Abstract
The goal of this study was to develop strategies to localize human collagen-based hydrogels within an infarcted mouse heart, as well as analyze its impact on endogenous extracellular matrix (ECM) remodeling. Collagen is a natural polymer that is abundantly used in bioengineered hydrogels because of its biocompatibility, cell permeability, and biodegradability. However, without the use of tagging techniques, collagen peptides derived from hydrogels can be difficult to differentiate from the endogenous ECM within tissues. Imaging mass spectrometry is a robust tool capable of visualizing synthetic and natural polymeric molecular structures yet is largely underutilized in the field of biomaterials outside of surface characterization. In this study, our group leveraged a recently developed matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) technique to enzymatically target collagen and other ECM peptides within the tissue microenvironment that are both endogenous and hydrogel-derived. Using a multimodal approach of fluorescence microscopy and ECM-IMS techniques, we were able to visualize and relatively quantify significantly abundant collagen peptides in an infarcted mouse heart that were localized to regions of therapeutic hydrogel injection sites. On-tissue MALDI MS/MS was used to putatively identify sites of collagen peptide hydroxyproline site occupancy, a post-translational modification that is critical in collagen triple helical stability. Additionally, the technique could putatively identify over 35 endogenously expressed ECM peptides that were expressed in hydrogel-injected mouse hearts. Our findings show evidence for the use of MALDI-IMS in assessing the therapeutic application of collagen-based biomaterials.
Keywords: collagen, biomaterials, peptides, imaging mass spectrometry, MALDI, myocardial infarction
Graphical Abstract
INTRODUCTION
Collagen is a natural, degradable polymer found in the extracellular matrix (ECM) that is widely used in therapeutic biomaterial applications because of its biocompatibility, cell adhesion promotion, and cell permeability.1-3 One application of biomaterials is in the treatment of myocardial infarction (MI), which if left untreated leads to the formation of noncontractile scar tissue and potentially heart failure.4-7 Collagen has been studied as a biomaterial for injectable hydrogels to repair heart tissue damage post-MI, with the goal of limiting scar formation.8 A previous study utilized two of the most abundant collagen subtypes within the myocardium, types I and III, to develop human-derived collagen hydrogels that could be used therapeutically to prevent adverse remodeling postinfarction.8 However, recent literature suggests that peptides, as opposed to whole proteins, provide a promising therapeutic advantage, as full-length human collagen is difficult to synthesize and stabilize because of extensive post-translational modifications.9,10 For this reason, determining which peptides within the protein used in purified collagen protein hydrogels are most efficacious may contribute to more applicable therapeutics. Recent studies show that peptide-based biomaterials can mimic the function of their full-length protein counterparts; are easier to produce; can be derived from human origin, reducing autoimmune effects; and can similarly contribute to structural cell-recognition motifs.11,12
Several imaging techniques have been developed over the years to analyze biomaterials in tissues. Fluorescence and MRI imaging techniques have been used to trace injected collagen ex vivo as well as in vivo at low penetrative depths.13,14 These techniques, however, require labeling of the injected material. Imaging mass spectrometry (IMS) can analyze the spatial distribution of molecules without labeling. Recent advances in this field have been applied to biomaterials research, using ionization techniques such as secondary ion MS (SIMS), matrix-assisted laser desorption/ionization (MALDI), and desorption electrospray ionization (DESI), among others.15 These IMS techniques have been used to detect synthetic polymeric biomaterial and nanoparticles in vitro.15-18 IMS is capable of analyzing not only the biomaterial itself but also potential interactions with the treated tissue.19 To date, however, the majority of studies have been on biomaterial surface analysis.15 A soft ionization technique, such as MALDI IMS, is arguably one of the most appropriate methods for natural polymer detection in situ due to minimal fragmentation, allowing for more information from a complex substrate such as tissue.
MALDI IMS is a robust technique that is able to identify endogenous or enzymatically derived peptides in tissue sections.20 However, proteomics-based techniques to evaluate ECM-based biomaterial interactions in surrounding tissues have been limited because of poor detection of ECM proteins. This is due to their low abundance and inter- and intramolecular cross-linking. Recently, our lab has developed a collagenase-based ECM-targeted MALDI imaging mass spectrometry (ECM-IMS) methodology that may be used sequentially with other enzymatic imaging strategies.21-27 In the current study, we utilize this method to elucidate the distribution and therapeutic effects of collagen-based biomaterials on extracellular matrix remodeling in the infarcted heart, as well as identify potential therapeutic collagen peptides to be evaluated in future studies.
■ METHODS
Materials and Chemicals.
All solutions were prepared using double distilled or HPLC-grade water, following all necessary safety and waste disposal regulations. Xylenes, 200 proof ethanol were purchased from Fisher Scientific (Pittsburgh, PA, U.S.A.). Acetonitrile, ammonium bicarbonate, calcium chloride, formic acid, and Trizma base were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). PNGase F PRIME was purchased from N-Zyme scientific (Charleston, SC, U.S.A.). Collagenase type III (COLase3) (C. histolyticum) was purchased from Worthington Biochemical (Lakewood, NJ, USA).
Preparation and Characterization of Collagen Hydrogel.
Collagen hydrogels were prepared as previously described by McLaughlin et al.8 Briefly, a 1% collagen solution was prepared by dissolving lyophilized recombinant human collagen type I or type III (rhCI and rhCIII, from Fibrogen) in 10 mL of ultrapure ddH2O. Chondroitin sulfate (CS; Wako), N-ethyl-N-(3-(dimethylamino)propyl) carbodiimide (EDC), and N-hydroxysuccinimide (NHS) were added to produce a final mixture with a mass ratio of 1:4:0.5:0.3 for collagen:CS:NHS:EDC. Before the final step of the hydrogel preparation, after EDC/NHS cross-linkers have been added but before NaOH addition to a pH of 7.4, 20 μL of AlexaFluor 594-NHS conjugate (Thermofisher) was added to label the hydrogels (25 nmol of dye/gel). The materials were prepared on ice using an enclosed system that allows homogeneous mixing.
