Abstract
DNA methylation is associated with transcriptional repression of eukaryotic genes and transposons, but the downstream mechanism of gene silencing is largely unknown. Here we describe two Arabidopsis methyl-CpG binding domain proteins, MBD5 and MBD6, that are recruited to chromatin by recognition of CG methylation, and redundantly repress a subset of genes and transposons without affecting DNA methylation levels. These methyl-readers recruit a J-domain protein, SILENZIO, that acts as a transcriptional repressor in loss-of-function and gain-of-function experiments. J-domain proteins often serve as co-chaperones with HSP70s. Indeed, we found that SILENZIO’s conserved J-domain motif was required for its interaction with HSP70s and for its silencing function. These results uncover an unprecedented role of a molecular chaperone J-domain protein in gene silencing downstream of DNA methylation.
One Sentence Summary:
Two CG specific DNA methyl-readers redundantly repress methylated genes and transposons by recruiting a J-domain protein.
Cytosine DNA methylation (mC) in eukaryotes is typically associated with transcriptional silencing of genes and transposable elements (TEs), however relatively little is known of the mechanism (1, 2). Mammalian genomes encode for several Methyl-CpG-Binding Domain (MBD) proteins that are recruited to chromatin in part by recognition of methylated CG dinucleotides, but they also play methylation-independent roles in gene regulation (3–7). One prevailing model is that MBDs recruit histone deacetylase complexes to methylated DNA, causing chromatin compaction and gene silencing (5–7). In plants, loss of DNA methylation causes derepression of many transposons and genes (8), but no evidence has been found for a role of methyl-readers in this process, leaving unresolved the question of what acts downstream of the methyl mark.
We recently identified two proteins named MBD5 and MBD6 from a mass spectrometry screen for methyl-readers in Arabidopsis thaliana (9). MBD5 and MBD6 belong to a family of 13 members that have been identified by sequence similarity with human MBD domains (10–12). Outside of this domain there is no sequence conservation between plants and animals. MBD5 and MBD6 are close relatives (10–12), they can interact with each other in vivo (13, 14), and were shown to bind methylated probes in electrophoretic mobility shift assays (10, 15, 16). While a function has not been assigned to MBD5, MBD6 was shown to be required for ribosomal RNA gene regulation in allotetraploid genetic hybrids (17).
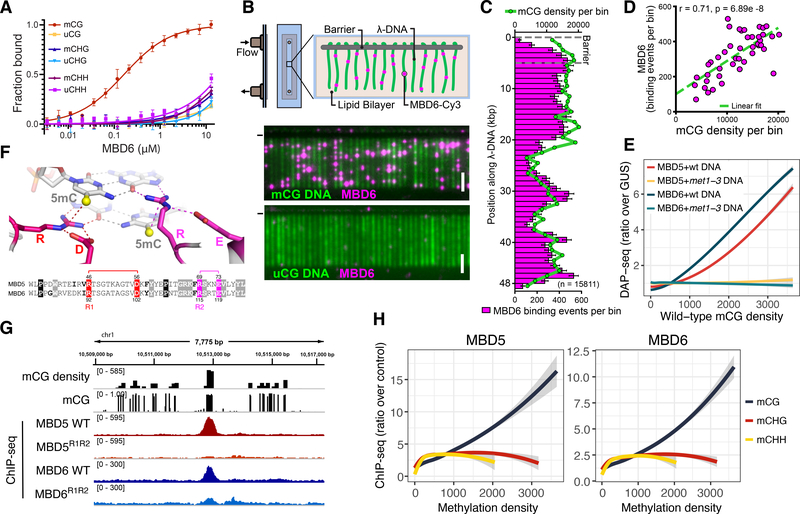
In plants, 5-Methylcytosines are common in CG, CHG, and CHH sequence contexts (18). The MBD typically recognizes symmetrically methylated CG dinucleotides (19), but exceptions have been reported such as MeCP2, that can also bind mCA sites (20). We tested the ability of MBD5 and MBD6 to bind CG, CHG, or CHH methylation by performing fluorescence polarization (FP) assays with oligonucleotides methylated in different contexts. Both MBD5 and MBD6 showed a strong preference for CG methylated oligonucleotides as compared to unmethylated controls, but little preference was observed for CHG or CHH methylation (Fig.1A, Fig. S1). We also employed DNA curtains, a single-molecule fluorescence microscopy assay, to visualize the interaction between MBD6 and flow-stretched bacteriophage λ DNA, which was methylated in vitro with the CG specific bacterial M.SssI methyltransferase. MBD6 bound methylated, but not unmethylated