments revealed that CSNsD1 and CSNsD2 are at least partially non-overlapping populations, we reasoned that this approach would allow us to distinguish large encoding differences between groups. We first analyzed neuronal activity in CSNs with confirmed synapses in the striatum, and found that it scales in duration with lever press sequences, similar to general CSNs (Figure 7F). Do CSNsD1 and CSNsD2 comprise similar proportions of ON, OFF, SUS, and SUPR neurons? We applied our classification scheme to rabies-labelled CSNs, and compared these data to unlabeled neurons from the same mice to control for potential differences in rabies expression across animals. Surprisingly, we found similar proportions of ON, OFF, SUS, and SUPR neurons in tdTomato+ neurons compared to tdTomato− CSNs, along with no apparent enrichment of any response type when comparing CSNsD1 and CSNsD2 (Figure 7G). These results support a model where the complex information encoded by CSNs during motor sequences is transmitted in a balanced fashion to both D1 and D2 SPNs, suggesting that the different encoding properties of D1 vs D2 SPNs emerge from differences in the synaptic properties, intrinsic connectivity, and neuromodulatory receptors of the target neurons.
Figure 7. Rabies-based in vivo identification of corticospinal neurons with striatal synapses.
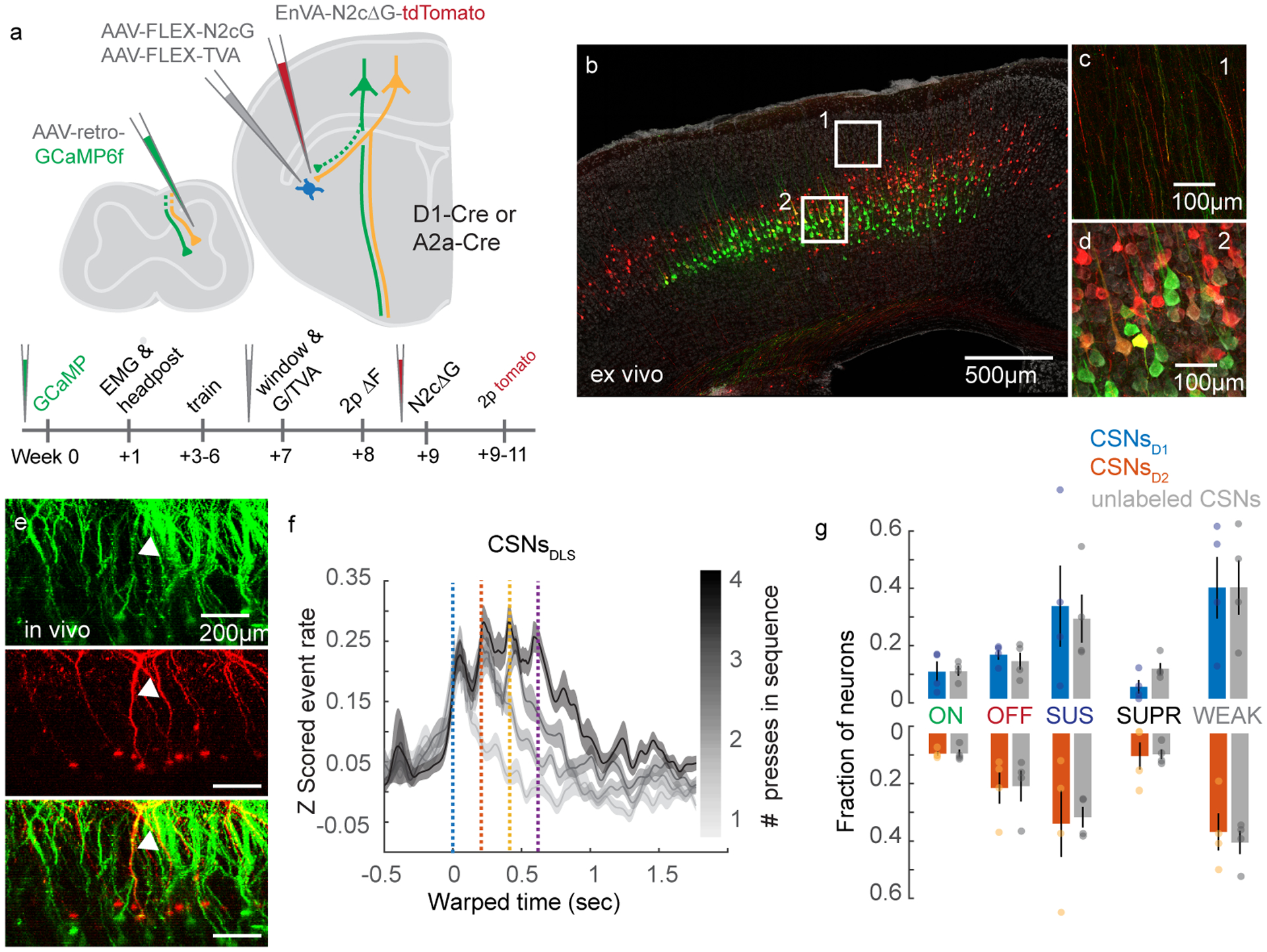
(A) The experimental strategy and timeline. (B) Confocal micrograph of GCaMP6f-tagged CSNs (green) and transynaptically-identified inputs to striatal SPNs (red). DAPI is grey. (C-D) High magnification images of the regions indicated in (B) showing green and red fluorescence in dendritic trunks (C) and somata (D). Double-labeled processes are yellow. (E) X-Z view of GCaMP- and tdTomato-expressing neurons in motor cortex, imaged in vivo. A double-labeled neuron is indicated with the arrowhead. (F) Average Z scored events of CSNsDLS aligned to the onset of lever press sequences, segregated by sequence length. n=347. (G) Fraction of ON, OFF, SUS, SUPR, and WEAK transynaptically-identified CSNsD1 and CSNsD2, compared to unlabeled CSNs. N=4 for each genotype. Error bars are SEM. Shaded area is SEM.
Discussion
The results presented here reveal that corticospinal neurons encode information that is diverse in its relationship to behavioral output, from muscle-related activity to higher order sequence-related information in the form of onset or offset responses. Our results further uncover synaptic and circuit principles governing the communication of these neural signals between spinal and basal ganglia circuits. Populations of CSNs preferentially synapsing onto D1 SPNs have different spinal projection patterns than CSNs preferentially synapsing onto D2 SPNs, and are biased in their connectivity to different spinal interneuron cell types. Surprisingly, these different CSN populations broadcast similar information to downstream circuits, suggesting that motor specificity emerges from the cell types innervated and differences in connectivity at these postsynaptic sites.
We first used a suite of anatomical tools to map the brain-wide organization of inputs to the spinal cord, and identified isocortical regions – particularly sensorimotor cortex – as containing the most spinal cord-projecting neurons across all mapped structures. In addition to sensorimotor cortex, we identified many midbrain and hindbrain inputs, including structures with well-defined roles in modulating premotor circuits32,33. Notably, the AAV-retro and rabies-based tracing methods we used do not necessarily reflect the strength of connectivity across these circuits, and differences in viral tropism may contribute to differences in labeling of brain structures30,52. Still, our approach to using these improved viral tools revealed details of corticospinal organization that have been elusive. First, we found that CSN collaterals project most prominently to the dorsolateral striatum, and that CSNsDLS comprise a diverse population originating in both motor and somatosensory cortex. An interesting future direction would be to characterize functional or anatomical differences between motor cortical and somatosensory cortical CSNsDLS.
Our results are at odds with some studies which have suggested that corticospinal and corticostriatal populations are largely non-overlapping, including antidromic stimulation-based experiments53. This may be due to the fact that individual CSNs have terminal fields that occupy a small region of striatum and form relatively few synapses, meaning that striatal stimulating electrode likely does not effectively drive antidromic action potentials in all CSNs that project to striatum20,54,55. And while some anatomical studies have deemphasized corticospinal innervation of supraspinal structures by CSNs, this may be due to less effective viral or histological methods. Indeed, Ramon y Cajal, using highly effective Golgi stains, made note of substantial supraspinal corticospinal innervation in his landmark studies21. Importantly, there may additionally be CSNs that do not collateralize in DLS and instead preferentially innervate other brain regions. Future studies will be useful in addressing this possibility. Within the striatum, CSNs drive larger responses in D1 SPNs than D2 SPNs, despite the fact that the lower input resistances of D1 SPNs should intuitively result in smaller postsynaptic currents24,40. Our strontium-based assay revealed that CSNs form nearly twice as many synapses on D1 SPNs, a postsynaptic dichotomy that may be amplified through the opposing influences that dopaminergic feedback have on D1 versus D2 SPN excitability56–58. This synaptic bias in the striatum is accompanied by an anatomical divergence in the spinal cord: CSNs that synapse on either D1 or D2 SPNs form distinct terminal fields in cervical spinal cord, revealing their capacity to differentially regulate spinal circuits through segregated interneuron populations. Consistent with this, we found CSNsD2 provide roughly twice as much input to GAD2 neurons compared to CSNsD1, an anatomical organization that may be important for the regulation of proprioceptive sensory feedback35,59,60.
