Abstract
Mechanisms underlying aspirin chemoprevention of colorectal cancer remain unclear. Prior studies have been limited due to the inability of preclinical models to recapitulate human normal colon epithelium or cellular heterogeneity present in mucosal biopsies. To overcome some of these obstacles we performed in vitro aspirin treatment of colon organoids derived from normal mucosal biopsies to reveal transcriptional networks relevant to aspirin chemoprevention. Colon organoids derived from 38 healthy individuals undergoing endoscopy were treated with 50μM aspirin or vehicle control for 72 hours and subjected to bulk RNA-sequencing. Paired regression analysis using DESeq2 identified differentially expressed genes (DEGs) associated with aspirin treatment. Cellular composition was determined using CIBERSORTx. Aspirin treatment was associated with 1,154 significant (q<0.10) DEGs prior to deconvolution. We provide replication of these findings in an independent population-based RNA-sequencing dataset of mucosal biopsies (BarcUVa-Seq), where a significant enrichment for overlap of DEGs was observed (P<2.2E−16). Single-cell deconvolution revealed changes in cell composition, including a decrease in transit-amplifying cells following aspirin treatment (P=0.01). Following deconvolution, DEGs included novel putative targets for aspirin such as TRABD2A (q=0.055), a negative regulator of Wnt signaling. Weighted gene co-expression network analysis identified 12 significant modules, including two that contained hubs for EGFR and PTGES2, the latter being previously implicated in aspirin chemoprevention. In summary, aspirin treatment of patient-derived colon organoids using physiologically relevant doses resulted in transcriptome-wide changes that reveal altered cell composition and improved understanding of transcriptional pathways, providing novel insight into its chemopreventive properties.
Keywords: aspirin chemoprevention, organoids, colorectal cancer, gene expression profiling, weighted gene co-expression network analysis
INTRODUCTION
While global rates of colorectal cancer (CRC) have seen a steady decrease, in part due to wider acceptance and implementation of endoscopic screening (1), CRC remains the third leading cause of cancer death. A multitude of studies have investigated aspirin use as a primary chemoprevention strategy to complement screening and further reduce incidence of CRC (2). In 2016, the United States Preventive Services Task Force (USPSTF) recommended aspirin use for the primary prevention of CRC among individuals between 50 and 69 years of age with a >10% ten-year risk of cardiovascular disease. Despite this landmark decision, the USPSTF cautioned that additional studies were required to clarify aspirin chemopreventive mechanisms before broadening these recommendations and suggested implementing precision prevention approaches to identify those most likely to benefit.
Determining aspirin’s mode of action has been complicated by a number of factors. Population-based cohort studies have examined how aspirin influences gene expression using transcriptome-wide RNA-sequencing (RNA-seq) in CRC tissue (3) or from bulk epithelial biopsies (4). While powerful, these studies are limited by factors such as cellular heterogeneity, and collection of variable and non-standardized aspirin data prior to tissue acquisition (3). Further, mechanisms derived from cancer tissues or immortalized cancer cell lines may not reflect modes of primary prevention relevant to normal tissue. Finally, mechanisms identified in preclinical mouse models have been limited in their translation to humans (5).
Despite these challenges, a framework of interrelated mechanisms centered on prostaglandin modulation has been proposed that requires further validation (2). This framework incorporates the combined effects of irreversible inhibition of Prostaglandin-Endoperoxide Synthase 1 (PTGS-1)-mediated platelet activation (6), PTGS-2 inhibition in epithelium (7), and aspirin’s anti-inflammatory properties. A number of PTGS-independent mechanisms, such as those involving the Epidermal Growth Factor (EGF) pathway (8) have also been proposed to contribute to the potent preventive properties of aspirin.
To overcome previous challenges in chemoprevention modeling, we aimed to interrogate aspirin-induced transcriptome-wide changes on healthy epithelial cells in vitro using human colon organoids derived from normal mucosal biopsy specimens. Previously, our group has used organoids to determine the transcriptomic effect of ethanol exposure, an established CRC risk factor, in PDO that demonstrated the capacity for this modeling system to recapitulate exogenous exposures inducing pro-tumorigenic signaling in otherwise normal cells (9,10).
Through the use of a large prospective biorepository of organoids, we identified a number of significant differentially expressed genes (DEG)s following aspirin treatment in vitro. We find that a subset of the genes identified have been previously associated with known cell markers (11), indicating that aspirin treatment affects cellular composition in a model of the colonic crypt. Using single-cell deconvolution approaches, we confirm this finding and extend our results to the identification of genes after accounting for cellular composition. We have previously employed this method to address the potential confounding effects of cellular composition in a largely overlapping cohort of colon organoids treated with ethanol (10). Here, we also take advantage of the unique size of this normal organoid dataset to perform weighted gene co-expression network analysis (WGCNA), and identify modules of co-expression that are altered following aspirin treatment. To the best of our knowledge, this study represents the first use of WGCNA in normal organoids/colon epithelial cells, shedding additional light upon the coordinated mechanisms by which aspirin exerts its chemopreventive effects.
MATERIALS AND METHODS
Patient Population
The majority of subjects (n=34) undergoing standard of care colonoscopy were recruited to a biopsy study at the University of Virginia (UVA). For the purpose of UVA recruitment, organoids were only generated from healthy individuals, determined as subjects presenting with 3 or fewer polyps and no personal or immediate family history of CRC. Four organoid lines were also included from patients that were recruited to a clinical trial (“ASPirin Intervention for the REDuction of colorectal cancer risk [ASPIRED]; NCT02394769) at Massachusetts General Hospital (MGH) (12). A total of 38 independent organoid lines were included in this study. Selected lines were balanced for sex (female = 20, male = 18), colon location (right = 18, left = 20), a broad age (22–74; median = 59) and Body Mass Index (19.7 – 50.4; median = 27.6). Participants in both studies provided written informed consent prior to participating in the research study and all research procedures were approved by the Institutional Review Boards (IRB) of UVA (IRB-HSR # 19439 and IRB-HSR#15274) and MGH through the Dana-Farber/Harvard Cancer Center IRB (DF/HCC IRB Protocol #14–496).
Biopsy Collection and Organoid Culture Derivation
Normal human colon organoids were established using a minimal modification of the method described by Sato et al (13). At UVA, biopsies (4 each from right and left colon) were taken at the time of colonoscopy using standard forceps and obtained immediately distal to the hepatic flexure (right colon) or immediately distal to the splenic flexure (left colon). At MGH, the ASPIRED trial protocol has been previously published (12). Biopsies were taken from grossly normal appearing distal colorectum mucosa (>15cm from the anal verge). While ASPIRED included an aspirin intervention, all organoids included in this study were derived from baseline, pre-treatment visits to remain consistent with UVA samples. Biopsies were placed immediately into DMEM/F12 media with 10% fetal bovine serum (FBS) and 100U/ml penicillin, 100μg/ml streptomycin on ice and transported to the laboratory where whole crypts were immediately isolated using methods adapted from Miyoshi and Stappenbeck (14) and Sato et al. (13) (Supplemental Methods).
