Abstract
The SH3-SH3-SH3-SH2 adapter Nck represents a two-gene family that includes Nckα (Nck) and Nckβ (Grb4/Nck2), and it links receptor tyrosine kinases to intracellular signaling networks. The function of these mammalian Nck genes has not been established. We report here a specific role for Nckβ in platelet-derived growth factor (PDGF)-induced actin polymerization in NIH 3T3 cells. Overexpression of Nckβ but not Nckα blocks PDGF-stimulated membrane ruffling and formation of lamellipoda. Mutation in either the SH2 or the middle SH3 domain of Nckβ abolishes its interfering effect. Nckβ binds at Tyr-1009 in human PDGF receptor β (PDGFR-β) which is different from Nckα's binding site, Tyr-751, and does not compete with phosphatidylinositol-3 kinase for binding to PDGFR. Microinjection of an anti-Nckβ but not an anti-Nckα antibody inhibits PDGF-stimulated actin polymerization. Constitutively membrane-bound Nckβ but not Nckα blocks Rac1-L62-induced membrane ruffling and formation of lamellipodia, suggesting that Nckβ acts in parallel to or downstream of Rac1. This is the first report of Nckβ's role in receptor tyrosine kinase signaling to the actin cytoskeleton.
Src homology (SH) domains, including SH2 and SH3 domains, are protein modules found in many otherwise functionally distinct molecules (25). The ligands for SH2 and SH3 domains are phosphotyrosine-containing peptides (pY-X-X-X) and proline-rich peptides (P-X-X-P), respectively. The C-terminal amino acid residues of the phosphotyrosines and the flanking amino acid residues of proline-rich segments determine binding affinity and specificity (26, 35). A family of SH2 and SH3 domain-containing proteins, including Crk, Grb2, and Nck, contain only SH2 and SH3 domains and have no other functional motifs (2, 19). They are therefore regarded as adapters. They act by binding to tyrosine-phosphorylated proteins via SH2 domains and associating with P-X-X-P motif-containing proteins through SH3 domains. SH3-associated proteins often get translocated to the proximity of phosphotyrosine proteins (32). Thus, SH2 and SH3 domains act as a second messenger connecting protein tyrosine phosphorylation to a variety of intracellular signaling networks. The best-characterized adapter is the SH3-SH2-SH3 protein Grb2. Grb2 binds to two separate pY-X-N-V motifs in the epidermal growth factor receptor (EGFR) via its SH2 domain and associates through its two SH3 domains to the PPPVPPRRR motifs in Sos, a guanine nucleotide exchange factor for Ras. As a result, Sos is translocated to the plasma membrane and activates Ras (32).
Nck contains three consecutive SH3 domains and one SH2 domain, which together occupy more than 70% of Nck's 377 amino acids (16). Similar to Grb2, Nck is widely expressed in various types of cells and acts as an adapter by linking receptor tyrosine kinases to downstream signaling networks. It has also been reported that there is a fraction of Nck that is associated with Sam68 in the nucleus (14), although its function remains unknown. The SH2 domain of Nck has been shown to bind either directly or indirectly to EGFR, platelet-derived growth factor receptor (PDGFR), Eph receptor, insulin receptor substrate 1, p130cas, and p62Dok (16, 20). For example, tyrosine 751 (Y751) in human PDGFR-β was identified as the binding site for Nckα. Since Y751 is also one of the two binding sites in the PDGFR for the p85 subunit of phosphatidylinositol 3-kinase (PI3-K) (the p85 subunit has two SH2 domains), Nckα and PI3-K may either compete with each other for binding to the PDGFR and thereby antagonize each other's function or bind to different pools of the cell surface PDGFR (22). There has been no evidence so far for or against either of these hypotheses. Stein et al. identified a binding site for Nckα in the Eph family receptor, Eph1 (ELK). They showed that Y-594 in the juxtamembrane region of Eph1 recruits Nck to the plasma membrane. The membrane-bound Nck in turn causes, apparently via Nck-interacting kinase (NIK), activation of the JNK/SAPK pathway (1, 36, 37). The current list of Nck (α, β, or both) SH3-binding molecules includes the Abl protein tyrosine kinase, Sos, Nck-associated kinase (NAK), p21cdc42/rac-activated kinases (PAK), Rho effector PKN-related kinase PRK2, protooncogene c-cbl, human Wiskott-Aldrich syndrome protein (WASp), the novel serine threonine kinase NIK, casein kinase 1 gamma-2, Sam-68, Nap1 (Nck-associated protein 1), and NAP4 (Nck-, Ash- and PLCγ-binding protein 4) (16). We and others recently reported that Nck represents a family of genes including two human (Nck/hNckα and hNckβ/Nck2) and two mouse (mNckα and mNckβ/Grb4) Nck genes (3, 4, 25, 40). hNckα and hNckβ reside in different chromosomes (4, 10, 41) and are coexpressed in most but not all cells (3). The newly identified Nckβ binds significantly better than Nck (Nckα) to both receptor and nonreceptor tyrosine kinases (3, 4, 40). Moreover, Nckα and Nckβ appear to have distinct functional assignments in the same cells (4).
Recently, a growing number of studies have suggested that Nck plays an important role in mediating receptor tyrosine kinase signaling to the actin cytoskeleton. Rockow et al. showed that overexpression of Nckα blocks nerve growth factor- and basic fibroblast growth factor-induced neurite outgrowth, a Rac1/Cdc42 GTPase-dependent actin cytoskeletal change, in rat adrenal pheochromocytoma cells PC12, through an extracellular signal-regulated kinase-independent mechanism (29). Two Nck-SH3-binding proteins, WASp and PAK1, have clear roles in regulation of the actin cytoskeleton through either Cdc42- and Rac-dependent or -independent mechanisms. Symons et al. showed that WASp binds to GTP-bound Cdc42 and clusters in polymerized actin (38). N-WASp (richest in neural tissues) is also involved in Cdc42 signaling to the actin cytoskeleton (21). Sells et al. reported that PAK1-induced actin organization depends upon binding to Nck but not upon PAK1 kinase activity or binding to Rac1 and Cdc42 (33). They showed that a kinase-dead PAK1 could mimic the effect of Rac and induce lamellipodia formation (33). Consistent with their observations, Obermeier et al. showed that brain-specific PAK (γPAK/PAK3) induces cell spreading, membrane ruffling, and increased lamellipodia formation (24). The strongest support of the notion that Nck links tyrosine kinases to the actin cytoskeleton comes from a genetic study of Drosophila melanogaster. Each of the eight R cells (R1 to R8) of the Drosophila compound eye is a distinct neuron and acts as a photoreceptor. Guidance and target recognition of these R cells toward axons are believed to be regulated by receptors at the surface of the growth cone, which resides at the leading edge of the axon. The growth cones receive extracellular cues and in turn control the intracellular actin cytoskeletol rearrangement. Zipursky and his colleagues found that the gene called Dreadlocks, or Dock, was concentrated in the R-cell growth cone and essential for R-cell guidance and target recognition. Mutations in the Dock gene disrupted signaling from the surface of the growth cone to the intracellular actin cytoskeleton, resulting in defects in R-cell fasciculation, targeting, and retinotopy (5). Dock is structurally related to and has an overall 40% amino acid identity with the mammalian Nck genes (4). Rao and Zipursky showed that the first and third SH3 domains and the SH2 domain are functionally redundant, whereas the middle SH3 domain is always required (27). Depending upon the specific neuron type, the middle SH3 domain could act with the SH2 domain or with the first and third SH3 domains. The critical downstream pathways for Dock include the Cdc42-Pak1 pathway (9) and the Ste20-like kinase misshapen pathway (30). In contrast, important questions regarding the function of the mammalian Nck genes remain unanswered. Does Nck play roles in mammals similar to that of Dock in Drosophila? Are both Nckα and Nckβ involved in regulation of the actin cytoskeleton? It has previously been demonstrated that PDGF induces actin polymerization at the plasma membrane of fibroblast cells to produce edge ruffles and lamellipodia (6, 28). This signaling event appears to involve the Rho family GTPase Rac1 (23, 28) and PI3-K (23, 42, 43). PI3-K appears to act upstream of Rac1 (8). Because Nck is a direct target for PDGFR, we set out to compare the roles of Nckα and Nckβ in PDGF-stimulated actin cytoskeletal rearrangement in NIH 3T3 cells. We found that Nckβ is specifically involved in PDGF-induced membrane ruffling and formation of lamellipodia. Our results suggest that Nckβ acts either parallel to or downstream of Rac1, a mediator between PDGFR and the actin cytoskeleton.
MATERIALS AND METHODS
Site-directed mutagenesis.
