Abstract
Background
Pathogenic variants of GNB5 encoding the β5 subunit of the guanine nucleotide-binding protein cause IDDCA syndrome, an autosomal recessive neurodevelopmental disorder associated with cognitive disability and cardiac arrhythmia, particularly severe bradycardia.
Methods
We used echocardiography and telemetric ECG recordings to investigate consequences of Gnb5 loss in mouse.
Results
We delineated a key role of Gnb5 in heart sinus conduction and showed that Gnb5-inhibitory signalling is essential for parasympathetic control of heart rate (HR) and maintenance of the sympathovagal balance. Gnb5−/− mice were smaller and had a smaller heart than Gnb5+/+ and Gnb5+/− , but exhibited better cardiac function. Lower autonomic nervous system modulation through diminished parasympathetic control and greater sympathetic regulation resulted in a higher baseline HR in Gnb5−/− mice. In contrast, Gnb5−/− mice exhibited profound bradycardia on treatment with carbachol, while sympathetic modulation of the cardiac stimulation was not altered. Concordantly, transcriptome study pinpointed altered expression of genes involved in cardiac muscle contractility in atria and ventricles of knocked-out mice. Homozygous Gnb5 loss resulted in significantly higher frequencies of sinus arrhythmias. Moreover, we described 13 affected individuals, increasing the IDDCA cohort to 44 patients.
Conclusions
Our data demonstrate that loss of negative regulation of the inhibitory G-protein signalling causes HR perturbations in Gnb5− /− mice, an effect mainly driven by impaired parasympathetic activity. We anticipate that unravelling the mechanism of Gnb5 signalling in the autonomic control of the heart will pave the way for future drug screening.
Keywords: GNB5variants, Gnb5-null mouse models, IDDCA, cardiac conduction anomalies
Introduction
Intellectual developmental disorder with cardiac arrhythmia (IDDCA, OMIM (Online Mendelian Inheritance in Man): #617173) is an autosomal recessive neurodevelopmental disorder with onset in early childhood. Inactivating and hypomorphic mutations in the β5 subunit of guanine nucleotide-binding protein (GNB5), respectively, cause severe and mild forms of the disorder.1 The former is associated with cognitive disability, poor or absent speech and/or severe cardiac arrhythmias. The moderate manifestation of the syndrome, also named language delay and ADHD/cognitive impairment with or without cardiac arrhythmia (LADCI) syndrome (OMIM: #617182), consists of mild intellectual impairment, language delay, attention deficit hyperactivity disorder (ADHD) and, in about half the cases, severe cardiac arrhythmia.1 2 Some patients with IDDCA also showed retinal dysfunction and nystagmus, epilepsy, hypotonia and gastrointestinal problems.1–10 The GNB5 retinopathy is a unique combination of dual retinal signalling defects reminiscent of features of both bradyopsia and rod ON-bipolar dysfunction,5 while the IDDCA epilepsy is characterised by early seizure onset (~3 months of age) with focal seizures rapidly evolving into epileptic spasms and consequent generalised multifocal discharges.4
The heart rate (HR) is established by the sinoatrial node, the pacemaker of the cardiac muscle, and controlled by the autonomic nervous system. This autonomic nervous system consists of two anatomically and functionally distinct divisions: the sympathetic and the parasympathetic branches, whose functions are often antagonistic but work together to maintain balance. In the heart, the postganglionic fibres of the sympathetic trunk stimulate the β-adrenoreceptors, thereby increasing HR and force of contraction. The parasympathetic modulation of the heart is primarily mediated by acetylcholine release, which activates the M2-muscarinic receptors (M2R) present on cells innervated by parasympathetic postganglionic neurons, including sinoatrial node cells. The activation of M2R triggers Gi/o subfamily G-proteins, which turn on G-protein-gated inwardly rectifying K+ channels (GIRK) resulting in membrane hyperpolarisation and decrease in HR. Regulator of G-protein signalling (RGS) proteins negatively regulate the timing of this M2R-GIRK signalling. GNB5, a divergent member of the Gβ family, has the unique property of forming complexes with R7-RGS proteins.11–16 In particular, the GNB5-RGS6 complex is involved in cardiac GIRK deactivation kinetics. Rgs6-null mice manifested heart conduction anomalies and hypersensitivity to parasympathomimetics.17 Zebrafish model defective for gnb5 gene correspondingly showed reduced heartbeat on reinforced parasympathetic stimulation, eye movement defects and altered swimming behaviour1 and cardiomyocytes differentiated from human induced pluripotent stem cells (iPSCs) edited to engineer the GNB5-Ser81Leu missense variant associated with LADCI showed a decrease in spontaneous activity on stimulation with carbachol compared with normal cells.7
Whereas homozygous Gnb5-null mice recapitulated many of the corresponding human disease phenotypes such as learning deficiencies, hyperactivity, impaired motor coordination and perturbed vision,18–22 a systematic cardiac evaluation has never been performed in a mammalian model. Here, we assessed heart electrophysiology of Gnb5 mice models. We detected an increased frequency of sinus arrhythmias in Gnb5−/ − animals, which have a smaller heart than wild-type and Gnb5+/− , but exhibited better cardiac function. Gnb5−/− mice also displayed enhanced parasympathetic sensitivity on stimulation with a cholinergic agonist. Consistent with this, transcriptome profiling of atria and ventricles revealed overexpression of genes involved in cardiac muscle contractility, along with reduced ventricular expression of genes required for development of pacemaker cells in Gnb5−/− mice. Finally, we expanded the number of ascertained IDDCA individuals and the GNB5 mutational spectrum.
Materials and methods
Enrolment
All affected individuals and their family members were recruited in Pakistan (families R–V), France (family W), Egypt (families X and Y) and Iran (family Z) after signing a written informed consent according to ethical review boards policies. Clinical ascertainment included physical examinations, medical history interviews and specialised consultations by a certified neurologist and cardiologist as appropriate. Venous blood was collected in EDTA for DNA extraction according to standard procedures.
Exome sequencing
Whole-exome sequencing of families R–V and Z was performed by Macrogen, Korea, as described in reference.23 Briefly, target enrichment was performed with 2 µg genomic DNA using the SureSelectXT Human All Exon Kit version 6 (Agilent Technologies, Santa Clara, California, USA) to generate barcoded whole-exome sequencing libraries. Libraries were sequenced on the HiSeqX platform (Illumina, San Diego, California, USA) with 50× coverage. Quality assessment of the sequence reads was performed by generating QC statistics with FastQC.24 The filtering strategy included screening for only exonic and donor/acceptor splicing variants. In accordance with the pedigree and phenotype, priority was given to variants rare or absent in public databases (1000 Genomes project, National Heart, Lung, and Blood Institute Exome Variant Server, Complete Genomics 69 and Exome Aggregation Consortium V.0.2).
Trio exome sequencing was performed in proband 37 (family W) and her parents using SeqCap EZ Medexome library preparation kit following manufacturer’s recommendations (Roche, Indiana, Indianapolis, USA). Libraries were sequenced on a NextSeq500 (Illumina) at a mean depth coverage of 73× with 93.3% of the target bases above 30×. Genomic alignment against the hg19/GRCh37 assembly and variant calling were, respectively, done with BWAMEM V.0.7.12 and GATK HaplotypeCaller V.3.4 (Broad Institute, Boston, Massachusetts, USA). Only highly confident variants were kept for analysis (total depth >9, alternative allele depth >4, no strand bias, mosaicism >10%). Rare variants were considered as having a frequency of <1% in GnomAD v2 dataset. Whole-exome sequencing of families X and Y was performed as described in Makrythanasis et al.25 Sanger sequencing in each family confirmed the segregation of GNB5 variants with the phenotype.
Mouse husbandry
The Gnb5 mouse line was recovered from cryopreserved sperm using in vitro fertilisation. The knockout allele was engineered in a C57BL/6J inbred genetic background by heterozygous deletion of exon 3 in the germline, as previously described.18 22 Genetically modified animals were born and housed in the Animal Facility of the Centre for Integrative Genomics, under controlled temperature conditions and a 12-hour light–dark cycle with free access to water, normal chow and nest building material. Mouse genomic DNA was extracted from ear biopsies using the hot shot protocol26 and used for genotyping as described.22 To prevent the previously documented high mortality of Gnb5− /− pups at weaning,18 heterozygous breeding couples used to obtain knockout pups were given breeding food pellet enriched for proteins and vitamins (Kliba 3336, extrudate). Additionally, litters including Gnb5 −/− pups were fed from 14 to 28 days of age, that is, starting 1 week before weaning, with powdered wet maintenance food (Kliba 3436) in a Petri dish placed directly onto the floor of the cages, an expedient that should provide easier access to the food for the pups.
In vivo transthoracic ultrasound imaging protocol
Transthoracic echocardiography was performed using a 30 MHz probe and the Vevo 2100 Ultrasound machine (VisualSonics, Toronto, Ontario, Canada). A light anaesthesia was achieved with 1%–1.5% isoflurane, maintaining HR at 400–500 beats/min. The mice were placed in decubitus dorsal on a heated 37°C platform to maintain body temperature. The heart was imaged in the 2D mode in the parasternal long-axis view. From this view, an M-mode cursor was positioned perpendicular to the interventricular septum and the posterior wall of the left ventricle, at the level of the papillary muscles. Diastolic and systolic interventricular septa, left ventricular posterior wall thickness and left ventricular internal end-diastolic and end-systolic chamber dimensions were measured. Three separate M-mode images were measured and averaged. Left ventricular fractional shortening and ejection fraction were also calculated. Fractional shortening and ejection fraction were assessed from M-mode based on the percentage changes of left ventricular end-diastolic and end-systolic diameters and volumes, respectively. We used male mice at 9 weeks of age of three different genotypes (Gnb5+/+ , Gnb5+/ − and Gnb5−/− ).
In vivo electrocardiography measurements
For in vivo electrocardiography monitoring we have subcutaneously implanted biopotential telemetric transponders (ETA-F10, Data Sciences International) allowing continuous monitoring in conscious freely moving animals at 12 weeks of age. The negative electrode was implanted at the top of the right pectoral muscle, and the positive one was anchored at the level of the last left rib (at about 1 cm of the xiphoid appendix), thus leading to a normal lead II trace. Baseline ECG was recorded 10 days after device implantation, over a period of 86 hours. After 30 min of basal measurements, we injected mice with 0.9% saline solution (intraperitoneal NaCl, 10 mL/kg) as a vehicle control; next, the following compounds were administered one at a time: atropine (PubChem CID: 174174, intraperitoneal, 1 mg/kg), carbachol (PubChem CID: 5831, intraperitoneal 0.1 mg/kg) and in a subset of mice isoprenaline (PubChem CID: 3779, intraperitoneal 4 mg/kg) and atenolol (PubChem CID: 2249, intraperitoneal 2 mg/kg) as well, with a one night interval between each injection. The amounts of isoprenaline and atenolol were chosen after testing 10 doses ranging from 4 µg/kg to 4 mg/kg and 2 µg/kg to 2 mg/kg, respectively. While atropine and carbachol respectively inhibit and activate the parasympathetic system atenolol and isoprenaline respectively block and promote the sympathetic response.
At the end of the experiment, mice were sacrificed by CO2 inhalation; the heart was digitally imaged both within its thoracic position and after excision. Heart weight was recorded and tibia length was measured to normalise the heart weight to body size. Images were taken by a Leica DCF295 digital colour camera with 3M pixels mounted on a MZ6 stereomiscroscope (Leica, Switzerland). The length of the tibia was measured using a precision calliper after removal from the left leg (without patella nor articular cartilage).
Baseline ECG traces were analysed as follows: 10 min of recording were analysed using ECG-Auto software in shape recognition mode (EMKA Technology, France) every 30 min, during night and day phases. Mean values were reported for each analysed parameter. During pharmacological challenges, ECG recording was analysed continuously, with 10 min steps; thus, one mean of each parameter was calculated every 10 min. On ECG traces we analysed: (1) RR interval (measured at R peak, expressed in millisecond; (2) HR: heart beating rate, calculated as 60/(RR/1000), expressed in beats/min; (3) PR interval (interval between beginning of P wave and R peak, expressed in millisecond); (4) QT duration: duration of the QT complex; (5) QTc (corrected QT, calculated from QT/√(RR/100)27 (all expressed in millisecond) that allows correction of QT from HR variations).
Temperature and activity were also recorded. Activity was estimated by displacement of the telemetric device from the antennas of the recording plate. This measurement was only used as a qualitative index of mouse activity/movement. Temperature and activity values are mean values of 1-hour interval.
Time-domain heart rate variation (HRV) analysis
Twelve-hour segments of day/night phases were selected from the baseline recording period and analysed separately for the HRV analysis. Specifically, R wave detection was performed and R-R interval time series were obtained. To ensure inclusion of sinus beats only, values not included between R–R intervals±2 SD were excluded as reported in Thireau et al.28 The analysed time-domain HRV parameters, computed using Kubios HRV Standard software V. 3.2.0, were: (1) mean R–R intervals (NN, in millisecond); (2) SD of all normal R–R intervals (SDNN, in millisecond); (3) square root of the mean square differences (RMSSD) between successive normal intervals (in millisecond); and (4) percentage of normal consecutive R–R intervals differing by >xms (pNNx, in %, in this study x=6 ms).
