Abstract
The striatum comprises multiple subdivisions and neural circuits that differentially control motor output. The islands of Calleja (IC) contain clusters of densely-packed granule cells situated in the ventral striatum, predominantly in the olfactory tubercle (OT). Characterized by expression of the D3 dopamine receptor, the IC are evolutionally conserved, but have undefined functions. Here we show that optogenetic activation of OT D3 neurons robustly initiates self-grooming in mice while suppressing other ongoing behaviors. Conversely, optogenetic inhibition of these neurons halts ongoing grooming, and genetic ablation reduces spontaneous grooming. Furthermore, OT D3 neurons show increased activity before and during grooming and influence local striatal output via synaptic connections with neighboring OT neurons (primarily spiny projection neurons, SPNs), whose firing rates display grooming-related modulation. Our study uncovers a novel role of the ventral striatum’s IC in regulating motor output and has important implications for the neural control of grooming.
Introduction
The striatum is an evolutionarily conserved brain structure essential for motor control1. By integrating brain-wide inputs, distinct striatal circuits facilitate the selection of appropriate behaviors through their downstream outputs. The striatum is divided into several subregions, each with distinct anatomical connectivity and putative functions2, 3. These striatal functions are topographically organized, progressing from motivational and reward-related behaviors in the ventral striatum to procedural habit learning in the dorsolateral striatum. The Islands of Calleja (IC) are a cluster of densely-packed granule cells in the ventral striatum (predominantly in the olfactory tubercle, OT)4–12, which are conserved across many species including rodents and humans7, 13, 14. It remains unclear how the enigmatic IC relates to the functional organization of striatal compartments.
The striatum, including the OT, has a fairly uniform cellular composition. The principal neurons are GABAergic, spiny projection neurons (SPNs, also called medium spiny neurons), classified as D1- or D2-type according to the dopamine receptors they express. They are intermingled with several subtypes of interneurons which are relatively scarce2, 3, 15, 16. One exception to this general cellular architecture are the IC, aggregations of GABAergic granule cells4–12. These neurons are characterized by expression of the D3 dopamine receptor17–22, even though the majority of IC D3 neurons also coexpress either the D1 or D2 receptor21, 22. The rodent IC start to form in early postnatal days and the IC neurons derive from the subventicular zone10, 11. While there are a few reports on projections to the IC6, 23, 24 and potential roles of the IC in neuropsychiatric disease25, 26 and cardiovascular regulation27, 28, the neural circuits in which IC neurons are embedded and the functions they support remain largely unknown.
Self-directed grooming, an evolutionally-conserved, stereotyped motor pattern, plays critical roles in hygiene maintenance, thermoregulation, de-arousal, and stress reduction29, 30. Notably, excessive grooming is an indication of compulsive repetitive behaviors in animal models of neurological and neuropsychiatric disorders29–32. Grooming can occur spontaneously or be triggered by external stimuli (e.g., water spraying on the face). In rodents, a complete grooming bout consists of a syntactic chain that progresses sequentially from nose (phase I) to face (phase II) and head (phase III) grooming and ends with body licking (phase IV). Although grooming typically starts with phase I, most grooming bouts (85–90%) in mice contain only a partial chain33, 34.
Neurobiological control of grooming behavior involves multiple brain regions. In addition to the brainstem35–37, amygdala38, 39 and hypothalamus40–45, the striatum is also implicated in patterning and execution of this behavior32, 46–51. Different parts of the striatum and their downstream targets play distinct roles in grooming behavior. Lesions to the dorsolateral striatum significantly reduce syntax completion without affecting total grooming time, while lesions to the ventral pallidum, a downstream target of the ventral striatum, reduce total grooming time without affecting syntax completion46.
In this study, we use a transgenic mouse line expressing Cre recombinase under the D3 receptor promoter to gain genetic access to the IC, an irregular and deep neural structure. Whole-brain imaging demonstrates that the IC form a continuous, branched network in the ventral striatum, largely within the OT. Optogenetic activation of OT D3 neurons induces robust grooming even in competition with other ongoing behaviors, while inactivation of these neurons halts ongoing grooming. Genetic ablation of OT D3 neurons reduces the total spontaneous grooming time by reducing both the number of grooming bouts and grooming duration per bout. Retrograde tracing reveals that OT D3 neurons receive direct inputs from other brain regions implicated in grooming (e.g., the ventral pallidum, lateral hypothalamus, and amygdala) while anterograde tracing and ex vivo electrophysiological recordings indicate that OT D3 neurons influence striatal output by inhibiting neighboring OT neurons including SPNs. In vivo calcium imaging via fiber photometry demonstrates increased OT D3 neuron activity before and during grooming. Moreover, single-unit recordings reveal that the firing rates of OT units display grooming-related modulation in freely behaving mice. Taken together, this study uncovers a novel role of ventral striatal circuitry involving OT D3 neurons in grooming control.
Results
Islands of Calleja contain densely-packed D3 neurons
To dissect the neural circuitry and function of the IC, we took advantage of a BAC transgenic D3-Cre line (see Methods for details), which allows genetic access to IC neurons characterized by expression of the D3 dopamine receptor. When crossed with a Cre-dependent tdTomato reporter line (Ai9) to generate D3-Cre/tdTomato mice, the IC are readily visible via whole-brain CLARITY imaging (Fig. 1a, Extended Data Fig. 1a–c, and Supplementary Video 1). In coronal sections, the IC are identifiable as clusters (“islands”) of D3-Cre/tdTomato granule cells in the ventral striatum (Fig. 1d, e). Interestingly, in whole brains, these “islands” appear as a contiguous structure with sparser cell bodies and neurites of isolated D3 neurons in between islands (Fig. 1a, Extended Data Fig. 1, and Supplementary Video 2). With the exception of the “major island” in the nucleus accumbens (NAc), all other islands are situated in the OT (Extended Data Fig. 1b). The shape and spatial distribution of the IC showed some variations between the two hemispheres of the same mouse and across different mice (Extended Data Fig. 2). We also observed tdTomato-positive neurons in the piriform cortex, hypothalamus, and hippocampus (Extended Data Fig. 3).
Figure 1. The Islands of Calleja (IC) contain densely-packed granule cells expressing the D3 dopamine receptor.
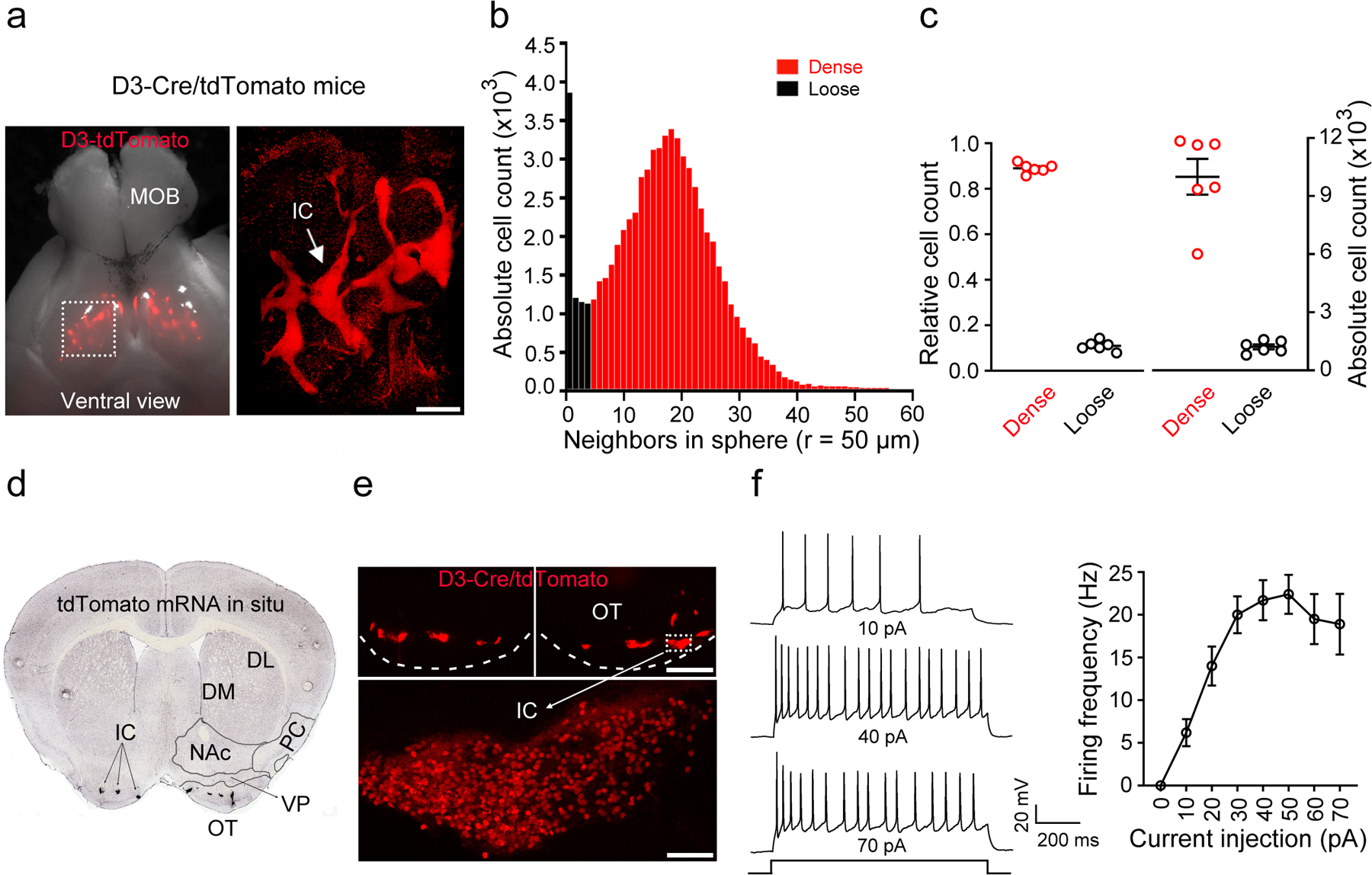
a, Ventral view of the brain from a D3-Cre/tdTomato mouse. Right, an enlarged image of the OT (dotted rectangle in left panel) after the brain was made transparent via CLARITY. Similar results were obtained from 6 OTs of 3 mice. MOB, main olfactory bulb. OT, olfactory tubercle. Scale bar = 500 μm. b, Histogram depicting the number of neighboring D3 neurons within the vicinity of a given cell (50 μm sphere radius). Two populations of neurons (dense versus loose) emerge with a cut-off of 5 neighboring cells. c, Relative and absolute numbers of dense versus loose D3 neurons. Data were from 6 OTs of 3 mice for b,c. d, A coronal section across the ventral striatum showing in situ hybridization against tdTomato mRNA in a D3-Cre/tdTomato mouse (image from Allen Mouse Brain Connectivity Atlas: http://connectivity.brain-map.org/transgenic/experiment/304168043)60. DL/DM, dorsolateral/dorsomedial striatum. NAc, nucleus accumbens. PC, piriform cortex. VP, ventral pallidum. OT, olfactory tubercle. e, D3-Cre/tdTomato neurons visualized as clusters (IC) in the OT (upper) and in a single island (lower) (similar observations in 10 mice used in f). Scale bars = 100 μm (upper) and 50 μm (lower). f, Left, firing of an IC D3-Cre/tdTomato neuron upon varied current injections. Right, average firing frequencies versus injected currents (n = 10 neurons selected from 10 mice). Holding potential = −60 mV. All averaged data are shown as mean ± SEM.
To determine whether we can specifically target the IC using this D3-Cre mouse line, we quantified D3-Cre/tdTomato neurons in the OT, NAc and ventral pallidum (Extended Data Fig. 1b), which account for 83.3%, 11.3% and 5.4% of the total tdTomato+ neurons in this region, respectively (Extended Data Fig. 1d,e). Within the OT, although there are some “loose” D3-Cre/tdTomato neurons, the vast majority (~90%) of OT D3-Cre/tdTomato neurons can be categorized as “dense” clusters belonging to the IC network (Fig. 1b,c; see Methods for more details). Overall, this transgenic D3-Cre line provides genetic access to OT D3 neurons, which predominantly reside in the IC.
We characterized intrinsic electrophysiological properties of OT D3 neurons using whole-cell patch-clamp recordings in acute brain slices from D3-Cre/tdTomato mice. We recorded IC D3-Cre/tdTomato neurons (Fig. 1e), which had small cell bodies (6–8 μm in diameter) and high input resistances (2.10 ± 0.28 GΩ, n = 10 cells). These neurons displayed no spontaneous action potentials, suggesting that they are under tonic inhibition and/or need external excitatory inputs to fire. Upon current injections, 29.1% (16 out of 55) of IC D3 neurons fired a single spike, but the majority (70.9%: 39 out of 55) fired persistently with a maximal firing rate of ~20 Hz (Fig. 1f), which guided the stimulation parameters in the initial optogenetic experiments described below.
Optogenetic activation of OT D3 neurons induces grooming
In order to manipulate the activity of OT D3 neurons, we generated D3-Cre/ChR2 mice by crossing the D3-Cre line with the Cre-dependent channelrhodopsin-2 (ChR2) line (Ai32). We verified that OT D3-Cre/ChR2 neurons in acute brain slices were reliably activated by blue laser stimulation (latency < 1 ms) (Fig. 2a). The persistent firing neurons (as in Fig. 1f) responded with high fidelity to stimulations up to 20 Hz, consistent with our acute slice analysis, and maintained robust firing for up to 20 s. These stimulation parameters were used in the initial optogenetic experiments in D3-Cre/ChR2 mice with an optical fiber unilaterally implanted in the OT (Fig. 2b). Upon blue light stimulation, mice immediately stopped exploration in the open field and started grooming. During 20 s stimulations, mice groomed for ~10 s before resuming other activities (walking, exploring, etc.) (Fig. 2b).
Figure 2. Activation of OT D3-Cre/ChR2 neurons induces grooming.
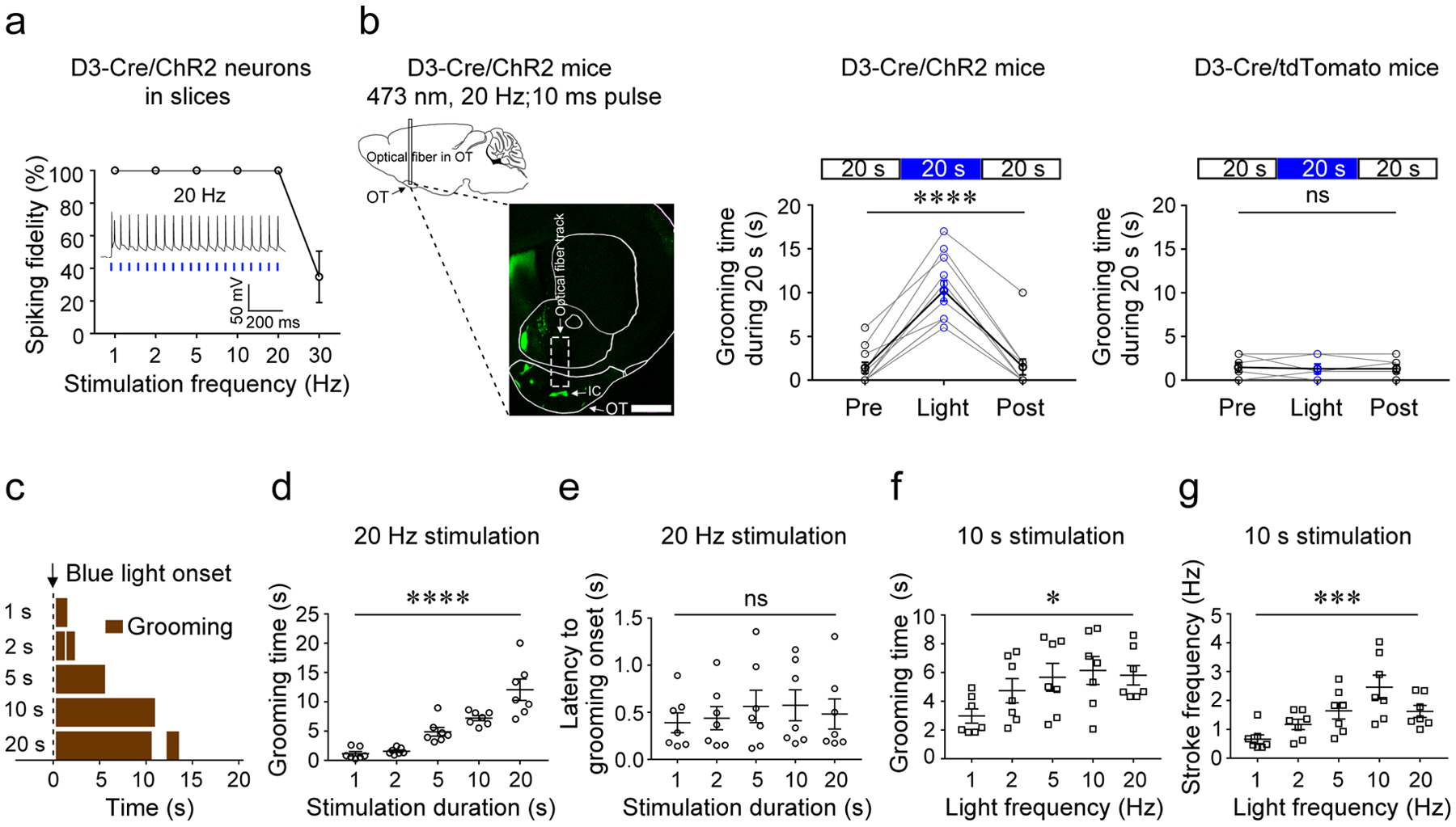
a, Spiking fidelity of IC D3-Cre/ChR2 neurons (n = 5 from 3 mice) upon blue light stimulation at different frequencies. The inset shows firing of an IC D3-Cre/ChR2 neuron upon laser stimulation at 20 Hz. b, Left, schematic of the optogenetic stimulation strategy and a representative image (coronal section) showing the optical fiber near the IC in the OT. Scale bar = 1 mm. Middle, the total grooming time in 20 s before, during, and after blue light stimulation in D3-Cre/ChR2 mice (n = 11; 3 trials/mouse; Friedman test, p = 1.69E-5). Right, the total grooming time in 20 s before, during, and after blue light stimulation in D3-Cre/tdTomato control mice (n = 6; 3 trials/mouse; Friedman test, p = 1.000). OT, olfactory tubercle. IC, islands of Calleja. c, Grooming behavior induced by light stimulation of varying lengths from a single mouse. d, The total time of grooming (within 20 s from the light onset) induced by varying stimulation durations (n = 7; F (4, 30) = 46.54; p = 1.90E-12). e, Latency from light onset to grooming onset (n = 7; F (4, 30) = 0.21; p = 0.933). f, The total time of grooming (within 10 s from the light onset) induced by varying stimulation frequencies (n = 7; F (4, 30) = 2.85; p = 0.0410). g, The stroke frequency of grooming induced by varying stimulation frequencies (n = 7; F (4, 30) = 6.45; p = 0.0007). Aligned Rank Transformation one-way ANOVA was used in (d)-(g). Each data point is an average of five trials from a single animal for d-g. *p < 0.05, ***p < 0.001, ****p < 0.0001, and ns, not significant. All averaged data are shown as mean ± SEM.
