Abstract
Ubiquitin-Specific Peptidase 7 (USP7), or herpes virus-associated protease (HAUSP), is the largest family of the deubiquitinating enzymes (DUBs). Recent studies have shown that USP7 plays a vital role in regulating various physiological and pathological processes. Dysregulation of these processes mediated by USP7 may contribute to many diseases, such as cancers. Moreover, USP7 with aberrant expression levels and abnormal activity are found in cancers. Therefore, given the association between USP7 and cancers, targeting USP7 could be considered as an attractive and potential therapeutic approach in cancer treatment. This review describes the functions of USP7 and the regulatory mechanisms of its expression and activity, aiming to emphasize the necessity of research on USP7, and provide a better understanding of USP7-related biological processes and cancer.
Keywords: ubiquitin-specific peptidase 7, deubiquitinase, cancer, neoplasms, protein homeostasis, proteostasis
1 Introduction
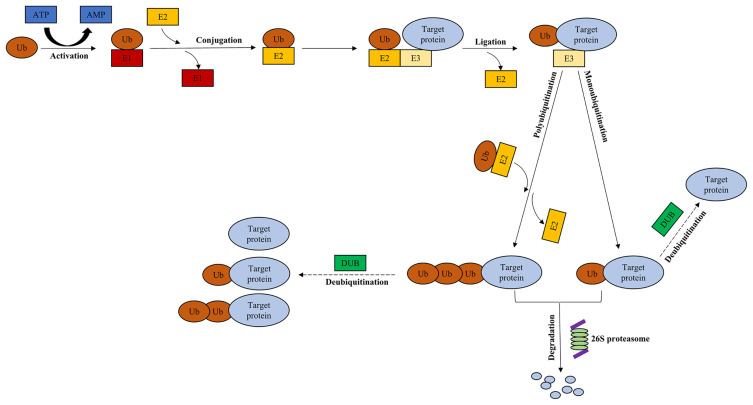
The ubiquitin-proteasome system (UPS), comprising ubiquitin, 26S proteasome, and four families of enzymes [ubiquitin‐activating enzyme E1, ubiquitin‐conjugating enzyme E2, ubiquitin ligase enzyme E3, and deubiquitinating enzymes (DUBs)] (1), regulates most intracellular protein degradation, cellular functions and maintains protein homeostasis (2) ( Figure 1 ). The UPS is involved in many biological processes, including immune response, cell cycle progression, signal transduction, and tumorigenesis (3). Ubiquitination, one of the most important posttranslational modifications (PTMs), takes part in regulating the stability, localization, and activity of proteins (4). The balance between protein synthesis and degradation is essential for homeostasis in cells (5). Ubiquitin is conjugated to the target protein via an enzymatic cascade, including E1, E2, and E3 (1). First, E1 activates ubiquitin, which is mediated by ATP (4). Subsequently, ubiquitin is transferred to E2 through trans-thiolation (4). Then, an isopeptide bond is formed following the conjugation between ubiquitin and a lysine residue of the target protein (6), subsequently recognized by 26S proteasome (5). Finally, the isopeptide bond is hydrolyzed by deubiquitinating enzymes (1).
Figure 1.
Ubiquitin-proteasome system. Ubiquitination of the target protein is catalyzed through E1, E2, and E3. Ub is activated by E1 in an ATP-dependent manner, which is subsequently transferred to E2. Then, an isopeptide bond is formed following the conjugation between ubiquitin and a lysine residue of the target protein via E3. Finally, the target protein is recognized by 26S proteasome, leading to the degradation of protein. The presence of DUBs rescues the target protein from being degraded, and contributes to its stabilization through deubiquitination. Dotted arrow: deubiquitination. Ub, ubiquitin; E1, ubiquitin‐activating enzyme; E2, ubiquitin‐conjugating enzyme; E3, ubiquitin ligase enzyme; DUB, deubiquitinating enzyme.
As one of the parts within UPS, the functions of DUBs are to reverse and antagonize the ubiquitination effect on the target protein, and participate in the balance between ubiquitination and deubiquitination. Recent studies have shown that, the aberrant activity and expression levels of DUBs correlates to numerous diseases, such as cancer.
DUBs mainly consist of 7 families: ubiquitin-specific proteases (USPs) (the largest family), ovarian tumor proteases (OTUs), Ub C-terminal hydrolases (UCHs), Machado–Joseph disease protein domain (MJD) proteases (MJDs), Jab1/MPN domain-associated metallo-isopeptidases (JAMM/MPM+), monocyte chemotactic protein-induced proteases (MCPIPs), and Zinc finger UB-specific proteases (ZUP/ZUFSP) (4).
