Abstract
We isolated mitochondria from Saccharomyces cerevisiae to selectively study polysomes bound to the mitochondrial surface. The distribution of several mRNAs coding for mitochondrial proteins was examined in free and mitochondrion-bound polysomes. Some mRNAs exclusively localize to mitochondrion-bound polysomes, such as the ones coding for Atm1p, Cox10p, Tim44p, Atp2p, and Cot1p. In contrast, mRNAs encoding Cox6p, Cox5a, Aac1p, and Mir1p are found enriched in free cytoplasmic polysome fractions. Aac1p and Mir1p are transporters that lack cleavable presequences. Sequences required for mRNA asymmetric subcellular distribution were determined by analyzing the localization of reporter mRNAs containing the presequence coding region and/or the 3′-untranslated region (3′UTR) of ATM1, a gene encoding an ABC transporter of the mitochondrial inner membrane. Biochemical analyses of mitochondrion-bound polysomes and direct visualization of RNA localization in living yeast cells allowed us to demonstrate that either the presequence coding region or the 3′UTR of ATM1 is sufficient to allow the reporter mRNA to localize to the vicinity of the mitochondrion, independently of its translation. These data demonstrate that mRNA localization is one of the mechanisms used, in yeast, for segregating mitochondrial proteins.
The biogenesis of mitochondria is a complex cellular process that involves the concerted expression of the two genomes in which molecular components of the organelles are encoded. The majority of mitochondrial proteins are encoded by the nucleus, synthesized on cytoplasmic polysomes as precursor proteins with short N-terminus extensions. Detailed knowledge of how polypeptides are sorted and imported to mitochondria has been elucidated in Saccharomyces cerevisiae and Neurospora crassa (43, 49). While mitochondrial protein import can occur posttranslationally, some precursors in vivo may also be imported cotranslationally (43). The surface of mitochondria isolated from yeast treated with protein synthesis inhibitors is coated with cytoplasmic polysomes. These polysomes are enriched in mRNAs encoding mitochondrially destined polypeptides and are clustered around contact sites between inner and outer mitochondrial membranes (29–31). In the early 1980s, two independent groups confirmed that mRNA of polysomes which coisolate with mitochondria is enriched in transcripts encoding mitochondrial proteins. Polysomes that cover the mitochondrial surface contain many more mRNAs for the α-, β-, and γ-subunits of F1 ATPase than free polysomes (2). Accordingly, cytoplasmic pools of these F1 ATPase subunits are small, arguing in favor of the possibility that the majority of their transcripts are attached to the outer mitochondrial membrane (3). Further, Suissa and Schatz measured the distribution of mRNAs for 12 imported mitochondrial polypeptides between free and mitochondrion-bound polysomes. These authors also showed the existence of a specific polysomal subpopulation bound to mitochondria, polysomes which could then insert cotranslationally some nuclearly encoded mitochondrial polypeptides (57). More recently, Pon et al. purified mitochondrial membrane components that were associated with cytoplasmic polysomes and retained the capacity to transport proteins across membranes (46). In vitro, a tight coupling between polypeptide synthesis and membrane translocation was observed when the translation of several precursors was performed in the presence of isolated yeast mitochondria. Such tight coupling between protein translation and membrane transport suggests a cotranslational import mechanism (19, 20, 59).
Interestingly, the nascent polypeptide-associated complex (NAC) was found on both ribosomes isolated from the cytosol and ribosomes associated with mitochondria (23). Moreover, a homologous in vitro system has recently been described, in which ribosome-attached nascent chains initiate import into mitochondria in the presence of NAC (21). Therefore, as for the endoplasmic reticulum (40), protein translation, targeting, and translocation across mitochondrial membranes are, at least in some cases, closely related processes.
While searching for new components of the mitochondrial import machinery, we found that the overexpression of KAP121 and KAP123, encoding two karyopherins involved in nucleocytoplasmic traffic of proteins and/or mRNAs (47, 50, 53), facilitates the mitochondrial import of highly hydrophobic proteins. This process was mediated by the specific targeting of corresponding mRNAs to mitochondrion-bound polysomes, which may lead to an enhancement of the cotranslationally import of such hydrophobic proteins (14). We then hypothesized that under physiological conditions the initial selection event for some mitochondrial proteins may be the localization of their transcripts to the mitochondrial periphery, where translation and import could be tightly coupled. We report here evidence for the asymmetric distribution of mRNAs encoding mitochondrial proteins, and we describe the mRNA elements involved in the molecular mechanism. Transcripts coding for Atm1p, Cox10p, Tim44p, Atp2p, and Cot1p are exclusively localized to mitochondrion-bound polysomes, while mRNAs encoding Cox6p, Cox5ap, Aac1p, and Mir1p are enriched in free cytoplasmic polysomes. The ability of the ATM1 untranslated regions (UTRs) or/and presequence to address the green fluorescent protein (GFP) coding sequence to mitochondrion-bound polysomes was analyzed. Interestingly, not only the 3′UTR of ATM1 but also 48 bp in the N-terminal coding region for amino acids 1 to 16 of Atm1p are sufficient to allow the hybrid mRNA to behave as the ATM1 transcript. To confirm these data, we used the recently described RNA-labeling system (7), which allows the direct visualization of RNA dynamics in live cells by microscopy of GFP fluorescence. Both the presequence coding region and the 3′UTR of ATM1 are independently sufficient to allow the localization of the reporter RNA to the vicinity of mitochondria. Therefore, in yeast, mRNA localization is involved in mitochondrial protein targeting, and consequently may play an essential role in the organelle biogenesis.
MATERIALS AND METHODS
Yeast strain and growth conditions.
The CW04 strain of S. cerevisiae was used in this study. This strain is isonuclear with the strain W303-1B (MATa leu2-3 ura 3-1 trp1-1 ade1-2 his3-11 can1-100) and was obtained as described earlier (12, 17).
CW04 cells were grown at 30°C in rich medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose, 30 μg of adenine per ml), galactose-rich medium [1% Bacto yeast extract, 1% Bacto peptone, 0.1% KH2PO4, 0.12% (NH4)2SO4, 2% galactose], or synthetic medium containing either 2% glucose or 2% galactose supplemented with the appropriate nutritional requirements. For solid medium, 2% agar was added. Yeast cells were transformed using a simplified lithium method (24).
Living cells expressing the GFP chimeras or green RNAs were observed by fluorescence microscopy with a chilled charge-coupled-device camera (Hamamatsu Photonics, Hamamatsu City, Japan) as previously described (14, 16). For all microscopic observations, cells were harvested in the early log phase.
Plasmids and DNA manipulations.
All bacteriological analyses and DNA manipulations were performed as previously described (48).
Sequences in the ATM1 gene were obtained by 16-cycle PCR amplifications using as template yeast genomic DNA (100 ng) and the recombinant Tth DNA polymerase XL (Perkin-Elmer Cetus) (5). For the promoter and the 5′UTR, the region spanning between the last 50 amino acids of the PRP12 gene (contiguous to ATM1) and either the first 3, 16, or 53 amino acids of Atm1p was amplified and cloned in frame with the GFP. For the 3′UTR, 507 bp between the Atm1p stop codon and the first 14 amino acids of the ADE4 gene (contiguous to ATM1) were amplified and cloned in frame with the GFP stop codon. To obtain deletions in the ATM1 3′UTR, the 362-bp region that separates the stop codon from the AATAAA signal was shortened by either 265 or 97 nucleotides. Specific oligonucleotides used to generate these PCR fragments are shown in Table 1. The 3′UTR of the PGK1 gene was obtained by HindIII digestion of the YepJB1-21-1 plasmid (4). The GFP coding sequence was obtained from the pRH431 plasmid (A. Delahhode et al., unpublished data). DNAs from PCR products and the GFP were cloned in the high-copy-number pRS426 vector (11) containing the URA3 marker. Probes used for Northern blot analyses represented fragments within each coding region obtained by PCR using specific oligonucleotides (Table 2) and purified from agarose gels with the Qiaquick gel extraction kit (Qiagen). Labeling of DNA fragments was performed using the NonaPrimer kit from Appligen according to the manufacturer's instructions.
TABLE 1.