To further analyze the light-induced grooming behavior, we videotaped D3-Cre/ChR2 mice in a clean cage from a side view. Blue light-induced grooming always started with phase I (paw and nose grooming with elliptical bilateral strokes) and about 10% (n = 70 trials from 10 and 20 s stimulation in Fig. 2d) progressed to phase II (face grooming with unilateral strokes) or phase III (head grooming with bilateral strokes) (Supplementary Video 3). We never observed blue light-induced grooming ending with body licking (phase IV), even though the mice exhibited spontaneous grooming bouts with complete four-phase syntax. In this study, if not otherwise stated, “grooming” thus refers to nose-face-head grooming (phase I to III) excluding body licking (phase IV). The beginning of a grooming bout was defined as when both paws were lifted to reach the face and the ending as when both paws returned to the cage floor. Notably, optogenetically induced grooming had a side bias: the forepaw on the stimulated side always lifted first, even though spontaneous grooming could start with lifting of either forepaw (40 bouts examined from the same mice in Fig. 2d).
Next, we quantified grooming behavior upon blue light stimulation of the OT by varying stimulation parameters. The latency to grooming onset upon light stimulation was short (0.49 ± 0.13 s, n = 175 trials from 7 mice) and independent of stimulation duration (Fig. 2c,e). Grooming time gradually increased with stimulation duration (Fig. 2c,d; Supplementary Video 4). For stimulation >10 s, grooming typically stopped before the light turned off, suggesting an internally programmed upper limit of grooming duration per bout (see below). When the frequency of light pulses increased from 1 to 20 Hz, mice showed a significant increase in grooming duration (Fig. 2f). Strikingly, the animals displayed faster strokes at higher stimulation frequencies (Fig. 2g and Supplementary Video 5). Taken together, these results suggest that OT D3 neurons may function in initiation, maintenance, and execution of grooming behavior.
We performed several controls to test if blue light-induced grooming indeed results from optogenetic activation of OT D3-Cre/ChR2 neurons. First, green laser with the same parameters did not elicit grooming behavior in the same cohort of D3-Cre/ChR2 mice (Supplementary Video 3). Green light should excite ChR2 with less efficiency although we did not validate this here. Second, the same blue laser stimulation did not elicit grooming behavior when the optical fiber was implanted in the OT of D3-Cre/tdTomato mice (n = 6) (Fig. 2b and Supplementary Video 6). Third, the same blue laser stimulation did not elicit grooming behavior when the optical fiber was implanted in either the NAc (n = 5) or hippocampus of D3-Cre/ChR2 mice (n = 3 in dentate gyrus and n = 2 in CA3; Supplementary Video 6). Finally, to rule out that blue light-induced grooming results from stimulation of en passant ChR2+ fibers originating elsewhere (such as the hippocampus, Extended Data Fig. 3c,c’), we focally injected Cre-dependent AAV1-DIO-ChR2-EYFP virus in the OT of D3-Cre/tdTomato mice. Here, blue light stimulation in the OT also induced grooming (n = 3). By contrast, we never observed grooming upon green light stimulation or blue light stimulation in a different brain region (Supplementary Video 7).
We next asked whether optogenetic activation of OT D3 neurons triggers grooming even when the animals are engaged in other highly motivated or compulsive behaviors (Fig. 3). In a cohort of D3-Cre/ChR2 mice (an optical fiber unilaterally implanted in the OT), we introduced an unfamiliar mouse into the test cage (both sexes used in separate trials). When the resident D3-Cre/ChR2 mouse was engaged in social investigation, blue light stimulation was triggered. Blue light induced grooming (with a similar latency as described in Fig. 2e) while suppressing social investigation, which was resumed after grooming termination. By contrast, green light, which should cause weaker activation of ChR2, had little effect on social investigation behavior (Fig. 3a,a’ and Supplementary Video 8). Similarly, blue (but not green) light induced grooming during both feeding behavior in food-deprived mice (Fig. 3b,b’ and Supplementary Video 9) and itch-induced scratching, a compulsive behavior52 (Fig. 3c,c’ and Supplementary Video 10). Because a complete grooming bout starts from nose grooming (phase I) and ends with body licking (phase IV), we examined the effect of OT D3 neuron activation on ongoing episodes of body licking. Blue light stimulation of the OT in D3-Cre/ChR2 mice terminated the ongoing body licking in all trials and reinitiated phase I grooming in more than 70% of the trials (17 out of 24) while green light had minimal effects (2 out of 24) (Fig. 3d,d’ and Supplementary Video 11). Taken together, these findings indicate that optogenetic activation of OT D3 neurons is sufficient to induce robust grooming behavior even in competition with alternative ongoing behaviors.
Figure 3. Activation of OT D3-Cre/ChR2 neurons induces grooming while suppressing alternative ongoing behaviors.
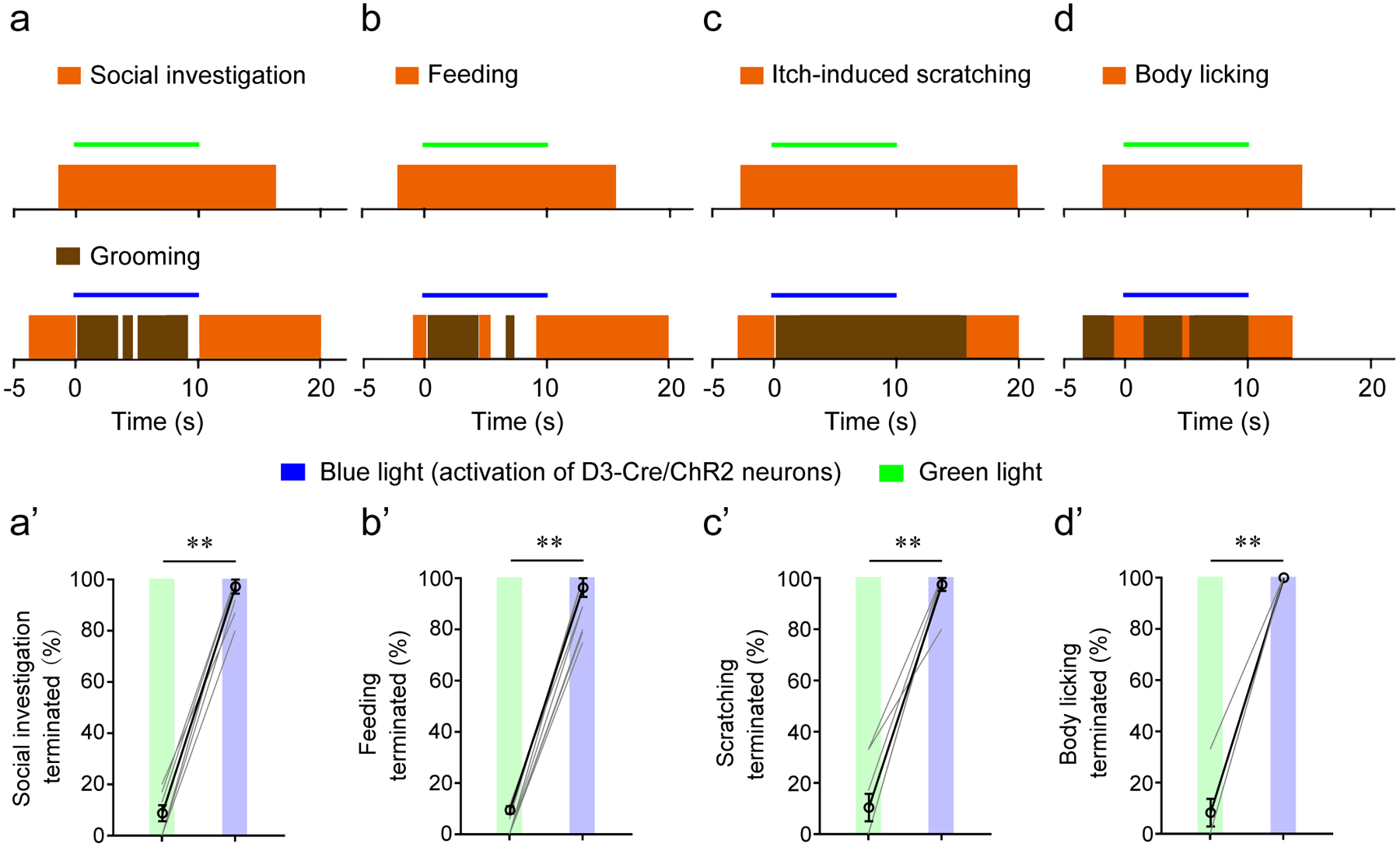
a-c, Blue light (activation of D3-Cre/ChR2 neurons) or green light (weaker activation of ChR2) were delivered during three ongoing behaviors: social investigation (green light, 74 trials; blue light, 102 trials) (a), feeding after 20-hr food deprivation (green light, 69 trials; blue light, 84 trials) (b), and itch-induced scratching (green light, 33 trials; blue light, 42 trials) (c). a’-c’, Summary data from 8 mice. Each mouse was tested in 8–15 trials for (a’), 6–18 trials for (b’), and 4–10 trials for (c’), and a percentage was calculated. d, d’, Blue light, but not green light, stopped body licking (n = 24 trials; 3 trials/mouse in 8 mice). Wilcoxon signed-rank test: p = 0.008 for a’-d’. **p < 0.01. Both blue and green laser stimulations were at 20 Hz with 10 ms pulses. All averaged data are shown as mean ± SEM.
OT D3 neuronal loss-of-function effects on grooming
Since phasic optogenetic activation of OT D3 neurons may impose unusually strong network activity in striatal circuits, we asked whether inactivation of OT D3 neurons affects ongoing grooming behavior. We injected Cre-dependent AAV2/9-DIO-eArchT-EGFP virus unilaterally into the OT of D3-Cre/tdTomato mice so that infected D3 neurons express the green light-activated outward proton pump archaerhodopsin and are thus inhibited upon green light exposure (Fig. 4a). We verified that D3-Cre/eArchT neurons in acute brain slices were hyperpolarized by green laser pulses without rebound firing (Fig. 4a). For in vivo testing, mice were monitored in clean cages four to six weeks after virus injection (to allow development of significant photocurrents upon modest light stimulation53) and fiber implantation. When mice started spontaneous grooming, green light was delivered to inhibit neuronal activity of D3-Cre/eArchT neurons (Fig. 4b and Supplementary Video 12). To rule out that behavioral changes resulted from thermal effects of continuous green light stimulation, we always subjected the same mice to continuous blue light exposure at the same intensity. Green light inactivation of OT D3-Cre/eArchT neurons halted spontaneous grooming within 3 s in 69% of the trials, while blue light with the same parameters, which should excite eArchT with less efficiency, only coincided with halted grooming in 12% of the trials (Fig. 4b and Supplementary Video 12). As expected, grooming duration in green light trials was significantly shortened compared to blue light trials (Fig. 4c,d). Similarly, green light inactivation of OT D3-Cre/eArchT neurons stopped water spray-induced grooming in 50% of the trials, significantly higher than the 3% in the blue light control group (Fig. 4e and Supplementary Video 13). These results suggest that OT D3 neuron activity is necessary to maintain grooming behavior.
Figure 4. Inactivation of D3-Cre/eArchT neurons halts ongoing grooming.
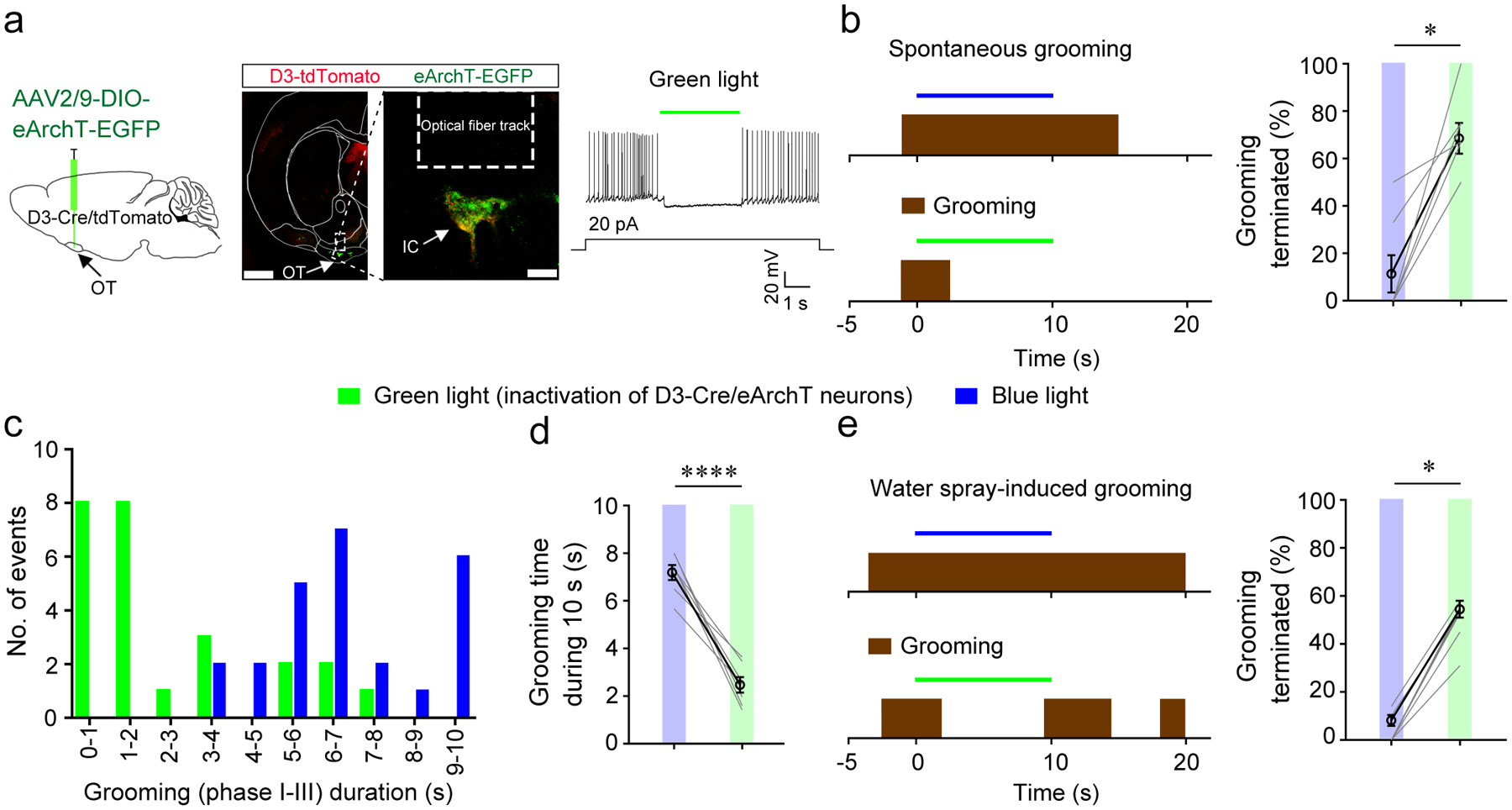
a, Left, viral injection strategy. Middle, post-mortem verification of viral infection and optical fiber implantation (similar results were observed in 7 mice). Scale = 1000 μm (left image) and 100 μm (right image). Right, green light effectively inhibited OT D3-Cre/eArchT neurons in acute brain slices (n = 3 out of 3 neurons). OT, olfactory tubercle. IC, islands of Calleja. b, Green light (inactivation of OT D3-Cre/eArchT neurons), but not blue light (which potentially excites eArchT with much less efficiency) shortened spontaneous grooming bouts. Left, representative trials. Right, the percentage of spontaneous grooming terminated within 3 s upon green or blue light stimulation. Wilcoxon signed-rank test, p = 0.016. c, Distribution of grooming durations upon 10 s stimulation of OT D3-Cre/eArchT neurons by green or blue light. d, Average grooming durations upon green light or blue light stimulation of OT D3-Cre/eArchT neurons. Student’s t test, t = 25.505 and p = 2.4E-07. The same dataset is analyzed for b-d: n = 25 trials from 7 mice with 3–6 trials/mouse. e, Green light (inactivation of OT D3-Cre/eArchT neurons), but not blue light stopped water spray-induced grooming. Left, representative trials. Right, the percentage of water spray-induced grooming terminated within 3 s upon green or blue light stimulation of OT D3-Cre/eArchT neurons (n = 7 mice; green light: n = 90 trials with 11–15 trials/mouse; blue light: n = 69 trials with 5–16 trials/mouse). Wilcoxon signed-rank test, p = 0.016. Both green and blue light were delivered continuously with the same intensity for a single mouse. *p < 0.05 and ****p < 0.0001. All averaged data are shown as mean ± SEM.
As multiple brain regions have been previously implicated in grooming29, we asked whether OT D3 neurons play a role in generating spontaneous grooming under normal conditions. For genetic ablation of OT D3 neurons, D3-Cre/ChR2 mice were unilaterally or bilaterally injected with Cre-dependent DTA (diphtheria toxin subunit A) virus (AAV8-mCherry-FLEX-DTA) or control AAV8-TurboRFP virus. DTA-dependent ablation of D3-Cre/ChR2 neurons was verified after completion of behavioral tests (Fig. 5a, b). Note that ChR2 was never stimulated but instead the fused EYFP was used to gauge effectiveness of the genetic ablation. EYFP fluorescent signal was significantly reduced in the ablated side compared to the control side in unilateral ablation mice (Fig. 5b), and similar efficacy was verified in bilateral ablation mice (Extended Data Fig. 5). The grooming behavior within 30 min was compared at different time points after viral injection (1 to 4 weeks). Compared to the control group, the total grooming time was reduced by 50% (Fig. 5c), arising from significant decreases in both the number of grooming bouts and the grooming duration per bout (Fig. 5d, e). Consistent with the results of unilateral optogenetic inactivation, unilateral ablation of OT D3 neurons caused similar effects as bilateral ablation. These findings suggest that OT D3 neurons play a critical role in grooming initiation as well as maintenance under normal conditions.
Figure 5. Ablation of OT D3 neurons reduces spontaneous grooming.