Among the USPs family, ubiquitin-specific protease 7 (USP7), or herpes virus-associated protease (HAUSP), is well- characterized. USP7 is a multi-domain protein that contains 1102 amino-acid residues (5), including (i) a catalytic domain (CD) with finger, thumb, and palm domain (7), (ii) a C-terminal domain that contains ubiquitin like domains (UBL) 1-5 (4), and (iii) a tumor necrosis factor (TNF) receptor associated factors (TRAFs) like domain in its N-terminal domain (4). The catalytic domain participates in cleaving the peptide bond between the substrates of USP7 and ubiquitin (8). Unlike other USPs, the conserved catalytic domain of USP7 has no catalytic capacity (4), which makes USP7 quite unique compared with other USPs. Both of the UBL domains in the C-terminal domain and the TRAFs like domain in the N-terminal domain are able to recognize multiple substrates of USP7 (8). The TRAFs like domain has been proved to interact with p53 and MDM2/MDMX (1, 9). A recent study also shows that, deleting the TRAF-domain effects nuclear localization of USP7 (4). Different UBL domains recognize different substrates: UBL1-2 has the binding sites of ring finger protein (RNF) 168 and 169 (the negative regulator of RNF168), xeroderma pigmentosum complementation group C (XPC), DNA methyltransferases 1 (DNMT1), and ICP0 (10–14). HDM2 is recognized by UBL3 (8, 15), and UBL4-5 binds the tumor suppressor p53 (8, 15) and forkhead box O4 (FOXO4) (8, 16). In addition, the C-terminal domain also plays a key role in modulating the catalytic activity of USP7 through regulating the activity or localization of the enzyme (17), the incomplete structure of which greatly impairs USP7 activity (15, 18).
USP7 functions as an essential role in different cell processes through interacting with its multiple substrates ( Table 1), such as DNA damage and repair, immune responses, epigenetic control, and tumor progression (4). The expression and activity of USP7 are also regulated by multiple regulators through gene transcription and PTMs. Recently, many studies have shown that dysregulated cell processes mediated by USP7 may contribute to numerous diseases, such as cancers. Moreover, USP7 with aberrant expression levels and abnormal activity also correlates with cancers. Therefore, given the association between USP7 and cancers, targeting USP7 could provide a potential therapeutic strategy in cancer treatment.
Table 1.
Substrates or related proteins of USP7.
| Substrates or related proteins | Processes | References |
|---|---|---|
| NF-κB | Inflammatory signaling | (19) |
| HSCARG | NF-κB signaling | (20, 21) |
| IKK-γ
a
TRAF6 b |
Innate immunity | (22) |
| ICP0 | Innate immune response Viral infection |
(22, 23) |
| LANA c | Viral infection | (24) |
| vIRF d | (25, 26) | |
| RTA e | (27) | |
| E1B‐55 K | (28) | |
| UL35 | (29) | |
| EBNA1 f | (30) | |
| Foxp3 g | Tumor evasion | (31) |
| VP24 protein | Innate immune evasion | (32) |
| Tip60 | DNA damage responses Cell apoptosis Tumor evasion |
(21, 31, 33, 34) |
| P53 | DNA damage repair DNA damage responses Cell proliferation, migration, and apoptosis Tumor progression Tumor suppression |
(1, 8, 31, 35–40) |
| MDM2 | P53 degradation Cell proliferation Cell apoptosis |
(31) |
| HDM2, HDMX | P53 destabilization and downregulation | (8) |
| CCDC6 h | DNA damage responses DNA repair |
(1) |
| CHK1 i | Double-strand DNA breaks Cell cycle Tumor resistance |
(1, 8, 41–43) |
| XPC j | Nucleotide excision repair (NER) | (10) |
| CSB k /ERCC6 | Transcription‐coupled nucleotide excision repair (TC‐NER) | (1, 44, 45) |
| HLTF l | DNA damage bypass Damaged gene replication Cell cycle |
(1, 46) |
| CHFR m | Cell cycle | (47) |
| Cyclin A2 | (48) | |
| Ki-67 antigen | (49) | |
| UHRF1 n | Replication | (1, 50) |
| Histone LSD1 | Cell proliferation and migration | (4) |
| MDC1° | Double-strand DNA breaks Homologous recombination |
(51) |
| PHF8 p | DNA damage responses Cell cycle |
(1, 4, 48, 52) |
| ANXA1 q | DNA damage responses Cell apoptosis |
(53) |
| Rad18 | DNA damage tolerance Cell cycle |
(46) |
| RNF168
r
RNF169 s |
DNA damage responses | (4, 8, 11, 12) |
| DNA polymerase polη | DNA damage | (54, 55) |
| RNA polymerase II | CSB stabilization Nucleotide excision repair (NER) |
(1) |
| UVSSA t | CSB stabilization Transcription‐coupled nucleotide excision repair (TC‐NER) |
(1, 44, 45) |
| DNMT1 u | Replication Cell proliferation and migration |
(1, 4, 56) |
| PTEN | Tumor metastasis Cell apoptosis Tumor suppression |
(1, 57, 58) |
| Claspin | Tumor suppression | (43) |
| FOXO v | (16) |
IκB kinase γ.
