Abstract
The baker's yeast Saccharomyces cerevisiae expresses three homologues of the Nramp family of metal transporters: Smf1p, Smf2p, and Smf3p, encoded by SMF1, SMF2, and SMF3, respectively. Here we report a comparative analysis of the yeast Smf proteins at the levels of localization, regulation, and function of the corresponding metal transporters. Smf1p and Smf2p function in cellular accumulation of manganese, and the two proteins are coregulated by manganese ions and the BSD2 gene product. Under manganese-replete conditions, Bsd2p facilitates trafficking of Smf1p and Smf2p to the vacuole, where these transport proteins are degraded. However, Smf1p and Smf2p localize to distinct cellular compartments under metal starvation: Smf1p accumulates at the cell surface, while Smf2p is restricted to intracellular vesicles. The third Nramp homologue, Smf3p, is quite distinctive. Smf3p is not regulated by Bsd2p or by manganese ions and is not degraded in the vacuole. Instead, Smf3p is down-regulated by iron through a mechanism that does not involve transcription or protein stability. Smf3p localizes to the vacuolar membrane independently of metal treatment, and yeast cells lacking Smf3p show symptoms of iron starvation. We propose that Smf3p helps to mobilize vacuolar stores of iron.
Nramp (for natural resistance-associated macrophage protein) represents a family of evolutionarily conserved membrane proteins that facilitate the transport of heavy metal ions (5, 6, 15, 17, 31). Members of the Nramp family have been found in mammals, birds, insects, plants, fungi, and bacteria (3, 5, 9, 15, 16, 22, 40). Among the best studied are the Nramp1 and Nramp2 transporters of rodents. Although these proteins share 61% homology at the amino acid level, they exhibit distinct functions. Mouse Nramp1 plays an important role in the control of infection against intracellular parasites and is exclusively expressed in monocytes/macrophages and polymorphonuclear leukocytes (2, 15). Nramp2 (also known as DCT1 or DMT1) is more ubiquitously expressed in most tissues (17) and acts as a divalent metal transporter capable of transporting iron, manganese, copper, zinc, cadmium, and lead (17). Mutations in Nramp2 have been associated with defects in duodenal iron uptake and cellular iron utilization in the mk mouse and the Belgrade rat models of anemia (13, 14).
The baker's yeast Saccharomyces cerevisiae expresses three closely related Nramp homologues, Smf1p, Smf2p, and Smf3p, encoded by SMF1, SMF2, and SMF3, respectively (6, 7, 29, 43). Like mammalian DMT1, the Smf proteins exhibit a somewhat broad substrate specificity. Smf1p was originally defined as a high-affinity manganese transporter (39) and was later shown to contribute to cellular accumulation of cadmium and copper (28). Smf2p can affect cobalt levels in yeast (28) and may also participate in manganese trafficking (43). In more recent studies, Chen and coworkers demonstrated that both Smf1p and Smf2p can stimulate iron uptake into Xenopus oocytes (7). The role of Smf3p in metal homeostasis has not been defined.
Studies on Smf1p have revealed a novel method of regulating Nramp transport activity in response to metals. Specifically, treatment of yeast cells with manganese triggers the rapid degradation of the Smf1 protein. When cells are replete with manganese, the bulk of Smf1p is targeted to the yeast vacuole for degradation, and this vacuolar targeting involves the S. cerevisiae BSD2 gene product (26). Bsd2p is a membrane protein localized to the endoplasmic reticulum that helps direct Smf1p to the vacuole in response to manganese treatment (26, 28). When cells are starved for manganese, Smf1p fails to enter the vacuole, and the transporter arrives at the plasma membrane, where metal ion uptake is thought to occur (25, 26). This plasma membrane localization of Smf1p is independent of Bsd2p (26). Such a simple switch in localization of the metal transporter allows for rapid changes in metal uptake without the need for new protein synthesis.
By comparison, nothing is known regarding the cellular localization or regulation of the other yeast Nramp homologues, Smf2p and Smf3p. Are these proteins functionally redundant with Smf1p, or do they act in unique pathways of metal transport? To address this issue, we have comparatively analyzed the three yeast Nramp homologues at the levels of cellular localization, transporter regulation, and function. We report here that like Smf1p, Smf2p is regulated at the posttranslational level by manganese ions and the BSD2 gene. By comparison, Smf3p shows no regulation by manganese; however, Smf3 protein levels are controlled by iron through a posttranscriptional mechanism. We additionally found that the three Smf proteins are housed at distinct cellular locations: Smf1p at the cell surface, Smf2p in intracellular vesicles, and Smf3p at the vacuolar membrane. Hence, as is the case with mammals, fungi have evolved with diverse Nramp isoforms that perform unique functions in metal metabolism.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The S. cerevisiae strains used in this study are presented in Table 1. The isogenic set of wild-type (YR98), bsd2Δ (XL115), pep4Δ (XL126), bsd2Δ pep4Δ (XL125), smf1Δ(XL112), and smf2Δ (XL117) strains has been previously described (26, 28). The smf1Δ smf2Δ strain XL131 was created by replacing the SMF2 gene of XL112 with HIS3 using the pSMF2Δ-HIS3 plasmid (28). The smf3Δ (MP112) and smf1Δ smf2Δ smf3Δ (MP113) strains were constructed by disrupting SMF3 in YR98 and XL131 using the smf3Δ::LEU2 plasmid pJS409. The aft1Δ (YPH250Δaft1), AFT1-up (M2P), ubc7Δ (SM3397), and pre1-1ts pre2-1ts (SM2899) strains and their corresponding wild-type parents (YPH250, CM3260, SM2561, and SM2898, respectively) were kind gifts of A. Dancis, D. Kosman, and S. Michaelis. Stocks of strains were maintained on standard yeast extract-peptone-dextrose (YPD) media. Cultures for experimental analysis were obtained by growth in a synthetic minimal medium containing dextrose (SD) (36) or in a metal-depleted minimal defined medium (MDM) prepared through the use of an ion-exchange resin (10, 26). As needed, 10 μM ZnCl2, 10 μM Fe(NH4)2(SO4)2, 1.0 μM CuSO4, 10 μM MnSO4, 1.0 μM CoSO4, or a combination of these metals was added to the MDM.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YR98 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 | 27 |
| XL115 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 bsd2Δ::HIS3 | 27 |
| XL126 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 pep4Δ::URA3 | 27 |
| XL125 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 pep4::URA3, bsd2Δ::HIS3 | 27 |
| XL112 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 smf1Δ::URA3 | 27 |
| XL117 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 smf2Δ::HIS3 | 27 |
| XL131 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 smf1Δ::URA3 smf2Δ::HIS3 | This study |
| MP112 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 smf3Δ::LEU2 | This study |
| MP113 | MATα ade2 his3Δ200 leu2-3 112 lys2Δ201 ura3-52 smf1Δ::URA3 smf2Δ::HIS3 smf3Δ::LEU2 | This study |
| YPH250 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 | 23 |
| YPH250Δaft1 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 aft1Δ::TRP1 | 23 |
| CM3260 | MATa ura3-52 trp1-63 leu2-3 gcn4-101 his3-609 | 23 |
| M2P | MATa ura3-52 trp1-63 leu2-3 gcn4-101 his3-609 AFT1-up | 23 |
| SM2561 | MATa his3 leu2 ura3 lys2 trp1 | S. Michaelis |
| SM3397 | MATa his3 leu2 ura3 lys2 trp1 ubc7Δ::LEU2 | S. Michaelis |
| SM2898 | MATa ura3 leu2-3,113 his3-11,15 | S. Michaelis |
| SM2899 | MATa ura3 leu2-3,113 his3-11,15 pre1-1 pre2-1 | S. Michaelis |
Molecular biology.