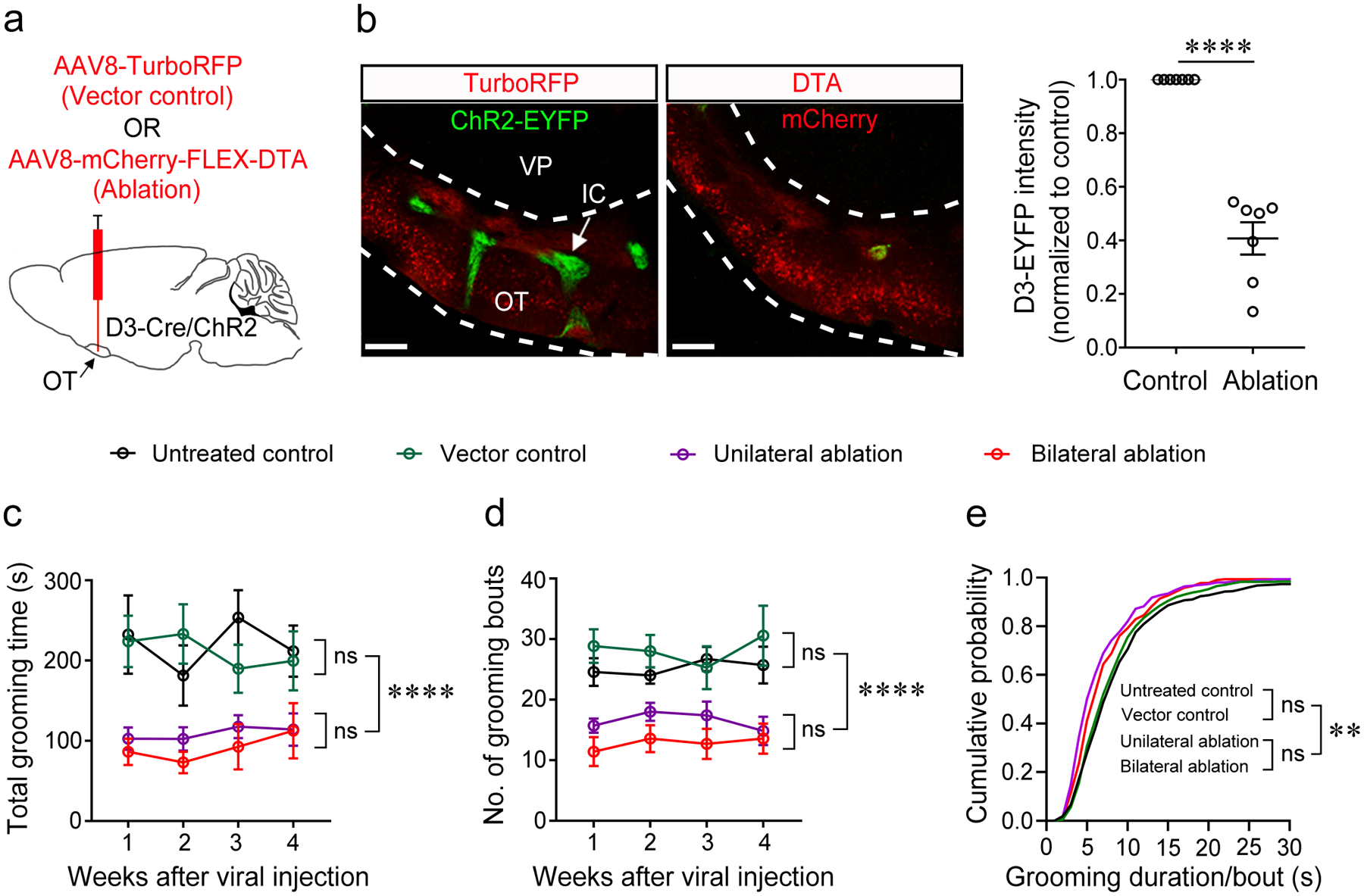
a, Strategy of genetic ablation of OT D3-Cre/ChR2-EYFP neurons via injection of Cre-dependent AAV-mCherry-FLEX-DTA with AAV-TurboRFP as control (800 nl). b, Left, injection of the DTA virus led to absence of the IC visualized by D3-Cre/ChR2-EYFP neurons (imaged four weeks post injection). Scale bars = 200 μm. Right, ablation efficiency of AAV8-DTA virus. Each data point is an average from three individual slices from one mouse. n = 7 mice. Wilcoxon signed-rank test, p = 6.10E-5. OT, olfactory tubercle. IC, islands of Calleja. VP, ventral pallidum. c-e, Summary of the total grooming time (c), number of grooming bouts (d), and cumulative probability of grooming duration/bout (e) during 30 min in the four groups of mice (n = 7 mice/group). Aligned Rank Transformation two-way ANOVA test was used in c and d. Grooming time: Ftime × treatment (9, 96) = 0.63 and p = 0.7696, Ftime (3, 96) = 0.48 and p = 0.6942, Ftreatment (3, 96) = 31.56 and p = 2.80E-14; Bout number: Ftime × treatment (9, 96) = 0.54 and p = 0.8435, Ftime (3, 96) = 0.17 and p = 0.9132, Ftreatment (3, 96) = 37.55 and p = 1.28E-15. In e, Kruskal-Wallis test was used to compare the median of the four groups, p = 0.0025. Two-sample Kolmogorov-Smirnov test was used to compare any two distributions: Untreated control vs Vector control, p = 0.512; Untreated control vs Unilateral ablation, p = 6.54E-9; Untreated control vs Bilateral ablation, p = 1.0E-4; Vector control vs Unilateral ablation, p = 6.29E-7; Vector control vs Bilateral ablation, p = 0.004; Unilateral vs Bilateral ablation, p = 0.054. **p < 0.01, ****p < 0.0001, and ns, not significant. All averaged data are shown as mean ± SEM.
OT D3 neurons receive inputs from other grooming centers
To define the brain regions that exert control over OT D3 neurons, we used the pseudotyped rabies virus (RV) system to retrogradely label neurons that are monosynaptically connected to this population54, 55. We unilaterally injected Cre-dependent AAVs that bicistronically express TVA-mCherry, a required receptor for EnvA-pseudotyped RV, and RV glycoprotein, required for transsynaptic spread, into the OT of D3-Cre mice. After 10 days, EnvA-pseudotyped RV-EGFP virus was injected into the OT. Brains were fixed 7 days later and sectioned coronally (100 μm thickness) for confocal microscopy imaging (Fig. 6a). We first verified that the “yellow” D3 neurons (TVA-mCherry and EGFP double-positive; presumptive “starter” cells) were located within the OT, predominantly within the IC (Fig. 6a, b). We then quantified 40, 55 and 68 starter cells in 3 mice (see Methods for details), and counted presynaptic (EGFP+ only) cells from every other section throughout the brain to calculate the percentage of retrogradely labeled cells in each brain region (Fig. 6c). Interestingly, presynaptic partners of OT D3 neurons were found in several brain regions (i.e., the ventral pallidum, lateral hypothalamic area, and amygdala) (Fig. 6c and Extended Data Fig. 4), which have been implicated in mediating grooming behavior35–45.
Figure 6. Whole-brain mapping of presynaptic partners of OT D3 neurons.
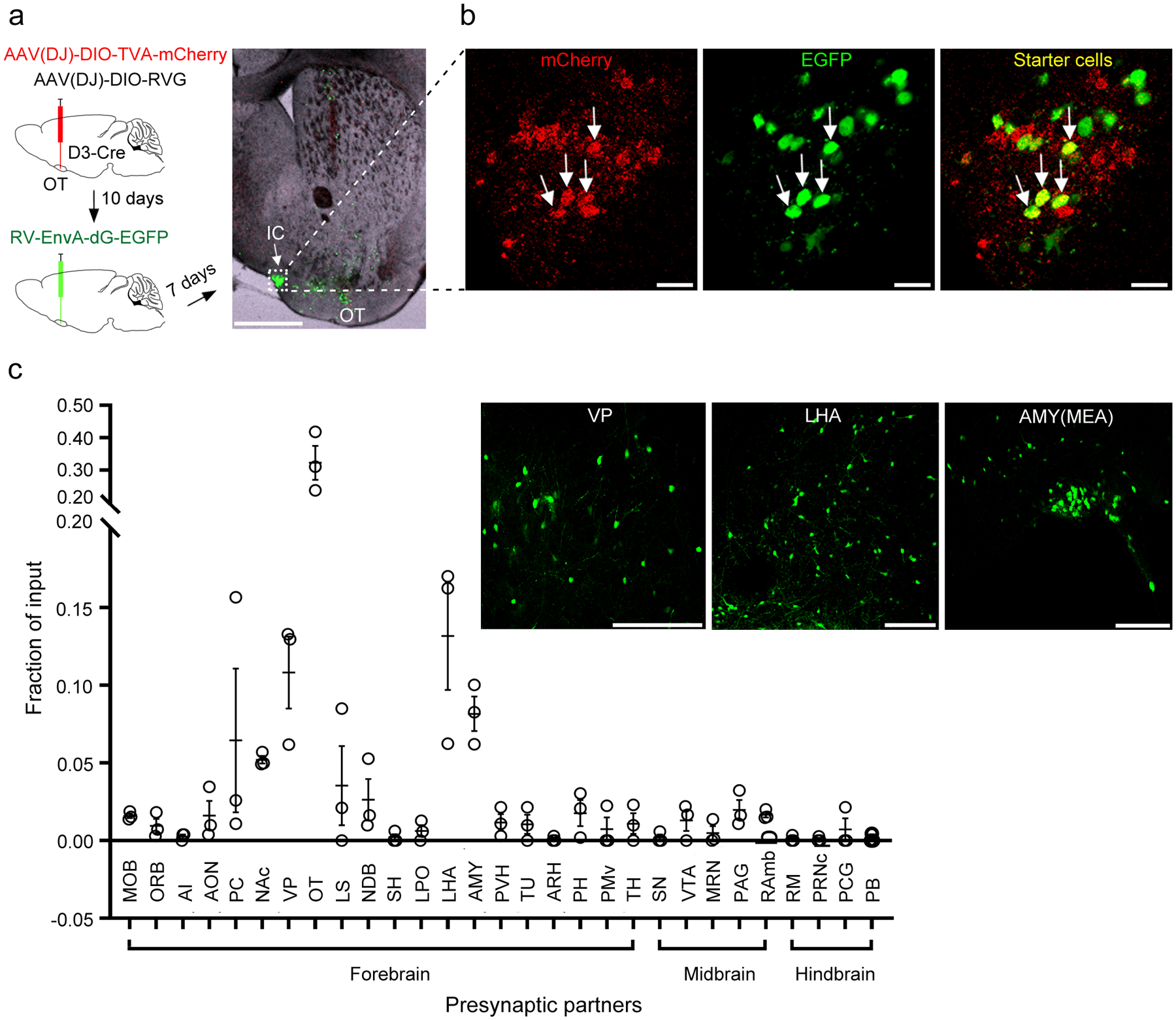
a, Left, schematic of retrograde tracing from OT D3-Cre neurons. Right, post-mortem verification of the viral injection site near one island in the OT (similar observations in 3 mice). Scale bar = 1 mm. b, Enlarged images of the island shown in a, revealing yellow “starter cells” (both mCherry and EGFP positive). Scale bars = 20 μm. c, Quantification of presynaptic inputs to OT D3 neurons in various brain regions (identified according to Allen Brain Atlas; more details in Methods and Extended Data Fig. 4). Data are averaged from 3 mice (mean ± SEM). Insets, representative images showing EGFP+ presynaptic cells in the ventral pallidum (VP), lateral hypothalamic area (LHA) and medial amygdala (AMY(MEA)). Scale bars = 200 μm. MOB, main olfactory bulb. ORB, orbital area. AI, agranular insular area. AON, anterior olfactory nucleus. PC, piriform cortex. NAc, nucleus accumbens. VP, ventral pallidum. OT, olfactory tubercle. LS, lateral septal nucleus. NDB, diagonal band nucleus. SH, septohippocampal nucleus. LPO, lateral preoptic area. LHA, lateral hypothalamic area. AMY, amygdala. PVH, paraventricular hypothalamic nucleus. TU, tuberal nucleus. ARH, arcuate hypothalamic nucleus. PH, posterior hypothalamic nucleus. PMv, ventral premammillary nucleus. TH, thalamus. SN, substantia nigra. VTA, ventral tegmental area. MRN, midbrain reticular nucleus. PAG, periaqueductal gray. RAmb, midbrain raphe nuclei. RM, nucleus raphe magnus. PRNc, pontine reticular nucleus, caudal part. PCG, pontine central gray. PB, parabrachial nucleus.
OT D3 neurons make local synaptic connections
To identify downstream targets of OT D3 neurons, we used multiple parallel approaches. First, we traced the output of OT D3-Cre/tdTomato neurons via CLARITY imaging and found no projections outside the ventral striatum (Extended Data Fig. 1), even though we were able to trace fine axons (Supplementary Video 2). It is unlikely that technical limitations precluded identification of such projections as we were able to trace fine projections of D3 neurons in the hypothalamus (Extended Data Fig. 3b,b’). Second, we performed anterograde tracing by injecting Cre-dependent AAV(DJ/8)-FLEX-Synaptophysin::EGFP unilaterally into the OT of D3-Cre/tdTomato mice (n = 7) (Fig. 7a). Four weeks later, coronal sections (100 μm thickness) were obtained from the entire brain and examined for fluorescent signals. Even though the expression of both tdTomato and synaptophysin-EGFP is driven by D3-Cre, the red and green signals do not show much overlap (Fig. 7a) as tdTomato strongly labels cell bodies and neurites while synaptophysin-EGFP mainly targets synaptic terminals. Third, a subset of mice injected with AAV1-DIO-ChR2-EYFP (n = 3) or AAV2/9-DIO-eArchT-EGFP (n = 4) in the OT for optogenetic behavioral experiments were additionally used for anterograde tracing. In all 14 mice used in viral tracing, the processes of OT D3 neurons were predominantly within the OT and no axonal projections were identified outside the ventral striatum. As shown in Fig. 7a, synaptophysin-EGFP axonal terminals are mainly observed in the OT, both within and outside the IC. We then used patch-clamp recording pipettes to fill individual D3-Cre/tdTomato neurons with a fluorescent dye and all 10 cells (7 within and 3 outside the IC) had neurites up to 200 μm in length within the ventral striatum (Fig. 7b). These anterograde tracing results, combined with previous reports that IC neurons are GABAergic and derive from the subventricular zone10, 11, led to the conclusion that these D3 neurons are a unique subtype of local striatal interneuron.
Figure 7. OT D3 neurons make local synaptic contacts.
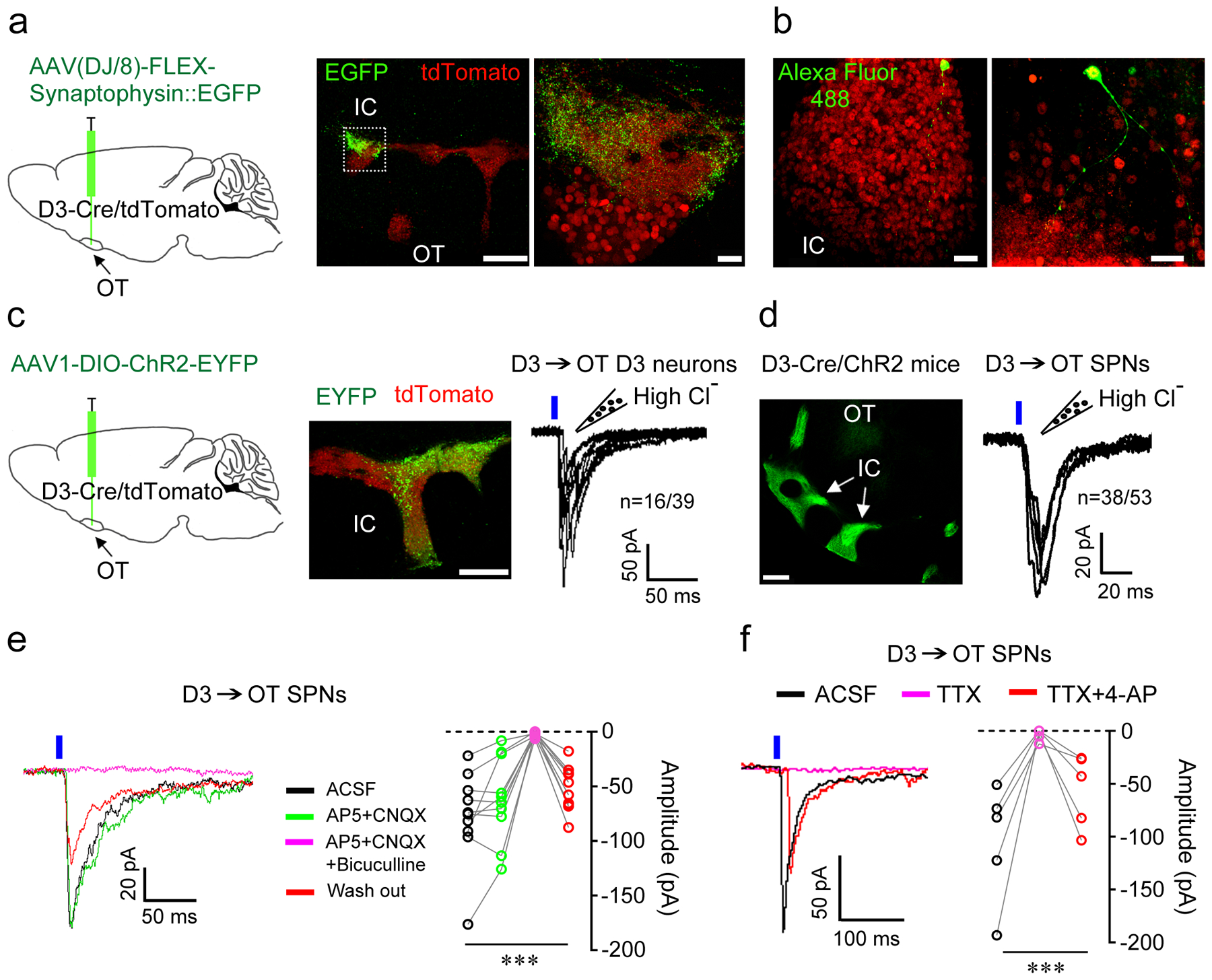
a, Left, experimental design of anterograde tracing from OT D3-Cre/tdTomato neurons. Middle, EGFP+ axonal terminals were visible after four weeks within the IC and OT. Right, enlarged image from the dashed rectangle. Note clustered D3 neuron cell bodies (bottom) and neuropils with numerous labeled synapses (top). Similar observations in 7 mice. Scale bars = 200 μm (middle) and 20 μm (right). b, Alexa Fluor 488 filled D3 neurons within an island (left; 7 neurons) or in between two islands (right; 3 neurons) of D3-Cre/tdTomato mice. Scale bars = 20 μm. c, OT D3 neurons inhibit each other. Left, experimental design. Middle, post-mortem verification of the viral injection site in the OT. Scale bar = 200 μm. Right, repeated 10 ms blue light-evoked postsynaptic currents (PSCs) in IC neurons (tdTomato+ but ChR2-EYFP−). d, D3 neurons inhibit neighboring OT SPNs. Left, IC with densely-packed D3-Cre/ChR2 neurons in the OT. Scale bar = 500 μm. Right, repeated 10 ms blue laser pulses evoked PSCs in OT SPNs near the IC. e, Light-evoked PSCs in OT SPNs were blocked by GABAA receptor antagonist 10 μM bicuculline but not changed by glutamate receptor antagonists (50 μM AP5+20 μM CNQX). Friedman test: F(3,36) = 18.840, p = 2.95E-4. f, Light-evoked PSCs in OT SPNs were blocked by TTX (1 μM) and reappeared after co-application of TTX + 4-AP (1 mM, potassium channel blocker), supporting monosynaptic connection. Friedman test: F(2,12) = 10.000, p = 7.72E-4. n = 10 and 5 cells from 5 mice in e and f, respectively. Each data point in e and f is averaged from 6–10 traces. ***p < 0.001. Holding potential = −60 mV.
To verify that anatomically identified synaptic connections are functional, we performed whole-cell patch-clamp recordings in acute brain slices. We first examined synaptic connections among OT D3 neurons by injecting Cre-dependent AAV1-DIO-ChR2-EYFP virus into the OT of D3-Cre/tdTomato mice (Fig. 7c). Three to four weeks later, OT D3 neurons were recorded in brain slices. As expected, in ChR2-EYFP+ neurons, blue laser pulses evoked high-fidelity action potentials under current clamp mode (similar to Fig. 2a) and inward somatic optical currents with extremely short latency (< 1 ms) under voltage clamp mode. To enhance our detection of GABAA-mediated inhibitory postsynaptic currents (PSCs), we used a high Cl− internal solution to set the reversal potential of GABAA-mediated currents at ~0 mV and recorded inward currents from a holding potential of −60 mV (see Methods for more details). In a subset (41.0%) of D3-Cre/tdTomato positive, but ChR2-EYFP negative neurons, blue light pulses evoked inward PSCs (latency: ~4.7 ms and jitter: ~1.3 ms; Fig. 7c and Extended Data Fig. 6a,b). The variation in latency and polyphasic appearance in some traces during repeated stimuli were likely due to the fact that some presynaptic D3 neurons fired more than one spike upon light stimulation (e.g., the first light pulse induced two spikes in the example shown in Fig. 2a) and that not all presynaptic D3 neurons fired/released neurotransmitters at exactly the same time. We next examined synaptic connections from D3-ChR2 neurons to neighboring SPNs (within the territory of ChR2-EYFP+ axons) in D3-Cre/ChR2 mice (Fig. 7d). In >70% of SPNs, identified by their low input resistance < 200 MΩ and characteristic firing patterns56, blue light pulses evoked inward PSCs (latency: ~4.6 ms and jitter: ~1.1 ms; Fig. 7d and Extended Data Fig. 6c,d), which were blocked by GABAA receptor antagonist bicuculline but not changed by glutamate receptor antagonists, AP5 and CNQX (Fig. 7e). Furthermore, light-evoked PSCs were blocked by tetrodotoxin (TTX) and reappeared after coapplication of TTX and 4-AP (a potassium channel blocker, which enables action potential-independent, ChR2-mediated neurotransmitter release), suggesting monosynaptic connections (Fig. 7f). To investigate whether there is biased innervation from D3 neurons onto neighboring D1 or D2 SPNs, we bred double transgenic D3-Cre and D1-tdTomato mice (see Methods) and injected Cre-dependent AAV1-DIO-ChR2-EYFP virus into the OT (similar to Fig. 7c). Four weeks later, we recorded D1-tdTomato and non-tdTomato SPNs (putative D2-type) alternately in OT slices. Blue light activation of D3-ChR2 neurons evoked inward currents in both D1-type (7 out of 10) and D2-type SPNs (6 out of 9) without evident innervation bias. These experiments reveal that OT D3 neurons make monosynaptic connections onto other OT neurons including both D3 neurons and SPNs, thus influencing striatal output.
