Abstract
A common approach to investigate oxidant-regulated intracellular pathways is to add exogenous H2O2 to living cells or tissues. However, the addition of H2O2 to the culture medium of cells or tissues approach does not accurately replicate intracellular redox-mediated cell responses. D-amino acid oxidase (DAAO)-based chemogenetic tools represent informative methodological advances that permit the generation of H2O2 on demand with a high spatiotemporal resolution by providing or withdrawing the DAAO substrate D-amino acids. Much has been learned about the intracellular transport of H2O2 through studies using DAAO, yet these valuable tools remain incompletely characterized in many cultured cells. In this study, we describe and characterize in detail the features of a new modified variant of DAAO (termed mDAAO) with improved catalytic activities. We tested mDAAO functionality in several cultured cell lines employing live-cell imaging techniques. Our imaging experiments show that mDAAO is suitable for the generation of H2O2 under hypoxic conditions imaged with the novel ultrasensitive H2O2 sensor (HyPer7). Moreover, this approach was suitable for generating H2O2 in a reversible and concentration-dependent manner in subcellular locales. Furthermore, we show that the choice of D-amino acids differentially affects mDAAO-dependent intracellular H2O2 generation. When paired with the hydrogen sulfide (H2S) sensor hsGFP, administration of the sulfur-containing amino acid D-cysteine to cells expressing mDAAO generates robust H2S signals. We also show that chemogenetic H2O2 generation in different cell types yields distinct HyPer7 profiles. These studies fully characterize the new mDAAO as a novel chemogenetic tool and provide multiparametric approaches for cell manipulation that may open new lines of investigations for redox biochemists to dissect the role of ROS signaling pathways with high spatial and temporal precision.
Keywords: D-amino Acid Oxidase (DAAO) catalysis and targeting, D-amino acids, Hydrogen Peroxide, Hydrogen Sulfide, Genetically Encoded Biosensors, hsGFP, HyPer7, Multiparametric imaging
Graphical Abstract
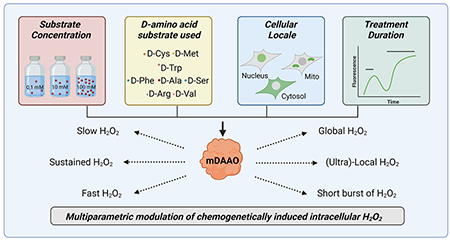
Introduction
Hydrogen peroxide (H2O2) is one the most studied reactive oxygen species (ROS) in biology both because of its involvement in pathological oxidative distress as well as its roles as a critical cell signaling molecule in redox-regulated pathways [1]. H2O2 causes oxidative distress at higher concentrations, while lower H2O2 concentrations lead to oxidative eustress that can drive physiological redox-signaling [2]. Natural sources for H2O2 include the enzymatic dismutation of superoxide anion (O2·−) generated by the mitochondrial respiration chain, as well as intracellular enzymes capable of directly synthesizing H2O2, such as NADPH oxidase 4 (Nox4) [3]. Under physiological conditions, these biochemical reactions are restricted to specific intracellular sites modulating redox-sensitive responses [4]. Yet, for many decades, H2O2 responses were typically studied by administering high concentrations of exogenous H2O2 to cells and tissues, which often led to contradictory results [5–7]. This approach may be informative in some experimental applications, yet in general, the practice of exposing cells to exogenous H2O2 fails to model the complexities of spatially-restricted redox-mediated signaling pathways in cells [6]. Owing in part to the lack of suitable experimental tools, the intracellular roles of endogenous H2O2 remain incompletely understood.
Over the last few years, a chemogenetic approach to generate H2O2 using recombinant yeast-derived D-amino acid oxidases (DAAO) has emerged as a novel technique that permits dynamic control of intracellular H2O2 [8–11]. By differentially targeting genetically encoded recombinant DAAO constructs, H2O2 can be generated in different subcellular locales simply by supplying the enzyme’s D-amino acid substrate [6,8]. The provision or withdrawal of D-amino acids can thereby modulate oxidant-regulated pathways in particular intracellular sites in living cells and tissues. The use of genetically-encoded H2O2 biosensors, such as HyPer [12–14], allows simultaneous production of H2O2 (by DAAO) and real-time detection (by HyPer) using high-resolution live-cell fluorescence imaging [15].
Contemporaneous advances in other genetically encoded biosensors have allowed the precise quantitation of diverse second messenger molecules, including other reactive oxygen species (ROS) and reactive nitrogen species (RNS) [16–21]. Combining chemogenetic approaches with next-generation biosensor methodologies has opened up new lines of investigation in complex model systems. Multispectral and multiparametric imaging allow the analysis of intracellular redox dynamics with high precision [22–26]. These approaches can be highly informative and enable the dynamic monitoring of oxidant-modulated cellular responses both in physiological conditions and in disease states characterized by pathological oxidative stress.
The experimental conditions optimal for exploiting chemogenetic tools paired with genetic biosensors are still being developed and validated in diverse cell systems [22]. In this paper, we characterize a mutated version of DAAO (“mDAAO”) and use this new chemogenetic construct in multiparametric imaging approaches exploiting recently developed informative biosensors [27,28]. We also provide new insights in the effects of different D-amino acid substrates on mDAAO-modulated H2O2 production. Our methods and multiparametric imaging techniques may provide new technological strategies to apply chemogenetic tools more precisely across a broad range of experimental conditions.
Material and Methods
Chemicals
Dulbecco’s modified Eagle’s medium (DMEM), phenol-free DMEM, penicillin and streptomycin, trypsin, and fetal bovine serum (FBS) were purchased from Pan Biotech (Aidenbach, Germany). High glucose DMEM without methionine and cysteine were purchased from Sigma Aldrich (Istanbul, Turkey). Cell culture supplements such as normocin were purchased from Invivogen (Toulouse – France) and hypoxanthine (HAT) supplements from LGC Standards (Istanbul, Turkey). Transfection reagent Polyjet was purchased from Signagen (Maryland, USA). D-amino acids D-alanine, D-arginine, D-phenylalanine, D-methionine, and D-tryptophan were purchased from Alfa Aesar (Landau, Germany). D-serine, D-valine, were purchased from Sigma Aldrich (Istanbul, Turkey). D-cysteine was purchased from ChemCruz (Heidelberg, Germany). P-Azido-L-phenylalanine (pAzF) was purchased from Bachem (Bubendorf, Swiss). L-Alanine and all chemicals to prepare imaging media and buffer solutions were purchased from neoFroxx (Einhausen, Germany).
Cell culture
Human embryonic kidney cells (HEK293), HeLa cells, human colon carcinoma cells (HCT116), and Uppsala 87 malignant glioma cells (U-87 MG) were grown in a high-glucose (4.5 g/L) complete medium including 10% FBS and 100 μg/ml streptomycin and 100 U/ml penicillin in a humidified incubator (37 °C, 5% CO2). Immortalized human umbilical vein endothelial cells (EA.hy926) were cultured in the same complete media as the cell lines described above except for additional supplements, including 2% HAT supplement (consisting of sodium hypoxanthine (5 mM), aminopterin (20 μM), and thymidine (0.8 mM)) and 100 μg/ml normocin. One day before transfection, cells were seeded (~3 x 105 cells per well) on a 30 mm glass coverslips No.1 (Glaswarenfabrik Karl Knecht Sondheim, Germany). At ~70–80% confluency, cells were co-transfected with CMV-driven mammalian expression vectors (AAV-MSC (Stratagene) or pCS2 (RZPD)) encoding the chemogenetic DAAO enzymes or genetically encoded biosensors using PolyJet transfection reagent according to the manufacturer’s instructions. All imaging experiments were performed 24 h after transfection. HEK293 cells transfected with hsGFP were cultured in a complete medium without L-methionine and L-cysteine containing 1 mM p-Azido-L-phenylalanine (pAzF) for 36-48 h. After this incubation, the culture medium was replaced by a fresh completemedium without L-methionine, L-cysteine, and pAzF for an additional 12 h to deplete the remaining pAzF before imaging experiments. All cell lines were cultured up to passage 30.
Molecular Cloning
DAAO constructs were differentially targeted to the cell nucleus [29], mitochondria matrix [30], and cytosol [31] using signal sequences, and were tagged with the red fluorescent protein mCherry2. The cDNA for mCherry2-mDAAO-NLS was synthesized and subcloned in the CMV-driven mammalian expression vector AAV-MSC (Stratagene) using the flanking restriction sites EcoRI and HindIII. The mitochondria-targeted constructs were generated by introducing a double tandem signal sequence of COXVIII by introducing the restriction site AgeI.
Buffer solutions and imaging media
Unless otherwise stated, all chemicals were purchased from neoFroxx (Einhausen, Germany). Prior to imaging experiments, cells were incubated at room temperature in a cell storage buffer consisting of 138 mM NaCl, 2 mM CaCl2, 5 mM KCl, 1 mM MgCl2, 1 mM HEPES, 2.6 mM NaHCO3, 0.44 mM KH2PO4, 0.34 mM Na2HPO4, 10 mM D-glucose, 0.1% vitamins, 0.2% essential amino acids, and 1% penicillin/streptomycin, pH 7.4 for 40 min. Imaging media for superfusing cells was a HEPES-buffered physiological salt solution consisting of 138 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10mM D-glucose, 10 mM HEPES. All imaging buffers were adjusted to pH 7.4. All D-amino acids were prepared fresh on the day of the experiment and dissolved in physiological buffer. For hypoxia experiments, physiological salt solution was bubbled for 1 hour at room temperature with nitrogen (N2) gas, and pH was subsequently adjusted. N2-mediated hypoxia was maintained during the imaging experiments.