Because of their widespread projections to DLS, we sought to characterize what information CSNs relate to basal ganglia. An important feature of the basal ganglia is that it is necessary for the performance of learned sequences of body movements25,28,29. Striatal SPNs encode features of movement sequences in their spiking activity, including neurons that encode the onset or offset of sequences, as well as neurons that are active around each individual movement in a sequence. Indeed, more D1 than D2 SPNs show activity that is sustained through sequence execution, while more D2 than D1 SPNs are suppressed during sequences. Because CSNs provide input to striatum, we reasoned that the activity properties of SPNs may me mirrored in the activity properties of their presynaptic CSN inputs. This led us to measure the sequence-related activity of CSNs, and then to measure whether CSNs that synapse on D1 or D2 SPNs differentially encode sequence activity. Using calcium imaging during a skilled head-fixed lever press sequence behavior, we first showed that the activity of many CSNs is closely related to muscle activity, which we measured use high resolution EMG techniques. Remarkably, a substantial proportion of both CSNs showed activity that was not highly correlated with individual muscle activity, but instead was correlated with either sequence onset or offset. Finally, a novel combination of 2p calcium imaging and transsynaptic rabies tracing revealed CSNsDLS encode lever press sequences in a similar fashion to the broader CSN population. Moreover, onset, offset, and sustained activity was found in equal proportions of CSNsD1 and CSNsD2. While some fraction of CSNs may synapse on both D1 and D2 SPNs, we found there are at least partially non-overlapping populations of CSNsD1 and CSNsD2. Because of this, we reason that our experimental approach would likely detect large differences in how these populations encode movement. These results raise the question how D1 and D2 SPNs differentially represent sequence information, particularly why more D1 SPNs are sustained, while more D2 SPNs are suppressed. One possibility is that CSNs act in a broadcasting capacity, transmitting efference copies of sequence-related performance to both the spinal cord the striatum. The bias of CSNs to synapse on D1 SPNs over D2 SPNs, combined with the state-dependent effects of dopaminergic feedback58 and local connectivity of striatal circuits, could then amplify sustained activity in D1 and suppress sustained activity in D2 SPNs. Another possibility is that there are differences in sequence-encoding in other populations of striatal inputs such as IT neurons or thalamic neurons. An interesting question is whether more complex behavioral information that is encoded in striatal SPNs is similarly encoded in CSNs presynaptic to those neurons. Future studies using our transsynaptic rabies tagging approach will be useful to address such possibilities.
In summary, our results unravel an organizational logic where separate populations of CSNs that synapse onto D1 and D2 SPNs form biased projection patterns and cell type-specific connectivity in spinal cord. However, CSNs targeting different striatal and spinal circuits broadcast similar movement-related information, including both neural signals closely related to motor output as well as signals related to higher-order features of behavior. These anatomical and functional circuit features suggest that motor specificity arises from the translation of corticospinal movement-related information by divergent circuits in the basal ganglia and spinal cord33. These differences in postsynaptic connectivity presumably act alongside various other neuronal inputs and downstream executive circuits to ultimately translate behavioral intent to action.
METHODS
A full list of resources and reagents can be found in Supplementary Information Table 1.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code Availability
The custom code used in this study is available from the corresponding author upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments and procedures were performed according to NIH guidelines and approved by the Institutional Animal Care and Use Committee of Columbia University.
Experimental Animals
Adult mice of both sexes, aged between 2–6 months were used for all experiments, including slice electrophysiology. The strains used were: C57BL6/J, Jackson Laboratories #000664, B6.Cg-Tg(Drd1a-tdTomato)6Calak/J, Jackson Laboratories #016204, Tg(Drd2-EGFP)S118Gsat/Mmnc, MMRRC #000230, Tg(Drd1a-cre)EY217Gsat/Mmucd, Jackson Laboratories #030778, B6.FVB(Cg)-Tg(Adora2a-cre)KG139Gsat/Mmucd, MMRRC #036158, Chx10-Cre, Custom Jessell Laboratory, B6J.Cg-Ssttm2.1(cre)Zjh/MwarJ, Jackson Laboratories #028864, Gad2tm2(cre)Zjh/J, Jackson Laboratories #010802, GAD2-GFP (GAD65-GFP), Gábor Szabó. Mice used for behavioral experiments were individually housed, and all mice were kept under a 12 hour light/dark cycle.
Methods Detail
Stereotaxic Viral Injections
Analgesia in the form of subcutaneous injection of carprofen (5 mg/kg) or buprenorphine SR (0.5–1mg/kg) was administered the day of the surgery, along with bupivacaine (2mg/kg). Mice were anesthetized with isoflurane and placed in a stereotaxic holder (Leica). A midline incision was made to expose the skull, and a craniotomy was made over the injection site. To label CSNs with GFP, 100nL of AAV-FLEX-GFP was injected into each of two sites of motor cortex, 1.5mm lateral to the midline and 0.5 and 1.0mm rostral to bregma, approximately 700μm below the pial surface. Care was made to ensure there was no efflux of virus by stabilizing the skull and waiting 10 minutes after penetration before injecting. AAV-retro-Cre.mCherry was then injected into spinal cord (see below). For rabies-based transsynaptic tracing from striatal SPNs, 40nL of a 1:1 mixture of AAV-FLEX-N2cG and AAV-FLEX-TVA.mCherry was injected into DLS at 0.5mm rostral, 2.65mm lateral, and 3.5mm ventral to bregma. To label CSN axons in spinal cord, a vertical approach was taken to target DLS, and 300nL of EnVA-N2cΔG-tdTomato was injected. For transsynaptic tracing following 2p imaging, a craniotomy was made just caudal to the cranial window. The injection pipette was angled along the rostrocaudal axis, and the same region of DLS targeted for injections of rabies helper viruses was injected with 300nL of pseudotyped deficient rabies virus. To express ChR2 in intratelencephalic neurons, 100nL of AAV-retro-ChR2.tdTomato was injected into either motor cortex or DLS contralateral to the hemisphere targeted for whole cell recording. For transsynaptic rabies tracing experiments to label inputs to CSNsDLS, 100nL of AAV-retro-FRT-Cre was injected into DLS, AAV-retro-FlpO was injected into spinal cord (see below), and 100nL of a 1:1 mixture of AAV-FLEX-N2cG and AAV-FLEX-TVA.mCherry was injected into forelimb motor cortex. Two weeks later, 300nL of EnVA-N2cΔG-tdTomato was injected into motor cortex. To tag CSNsDLS with GFP, AAV-retro-FLEX-GFP was injected into DLS, and AAV-retro-Cre.mCherry was injected into spinal cord (see below). To tag subpopulations of CSNs, 250nL of AAV-FRT-EYFP was injected into forelimb motor cortex at each of two sites. AAV-FLEX-N2cG and AAV-FLEX-TVA.mCherry was then injected into spinal cord, followed two weeks later by injections of EnVA-N2cΔG-FlpO.mCherry into spinal cord (see below).
Spinal Cord Viral Injections
Analgesia in the form of subcutaneous injection of carprofen (5 mg/kg) or buprenorphine SR (0.5–1mg/kg) was administered the day of the surgery, along with bupivacaine (2mg/kg). Mice were anesthetized with isoflurane and placed in a stereotaxic holder (Leica). A midline incision was made to expose the spinal column. The muscular overlying the column was resected, and a metal clip attached to a spinal clamp was used to secure the T2 process and minimize spinal cord movement. The tail was gently stretched with another spinal clamp to separate the vertebrae. A surgical microknife and fine forceps were used to sever the meninges, exposing the spinal cord. A pulled glass pipette was filled with virus, and a Nanoject III was used to make multiple small volume injections across into the spinal cord, with parameters that depended on the experiment and reagents used. For injections of AAV-retro-GCaMP6f, AAV-retro-Cre.mCherry, AAV-retro-ChR2.tdTomato, or AAV-retro-FlpO, one penetration was made into each segment of the spinal cord between C3 and C7. Twenty injections of 10nL each were made into the center of the spinal grey, for a total volume of 200nL per spinal segment. For injections of AAV-FLEX-N2cG or AAV-FLEX-TVA.mCherry, two penetrations were made into each segment of the spinal cord between C3 and C7. 10nL of virus was injected along the dorsoventral axis every 50μm between 1.2mm and 0.1mm below the surface of the cord, totaling 460nL per segment. For injections of EnVA-N2cΔG-FlpO.mCherry, three penetrations were made into each segment of the spinal cord between C3 and C7. 15nL of virus was injected along the dorsoventral axis every 50μm between 1.2mm and 0.1mm below the surface of the cord, totaling 1035nL per segment. Following all injections, the skin was sutured closed and animals were closely monitored during recovery.
Slice Electrophysiology and Optogenetic Photostimulation
Mice were deeply anesthetized with isoflurane and transcardially perfused with an ice-cold carbogenated high magnesium (10mM) ACSF. The brain was removed from the skull, and glued to the stage of a vibrating microtome (Leica). 300μm coronal brain slices were cut in a bath of ice-cold, slushy, carbogenated low calcium ACSF. Slices were incubated for 15–30 minutes in a 37°C bath of normal ACSF containing (in mM): 124 NaCl, 2.7 KCl, 2 CaCl2, 1.3 MgSO4, 26 NaHCO3, 1.25 NaH2PO4, 18 glucose, 0.79 sodium ascorbate. Slices were then transitioned to room temperature, where they remained for the duration of the experiment. Patch electrodes (3–6MΩ) were filled with either a potassium gluconate based internal solution (135 mM K-gluconate, 2 mM MgCl2, 0.5 mM EGTA, 2 mM MgATP, 0.5 mM NaGTP, 10 mM HEPES, 10 mM phosphocreatine, 0.15% Neurobiotin) or a cesium/QX-314 based internal solution (5 mM QX-314, 2 mM ATP Mg salt, 0.3 mM GTP Na salt, 10 mM phosphocreatine, 0.2 mM EGTA, 2 mM MgCl2, 5 mM NaCl, 10 mM HEPES, 120 mM cesium methanesulfonate, and 0.15% Neurobiotin). All recordings were made using a Multiclamp 700B amplifier, the output of which was digitized at 10 kHz (Digidata 1440A). Series resistance was always <35 MΩ and was compensated up to 90%. Neurons were targeted with differential interference contrast (DIC) and epifluorescence when appropriate. For simultaneous recordings, pairs of neighboring SPNs (within 50 μm of each other) were identified first by morphology using DIC imaging. The cellular identity of targeted neurons was confirmed through expression or lack of expression of transgenically-targeted fluorescent reporters. For experiments exploiting potassium gluconate based internal solutions, neurons were further identified through intrinsic electrophysiological properties, including excitability and current/voltage transformation. In a subset of experiments, cell morphology was visualized through internal dialysis of 0.1 mM Alexa Fluor 594 cadaverine or 0.1 mM Alexa Fluor 488 Na salt. ChR2-expressing axons were photostimulated using 10ms pulses of 473nm LED light (CoolLED) delivered through a 10x objective centered over the recording site. Brain slices were histologically processed to visualize Neurobiotin-filled cells through streptavidin-Alexa Fluor processing.