Tumor necrosis factor (TNF) receptor associated factor 6.
Latency‐associated nuclear antigen.
Viral interferon regulatory factor.
RNA transcriptional activator.
Epstein‐Barr nuclear antigen 1.
Forkhead box P3.
Coiled-coil domain containing 6.
Checkpoint kinase 1.
Xeroderma pigmentosum complementation group C.
Cockayne syndrome B.
Helicase-like transcription factor.
Checkpoint protein with FHA and RING domains.
Ubiquitin‐like, containing PHD and RING finger domains 1.
° Mediator of DNA damage checkpoint 1.
Plant homeodomain finger‐containing protein 8.
Annexin‐1.
Ring finger protein 168.
Ring finger protein 169.
UV-stimulated scaffold protein A.
DNA methyltransferases 1.
Forkhead box O.
This review summarizes various functions of USP7 in diverse cellular processes and cancers, as well as the regulatory mechanisms of its expression levels and activity, aiming to emphasize the necessity of research on USP7, and provide a better understanding of USP7-related biological processes and cancer.
2 Physiological and Pathological Roles of USP7
USP7 has been shown to be involved in a variety of biological processes in cells, and is associated with many cancers through the regulation of its downstream substrates.
2.1 USP7 and Immune Signaling
Multiple proteins have been shown to be the substrates of USP7 in immune signaling. USP7 may promote inflammatory signaling by stabilizing NF-κB (19). IκB kinase γ (IKK-γ) polyubiquitination and IκB degradation can be suppressed when USP7 interacts with HSCARG, which subsequently inhibits NF-κB signaling (20). In the innate immune response, USP7 promotes deubiquitination of TRAF3 and TRAF6 by binding to vIRF2, resulting in extended transactivation of TRAF3 and TRAF6, thereby regulating the interferon response (21). Moreover, USP7 reduces NFκB -mediated innate immune responses through TRAF6 and IKK-γ deubiquitination, which appears to be directed by the USP7-ICP0 complex (22, 23). This may be the mechanism by which USP7 prevents HSV from innate host immunity (22).
During the adaptive immune response, USP7 contributes to the evasion of tumors by promoting the deubiquitination and stability of proteins, such as Tip60 and Forkhead box P3 (Foxp3) (31), which act a pivotal part in promoting the suppressive functions of both Treg and Teff (59). In mice treated by DSS, it appears that a novel small molecule cambogin relieves the symptoms of enteritis and reduces the production of pro-inflammatory cytokines (60). This may be due to the effect of cambogin on the promoter of USP7, which promotes the deubiquitination and expression of Foxp3, and reduces inflammation by affecting the immune response mediated by Treg cells (61). DNMT1 and Ubiquitin‐like, containing PHD and RING finger domains 1 (UHRF1) play essential roles in modulating Treg function and development (62), and the stability of the former is also regulated by USP7 (62). In mouse models, USP7 deletion in Treg cells has been found to induce lethal systemic autoimmunity (63). In addition, deleting USP7 also down-regulates the expression of many transcription factors that are essential to Treg cell development (48), and is correlated to the increased-level of IFNγ and IL-2 expression (62), resulting in the impairment of Treg suppressive functions (31), which seems to promote antitumor immunity (62) and break immunotolerance (31). Therefore, given the suppressive functions of USP7 inhibitors in regulating Foxp3+Treg cells, USP7 could be a potential target in cancer immunotherapy.
2.2 DNA Damage Responses
In response to DNA damage, USP7 regulates multiple proteins in double-strand DNA breaks (ATR-CHK1 and ATM-CHK2 signaling cascade), homologous recombination repair (HRR), cell cycle checkpoint activation, nucleotide excision repair (NER), and DNA damage bypass, where p53 functions as one of the key factors ( Figures 2 and 3 ).
