Abstract
The prevalence of staphylococcal infection and the emergence of multidrug resistance of Staphylococcus aureus (S. aureus) are major concerns in food safety and public health. This study aimed to investigate the prevalence of S. aureus isolated from traditional Chinese Rubing and Rushan cheese, antimicrobial resistance profiles, genomic characteristics, and predict antimicrobial resistance genes (ARGs). From 124 samples, 18 of 62 (29.03%) of Rubing and 5 of 62 (8.06%) of Rushan cheese were confirmed to be S. aureus positive by standard culture-based methods. Twenty-three coagulase-positive staphylococci isolates were grouped into 16 clusters by pulsed-field gel electrophoresis and subjected to routine susceptibility testing to 12 antibiotics. Those isolates exhibited high resistance to penicillin (100%), erythromycin, trimethoprim-sulphamethoxazole (34.78%), oxacillin, clindamycin, and cefoxitin (21.74%). Multidrug-resistant (MDR) S. aureus was found in 34.78% (8 of 23) of isolates. Further, S. aureus strain DC.RB_015 isolated from Rubing cheese, recognized as the most resistant to six antibiotics, was selected for whole-genome sequencing (WGS), continued with in silico approaches. S. aureus DC.RB_015 had a single chromosome size of 2,794,578 bp and a plasmid size of 22,961 bp. The strain harbored 18 predicted ARGs, including eight efflux pump genes (mepA, tet(K), arlR, arlS, norA, mgrA, tet(38), LmrS), one peptidoglycan biosynthesis gene (bacA), two β-lactams resistance genes (mecA, blaZ), and seven genes conferring other antimicrobial resistance (APH(3′)-IIIa, aad(6), ErmB, SAT-4, mecR1, GlpT, murA). The results of this study expand the knowledge of S. aureus strain DC.RB_015, increase food safety awareness, and will be helpful in establishing therapeutic therapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-03072-4.
Keywords: Staphylococcus aureus, Traditional cheese, Antimicrobial susceptibility, In silico approaches, Antimicrobial resistance genes
Introduction
Staphylococcus aureus contamination has been investigated as one of the leading causative agents of various severe clinical infections in humans due to their high occurrence of foodborne illness outbreaks worldwide (Johler et al. 2016; Le et al. 2021; Nouws et al. 2021). S. aureus is a type of coagulase-positive, facultative anaerobic Gram-positive bacteria, drug-resistant pathogen, which secretes heat-stable enterotoxins, and is capable of causing cross-contamination on food processing facilities, primarily through food handlers and food contact surfaces (Dai et al. 2019; Naorem et al. 2021). S. aureus may enter the food chain and has the potential to contaminate food materials, particularly milk, meat, mutton, beef, fish, poultry, eggs, and their byproducts during farming, slaughtering, processing, cooking, and storage (Wang and Ruan 2017). If these food materials are contaminated by staphylococcal enterotoxins (SEs), it may trigger an immediate foodborne illness. S. aureus proliferation above 105–106 CFU/g is the main factor of foodborne toxicity level and illness outbreak (Ertas et al. 2010). Based on a survey during 2006–2015 in China, microbiological hazards, including S. aureus contamination, were responsible for 61.3% of food safety issues (Wang 2018).
S. aureus proliferation leads to fatal skin and soft tissue infections (SSTI), furunculosis, carbuncles, cellulitis, impetigo, scalded skin syndrome, folliculitis, nosomical and osteoarticular infections, and pleuropulmonary complications. These are major causes of chronic community- and hospital-acquired infections (Balachandra et al. 2015; Pereira 2014; Sasirekha et al. 2014; Tong et al. 2015; Alni et al. 2018; Troxell and Hall 2020). The pathogen releases various toxins in both methicillin-resistance S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) strains, including toxic shock syndrome toxin (TSST-1), Panton-Valentine leukocidin (PVL), staphylococcal enterotoxins (SEs), and staphylococcal enterotoxin-like proteins (SEls). Moreover, more than 20 types of SEs or SEl have also been determined recently (Bien et al. 2011; Dai et al. 2019; Shallcross et al. 2013; Spanu et al. 2012; Vitale et al. 2018). In a case when S. aureus penetrates the human digestive system, it can multiply unfavorable condition of food poisoning symptoms, e.g., staphylococcal pulmonary disease in cystic fibrosis, gastrointestinal syndromes, including abdominal cramps, diarrhea, vomiting, nausea, and serious food safety incidents (Kümmel et al. 2016; Shi et al. 2021; Wei et al. 2018).
Recent regional surveillance of S. aureus conducted by Li et al. (2021) reported that 16.7% of ready-to-eat sushi and 25.0% of sliced raw pork products from 50 food outlets and supermarkets in Beijing were positive for S. aureus, including MDR in MRSA strains identified as ST8, ST15, ST25, ST59, ST398, and ST2631. Wang et al. (2017) also reported that the prevalence of S. aureus contamination in retail foods in China was 4.3%, with 7.9% mecA-positive, and 57.5% of S. aureus isolates were confirmed as MDR. In the past few decades, MDR staphylococci, particularly MRSA, have shown the capability for zoonotic transmission, and their prevalence has increased dramatically due to bacterial evolution and antibiotic overuse for prophylaxis or therapy (Crespo-Piazuelo and Lawlor 2021; Khan et al. 2021). The strict implementation of a code of practice for control of the use of veterinary antimicrobial agents and public health programs in the context of a One Health perspective is strongly needed to control the potential risks of MDR S. aureus (Ali et al. 2021a, b; Rodríguez-Lázaro et al. 2017).
The resistance profile to β-lactam antibiotics in S. aureus strains is mediated by exogenous mecA gene expression, which ascertains that the host is confirmed to fail in recognizing β-lactams (Otarigho and Falade 2018; Zuo et al. 2021). The mecA gene, which is conveyed on a mobile genetic element (MGE) embedded in a chromosome specified as staphylococcal cassette chromosomal mec (SCCmec), encoding for the production of a 78-kDa monofunctional transpeptidase penicillin-binding protein 2a (PBP2a) (Naorem et al. 2020; Parvin et al. 2021; Shahkarami et al. 2014). The SCCmec cassette also plays an essential regulatory role in the pathogenicity of MRSA strains (Dehbashi et al. 2021). Technological advances in high-throughput WGS and bioinformatics tools have developed a rapid and cost-effective framework for detecting and analyzing ARGs in isolated bacterial genomes and complex metagenomes obtained from food and environmental samples (Ali et al. 2021b; Gupta et al. 2020; Li et al. 2021; Saima et al. 2020).
Rubing and Rushan cheese, the traditional Chinese foods, have been made for over a thousand years by the Bai minority inhabiting Southwestern China based on their culture, heritage, and local ingredient knowledge. Milk and other dairy products are suitable growing substances and production mediums for S. aureus, with the biological activity of SEs remaining present after pasteurization (Dai et al. 2019). In this present study, S. aureus isolation and detection, molecular typing, antimicrobial susceptibility test, WGS, and ARGs prediction using in silico approaches were systematically performed to investigate the emergence of staphylococcal contamination in traditional Chinese Rubing and Rushan cheese samples obtained from different markets and geographic locations in Yunnan province, China.
Materials and methods
Study area
In this study, a total of 124 traditional cheese samples of Rubing (n = 62) and Rushan (n = 62) were purchased from different local markets, retailer shops, online shops, and farmers in Kunming, Dali, Lijiang, and Dehong city in Yunnan province, China. All samples were directly stored in a cold box at a temperature of 4 °C, wrapped in sterile plastic-packing, and delivered to the food safety laboratory in Yunnan Agricultural University, Kunming. Those samples were subjected to further screening for foodborne contamination of S. aureus within 10 h.
Isolation and detection
Qualitative analysis was performed to detect the presence of S. aureus in observed samples. S. aureus contamination in Rubing and Rushan cheese samples was detected according to Chinese National Food Safety Standards document GB 4789.10-2016, with slight technical adjustment (Chinese National Standard 2016). A total of 25 g of Rubing and Rushan cheese samples were prepared in separate parts and subjected to homogenization for 2 min at 10,000 rpm, with 225 ml of 7.5% sodium chloride broth (Guangdong Huankai Microbial Science and Technology Co., Ltd., China). With the homogeneous sample solution incubated at 37 °C for 18 h, S. aureus was observed to show turbid growth in the medium. The culture was enriched and incubated on Baird-Parker agar and blood plates at 37 °C for 20 h. Suspected colonies were also collected for Gram staining microscopic examination, accompanied by a plasma coagulase test. They were inoculated on 5 ml of brain heart infusion (BHI) and nutrient agar slant at 37 °C for 20 h. A total of 0.2 ml of BHI culture was added into a small tube and mixed well with 0.5 ml of fresh rabbit plasma (Guangdong Huankai Microbial Science and Technology Co., Ltd., China). Those samples were then incubated at 37 °C in a water bath and observed every half an hour for the next 6 h. Both broth cultures of plasma coagulase-positive and also Staphylococcus strains were used as control references. The isolated colonies were preserved in 40% glycerol stocks at − 20 °C for further purposes (Manukumar and Umesha 2017). Positive colonies with the confirmed staphylococcal morphology on Baird-Parker agar plates were picked for further evaluation.