DNA curtains, and its enrichment profile correlated strongly with the local density of methylated CG sites (Fig. 1B–D). To test the ability of MBD5 and MBD6 to bind methylation in natural Arabidopsis genomic sequences, we performed DNA affinity purification sequencing (DAP-seq) (21) by incubating Halo-tagged recombinant proteins with DNA extracted from wild-type plants or from met1–3 mutant plants. The met1–3 mutant is almost completely lacking in CG methylation due to mutation of the maintenance DNA METHYLTRANSFERASE 1 (MET1) gene, but retains substantial levels of CHG and CHH methylation (22). We observed a strong genome-wide correlation between MBD5/6 DAP-seq enrichment and CG methylation density with DNA from a wild-type background, and an almost complete loss of binding to DNA in the met1–3 background (Fig. 1E). Only a few small peaks were retained in regions that did not completely lose CG methylation (Fig. S2). Overall, these results strongly support the specificity of MBD5 and MBD6 for CG methylation in vitro.
Figure 1: MBD5 and MBD6 are CG specific methyl-readers in vitro and in vivo.
A) Binding curves of MBD6 with DNA oligos methylated (m) or unmethylated (u) in the indicated contexts, measured by fluorescence polarization (N=3, standard error of the mean [SEM]). B) Diagram of DNA curtain assay and representative image of YOYO-1 stained methylated (mCG) and unmethylated (uCG) DNA (green) bound by Cy3-labeled MBD6 (magenta). (−) chrome diffusion barriers. Scale bar - 5 μm. C) Distribution of MBD6 binding events along mCG DNA overlayed with the distribution of mCG density (green line). Error bars: 95% confidence intervals (CI) by bootstrap. D) Correlation scatterplot of MBD6 binding to methylated curtains and mCG density (1 kb bins). R: Pearson. E) Genome-wide correlation between DAP-seq and mCG density (400 bp bins). Trend lines calculated by locally weighted polynomial regression (loess curves). F) Homology models of MBD5 and MBD6. The two arginine residues of the 5mC–Arg–G triads (R1 and R2) are shown in the sequence alignment. G) Example ChIP-seq peaks at regions of dense CG methylation. H) Loess curves of ChIP-seq enrichment and methylation density (400 bp bins overlapping Pol V ChIP-seq peaks). E,H) Shaded area: 95% CI.
We generated homology models of Arabidopsis MBD domains based on known mammalian MBD structures. High confidence models could be determined except for the most divergent protein MBD9, which is known not to bind methylated DNA in vivo (23) (Fig. S3). The MBD5 and MBD6 structural models highlighted two arginine residues (R1 and R2) that are predicted to directly interact with methylated CGs by forming the previously described “methyl-Arg-G triad” (19) (Fig. 1F, Fig. S3). We tested the importance of these residues by mutating them to alanine (MBD5R1R2, MBD6R1R2), and indeed we observed a loss of specificity for binding to CG methylation in FP assays (Fig. S1B).
We next investigated the genomic localization of MBD5 and MBD6 in vivo by chromatin immunoprecipitation-sequencing (ChIP-seq) using FLAG-tagged transgenic lines. MBD5 and MBD6 bound methylated chromatin, with a clear preference for mCG density as opposed to mCHG and mCHH density (Fig. 1G–H). Importantly, no correlation was found with the density of unmethylated CG sites (Fig. S4). The MBD5R1R2 and MBD6R1R2 mutants showed a strong reduction of ChIP-seq enrichment (Fig. 1G, Fig. S5), demonstrating that recognition of mCGs is required for recruitment of MBD5 and MBD6 to chromatin.