An SMF3 deletion plasmid, pJS409, was constructed by amplifying SMF3 sequences from −493 to +116 and from +1313 to +1862 by PCR using primers designed to introduce HindIII and BamHI sites or SalI and HindIII sites on the upstream and downstream fragments, respectively. After digestion at these sites, the fragments were simultaneously ligated into the BamHI and SalI sites of pRS305 (LEU2) (37). Linearization of this plasmid with HindIII and transformation of yeast cells resulted in a deletion in chromosomal SMF3 sequences from +116 to +1313 that was verified by PCR. The CEN Smf1-hemagglutinin (HA)-expressing plasmid pSF4 was previously described (26). A multicopy Smf1-HA plasmid, pSF5, was constructed by inserting the ApaI-to-NdeI fragment of pSF4 into the 2μ LEU2 plasmid pVC36 (23). The Smf2-HA-expressing plasmid pSF6 was constructed by amplifying SMF2 sequences from −257 to the stop codon employing a primer that replaced the termination sequence with NdeI and by using these sequences to replace the NdeI-ApaI BSD2-containing fragment of plasmid pXL36 (CEN LEU2) (27). As a result, Smf2p was fused at the C terminus to two copies of the HA epitope. A multicopy Smf2-HA plasmid, pSF7, was obtained by inserting the ApaI-NdeI fragment of pSF6 into the 2μ LEU2 plasmid pVC36 (23).
Two Smf3-HA plasmids were constructed: one whose product contained the HA tag at the C terminus and the other whose product had HA integrated at amino acid position 424. The SMF3 construct (pMP043) whose product had a C-terminal tag was obtained in a manner identical to that of pSF6 using SMF3 sequences from −578 to the stop codon. The internally tagged Smf3-HA plasmid (pMP054) was constructed by first amplifying SMF3 sequences from −578 to +1824 and inserting these sequences into the BamHI and XhoI sites of pRS316 (CEN URA3) (37). Dual tandem copies of the sequence encoding the HA epitope were then introduced in frame at position +1272 by two successive rounds of site-directed mutagenesis (Quikchange; Stratagene).
For RNA blot analysis of SMF2 and SMF3 expression, 30 and 10 μg, respectively, of total RNA was subjected to formaldehyde-agarose gel electrophoresis, followed by transfer to a nylon membrane as described previously (35). Detection of specific RNA transcripts employed SMF2 (sequences from −257 to +1664) and SMF3 (+418 to +1429) DNA probes amplified by PCR and radiolabeled with 32P.
Biochemical analyses and immunodetection techniques.
Measurements of manganese ion accumulation were obtained through atomic absorption spectrophotometry. Cells were grown to an optical density at 600 nm (OD600) of 1.0 in 10 ml of MDM supplemented with 150 nM CuSO4 and 10 μM ZnCl2. Cells were then harvested, washed, resuspended in 500 μl of distilled water, and subjected to atomic absorption spectrophotometry as described earlier (27).
To monitor iron regulation of the FET3 promoter, cells transformed with a 2μ URA3 FET3-lacZ reporter plasmid (kind gift of D. Winge) were grown overnight in selecting SD medium, diluted into YPD to an OD600 of 0.5, and grown for an additional 5 h until mid-log phase. Cells were harvested and washed, and crude extracts were prepared by glass bead homogenization in Z buffer (0.06 M Na2HPO4, 0.04 M Na2H2PO4, 0.01 M KCl, 0.001 M MgSO4 [pH 7.0]). A total of 100 μg of whole-cell extract in 400 μl of Z buffer was then combined with 100 μl of a 4-mg/ml solution of o-nitrophenyl-β-d-galactopyranoside. Following a 1-h incubation at 30°C, lacZ activity was measured as a function of absorption at 420 nm.
For Western blot analysis, yeast cells expressing the Smf1-HA, Smf2-HA, or Smf3-HA fusion protein were grown to mid-logarithmic phase (OD600 = 1.0) in selecting SD medium or MDM as needed. Extracts were prepared by either alkaline lysis (30) or glass bead homogenization (32), with similar results. Samples (10 to 20 μg) were subjected to sodium dodecyl sulfate gel electrophoresis on precast 12% polyacrylamide gels (Invitrogen) and were analyzed by Western blotting using a mouse anti-HA antibody (BABCO) as previously described (27).
Immunofluorescence microscopy analysis was conducted essentially as described previously (26), using strains transformed with a CEN plasmid for the expression of either Smf3-HA, Smf2-HA, or Smf1-HA. Cells were grown to a mid-logarithmic stage in selecting SD medium or MDM, fixed with formaldehyde, digested with Zymolyase, and stained with a mouse anti-HA antibody for 2 h. The secondary antibody consisted of a goat anti-mouse antibody coupled to fluorescein isothiocyanate (FITC) (Boehringer Mannheim), and staining proceeded for 1 h. Nucleic acids were stained by incubation with 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) per ml for 5 min. FITC and DAPI staining were monitored by fluorescence microscopy, whereas visualization of yeast vacuoles used Nomarski optics.
RESULTS
Regulation of Smf2p by the BSD2 gene and metal ions.