Human Nckα and Nckβ genes have been described previously (4). Mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene). PCRs were carried out using an overlap extension by Pfu DNA polymerase (Boehringer Mannheim) in order to generate both the SH2 domain mutant, in which the conserved arginine residue (R) in the FLVRES motif was changed to lysine (K), and the SH3 domain mutants, in which the first tryptophan residue (W) in the conserved WW motif was changed to lysine (K). These residues have previously been identified as being essential for binding to their ligands (17). The oligonucleotides used in the mutageneses were GCTTCTGGATGATTCTAAGTCCAAGTGGCGAGTTCGAAATTCC for Nckα (W38K), GGAGAAATCGAGTGATGGGAAGTGGCGTGGTAGCTACAATGG for Nckα (W143K), CCTGAAAATGACCCAGAGAAGTGGAAATGCAGGAAGATCAATGG for Nckα (W229K), GGGGATTTCCTCATTAAGGATAGTGAATCTTCGCC for Nckα (R308K), GGACGACTCCAAGACGAAGTGGCGGGTGAGGAACGCG for Nckβ (W39K), GGAGAAGTGCAGCGACGGTAAGTGGCGGGGCAGCTACAACG for Nckβ (W149K), CCGGAGAACGACCCCGAGAAGTGGAAATGCAAAAATGCC for Nckβ (W235K), and GGCGACTTCCTCATTAAGGACAGCGAGTCCTCG for Nckβ (R312K). After DpnI digestion of the parental cDNA templates, mutant DNA clones were subcloned into the pRK5 mammalian expression vector (15) and transformed into XL1-Blue competent bacteria. Plasmids were isolated and purified. Mutations were confirmed by nucleotide sequencing analysis (at the DNA Sequencing Facility of the University of Chicago). Double or triple mutations were carried out by sequentially repeating the above procedure.
Construction of membrane-attached Nckα and Nckβ.
Wild-type Nck genes were linked in frame with the Ras farnesylation sequence, KLNPPDESGPGCMSCKCVLS, encoded by AAACTTAATCCTCCTGATGAATCTGGTCCTGGTTGTATGTCTTGTAAATGTGTTCTTTCT, at their carboxyl termini through three sequential PCRs. The sequence of the 5′-end oligonucleotide was AGCTGGTACCAAGCTTGGCACCATGTTTTACCCATACGAT for all three PCRs. The sequences of the 3′-end oligonucleotides for the three sequential PCRs were AGATTCATCAGGAGGATTAAGTTTCTGCAGGGCCCTGACGAGGTA, TTTACAAGACATACAACCAGGACCAGATTCATCAGGAGGATTAAGTTT, and GGATCCGAATTCGTCATCAAGAAAGAACACATTTACAAGACATACAACCAGGACC. The hemagglutinin (HA)-tagged wild-type Nck genes were used as templates for the PCRs. Two percent (vol/vol) of each PCR was used as the template for the following PCR. Products of the final PCR were purified and subcloned into pRK5 at the XbaI site, and sequences were confirmed by DNA sequencing analyses. The constructs were transfected into NIH 3T3 cells, and membrane attachment was confirmed by a cell fractionation study (see below).
Cell fractionation, immunoprecipitation, and immunoblotting.
Transfected NIH 3T3 cells in 15-cm tissue culture dishes were scraped off in 2 ml of ice-cold phosphate-buffered saline (PBS) containing 1 mM iodoacetate, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.1 mM orthovanadate. Cell pellets, obtained through centrifugation at 400 × g for 5 min, were swollen in 8 volumes of hypotonic buffer (5 mM Tris-HCl [pH 7.4], 10 mM sodium pyrophosphate, 10 mM sodium fluoride, 1 mM sodium iodoacetate, 0.5 mM sodium orthovanadate, 1 mM PMSF) for 15 min and Dounce homogenized on ice until over 90% of the cells were broken (the percentage of broken cells was monitored under the microscope). Then 0.25 volume of compensation buffer (20 mM Tris-HCl [pH 6.7], 0.95 M sucrose, 0.1 M sodium chloride, 30 mM sodium pyrophosphate, 100 μM sodium fluoride, 0.5 mM sodium orthovanadate, 0.025 mM zinc chloride) was added to bring back isotonicity. Nuclei were separated from the rest of the cell extract by centrifugation at 2,200 rpm for 1 min. The pellet was the nucleus fraction. The supernatant was further centrifuged at 30,000 rpm (100,000 × g) in a Beckman SW60.1 for 30 min at 4°C. The supernatant was the postnuclear cytosol fraction, and the pellet was the membrane fraction. Equal portions of the cytosol fraction and Triton- X-100 (in lysis buffer)-soluble fractions of the nuclei and membrane pellets were either directly analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting (Western) or immunoprecipitated with anti-HA monoclonal antibody 12CA5. Immunoprecipitates were then analyzed through SDS-PAGE and immunoblotting. Results were visualized through enhanced chemiluminescence (ECL) reactions according to the manufacturer's instructions (Amersham).
For immunoprecipitation, cells extracts were incubated with the corresponding primary antibody for 3 h, followed by secondary antibody incubation for an additional 1 h at 4°C. Immune complexes were precipitated by incubation with protein A-Sepharose beads for 1 h at 4°C. Beads were washed three to five times with lysis buffer without bovine serum albumin (BSA) and heated at 95°C for 5 min in sample buffer containing 0.2 M β-mercaptoethanol. Supernatants were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and blotted with the corresponding antibodies.
Identification of Nckβ binding site in the human PDGFR-β.
Dog kidney epithelial cells (TRMP) expressing wild-type or mutant human PDGF β-receptors were maintained as described previously (12). TRMP cells were grown to 80% confluence in 6-cm tissue culture dishes and incubated in medium containing 0.5% fetal calf serum for 16 h. Cells were then treated or not with human recombinant PDGF (400 ng/ml; Sigma) at 37°C for 5 min, at which time maximum protein tyrosine phosphorylation was detected (15). Cells were washed three times with ice-cold PBS buffer and solubilized in lysis buffer. The supernatants of the clarified cell lysates were resolved in an SDS gel, transferred to a nitrocellulose membrane, and incubated with purified glutathione S-transferase (GST) alone or GST-Nck (3 μg/ml) for 2 h at 4°C. The GST-Nckβ-bound PDGFR was visualized by further immunoblotting with anti-GST antibody, followed by ECL.
Generation of anti-Nck-specific antibodies.
Full-length Nckα and Nckβ cDNAs were fused with the bacterial GST gene in the pGEX vector and expressed in the bacterial strain XL-1. Fusion proteins were purified and used to immunize three rabbits per antigen. All the initial antisera cross-reacted to various degrees with both Nckα and Nckβ. To further isolate anti-Nckα-specific antibodies, each of the anti-Nckα antisera was passed through a GST-Nckβ fusion protein column to remove Nck-β-binding immunoglobulin G (IgG) molecules. The leftover supernatant was subjected to purification by a GST-Nckα fusion protein affinity column, in accordance with a previously published procedure (7). Likewise, to isolate anti-Nckβ-specific antibodies, each of the anti-Nckβ antisera was passed through a GST-Nckα fusion protein column to remove the portion of Nckα-interacting antibodies. Anti-Nckβ antibodies were further purified from the leftover supernatant with a GST-Nckβ protein affinity column. Antibodies were washed and concentrated in 0.9% NaCl by Cetricon-50. These antibodies were tested for recognizing and neutralizing the function of the cellular Nck proteins. To test whether the antibodies recognize native Nck proteins, increasing concentrations of each antibody were added to a fixed amount of cellular extract and tested for complete depletion of the cellular Nckα or Nckβ proteins. The antibodies were removed by protein A-Sepharose, and the lysates were examined by immunoblotting with the same anti-Nck antibody. To test whether the antibodies block interactions between Nck and activated EGFR or PDGFR, lysates of EGF- or PDGF-stimulated cells were added simultaneously with GST-Nck on agarose beads and increasing amounts of the purified antibodies. The bead-bound EGFR or PDGFR was analyzed by immunoblotting analysis with anti-EGFR or anti-PDGFR and antiphosphotyrosine antibodies.
Cell culture, transfection, and microinjection.
NIH 3T3 cells expressing endogenous PDGF receptors (105 binding sites/cell) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), streptomycin, and penicillin (50 U/ml). Cells were transfected with the individual DNA constructs for 2 h using SuperFect reagent (Qiagen), according to the manufacturer's instructions. The ratio of DNA to SuperFect reagent was 1:2. Three hours following transfection, medium was changed to fresh DMEM with 10% FCS and incubated for 48 h prior to further analysis. As a control for detecting doubly transfected cells for cotransfection experiments, duplicate cells were transfected with one of the two DNA constructs, using vector DNA to replace the other DNA construct so that the total amounts of DNA were equal.
For microinjection, cells were cultured on coverslips at ∼400 cells/well. After 2 days of culture, cells were serum starved for 18 h in DMEM containing 0.2% FCS. Immediately prior to microinjection, cells were changed to specially prepared DMEM with 10% of the normal amount of NaHCO2 in order to maintain neutral pH during the microinjection period. Rabbit IgG (as a control), purified rabbit anti-Nckα, and anti-Nckβ antibody at a concentration of 500 ng/μl were independently microinjected into the cytoplasm of cells together with fluorescein isothiocyanate (FITC)-labeled dextran as a marker protein to identify injected cells. (The whole procedure was carried out at the Transgenic Facility of the University of Southern California.) Usually, 25 to 30 cells were successfully injected within 10 min for each condition (in each well) in one experiment. Cells were then returned to the incubator for 2 h and treated or not with PDGF (Sigma; 100 ng/ml) for 15 min before fixation, as described below. Four independent experiments were carried out under similar conditions.