Methylated DNA is associated with three different chromatin states in Arabidopsis: euchromatic patches of RNA-directed DNA methylation (RdDM) which contain CG and non-CG methylation, peri-centromeric heterochromatin which is enriched in H3K9me2 as well as CG and non-CG methylation, and expressed genes containing Gene Body Methylation (GbM) that are exclusively marked by CG methylation (18). We observed MBD5 and MBD6 ChIP-seq enrichment at a large fraction of sites in all three chromatin states, but the extent of enrichment was higher at RdDM sites compared to heterochromatin or GbM sites (Fig. S6). Interestingly, the preference for RdDM sites was not observed by DAP-seq, which tests the ability of proteins to bind naked genomic DNA (Fig. S6C–D). These observations suggest that recruitment of MBD5 and MBD6 to chromatin in vivo may be influenced by histones or other chromatin components.
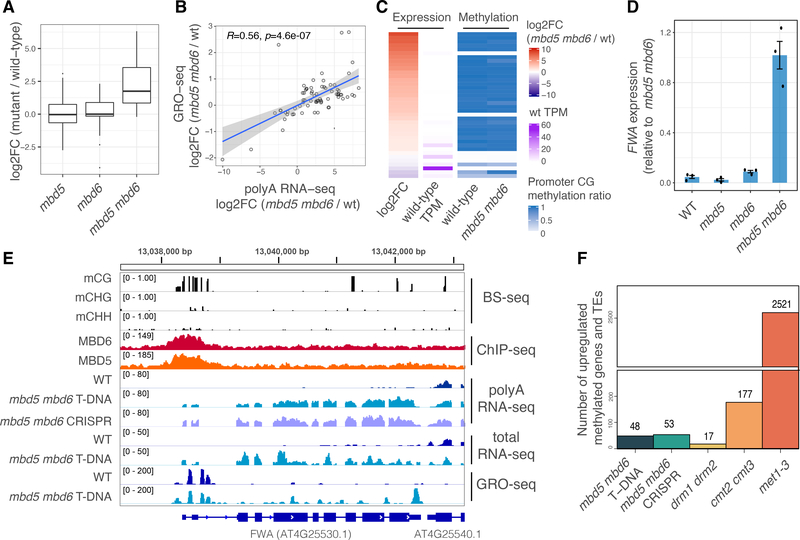
To test if MBD5 and MBD6 regulate transcription at their targets we performed RNA sequencing (RNA-seq) of mbd5 and mbd6 T-DNA mutants and of a double mutant generated by crossing (mbd5 mbd6). A number of transposons and protein coding genes were derepressed only in the double mutant, indicating genetic redundancy of MBD5 and MBD6 (Fig. 2A, Fig. S7). We confirmed this with an independent mbd5 mbd6 double mutant generated by CRISPR/Cas9 (Fig. S7, Fig. S8). Global run-on sequencing (GRO-seq) showed a similar pattern of changes, indicating that the derepression in mbd5 mbd6 occurs at the transcriptional level (Fig. 2B). Most upregulated genes and transposons were not expressed in wild-type and showed high levels of promoter CG methylation, suggesting that they are direct targets (Fig. 2C). DNA methylation levels were not altered in mbd5 mbd6 (Fig. 2C, Fig. S9), indicating that the methyl-readers act strictly downstream of DNA methylation. One of the derepressed genes was FWA, a well characterized imprinted gene that is silenced by promoter methylation (24) (Fig. 2D–E). Reintroduction into mbd5 mbd6 mutant plants of FLAG-tagged versions of wild-type MBD5 or MBD6, but not their R1R2 mutant counterparts, was sufficient to largely rescue the derepression of FWA and of other genes and transposons (Fig. S10). Overall, these results suggest that MBD5 and MBD6 are recruited to DNA by methylation and translate the methyl mark into gene repression at a subset of methylated sites.
Figure 2: MBD5 and MBD6 redundantly repress a subset of genes and transposons downstream of DNA methylation.
A) Boxplot of polyA RNA-seq for different mutants. Shown are the transcripts (genes and transposons) upregulated in mbd5 mbd6. B) Scatterplot comparing polyA RNA-seq with GRO-seq data at mbd5 mbd6 T-DNA differential transcripts. R and p-value: Spearman. Shaded area: 95% CI. C) Heatmap of mbd5 mbd6 T-DNA differential transcripts, showing polyA RNA-seq and BS-seq data (average methylation ratio at 400 bp windows around the TSS). D) RT-qPCR analysis of FWA expression normalized to IPP2. Dots: individual plants. Error bars: SEM. E) Genome browser tracks at FWA. The GRO-seq enrichment at the FWA promoter likely corresponds to Pol V transcription. F) Number of promoter methylated genes and TEs, upregulated in different mutants.