Molecular subtyping analysis of S. aureus isolates
Pulsed-field gel electrophoresis (PFGE) was used to analyze molecular typing bands of S. aureus isolates obtained from Rubing and Rushan cheese, according to the standard procedure for PFGE operation issued by Yunnan Centers for Disease Control and Prevention (CDC) and He et al. (2013), with slight modification. A 2–2.5 mm single-bacteria colony was subjected to inoculation in 5 ml BHI broth, incubated at 37 °C for 14–18 h, harvested, and suspended in 200 μl 1X Tris–EDTA (TE) buffer containing 10 mM Tris Hydrochloride (Tris HCl), 1 mM Ethylenediaminetetraacetic acid (EDTA), and a pH of 8. A total of 400 μl of bacterial suspension was transferred into a 1.5-ml microcentrifuge tube, and 4 μl staphylococcal lysozyme was added to each. The mixture was processed under incubation in a water bath at 55–60 °C for 10–20 min. A total of 400 μl dissolved 1% SeaKem Gold (SKG) agarose was filled into each tube. The mixture was immediately placed into a plug mold (Bio-Rad Laboratories, Inc, Hercules, CA) to solidify at room temperature. In each tube, a total of 10–15 ml ultrapure water was added and pre-heated to 54–55 °C, then oscillated in a water bath from 55 to 60 °C for 15 min at 170 rpm.
A 2.0- to 2.5 mm-wide gel block containing S. aureus was transferred into a 200-μl 1X TE buffer in 1.5 ml microcentrifuge tubes and incubated in a water bath at 30 °C for 5–10 min. Genomic DNA of S. aureus was then digested at 30 °C for 4 h with 20U SmaI restriction enzyme (New England Biolabs Ltd., UK). The DNA fragment was separated using the BioRad CHEF Mapper XA System in a 1% SKG gel (Guangdong Huankai Microbial Science and Technology Co., Ltd., China) stained with ethidium bromide (0.5 g/ml). After that, the gel was exposed to 0.5X Tris–borate EDTA buffer at 200 V for 15 min. A PFGE standard was used as a DNA size ladder to identify electrophoresis results. The gel was observed under a UV transilluminator, with its image documented by Molecular Imager Gel Doc XR (Bio-Rad Laboratories, Inc, Hercules, CA). The relatedness of S. aureus isolates was determined by the patterns of the DNA fragment, and PFGE profiles with 100% similarity were recognized as a pulsotype. Gel pictures were analyzed using BioNumerics bioinformatic software version 5.1 (Applied Math NV, Ghent, Belgium) to obtain high reproducibility normalization of PFGE fingerprints comparison.
Antimicrobial susceptibility test
Antimicrobial susceptibility testing of S. aureus isolates to different antibiotic classes was performed by Kirby-Bauer disk diffusion method on Mueller–Hinton agar (MHA). The method used MHA as a medium to spread bacterial suspensions in BHI broth, with an equivalent of colony suspension closer to a 0.5 McFarland standard. Minimum inhibitory concentration (MIC) testing was carried out by broth microdilution method by Cation-Adjusted Mueller–Hinton Broth (CAMHB), which was enriched using 2% NaCl for oxacillin and 50 μg/ml calcium for daptomycin, in order to determine MIC values in detail. MICs were observed visually and interpreted according to Clinical and Laboratory Standards Institute (CLSI) M100-S30 performance standards (CLSI 2020). S. aureus American Type Culture Collection (ATCC) 29213 was included as quality control and reference strain. Twelve-panel antibiotic compounds (Oxoid Ltd., UK) were tested to evaluate S. aureus antimicrobial susceptibility, including penicillin (5 μg), erythromycin (15 μg), trimethoprim-sulfamethoxazole (25 μg), oxacillin (1 μg), clindamycin (1 μg), cefoxitin (30 μg), tetracycline (30 μg), ciprofloxacin (30 μg), vancomycin (30 μg), chloramphenicol (10 μg), gentamicin (10 μg), and daptomycin (10 μg). Inoculated plates were then incubated at 35–37 °C for 17 h before measuring inhibition zone diameters. The interpretation of each isolate’s antimicrobial susceptibility criteria (susceptible, intermediate, and resistant) was established according to the inhibition zone diameter described in CLSI guidelines. MDR isolates were defined as those that were resistant to at least one agent in three or more antibiotic classes (Parvin et al. 2021). Furthermore, the phenotype of S. aureus strain with the highest resistance against antibiotics was selected for WGS.
Genomic DNA isolation and sequencing
For genomic DNA isolation, S. aureus cultured in BHI medium were prepared by re-suspending pure colonies in 300 μl of Tris-EDTANaCl-TritonX100 (TENT) buffer (10 mM Tris–HCl, 1 mM EDTA, 0.1 M NaCl, 5% [v/v] Triton X100, pH 8.0) as described elsewhere (Hassanzadeh et al. 2016). The cell suspension was boiled at 100 °C, centrifuged at 3450 rpm for 10 min, and then the supernatants were replaced into new sterile tubes. Total genomic DNA extraction was performed by purifying the supernatant using Qiagen assay kit 13323 (Qiagen Co. Ltd., China), following the manufacturer’s protocols. Extracted DNA samples were stored at − 20 °C for further complete genome analysis. DNA concentration was measured using NanoDrop 2000 spectrophotometer at OD 260/280 ratio (Thermo Fisher Scientific Inc., Waltham, MA, USA), and the quality was evaluated using 0.8% gel electrophoresis. A total of 1 ng of genomic DNA was subjected to WGS, with the purity and concentration of DNA was determined using Qubit 1X dsDNA HS (High-Sensitivity) assay kit in a Qubit 3.0 Fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Sequencing-ready libraries were built using Nextera XT DNA sample preparation kit, following the manufacturer’s guidelines (Illumina, San Diego, CA, USA). Library validation to control DNA fragmented distribution was executed on a 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA). Illumina high-throughput Nova-Seq sequencing platform (Illumina, San Diego, CA, USA) was used to perform WGS to obtain 150-bp paired-end reads, following the manufacturer’s instructions. Genomes were sequenced to effective average coverage of a minimum of 30X (Durand et al. 2018).
Genome annotation and ARGs prediction
After sequencing, all short-read sequencing data were assembled, and hybrid assemblies under default parameters were generated with Unicycler v0.4.8 software (Wick et al. 2017). In analyzing genome-wide annotation, Rapid Annotation using Subsystem Technology (RAST) online was also utilized. The same source ratio was performed using Basic Local Alignment Search Tool (BLAST) local ratio, combined with a functional database’s characteristics. InterPro database and InterScanPro software (http://www.ebi.ac.uk/interpro/) were used to annotate the functional domain of all identified proteins gene ontology (GO) based on sequence homology (Blum et al. 2020).
National Center for Biotechnology Information (NCBI) clusters of orthologous groups (COG) database was utilized to classify protein families encoded in completely sequenced genomes. Kyoto Encyclopedia of Genes and Genomes (KEGG) database description was obtained using KEGG Automatic Annotation Server (KAAS). The annotation was mapped to KEGG pathway database using KEGG mapper. Subsequently, RNAmmer was used to predict rRNA coding genes. The tRNAscan-SE was used to identify tRNA genes, as Guanine-Cytosine (GC)-profile was utilized to observe genomic GC-content, while Circular Genome Viewer (CGView) (https://paulstothard.github.io/cgview/) generated circular maps of chromosomes and plasmids (Stothard and Wishart 2005). The predicted toxicity factor was determined using Virulence Factor Database (VFDB), with insertion sequence (IS) element ascertained using ISfinder. Genomic sequence of S. aureus DC.RB_015 in FASTA format was uploaded on Resistance Gene Identifier (RGI) tool in antibiotic resistance gene database platform for ARGs analysis (https://card.mcmaster.ca/analyze/rgi) (Alcock et al. 2020). All default criteria and settings, which only filtered genes base on bitscore cut-off perfect or strict algorithms, were retained.
Sequence supporting data
The genomic data of S. aureus DC.RB_015 isolated from Rubing cheese used in this study were deposited in the NCBI GenBank database under accession numbers: BioProject PRJNA762640, BioSample SAMN21399875, Nucleotide CP083434 (DC_RB_015_chr), and Nucleotide CP083435 (DC_RB_015_plas1).
Results
Prevalence of S. aureus contamination
During this current study, 18 out of 62 isolates (29.03%) in Rubing cheese and 5 out of 62 isolates (8.06%) in Rushan cheese were detected with contamination of coagulase-positive staphylococci (CPS) (Table S2). The highest prevalence of CPS in Rubing and Rushan cheese was found in Dali city, which was 38.24% and 9.52%, respectively (Table S1). Across all isolates, the prevalence of CPS in Rubing collected from local agricultural markets (66.67%) was higher than in other outlets, followed by samples from retailers and farmers. Rushan cheese from retailer shops showed the highest prevalence of CPS (60.00%) (Table S2). These findings confirmed the presence of S. aureus contamination in Rubing and Rushan cheese and brought awareness that corrective actions need to be taken to control the pathogen in local agricultural markets and retailer shops. The disease-causing bacteria can be transmitted through the use of a contaminated facility or via direct physical contact with an infected individual.