Real-time fluorescence imaging and high–resolution confocal microscopy
Real-time cell imaging experiments were performed on inverted wide-field epifluorescent microscopes, either the Axio Observer.Z1/7 or the Axio Vert.A1 (Zeiss, Germany). These microscopes are equipped with a Plan-Apochromat 20x/0.8 dry objective, a Plan-Apochromat 40x/1.4 DIC (UV) VIR-IR oil immersion objective, and monochrome CCD cameras Axiocam 503. Axio Observer.Z1/7 was equipped with an LED light source Colibri 7 R[C/Y] CBV-UV and Axio Vert.A1 with an LED light source Colibri 2 (365nm, 470 nm, 625nm). For ratiometric imaging of HyPer7, the Axio Observer.Z1/7 was used. Probes were alternately excited with the excitation wavelength of 423/44 nm and 469/38 nm using LED modules of Colibri 7, and emission was alternately collected using a motorized dual filter wheel equipped with the filter combinations: beam splitter FT455 (for HyPer low), FT495 (for HyPer high) and emission filter BP 525/50 for both channels. For the HyPer ratio, the GFP emission excited in the GFP channel was divided by the GFP emission excited in the CFP channel (F469/F423). Intensiometric HyPer 7 imaging was performed using Axio Observer.Z1/7 by exciting with 469/38 nm, and the emission was collected with the filter combinations FT495 (BS) and emission filter BP 525/50. Differentially targeted mCherry-mDAAO imaging with Axio Observer.Z1/7 was excited with 555/30 nm, and emission was collected with the filter combinations FT570 (BS) and emission filter BP 605/70. Intensiometric HyPer 7 imaging on Axio Vert.A1 was excited with the LED module 470 nm, and emission was collected with the filter cube combination number 38 (GFP). mCherry-mDAAO images were taken by excitation with the LED module white, and emission was collected with the filter cube combination 43 (DsRed). Data acquisition and control on both imaging systems was carried out using the Zen Blue 3.1 software (Zeiss, Germany). All imaging experiments were performed using a gravity-based or pump-driven custom perfusion system to supply or withdraw drugs or substrates to the cells. The perfusion chamber was purchased from NGFI (Graz, Austria). Camera binning for real-time imaging was set to 4x4, light intensities were adjusted between 1.5% and 5%, and exposure times were adjusted according to the expression rate set between 30 ms and 300 ms.
High-resolution confocal images were taken using a laser scanning confocal microscope LSM 800 (Zeiss, Germany) equipped with a Plan-Apochromat 40x/1.3 DIC (UV) VIS-IS oil immersion objective. Differentially targeted HyPer7 were excited with a 488 nm and 405 nm laser, and emissions were collected using a 509 nm filter system. Fluorescence detection was acquired with an A GaAsP-PMT detector and 400-565 nm filter using a Multialkali-PMT detector. mCherry-mDAAO constructs were excited using a 561 nm laser, and emission wavelength was captured between 616-700 nm. The digital detector gain for all channels was set at 1; detector gain was applied between 500-1000 V. Laser intensities were set between 0.90% to 0.95%, and the pinhole was set between 29-32 μm according to the expression level of the fluorescent proteins. Bright-field images were taken using a photodiode detector. Regions of interest were selected and processed using Zen Blue 3.1 software (Zeiss, Germany). All experiments have been conducted under normal air conditions (18 kPa) unless stated otherwise in the figure legends (see Figure 2).
Figure 2: Hypoxia and HyPer7 responses to DAAO and mDAAO activation:
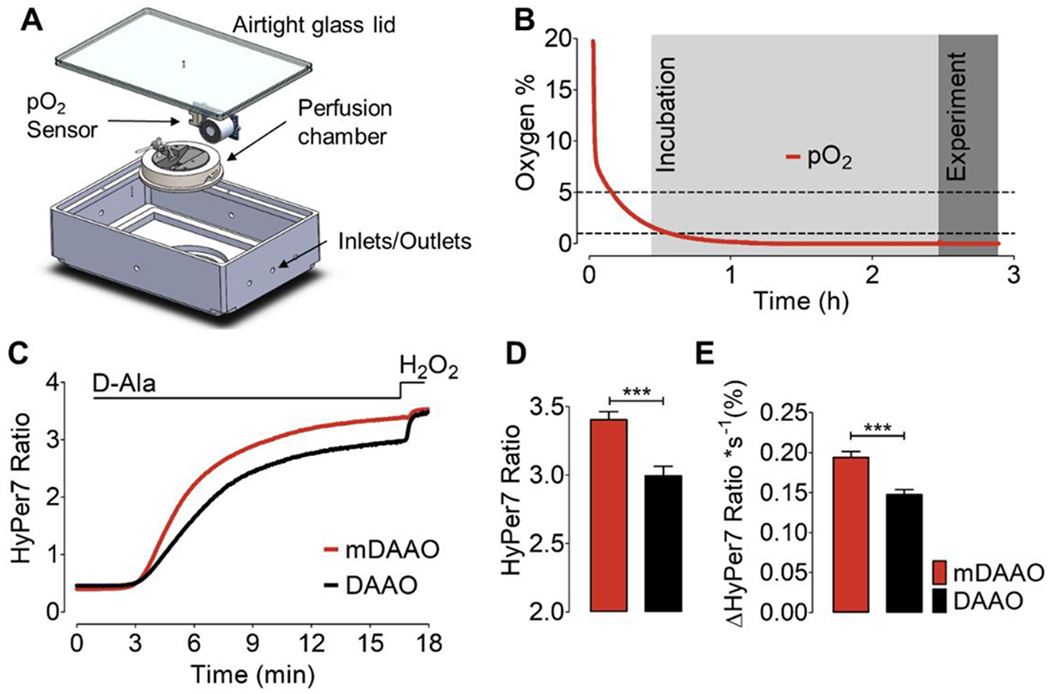
(A) This panel shows our design of a 3D-printed airtight hypoxia chamber equipped with an integrated oxygen sensor and perfusion chamber. (B) This panel shows a representative time course of pO2 levels measured in the sealed hypoxia chamber following continuous exposure to N2 gas. Light gray shows the duration of incubation of cells at ~1% pO2 for 2 hours before real-time imaging experiments. highlighted in dark grey. (C) This panel shows the HyPer7 ratio measured in HEK293 cells co-expressing the untargeted version of HyPer7 along with cytosol-targeted mCherry-mDAAO-NES or DAAO-NES in response to 10 mM D-Alanine, followed by addition of 100 μM H2O2 under hypoxic conditions. Cells were kept at ~1% O2 for 2 hours before the experiment. (D) This bar graph shows quantitative analysis of the maximum HyPer7 ratio as derived from the experiments shown in panel C. Bars show maximum HyPer7 ratio signals of HEK293 cells co-expressing either mDAAO (red bar, n=3/122) or DAAO (black bar, n=3/165) in response to 10 mM D-Alanine. (E) Bars represent kinetic analysis showing the maximum slope of HyPer7 ratio signals derived from the experiment shown in panel C. Unpaired t-test, two-tailed = p < 0.0001 P-value summary ***. All values are given as ±SEM.
Design and generation of an oxygen-controlled on-stage chamber and experimental procedure
The oxygen-controlled on-stage chamber was designed in computer-aided design (CAD) to fit the microscope stage of an Axio Observer.Z1/7. The chamber was 3D printed using stereolithography methods (Formlabs Form 3, Cat No: PKG-F3-WSVC-BASIC, USA) using white resin (Formlabs White Resin, Cat No: RS-F2-GPWH-04, USA). After 3D printing, the chamber was washed with isopropanol and cured by exposing the construct to 405 nm light at 65°C for 1h (Form Cure, Cat No: FH-CU-01, USA). For live-cell imaging under oxygen-controlled conditions, cells were placed in a perfusion chamber (NGFI, Austria) which was placed into an airtight hypoxia chamber sealed with a Plexiglas. Air in the chamber was displaced with 99.9% N2 gas using silicon tubing holes. O2 levels were measured using an Arduino IDE-based oxygen sensor integrated into the chamber (Grove Oxygen Sensor, Winsen, Cat No: 101020002, China). Buffers were also gassed with 99.9% N2 using a magnetic stirrer. When < 5% O2 levels were reached in the chamber, cells were incubated under low oxygen conditions (5% - 1% O2) for 2 hours before the experiment. After preincubation, real-time imaging experiments were performed using N2-gassed buffers.
Statistical analysis
All acquired imaging data were analyzed using GraphPad Prism software version 5.04 (GraphPad Software, San Diego, CA, USA). All experiments were repeated at least three times. The number of experiments is given as ‘N’, and the total number of cells imaged is indicated as ‘n.’ For instance: 3/18 indicates N=3 (triplicate) and n=18 (number of cells imaged in this particular experiment. All statistical data are presented as ±SD in addition to the representative real-time traces shown as curves (if not indicated otherwise). Statistical comparison of two groups was evaluated using a two-tailed Student t-test. Statistical significances were considered significant and indicated with “*”. Statistical comparisons of multiple groups, one-way ANOVA analyses of variances with post-test Bonferroni (comparison of all pairs of columns), or Dunnett (multiple comparison post test), or Mixed effect analysis (Tukey’s multiple comparison test) were performed. Concentration-response curves were calculated using non-linear regression (log agonist vs log response) and the application of the formula: Y=Bottom + (Top-Bottom)/(1+10^((LogEC50-X))). All single values for concentration-response were performed at least in triplicate.