Animal Experiments.
All procedures were approved by the University of Ottawa Animal Care Committee and performed according to the National Institute of Health Guide for the Care and Use of Laboratory Animals. Myocardial infarction (MI) was induced in 8- to 9-week-old female C57BL/6 mice (Charles River), and treatment delivery was performed using an established protocol.28,29 Briefly, mice were anesthetized (2% isoflurane), intubated, and the heart was exposed via fourth intercostal thoracotomy. At 1-week post-MI (baseline), mice were randomly assigned to receive treatment of PBS (control), rhCI or rhCIII matrices, delivered in 5 equivolumetric intramyocardial injections (10 μL each site, 50 μL total) through a 25G needle using an ultrasound-guided closed-chest procedure. Mice were sacrificed by terminal anesthesia at 2 days post-treatment (9 days post-MI), and hearts were collected and frozen in OCT for histological sectioning.
Tissue Preparation and Histology.
Frozen tissue sections of mouse hearts injected with AF594-labeled hydrogels or PBS (10 μm thick) were cut via cryostat. A section approximately 2.16 mm from the apex was used in the present study. Fresh frozen mouse heart tissue sections were fixed briefly in 4% paraformaldehyde and stained with DAPI counterstain (Sigma-Aldrich, St Louis, MO, U.S.A.). Sections were mounted with Dako Fluorescent mounting medium (Agilent Technologies, Santa Clara, CA, U.S.A.), coverslipped, and sealed with clear nail polish. High-resolution digital captures were taken of IHC-stained mouse heart sections before MALDI IMS studies of the same slide. Slides are imaged using a Leica Aperio Versa slide scanner with the 20× objective for DAPI and AF594 signals (λexcitation: 580 nm; λemission: 625 nm).
For N-glycan and ECM peptide MALDI IMS studies, FFPE tissues were prepared as previously described.21,30 Briefly, after staining, AF594-labeled hydrogel detection, and coverslip removal, tissues were cleared of potential remaining OCT and mounting media with the following washing steps: 1 min Xylene, 2 × 1 min 100% ethanol, 1 min 95% ethanol, 1 min 70% ethanol, 2 × 1 min HPLC water. Tissues were then antigen retrieved in Citraconic Buffer pH 3 and 10 mM Tris pH 9.21,30 Tissue sections were then digested with Collagenase Type III (COLase3) after deglycosylation with PNGaseF (N-Zyme Scientifics) for MALDI IMS experiments.24,25 Enzyme solutions were sprayed with a TM Sprayer M3 (HTX Imaging, Chapel Hill, NC, U.S.A.) under the following parameters: 15 passes, crisscross pattern, 3.0 mm track spacing, velocity of 1200 mm/min, and a dry time of zero.
COLase3 treated samples were digested in ≥80% relative humidity at 37.5 °C for 5 h. Matrix was 7 mg/mL solution of alpha-cyano-4-hydroxycinnamic acid (CHCA) solution, prepared in 50% acetonitrile and 1.0% TFA, spiked with a standard of 200 femtomol/L [Glu1]-fibrinopeptide B human (GluFib) (Sigma-Aldrich, St Louis, MO, U.S.A.). CHCA automatic spray conditions included: 79 °C, 10 psi, 70 μL/min, 1300 velocity, and 14 passes with a 2.5 mm offset. Slides for peptide imaging were rapidly dipped (<1 s) in cold 5 mM ammonium phosphate and immediately dried in a desiccator.
MALDI Imaging Mass Spectrometry.
MALDI IMS analysis was via a 7.0 T solariX Legacy FT-ICR (Bruker Scientific, LLC) operated in positive ion broadband mode over m/z range 600–2500. A transient length of 0.8389 ms was used with a resolving power of 29 000 calculated at 1400 m/z. Laser settings used were 200 shots/pixel with a 75 μm stepsize. A lock mass of 1570.677 (matrix-spiked GluFib peptide, see above) was used. Images were visualized in FlexImaging v5.0 and analyzed using SCiLS (Bruker Scientific). All images shown are normalized to total ion current. Spectra shown were analyzed using mMass version 5.5.0. A filter of 10 ppm mass error was used for this study. Target peptides were were further fragmented by on-tissue MALDI MS/MS and analyzed for ion mobility to confirm post-translational modification site occupancy via CID on a timsTOF fleX as previously described (Bruker Scientific, LLC).25
Proteomic Tissue Preparation.
Tissue was prepared as previously described.26 Briefly, tissues were antigen retrieved and deglycosylated as described above. One slide with 6 serial sections per slide was used per treatment group. Tissue was scraped off the slide and ultrasonicated at 50% energy (Fisherbrand 120 sonic dismembrator; Fisher Scientific, Pittsburgh, PA, U.S.A.) for 2 min each in 10 mM ammonium bicarbonate 1 mM CaCl2 (pH 7.4). Tissues were incubated with 2 μg of Collagenase Type III overnight and centrifuged to collect supernatant for proteomic analysis. Samples were purified by C18 STAGE tip (Pierce Biotechnology, Waltham, MA) according to manufacturer’s protocol. STAGE-tip eluate was dried down via speed vac and resuspended in mobile phase A (HPLC water).
Proteomics.
Peptides were analyzed by data-dependent acquisition on a timsTOF fleX equipped with a nano-HPLC (nano-Elute, Bruker Daltonics). Peptides were loaded onto a trap column and separated on a 75 μm × 25 cm classic pulled tip column (Aurora C18 1.6um particles) at 50 °C. The gradient was from 2 to 80% solvent B over 105 min, where solvent A was HPLC water and solvent B was acetonitrile. Parallel accumulation serial fragmentation (PASEF) scan mode and TIMS were enabled in positive ion mode. TIMS accumulation time was fixed to 2 ms with a cycle time of 100 ms, covering a mass range of 100–1700 m/z and a 1/K0 range of 0.6–1.6 Vs/cm2.