Histology and Confocal Imaging
Mice were deeply anesthetized with isoflurane and transcardially perfused with phosphate buffered saline (PBS) followed by ice cold 4% paraformaldehyde. Brains and spinal cords were post-fixed overnight in 4% paraformaldehyde, and then cryopreserved in a 30% sucrose solution for 3 days at 4°C. Brains and spinal cords were embedded in Optimum Cutting Temperature Compound (Tissue-Tek), and 70μm coronal sections were cut on a cryostat. Tissue was rinsed several times in PBS, then permeabilized in PBS containing 0.2% Triton X-100 (PBST). For imaging synapses in spinal cord, tissue sections were first permeabilized in 1% PBST to aid in antibody penetration. Immunostaining was performed with primary antibodies diluted at 1:1000 for 3 days at 4°C, and with secondary antibodies at 1:000 overnight at 4°C. Counterstains of DAPI or Neurotrace were included in the secondary antibody incubation at 1:1000. Brain and spinal cord slices mounted to slides were briefly incubated with TrueBlack diluted in 70% ethanol to quench lipofuscin and background autofluorescence. Confocal imaging was performed on a Zeiss 710 or Zeiss 880 using 10x, 20x, 40x, 63x, or 100x objectives. For mapping the distribution of spinal synapses arising from CSNsDLS, high XYZ resolution stitched images were acquired overnight using a 40x water immersion high NA objective. Imaris was used to identify tdTomato+ axons that colocalize with vGlut1 expression. Synaptic boutons were then marked with spots, and the coordinates of these spots were measured relative to the center of the central canal.
Slide Scanning and Anatomical Reconstructions
70μm coronal sections were serially mounted on slides, and were treated with TrueBlack diluted in 70% ethanol to quench lipofuscin and background autofluorescence. Sections were imaged using an AZ100 automated slide scanning microscope equipped with a 4X 0.4NA objective. (Nikon). Image processing and analysis using BrainJ proceeded as previously described61. Briefly, brain sections were aligned and registered using 2D rigid body registration62. A 7-pixel rolling ball filter was used on all images to reduce background signal and a machine learning pixel classification approach using Ilastik was employed to identify cell bodies and neuronal processes63. To this end, several background-subtracted images were imported into Ilastik (separately for each fluorescence channel) to generate a large sample of neurons with variable morphological characteristics. Representative cell bodies, neurites, and background fluorescence features were selected to train the algorithm across all images. Probabilistic assignment of image features was continuously checked with a live preview feature to ensure accuracy. The algorithms were then use to generate probability images for each fluorescence channel of each brain section image, and the resulting images were processed for segmentation of cell bodies and neurites. To map the location of these structures to an annotated brain atlas, 3D image registration was performed using Elastix relative to a reference brain64. The coordinates of detected cells and processes were then projected into the Allen Brain Atlas Common Coordinate Framework65. Visualizations of the data were performed in ImageJ and Imaris, and subsequent analyses were performed in MATLAB using custom software.
Electromyographic Electrode and Headpost Implantation
Electromyographic electrodes were fabricated as previously described66. Two pieces of insulated braided stainless-steel wire were knotted, and half-millimeter portions of insulation were stripped from each wire just below the knot, so that exposed contact sites were separated by 0.5 millimeter. The portions of wire with contact sites were twisted, and the ends secured in a crimped hypodermic needle to permit easy insertion into targeted muscle groups. The opposing strands were soldered to a miniature connector. This process was repeated three times to produce a total of four differential recording electrodes that could be implanted into four muscles.
Analgesia in the form of subcutaneous injection of carprofen (5 mg/kg) or buprenorphine SR (0.5–1mg/kg) was administered the day of the surgery, along with bupivacaine (2mg/kg). Mice were anesthetized with isoflurane and placed in a stereotaxic holder (Leica). Hair was carefully shaved from the right forelimb, neck, and head, and the skin was thoroughly cleaned. Incisions were made over the neck and forelimb, and the electrode assemblage was snaked through these sites so that the miniature connector was positioned near the head and the individual recording electrodes positioned near biceps, triceps, extensor digitorum communis, and palmaris longus. Electrodes were implanted in each muscle by passing each needle and wire through targeted muscle groups until the knot was abutted to the muscle entry point. The tag ends of wire were then knotted by the exit point, thus securing the contact sites within the muscle. Forelimb incisions were closed with sutures, and the headpost implantation proceeded. The scalp was removed to expose the cranium, and facia was cleared using a scalpel and saline irrigation. A custom, 3D printed plastic headpost was affixed to the cranium using Metabond dental cement (Parkell), and reference points were marked to facilitate the implantation of a cranial window. Finally, the miniature connecter for the EMG electrode assemblage was cemented to the caudal edge of the headpost, and the skin overlying the neck was closed with sutures.
Cranial Window Implantation
Analgesia in the form of subcutaneous injection of buprenorphine SR (0.5–1mg/kg) and was administered the day of the surgery, along with bupivacaine (2mg/kg) and the anti-inflammatory dexamethasone (2mg/kg). Mice were secured in a stereotaxic frame (Leica) and the head was secured using 3D printed forks designed to clamp the custom headpost. The custom cranial window was composed of two semicircular pieces of glass coverslip (200 μm thick, Tower Optical Corp.), fused together and then to a 4mm round #1 coverslip (Warner Instruments) with optical cement (Norland Optical Adhesive 61). A craniotomy the shape of the insertable coverslips was made over forelimb motor cortex, and the window was implanted so that the semicircular plug was gently pressing on the brain. The entire assemblage was secured using Metabond.
Behavior
Behavioral training occurred in parallel using behavioral chambers equipped with custom-made and assembled components. Mice were head-fixed using 3D printed hard plastic forks that clamped around a custom plastic headpost cemented to the cranium. The body rested in an opaque plastic tube, and the left forelimb was allowed to rest on a moveable perch. The right forelimb was positioned over a milled plastic lever that had a small counterweight. Lever presses were reported as the counterweighted arm passed through an infrared beam. Water rewards were dispensed through a blunt needle positioned ~3mm from the mouth so that beads of water reward were reachable by licking. Water reward was calibrated regularly by adjusting the length of the TTL pulse sent to a solenoid valve. Behavioral assays were controlled using software written for and deployed with pyControl (https://pycontrol.readthedocs.io/en/latest/). Performance was continuously monitored and recorded with webcams.
Mice were accustomed to handling for several days, and then placed on a water restriction schedule using established guidelines67. Weight, appearance, and general health was monitored daily, and supplemental water was administered when necessary. Water-restricted mice were acclimated to the custom-made behavioral apparatus for two days, where they received water reward (5μL) at sporadic intervals for 15 minutes (day 1) or 30 minutes (day 2). For the first phase of 7 days of training, mice were required only to press the lever once to receive reward. A timeout period of 3 seconds following reward was imposed to discourage continuous pressing, and the session ended when reward volume totaled 1000μL or one hour passed. Supplemental water was given to ensure an adequate daily volume. For the second phase of training, reward was delivered after every forth lever press, regardless of the inter-press interval duration. After 3 days of this initial sequence training, the maximum sequence length that issued reward was lowered first to 3, then 2 seconds. The countdown was reset after reward delivery, and a 3 second timeout was imposed.
Two-photon Imaging
Calcium imaging experiments were performed using a modified two-photon microscope (Bruker) outfitted with a 25× 1.0NA water immersion objective (Olympus) and a mode locked Ti:sapphire laser (Verdi 18W, Coherent) at 940nm. A custom-made computerized, motorized goniometer was used to subtly and reproducibly angle the head so that the cranial window was orthogonal to the beam path. Images were acquired using Prairie View software (Bruker) at 64Hz, and every 4 images were averaged, yielding an effective sampling rate of 16Hz. Data was acquired from an area approximately 430μm × 430μm with 256 × 256 pixels. Multiple non-overlapping field of view were imaged from each mouse over ~7 days. Following injections of EnVA-N2cΔG-tdTomato, fields of view from functional imaging sessions were identified by first aligning surface vasculature, then carefully aligning basal GCaMP fluorescence signals to reference images taken during functional imaging. Z stacks and 2D images of tdTomato fluorescence were acquired at a wavelength of 1040nm.
Electromyographic Recordings
EMG signals were amplified and filtered (250–20,000 Hz) with a differential amplifier (MA102 with MA103S preamplifiers, University of Cologne electronics lab). These signals were acquired at 10kHz alongside two-photon imaging data using Prairie View. EMG signals were down-sampled to 1kHz, high-pass filtered at 40Hz, rectified, and convolved with a Gaussian that had 10 ms standard deviation.
Statistics and Reproducibility
Below are details of experimental design and statistical analyses. No statistical methods were used to predetermine sample size, but our sample sizes are similar to those reported in previous publications61,68. No data were excluded from the analyses. Data for measuring the distribution of spinal synapses were subsampled and randomized. Randomization was not used for other experiments. Investigators were blinded to genotype when quantifying spinal synapses. Blinding was not used for other experiments, but automated analyses were used to limit experimenter bias. Data distribution was assumed to be normal but this was not formally tested. Animals (within genotype pools) were randomly assigned to experimental groups.
Automated Anatomical Reconstruction
Analysis of slide scanning data was performed using MATLAB. Data was output from the BrainJ pipeline in the form of CSV files containing measurements of neurite labeling and cell body count from each region in the Allen Brain Atlas Common Coordinate Framework. These measurements were hierarchically organized so that analyses from sub-regions (i.e. layers of primary motor cortex) could be performed alongside more general annotations (i.e. primary motor cortex), referred to as “summary structures” in the Allen Institute Mouse Common Coordinate Framework69. For measurements from high order brain regions regions (i.e. isocortex), measurements from descendent regions identified by Allen Brain Atlas application programming interface were grouped. In the Allen Institute Mouse Common Coordinate Framework, these are referred to as “major divisions”.
Slice Electrophysiology
Analysis of slice electrophysiology data was performed in MATLAB and in Clampfit (Molecular Devices). Tests of significance were performed using paired two-tailed t-tests with an alpha of 0.05. Amplitude and charge were measured from a 200ms window following stimulus onset relative to a baseline period 250ms before the onset of stimulus. To measure the amplitude of miniature EPSCs evoked through optogenetic stimulation of CSNs using strontium-containing ACSF, a mEPSC template was created in Clampfit. That template was used to search for mEPSCs in the tail response following the early synchronous release of neurotransmitter. Each mEPSC was manually reviewed, misidentified events were excluded from analysis, and the resulting mEPSCs were averaged for each cell.