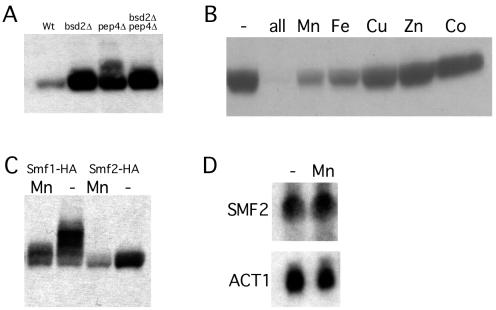
Yeast Smf1p is known to be regulated at the posttranslational level by manganese ions and Bsd2p; however, the regulation of other yeast Nramp homologues had not been explored. To monitor expression and regulation of Smf2p, the protein was tagged at the C terminus with an HA epitope and was expressed from its native promoter on a CEN vector. The corresponding protein complemented a manganese trafficking defect associated with loss of SMF2 (E. Luk and V. C. Culotta, unpublished data). To examine whether Smf2-HA is subject to the same pathway of protein turnover seen with Smf1p, we used a set of isogenic strains containing mutations in BSD2 or in PEP4. PEP4 is necessary for vacuolar proteolysis (19). As shown in Fig. 1A, Smf2-HA protein levels showed a dramatic increase in the bsd2Δ strain compared to those of the isogenic wild-type strain. Smf2-HA also accumulated to a high level in the pep4Δ mutant, and there was no additive effect seen with bsd2 and pep4 mutations. Thus, as is the case with Smf1p (26), Smf2p is subject to degradation by vacuolar proteases in a Bsd2p-dependent fashion. We additionally examined whether Smf2p falls under the negative control of metal ions. As shown by immunoblot analysis, depletion of the heavy metals zinc, copper, iron, and manganese from the growth medium resulted in a dramatic increase in Smf2-HA protein levels in a wild-type strain (Fig. 1B). The individual addition of manganese and iron back to the medium partially suppressed Smf2-HA levels, whereas supplementation with zinc, copper, or cobalt had no effect (Fig. 1B). This pattern of negative regulation by metal ions is very similar to what has been previously reported for Smf1p (26). As shown in Fig. 1C, both Smf1p and Smf2p are repressed by the addition of manganese to the growth medium, although the accumulation of Smf1-HA under metal starvation conditions appears to be approximately two- to threefold higher than that obtained with Smf2-HA.
FIG. 1.
Role of BSD2, vacuolar degradation, and metal ions in the control of Smf2p expression levels. (A) The indicated yeast strains were transformed with pSF6 expressing Smf2-HA, and the corresponding cell lysates were analyzed by Western blot analysis using an anti-HA antibody. Strains used: wild type (Wt), YR98; bsd2Δ, XL115; pep4Δ, XL126; pep4Δ bsd2Δ, XL125. (B) Strain YR98 transformed with the Smf2-HA plasmid pSF6 was grown in MDM depleted of metal ions (−) or in the same medium supplemented with the individual metals [i.e., 10 μM ZnCl2, 10 μM Fe(NH4)2(SO4)2, 1.0 μM CuSO4, 10 μM MnSO4, or 1.0 μM CoSO4] or a combination of all the metals. Smf2-HA expression was monitored by Western blotting as for panel A. (C) Expression of Smf1-HA (pSF4) and Smf2-HA (pSF6) proteins in strain YR98 was analyzed by Western blotting as for panel A. (D) YR98 cells grown in MDM (−) or in MDM supplemented with 10 μM MnSO4 were subjected to Northern blot analysis with sequential hybridization to an SMF2 probe and an ACT1 probe as a control.
To test whether gene transcription played any role in metal regulation of SMF2, RNA prepared from cells grown in metal-depleted or manganese-replete medium was analyzed by Northern blotting. As shown in Fig. 1D, SMF2 mRNA levels were not changed by metal depletion, demonstrating that metal ions increase Smf2p expression only through an increase in protein accumulation, as has been reported previously for Smf1p (26).
Localization of Smf2p.
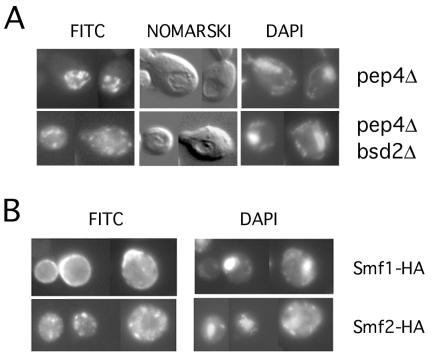
Indirect immunofluorescence microscopy was utilized to determine the subcellular localization of Smf2p. Cells expressing Smf2-HA from its native promoter were probed with a mouse anti-HA antibody and with a secondary antibody coupled to FITC. Under metal-replete conditions, the steady-state levels of Smf2p were too low to be detected by immunofluorescence microscopy (not shown). Yet since the bulk of Smf2p is degraded by vacuolar proteases under these conditions, we could readily detect Smf2-HA localization in a pep4Δ mutant. In this case, very intense staining of Smf2-HA was observed within the lumen of the vacuole identified by Nomarski optics (Fig. 2A). A quite different staining pattern was found in an isogenic pep4Δ bsd2Δ double mutant. In this case, there was no vacuolar staining, and the Smf2-HA localization was restricted to punctate bodies reminiscent of Golgi-like vesicles. Hence, as has been reported for Smf1p (26), Smf2p is normally targeted to the vacuole for degradation by PEP4-dependent proteases, and this delivery of Smf2p to the vacuole is dependent on the BSD2 gene product. However, contrary to what has been observed for Smf1p (26), we failed to detect any cell surface staining of Smf2p in bsd2 mutants.
FIG. 2.
Immunofluorescence microscopy of Smf2-HA. (A) Strains XL126 (pep4Δ) and XL125 (pep4Δ bsd2Δ) transformed with pSF6 expressing Smf2-HA were grown in MDM supplemented with all five essential metal ions as described in the legend to Fig. 1B. Cells were probed with anti-HA and an FITC-conjugated anti-mouse antibody and doubly stained with DAPI for nucleic acid detection. Cells were analyzed by epifluorescence at a magnification of ×1,000 or by Nomarski optics for visualization of vacuoles seen as indentations. (B) Strain YR98 expressing either Smf1-HA (from pSF4) or Smf2-HA (pSF6) grown in MDM without metal supplementation was subjected to immunofluorescence microscopy as for panel A.
We additionally compared the localization of Smf1p and Smf2p under metal starvation conditions. Consistent with our earlier studies, the majority of Smf1-HA exhibited a cell surface staining pattern when cells were starved for manganese (Fig. 2B). However, under precisely the same conditions, Smf2-HA localization was restricted to intracellular punctate bodies. Therefore, although Smf1p and Smf2p are coregulated by manganese ions and Bsd2p, they exhibit distinct cellular localizations.
The SMF3 gene.