Data was searched using MSFragger with IonQuant31 (v3.2; Fragpipe v15.0) against the mouse extracellular matrix database (downloaded February 26, 2021), a subset database of 2976 entries with keywords used (collagen, elastin, aggrecan, gelatin, osteonectin, perlecan, plasminogen, and fibronectin). Parameters included unspecified proteolytic enzyme, precursor mass tolerance of ±20 ppm, and fragment mass tolerance ±0.8 Da. Methionine oxidation and proline hydroxylation were included as variable modifications. Proteins were identified with FDR ≤ 0.05 and at least two peptides. Scaffold v5 was used for protein and peptide level analyses.
Bioinformatics.
Normalized peak intensities were evaluated by ANOVA and Mann–Whitney U test, correcting for multiple comparisons (IBM SPSS Statistics, version 25). A Type I error probability of ≤0.1 was used to evaluate the significance of the result. For this proof of principle study, p-values of ≤0.1 are reported as trending to significance and p-values of ≤0.05 were used determine significant results. AUC p-values are reported as the maximal discriminative power at the centroid. Peaks reported as significantly regulated were identified via SCiLS (v. 2017a). Exported peak intensities are visualized as heatmaps after natural log transformation with MultiExperiment Viewer (http:/mev.tm4.org).32
Peptide Synthesis.
GRPoxGEVGPoxP, GRPGEVGPoxPox, and GRPoxGEVGPPox peptides were synthesized using the Liberty Blue (CEM) automated microwave peptide synthesizer (where Pox is HYP). Fmoc-protected amino acids were purchased from CEM. HYP were purchased from BACHEM. To 2-chloro trityl resin (ProTideCl-TCP(Cl) resin, CEM), the first amino acid was loaded under microwave (90 °C), DIEA (1M), and KI (0.125M) for 10 min with twice coupling. Next, Fmoc deprotection was carried out with 20% piperidine at 90 °C for 60 s, while standard coupling cycles using DIC/Oxyma Pure were run at 90 °C for 240 s. Peptides were cleaved from the resin and deprotected with 92.5/2.5/2.5/2.5% v/v TFA/TIS/EDT/H2O at 42 °C for 30 min and then precipitated in −20 °C diethyl ether. Crude peptides were dried under vacuum overnight and purified by RP-HLPC in a Waters 1525EF semipreparative system with a 21.6 × 250 mm C18 column at 20 mL/min. Peptide purity and identity was confirmed via RP-UPLC-UV/MS in a Waters Acquity UPLC Xevo TQD using a 2.1 × 100 mm UPLC BEH C8 column. A purity of 95% was determined through UV peak analysis. The theorical monoisotopic mass for the 3 peptides is equal to 896.4 Da. For the peptide GRPoxGEVGPoxP, mass spectrometry results gave the most abundant peaks of 449.3 m/z [M+2H+]2+ and 897.4 m/z [M+H+]+ for an experimental mass of 896.4 Da. For the peptide GRPGEVGPoxPox, mass spectrometry results gave the most abundant peaks of 449.4 m/z [M+2H+]2+ and 897.6 m/z [M+H+]+ for an experimental mass of 896.6 Da. For the peptide GRPoxGEVGPPox mass spectrometry results gave the most abundant peaks of 449.3 m/z [M+2H+]2+ and 897.4 m/z [M+H+]+ for an experimental mass of 896.4 Da (Figure S4a-c).
■ RESULTS AND DISCUSSION
Study Design and Hydrogel Region of Interest Analysis.
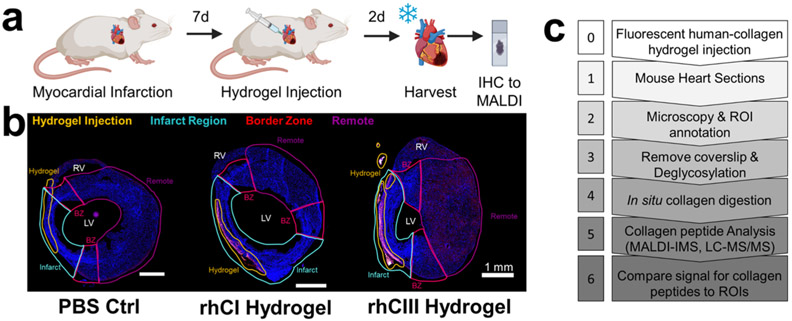
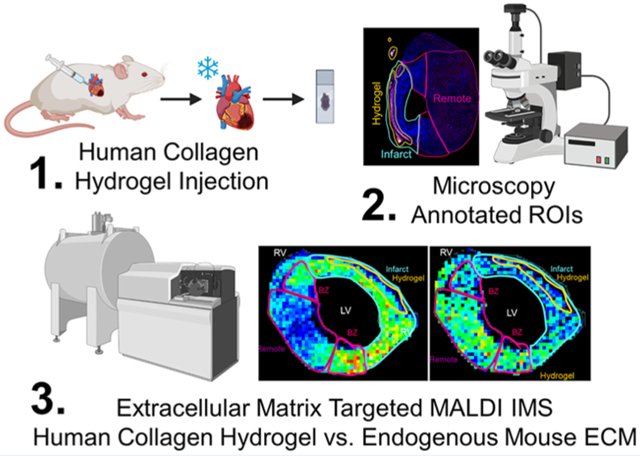
In this study, mice with myocardial infarction (MI) were treated at 7-days post-MI with either a collagen type I (rhCI) or a collagen type III (rhCIII) hydrogel.8 Two days after hydrogel treatment, mouse hearts were harvested, frozen, and sectioned for tandem fluorescent-microscopy-based histology and MALDI IMS studies (Figure 1a). Injected human collagen hydrogels were fluorescently labeled prior to injection and analyzed via immunofluorescence microscopy (Figure 1b). Microscopy-based analysis of tissues identified the potential site of hydrogel injection as well as the infarcted region, the border zone (area immediately adjacent to the infarct), and the remote region (intact myocardium) (Figure 1b, Figure S1). These microscopy-based observations were used to create regions of interest (ROIs) for downstream MALDI IMS analysis of collagen and other ECM peptides (ECM-IMS) (Figure 1c). While downstream peak-picking was performed unbiased of microscopy-based observations, ROIs were used to further evaluate collagen-type peaks of interest in relation to hydrogel specificity.