Immunofluorescence microscopy.
Serum-starved parental cells, transfected cells, and microinjected cells on coverslips were either untreated or treated with PDGF-bb (100 ng/ml) for 15 min at 37°C. Cells were rinsed with PBS and fixed in freshly prepared 4% (wt/vol) paraformaldehyde in PBS for 10 min. Cells were rinsed twice with PBS and permeabilized in PBS containing 0.2% Triton X-100 for 5 min. Following a PBS rinse, cells were incubated with primary antibodies anti-HA (12CA5, 2 μg/ml) or rabbit anti-Myc antibody (N-262; Santa Cruz; 4 μg/ml) or both in PBS containing 1% BSA for 2 h. Cells were rinsed and incubated with secondary antibody mixtures containing FITC-conjugated rabbit anti-mouse IgG (Jackson Laboratory; 10 μg/ml) and/or AMCA (coumarin)-conjugated goat anti-rabbit IgG (Sigma, 10 μg/ml) together with TRITC (rhodamine)-conjugated phalloidin (0.1 μg/ml; Molecular Probes) for 45 min. Therefore, in cells simultaneously transfected with Myc-tagged Rac/Cdc42 and HA-tagged Nck, expression of these genes and changes in actin polymerization in a single cell could be visualized by triple (green [FITC], blue [AMCA], and red [TRITC]) staining. Cells were rinsed three times with PBS (10 min of incubation each time) and air dried. The coverslips were mounted with antifade reagent (Molecular Probes). Expression of transfected genes and actin polymerization in the cells was examined by confocal microscopy (at the University of Southern California Confocal Core Facility), using ZEISS 100X 1.0 oil immersion objectives. Thirty to 120 randomly selected cells from either vector-transfected population or gene-transfected double (FITC and TRITC)- or triple-stained (FITC, AMCA, and TRITC) populations were analyzed for peripheral filamentous actin in membrane ruffles. Images shown are representative of significantly responding cells under each condition. The percentage of cells that had undergone membrane ruffling was calculated as responding cells over total positively stained cells.
RESULTS
Overexpression of Nckβ but not Nckα blocked PDGF-induced actin polymerization.
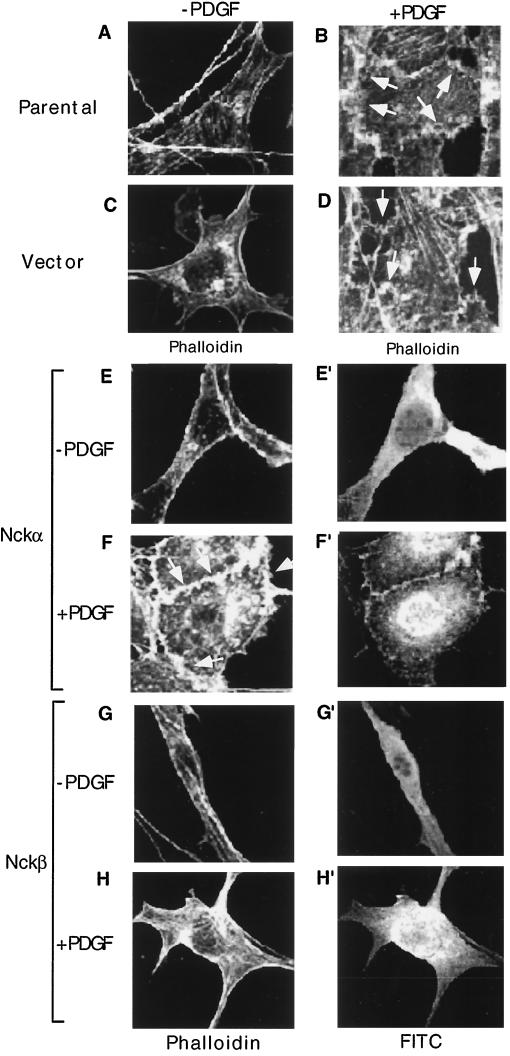
Prompted by the finding that the Drosophila Nck-like gene, Dock, plays a critical role in mediating extracellular cues to intracellular actin cytoskeleton at the growth cone during axon guidance and targeting (5, 27), we were interested in understanding whether or not Nck has a similar function in mammalian cells. We chose PDGFR signaling in fibroblasts as the biological system, because it has been well established that in these cells PDGF stimulates, via Rac, actin polymerization, which leads to formation of membrane ruffles and lamellipodia (23, 28), and Nck is a direct target for the PDGFR (15, 22). We started out by confirming the PDGF effect in NIH 3T3 cells and by testing whether or not Nck regulates PDGFR signaling to the actin cytoskeleton. It is shown in Fig. 1 that in quiescent (serum-starved) cells, a fine ring of polymerized actin at the periphery of the cell was seen by staining with rhodamine-labeled phalloidin (Fig. 1A). Following PDGF treatment for 15 min, a dramatic alteration in the actin cytoskeleton of the cell, including accumulation of polymerized actin in the peripheral plasma membrane and formation of lamellipodia and membrane edge ruffles, could be visualized (Fig. 1B) under the fluorescent microscope. Of the 212 parental cells examined, 208 showed this phenotype (see statistical analyses in Fig. 5). Similar results were observed in cells transfected with an empty pRK5 expression vector (Fig. 1D versus C). These results have established NIH 3T3 cells as an adequate cell culture system for studying PDGFR signaling to the actin cytoskeleton.
FIG. 1.
Overexpression of Nckβ but not Nckα blocks PDGF-stimulated membrane ruffling. NIH 3T3 cells, cultured in fibronectin-coated (10 μg/ml, 2 h) eight-chamber culture slides, were either untransfected (A and B), transfected with vector alone mixed with a GFP-containing vector (C and D), or transfected with the wild-type Nckα (E to F′) or Nckβ (G to H′) construct at 0.5 μg/well. After 48 h, cells were starved in low-serum medium for an additional 18 h and treated (B, D, F, and H) or not (A, C, E, and G) with PDGF-bb (100 ng/ml) at 37°C for 15 min. Expression of the Nck proteins was monitored by anti-HA antibody blotting, followed by a secondary antibody conjugated with FITC. The actin cytoskeleton was revealed by rhodamine-labeled phalloidin staining. Eighty to 100 cells which showed positive FITC staining were selected and analyzed in each experiment. Vector-transfected cells were identified as GFP positive. Images were recorded with a Zeiss confocal microscope. Magnifications, × 150. This experiment was repeated four times.
FIG. 5.
Statistical analysis of data shown in Fig. 1 and 4. FITC-staining cells (80 to 100 cells for each of the conditions) were randomly selected and analyzed for membrane ruffling and lamellipodium formation in response to PDGF stimulation.Values are [(number of actin-polymerized cells)/(total number of cells)] × 100. Due to variations in exogenous expression levels of any given HA-tagged Nck construct in different cells, degrees of PDGF-stimulated actin polymerization vary. Nckβ-WTK, Nckβ-W38/143/229K triple mutant.
We have previously shown that Nckα binds directly to human PDGFR at Y751 (22), and Nckβ binds 10 times better than Nckα to the PDGFR, via an unknown site (4). We tested if overexpression of HA-tagged wild-type Nckα and Nckβ would interfere with (enhance and suppress) PDGF-stimulated actin polymerization. To examine expression of transfected HA-Nck and changes of the actin cytoskeleton in the same cells, HA-Nck-positive cells were identified by anti-HA antibody blotting, followed by FITC-conjugated secondary antibody staining, whereas the actin cytoskeleton was visualized by staining with rhodamine-labeled phalloidin, as previously used. It is also shown in Fig. 1E to H′ that enforced overexpression of wild-type Nckα or wild-type Nckβ had no significant effect on the actin structure in serum-starved, unstimulated cells (Fig. 1E and G). Expression of HA-Nckα or HA-Nckβ protein in the same cells was indicated by FITC staining (Fig. 1E′ and G′). In the PDGF-stimulated cells, cells transfected with wild-type Nckα exhibited a pattern of actin assembly similar to that in the cells transfected with the vector alone (Fig. 1F versus D), i.e., polymerized actin assembly at the leading edge of the plasma membrane and formation of membrane ruffles. Expression of HA-Nckα protein in the same cell was indicated by FITC staining (Fig. 1F′). Surprisingly, in cells transfected with the wild-type Nckβ, the PDGF-stimulated accumulation of actin in membrane ruffles was dramatically inhibited (Fig. 1H) in more than 80% of HA-Nckβ-positive cells (71 of 87) examined (see statistical analysis in Fig. 5). Expression of transfected HA-Nckβ in the same cell was indicated by FITC staining (Fig. 1H′). These observations suggest that Nckβ but not Nckα participates in PDGF signaling to the actin cytoskeleton.