Distribution of spinal synapses
For resampling analysis, subsets of dorsoventral and mediolateral synaptic position coordinates (from 100 samples up to 8000 samples) were randomly selected from the population, and this was repeated to generate 10,000 random datasets for each subsampled group. Statistical significance was measured for each group across genotype using a two-tailed unpaired t-test. The results were plotted in a histogram to illustrate the frequency of calculated p-values measured from each subsample size. To measure statistical differences in the spatial distribution (i.e. clusters) of spinal synapses from each genotype, the coordinates of identified synapses were binned to 20 microns for each mouse. Sets of nine (3 by 3) bins were selected using a sliding window of 1 bin in each mediolateral and dorsoventral direction, and the values of those bins were concatenated across mice for each genotype. For each sliding window position, an unpaired two-tail t-test with an alpha of 0.05 was used to measure significance, and the resulting p-value was assigned to the center of that sliding window. The sliding window was then shifted by one bin, and the process was repeated. To quantify corticospinal appositions on GAD2 or CB neurons, high resolution tiled Z stacks acquired using 40x or 63x high NA objectives with pinholes set at 1AU were imported into Imaris. Labeled GAD2 or CB neurons were identified by measuring colocalization between cell type labeling and Neurotrace fluorescence. The Spots function was used to identify the centroids of those neurons, and the image was manually inspected to eliminate false positives and add missed somata. Corticospinal varicosities were identified by colocalization of tdTomato and vGlut1+ staining. The Spots function was then used to identify the centroids of those structures. RFP+/vGlut1+ varicosities that formed close appositions to cell bodies or clearly identifiable proximal dendrites were marked with a Spot. The number of close appositions was then divided by the total number of GAD2 or CB cell bodies in the region of interest, and that number was then divided by the number of RFP+/vGlut1+ varicosities in the imaging field, in order to normalize to variable axonal labeling densities.
Calcium Imaging
Calcium imaging analysis was performed using constrained non-negative matrix factorization (CNMF). First, raw imaging datasets (~10 minutes each) were motion corrected using rigid, then non-rigid registration. Registered datasets were then processed in CNMF using an autoregressive process p of 2. Analysis was also performed using a p of 0 to replicate results, although this data is not included in this study. Output of the CNMF was in the form of ΔF/F and deconvolved events. Signals were up-sampled to match the sampling rate of EMG data, and Z-scored for further analysis. Time warping was used to standardize the inter-press interval of lever press trials. To this end, a lever press sequence time series template was created that comprised 6 time anchors, four of which correspond to a lever press sequence as well as 2 anchors for pre- and post-trial time periods. The inter-press interval for this template was standardized to 200ms. For each behavioral trial, the real time interval was measured between lever press, and linear resampling was performed as needed to either increase or decrease the number of samples within this window to match the 200ms template. Neurons were classified by their response properties as follows. For each neuron, trials of 4 lever press sequences were identified and time warped. A baseline period was defined as the first 250ms of each trial (beginning 1.5sec before the first lever press). Each trial (excluding the baseline period) was segmented into bins 10 samples in length, and the bins with mean activity significantly different than baseline (measured using within-trial paired two-tail t-test with an alpha of 0.05) were marked. Within this group, bins with mean activity greater than 2.5 standard deviations of baseline were then identified as positively modulated, and bins with mean activity less than that of baseline were identified as negatively modulated. We then identified significantly modulated bins in each of 9 time periods that spanned the trial (excluding the baseline period). The rationale for analyzing short bins was that brief deviations in activity could be overlooked or diluted if averaging across longer time windows. Neurons with significant and positively modulated activity in one or more of periods 1–4, and zero in periods 6–9 were classified as ON. Neurons with significant and positively modulated activity in one or more of periods of 6–9, and zero in periods 1:4 were classified as OFF. Neurons with significant and positively modulated activity in two or more of periods 3–7 were marked as SUS. Neurons with significant and negatively modulated activity in two or more of periods 3:7 were classified as SUPR. Neurons that met none of these criteria were marked as WEAK.
To mark CSNs co-labeled with tdTomato through rabies infection, we used a 3D reconstruction approach to improve identification of red fluorescent neurites. Around one week to ten days after rabies injection, Z stacks of tdTomato fluorescence were acquired at 1040nm. These tdTomato Z stacks were imported into Imaris, and binary masks were generated using the surfaces function. The binary stack was then resliced to generate one binary mask at the same Z plane used for functional imaging of GCaMP. This mask was registered to the functional data set using shift parameters derived from registration of reference GCaMP fluorescence images. We then identified tdTomato pixels that fell within the spatial boundaries of GCaMP ROIs, and summed these pixels, which were weighted by how close they were to the center of the ROI. This number was divided by the total tdTomato pixels within that structure, yielding a value that reflected 1) the proximity of the tdTomato process to the center of the GCaMP ROI, and 2) the degree to which the tdTomato structure was overlapping with the GCaMP ROI. If this value was greater than 60% of the sum of weighted GCaMP ROI pixels divided by the total number of those pixels, that ROI was marked as tdTomato+.
Extended Data
Extended Data Fig. 1. Mapping brainwide inputs to the spinal cord.

(A) Illustration of approach to visualize inputs to cervical spinal cord. (B) 3D reconstruction of inputs to spinal cord. Colors correspond to major brain divisions. (C) The top brain regions that project to spinal cord, determined by the relative fraction of total somata. Notable regions are indicated by colored bars. Insets illustrate exemplar brain regions with substantial labeling. Dashed boxes are colored to correspond to notable brain regions. The inset pie chart shows the major brain structures (“major divisions” as classified by the Allen Institute Mouse Common Coordinate Framework; see Methods) projecting to cervical spinal cord. N=3. (D–F) Micrographs of CSN axons expressing GFP (green) in transverse cross-sections of cervical (D), thoracic (E), and lumbar (F) spinal cord. The insets are high magnification images of GFP+ bulbous varicosities from different laminae of cervical (7Sp/8Sp), thoracic (7Sp/ICl), and lumbar (4Sp) segments. Neuronal processes expressing Cre.RFP are in red. Representative of N=3. (G) Quantification of cortical inputs to spinal cord (RFP+), divided by cortical region and laminae. The inset photomicrograph illustrates the L5b positioning of CSNs. Note that the Allen Brain Atlas classification did not subdivide L5 into L5a and L5b, and the position of corticospinal somata fell around the boundary between L5 and L6a. N=3. (H) Experimental strategy, same as Figure 1A. (I) The cortical regions giving rise to corticospinal somata (GFP+). Dark green bars represent the major regions; light green bars represent subdivisions of cortical regions. N=3. (J) Major brain regions containing GFP+ neurites. This includes dendritic processes in sensorimotor cortex. Grouping the many brain regions comprising these major structures reveals the intense innervation of several subcortical structures, including brainstem. (K) Photomicrograph of brainstem labeling by CSNs. N=3. (L) Experimental strategy to label synapses arising from CSNs. (M) Synaptophysin GFP (green) labeling in the brain. N=3. (N) Synaptophysin GFP (green) and FlpO (red) labeling in motor cortex. N=3. (O) Synaptophysin GFP (green) labeling in DLS. N=3. (P) Top brain regions to which CSNs project, excluding sensorimotor cortex and fiber tracts. N=2. Error bars are SEM.
Extended Data Fig. 2. Mapping the brainwide targets of CSNsDLS.

(A) Experimental strategy to label corticospinal neurons that project to striatum (CSNsDLS). (B) Photomicrographs exemplifying the cortical distribution CSNsDLS. Representative of N=3. (C) Sagittal Z projection of raw fluorescence aligned to atlas space. Representative of N=3. (D) 3D reconstruction of CSNsDLS cell bodies. Representative of N=3. (E) Quantification of cortical regions contributing to the total population of CSNsDLS, compared to experiments from Figure 1 targeting primarily the motor cortical population of CSNs (M-CSNs). Asterisks indicate statistically significant differences in innervation between M-CSNs and CSNsDLS (*: p<0.05, **: p<0.005, 2-way ANOVA with post-hoc t-test). (F) Quantification of brain regions targeted by CSNsDLS, compared to data from Figure 1. Note that – despite the differences in experimental strategy – DLS is a primary target of CSNsDLS. N=3. (G) 3D reconstruction of CSNsDLS projections throughout the brain, colored by targeted brain region. (H) Photomicrograph showing CSNsDLS axon labeling in the brainstem. Representative of N=3. (I) Higher magnification inset from (H). Representative of N=3. (J) Experimental strategy to drive expression of GFP in corticospinal neurons that form synapses on identified spinal cell types. (K) 3D reconstruction of axons from CSNsChx10, color coded by brain region. (L) Confocal micrograph exemplifying CSNChx10 axon labeling in DLS. (M) Quantification of brain structures that receive substantial input from CSN subtypes. N=4, Chx10, N=2, SST, N=3, GAD2. Error bars are SEM.
Extended Data Fig. 3. Controls for transsynaptic tracing experiments.
(A) Experimental strategy to confirm Cre-dependent expression of AAVs-FLEX encoding rabies glycoprotein and TVA.mCherry in DLS, as well as dependence of EnVA-N2cΔG-tdTomato infection on expression of TVA. The AAVs were injected into wild type mice, followed by injection of rabies. (B-D) Photomicrographs illustrating the absence of any mCherry or tdTomato labeling in the brain. Representative of N=3. (E) Experimental strategy to confirm Cre-dependent expression of AAVs-FLEX encoding rabies glycoprotein and TVA.mCherry in the spinal cord, as well as dependence of EnVA-N2cΔG-FlpO.mCherry infection on expression of TVA. (F-G) Photomicrographs illustrating the absence of any mCherry or tdTomato labeling in the spinal cord or brain. Representative of N=3. (H) Experimental strategy to confirm dependency of transsynaptic spread on rabies glycoprotein, in the DLS. AAV-FLEX-TVA.mCherry was injected into DLS of D1-Cre mice. AAV-FLEX-N2cG was omitted from the injection. (I-K) Injecting EnVA-N2cΔG-tdTomato led to local tdTomato expression, but no expression in presynaptic inputs to DLS. Representative of N=1. (L) Experimental strategy to confirm dependency of transsynaptic spread on rabies glycoprotein, in the spinal cord. AAV-FLEX-TVA.mCherry was injected into spinal cord of GAD2-Cre mice. (M-O) Injecting EnVA-N2cΔG-FlpO.mCherry led to local mCherry expression, but no expression in presynaptic inputs to spinal cord. Representative of N=3.