Oligonucleotides used in this study to generate GFP chimeric constructs
| Oligonucleotidea | Sequence
|
PCR product length (bp) | Cloning sites in pRS 426 | |
|---|---|---|---|---|
| 5′ primer (5′–3′) | 3′ primer (5′–3′) | |||
| 5′UTR + 3 aa of ATM1 | TGCTCTAGAGCATGGTAGAATCAAGAAATGGGAAGAAGA | CGCGGATCCGCGAAGCAGCATTTGCAGGTTGGAGTGGAA | 613 | XbaI-BanHI |
| 5′UTR + 53 aa of ATM1 | TGCTCTAGAGCATGGTAGAATCAAGAAATGGGAAGAAGA | CGCGGATCCGCGTTGATTCCATAAATTCACAGCTGAATT | 763 | XbaI-BanHI |
| 5′UTR + 16 aa of ATM1 | TGCTCTAGAGCATGGTAGAATCAAGAAATGGGAAGAAGA | CGCGGATCCGCGTGATCTTACAATTCGCCCAATTACAGG | 652 | XbaI-BanHI |
| Stop codon + 3′UTR of ATM1 | CCGCTCGAGCGGTGAACGCTCGTAAGTAAATATTGATTT | CGGGGTACCCCGAGTGGTTTGGTTTGCTAATACAATACC | 507 | XhoI-KpnI |
| 97-bp deletion in ATM1's 3′UTR | CCATCGATGGACCCGATGTATTGACTCTTCCTGACCGAA | CGGGGTACCCCGAGTGGTTTGGTTTGCTAATACAATACC | 410 | ClaI-KpnI |
| 238-bp deletion in ATM1's 3′UTR | CCATCGATGGCTTACTTACAATTTCAATAATAGGTTTGT | CGGGGTACCCCGAGTGGTTTGGTTTGCTAATACAATACC | 269 | ClaI-KpnI |
aa, amino acids.
TABLE 2.
Oligonucleotides used in this study to generate radiolabelled probes.
| Oligonucleotide | Sequence
|
PCR product length (bp) | |
|---|---|---|---|
| 5′ Primer | 3′ Primer | ||
| ATM1 | CAATGACACTATCTGGGAAAATGT | TCATAGTTCTTGCTGGTCTTTTAG | 507 |
| COX10 | AATCATGAATAAAGACATTGATGTTATGTA | GGAAGCTATTGGGGCGAGGGTTTTTGAATT | 2,000 |
| TIM44 | TGAACAGACCTACTTTGGTAATTTTATATG | AAGAAGTATTAGTTATAGTACTATTAGTAC | 2,030 |
| ATP2 | ATGGTTTTGCCAAGACTATATACTGCT | CTAGTTGGCTTCACGGGCTTTGAAAAGTCG | 1,536 |
| COT1 | GTGTTCTTCGGGATCGAGATAACT | ATTTCCGCCGCTGGCAATCGTATG | 606 |
| ABF2 | ATGAACAGTTACAGCCTATTAACT | CTAGTTGAGAGGGTAGCGAGCATT | 552 |
| TOM20 | ATGCTTCTATATTCAATGGATTTCCTGTCT | TTCGTTAATGGTCGTCCACGTGTGTATACT | 1,025 |
| SMF2 | ATGACGTCCCAAGAATATGAACCT | CTTGGAAAGAGTACTACAGAGTAA | 1,080 |
| COX6 | ATGTTATCAAGGGCCATATTCAGAAAT | TTAGTTCTGAGGTGGATGAATATCCCA | 638 |
| COX5a | ATGTTACGTAACACTTTTACTAGA | GATTGGACCTGAGAATAACCACCC | 431 |
| AAC1 | ATGTCTCACACAGAAACACAGACT | TTGCAACTGATCGTACAATGAGAT | 900 |
| MIR1 | GTCTGCTGCTCCTGCTATTCCACA | CCTCCGACGAGTCCATTAGCGAAC | 458 |
| COX3 | ATGACACATTTAGAAAGAAGTAGACATCAA | TTAAACACCTCATCAATAAAATAGTACGTA | 810 |
| ACT1 | GAGGTTGCTGCTTTGGTTATTG | GTGGTGAACGATAGATGGACC | 500 |
Purification of free and mitochondrion-bound polysomes.
Purification of mitochondrion-bound polysomes and free cytoplasmic polysomes was performed as described elsewhere, with few modifications (2, 57). To convert cells to spheroplasts, a 60-min treatment with Quantazyme (Quantum; Appligen) at 30°C with shaking was performed; 1,000 U per g (wet weight) of cells was used. To produce EDTA-washed mitochondria, spheroplast preparations were homogenized in breaking buffer containing 0.6 M mannitol, 30 mM Tris-HCl (pH 7.4), 10 mM EDTA, 5 mM 2-mercaptoethanol, 200 μg of cycloheximide per ml, and 500 μg of heparin per ml. EDTA-free buffers were used for all the other steps. To obtain mitochondria coated with polysomes, the breaking buffer contained 5 mM MgCl2 and 100 mM KCl instead of 10 mM EDTA.
Polysomes bound to mitochondria were released by incubation in polysome buffer (30 mM Tris-HCl [pH 7.4], 10 mM EDTA, 5 mM 2-mercaptoethanol, 200 μg of cycloheximide per ml, and 500 μg of heparin per ml) with 1.5% of Triton X-100 for 20 min on ice. After that, a centrifugation at 15,000 rpm for 20 min was performed; the supernatant contained released polysomes. To analyze polysome profiles, 50 A260 units of polysomes released from mitochondria were layered over an 11.4-ml linear sucrose gradient (10 to 50% sucrose in 200 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 2.1 mM magnesium acetate, and 100 mM KCl). The gradients were centrifuged in a Kontron TST41 Ti rotor at 40,000 rpm for 3 h and scanned at 260 nm with a Pharmacia spectrophotometer connected to a turbulence-free flow cell.
RNA extraction and Northern blot analyses.
Total RNA extraction from mitochondrion-bound polysomes, from free cytoplasmic polysomes, and from whole cells was performed by a hot-phenol method (51). For Northern blots, 10 μg (subcellular polysome fractions) or 30 μg (whole cells) of RNA was separated by electrophoresis through denaturing formaldehyde-agarose gels after the transfer nylon membranes were stained with methylene blue solution (48). Hybridizations and washings were performed as described elsewhere (48). The PhosphorImager system and TINA software were used to compare the relative abundance of each individual mRNA specie.
Direct visualization of RNA in live cells.
The coat protein (CP) of bacteriophage MS2 was fused to the GFP and expressed from the plasmid pCP-GFP, generously provided by D. L. Beach and K. Blomm (7). The pCP-GFP is a low-copy HIS3 selectable plasmid that produced CP-GFP regulated by the MET25 promoter. Cells grown in the presence of methionine produced no detectable CP-GFP protein product, as determined from the fluorescence signal intensity of imaged cells (data not shown). To induce CP-GFP production, methionine starvation was performed by switching the cells to a synthetic medium with 2% glucose or 2% galactose for 2 h.
To obtain reporter RNA, we used the pIIIA/MS2-2 plasmid (7), which contains two tandem copies of the CP-binding site. This plasmid can express a transcript tagged by the two tandem copies of the CP-binding site using the RNA seP promoter; this constitutive RNA polymerase III promoter maintains RNA levels throughout the cell cycle. The single SmaI site was used to introduce the following nucleotide sequences: the complete 3′UTR of ATM1, the complete 3′UTR of PGK1, and the sequence encoding the first 53 amino acids of Atm1p. In all the cases the binding site precedes the sequence examined. For each inserted sequence, we studied both possible orientations. The cells expressing the constructions where the inserted sequence was placed in the opposite orientation, such that the noncoding sequence was transcribed, were examined by fluorescence microscopy. These cells, like those expressing the empty pIIIA/MS2-2 plasmid, showed a low amount of fluorescence that was distributed throughout the cytoplasm and was excluded only from the vacuole (data not shown), as previously shown (7).
RESULTS
Asymmetric distribution of mRNAs coding for imported mitochondrial proteins between free and mitochondrion-bound polysomes.
