Extended Data Figure 6 |. The PXL region is necessary for ALK signaling.
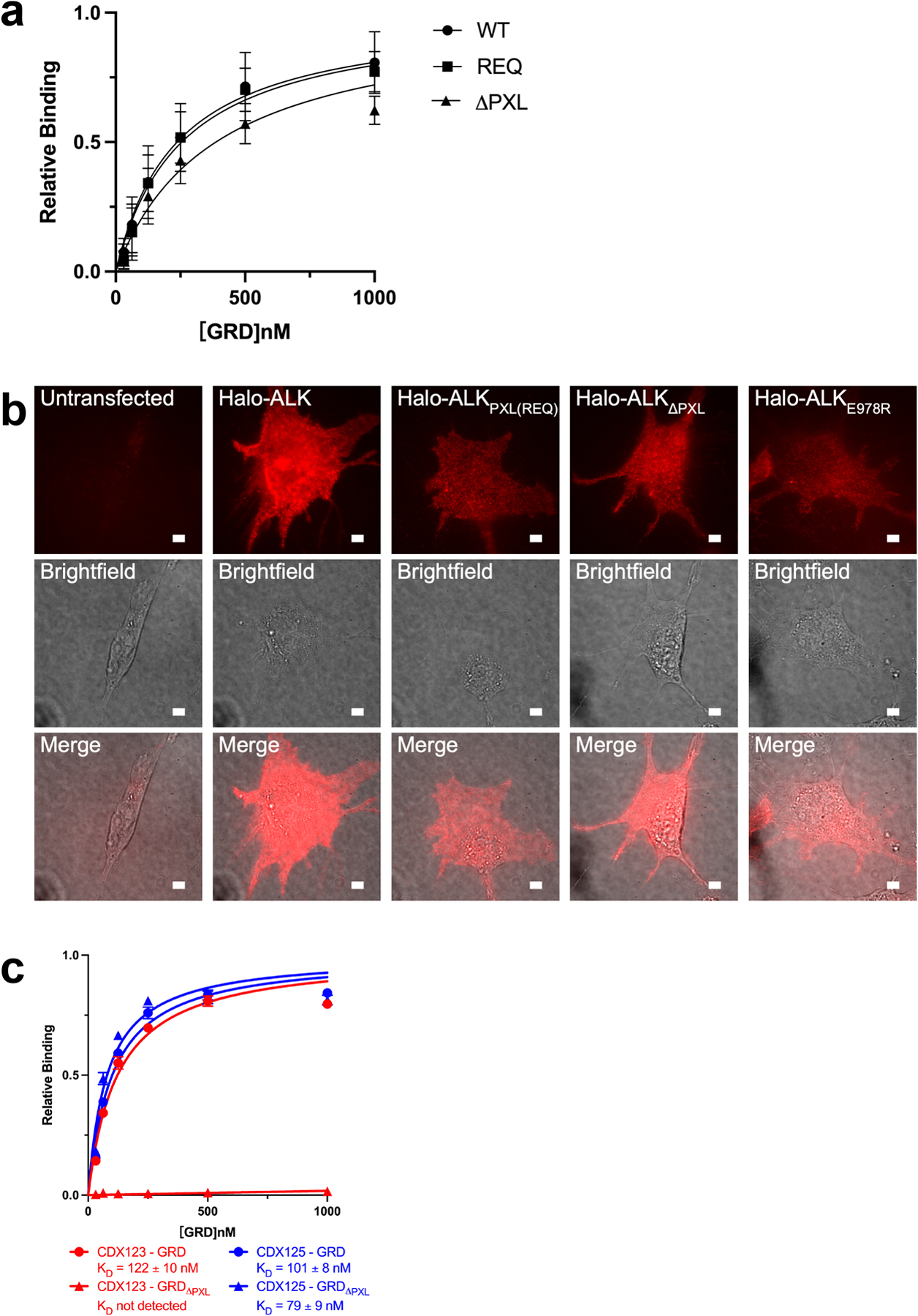
a, Binding of ALKAL2-AD to GRD PXL mutations detected by BLI. The sensors were loaded with ALKAL2-AD. REQ is a mutant that alters the interface residues 795-“IGE”−797 (shown in Fig. 3b, c) to REQ. ΔPXL is a mutant that removes the entire “disulfided helix” 783–797 (shown in Fig. 3b, c). The relative binding is the binding normalized to the maximum responses. Data represent mean ± SD of four measurements. b, Expression of Halo-ALK on NIH/3T3 cell membranes. NIH/3T3 cells were untransfected, transfected with wildtype Halo-ALK or ALK mutants. Cells were stained with membrane impermeable dye, JF549i, before imaging. For each construct fluorescence (top row), brightfield (middle row), and a merge (bottom row) were shown. Scale bar 5 μm. The experiment was repeated two times with similar results. c, Binding of CDX123 or CDX125 to GRD or GRDΔPXL detected by BLI. Data represent mean ± SD from three independent experiments.