Arrhythmia assessment
Arrhythmias were identified based on ECG trace (RR interval) and counted. Observed arrhythmias were defined as follows: (1) escape atrial beat, with a P wave morphology different from that of the sinus P wave, classified as ‘long’ and ‘short’ according to the location of the P wave on the ECG trace (specifically, long was an escape beat whose duration was longer than two normal PP intervals, and short were those escape beats that lasted less than two normal PP intervals); (2) atrioventricular block defined on ECG by more than one P wave for one QRS complex; (3) premature beats; and (4) episodes of tachycardia followed by bradycardia, with HR oscillation between high and low values within a few seconds, independently of other type of arrhythmias.
Statistical tests
Each parameter measured was reported as mean±SD. Standard t-test was used to assess differences between two groups. Analysis of variance tests, followed by Tukey post hoc tests, were also calculated and are displayed. A p value of <0.05 was considered significant, and stars on the plots represent the level of significance (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001; p>0.05 was considered not significant).
Transcriptome profiling, data processing and differential expression analysis
A total of 72 RNA sequencing libraries were generated from two heart tissues, atria and ventricles, and three brain regions, cerebellum, hippocampus and cerebral cortex. Both the entire ventricles (left and right) or atria (left and right) were used for the sample processing; therefore, the atrial tissue represents a mixture of heart muscle cells, as well as sinoatrial and atrioventricular cells. The whole cerebellum, the hippocampi from each hemisphere of the brain and the whole cerebral cortex were dissected for RNA extraction. For this experiment, we created a cohort of adult (18 weeks) male mice including 6 Gnb5−/− , 6 Gnb5+/− and 6 Gnb5+/+ animals. Brain transcriptome was only assessed in Gnb5−/− and Gnb5+/+ animals. Tissue collection and processing procedures were designed to minimise biological and technical variation. Specifically, tissues were dissociated in QIAzol Lysis Reagent (Qiagen) using the gentleMACS Dissociator (Miltenyi Biotec). Cell suspension was used to obtain total RNA. Genomic DNA contamination was removed by digestion with RNase-free Deoxyribonuclease I (Qiagen). RNA concentration and purity were measured by ND-1000 spectrophotometer (Thermo Scientific, Wilmington, North Carolina, USA), and RNA integrity was verified by fragment analyser automated CE system (Advanced Analytical Technologies) according to manufacturer’s instructions. Libraries were then prepared with TruSeq Stranded RNA Library Prep Kit (Illumina) and sequenced on multiple lanes of an Illumina HiSeq4000 platform, generating an average of 50M single-end 125-cycle reads for each sample. Quality of sequence was assessed by FastQC V.0.11.4.24 Purity-filtered reads were adapters-trimmed and quality- trimmed with Cutadapt V.1.8.29 Reads matching to ribosomal RNA sequences were removed with fastq_screen V.0.11.1. Remaining reads were further filtered for low complexity with reaper V.15–065.30 Reads were then aligned against Mus Musculus.GRCm38.92 genome using STAR V.2.5.3a.31 The number of read counts per gene locus was summarised with htseq-count V.0.9.132 using Mus Musculus.GRCm38.92 gene annotations. Quality of the RNA-seq data alignment was assessed using RSeQC V. 2.3.7.33 Reads were also aligned to the Mus Musculus.GRCm38.92 transcriptome using STAR V. 2.5.3a,31 and the estimation of the isoforms abundance was computed using RSEM V.1.2.31.34 To assess differential expression between genotypes within each tissue, we compared Gnb5−/− and Gnb5+/− ± to wild-type specimens. Data analysis was performed with the R Bioconductor package DESeq2 V.1.14.1.35 Differentially expressed genes (DEGs) were identified at the Benjamini-Hochberg adjusted p<0.05 level, using Wald test under design ~genotype. For gene set enrichment analysis, no direction criterion on fold change was applied. Enriched Gene Ontology (GO) categories were identified using the enrichment analysis package in R/Bioconductor, clusterProfiler,36 considering only categories with at least 10 and maximum 500 annotated genes. Nominally significant enriched terms were retained for results interpretation.
Western blotting analysis
Tissue lysates, including atria, ventricles, cerebral cortex, cerebellum and hippocampi, were prepared in RIPA buffer (Millipore) supplemented with protease inhibitors (Thermo Fisher Scientific). Tissues were homogenised using the gentleMACS Dissociator (Miltenyi Biotec). After SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transfer to nitrocellulose membrane, blots were incubated with anti-Gnb5, anti-Rgs7 (both a generous gift from Dr William F Simonds) and anti-Gnb3 (Cell Signalling Technology) antibodies, separately, and with an antiactin antibody (Sigma), used for loading control. Horseradish peroxidase-conjugated anti-rabbit antibody (Santa Cruz) and the ECL chemiluminescence system (Millipore) were used for detection.
Results
Clinical and molecular features of thirteen novel patients with IDDCA
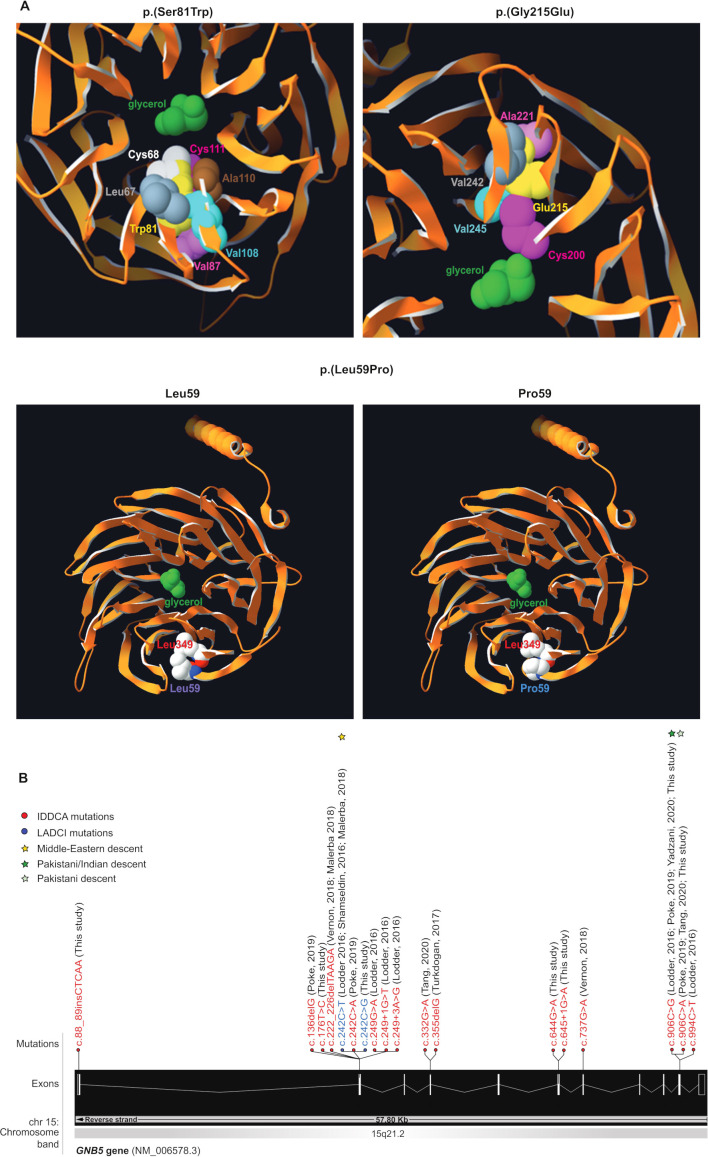
We identified 13 additional IDDCA cases (online supplemental figure S1, table S1 and table 1) through exome sequencing of nine consanguineous families and data aggregation of multiple laboratories and clinical centres via GeneMatcher37 38 or direct contacts. Consistent with previous reports,1–10 the carrier of a homozygous GNB5 missense variant on Ser81 presented with LADCI, the mild form of IDDCA (family W), while the nine individuals with biallelic loss-of-function (LoF) alleles due to truncating or splicing mutations displayed phenotypes corresponding to the severe end of the disease spectrum (families R and S, U and V, and X and Z). The remaining three individuals (families T and Y) carrying novel homozygous missense variants on Gly215 (family T) and on Leu59 (family Y) similarly presented with the severe IDDCA phenotypical spectrum. Clinical features of affected individuals are summarised in table 1 and detailed in the online supplemental note and table S1. The three missense variants (c.644G>A, p.(Gly215Glu), family T, c. 242C>G, p.(Ser81Trp), family W and c.176T>C, p.(Leu59Pro), family Y; transcript NM_006578.3) were not reported before (figure 1). They are predicted by a majority of prediction tools to be likely damaging to protein function (online supplemental table S2) and are absent from GnomAD V.3.39 To assess possible impact, we modelled these three substitutions using the crystal structure of the GNB5-RGS9 complex40 and found that each variant could impact the potential binding properties of the GNB5 central pore (p.(Ser81Trp) and p.(Gly215Glu)) or the protein folding (p.(Leu59Pro)) (figure 1A). Specifically, Serine 81 is buried inside a β strand of the first WD40 repeat close to the central pore of the β propeller structure1 2 where a glycerol molecule is observed in pbi structure 2pbi (figure 1A, top left panel). Reminiscent of the Ser81Leu variant previously documented,1 a tryptophan at position 81 cannot be accommodated without disrupting the structure and potential binding properties of the pore. Our model suggests that the rearrangements necessary to settle such a bulky sidechain will change the channel characteristics. We have investigated the related rotamers, emphasising the steric hindrance-induced local rearrangements associated with the tryptophan replacement, whose perturbations were evaluated using the backbone-dependent rotamer library implemented in the Swiss-PdbViewer.41 Depending on the rotamer, tryptophan 81 will severely encroach with leucine 67, cysteine 68, valine 87, cysteine 111, valine 108 and/or alanine 110 (figure 1A, top left panel).
Table 1.
Overlapping clinical features of individuals with IDDCA and LADCI syndromes
| Individuals with phenotype/individuals (total n) | Phenotype severity (IDDCA) |
Phenotype severity (LADCI) |
Phenotype severity (intermediate) |
|
| Gender | 23F, 21M | 15F, 19M | 7F, 2M | 1F |
| Clinical examination | ||||
| Dysmorphic features | 11/44 (25%) | 10/34 | – | 1/1 |
| Congenital malformations | 4/44 (9%) | 4/34 (heart) | – | – |
| Neurological manifestations | ||||
| Intellectual disability | 37/44 (84%) | 32/34 | 4/9 | 1/1 |
| Speech delay | 34/44 (77%) | 27/34 | 6/9 | 1/1 |
| Hypotonia | 33/44 (75%) | 29/34 | 3/9 | 1/1 |
| Seizures | 25/44 (57%) | 25/34 | – | – |
| Behavioural disorders | 6/44 (14%) | 3/34 (ASD) | 3/9 (ADHD) | – |
| MRI anomalies | 8/44 (18%) | 8/34 | – | – |
| Sleep disturbance | 2/44 (5%) | 2/34 | – | – |
| Cardiac manifestations | ||||
| Sinus sick syndrome | 27/44 (61%) | 22/34 | 4/9 | 1/1 |
| Pacemaker implantation | 6/44 (14%) | 4/34 | 1/9 | 1/1 |
| Ophthalmological findings | ||||
| Nystagmus | 26/44 (59%) | 26/34 | – | – |
| Strabismus | 6/44 (14%) | 5/34 | – | 1/1 |
| Retinal disease | 14/44 (32%) | 14/34 | – | – |
| Gastrointestinal problems | ||||
| Pathological gastric reflux | 18/44 (41%) | 18/34 | – | – |
Pedigree charts and variants of single individuals are detailed in the corresponding published reports1–4 6 8–10 81 and in online supplemental figure S1. Detailed phenotypical information of each affected individuals is reported in online supplemental table S1 and supplemental note.
ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; F, female; IDDCA, Intellectual developmental disorder with cardiac arrhythmia; LADCI, language delay and ADHD/cognitive impairment; M, male.
Figure 1.