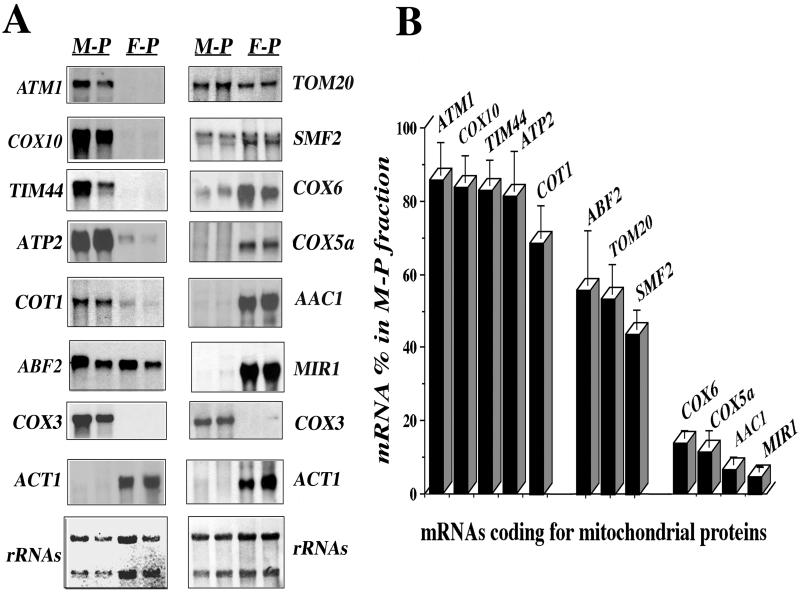
We purified mitochondria with cytoplasmic polysomes bound to the outer membrane from growing yeast spheroplasts in the presence of Mg2+ and cycloheximide containing buffers, as previously described (2, 57). RNA was extracted from free and mitochondrion-bound polysomes and submitted to Northern blot analysis. We used two genes as typical markers of either mitochondrial or cytoplasmic fractions: COX3, which is encoded by the mitochondrial DNA, and ACT1, whose mRNA is exclusively found in cytoplasmic fractions (Fig. 1A). Mitochondrial preparations appeared to be devoid of other cellular fractions. Only probes related to rough endoplasmic reticulum proteins, such as SSH1, revealed the presence of irrelevant mRNA species (data not shown). This is in agreement with recent observations, showing a tight association between both organelle membranes (1). Twelve nuclear genes, covering an array of functions and localizations within the organelle, were chosen for mRNA localization analysis. These experiments consistently showed that the transcripts examined are asymmetrically distributed in the two subcellular polysome fractions examined (Fig. 1). Three families of mRNAs according to their localization can be defined.
FIG. 1.
Asymmetric distribution of mRNAs coding for mitochondrial imported proteins among free and mitochondrion-bound polysomes. (A) CW04 cells were grown aerobically (rich galactose medium) and harvested in early log phase; free polysomes (F-P) and mitochondrion-bound polysomes (M-P) were prepared at pH 7.4 as described in Materials and Methods. Their RNA was extracted and subjected to Northern blot analysis, using probes for different genes encoding mitochondrial proteins. Cross-contamination of the fractions was checked with the ACT1 gene as a cytoplasmic protein control and the COX3 gene as a mitochondrial protein control. Results obtained for two individual RNA preparations are shown; at the bottom, methylene blue staining of the filters is shown. The approximate sizes measured for individual mRNAs were as follows: ATM1, 2.2 kb; COX10, 1.7 kb; TIM44, 1.5 kb; ATP2, 1.6 kb; COT1, 1.5 kb; ABF2, 0.6 kb; COX3, 3.7 kb; ACT1, 1.7 kb; TOM20, 0.8 kb; SMF2, 1.9 kb; COX6, 0.6 kb; COX5a, 0.6 kb; AAC1, 1.1 kb; and MIR1, 1.2 kb. (B) Densitometric analyses of the results obtained with 12 independent polysome preparations were performed. A signal obtained for an individual transcript in the RNA preparation from mitochondrion-bound polysomes was normalized with the COX3 mRNA signal. The normalization for the signal in the free-polysome fraction was performed with the ACT1 mRNA signal. For a given mRNA, addition of specific signals measured in mitochondrion-bound polysomes and in free polysomes after normalization was considered as 100%. The percentage of mRNA signal found in mitochondrion-bound polysomes is shown for the 12 genes examined.
The first family includes mRNAs exclusively localized to polysomes bound to mitochondria (Fig. 1A, lanes M-P), such as ATM1, COX10, TIM44, ATP2, and COT1 mRNAs. Consistently, between 70 and 85% of the total signal measured for these five transcripts was found in polysomes bound to mitochondria (Fig. 1B). Surprisingly, Cot1p, when overproduced, was found to be localized to the vacuole (37). We found that approximately 70% of its mRNA was present in mitochondrion-bound polysomes, suggesting that Cot1p might also be addressed to mitochondria, a result in agreement with previous studies (13).
The second family includes mRNAs equally divided between free and mitochondrion-bound polysomes, such as ABF2, TOM20, and SMF2 (Fig. 1A). For these transcripts, between 58 and 42% of the overall signal was consistently measured in mitochondrion-bound polysomes (Fig. 1B).
The third family mRNAs were those predominantly associated with free cytoplasmic polysomes (Fig. 1A, lanes F-P), such as COX6, COX5a, AAC1, and MIR1. Remarkably, <15% of the overall signal for these mRNAs was found in polysomes bound to mitochondria (Fig. 1B).
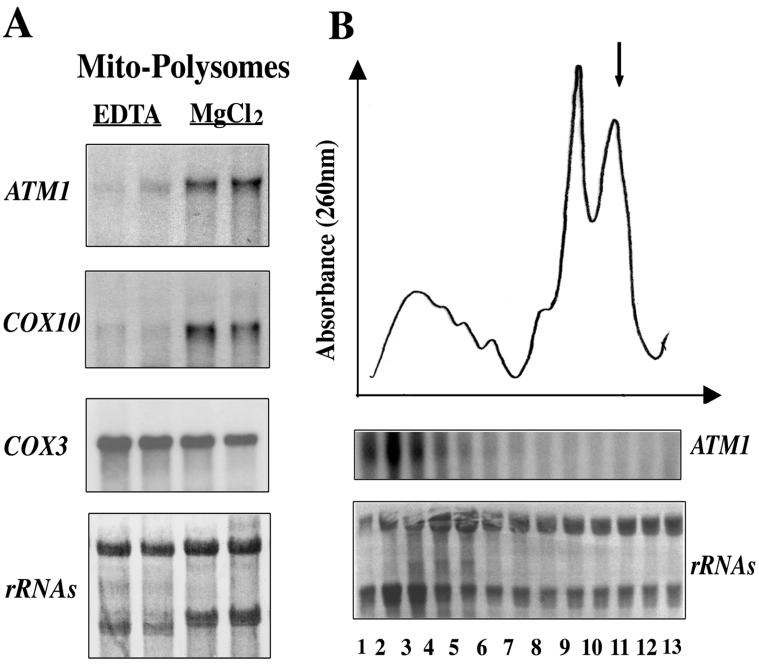
As expected, EDTA treatment of mitochondrion-bound polysome fractions (15, 46) led to the complete release of mitochondrion-bound mRNAs, such as ATM1 or COX10 (Fig. 2A). Polysomes bound to mitochondria were separated into size classes by sucrose gradient centrifugation, and each fraction was analyzed by Northern blotting. The ATM1 mRNA signal was enriched in fractions 1 to 4, corresponding to polysomes with four or more ribosomes. In contrast, a very weak signal was detected in the 80S monosome fraction (Fig. 2B).
FIG. 2.
The ATM1 transcript is specifically associated with mitochondrion-bound polysomes. (A) Northern blots were performed with RNA prepared from EDTA-treated mitochondria in MgCl2-free buffers (EDTA) or from untreated mitochondria in buffers containing 5 mM MgCl2 and 100 mM KCl (MgCl2). In the presence of EDTA, polysomes attached to mitochondria were washed off. This finding is visible after staining of the filter with methylene blue (shown at the bottom): the 18S rRNA band is strongly diminished, and the 16S rRNA band representing the mitochondrial specie is clearly distinguished. In these preparations, approximately 90% of the ATM1 and COX10 mRNA signal disappeared. (B) Fifty A260 units of cytoplasmic polysomes released from mitochondria, after Triton X-100 treatment, were sedimented in a linear sucrose gradient. The gradients were scanned at 260 nm, and fraction 1 is the bottom of the gradient. An arrow (upper panel) indicates the position of the 80S monosomes. Thirteen fractions were collected from each gradient to prepare RNA and then subjected to Northern blot analysis using ATM1 radiolabeled DNA (lower panel). The patterns obtained for polysomes released from mitochondria are very similar to the ones described by Suissa and Schatz (57). Furthermore, the profile of ATM1 mRNA is consistent with its almost exclusive association with polysomes containing four or more ribosomes. Very low levels of ATM1 signal were found in the 80S monosome fraction. Methylene blue staining of the filter is shown at the bottom.