We compared mbd5 mbd6 gene expression data with that of mutants affecting different methylation pathways: drm1 drm2 and cmt2 cmt3 lose non-CG methylation at euchromatic RdDM sites and heterochromatic regions respectively, while met1–3 loses CG methylation genome-wide (22, 25). Most of the loci upregulated in mbd5 mbd6 were also derepressed in met1–3, indicating that they are silenced by CG methylation (Fig. S11A). The MBD5/6 derepressed TEs were also longer than average and more enriched in H3K9me2, indicating that they are mostly heterochromatic TEs (Fig. S11B–C). A small number of loci were also derepressed in cmt2 cmt3, but none were derepressed in drm1 drm2 (Fig. S11A). Thus, while MBD5 and MBD6 are enriched at a wide range of CG methylated sites, their repressive role is strongest at a subset of MET1-dependent sites. Furthermore, the number of derepressed transposons and the amplitude of derepression in mbd5 mbd6 was much smaller than in met1–3 (Fig. 2F, Fig. S11), suggesting that MBD5 and MBD6 are not the only factors mediating repression downstream of DNA methylation.
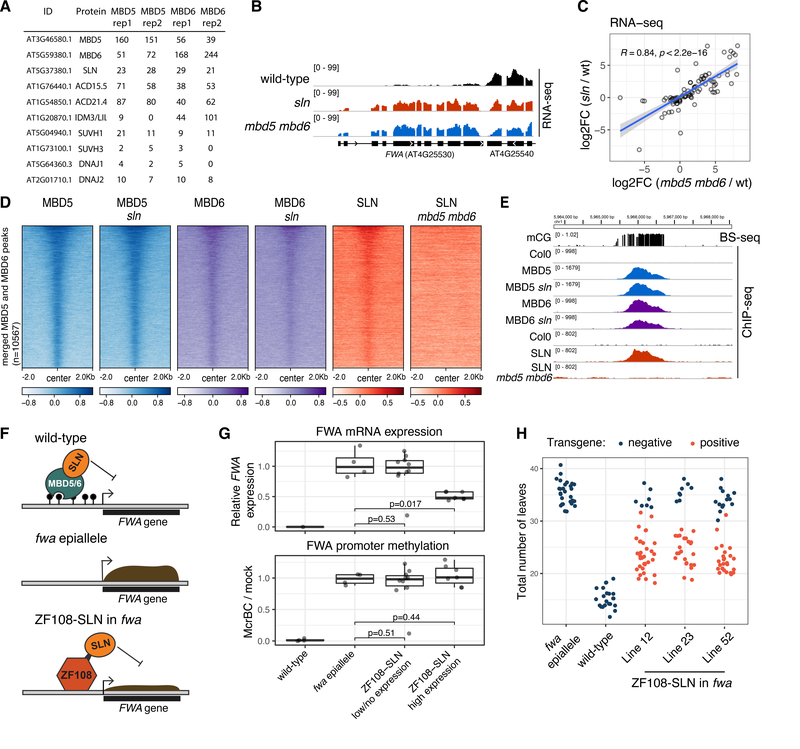
To investigate the mechanism of action of MBD5 and MBD6, we performed immunoprecipitation–mass spectrometry (IP-MS) utilizing the FLAG-tagged transgenic lines. Both proteins pulled-down each other and three small heat shock proteins (ACD15.5, ACD21.4 and IDM3/LIL) that were previously found to interact with MBD5 and MBD7 (13). In addition, we detected an uncharacterized class C J-domain protein (AT5G37380) (26, 27) which we have named SILENZIO (SLN) (Fig. 3A, Table S1). MBD5 and MBD6 also pulled down a smaller number of peptides of SUVH1, SUVH3, DNAJ1 and DNAJ2, which are components of a methyl-reader complex known to bind at RdDM sites and upregulate nearby protein coding genes (9, 28).
Figure 3: SLN represses transcription downstream of MBD5 and MBD6.