Molecular fingerprints analysis
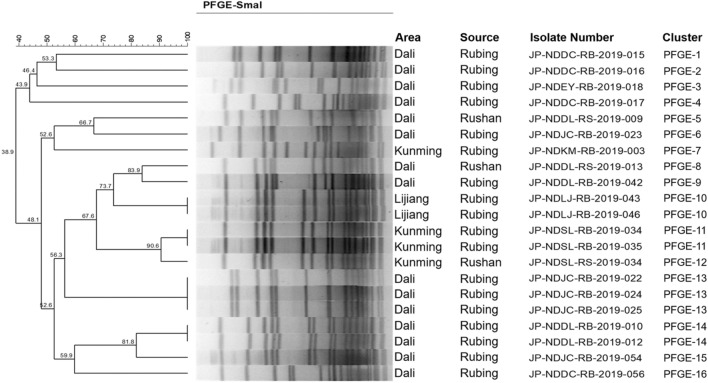
Of 23 S. aureus isolates obtained from Rubing and Rushan cheese, 21 PFGE pulsotypes were grouped into 16 clusters with 10–18 DNA fragment bands and differed using SmaI macro-digestion enzyme, which was allied to geographic resources (Fig. 1). The PFGE clusters differentiated based on 80% similarity of coefficient cut-off point showed 15 pulsotypes within samples collected from Dali, four from Kunming, and two from Lijiang. In this screening test, pulsotype of observed S. aureus isolates indicated 38.9–90.6% similarity, demonstrated significant genetic characteristics diversity and was dominated by PFGE-13. It was important to note that molecular similarity among S. aureus pulsotypes obtained from Rubing and Rushan cheese indicated that the products might have been processed by the same manufacturer, although sold through different outlets. The PFGE subtypes demonstrating identical DNA band patterns were observed in the same manufacturer, such as PFGE-10 (JP-NDSL-RB-2019-043 and -046) from Lijiang and PFGE-13 (JP-NDSL-RB-2019-022, -024, and -025) from Dali, demonstrated identical pulsotypes, with 100% similarity of homology. PFGE genotyping technique demonstrated promising results to identify foodborne illness isolates and their origin based on DNA fingerprints for subtyping pathogens (Machado et al. 2020; Sandt et al. 2020). Genetic similarity among S. aureus isolates was also explained due to the pathogen’s difference in food source origins (Dorneles et al. 2019; Neyaz et al. 2020).
Fig. 1.
Dendrogram depicting S. aureus pulsotypes isolates in Dali, Kunming, and Lijiang city isolated from traditional Chinese Rubing and Rushan cheeses, established used pulsed-field gel electrophoresis (PFGE) with SmaI macro-digestion enzyme. The corresponding area, source, isolate number, and cluster are demonstrated
Antimicrobial resistance profile
A high resistance level ranging from 34.78 to 100% was found among S. aureus isolates to erythromycin, trimethoprim-sulphamethoxazole, and penicillin. On the contrary, a comparably lower resistance was identified to oxacillin, clindamycin, cefoxitin, and tetracycline at 21.74%. None of the isolates presented resistance to ciprofloxacin, vancomycin, chloramphenicol, gentamicin, and daptomycin (0%). Antimicrobial susceptibility profiles are shown in Table 1 and S3. Notably, all isolates showed susceptibility to vancomycin, gentamicin, and daptomycin (100%). A total of 21.74%, 8.70%, and 4.35% of S. aureus isolates demonstrated intermediate susceptibility to tetracycline, chloramphenicol, and ciprofloxacin, respectively. Overall, 8 out of 23 S. aureus isolates (34.78%) showed multidrug resistance, including three or more antimicrobial resistance (Table 2, S3, and S4). Of 8 MDR S. aureus, five isolates (NDDC-RB-2019-015, -016, -022, -024, and -025) demonstrated high-level resistance against penicillin, oxacillin, erythromycin, clindamycin, and cefoxitin with high MICs (> 4, ≥ 16, > 16, > 8, and > 8 μg/ml, respectively) (Table S4). Moreover, S. aureus isolate JP-NDDC-RB-2019-015 (strain DC.RB_015) had the highest multidrug resistance to six different antibiotics’ applications, including penicillin, oxacillin, erythromycin, clindamycin, tetracycline, and cefoxitin. S. aureus DC.RB_015 was then selected to be subjected to WGS to obtain further knowledge regarding genome component characteristics, annotations, and in silico profiles to predict ARGs.
Table 1.
Antimicrobial susceptibility profile of S. aureus isolates to 12 antibiotics (n = 23)
| Antibiotics | Class | Resistant (R) | Intermediate (I) | Sensitive (S) |
|---|---|---|---|---|
| Penicillin (PEN) | Penicillins | 100.00% (n = 23) | 0.00% (n = 0) | 0.00% (n = 0) |
| Erythromycin (ERY) | Macrolides | 34.78% (n = 8) | 0.00% (n = 0) | 65.22% (n = 15) |
| Trimethoprim-sulphamethoxazole (SXT) | Sulfonamides | 34.78% (n = 8) | 0.00% (n = 0) | 65.22% (n = 15) |
| Oxacillin (OXA) | Penicillins | 21.74% (n = 5) | 0.00% (n = 0) | 78.26% (n = 18) |
| Clindamycin (CLI) | Lincosamides | 21.74% (n = 5) | 0.00% (n = 0) | 78.26% (n = 18) |
| Cefoxitin (CFX) | β-lactam | 21.74% (n = 5) | 0.00% (n = 0) | 78.26% (n = 18) |
| Tetracycline (TET) | Tetracyclines | 21.74% (n = 5) | 21.74% (n = 5) | 56.52% (n = 13) |
| Ciprofloxacin (CIP) | Fluoroquinolones | 0.00% (n = 0) | 4.35% (n = 1) | 95.65% (n = 22) |
| Vancomycin (VAN) | Glycopeptides | 0.00% (n = 0) | 0.00% (n = 0) | 100.00% (n = 23) |
| Chloramphenicol (CHL) | Chloramphenicols | 0.00% (n = 0) | 8.70% (n = 2) | 91.30% (n = 21) |
| Gentamicin (GEN) | Aminoglycoside | 0.00% (n = 0) | 0.00% (n = 0) | 100.00% (n = 23) |
| Daptomycin (DAP) | Bactericidals | 0.00% (n = 0) | 0.00% (n = 0) | 100.00% (n = 23) |
CLSI guidelines were followed to make the breakpoints of each antibiotic
Table 2.
Multidrug-resistance spectrum of S. aureus isolates to 12 antibiotics (n = 23)
| Antibiotics combination | Number of resistant strains |
|---|---|
| PEN | 30.43% (n = 7) |
| PEN-SXT | 26.09% (n = 6) |
| PEN-TET | 8.70% (n = 2) |
| PEN-ERY-TET | 4.35% (n = 1) |
| PEN-ERY-SXT | 4.35% (n = 1) |
| PEN-ERY-SXT-TET | 4.35% (n = 1) |
| PEN-OXA-ERY-CLI-CFX | 17.39% (n = 4) |
| PEN-OXA-ERY-CLI-TET-CFX | 4.35% (n = 1) |
Genome component prediction and gene function profile of S. aureus DC.RB_015
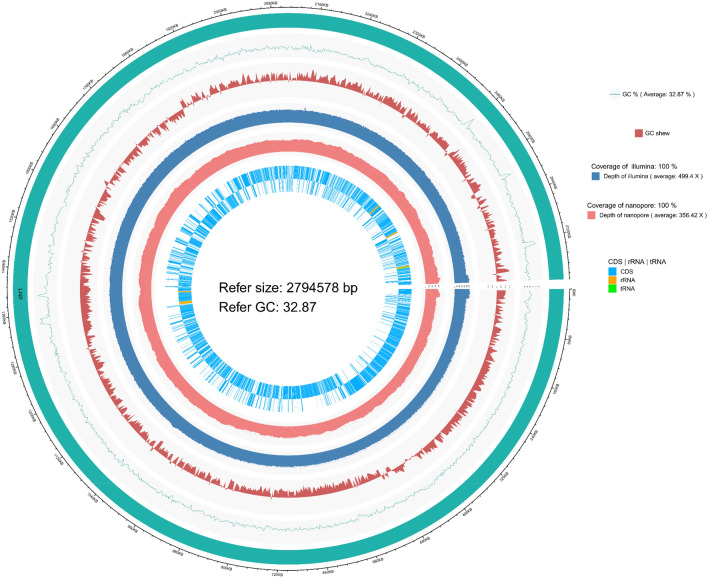
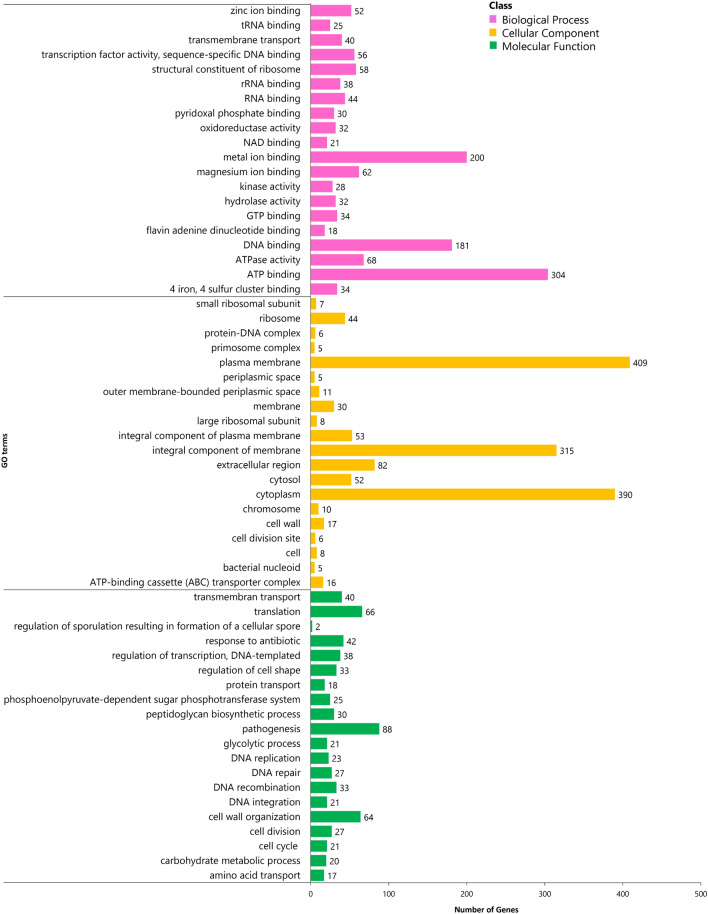
The concentration of genomic DNA of S. aureus isolates was measured within the range of 150–180 ng/µl at OD 260/280. One contig N50 genome of S. aureus DC.RB_015 with a circular chromosome of 2,794,578 bp, a plasmid size of 22,961 bp, and 32.87% GC-content were observed from the reads of the hybrid assembly of Illumina DNAPlotter (Fig. 2). A total of 2770 protein-encoding genes, 2576 annotation protein-encoding genes, 60 tRNA, 19 rRNA, 1 tmRNA, and 3 miscRNA genes were predicted and verified using publicly available genome databases including GO, COG, KEGG, Swiss-Prot, RefSeq, and other functional databases to obtain gene function annotation results. According to GO analysis, 1783 predicted proteins accounting for 64.37% of the whole genome were determined and classified into three major functional classes, including biological process (2189 genes), cellular components (1587 genes), and molecular function (1357 genes) (Fig. 3 and Table S6).