These data provide strong evidence that the 12 mRNA species tested are partitioned quite differently between free and mitochondrion-bound polysomes. Among them, at least the ATM1 mRNA is a genuine, efficiently translated component of the mitochondrion-bound polysome fraction.
Characterization of ATM1 sequences required for mRNA localization.
Atm1p is an ABC transporter of the mitochondrial inner membrane, is involved in mitochondrial iron metabolism, and is required for the generation of cytosolic Fe/S proteins (33, 34, 36). Atm1p is a highly hydrophobic protein that contains six putative transmembrane fragments. ATM1 mRNA localizes exclusively to mitochondrion-bound polysomes (Fig. 1 and 2). To characterize the sequences responsible for the asymmetric mRNA localization, we constructed chimeras in which the GFP was expressed under the control of the ATM1 promoter. Two regions within ATM1 were examined: the mitochondrial targeting sequence (mts) coding region, corresponding to the first 53 amino acids of the precursor protein (36), and the 3′UTR of 507 bp encompassing the region between the Atm1p stop codon and the first 14 amino acids of the adjacent gene, ADE4.
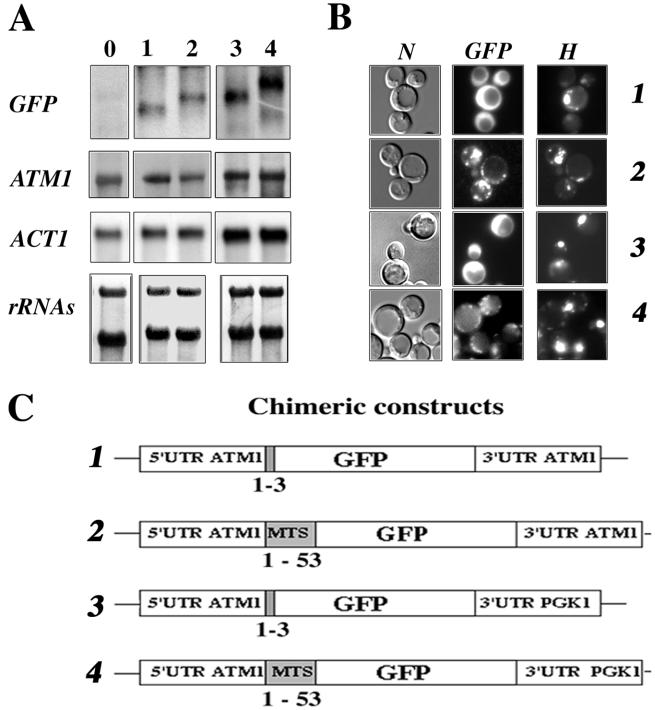
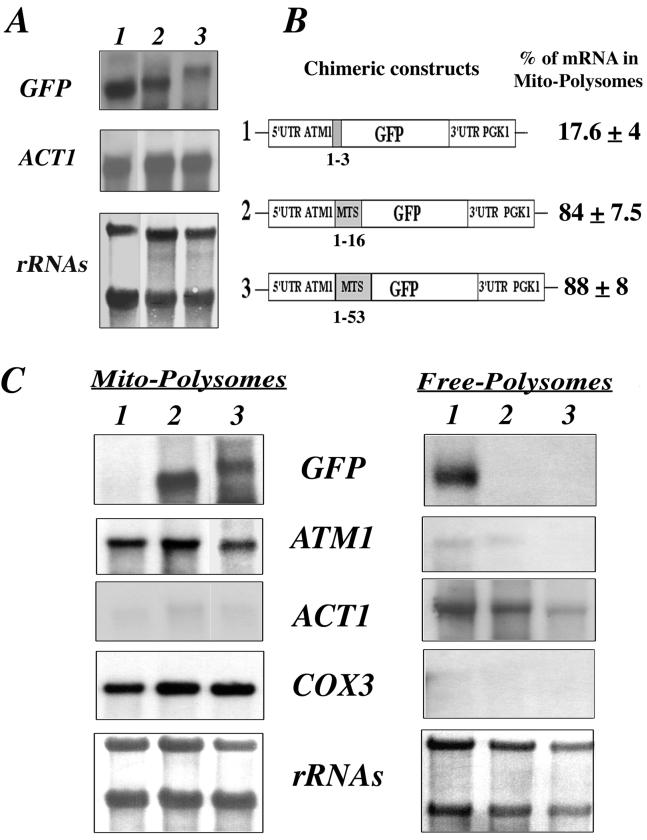
Four plasmids were obtained (Fig. 3C): in plasmids 2 and 4, the GFP was fused in frame with the complete mts, while in plasmids 1 and 3 only the first three amino acids were fused to the GFP (Fig. 3C). Additionally, in plasmids 3 and 4, the 3′UTR of ATM1 was replaced by 400 bp corresponding to the 3′UTR of PGK1, a gene coding for a cytoplasmic protein and usually used as a marker of soluble cytosolic fractions (52) (Fig. 3C).
FIG. 3.
Expression and localization of GFP-Atm1p chimeras. (A) A portion (30 μg) of RNA prepared from whole cells was separated on formaldehyde-agarose gels and subjected to Northern blot analysis using specific radiolabeled probes. Lane 0 represents CW04 wild-type cells devoid of recombinant plasmids, and lanes 1 to 4 represent CW04 cells expressing plasmids 1 to 4, respectively, shown in panel C. The approximate sizes of each individual GFP hybrid mRNA were as follows: plasmid 1, 0.75 kb; plasmid 2, 0.9 kb; plasmid 3, 0.9 kb; and plasmid 4, 1.1 kb. (B) Direct fluorescence microscopy of CW04 cells carrying plasmids expressing the GFP chimeras. Panel N represents the cells viewed by Nomarski optics, panel GFP represents the GFP signal, and panel H shows cells stained with the DNA dye reagent Hoechst. Cells carrying plasmids 1 and 3 had homogeneous cytosolic staining, in contrast to cells carrying plasmids 2 and 4, which gave a cytoplasmic GFP signal localized to discrete spots, which were also stained by the DNA reagent Hoechst. (C) Representation of the four chimeric constructs tested. GFP protein is expressed under the control of ATM1 promoter. All of the plasmids shared the complete 5′UTR of ATM1, and translation for all them was initiated at the authentic Atm1p AUG. For plasmids 1 and 3, the GFP AUG is in the fourth position of the chimeras. In contrast, fusion proteins translated from plasmids 2 and 4 possess the entire Atm1p 53-amino-acid presequence in frame with the GFP AUG codon. In plasmids 3 and 4, the 3′UTR of ATM1 was replaced with the 3′UTR of PGK1, a gene coding for a cytoplasmic protein and usually used as a marker of soluble cytosolic fractions (52).
Total RNAs were purified from cells expressing GFP chimeras or wild-type cells and analyzed by Northern blot analysis (Fig. 3A). No signal was detected with the GFP probe in wild-type cells (Fig. 3A, lane 0). In cells expressing chimeras 1 and 2, two mRNAs of 0.75 and 0.9 kb, respectively, were revealed (Fig. 3A, lanes 1 and 2). In cells expressing chimeras 3 and 4, we detected two slightly longer mRNAs of 0.9 and 1.1 kb, respectively; the difference observed was probably related to the presence in these plasmids of the PGK1 3′UTR (Fig. 3A, lanes 3 and 4). It appears clearly from these results that the steady-state levels of hybrids mRNAs tested are similar (Fig. 3A).