A) IP-MS spectral counts of FLAG-tagged MBD5 and MBD6. All proteins displayed were not detected in the no-FLAG negative control (see Table S1). B) RNA-seq data at FWA. C) Scatterplot of the union of mbd5 mbd6 CRISPR and sln differential transcripts. R and p-value: Spearman. Blue line: linear model fit. Shaded area: 95% CI. D) Heatmap of ChIP-seq data (log2 fold change over no-FLAG control). E) Example methylated site bound by MBD5, MBD6, and SLN, in the indicated backgrounds. F) Cartoon showing SLN’s ectopic recruitment to unmethylated FWA via fusion to ZF108. G) RT-qPCR analysis of FWA expression and McrBC-qPCR analysis of FWA promoter methylation in T1 lines expressing low or high levels of ZF108-SLN (western blot in Figure S15A). Dots: individual plants. P-value: t-test. RT-qPCR data (normalized to IPP2) is relative to fwa epiallele plants. H) Flowering time (number of leaves produced before flowering) of segregating T2 populations from three transgenic lines expressing high levels of ZF108-SLN, comparing transgene positive to null segregant (negative) plants.
We focused our further investigation on SILENZIO because of the recently described role of the J-domain proteins DNAJ1 and DNAJ2 in gene activation downstream of DNA methylation (9, 28). SILENZIO homologs were found to be present widely throughout the plant kingdom, but only the J-domain was conserved in animals (Figure S12). To determine whether SILENZIO was involved in gene silencing, we performed RNA-seq on an sln T-DNA mutant line. Strikingly, we found a strong correlation between the sln and the mbd5 mbd6 RNA-seq data, with a similar extent of derepression of transposable elements and genes, including FWA (Fig. 3B–C). We performed ChIP-seq with a complementing FLAG-tagged SLN line (Fig. S13) and observed localization to the same sites as MBD5 and MBD6, but this localization was abolished in mbd5 mbd6 mutants, suggesting that SLN is recruited to chromatin by the methyl-readers (Fig. 3D–E, Fig. S14). Conversely, MBD5 and MBD6 ChIP-seq signal was unaffected in sln, indicating that their recruitment to chromatin does not require SLN (Fig. 3D–E, Fig. S14). Overall, these results suggest that SLN acts as a gene repressor downstream of MBD5 and MBD6.
To further test the role of SLN as a repressor, we created a fusion of SLN with ZF108, an artificial zinc finger that allows ectopic targeting of proteins to the FWA promoter (Fig. 3F) (29, 30). We transformed this fusion construct driven by the constitutive UBIQUITIN10 promoter (pUBQ10::ZF108-SLN) into fwa epiallele mutant plants (24), in which the FWA gene has heritably lost DNA methylation, leading to FWA overexpression and a late-flowering phenotype. Transgenic (T1) plants that expressed high levels of the fusion protein displayed downregulation of FWA, thus supporting a role of SLN as transcriptional repressor (Fig. 3G, Fig. S15A). Importantly, FWA repression was not accompanied by promoter methylation (Fig. 3G, Fig. S15B), demonstrating that SLN’s ability to repress transcription can be uncoupled from DNA methylation. Indeed, in the T2 segregant population, the null segregants recovered FWA overexpression and the corresponding late flowering time (Fig. 3H, Fig. S15C). ZF108 was designed to bind FWA, but it also binds to thousands of off-target sites in the genome (30), allowing us to examine gene expression changes at these sites by performing RNA-seq in the pUBQ10::ZF108-SLN lines. We observed that genes with a ZF108 peak near their promoter showed a tendency to be downregulated (Fig. S16), demonstrating that ectopic recruitment of SLN can repress many genes in addition to FWA.