Results
Expression and functional characterization of mDAAO in single cells
Figure 1A shows a schematic of DAAO catalysis, in which the yeast-derived enzyme oxidizes D-amino acids to their corresponding alpha-keto acids in the presence of FAD and molecular oxygen (O2), producing equimolar concentrations of H2O2, pyruvate, and ammonia (NH3). Due to the high intracellular concentrations of pyruvate and NH3, the generation of these DAAO co-products yields only a negligible effect on pyruvate and ammonia levels in cells, whereas there is a striking fold increase in levels of intracellular H2O2 following DAAO activation [32]. A mutant DAAO variant (termed mDAAO) has been developed that was found to have a ten-fold lower Km for the DAAO co-substrate O2 [33]. We hypothesized that this more O2-sensitive mDAAO mutant might have advantages over the original DAAO construct. To our knowledge, the mDAAO mutant has not previously been reported in chemogenetic applications. We generated mDAAO by introducing five distinct amino acid substitutions into the wild-type DAAO, which include S19G, S120P, Q144R, K321M, and A345V mutations as previously described [33]. We then made a chimeric construct in which the mDAAO was fused to a red fluorescent protein (mCherry) at the N-terminus of the mDAAO enzyme. For our initial characterizations of the mCherry-mDAAO constructs, we studied HEK293 cells, a cell line that can be efficiently transfected and imaged. To confirm the functionality of mDAAO, we employed real-time imaging of H2O2 using the recently developed genetically encoded biosensor HyPer7 [27]. HyPer7 permits the detection of chemogenetically-produced H2O2 with high spatial and temporal resolution as it reversibly and rapidly increases its fluorescence when oxidized by H2O2 (Figure 1A). D-alanine yields robust and reliable chemogenetic H2O2 signals in cells and tissues expressing mDAAO and HyPer7. To date, most of the cell- and tissue-based chemogenetic reports on DAAO have utilized D-alanine as the DAAO substrate to generate intracellular H2O2. We tested mDAAO functionality in HEK293 cells co-expressing mDAAO and HyPer7 by providing D-alanine (1 mM) to cells. As expected, D-alanine treatment led to a rapid increase of the HyPer7 ratio signal that plateaued within minutes, while L-alanine treatment did not affect the HyPer7 signal (Figure 1B–D), confirming the stereoselectivity of mDAAO for D-amino acids. Subsequent addition of 50 μM H2O2 directly to the cells in culture significantly increased the HyPer7 ratio in cells treated with L-alanine, but led to no further increase in the HyPer7 signal in cells that had been treated with D-alanine. These observations indicate that 1 mM D-alanine is sufficient to robustly and rapidly generate intracellular H2O2 to a level similar to that seen when 50 μM H2O2 is added exogenously.
Figure 1: Mechanism and stereoselectivity of DAAO:
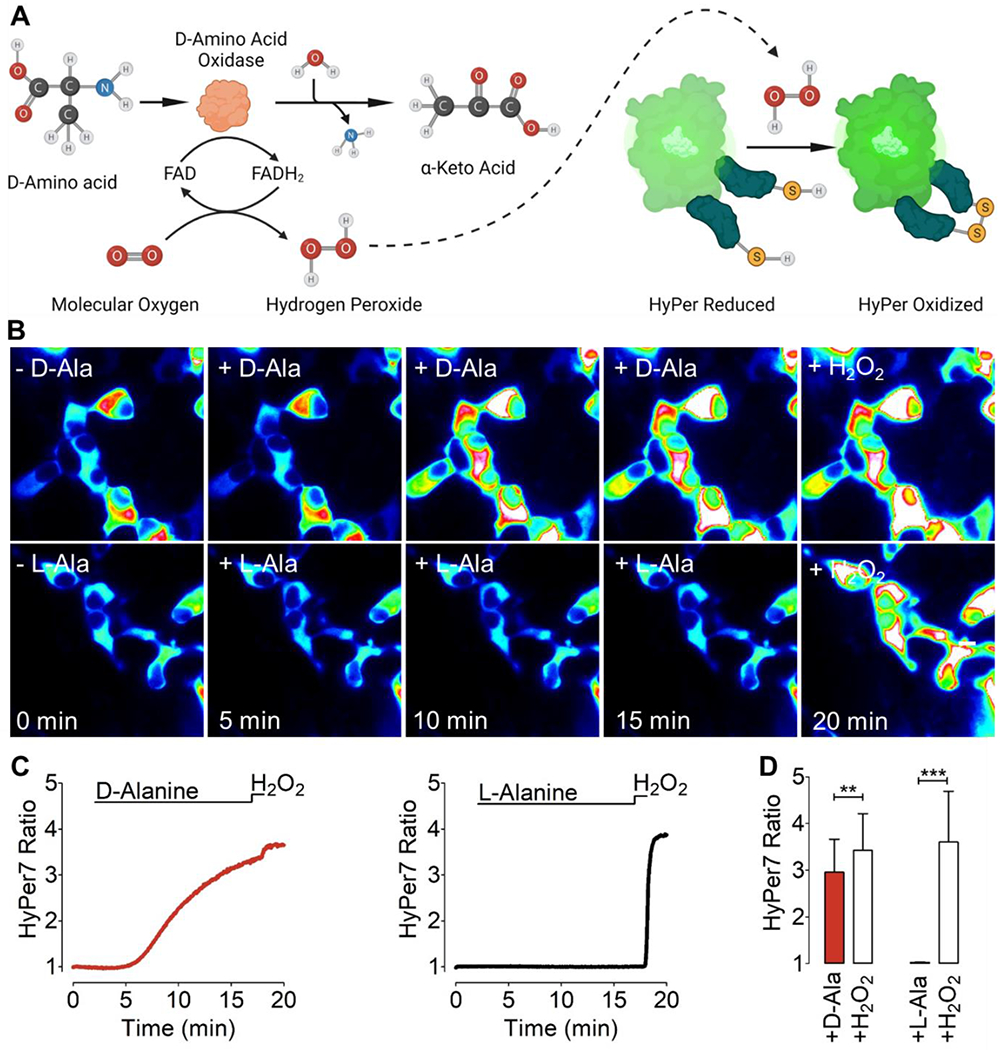
(A) The left part of the schematic in Panel A shows the generation of H2O2 following the DAAO-catalyzed oxidation of D-amino acids to the corresponding a-keto acid plus equimolar ammonia, with FAD serving as a cofactor and molecular O2 as the co-substrate. The right side of Panel A is a schematic showing that the H2O2 generated by DAAO can be detected by the genetically encoded biosensor HyPer7. (B) This panel shows representative intensiometric pseudo-colored wide-field images of HEK cells imaged over time in response to addition of 1 mM D-alanine (D-Ala, upper row) or L-alanine (L-Ala, lower row), followed by the addition of 50 μM H2O2. (C) The left panels show representative real-time traces quantitating the H2O2 signals (red curve) generated by cytosol-targeted mCherry-mDAAO-NES and visualized with cytosol-targeted HyPer7 in HEK293 cells following addition of 1 mM D-Alanine. The right panel shows the same treatment in response to 1 mM L-Alanine (black curve). In both experiments, cells were subsequently exposed to 50 μM exogenous H2O2. (D) The bar graph on the left show the maximum HyPer7 ratio signals that are observed in response to 1 mM D-Alanine (red bar, n=3/36) or to exogenous 50 μM H2O2 (white bar, n=3/36). The bar graph on the right shows the maximum HyPer7 ratio signals of in response to 1 mM L-Alanine (black bar, n=3/42) or exogenous H2O2 (white bar, n=3/42). Unpaired t-test, two-tailed = p < 0.0098 P-value summary **, p < 0.0001 P-value summary ***. All values are given as ±SD.
mDAAO is suitable for chemogenetic H2O2 generation under hypoxic conditions
Previous studies have shown that the mDAAO enzyme has significantly higher enzymatic activity at low O2 and low D-alanine concentrations compared to wild-type DAAO [33]. We compared the wild-type DAAO and mDAAO under hypoxic conditions utilizing a hypoxia chamber with an integrated oxygen sensor to control the pO2 levels in the chamber during imaging (Figure 2A). Before the imaging experiments, cells were placed in a metal superfusion chamber that was fixed in the hypoxia chamber under airtight conditions and continuously gassed with N2 for 2 hours to establish hypoxic conditions in the chamber and to allow cells to adapt to low oxygen (Figure 2B). We used a pump-driven custom-made superfusion system to provide and withdraw the imaging media and continuously purged the buffers and the hypoxia chamber with N2 gas. The rate of the maximum amplitude of the HyPer7 ratio in response to D-alanine administration to cells expressing mDAAO was significantly higher than for wild-type DAAO, indicating increased levels of H2O2 derived from mDAAO under hypoxic conditions (Figure 2C+D). Kinetic analyses showed that the rate of the HyPer ratio increase over time was also significantly higher in cells expressing mDAAO than with wild-type DAAO, further confirming the greater activity of mDAAO under hypoxic conditions (Figure 2E). Our observations demonstrate that both mDAAO and HyPer7 are suitable for live-cell imaging studies under hypoxic conditions, and show that mDAAO yields a more robust HyPer7 signal compared to wild-type DAAO.
Substrate and locale-dependent kinetic activity of mDAAO’s
We next generated differentially-targeted mCherry-mDAAO constructs to test the performance of these constructs in three specific subcellular locales: mitochondria, cytosol, and nucleus. We again used HEK293 cells, and co-transfected cells with the differentially-targeted mCherry tagged mDAAO and HyPer7 plasmid constructs. High-resolution laser scanning confocal microscopy images confirmed correct localization and similar expression levels of both constructs in a given cell (Figure 3A–C). Addition and withdrawal of D-alanine (10 mM) for a short period (300 seconds) evoked a rapid and strong H2O2 signal that plateaued within a few minutes in all three compartments tested. The recovery phase of the HyPer7 signal lasted for approximately 60 more minutes until the HyPer7 ratio signal returned to baseline. Subsequent D-alanine treatment with the same concentration and duration evoked similar HyPer7 profiles and levels as were observed following the initial addition of substrate (left panels of Figure 3D–F). These experiments verify the reversibility and functionality of both the H2O2 generator (mDAAO) and the H2O2 biosensor (HyPer7) in different subcellular locales. We next performed D-alanine concentration-response curves in the cytosol, nucleus, and mitochondria, and recorded the HyPer7 ratios. We observed a dose-dependent relationship between the concentration of added D-alanine and the HyPer7 ratio response in all three compartments. The half-maximal response (EC50) values in the three cell compartments were comparable, indicating that the substrate D-alanine can reach different subcellular locales to activate mDAAO and generate H2O2 (middle panels of Figure 3D–F). Kinetic analyses documented that the rate of increase in the HyPer7 ratio response to D-alanine also increased in a concentration-dependent manner (right panels of Figure 3). Overall, our data confirm that the mDAAO-dependent production of H2O2 in different subcellular locales can be modulated by varying the concentration of D-alanine provided to the transfected cells.
Figure 3: D-alanine dependent H2O2 generation by mDAAO targeted to subcellular locales.