Figure 1.
Mouse model study design and multimodal imaging mass spectrometry workflow. (a) Timeline of mouse model. 9-week old mice were induced with myocardial infarction. 7-days postsurgery, collagen hydrogel or PBS control was injected into the left ventricle. 2-days post injection, hearts were harvested. (b) Fluorescence microscopy images showing region of interest (ROI) annotations of heart sections in relation to fluorescently labeled human hydrogel signal. BZ: border zone; LV: left ventricle; RV: right ventricle. Scale bar shown is 1 mm. (c) Simplified workflow of tandem high-resolution fluorescent microscopy and collagenase-based MALDI IMS and LC MS/MS experiments.
Fibrillar Collagen Homology Sequences Are More Abundant in Hydrogel Injection Sites.
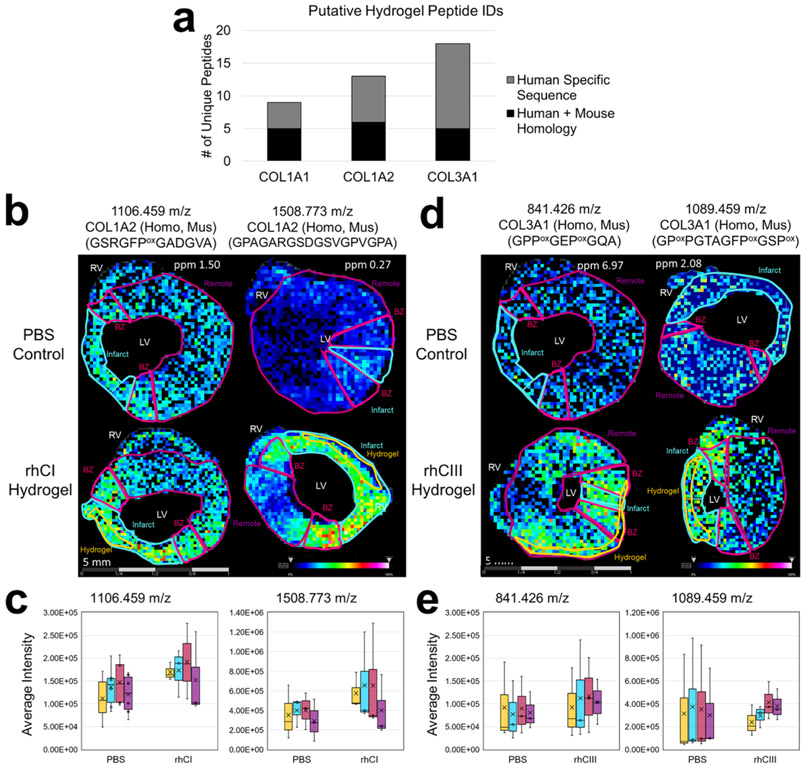
A challenge of this study is that humans and mice have 93–94% homology between collagen sequences (for the collagen subtypes used for hydrogel injections: COL1A1, COL1A2, and COL3A1). To determine specificity of hydrogel-derived human collagen sequences, the localization capability of MALDI IMS was leveraged with multiplexed histopathology-based studies in conjunction with on-tissue peptide sequencing via MALDI MS/MS. Overall, 22 collagen type 1 putative peptides (9 COL1A1, and 13 COL1A2) were identified; 11 of all collagen type 1 putative peptides identified contained sequences homologous between human and mouse (Figure 2a, Supplemental Table S1). Additionally, ECM-IMS analysis identified 18 human COL3A1 peptides, with only 5 being homologous between human and mouse sequences (Figure 2a, Supplemental Table S1). Among these, pathological ROI analysis identified four collagen type 1 peaks that were differentially regulated in rhCI-injected hearts, specifically at the infarct ROI (Supplemental Table S2). Similarly, two COL3A1 peaks were differentially expressed between PBS-control and rhCIII-injected hearts (Supplemental Table 2). Two representative images of collagen type 1 in rhCI hearts and COL3A1 in rhCIII hearts are shown as compared to PBS injected controls (Figure 1b,d respectively). Average intensity levels of peaks shown in representative images were quantified across all tissue sections studied (Figure 1c,e). Overall, ROI annotation and differential intensity distributions were able to identify putative peptides corresponding to the site of hydrogel injections.
Figure 2.
Human collagen peptides with mouse homology are more abundant in hydrogel injection sites. (a) Total number of putative human peptide identifications from hydrogel injected tissues as a function of collagen subtype for collagens injected. (b) Representative collagen type 1 peaks as visualized in rhCI injected hearts as compared to PBS controls. Average intensity levels measured between all samples studied are shown in (c). (d) Representative collagen type 3a1 peaks as visualized in rhCIII injected hearts as compared with PBS controls. Average intensity levels measured between all samples studied are shown in (e). n = 3. Six tissue sections were visualized for each biological replicate. Different representative samples are used to show reproducibility across n’s. Peptides shown in panel d (left) represent nearest identified putative peptides from previously acquired human databases.
ECM-IMS Identifies Human-Specific Collagen Sequences in Hydrogel Injected Mouse Hearts.