Fig. 2.
Genomic circular map of S. aureus DC.RB_015. From inner to outer circle: (1) reference line, with each scale being 5 kb; (2) reference species genome sequence, informed the total length of gene; (3) statistical average GC-content of reference genome sequence, with a sliding window of 2,000 bp; (4) GC skew curve of reference genome sequence; (5) Illumina sequencing depth and coverage information; (6) nanopore sequencing depth and coverage information, and (7) gene coding and non-coding RNA sequences, including rRNA and tRNA in reference genome, by two inner and outer layers representing positive and negative circles, respectively. The Circos genomic circular map and data visualization were generated using a hybrid assembly of Illumina DNAPlotter
Fig. 3.
GO classification of S. aureus DC.RB_015 core genome with three functional classes, including biological process, cellular component, and molecular function
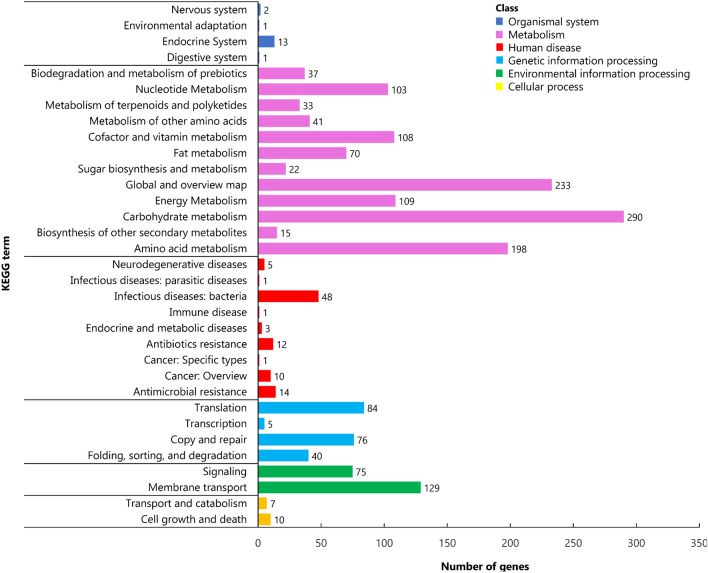
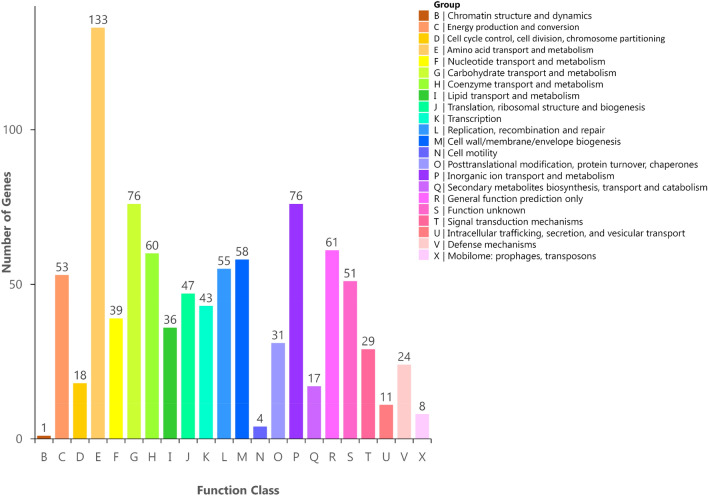
To further characterize the gene function of S. aureus DC.RB_015, KEGG function classification was performed. A total of 1486 putative proteins, accounting for 53.65% of a total number of predicted genes, were assigned to their functional orthologues in KEGG database enriched in 156 metabolic pathways and classified into six functional classes. They included 17, 204, 205, 95, 1259, and 17 genes of cellular process, environmental information processing, genetic information processing data, human diseases, metabolism, and organic systems process, respectively (Fig. 4 and Table S7). It was also found that 22 and 14 genes in S. aureus strain DC.RB_015 were involved in peptidoglycan biosynthesis and beta-lactam resistance, which further confirmed that the strain is capable of antimicrobial resistance (Table S7). COG mapping in NCBI obtained 891 proteins (32.17%), accounting for 88.99% of a total number of predicted coding genes, were assigned to COG functional categories. Twenty-two functional classes were grouped, with the amino acid transport and metabolism category having the highest number of genes (133), followed by carbohydrate transport and metabolism (76) and inorganic ion transport and metabolism (76) (Fig. 5 and Table S8).
Fig. 4.
KEGG classification of S. aureus DC.RB_015 core genome with six functional classes, including organismal systems, metabolism, human disease, genetic information processing, environmental information processing, and cellular processing
Fig. 5.
COG classification of S. aureus DC.RB_015 core genome, which is a total of 931 genes associated with 22 functional classes
In silico characterization of ARGs
In silico analysis of ARGs using Comprehensive Antibiotic Resistance Database (CARD) showed the presence of 18 antibiotics resistance genes predicted in S. aureus strain DC.RB_015, including two resistance genes encoding for resistance to methicillin and β-lactam (mecA) and penicillin (blaZ) involved with a reference pathway ko01501, one gene encoding for peptidoglycan biosynthesis (bacA) involved with a reference pathway ko00550, eight genes regulate efflux pumps providing resistance against tigecycline (mepA), tetracycline (tet(K), tet(38)), fluoroquinolone (arlR, arlS, norA, mgrA), and macrolide, aminoglycoside, oxazolone, diaminopyrimidine, fenicol (LmrS), and seven genes related to resistance against other antibiotic classes, including aminoglycosides (APH(3′)-IIIa, aad(6)), macrolides (ErmB), nucleosides (SAT-4), penicillins (mecR1), and fosfomycin (GlpT, murA). These predicted genes were also found in reference genomes of type strains of lactobacilli and bifidobacteria (Duranti et al. 2017; Rodríguez et al. 2019). The functional classification and antibiotic types of the predicted genes are outlined in Table 3 and S5. Additionally, according to an arrangement in performance of antimicrobial susceptibility test and ARGs detection, the presence of mecA gene using WGS and CARD database revealed conformable results, thus S. aureus DC.RB_015 was also identified as MRSA.
Table 3.