Extended Data Fig. 4. Synaptic organization of intratelencephalic corticostriatal projections.
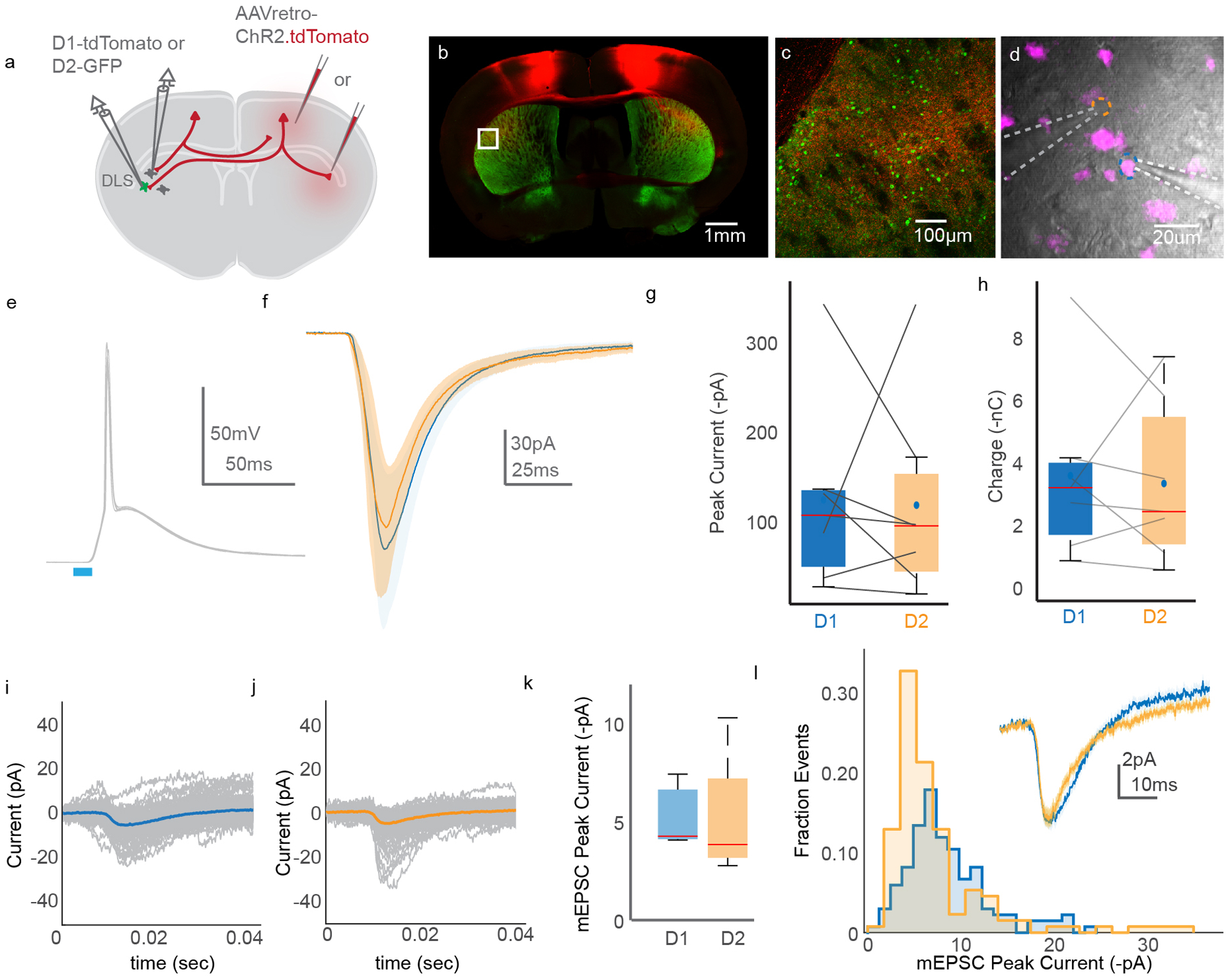
(A) Schematic illustrating the experimental strategy. Retrogradely-transported and expressed AAV encoding ChR2.tdTomato was injected into contralateral DLS or M1. D1 and D2 SPNs were targeted for simultaneous recording. (B) Photomicrograph of ChR2.tdTomato (red) and D2-GFP (green) labeling in a brain slice. (C) High magnification image of the boxed region from (B). Note the expansive axonal plexus. (D) DIC image of a D1+ (magenta) and D1− SPNs targeted for simultaneous whole cell recording. The dashed lines indicate the location of recording electrodes. (E) Superimposed current-clamp voltage recordings from an SPN following optogenetic stimulation of IT corticostriatal axons, highlighting the potency of this projection. (F) Grand average response of all D1 (blue) and D2 (orange) SPNs to optogenetic stimulation of IT corticostriatal neurons. (G-H) Pairwise comparison of ChR2-evoked amplitude (G) and charge (H) in D1 versus D2 SPNs. N=3 animals, n=7 cells. Paired t-test. (I-J) Trial average of mEPSCs evoked from an example D1 (I) and D2 (J) SPN. Individual trials are in grey. (K) Average mEPSC amplitudes in D1 versus D2 SPNs. N=2 animals, n=7 cells. (L) Distribution of all mEPSCs ordered by mEPSC peak current, recorded in D1 (blue) or D2 (orange) SPNs. The inset is an overlay of the average mEPSC from D1 and D2 SPNs. Error bars are SEM. Shaded areas are SEM. The horizontal dashes in the box plots in (G), (H), and (K) represent the median. The dots in (G) and (H) indicate the means. The bottom and top edges of all boxes indicate the 25th and 75th percentiles, respectively, while the whiskers indicate the minima and maxima, excluding outliers.
Extended Data Fig. 5. Distribution of spinal synapses arising from CSNsDLS.
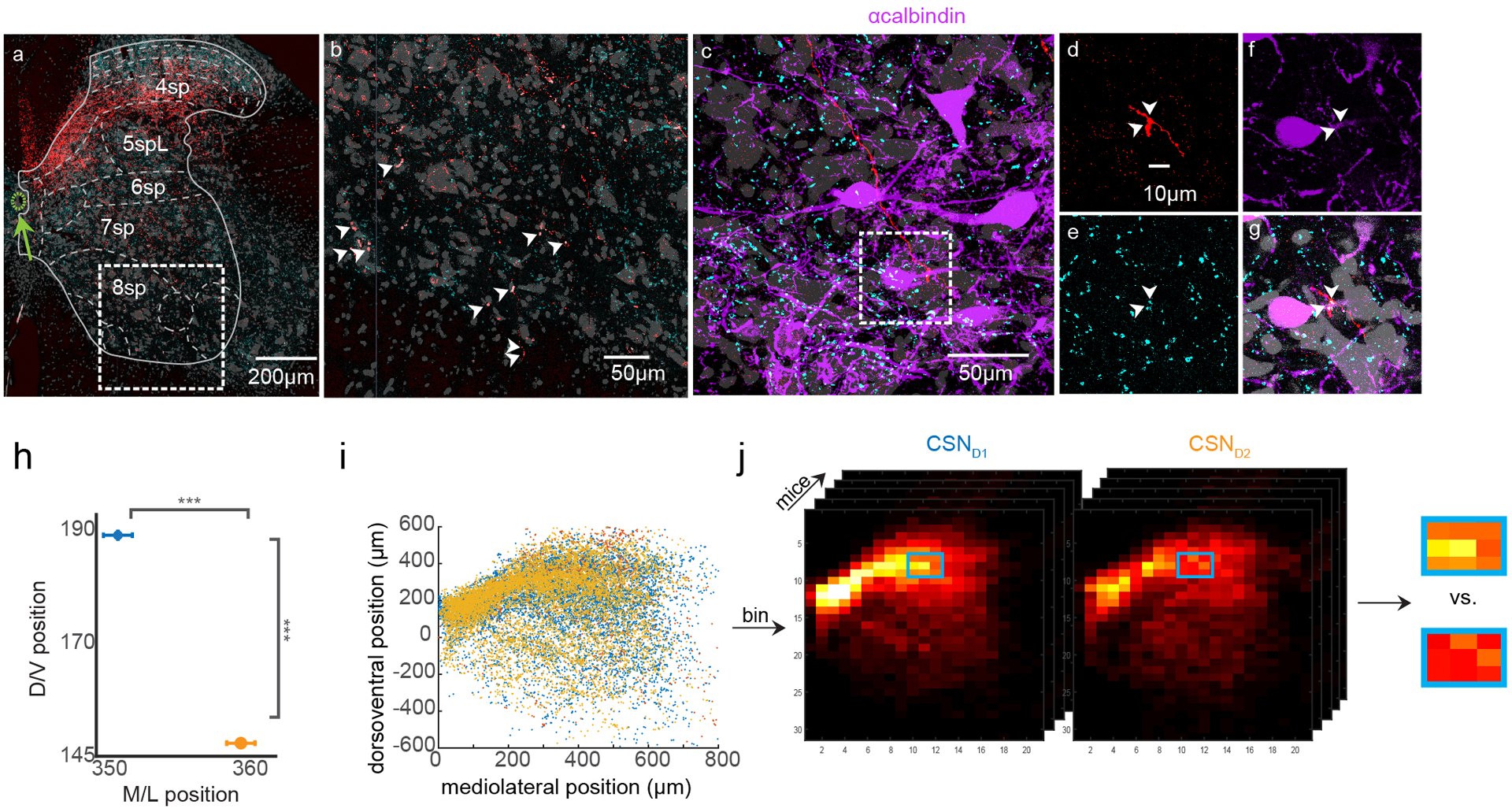
(A) The same confocal micrograph from Figure 3B. The ventral horn inset is shown in (B), with vGlut1+ varicosities indicated with arrowheads. (C) Confocal micrograph of immunolabeled calbindin-expressing interneurons located below motor pools in cervical spinal cord (putative Renshaw neurons). (D–G) High magnification 63x single optical section images showing two vGlut1+ synapses from a rabies-labeled CSN in close apposition to the proximal dendrite belonging to a calbindin+ interneuron. (H) Quantification of the mean mediolateral and dorsoventral position of CSNsD1 (blue) and CSNsD2 (orange). N=5 each, unpaired two-sided t-test, p=1.08×10−293. (I) Raw CSN spinal synapse data from three example mice. The position of each dot corresponds to a vGlut1+ axonal varicosity. (J) Raw data is spatially binned for each mouse. A sliding window is used to group local bins, and the density of labeling within these groups is compared across genotypes of mice.