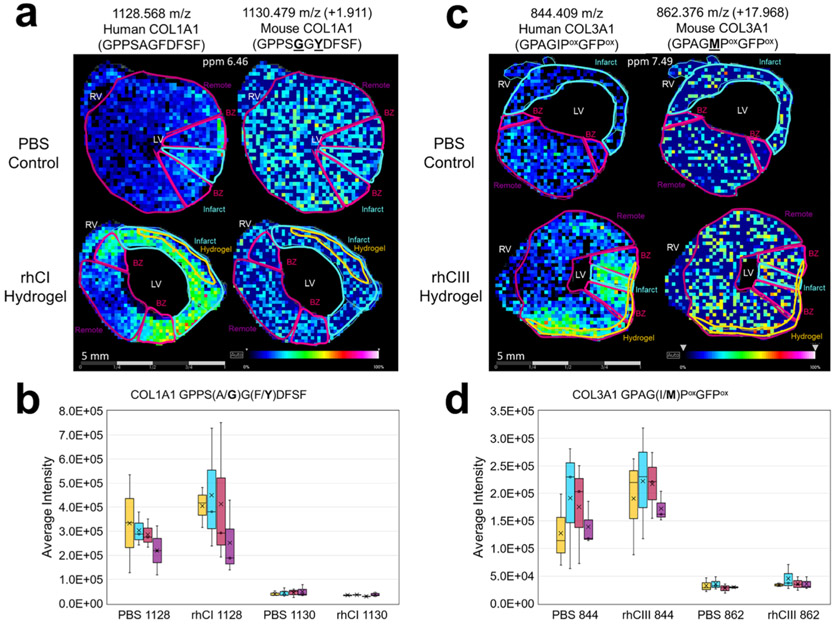
An example of a significantly increased peptide identified that aligned uniquely to the COL1A1 human sequence is shown in Figure 3 (rhCI 1128.568 m/z; GPPSAGFDFSF). ECM-IMS analysis was also able to identify the mouse peptide corresponding to this human sequence (1130.479 m/z, + 1.911 m/z; GPPSGGYDFSF) (Figure 3a,b). As can be seen in Figure 3, the peak corresponding to the mouse peak is equally present in the PBS control and rhCI hydrogel tissue. However, the corresponding human-specific peptide is significantly more abundant in the rhCI-injected heart (AUC at infarct 0.689, p-value 0.013). A similar result is seen for a COL3A1 peptide sequence in the rhCIII-treated heart (AUC at infarct, 0.923, p-value 0.008) (Figure 3c,d). Importantly, m/z 1128 and m/z 844 were not identified in the mouse-database identified peptides via LC-MS/MS (Figure 4, Supplemental Table S5). Finding these human-proteome unique peptides provides further evidence of biomaterial localization via MALDI ECM IMS. While fluorescent label-based microscopy techniques may be able to identify the site of injection, ECM-IMS identifies unique peptides within the hydrogel that may be contributing to endogenous ECM remodeling and improved cardiac function, as seen in tandem studies on the same mice.8
Figure 3.
Collagen peptides unique to human proteome have differential distribution to corresponding mouse peptides. Species differentiating amino acids are underlined in bold within the sequence. (a) Representative collagen type 1 peaks as visualized in rhCI injected hearts as compared with PBS controls are shown for the human-specific collagen sequence (left) and corresponding mouse sequence (right). Average intensity levels measured between all samples studied are shown in (b). (c) Representative collagen type 3a1 peaks as visualized in rhCIII injected hearts as compared to PBS controls are shown for the human-specific collagen sequence (left) and corresponding mouse sequence (right). Average intensity levels measured between all samples studied are shown in (d). n = 3. Six tissue sections were visualized for each biological replicate. Different representative samples are used to show reproducibility across n’s. Note: Location of hydroxyproline (Pox) are ambiguous and require further validation. Peptides shown in a–d represent nearest identified putative peptides from previously acquired human databases.
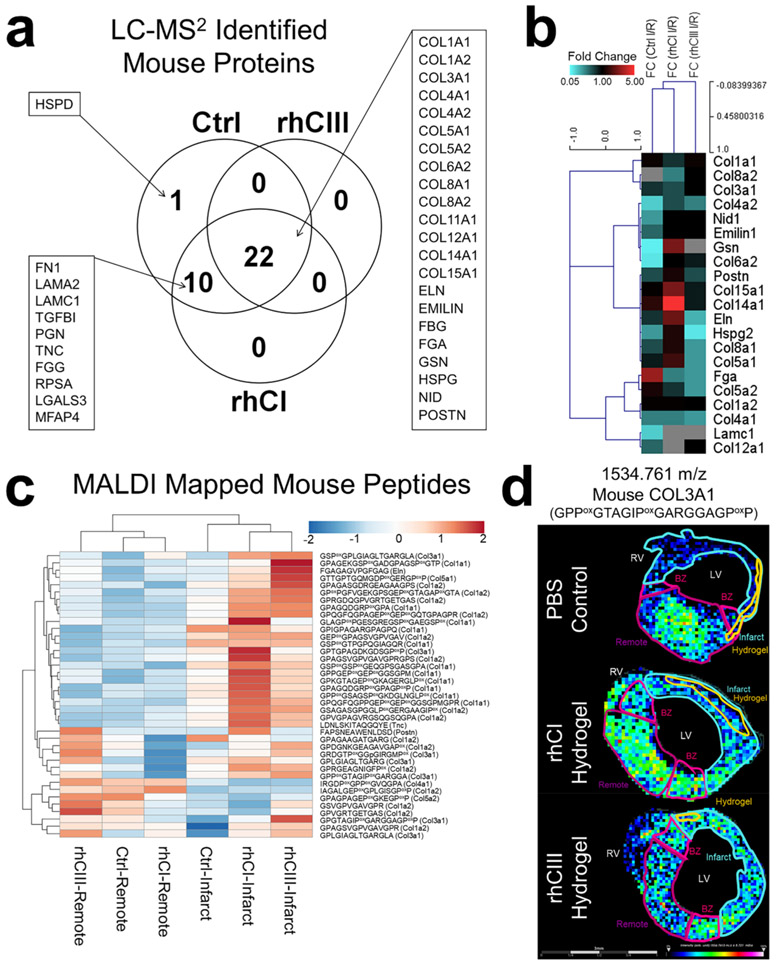
Figure 4.