Prediction of antimicrobial resistance genes, functional classification, antibiotic types, and biosynthesis pathway of S. aureus DC.RB_015
| Types of resistance genes | Functional classification | Antibiotic types | Pathway ID |
|---|---|---|---|
| APH(3’)-IIIa (Aminoglycosides) |
Aminoglycoside 3′-O-phosphotransferase, which modifies the amino group by phosphorylation Glycosides |
Kanamycin, Barrotomycin, Isapamycin, Butirosine, Gentamicin, Penicillin, Ribomycin, Neomycin, Amikacin | – |
| ErmB (Macrolides) | rRNA adenine N-6-methyltransferase can methylate adenine at position 2058 of 23S rRNA, giving erythromycin resistance | Streptomycin B, Erythromycin, Lincosamide | – |
| aad(6) (Aminoglycosides) | Aminoglycoside 6′-adenosyltransferase | Aminoglycoside | – |
| SAT-4 (Nucleosides) | Streptomycin acetyltransferase A | Nucleoside | – |
| mecA (β-lactams) | Penicillin-binding proteins have low affinity for β-lactam and catalyzes penicillin-insensitive transpeptidase | β-lactam, Methicillin | Ko01501 |
| mepA (Glycylcyclines) | Multiple antibacterial extrusion (MATE) family proteins, multi-drug resistant efflux pump | Tigecycline | – |
| tet(K) (Tetracyclines) | Superfamily transporter, tetracycline efflux pump | Tetracycline | – |
| arlR (Fluoroquinolones) | Major Facilitator Superfamily (MFS) antibiotic efflux pump | Fluoroquinolone | – |
| arlS (Fluoroquinolones) | Major Facilitator Superfamily (MFS) antibiotic efflux pump | Fluoroquinolone | – |
| norA (Fluoroquinolones) | Major Facilitator Superfamily (MFS) antibiotic efflux pump | Fluoroquinolone | – |
| mgrA (Fluoroquinolones) | ATP binding cassette (ABC) antibiotic efflux pump; the main facilitator superfamily (MFS) antibiotic efflux pump | Fluoroquinolones, Cephalosporins, Penicillins, Tetracyclines, Peptides | – |
| tet(38) (Tetracyclines) | Major Facilitator Superfamily (MFS) antibiotic efflux pump | Tetracycline | – |
| LmrS (Macrolides) | Major Facilitator Superfamily (MFS) antibiotic efflux pump | Macrolide, Aminoglycoside, Oxazolone, Diaminopyrimidine, Fenicol | – |
| glpT (Fosfomycin) | Mutant glpT, resistant to fosfomycin | Fosfomycin | – |
| murA (Fosfomycin) | Mutant MurA, resistant to fosfomycin | Fosfomycin | – |
| BacA (Polypeptides) | Undecenyl pyrophosphate phosphatase leads to the separation of base pyrophosphate | Bacitracin | Ko00550 |
| blaZ (Penicillins) | Penicillinase extrudes the β-lactam antibiotic ring and invalidates the antibacterial properties of the molecule | Penicillin | Ko01501 |
| mecR1 (Penicillins) | Methicillin-resistance mecR1 proteins | Methicillin | – |
The antimicrobial resistance genes family, resistance mechanism, and predicted protein sequence are separately outlined in Table S5
Discussion
The rapid spread of MDR S. aureus harboring ARGs had become a major problem in veterinary and human medicine worldwide due to the difficulty of treating them. Most infections that lead to severe clinical implications for human health occur via pathogenic potential lineages and are classified as high-risk microbiological hazards in Hazard Analysis Critical Control Point (HACCP) (Ali et al. 2021a; Dehbashi et al. 2021; Fiaz et al. 2021; Rodríguez-Lázaro et al. 2017; Saima et al. 2020). This present study is the first investigation to evaluate the spread of AMR and ARGs of S. aureus in Rubing and Rushan cheese in Yunnan province, China, through the integration of phenotypic approach and in silico genome-based methods using publicly available databases. Recently, only a few studies that correlate the prevalence of phenotype of antibiotic-resistant S. aureus isolated from traditional cheese types have been reported elsewhere. The findings of this research expanded the understanding of antimicrobial resistance prevalence and profiles of S. aureus in traditional cheese products, particularly strain DC.RB_015.
In the current study, 18.55% (23 of 124) of observed Rubing and Rushan cheese samples were positively contaminated with S. aureus, with the majority of contaminations found in agricultural markets (12 of 18 isolates for Rubing) and retail shops (2 of 5 for Rushan). These prevalence results were also similar to the previous investigation conducted in Tabriz, Iran, where 27% of all dairy product samples (milk and cheese) had been confirmed with the presence of S. aureus (Saadat et al. 2014). Furthermore, the presence of S. aureus contamination in cheese has raised serious concerns in Europe, as it has the potential to increase the prevalence of human illnesses and outbreaks (Verraes et al. 2015). Thus far, unpasteurized milk cheeses were responsible for about 70% of foodborne illness outbreaks caused by cheese consumption (Donnelly 2018; Martínez-Vasallo et al. 2019). Food handlers who work in cheese manufacturers and have frequent contact with food chains and facilities should be trained on the potential risks of spreading MDR S. aureus from animals and the environment in order to eliminate staphylococcal bacterial contamination.
According to the phenotypic test results of antimicrobial resistance, we detected resistance to penicillin with MIC ≥ 0.25 μg/ml was the most frequent than other antibiotics tested (100%) among the isolates, while resistance against ciprofloxacin (MIC ≤ 2 μg/ml), vancomycin (MIC ≤ 2 μg/ml), chloramphenicol (MIC ≤ 16 μg/ml), gentamicin (MIC ≤ 1 μg/ml), and daptomycin (MIC ≤ 1 μg/ml) showed the lowest resistance (0%). Besides, erythromycin and trimethoprim-sulphamethoxazole resistance was found in 34.78% of isolates. Due to the widespread and inappropriate use of penicillin and other antibiotics in infection treatment in dairy farming and healthcare, the majority of S. aureus strains are resistant to these agents (de los Santos et al. 2017; Haulisah et al. 2021). In this experiment, the Kirby-Bauer method using MHA standard medium demonstrated high performance to support better growth conditions to spread the suspension of S. aureus in BHI broth. It was indicated by the growth ability in a broth of 23 S. aureus isolates, which is consistent with other studies performed by Nassar et al. (2019).
Previously, Liu et al. (2017) reported that penicillin, ampicillin, streptomycin, gentamicin, ciprofloxacin, and sulfamethoxazole-trimethoprim were commonly used for dairy mastitis treatment in northern China. Those antibiotic applications were designed to target the mitochondrial ribosome in the bacterial cell to control the infection through a protein synthesis inhibition mechanism (Chellat et al. 2016; Wang et al. 2015). Penicillin resistance in S. aureus strains isolated from milk and its byproducts has been detected in several previous studies worldwide (Dittmann et al. 2017; Kalayu et al. 2020; Liao et al. 2018; Mohammed et al. 2018; Pekana and Green 2018; Rodrigues et al. 2017). A high prevalence of S. aureus resistance to penicillin, ampicillin, erythromycin, tetracycline, and clindamycin was also screened in Brazil and Turkey, which was 77.9%, 68.3%, 65.4%, 24.0%, 19.2% (in milk and Brazilian Coalho cheese samples), and 84.71%, 72.94%, 22.35%, 5.88%, 50.59% (in soft and semi-soft ripened Turkish cheese samples), respectively (Kayili and Sanlibaba 2020; Pereira et al. 2018). Rodrigues et al. (2017) studied the antimicrobial resistance profiles of Staphylococcus spp. isolated from one hundred traditional Brazilian Minas Frescal cheese samples and reported 18 resistance profiles, with the most considerable being resistance against penicillin (23%), followed by oxacillin, cefoxitin, erythromycin, and clindamycin (3%). The resistance profile to erythromycin, tetracycline, vancomycin, and ciprofloxacin was caused by the presence of mgrA gene, classified into the staphylococcal accessory regulator A (SarA) protein family, and tetK gene (Leroy et al. 2019; Sun et al. 2011).
Herein, the prevalence of MDR S. aureus in this study was lower (34.78%) when compared to a previous study reported in Malaysia that observed a high proportion of MDR (96.15%) in all tested MRSA isolates from milk and nasal swabs of dairy cattle (Aklilu and Chia 2020). Comparing the findings of this study with other studies regarding the occurrence of AMR S. aureus, a Turkish study reported the investigation of S. aureus isolates from 387 traditional cheese samples (White, Tulum, Ezine, and raw sheep milk cheese) showed a high rate of MDR (72.94%). Meanwhile, a Brazilian study found a lower prevalence of S. aureus isolates (29%) (Kayili and Sanlibaba 2020; Pereira et al. 2018). Several efflux pump genes are identified as responsible for MDR mechanisms (Mahmood et al. 2016; Mbindyo et al. 2021). The presence of a non-native gene involved in the peptidoglycan biosynthesis pathway and producing penicillin-binding proteins (PBPs), a mesh-like polymer surrounding the bacterial cell, has also been suggested as a possible explanation for the high prevalence of S. aureus resistance occurrences (Peacock and Paterson 2015; Stapleton and Taylor 2002).
Moreover, the expression of transpeptidase penicillin-binding protein 2a (PBP2a), a modified PBP, leads to a change in S. aureus to become more resistant to methicillin and various β-lactam antibiotics. In our approach, which used several tools and publicly available databases for antimicrobial resistance, we detected genes encoding resistance to methicillin, β-lactam, and penicillin, peptidoglycan biosynthesis, efflux pumps, and resistance to other antibiotic classes (e.g., aminoglycosides, macrolides, nucleosides, and fosfomycin). We observed S. aureus DC.RB_015 harbored mecA gene herein and was classified as an MRSA strain. Methicillin resistance in MRSA strains can be conferred by its ability to conduct peptidoglycan biosynthesis mediated by PBP2a, which cannot be bonded by β-lactam (Ali et al. 2021c; Chiang et al. 2020). In another work related to this current research project, Shi et al. (2021) identified 440 lysine-malonylated sites in 281 proteins as novel post-translational modifications (PTMs) in key rate-limiting enzymes, such as 6-phosphofructokinase, F1F0-ATP synthase, pyruvate kinase (PYK), and dihydrolipoyl dehydrogenase, which were involved in regulating energy-yielding metabolic pathways and indirectly affected the pathogenicity and susceptibility to antibiotics in MRSA S. aureus DC.RB_015.