Aspirin Treatment Experiments, Nucleic Acid Extraction, and RNA-seq
A total of 38 independent organoid lines were used in the study. Organoid lines were grown in 48-well plates, and seeded at a density of approximately 105 cells per well and allowed to grow for approximately 3 days prior to treatment with aspirin. Cells within the treatment group were treated with 0.5μL aspirin (100mM Sigma A5376) per 1mL complex media (Supplemental Methods), equating to a 50μM dose of aspirin. Vehicle control samples were treated with an equal amount of 1M Tris (pH=7.2) with a final concentration of 50μM in complex media. In vitro dose selection was extrapolated for standard (325–400 mg) dose aspirin based on pharmacodynamic data supporting that the physiologically-relevant concentration [Cmax] for low-dose (81–100 mg) aspirin in humans is approximately 10μM (15) (see Supplemental Methods; Supplemental Figure 1). RNA was extracted 72 hours after the initial treatment (see Supplemental Methods). RNA was checked for quality using the Agilent 4200 Tapestation. Only samples with a RIN score > 8.5 were used for library preparation and sequencing using the Illumina NovaSeq 6000 (Northwest Genomics Center at the University of Washington, Seattle).
Bioinformatic Analysis
A detailed description of bioinformatic methods used in this study is provided in Supplemental Methods. Briefly, 100bp paired-end reads were trimmed and aligned to the GENCODE reference transcriptome using STAR and reads were quantified using HT-Seq (16,17). An average of 49.82 million reads per sample uniquely mapped to the reference GENCODE transcriptome. Gene counts were generated for each sample and collated into a singular matrix for paired regression analysis in DESeq2 (18). Significant DEGs were defined as genes surviving Benjamani-Hochberg False Discovery Rate (FDR) correction (q=0.10) using DESeq2 default parameters. Pathway analysis was performed in ToppFun (19).
For the purpose of single-cell deconvolution, single-cell RNA-sequencing (scRNA-seq) data was downloaded from a large, publicly available dataset of colon tissue (11) in which only cells derived from healthy donors were considered. We used Seurat V3 (20) to filter cells and pre-process cells before transcripts per million were uploaded to CIBERSORTx (21) for deconvolution. Four main cell populations were defined: transit-amplifying (TA), immature colonocytes, immature goblet cells and stem cells. Cell scores generated using this approach were centered to the mean and scaled prior to incorporation into regression models. Single-cell deconvolution was also applied to the BarcUVa-Seq dataset. Previously reported cell identities were considered for the current analysis (11), consistent with our prior study (22) except that here, endothelial cell subsets were grouped and that innate lymphoid cells were removed. This adjustment accounted for context of normal colon tissue used here rather than tumor tissue previously used for deconvolution (Supplemental Methods).
Given that small but consistent fold changes were identified at the single gene level, WGCNA (23) was performed across all PDO following correction for cell composition and sample pairing (24). Resulting modules were correlated with aspirin treatment.
Independent Validation Using BarcUVa-Seq Data
Details of pre-processing and sample acquisition have previously been reported (25). Regular aspirin users were defined as those who responded “yes” to whether they had taken more than 30 doses, low (81–100mg) or standard (325+mg/day) of aspirin over their lifetime (Supplemental File 1). Samples were filtered to exclude individuals with missing phenotypic information for adjustment covariates. Only samples derived from white individuals were included to minimize the effects of potential population stratification in the analysis. The final cohort consisted of RNA-seq data from 387 individuals, of which 31 were considered regular aspirin users. A regression model for aspirin use (regular vs no) was performed in DESeq2 (18). Sequencing batch, colon location (left/right/transverse), smoking status (never/former/current) and gender were included as adjustment covariates.
Annotation of DEGs with CRC-related genes
CRC-related loci identified through genome-wide association studies (GWAS) were downloaded from the GWAS catalog (26). Genes were annotated to a SNP if they were found to have at least one nucleotide of one exon overlapping a 1Mb window centered on the SNP. SNP locations were generated according to their hg38 location. BiomaRt was used to determine respective GrCH38 gene coordinates (27).
Data Availability
Raw fastq files and pre-processed count matrix data have been uploaded to Gene Expression Omnibus(28), accession number: GSE163282.
RESULTS
Widespread Transcriptomic Changes in Normal Colon PDO following Aspirin Exposure.
RNA-seq was performed on a large prospective organoid cohort (n=38 patients; Supplemental Table 1) subsequently treated in vitro with aspirin or vehicle control. One sample pair was considered to be an outlier and was removed (Supplemental Figure 2A). Recruitment center had no obvious effect on clustering (Supplemental Figure 2B), therefore, MGH and UVA organoids were analyzed together. Principal component analysis (PCA) was performed across all genes (Supplemental Figure 3). Overall, the nearest neighbor of each control organoid was its aspirin-treated pair. This not only demonstrated unique, intrinsic organoid properties, but measurable effect specifically attributable to aspirin treatment. No obvious clustering for age, body mass index (BMI) or sex were observed.
A paired regression analysis was performed in DESeq2, which identified 1,154 FDR corrected DEGs (Figure 1A). We confirmed the direction of effect for the majority of genes considered (7/9) using a subset of organoids (n=4) (Supplemental Methods; Supplemental Table 2). Importantly, pathway analysis of DEGs whose expression increased following aspirin treatment identified relevant enrichments for genes previously associated with aspirin (ctd:D001241; q=1.63E−14), as well as a number of other non-steroidal anti-inflammatory drugs (Figure 1B).
Figure 1: Differential expression analysis of aspirin treated colon organoids.
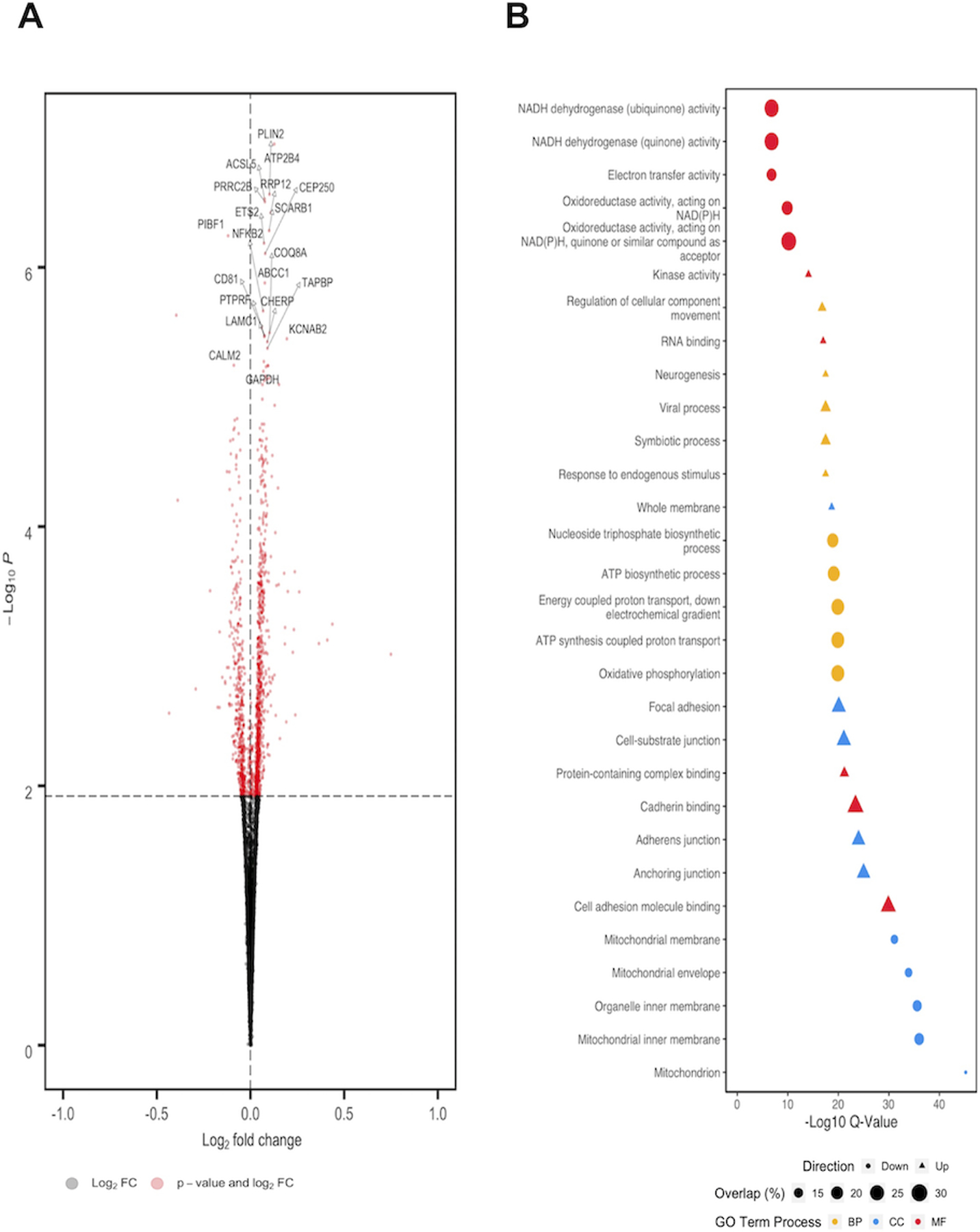
A) Volcano plot of DEGs. DEGs with FDR corrected q-values that met significance (red dots) appear above the FDR threshold (horizontal hashed line). Positive log2fold changes (right of vertical hashed line) indicate higher expression in aspirin treated samples compared to vehicle controls, whereas negative log2fold changes (left of vertical hashed line) indicate lower expression compared to vehicle controls. Connectors were placed to highlight the most significant 20 DEGs associated with aspirin exposure. B) Plot of five most significantly enriched Gene Ontology terms for each category: Biological Process (BP); Cellular Component (CC) and Molecular Function (MF). Direction corresponds to whether nominally over-expressed (triangle) or under-expressed (circle) DEGs were used for pathway enrichment analysis. Size of each pathway corresponds to the overlap (%) of DEGs identified in aspirin response versus the total number of genes in the pathway annotation.