Variants modelling and IDDCA mutational spectrum. (A, top left) Top view of the Gnb5 (orange, PDB entry 2pbi) 3D protein model, showing the mutated Trp81 (yellow) and the glycerol molecule (green) in the centre of the pore. The rearrangements necessary to accommodate a tryptophan residue at position 81 will change the channel characteristics. The rotamer displayed here highlights clashes of Trp81 with Cys68 (white) and Cys111 (magenta). In additional rotamers, the bulky tryptophan sidechain will severely bump into Leu67 (grey), Val87 (pink), Val108 (cyan) and Ala110 (brown). (A, top right) As shown in this view of the beta propeller from above, the ‘wild-type’ Gly215 (not shown) lays in a beta-sheet, with on top Ala221 (pink), at the bottom Cys200 (magenta), and in front a beta-strand harbouring Val242 (grey) and Val245 (cyan). The presence of Glu215 cannot be tolerated, as it will encroach into one of the residues previously enumerated. Another rotamer shows clashes into Val245 (cyan). Overall, all rotamers may also force the sidechain of Cys200 (magenta) to reorient itself toward the internal part of the channel to provide space to accommodate glutamine at position 215 (yellow). In this position, the Cys200 sidechain will occupy the space dedicated to the glycerol (green), thus changing the properties of the channel. (A, bottom) The Leu59 (purple label, left panel) is positioned closely to the Leu349 residue just above (red label) in an antiparallel beta-sheet. This secondary structure will likely be broken in presence of a proline at that position (blue label, right panel), which in turn will disrupt the protein folding as the proline sidechain will collide into Leu349 (red label). (B) Distribution of the IDDCA published and novel variants along the schematically represented 11 exons of the human GNB5 gene (transcript NM_006578.3; Ensembl (release 98, September 2019). The variants of IDDCA affected individuals are represented in red (LoF) and the missense LADCI variants in blue. The yellow star marks the variant of Middle Eastern descent, while green stars indicate the amber and ochre variants from the Indian subcontinent (dark green) and of Pakistani descent (light green), respectively. IDDCA, intellectual developmental disorder with cardiac arrhythmia; LADCI, language delay and ADHD/cognitive impairment; LoF, loss of function.
jmedgenet-2020-107015supp003.pdf (5.8MB, pdf)
jmedgenet-2020-107015supp004.pdf (938.6KB, pdf)
jmedgenet-2020-107015supp002.pdf (49.9KB, pdf)
Pedigree charts and variants of single individuals are detailed in the corresponding published reports1–4 6 8–10 43 and in (online supplemental figure S1). Detailed phenotypic information of each affected individuals is reported in (online supplemental table S1 and Supplemental Note). Abbreviations are as follows: M: male; F: female, ASD: Autism Spectrum Disorder, ADHD: Attention Deficit Hyperactivity Disorder.
Glycine 215 (figure 1A, top right panel) lies in a beta-sheet, between alanine 221 and cysteine 200. It faces a beta-strand harbouring valine 242 and valine 245. There is not enough space to accommodate the glutamic acid sidechain as it would encroach into one or more sidechains of the above-enumerated residues. All rotamers would also probably force the sidechain of cysteine 200 to reorient itself toward the interior of the channel to accommodate the glutamic acid 215 sidechain. This will infringe on the glycerol molecule space40 changing the channel characteristics. Additionally, the c.644G>A, p.(Gly215Glu) variant that affects the second to last nucleotide of exon 6 might alter the activity of the donor splice site as well as create cryptic exonic splicing enhancers or silencers according to the NNSplice, NetGene2 and Splicing Finder prediction tools (online supplemental table S2).
Leucine 59 (figure 1A, bottom left panel) is in close contact with leucine 349, and both residues, linked by a hydrogen bond, belong to an antiparallel beta-sheet. A proline at position 59 (figure 1A, bottom right panel) will likely destabilise (break) this hydrogen bond, thus representing a structural conundrum. Locally, the presence of a proline might disrupt the overall protein fold as there will not be enough space to accommodate this residue, its cyclic sidechain clashing into Leu349.
To date a total of 18 pathogenic GNB5 variants and one homozygous deletion at 15q21.2 encompassing GNB5 gene have been identified in 44 individuals with IDDCA (online supplemental ffigure 1S 1B and table S1). Suggestive of a founder effect, the eight affected individuals from three families (families E–G) carrying the Ser81Leu variant all originate from Arab countries (Morocco, Algeria and Saudi Arabia). Of note, the Greater Middle East Variome Project43 (http://igm.ucsd.edu/gme/) did not identify this variant within 2497 individuals. Similarly, the patients from six families (families D, L, P, R, S and V) harbouring the amber nonsense c.906C>G (Tyr302*) variant are from the Indian subcontinent (one from India and five from Pakistan). Another variant modifying the Tyr302 codon in an ochre codon (c.906C>A) was found in two additional Pakistani families (families N and U), suggesting again a possible founder effect. Of note, the same ochre variant was shown to be de novo on the paternal allele of the proband of the Chinese descent family O.9 The 34 IDDCA individuals present with the severe end of the disease spectrum, which is characterised by severe ID (32 out of 34) with poor or absent speech (27/34), early onset sinus node dysfunction (22/34) with 4/22 who had a pacemaker implanted, variable visual abnormalities (26/34), seizures (25/34), hypotonia (29/34) and gastrointestinal problems (18/34). Additionally, 9/34 individuals showed different types of dysmorphic features (table 1 and online supplemental table 1), and MRI evaluation revealed altered brain structure in 8/34 children, with four having thinner corpus callosum, two having long posterior and hypogenesis of corpus callosum, respectively, one cerebral atrophy and one cerebral and cerebellar cortical atrophy. Three individuals (individual 16, family H; individual 39, family X; and individual 44, Family Z) showed autistic features, and the other two displayed sleep disturbances (individual 27, family N, and individual 29, family P). All severely affected individuals carry either biallelic truncation mutations or biallelic missense variants that probably result in LoF. Nine patients displayed the milder LADCI syndrome and biallelic missense variants at position 81: 4/9 presented with mild ID with 6/9 showing language deficits; 4/9 were noted to have sinus node dysfunction (one of which with pacemaker implantation); and 3/9 were reported with impaired fine motor skills. Behaviorally, 3/9 patients exhibited ADHD. The remaining patient (family J) is compound heterozygous for the LoF p.Asp74Glufs52* and the Ser81Leu variants.3 She presented with an intermediate manifestation of the symptoms with mild ID accompanied by speech delay, hypotonia and sinus bradycardia (table 1). Like patient 21 from family I who carries the same LoF p.Asp74Glufs52* combined with a different missense, p.(Arg246Gln),8 she is affected by hearing loss (online supplemental table S1).
Gnb5 knockout mouse cohort
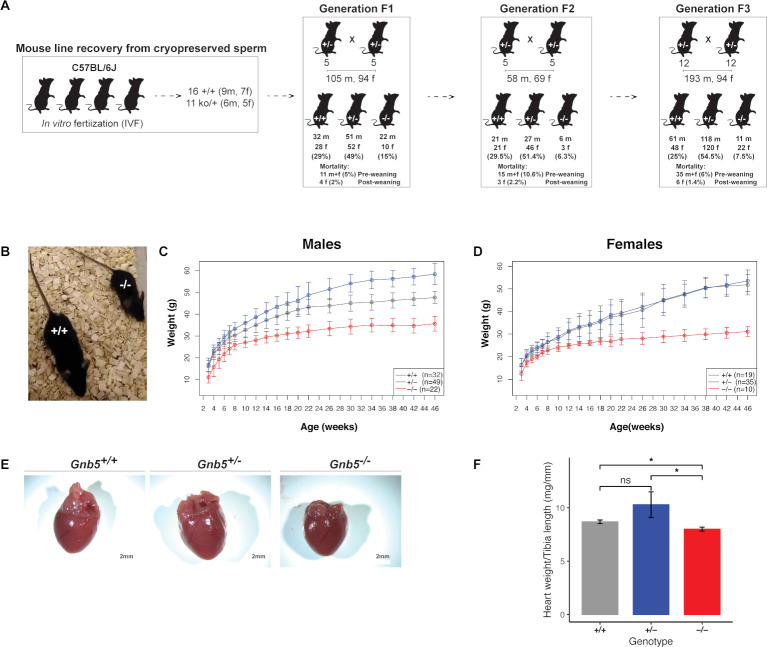
To model the cardiac manifestations occurring in IDDCA syndrome, we used the mouse model knockout for Gnb5 (Gnb5–/ –), thus mimicking a complete LoF. Whereas ~66% of the pups carrying the homozygous null allele were previously reported to die prior to or at weaning,18 preweaning mortality was very low in our husbandry setting (see Materials and methods). We experienced only 5%, 11% and 6% of losses in the three cohorts we generated by mating heterozygous parents (figure 2A). We recorded two to eight breeding events during three generations of husbandry with an average litter size of 4–10 pups. The vast majority of preweaning lethality appears to be associated with Gnb5–/– as shown by the quasi-Mendelian distribution of genotypes (figure 2A). We longitudinally monitored the body weight of Gnb5+/+ (wild type), Gnb5+/– and Gnb5–/– male and female mice from 3 to 46 weeks of age (figure 2C, D). We confirmed previous reports18 22 that showed that female and male knockout animals are smaller (figure 2B–D) and that heterozygote male mice are heavier (figure 2). Importantly, knockout mice had a smaller heart (figure 2E), even smaller than expected when the heart weight was normalised to the tibia length, a proxy for animal size (figure 2F). During animal handling, no obvious gender differences were observed regarding development, behaviour or other gross phenotypes.
Figure 2.
Gnb5 mouse line features. (A) Mouse mating strategy and gender and genotype distribution over three successive generations. Preweaning and postweaning mortality is reported for each colony. (B) Size of Gnb5+/+ and Gnb5−/ − mice. (C, D) Body weights profile monitored from 3–46 weeks of age. All mice were weaned on week 3. Data are shown as mean±SD. (C, D) Panels separate body weights according to sex. Gnb5+/+ is depicted in grey, Gnb5+/ − in blue and Gnb5−/ − in red. (E) At sacrifice, neither significant morphology difference nor thoracic position of the heart were observed among groups. (F) Bar plot showing that Gnb5−/ − hearts (red, n=16) are smaller compared with the other genotypes (Gnb5+/ −, blue (n=8) and Gnb5+/+, grey (n=16)). Data are shown as mean of the ratio between heart weight and tibia length (used to normalise for animal size)±SEM. Asterisks on the plots represent the level of significance: *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001; p>0.05 was considered not significant.
Ultrasound scans pinpointed increased cardiac function in Gnb5 knockout mice
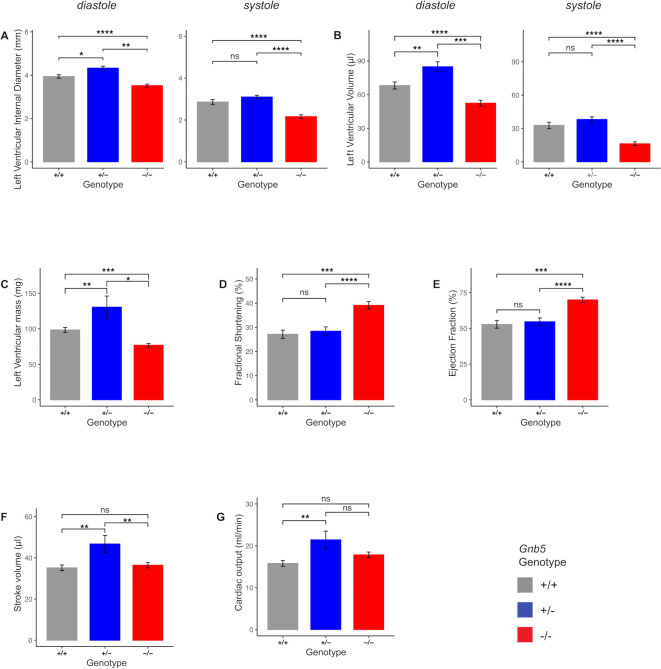
To characterise the Gnb5 knockout mouse line at cardiac level, we first performed ultrasound scans in baseline conditions. We used 16 Gnb5+/+ , 8 Gnb5+/– and 16 Gnb5–/– male mice (9 wo (weeks of age)) and analysed heart morphology and function (see Materials and methods section). Echocardiography confirmed that Gnb5–/– animals had smaller hearts, as demonstrated by reduced ventricular chambers both in diastole and systole (figure 3A). Consequently, ventricular volume was also smaller (figure 3B). Estimated left ventricular weight was significantly lower in Gnb5–/– compared with Gnb5+/– and wild-type (figure 3C). Left posterior ventricular wall and interventricular septum thickness were not substantially modified (online supplemental figure S2A, B). Interestingly, Gnb5–/– mice demonstrated improved cardiac function, as judged by increased fractional shortening (figure 3D) and ejection fraction (figure 3E). However, stroke volume (figure 3F) and cardiac output (figure 3G) were similar to those measured in wild-type mice. Notably, and unexpectedly, Gnb5+/– have a bigger heart (figure 3A–C) compared with wild type, while their cardiac function remains unchanged (figure 3D–E). Therefore, the increased stroke volume (figure 3F) and cardiac output (figure 3G) reflected an increased volume of blood pumped by the ventricle.
Figure 3.
Morphological and functional parameters measured by ultrasound scan in Gnb5+/+ , Gnb5+/ − and Gnb5−/ − male mice. (A) Left ventricular internal diameter at diastole (left) and systole (right). (B) Left ventricular volume in diastole (left) and systole (right). (C) Mass of the left ventricle. (D–G) Cardiac function expressed as fractional shortening (D), ejection fraction (E), cardiac output (F) and stroke volume (G). Parameters unchanged between the three genotypes are shown in online supplemental figure S2A, B.
Taken together, these results indicated that Gnb5–/– mouse hearts were smaller than that of the other two genotypes but compensated their smaller size by increased cardiac efficiency.