Upon inspection of the S. cerevisiae genome sequence, we identified an open reading frame whose product exhibited extensive homology to yeast Smf1p and Smf2p (YLR034C); this gene was entered in the Saccharomyces cerevisiae Genome Database as SMF3. Overall, the three yeast Smf proteins show ∼26% identity to human Nramp2 (data not shown) and ∼48% identity to each other at the amino acid level (Fig. 3). This homology extends over most of the proteins' sequences, with three notable exceptions: Smf3p lacks an amino-terminal extension present on Smf1p and Smf2p, Smf1p harbors additional intervening sequence separating transmembrane domains 6 and 7, and Smf3p has a comparatively short sequence separating transmembrane domains 10 and 11 (Fig. 3).
FIG. 3.
Amino acid compositions of the S. cerevisiae Nramp homologues. Amino acid alignment of Smf1p, Smf2p, and Smf3p using the ClustalW alignment program. Asterisks represent amino acid identity and dots represent amino acid similarity. Bold lines above sequences represent predicted transmembrane domains based on hydropathy analysis and on homology to other members of the Nramp family (6, 17). The arrow represents the position in Smf3p where two copies of HA were introduced by site-directed mutagenesis (see Materials and Methods).
Regulation of SMF3 by iron.
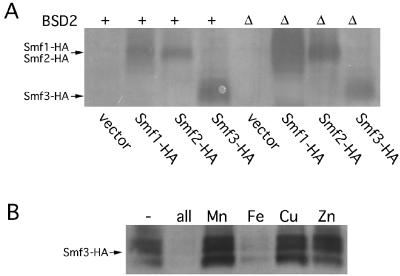
Expression of Smf3p was monitored through introduction of an HA epitope either at the C terminus of the protein (as was used for Smf1p and Smf2p in the experiment shown in Fig. 1) or at an internal region between transmembrane segments 10 and 11 (as was used for Smf1p in other studies [43]). To address whether Smf3p is negatively regulated by BSD2, the epitope-tagged versions of Smf3-HA were expressed in an isogenic set of wild-type and bsd2Δ mutant strains. As seen in Fig. 4A, loss of BSD2 increased Smf1-HA and Smf2-HA protein expression, but no similar increase was observed with Smf3-HA. Identical results were obtained with Smf3p regardless of whether the HA tag was placed at the C terminus (Fig. 4A) or at the internal site (not shown). We next tested whether SMF3 is regulated by metals. As seen in Fig. 4B, levels of the Smf1-HA protein were greatly increased in medium depleted of manganese, iron, copper, and zinc, demonstrating that like Smf1p and Smf2p, the Smf3 polypeptide is negatively regulated by heavy metals. However, while Smf1p and Smf2p are down-regulated by manganese (Fig. 1C), addition of manganese ions to the growth medium had no effect on Smf3-HA (Fig. 4B). Instead, Smf3-HA was strongly repressed by iron (Fig. 4B). Again, the results with Smf3p were independent of the position of the HA epitope tag.
FIG. 4.
Regulation of Smf3 protein levels. (A) The isogenic wild-type YR98 (+) and bsd2Δ mutant strain XL115 (Δ) were transformed with plasmids for the expression of Smf1-HA (pSF5), Smf2-HA (pSF7), or Smf3-HA (pMP043) or with the empty vector pRS425. Cells were grown in selecting SD medium, and lysates were prepared and analyzed by Western blotting as described for Fig. 1A. Arrows mark the positions of the 65-kDa Smf1-HA, 62-kDa Smf2-HA, and 54-kDa Smf3-HA polypeptides; molecular masses were confirmed by comigration of standards (not shown). (B) Strain YR98 expressing Smf3-HA from plasmid pMP054 was grown in MDM that was supplemented with the indicated metal ions as described for Fig. 2B. Cells lysates were prepared and subjected to Western analysis as for panel A.
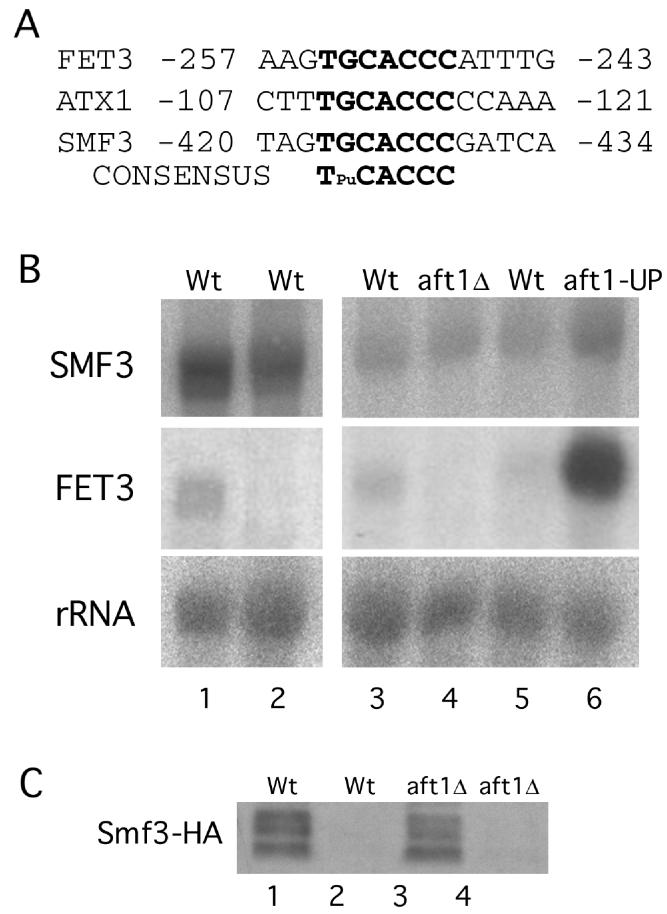
Upon inspection of the 5′ flanking region of SMF3, we noted a consensus sequence for Aft1p, an iron-sensing transcription factor that activates iron metabolism genes in yeast (44, 45) (Fig. 5A). No such sequence was noted for SMF1 or SMF2 (data not shown), suggesting that iron regulation of SMF3 may reflect transcription control by Aft1p. To address this, SMF3 mRNA levels were monitored by Northern blotting. Under the same iron starvation and iron-replete conditions that modulate Smf3 protein (Fig. 4B), there was no substantial effect on SMF3 mRNA (Fig. 5B, lanes 1 and 2). By comparison, transcription of the FET3 gene, involved in iron uptake (1, 11, 21), was completely repressed by iron. FET3 is Aft1 regulated (4, 45), and accordingly the gene was strongly induced in a strain expressing a constitutively active AFT-up allele (Fig. 5B, lanes 5 and 6) and repressed in an aft1Δ null strain (Fig. 5B, lanes 3 and 4). By contrast, SMF3 transcription was not decreased by the aft1Δ mutation and was only modestly increased in the AFT-up strain (Fig. 5B, lanes 3 through 6). Moreover, a null mutation in AFT1 had no effect on the dramatic iron repression of Smf3p seen at the level of protein expression (Fig. 5C). Therefore, iron regulation of Smf3p does not involve either Aft1p or SMF3 gene transcription.