Examination of Aspirin-associated DEGs in Large Colon Biopsy Cohort
We extended our in vitro findings to an in vivo human setting by performing a similar analysis in an independent population-based RNA-seq dataset (BarcUVa-Seq) of normal colon biopsies taken at colonoscopy with available questionnaire-derived data for aspirin use (25). One-way Fisher’s exact test confirmed an enrichment for overlap between organoid and BarcUVa-Seq DEGs (Odds Ratio = 3.09; P<2.2E−16) (Supplemental Table 3). These results indicate that in vitro aspirin treatment of PDO results in broadly similar transcriptomic profiles observed in mucosal biopsies from individuals reporting aspirin use, providing independent replication of our findings.
Single-cell Deconvolution of Bulk RNA-seq Data.
We identified patterns of expression in nominal (P<0.05) and significant DEGs following FDR correction (q<0.10) indicative of a reduction in TA cell content and an increase in more differentiated cell populations in organoid (Figure 2A). Similar results were also observed when cross referencing DEGs identified in the BarcUVA-Seq cohort with potential markers of colon epithelial cell types, where a general reduction in expression of TA cell markers and an increase in expression of more differentiated cell markers was also observed in aspirin users (Figure 2B) (11). We therefore hypothesized that aspirin may drive cellular transcriptomic programs of epithelial cells in organoid towards specific cell states.
Figure 2: Single-cell deconvolution of bulk RNA-seq from aspirin exposed colon organoids.
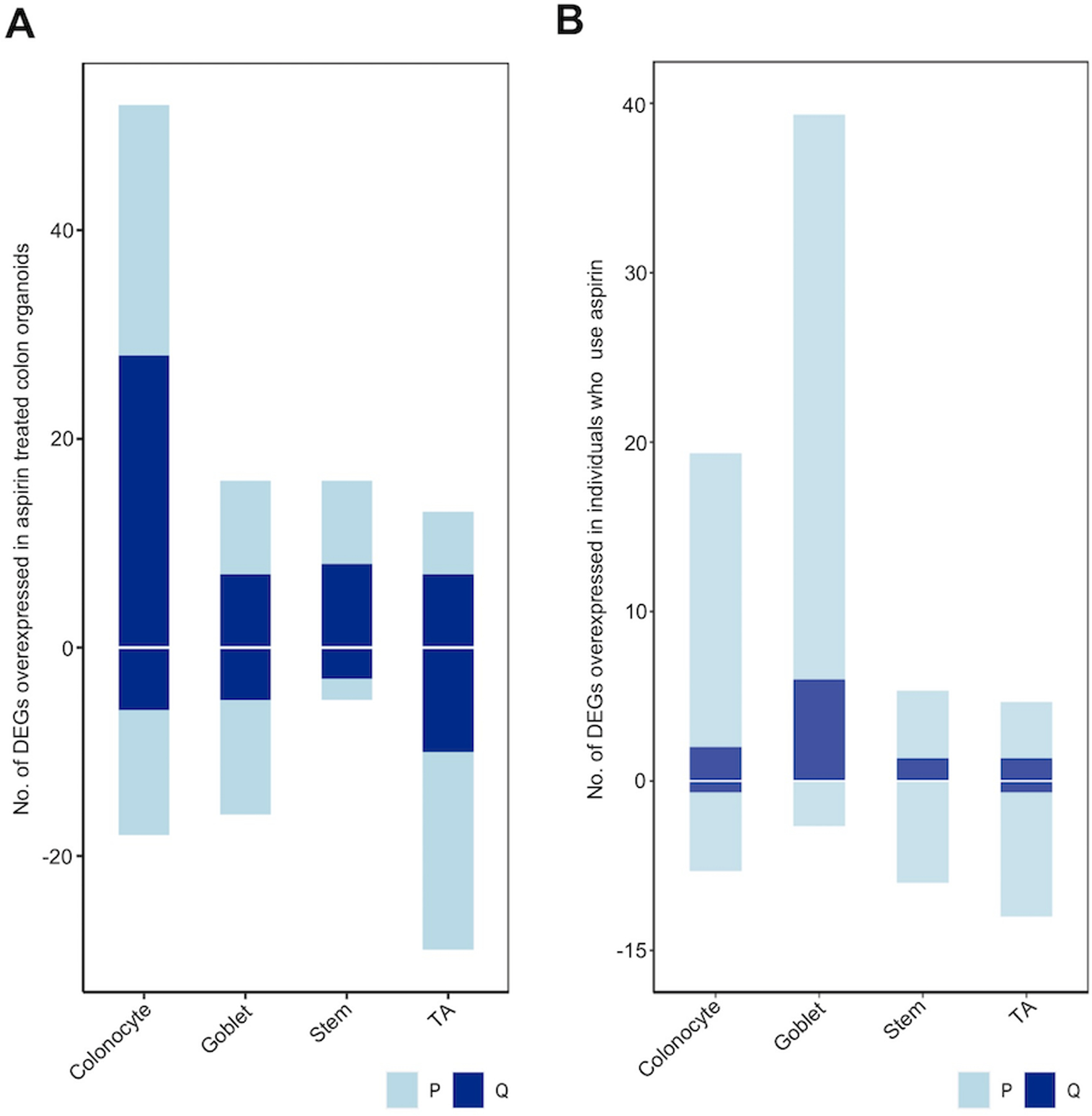
Nominally significant (P=0.05; light blue) and FDR corrected (q=0.10; dark blue) DEGs were overlaid with known cell markers of colon cell types as identified through scRNA-seq. Positive values indicate the number of genes displaying increased expression, while negative values indicate number of genes with reduced expression in A) aspirin treated organoids or B) regular users of aspirin.