Loss of functional Gnb5 determines the onset of sinus arrhythmias
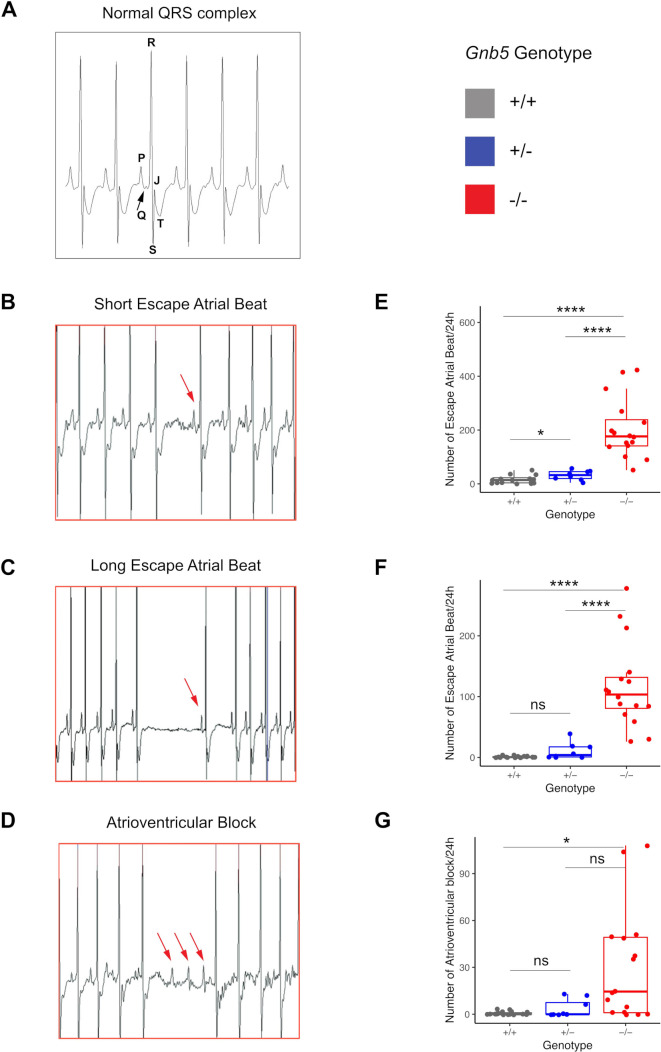
As cardiac arrhythmia in the form of bradycardia and ectopic beats is one of the core symptoms in IDDCA, we examined HRVs in Gnb5 mouse models with in vivo ECG monitoring. ECG was performed at 12 weeks on the same male mice used for echocardiography. Baseline ECG parameters were not different among Gnb5+/+ , Gnb5+/– and Gnb5–/– animals, except for HR in knocked-out animals that showed a trend toward higher values (online supplemental figure S2C, minimum and maximum HR values registered in wild-type were 313 and 741 beats/min, while lowest and greatest HR values in knockout were 349 and 744 beats/min, significance varying between p=1.28E-04 and p=0.9971, over 36 daylight time points).
Close inspection of baseline ECG over a 24-hour window allowed the quantification and characterisation of small changes in the intervals between successive heartbeats (RR interval) corresponding to cardiac arrhythmias (Materials and methods). The 24-hour ECGs unearthed a significant increase in arrhythmic events in Gnb5–/– mice compared with heterozygous and wild-type littermates. We counted on average 53 short atrial escape beats in Gnb5+/+ and 204 in Gnb5–/– animals over 24 hours (figure 4B–E, p=4.936e-06). Long atrial escape beats (figure 4C–F; 1 vs 117 events, p=7.792e-06) and atrioventricular blocks (figure 4D–G; 0.6 vs 30 events, p=0.04078) were similarly observed significantly more frequently in knockout animals. Few episodes of tachycardia followed by bradycardia and premature beats were also observed in homozygous knockouts.
Figure 4.
Cardiac arrhythmias recorded in Gnb5 mouse line. (A) Normal ECG trace recorded in wild-type male mice with the main spikes specified as used in the text. (B, C) Gnb5−/− male mice ECG traces showing escape atrial beats classified as short (B) and long (C) and characterised by the occurrence of a late P-wave (red arrow). (D) Gnb5−/ − male mice ECG trace demonstrating atrioventricular blocks, with more than one P-wave per QRS complex (consecutive red arrows). (E–G) Respective box plots indicating the number of arrhythmias, that is, the number of short (E) and long (F) escape atrial beats and atrioventricular blocks (G) per 24 hours. ns, not significant.
These results demonstrated that Gnb5–/– mice have a defect at the level of sinus node as well as cardiac conduction anomalies linked to the atrioventricular node. Of note, we did not assess for other anomalies.
Gnb5-deficient mice exhibit higher cholinergic sensitivity and normal sympathetic activity
Human homozygote carriers of GNB5 pathogenic variants show severe bradycardia at rest with a maximal HR unchanged during exercise. Zebrafish and human cell modelling supported predominantly parasympathetic modulation in the aetiology of HR disturbances in IDDCA individuals.1 7 To better assess the possible involvement of the autonomic innervation, we used an in vivo mammalian model system whose physiology is closer to humans. We used ECG telemetry to monitor HR and observed higher HR in baseline conditions (online supplemental figure S2C) possibly reflecting a higher rate of activity of the Gnb5–/– mice, as measured qualitatively here and reported previously22 (online supplemental figure S2D, E), or alternatively differences in the HR regulation.
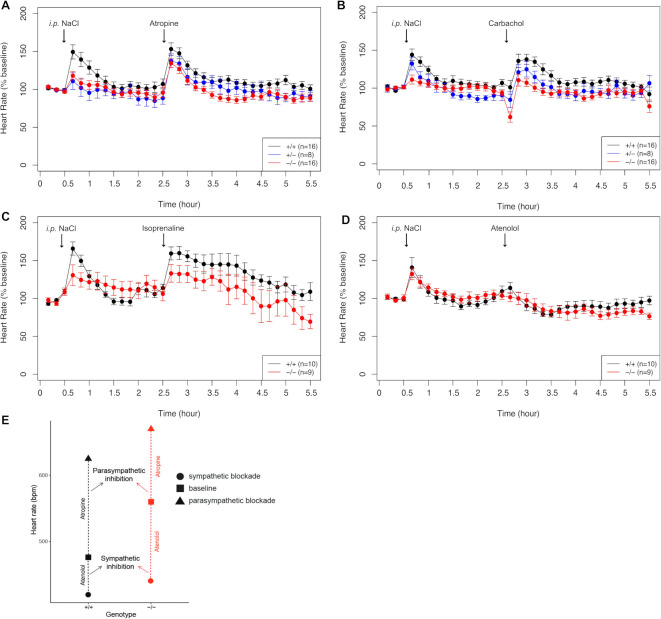
Parasympathetic blockade with atropine (1 mg/kg) had a positive chronotropic effect; that is, HR increased (HR Gnb5+/+ =690 beats/min±68, HR Gnb5+/– =700 beats/min±34, HR Gnb5–/– =758 beats/min±28 (p+/+ vs. +/–=0.63, p+/+ vs –/–=2.62E-03, p+/– vs –/–=2.38E-03)) (online supplemental figure S3A). In contrast, carbachol administration (0.1 mg/kg) triggered a rapid decrease of the HR in the three groups, with a significant effect in Gnb5–/– mice, whose HR dropped until 335 beats/min (HR Gnb5+/+ =448 beats/min±147, HR Gnb5+/– =415 beats/min±157, HR Gnb5–/– =335 beats/min±141 (p+/+ vs –/–=3E-02; online supplemental figure S3B). HR quickly recovered in all genotypes. The duration of the bradycardia was similar in the three groups (~1 hour). Moreover, since the baseline HR of knockout animals was higher, when expressed in relation to the basal values, the effect of atropine was not different in the three groups of mice (p+/+ vs +/–=0.22, p+/+ vs –/–=0.07, p+/– vs –/–=0.77; figure 5A), while the carbachol-induced bradycardia was more severe in Gnb5–/– (p=1.79E-03, figure 5B).
Figure 5.
Pharmacological administration of compounds mimicking parasympathetic and sympathetic stimulation. (A) HR monitoring after injection of atropin (intraperitoneal 1 mg/kg). (B) Bradycardia measured in response to carbachol (intraperitoneal 0.1 m/kg). (C) HR variation in response to atenolol (intraperitoneal 2 mg/kg). (D) Increased HR after isoprenaline administration (intraperitoneal 4 mg/kg). Data points are expressed as percentage of the baseline values. (E) Smaller parasympathetic and bigger sympathetic blockade in Gnb5−/− mice (red), compared with wild-type littermates (black), indicative of lower parasympathetic and higher sympathetic tones in basal conditions. HR, heart rate.
To mimic the sympathetic response and investigate a possible role of the β-adrenergic response in the heart rhythm perturbations of IDDCA syndrome, we challenged Gnb5+/+ and Gnb5–/– animals with either isoprenaline or atenolol. Injection of the sympathetic agonist isoprenaline (4 mg/kg) resulted in a prolonged (~1 hour) increase of HR, with values comparable in both genotypes (p=0.41, online supplemental figure S3C). HR slowly decreased to baseline; this reduction reached lower than baseline values in Gnb5–/– mice (online supplemental figure S3C). However, when expressed in percentage of the baseline, the tachycardia seemed stronger in wild-type (p=0.09, figure 5C).
The sympathetic antagonist atenolol (2 mg/kg) induced a similar decrease of HR in both groups (p=0.73, p=0.22 when compared with baseline (online supplemental figure S3D and figure 5D).
These results indicate that the Gnb5–/– mice bradycardia results from enhanced parasympathetic (cholinergic) stimulation/reflex. Our data also suggest an increased sympathetic activation when animals are under stress, that is, when they are challenged by the injection of drugs.
In contrast to individuals affected with IDDCA, Gnb5–/– mice showed a higher HR in baseline conditions (online supplemental figure S2C and S3A-D, first three data points). We hypothesised that such discrepancy could be linked to different autonomic nervous system tone between human and mouse.42 We therefore analysed the response to the administration of drugs to determine which of the parasympathetic and sympathetic system mostly influences baseline HR. Atropine-mediated parasympathetic inhibition induced an increase in HR with a variation from the baseline, which was smaller in knockout animals than in wild types (figure 5E), pinpointing that the parasympathetic tone is lower in Gnb5–/– mice. Conversely, the sympathetic blockade by atenolol induced a greater reduction of the HR in Gnb5–/– than control littermates (figure 5E) evocative of the greater sympathetic tone observed in basal conditions. Of note, the control responses mediated by NaCl injection also showed a difference in HR attributable to increased parasympathetic/sympathetic balance; the HR increase, due to the stress caused by the injection and animal handling, was less pronounced in Gnb5–/– mice. Our results suggest that higher basal HR in Gnb5–/– mice could be due to lower parasympathetic tone and higher sympathetic tone.
To further assess the functioning of cardiac autonomic regulation, we performed time domain analysis of HRV in Gnb5+/+ and Gnb5–/– mice. We measured NN, SDNN, RMSSD and pNN6 during light (day) and dark (night) phases in each genotype. Whereas all parameters differ in the wild-types, the knockouts showed no significant HRV differences between light and dark phases (table 2) confirming that differences in RR intervals (NN), and hence in HR, were greater in Gnb5+/+ (pNN=1.25E-08, table 2) than in Gnb5–/– mice (pNN=0.72, table 2). This suggests abnormal autonomic regulation, in particular via the parasympathetic system in the Gnb5 –/– mouse. Concordantly, RMSSD, which reflects short-term variations in HR, and pNN6, both measures of the parasympathetic nervous system regulation, were significantly different between night and day in Gnb5+/+ mice (pRMSSD=4.38E-05, ppNN6=9.71E-06; table 2), while they were almost unchanged in Gnb5 –/– mice (pRMSSD=0.65, ppNN6=0.9, table 2). Similarly, SDNN that indicates the total autonomic variability, fluctuated in Gnb5+/+ mice (pSDNN=1.16E-02, table 2) but not in Gnb5 –/– mice (pSDNN=0.61, table 2) at night.
Table 2.
Results of HRV analysis
| Genotype | Gnb5+/+ | Gnb5−/− | Gnb5+/+ vs Gnb5−/− | |||||||||||||
| Night/day | Night | Day | P value | Night | Day | P value | P value | P value | ||||||||
| Mean | SD | SEM | Mean | SD | SEM | Night versus day | Mean | SD | SEM | Mean | SD | SEM | Night versus day | Night | Day | |
| Mean RR (NN) | 112.27 | 10.32 | 2.66 | 136.81 | 13.25 | 3.31 | 1.25E-08 | 110.00 | 7.45 | 1.92 | 111.63 | 24.18 | 6.04 | 0.72 | 0.21 | 3.45E-03 |
| SDNN | 23.47 | 3.44 | 0.89 | 26.56 | 5.02 | 1.25 | 0.012 | 24.05 | 4.16 | 1.11 | 23.17 | 3.31 | 0.83 | 0.61 | 0.94 | 4.76E-02 |
| RMSSD | 4.77 | 1.74 | 0.45 | 6.54 | 2.10 | 0.53 | 4.38E-05 | 5.05 | 2.49 | 0.64 | 4.69 | 1.05 | 0.26 | 0.65 | 0.86 | 9.07E-03 |
| pNN6 | 13.81 | 8.96 | 2.31 | 24.13 | 10.49 | 2.62 | 9.71E-06 | 13.33 | 7.98 | 2.06 | 13.21 | 5.00 | 1.25 | 0.90 | 0.88 | 7.34E-04 |
HRV parameters measured in time-domain HRV analysis over 12-hour nocturnal and 12-hour diurnal intervals. Values are reported as mean; SD and SEM were calculated and reported. Standard t-test was used to assess differences between Gnb5+/+ and Gnb5−/− groups, as well as for differences between day and night. Description of the parameters used can be found in the Materials and Methods section.