FIG. 5.
AFT1 and SMF3 gene expression. (A) Marked in bold are the Aft1p consensus binding sequences for FET3 (45), ATX1 (24), and SMF3. Numbers indicate sequences with respect to the translational start site. (B) The indicated strains were grown in MDM that was depleted of heavy metals (lane 1), MDM supplemented with 10 μM Fe(NH4)2(SO4)2 (lane 2), or YPD medium (lanes 3 through 6). Total RNA was subjected to Northern analysis with sequential hybridization to SMF3 and FET3 probes. The following strains were utilized: wild type (Wt), YR98 (lanes 1 and 2), YPH250 (lane 3, and CM3260 (lane 5); aft1Δ, YPH250aft1Δ; and aft1-UP, M2P. (C) The indicated strains expressing Smf3-HA (from pMP054) were grown in MDM that was either iron depleted (lanes 1 and 3) or supplemented with iron (lanes 2 and 4) as for panel B. Cell lysates were subjected to Western blot analysis as for Fig. 4. Wt, YPH250; aft1Δ, YPH250aft1Δ.
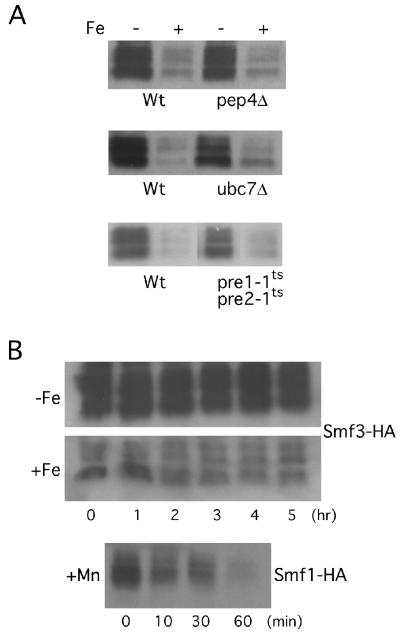
We next tested whether iron regulates Smf3p at the level of protein turnover. A null mutation in PEP4 did not alter the levels of Smf3p tagged with HA at amino acid 424 (Fig. 6A), demonstrating that this protein is not subject to vacuolar degradation. (While Smf3p tagged with HA at position 424 is resistant to proteolysis, the corresponding tag at the C terminus is degraded in a PEP4-dependent manner, as this peptide is predicted to lie within the lumen of the vacuole (see Fig. 10). It is likely that the short hydrophilic C terminus of native Smf3p (≈6 amino acids) is normally resistant to vacuolar proteases, but addition of the 18-amino-acid HA tag exposes the C terminus to proteolytic digestion.) Another major protein degradation pathway in yeast involves the 26S proteasome, where proteins destined for degradation are tagged with ubiquitin via the action of ubiquitin-conjugating enzymes, such as Ubc7p (8, 20). It has previously been shown that Smf1p is partially stabilized in a ubc7 mutant (26). However, as seen in Fig. 6A, the steady-state levels of Smf3-HA were only modestly affected by a ubc7Δ mutation, and the same results were obtained regardless of the position of the HA epitope. To more definitively address the role of the proteasome in Smf3p degradation, we monitored Smf3-HA expression in a strain containing temperature-sensitive mutations in PRE1 and PRE2, two essential genes encoding components of the proteasome (18). In this experiment, cells were grown at the permissive temperature prior to shifting to the nonpermissive temperature for 3 h to inactivate the proteasome. As seen in Fig. 6A, Smf3-HA levels were not substantially affected by the pre1-1 pre2-1 mutation, indicating that proteasome degradation does not account for the large effects of iron on Smf3p expression. In parallel with these genetic studies, the effect of iron on Smf protein turnover was also tested by monitoring degradation rates of the protein under metal-starved or iron-replete conditions. For these studies, the time course of Smf3 stability was examined in wild-type cells treated with cycloheximide to inhibit new protein synthesis. As shown in Fig. 6B, Smf3-HA was extremely stable. The protein levels remained largely constant over 5 h, even in iron-treated cells. By comparison, the same cycloheximide treatment resulted in rapid degradation of Smf1-HA in cells replete with manganese (Fig. 6B, bottom). Overall, these studies strongly indicate that iron regulates Smf3 protein levels by a novel mechanism that does not appreciably involve either transcription or protein stability.
FIG. 6.
Stability of the Smf3 polypeptide. (A) The designated strains were transformed with the Smf3-HA-expressing vector pMP054 and were grown in MDM that was supplemented with iron where indicated as for Fig. 5B. In the experiment with the pre1-1ts pre2-1ts mutant, cells were cultured at 25°C prior to shifting to 37°C for 3 h. All cells were subjected to Western blot analysis as for Fig. 4. The following strains were utilized: wild type (Wt), YR98 (upper panel), SM2561 (middle panel), and SM2898 (lower panel); pep4Δ, XL126; ubc7Δ, SM3397; and pre1-1ts pre2-1ts, SM2899. (B) Strain YR98 expressing either Smf3-HA (on pMP054) or Smf1-HA (on pSF5) were grown in MDM alone (−Fe) or in MDM supplemented either with 10 μM iron (+Fe) or with 10 μM manganese (+Mn) as described in the legend to Fig. 1B. Following growth to an OD600 of 0.6, 100 μg of cycloheximide per ml was added and aliquots of cells were removed at the indicated time points for Western analysis.
FIG. 10.