Extended Data Fig. 6. Analysis of behavior and deconvolution.
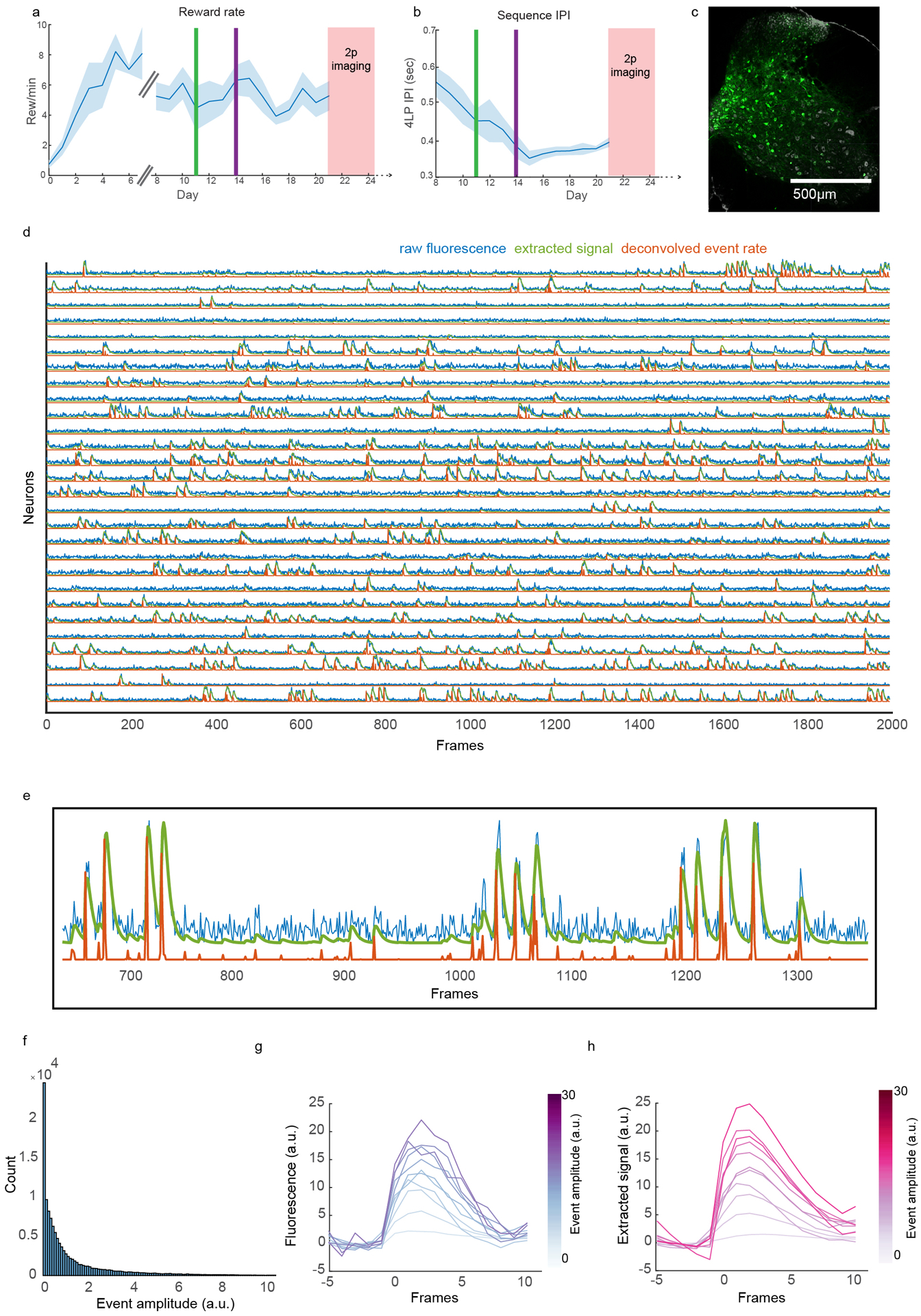
(A) Rate of reward across training. The broken X axis indicates where training changed from Phase 1 (one press issues reward) to Phase 2 (four press sequences issue reward). The green vertical line indicates the day at which the maximum rewarded inter-press interval (IPI) for four lever press sequences is limited to 3 seconds. Before this line (days 8–10), reward was issued every four presses, regardless of IPI. The purple vertical line indicates the day at which the maximum IPI for four lever press sequences to lead to reward is limited to 2 seconds. (B) The inter-press interval of lever press sequences across Phase 2 of training. (C) Confocal micrograph of the spinal cord injection site. (D) Signals extracted from CSNs from one mouse for a portion of a session. Raw fluorescence signal in blue. Calcium signal derived using CNMF is in green. Deconvolved event rate is in orange. Units are a.u. (E) A higher magnification view of signals extracted from one neuron. (F) Histogram depicting the number of deconvolved events as a function of their amplitude. (G) Average of raw fluorescence signal triggered by deconvolved events, aligned to event time. The shade of purple corresponds to the size of the associated deconvolved event. (H) Average of CNMF-derived calcium signal triggered by deconvolved events, aligned to event time. The shade of red corresponds to the size of the associated deconvolved event. Shaded area is SEM.
Extended Data Fig. 7. Imaging the activity of CSNs during behavior.

(A) Trial-averaged calcium activity aligned to lever press for neurons from a single mouse. (B) Same data as (A), but for deconvolved events. (C) Z scored deconvolved events of neurons at baseline versus at lever press. (D) Z scored activity of neurons aligned to single lever press events. (E) Histogram of the times of peak activity relative to lever press, for all neurons. (F) Average activity traces for neurons with peak activity that falls within different bins of time relative to lever press. (G-I) Illustration of time warping procedure for four press sequences. Dots indicate lever press times, as well as time anchors used for pre- and post-trial alignment (six time points per trial). (H) Illustration of how time is either dilated or contracted to match a template sequence. (J-K) Z scored calcium activity before (J) and after (K) time warping, to illustrate the utility of time warping for resolving latent features in unaligned activity. Note the emergence in (K) of peaks in activity corresponding to individual lever press events. This is the same dataset as Figure 6J, but is calcium signal, instead of deconvolved events. Shaded area is SEM.
Extended Data Fig. 8. Classification of CSN activity during lever press sequences.
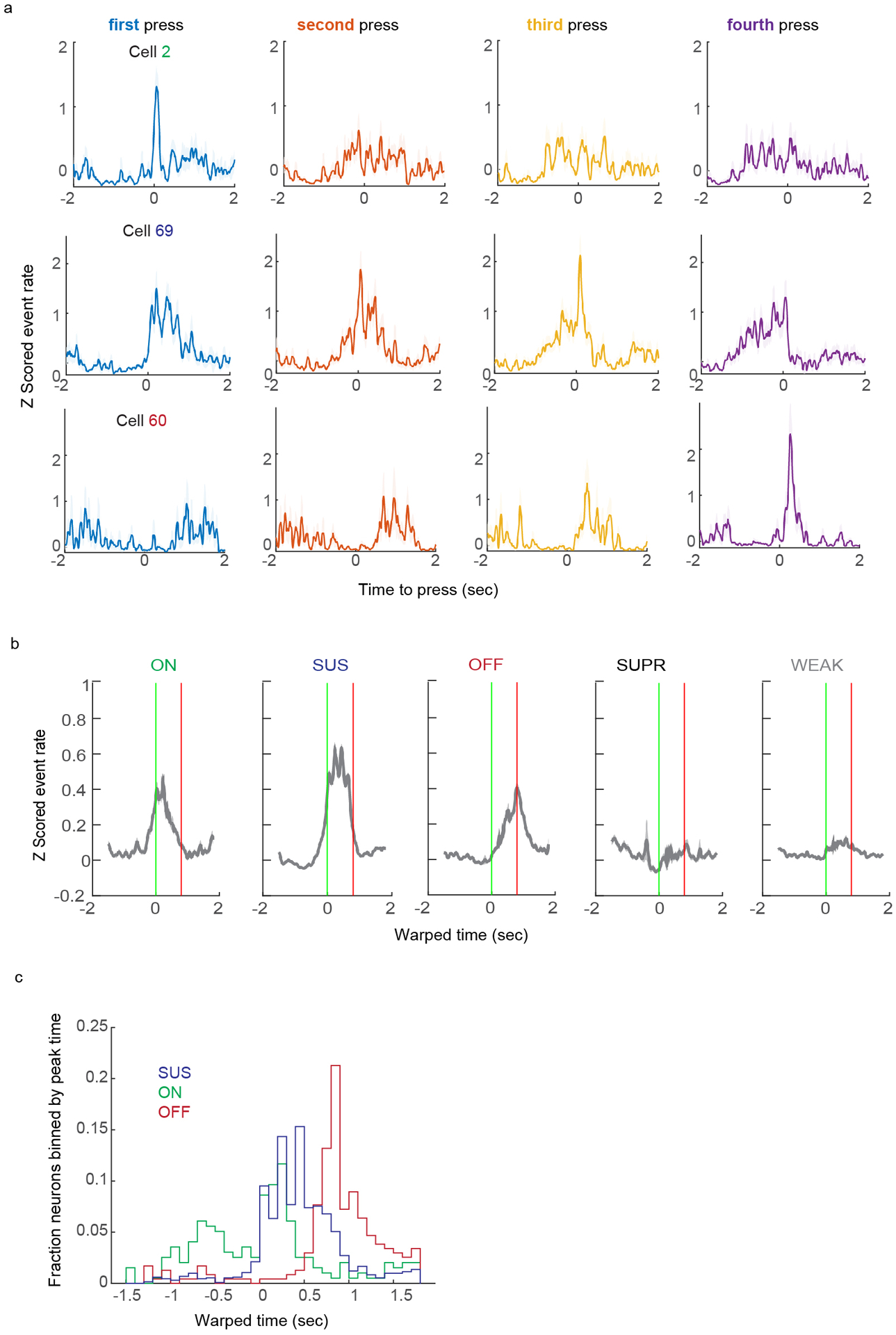
(A) The same neurons as (Figure 6L–N), instead displaying the average of events aligned to first, second, third, or forth press in the sequence. (B) The average activity of neurons belonging to each activity profile, aligned to four lever press sequence onset. (C) Histogram of the times of peak activity for CSNs with categorized activity profiles, aligned to lever press sequence onset. Shaded area is SEM.