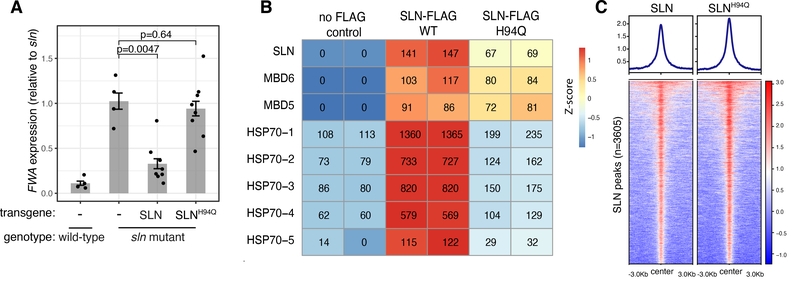
IP-MS analysis of SLN-FLAG identified peptides corresponding to MBD5 and MBD6 as expected, but also showed a strong enrichment of five HEAT SHOCK PROTEIN 70 (HSP70s) known to be constitutively expressed and localized in the nucleus (31) (Fig. 4B, Table S1). Enrichment for HSP70s was also detected in the MBD5 and MBD6 IP-MS datasets, and was lost in sln mutant plants (Figure S17, Table S1). This suggests that SLN mediates the interaction between the methyl-readers and the HSP70s.
Figure 4: SLN silencing function requires the conserved HPD tripeptide.
A) RT-qPCR analysis of FWA expression (normalized to IPP2) in T1 lines expressing SLN or SLNH94Q in the sln mutant background. p-values: t-test. Error bars: SEM. Dots: individual plants. B) IP-MS spectral counts of wild-type and H94Q mutant SLN-FLAG (representative of two independent experiments, see Table S1). C) ChIP-seq of FLAG-tagged SLN and SLNH94Q (log2 fold change over the no-FLAG control).
The canonical function of J-domain proteins is to bind clients, recruit HSP70 chaperones utilizing a conserved HPD tripeptide, and stimulate the ATPase activity of HSP70s to increase their affinity for substrates. The HSP70-substrate interaction can induce folding, disaggregation, or assembly/disassembly of complexes involving client proteins (32). Mutating the histidine of the HPD tripeptide to glutamine can abrogate the J-domain-HSP70 interaction (32). To test if SLN’s binding to HSP70s was associated with its gene silencing function, we generated an HPD mutant version of SLN by mutating the histidine to glutamine (SLNH94Q) and transformed this into sln mutant plants. The SLNH94Q mutant failed to rescue the derepression of FWA and of the other genes and transposons, suggesting that the gene silencing function of SLN requires the J-domain and HSP70 interaction (Fig. 4A, Fig. S18A–F). Indeed, IP-MS of SLNH94Q showed greatly reduced enrichment of HSP70s, while the interaction with MBD5 and MBD6 was retained (Fig. 4B, Table S1). Furthermore, ChIP-seq enrichment of SLN on chromatin was not affected by the H94Q mutation (Fig. 4C, Fig. S18G–H). These results suggest that recruitment of SLN by the methyl-readers may serve as a tether to bring the chaperone activity of SLN-HSP70s to CG dense methylated chromatin to enforce gene silencing. The interaction between chaperones and their clients is often transient and difficult to detect by IP-MS (32), meaning that SLN might exert its repressive activity via recruitment, stabilization, or assembly of currently unknown repressive complexes, or via targeted inhibition or disassembly of activators.
In conclusion, this work identifies a novel pathway that links DNA methylation to silencing of sites marked by CG methylation. The characterization of the methyl binding proteins MBD5 and MBD6 shows that they likely act via a mechanism distinct from that of known MBD proteins in animals. The identification of the novel J-domain protein SILENZIO as a silencing effector further suggests that gene repression downstream of methylation is linked to chaperone activity, and this new pathway is likely to be conserved among divergent plant lineages.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mahnaz Akhavan and the Broad Stem Cell Research Center BioSequencing core for high throughput sequencing, Ece Ograg for help with plant care, Colette Picard, Zhenhui Zhong and Giuseppe Barisano for bioinformatic scripts, Vijay Ramani, Nour J. Abdulhay and Aidan Keith for their guidance with the EM-seq of in vitro methylated DNA, Barbara Panning and members of the Jacobsen lab for helpful advice and discussions.
Funding: This work was supported by NIH R35 GM130272 to S.E.J., the UCSF Program for Breakthrough Biomedical Research and the Sandler Foundation to S.R., NIH R01 GM089778 to J.A.W., NIH R35GM134744 and CPRIT RR160029 to X.C. (who is a CPRIT Scholar in Cancer Research), NIH/NIGMS K99GM135515 to S.H.D, the Philip Whitcome Pre-Doctoral Fellowship in Molecular Biology to L.I. and the Ruth L. Kirschstein National Research Service Award GM007185 to B.A.B.. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: The high-throughput sequencing data generated in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession GSE165095).