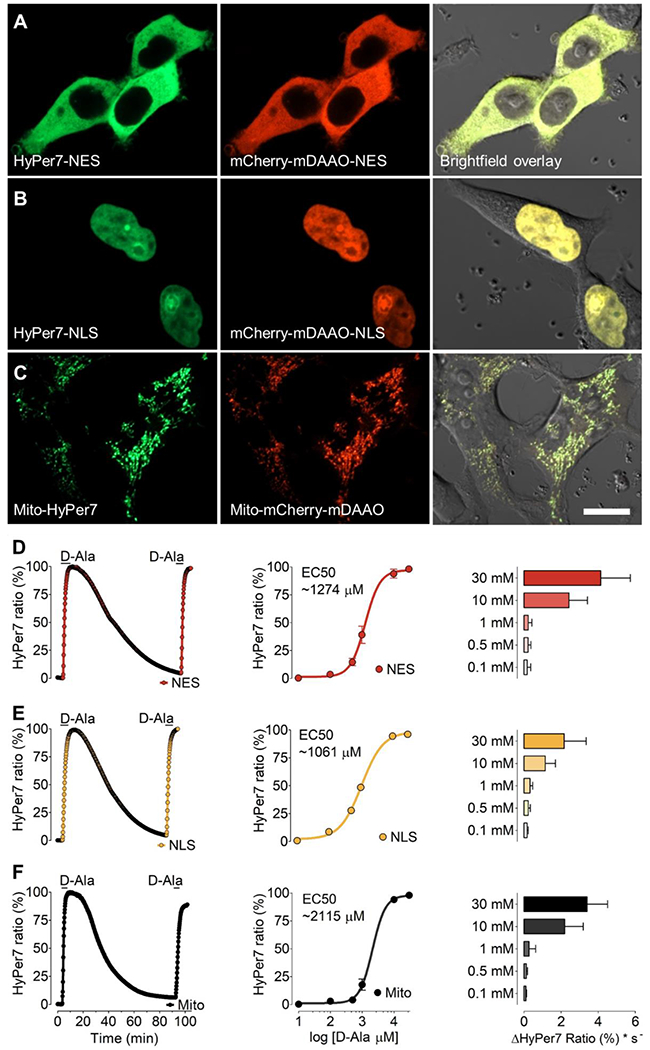
Representative confocal images of HEK293 cells co-expressing HyPer7 (left panels) and mCherry-mDAAO (middle panels) are shown in the cytosol (A) nucleus (B) or mitochondria (C). Right panels show phase-contrast images overlayed with the GFP and RFP channel. Scale bar represents 20 μm. (D) The left panel shows representative real-time traces of HyPer signals in response to the addition of 10 mM D-Alanine in HEK293 cells expressing the cytosolic mCherry-mDAAO-NES and HyPer7-NES. Middle panel shows normalized changes in maximum HyPer7-NES ratio signals in response to D-Alanine concentrations as shown, including 100 μM (n=3/61), 500 μM (n=3/56), 1 mM (n=3/44), 10 mM (n=3/45) and 30 mM (n=3/54). The right panel shows the initial maximum slope of the HyPer ratio in response to different D-Alanine concentrations derived from the data shown in the middle panel. Panel (E) shows the same experimental setup as presented in panel A in HEK293 cells expressing the nuclear-targeted mCherry-mDAAO-NLS and HyPer7-NLS. Normalized concentration response curve: 100 μM (n=3/65), 500 μM (n=3/61), 1 mM (n=3/54), 10 mM (n=3/58) and 30 mM (n=3/53). Experiments in panel (F) show results in HEK293 cells expressing mitochondria-targeted Mito-mCherry-mDAAO and Mito-HyPer7. Normalized concentration response curve: 100 μM (n=3/54), 500 μM (n=3/46), 1 mM (n=3/55), 10 mM (n=3/42) and 30 mM (n=3/57).One-way ANOVA and Bonferroni’s multiple comparison post-test were applied to compare all columns with each other. Concentration-response curves were calculated using non-linear regression (log agonist vs log response) and the application of the formula: Y=Bottom + (Top-Bottom)/(1+10^((LogEC50-X))). Concentration-response curves are shown as ±SEM values and slope analysis as ±SD.
Different D-amino acids allow temporal manipulation of H2O2 levels in cells
In mammalian cells, amino acid transporters appear to broadly distributed in different subcellular locales, including the plasma membrane [34], the inner mitochondrial membrane [35], and lysosomal membranes [36]. We next sought to test mDAAO-dependent H2O2 generation following the addition of different D-amino acids to the transfected cells. We again used HEK293 cells and tested the kinetics of H2O2 generation – measured by HyPer7 ratio – in response to different D-amino acids, each at a concentration of 1 mM. We chose amino acids from chemically distinct classes, including polar (D-serine), charged (D-arginine), hydrophobic (D-alanine, D-phenylalanine, D-tryptophan and D-valine), and sulfur-containing (D-cysteine and D-methionine) amino acids. Considering D-alanine as the standard, treatment with 1 mM D-alanine evoked a robust H2O2 signal and plateaued with a rate of 3.58 ± 2.11 ΔF*s−1 (Figure 4A). The hydrophobic amino acid D-valine yielded a much lower HyPer7 signal (0.9 ± 0.31 ΔF*s−1). The aromatic amino acids D-phenylalanine and D-tryptophan yielded a robust HyPer7 signal (6.71 ± 3.72 ΔF*s−1 and 4.46 ± 2.41 ΔF*s−1, respectively) compared to D-alanine. The positively charged amino acid D-arginine (1.24 ± 0.82 ΔF*s−1) elicited only a weak HyPer7 response while polar uncharged amino acid D-serine (3.87 ± 1.71 ΔF*s−1) displayed comparable on-kinetics to D-alanine. Much to our surprise, the sulfur-containing amino acids D-cysteine (25.10 ± 10.34 ΔF*s−1) and D-methionine (52.51 ± 20.37 ΔF*s−1) produced extremely robust H2O2 signals that plateaued within a few seconds (Figure 4A). The kinetics and magnitude of HyPer signals that are observed following addition of different D-amino acids (Figure 4A) reflect multiple independent parameters, including the rates of D-amino acid uptake transport into the cells, the differential distribution of D-amino acid into cellular compartments, as well the rates of enzyme catalysis for different D-amino acid substrates. As shown in Figure 4B, 1 mM D-alanine treatment in HEK293 cells induced a robust HyPer7 signal. However, under the same experimental setup, a mixture of 1 mM D-alanine and 1 mM L-alanine yielded a significantly lower HyPer7 signal. Increasing the D-alanine concentration relative to the L-alanine concentration yielded considerably higher HyPer7 signals. The subsequent withdrawal of L-alanine from the cell media while maintaining the same D-alanine concentration led to a stable increase of the HyPer7 signal (Figure 4C). These observations suggest there is competition between D- and L-amino acids at the level of amino acid transporters - even if the mDAAO enzyme itself is highly stereoselective for the D enantiomer.
Figure 4: H2O2 generation by mDAAO in response to different D-amino acids.
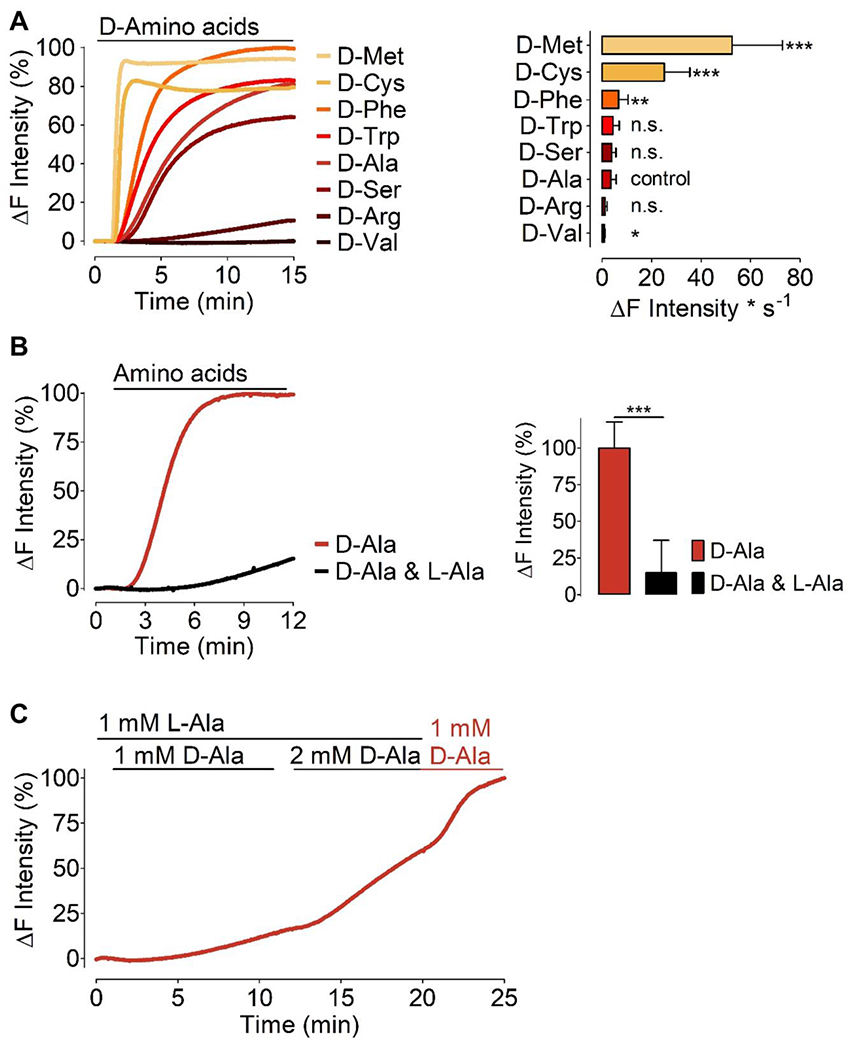
(A) Left panel shows normalized average curves of HyPer7 signals in HEK293 cells expressing HyPer7-NLS and mCherry-mDAAO-NLS in response to different D-amino acids as indicated in the figure. Bars in the right panel show maximum initial slope in response to various D-amino acids (all added at 1 mM) as indicated: D-Methionine (n=3/89), D-Cysteine (n=3/71), D-Phenylalanine (n=3/89), D-Tryptophan (n=3/81), D-Serine (n=3/75), D-Alanine (n=10/254), D-Arginine (n=3/89), and D-Valine (n=3/87). D-alanine has been defined as the control group, and all D-amino acids were compared to the kinetics of D-alanine using One-way ANOVA analysis, Dunnett’s multiple comparison post-test comparison of all columns vs. control column (D-alanine) p < 0.05 P-value summary *** or ** or *. (B) Left panel shows normalized average curves of H2O2 signals in HEK293 cells co-expressing mCherry-mDAAO-NLS and HyPer7-NLS in response to 1 mM D-alanine (red curve, n=3/70) or 1 mM D-alanine and 1 mM L-alanine (black curve, n=3/68). The right panel shows statistical analysis of the experiments shown in the left panel. Unpaired t-test, two-tailed = p < 0.0001 P-value summary ***. All values are given as ±SD. (C) Average curve (n=20 cells) of real-time HyPer traces shows results obtained in HEK cells co-expressing nuclear-targeted mCherry-mDAAO-NLS and HyPer7-NLS. Cells were pre-incubated in 1 mM L-Alanine for 1 hour prior to imaging and during imaging experiments consecutively provided with L-Ala or D-Ala at the concentrations indicated in the figure.