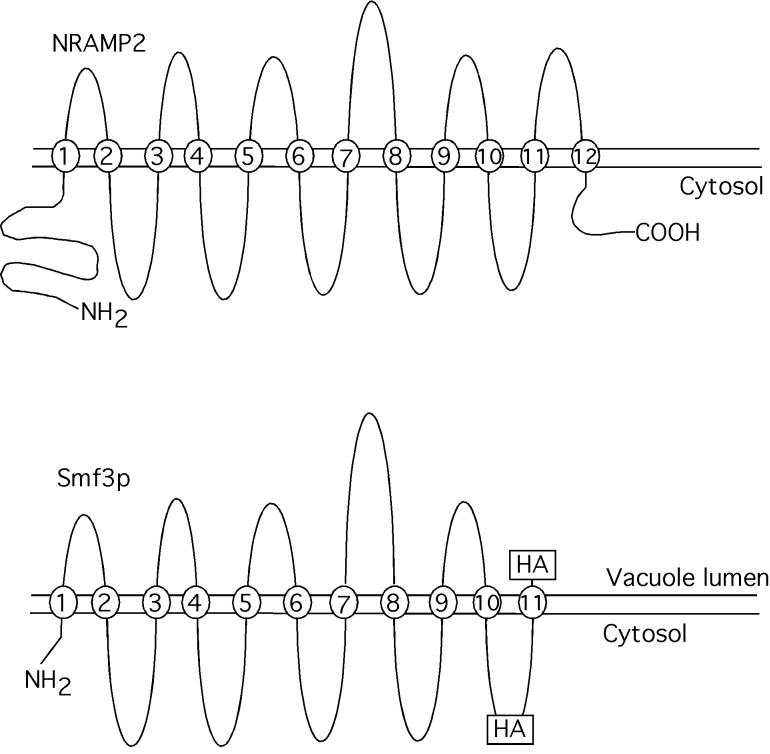
Membrane topology of Smf3p. Shown are the predicted membrane topologies for mammalian DMT1 (6, 17) and yeast Smf3p. Positions at which HA epitopes were introduced into Smf3p are indicated.
Localization of Smf3p.
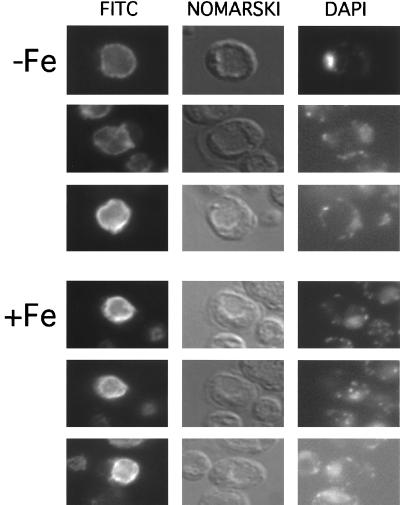
The subcellular localization of Smf3p was examined by indirect immunofluorescence microscopy. Since Smf1p (26) and Smf2p (Fig. 2B) exhibit shifts in localization with metal treatment, we analyzed Smf3-HA localization under iron-replete and iron starvation conditions. As seen in Fig. 7, Smf3-HA exhibited rim staining coincident with the vacuolar membrane. The vacuoles were identified as indentations under Nomarski optics. It is noteworthy that this rim or membrane staining of the vacuole is quite distinct from the luminal or internal staining of the vacuole seen with Smf2p (Fig. 2A) or Smf1p (26) under metal-replete conditions. Furthermore, unlike that of Smf1p and Smf2p, Smf3p localization did not change under metal starvation conditions, and these results were obtained with Smf3p containing the HA epitope either at the C terminus (not shown) or at position 424 (Fig. 7). It thus appears that Smf3p is a constant resident of the vacuolar membrane.
FIG. 7.
Immunofluorescence microscopy localization of Smf3p. Strain YR98 expressing Smf3-HA (from pMP054) was grown in MDM supplemented with 10 μM iron where indicated. Cells were fixed and subjected to immunofluorescence microscopy as for Fig. 2.
Role of Smf3p in iron homeostasis.
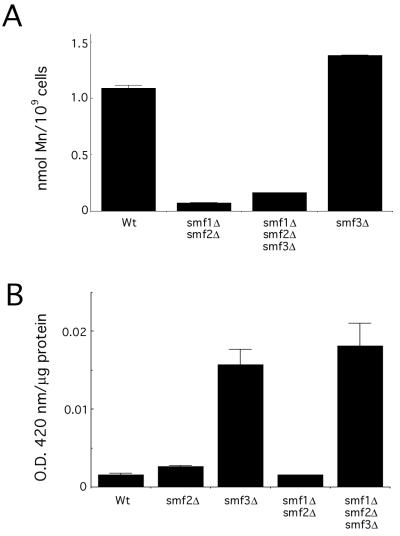
Since yeast Smf1p and Smf2p are specifically regulated by manganese, it is not surprising that these transporters participate in manganese uptake and distribution (39, 43; Luk and Culotta, unpublished). As expected, a combined deletion of SMF1 and SMF2 results in a dramatic decrease in steady-state accumulation of cellular manganese as measured by atomic absorption spectroscopy (Fig. 8A). However, a corresponding mutation in SMF3 had no effect on manganese accumulation. A single smf3Δ mutant displayed wild-type cellular levels of manganese, and there was no further decrease in manganese accumulation in the smf1Δ smf2Δ smf3Δ triple mutant compared to that of the smf1Δ smf2Δ strain (Fig. 8A).
FIG. 8.
Effects of smf null mutations. (A) The indicated strains were grown in MDM supplemented with 150 nM CuSO4 and 10 mM ZnCl2 (the medium was depleted of iron and manganese to maximize expression of the Smf proteins). Cells were washed and prepared for atomic absorption spectroscopy analysis of manganese content. The following strains were analyzed: wild type (Wt), YR98; smf2Δ, XL117; smf3Δ, MP112; smf1Δ smf2Δ, XL131; and smf1Δ smf2Δ smf3Δ, MP113. (B) The indicated strains were tested for expression of an FET3-lacZ reporter plasmid as described in Materials and Methods. Beta-galactosidase activity is reported as units of absorption at 420 nm per microgram of lysate protein. Strains utilized are as described in the legend to panel A.
Based on the tight regulation of Smf3p by iron (and not manganese), it seemed possible that this Nramp isoform functions in iron homeostasis. To address whether Smf3p affects intracellular iron stores, we used a reporter construct in which the FET3 gene promoter was fused to lacZ. Through regulation involving Aft1p, this promoter is strongly induced by iron starvation conditions (4, 45). As shown in Fig. 8B, the smf3Δ strain exhibited a large induction of lacZ reporter activity suggestive of an iron starvation status. By comparison, an smf2Δ strain and an smf1Δ smf2Δ strain showed no induction of reporter activity, and an smf1Δ smf2Δ smf3Δ strain exhibited the same level of induction as seen with the single smf3Δ mutant (Fig. 8B). Therefore, Smf3p appears to play a role in controlling the intracellular availability of iron.