Multidrug resistance phenotype among eight observed isolates suggested the possible presence of a multidrug transporter protein mepA gene from the multi-antimicrobial extrusion (MATE) protein family, which is available in the mepRAB operon (Costa et al. 2013). Tetracycline-resistant tetK, frequently located on small transmissible plasmids, and tet(38) gene included in the Major Facilitator Superfamily (MFS) efflux pump genes were also detected in this study. The LmrS efflux pump gene, a lincomycin resistance protein of S. aureus, was responsible for gentamicin (aminoglycoside) and erythromycin (macrolide) resistance. Through in silico analysis, LmrS gene showed potential resistance to other antibiotics, including oxazolone, diaminopyrimidine (trimethoprim), and fenicol. Besides, Andersen et al. (2015) reported that the activity of LmrS gene effectively extruded fusidic acid, kanamycin, lincomycin, and linezolid with the use of a predicted 47 kDa protein product, which comprises 14 transmembrane alpha-helices (TMH). A chromosomally encoded resistance mechanism to ciprofloxacin (quinolone) was associated with the expression of the norA efflux pump gene, which modulates ferric uptake regulator (Fur), and mutational alterations occurring in norA promoter region (Leroy et al. 2019).
The bacA gene encoding for undecaprenyl-diphosphatase-related proteins and the expression of the PC1 β-lactamase enzyme encoded by blaZ gene also contributed to MDR profiles among observed isolates, where bacA demonstrates drug resistance through a target alteration mechanism to a peptide class involved in peptidoglycan biosynthesis in the ko00550 pathway (Fig. S1), and blaZ regulates the hydrolyzing of β-lactam ring and renders the antibiotic inactivation. Our findings were consistent with previous results reported by Naorem et al. (2020, 2021) that also found 18 antimicrobial resistance-associated genes in S. aureus obtained from four MRSA clinical isolates in Germany and Hungary. Among these genes, 16 genes were responsible for resistance to methicillin (mecA), β-lactams (blaZ), tetracycline (tet-38, mgrA), daptomycin (clsA), fluoroquinolone and acridine dye (arlS and arlR), rifamycin (rpoB32), nitroimidazole (msbA), defensin (mprF), glycylcycline (mepA), multidrug and toxic compound (mepR), fluoroquinolone (norA, gyrA, gyrB), and diaminopyrimidine (dfrC).
In addition, based on GO analysis results for MRSA S. aureus DC.RB_015, we found a total of 88, 66, 409, 390, and 304 predicted genes (5.88%, 2.38%, 14.77%, 11.16%, and 10.97%) encoding for pathogenesis, translation, plasma membrane, cytoplasm, and ATP-binding, respectively. Considering the clinical treatment of MDR S. aureus and MRSA strains spread in cheese and other dairy products is challenging, it is imperative to monitor the spread of AMR and resistance mechanisms of this bacterium along its food chains. Practical control measures, including proper pasteurization, HACCP implementation, good hygiene practices (GHP), good manufacturing practices (GMP) from on-farm to off-farm, maintaining time/temperature control for safety (TCS), microbial risk assessment, and performing foodborne illness surveillance, are urgently needed (Choi et al. 2016; Liu et al. 2017; Regecová et al. 2021).
Conclusion
This current study provides fundamental knowledge into the antimicrobial resistance of S. aureus isolated from artisanal Chinese traditional Rubing and Rushan cheese, with particular attention on penicillin resistance. The results contribute to a baseline for controlling the process flow of cheese production and distribution practices. A pathogen monitoring program (PEM) should be conducted to evaluate the spread and emergence of AMR in food chains. Hence, HACCP and other food safety risk assessments in the context of AMR should always be promoted straightforwardly to prevent potential hazards. WGS and CARD analysis could extend the knowledge of comprehensive molecular pathogenesis and predict antimicrobial resistance profiles in real-time and rapid detection. Putative antimicrobial resistance proteins and ARGs were successfully predicted in S. aureus DC.RB_015. The performance of the antimicrobial susceptibility test and in silico CARD analysis showed that phenotypic MDR to six different antibiotics in S. aureus DC.RB_015 corresponded with the expression of the 18 resistance-associated genes. The presence of mecA gene detected in observed strain confirmed the phenotypic expression of methicillin resistance S. aureus (MRSA). To meet the basic compliance of the suitable presumption of food safety standards that requires the absence of common ARGs encoding for resistance to specific antimicrobials, future research of the transferability mechanisms of MGEs, biofilm-forming ability, expression profiling, and phylogenetic analysis is needed to gain insight into the potential risk of the presence of the ARGs in the S. aureus strains of traditional Chinese cheeses or other foods.
Supplementary Information
Below is the link to the electronic supplementary material.
The online version contains supplementary material for this article (Table S4, S5, S6, S7, and S8 dataset) can be accessed at https://doi.org/10.6084/m9.figshare.14479809 (DOCX 204 KB)
Acknowledgements
We would like to extend our heartful gratitude to the anonymous reviewers and colleagues for their helpful comments and suggestions that improved the content quality of the manuscript. Financial support for the research was provided by the Key Research and Development Program of Yunnan Province entitled “Study on Traceability Technology and Risk Assessment of Foodborne Pathogens in Yunnan Province” (Grant No. 2018BC006), and Dairy Processing and Food Safety Program of Yunnan Modern Agricultural Cattle Milk Industrial Technology System (Grant No. 2019KJTX0014). We also thank Mr. Ren Xiang for his kind help in the preservation of Staphylococcus aureus strain DC.RB_015, which provides support to the Yunnan Center for Disease Control and Prevention.
Author contributions
AH, ZY, and YX designed, developed, financed, supervised the experiment, and approved the final draft. ASP and JZ performed the experiments, analyzed the experimental data, drafted the final report, interpretation, documentation, prepared tables and/or figures, and authored drafts of the manuscripts. YS, QM, and QZ prepared samples, reagents, equipment, submitted genomic data to GeneBank, and approved the final draft.
Declarations
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Adhita Sri Prabakusuma and Jingjing Zhu have contributed equally to this work.
References
- Aklilu E, Chia HY. First mecC and mecA positive livestock-associated methicillin resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from dairy cattle in Malaysia. Microorganisms. 2020 doi: 10.3390/microorganisms8020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Liaqat S, Tariq H, Abbas S, Arshad M, Li WJ, Ahmed I. Neonatal calf diarrhea: a potent reservoir of multi-drug resistant bacteria, environmental contamination and public health hazard in Pakistan. Sci Total Environ. 2021;799:149450. doi: 10.1016/j.scitotenv.2021.149450. [DOI] [PubMed] [Google Scholar]
- Ali A, Tariq H, Abbas S, Arshad M, Li S, Dong L, Li L, Li WJ, Ahmed I. Draft genome sequence of a multidrug-resistant novel candidate Pseudomonas sp. NCCP-436T isolated from faeces of a bovine host in Pakistan. J Glob Antimicrob Resist. 2021;27:91–94. doi: 10.1016/j.jgar.2021.08.011. [DOI] [PubMed] [Google Scholar]
- Ali T, Basit A, Karim AM, Lee J-H, Jeon J-H, Rehman SU, Lee S-H. Mutation-based antibiotic resistance mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. Pharmaceuticals. 2021;14:420. doi: 10.3390/ph14050420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alni RH, Mohammadzadeh A, Mahmoodi P. Molecular typing of Staphylococcus aureus of different origins based on the polymorphism of the spa gene: characterization of a novel spa type. 3Biotech. 2018;8(1):58. doi: 10.1007/s13205-017-1061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, He GX, Kakarla P, Ranjana KC, Kumar S, Lakra WS, Mukherjee MM, Ranaweera I, Shrestha U, Tran T, Varela MF. Multidrug efflux pumps from enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int J Environ Res Public Health. 2015;12(2):1487–1547. doi: 10.3390/ijerph120201487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandra S, de la Gandara MP, Salvato S, Urban T, Parola C, Khalida C, Kost RG, Evering TH, Pastagia M, D’Orazio BM, Tomasz A, de Lencastre H, Tobin JN. Recurrent furunculosis caused by a community-acquired Staphylococcus aureus strain belonging to the USA300 Clone. Microb Drug Resist. 2015;21(2):237–243. doi: 10.1089/mdr.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien J, Sokolova O, Bozko P. Characterization of virulence factors of Staphylococcus aureus: novel function of known virulence factors that are implicated in activation of airway epithelial proinflammatory response. J Pathog. 2011;11:601905. doi: 10.4061/2011/601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Chang H, Chuguransky S, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020;49:D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellat MF, Raguž L, Riedl R. Targeting antibiotic resistance. Angew Chem Int Ed Engl. 2016;55(23):6600–6626. doi: 10.1002/anie.201506818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YC, Wong MTY, Essex JW. Molecular dynamics simulations of antibiotic ceftaroline at the allosteric site of Penicillin-Binding Protein 2a (PBP2a) Isr J Chem. 2020;60:754–763. doi: 10.1002/ijch.202000012. [DOI] [Google Scholar]