HRV, heart rate variation; NN, mean R–R interval; RMSSD, square root of the mean square difference; SDNN, SD of all normal R–R intervals.
All parameters were significantly reduced during the light phase when comparing Gnb5+/+ and Gnb5 –/– mice (pNN=3.45E-03, pSDNN=4.76E-02, pRMSSD=9.07E-03, ppNN6=7.34E-04; table 2).
Taken together, these results demonstrated an impaired autonomic regulation in Gnb5 –/– mice, in particular a lower modulation of the parasympathetic activity.
Transcriptome analysis of Gnb5 mice
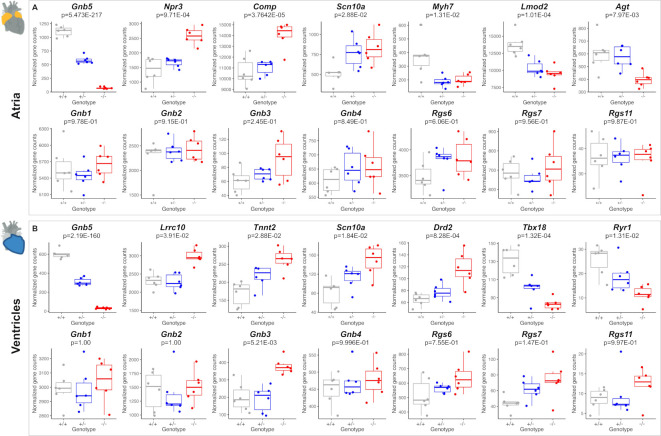
To explore the transcriptional consequences of Gnb5 loss in the heart, we profiled the transcriptomes of atria and ventricles of Gnb5 –/– , Gnb5+/– and wild-type male mice at 18 weeks of age using RNA-sequencing (RNA-seq) (online supplemental table S3). RNA-seq libraries were sequenced to a median depth of ~50 000 000 single-end reads per samples. Samples clustering (using Poisson model44) to define the global relationship among all samples, showed a very clear separation of atria and ventricles (online supplemental figure S4A) as well as of hippocampi, cerebellum and cerebral cortex (online supplemental figure S4B). Transcripts quantification confirmed that Gnb5, the orthologue of GNB5, is expressed in the brain (online supplemental figure S5) and cardiac tissues (atria and ventricles; figure 6A, B, top) of adult mice and that its expression levels correlated with gene dosage. We identified 98 significantly DEGs in atria (online supplemental table S4) and 63 in ventricles (online supplemental table S5) applying a false discovery rate method for multiple testing with a 5% threshold. Consistent with the phenotype described in Gnb5 –/– mice, we found altered expression of genes involved in cardiac muscle contractility, HR regulation and cardiac conduction. Among the upregulated genes falling into these categories in atria were Npr3 (p=9.71E-04), Comp (p=3.7E-05) and Scn10a (p=2.88E-02), while downregulated transcripts were Myh7 (p=1.31E-02), Lmod2 (p=1.04E-04) and Agt (p=7.97E-03) (figure 6A, top). Profiling of the ventricles demonstrated significant upregulation of Lrrc10 (p=3.91E-02), Tnnt2 (cardiac troponin, p=2.88E-02), Scn10a (p=1.84E-02) and Drd2 (p=8.28E-04) (figure 6B, top), and reduced expression of Tbx18 (p=1.32E-04). Notably, the expression level of TNNT2 gene was not modified in GNB5-Ser81Leu hiPSC.7 Correspondingly, nominally enriched gene sets were associated with development of the cardiac conduction system (GO:0003161, online supplemental table S6, S7), regulation of HR (GO:0002027, online supplemental table S6, S7) and cardiac muscle contraction (GO:0060047, GO:0060048; online supplemental table S6, S7). MYH7 and TNNT2 encode sarcomeric proteins, essential for contraction and relaxation of the heart muscle, and mutations in these genes have been linked to cardiomyopathy.45
Figure 6.
Comparison of transcriptome profiles of atria and ventricles in Gnb5+/+ , Gnb5+/ and Gnb5−/− male mice. (A) Atrial expression profiles of Gnb5 (top, left) and other DEGs (top Lane); Gnb and Rgs transcripts quantification in atria (bottom lane). (B) Ventricular expression profiles of Gnb5 gene (top, left) and other DEGs (top lane); Gnb and Rgs transcripts quantification in ventricles (bottom). DEG, differentially expressed gene.
Scn10a encodes the voltage-gated sodium channel NaV1.8. Its human orthologue was associated with HR regulation and genome-wide association studies suggested it as a modulator of PR interval duration, that is, atrial conduction time.46 Lrrc10 and Lmod2 are associated with dilated cardiomyopathy in both human and mice47–50; while Lrrc10 functions in the sarcomeric Z-disc and the T-tubule components, involved in muscle contraction,47 loss of functional Lmod2 has been linked to short thin filaments and reduction in maximum calcium-activated force production.48 Rodent ventricular cardiomyocytes are converted into spontaneous firing cells (eg, sinoatrial-node pacemaker cells) by expression of the transcription factor Tbx18,51 whereas a polymorphism in DRD2 has been associated with motor learning and HR.52 RYR1 has a dominant role in muscles contractility, even though it is more prominently associated with skeletal muscle.53 Common variants in RYR1 are associated with left ventricular hypertrophy.54
Given that (1) Gnb5 has four paralogous genes, with ~50% sequence identity55 56; (2) the respective encoded proteins may have redundant function; (3) the propagation of the Gnb5-mediated signal is controlled by RGS15 16 57; and (4) Gnb5 and Rgs (ie, Rgs6 and Rgs7) are coexpressed both at RNA (online supplemental figure S6A, B) and protein levels,17 we investigated the expression patterns of these two families of genes in our transcriptome profiles to investigate possible compensatory mechanisms. We found significant upregulation of Gnb3 in ventricles (figure 6B, bottom, p=5.21E-03) and a trend toward increased expression level in atria (figure 6A, bottom, p=0.24). The genes encoding the other Gβ subunits, named Gnb1, Gnb2 and Gnb4, did not change expression (figure 6A, B, bottom). In ventricles, the three R7-Rgs expressed in heart, Rgs6, Rgs7 and Rgs11, showed a trend towards increased expression when Gnb5 was knocked out (figure 6B, bottom). Only Rgs6 showed a similar trend in atria (figure 6A, bottom). Gnb5+/− mice showed no or very subtle changes of their transcriptome compared with controls; we detected only six DEGs in atria (online supplemental table S8) and one in ventricles (online supplemental table S9) beside the engineered Gnb5. Profiling Gnb5 protein expression revealed detectable levels in the heart of wild-type mice, along with abundant presence in three brain regions (cortex, cerebellum and hippocampi), while immunoblotting of Gnb5−/− tissues verified the complete absence of the protein (online supplemental figure S6C). Notably, we detected no compensatory changes in either Gnb3 or Rgs7 levels; the two proteins were undetectable in Gnb5−/− atria and ventricles (online supplemental figure S6C). This suggests that Gnb3 protein stability may be decreased and further demonstrates that the physical association with Gnb5 is critical for the stability of the Gnb5/R7-Rgs complex, as previously described for Rgs6.17 Gnb5 and Rgs7 proteins were enriched in atria compared with ventricles in wild-type mice (online supplemental figure S6C).
We further investigated transcriptome signatures in three brain regions relevant to the IDDCA pathology: the cortex because of its role in higher cognitive function, the hippocampus as it participates in the formation of long-term and spatial memories and the cerebellum for its motor control and language processing. Cerebellar and hippocampal anomalies were documented in Gnb5 knockout pups.22 Overall, we observed 206 DEGs in cerebellum, 105 in hippocampus and 48 in cerebral cortex (at false discovery rate (FDR) 5%) (online supplemental table S10-S12). These genes resulted in over-representation of gene ontology (GO) terms involved in regulation of excitatory postsynaptic membrane potential (GO:0060079), regulation of neurotransmitters secretion (GO:0046928), learning (GO:0007612) and synapse organisation (GO:0050808), regulation of inositol 3-phosphate (GO:0014065), regulation of cAMP-mediated signalling (GO:0043951) (in cerebellum, online supplemental table S13); visual perception (GO:0007601), phototransduction (GO:007602), guanylate cyclase activity (GO:0031282) and regulator of signalling receptors (GO:2000272) (in hippocampus, online supplemental table S14); and sensory perception of light stimuli (GO:0050953) and lens development (GO:0002088) (in cortex, online supplemental table S15).
Quantitative RT-PCR and microarray data previously indicated altered expression levels of genes implicated in neuronal development and function, such as down-regulation of Grid2 (glutamate ionotropic receptor, delta 2) and upregulation of Synpo (synaptopodin),22 as well as increased expression of Guca1a and Guca1b, calcium-binding protein activating photoreceptor guanylate cyclases. Guca1a (phippocampus=1.81E-03) and Guca1b (pcerebellum=1.56E-302, phippocampus=1.9589E-196, pcortex=2.209E-178) were DEGs in the brain of Gnb5 knockout mice; they showed significant upregulation (online supplemental figure S5). Synpo showed a trend of increased expression in the hippocampus (online supplemental figure S5B, p=0.55).
A subset of additional DEGs included Grin2a (p=1.044E-06), Snca (p=7.96E-03), Gnai2 (p=7.79E-03) and Kcnj2 (p=0.04), in the cerebellum (online supplemental figure S5A), Hes5 (p=0.04) and Sox6 (p=0.02) in the hippocampus (online supplemental figure S5B) and Ntrk3 (p=9.69E-04) and Cacna2d4 (p=0.03) in the cortex (online supplemental figure S5C). The glutamate-gated ion channel Grin2a protein plays important roles in long-term potentiation and in efficient synaptic transmission. Disruption of this gene is associated with focal epilepsy and speech disorder with or without cognitive disability.58 Hes5 and Sox6 genes, respectively, are a transcriptional repressor and activator, required for the regulation of transition timing of neurogenesis and gliogenesis in mammalian neocortical development59 and in the normal development of the central nervous system.60 Cacna2d4 encodes for a calcium channel whose mutations are associated with retinal dysfunction in human.61 The expression of transcripts encoding for different Gβ subunits and R7-RGS genes remained unchanged (online supplemental figure S5 A-C, bottom), suggesting that the brain could be less proficient than the heart in compensating dysregulated pathways. Rgs7 protein level was greatly reduced in Gnb5−/− brain, further emphasising the Gnb5/R7-Rgs codependence (online supplemental figure S6C).
Discussion
The advent of high-throughput sequencing allowed identification of the cause of hundreds of Mendelian diseases,62 in particular those involving intellectual disability.63 For example, data aggregation of exome sequencing uncovered a link between variants in genes encoding the Gβ subunits of the heterotrimeric G-proteins with a group of neuropsychiatric conditions with cardiac manifestations and ophthalmic pathologies.1–4 6 8–10 64–79 Mutations in GNB5, encoding the divergent Gβ5 subunit of the guanine–nucleotide binding protein family, were recently shown to be causative of the autosomal recessive IDDCA syndrome. Differently from Gβ1–4, Gβ5 forms irreversible dimer with the G-protein γ-like domain80 present in the R7 regulator group of G-protein signalling proteins (R7 RGS). Dimerisation with Gβ5 is absolutely required for the stability of the R7 RGS proteins16.
Genotype–phenotype correlation showed that carriers of truncating GNB5 variants present with the severe form of this syndrome, while missense alleles are associated with milder phenotypical manifestations. Recently, an individual carrying a homozygous deletion spanning three genes, BCL2L, GNB5 and MYO5C, was reported to display a phenotype which overlaps significantly with the IDDCA manifestations,81 further confirming that this phenotype is associated with loss of function of GNB5. In this work, we increased the number of reported individuals with IDDCA and associated variants, through identification of nine additional families and five novel causative variants. The core symptoms of IDDCA include cognitive disability, epilepsy, retinopathy and cardiac arrhythmia.1–10 This is a life-long condition with a risk of sudden death. Six of the seven deceased IDDCA individuals are suspected to have died from a sudden cardiac arrest secondary to an arrhythmia. Although the mutational spectrum of this syndrome has already been partly defined, the overall molecular mechanism by which perturbations of GNB5 translate into IDDCA phenotypical manifestations remains unclear. Modelling in zebrafish and cardiomyocytes differentiated from human iPSCs provided some initial answers1 7; however, we thought that the mouse would be a more appropriate tool to investigate consequences of this human syndromic neurodevelopmental condition. As little was known about possible cardiac conduction anomalies in mice, we characterised both the cardiac phenotype and molecular outcome of knocking-out Gnb5. Gnb5−/− animals were smaller and had smaller ventricles, and therefore the chambers would contain less blood as a consequence. However, they exhibited an increase in fractional shortening and ejection fraction, a sign of compensatory cardiac function. The quantity of blood ejected by the ventricles, as defined by the cardiac output and stroke volume, remained unchanged between Gnb5−/− and Gnb5+/+ mice, indicating that Gnb5−/− heart has adapted to ensure that it efficiently meets the body’s demands for perfusion.