Cells were visualized by fluorescence microscopy in order to determine the subcellular localization of GFP chimeras (Fig. 3B). When the GFP protein was fused with the first three amino acids of Atm1p, we consistently observed a homogeneous cytosolic staining, suggesting that the protein was localized to the cytoplasm (Fig. 3B, GFPs 1 and 3). In contrast, when the GFP was fused to the functional mts of Atm1p, the chimera localized to discrete spots in the cytoplasm, a result reminiscent of the mitochondrial distribution (41) (Fig. 3B, GFP 2 and 4). These discrete fluorescent spots were also stained with the specific DNA labeling reagent, Hoechst, confirming that they contain DNA (Fig. 3B, H 2 and 4). Thus, GFP fused to the first 53 amino acids of Atm1p is addressed to mitochondria.
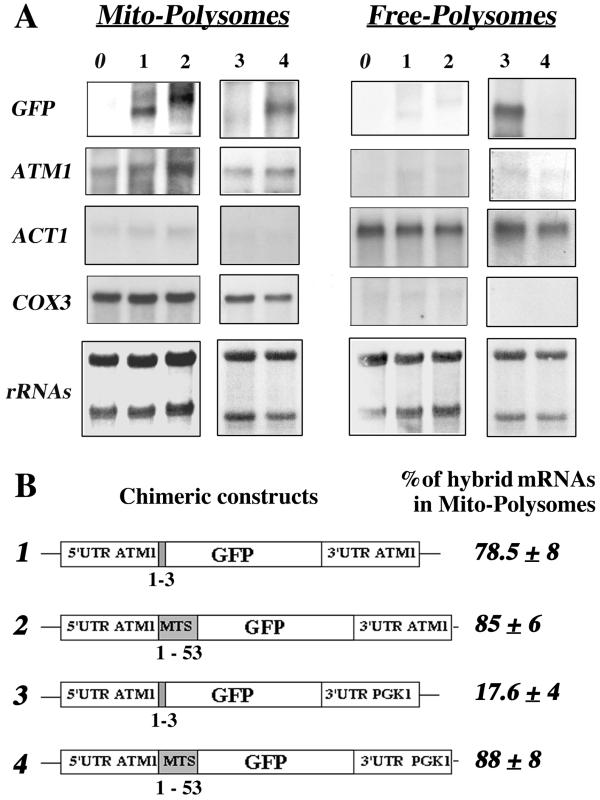
Fractionation experiments were performed to obtain free and mitochondrion-bound polysomes from wild-type cells and cells expressing GFP chimeras (Fig. 4B). RNAs were purified and subjected to Northern blot analysis. As shown in Fig. 1, very little cross-contamination of the subcellular fractions was measured by hybridization with ACT1 and COX3 probes (Fig. 4A). In addition, no signal was detected with the GFP probe in cells devoid of GFP plasmids (Fig. 4A, lane 0). When the chimeras containing ATM1 3′UTR were expressed, we detected, as expected, two transcripts of 0.75 and 0.9 kb. These transcripts were remarkably enriched in mitochondrion-bound polysomes. Very few of the messengers were detected in free cytoplasmic polysomes (Fig. 4A, lanes 1 and 2). Based on six independent experiments, we deduced that in the presence of the 3′UTR ATM1 approximately 80% of hybrid mRNAs was localized to the mitochondrial vicinity, independently of the mts coding sequence (Fig. 4B). This distribution is very similar to the one detected in the same cells for the endogenous ATM1 mRNA, where approximately 85% of the ATM1 mRNA signal was found in mitochondrion-bound polysomes (Fig. 4A). When the 3′UTR of ATM1 was replaced by the 3′UTR of PGK1, the mRNA distribution dramatically changed. In the absence of the mts coding sequence, the 0.9-kb transcript was almost exclusively localized to free cytoplasmic polysomes (Fig. 4A, lane 3); approximately 82% of the overall signal was found in this fraction (Fig. 4B). In contrast, 88% of the 1.1-kb hybrid mRNA containing the mts but not the 3′UTR ATM1 localized predominantly to mitochondrion-bound polysomes, as did ATM1 mRNA (Fig. 4A, lane 4, and Fig. 4B).
FIG. 4.
ATM1 sequences required for the asymmetric subcellular distribution of GFP hybrid mRNAs. (A) Wild-type CW04 cells (lane 0) or cells carrying the four plasmids (lanes 1 to 4) described in Fig. 3 C were grown until early log phase and harvested in order to purify free and mitochondrion-bound polysomes (see Materials and Methods). Then, 10 μg of RNA extracted from each polysomal fraction was separated on formaldehyde-agarose gels, subjected to Northern blot analysis, and hybridized successively with GFP, ATM1, ACT1, and COX3 probes. Methylene blue staining of the filters is shown at the bottom. Results obtained for RNA prepared from mitochondrion-bound polysomes (Mito-Polysomes) are shown in the left panel, and those obtained from free cytoplasmic polysomes (Free-Polysomes) are shown in the right panel. The autoradiograms represent exposure times of between 2 and 4 h at −80°C, with Amersham intensifying screens, for all the probes except ATM1, which required an exposure time of approximately 16 h. (B) Chimeric constructs are represented, as well as the percentage of the hybrid mRNA signal measured in polysomes bound to mitochondria, given as a mean of six independent experiments. In all cases, the presence of the ATM1 mts or its 3′UTR allowed the GFP hybrid mRNA to behave, with respect to its final subcellular localization, as the endogenous ATM1 mRNA.
Thus, at least two regions in the ATM1 gene can act independently to address the reporter mRNA to the mitochondrial vicinity: the mts coding sequence and the 3′UTR.
Only 48 bp of the Atm1p's mts coding sequence are sufficient to guide the reporter mRNA to the vicinity of mitochondria.
To better define the region within the mts coding sequence involved in the targeting of the reporter mRNA to the mitochondrial vicinity, plasmids were obtained in which 3, 16, or 53 amino acids of the Atm1p N-terminal region were fused to the GFP (Fig. 5B). Cells expressing the different chimeras under the transcriptional control of the ATM1 promoter and possessing the PGK1 3′UTR were examined. Transcripts synthesized from these plasmids were expressed at similar levels, and the sizes visualized for each individual mRNA were in agreement with theoretical calculations (Fig. 5A).
FIG. 5.
Minimal region within the mts of Atm1p required for the correct localization of GFP hybrid mRNAs. (A) Three plasmids in which the GFP was fused in frame either with the complete mts of Atm1p or with shortened mts versions were obtained. RNA extracted from whole cells was checked by Northern blot analysis. The steady-state levels of each hybrid mRNA were very similar, and the sizes measured were as expected. The approximate size for each individual mRNA was as follows: plasmid 1, 0.9 kb; plasmid 2, 1 kb; and plasmid 3, 1.1 kb. (B) Chimeric constructs examined are represented. In plasmid 1 the GFP was fused in frame to the first three amino acids of Atm1p, in plasmid 2 the first 16 amino acids of Atm1p were fused to the GFP, and in plasmid 3 the complete mts was present. In all of the plasmids, the stop codon was associated with the 3′UTR of the PGK1 gene. Calculations of the relative abundance of each mRNA in mitochondrion-bound polysomes, for six independent experiments, were obtained after the normalization of each signal with the internal markers COX3 or ACT1. (C) Northern blots performed with RNAs purified from mitochondrion-bound polysomes (Mito-Polysomes) and free cytoplasmic polysomes (Free-Polysomes) are shown. Methylene blue staining of the filters is shown at the bottom. The autoradiograms represent exposures times of between 2 and 4 h at −80°C, with Amersham intensifying screens, for all of the probes except ATM1, which required an exposure time of approximately 16 h.
RNA for mitochondrion-bound polysomes and free cytoplasmic polysomes were obtained and subjected to Northern blot analysis (Fig. 5C). Deletion of the region coding for amino acids 17 to 53 of Atm1p had no effect on the localization of the hybrid mRNA. This mRNA behaves as the mRNA containing the full-length presequence and as endogenous ATM1 mRNA; 84% of the overall mRNA signal was found in mitochondrion-bound polysomes (Fig. 5B and C, lanes 2 and 3). As shown in Fig. 4, we confirmed that the mitochondrial localization of the reporter mRNA was completely abolished when the sequence coding for the first three amino acids of Atm1p were fused to the GFP in the absence of the ATM1 3′UTR (Fig. 5C, lane 1). Hence, one of the necessary and sufficient addressing components of the ATM1 mRNA encompasses the sequence coding for amino acids 1 to 16 of the precursor protein.