- Chinese National Standard (2016) GB 4789.10-2016 National food safety standard food microbiological examination: Staphylococcus aureus. National Standard of The People’s Republic of China. http://tradechina.dairyaustralia.com.au/wp-content/uploads/2018/08/GB-4789.10-2016-Safety-Standard-Food-Microbiological-Examination-Staphylococcus-Aureus.pdf. Accessed 24 Dec 2020
- Choi KH, Lee H, Lee S, Kim S, Yoon Y. Cheese microbial risk assessments—a review. Asian–aust J Anim Sci. 2016;29(3):307–314. doi: 10.5713/ajas.15.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2020) performance standards for antimicrobial susceptibility testing, CLSI supplement M100 30th edition. Clinical and Laboratory Standards Institute, Wayne, PA, USA
- Costa SS, Viveiros M, Amaral L, Couto I. Multidrug efflux pumps in Staphylococcus aureus: an update. Open Microbiol J. 2013;7:59–71. doi: 10.2174/1874285801307010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Piazuelo D, Lawlor PG. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir Vet J. 2021;74:21. doi: 10.1186/s13620-021-00200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wu S, Huang J, Wu Q, Zhang F, Zhang J, Wang J, Ding Y, Zhang S, Yang X, Lei T, Xue L, Wu H. Prevalence and characterization of Staphylococcus aureus isolated from pasteurized milk in China. Front Microbiol. 2019;10:641. doi: 10.3389/fmicb.2019.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos RI, Zunino PM, Gil AD, Laport A, Hirigoyen DJ. Antibiotic resistance of Staphylococcus aureus associated with subclinical and clinical mastitis in Uruguay during an eight-year period. Austral J Vet Sci. 2017 doi: 10.4067/S0719-81322017000300191. [DOI] [Google Scholar]
- Dehbashi S, Tahmasebi H, Zeyni B, Arabestani MR. Regulation of virulence and β-lactamase gene expression in Staphylococcus aureus isolates: cooperation of two-component systems in bloodstream superbugs. BMC Microbiol. 2021;21:192. doi: 10.1186/s12866-021-02257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann KK, Chaul LT, Lee SHI, Corassin CH, de Oliveira CAF, De Martinis ECP, Alves VF, Gram L, Oxaran V. Staphylococcus aureus in some Brazilian dairy industries: changes of contamination and diversity. Front Microbiol. 2017;8:2049. doi: 10.3389/fmicb.2017.02049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C (2018) Control of pathogens in cheeses made from unpasteurised milk. Food Standards Scotland. https://www.foodstandards.gov.scot/publications-and-research/publications/control-of-pathogens-in-cheeses-made-from-unpasteurised-milk. Accessed 6 May 2021
- Dorneles EMS, Fonseca MDAM, Abreu JAP, Lage AP, Brito MAVP, Pereira CR, Brandão HM, Guimarães AS, Heinemann MB. Genetic diversity and antimicrobial resistance in Staphylococcus aureus and coagulase-negative Staphylococcus isolates from bovine mastitis in Minas Gerais Brazil. Microbiologyopen. 2019;8(5):e00736. doi: 10.1002/mbo3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand G, Javerliat F, Bes M, Veyrieras JB, Guigon G, Mugnier N, Schicklin S, Kaneko G, Santiago-Allexant E, Bouchiat C, Martins-Simões P, Laurent F, Van BA, Vandenesch F, Tristan A. Routine whole-genome sequencing for outbreak investigations of Staphylococcus aureus in a national reference center. Front Microbiol. 2018;9:511. doi: 10.3389/fmicb.2018.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti S, Lugli GA, Mancabelli L, Turroni F, Milani C, Mangifesta M, Ferrario C, Anzalone R, Viappiani A, van Sinderen D, Ventura M. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl Environ Microbiol. 2017;83:e02894–e2916. doi: 10.1128/AEM.02894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertas N, Gonulalan Z, Yildirim Y, Kum E. Detection of Staphylococcus aureus enterotoxins in sheep cheese and dairy desserts by multiplex PCR technique. Int J Food Microbiol. 2010;142(1–2):74–77. doi: 10.1016/j.ijfoodmicro.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Fiaz M, Ahmed I, Riaz R, Nawaz U, Arshad M. Prevalence of antibiotic-resistant bacterial strains in wastewater streams: molecular characterization and relative abundance. Folia Microbiol (praha) 2021 doi: 10.1007/s12223-021-00902-z. [DOI] [PubMed] [Google Scholar]
- Gupta CL, Tiwari RK, Cytryn E. Platforms for elucidating antibiotic resistance in single genomes and complex metagenomes. Environ Int. 2020;138:105667. doi: 10.1016/j.envint.2020.105667. [DOI] [PubMed] [Google Scholar]
- Hassanzadeh S, Pourmand MR, Afshar D, Dehbashi S, Mashhadi R (2016) TENT: a rapid DNA extraction method of Staphylococcus aureus. Iran J Public Health 45(8):1093–1095. https://pubmed.ncbi.nlm.nih.gov/27928541 [PMC free article] [PubMed]
- Haulisah NA, Hassan L, Bejo SK, Jajere SM, Ahmad NI. High levels of antibiotic resistance in isolates from diseased livestock. Front Vet Sci. 2021;8:652351. doi: 10.3389/fvets.2021.652351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xie Y, Reed S. Pulsed-field gel electrophoresis typing of staphylococcus aureus isolates. In: Ji Y, editor. Methicillin-resistant Staphylococcus aureus (MRSA) protocols. Berlin: Springer Science+Business Media; 2013. pp. 103–111. [Google Scholar]
- Johler S, Zurfluh K, Stephan R. Tracing and inhibiting growth of Staphylococcus aureus in barbecue cheese production after product recall. J Dairy Sci. 2016;99:3345–3350. doi: 10.3168/jds.2015-10689. [DOI] [PubMed] [Google Scholar]
- Kalayu AA, Woldetsadik DA, Woldeamanuel Y, Wang SH, Gebreyes WA, Teferi T. Burden and antimicrobial resistance of S. aureus in dairy farms in Mekelle Northern Ethiopia. BMC Vet Res. 2020;16:20. doi: 10.1186/s12917-020-2235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayili E, Sanlibaba P. Prevalence, characterization and antibiotic resistance of Staphylococcus aureus isolated from traditional cheeses in Turkey. Int J Food Prop. 2020;23(1):1441–1451. doi: 10.1080/10942912.2020.1814323. [DOI] [Google Scholar]
- Khan J, Tarar SM, Gul I, Nawaz U, Arshad M. Challenges of antibiotic resistance biofilms and potential combating strategies: a review. 3Biotech. 2021;11:169. doi: 10.1007/s13205-021-02707-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmel J, Stessl B, Gonano M, Walcher G, Bereuter O, Fricker M, Grunert T, Wagner M, Ehling-Schulz M. Staphylococcus aureus entrance into the dairy chain: tracking S. aureus from dairy cow to cheese. Front Microbiol. 2016;7:1603. doi: 10.3389/fmicb.2016.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HHT, Dalsgaard A, Andersen PS, Nguyen HM, Ta YT, Nguyen TT. Large-scale Staphylococcus aureus foodborne disease poisoning outbreak among primary school children. Microbiol Res. 2021;12:43–52. doi: 10.3390/microbiolres12010005. [DOI] [Google Scholar]
- Leroy S, Christieans S, Talon R. Tetracycline gene transfer in Staphylococcus xylosus in situ during sausage fermentation. Front Microbiol. 2019;10:392. doi: 10.3389/fmicb.2019.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tang T, Stegger M, Dalsgaard A, Liu T, Leisner JJ. Characterization of antimicrobial-resistant Staphylococcus aureus from retail foods in Beijing China. Food Microbiol. 2021;93:103603. doi: 10.1016/j.fm.2020.103603. [DOI] [PubMed] [Google Scholar]
- Liao F, Gu W, Yang Z, Mo Z, Fan L, Guo Y, Fu X, Xu W, Li C, Dai J. Molecular characteristics of Staphylococcus aureus isolates from food surveillance in southwest China. BMC Microbiol. 2018;18:91. doi: 10.1186/s12866-018-1239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li S, Meng L, Dong L, Zhao S, Lan X, Wang J, Zheng N. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J Dairy Sci. 2017;100:8796–8803. doi: 10.3168/jds.2017-13370. [DOI] [PubMed] [Google Scholar]
- Machado V, Pardo L, Cuello D, Giudice G, Luna PC, Varela G, Camou T, Schelotto F. Presence of genes encoding enterotoxins in Staphylococcus aureus isolates recovered from food, food establishment surfaces and cases of foodborne diseases. Rev Inst Med Trop Sao Paulo. 2020;62:e5. doi: 10.1590/s1678-9946202062005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood HY, Jamshidi S, Sutton JM, Rahman KM. Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr Med Chem. 2016;23(10):1062–1081. doi: 10.2174/0929867323666160304150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manukumar HM, Umesha S. MALDI-TOF-MS based identification and molecular characterization of food associated methicillin-resistant Staphylococcus aureus. Sci Rep. 2017;7:1–16. doi: 10.1038/s41598-017-11597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Vasallo A, Ribot-Enríquez A, Riverón-Alemán Y, Remón-Díaz D, Martínez-García YA, Jacsens L, Uyttendaele M (2019) Staphylococcus aureus in the production chain of artisan fresh cheese. Rev Salud Anim 41(1):1–9. http://scielo.sld.cu/pdf/rsa/v41n1/2224-4700-rsa-41-01-e03.pdf