To address this hypothesis, we performed in silico single-cell deconvolution to determine the abundance of four major cell populations (TA cells, stem cells, immature goblets and immature colonocytes) (11,21). We validated our cell abundance scores by confirming that they accurately captured expression differences of known cell markers (Supplemental Methods; Supplemental Figure 4A–B). Linear mixed effect regressions of cell scores validated our hypothesis, identifying a significant decrease in TA cells (P=0.01; 25 of 37 organoid) in aspirin treated organoid compared to controls. We also identified a significant increase in stem cells (P=0.03; 24 of 37 organoid) and a trend for increase in immature goblets (P=0.07; 20 of 37 organoid) in aspirin treated organoid. We similarly measure an increase in goblet cells using BarcUVa-Seq (P=0.042) and a trend towards reduction of TA cell populations in aspirin users (P=0.12). Taken together, these data imply that aspirin treatment results in transcriptomic programming favoring a less overall proliferative state highlighted by a reduction in the TA cells.
Controlling for Cellular Composition in Analysis of Colon Organoids
Given the change to cell type markers, we next aimed to define cell type agnostic DEGs in organoids, those universally impacted by aspirin treatment, by incorporating cell scores as covariates into our paired regression analysis. After adjusting for cell composition, we identified 334 additional DEGs (P<0.05) that were only apparent following this correction (Figure 3A, red dots, 334 of 1,257 nominally significant DEGs). The remaining 73.4% (923) were consistent with DEGs identified in the primary analysis prior to adjustment for cell composition (Figure 3A, blue dots). Importantly, several candidate genes within pathways postulated to be central to aspirin chemopreventive effects emerged following adjustment for cell composition. For example, decreased Prostaglandin E Synthase (PTGES2; P=0.02), as well as increased expression ATP-binding cassette sub-family C member 4 (ABCC4; P=0.03) (29). An enrichment for overlap with BarcUVa-Seq biopsy data was still present in DEGs following cell type adjustment, though reduced (Odds Ratio = 1.44, P=4.31E−05). Following FDR correction, 46 DEGs persisted (q<0.10), suggesting this subset of DEGs are the most likely to be associated with aspirin treatment across a range of cell types (Supplemental Table 4).
Figure 3: Differential expression analysis of aspirin-treated colon organoids following adjustment for cell composition.
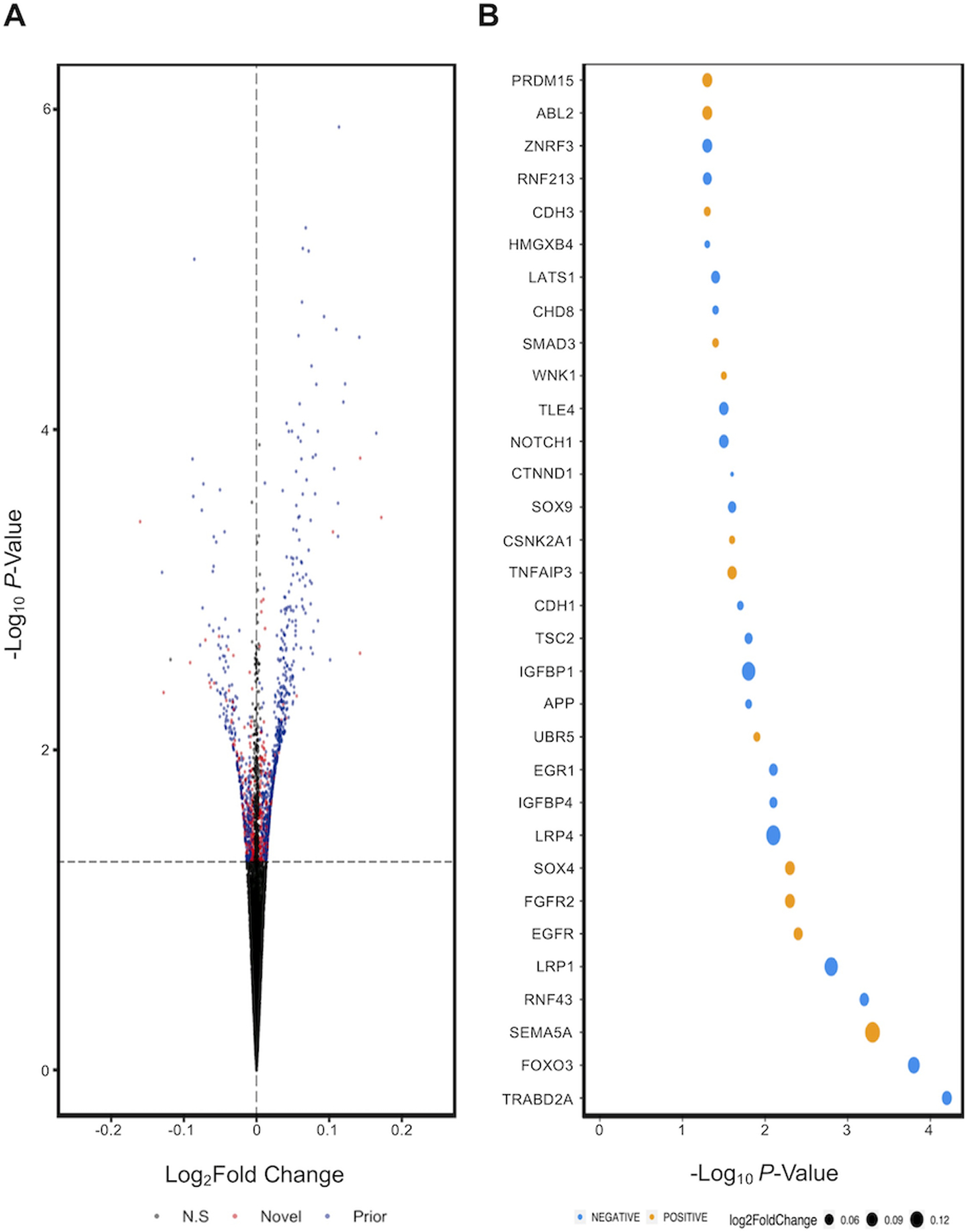
A) Volcano plot of significant DEGs that were also identified in original analysis (blue) and novel (red). B) Inspection of fold change of nominal DEGs that are positively (orange) or negatively (blue) correlated with Wnt signaling, ranked from left to right in order of significance.
Pathway analysis of genes upregulated in aspirin treated organoids following adjustment for cell composition revealed enrichment of several chemoprevention related pathways, including those associated with aspirin (ctd:D001241; q=1.46E−10), positive regulation of cell differentiation (q=1.34E−08), 8-iso-prostaglandin E2 (CID000000158; q=4.33E−06) and Wnt signaling (q=2.31E−05). To better determine the effect of aspirin on Wnt, genes were mapped to GO pathways with increased granularity (GO:0030178, GO:0090090, GO:0090263, GO:0030177), such as positive or negative regulation of Wnt (Figure 3B). TraB Domain Containing 2A (TRABD2A; q=0.055) and Forkhead Box O3 (FOXO3; q=0.065) both were upregulated in aspirin treated PDOs and both are negative regulators of Wnt. To the best of our knowledge, TRABD2A has not previously been implicated in the mechanism of action of aspirin.