Extended Data Fig. 9. Analysis of EMG during behavior and CSN activity correlations to EMG.
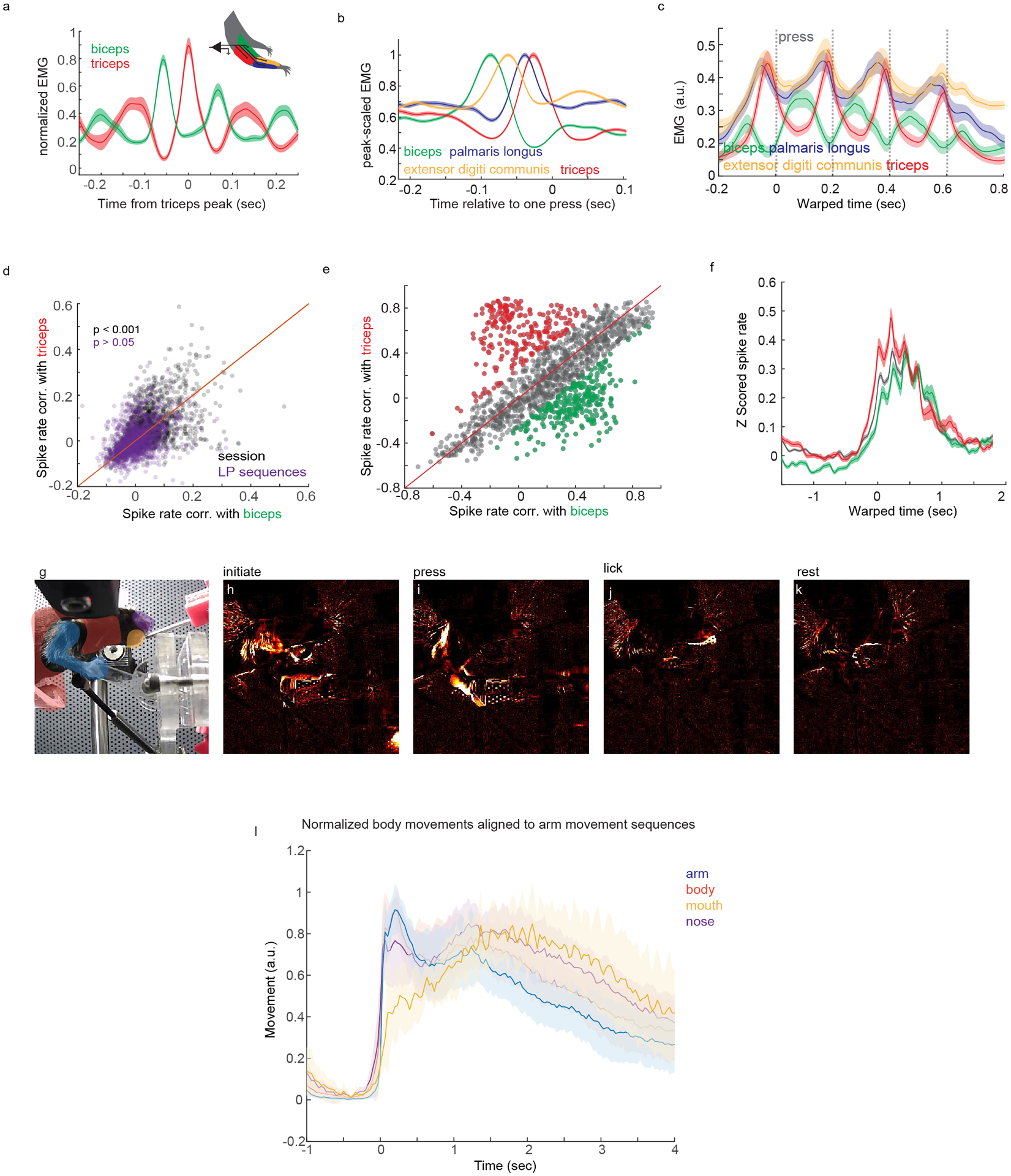
(A) Example recording of biceps and triceps muscle activity from one mouse. Biceps EMG is aligned to peaks in triceps EMG. (B) Average EMG activity for four forelimb muscles aligned to single lever presses. (C) Average EMG activity for four forelimb muscles aligned to lever press sequences. (D) Correlation of CSN activity to biceps versus triceps EMG during concatenated random segments of behavior and rest (grey) or concatenated lever press sequences (purple), matched in duration. Paired two-sided t-tests, concatenated random segments: p=6.66×10−7; concatenated lever press sequences: p=0.722, N=8, n=2252. (E) Correlation of trial-averaged CSN activity with biceps or triceps EMG. Neurons with correlations biased to triceps or biceps are colorized in red or green, respectively. (F) Average lever press sequence-related activity of CSNs highly correlated to triceps (red) or biceps (green) EMG. Activity from neurons with similar correlation coefficients is in grey. (G-L) Analysis of body movements during behavior. (G) A video still with highlighted regions of interest used for quantifying body movements. (H-K) Images of variance from the same vantage as (G) revealing different body parts moving during the behavior. (L) Variance from different body regions of interest aligned to the onset of arm movement sequences. Notice the absence of overt body movements before arm movement. Shaded area is SEM.
Extended Data Fig. 10. Method to identify CSNs with identified striatal synapses.

(A) Exemplar photomicrograph of CSNs expressing GCaMP (green), and corticostriatal neurons marked with tdTomato (red). Representative of N=8. (B) Cartoon depiction of fluorescent expression possibilities, viewed from an X-Z perspective. (C) Cartoon depiction of fluorescent expression possibilities, viewed from an X-Y perspective. (D) Two example possibilities for overlapping green and red fluorescence, one constituting a double-positive (top) and one rejected from being a double-positive (bottom).
Supplementary Material
Acknowledgements
We thank K. Fidelin and V. Athalye for feedback on this manuscript. We thank H. Rodrigues for designing and constructing behavioral equipment. We thank S. Brenner-Morton for custom antibodies, and S. Fageiry & K. Ritola for custom viral constructs. We would like to thank Zuckerman Institute’s Cellular Imaging platform for instrument use and technical advice. We are grateful for technical assistance from L. Hammond, G. Martins, M. Correia, C. Warriner, A. Miri, and K. MacArthur. We thank I. Marcelo for time warping code. Imaging was performed with support from the Zuckerman Institute’s Cellular Imaging platform. We thank T. Jessell for inspiring this research and for his critical feedback. R.M.C was funded by the National Institute of Health (5U19NS104649) and the Simons-Emory International Consortium on Motor Control. A.N. was a Howard Hughes Medical Institute Fellow of the Helen Hay Whitney Foundation and is currently supported by NIH Pathway to Independence Award 1K99NS118053-01.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
References
- 1.Shinoda Y, Arnold AP & Asanuma H Spinal branching of corticospinal axons in the cat. Exp Brain Res 26, 215–234 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Ueno M et al. Corticospinal Circuits from the Sensory and Motor Cortices Differentially Regulate Skilled Movements through Distinct Spinal Interneurons. Cell Rep 23, 1286–1300 e1287, doi: 10.1016/j.celrep.2018.03.137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter R & Lemon R Corticospinal function and voluntary movement. (Clarendon Press; Oxford University Press, 1993). [Google Scholar]
- 4.Lloyd DPC The spinal mechanism of the pyramidal system in cats. J Neurophysiol 4, 525–546 (1941). [Google Scholar]
- 5.Wang X et al. Deconstruction of Corticospinal Circuits for Goal-Directed Motor Skills. Cell 171, 440–455 e414, doi: 10.1016/j.cell.2017.08.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tennant KA et al. The organization of the forelimb representation of the C57BL/6 mouse motor cortex as defined by intracortical microstimulation and cytoarchitecture. Cerebral cortex 21, 865–876, doi: 10.1093/cercor/bhq159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamiyama T et al. Corticospinal tract development and spinal cord innervation differ between cervical and lumbar targets. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 1181–1191, doi: 10.1523/JNEUROSCI.2842-13.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivares-Moreno R et al. Mouse corticospinal system comprises different functional neuronal ensembles depending on their hodology. BMC Neurosci 20, 50, doi: 10.1186/s12868-019-0533-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemon RN Descending pathways in motor control. Annu Rev Neurosci 31, 195–218, doi: 10.1146/annurev.neuro.31.060407.125547 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Evarts EV Relation of pyramidal tract activity to force exerted during voluntary movement. J Neurophysiol 31, 14–27, doi: 10.1152/jn.1968.31.1.14 (1968). [DOI] [PubMed] [Google Scholar]
- 11.Sherrington CS Flexion-reflex of the limb, crossed extension-reflex, and reflex stepping and standing. J Physiol 40, 28–121, doi: 10.1113/jphysiol.1910.sp001362 (1910). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overduin SA, d’Avella A, Carmena JM & Bizzi E Microstimulation activates a handful of muscle synergies. Neuron 76, 1071–1077, doi: 10.1016/j.neuron.2012.10.018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheney PD & Fetz EE Functional classes of primate corticomotoneuronal cells and their relation to active force. J Neurophysiol 44, 773–791, doi: 10.1152/jn.1980.44.4.773 (1980). [DOI] [PubMed] [Google Scholar]
- 14.Fetz EE, Cheney PD, Mewes K & Palmer S Control of forelimb muscle activity by populations of corticomotoneuronal and rubromotoneuronal cells. Progress in brain research 80, 437–449; discussion 427–430, doi: 10.1016/s0079-6123(08)62241-4 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Peters AJ, Lee J, Hedrick NG, O’Neil K & Komiyama T Reorganization of corticospinal output during motor learning. Nature neuroscience 20, 1133–1141, doi: 10.1038/nn.4596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraskov A et al. Corticospinal mirror neurons. Philos Trans R Soc Lond B Biol Sci 369, 20130174, doi: 10.1098/rstb.2013.0174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canedo A Primary motor cortex influences on the descending and ascending systems. Prog Neurobiol 51, 287–335, doi: 10.1016/s0301-0082(96)00058-5 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Lemon RN & Griffiths J Comparing the function of the corticospinal system in different species: organizational differences for motor specialization? Muscle Nerve 32, 261–279, doi: 10.1002/mus.20333 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kita T & Kita H The subthalamic nucleus is one of multiple innervation sites for long-range corticofugal axons: a single-axon tracing study in the rat. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 5990–5999, doi: 10.1523/JNEUROSCI.5717-11.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooks BM et al. Topographic precision in sensory and motor corticostriatal projections varies across cell type and cortical area. Nat Commun 9, 3549, doi: 10.1038/s41467-018-05780-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramón y Cajal S Histologie du système nerveux de l’homme & des vertébrés. Ed. française rev. & mise à jour par l’auteur, tr. de l’espagnol par Azoulay L. edn, Vol. v. 1 (Maloine, 1909). [Google Scholar]
- 22.Graybiel AM Neurotransmitters and neuromodulators in the basal ganglia. Trends in neurosciences 13, 244–254, doi: 10.1016/0166-2236(90)90104-i (1990). [DOI] [PubMed] [Google Scholar]