Collagen targeted proteomics investigates endogenous mouse ECM remodeling. (a) Venn Diagram showing the number of proteins identified in PBS control, rhCI hydrogel, and rhCIII hydrogel samples. (b) Hierarchical clustering of protein level fold change (infarct/remote) values for each treatment group. (c) Hierarchical clustering of LC-MS/MS identified mouse peptides mapped via MALDI IMS studies. ROI intensity for infarct and remote is shown for each treatment group. (d) Representative image of a COL3A1 peptide that was differentially abundant in PBS Ctrl vs hydrogel treatment, mapped via MALDI IMS.
On-Tissue MALDI Tandem MS Peptide Sequencing Provides Evidence toward Identifications and PTM Site Occupancy.
Within the differentially expressed COL1A1 peptides identified, one peptide had potential for differential hydroxyproline (HYP) sites. HYP is a post-translational modification necessary for collagen triple helix structural stability. The putative HYP containing peptide, with a COL1A1 peak at m/z 897.445 (GRPoxGEVGPoxP, where Pox is HYP; AUC = 0.686, p-value 0.018); this peptide has one hydroxylated proline, with three potential variants at isobaric peaks. To identify fragments associated with each HYP variant, on-tissue fragmentation was performed MALDI MS/MS to identify specific fragments relating to each variant (Figure S2a,b). On-tissue, the primary HYP variant identified was in proline positions 1 and 2 (1,2; GRPoxGEVGPoxP), with 17 unique fragment peaks out of the 30 peaks identified. However, this experiment showed a mix of peptide variants, with 1,3 and 2,3 variants also being identified (Supplementary Figure S2b)
HYP site occupancy in the hydrogel-derived 897 m/z peptide were putatively identified but were investigated further using synthesize peptide standards (Figure S2c-e). The three synthesized peptide variant standards were also investigated via trapped ion mobility, to distinguish the three isoforms of the 897 m/z peptide by their collisional cross-section (Figure S2f). An additional unmodified COL1A2 peptide m/z 1508.773 (GPAGARGSDGSVGPVGPA; AUC 0.690) was also targeted for on-tissue fragmentation (Figure S3). These preliminary results have tentatively identified putative peptides, as validated by on-tissue MALDI MS/MS, that may be promising candidates for use in developing therapeutic peptide-based biomaterials.
Collagen-Based Biomaterials Effects on Endogenous Mouse ECM Remodeling.
To further identify endogenous mouse ECM proteins that were differentially regulated with hydrogel treatment, we performed parallel chromatographic proteomic experiments on macro dissected regions, infarct and remote. In these studies, created mouse ECM databases were used to earmark endogenous mouse peptides in serial sections of tissue used in the ECM-IMS studies. A total of 33 proteins were identified, with 42% (14 out of 33) proteins being collagen subtypes, varied across families of fibril, network-associated, FACIT (fibril-associated collagen with interrupted triple helices) and multiplexin (Figure 4a,b, Figure S5, Supplemental Table S3). Peptides putatively identified from noncollagen type proteins were implicated in pathways related to collagen biosynthesis and chain trimerization, GAG biosynthesis, and ECM degradation (Supplemental Table S4). Proteins were investigated as a function of fold change between each treatment’s infarct ROI vs remote ROI (Figure 4b).
These proteomic experiments were then used to create a mouse ECM peptide database to apply to the MALDI IMS data set. Of the 62 mouse peptides mapped in the MALDI data set, 39 peptides contained sequences unique to mouse and were used for further analysis (Figure 4c,d, Supplemental Table S5). Figure 4c shows that peptide intensities as measured between infarct ROIs (closest to hydrogel injection site) and remote ROIs (furthest from hydrogel injection site) can be unbiased clustered apart from one another based on ECM peptide expression (Figure 4c). Within the infarcted region, rhCI and rhCIII hydrogel treated mice show similar endogenous peptide expression as compared with PBS-treated control mice (Figure 4c).
Of these 39 endogenous mouse peptides shown in Figure 4c, 8 were found to be differentially expressed among treatment groups (Supplemental Table S6). Figure 4d shows a representative image of one of these peptides (m/z 1534.761; COL3A1 GPPoxGTAGIPoxGARGGAGPoxP) via MALDI-IMS mapping. Here, the unique spatial distribution of this endogenous mouse peptide is seen as compared to hydrogel distribution in previous figures. In previous figures, ROI analysis shows that peptides’ sharing homology sequence between human and mouse (Figure 2) have greater signal intensities within the hydrogel injection site (HIS) and infarct ROIs, as compared to the more distant remote region, corroborating microscopy-derived annotations of injection sites. This data may further suggest increased sensitivity of MALDI IMS data compared with microscopy annotated ROIs, as diffusion of hydrogel-derived peptides can be visualized via MALDI IMS outside of the hydrogel injection site ROI, toward the next-nearest infarct ROI and in some instances toward the border zone ROI. This trend is even more apparent when analyzing putatively identified peptides that are unique to the human sequence (Figure 3). Importantly, MALDI IMS maps of putatively identified peptides unique to the mouse proteome (Figure 4d) are homogeneously distributed throughout the tissue, with no preferential distribution corresponding to hydrogel-dependent ROIs. Interestingly, the COL3A1 peptide shown appears to be localized only to healthy remote tissue in PBS-treated control, while being more homogeneously distributed in hydrogel-treated samples. Together, these studies show the utility of MALDI-IMS to localize both human hydrogel-specific peptides and endogenous mouse extracellular matrix peptides within a site of therapeutic biomaterial injection.