Nckβ binds to a distinct site in the PDGFR and does not compete with PI3-K binding.
One could argue that the inhibitory effect of Nckβ was due to nonspecific binding competition for PDGFR, occupying the binding sites of other signaling proteins such as PI3-K and Nckα, which have a common binding site, Y751 (22). To address this problem, we set out to identify the Nckβ binding site in human PDGFR-β. TRMP cells expressing all the possible PDGFR phosphotyrosine mutants, previously described (12), were used for the experiment. Lysates of these cells either untreated or treated with PDGF were resolved in duplicate in SDS gels, transferred to a nitrocellulose membrane, and blotted either with an antiphosphotyrosine antibody or with purified GST-Nckβ proteins. The GST-Nckβ-bound PDGFR was further visualized by anti-GST antibody immunoblotting, followed by ECL. The advantage of this technique is that it allows determination of direct interaction between Nckβ and PDGFR. It is shown in Fig. 2Aa that comparable amounts of PDGFR in various cell lines were subjected to the binding study. While GST-Nckβ was able to bind the wild type and most of the PDGFR mutants (Fig. 2Ab, lanes 2 to 8 lane 11), its binding to the PDGFR with a single mutation at Y1009 or double mutations at Y1009 and Y1021 was dramatically reduced (lanes 9 and 10). The slightly reduced binding to the Y740/751F mutant was not always reproducible.
FIG. 2.
Nckβ binds at Y1009 in the PDGFR and its overexpression did not affect PI3-K binding to the PDGFR. (A) TRMP cells expressing wild-type (wt) or mutant PDGFR were serum starved and either untreated or treated with PDGF-bb (400 ng/ml) for 5 min. Total lysates (50 μg of protein per lane) of the cells were resolved in an SDS gel, transferred to a nitrocellulose membrane, and blotted with either antiphosphotyrosine (anti-PY) (a) or purified GST-Nckβ (3 μg/ml), followed by anti-GST antibody blotting (b) at 4°C for 2 h. Membranes were washed, and the results were visualized with ECL. (B) Lysates of cells (2 × 106 cells/dish), untransfected (lanes 1 and 2) or transfected with 1.5 μg (lanes 3 and 4) or 5 μg (lanes 5 and 6) of Nckβ cDNA, and either untreated or treated with PDGF (5 min, 37°C), were immunoprecipitated (IP) with an anti-p85 antibody (Z-8; Santa Cruz). The immunoprecipitates were resolved in an SDS gel, transferred to a nitrocellulose membrane, and immunoblotted with either monoclonal anti-PDGFR antibody (61520.11; R&D Systems) (a) or the anti-p85 antibody (b), or the same set of total lysates (30 μg of protein/lane) were directly analyzed by Western blot using anti-HA (12CA5) antibody (c). Results were visualized by ECL. (C) Schematic representation of binding of the two Ncks to human PDGFR-β together with their shared binding partners.
Similar results were observed in co-immunoprecipitation experiments using our newly developed anti-Nckβ-specific antibodies (data not shown). These results demonstrate that Nckβ binds to Y1009 on human PDGFR-β. Since this site has previously been shown to be the binding site for the SH2 domain of SHP2 (13), Nckβ, similar to Nckβ, shares a binding site with another SH2-containing protein. We then tested whether or not overexpressed Nckβ would cause nonspecific competition for other binding sites on PDGFR. We compared the binding of PI3-K's p85 subunit to PDGFR in control and Nckβ-overexpressing cells, particularly because PI3-K has been shown to play an important role in PDGF-stimulated actin polymerization (8, 42, 43) and shares the binding site Y-751 with the SH2 domain of Nckα. Figure 2B clearly shows that increasing concentrations of Nckβ expression in cells (c, lanes 3 to 6 versus lanes 1 and 3) did not affect the amount of p85-coimmunoprecipitated PDGFR (Fig. 2Ba, lanes 4 and 6 versus lane 2). Similar amounts of p85 were recovered by anti-p85 antibody immunoprecipitation (Fig. 2Bb, lanes 1 to 6). These results suggest that the observed inhibitory effect of Nckβ on PDGFR signaling to the actin cytoskeleton was probably not due to nonspecific binding competition, although we did not test this for each of the dozen previously shown PDGFR-binding proteins.
Unfortunately, because Y1009 is also shared by the SH2 domain of SHP2, the PDGFR-Y1009F mutant cannot be used to evaluate the specificity of Nckβ's effect. Instead, another approach has been used; see below. A schematic representation of the binding of the two Ncks to human PDGFR is shown in Fig. 2C, in which both Nckα and Nckβ share a binding site with another PDGFR-interacting protein(s).
Mutations in the SH2 and SH3 domains of Nckβ abolish its interfering effect.
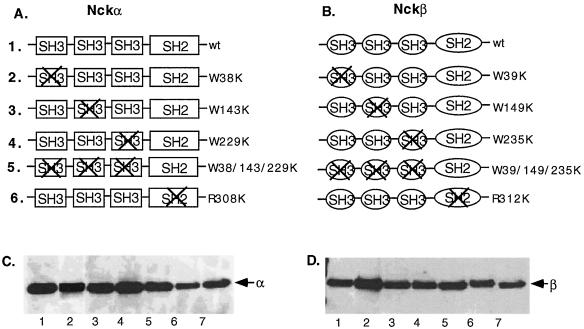
To study the possible mechanism of Nckβ's action, we generated HA-tagged SH2 and SH3 mutants of these two genes, as schematically shown in Fig. 3. The highly conserved arginine (R) of the FLVRES motif in the SH2 domains and the first tryptophan (W) of the characteristic double tryptophans in the SH3 domains were replaced with lysine residues (K). Figure 3A and B show the list of HA-tagged wild-type and SH2 mutants and SH3 mutants of Nckα and Nckβ, respectively. To confirm the expression of these transgenes, pRK5-cDNA constructs were transfected into NIH 3T3 cells, and lysates of the transfected cells were immunoblotted with anti-HA-tagged antibody (the transfection efficiency by Superfect reagent was around 35% for NIH 3T3 cells). It is shown in Fig. 3C and D that a similar level of protein expression of the various forms of Nckα (C) and Nckβ (D) genes was achieved. When the same samples were immunoblotted with an anti-Nckβ or anti-Nckβ antibody (71-2800; Zymed), which recognize both HA-tagged and endogenous Nck, five- to sevenfold-higher expression of HA-Nck over endogenous Nck was observed (data not shown).
FIG. 3.
Schematic representation of Nckα (A) and Nckβ (B) mutant constructs and their expression. A nucleotide fragment encoding three repeats of the HA peptide YPYDVPDY was linked in frame to the N termini of the Nck genes. Site-directed mutagenesis was carried out to generate point mutations in the previously established conserved sites within the SH2 and SH3 domains (see text). (C and D) Expression of the transgenes in NIH 3T3 cells following transfection is indicated by Western blot analysis (30 μg of total cellular protein/lane) using anti-HA monoclonal antibody 12CA5. The results were visualized by ECL.
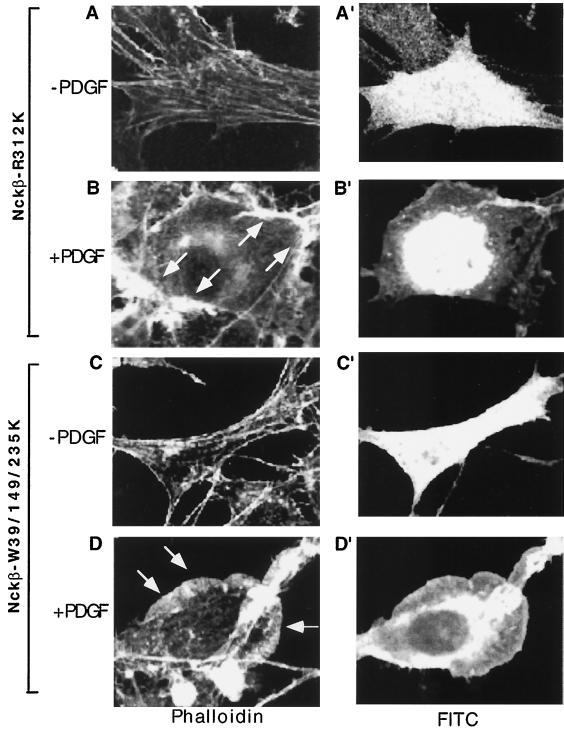
We first investigated whether or not the SH2 domain of Nckβ was required for its dominant interfering effect. Cells were transfected with the Nckβ SH2 mutant, Nckβ-R312K, and either untreated or treated with PDGF. It is shown in Fig. 4 that expression of HA-Nckβ-R312K was indicated by anti-HA antibody blotting followed by FITC antibody staining (A′ and B′). Rhodamine-labeled phalloidin staining of the same cells revealed that Nckβ-R312K had little effect on actin polymerization in the absence of PDGF (Fig. 4A). However, in contrast to the effect of wild-type Nckβ, Nckβ-R312K was no longer able to block PDGF-induced membrane ruffling (Fig. 4B). These results suggest that binding to PDGFR is essential for the function of Nckβ. We then tested the role of the three SH3 domains by using an SH3 triple mutant of Nckβ, Nckβ-W39/149/235K. Nckβ-W39/149/235K also failed to block PDGF-stimulated membrane ruffling (Fig. 4D versus C). Expression of HA-Nckβ-W39/149/235K was indicated by anti-HA antibody blotting followed by FITC antibody staining (Fig. 4C′ and D′). In 89 cells examined, all of which positively expressed Nckβ-W39/149/235K, we did not detect any significant inhibition of PDGF-induced membrane ruffling and lamellipodium formation (see statistical analysis in Fig. 5).