Effects of deletions in the ATM1 3′UTR on mRNA subcellular distribution.
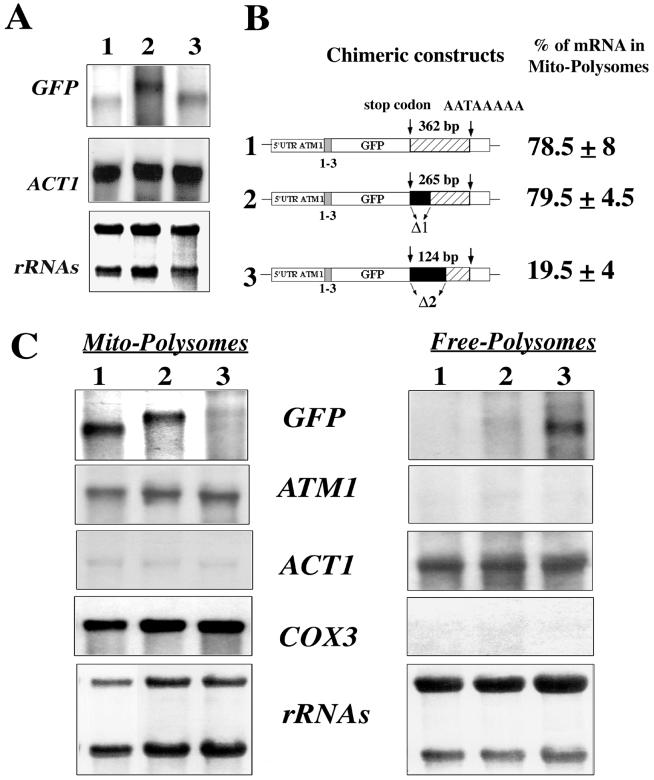
To better characterize the nucleotide sequence within the 3′UTR of ATM1 responsible for mRNA transport to mitochondrion-bound polysomes, the regions between the stop codon and the canonical polyadenylation signal sequence AATAAA (28) were shortened by 97 and 238 bp, respectively (Fig. 6B). Total RNA was prepared from cells expressing either of these plasmids and submitted to Northern blot analysis. To our surprise, mRNAs transcribed from plasmids bearing deletions in their 3′UTR are slightly longer than those from the original plasmid; the cleavage at the polyadenylation site may be modified in these constructs. However, the steady-state levels of each hybrid mRNA were quite similar in all the tested cells (Fig. 6A). To examine the distribution of transcripts produced from the new set of chimeric constructs, RNA was extracted from mitochondrion-bound polysomes and free cytoplasmic polysomes and submitted to Northern blot analysis (Fig. 6C). The hybrid mRNA, bearing the 238-bp deletion, was almost exclusively found in free cytoplasmic polysomes; only 19.5% of the overall signal was measured in mitochondrion-bound polysomes (Fig. 6B and C, lane 3). In contrast, the 97-bp deletion had little effect on mRNA targeting; the transcript synthesized from the corresponding plasmid behaves roughly like ATM1 mRNA. Consistently, 80% of the hybrid mRNA localized to mitochondrion-bound polysomes (Fig. 6B and C, lane 2). Therefore, the region between the stop codon and the AATAAA signal plays a critical role in the correct cellular localization of the reporter mRNA.
FIG. 6.
Minimal region within the 3′UTR of ATM1 required for the correct localization of GFP hybrid mRNAs. (A) Northern blots were performed using RNA extracted from whole cells carrying plasmids in which the regions between the stop codon and the canonical polyadenylation signal sequence AATAAA was shortened by 238 and 97 nucleotides, respectively. The steady-state levels of these individual mRNAs were quite similar in all of the cells tested. The approximate sizes for each hybrid mRNA were as follows: plasmid 1, 0.7 kb; plasmid 2, 1 kb; and plasmid 3, 0.8 kb. (B) Two plasmids were constructed in which deletions of the ATM1 3′UTR were performed. In plasmid 1 the complete 507 bp region of the 3′UTR was associated with the stop codon; the stop codon and the canonical polyadenylation signal sequence AATAAA are separated in this plasmid by 362 nucleotides. In plasmids 2 and 3, 97 and 238 nucleotides, respectively, shortened the region between the stop codon and the AATAAA signal. In all three chimeric constructs, the GFP is associated in frame with the first three amino acids of Atm1p. Calculations of the relative abundance of each transcript in mitochondrion-bound polysomes, for six independent experiments, were obtained after the normalization of each signal with the internal markers COX3 or ACT1. (C) Northern blots performed with RNAs purified from mitochondrion-bound polysomes (Mito-Polysomes) and free cytoplasmic polysomes (Free-Polysomes). Methylene blue staining of the filters is shown at the bottom. The autoradiograms represent exposures times of between 2 and 4 h at −80°C, with Amersham intensifying screens, for all of the probes except ATM1, which required an exposure time of approximately 16 h.
Imaging gRNAATM1 localization in live cells.
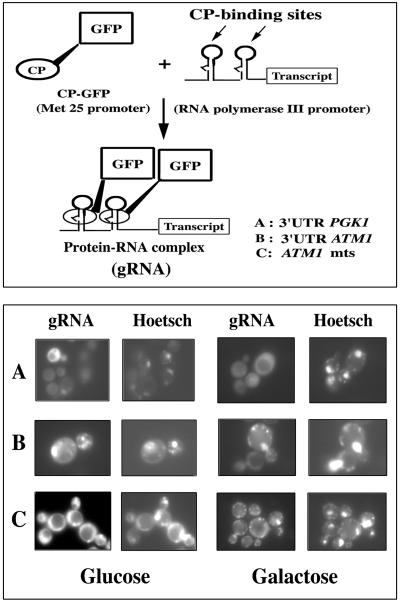
The RNA-labeling system recently described (7) to visualize native RNA movement in living cells was used to confirm the mRNA addressing information included in the mts coding sequence and the 3′UTR of ATM1. A regulated cytoplasmic coat protein (CP)-GFP was used to visualize the transcript without high levels of background fluorescence. Additionally, the plasmid contains only two CP-binding sites, which was sufficient to visualize the mRNA (Fig. 7, upper panel). Three sequences were studied as possible regulators of the RNA subcellular distribution: (i) the 3′UTR of the PGK1 gene, which encodes a cytoplasmic protein and which directs the localization of hybrid GFP mRNAs to free cytoplasmic polysomes (Fig. 4 and 5); (ii) the 507 bp of the complete ATM1 3′UTR, which was able to address the hybrid GFP mRNA to mitochondrion-bound polysomes (Fig. 4 and 6); and (iii) the nucleotide sequence corresponding to the first 53 amino acids of Atm1p, which we know to be sufficient to allow the reporter mRNA localization to the vicinity of mitochondria (Fig. 4 and 5).
FIG. 7.
gRNAATM1 transcripts localized to the mitochondrial vicinity in living yeast cells. (Upper panel) The RNA-labeling system contains two components: the RNA binding MS2 CP fused to the GFP (CP-GFP) and an RNA transcript containing two CP-binding sites (7). The CP-binding sites were fused to the complete 3′UTR of PGK1 (A), the complete 3′UTR of ATM1 (B), and the nucleotide sequence coding for the first 53 amino acids of Atm1p (C). When coexpressed in the cell, the two components interact to form a GFP-labeled RNA (gRNA) which can be visualized using common fluorescence microscopy techniques. (Lower panel) We generated gRNAATM1 by placing either the entire 3′UTR of ATM1 (B) or the sequence corresponding to the Atm1p's mts (C) downstream of the CP-binding sites. Coexpression of CP-GFP and ATM1 reporter RNAs produces spots of fluorescence, which colocalize with the Hoechst cytoplasmic labeling. The discrete GFP fluorescent spots distinguished are very similar to the chondriolites, previously described as representing mitochondrial DNA (41, 55, 60). We were able to visualize arrangement changes of chondriolites by using Hoechst staining in cells that had grown in 2% galactose (galactose) instead of 2% glucose (glucose); gRNAATM1 remarkably followed the same cytoplasmic distribution as mitochondrial DNA. In contrast, the gRNA containing the PGK1 3′UTR consistently showed a homogeneous staining all over the cytoplasm (A).