■ CONCLUSIONS
ECM-IMS of collagen-based biomaterial therapeutics enabled complex protein and PTM expression data analysis, which were localized to surrounding pathological regions and within the biomaterial injection site. The use of MALDI-IMS in this study was able to localize and identify peptides within hydrogel injected tissues, as well as identify potential endogenously differentiated extracellular matrix proteins. While this study focused on imaging of the collagenous component of the injected biomaterials, glycosaminoglycans such as chondroitin sulfate (as used in this study) and recombinant elastin proteins are other common components of hydrogels. Serial-enzyme ECM-targeted strategies used on-tissue may be beneficial in hydrogel studies to understand therapies involving complex hydrogel mixtures.25 These and future studies may provide evidence and insight for which hydrogel-derived peptides are most efficacious at promoting healing in the initial wound environment before therapeutic hydrogel degradation.
Supplementary Material
ACKNOWLEDGMENTS
Work by C.L.C. is supported by a T32 from NHLBI (HL007260) and an F31 from NHLBI (HL156524). Additional support was provided by the South Carolina Centers of Economic Excellence SmartState Program to R.R.D. and A.S.M. Additional support to R.R.D., A.S.M., and P.M.A. was supplied by U01CA242096 (NCI). P.M.A. appreciates support by 16GRNT31380005 (American Heart Association), R21CA240148 (NCI), P20GM103542 (NIH/NIGMS). The authors appreciate the guidance of Grace Grimsley for proteomic analysis using the timsTOF fleX. E.I.A. and E.J.S. thanks the funding of a Collaborative Health Research Program grant from the Canadian Institutes of Health Research (CPG-158280) and the Natural Sciences and Engineering Research Council (CHRP 523794-18). E.I.A. thanks the Government of Ontario for an Early Career Research Award.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.1c00189.
MALDI IMS mapped human hydrogel peptides; significant human hydrogel peptides in infarct; LC-MS/MS identified proteins; pathway annotation for noncollagen proteins; MALDI IMS mapped mouse ECM peptides; significant mouse ECM peptides; ROI annotations of all biologic replicates; 897 m/z peptide HYP mapping via MS/MS; 1508 m/z peptide MS/MS; MS of synthesized peptides; LC-MS/MS determined fold change (infarct/remote); representative collagenase spectra (PDF)
The authors declare no competing financial interest.
Contributor Information
Cassandra L. Clift, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States.
Sarah McLaughlin, Division of Cardiac Surgery, University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada.
Marcelo Muñoz, Division of Cardiac Surgery, University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada.
Erik J. Suuronen, Division of Cardiac Surgery, University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada
Benjamin H. Rotstein, Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Ottawa, Ontario K1H 8M5, Canada.
Anand S. Mehta, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States.
Richard R. Drake, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States
Emilio I. Alarcon, Division of Cardiac Surgery, University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada; Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Ottawa, Ontario K1H 8M5, Canada.
Peggi M. Angel, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States.
REFERENCES
- (1).Chattopadhyay S; Raines RT Review collagen-based biomaterials for wound healing. Biopolymers 2014, 101, 821–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cen L; Liu W; Cui L; Zhang W; Cao Y Collagen tissue engineering: Development of novel biomaterials and applications. Pediatr. Res 2008, 63, 492–496. [DOI] [PubMed] [Google Scholar]
- (3).Parenteau-Bareil R; Gauvin R; Berthod F Collagen-based biomaterials for tissue engineering applications. Materials 2010, 3, 1863–87. [Google Scholar]
- (4).Rane AA; Christman KL Biomaterials for the treatment of myocardial infarction: A 5-year update. J. Am. Coll. Cardiol 2011, 58, 2615–29. [DOI] [PubMed] [Google Scholar]
- (5).Zhu Y; Matsumura Y; Wagner WR Ventricular wall biomaterial injection therapy after myocardial infarction: Advances in material design, mechanistic insight and early clinical experiences. Biomaterials 2017, 129, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ungerleider JL; Christman KL Concise Review: Injectable Biomaterials for the Treatment of Myocardial Infarction and Peripheral Artery Disease: Translational Challenges and Progress. Stem Cells Transl. Med 2014, 3, 1090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Venugopal JR; Prabhakaran MP; Mukherjee S; Ravichandran R; Dan K; Ramakrishna S Biomaterial strategies for alleviation of myocardial infarction. J. R. Soc., Interface 2012, 9, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).McLaughlin S; McNeill B; Podrebarac J; Hosoyama K; Sedlakova V; Cron G; Smyth D; Seymour R; Goel K; Liang W; Rayner KJ; Ruel M; Suuronen EJ; Alarcon EI Injectable human recombinant collagen matrices limit adverse remodeling and improve cardiac function after myocardial infarction. Nat. Commun 2019, 10, 4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Koide T Triple Helical Collagen-Like Peptides: Engineering and Applications in Matrix Biology. Connect. Tissue Res 2005, 46, 131–41. [DOI] [PubMed] [Google Scholar]