Real-time imaging of H2S generation with mDAAO
Hydrogen sulfide (H2S) is a gaseous second messenger involved in many biological functions, and may have cytoprotective effects in oxidative stress [37]. H2S can be produced from L-cysteine by cystathionine β-synthase, cystathionine-γ-lyase, and 3-mercaptopyruvate sulfurtransferase. mDAAO’s can also generate H2S from D-cysteine when the 3-mercaptopyruvate produced by mDAAO is converted into pyruvate plus H2S [38]. Figure 5A shows a schematic of the D-cysteine-dependent pathway. In analogy to H2O2 generation with mDAAO and detection with HyPer7, we used the genetically encoded H2S biosensor hsGFP to visualize H2S [28]. hsGFP contains a synthetic amino acid in the chromophore that becomes reduced in the presence of H2S, which leads to an increase in fluorescence (Figure 5A, right part). To test the generation of H2S by mDAAO in response to sulfur-containing amino acids, we again used HEK293 cells and sequentially added 1 mM D-alanine, D-methionine, and D-cysteine. As expected, D-cysteine but not D-alanine nor D-methionine generated robust H2S signals (Figure 5B). It should be noted that the oxidation of D-cysteine by mDAAO generates both H2O2 as well as H2S, which confounds the interpretation of experiments using D-cysteine to selectively probe the roles of H2S.
Figure 5: Mechanism of mDAAO-dependent H2S generation and detection in response to D-cysteine.
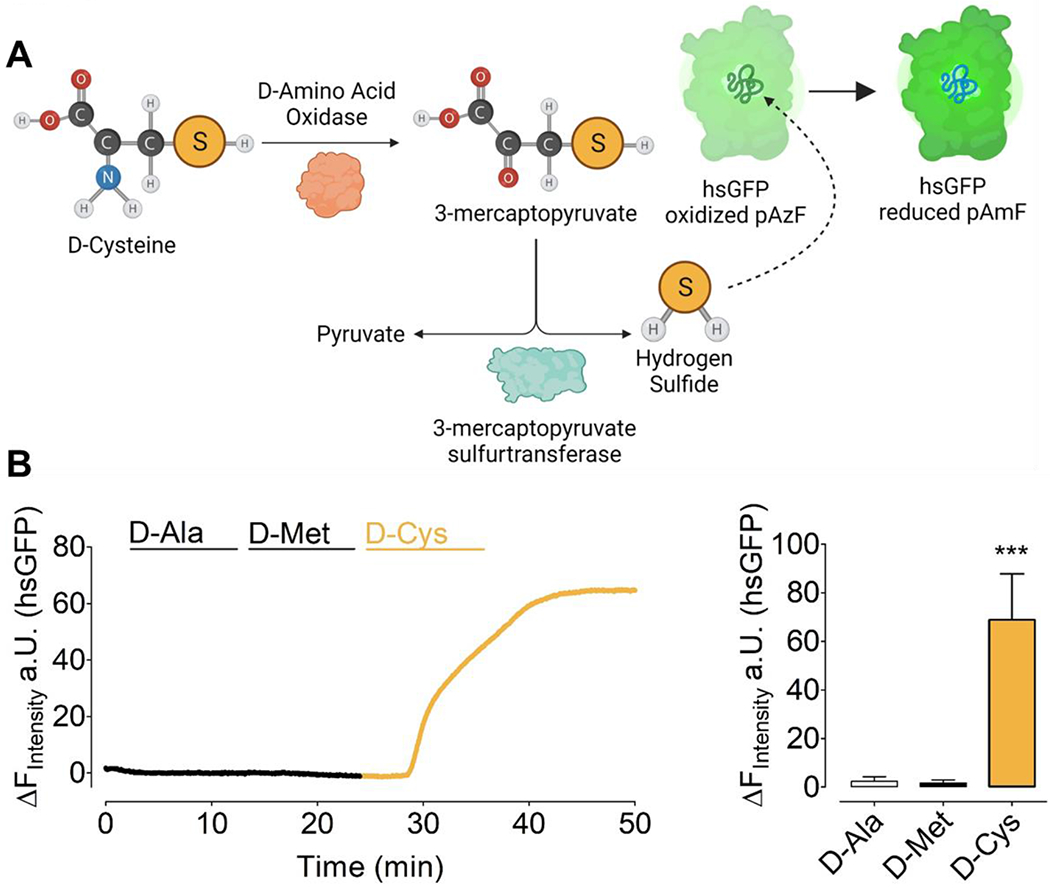
(A) This left hand portion of this schematic represents the simplified biochemical pathway for the oxidation of D-Cysteine by mDAAO to 3-mercaptopyruvate, leading to formation of H2S.The right hand portion of the schematic shows reduction of the genetically encoded hsGFP biosensor upon its interaction with H2S. (B) The left panel shows representative real-time traces of chemogenetically-produced H2S in response to 1 mM D-Alanine, D-Methionine, and D-Cysteine, respectively, in HEK293 cells co-expressing untargeted hsGFP and mCherry-mDAAO-NLS. The bar graph shows statistical analysis of the maximum response of hsGFP in response to 1 mM D-Alanine (n=27, white bar), 1 mM D-Methionine (n=27, black bar), 1 mM D-Cysteine (n=27, yellow bar). One-way ANOVA and Bonferroni’s multiple comparison post-test were applied to compare all columns with each other, p < 0.0001 P-value summary ***. All values are given as ±SD.
Chemogenetically induced H2O2 in different cell types
We next extended these experimental approaches from HEK293 cells to other cultured cell lines. We co-transfected cancer and endothelial cell lines with nucleus-targeted HyPer7 and mCherry-mDAAO, including HCT116, HeLa, U-87 MG, EA.hy926, or HEK293 cells. As shown in Figure 6A, transfected HEK293 cells treated with 1 mM D-alanine yielded a rapid HyPer7 signal (0.77 ± 0.33 ΔF*s−1), while the other cell lines responded more slowly; HCT116 (0.37 ± 0.10 ΔF*s−1), EA.hy926 (0.26 ± 0.28 ΔF*s−1), U-87 MG (0.24 ± 0.12 ΔF*s−1), and HeLa cells (0.21 ± 0.05 ΔF*s−1). These data suggest that D-alanine treatment of different cell lines transfected with mDAAO leads to variable rates of intracellular H2O2 responses, although all cell types showed similar mDAAO expression (Figure 6A, inset). We next tested the effects of D-methionine on transfected HEK293 cells, and found that 1 mM D-methionine led to a robust HyPer7 response that plateaued within several seconds (6.60 ± 2.27 ΔF*s−1). In contrast, the immortalized endothelial cell line EA.hy926 showed significantly slower on kinetics (1.44 ± 0.89 ΔF*s−1), while the cancer cell lines U-87 MG (7.20 ± 3.99 ΔF*s−1), HeLa (12.13 ± 5.70 ΔF*s−1) and HCT116 (13.75 ± 4.20 ΔF*s−1) rapidly generated H2O2 following the addition of 1 mM D-methionine but not to D-alanine (Figure 6B). As shown in Figure 6C 1 mM D-methionine yielded significantly higher HyPer7 amplitudes in all cell lines compared to 1 mM D-alanine. Moreover, D-methionine derived HyPer7 signals appeared to be significantly faster compared to 1 mM D-alanine (Figure 6D).
Figure 6: Expression of mDAAO and H2O2 generation in different cell types.