We stratified our dataset according to colon biopsy location before performing independent regression analyses. Initially, more significant DEGs (q<0.10) were observed in proximal (206 DEGs) than distal organoids (13 DEGs). Following adjustment for cell composition, 35 DEGs and 8 DEGs remained, respectively, with no overlap (Supplemental Table 5). Pathway analysis revealed that 114 of the 326 GO-terms enriched in DEGs overexpressed in distal organoid following aspirin treatment were identical to those enriched in proximal colon. These findings suggest that while some pathways may be commonly impacted by aspirin treatment, colon location may be relevant to consider with regard to aspirin effects. Future studies specifically powered to examine location, while accounting for cellular composition will be needed to confirm these findings.
Combined, these data underscore the potential importance of cell context for future aspirin mechanism studies. Relevant DEGs may be lost in the noise of cellular heterogeneity, and single-cell studies may help explain why novel targets have yet to emerge from, and strong putative mechanisms have yet to be validated by, prior RNA-seq studies of bulk tissue samples.
Aspirin associated DEGs and CRC related Genes
To better understand the potential relationship of identified DEGs with CRC risk and chemoprevention, we cross referenced nominally significant DEGs in aspirin treated organoids with genes previously associated with inherited CRC risk loci identified through GWAS (Supplemental Table 6) (26). We identified 90 DEGs that are within the vicinity of CRC-related GWAS loci (see Methods), ten of which were also present in BarcUVa-Seq, with the same direction of effect. Of the 90, six survived FDR correction, including increased expression of Laminin Subunit Gamma-1 (LAMC1) (q=0.095), CEA Cell Adhesion Molecule 7 (CEACAM7) (q=0.059) and Acyl-COA Synthetase Long Chain Family Member 5 (ACSL5) (q=0.02).
WGCNA of aspirin treatment in PDOs
WGCNA was performed to identify networks of genes impacted by aspirin treatment (see Methods, Supplemental Figures 5 and 6, Supplemental File 2). WGCNA identifies correlations between gene expression without bias from prior knowledge of function. Strongly correlated genes are then grouped into modules and these modules are subsequently tested for their association to a trait (aspirin response). We identified 32 modules of co-expression in our organoid dataset. Module gene lists were cross-referenced with genes in the vicinity of CRC GWAS loci (Supplemental File 3). Of these, 13 were nominally associated with treatment condition (P<0.05), of which 12 survived FDR correction (q<0.05). Hub genes were determined for each module (Figure 4). The tan and skyblue2 modules emerged as potentially the most relevant for aspirin chemoprevention based upon the overall significance of a module to the treatment conditions, and on a priori knowledge of aspirin mechanism.
Figure 4: Bar plot to show significance of modules defined through WGCNA with relation to aspirin treatment status.
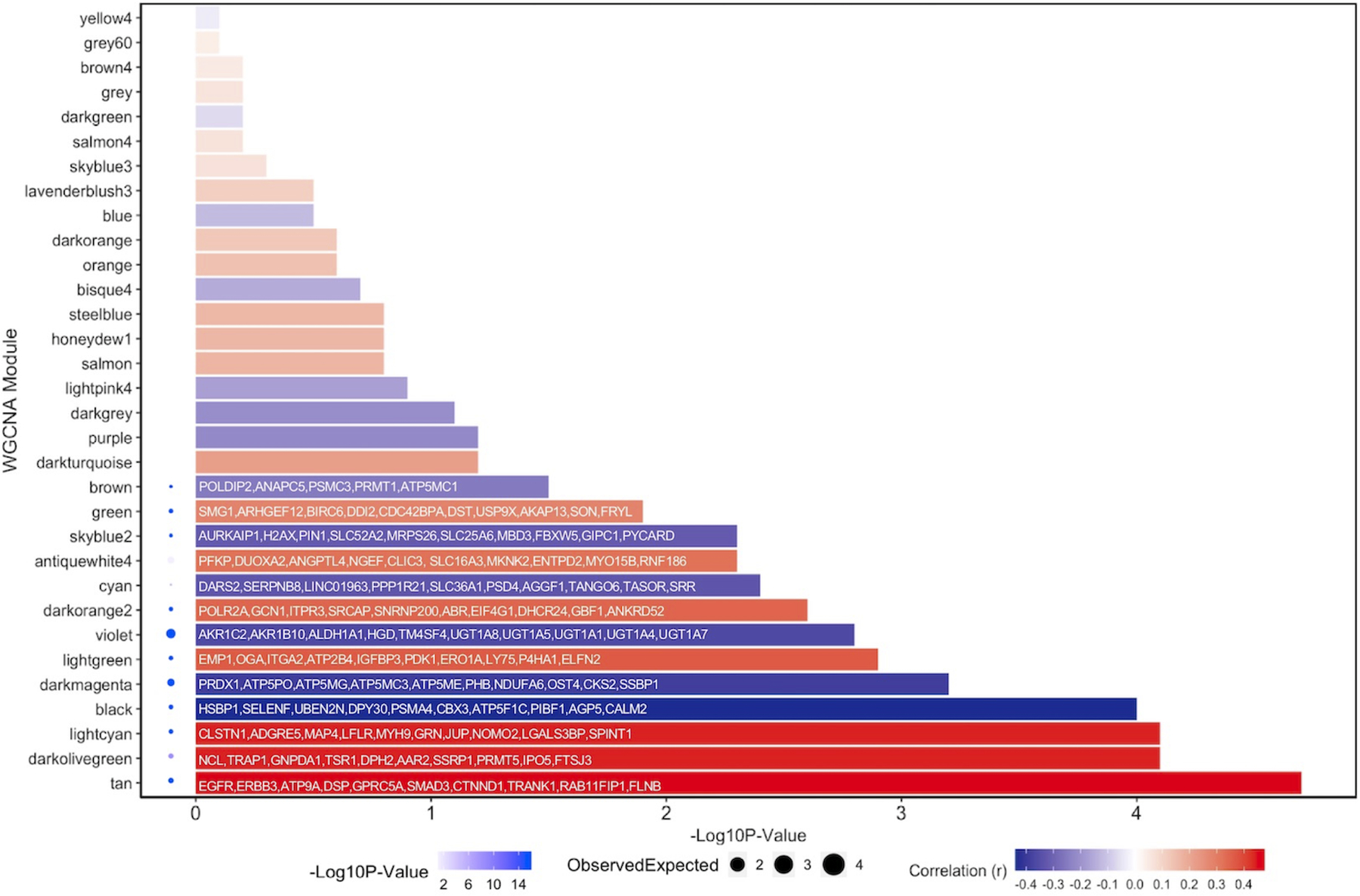
Extent of positive correlation to aspirin treatment can be visualized as deepening red hue. For each significant module, the genes with the highest module memberships were annotated. PPI enrichment analysis was performed for each significant module by uploading genes found in each module to STRING. Significance of PPI enrichment can be observed by a deepening blue hue next to the relevant module. Size of circle refers to the number generated when dividing the number of observed edges in the PPI network by the number of expected edges given the size of the network analyzed.
The tan module contained 13 genes previously found to be associated with aspirin response, including EGFR (30) (Supplemental Figure 7). Surprisingly, we find that EGFR expression positively correlates with aspirin treatment and is a central hub to the tan module, despite data showing evidence that at high doses, aspirin leads to EGFR inhibition in colon epithelial cells (31). Interestingly, the sixth most representative hub gene within this module was SMAD Family Member 3 (SMAD3) (26,32). This module was also enriched for pathways associated with cell adhesion (q=6.02E−07), epithelial cell development (q=4.87E−05) and positive regulation of cell differentiation (q=0.02), further supporting a potential role for aspirin in promoting differentiation.