Yeast cells were transformed with both plasmids directing the production of the CP-GFP fusion protein and the reporter RNA transcripts (gRNA), respectively. Two conditions of cell growth were used to examine the gRNA subcellular distribution. Cells were grown either in 2% glucose or 2% galactose. In yeast, the effects of growth conditions on mitochondrial morphology have been well documented (41, 55, 60). Yeast cells expressing both CP-GFP chimera and the ATM1 reporter RNAs, called gRNAATM1 (Fig. 7, lower panel, rows B and C) contained small discrete fluorescence spots, which remarkably colocalized with Hoechst cytoplasmic staining. These discrete speckles are reminiscent of mitochondrial nucleoids, termed chondriolites, described as mitochondrial DNA visualized by the DAPI (4′,6′-diamidino-2-phenylindole) or DASPMI staining techniques (41, 55, 60). Cells expressing the gRNA transcript containing the 3′UTR of PGK1 consistently presented no organized subcellular distribution, with a general staining in the cytoplasm (Fig. 7, lower panel, row A). Comparison of the fluorescence labeling in cells grown in glucose to cells grown in galactose showed significant differences in the punctuate distribution of GFP spots, which are consistently localized to cytoplasmic regions also stained by the Hoechst reagent. Hence, gRNAATM1 specifically localizes to the vicinity of mitochondria in living cells.
DISCUSSION
Asymmetric cell division contributes to the generation of different cell types during the development of multicellular organisms. In Drosophila and Xenopus, mRNAs rather than proteins are often asymmetrically distributed (35, 42). Segregating mRNAs instead of proteins has the advantage of localizing protein synthesis to the site of the protein's action (6, 27, 32), which could be an advantage for hydrophobic proteins. In yeast, regulation of mating-type switching requires the concentration of Ash1p within the daughter nucleus (9, 54). Recent work demonstrated that the asymmetric distribution of Ash1p to daughter nuclei requires localization of its mRNA at the bud tip (38, 58).
While analyzing the mRNA composition of mitochondrion-bound polysomes in yeast, three classes of mRNAs coding for mitochondrially destined proteins were distinguished: (i) those like ATM1 and COX10, which are exclusively associated to mitochondria; (ii) those which are equally distributed between mitochondrial and nonmitochondrial fractions (TOM20 and ABF2); and (iii) those that are weakly found in the mitochondrial fraction (COX6, AAC1, and MIR1).
Unexpectedly, this distribution does not overlap the hydrophobic properties of the corresponding proteins; indeed, they are found in both the first and third categories. However, it is worth noting that the hydrophobic proteins of the first group, Atm1p and Cox10p, have an N-terminal targeting sequence, whereas the two hydrophobic proteins of the third class, Aac1p and Mir1p, are carriers possessing internal targeting signals. This prompted us to examine the mRNA addressing information contained in the N-terminal targeting sequence, taking ATM1 as a model system. Atm1p is an ABC transporter of the mitochondrial inner membrane, involved in mitochondrial iron metabolism and required for generation of cytosolic Fe/S proteins (33, 34, 36). Atm1p is a highly hydrophobic protein that, when overproduced, is poorly translocated inside the organelle (14). Furthermore, we demonstrated here that ATM1 mRNA localizes exclusively to mitochondrion-bound polysomes. Chimeric plasmids were produced using the GFP expressed under the control of the ATM1 promoter. The sequence coding for the mts of Atm1p, which is 53 amino acids long (36), was tested for its ability to address GFP mRNA to the mitochondrial vicinity. The reporter mRNA possessing this sequence is exclusively localized to mitochondrion-bound polysomes, just like the endogenous ATM1 mRNA. Therefore, the Atm1p mts coding sequence is able to deliver ATM1 mRNA to its final destination at the mitochondrial vicinity. More precisely, we were able to show that the cis-acting information that guides the mRNA to the mitochondrial vicinity is included in the nucleotide region coding for the first 16 amino acids of Atm1p. Several examples in mammalian cells demonstrated unambiguously that translation is tightly coupled with mitochondrial import. This is the case for aldehyde dehydrogenase and adenylate kinase enzymes, both of which use a cotranslational mitochondrial import pathway (44, 45). Our data suggest that the ATM1 mts coding region could play a role in connecting the mRNA specific localization and the translation processes. However, using a novel approach to visualize RNA movement in real time in living cells (7, 8), we demonstrated that the mRNA targeting process does not require the translation of the mts coding region. Indeed, the green RNA, which colocalizes with mitochondria (Fig. 7), is unlikely to be translated. These experiments confirm the biochemical studies of mitochondrion-bound polysomes and strengthen the idea that, under physiological conditions, the ATM1 mRNA when exported from the nucleus is quickly sorted to mitochondria.
Moreover, the process of specific intracellular compartment mRNA localization is often mediated by cis-acting elements within the 3′UTR of the transcripts (27, 56). In yeast, ASH1 mRNA is localized to the distal tip of daughter buds in postanaphase cells. To achieve this localization, the transcript must have its 3′UTR and the actin cytoskeleton must be intact (38, 58). In our study, the entire 3′UTR region of ATM1 was able to address a GFP mRNA to mitochondrion-bound polysomes in the absence of the Atm1p mts coding sequence. Interestingly, the region between the stop codon and the polyadenylation signal is important in this process. Indeed, a deletion of 238 nucleotides within the region spanning the codon stop and the AATAAA signal, normally separated by 362 bp, abolishes the localization of the reporter mRNA to mitochondrion-bound polysomes. Besides, our observation that green RNA containing the ATM1 3′UTR exclusively localized to the vicinity of mitochondria in living cells (Fig. 7) is in agreement with our biochemical studies. Since this gRNA is not translated, it appears clearly that the sorting process is not the consequence of the messenger tethering by the translation machinery located in the vicinity of the mitochondria. The secondary structure for the 3′UTR of ATM1 and other nuclearly encoded mitochondrial genes predicted several long and stable double-stranded regions. Additionally, for all of them the AU content is approximately 70%, a finding which is reminiscent of the highly conserved 3′UTR of nucleus-encoded mitochondrial proteins of rat liver (28). It has been shown that the 3′UTR of the β1-F1 ATPase transcript is involved in controlling the translation, stability, and subcellular localization of the mRNA during rat liver development (18).
We demonstrated here that, for the ATM1 gene, either the mts coding sequence or the 3′UTR is independently sufficient to localize the mRNA to mitochondrion-bound polysomes. The redundancy of localizing elements has been observed in only three cases: ASH1 mRNA in yeast (10, 25), in the Drosophila bicoid mRNA (39), and in the Xenopus Vg1 mRNA (22). The coding sequence for the mts and the 3′UTR of the ATM1 mRNA could interact to achieve the efficient translation and mitochondrial translocation of the precursor. This is reminiscent of the β1-F1 ATPase translation process, which is mediated by the cross-talk between the 5′ and 3′ ends of its mRNA (26).
In conclusion, this study presents compelling evidence that, in yeast, translation of a subset of mitochondrion-destined precursors occurs subsequent to mRNA localization to the mitochondrial vicinity. A study using DNA micro-array is currently in progress to find consensus targeting motifs based on different mRNA species exclusively localizing to mitochondrion-bound polysomes. Finally, the GFP chimera coupled to gRNAATM1 reporters will provide a rapid and convenient way to characterize mutants affecting RNA localization (8, 38) and mitochondrial function. These two complementary approaches will help us to elucidate some of the molecular mechanisms involved in yeast mitochondrial biogenesis.
ACKNOWLEDGMENTS
We thank A. Delahodde, E. Carvajal, and M. G. Claros for useful discussions and comments on the manuscript. We thank D. Beach and K. Bloom for the kind gift of plasmids required for the RNA-labeling system. We are also grateful to D. Ojcius for proofreading the manuscript.
This study was supported by funds from the CNRS (UMR 8541), the Ecole Normale Supérieure, the AFM (grant 586073), and the French Association Against Cancer (ARC grant 5691).