References and Notes
- 1.Goll MG, Bestor TH, Eukaryotic Cytosine Methyltransferases. Annual Review of Biochemistry. 74, 481–514 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Zemach A, McDaniel IE, Silva P, Zilberman D, Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 328, 916–919 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Baubec T, Ivánek R, Lienert F, Schübeler D, Methylation-Dependent and -Independent Genomic Targeting Principles of the MBD Protein Family. Cell. 153, 480–492 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Shimbo T, Wade PA, Proteins that read DNA methylation. Advances in Experimental Medicine and Biology. 945, 303–320 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Boxer LD, Renthal W, Greben AW, Griffith EC, Bonev B, Greenberg ME, MeCP2 Represses the Rate of Transcriptional Initiation of Highly Methylated Long Genes. Molecular Cell. 77, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, De Lima Alves F, Rappsilber J, Greenberg ME, Bird A, Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nature Neuroscience. 16, 898–902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A, Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 393, 386–389 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW-L, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR, Genome-wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell. 126, 1189–1201 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z, Yen L, Barshop WD, Rayatpisheh S, Gallego-Bartolome J, Groth M, Wang Z, Wohlschlegel JA, Du J, Rothbart SB, Butter F, Jacobsen SE, A DNA methylation reader complex that enhances gene transcription. Science. 362, 1182–1186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zemach A, Grafi G, Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. The Plant Journal. 34, 565–572 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Berg A, Meza TJ, Mahić M, Thorstensen T, Kristiansen K, Aalen RB, Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial for normal development. Nucleic Acids Research. 31, 5291–5304 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Springer NM, Kaeppler SM, Evolutionary Divergence of Monocot and Dicot Methyl-CpG-Binding Domain Proteins. Plant Physiology. 138, 92–104 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Palanca AMS, Won SY, Gao L, Feng Y, Vashisht AA, Liu L, Zhao Y, Liu X, Wu X, Li S, Le B, Kim YJ, Yang G, Li S, Liu J, Wohlschlegel JA, Guo H, Mo B, Chen X, Law JA, The MBD7 complex promotes expression of methylated transgenes without significantly altering their methylation status. eLife. 6, e19893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zemach WA, Li Y, Wayburn B, Ben-Meir H, Kiss V, Avivi Y, Kalchenko V, Jacobsen SE, Grafi G, DDM1 Binds Arabidopsis Methyl-CpG Binding Domain Proteins and Affects Their Subnuclear Localization. The Plant cell. 17, 1549–1558 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scebba F, Bernacchia G, De Bastiani M, Evangelista M, Cantoni RM, Cella R, Locci MT, Pitto L, Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Molecular Biology. 53, 755–771 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Ito M, Koike A, Koizumi N, Sano H, Methylated DNA-Binding Proteins from Arabidopsis. Plant Physiology. 133, 1747–1754 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, Pikaard CS, Multimegabase Silencing in Nucleolar Dominance Involves siRNA-Directed DNA Methylation and Specific Methylcytosine-Binding Proteins. Molecular Cell. 32, 673–684 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law JA, Jacobsen SE, Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Zhang X, Blumenthal RM, Cheng X, A common mode of recognition for methylated CpG. Trends in Biochemical Sciences. 38, 177–183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME, Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 522 (2015), doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett A, Malley RCO, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR, Mapping genome-wide transcription-factor binding sites using DAP-seq. Nature Protocols. 12, 1659–1672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroud H, Greenberg MVC, Feng S, V Bernatavichute Y, Jacobsen SE, Comprehensive Analysis of Silencing Mutants Reveals Complex Regulation of the Arabidopsis Methylome. Cell. 152, 352–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potok ME, Wang Y, Xu L, Zhong Z, Liu W, Feng S, Naranbaatar B, Rayatpisheh S, Wang Z, Wohlschlegel JA, Ausin I, Jacobsen SE, Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nature Communications. 10, 3352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM, The Late Flowering Phenotype of fwa Mutants Is Caused by Gain-of-Function Epigenetic Alleles of a Homeodomain Gene. Molecular Cell. 6, 791–802 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE, Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nature Structural & Molecular Biology. 21, 64–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finka A, Mattoo RUH, Goloubinoff P, Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress and Chaperones. 