Examining the skyblue2 module we find a node containing PTGES2, a gene responsible for PGE2 synthesis (Supplemental Figure 8). PTGES2 was nominally downregulated in our adjusted model at a single gene level (P=0.021) and displayed a high module membership (r=0.75, P=1.26E−14) with the skyblue2 module. This module also contained 13 of the top 20 genes that were identified through other gene-gene networks investigating the interactions of PTGES2 (33), indicating some reproducibility of this co-expression module in prior research. This module was enriched for prostaglandin E2 related processes (q=7.60E−03), as well as for processes such as apoptotic mitochondrial changes (q=9.96E−03) and positive regulation of apoptotic signaling pathway (q=0.05), highlighting the translational potential of the use of colon organoids.
We used an independent method to validate the co-expression of significant modules. Gene lists were imported into STRING (34) to construct protein-protein interaction (PPI) networks. For each significant module, a PPI enrichment P-value was calculated (Figure 4). Intuitively, if co-expression within modules exists, gene lists should be enriched for PPI. Indeed, all but one significant module (AntiqueWhite4) was enriched for PPI (P=0.087). Further, when considering STRING analysis of the tan module, EGFR appeared central to the generated PPI (Supplemental Figure 9), concordant with its relationship to the tan module. For comparison, this approach was then applied to the grey module, the default color assigned to a module comprised of genes with limited, if any co-expression. This module was not significantly enriched for PPI (P=0.698). These findings confirm that the modules identified in our network analysis are plausibly correlated. Further inspection of all 32 modules identified in our WGCNA analysis revealed that modules significantly associated with aspirin response appeared more likely to be enriched for aspirin-related pathways. In total, five of 12 (41.7%) significant modules (light cyan, light green, darkorange2, green and violet) as well as the nominally significant brown module were all enriched for aspirin-related processes. In contrast, only three of 19 (15.8%) non-significant modules had evidence for a relationship with aspirin: darkorange, blue and skyblue3. Taken together, these findings suggest that WGCNA analysis employed here captured plausible modules, and that differences occurring across significant modules are likely direct measured effects of aspirin treatment, which may have novel and significant functional consequences.
DISCUSSION
Numerous observational studies and randomized control trials have reported positive associations between the routine administration of aspirin and reduced CRC risk (7,12,35–38). However, while a number of known targets for aspirin response have been identified (2), transcriptome-wide analysis of population-based studies have typically identified few, if any, consistently modulated genes associated with aspirin use (3,4). Here, we identify DEGs associated with in vitro aspirin treatment of normal colon organoids including novel and previously proposed genes related to aspirin chemoprevention that particularly emerge following adjustment for cell composition. Aspirin also induces transcriptomic profiles that favor the reduction of rapidly cycling TA cells for increased goblet or stem cell states, and modulation of several gene networks that may be central to aspirin’s pleiotropic protective effects. These findings were validated in a novel analysis of an independent large observational cohort with RNA-seq of mucosal biopsies and aspirin use data.
Previous inconsistencies in transcriptomic signatures associated with aspirin use in normal colon epithelium may have been impacted by limitations of study design. In combination with factors such as small sample size, inconsistent dosing, and cellular heterogeneity, the subtle effects of aspirin on colon epithelial cells may be one reason why previous studies have been unable to identify gene expression responses in studies of aspirin in bulk epithelium (4), despite strong clinical and epidemiologic evidence for its chemopreventive capacity (2). Specifically, our results underscore the importance of cell context in understanding aspirin’s transcriptomic effects and disentangling putative chemopreventive mechanisms in normal epithelial cells derived from organoids or mucosal biopsies. For example, modulated expression of PTGES2 and ABCC4, two genes central to the hypothesized mechanism of action of aspirin (2), only emerged following single-cell deconvolution and adjustment for cell composition in PDOs.
Basic cell composition and individual-level factor differences also appear to persist following organoid derivation. Additional sources of variation, including individual (i.e., age, sex, BMI, etc.) or technical (i.e., colon biopsy location or clinical site) factors, may also contribute to cellular heterogeneity and/or noise in transcriptome-wide profiling studies. In the current study, we have attempted to account for possible interindividual differences using several approaches, especially age and BMI which have previously been hypothesized to significantly modify aspirin’s chemopreventive effects (2). First, every individual served as their own experimental control. We used a large, paired study design, which reduces the effects of subject-level confounding (39). By comparison, in vitro studies typically use a small (n=3–5) number of lines. Second, we explored and found no obvious effect for age, gender, BMI, or clinical site (UVA or MGH). However, we do note a potential differential response to aspirin in proximally-derived organoids compared to distal organoids that persisted, but was substantially attenuated following adjustment for cell composition. The identification of this finding is intriguing, and may allude to biological differences across the colon that affect organoids response to aspirin. Future studies should consider matching proximal and distal lines from the sane individual to account for potential genetic heterogeneity that may, in part, account for this finding Combined, these results highlight that despite uniform in vitro conditions, additional adjustment for cell composition is important for mechanistic interrogation using bulk RNA-seq of normal epithelial cells to clarify cell context, especially when biopsies are derived from different colon locations within the same study. Data generated by clinical trials with available scRNA-seq data paired with epithelial cell RNA-seq data may be able to specifically validate these differences. Further, the application of single-cell deconvolution approaches to other existing datasets may provide new insight and replication.
Our results also extend previous findings that aspirin reduces Wnt signaling in mice and small intestinal organoids (40), a mechanism previously associated with both PTGS-dependent and independent mechanisms of aspirin response (41), by using human colon organoids, more physiologically relevant doses of aspirin, and a shorter window from initial treatment. Further, Wnt signaling is crucial for early progenitor cell proliferation and dysregulation is a hallmark of CRC (42). Our network analysis identified an enrichment for DEGs in Wnt signaling pathways. Both highly significant increased DEGs (TRABD2A and FOXO3) are negative regulators of Wnt, supporting that aspirin promotes Wnt inhibition. This relationship also extended to nominal DEGs, where 62.5% of overexpressed DEGs were negative regulators of Wnt, including E-Cadherin (CDH1). High doses of aspirin have previously been shown to increase E-Cadherin expression in CRC cell lines (40). However, to the best of our knowledge, this is the first report describingTRABD2A as an aspirin target.
The identification of multiple novel aspirin associated DEGs that map to CRC GWAS loci presents an interesting avenue to explore interplay between genotype and environment. Environmental risk factors play a major role in contributing to CRC risk and protection. Further, GWAS associations between risk SNPs and reported genes are often limited by a lack of functional follow up. Here we identify an aspirin associated increased expression of CEACAM7, which is found to be decreased in rectal neoplasia compared to normal mucosa (43). Further, we also find that aspirin treatment increases ACSL5 and LAMC1 expression. Low expression of ACSL5 has previously been associated with CRC tumor recurrence (44). Lamc1 has previously led to epithelial hyperplasia in mice (45). In the context of CRC, LAMC1 has been previously shown to be elevated in tumor versus normal adjacent tissue (46). Increased cellular heterogeneity of tumors compared to early neoplasia may contribute to this relationship, as duodenal levels of Lamc1 were shown to be greater in mesenchymal than epithelial tissues (45), thus the epithelial to mesenchymal transition of cancer cells may in part account for increased LAMC1, further supporting the important context cell composition can have in interpreting preventive mechanisms.