FIG. 4.
SH2 and middle SH3 domains of Nckβ are required for the regulatory effect of Nckβ on PDGFR signaling. Cells were transfected with either the SH2 mutant Nckβ-R312K (A, A′, B, and B′), with the triple SH3 mutant Nakβ-W39/149/235K (C, C′, D, and D′), or with the individual SH3 mutants indicated (E to J′). The rest of the experimental procedures were identical to those described for Fig. 1. Three independent experiments were carried out, and they showed similar results.
The effect of Nckβ-W39/149/235K on PDGFR signaling was unexpected. We initially had predicted that this mutant should have a strong dominant negative effect because its SH2 domain was still intact and could compete with endogenous Nckβ for binding to PDGFR. A possible explanation is that mutations in SH3 domains might have weakened SH2 binding to phosphotyrosine. Interestingly, while there is currently no evidence either for or against this hypothesis, similar results were previously reported for the Drosophila Nck-like gene Dock, for which it was shown that a similar mutant had no dominant negative effect (27).
To identify the specific SH3 domain(s) which is required for the interfering action of Nckβ, we tested the effects of each of the individual SH3 domain mutations of Nckβ, W39K, W149K, and W235K. It is also shown in Fig. 4 that Nckβ-W39K (E and F) and Nckβ-235K (I and J) were still able to block PDGF-stimulated membrane ruffling (F versus E and J versus I). Interestingly, the Nckβ-W149K mutant failed to inhibit PDGF-induced actin polymerization in the cell (Fig. 4G and H), resulting in clearly detectable PDGF-induced membrane ruffling (Fig. 4H versus G). These results indicated that the middle SH3 domain of Nckβ plays a critical role. The statistical analysis of these data is summarized in Fig. 5.
Microinjection of anti-Nckβ-specific antibody inhibits PDGF-stimulated actin polymerization.
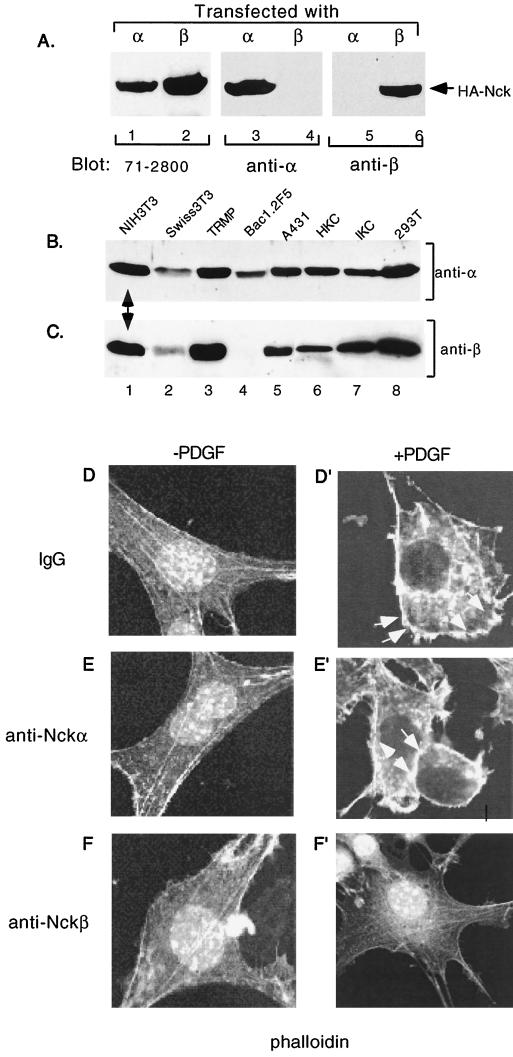
As mentioned previously, since Y1009 is also the binding site for SHP2, the PDGFR-Y1009F mutant became less useful for determining the specific effect of Nckβ on PDGFR-mediated actin polymerization. Therefore, we undertook a microinjection approach. We first generated anti-Nckα and anti-Nckβ antibodies that recognize the native forms of Nckα and Nckβ, respectively. It is shown in Fig. 6A that a commercial anti-Nck antibody (71-2800; Zymed) recognized both HA-tagged Nckα and Nckβ proteins (lanes 1 and 2). In contrast, our anti-Nckα and anti-Nckβ antibodies only recognized HA-tagged Nckα (lanes 3 versus 4) and HA-tagged Nckβ (lanes 5 versus 6), respectively. To confirm that both Nckα and Nckβ are expressed in NIH 3T3 cells, total lysates of NIH 3T3 and seven other cell lines were immunoblotted with either anti-Nckα (Fig. 6B) or anti-Nckβ (Fig. 6C) antibody. It is clearly shown that Nckα is expressed in all the cells tested (Fig. 6B), whereas Nckβ is expressed in most but not all of the eight cell lines tested (Fig. 6C). Nonetheless, Nckα and Nckβ are coexpressed in NIH 3T3 cells (indicated by arrows). The anti-Nckα and anti-Nckβ antibodies showed neutralizing effects in vitro, since they blocked GST-Nckα and GST-Nckβ binding to PDGFR in a concentration-dependent fashion (data not shown).
FIG. 6.
Microinjection of anti-Nckβ but not anti-Nckα antibodies blocks PDGF-stimulated actin polymerization. (A) Lysates of HA-Nckα-transfected (lanes 1, 3, and 5) or HA-Nckβ-transfected (lanes 2, 4, and 6) cells were resolved in an SDS gel, transferred to a nitrocellulose membrane, and blotted with either 71-2800 (Zymed; cross-reacting with α and β) (lanes 1 and 2), anti-Nckα (lanes 3 and 4), or anti-Nckβ (lanes 5 and 6) antibody. Results were visualized by ECL. (B and C) Total lysates of the eight indicated cell lines were resolved in duplicate SDS gels and subjected to Western blotting using either anti-Nckα (B) or anti-Nckβ (C) antibody, followed by ECL. (D to F′) Serum-starved NIH 3T3 cells, cultured in eight-chamber coverslips, were microinjected with either control IgG or antibodies (500 ng/μl), together with FITC-dextran as a marker protein to identify injected cells. Cells were then stimulated with PDGF-bb (100 ng/ml) for 15 min at 37°C. The actin cytoskeleton was revealed by rhodamine-labeled phalloidin staining as described in the text. Images were recorded with a Zeiss confocal microscope (magnification, × 150). For one experiment, 25 to 50 cells were injected with each antibody, and the experiment was repeated three times.
These antibodies were further purified and used for microinjection. Figures 6D to F′ show that microinjection of either an irrelevant rabbit immunoglobulin (D and D′) or anti-Nckα (E and E′) antibody did not affect PDGF-induced membrane ruffling (D′ versus D and E′ versus E). In contrast, microinjection of the anti-Nckβ antibody significantly, albeit not completely, inhibited the effect of PDGF (Fig. 6F′ versus F). These results were reproducible in three independent microinjection experiments. We conclude that Nckβ regulates PDGFR signaling to the actin cytoskeleton.
Membrane-bound Nckβ inhibits Rac signaling.
To gain further insights into the mechanisms of Nckβ action, we tested whether or not Nckβ interferes with Rac1 signaling, which is known to mediate PDGF-induced formation of lamellipodia and membrane ruffles (28). A Myc-tagged, constitutively active Rac1 (Rac1-L61) was introduced into NIH 3T3 cells with and without cotransfection with Nckα or Nckβ. Constitutively active Cdc42 (Cdc42-L61) and Rho (Rho-L63) were included as controls. Previous studies indicate that membrane localization is the key step for Nck to activate PAK (17, 34). Therefore, we speculated that if the binding of Nckβ to PDGFR, i.e., relocation from the cytoplasm to the plasma membrane, is an essential step for Nckβ to execute its interfering effect on PDGFR signaling, one would need to construct a constitutively membrane-bound Nckβ to mimic the “active stage” (PDGFR bound) of Nckβ.