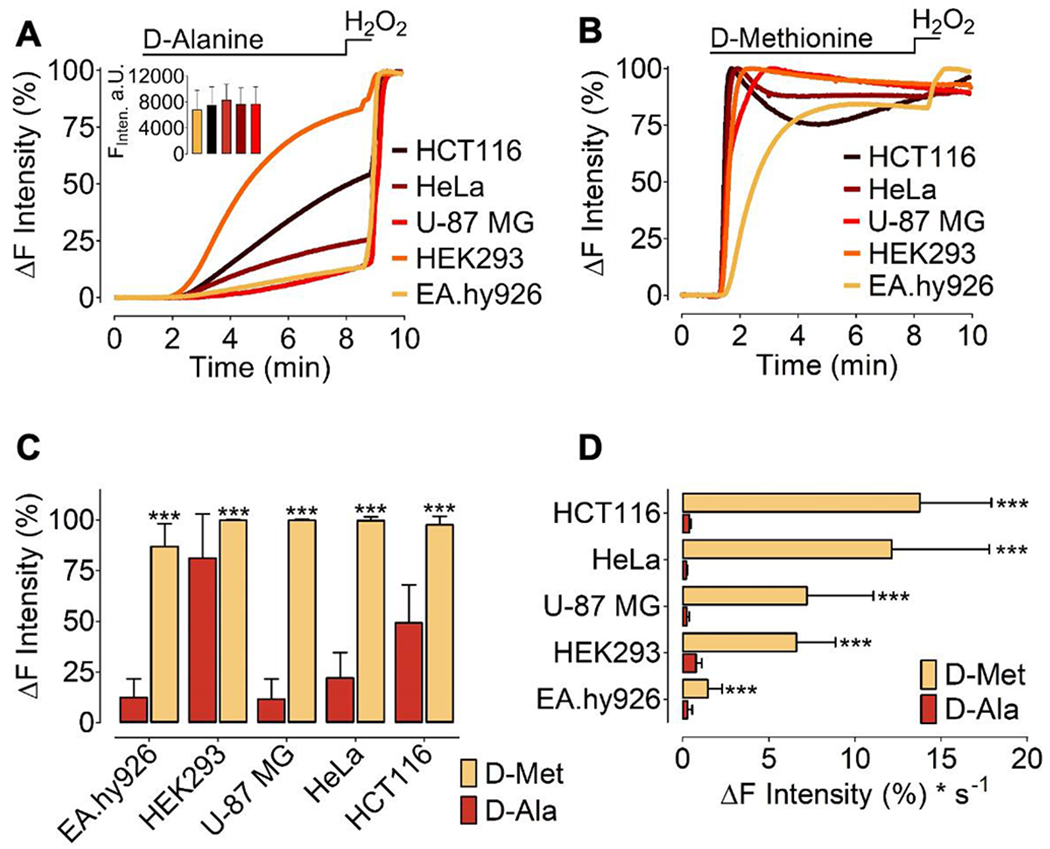
(A) Normalized average curves of HyPer7 signals measured in different cell types (as indicated in the figure) expressing mCherry-mDAAO-NLS and HyPer7-NLS in response to 1 mM D-alanine or (B) 1 mM D-methionine. (A) The inset shows statistical analysis of the mean fluorescence intensity of mCherry-mDAAO-NLS expressed in theEA.hy926 (n=60), HCT116 (n=59), HEK293 (n=61), HeLa (n=59), U87-MG (n=26). Mixed-effects analysis with Tukey’s multiple comparison post-test was performed p>0,9999 (n.s.) (C) Normalized maximum HyPer7 signals in response to 1 mM D-alanine or 1 mM D-methionine in different cell lines as indicated in the figure. Shown are the cell lines HTC116 (D-met, n=4/50; D-ala, n=3/44); HeLa (D-met, n=4/67; D-ala, n=3/53); U-87 MG (D-met n=2/16; D-ala, n=5/19); HEK293 (D-met, n=3/62; D-ala, n=3/83); and EA.hy926 cells (D-met, n=4/9; D-ala, n=5/26). (D) Bars show kinetics of the HyPer7 signal comparing signals seen following addition of 1 mM D-alanine or 1 mM D-methionine in different cell types. HTC116 (D-met, n=4/50; D-ala, n=3/44), HeLa (D-met, n=4/67; D-ala, n=3/53), U-87 MG (D-met n=2/16; D-ala, n=5/19), HEK293 (D-met, n=3/62; D-ala, n=3/83), EA.hy926 (D-met, n=4/9; D-ala, n=5/26). Unpaired t test was applied: p < 0.0001 (Two-tailed) P-value summary ***. All values are shown as ±SD.
Discussion
In these studies, we have pursued multiparametric imaging approaches using the mutated DAAO variant mDAAO, and we have expanded the toolbox and the possible experimental approaches that exploit chemogenetic tools for analyses of intracellular redox balance. To characterize intracellular H2O2 signals derived from mDAAO in single cells, we paired this enzyme with the novel ultrasensitive genetically encoded biosensor HyPer7 [27] and have shown the chemogenetic enzyme’s functionality and stereoselectivity for D-amino acids (Figure 1). The various HyPer biosensors have been well established as probes of intracellular H2O2[17] in many experimental systems [6,9–11,15,32]. In addition, many other redox probes such as NeonOxIrr [39], probes of the roGFP family [18,40,41], UFP-Tyr66pBoPhe [42], OxyFRET [43], Apollo NADP+ [44], NADP-Snifit [45], or the iNAP biosensors [46] family are also useful for detection of intracellular redox responses in single cells. Here we have exploited the ultrasensitive HyPer7 biosensor to investigate kinetic properties of mDAAO in chemogenetically generated H2O2 levels in subcellular locales of cultured cells (Figures 3–6).
Previous studies of mDAAO catalysis have shown that the mDAAO variant is more active at lower substrate concentrations than the wild-type enzyme, both for molecular O2 and for various D-amino acid substrates [33]. The lower substrate requirements of mDAAO represent a critical feature that may have advantages over the original DAAO construct for many in vivo and in vitro applications. Such experimental model systems include long-term cultured cells under oxygen-controlled conditions [47], ischemia-reperfusion injury model systems in cultured cells [48], or solid (cancer) organoid model systems [49]. Employing real-time imaging experiments in cultured cells using a custom-made hypoxia chamber, we confirmed the first time in live cells that the mDAAO variant indeed performed better than wild-type DAAO under hypoxic conditions (Figure 2). Also, these experiments demonstrate the functionality of the HyPer7 biosensor under hypoxic conditions. However, we acknowledge several limitations to this hypoxia imaging system. We preconditioned cells for only 2 hours before the imaging experiment in this setup representing acute hypoxic conditions. Future studies with cells adapted to long-term normoxia/hypoxia conditions will help us to more completely understand the functionality of mDAAO combined with HyPer (or other relevant biosensors for detecting H2O2) and the chemogenetic enzymes. Notably, many fluorescent protein-based genetically encoded biosensors rely on O2 to adopt the fluorescent state [50]. Thus, oxygen-dependent maturation of the chromophore and functionality of the probe may be affected under hypoxic conditions, and this may alter the signal observed. Despite the differences in the redox states that are found in different subcellular organelles [51] and the existence of intracellular membrane barriers with distinct biochemical properties, chemogenetic approaches can be devised to generate and control H2O2 generation in different subcellular locales (Figure 3). Our data confirm that the mCherry-mDAAO chimera can be readily expressed and remains fully functional in different organelles. This is a critical finding, as the genetic encodability of mDAAO permits its subcellular localization to virtually any cellular compartment using appropriately designed recombinant targeting constructs. More importantly, D-alanine appears to reach these various subcellular locales with similar kinetics, and mDAAO can generate H2O2 in different organelles in a concentration-dependent manner (Figure 3). These important features of the mDAAO-HyPer7 approach permit experimenters to control oxidative eustress or distress with high spatial and temporal resolution.
Another parameter that allows the dynamic control of chemogenetic H2O2 generation in cells is the choice of specific D-amino acids as substrates for recombinant mDAAO, which represents a powerful tool to modulate intracellular H2O2 either for acute or more sustained ROS stress (Figure 4). D-alanine is currently the gold standard as a DAAO substrate because it can readily be metabolized to its final product and does not affect cellular pH levels; moreover, D-alanine is quite inexpensive [8]. In addition, pyruvate is present in cells in high concentrations, and thus there are only nominal changes in intracellular pyruvate following the oxidation of D-alanine to pyruvate by DAAO. Our findings also document for the first time that the substrate type dependent chemogenetic H2O2 generation in single cells is a critical parameter that can be exploited to more accurately control oxidative stress generation. However, one needs to consider that different D-amino acid-derived byproducts yield different α-keto acids upon oxidative deamination by DAAO. For example, D-methionine is converted to 2-keto-4 (methylthio) butanoic acid [52]. D-cysteine administration to DAAO yields 3-mercaptopyruvate [38], instead of the pyruvate that is produced from D-alanine. The DAAO-catalyzed oxidation of D-phenylalanine produces phenyl pyruvic acid [53]. With D-tryptophan as its substrate, DAAO generates indole-3-pyruvic acid [54]. In addition, other amino acids used in this study may affect local pH [8], which can confound experiments using genetically-encoded biosensors since most of these biosensors are pH-sensitive [55,56].
Here we employed the relatively pH stable biosensor HyPer7; however, attention and awareness are required when using different D-amino acids. We found that the aromatic amino acids D-tryptophan and D-phenylalanine allow for the generation of rapid intracellular H2O2 even in the cell nucleus within seconds after providing the substrate to cells (Figure 4A). In contrast, D-valine, and D-arginine show slower kinetics of intracellular H2O2 production from mDAAO, and these amino acids may be more suitable to yield chemogenetic H2O2 generation over a more extended period. Taken together, these findings indicate that the choice of the specific D-amino acid used for chemogenetic generation of H2O2 can affect the cellular consequences of mDAAO activation. We conducted these experiments in a simplified cell media consisting of physiologically essential salts and lacking any amino acids except for D-amino acids. Indeed, under cell culture conditions, the nutrient medium often consists of L- amino acids, macromolecules, and other biochemicals to maintain cell functionality- which may affect the amino acid uptake machinery or even DAAO activity. Here we tested the consequences of adding L-alanine together with D-alanine (Figure 4B). Our findings confirm that mDAAO-generated H2O2 level was significantly reduced when cells were provided with equal amounts of D- and L-alanine (Figure 4C), suggesting that these two enantiomers compete with one another. The mDAAO enzyme does not act on L-alanine as a substrate, exhibiting exquisite stereoselectivity. Thus, the inhibitory effects of L-alanine on D-alanine-dependent mDAAO activation probably reflects competition at the level of amino acid transport into the cells. Indeed, most amino acid transporters exhibit only modest stereoselectivity [57]. This hypothesis is in line with former studies with recombinant DAAO’s demonstrating that L-amino acids are neither substrates nor inhibitors for the enzyme [58,59]. Overall, we conclude from these observations that the presence of L-amino acids under some experimental conditions can importantly influence the intracellular generation of H2O2 by mDAAO by interfering with D-alanine transport.
In 2013 a new biosynthetic pathway was identified for the generation of H2S from D-cysteine involving DAAO in mice brains and kidneys [38]. Shibuya et al. demonstrate that D-cysteine-derived H2S has cytoprotective effects on cultured cerebellar neurons against H2O2. Our results employing mDAAO paired with the H2S sensor hsGFP confirm the enzyme’s capability to robustly generate H2S from the substrate D-cysteine, but not when D-methionine or D-alanine are provided (Figure 5). However, our data also suggest that interpretation of cellular responses using D-cysteine as a mDAAO substrate can be confounded due to the generation of both species, H2S and H2O2 (Figures 4 and 5). Future studies employing multispectral co-imaging of H2O2 and H2S using optimized and spectrally distinct biosensors may provide new information on the cellular metabolism and signaling properties of these two important biomolecules.