16, 15–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babu V, Rajan V, D’silva P, Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Functional & Integrative Genomics. 9, 433–446 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Zhao QQ, Lin RN, Li L, Chen S, He XJ, A methylated-DNA-binding complex required for plant development mediates transcriptional activation of promoter methylated genes. Journal of Integrative Plant Biology. 61, 120–139 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, Patel DJ, Jacobsen SE, SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 507, 124–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallego-Bartolomé J, Liu W, Kuo PH, Feng S, Ghoshal B, Gardiner J, Zhao JM-C, Park SY, Chory J, Jacobsen SE, Co-targeting RNA Polymerases IV and V Promotes Efficient De Novo DNA Methylation in Arabidopsis. Cell. 176, 1068–1082.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leng L, Liang Q, Jiang J, Zhang C, Hao Y, Wang X, Su W, A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. Journal of Plant Research. 130, 349–363 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B, The Hsp70 chaperone network. Nature Reviews Molecular Cell Biology. 20, 665–680 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Pulido P, Leister D, Novel DNAJ-related proteins in Arabidopsis thaliana. New Phytologist. 217, 480–490 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q, High-Efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Molecular Plant. 8, 1820–1823 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Greene EC, Wind S, Fazio T, Gorman J, Visnapuu ML, DNA curtains for high-throughput single-molecule optical imaging. Methods in enzymology. 472, 293–315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The UniProt Consortium, UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. Journal of Molecular Biology. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 38.NCBI Resource Coordinators, Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 46, D8–D13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R, The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moissiard G, Bischof S, Husmann D, Pastor WA, Hale CJ, Yen L, Stroud H, Papikian A, Vashisht AA, Wohlschlegel JA, Jacobsen SE, Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. PNAS. 111, 7474–7479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hetzel J, Duttke SH, Benner C, Chory J, Nascent RNA sequencing reveals distinct features in plant transcription. Proceedings of the National Academy of Sciences of the United States of America. 113, 12316–12321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Link VM, Duttke SH, Chun HB, Holtman IR, Westin E, Hoeksema MA, Abe Y, Skola D, Romanoski CE, Tao J, Fonseca GJ, Troutman TD, Spann NJ, Strid T, Sakai M, Yu M, Hu R, Fang R, Metzler D, Ren B, Glass CK, Analysis of Genetically Diverse Macrophages Reveals Local and Domain-wide Mechanisms that Control Transcription Factor Binding and Function. Cell. 173, 1796–1809.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villar CBR, Köhler C, Plant Chromatin Immunoprecipitation. Methods in Molecular Biology. 655, 401–411 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Li X, Jake Harris C, Zhong Z, Chen W, Liu R, Jia B, Wang Z, Li S, Jacobsen SE, Du J, Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proceedings of the National Academy of Sciences of the United States of America. 115, E8793–E8802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krueger F, Andrews SR, Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang X, Zhang S, Li K, Thimmapuram J, Xie S, ViewBS: A powerful toolkit for visualization of high-throughput bisulfite sequencing data. Bioinformatics. 34, 708–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nature Methods. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T, deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic acids research. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Shirley XS, Model-based analysis of ChIP-Seq (MACS). Genome Biology. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Harris CJ, Liu Q, Liu W, Ausin I, Long Y, Xiao L, Feng L, Chen X, Xie Y, Chen X, Zhan L, Feng S, Jingyi JL, Wang H, Zhai J, Jacobsen SE, Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc Natl Acad Sci U S A. 115, E1069–E1074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W, Duttke SH, Hetzel J, Groth M, Feng S, Gallego-Bartolome J, Zhong Z, Kuo HY, Wang Z, Zhai J, Chory J, Jacobsen SE, RNA-directed DNA methylation involves co-transcriptional small-RNA-guided slicing of polymerase V transcripts in Arabidopsis. Nature plants. 4, 181–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Pyl PT, Huber W, HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 15 (2014), doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Gu Z, Eils R, Schlesner M, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Schmitz RJ, Schultz MD, Lewsey MG, O’Malley R. c., Urich MA, Libiger O, Schork NJ, Ecker JR, Transgenerational epigenetic instability is a source of novel methylation variants. Science. 334, 369–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlan AR, Hall IM, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.