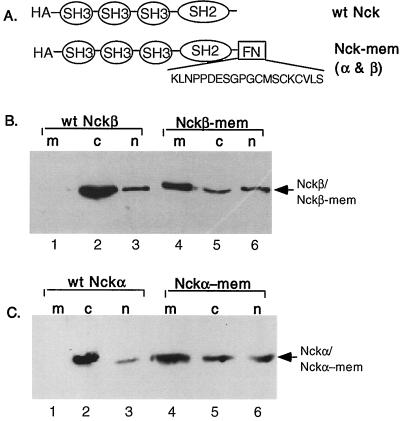
Figure 7 shows that the farnesylation signal sequence of Ras, KLNPPDESGPGCMSCKCVLS, was fused to the carboxyl termini of Nckβ and Nckα to create Nckβ-mem and Nckα-mem, repectively (Fig. 7A). To verify the effectiveness of the farnesylation signal sequence, transfected cells were fractionated into membrane, cytosol, and nucleus fractions. Equal portions of the cellular fractions were resolved by an SDS gel, transferred to a nitrocellulose membrane, and immunoblotted with anti-HA antibody. It can be seen that the majority of wild-type Nckβ was detected in the cytosol fraction (Fig. 7B, lane 2 versus lanes 1 and 3), and a small amount was detected in the nuclear fraction (lane 3). However, over 50% of the HA-Nckβ-mem was found in the membrane fraction (lane 4 versus lanes 5 and 6). The small amount of Nckβ-mem that still remained in the cytosol fraction (lane 5) is most likely the unfarnesylated portion of Nckβ-mem. Similar results were observed for Nckα-mem (Fig. 7C). The majority of the Nckα-mem was found in the membrane fraction (lane 4 versus lane 1). The amounts of membrane-associated Nckβ (Nckβ-mem) and Nckα (Nckα-mem) should be regarded as highly significant, because even in PDGF-stimulated cells, only a small percentage (∼5 to 7%) of Nck binds to the activated PDGFR (15, 22). These membrane-bound Nck gene constructs were cotransfected with the Rho GTPases, and their effects on the GTPases' signaling were investigated. In these experiments, coexpression of HA-Nck and Myc-Rac1 in the same cells was differentiated by double staining with FITC-conjugated (green) rabbit anti-mouse IgG (against anti-HA monoclonal antibody) and AMCA-conjugated (blue) goat anti-rabbit IgG (against rabbit anti-Myc antibody), while the changes in actin polymerization was again visualized by TRITC-conjugated phalloidin.
FIG. 7.
Construction of membrane-bound Nckα and Nckβ. The Ras farnesylation sequence, KLNPPDESGPGCMSCKCVLS, was linked in frame to the C termini (immediately following the last amino acid residues) of the Nck genes (A). Wild-type (wt) HA-Nckβ and HA-Nckβ-mem gene constructs (B) or HA-Nckα and HA-Nckα-mem and constructs (C) were transfected into NIH 3T3 cells. After 48 h, cells were subjected to a cellular fractionation procedure (see text) to obtain the membrane (m), cytosol (c), and nuclear (n) factions. Equal portions of each of the fractions were analyzed by Western blot analysis using anti-HA monoclonal antibody 12CA5. The results were visualized by ECL.
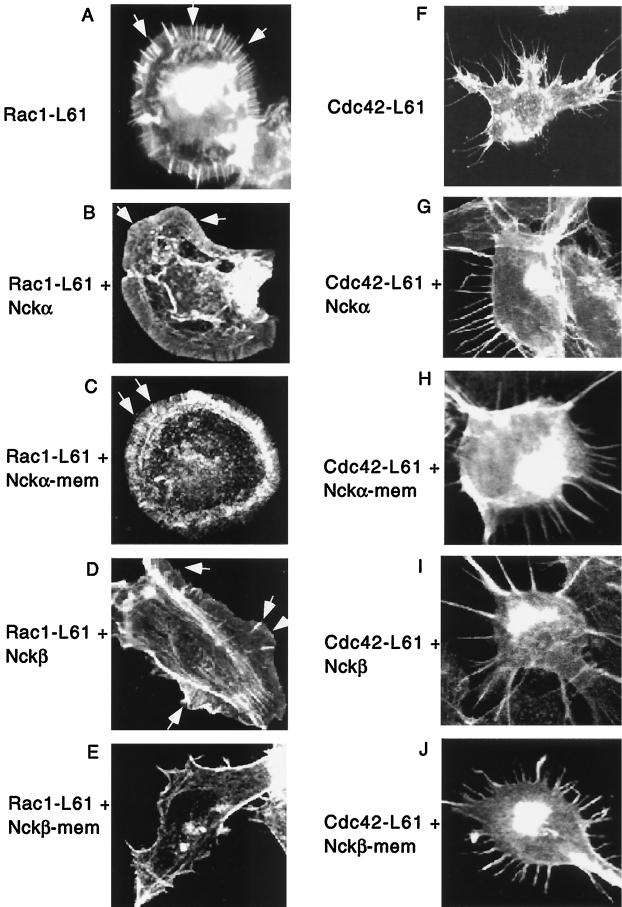
Consistent with previously published studies (6), expression of Rac1-L61 induced dramatic lamellipodia and membrane ruffles as well as filopodia in NIH 3T3 cells (Fig. 8A). The filopodium formation was likely due to activation of Cdc42 by Rac-L61 in these cells. Expression of Cdc42-L61 strongly induced filopodium formation (Fig. 8F). For unknown reasons, Rho-L63 did not cause clear actin stress fiber formation in NIH 3T3 cells (data not shown). Cells cotransfected with wild-type Nckα showed little inhibition of Rac1-L61-induced lamellipodium formation and membrane ruffling, although filopodia no longer appeared (Fig. 8B). Even the membrane-bound Nckα produced no effect (Fig. 8C). Cells cotransfected with wild-type Nckβ exhibited a moderate inhibition of lamellipodium formation and membrane ruffling, although thickness of the ruffled membrane was still evident (Fig. 8D versus 8A). Moreover, this moderate inhibition occurred in only 15% of the positively stained cells examined.
FIG. 8.
Nckβ-mem but not Nckα-mem inhibits Rac1-induced membrane ruffling and lamellipodium formation. Cells were transfected with Rac1-L61 (A) or Cdc42-L61 (F) (0.3 mg/well) alone or cotransfected with Rac1-L61 plus wild-type Nckα (B), Rac1-L61 plus Nckα-mem (C), Rac1-L61 plus wild-type Nckβ (D), Rac1-L61 plus Nckβ-mem (E), Cdc42-L61 plus wild-type Nckα. (G), Cdc42-L61 plus Nckα-mem (H), Cdc42-L61 plus wild-type Nckβ (I) or Cdc42-L61 plus wild-type Nckβ-mem (J) (Rac/Cdc42:Nck ratio, 0:3:2.5). To identify the double-transfected Rac1/Cdc42 plus Nck cells, staining with a combination of mouse anti-HA antibody 12CA5 followed by FITC-conjugated rabbit anti-mouse IgG and rabbit anti-Myc antibody followed by AMCA-conjugated goat anti-rabbit IgG was used. Changes in actin polymerization were detected by TRITC-conjugated phalloidin. Statistical analysis of Rac1 (K) and Cdc42 (L) was made from 80 to 100 randomly selected FITC and AMCA double-stained positive cells. Values represent [(number of actin-polymerized cells)/(total number of cells selected)] × 100. Four independent experiments were carried out, and they showed similar results.
Interestingly, cotransfection with Nckβ-mem resulted in dramatic inhibition of Rac1-L61-induced lamellipodium formation (Fig. 8E) in more than 50% of the Nckβ-mem-positive cells, in which membrane ruffling was almost completely gone and a rather thin and smooth membrane appeared. The statistical analysis of Nckβ-mem's effect on Rac1-L61 is summarized in Fig. 8K.
In contrast, neither Nckβ nor Nckβ-mem showed any inhibitory effect on Cdc42-L61-induced filopodium formation (Fig. 8I and J), suggesting that the effect of Nckβ-mem on Rac1-L61 was specific. Similarly, neither Nckα nor Nckα-mem had any effect on Cdc42-L61-induced filopodium formation (Fig. 8G and H). As previously mentioned, since constitutively active Rho1, Rho1-L63, did not cause significant stress fiber formation in NIH 3T3 cells, we were not able to assess the effect of Nckα-mem or Nckβ-mem on Rho1-L63 signaling. The statistical analysis of Nckβ's effect on Cdc42-L61 is summarized in Fig. 8L. The above results suggest that Nckβ participates, via an unknown mechanism, in Rac1 signaling in response to PDGF. A likely possibility is that Nckβ relocates cytoplasmic signaling proteins such as PAK to the plasma membrane and presents them to activated Rac.