OT neurons display grooming-related activity
We next addressed the question how OT D3 neuron activity relates to grooming using fiber photometry-based in vivo calcium imaging (Fig. 8a). We injected Cre-dependent AAV9-FLEX-GCaMP7s (a genetically encoded Ca2+ indicator) virus unilaterally into the OT of D3-Cre/tdTomato mice and implanted an optical fiber above the injection site. After allowing 3–4 weeks for GCaMP7s expression, we recorded temporally demodulated 490 nm and 405 nm fluorescent signals (Ca2+-dependent and -independent, respectively, Fig. 8b) in freely behaving mice. Given the IC irregular shape and deep location in the brain (Fig. 1 and Extended Data Figs. 1, 2), we expected some variations in fiber location relative to GCaMP7s+ “islands”. Indeed, based on post-mortem histology (Fig. 8a), mice were categorized according to optical fiber position, as either “near IC” or “away from IC” (see Methods for more details). Mice with “near IC” fiber position showed significantly elevated calcium levels (as reflected in ΔF/F) a few seconds before and after grooming onset for both spontaneous and water spray-induced grooming compared to non-grooming periods (Fig. 8c,d). By contrast, mice with “away from IC” fiber placement did not show grooming-related signals (Fig. 8a,c,d). The peak ΔF/F value (within ± 1 s relative to grooming onset) was negatively correlated with the distance from the fiber tip to the closest GCaMP7s+ island (Fig. 8e). These results indicate that OT D3 neurons were active before and during grooming, supporting their contribution to grooming initiation and possibly maintenance.
Figure 8. OT neurons show grooming-related activity.
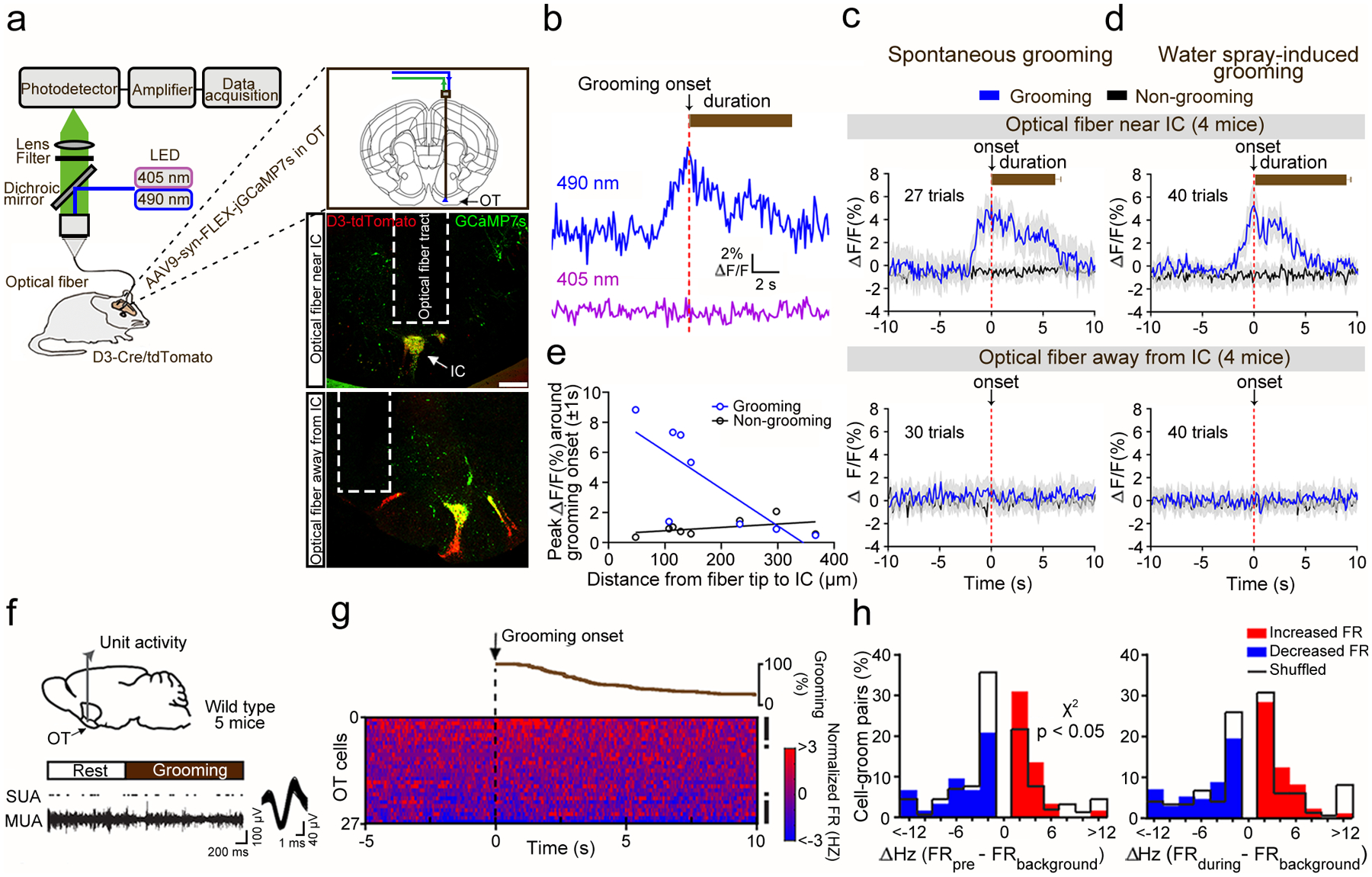
a, Schematic of fiber photometry recording from OT D3 neurons (left) and viral injection/fiber implantation sites (right). Scale bars = 200 μm. b, Fluorescent signals during a single spontaneous grooming bout. c-d, Ca2+-dependent fluorescent changes during grooming (blue) and non-grooming bouts (black) from mice with optical fiber near (upper) or away from the IC (lower). Bar graphs indicate grooming durations (mean ± SEM). Randomization test was used to compare grooming versus non-grooming events: p = 1.84E-3 (spontaneous) and 5.45E-3 (water spray-induced) for “Optical fiber near IC” mice, and p = 0.388 (spontaneous) and 0.527 (water spray-induced) for “Optical fiber away from IC” mice. e, Correlation between peak ΔF/F around grooming onset (± 1 s) and distance from fiber tip to closest GCaMP7s+ “island”. Pearson’s linear regression fitting: r = −0.784, p = 0.021 for grooming, and r = 0.432, p = 0.285 for non-grooming events. f, Schematic of OT unit recording in freely moving mice (upper) and example single unit activity (SUA; overlaid waveform in Inset) raster isolated from multi-unit activity (MUA) trace from an OT neuron during a spontaneous grooming bout (lower). g, Cumulative percentage of grooming bout durations aligned to onset (upper) and 2D histogram (50 ms bins) displaying normalized firing rates (FR) of all OT single units averaged across all grooming bouts (lower). Each row represents one unit. Rows are arranged from highest to lowest average change in normalized FRs ± 1s relative to groom onset. Right, black rasters denote significantly modulated units. h, Cell-groom pairs’ distributions of FR changes (ΔHz) pre- (left) or during (right) grooming compared to background, overlaid with ‘shuffled’ data (black staircase plots). More cell-groom pairs displayed an FR increase prior to grooming onset (χ2 (1, N = 334) = 3.842, p = 0.029; real vs shuffled).
To test how OT neuron firing is modulated during grooming behavior, we implanted 5 wild-type mice with tungsten multiwire electrode arrays in their OT (Fig. 8f) and analyzed single unit activity during a total of 117 spontaneous grooming events (Fig. 8g; see Extended Data Fig. 7 for single unit verification). As D3 neurons are small granule cells, we assume their activity will not be directly acquired by wire electrode arrays and thus the activity most likely stems from SPNs, the predominant type of striatal neurons. Further supporting this, from the 27 isolated OT single units, most of them had relatively low background firing rates (mean ± SEM, 2.0 ± 1.5 Hz; Extended Data Fig. 7), which is characteristic of SPNs. We compared the averaged firing rates within ± 1 s relative to grooming onset to their background activity. Among all neurons, 14 out of 27 were significantly modulated by grooming: 7 displayed significant decreases and 7 displayed increases in firing rates (Fig. 8g; see Methods for details on statistical analysis). Since each grooming bout displayed by a mouse may uniquely influence and/or be influenced by local OT neural activity, we generated individual “cell-groom pairs” (n = 621) derived from significantly modulated neurons in relation to each bout and classified them as being decreased or increased during the 1 s before grooming (“ΔHz FRpre – FRbackground”), or within 1 s following the start of grooming (“ΔHz FRduring – FRbackground”). This allowed us to identify both the magnitude and direction of firing changes from all neurons monitored in a given bout relative to grooming. In both time windows, some OT cell-groom pairs (87 and 82 in “pre” and “during”, respectively) displayed decreases in firing rates while others (90 and 89 in “pre” and “during”, respectively) displayed increases (Fig. 8h). This bidirectional change in firing relative to grooming differed from what was observed when sampling from the same neurons yet during pseudorandomly selected grooming-free ‘shuffled’ periods of time, which revealed statistically more cell-groom pairs with increased firing in the period of time before grooming onset (Fig. 8h; see Discussion). Together these findings indicate that firing of OT neurons (primarily SPNs) are altered during grooming.
Discussion
In the present study, we investigated the neural circuitry and function of OT D3 neurons mostly concentrated in the IC, a previously understudied cell population of the ventral striatum. By combining optogenetics, ex vivo and in vivo electrophysiology, whole-brain CLARITY imaging, viral circuit tracing, and in vivo fiber photometry, we discovered that this neuronal population is involved in controlling grooming behavior, highlighting a novel function of the IC striatal circuits.
Our study strongly supports that OT D3 neurons play a role in grooming initiation, maintenance, and execution. Supporting a role in grooming initiation, optogenetic activation of these neurons was sufficient to elicit robust grooming with a short latency of ~0.5 sec (Fig. 2), even when the animals were engaged in other highly motivated or compulsive behaviors including social investigation, feeding, and itch-induced scratching (Fig. 3). In addition, genetic ablation of OT D3 neurons reduced the total grooming time and the total grooming bouts by ~ 50% (Fig. 5), suggesting that these neurons contribute to grooming initiation under normal conditions. Moreover, in vivo calcium recordings from populations of OT D3 neurons via fiber photometry reveals elevated neuronal activity before grooming onset, consistent with the finding that significantly more OT neurons show firing changes before the grooming onset (Fig. 8).
Several lines of evidence support the contribution of OT D3 neurons to grooming maintenance. When OT D3-Cre/ChR2 neurons were optogenetically activated, the duration of induced grooming increased with the stimulation duration from 1 to 20 sec (Fig. 2). Furthermore, optogenetic inactivation of these neurons halted ongoing grooming (Fig. 4) and genetic ablation of these neurons significantly decreased the grooming duration per bout (Fig. 5). Finally, OT D3 neurons were active before the grooming onset and the elevated activity remained for a few seconds during grooming (Fig. 8), although we note the limited temporal resolution of this approach.
The potential involvement of OT D3 neurons in grooming execution is supported by optogenetically induced grooming experiments, wherein stroke frequency increased with optogenetic stimulation frequency from 1 to 10 Hz (Fig. 2). Moreover, initiation of grooming exhibited a side bias (i.e., the forepaw on the stimulated side lifted first), suggesting that activation of OT D3 neurons produces a motor rather than motivational signal. Taken together, the current dataset is consistent with involvement of OT D3 neurons in grooming initiation, maintenance, and execution.
While OT D3 neurons are intimately linked to grooming, our data also suggest additional mechanisms for grooming control29. For example, during 20 s optical stimulation of OT D3-Cre/ChR2 neurons, the induced grooming typically stops before the light turns off (Fig. 2), suggesting other pathways involved in cessation of grooming. Animals with bilateral ablation of OT D3 neurons still display spontaneous grooming (Fig. 5). Since unilateral ablation had similar effects as bilateral ablation, this finding likely results from involvement of other grooming centers rather than incomplete ablation. It is worth mentioning that OT D3 neurons unlikely contribute to programming of the grooming syntax. Although optogenetic activation of OT D3 neurons robustly initiated grooming, the induced grooming was limited to nose-face-head grooming (phase I to III) and almost never progressed to body licking (phase IV). Furthermore, activation of OT D3 neurons during phase IV (resulted from a spontaneous grooming bout) stopped body licking, and in most trials, reinitiated nose grooming (phase I) (Fig. 3). These results further support a role of OT D3 neurons in grooming initiation.
While the IC connectivity had been previously probed6, our study directly assesses the inputs and outputs of OT/IC D3 neurons via viral tracing tools. Our findings not only confirm previously identified presynaptic areas (e.g., the nucleus accumbens, piriform cortex, amygdala, and ventral tegmental area)6, but also extend to new regions (e.g., the ventral pallidum, hypothalamus, and midbrain structures) (Fig. 6). Some of these regions have previously been implicated in grooming control, including the ventral pallidum46, lateral hypothalamus40–44 and medial amygdala38, 39. Interestingly, optogenetic activation of glutamatergic neurons in lateral hypothalamus and medial amygdala also induce self-grooming, but with a latency of several seconds39, 44, longer than the latency period observed upon OT D3 neuron stimulation (Fig. 2). The IC/OT D3 neuron network may thus coordinate with other grooming centers in the brain to generate grooming under different conditions. Dissecting the distinct contributions from each region warrants future investigations.
IC/OT D3 neurons likely influence striatal output and behavior by inhibiting other neurons in the ventral striatum. In contrast to the “Islands of Calleja” nomenclature, whole-brain imaging of D3-Cre/tdTomato neurons reveals that these islands actually form a continuous, branched structure throughout the ventral striatum (Fig. 1, Extended Data Figs. 1, 2). Via anterograde tracing from OT D3-Cre/tdTomato neurons, we did not find projections outside the ventral striatum, suggesting that these neurons act as local interneurons and exert their function through modulation of other neighboring neurons including both D3 neurons and OT SPNs (Fig. 7). During in vivo optogenetic experiments, presumably only a subset of OT D3 neurons can be directly activated or inactivated optically via a 400 μm fiber, but such manipulations reliably triggered or halted grooming, respectively (Figs. 2–4). In addition, unilateral ablation of OT D3 neurons had a similar effect as bilateral ablation in reducing the number of grooming bouts and grooming duration per bout (Fig. 5), suggesting that loss-of-function of a partial IC network is sufficient to interfere with the grooming behavior. The effectiveness of these manipulations suggests that either a small subset of D3 neurons are sufficient to mediate grooming or that IC neurons function as a unified network, with local manipulations propagating to distant parts of the structure. The unique geometry and connectivity may enable IC neurons to act as a unified network to simultaneously modulate widespread SPNs in the ventral striatum. In addition to chemical synapses (Fig. 7), densely-packed IC D3 neurons may also communicate with each other via gap junctions57 and/or ephaptic coupling58, which may facilitate synchronized activity among the densely-packed D3 neurons. Since single-cell recordings from tiny IC neurons situated in the most ventral part of the brain are extremely challenging, we used in vivo fiber photometry to demonstrate elevated neuronal activity from populations of OT D3 neurons during grooming (Fig. 8). Given that OT D3 neurons provide direct GABAergic inputs onto neighboring neurons including SPNs (Fig. 7), it may seem counterintuitive that more OT units show increased firing before the grooming onset (Fig. 8). Several factors possibly contribute to this finding. As all OT neurons are embedded in a largely inhibitory local network, activation of a subpopulation of GABAergic neurons (e.g., IC D3 neurons) inhibits monosynaptically connected cells while leading to disinhibition of others. Additionally, D1- and D2-type SPNs, which are not distinguished in our in vivo recordings, may be differentially modulated during grooming behavior. Moreover, the average firing rate of OT neurons is relatively low (~2 Hz), which makes it challenging to identify decreased firing. Nevertheless, the results support the notion that IC neurons are uniquely positioned to change striatal output and behavior.
One question remaining is what specific role(s) the D3 receptor in IC neurons may play in grooming control and/or other physiological functions in health and disease25–28. Genetic knockout of the D3 receptor increases the basal level of grooming behavior59. However, since the majority (~80%) of IC D3 neurons coexpress D1 receptor, whereas the remaining likely coexpress D2 receptor21, 22, specific pharmacological reagents targeting the D3 receptor or genetic knockdown or knockout of this receptor in defined cell populations would be required to tease out the function of the D3 receptor in IC D3 neurons.
As the most ventral part of the striatum, the OT receives sensory inputs from the olfactory system among others, and has been implicated in associative learning, reinforcement and reward-related behaviors15, 16. The current study reveals a surprising role of IC and OT circuitry in motor control, specifically in mediating grooming behavior, expanding our current understanding of distinct roles of different subdivisions of the striatum. Interestingly, abnormal grooming is frequently observed in animal models of neurological and neuropsychiatric disorders (Tourette syndrome, obsessive compulsive disorder, and autism spectrum disorder), which are often accompanied by impairment of striatal circuits29–32. Our study highlights a novel role of the IC network in grooming control under normal conditions and suggests their dysfunction in pathological states.
Methods
Animals
The bacterial artificial chromosome (BAC) transgenic D3-Cre line (STOCK B6.FVB(Cg)-Tg(Drd3-cre)KI198Gsat/Mmucd, RRID:MMRRC_031741-UCD) line was obtained from the Mutant Mouse Resource and Research Centers (MMRRC) at University of California at Davis, an NIH-funded strain repository, and was donated to the MMRRC by Nathaniel Heintz, Ph.D., The Rockefeller University, GENSAT and Charles Gerfen, Ph.D., National Institutes of Health, National Institute of Mental Health. The D3-Cre line was crossed with the Cre-dependent tdTomato reporter line (JAX Stock No: 007909 or Ai9 line: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/) or Cre-dependent channelrhodopsin 2 (ChR2)-EYFP line (JAX Stock No: 024109 or Ai32 line: B6.Cg-Gt(ROSA)26Sortm32(CAG-COP4*H134R/EYFP)Hze)61 to generate D3-Cre/tdTomato or D3-Cre/ChR2 mice, respectively. Double transgenic D3-Cre and D1-tdTomato mice (JAX Stock No:016204, B6.Cg-Tg(Drd1a-tdTomato)6Calak)62 were bred to achieve identification of D1-type and presumptive D2-type SPNs in slice recordings. Wild-type C57bl/6j mice were offsprings from breeders purchased from the Jackson Laboratory. Approximately equal numbers of male and female mice (8–12 weeks old) were used for all experiments except for in vivo unit recordings. Since no sex difference was observed, the data from both sexes were combined. Mice were housed in temperature- and humidity-controlled facilities on a 12 h light/dark cycle with food and water available ad libitum unless otherwise stated. Mice were group-housed until the surgery of receiving virus injection and intra-cranial implantation and singly-housed afterwards. All experimental procedures were performed in accordance with the guidelines of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committees of the University of Pennsylvania and the University of Florida. The number/sex of mice used in each set of experiments are summarized in Supplementary Table 1.