We also explored the features of mDAAO expressed in different cell types, including cancer cell lines, immortalized endothelial cells, and HEK cells (Figure 6). To our surprise, we observed striking differences in the kinetics of the HyPer responses between different cells in response to the same D-alanine concentration. Although the expression levels for mDAAO was similar in all cell types investigated, only HEK cells showed robust chemogenetic H2O2 generation in response to D-alanine (Figure 6A). But in contrast to the variable mDAAO responses to D-alanine in different cells, D-methionine evoked a reproducibly robust and rapid intracellular H2O2 signal in all cell types (Figure 6B). Thus, the addition of D-methionine to all cells expressing mDAAO generated significantly higher levels of H2O2 with more rapid activation kinetics compared to D-alanine (Figure 6C+D). Variations in the HyPer response to D-alanine might be caused by differences in the expression levels of amino acid transporters [15].
Our data demonstrate that there is no absolute general rule in the application of chemogenetic tools to induce oxidative eustress or distress in different cell types and under different experimental conditions. In order to most effectively validate and compare cellular responses in different models, we recommend a systematic characterization of key experimental parameters–the identity and concentration of the D-amino acid used, the treatment period and culture conditions, and the subcellular locale of the recombinant DAAO– for any given model system.
Conclusion
Multiparametric manipulation and imaging of intracellular redox-pathways exploiting chemogenetic enzymes paired with informative biosensors represent novel and powerful techniques that have recently been added to the experimental toolbox of redox scientists. Here we have characterized the variant D-amino acid oxidase mDAAO in the context of genetically encoded biosensors for H2O2 and H2S. We provide detailed information on how to manipulate single cells controlling multiple parameters that need consideration when investigating redox pathways with the aid of chemogenetic tools and redox biosensors. We found striking differences in the D-alanine activation kinetics of mDAAO between different cultured cell lines, yet we found that D-methionine led to robust mDAAO activation in all the cell types we studied. Our findings help to demonstrate that multiparametric real-time imaging methods are valuable tools to manipulate and dissect redox signaling pathways in cells. We anticipate that the work presented here will be helpful to scientists who are seeking to apply chemogenetic tools in different experimental conditions and to understand the roles of H2O2.
Highlights.
mDAAO catalyzes H2O2 generation even under relatively hypoxic conditions
Different D-amino acids differentially promote H2O2 generation by recombinant mDAAO
Targeted mDAAO can reversibly generate H2O2 in discrete subcellular locales
H2O2 generated by mDAAO in different cell types yields distinct cellular responses
mDAAO catalyzes the oxidation of D-cysteine to yield H2S, which can be visualized with hsGFP in single cells
Acknowledgment
This research was supported by funds from the Scientific and Technological Research Council of Turkey Grant 118C242 (to E.E., T.A.C., H.Y.A., M.S., G.S., and B.N.A) the BAGEP award of the Science Academy Turkey with funds of the Integration Projects of Sabanci University EPD-2019-1 (to E.E., Y.C.E., Z.C., and A.G.Z.), Austrian Research Fund J-4113 (to E.E.); grant 075-15-2019-1789 from the Ministry of Science and Higher Education of the Russian Federation (to V.V.B.); and by National Institutes of Health Grants R21 AG063073 R01 HL152173, and R33 HL157918, and by the Brigham Biomedical Research Institute Fund to Sustain Research Excellence (to TM). Figures 1, 5, and graphical abstract were adapted from images created with BioRender.com (Agreement number: Figure 1: YI22Z9CJQI; Figure 5: ED22Z9C8JK; graphical abstract: EC22Z9C68G)
Nonstandard Abbreviations and Acronyms
- DAAO
D-amino acid oxidases
- mDAAO
mutated D-amino acid oxidase
- HyPer
genetically-encoded hydrogen peroxide biosensor
- hsGFP
genetically-encoded hydrogen sulfide biosensor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sies H, Jones DP, Reactive oxygen species (ROS) as pleiotropic physiological signalling agents, Nat. Rev. Mol. Cell Biol 21 (2020) 363–383. 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- [2].Sies H, Oxidative eustress and oxidative distress: Introductory remarks, Academic Press, (2020) 3–12. 10.1016/B978-0-12-818606-0.00001-8. [DOI] [Google Scholar]
- [3].Serrander L, Cartier L, Bedard K, Banfi B, Lardy B, Plastre O, Sienkiewicz A, Schlegel W, Krause K-H, NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation, Biochem. J 406 (2007) 105–114. 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schieber M, Chandel NS, ROS function in redox signaling and oxidative stress, Curr. Biol 24 (2014) R453–R462. 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sorrentino A, Michel T, Redox à la carte: Novel chemogenetic models of heart failure, Br. J. Pharmacol 177 (2020) 3162–3167. 10.1111/bph.15093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Saeedi Saravi SS, Eroglu E, Waldeck-Weiermair M, Sorrentino A, Steinhorn B, Belousov V, Michel T, Differential endothelial signaling responses elicited by chemogenetic H2O2 synthesis, Redox Biol. 36 (2020) 101605. 10.1016/j.redox.2020.101605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Forman HJ, Use and abuse of exogenous H2O2 in studies of signal transduction, Free Radic. Biol. Med 42 (2007) 926. 10.1016/j.freeradbiomed.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bogdanova YA, Schultz C, Belousov VV, Local Generation and Imaging of Hydrogen Peroxide in Living Cells, Curr. Protoc. Chem. Biol 9 (2017) 117–127. 10.1002/cpch.20. [DOI] [PubMed] [Google Scholar]
- [9].Steinhorn B, Sorrentino A, Badole S, Bogdanova Y, Belousov V, Michel T, Chemogenetic generation of hydrogen peroxide in the heart induces severe cardiac dysfunction, Nat. Commun 9 (2018) 1–10. 10.1038/s41467-018-06533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sorrentino A, Steinhorn B, Troncone L, Saravi SSS, Badole S, Eroglu E, Kijewski MF, Divakaran S, Di Carli M, Michel T, Reversal of heart failure in a chemogenetic model of persistent cardiac redox stress, Am. J. Physiol. Heart Circ. Physiol 317 (2019) H617–H626. 10.1152/ajpheart.00177.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eroglu E, Saeedi Saravi SS, Sorrentino A, Steinhorn B, Michel T, Discordance between eNOS phosphorylation and activation revealed by multispectral imaging and chemogenetic methods, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 20210–20217. 10.1073/pnas.1910942116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S, Genetically encoded fluorescent indicator for intracellular hydrogen peroxide, Nat. Methods 3 (2006) 281–286. 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- [13].Bilan DS, Pase L, Joosen L, Gorokhovatsky AY, Ermakova YG, Gadella TWJ, Grabher C, Schultz C, Lukyanov S, Belousov VV, HyPer-3: A genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging, ACS Chem. Biol 8 (2013) 535–542. 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- [14].Ermakova YG, Bilan DS, Matlashov ME, Mishina NM, Markvicheva KN, Subach OM, Subach FV, Bogeski I, Hoth M, Enikolopov G, Belousov VV, Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide, Nat. Commun 5 (2014). 10.1038/ncomms6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Matlashov ME, Belousov VV, Enikolopov G, How much H2O2 is produced by recombinant D-Amino acid oxidase in mammalian cells?, Antioxidants Redox Signal. 20 (2014) 1039–1044. 10.1089/ars.2013.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eroglu E, Charoensin S, Bischof H, Ramadani J, Gottschalk B, Depaoli MR, Waldeck-Weiermair M, Graier WF, Malli R, Genetic Biosensors for Imaging Nitric Oxide in Single Cells, Free Radic. Biol. Med 128 (2018) 50–58. 10.1016/j.freeradbiomed.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bilan DS, Belousov VV, HyPer Family Probes: State of the Art, Antioxidants Redox Signal. 24 (2016) 731–751. 10.1089/ARS.2015.6586. [DOI] [PubMed] [Google Scholar]
- [18].Morgan B, Van Laer K, Owusu TNE, Ezerina D, Pastor-Flores D, Amponsah PS, Tursch A, Dick TP, Real-time monitoring of basal H 2 O 2 levels with peroxiredoxin-based probes, Nat. Chem. Biol 2016 126. 12 (2016) 437–443. 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
- [19].Eroglu E, Gottschalk B, Charoensin S, Blass S, Bischof H, Rost R, Madreiter-Sokolowski CT, Pelzmann B, Bernhart E, Sattler W, Hallstrom S, Malinski T, Waldeck-Weiermair M, Graier WF, Malli R, Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics, Nat. Commun 7 (2016) 1–11. 10.1038/ncomms10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Eroglu E, Hallström S, Bischof H, Opelt M, Schmidt K, Mayer B, Waldeck-Weiermair M, Graier WF, Malli R, Real-time visualization of distinct nitric oxide generation of nitric oxide synthase isoforms in single cells, Nitric Oxide. 70 (2017) 59–67. 10.1016/j.niox.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen Z, Zhang S, Li X, wang Ai H, A high-performance genetically encoded fluorescent biosensor for imaging physiological peroxynitrite, Cell Chem. Biol (2021). 10.1016/j.chembiol.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kostyuk AI, Panova AS, Bilan DS, Belousov VV, Redox biosensors in a context of multiparameter imaging, Free Radic. Biol. Med 128 (2018) 23–39. 10.1016/j.freeradbiomed.2018.04.004. [DOI] [PubMed] [Google Scholar]