DISCUSSION
Nck has been implicated to play a role in cell mitogenesis and morphogenesis. Recent genetic studies in Drosophila suggest that Nck links cell surface tyrosine phosphorylation to the actin cytoskeleton during neuronal guidance and targeting (5). Whether or not Nck has a similar function in mammalian cells remained unclear. We and others have recently shown that Nck represents a two-gene family including Nckα (formerly Nck) and Nckβ (also known as Grb4/Nck2) (3, 4, 38). It is of interest to understand whether or not different Nck species have their own specific functions in cells. In the current study, we have investigated the roles of Nckα and Nckβ in PDGF-stimulated actin polymerization and subsequent membrane ruffling in NIH 3T3 cells. The results of these experiments show that Nckβ but not Nckα plays a specific role in PDGFR signaling to the actin cytoskeleton. This function of Nckβ requires binding to PDGFR, because the SH2 domain mutant of Nckβ failed to act in a dominant negative fashion and membrane-bound Nckβ showed a constitutive interfering effect. Mutations in the SH3 domains of Nckβ also abolished the interfering effect of Nckβ, and the middle SH3 domain of Nckβ appeared to play the most important role. Interestingly, only membrane-attached, not cytoplasmic, Nckβ blocked the function of Rac1, a mediator between PDGFR and membrane ruffling and formation of lamellipodia. Under similar conditions, membrane-bound Nckβ had no inhibitory effect on Cdc42-induced formation of filopodia. In comparison, Nckα, either cytoplasmic or membrane bound, had no effect on either Rac1 or Cdc42 signaling. These results suggest that Nckβ acts either downstream of or in parallel with Rac1 signaling in response to PDGF. We hypothesize that following PDGF stimulation, Nckβ associates, via its SH2 domain, with the activated PDFGR, thereby relocating its SH3-bound molecules to the plasma membrane. These Nckβ-SH3-associated molecules then participate in Rac1 signaling to the actin cytoskeleton.
The observation that overexpression of wild-type Nckβ produced a dominant negative instead of an enhancing effect on PDGFR signaling and that, in comparison, overexpression of wild-type Nckα had no such effect was somewhat unexpected. A likely explanation is that Nckβ orchestrates a number of SH3-binding proteins and maintains them in a certain stoichiometry in order to execute its function. Increasing the cellular concentration of Nckβ alone would disrupt or titrate the ratio between Nckβ and its SH3-interacting proteins. For instance, if the middle SH3-binding protein plays a critical role, the overexpressed Nckβ would have its middle SH3 domain unoccupied due to lack of free middle-SH3-binding proteins in the cytoplasm. Rao and Zipursky showed that in Drosophila, Dock requires multiple domains acting in cis. Either a combination between the middle SH3 domain and the SH2 domain or a combination between the middle SH3 domain and the first and the third SH3 domains could mediate the signaling events (27). In Dock, it was the middle SH3 domain, not the SH2 domain, that was always required. Furthermore, none of the domain mutations in the Dock gene could act in a dominant negative fashion either by itself or in combination (27). We made similar observations. We initially predicted that the middle SH3 mutation and the SH3 triple mutations should act in a strong dominant negative fashion, but they did not. It is possible that the binding of the middle SH3 domain to its target molecule plays a role in stabilizing the binding of the SH2 domain to PDGFR. The second possible explanation for the dominant negative effect of wild-type Nckβ is that Nckβ plays a negative role in the PDGFR signaling to the actin cytoskeleton. Overexpression of Nckβ, similar to overexpression of a negative regulator such as a protein tyrosine phosphatase, would enhance its endogenous inhibitory effect. In fact, the results of our mutagenesis studies favor this hypothesis, in which both the SH2 and the triple and middle SH3 mutants are no longer able to block PDGFR signaling, or the negative signal can no longer be propagated through these mutants. While future studies will be required to further distinguish between these possibilities, the results of our microinjection experiments strongly argue that Nckβ plays a direct role in PDGFR signaling to the actin cytoskelton.
During the course of this study, a critical issue was the specificity of Nckβ action. We initially argued that overexpressed Nckβ may have had nonspecific competition for binding to other phosphotyrosine sites in addition to binding to its own site in the activated PDGFR. In this case, overexpressed Nckβ could prevent other PDGFR-binding molecules from getting into their sites, by which PDGFR signaling to the actin cytoskeleton was indirectly blocked. This argument has since been challenged by three lines of evidence that strongly suggest that the interfering effect of the overexpressed Nckβ was specific for Nckβ. First, overexpression of the other Nck family member Nckα, which has previously been shown to share a phosphotyrosine binding site with one of the two SH2 domains of the p85 subunit of PI3-K (22), did not show any interfering effect on either PDGFR or Rac-L61 signaling to the actin cytoskeleton, even though the SH2 domains of Nckβ and Nckα have a high degree (85%) of homology. In particular, since PI3-K has been reported to play a role in PDGF signaling to the actin cytoskeleton (6, 8), Nckα, not Nckβ, would be considered more likely to block PDGF-stimulated actin polymerization. The fact that Nckα did not inhibit PDGFR/PI3-K signaling to the actin cytoskeleton can be explained by the fact that the p85 subunit has two SH2 domains and its binding to Y740 has a much higher affinity than the binding to Y751 (11). p85 could even bind PDGFR with a mutation at Y751, where Nckα binds. It has recently been shown that tyrosine-778 (its binding protein remains unknown) in PDGFR-β plays an important role in PDGFR signaling to the actin cytoskeleton (31). Thus, multiple PDGFR-binding proteins may be involved in regulation of the actin cytoskelton. Second, overexpression of membrane-bound Nckβ inhibited the constitutively active Rac-L61-induced membrane ruffling and lamellipodium formation, in which SH2 domain binding was apparently not involved because of a lack of PDGFR activation. This observation suggests that Nckβ acts either downstream of or in parallel with Rac1. Nckβ could play such a role as “feeding” (i.e., relocation of critical effector molecules to the GTP-bound Rac1) Rac1 with cytoplasmic targets such as PAK1 kinase. Again, under similar conditions, membrane-bound Nckα had no effect. Third, we have recently generated anti-Nckα and anti-Nckβ antibodies which recognize the native forms of the gene products. Microinjection of anti-Nckβ antibody but not anti-Nckα antibody or irrelevant immunoglobulin molecules significantly blocked PDGF-induced actin polymerization. The exact mechanism by which the microinjected anti-Nckβ antibodies blocked the function of the endogenous Nckβ in the cells remains unknown. Assuming that the antibodies block PDGFR signaling by binding to Nckβ and preventing it from interacting with PDGFR, based on the fact that they had a neutralizing effect in an in vitro test by blocking GST-Nckβ binding to PDGFR, the microinjection results strongly support the hypothesis that Nckβ acts between PDGFR and the actin cytoskeleton.
The specific function of Nckα remains to be further studied. Both Northern and Western analyses showed that Nckα is expressed in all the cell lines so far tested, in comparison to Nckβ, whose expression is absent in certain cell types. It is possible that Nckα has a similar function to Nckβ but mediates signaling by a different cell surface receptor(s). For instance, Nckα may mediate Eph receptor signaling in the pathway of Eph/Nck/NIK/JNK (1). It is also possible that Nck in different cell types has different functions. In T lymphocytes, Nck (whether it is Nckα or Nckβ remains unknown) is required for T-cell receptor-mediated interleukin-2 gene expression and, therefore, cell proliferation (44) and cytoskeletal assembly (3a). Now, having recognized Nck as a multiple gene family, we have begun to reveal the cellular function and specificity of different Nck adapters. Mice deficient in either Nckα or Nckβ have been made available (T. Pawson, personal communication). Cell lines derived from these Nck-knockout mice or embryos will provide powerful tools for better understanding Nck signaling and function. Continued genetic studies of Drosophila and Caenorhabditis elegans will provide more guidance for studying the mammalian Nck genes. Lastly, the chromosomal locations of the Nck genes coincide with the locations of mutations which are associated with a number of human diseases, including cancer (9, 38). It would be interesting to study whether Nck gene mutations influence the occurrence or frequency of human diseases.
ACKNOWLEDGMENTS
We are very grateful to John Cooper for PDGFR-expressing TRMP cell lines and to Alan Hall for the Rho GTPase constructs. We thank Andrius Kazlauskas for his useful advice and suggestions and Elaine Fuchs for allowing us to access her confocal facility. We thank Nancy Wu of the USC Transgenic Facility for her help in microinjection and Ernie Brown at the USC Confocal Core Facility. The DNA Sequencing Facility at the University of Chicago is also acknowledged.
This work was supported in part by NCI grant R01 CA65567 (to W.L.) and by NIH grant R01 AR46538 (to D.T.W.). W.L. was a recipient of the American Cancer Society Junior Faculty Research Award.
The first two authors contributed equally to this work.