Viruses
Cell type-specific trans-synaptic rabies virus (RV) labeling requires expression of Cre-dependent AAVs expressing TVA-2a-mCherry, the receptor for EnvA-pseudotyped RV, and RV glycoprotein to enable transsynaptic spread. These plasmids (pAAV.CAG.DIOloxP.TVA66T.2a.mCherry, pAAV.CAG.DIOloxP.G, and (EnvA).SAD.ΔG.EGFP) were obtained as kind gifts from Dr. Liqun Luo and the viruses were packaged in house with the DJ serotype. EnvA-pseudotyped RV-EGFP was generated both in-house (expanded RV-EGFP virus derived from stock kindly provided by Byungkook Lim63) as well as purchased from the Salk viral core. AAV(DJ/8)-FLEX-Synaptophysin::EGFP (AAV(DJ/8)-EF1a-FLEX-Synaptophysin::EGFP-WPRE-hGHpA)64 and AAV2/9-DIO-eArchT-EGFP (AAV2/9-CAG-DIO-eArchT3.0-EGFP, 1012 GC/ml)44 were from the Neuroconnectivity Core, Baylor College of Medicine, AAV1-DIO-ChR2-EYFP (AAV1-EF1a-DIO-hChR2 (H134R)-eYFP-WPRE-hGH, ≥7×1012 GC/ml; a gift from Karl Deisseroth, Addgene viral prep # 20298-AAV1) and AAV9-FLEX-GCaMP7s (pGP-AAV9-syn-FLEX-jGCaMP7s-WPRE, > 1×10¹³ vg/mL)65 from ADDGENE, AAV9-EGFP virus (AAV9.CMV.PI.EGFP.WPRE.bGH, ≥1×10¹³ μg/ml; a gift from James M. Wilson, Addgene viral prep # 105530-AAV9) from Penn Vector Core, and AAV8-CMV-TurboRFP-WPRE-rBG (2.94×1010 GC/ml) and AAV8-EF1α-mCherry-FLEX-DTA (3.3×109 viral units/ml) from the University of North Carolina Viral Vector Core (Chapel Hill, NC).
Viral Injection and Optical Fiber Implantation
For viral injection, mice were anesthetized with isoflurane (~3% in oxygen) and secured in a stereotaxic system (Model 940, David Kopf Instruments). Isoflurane levels were maintained at 1.5–2% for the remainder of the surgery. Body temperature was maintained at 37 °C with a heating pad connected to a temperature control system (TC-1000, CWE Inc.). Local anesthetic (bupivacaine, 2 mg/kg, s.c.) was applied before skin incision and hole drilling on the dorsal skull. In order to target the IC in the OT, a relatively large structure, we tested two sets of coordinates from bregma: anteroposterior (AP) 1.2 (or 1.54) mm; mediolateral (ML) ±1.1 (or 1.15) mm; dorsoventral (DV), −5.5 (or −5.0) mm for both viral injections and optical fiber implantations (Extended Data Fig. 5). The results were combined as we did not observe significant differences. Viral injection (300–500 nl except for genetic ablation experiments in Fig. 5 at 800 nl) was conducted via a Hamilton syringe (5 μl) with a flow rate of 40 nl/min controlled by an Ultra Micro Pump III (UMP3) with a SYS-micro4 controller attachment (World Precision, Sarasota, USA). The tip of the syringe was left for 10–15 min after the injection. For implantation of the optical fiber, a cannula (CFMC14L10-Fiber Optic Cannula, Ø2.5 mm Ceramic Ferrule, Ø400 μm Core, 0.39 NA; Thorlabs, Newton, NJ), customized to 6 mm length, was placed in the OT at the same coordinates as described above and fixed on the skull with dental cement. In viral injected mice, the optical fiber was implanted right above the injection site. D3-Cre/ChR2 mice were returned to home cage for recovery for one week before behavioral tests and mice with viral injection had at least three-week waiting period before tests. In a subset of D3-Cre/ChR2 mice, the optical fiber cannula was unilaterally placed in the nucleus accumbens (NAc) (AP 1.45 or 1.54 or 1.78 mm, ML ± 1.6 or ±1.13 or ± 0.62 mm, and DV −4.45 or −4.85 or −4.33 mm), hippocampus (dentate gyrus coordinates: AP −1.7 mm, ML ± 0.7 mm, and DV −2.04 mm and CA3 coordinates: AP −1.7 mm, ML ± 2.0 mm, and DV −1.85 mm). In a subset of D3-Cre/tdTomato mice with unilateral AAV1-DIO-ChR2-EYFP virus injection, the optical fiber was unilaterally placed in the paraventricular hypothalamic nucleus (PVH) (AP −0.58 mm, ML ± 0.2 mm, and DV −4.75 mm). All viral injection/expression sites and optical fiber locations were verified post-mortem (summarized in Extended Data Fig. 5) and only mice with the intended targeted site were included in data analysis except for in vivo fiber photometry experiments (see below).
In Vivo Optical Stimulation and Behavioral Assays
All behavioral procedures were performed during the light cycle between 9:00 am and 12:00 pm. The experimental mice were transferred to the testing room at least 1 h before the tests to acclimatize to the environment. Before each test, the mouse was briefly anesthetized via isoflurane and the laser fiber was coupled to the implanted fiber stud with a mating sleeve (Thorlabs Inc.). The mice were placed in an open field (40 cm × 40 cm) or a clean new cage when blue (473 nm, 10–15 mW/mm2, 1–20 Hz of 10 ms pulses for optogenetic activation) or green laser (532 nm, continuous for optogenetic inactivation) were delivered. The interval between two successive optogenetic stimulations was 3–5 min to avoid cumulative effects of the light stimulation. Mice were videotaped using a webcam at 30 frames/sec or an iPhone 6 at 240 frames/sec (for accurate measurement of the latency to grooming onset and stroke frequency).
The water spray-induced grooming was produced in D3-Cre/tdTomato mice with AAV2/9-DIO-eArchT-EGFP viral injection in the OT using a spray bottle filled with room-temperature pure water33. The mice were faced toward the direction of the nozzle (~ 20 cm away), and water mist was sprayed three times to adequately coat the mouth and facial parts. The effects of green (optogenetic inactivation) and blue laser (exciting eArchT with less efficiency) were tested for 3–5 times/day for three days. For spontaneous grooming, the effects of blue and green laser were tested for 1–3 times/day for three days.
The effects of optogenetic activation of OT D3 neurons in D3-Cre/ChR2 mice with unilateral optical fiber implantation were tested in alternative ongoing behaviors. To induce social investigation behavior, the mice were placed in a new cage with fresh bedding for 10 min of habituation. Then a stranger mouse (either male or female at the same age) was introduced into the cage. To induce feeding behavior, the mice were starved for 20 h, and then were individually placed in a new cage with fresh bedding with ad libitum access to food pellets. To elicit itch-induced scratching behavior, chloroquine (40 mM; 15 μl) was unilaterally injected into the mouse cheek66, 67. In a subset of animals, the alternative behavior included spontaneous grooming that progressed to phase IV (body licking). While the mice were engaged in these behaviors, blue (optogenetic activation) or green laser (exciting ChR2 with less efficiency) was delivered.
Anatomical Tracing and Confocal Imaging
For viral retrograde trans-synaptic tracing experiments, a mixture virus of AAV5.CAG.DIOloxP.TVA66T.2a.mCherry and AAV8.CAG.DIOloxP.G (1:1; 300 nl) was unilaterally injected into the OT, and after a recovery of 10 days, the (EnvA).SAD.ΔG.EGFP virus (300 nl) was injected into the same site to initiate trans-synaptic tracing. Seven days later, the mice were sacrificed, and the brain was dissected for fluorescent imaging.
Mice were perfused transcardially with 4% paraformaldehyde (PFA) in fresh phosphate buffered saline (PBS). The brain was dissected out and post fixed in 4% PFA overnight at 4 °C, then transferred into PBS. Coronal slices (100 μm thick) were prepared using a Leica VT 1200S vibratome. The slices were treated with glycerol in PBS (volume ratio 1:1) for 30 min followed by glycerol in PBS (volume ratio 7:3) for 30 min before being mounted onto superfrost slides for imaging. A SP5 Leica confocal microscope equipped with LAS AF Lite software and 473 nm, 599 nm, and 635 nm lasers was used to obtain images at 10x (for coarse location of fluorescent regions as in Extended Data Fig. 4) and 40x (for cell counting in fluorescent regions as in Fig. 6). For cell counting, we acquired z-stack confocal images for 40 μm (with a z step of 1 μm) and obtained a projected image for that region. At the injection site, we identified “starter” cells (mCherry and EGFP double positive) from 5 to 6 sections/mouse and counted 16, 22, and 27 cells from the three mice. Since we only imaged 40 μm out of 100 μm sections, we estimated the total number of starter cells from the three mice to be 40, 55 and 68, respectively, assuming relative homogeneity throughout. For quantification of presynaptic cells, we counted EGFP positive (but mCherry negative) cells from every other section throughout the brain and calculated the percentage of cells in each brain region. Although this approach underestimated the number of labeled cells, it allowed consistent identification of the major input areas in all three mice.
For anterograde tracing, Cre-dependent AAV(DJ/8)-FLEX-Synaptophysin::EGFP virus (300 nl) was unilaterally injected into the OT, and four weeks later, the brain was dissected out for fluorescent imaging as described above. A subset of mice injected with Cre-dependent AAV1-DIO-ChR2-EYFP or AAV2/9-DIO-eArchT-EGFP (300–500 nl) were also used for anterograde tracing. All brain structures in coronal sections are outlined based on the Allen Mouse Brain Common Coordinate Framework version 3 (https://scalablebrainatlas.incf.org/mouse/ABA_v3)68.
For the efficiency of DTA viral ablation of OT D3 neurons, the EYFP fluorescence intensity (ChR2-EYFP as a marker for D3-Cre/ChR2 neurons) between the two sides of the OT on the same coronal sections in the unilateral ablation animals was compared using the ImageJ software (https://imagej.nih.gov/ij/).
Patch-clamp Recording
For whole-cell patch-clamp recordings, mice were deeply anesthetized with ketamine-xylazine (200 and 20 mg/kg body weight, respectively) and decapitated. The brain was dissected out and immediately placed in ice-cold cutting solution containing (in mM) 92 N-Methyl D-glucamine, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 Sodium L-ascorbate, 2 Thiourea, 3 Sodium Pyruvate, 10 MgSO4, and 0.5 CaCl2; osmolality ~300 mOsm and pH ~7.3, bubbled with 95% O2-5% CO2. Coronal sections (250 μm thick) containing the OT were cut using a Leica VT 1200S vibratome. Brain slices were incubated in oxygenated artificial cerebrospinal fluid (ACSF in mM: 124 NaCl, 3 KCl, 1.3 MgSO4, 2 CaCl2, 26 NaHCO3, 1.25 NaH2PO4, 5.5 glucose, and 4.47 sucrose; osmolality ~305 mOsm and pH ~7.3, bubbled with 95% O2-5% CO2) for ~30 min at 31°C and at least 30 minutes at room temperature before use. For recordings, slices were transferred to a recording chamber and continuously perfused with oxygenated ACSF. Fluorescent cells were visualized through a 40X water-immersion objective on an Olympus BX61WI upright microscope equipped with epifluorescence.
Whole-cell patch-clamp recordings were controlled by an EPC-10 amplifier combined with Pulse v8.74 (HEKA Electronik) and analyzed using Igor Pro 6 (Wavematrics). Recording pipettes were made from borosilicate glass with a Flaming-Brown puller (P-97, Sutter Instruments; tip resistance 5–10 MΩ). The pipette solution contained (in mM) 120 K-gluconate, 10 NaCl, 1 CaCl2, 10 EGTA, 10 HEPES, 5 Mg-ATP, 0.5 Na-GTP, and 10 phosphocreatine. For light-evoked inhibitory postsynaptic currents (IPSCs), a high Cl− pipette solution (120 mM KCl instead of K-gluconate) was used so that the reversal potential of [Cl−] was at ~ 0 mV and GABAA receptor-mediated currents would be inward from a holding potential of −60 mV. Holding the cells near the resting membrane potential kept them healthy for a longer time during recording. Light stimulation was delivered through the same objective via pulses of blue laser (473 nm, FTEC2473-V65YF0, Blue Sky Research, Milpitas, USA) with varying lengths and frequencies. Viral infection in the OT was confirmed in brain slices during recording. Pharmacological drugs were bath perfused during recording: (2R)-amino-5-phosphonovaleric acid (AP5), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), bicuculline, tetrodotoxin (TTX) citrate and 4-Aminopyridine (4-AP).
CLARITY: Tissue Clearing, Brain Imaging, and Data Analysis
CLARITY-based tissue clearing69, 70 and whole-brain microscopy were performed as previously described71, 72 with minor adjustments. Briefly, perfusion-fixed brains from adult D3-Cre/tdTomato mice of both sexes were removed from the skull. To maintain structural integrity, tissue was fixed overnight at 4°C in hydrogel fixation solution containing 4% acrylamide, 0.05% bis-acrylamide, 0.25% VA-044 initiator, 4% PFA in Ca2+- and Mg2+-free phosphate buffered saline (PBS−/−). After acrylamide polymerization at 37°C, samples were washed and incubated in 4% SDS and 20 mM boric acid (pH 8.5) at room temperature for 24 h and, subsequently, at 37°C for two more months. Next, samples were incubated for 24 h in refractive index matching solution containing 80 g Nycodenz (nRIMS80), 20 mM PS, 0.1% Tween 20, and 0.01% sodium acid. Specimen were kept in a custom-designed imaging chamber that was mounted on the stage of a Leica DMi8 inverted microscope. Imaging was performed with an Andor Dragonfly 500 spinning-disk confocal unit using a 561 nm laser and a HC PL FL 10x/0.30 objective.
For anatomical mapping of D3-positive neurons, data were preprocessed using IMARIS™ 9.5.1 3D rendering software (bitplane, Zurich, Switzerland). Surface objects were generated and exported to MeshLab73 to generate STL files. Next, Allen Brain Atlas version 3 (Allen Institute for Brain Science) was merged and manually adjusted to match the specimen surface objects using the mesh deform modifier in Blender 2.79b-2.80 (www.blender.org). The Iterative Closest Point (ICP) Registration add-on for Blender by Niels Klop was used for accurate mesh alignment. Importantly, we exclusively transformed the original 3D brain atlas’ bounding box to match the specimen structure excluding somewhat biased deformations. The resulting sample-matched brain atlas was then sliced into a virtual z-stack that we superimposed onto the imaging data in IMARIS. Next, OT surfaces were generated and exported using IMARIS and Microsoft 3D Builder 18.0.1931. Virtual volumes were generated using PrusaSlicer 2.1 (Prusa Research, Praha, Czech Republic). In Blender the optimal overlay was iteratively determined by optimizing the overlap of both volumes.
For detection and registration of individual tdTomato-labeled cells, we first applied a Laplacian-of-Gaussian Scale-Space Maximum Projection (LoGSSMP), corresponding to the seed detection stage of the nucleus detection algorithm TWANG (using the ITK-based C++ application XPIWIT)74, 75. In brief, the original images were filtered using differently scaled Laplacian-of-Gaussian filters and the kernel standard deviations were matched to the manually measured minimum and maximum cell sizes (σmin = 2, σmax = 4 pixels). The 4D scale-space was reduced to a 3D image by a maximum intensity projection of the individual LoG-filtered images. Local maxima were then identified in the 3D LoGSSMP image. To minimize false negatives (i.e., missed cells), we additionally allowed to detect multiple seed points on intensity plateaus where no single maximum pixel was present. To coarsely reduce false positive detections in background regions, only detections with an intensity larger than the global mean plus one standard deviation of the LoGSSMP image intensities were considered. Redundant detections with a distance of less than three voxels in the image space were locally fused to a single detection.
The detection module was parameterized to be very sensitive, i.e., to avoid false negatives. This, however, results in increased false positive detections. Given different signal intensity levels at different tissue depths, using a simple threshold for false positive rejection was not possible. Instead, we trained a classifier to suppress erroneous detections, i.e., to distinguish false positives from true cells. Training data were obtained by manually labeling a set of representative cells using EmbryoMiner for ground truth generation and visual result confirmation76. This interactive software allows superimposing automatically obtained detections on raw images. Using freehand selection tools we assigned cells to different groups. For each manually classified detection, we cropped a 32×32 2D patch with the detection located in the center from the z-slice running through the center of the cell. The resulting training snippets (>5000 per class) were normalized to zero mean and unit standard deviation and then used to train a small classification CNN with the following architecture: Input-Layer (32×32×1), Conv2D-Layer (32×32×32), Conv2D-Layer (32×32×64), Max Pooling-Layer (16×16×64), Conv2D (16×16×64), Conv2D-Layer (16×16×128), Dropout-Layer (p = 0.5), Fully-Connected-Layer (1024), Dropout-Layer (p = 0.5), Fully-Connected Output-Layer75. The convolutional neural network was implemented in MATLAB using the Deep Learning Toolbox. All convolutions used a stride of 1, zero padding and a receptive field of 3×3. Max pooling was implemented with a receptive field of 2×2 and a stride of 1. All layers use ReLU activations except of the output layer, where softmax activation was used to obtain class probabilities. With a batch size set to 100, we trained for 100 epochs using the adam optimizer. We assessed the performance of the classifier using a five-fold cross validation and achieved an average classification accuracy of 96.18% on an independent test set. The trained model was then used to classify all remaining cells and to remove all cells that were classified as false positive in the background.
For categorization of OT D3 neurons into either dense or loose populations, we computed a cell density measure by counting the number of detections located in a sphere of radius r = 50 μm surrounding each detection. This measure yields high values if the cell is located in a dense region and low values if few cells are residing in the vicinity. Distribution analysis based on histogram plots of various bin widths identified a threshold of t = 5 to separate loose from dense cell. Thus, cells with >5 neighbors were considered to reside in a dense region. This criterion, however, might not apply to few cells at the edges of dense regions. Consequently, the percentage of truly loose cells is likely an overestimate.