- 23.Gerfen CR et al. D1 and D2 Dopamine Receptor Regulated Gene-Expression of Striatonigral and Striatopallidal Neurons. Science 250, 1429–1432, doi:DOI 10.1126/science.2147780 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Gertler TS, Chan CS & Surmeier DJ Dichotomous anatomical properties of adult striatal medium spiny neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 10814–10824, doi: 10.1523/JNEUROSCI.2660-08.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyachi S, Hikosaka O, Miyashita K, Karadi Z & Rand MK Differential roles of monkey striatum in learning of sequential hand movement. Exp Brain Res 115, 1–5, doi: 10.1007/pl00005669 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Pisa M Motor functions of the striatum in the rat: critical role of the lateral region in tongue and forelimb reaching. Neuroscience 24, 453–463, doi: 10.1016/0306-4522(88)90341-7 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Cui G et al. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature 494, 238–242, doi: 10.1038/nature11846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X, Tecuapetla F & Costa RM Basal ganglia subcircuits distinctively encode the parsing and concatenation of action sequences. Nature neuroscience 17, 423–430, doi: 10.1038/nn.3632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin X & Costa RM Start/stop signals emerge in nigrostriatal circuits during sequence learning. Nature 466, 457–462, doi: 10.1038/nature09263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tervo DG et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382, doi: 10.1016/j.neuron.2016.09.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botta P et al. An Amygdala Circuit Mediates Experience-Dependent Momentary Arrests during Exploration. Cell, doi: 10.1016/j.cell.2020.09.023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esposito MS, Capelli P & Arber S Brainstem nucleus MdV mediates skilled forelimb motor tasks. Nature 508, 351–356, doi: 10.1038/nature13023 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Arber S & Costa RM Connecting neuronal circuits for movement. Science 360, 1403–1404, doi: 10.1126/science.aat5994 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Bikoff JB et al. Spinal Inhibitory Interneuron Diversity Delineates Variant Motor Microcircuits. Cell 165, 207–219, doi: 10.1016/j.cell.2016.01.027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fink AJP et al. Presynaptic inhibition of spinal sensory feedback ensures smooth movement. Nature 509, 43–+, doi: 10.1038/nature13276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azim E, Jiang J, Alstermark B & Jessell TM Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature 508, 357–+, doi: 10.1038/nature13021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan B et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell 159, 1417–1432, doi: 10.1016/j.cell.2014.11.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reardon TR et al. Rabies Virus CVS-N2c(Delta G) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron 89, 711–724, doi: 10.1016/j.neuron.2016.01.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kha HT et al. Projections from the substantia nigra pars reticulata to the motor thalamus of the rat: single axon reconstructions and immunohistochemical study. The Journal of comparative neurology 440, 20–30, doi: 10.1002/cne.1367 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Kress GJ et al. Convergent cortical innervation of striatal projection neurons. Nature neuroscience 16, 665–667, doi: 10.1038/nn.3397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuypers HG An anatomical analysis of cortico-bulbar connexions to the pons and lower brain stem in the cat. J Anat 92, 198–218 (1958). [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey DR & Corrie WS Properties of pyramidal tract neuron system within a functionally defined subregion of primate motor cortex. J Neurophysiol 41, 216–243, doi: 10.1152/jn.1978.41.1.216 (1978). [DOI] [PubMed] [Google Scholar]
- 43.Johansson Y & Silberberg G The Functional Organization of Cortical and Thalamic Inputs onto Five Types of Striatal Neurons Is Determined by Source and Target Cell Identities. Cell Rep 30, 1178–1194 e1173, doi: 10.1016/j.celrep.2019.12.095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu-Friedman MA & Regehr WG Probing fundamental aspects of synaptic transmission with strontium. The Journal of neuroscience : the official journal of the Society for Neuroscience 20, 4414–4422 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franks KM et al. Recurrent circuitry dynamically shapes the activation of piriform cortex. Neuron 72, 49–56, doi: 10.1016/j.neuron.2011.08.020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Acunzo P et al. A conditional transgenic reporter of presynaptic terminals reveals novel features of the mouse corticospinal tract. Front Neuroanat 7, 50, doi: 10.3389/fnana.2013.00050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carr PA, Alvarez FJ, Leman EA & Fyffe RE Calbindin D28k expression in immunohistochemically identified Renshaw cells. Neuroreport 9, 2657–2661, doi: 10.1097/00001756-199808030-00043 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Lopez-Bendito G et al. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cerebral cortex 14, 1122–1133, doi: 10.1093/cercor/bhh072 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Beaulieu-Laroche L, Toloza EHS, Brown NJ & Harnett MT Widespread and Highly Correlated Somato-dendritic Activity in Cortical Layer 5 Neurons. Neuron 103, 235–241 e234, doi: 10.1016/j.neuron.2019.05.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittmann W et al. Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nature neuroscience 14, 1089–1093, doi: 10.1038/nn.2879 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Pnevmatikakis EA et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299, doi: 10.1016/j.neuron.2015.11.037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nectow AR & Nestler EJ Viral tools for neuroscience. Nat Rev Neurosci 21, 669–681, doi: 10.1038/s41583-020-00382-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauswein E, Fromm C & Preuss A Corticostriatal cells in comparison with pyramidal tract neurons: contrasting properties in the behaving monkey. Brain Res 493, 198–203, doi: 10.1016/0006-8993(89)91018-4 (1989). [DOI] [PubMed] [Google Scholar]
- 54.Morita K, Im S & Kawaguchi Y Differential Striatal Axonal Arborizations of the Intratelencephalic and Pyramidal-Tract Neurons: Analysis of the Data in the MouseLight Database. Front Neural Circuits 13, 71, doi: 10.3389/fncir.2019.00071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kincaid AE, Zheng T & Wilson CJ Connectivity and convergence of single corticostriatal axons. The Journal of neuroscience : the official journal of the Society for Neuroscience 18, 4722–4731 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albin RL, Young AB & Penney JB The functional anatomy of basal ganglia disorders. Trends in neurosciences 12, 366–375 (1989). [DOI] [PubMed] [Google Scholar]
- 57.DeLong MR Primate models of movement disorders of basal ganglia origin. Trends in neurosciences 13, 281–285 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Surmeier DJ, Ding J, Day M, Wang Z & Shen W D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends in neurosciences 30, 228–235, doi: 10.1016/j.tins.2007.03.008 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Akay T, Tourtellotte WG, Arber S & Jessell TM Degradation of mouse locomotor pattern in the absence of proprioceptive sensory feedback. Proceedings of the National Academy of Sciences of the United States of America 111, 16877–16882, doi: 10.1073/pnas.1419045111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crapse TB & Sommer MA Corollary discharge across the animal kingdom. Nat Rev Neurosci 9, 587–600, doi: 10.1038/nrn2457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Botta P et al. An Amygdala Circuit Mediates Experience-Dependent Momentary Arrests during Exploration. Cell, doi: 10.1016/j.cell.2020.09.023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thevenaz P, Ruttimann UE & Unser M A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7, 27–41, doi: 10.1109/83.650848 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Sommer C, Straehle C, Köthe U & Hamprecht FA in 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 230–233.
- 64.Klein S, Staring M, Murphy K, Viergever MA & Pluim JPW elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Transactions on Medical Imaging 29, 196–205, doi: 10.1109/TMI.2009.2035616 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Ragan T et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature methods 9, 255–258, doi: 10.1038/nmeth.1854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akay T, Acharya HJ, Fouad K & Pearson KG Behavioral and electromyographic characterization of mice lacking EphA4 receptors. J Neurophysiol 96, 642–651, doi: 10.1152/jn.00174.2006 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Guo ZV et al. Procedures for behavioral experiments in head-fixed mice. PLoS One 9, e88678, doi: 10.1371/journal.pone.0088678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson A, Mooney R. The Basal Forebrain and Motor Cortex Provide Convergent yet Distinct Movement-Related Inputs to the Auditory Cortex. Neuron. 2016. May 4;90(3):635–48. doi: 10.1016/j.neuron.2016.03.031. Epub 2016 Apr 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953 e920, doi: 10.1016/j.cell.2020.04.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