ECG measurements showed that, conversely to IDDCA affected individuals who present with severe bradycardia at rest, Gnb5−/− mice have higher HRs in basal condition, especially during the day, the mouse sleeping phase. Zebrafish knocked-out for all gnb5a and gnb5b copies present in the fish genome, showed a similar trend of increased basal HR.1 It is plausible that these differences are linked to a lower parasympathetic tone in fish and rodents compared with humans. Of note, we showed that the parasympathetic and sympathetic tones of Gnb5 −/− mice were, respectively, lower and higher than those of controls (figure 5E), thus possibly influencing the basal HR. Whereas we cannot conclude that this higher basal HR is exclusively due to changes in HR regulation as we and others observed an increase in activity of Gnb5 −/− mice21 22 (online supplemental figure S2D), the zebrafish model showed no sign of hyperactivity.1 In normal conditions, HR is balanced by the synergic interactions between the sympathetic and parasympathetic nervous systems. Our HRV analysis, aimed at assessing such sympathovagal balance, confirmed a reduced modulation of the parasympathetic stimulation in the heart of Gnb5 −/− mice (table 2).
ECG recordings additionally revealed a high number of arrhythmias in knockout animals, including escape beats, tachycardia/bradycardia episodes and atrioventricular block. These sinus arrhythmias and conduction problems, reminiscent of arrhythmias observed in IDDCA subjects, further corroborate the involvement of Gnb5 gene in altered cardiac function and irregular heartbeat. Finally, our mice study showed that Gnb5-inhibitory signalling is essential for the parasympathetic control of the HR as suggested by previous studies in other models.1 7 Knockout mice treated with a parasympathomimetic presented with bradycardia, while injection of an antiparasympathetic drug atropine had the same effect as in wild type. The β-adrenergic activity of Gnb5 −/− mice was unaltered, suggesting a normal sympathetic modulation of the cardiac stimulation, and further confirming that the more efficient cardiac function is an adaptation to counteract the reduced size of the Gnb5 −/− heart.
Consistent with this observation, transcriptome profiling provided insights into modifications of cardiac contraction properties, along with reduced ventricular expression of genes required for development of pacemaker cells. Instead of specific signals (eg, pacemaker function genes in atria), genes related to heart muscle contraction (eg, Myh7 and Lmod2 in atria, Tnnt2 and Lrrc10 in ventricles) and conduction system functions (eg, Agt and Npr3 in atria, Tbx18 in ventricles and Snc10a in both) were identified as DEGs in both atria and ventricles in Gnb5 −/− animals. These transcriptome alterations may shed light on the mechanisms that result in the pathology of IDDCA. Approaches like single-cell RNA-sequencing combined with the isolation of sinoatrial node cells are warranted to complement our findings in the future. Globally, these data further challenge our study, since we cannot unravel whether the transcriptome modifications are cause or consequence of the augmented contractility. We argue, in line with the aforementioned results, that we are possibly investigating a compensatory mechanism. Interestingly, mice lacking Gnb5 overexpress Gnb3, a gene encoding a different Gβ subunit and involved in the activation of GIRK channels.82 In contrast to Gnb1 and Gnb4 that have a well-documented role in the nervous system,76 83 Gnb3-null mice presented with cardiac manifestations, including slower HRs. However, their isolated hearts responded equivalently to muscarinic receptor-adrenergic and β-adrenergic receptor-stimulations, thus suggesting that Gnb3 is unlikely to be involved directly in the G-protein signalling controlling heart pacemaker activity.84 Nonetheless, its higher expression may play a role in coping with the loss of Gnb5; further studies are warranted to elucidate such compensatory mechanisms. The expression of Gnb2 is unchanged in the cardiac tissue of Gnb5 knockouts; however, it was also linked to heart functions in mouse and human66 69 with Gnb2 knockout mice having increased HR, and ECG trace revealing shortened RR and PQ intervals and ST segment (https://www.mousephenotype.org/data/genes/MGI:95784).85
The transcriptome data provided molecular data supporting an increased cardiac function of Gnb5 −/− mice. It probably results from both a compensatory mechanism, as shown by misexpression of genes involved in cardiac muscle contractility, and an alteration of the expression of genes involved in the regulation of the HR. We also documented transcriptional changes in three brain regions of knockout mice which correlate with the IDDCA neuropsychiatric disease spectrum.
HR regulation by the parasympathetic and sympathetic branches of the autonomic nervous system takes place in the pacemaker cells of the sinoatrial node. While sympathetic modulation increases pacemaker cell firing rate, the vagal parasympathetic activity decreases the HR. Both activities are mediated by G-protein-coupled signalling on β-adrenergic (sympathetic) and cholinergic M2 muscarinic (parasympathetic) receptors, respectively. Autonomic innervation is regulated in vivo by RGS proteins. In particular, in the heart, the M2 receptor signalling is mediated by the Rgs6–Gnb5 complex.17 As Gnb5 and Rgs genes are coexpressed and as the physical association of the encoded proteins is critical for complex stability, we expected to observe reduced Rgs expression levels. In a previous report,17 no Rgs6 was detected in the atria and in the brain of Gnb5 knockout mice. Surprisingly, we detected a trend of increased expression of Rgs subunits, in particular Rgs6, contrary to findings in reference7 where expression of RGS6 was not different between GNB5-Ser81Leu and wild-type hiPSCs. These discrepancies may stem from the fact that we are investigating different model systems, that is, mouse tissues versus human cells, as well as by the use of different technologies to detect mRNA abundance; that is, RNA-sequencing versus qRT-PCR and that in case of hiPSCs data were generated from one clone per genotype.
Altogether, our results unveil that potentially compensatory changes occur at a transcriptional level in Gnb5 −/− mice through higher expression of Gnb3 and Rgs genes in heart tissues. Additional evidence at protein level did not confirm compensatory changes. The previously cited link between GNB5 and the R7-RGS RGS6 and RGS11 suggests that they are part of the same pathway. Consistent with this hypothesis, the members of a Tunisian family presenting with cataract, mental retardation and microcephaly were carriers of biallelic mutations in RGS6 86. Another unpublished case harbouring mutation in RGS11 presented with overlapping neuropsychiatric phenotype (Sarah Montgomery, in litt). No specific cardiac evaluation is reported in these individuals, but our data suggest that this should be considered. Co-occurrence of neuropsychiatric symptoms with visual and cardiac manifestations could represent a unique combination associated with Gβ and the R7-RGS related proteins. We hypothesise that the disease mechanism responsible for HR perturbations is the reduction or the loss of negative regulation by Rgs6 on the inhibitory Gβ5 signalling, resulting in enhanced parasympathetic activity.
Overall, our work highlights that Gnb5 −/− mice not only recapitulate IDDCA’s neurological manifestations previously outlined elsewhere22 but also mimic its cardiac perturbations, especially regarding heart arrhythmias and autonomic nervous system control, allowing for future screening of drugs modulating the parasympathetic branch of the autonomic nervous system, in view of the development of patients’ therapy.
Web resources
GnomAD: https://gnomad.broadinstitute.org/about
IMPC: http://www.mousephenotype.org/MutationTaster2: http://www.mutationtaster.org/
PolyPhen-2: http://genetics.bwh.harvard.edu/pph2/index.shtml
PROVEAN: http://provean.jcvi.org/index.phpSIFT: http://sift.jcvi.org/
UMD Predictor: http://umd-predictor.eu/
Functional Analysis through Hidden Markov Models V.2.3: http://fathmm.biocompute.org.uk/
Splice Site Prediction by Neural Network (NNSplice): https://www.fruitfly.org/seq_tools/splice.html
NetGene2 server: http://www.cbs.dtu.dk/services/NetGene2/
Human Splicing Finder: http://www.umd.be/HSF/
CADD: https://cadd.gs.washington.edu/
Greater Middle East Variome Project: http://igm.ucsd.edu/gme/
Accession Numbers
The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE156898.
jmedgenet-2020-107015supp001.pdf (66.5KB, pdf)
jmedgenet-2020-107015supp005.pdf (60.8KB, pdf)
Acknowledgments
We thank the probands and their families for their participation in this study and Jacques S Beckmann for comments. We are grateful to the Genomics Technologies Facility of the University of Lausanne and the Transgenic Core Facility of the EPFL.
Footnotes
Twitter: @EDN_88, @NicoChatron
Collaborators: SYNAPS Study Group: Stanislav Groppa, Blagovesta Marinova Karashova, Wolfgang Nachbauer, Sylvia Boesch, Larissa Arning, Dagmar Timmann, Bru Cormand, Belen Pérez-Dueñas, Jatinder S Goraya, Tipu Sultan, Jun Mine, Daniela Avdjieva, Hadil Kathom, Radka Tincheva, Selina Banu, Mercedes Pineda-Marfa, Pierangelo Veggiotti, Michel D. Ferrari, Arn M. J. M. van den Maagdenberg, Alberto Verrotti, Giangluigi Marseglia, Salvatore Savasta, Mayte García-Silva, Alfons Macaya Ruiz, Barbara Garavaglia, Eugenia Borgione, Simona Portaro, Benigno Monteagudo Sanchez, Richard Boles, Savvas Papacostas, Michail Vikelis, Eleni Zamba Papanicolaou, Efthymios Dardiotis, Shazia Maqbool, Shahnaz Ibrahim, Salman Kirmani, Nuzhat Noureen Rana, Osama Atawneh, George Koutsis, Salvatore Mangano, Carmela Scuderi, Eugenia Borgione, Giovanna Morello, Tanya Stojkovic, Massimo Zollo, Gali Heimer, Yves A. Dauvilliers, Pasquale Striano, Issam Al-Khawaja, Fuad Al-Mutairi, Hamed Sherifa.
Contributors: PDN and AR conceived and directed the study. SE, AP, TS, JRA, ZuR, FZ, NR, FR, NA, SM, MSZ, JGG, DM, HG, GS, NM, ASe, GL, NC, VDS, MC, RM and HH recruited the patients, gathered clinical information, prepared samples, and performed whole-exome and mutational analysis. PDN, ASa and JC carried out the other experiments. NG performed structural modeling of GNB5 variants. ASa, TP, PDN and AR designed the mouse experiments. ASa, with the help of PDN, conducted the mouse experiments. PDN and ASa performed statistical analysis and analysed the data. WFS provided Gnb5 knockout mouse model. PDN and AR wrote the manuscript. All authors reviewed and approved the manuscript.
Funding: This work was supported by grants from the Swiss National Science Foundation (31003A_182632) and the Jérôme Lejeune Foundation to AR, the MRC (MR/S01165X/1, MR/S005021/1, G0601943), Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK) and Muscular Dystrophy Association (MDA USA) to HH. The families (R-V and Z) were collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA). Their assessment was conducted as part of the Queen Square Genomics group at University College London, with support from the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Author note: The Twitter account of Pasquelena De Nittis is wrong it is "@elena_denittis"
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
Collaborators: SYNAPS Study Group, Stanislav Groppa, Blagovesta Marinova Karashova, Wolfgang Nachbauer, Sylvia Boesch, Larissa Arning, Dagmar Timmann, Bru Cormand, Belen Pérez-Dueñas, Jatinder S Goraya, Tipu Sultan, Jun Mine, Daniela Avdjieva, Hadil Kathom, Radka Tincheva, Selina Banu, Mercedes Pineda-Marfa, Pierangelo Veggiotti, Michel D Ferrari, Arn M J M van den Maagdenberg, Alberto Verrotti, Giangluigi Marseglia, Salvatore Savasta, Mayte García-Silva, Alfons Macaya Ruiz, Barbara Garavaglia, Eugenia Borgione, Simona Portaro, Benigno Monteagudo Sanchez, Richard Boles, Savvas Papacostas, Michail Vikelis, Eleni Zamba Papanicolaou, Efthymios Dardiotis, Shazia Maqbool, Shahnaz Ibrahim, Salman Kirmani, Nuzhat Noureen Rana, Osama Atawneh, George Koutsis, Salvatore Mangano, Carmela Scuderi, Eugenia Borgione, Giovanna Morello, Tanya Stojkovic, Massimo Zollo, Gali Heimer, Yves A Dauvilliers, Pasquale Striano, Issam Al-Khawaja, Fuad Al-Mutairi, and Hamed Sherifa
Data availability statement
Data are available in a public, open access repository. The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE156898.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Maintenance and mouse experimental procedures were approved by institutional and Swiss Federal Veterinary Office (experimental protocol VD3289.c).