In vivo Fiber Photometry
The AAV9-FLEX-GCaMP7s virus (500 nl) was unilaterally injected into the OT of D3-Cre/tdTomato mice and a 400 μm optical fiber cannula was implanted above the injection site. Based on post-mortem histology, the mice were categorized into two groups: (1) “Optical fiber near IC”: the optical fiber was right above a GCaMP7s+ island and the vertical distance between the fiber tip and the island was < 200 μm, and (2) “Optical fiber away from IC”: there was no GCaMP7s+ island right below the fiber tract. In this case, the distance from the fiber tip to the closest GCaMP7s+ island was measured and used in Fig. 8e. After allowing 3–4 weeks for GCaMP7s expression, mice were attached via an optical fiber (400 ìm core, 0.48 NA; Thor Labs, Model M76L01) to a Doric 4-port minicube (FMC4, Doric Lenses). Blue (490 nm wavelength for GCaMP7s stimulation, ThorLabs #M490F3) and violet (405 nm wavelength for artifact control fluorescence, ThorLabs #M405FP1) LED light was delivered to the brain at 8–15 μW (LED driver: Thor Labs, Model DC4104). Mice were hooked up to the optical fiber with the LEDs on for 10 – 15 min prior to recording start. Emissions passed through a dichroic mirror, a 500–550 nm cut filter and were then detected by a femtowatt silicon photoreceiver (Newport, Model 2151). Analog signals were demodulated and recorded using the RZ5 processor and Synapse Software (Tucker Davis Technologies). Time-locked behavioral videos were obtained via a webcam (30 frames/sec) and grooming events were scored. Grooming bouts within 5 s from a previous bout were excluded in the analysis to avoid cumulative effects on the fluorescent signals. A single non-grooming period right before each grooming bout was included as a control. To calculate the fluorescence change ratios (ΔF/F), the raw data were analyzed using MATLAB and downsampled to 9 Hz. For each wavelength, the fluorescence change (ΔF/F490 or ΔF/F405) was calculated as (F-F0)/F0, where F0 was the baseline fluorescence signal averaged from periods without grooming. Then ΔF/F was obtained by subtracting ΔF/F405 from ΔF/F490, and the peak ΔF/F measured around grooming onset (± 1 s). Randomization test was performed in R package by shuffling the data 100,000 times to compare the difference between the grooming and non-grooming events.
In vivo Unit Recordings
We analyzed electrophysiological recordings of OT neural activity from five adult male C57BL/6J mice which were stereotactically implanted with chronic tungsten electrode arrays as described previously77. Briefly, surgeries occurred under isoflurane anesthesia, with the local anesthetic Marcaine (Patterson Veterinary, Greely, CO) applied at the wound margin and the analgesic meloxicam delivered for 3 days following surgery. To target the OT, 8 electrode wires were installed +1.1 mm medial and +0.6 or +1.1–1.2 mm anterior to bregma and lowered to 4.9 mm ventral to the brain surface. These implanted mice contributed single unit data to a prior study77. For the present study, we utilized single unit data in which timestamped grooming events were completely separated in time from behaviors investigated in the prior study involving reward behavior77.
Spontaneous grooming events were captured via a camera as mice explored a familiar 15 × 15 × 30 cm chamber in 1 h sessions. They were connected to a flexible tether and the output of the electrode arrays was amplified with a digital headstage (Intan Technologies). Recordings were acquired at 24.4 kHz (0.3–5 kHz band pass) using SYNAPSE (Tucker-Davis Technologies) with one of the electrodes in the OT of each mouse used as a local reference. Following behavior, mice were overdosed with sodium pentobarbital (Fatal Plus, Patterson Veterinary) and transcardially perfused with cold 0.9% saline and 10% phosphate-buffered formalin. Brains were sectioned at 40 μm and counterstained with DAPI for post-mortem histological verification of electrode recording sites (Extended Data Fig. 5h).
We used offline spike sorting (Spike2, Cambridge Electronic Design) with a combination of waveform template matching and K-means assisted cluster cutting based on principal component analysis (PCA) to establish a population of putative single units. Next, among these putative single units, we used a conservative inter-spike interval threshold to eliminate possible multi-unit data from the population. To accomplish this, we eliminated any putative single units which displayed >2% of their spikes within a 2 ms refractory period78 to generate a population of confirmed single units (Extended Data Fig.7). Any instances of a single unit whose activity was observed across more than one channel was removed to prevent oversampling.
Among confirmed single units, spike times aligned to grooming onset events or aligned to pseudorandomly identified (for shuffled comparison) time markers that did not include grooming events were extracted and imported into MATLAB (MathWorks, R2020a). Spike density functions in 50 ms bins were calculated based on convolving spike trains with a function resembling a postsynaptic potential79. Each neuron’s background firing rate was calculated based on its activity during numerous 2 s windows (mean = 53.5), equally sampled throughout the 1 h sessions that did not include any grooming. Spike density functions for each analysis window were z-score normalized to each neuron’s average background firing.
To identify task-modulated neurons, we compared each neuron’s averaged background firing to its averaged firing rate ± 1 sec relative to grooming onsets via t-tests with a False Discovery Rate (FDR) Q of 1% using the Benjamini and Hochberg method80, 81. To examine and identify cell-groom pairs, among significantly modulated neurons defined above, we subtracted background activity from their firing rates during individual groom bouts and averaged their firing rates across time bins within 1 s prior to grooming onset (“pre”), and within 1 s following grooming onset (“during”). This gave the change in firing rates: ΔHz = FRpre – FRbackground or FRduring – FRbackground. Only cell-groom pairs with ΔHz ± 1 Hz were included. The same approach was used to determine the distribution of firing rate changes for shuffled, non-grooming events, including ΔHz values within ± 1 Hz. Analyses and figures were generated using MATLAB and/or Prism 7 (Graph Pad).
Statistics & Reproducibility
Sample sizes for individual experiments were determined according to the NIH “Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research” (Sample Size Determination: https://www.ncbi.nlm.nih.gov/books/NBK43321/). Shapiro-Wilk normality tests were used to verify normal distribution of each dataset. Parametric statistical tests were used for normally distributed datasets; otherwise, non-parametric tests were used. All statistical tests and results are reported in Supplementary Table 2. Various controls (genotype, virus, laser wavelength, brain regions, etc) were included in experimental design, and the number of replications for each experiment was included in figure legends as well as in Supplementary Table 1. Animals were allocated randomly in all experiments and various stimulating conditions (e.g., blue versus green laser, stimulation duration and frequency) for individual mice were randomized. All animals with verified optical fiber implantation/viral injection sites were included in data analysis. Blinding in data collection and analysis were carried out as much as possible with the following exceptions. For CLARITY imaging and patch clamp recordings, only mice with desired genotype were used. For optogenetic experiments in behaving mice, blue light activation of OT D3-Cre/ChR2 neurons always induced grooming behavior, which would reveal the genotype or virus type of the mice, thus preventing complete blinding in data collection and analysis.
Extended Data
Extended Data Fig. 1. Quantification of D3-Cre/tdTomato neurons in the ventral striatum.
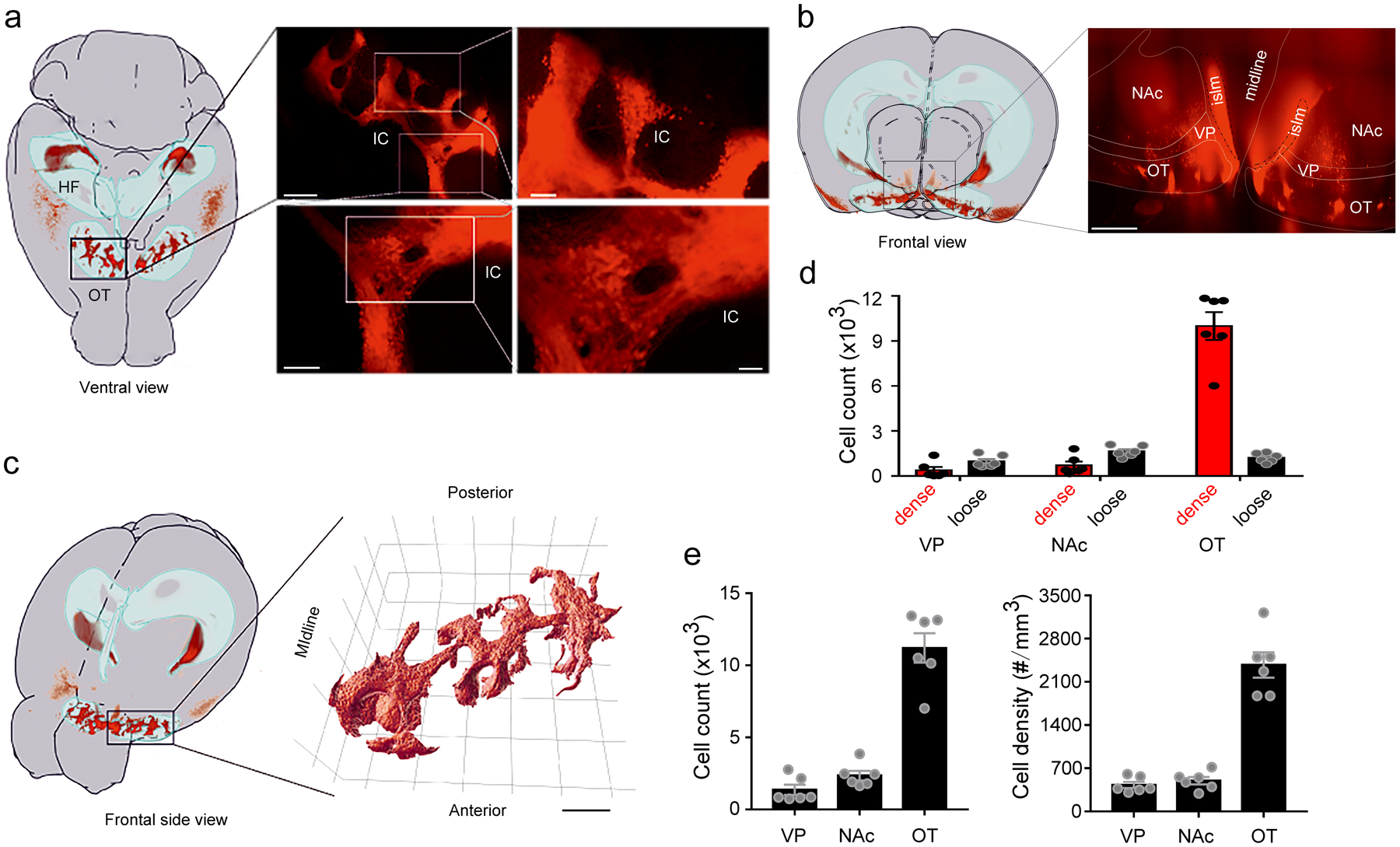
a, Left, ventral view of location(s) of D3-Cre/tdTomato neurons within a mouse brain mapped onto a sample-adjusted version of the Allen Mouse Brain Atlas. The OT and hippocampus are outlined as light blue volumes. Right, 3D projection of the OT region outlined (black rectangle). Scale bars = 400 μm (upper left), 100 μm (upper right), 150 μm (lower left), and 50 μm (lower right). b, Left, frontal view projection of the location(s) of D3-Cre/tdTomato neurons. Right, 3D projection of the ventral striatum. Scale bar = 300 μm. c, Left, frontal side view projection of the location(s) of D3-Cre/tdTomato neurons. Right, 3D surface rendering of the IC network. Scale bar = 500 μm. d-e, Quantification of D3 neurons in the ventral striatum and ventral pallidum. Absolute numbers of dense versus loose neurons in d, total numbers of neurons in e (left) and cell density in e (right) in the VP, the NAc and the OT. n = 6 hemispheres from 3 mice. All averaged data are shown as mean ± SEM. OT, olfactory tubercle. HF, hippocampal formation. IC, islands of Calleja. islm, major island. VP, ventral pallidum. NAc, nucleus accumbens.
Extended Data Fig. 2. Quantification of the IC between hemispheres and among individuals.
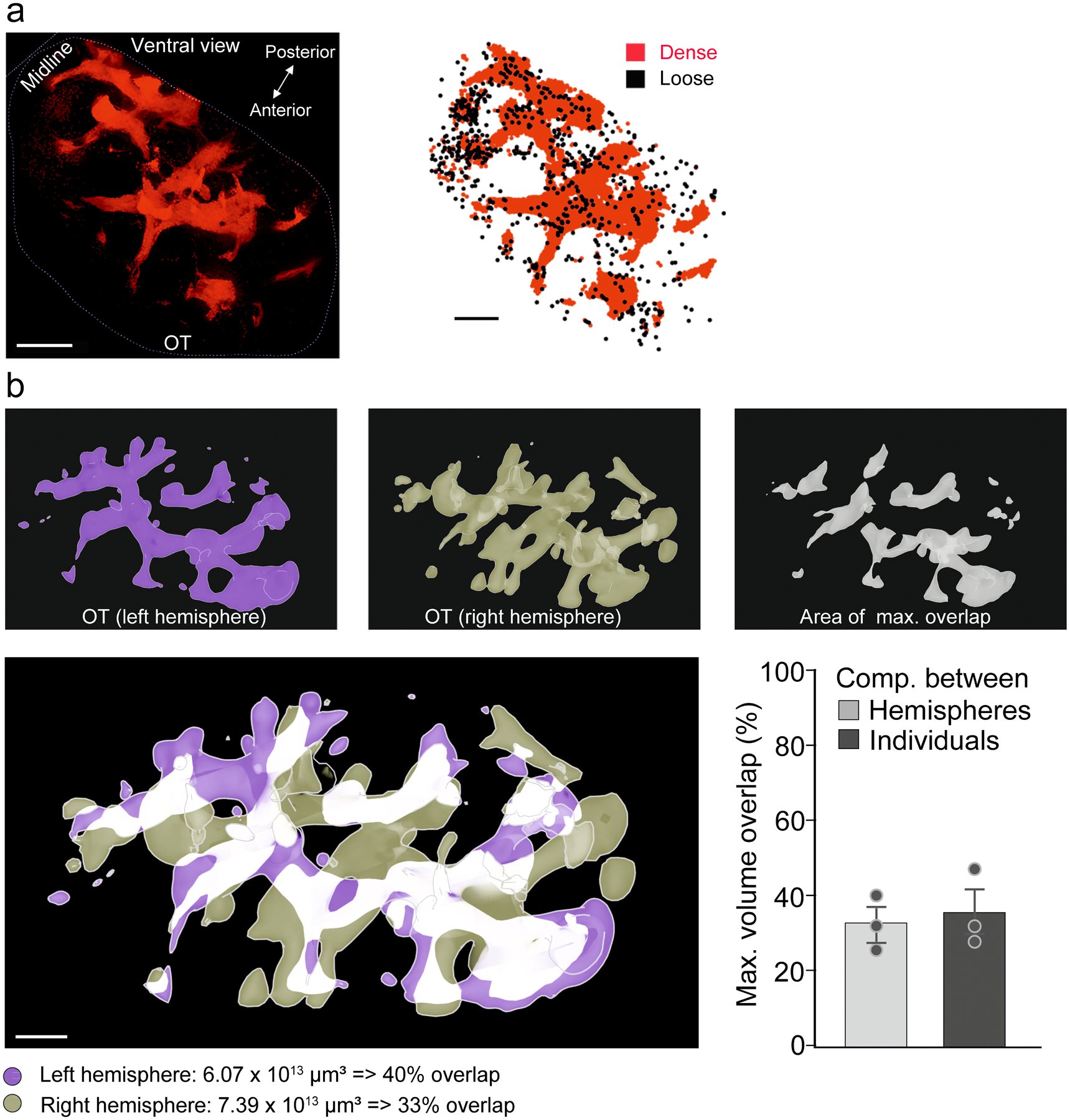
a, Left, 3D reconstruction of D3-Cre/tdTomato neurons in the OT demonstrating that the IC form a continuous branched network. Similar results were observed in 6 OTs from 3 mice. Right, map of registered neurons categorized as dense or loose neurons, respectively. Scale bars = 600 μm (left) and 500 μm (right). b, Top, 3D reconstructions of OT IC structures in individual hemispheres (left and middle panel), and the area of maximal overlap (right panel). Bottom left, IC overlap between the two hemispheres from the same mouse. Mirrored 3D objects were merged and aligned to create maximum overlap (white voxels). Scale bar = 500 μm. Bottom right, maximum volume overlap of the IC network between two hemispheres and among individuals. All averaged data are shown as mean ± SEM.
Extended Data Fig. 3. D3-Cre/tdTomato neurons in the piriform cortex, the hypothalamus and the hippocampus.
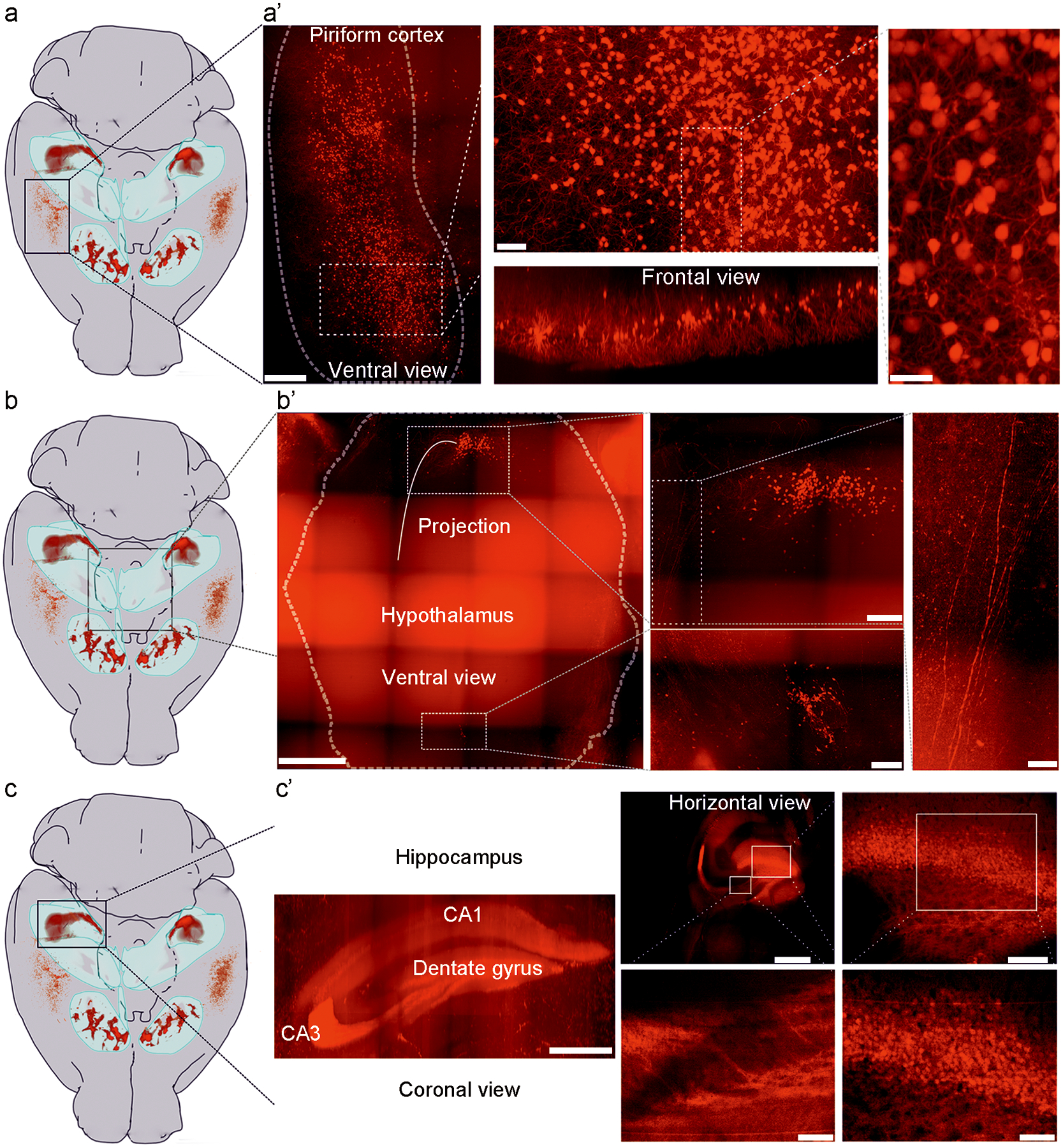
Location(s) of D3 neurons within the mouse brain are mapped onto a sample-adjusted version of the Allen Mouse Brain Atlas. The OT and hippocampal formation (HF) are outlined as light blue volumes. a, D3 neurons in the piriform cortex do not project to targets outside this region. a’, 3D projection of the piriform cortex region outlined in a (black rectangle). Different areas are shown at higher magnification. Note that no projection fibers are evident; in frontal view, D3 neurons appear to adhere to a layer-specific organization. Scale bars = 500 μm (left), 200 μm (middle), and 100 μm (right). b, Two areas in the hypothalamus harbor D3 neurons. Of these, neurons in the caudal aspect of the hypothalamus exhibit some projections. b’, 3D maximum projection of the hypothalamic region outlined in b (black rectangle). Different areas are shown at higher magnification. Relatively sparse, but consistent tdTomato expression is observed at both a relatively caudal and rostral region within the hypothalamus (close to midline). Note that few fibers are evident at the caudal site, whereas no fibers are found at the rostral site. Scale bars = 1000 μm (left), 300 μm (middle), and 100 μm (right). c, D3 neurons in the hippocampus. c’, 3D projection of the hippocampal region outlined in c (black rectangle). Different areas are shown at higher magnification. Scale bars = 1000 μm (left), 1000 μm (middle, upper panel), 100 μm (middle, lower panel), 200 μm (right, upper panel) and 100 μm (right, lower panel). Similar results were observed in 3 mice for a-c.