- [23].Eroglu E, Rost R, Bischof H, Blass S, Schreilechner A, Gottschalk B, Depaoli MR, Klec C, Charoensin S, Madreiter-Sokolowski CT, Ramadani J, Waldeck-Weiermair M, Graier WF, Malli R, Application of Genetically Encoded Fluorescent Nitric Oxide (NO•) Probes, the geNOps, for Real-time Imaging of NO• Signals in Single Cells, JoVE (Journal Vis. Exp. 2017 (2017) e55486. 10.3791/55486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Depaoli MR, Bischof H, Eroglu E, Burgstaller S, Ramadani-Muja J, Rauter T, Schinagl M, Waldeck-Weiermair M, Hay JC, Graier WF, Malli R, Live cell imaging of signaling and metabolic activities, Pharmacol. Ther 202 (2019) 98–119. 10.1016/j.pharmthera.2019.06.003. [DOI] [PubMed] [Google Scholar]
- [25].Charoensin S, Eroglu E, Opelt M, Bischof H, Madreiter-Sokolowski CT, Kirsch A, Depaoli MR, Frank S, Schrammel A, Mayer B, Waldeck-Weiermair M, Graier WF, Malli R, Intact mitochondrial Ca2+ uniport is essential for agonist-induced activation of endothelial nitric oxide synthase (eNOS), Free Radic. Biol. Med 102 (2017) 248–259. 10.1016/j.freeradbiomed.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Waldeck-Weiermair M, Bischof H, Blass S, Deak AT, Klee C, Graier T, Roller C, Rost R, Eroglu E, Gottschalk B, Hofmann NA, Graier WF, Malli R, Generation of Red-Shifted Cameleons for Imaging Ca2+ Dynamics of the Endoplasmic Reticulum, Sensors 2015, Vol. 15, (2015) 13052–13068. 10.3390/S150613052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pak VV, Ezeriņa D, Lyublinskaya OG, Pedre B, Tyurin-Kuzmin PA, Mishina NM, Thauvin M, Young D, Wahni K, Martínez Gache SA, Demidovich AD, Ermakova YG, Maslova YD, Shokhina AG, Eroglu E, Bilan DS, Bogeski I, Michel T, Vriz S, Messens J, Belousov VV, Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function, Cell Metab. 31 (2020) 642–653.e6. 10.1016/j.cmet.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen ZJ, Ai HW, A Highly Responsive and Selective Fluorescent Probe for Imaging Physiological Hydrogen Sulfide, Biochemistry. 53 (2014) 5966–5974. 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- [29].Lanford RE, Kanda P, Kennedy RC, Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal, Cell. 46 (1986) 575–582. 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- [30].Rizzuto R, Simpson AWM, Brini M, Pozzan T, Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin, Nature. 358 (1992) 325–327. 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- [31].Wen W, Meinkotht JL, Tsien RY, Taylor SS, Identification of a signal for rapid export of proteins from the nucleus, Cell. 82 (1995) 463–473. 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- [32].Sorrentino A, Eroglu E, Michel T, In vivo applications of chemogenetics in redox (patho)biology, Academic Press. (2020) 97–112. 10.1016/B978-0-12-818606-0.00007-9. [DOI] [Google Scholar]
- [33].Rosini E, Pollegioni L, Ghisla S, Orru R, Molla G, Optimization of d-amino acid oxidase for low substrate concentrations - Towards a cancer enzyme therapy, FEBS J. 276 (2009) 4921–4932. 10.1111/j.1742-4658.2009.07191.x. [DOI] [PubMed] [Google Scholar]
- [34].Palacín M, Estévez R, Bertran J, Zorzano A, Molecular Biology of Mammalian Plasma Membrane Amino Acid Transporters, Physiol Rev. 78 (1998) 969–1054. 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- [35].Monné M, Vozza A, Lasorsa FM, Porcelli V, Palmieri F, Mitochondrial Carriers for Aspartate, Glutamate and Other Amino Acids: A Review, Int. J. Mol. Sci 20 (2019). 10.3390/ijms20184456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pisoni RL, Thoene JG, The transport systems of mammalian lysosomes, Biochim. Biophys. Acta - Rev. Biomembr 1071 (1991) 351–373. 10.1016/0304-4157(91)90002-E. [DOI] [PubMed] [Google Scholar]
- [37].Xie ZZ, Liu Y, Bian JS, Hydrogen Sulfide and Cellular Redox Homeostasis, Oxid. Med. Cell. Longev 2016 (2016). 10.1155/2016/6043038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shibuya N, Koike S, Tanaka M, Ishigami-Yuasa M, Kimura Y, Ogasawara Y, Fukui K, Nagahara N, Kimura H, A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells, Nat. Commun 4 (2013) 1–7. 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
- [39].Subach OM, Kunitsyna TA, Mineyeva OA, Lazutkin AA, Bezryadnov DV, Barykina NV, Piatkevich KD, Ermakova YG, Bilan DS, Belousov VV, Anokhin KV, Enikolopov GN, Subach FV, Slowly Reducible Genetically Encoded Green Fluorescent Indicator for In Vivo and Ex Vivo Visualization of Hydrogen Peroxide, Int. J. Mol. Sci 20 (2019) 3138. 10.3390/IJMS20133138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gutscher M, Pauleau A-L, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP, Real-time imaging of the intracellular glutathione redox potential, Nat. Methods 2008 56. 5 (2008) 553–559. 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- [41].Gutscher M, Sobotta MC, Wabnitz GH, Ballikaya S, Meyer AJ, Samstag Y, Dick TP, Proximity-based Protein Thiol Oxidation by H2O2-scavenging Peroxidases *♦, J. Biol. Chem 284 (2009) 31532–31540. 10.1074/JBC.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Niu W, Guo J, Expanding the chemistry of fluorescent protein biosensors through genetic incorporation of unnatural amino acids, This J. Is c R. Soc. Chem 9 (2013) 2961. 10.1039/c3mb70204a. [DOI] [PubMed] [Google Scholar]
- [43].Enyedi B, Zana M, Donkó Á, Geiszt M, Spatial and Temporal Analysis of NADPH Oxidase-Generated Hydrogen Peroxide Signals by Novel Fluorescent Reporter Proteins, Antioxid Redox Signal. 19 (2013) 523–534. 10.1089/ARS.2012.4594. [DOI] [PubMed] [Google Scholar]
- [44].Cameron WD, V Bui C, Hutchinson A, Loppnau P, Gräslund S, V Rocheleau J, Apollo-NADP + : a spectrally tunable family of genetically encoded sensors for NADP +, Nat. Methods 13 (2016) 352–358. 10.1038/nmeth.3764. [DOI] [PubMed] [Google Scholar]
- [45].Sallin O, Reymond L, Gondrand C, Raith F, Koch B, Johnsson K, Semisynthetic biosensors for mapping cellular concentrations of nicotinamide adenine dinucleotides, Elife. 7 (2018). 10.7554/ELIFE.32638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, Zou Y, Shi M, Liu R, Su N, Du J, Zhou H-M, Zhu L, Qian X, Liu H, Loscalzo J, Yang Y, Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism, Nat. Methods 14 (2017) 720. 10.1038/NMETH.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Burr S, Caldwell A, Chong M, Beretta M, Metcalf S, Hancock M, Arno M, Balu S, Kropf VL, Mistry RK, Shah AM, Mann GE, Brewer AC, Oxygen gradients can determine epigenetic asymmetry and cellular differentiation via differential regulation of Tet activity in embryonic stem cells, Nucleic Acids Res. 46 (2018) 1210–1226. 10.1093/NAR/GKX1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].G W, MJ S, S S, TP K, RCM S, PA F, GE M, Nrf2-regulated redox signaling in brain endothelial cells adapted to physiological oxygen levels: Consequences for sulforaphane mediated protection against hypoxia-reoxygenation, Redox Biol. 37 (2020). 10.1016/j.redox.2020.101708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, Mclendon RE, Sloan AE, Rich JN, Tumor and Stem Cell Biology A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo, (2016). 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Erapaneedi R, V Belousov V, Schäfers M, Kiefer F, A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level, EMBO J. 35 (2016) 102. 10.15252/EMBJ.201592775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jones DP, Go Y-M, Redox compartmentalization and cellular stress, Diabetes. Obes. Metab 12 (2010) 116. 10.1111/J.1463-1326.2010.01266.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhang S, Gilbert ER, Noonan KJT, Saremi B, Wong EA, Gene expression and activity of methionine converting enzymes in broiler chickens fed methionine isomers or precursors, Poult. Sci 97 (2018) 2053–2063. 10.3382/ps/pey037. [DOI] [PubMed] [Google Scholar]
- [53].Fernández-Lafuente R, Rodriguez V, Guisán JM, Guisán JM, The coimmobilization of d-amino acid oxidase and catalase enables the quantitative transformation of d-amino acids (d-phenylalanine) into α-keto acids (phenylpyruvic acid), Enzyme Microb. Technol 23 (1998) 28–33. 10.1016/S0141-0229(98)00028-3. [DOI] [Google Scholar]
- [54].Nguyen LP, Hsu EL, Chowdhury G, Dostalek M, Guengerich FP, Bradfield CA, d-Amino Acid Oxidase Generates Agonists of the Aryl Hydrocarbon Receptor from d-Tryptophan, Chem. Res. Toxicol 22 (2009) 1897–1904. 10.1021/TX900043S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Weller J, Kizina KM, Can K, Bao G, Müller M, Response properties of the genetically encoded optical H2O2 sensor HyPer, Free Radic. Biol. Med 76 (2014) 227–241. 10.1016/j.freeradbiomed.2014.07.045. [DOI] [PubMed] [Google Scholar]
- [56].Burgstaller S, Bischof H, Gensch T, Stryeck S, Gottschalk B, Ramadani-Muja J, Eroglu E, Rost R, Balfanz S, Baumann A, Waldeck-Weiermair M, Hay JC, Madl T, Graier WF, Malli R, PH-Lemon, a Fluorescent Protein-Based pH Reporter for Acidic Compartments, ACS Sensors 4 (2019) 883–891. 10.1021/acssensors.8b01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sahoo S, Aurich MK, Jonsson JJ, Thiele I, Membrane transporters in a human genome-scale metabolic knowledgebase and their implications for disease, Front. Physiol 5 (2014) 1–24. 10.3389/fphys.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pollegioni L, Falbo A, Pilone MS, Specificity and kinetics of Rhodotorula gracillis d-amino acid oxidase, Biochim. Biophys. Acta (BBA)/Protein Struct. Mol 1120 (1992) 11–16. 10.1016/0167-4838(92)90418-D. [DOI] [PubMed] [Google Scholar]
- [59].Molla G, Motteran L, Piubelli L, Pilone MS, Pollegioni L, Regulation of D-amino acid oxidase expression in the yeast Rhodotorula gracilis, Yeast. 20 (2003) 1061–1069. 10.1002/yea.1023. [DOI] [PubMed] [Google Scholar]