Moreover, many genes that may have clinical relevance and key roles in anti-cancer effects in colon epithelial cells may be overlooked in single-gene approaches. To the best of our knowledge, this study represents the first WGCNA of normal organoids and our findings highlight several possible modules encompassing existing and novel networks through which aspirin may exert its anti-cancer effects that are in need of future validation and deeper mechanistic interrogation. The skyblue2 module emerged as one characterized by PTGS-dependent genes including PTGES2, which has been shown in preclinical models to be critical to PGE2-mediated tumorigenesis (47). In addition, we identify several other modules of coordinated gene expression changes associated with aspirin treatment, the most significant of which was the tan module. The central hub gene for the tan module was EGFR. Previous preclinical studies in immortalized or transformed cell lines have found that aspirin leads to reduced EGFR expression, but these studies typically required high doses of aspirin that often ranged beyond therapeutic dose (31), perhaps a consequence of the model selection. While our results suggest that EGFR may play a significant role in potentiating the cancer protective properties of aspirin, this mechanism requires further validation. While the tan module appeared primarily driven by EGFR, SMAD3 was the sixth most representative hub gene identified, indicating an important role for SMAD3 in this module. Further, 33 of the 365 genes identified within the tan module were related to CRC through GWAS loci, including SMAD3 and CEACAM7. WGCNA performance benefits from increased sample sizes, and confirmation of these modules in larger studies may provide novel insight into how well-characterized GWAS-related genes may play an important role in defining a coordinated gene expression response to aspirin.
Our study does have limitations. organoid models enrich for stem cells and progenitor cell populations. This model lacks both immune and stromal cell types, which may play important additional roles in aspirin chemoprevention. However, as intestinal stem cells and their progenitors are the most recognized tumor initiating cell population (48), these populations may provide the best model for chemoprevention studies. Further, we exposed organoids to a single dose of aspirin for 72 hours to observe the prolonged effects of single dose of aspirin that is consistent with pharmacodynamically expected and physiologically relevant concentrations in the colon. While we note this limitation in the modelling of aspirin, we are able to provide strong replication of our findings in BarcUVa-Seq for regular aspirin use. Similarly, an important strength of the current study is the validation of in vitro findings in this real-world data. However, the measurement recorded for regular aspirin use is not as rigorous as the doses employed in our in vitro work or other more rigorous human studies. Future studies would benefit from the collection of more precise measurements and/or large controlled trials of aspirin. Future studies should also consider repeat, daily dosing, at a larger range of doses, including lower doses representative of 81–100 mg dose, so as to better mimic the widespread use of low dose aspirin in the population for primary prevention. A range of doses may also unveil possible relationships with additional factors, such as height, weight, or body mass index, which have been determined to influence bioavailability (49,50). Nonetheless, while dose-dependent effects of aspirin have been previously documented, studies should consider limiting in vitro doses to those that remain clinically relevant (micromolar range).
In conclusion, we identify several genes and pathways differentially expressed in response to aspirin administered in vitro at a therapeutic dose in a large biorepository of patient-derived human colon organoids. Single-cell deconvolution revealed cellular composition changes, and adjustment for cellular composition identified novel targets related to chemoprevention. WGCNA further elucidated networks of genes, especially those previously hypothesized to be central to the mode of action of aspirin including prostaglandin inhibition. Overall, our results demonstrate that the use of organoids from normal mucosal specimens along with single-cell deconvolution methods may overcome limitations associated with prior cancer prevention studies and provide a suitable experimental model system in which to interrogate the chemopreventive mechanism of aspirin.
Supplementary Material
Prevention Relevance:
Numerous studies have highlighted a role for aspirin in colorectal cancer chemoprevention, though the mechanisms driving this association remain unclear. We addressed this by showing that aspirin treatment of normal colon organoids diminished the transit-amplifying cell population, inhibited prostaglandin synthesis, and dysregulated expression of novel genes implicated in colon tumorigenesis.
Acknowledgments
Grant Support
This work was supported by funding through National Institutes of Health (NIH) grants: NIH/NCI R01 CA201407 (UP and GC), NIH/NCI R01 CA143237 and NIH/NCI R01 CA204279 (GC), NIH/NIDDK K01 DK120742 and NIH/NCI L30 CA209764 (DAD), NIH/NCI R35 CA253185 and R01 CA137178 and the Stuart and Suzanne Steele MGH Research Scholar Award (ATC). NIH/NCI R01 CA211184 and NIH/NCI CA254314 (ÖHY). Study sponsors had no role in the study design, collection, analysis, or interpretation of data.
Abbreviations
- ABCC4
ATP-binding cassette sub-family C member 4
- ACSL5
Acyl-COA Synthetase Long Chain Family Member 5
- ASPIRED
ASPirin Intervention for the REDuction of colorectal cancer risk
- BMI
body mass index
- CDH1
E-Cadherin
- CEACAM7
CEA Cell Adhesion Molecule 7
- CRC
colorectal cancer
- DEG
differentially expressed gene
- EGF
Epidermal Growth Factor
- EGFR
Epidermal Growth Factor Receptor
- eQTL
expression quantitative trait loci
- FBS
fetal bovine serum
- FDR
false discovery rate
- FOXO3
Forkhead Box O3
- GWAS
Genome Wide Association Studies
- IRB
institutional review boards
- LAMC1
Laminin Subunit Gamma-1
- LGR5
Leucine-rich-repeat-containing G protein coupled receptor 5
- MGH
Massachusetts General Hospital
- PCA
principal component analysis
- PPI
protein-protein interaction
- PTGES1/COX-1
Prostaglandin-Endoperoxide Synthase 1
- PTGES2
Prostaglandin E Synthase 2
- qPCR
quantitative PCR
- RNA-seq
RNA-sequencing
- scRNA
single-cell RNA-sequencing
- SERPINB8
Serpin Family B Member 8
- SMAD3
SMAD Family Member 3
- TA
transit-amplifying
- TRABD2A
TraB Domain Containing 2A
- USPSTF
United States Preventive Services Task Force
- UVA
University of Virginia
- WGCNA
Weighted Gene Co-expression Network Analysis
Footnotes
Disclosures
The authors declare that there are no conflict of interests.
Transcript Profiling
Raw data for this manuscript has been uploaded to Gene Expression Omnibus and is available for download using accession number: GSE163282.