Extended Data Fig. 4. Retrograde tracing of presynaptic partners of OT D3 neurons.
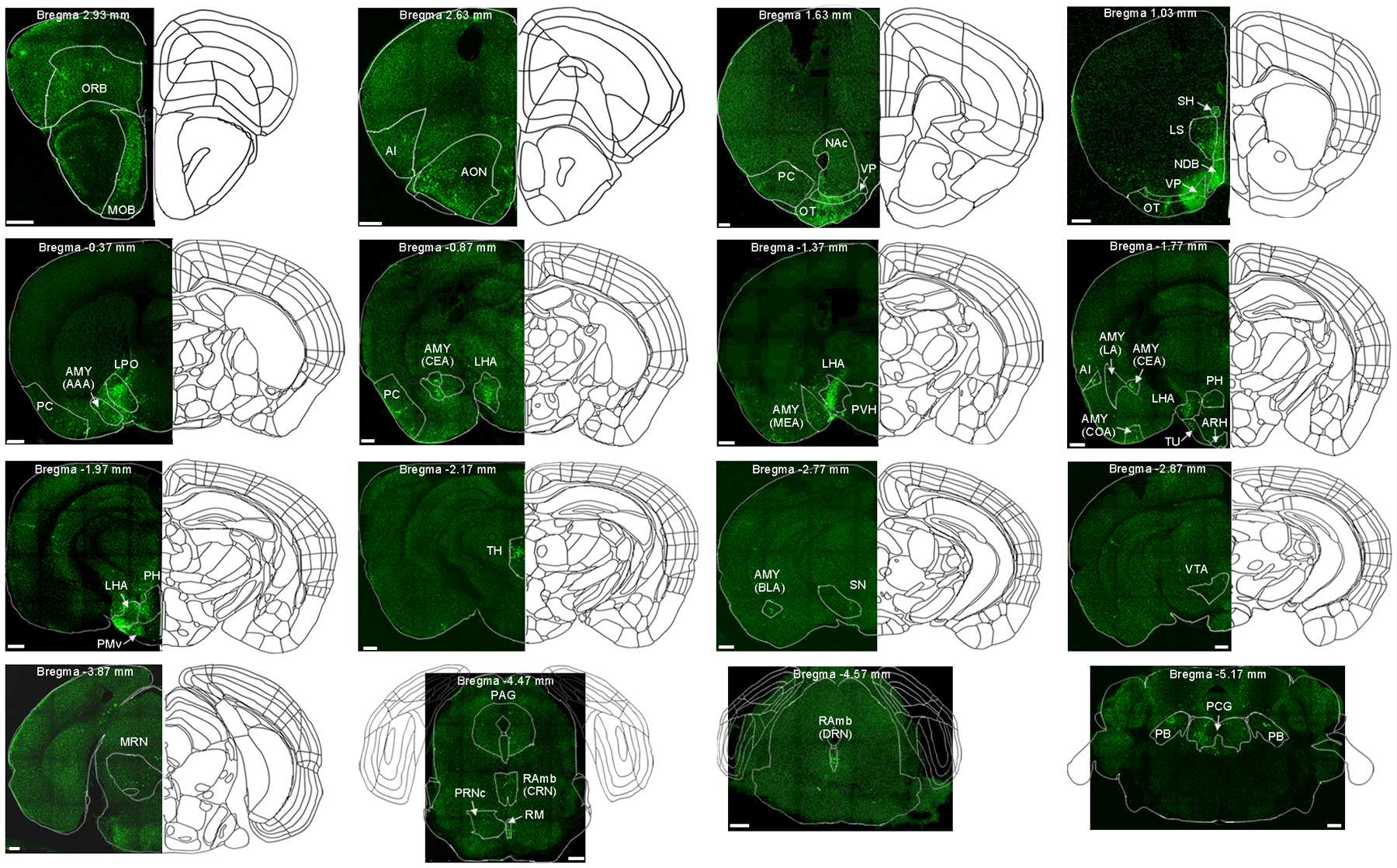
Representative images showing labeled presynaptic partners of OT D3 neurons from the anterior (upper left) to posterior brain sections (lower right). Similar results were observed in 3 mice. MOB, main olfactory bulb. ORB, orbital area. AI, agranular insular area. AON, anterior olfactory nucleus. PC, piriform cortex. NAc, nucleus accumbens. VP, ventral pallidum. OT, olfactory tubercle. LS, lateral septal nucleus. NDB, diagonal band nucleus. SH, septohippocampal nucleus. LPO, lateral preoptic area. LHA, lateral hypothalamic area. AMY (AAA), anterior amygdalar area. AMY (CEA), central amygdalar nucleus. AMY (MEA), medial amygdalar nucleus. AMY (LA), lateral amygdalar nucleus. AMY (COA), cortical amygdalar area. PVH, paraventricular hypothalamic nucleus. TU, tuberal nucleus. ARH, arcuate hypothalamic nucleus. PH, posterior hypothalamic nucleus. PMv, ventral premammillary nucleus. TH, thalamus (mostly in the subparafascicular area). AMY (BLA), basolateral amygdalar nucleus. SN, substantia nigra. VTA, ventral tegmental area. MRN, midbrain reticular nucleus. PAG, periaqueductal gray. RAmb (CRN), medbrain raphe nuclei, central part. RM, nucleus raphe magnus. PRNc, pontine reticular nucleus, caudal part. RAmb (DRN), medbrain raphe nuclei, dorsal part. PCG, pontine central gray. PB, parabrachial nucleus. Brain atlas images are modified from Allen Mouse Brain Common Coordinate Framework version 3 (https://scalablebrainatlas.incf.org/mouse/ABA_v3). Scale bars = 500 μm.
Extended Data Fig. 5. Viral injection/expression sites and optical fiber placements for experiments in Figs. 2–8.
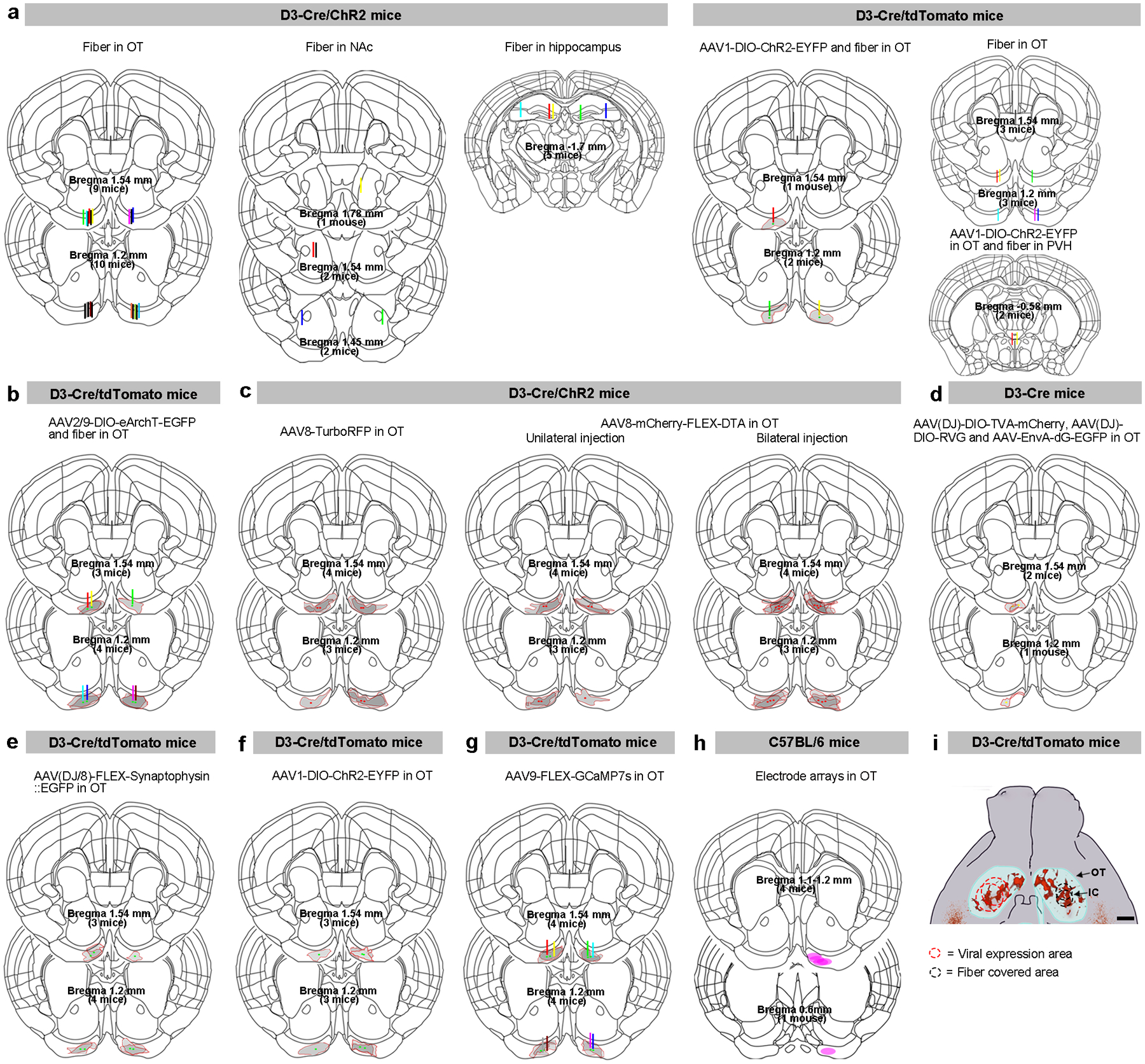
Coronal brain sections at the bregma levels showing viral injection sites (dots)/expression areas (gray shadow areas with red borders) and/or optical fiber tracts (vertical bars) for mice included. a-b, Optogenetic experiments (Figs. 2, 3 in a and Fig. 4 in b). c, DTA ablation experiments (Fig. 5). d-e, Retrograde (Fig. 6 in d) and anterograde (Fig. 7 in e) tracing experiments. f, Electrophysiological recordings (Fig. 7 in f), and (g) fiber photometry experiments (Fig. 8). For clarity, an optical fiber (400 μm) is shown as a thin vertical bar at the center of the tract. h, Electrode array locations from in vivo unit recording experiments (Fig. 8). i, Schematic showing viral expression and fiber covered areas for the two coordinates used in the OT. Scale bar = 500 μm. Each dot or line represents one animal except for bilateral AAV8-DTA injection in c. Brain atlas images are modified from Allen Mouse Brain Common Coordinate Framework version 3 (https://scalablebrainatlas.incf.org/mouse/ABA_v3). IC, islands of Calleja. OT, olfactory tubercle. NAc, nucleus accumbens. PVH, paraventricular hypothalamic nucleus.
Extended Data Fig. 6. Properties of postsynaptic currents (PSCs) upon optogenetic stimulation of D3-Cre/ChR2 neurons in the OT.
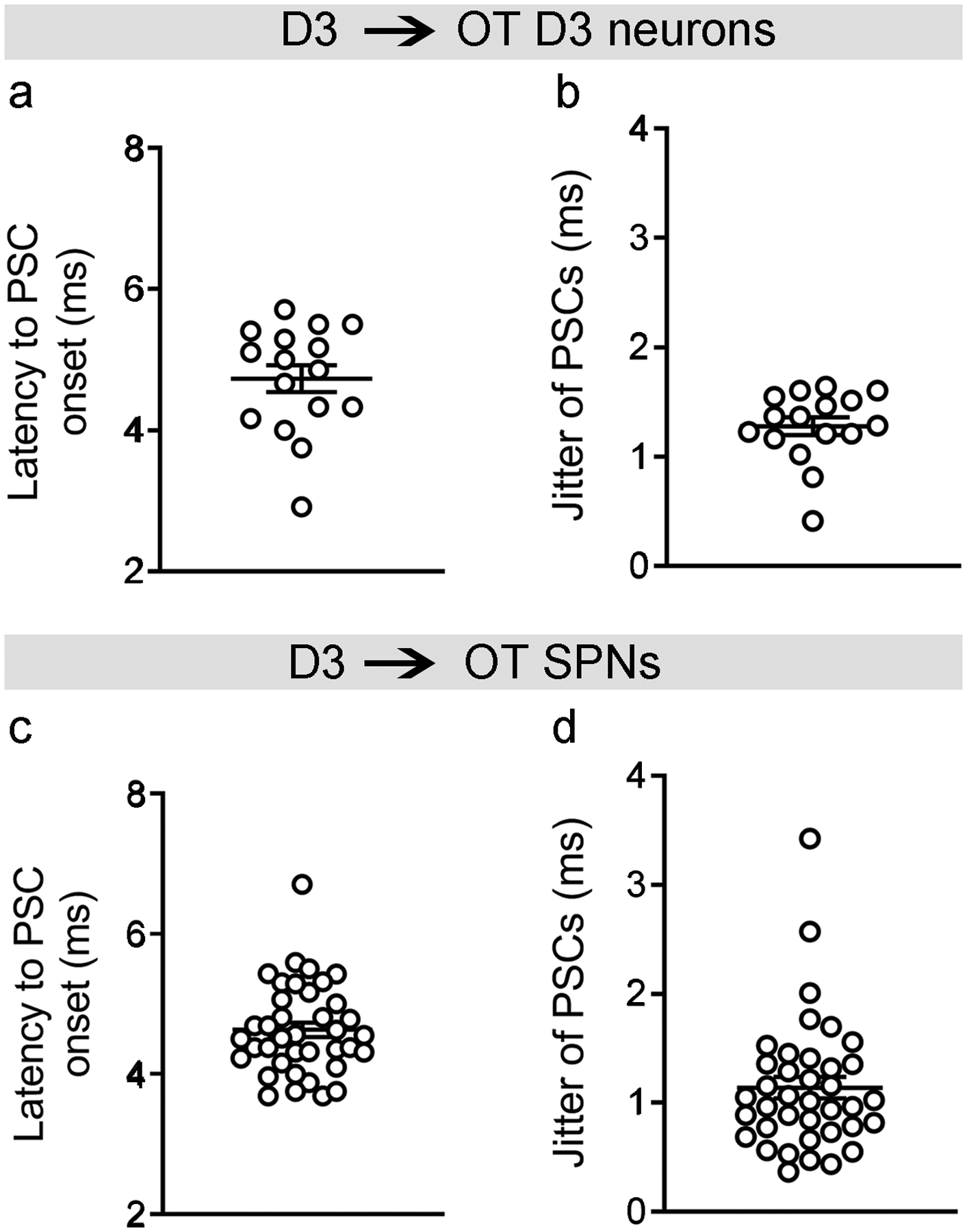
a and c, Latency to PSC onset in D3-Cre/tdTomato neurons (4.73 ± 0.19 ms) (a) and SPNs (4.63 ± 0.11 ms) (c). b and d, Jitter of PSCs (SD of latencies during repeated light stimuli) in D3-Cre/tdTomato neurons (1.28 ± 0.08 ms) (b) and SPNs (1.14 ± 0.10 ms) (d). Data are quantified in 16 D3-Cre/tdTomato neurons and 38 SPNs (6–10 traces/neuron) showing light-evoked PSCs. All averaged data are shown as mean ± SEM.
Extended Data Fig. 7. Single unit quality control metrics.
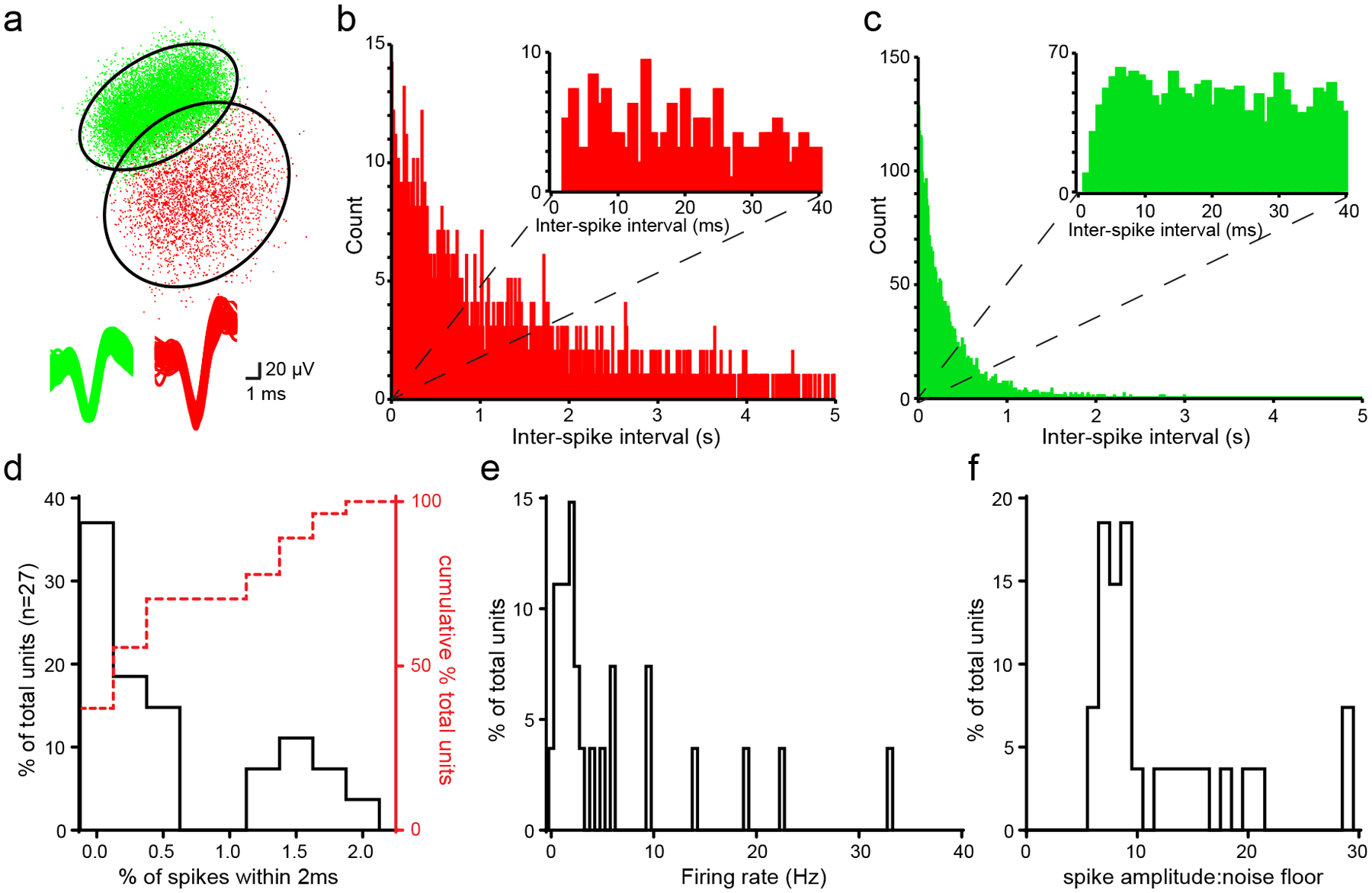
a, Upper panel, PCA plot of two putative single units recorded from the same electrode. Ellipses denote 2.5x SD of each K-means cluster. Lower panel, overlaid waveforms of the same neurons. b-c, Inter-spike intervals (ISIs, 2 ms bins) for the same two neurons as in (a) indicating significantly different distributions (two-sample Kolmogorov-Smirnov test D (2493) = 0.22, p <0.0001). Insets, ISI distributions (1 ms bins) showing limited numbers (< 2%) of ISI events < 2ms. d, Distribution of the proportion of ISI violations (< 2ms between spikes) among all single units. 100% of units had <2% of their spikes occurring within 2ms of each other. e, Distribution of mean firing rates during entire recording session of all single units (median: 1.97 Hz, typical of spiny projection neurons). f, Distribution of spike amplitude: noise floor values for all single units.