- (10).Tanrikulu IC; Forticaux A; Jin S; Raines RT Peptide tessellation yields micrometre-scale collagen triple helices. Nat. Chem 2016, 8, 1008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hosoyama K; Lazurko C; Muñoz M; McTiernan CD; Alarcon EI Peptide-based functional biomaterials for soft-tissue repair. Front. Bioeng. Biotechnol 2019, 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Swartzlander MD; Blakney AK; Amer LD; Hankenson KD; Kyriakides TR; Bryant SJ Immunomodulation by mesenchymal stem cells combats the foreign body response to cell-laden synthetic hydrogels. Biomaterials 2015, 41, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Artzi N; Oliva N; Puron C; Shitreet S; Artzi S; Bon Ramos A; Groothuis A; Sahagian G; Edelman ER In vivo and in vitro tracking of erosion in biodegradable materials using non-invasive fluorescence imaging. Nat. Mater 2011, 10, 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mertens ME; Hermann A; Bühren A; Olde-Damink L; Möckel D; Gremse F; Ehling J; Kiessling F; Lammers T Iron oxide-labeled collagen scaffolds for non-invasive MR imaging in tissue engineering. Adv. Funct. Mater 2014, 24, 754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Paine MRL; Kooijman PC; Fisher GL; Heeren RMA; Fernández FM; Ellis SR Visualizing molecular distributions for biomaterials applications with mass spectrometry imaging: A review. J. Mater. Chem. B 2017, 5, 7444–60. [DOI] [PubMed] [Google Scholar]
- (16).Kokesch-Himmelreich J; Woltmann B; Torger B; Rohnke M; Arnhold S; Hempel U; Müller M; Janek J Detection of organic nanoparticles in human bone marrow-derived stromal cells using ToF-SIMS and PCA. Anal. Bioanal. Chem 2015, 407, 4555–65. [DOI] [PubMed] [Google Scholar]
- (17).Ulrich TA; Lee TG; Shon HK; Moon DW; Kumar S Microscale mechanisms of agarose-induced disruption of collagen remodeling. Biomaterials 2011, 32, 5633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chen S; Xiong C; Liu H; Wan Q; Hou J; He Q; Badu-Tawiah A; Nie Z Mass spectrometry imaging reveals the sub-organ distribution of carbon nanomaterials. Nat. Nanotechnol 2015, 10, 176–82. [DOI] [PubMed] [Google Scholar]
- (19).Klerk LA; Dankers PYW; Popa ER; Bosman AW; Sanders ME; Reedquist KA; Heeren RMA TOF-secondary ion mass spectrometry imaging of polymeric scaffolds with surrounding tissue after in vivo implantation. Anal. Chem 2010, 82, 4337–43. [DOI] [PubMed] [Google Scholar]
- (20).Groseclose MR; Andersson M; Hardesty WM; Caprioli RM Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom 2007, 42, 254–62. [DOI] [PubMed] [Google Scholar]
- (21).Angel PM; Comte-Walters S; Ball LE; Talbot K; Mehta AS; Brockbank KGM; Drake RR Brockbank KGMM, Drake RR. Mapping Extracellular Matrix Proteins in Formalin-Fixed, Paraffin-Embedded Tissues by MALDI Imaging Mass Spectrometry. J. Proteome Res 2018, 17, 635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Angel PM; Schwamborn K; Comte-Walters S; Clift CL; Ball LE; Mehta AS; Drake RR Extracellular Matrix Imaging of Breast Tissue Pathologies by MALDI Imaging Mass Spectrometry. Proteomics: Clin. Appl 2019, 13, 1700152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Angel PM; Bruner E; Bethard J; Clift CL; Ball L; Drake RR; Feghali-Bostwick C Extracellular matrix alterations in low-grade lung adenocarcinoma compared with normal lung tissue by imaging mass spectrometry. J. Mass Spectrom 2020, 55, e4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Clift CL; Mehta A; Drake RR; Angel PM Multiplexed Imaging Mass Spectrometry of Histological Staining, N-glycan and Extracellular Matrix from One Tissue Section: A Tool for Fibrosis Research. Methods Mol. Biol 2019, 2350, 313–329. [DOI] [PubMed] [Google Scholar]
- (25).Clift CL; Drake RR; Mehta A; Angel PM Multiplexed imaging mass spectrometry of the extracellular matrix using serial enzyme digests from formalin-fixed paraffin-embedded tissue sections. Anal. Bioanal. Chem 2021, 413, 2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Clift CL; Su YR; Bichell D; Jensen Smith HC; Bethard JR; Norris-Caneda K; Comte-Walters S; Ball LE; Hollingsworth MA; Mehta AS; Drake RR; Angel PM Collagen fiber regulation in human pediatric aortic valve development and disease. Sci. Rep 2021, 11, 9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Angel PM; Spruill L; Jefferson M; Bethard JR; Ball LE; Hughes-Halbert C; Drake RR Zonal regulation of collagen-type proteins and posttranslational modifications in prostatic benign and cancer tissues by imaging mass spectrometry. Prostate 2020, 80, 1071–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ahmadi A; McNeill B; Vulesevic B; Kordos M; Mesana L; Thorn S; Renaud JM; Manthorp E; Kuraitis D; Toeg H; Mesana TG; Davis DR; Beanlands RS; DaSilva JN; deKemp RA; Ruel M; Suuronen EJ The role of integrin alpha2 in cell and matrix therapy that improves perfusion, viability and function of infarcted myocardium. Biomaterials 2014, 35 (17), 4749–58. [DOI] [PubMed] [Google Scholar]
- (29).Blackburn NJ; Sofrenovic T; Kuraitis D; Ahmadi A; McNeill B; Deng C; Rayner KJ; Zhong Z; Ruel M; Suuronen EJ Timing underpins the benefits associated with injectable collagen biomaterial therapy for the treatment of myocardial infarction. Biomaterials 2015, 39, 182–92. [DOI] [PubMed] [Google Scholar]
- (30).Powers TW; Neely BA; Shao Y; Tang H; Troyer DA; Mehta AS; Haab BB; Drake RR MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 2014, 9, e106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yu F; Haynes SE; Ci Teo G; Avtonomov DM; Polasky DA; Nesvizhskii AI Fast quantitative analysis of timsTOF PASEF data with MSFragger and IonQuant. bioRxiv, July 3, 2020. DOI: 10.1101/2020.03.19.999334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Saeed AI; Bhagabati NK; Braisted JC; Liang W; Sharov V; Howe EA; Li J; Thiagarajan M; White JA; Quackenbush J TM4Microarray Software Suite. Methods Enzymol. 2006, 411, 134–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.