References
- 1.Zauber AG, Winawer SJ, O’Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med 2012;366(8):687–96 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drew DA, Chan AT. Aspirin in the Prevention of Colorectal Neoplasia. Annu Rev Med 2020. 10.1146/annurev-med-060319-120913. [DOI] [PMC free article] [PubMed]
- 3.Slattery ML, Pellatt DF, Mullany LE, Wolff RK. Differential Gene Expression in Colon Tissue Associated With Diet, Lifestyle, and Related Oxidative Stress. PLoS One 2015;10(7):e0134406 10.1371/journal.pone.0134406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas SS, Makar KW, Li L, Zheng Y, Yang P, Levy L, et al. Tissue-specific patterns of gene expression in the epithelium and stroma of normal colon in healthy individuals in an aspirin intervention trial. Genom Data 2015;6:154–8 10.1016/j.gdata.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y, Su ZY, Zhang CY, Gaspar JM, Wang R, Hart RP, et al. Mechanisms of colitis-accelerated colon carcinogenesis and its prevention with the combination of aspirin and curcumin: Transcriptomic analysis using RNA-seq. Biochem Pharmacol 2017;135:22–34 10.1016/j.bcp.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtenberger LM, Fang D, Bick RJ, Poindexter BJ, Phan T, Bergeron AL, et al. Unlocking Aspirin’s Chemopreventive Activity: Role of Irreversibly Inhibiting Platelet Cyclooxygenase-1. Cancer Prev Res (Phila) 2017;10(2):142–52 10.1158/1940-6207.CAPR-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. New Engl J Med 2007;356(21):2131–42 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 8.Bashir AIJ, Kankipati CS, Jones S, Newman RM, Safrany ST, Perry CJ, et al. A novel mechanism for the anticancer activity of aspirin and salicylates. Int J Oncol 2019;54(4):1256–70 10.3892/ijo.2019.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devall M, Jennelle LT, Bryant J, Bien S, Peters U, Powell S, et al. Modeling the effect of prolonged ethanol exposure on global gene expression and chromatin accessibility in normal 3D colon organoids. PLoS One 2020;15(1):e0227116 10.1371/journal.pone.0227116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devall M, Plummer SJ, Bryant J, Jennelle LT, Eaton S, Dampier CH, et al. Ethanol exposure drives colon location specific cell composition changes in a normal colon crypt 3D organoid model. Sci Rep 2021;11(1):432 10.1038/s41598-020-80240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019;178(3):714–30 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drew DA, Schuck MM, Magicheva-Gupta MV, Stewart KO, Gilpin KK, Miller P, et al. Effect of Low-dose and Standard-dose Aspirin on PGE2 Biosynthesis Among Individuals with Colorectal Adenomas: A Randomized Clinical Trial. Cancer Prev Res (Phila) 2020. 10.1158/1940-6207.CAPR-20-0216. [DOI] [PMC free article] [PubMed]
- 13.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141(5):1762–72 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 2013;8(12):2471–82 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patrignani P, Tacconelli S, Piazuelo E, Di Francesco L, Dovizio M, Sostres C, et al. Reappraisal of the clinical pharmacology of low-dose aspirin by comparing novel direct and traditional indirect biomarkers of drug action. J Thromb Haemost 2014;12(8):1320–30 10.1111/jth.12637. [DOI] [PubMed] [Google Scholar]
- 16.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12) doi ARTN 550 10.1186/s13059–014-0550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37(Web Server issue):W305–11 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177(7):1888–902 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37(7):773–82 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devall MAM, Casey G. Controlling for cellular heterogeneity using single-cell deconvolution of gene expression reveals novel markers of colorectal tumors exhibiting microsatellite instability. Oncotarget 2021;12(8):767–82 10.18632/oncotarget.27935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diez-Obrero V, Dampier CH, Moratalla-Navarro F, Devall M, Plummer SJ, Diez-Villanueva A, et al. Genetic Effects on Transcriptome Profiles in Colon Epithelium Provide Functional Insights for Genetic Risk Loci. Cell Mol Gastroenterol Hepatol 2021;12(1):181–97 10.1016/j.jcmgh.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47(D1):D1005–D12 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4(8):1184–91 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol 2016;1418:93–110 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung KH, Chu K, Lee ST, Yoon HJ, Chang JY, Nam WS, et al. Prolonged use of aspirin alters human and rat intestinal cells and thereby limits the absorption of clopidogrel. Clin Pharmacol Ther 2011;90(4):612–9 10.1038/clpt.2011.163. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Xie Y, Wang L, Zhang Y, Gu H, Chai Y. Significant Modules and Biological Processes between Active Components of Salvia miltiorrhiza Depside Salt and Aspirin. Evid Based Complement Alternat Med 2016;2016:3439521 10.1155/2016/3439521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21(11):1350–6 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet 2019;51(1):76–87 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim P, Cheng F, Zhao J, Zhao Z. ccmGDB: a database for cancer cell metabolism genes. Nucleic Acids Res 2016;44(D1):D959–68 10.1093/nar/gkv1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607–D13 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan AT, Giovannucci EL, Meyerhardt JA, Schernhammer ES, Curhan GC, Fuchs CS. Long-term use of aspirin and nonsteroidal anti-inflammatory drugs and risk of colorectal cancer. JAMA 2005;294(8):914–23 10.1001/jama.294.8.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drew DA, Cao Y, Chan AT. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat Rev Cancer 2016;16(3):173–86 10.1038/nrc.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez LAG, Soriano-Gabarro M, Bromley S, Lanas A, Soriano LC. New use of low-dose aspirin and risk of colorectal cancer by stage at diagnosis: a nested case-control study in UK general practice. Bmc Cancer 2017;17 10.1186/s12885-017-3594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosetti C, Santucci C, Gallus S, Martinetti M, La Vecchia C. Aspirin and the risk of colorectal and other digestive tract cancers: an updated meta-analysis through 2019. Ann Oncol 2020;31(5):558–68 10.1016/j.annonc.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Stevens JR, Herrick JS, Wolff RK, Slattery ML. Power in pairs: assessing the statistical value of paired samples in tests for differential expression. BMC Genomics 2018;19(1):953 10.1186/s12864-018-5236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunbar K, Valanciute A, Lima A, Vinuela PF, Jamieson T, RajasekaranV, et al. Aspirin rescues Wnt-driven stem-like phenotype inhuman intestinal organoids and increases the Wnt antagonist Dickkopf-1,. Cellular and MolecularGastroenterology and Hepatology 2020. 10.1016/j.jcmgh.2020.09.010. [DOI] [PMC free article] [PubMed]
- 41.Gala MK, Chan AT. Molecular pathways: aspirin and Wnt signaling-a molecularly targeted approach to cancer prevention and treatment. Clin Cancer Res 2015;21(7):1543–8 10.1158/1078-0432.CCR-14-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schatoff EM, Leach BI, Dow LE. Wnt Signaling and Colorectal Cancer. Curr Colorectal Cancer Rep 2017;13(2):101–10 10.1007/s11888-017-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messick CA, Sanchez J, Dejulius KL, Hammel J, Ishwaran H, Kalady MF. CEACAM-7: a predictive marker for rectal cancer recurrence. Surgery 2010;147(5):713–9 10.1016/j.surg.2009.10.056. [DOI] [PubMed] [Google Scholar]
- 44.Hartmann F, Sparla D, Tute E, Tamm M, Schneider U, Jeon MK, et al. Low acyl-CoA synthetase 5 expression in colorectal carcinomas is prognostic for early tumour recurrence. Pathol Res Pract 2017;213(3):261–6 10.1016/j.prp.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Fields B, DeLaForest A, Zogg M, May J, Hagen C, Komnick K, et al. The Adult Murine Intestine is Dependent on Constitutive Laminin-gamma1 Synthesis. Sci Rep 2019;9(1):19303 10.1038/s41598-019-55844-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou J, Gong J, Ke J, Tian J, Zhang Y, Li J, et al. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis 2017;38(2):177–83 10.1093/carcin/bgw204. [DOI] [PubMed] [Google Scholar]
- 47.Nakanishi M, Menoret A, Tanaka T, Miyamoto S, Montrose DC, Vella AT, et al. Selective PGE(2) suppression inhibits colon carcinogenesis and modifies local mucosal immunity. Cancer Prev Res (Phila) 2011;4(8):1198–208 10.1158/1940-6207.CAPR-11-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J 2012;31(14):3079–91 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrucci G, Zaccardi F, Giaretta A, Cavalca V, Capristo E, Cardillo C, et al. Obesity is associated with impaired responsiveness to once-daily low-dose aspirin and in vivo platelet activation. J Thromb Haemost 2019;17(6):885–95 10.1111/jth.14445. [DOI] [PubMed] [Google Scholar]
- 50.Rothwell PM, Cook NR, Gaziano JM, Price JF, Belch JFF, Roncaglioni MC, et al. Effects of aspirin on risks of vascular events and cancer according to bodyweight and dose: analysis of individual patient data from randomised trials. Lancet 2018;392(10145):387–99 10.1016/S0140-6736(18)31133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw fastq files and pre-processed count matrix data have been uploaded to Gene Expression Omnibus(28), accession number: GSE163282.