DISCUSSION
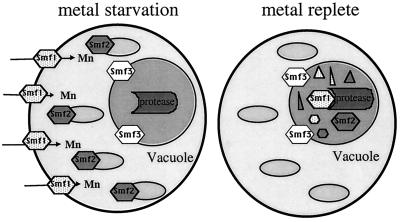
Here we have compared and contrasted the three isoforms of yeast Nramp proteins based on their regulation by metals, cellular localization, and function. Our major finding is that yeast Smf1p, Smf2p, and Smf3p are nonredundant. Smf1p and Smf2p are regulated by manganese and BSD2, and both are targeted to the vacuole for degradation when manganese is plentiful. However, Smf1p responds to manganese starvation by moving to the plasma membrane, whereas Smf2p redistributes to intracellular vesicles (Fig. 9). Smf3p is even more disparate. The Smf3 polypeptide is uniquely down-regulated by iron in a manner that does not involve protein turnover. Moreover, while Smf1p and Smf2p shift their localization upon metal starvation, Smf3p appears to be a constant resident of the vacuolar membrane (Fig. 9). We propose that Smf1p is primarily involved in the uptake of manganese (and possibly other metals) from the extracellular environment, whereas Smf2p's role may be to mobilize metals from vesicular stores. The nature of the vesicles in which Smf2p resides is unknown, but they appear to be important for delivering manganese to the mitochondria (Luk and Culotta, unpublished). We hypothesize that the primary role of Smf3p is to help mobilize iron stores of the vacuole (see below).
FIG. 9.
Model for yeast Nramp localization. Depicted is a diagram of a yeast cell, illustrating the distinct localizations of Smf1, Smf2, and Smf3. With metal starvation, Smf1 localizes to the cell surface (arrows indicate direction of manganese uptake), Smf2 localizes to intracellular vesicles (represented by small ovals), and Smf3 localizes at the vacuolar membrane (represented by inner circle). Upon metal-replete conditions (treatment with manganese and iron), both Smf1 and Smf2 are targeted to the vacuole lumen for degradation by proteases (small triangles and hexagons represent products of Smf degradation). Smf3 remains at the vacuolar membrane and is not degraded.
The divergent patterns of regulation and localization observed with Smf1p, Smf2p, and Smf3p presumably reflect unique features of the corresponding polypeptide sequences. Of the three Nramp isoforms, Smf1p and Smf2p are most closely related in overall topology, although Smf1p contains a somewhat extended N-terminal region compared to that of Smf2p, and this may facilitate a cell surface localization. The Smf3p sequence is the most different of the three proteins. Smf3p is completely devoid of the 5- to 7-kDa N-terminal extensions that are present in Smf1p and Smf2p and also exhibits a gap between transmembrane segments 10 and 11. It is possible that one or more of these missing sequences in Smf3p accounts for its lack of regulation by BSD2. Our preliminary studies suggest that the N-terminal regions of Smf1p and Smf3p are involved in the response to manganese versus iron; however, other, as-yet-unidentified sequences are needed for metal-specific regulation (data not shown). Work to identify these sequences is under way.
How is Smf3p regulated by iron? We found that the Smf3 polypeptide is not degraded in response to iron, and furthermore, iron does not appreciably regulate transcription of the SMF3 gene. While SMF3 does contain in its promoter a consensus site for the iron-sensing Aft1p transcription factor, Aft1p does not significantly modulate SMF3 expression. The effects seen are similar to the modest regulation of the S. cerevisiae ATX1 gene by AFT1 (24). Since mRNA and protein stability effects cannot account for the strong regulation of Smf3p by iron, Smf3p may be regulated at the level of protein translation. A limited degree of secondary structure in the 5′ untranslated region of SMF3 has been noted (data not shown), and work to determine the role of these sequences in modulating SMF3 expression is under way. Iron-mediated control of polypeptide translation has been described for mammalian proteins, such as ferritin (34, 41); however, the mechanism is likely to be different in the case of yeast Smf3p, as the relevant mammalian iron regulatory factor (i.e., cytosolic aconitase) is not expressed in this fungus.
Our data are consistent with a model in which Smf3p helps to mobilize iron from stores in the vacuole. Smf3p is always localized to the vacuolar membrane, whereas Smf1p and Smf2p are targeted to the vacuolar lumen for degradation. The vacuole in yeast has been hypothesized to contain a large reservoir of iron (33), and indeed smf3 mutants exhibit signs of iron starvation, as seen through induction of a FET3 reporter construct. A comparison of the predicted topology maps for Smf3p and the mammalian Nramp transporters is consistent with the transport of metal from the lumen of the vacuole to the cytosol (6) (Fig. 10). Smf3p lacks the most C-terminal transmembrane domain found in the mammalian Nramp transporters, and therefore, the C terminus of Smf3p is predicted to lie on the luminal face of the vacuole membrane. As evidence for the predicted topology, the HA epitope at the C terminus of Smf3p is removed by vacuolar proteases (susceptible to PEP4-dependent degradation), whereas the HA epitope placed at the predicted cytoplasmic side is resistant to PEP4 proteolysis (see above) (summarized in Fig. 10).
Recent work by Urbanowski and Piper (42) has identified a pair of proteins (Fth1p and Fet5p) on the vacuolar membrane that are homologous to the high-affinity iron uptake proteins, Ftr1p and Fet3p, at the cell surface. Based on topology analysis and homology to Ftr1p and Fet3p, the authors suggest that Fth1p and Fet5p may act to mobilize the iron stores of the vacuole (42). It is therefore possible that multiple pathways exist for moving iron out of the vacuole. Indeed, multiple systems operate at the cell surface for the uptake of iron (e.g., high-affinity Ftr1p and low-affinity Fet4p iron transporters [12, 38]), and Smf3p may be somewhat analogous to the Fet4p transporter, which operates on a variety of metals, including iron.
Although Nramp transporters can potentially recognize a broad range of metal substrates (e.g., Cu, Cd, Mn, and Fe) (17, 28, 40), biological specificity may come from their regulation. For example, SMF1 and SMF2 are expressed only under manganese or iron starvation conditions, whereas SMF3 is predominantly expressed under iron starvation. Overall, the Smf family of Nramp metal transporters in yeast is providing useful insight into metal-regulated gene expression and the complexity of metal ion homeostasis within a single cell.
ACKNOWLEDGMENTS
This work was supported by the JHU NIEHS center and by NIH grant ES 08996 to V.C.C. M.E.P. was supported by EPA STAR Fellowship U915646, and X.F.L. was supported by NIEHS training grant ES 07141.
We thank D. Winge for the FET3-lacZ reporter plasmid and S. Michaelis for yeast strains. We are also indebted to J. Strain for technical assistance and D. Sullivan for helpful discussions.