Supplementary Material
3D reconstruction of the Islands of Calleja in the OT. Red: D3-Cre/tdTomato neurons. Green: axonal fibers from the MOB projection neurons labeled via viral injection of AAV9-EGFP. Blue: cell nuclei labeled by DRAQ5TM, a far-red fluorescent DNA dye.
3D reconstruction of individual D3 neurons in and between two Islands of Calleja in the OT showing the neuronal processes of these neurons. Colors are the same as in Supplementary Video 1.
Blue light (but not green light) activation of OT D3-Cre/ChR2 neurons induces grooming.
Grooming behavior induced by blue light stimulation of OT D3-Cre/ChR2 neurons with different durations (1 to 20 s at 20 Hz).
Grooming behavior induced by blue light stimulation of OT D3-Cre/ChR2 neurons with different frequencies (1–20 Hz for 10 s).
Genotype and stimulation site controls. Blue light stimulation of OT D3-Cre/tdTomato neurons or blue light stimulation of D3-Cre/ChR2 neurons in the nucleus accumbens (NAc) and hippocampus (dentate gyrus and CA3 region) do not induce grooming.
Grooming behavior induced by blue light activation of the OT in D3-Cre/tdTomato mice injected with Cre-dependent AAV1-DIO-ChR2-EYFP virus. Additional conditions include green light or optical fiber implanted in the paraventricular hypothalamic nucleus (PVH).
Blue light (but not green light) activation of OT D3 neurons stops social investigation and initiates grooming in D3-Cre/ChR2 mice.
Blue light (but not green light) activation of OT D3 neurons stops feeding and initiates grooming in D3-Cre/ChR2 mice.
Blue light (but not green light) activation of OT D3 neurons stops itch-induced scratching and initiates grooming in D3-Cre/ChR2 mice.
Blue light (but not green light) activation of OT D3 neurons stops body licking and initiates grooming in D3-Cre/ChR2 mice.
Green light (but not blue light) inactivation of OT D3-Cre/eArchT neurons shortens spontaneous grooming.
Green light (but not blue light) inactivation of OT D3-Cre/eArchT neurons halts water spray-induced grooming.
Acknowledgement
We thank Acal BFi and Andor for providing access to the Dragonfly 500 spinning-disk confocal microscope platform. This work was supported by the National Institutes of Health (R01NS117061 to D.W.W., M.V.F, and M.M., R01DC006213 to M.M., R01DA049545 and R01DA049449 to M.M. and D.W.W, R01DC016519 and R01DC014443 to D.W.W., R01MH115030 to M.V.F., R21DC019193 to J.P.B., F32DC018452 to K.N.W., F31DC017054 to M. Schreck, and F31MH124372 to E.J.), by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) 368482240/GRK2416 to M. Spehr, 269953372/GRK2150 to J.M. and M. Spehr, and by the Whitehall Foundation and Foundation for OCD Research to M.V.F. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Data Availability
The raw data generated and/or analyzed during the current study are available from the corresponding authors on reasonable request. Source data are provided for all figures wherever applicable. The Allen Mouse Brain Connectivity Atlas (http://connectivity.brain-map.org/transgenic/experiment/304168043) was used for Fig. 1d and the Allen Mouse Brain Common Coordinate Framework version 3 (https://scalablebrainatlas.incf.org/mouse/ABA_v3) was used to outline brain structures in Figs. 2b, 4a, and Extended Data Figs. 4, 5.
References
- 1.Grillner S & Robertson B The Basal Ganglia Over 500 Million Years. Curr Biol 26, R1088–R1100 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Burke DA, Rotstein HG & Alvarez VA Striatal Local Circuitry: A New Framework for Lateral Inhibition. Neuron 96, 267–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox J & Witten IB Striatal circuits for reward learning and decision-making. Nat Rev Neurosci 20, 482–494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millhouse OE Granule cells of the olfactory tubercle and the question of the islands of Calleja. J Comp Neurol 265, 1–24 (1987). [DOI] [PubMed] [Google Scholar]
- 5.De Marchis S, Fasolo A & Puche AC Subventricular zone-derived neuronal progenitors migrate into the subcortical forebrain of postnatal mice. J Comp Neurol 476, 290–300 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Fallon JH, Riley JN, Sipe JC & Moore RY The islands of Calleja: organization and connections. J Comp Neurol 181, 375–395 (1978). [DOI] [PubMed] [Google Scholar]
- 7.Meyer G, Gonzalez-Hernandez T, Carrillo-Padilla F & Ferres-Torres R Aggregations of granule cells in the basal forebrain (islands of Calleja): Golgi and cytoarchitectonic study in different mammals, including man. J Comp Neurol 284, 405–428 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Adjei S & Wesson DW Laminar and spatial localization of the islands of Calleja in mice. Neuroscience 287, 137–143 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Fallon JH, Loughlin SE & Ribak CE The islands of Calleja complex of rat basal forebrain. III. Histochemical evidence for a striatopallidal system. J Comp Neurol 218, 91–120 (1983). [DOI] [PubMed] [Google Scholar]
- 10.Hsieh YC & Puche AC Development of the Islands of Calleja. Brain Res 1490, 52–60 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Hsieh YC & Puche AC GABA modulation of SVZ-derived progenitor ventral cell migration. Dev Neurobiol 75, 791–804 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Martin-Lopez E, Xu C, Liberia T, Meller SJ & Greer CA Embryonic and postnatal development of mouse olfactory tubercle. Mol Cell Neurosci 98, 82–96 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Suzuki M, Hurd YL, Sokoloff P, Schwartz JC & Sedvall G D3 dopamine receptor mRNA is widely expressed in the human brain. Brain Res 779, 58–74 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Neddens J & Buonanno A Expression of the neuregulin receptor ErbB4 in the brain of the rhesus monkey (Macaca mulatta). PLoS One 6, e27337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giessel AJ & Datta SR Olfactory maps, circuits and computations. Curr Opin Neurobiol 24, 120–132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesson DW The Tubular Striatum. J Neurosci 40, 7379–7386 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levant B Differential distribution of D3 dopamine receptors in the brains of several mammalian species. Brain Res 800, 269–274 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Bouthenet ML, et al. Localization of dopamine D3 receptor mRNA in the rat brain using in situ hybridization histochemistry: comparison with dopamine D2 receptor mRNA. Brain Res 564, 203–219 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Li Y & Kuzhikandathil EV Molecular characterization of individual D3 dopamine receptor-expressing cells isolated from multiple brain regions of a novel mouse model. Brain Struct Funct 217, 809–833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landwehrmeyer B, Mengod G & Palacios JM Dopamine D3 receptor mRNA and binding sites in human brain. Brain Res Mol Brain Res 18, 187–192 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Ridray S, et al. Coexpression of dopamine D1 and D3 receptors in islands of Calleja and shell of nucleus accumbens of the rat: opposite and synergistic functional interactions. Eur J Neurosci 10, 1676–1686 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Le Moine C & Bloch B Expression of the D3 dopamine receptor in peptidergic neurons of the nucleus accumbens: comparison with the D1 and D2 dopamine receptors. Neuroscience 73, 131–143 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Novejarque A, Gutierrez-Castellanos N, Lanuza E & Martinez-Garcia F Amygdaloid projections to the ventral striatum in mice: direct and indirect chemosensory inputs to the brain reward system. Front Neuroanat 5, 54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubeda-Banon I, et al. Projections from the posterolateral olfactory amygdala to the ventral striatum: neural basis for reinforcing properties of chemical stimuli. BMC Neurosci 8, 103 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joyce JN & Gurevich EV D3 receptors and the actions of neuroleptics in the ventral striatopallidal system of schizophrenics. Ann N Y Acad Sci 877, 595–613 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Inta D, Meyer-Lindenberg A & Gass P Alterations in postnatal neurogenesis and dopamine dysregulation in schizophrenia: a hypothesis. Schizophr Bull 37, 674–680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calaresu FR, Zhang J, Chitravanshi VC & Mckitrick DJ Cardiovascular and Single-Unit Responses Elicited by Stimulation of the Islands of Calleja and by Changes in Arterial-Pressure. Brain Res 655, 45–50 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Meyer G, Gonzalez-Hernandez T, Galindo-Mireles D, Carrillo-Padilla F & Ferres-Torres R NADPH-d activity in the islands of Calleja: a regulatory system of blood flow to the ventral striatum/pallidum? Neuroreport 5, 1281–1284 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Kalueff AV, et al. Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat Rev Neurosci 17, 45–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smolinsky AN, Bergner CL, LaPorte JL & Kalueff AV Analysis of grooming behavior and its utility in studying animal stress, anxiety, and depression. Mood and Anxiety Related Phenotypes in Mice, Characterirzation Using Behavioral Tests, Neuromethods 42, Chapter 42. (2009). [Google Scholar]
- 31.Fuccillo MV Striatal Circuits as a Common Node for Autism Pathophysiology. Front Neurosci 10, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burguiere E, Monteiro P, Mallet L, Feng G & Graybiel AM Striatal circuits, habits, and implications for obsessive-compulsive disorder. Curr Opin Neurobiol 30, 59–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalueff AV, Aldridge JW, LaPorte JL, Murphy DL & Tuohimaa P Analyzing grooming microstructure in neurobehavioral experiments. Nat Protoc 2, 2538–2544 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Berridge KC, Fentress JC & Parr H Natural syntax rules control action sequence of rats. Behav Brain Res 23, 59–68 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Berntson GG, Jang JF & Ronca AE Brainstem systems and grooming behaviors. Ann N Y Acad Sci 525, 350–362 (1988). [DOI] [PubMed] [Google Scholar]
- 36.Berridge KC Progressive degradation of serial grooming chains by descending decerebration. Behav Brain Res 33, 241–253 (1989). [DOI] [PubMed] [Google Scholar]
- 37.Spruijt BM, van Hooff JA & Gispen WH Ethology and neurobiology of grooming behavior. Physiol Rev 72, 825–852 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Alo R, Avolio E, Mele M, Di Vito A & Canonaco M Central amygdalar nucleus treated with orexin neuropeptides evoke differing feeding and grooming responses in the hamster. J Neurol Sci 351, 46–51 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Hong W, Kim DW & Anderson DJ Antagonistic control of social versus repetitive self-grooming behaviors by separable amygdala neuronal subsets. Cell 158, 1348–1361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunn AJ, Berridge CW, Lai YI & Yachabach TL CRF-induced excessive grooming behavior in rats and mice. Peptides 8, 841–844 (1987). [DOI] [PubMed] [Google Scholar]
- 41.Roeling TA, Veening JG, Peters JP, Vermelis ME & Nieuwenhuys R Efferent connections of the hypothalamic “grooming area” in the rat. Neuroscience 56, 199–225 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Kruk MR, et al. The hypothalamus: cross-roads of endocrine and behavioural regulation in grooming and aggression. Neurosci Biobehav Rev 23, 163–177 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Dunn AJ Studies on the neurochemical mechanisms and significance of ACTH-induced grooming. Ann N Y Acad Sci 525, 150–168 (1988). [DOI] [PubMed] [Google Scholar]
- 44.Mangieri LR, et al. A neural basis for antagonistic control of feeding and compulsive behaviors. Nat Commun 9, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mu MD, et al. A limbic circuitry involved in emotional stress-induced grooming. Nat Commun 11, 2261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cromwell HC & Berridge KC Implementation of action sequences by a neostriatal site: a lesion mapping study of grooming syntax. J Neurosci 16, 3444–3458 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapanelli M, Frick L, Bito H & Pittenger C Histamine modulation of the basal ganglia circuitry in the development of pathological grooming. Proc Natl Acad Sci U S A 114, 6599–6604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu X, et al. Reducing Astrocyte Calcium Signaling In Vivo Alters Striatal Microcircuits and Causes Repetitive Behavior. Neuron 99, 1170–1187 e1179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graybiel AM Habits, rituals, and the evaluative brain. Annu Rev Neurosci 31, 359–387 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Graybiel AM & Grafton ST The striatum: where skills and habits meet. Cold Spring Harb Perspect Biol 7, a021691 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldridge JW, Berridge KC & Rosen AR Basal ganglia neural mechanisms of natural movement sequences. Can J Physiol Pharmacol 82, 732–739 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Ikoma A, Steinhoff M, Stander S, Yosipovitch G & Schmelz M The neurobiology of itch. Nat Rev Neurosci 7, 535–547 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickersham IR, Sullivan HA & Seung HS Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat Protoc 5, 595–606 (2010). [DOI] [PubMed] [Google Scholar]
- 56.White KA, et al. Glutamatergic Neurons in the Piriform Cortex Influence the Activity of D1- and D2-Type Receptor-Expressing Olfactory Tubercle Neurons. J Neurosci 39, 9546–9559 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halliwell JV & Horne AL Evidence for enhancement of gap junctional coupling between rat island of Calleja granule cells in vitro by the activation of dopamine D3 receptors. J Physiol 506 ( Pt 1), 175–194 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribak CE & Fallon JH The island of Calleja complex of rat basal forebrain. I. Light and electron microscopic observations. J Comp Neurol 205, 207–218 (1982). [DOI] [PubMed] [Google Scholar]
- 59.Le Foll B, Diaz J & Sokoloff P Neuroadaptations to hyperdopaminergia in dopamine D3 receptor-deficient mice. Life Sci 76, 1281–1296 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shuen JA, Chen M, Gloss B & Calakos N Drd1a-tdTomato BAC transgenic mice for simultaneous visualization of medium spiny neurons in the direct and indirect pathways of the basal ganglia. J Neurosci 28, 2681–2685 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi K, Holly EN, Davatolhagh MF, Beier KT & Fuccillo MV Integrated anatomical and physiological mapping of striatal afferent projections. Eur J Neurosci 49, 623–636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herman AM, et al. A cholinergic basal forebrain feeding circuit modulates appetite suppression. Nature 538, 253–256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dana H, et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16, 649–657 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Liu Q, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 139, 1353–1365 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onigbogi O, Ajayi AA & Ukponmwan OE Mechanisms of chloroquine-induced body-scratching behavior in rats: evidence of involvement of endogenous opioid peptides. Pharmacol Biochem Behav 65, 333–337 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Chung K & Deisseroth K CLARITY for mapping the nervous system. Nat Methods 10, 508–513 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gretenkord S, et al. Coordinated electrical activity in the olfactory bulb gates the oscillatory entrainment of entorhinal networks in neonatal mice. PLoS Biol 17, e2006994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marom K, et al. The Vomeronasal System Can Learn Novel Stimulus Response Pairings. Cell Rep 27, 676–684 e676 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Pietroni N, Tarini M & Cignoni P Almost isometric mesh parameterization through abstract domains. IEEE Trans. Vis. Comput. Graph 16, 621–635 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Stegmaier J, et al. Fast segmentation of stained nuclei in terabyte-scale, time resolved 3D microscopy image stacks. PLoS One 9, e90036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bartschat A, Hubner E, Reischl M, Mikut R & Stegmaier J XPIWIT--an XML pipeline wrapper for the Insight Toolkit. Bioinformatics 32, 315–317 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Schott B, et al. EmbryoMiner: A new framework for interactive knowledge discovery in large-scale cell tracking data of developing embryos. PLoS Comput Biol 14, e1006128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wright KN & Wesson DW The tubular striatum and nucleus accumbens distinctly represent reward-taking and reward-seeking. J Neurophysiol 125, 166–183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gadziola MA, Tylicki KA, Christian DL & Wesson DW The olfactory tubercle encodes odor valence in behaving mice. J Neurosci 35, 4515–4527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson KG, Hanes DP, Bichot NP & Schall JD Perceptual and motor processing stages identified in the activity of macaque frontal eye field neurons during visual search. J Neurophysiol 76, 4040–4055 (1996). [DOI] [PubMed] [Google Scholar]
- 80.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57, 289–300 (1995). [Google Scholar]
- 81.Esmaeili V, et al. Rapid suppression and sustained activation of distinct cortical regions for a delayed sensory-triggered motor response. Neuron 109, 2183–2201 e2189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D reconstruction of the Islands of Calleja in the OT. Red: D3-Cre/tdTomato neurons. Green: axonal fibers from the MOB projection neurons labeled via viral injection of AAV9-EGFP. Blue: cell nuclei labeled by DRAQ5TM, a far-red fluorescent DNA dye.
3D reconstruction of individual D3 neurons in and between two Islands of Calleja in the OT showing the neuronal processes of these neurons. Colors are the same as in Supplementary Video 1.
Blue light (but not green light) activation of OT D3-Cre/ChR2 neurons induces grooming.
Grooming behavior induced by blue light stimulation of OT D3-Cre/ChR2 neurons with different durations (1 to 20 s at 20 Hz).
Grooming behavior induced by blue light stimulation of OT D3-Cre/ChR2 neurons with different frequencies (1–20 Hz for 10 s).
Genotype and stimulation site controls. Blue light stimulation of OT D3-Cre/tdTomato neurons or blue light stimulation of D3-Cre/ChR2 neurons in the nucleus accumbens (NAc) and hippocampus (dentate gyrus and CA3 region) do not induce grooming.
Grooming behavior induced by blue light activation of the OT in D3-Cre/tdTomato mice injected with Cre-dependent AAV1-DIO-ChR2-EYFP virus. Additional conditions include green light or optical fiber implanted in the paraventricular hypothalamic nucleus (PVH).
Blue light (but not green light) activation of OT D3 neurons stops social investigation and initiates grooming in D3-Cre/ChR2 mice.
Blue light (but not green light) activation of OT D3 neurons stops feeding and initiates grooming in D3-Cre/ChR2 mice.
Blue light (but not green light) activation of OT D3 neurons stops itch-induced scratching and initiates grooming in D3-Cre/ChR2 mice.
Blue light (but not green light) activation of OT D3 neurons stops body licking and initiates grooming in D3-Cre/ChR2 mice.
Green light (but not blue light) inactivation of OT D3-Cre/eArchT neurons shortens spontaneous grooming.
Green light (but not blue light) inactivation of OT D3-Cre/eArchT neurons halts water spray-induced grooming.
Data Availability Statement
The raw data generated and/or analyzed during the current study are available from the corresponding authors on reasonable request. Source data are provided for all figures wherever applicable. The Allen Mouse Brain Connectivity Atlas (http://connectivity.brain-map.org/transgenic/experiment/304168043) was used for Fig. 1d and the Allen Mouse Brain Common Coordinate Framework version 3 (https://scalablebrainatlas.incf.org/mouse/ABA_v3) was used to outline brain structures in Figs. 2b, 4a, and Extended Data Figs. 4, 5.