References
- 1. Lodder EM, De Nittis P, Koopman CD, Wiszniewski W, Moura de Souza CF, Lahrouchi N, Guex N, Napolioni V, Tessadori F, Beekman L, Nannenberg EA, Boualla L, Blom NA, de Graaff W, Kamermans M, Cocciadiferro D, Malerba N, Mandriani B, Coban Akdemir ZH, Fish RJ, Eldomery MK, Ratbi I, Wilde AAM, de Boer T, Simonds WF, Neerman-Arbez M, Sutton VR, Kok F, Lupski JR, Reymond A, Bezzina CR, Bakkers J, Merla G. GNB5 mutations cause an autosomal-recessive multisystem syndrome with sinus bradycardia and cognitive disability. Am J Hum Genet 2016;99:786. 10.1016/j.ajhg.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shamseldin HE, Masuho I, Alenizi A, Alyamani S, Patil DN, Ibrahim N, Martemyanov KA, Alkuraya FS. GNB5 mutation causes a novel neuropsychiatric disorder featuring attention deficit hyperactivity disorder, severely impaired language development and normal cognition. Genome Biol 2016;17:195. 10.1186/s13059-016-1061-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Malerba N, Towner S, Keating K, Squeo GM, Wilson W, Merla G. A NGS-Targeted Autism/ID panel reveals compound heterozygous GNB5 variants in a novel patient. Front Genet 2018;9:626. 10.3389/fgene.2018.00626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poke G, King C, Muir A, de Valles-Ibáñez G, Germano M, Moura de Souza CF, Fung J, Chung B, Fung CW, Mignot C, Ilea A, Keren B, Vermersch A-I, Davis S, Stanley T, Moharir M, Kannu P, Shao Z, Malerba N, Merla G, Mefford HC, Scheffer IE, Sadleir LG. The epileptology of GNB5 encephalopathy. Epilepsia 2019;60:e121–7. 10.1111/epi.16372 [DOI] [PubMed] [Google Scholar]
- 5. Shao Z, Tumber A, Maynes J, Tavares E, Kannu P, Heon E, Vincent A. Unique retinal signaling defect in GNB5-related disease. Doc Ophthalmol 2020;140:273-277. 10.1007/s10633-019-09735-1 [DOI] [PubMed] [Google Scholar]
- 6. Turkdogan D, Usluer S, Akalin F, Agyuz U, Aslan ES. Familial early infantile epileptic encephalopathy and cardiac conduction disorder: a rare cause of SUDEP in infancy. Seizure 2017;50:171–2. 10.1016/j.seizure.2017.06.019 [DOI] [PubMed] [Google Scholar]
- 7. Veerman CC, Mengarelli I, Koopman CD, Wilders R, van Amersfoorth SC, Bakker D, Wolswinkel R, Hababa M, de Boer TP, Guan K, Milnes J, Lodder EM, Bakkers J, Verkerk AO, Bezzina CR. Genetic variation in i>i>GNB5i>i> causes bradycardia by augmenting the cholinergic response via increased acetylcholine-activated potassium current (i>Ii>K, ACh). Dis Model Mech 2019;12. 10.1242/dmm.037994. [Epub ahead of print: 09 07 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vernon H, Cohen J, De Nittis P, Fatemi A, McClellan R, Goldstein A, Malerba N, Guex N, Reymond A, Merla G. Intellectual developmental disorder with cardiac arrhythmia syndrome in a child with compound heterozygous GNB5 variants. Clin Genet 2018;93:1254–6. 10.1111/cge.13194 [DOI] [PubMed] [Google Scholar]
- 9. Tang M, Wang Y, Xu Y, Tong W, Jin D, Yang X-A. IDDCA syndrome in a Chinese infant due to GNB5 biallelic mutations. J Hum Genet 2020;65:627-631. 10.1038/s10038-020-0742-x [DOI] [PubMed] [Google Scholar]
- 10. Yazdani S, Badjatiya A, Dorrani N, Lee H, Grody WW, Nelson SF, Dipple KM. Genetic characterization and long-term management of severely affected siblings with intellectual developmental disorder with cardiac arrhythmia syndrome. Mol Genet Metab Rep 2020;23:100582. 10.1016/j.ymgmr.2020.100582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nini L, Zhang J-H, Pandey M, Panicker LM, Simonds WF. Expression of the Gβ5/R7-RGS protein complex in pituitary and pancreatic islet cells. Endocrine 2012;42:214–7. 10.1007/s12020-012-9611-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sánchez-Blázquez P, Rodríguez-Díaz M, López-Fando A, Rodríguez-Muñoz M, Garzón J. The GBeta5 subunit that associates with the R7 subfamily of RGS proteins regulates mu-opioid effects. Neuropharmacology 2003;45:82–95. 10.1016/s0028-3908(03)00149-7 [DOI] [PubMed] [Google Scholar]
- 13. Sondek J, Siderovski DP, gamma-like G. Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem Pharmacol 2001;61:1329–37. 10.1016/s0006-2952(01)00633-5 [DOI] [PubMed] [Google Scholar]
- 14. Witherow DS, Slepak VZ. A novel kind of G protein heterodimer: the G beta 5-RGS complex. Receptor Channel 2003;9:205–12. [PubMed] [Google Scholar]
- 15. Xie K, Allen KL, Kourrich S, Colón-Saez J, Thomas MJ, Wickman K, Martemyanov KA. Gbeta5 recruits R7 RGS proteins to GIRK channels to regulate the timing of neuronal inhibitory signaling. Nat Neurosci 2010;13:661–3. 10.1038/nn.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie K, Ge S, Collins VE, Haynes CL, Renner KJ, Meisel RL, Lujan R, Martemyanov KA. Gβ5-RGS complexes are gatekeepers of hyperactivity involved in control of multiple neurotransmitter systems. Psychopharmacology 2012;219:823–34. 10.1007/s00213-011-2409-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Posokhova E, Wydeven N, Allen KL, Wickman K, Martemyanov KA. RGS6/Gβ5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ Res 2010;107:1350–4. 10.1161/CIRCRESAHA.110.224212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen C-K, Eversole-Cire P, Zhang H, Mancino V, Chen Y-J, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A 2003;100:6604–9. 10.1073/pnas.0631825100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krispel CM, Chen C-K, Simon MI, Burns ME. Prolonged photoresponses and defective adaptation in rods of Gbeta5-/- mice. J Neurosci 2003;23:6965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rao A, Dallman R, Henderson S, Chen C-K. Gbeta5 is required for normal light responses and morphology of retinal on-bipolar cells. J Neurosci 2007;27:14199–204. 10.1523/JNEUROSCI.4934-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Levay K, Chanturiya T, Dvoriantchikova G, Anderson KL, Bianco SDC, Ueta CB, Molano RD, Pileggi A, Gurevich EV, Gavrilova O, Slepak VZ. Targeted deletion of one or two copies of the G protein β subunit Gβ5 gene has distinct effects on body weight and behavior in mice. Faseb J 2011;25:3949–57. 10.1096/fj.11-190157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J-H, Pandey M, Seigneur EM, Panicker LM, Koo L, Schwartz OM, Chen W, Chen C-K, Simonds WF. Knockout of G protein β5 impairs brain development and causes multiple neurologic abnormalities in mice. J Neurochem 2011;119:544–54. 10.1111/j.1471-4159.2011.07457.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mencacci NE, Kamsteeg E-J, Nakashima K, R'Bibo L, Lynch DS, Balint B, Willemsen MAAP, Adams ME, Wiethoff S, Suzuki K, Davies CH, Ng J, Meyer E, Veneziano L, Giunti P, Hughes D, Raymond FL, Carecchio M, Zorzi G, Nardocci N, Barzaghi C, Garavaglia B, Salpietro V, Hardy J, Pittman AM, Houlden H, Kurian MA, Kimura H, Vissers LELM, Wood NW, Bhatia KP. De novo mutations in PDE10A cause childhood-onset chorea with bilateral striatal lesions. Am J Hum Genet 2016;98:763–71. 10.1016/j.ajhg.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andrews S. FastQC: a quality control tool for high throughput sequence data, 2010. [Google Scholar]
- 25. Makrythanasis P, Maroofian R, Stray-Pedersen A, Musaev D, Zaki MS, Mahmoud IG, Selim L, Elbadawy A, Jhangiani SN, Coban Akdemir ZH, Gambin T, Sorte HS, Heiberg A, McEvoy-Venneri J, James KN, Stanley V, Belandres D, Guipponi M, Santoni FA, Ahangari N, Tara F, Doosti M, Iwaszkiewicz J, Zoete V, Backe PH, Hamamy H, Gleeson JG, Lupski JR, Karimiani EG, Antonarakis SE. Biallelic variants in KIF14 cause intellectual disability with microcephaly. Eur J Hum Genet 2018;26:330–9. 10.1038/s41431-017-0088-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000;29:52–4. 10.2144/00291bm09 [DOI] [PubMed] [Google Scholar]
- 27. Mitchell GF, Jeron A, Koren G. Measurement of heart rate and Q-T interval in the conscious mouse. Am J Physiol 1998;274:H747–51. 10.1152/ajpheart.1998.274.3.H747 [DOI] [PubMed] [Google Scholar]
- 28. Thireau J, Zhang BL, Poisson D, Babuty D. Heart rate variability in mice: a theoretical and practical guide. Exp Physiol 2008;93:83–94. 10.1113/expphysiol.2007.040733 [DOI] [PubMed] [Google Scholar]
- 29. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 2011;17. [Google Scholar]
- 30. Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 2013;63:41–9. 10.1016/j.ymeth.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L, Wang S, Li W. RSeQC: quality control of RNA-Seq experiments. Bioinformatics 2012;28:2184–5. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 34. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat 2015;36:425–31. 10.1002/humu.22769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015;36:928–30. 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP. Variation across141,456 human exomes and genomes reveals the spectrum of loss-of-functionintolerance across human protein-coding genes. bioRxiv 2019;531210. [Google Scholar]
- 40. Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat Struct Mol Biol 2008;15:155–62. 10.1038/nsmb.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997;18:2714–23. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- 42. Kurhanewicz N, Ledbetter A, Farraj A, Hazari M. TRPA1 mediates the cardiac effects of acrolein through parasympathetic dominance but also sympathetic modulation in mice. Toxicol Appl Pharmacol 2018;347:104–14. 10.1016/j.taap.2018.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, Gabriel SB, Belkadi A, Boisson B, Abel L, Clark AG, Alkuraya FS, Casanova J-L, Gleeson JG, Greater Middle East Variome Consortium . Characterization of greater middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 2016;48:1071–6. 10.1038/ng.3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Witten DM. Classification and clustering of sequencing data using a poisson model. Ann Appl Stat 2011;5:2493–518. 10.1214/11-AOAS493 [DOI] [Google Scholar]
- 45. Frey N, Luedde M, Katus HA. Mechanisms of disease: hypertrophic cardiomyopathy. Nat Rev Cardiol 2011;9:91–100. 10.1038/nrcardio.2011.159 [DOI] [PubMed] [Google Scholar]
- 46. Chambers JC, Zhao J, Terracciano CMN, Bezzina CR, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi JS, Kooner MK, Deng G, Siedlecka U, Parasramka S, El-Hamamsy I, Wass MN, Dekker LRC, de Jong JSSG, Sternberg MJE, McKenna W, Severs NJ, de Silva R, Wilde AAM, Anand P, Yacoub M, Scott J, Elliott P, Wood JN, Kooner JS. Genetic variation in SCN10A influences cardiac conduction. Nat Genet 2010;42:149–80. 10.1038/ng.516 [DOI] [PubMed] [Google Scholar]
- 47. Brody MJ, Lee Y. The role of leucine-rich repeat containing protein 10 (LRRC10) in dilated cardiomyopathy. Front Physiol 2016;7:337. 10.3389/fphys.2016.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pappas CT, Mayfield RM, Henderson C, Jamilpour N, Cover C, Hernandez Z, Hutchinson KR, Chu M, Nam K-H, Valdez JM, Wong PK, Granzier HL, Gregorio CC. Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality. Proc Natl Acad Sci U S A 2015;112:13573–8. 10.1073/pnas.1508273112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brody MJ, Hacker TA, Patel JR, Feng L, Sadoshima J, Tevosian SG, Balijepalli RC, Moss RL, Lee Y. Ablation of the cardiac-specific gene leucine-rich repeat containing 10 (Lrrc10) results in dilated cardiomyopathy. PLoS One 2012;7:e51621. 10.1371/journal.pone.0051621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ahrens-Nicklas RC, Pappas CT, Farman GP, Mayfield RM, Larrinaga TM, Medne L, Ritter A, Krantz ID, Murali C, Lin KY, Berger JH, Yum SW, Carreon CK, Gregorio CC. Disruption of cardiac thin filament assembly arising from a mutation in LMOD2: A novel mechanism of neonatal dilated cardiomyopathy. Sci Adv 2019;5:eaax2066. 10.1126/sciadv.aax2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kapoor N, Liang W, Marbán E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol 2013;31:54–62. 10.1038/nbt.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huertas E, Bühler K-M, Echeverry-Alzate V, Giménez T, López-Moreno JA. C957T polymorphism of the dopamine D2 receptor gene is associated with motor learning and heart rate. Genes Brain Behav 2012;11:677–83. 10.1111/j.1601-183X.2012.00793.x [DOI] [PubMed] [Google Scholar]