REFERENCES
- 1.Askwith C, Eide D, Ho A V-, Bernard P S, Li L, Davis-Kaplan S, Sipe D M, Kaplan J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 1994;76:403–410. doi: 10.1016/0092-8674(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson P G, Blackwell J M, Barton C H. Nramp1 locus encodes a 65 kDa interferon-gamma-inducible protein in murine macrophages. Biochem J. 1997;325:779–786. doi: 10.1042/bj3250779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belouchi A, Kwan T, Gros P. Cloning and characterization of the OsNramp family from Oryza sativa, a new family of membrane proteins possibly implicated in the transport of metal ions. Plant Mol Biol. 1997;33:1085–1092. doi: 10.1023/a:1005723304911. [DOI] [PubMed] [Google Scholar]
- 4.Casas C, Aldea M, Espinet C, Gallego C, Gil R, Herrero E. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast. 1997;13:621–637. doi: 10.1002/(SICI)1097-0061(19970615)13:7<621::AID-YEA121>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 5.Cellier M, Belouchi A, Gros P. Resistance to intracellular infections: comparative genomic analysis of Nramp. Trends Genet. 1996;12:201–204. doi: 10.1016/0168-9525(96)30042-5. [DOI] [PubMed] [Google Scholar]
- 6.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nramp defines a family of membrane proteins. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X Z, Peng J B, Cohen A, Nelson H, Nelson N, Hediger M A. Yeast SMF1 mediates H(+)-coupled iron uptake with concomitant uncoupled cation currents. J Biol Chem. 1999;274:35089–35094. doi: 10.1074/jbc.274.49.35089. [DOI] [PubMed] [Google Scholar]
- 8.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 9.Curie C, Alonso J M, Jean M L, Ecker J R, Briat J F. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- 10.Dancis A, Klausner R D, Hinnebusch A G, Barriocanal J G. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De-Silva D M, Askwith C C, Eide D, Kaplan J. The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J Biol Chem. 1995;270:1098–1101. doi: 10.1074/jbc.270.3.1098. [DOI] [PubMed] [Google Scholar]
- 12.Dix D, Bridgham J T, Broderius M A, Byersdorfer C A, Eide D J. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 13.Fleming M D, Romano M A, Su M A, Garrick L M, Garrick M D, Andrews N C. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming M D, Trenor C C, Su M A, Foernzler D, Beier D R, Dietrich W F, Andrews N C. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 15.Govoni G, Gros P. Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm Res. 1998;47:277–284. doi: 10.1007/s000110050330. [DOI] [PubMed] [Google Scholar]
- 16.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1Gly169 allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunshin H, Mackenzie B, Berger U V, Gushin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 18.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones E. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- 20.Jungmann J, Reins H A, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan J, O'Halloran T V. Iron metabolism in eukaryotes: Mars and Venus at it again. Science. 1996;271:1510–1512. doi: 10.1126/science.271.5255.1510. [DOI] [PubMed] [Google Scholar]
- 22.Kehres D G, Zaharik M L, Finlay B B, Maguire M E. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 23.Lapinskas P J, Lin S J, Culotta V C. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol Microbiol. 1996;21:519–528. doi: 10.1111/j.1365-2958.1996.tb02561.x. [DOI] [PubMed] [Google Scholar]
- 24.Lin S J, Pufahl R, Dancis A, O'Halloran T V, Culotta V C. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem. 1997;272:9215–9220. [PubMed] [Google Scholar]
- 25.Liu X F, Culotta V C. Mutational analysis of Saccharomyces cerevisiae Smf1p, a member of the Nramp family of metal transporters. J Mol Biol. 1999;289:885–891. doi: 10.1006/jmbi.1999.2815. [DOI] [PubMed] [Google Scholar]
- 26.Liu X F, Culotta V C. Post-translational control of Nramp metal transport in yeast: role of metal ions and the BSD2 gene. J Biol Chem. 1999;274:4863–4868. doi: 10.1074/jbc.274.8.4863. [DOI] [PubMed] [Google Scholar]
- 27.Liu X F, Culotta V C. The requirement for yeast superoxide dismutase is bypassed through mutations in BSD2, a novel metal homeostasis gene. Mol Cell Biol. 1994;14:7037–7045. doi: 10.1128/mcb.14.11.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X F, Supek F, Nelson N, Culotta V C. Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J Biol Chem. 1997;272:11763–11769. doi: 10.1074/jbc.272.18.11763. [DOI] [PubMed] [Google Scholar]
- 29.Nelson N. Metal ion transporters and homeostasis. EMBO J. 1999;18:4361–4371. doi: 10.1093/emboj/18.16.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ooi C E, Rabinovich E, Dancis A, Bonifacino J S, Klausner R D. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 1996;15:3515–3523. [PMC free article] [PubMed] [Google Scholar]
- 31.Orgad S, Nelson H, Segal D, Nelson N. Metal ions suppress the abnormal taste behavior of the Drosophila mutant malvolio. J Exp Biol. 1998;201:115–120. doi: 10.1242/jeb.201.1.115. [DOI] [PubMed] [Google Scholar]
- 32.Portnoy M E, Rosenzweig A C, Rae T, Huffman D L, O'Halloran T V, Culotta V C. Structure-function analyses of the ATX1 metallochaperone. J Biol Chem. 1999;274:15041–15045. doi: 10.1074/jbc.274.21.15041. [DOI] [PubMed] [Google Scholar]
- 33.Raguzzi F, Lesuisse E, Crichton R R. Iron storage in Saccharomyces cerevisiae. FEBS Lett. 1988;231:253–258. doi: 10.1016/0014-5793(88)80742-7. [DOI] [PubMed] [Google Scholar]
- 34.Rouault T, Klausner R. Regulation of iron metabolism in eukaryotes. Curr Top Cell Regul. 1997;35:1–19. doi: 10.1016/s0070-2137(97)80001-5. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. 1 to 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sherman F, Fink G R, Lawrence C W. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1978. [Google Scholar]
- 37.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stearman R, Yuan D, Yamaguchi-Iwan Y, Klausner R D, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 39.Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomine S, Wang R, Ward J M, Crawford N M, Schroeder J I. Cadmium and iron transport by members of a plant metal transporter family in arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson A M, Rogers J T, Leedman P J. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int J Biochem Cell Biol. 1999;31:1139–1152. doi: 10.1016/s1357-2725(99)00080-1. [DOI] [PubMed] [Google Scholar]
- 42.Urbanowski J L, Piper R C. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J Biol Chem. 1999;274:38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- 43.West A H, Clark D J, Martin J, Neupert W, Hart F U, Horwich A L. Two related genes encoding extremely hydrophobic proteins suppress a lethal mutation in the yeast mitochondrial processing enhancing protein. J Biol Chem. 1992;267:24625–24633. [PubMed] [Google Scholar]
- 44.Yamaguchi-Iwai Y, Dancis A, Klausner R. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner R D. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]