REFERENCES
- 1.Becker E, Huynh-Do U, Holland S, Pawson T, Daniel T O, Skolnik E Y. Nck-interacting Ste20 kinase couples Eph receptors to c-Jun N-terminal kinase and integrin activation. Mol Cell Biol. 2000;20:1537–1545. doi: 10.1128/mcb.20.5.1537-1545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birge R B, Knudsen B S, Besser D, Hanafusa H. SH2 and SH3-containing adaptor proteins: redundant or independent mediators of intracellular signal transduction. Genes Cells. 1996;1:595–613. doi: 10.1046/j.1365-2443.1996.00258.x. [DOI] [PubMed] [Google Scholar]
- 3.Braverman L E, Quilliam L A. Identification of Grb4/Nck, a src homology 2 and 3 domain-containing adapter protein having similar binding and biological properties to Nck. J Biol Chem. 1999;274:5542–5559. doi: 10.1074/jbc.274.9.5542. [DOI] [PubMed] [Google Scholar]
- 3a.Bubeck Wardenburg J, Pappu R, Bu J Y, Mayer B, Chernoff J, Straus D, Chan A C. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- 4.Chen M, She H, Davis E M, Spicer C M, Kim L, Ren R, Le Beau M, Li W. Nck family genes, chromosomal location, expression and signaling specificity. J Biol Chem. 1998;273:25171–25178. doi: 10.1074/jbc.273.39.25171. [DOI] [PubMed] [Google Scholar]
- 5.Garrity P A, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky S L. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- 6.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 7.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 283–312. [Google Scholar]
- 8.Hawkins P T, Eguinoa A, Qiu R-G, Stokoe D, Cooke F T, Walters R, Wennström S, Claesson-Welsh L, Evans T, Symons M, Stephens L. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- 9.Hing H, Xiao J, Harden N, Lim L, Zipursky S L. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 10.Huebner K, Kastury K, Druck T, Salcini A E, Lanfrancone L, Pelicci G, Lowenstein E, Li W, Park S H, Cannizzaro L, Pelicci P G, Schlessinger J. Chromosome locations of genes encoding human signal transduction adapter proteins Nck, Shc and Grb2. Genomics. 1994;22:281–287. doi: 10.1006/geno.1994.1385. [DOI] [PubMed] [Google Scholar]
- 11.Kazlauskas A. Receptor tyrosine kinases and their targets. Curr Opin Genet Dev. 1994;4:5–14. doi: 10.1016/0959-437x(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 12.Kazlauskas A, Cooper J A. Autophosphorylation of the PDGF receptor in the kinase insert region regulates interaction with cellular proteins. Cell. 1989;58:1121–1133. doi: 10.1016/0092-8674(89)90510-2. [DOI] [PubMed] [Google Scholar]
- 13.Kazlauskas A, Feng G S, Pawson T, Valius M. The 64-kDa protein that associates with the PDGF receptor subunit via tyrosine 1009 is the SH2-containing phosphotyrosine phosphatase Syp/SHPTP2/PTP1D/SHPTP3. Proc Natl Acad Sci USA. 1993;90:6939–6942. doi: 10.1073/pnas.90.15.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawe D C, Hahn C, Wong A J. The Nck SH2/SH3 adapter protein is present in nucleus and associates with the nuclear SAM68. Oncogene. 1997;14:223–231. doi: 10.1038/sj.onc.1200821. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Hu P, Skolnik E Y, Ullrich A, Schlessinger J. The SH2 and SH3 domain-containing Nck protein is oncogenic and a common target for phosphorylation by different surface receptors. Mol Cell Biol. 1992;12:5824–5833. doi: 10.1128/mcb.12.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, She H. The SH2 and SH3 adapter Nck: a two-gene family and a linker between tyrosine kinases and multiple signaling networks. Histol Histopathol. 2000;15:947–955. doi: 10.14670/HH-15.947. [DOI] [PubMed] [Google Scholar]
- 17.Lu W, Katz S, Gupta R, Mayer B J. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 18.Margolis B. Proteins with SH2 domains: transducers in the tyrosine kinase signaling pathway. Cell Growth Differ. 1992;3:73–80. [PubMed] [Google Scholar]
- 19.Mayer B J, Baltimore D. Signaling through SH2 and SH3 domains. Trends Cell Biol. 1993;3:8–13. doi: 10.1016/0962-8924(93)90194-6. [DOI] [PubMed] [Google Scholar]
- 20.McCarty J H. The Nck SH2/SH3 adaptor protein: a regulator of multiple intracellular signal transduction events. Bioessays. 1998;20:913–921. doi: 10.1002/(SICI)1521-1878(199811)20:11<913::AID-BIES6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura R, Li W, Kashishian A, Zhou M, Mondino A, Hu P, Cooper J, Schlessinger J. Two signaling molecules share a phosphotyrosine-containing binding site in the PDGF receptor. Mol Cell Biol. 1994;13:6889–6896. doi: 10.1128/mcb.13.11.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobes C D, Hawkins P, Stephens L, Hall A. Activation of the small GTP-binding proteins rho and rac by growth factor receptors. J Cell Sci. 1995;108:225–233. doi: 10.1242/jcs.108.1.225. [DOI] [PubMed] [Google Scholar]
- 24.Obermeier A, Ahmed S, Mannser E, Yen S C, Hall C, Lim L. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 1998;17:4238–4339. doi: 10.1093/emboj/17.15.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park D. Cloning, sequencing, and overexpression of SH2/SH3 adaptor protein Nck from mouse thymus. Mol Cells (Korea) 1997;7:231–236. [PubMed] [Google Scholar]
- 26.Pawson T. Protein modules and signaling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 27.Rao Y, Zipursky S L. Domain requirements for the Dock adapter protein in growth cone signaling. Proc Natl Acad Sci USA. 1998;95:2077–2082. doi: 10.1073/pnas.95.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 29.Rockow S, Tang J, Xiong W, Li W. Nck inhibits NGF and basic FGF induced PC12 cell differentiation via mitogen-activated protein kinase-independent pathway. Oncogene. 1996;12:2351–2359. [PubMed] [Google Scholar]
- 30.Ruan W, Pang P, Rao Y. The SH2/SH3 adaptor protein dock interacts with the Ste20-like kinase misshapen in controlling growth cone motility. Neuron. 1999;24:595–605. doi: 10.1016/s0896-6273(00)81115-0. [DOI] [PubMed] [Google Scholar]
- 31.Ruusala A, Sundberg C, Arvidsson A K, Rupp-Thuresson E, Heldin C H, Claesson-Welsh L. Platelet-derived growth factor (PDGF)-induced actin rearrangement is deregulated in cells expressing a mutant Y778F PDGF beta-receptor. J Cell Sci. 1998;111:111–120. doi: 10.1242/jcs.111.1.111. [DOI] [PubMed] [Google Scholar]
- 32.Schlessinger J. SH2/SH3 signaling proteins. Curr Opin Genet. 1994;4:25–30. doi: 10.1016/0959-437x(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 33.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (PAK1) regulates actin organization of mammalian cells. Curr Biol. 1996;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 34.Smith J M, Katz S, Mayer B J. Activation of the Abl tyrosine kinase in vivo by Src homology 3 domains from the Src homology 2/Src homology 3 adaptor Nck. J Biol Chem. 1999;274:27956–27962. doi: 10.1074/jbc.274.39.27956. [DOI] [PubMed] [Google Scholar]
- 35.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 36.Stein E, Huynh-Do U, Lane A A, Cerretti D P, Daniel T O. Nck recruitment to Eph receptor, EphB1/ELK, couples ligand activation to c-Jun kinase. J Biol Chem. 1998;273:1303–1308. doi: 10.1074/jbc.273.3.1303. [DOI] [PubMed] [Google Scholar]
- 37.Su Y C, Han J, Xu S, Cobb M, Skolnik E Y. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Symons M, Derry J M, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka M, Gupta R, Mayer B J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu Y, Li F, Wu C. Nck-2, a novel src homology2/3-containing adapter protein that interacts with the LIM-only protein PINCH and components of growth factor receptor kinase-signaling pathways. Mol Biol Cell. 1998;9:3367–3382. doi: 10.1091/mbc.9.12.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorobieva N, Protopopov A, Protopopov M, Kashuba V, Allikmets R L, Modi W, Zabarovsky E R, Klein G, Kisselev L, Graphodatsky A. Localization of human AF2 and NCK genes and 13 other Notl-linking clones to chromosome 3 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1995;68:91–94. doi: 10.1159/000133898. [DOI] [PubMed] [Google Scholar]
- 42.Wennström S, Hawkins P, Cooke F, Hara K, Yonezawa K, Kasuga M, Jackson T, Claesson-Welsh L, Stephens L. Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr Biol. 1994;4:385–393. doi: 10.1016/s0960-9822(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 43.Wennström S, Siegbahn A, Yokote K, Arvidsson A K, Heldin C H, Mori S, Claesson-Welsh L. Membrane ruffling and chemotaxis transduced by the PDGF beta-receptor require the binding site for phosphatidylinositol 3′ kinase. Oncogene. 1994;9:651–660. [PubMed] [Google Scholar]
- 44.Yablonski D, Kane L P, Qian D, Weiss A. A Nck-Pak1 signaling module is required for T-cell receptor-mediated activation of NFAT, but not of JNK. EMBO J. 1998;17:5647–5657. doi: 10.1093/emboj/17.19.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]