- 53. Hernández-Ochoa EO, Pratt SJP, Lovering RM, Schneider MF. Critical role of intracellular RyR1 calcium release channels in skeletal muscle function and disease. Front Physiol 2015;6:420. 10.3389/fphys.2015.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hong K-W, Shin D-J, Lee S-H, Son N-H, Go M-J, Lim J-E, Shin C, Jang Y, Oh B. Common variants in RyR1 are associated with left ventricular hypertrophy assessed by electrocardiogram. Eur Heart J 2012;33:1250–6. 10.1093/eurheartj/ehr267 [DOI] [PubMed] [Google Scholar]
- 55. Watson AJ, Aragay AM, Slepak VZ, Simon MI. A novel form of the G protein beta subunit Gbeta5 is specifically expressed in the vertebrate retina. J Biol Chem 1996;271:28154–60. 10.1074/jbc.271.45.28154 [DOI] [PubMed] [Google Scholar]
- 56. Watson AJ, Katz A, Simon MI. A fifth member of the mammalian G-protein beta-subunit family. Expression in brain and activation of the beta 2 isotype of phospholipase C. J Biol Chem 1994;269:22150–6. [PubMed] [Google Scholar]
- 57. Xie K, Masuho I, Brand C, Dessauer CW, Martemyanov KA. The complex of G protein regulator RGS9-2 and Gβ(5) controls sensitization and signaling kinetics of type 5 adenylyl cyclase in the striatum. Sci Signal 2012;5:ra63. 10.1126/scisignal.2002922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strehlow V, Heyne HO, Vlaskamp DRM, Marwick KFM, Rudolf G, de Bellescize J, Biskup S, Brilstra EH, Brouwer OF, Callenbach PMC, Hentschel J, Hirsch E, Kind PC, Mignot C, Platzer K, Rump P, Skehel PA, Wyllie DJA, Hardingham GE, van Ravenswaaij-Arts CMA, Lesca G, Lemke JR, GAs group, GRIN2A study group . GRIN2A-related disorders: genotype and functional consequence predict phenotype. Brain 2019;142:80–92. 10.1093/brain/awy304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bansod S, Kageyama R, Ohtsuka T. Hes5 regulates the transition timing of neurogenesis and gliogenesis in mammalian neocortical development. Development 2017;144:3156–67. 10.1242/dev.147256 [DOI] [PubMed] [Google Scholar]
- 60. Lee KE, Seo J, Shin J, Ji EH, Roh J, Kim JY, Sun W, Muhr J, Lee S, Kim J. Positive feedback loop between Sox2 and SOX6 inhibits neuronal differentiation in the developing central nervous system. Proc Natl Acad Sci U S A 2014;111:2794–9. 10.1073/pnas.1308758111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wycisk KA, Zeitz C, Feil S, Wittmer M, Forster U, Neidhardt J, Wissinger B, Zrenner E, Wilke R, Kohl S, Berger W. Mutation in the auxiliary calcium-channel subunit CACNA2D4 causes autosomal recessive cone dystrophy. Am J Hum Genet 2006;79:973–7. 10.1086/508944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bamshad MJ, Nickerson DA, Chong JX. Mendelian gene discovery: fast and Furious with no end in sight. Am J Hum Genet 2019;105:448–55. 10.1016/j.ajhg.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 2016;17:9–18. 10.1038/nrg3999 [DOI] [PubMed] [Google Scholar]
- 64. Arno G, Holder GE, Chakarova C, Kohl S, Pontikos N, Fiorentino A, Plagnol V, Cheetham ME, Hardcastle AJ, Webster AR, Michaelides M, UK Inherited Retinal Disease Consortium . Recessive retinopathy consequent on mutant G-protein β subunit 3 (GNB3). JAMA Ophthalmol 2016;134:924–7. 10.1001/jamaophthalmol.2016.1543 [DOI] [PubMed] [Google Scholar]
- 65. Endo W, Ikemoto S, Togashi N, Miyabayashi T, Nakajima E, Hamano S-I, Shibuya M, Sato R, Takezawa Y, Okubo Y, Inui T, Kato M, Sengoku T, Ogata K, Hamanaka K, Mizuguchi T, Miyatake S, Nakashima M, Matsumoto N, Haginoya K. Phenotype-Genotype correlations in patients with GNB1 gene variants, including the first three reported Japanese patients to exhibit spastic diplegia, dyskinetic quadriplegia, and infantile spasms. Brain Dev 2020;42. 10.1016/j.braindev.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 66. Fukuda T, Hiraide T, Yamoto K, Nakashima M, Kawai T, Yanagi K, Ogata T, Saitsu H. Exome reports a de novo GNB2 variant associated with global developmental delay, intellectual disability, and dysmorphic features. Eur J Med Genet 2019;103804. [DOI] [PubMed] [Google Scholar]
- 67. Hemati P, Revah-Politi A, Bassan H, Petrovski S, Bilancia CG, Ramsey K, Griffin NG, Bier L, Cho MT, Rosello M, Lynch SA, Colombo S, Weber A, Haug M, Heinzen EL, Sands TT, Narayanan V, Primiano M, Aggarwal VS, Millan F, Sattler-Holtrop SG, Caro-Llopis A, Pillar N, Baker J, Freedman R, Kroes HY, Sacharow S, Stong N, Lapunzina P, Schneider MC, Mendelsohn NJ, Singleton A, Loik Ramey V, Wou K, Kuzminsky A, Monfort S, Weiss M, Doyle S, Iglesias A, Martinez F, Mckenzie F, Orellana C, van Gassen KLI, Palomares M, Bazak L, Lee A, Bircher A, Basel-Vanagaite L, Hafström M, Houge G, Goldstein DB, Anyane-Yeboa K, Ramey LV, C4RCD Research Group, DDD study . Refining the phenotype associated with GNB1 mutations: clinical data on 18 newly identified patients and review of the literature. Am J Med Genet A 2018;176:2259–75. 10.1002/ajmg.a.40472 [DOI] [PubMed] [Google Scholar]
- 68. Jones HF, Morales-Briceño H, Barwick K, Lewis J, Sanchis-Juan A, Raymond FL, Stewart K, Waugh M-C, Mahant N, Kurian MA, Dale RC, Mohammad SS. Myoclonus-dystonia caused by GNB1 mutation responsive to deep brain stimulation. Mov Disord 2019;34:1079–80. 10.1002/mds.27708 [DOI] [PubMed] [Google Scholar]
- 69. Kuß J, Stallmeyer B, Goldstein M, Rinné S, Pees C, Zumhagen S, Seebohm G, Decher N, Pott L, Kienitz M-C, Schulze-Bahr E. Familial sinus node disease caused by a gain of GIRK (G-Protein Activated Inwardly Rectifying K+ Channel) channel function. Circ Genom Precis Med 2019;12:e002238. 10.1161/CIRCGEN.118.002238 [DOI] [PubMed] [Google Scholar]
- 70. Laššuthová P, Šafka Brožková D, Neupauerová J, Krůtová M, Mazanec R, Seeman P. Confirmation of the GNB4 gene as causal for Charcot-Marie-Tooth disease by a novel de novo mutation in a Czech patient. Neuromuscul Disord 2017;27:57–60. 10.1016/j.nmd.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 71. Lohmann K, Masuho I, Patil DN, Baumann H, Hebert E, Steinrücke S, Trujillano D, Skamangas NK, Dobricic V, Hüning I, Gillessen-Kaesbach G, Westenberger A, Savic-Pavicevic D, Münchau A, Oprea G, Klein C, Rolfs A, Martemyanov KA. Novel GNB1 mutations disrupt assembly and function of G protein heterotrimers and cause global developmental delay in humans. Hum Mol Genet 2017;26:1078–86. 10.1093/hmg/ddx018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Malerba N, De Nittis P, Merla G. The emerging role of Gβ subunits in human genetic diseases. Cells 2019;8. 10.3390/cells8121567. [Epub ahead of print: 04 12 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miura S, Morikawa T, Fujioka R, Noda K, Kosaka K, Taniwaki T, Shibata H. A novel missense variant (Gln220Arg) of GNB4 encoding guanine nucleotide-binding protein, subunit beta-4 in a Japanese family with autosomal dominant motor and sensory neuropathy. Eur J Med Genet 2017;60:474–8. 10.1016/j.ejmg.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 74. Peng J, Wang Y, He F, Chen C, Wu L-W, Yang L-F, Ma Y-P, Zhang W, Shi Z-Q, Chen C, Xia K, Guo H, Yin F, Pang N. Novel West syndrome candidate genes in a Chinese cohort. CNS Neurosci Ther 2018;24:1196–206. 10.1111/cns.12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Petrovski S, Küry S, Myers CT, Anyane-Yeboa K, Cogné B, Bialer M, Xia F, Hemati P, Riviello J, Mehaffey M, Besnard T, Becraft E, Wadley A, Politi AR, Colombo S, Zhu X, Ren Z, Andrews I, Dudding-Byth T, Schneider AL, Wallace G, Rosen ABI, Schelley S, Enns GM, Corre P, Dalton J, Mercier S, Latypova X, Schmitt S, Guzman E, Moore C, Bier L, Heinzen EL, Karachunski P, Shur N, Grebe T, Basinger A, Nguyen JM, Bézieau S, Wierenga K, Bernstein JA, Scheffer IE, Rosenfeld JA, Mefford HC, Isidor B, Goldstein DB. Germline de novo mutations in GNB1 cause severe neurodevelopmental disability, hypotonia, and seizures. Am J Hum Genet 2016;98:1001–10. 10.1016/j.ajhg.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Soong B-W, Huang Y-H, Tsai P-C, Huang C-C, Pan H-C, Lu Y-C, Chien H-J, Liu T-T, Chang M-H, Lin K-P, Tu P-H, Kao L-S, Lee Y-C. Exome sequencing identifies GNB4 mutations as a cause of dominant intermediate Charcot-Marie-Tooth disease. Am J Hum Genet 2013;92:422–30. 10.1016/j.ajhg.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Steinrücke S, Lohmann K, Domingo A, Rolfs A, Bäumer T, Spiegler J, Hartmann C, Münchau A. Novel GNB1 missense mutation in a patient with generalized dystonia, hypotonia, and intellectual disability. Neurol Genet 2016;2:e106. 10.1212/NXG.0000000000000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Szczałuba K, Biernacka A, Szymańska K, Gasperowicz P, Kosińska J, Rydzanicz M, Płoski R. Novel GNB1 de novo mutation in a patient with neurodevelopmental disorder and cutaneous mastocytosis: clinical report and literature review. Eur J Med Genet 2018;61:157–60. 10.1016/j.ejmg.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 79. Vincent A, Audo I, Tavares E, Maynes JT, Tumber A, Wright T, Li S, Michiels C, Condroyer C, MacDonald H, Verdet R, Sahel J-A, Hamel CP, Zeitz C, Héon E, GNB3 Consortium . Biallelic mutations in GNB3 cause a unique form of autosomal-recessive congenital stationary night blindness. Am J Hum Genet 2016;98:1011–9. 10.1016/j.ajhg.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Patil DN, Rangarajan ES, Novick SJ, Pascal BD, Kojetin DJ, Griffin PR, Izard T, Martemyanov KA. Structural organization of a major neuronal G protein regulator, the RGS7-Gβ5-R7BP complex. Elife 2018;7. 10.7554/eLife.42150. [Epub ahead of print: 12 12 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sciacca FL, Ciaccio C, Fontana F, Strano C, Gilardoni F, Pantaleoni C, D'Arrigo S. Severe Phenotype in a Patient With Homozygous 15q21.2 Microdeletion Involving BCL2L10, GNB5, and MYO5C Genes, Resembling Infantile Developmental Disorder With Cardiac Arrhythmias (IDDCA). Front Genet 2020;11:399. 10.3389/fgene.2020.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lei Q, Jones MB, Talley EM, Garrison JC, Bayliss DA. Molecular mechanisms mediating inhibition of G protein-coupled inwardly-rectifying K+ channels. Mol Cells 2003;15:1–9. [PubMed] [Google Scholar]
- 83. Okae H, Iwakura Y. Neural tube defects and impaired neural progenitor cell proliferation in Gbeta1-deficient mice. Dev Dyn 2010;239:1089–101. 10.1002/dvdy.22256 [DOI] [PubMed] [Google Scholar]
- 84. Ye Y, Sun Z, Guo A, Song L-S, Grobe JL, Chen S. Ablation of the GNB3 gene in mice does not affect body weight, metabolism or blood pressure, but causes bradycardia. Cell Signal 2014;26:2514–20. 10.1016/j.cellsig.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu C-W, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LMJ, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A, MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon A-M, Henkelman RM, Brown SDM, Adams DJ, Lloyd KCK, McKerlie C, Beaudet AL, Bućan M, Murray SA, Consortium IMP, International Mouse Phenotyping Consortium, Jackson Laboratory, Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS), Charles River Laboratories, MRC Harwell, Toronto Centre for Phenogenomics, Wellcome Trust Sanger Institute, RIKEN BioResource Center . High-Throughput discovery of novel developmental phenotypes. Nature 2016;537:508-+–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chograni M, Alkuraya FS, Maazoul F, Lariani I, Chaabouni-Bouhamed H. RGS6: a novel gene associated with congenital cataract, mental retardation, and microcephaly in a Tunisian family. Invest Ophthalmol Vis Sci 2014;56:1261–6. 10.1167/iovs.14-15198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2020-107015supp003.pdf (5.8MB, pdf)
jmedgenet-2020-107015supp004.pdf (938.6KB, pdf)
jmedgenet-2020-107015supp002.pdf (49.9KB, pdf)
jmedgenet-2020-107015supp001.pdf (66.5KB, pdf)
jmedgenet-2020-107015supp005.pdf (60.8KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. The RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE156898.