REFERENCES
- 1.Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein S D, Perktold A, Zellnig G, Daum G. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–553. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 2.Ades I Z, Butow R A. The products of mitochondria-bound cytoplasmic polysomes in yeast. J Biol Chem. 1980;255:9918–9924. [PubMed] [Google Scholar]
- 3.Ades I Z, Butow R A. The transport of proteins into yeast mitochondria. J Biol Chem. 1980;255:9925–9935. [PubMed] [Google Scholar]
- 4.Banroques J, Delahodde A, Jacq C. A mitochondrial RNA maturase gene transferred to the yeast nucleus can control mitochondrial mRNA splicing. Cell. 1986;46:837–844. doi: 10.1016/0092-8674(86)90065-6. [DOI] [PubMed] [Google Scholar]
- 5.Barnes W M. PCR amplification of up to 35 kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassell G J, Oleynikov Y, Singer R H. The travels of mRNAs through all cells large and small. FASEB J. 1999;13:447–454. doi: 10.1096/fasebj.13.3.447. [DOI] [PubMed] [Google Scholar]
- 7.Beach D L, Salmon E D, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand E, Chartrand P, Schaefer M, Shenoy S M, Singer R H, Long R M. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 9.Bobola N, Jansen R-P, Shin T H, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 10.Chartrand P, Meng X-H, Singer R H, Long R M. Structural elements required for the localization of Ash1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 11.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 12.Conde J, Fink G. A mutant of S. cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conklin D S, McMaster J A, Culbertson M R, Kung C. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:3678–3688. doi: 10.1128/mcb.12.9.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corral-Debrinski M, Belgareh N, Blugeon C, Claros M G, Doye V, Jacq C. Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol Microbiol. 1999;31:1499–1511. doi: 10.1046/j.1365-2958.1999.01295.x. [DOI] [PubMed] [Google Scholar]
- 15.Crowley K S, Payne R M. Ribosome binding to mitochondria is regulated by GTP and the transient peptide. J Biol Chem. 1998;273:17278–17285. doi: 10.1074/jbc.273.27.17278. [DOI] [PubMed] [Google Scholar]
- 16.Doye V, Wepf R, Hurt E C. A novel nuclear pore protein Nup133p with distinct roles in poly(A+) RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dujardin G, Pajot P, Groudinsky O, Slonimski P. Long-range control circuits within mitochondria and between nucleus and mitochondria. Mol Gen Genet. 1980;179:469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- 18.Egea G, Izquierdo J M, Martin R C S, Cuezva J M. mRNA encoding the b-subunit of the mitochondrial F1-ATPase is a localized mRNA in rat hepatocytes. Biochem J. 1997;322:557–565. doi: 10.1042/bj3220557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiki M, Verner K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. J Biol Chem. 1993;268:1914–1920. [PubMed] [Google Scholar]
- 20.Fujiki M, Verner K. Coupling of protein synthesis and mitochondrial import in a homologous yeast in vitro system. J Biol Chem. 1991;266:6841–6847. [PubMed] [Google Scholar]
- 21.Fünfschilling U, Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol Biol Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautreau D, Cote C A, Mowry K L. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocyte. Development. 1997;124:5013–5020. doi: 10.1242/dev.124.24.5013. [DOI] [PubMed] [Google Scholar]
- 23.George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high-efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez I, Buonomo S B C, Nasmyth K, Ahsen U V. ASH1 mRNA localization in yeats involves multiple secondary elements and Ash1 protein translation. Curr Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- 26.Izquierdo J M, Cuezva J M. Control of the translational efficiency of b-F1-ATPase mRNA depends on the regulation of a protein that binds the 3′ untranslated region of the mRNA. Mol Cell Biol. 1997;17:5255–5268. doi: 10.1128/mcb.17.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston D S. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 28.Juretic N, Theus M. Analysis of the polyadenylation consensus sequence context in the genes of nuclear encoded mitochondrial proteins. FEBS Lett. 1991;290:4–8. doi: 10.1016/0014-5793(91)81212-q. [DOI] [PubMed] [Google Scholar]
- 29.Kellems R E, Allison V F, Butow R A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J Biol Chem. 1974;249:3297–3303. [PubMed] [Google Scholar]
- 30.Kellems R E, Allison V F, Butow R A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellems R E, Butow R A. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. III. Changes in the amount of bound ribosomes in response to changes in metabolic state. J Biol Chem. 1974;249:3304–3310. [PubMed] [Google Scholar]
- 32.King M L. Molecular basis for cytoplasmic localization. Dev Genet. 1996;19:183–189. doi: 10.1002/(SICI)1520-6408(1996)19:3<183::AID-DVG1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Kispal G, Csere P, Guiard B, Lill R. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 1997;418:346–350. doi: 10.1016/s0014-5793(97)01414-2. [DOI] [PubMed] [Google Scholar]
- 34.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lasko P. RNA sorting in Drosophila oocytes and embryos. FASEB J. 1999;13:421–433. doi: 10.1096/fasebj.13.3.421. [DOI] [PubMed] [Google Scholar]
- 36.Leighton J, Schatz G. An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 1995;14:188–195. doi: 10.1002/j.1460-2075.1995.tb06989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Kaplan J. Defects in the yeast high-affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem. 1998;273:22181–22187. doi: 10.1074/jbc.273.35.22181. [DOI] [PubMed] [Google Scholar]
- 38.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 39.MacDonald P M, Kerr K. Redundant RNA recognition events in bicoid mRNA localization. RNA. 1998;3:1413–1420. [PMC free article] [PubMed] [Google Scholar]
- 40.Martoglio B, Dobberstein B. Snapshots of membrane-translocation proteins. Trends Cell Biol. 1996;6:142–147. doi: 10.1016/0962-8924(96)10001-5. [DOI] [PubMed] [Google Scholar]
- 41.Miyakawa I, Aoi H, Sando N, Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis ans sporulation in the yeast, Saccharomyces cerevisiae. J Cell Sci. 1984;66:21–38. doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- 42.Mowry K L, Cote C A. RNA sorting in Xenopus eggs and oocytes. FASEB J. 1999;13:435–445. doi: 10.1096/fasebj.13.3.435. [DOI] [PubMed] [Google Scholar]
- 43.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:683–717. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 44.Ni L, Heard T S, Weiner H. In vitro mitochondrial import: a comparison of leader sequence charge and structural relationships with the in vitro model resulting in evidence for co-translational import. J Biol Chem. 1999;274:12685–12691. doi: 10.1074/jbc.274.18.12685. [DOI] [PubMed] [Google Scholar]
- 45.Nobumoto M, Yamada M, Song S, Inouye S, Nakasawa A. Mechanism of mitochondrial import of adenylate kinase enzymes. J Biochem. 1998;123:128–135. doi: 10.1093/oxfordjournals.jbchem.a021899. [DOI] [PubMed] [Google Scholar]
- 46.Pon L, Moll T, Vestweber D, Marshallsay B, Schatz G. Protein import into mitochondria: ATP-dependent translocation activity in submitochondrial fraction enriched membrane contact sites and specific proteins. J Cell Biol. 1989;109:2603–2616. doi: 10.1083/jcb.109.6.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 50.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Gorlich D, Ponstingl H, Bischoff F R. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmiit M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, Paltauf F, Wieland F T, Kohlwein S D. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sil A, Herskowitz I. Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 55.Stevens B J. Variation in number and volume of the mitochondria in yeast according to growth conditions. A study based on serial sectioning and computer graphics reconstitution. Biol Cell. 1977;28:37–56. [Google Scholar]
- 56.Steward O, Singer R H. mRNA metabolism and posttranscriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. The intracellular RNA sorting system: postal zones, zip codes, mail bags and mail boxes; pp. 127–146. [Google Scholar]
- 57.Suissa M, Schatz G. Import of proteins into mitochondria. J Biol Chem. 1982;257:13048–13055. [PubMed] [Google Scholar]
- 58.Takizawa P, Sil A, Swedlow J, Herskowitz I. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 59.Verner K. Co-translational protein import into mitochondria: an alternative view. Trends Biochem Sci. 1993;18:364–371. doi: 10.1016/0968-0004(93)90090-a. [DOI] [PubMed] [Google Scholar]
- 60.Williamson D H. Packing and recombination of mitochondrial DNA in vegetatively growing yeast cells. In: Bandlow R J S W, Thomas D Y, Wolf K, Kaudewitz F, editors. Genetics, biogenesis, and bioenergetics of mitochondria. Berlin, Germany: De Gruyter and Co.; 1976. pp. 117–123. [Google Scholar]