- Mbindyo CM, Gitao GC, Plummer PJ, Kulohoma BW, Mulei CM, Bett R. Antimicrobial resistance profiles and genes of Staphylococci isolated from mastitic cow’s milk in Kenya. Antibiotics. 2021;10:772. doi: 10.3390/antibiotics10070772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Ziwa MH, Hounmanou YMG, Kisanga A, Tuntufye HN. Molecular typing and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from bovine milk in Tanzania. Int J Microbiol. 2018;4287431:1–6. doi: 10.1155/2018/4287431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naorem RS, Urban P, Goswami G, Fekete C. Characterization of methicillin-resistant Staphylococcus aureus through genomics approach. 3Biotech. 2020;10:401. doi: 10.1007/s13205-020-02387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naorem RS, Blom J, Fekete C. Genome-wide comparison of four MRSA clinical isolates from Germany and Hungary. PeerJ. 2021;9:e10185. doi: 10.7717/peerj.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar MSM, Hazzah WA, Bakr WMK. Evaluation of antibiotic susceptibility test results: how guilty a laboratory could be? J Egypt Public Health Assoc. 2019 doi: 10.1186/s42506-018-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyaz L, Rajagopal N, Wells H, Fakhr MK. Molecular characterization of Staphylococcus aureus plasmids associated with strains isolated from various retail meats. Front Microbiol. 2020;11:223. doi: 10.3389/fmicb.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouws S, Bogaerts B, Verhaegen B, Denayer S, Laeremans L, Marchal K, Roosens NHC, Vanneste K, De Keersmaecker SCJ. Whole genome sequencing provides an added value to the investigation of staphylococcal food poisoning outbreaks. Front Microbiol. 2021;12:750278. doi: 10.3389/fmicb.2021.750278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otarigho B, Falade MO. Analysis of antibiotics resistant genes in different strains of Staphylococcus aureus. Bioinformation. 2018;14(3):113–122. doi: 10.6026/97320630014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin MS, Ali MY, Talukder S, Nahar A, Chowdhury EH, Rahman MT, Islam MT. Prevalence and multidrug resistance pattern of methicillin resistant S. aureus isolated from frozen chicken meat in Bangladesh. Microorganisms. 2021;9:636. doi: 10.3390/microorganisms9030636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- Pekana A, Green E. Antimicrobial resistance profiles of Staphylococcus aureus isolated from meat carcasses and bovine milk in abattoirs and dairy farms of the eastern Cape, South Africa. Int J Environ Res Public Health. 2018;15(10):2223. doi: 10.3390/ijerph15102223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LB. Impetigo—review. An Bras Dermatol. 2014;89:293–299. doi: 10.1590/abd1806-4841.20142283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CTM, de Oliveira DSV, Veloso VS, Silva SSP, Santos LS, Lima AF, Oliveira FAA, de Melo MCN, Soares MJS. Microbiology quality, detection of enterotoxin genes and antimicrobial resistance of Staphylococcus aureus isolated from milk and Coalho cheese. Semina Ciências Agrárias. 2018;39(5):1957–1967. doi: 10.5433/1679-0359.2018v39n5p1957. [DOI] [Google Scholar]
- Regecová I, Výrostková J, Zigo F, Gregová G, Kováčová M. Detection of antimicrobial resistance of bacteria Staphylococcus chromogenes isolated from sheep’s milk and cheese. Antibiotics. 2021 doi: 10.3390/antibiotics10050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues MX, Silva NCC, Trevilin JH, Cruzado MMB, Mui TS, Duarte FRS, Castillo CJC, Canniatti-Brazaca SG, Porto E. Molecular characterization and antibiotic resistance of Staphylococcus spp. isolated from cheese processing plants. J Dairy Sci. 2017;100:5167–5175. doi: 10.3168/jds.2016-12477. [DOI] [PubMed] [Google Scholar]
- Rodríguez C, Petrelli L, Ramirez MS, Centron D, Hebert EM, Saavedra L. Genomic overview of acquired antibiotic resistance mechanisms in Lactobacillus. In: Ruzal SM, editor. Lactobacillus genomics and metabolic engineering. Buenos Aires: Caister Academic Press; 2019. pp. 185–204. [Google Scholar]
- Rodríguez-Lázaro D, Oniciuc EA, García PG, Gallego D, Fernández-Natal I, Dominguez-Gil M, Eiros-Bouza JM, Wagner M, Nicolau AI, Hernández M. Detection and characterization of Staphylococcus aureus and methicillin-resistant S. aureus in foods confiscated in EU borders. Front Microbiol. 2017;8:1344. doi: 10.3389/fmicb.2017.01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat YR, Fooladi AAI, Shapouri R, Hosseini MM, Deilami KZ (2014) Prevalence of enterotoxigenic Staphylococcus aureus in organic milk and cheese in Tabriz, Iran. Iran J Microbiol 6:345–349. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4385576/pdf/IJM-6-345.pdf [PMC free article] [PubMed]
- Saima S, Fiaz M, Manzoor M, Zafar R, Ahmed I, Nawaz U, Arshad M. Molecular investigation of antibiotic resistant bacterial strains isolated from wastewater streams in Pakistan. 3Biotech. 2020;10:378. doi: 10.1007/s13205-020-02366-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandt CH, Krouse DA, Cook CR, Hackman AL, Chmielecki WA, Warren NG. The key role of pulsed-field gel electrophoresis in investigation of a large multiserotype and multistate foodborne outbreak of Salmonella infections centered in Pennsylvania. J Clin Microbiol. 2020;44(9):3208–3212. doi: 10.1128/JCM.01404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasirekha B, Usha MS, Amruta JA, Ankit S, Brinda N, Divya R. Incidence of constitutive and inducible clindamycin resistance among hospital-associated Staphylococcus aureus. 3Biotech. 2014;4(1):85–89. doi: 10.1007/s13205-013-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahkarami F, Rashki A, Ghalehnoo ZR. Microbial susceptibility and plasmid profiles of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. Jundishapur J Microbiol. 2014;7(7):e16984. doi: 10.5812/jjm.16984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallcross LJ, Fragaszy E, Johnson AM, Hayward AC. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13(1):43–54. doi: 10.1016/S1473-3099(12)70238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhu J, Xu Y, Tang X, Yang Z, Huang A. Malonyl-proteome profiles of Staphylococcus aureus reveal lysine malonylation modification in enzymes involved in energy metabolism. Proteome Sci. 2021;19:1. doi: 10.1186/s12953-020-00169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu V, Spanu C, Virdis S, Cossu F, Scarano C, de Santis EPL. Virulence factors and genetic variability of Staphylococcus aureus strains isolated from raw sheep’s milk cheese. Int J Food Microbiol. 2012;153(1–2):53–57. doi: 10.1016/j.ijfoodmicro.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002;85(1):57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhou L, Zhao BC, Deng X, Cho H, Yi C, Jian X, Song CX, Luan CH, Bae T, Li Z, He C. Targeting MgrA-mediated virulence regulation in Staphylococcus aureus. Chem Biol. 2011;18(8):1032–1041. doi: 10.1016/j.chembiol.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell T, Hall CA (2020) Carbuncle. Treasure Island (FL): StatPearls Publishing [Internet]. https://www.ncbi.nlm.nih.gov/books/NBK554459/. Accessed 6 May 2021
- Verraes C, Vlaemynck G, Van Weyenberg S, De Zutter L, Daube G, Sindic M, Uyttendaele M, Herman L. A review of the microbiological hazards of dairy products made from raw milk. Int Dairy J. 2015;50:32–44. doi: 10.1016/j.idairyj.2015.05.011. [DOI] [Google Scholar]
- Vitale M, Gaglio S, Galluzzo P, Cascone G, Piraino C, Presti DML, V, Alduina R, Antibiotic resistance profiling, analysis of virulence aspects and molecular genotyping of Staphylococcus aureus isolated in Sicily, Italy. Foodborne Pathog Dis. 2018;15(3):177–185. doi: 10.1089/fpd.2017.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Characteristics of food poisoning in mainland China, 2006–2015. Prev Med (baltim) 2018;25(3):257–260. doi: 10.3969/j.issn.1006-3110.2018.03.001. [DOI] [Google Scholar]
- Wang L, Ruan S. Modeling nosocomial infections of methicillin-resistant Staphylococcus aureus with environment contamination. Sci Rep. 2017;7:580. doi: 10.1038/s41598-017-00261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ryu D, Houtkooper RH, Auwerx J. Antibiotic use and abuse: a threat to mitochondria and chloroplasts with impact on research, health, and environment. BioEssays. 2015;37(10):1045–1053. doi: 10.1002/bies.201500071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Baloch Z, Jiang T, Zhang C, Peng Z, Li F, Fanning S, Ma A, Xu J. Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus Isolated from retail food in China. Front Microbiol. 2017;8:2256. doi: 10.3389/fmicb.2017.02256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Zhong J, Hu T, Zhao X. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3Biotech. 2018;8(1):76. doi: 10.1007/s13205-018-1086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H, Uehara Y, Lu Y, Sasaki T, Hiramatsu K. Genetic and phenotypic diversity of methicillin-resistant Staphylococcus aureus among Japanese inpatients in the early 1980s. Sci Rep. 2021;11:5447. doi: 10.1038/s41598-021-84481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version contains supplementary material for this article (Table S4, S5, S6, S7, and S8 dataset) can be accessed at https://doi.org/10.6084/m9.figshare.14479809 (DOCX 204 KB)