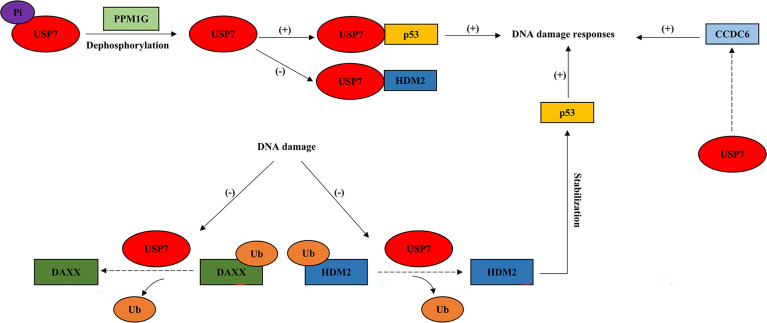
Figure 2.
USP7 interacts with tumor suppressors in response to DNA damage. This model describes how USP7 effects DNA damage responses by interacting with tumor suppressive proteins, p53 and CCDC6. Dotted arrow: deubiquitination; Brackets with a “+”: promotional effect; Brackets with a “-”: inhibitory effect. USP7, ubiquitin-specific peptidase 7; DAXX, death-associated protein 6; PPM1G, protein phosphatase 1G; CCDC6, coiled-coil domain containing 6.
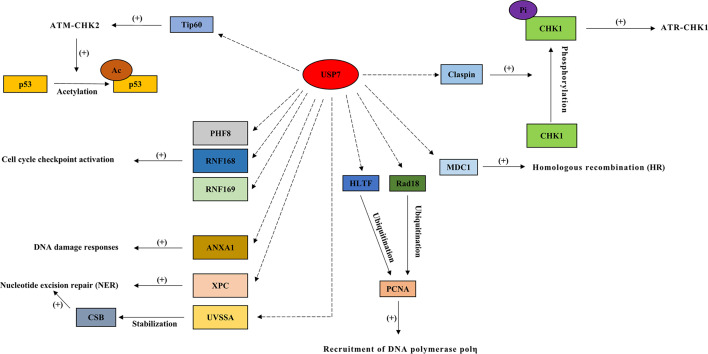
Figure 3.
Interactions between USP7 and its substrates in double-strand DNA breaks, homologous recombination repair (HRR), cell cycle checkpoint activation, nucleotide excision repair (NER), and DNA damage bypass. In response to DNA damage, USP7 regulates multiple proteins in double-strand DNA breaks (ATR-CHK1 and ATM-CHK2 signaling cascades), homologous recombination repair (HRR), cell cycle checkpoint activation, nucleotide excision repair (NER), and DNA damage bypass. Dotted arrow: deubiquitination; Brackets with a “+”: promotional effect; Brackets with a “-”: inhibitory effect. USP7, ubiquitin-specific peptidase 7; CHK1, checkpoint kinase 1; MDC1, mediator of DNA damage checkpoint 1; PHF8, plant homeodomain finger‐containing protein 8; RNF168, ring finger protein 168; RNF169, ring finger protein 169; XPC, xeroderma pigmentosum complementation group C; UVSSA, UV-stimulated scaffold protein A; HLTF, helicase-like transcription factor.
During DDR, USP7 acts as a pivotal role in the regulation of genomic stress and cell fate through deubiquitinating and stabilizing the key factor, tumor suppressor p53 ( Figure 2 ). The interaction between p53 and USP7 leads p53 to effect DNA damage repair at the level of gene transcription (35). DNA damage results in USP7 dephosphorylation via protein phosphatase 1G (PPM1G), which is in a way dependent on ATM (64). This causes lower USP7 affinity to HDM2 and increases its affinity towards p53 instead, driving p53-dependent DDR (8, 64). DNA damage also leads to the disrupted association between USP7 and DAXX, resulting in the ubiquitination of HDM2 and stabilization of p53 later (21). Moreover, USP7 is also required for DDR and DNA repair via its interaction with another tumor suppressor, coiled-coil domain containing 6 (CCDC6) (1), which has been shown to produce the pro‐apoptotic protein (65) ( Figure 2 ).
During double-strand DNA breaks (DSBs), USP7 acts as an essential part of the ATR-CHK1 branch (8) by regulating checkpoint kinase 1 (CHK1), thereby playing a vital role in DDR (41). USP7 promotes CHK1 phosphorylation mediated by ATR (1), and is required for the regulation of claspin that is key to CHK1 activation (66). Another crucial part of DDR, the ATM-CHK2 pathway, is also regulated by USP7 (67). USP7 indirectly activates p53 through deubiquitinating Tip60 (21), which activates the ATM-CHK2 signaling cascade through acetylating ATM, causing the acetylation and activation of p53 (33).
USP7 plays a role in recruiting DNA repair proteins towards DSB lesions, and influences homologous recombination (HR) through regulating the mediator of DNA damage checkpoint 1 (MDC1) (51). In cells depleted of MDC1, the failure of DNA repair proteins recruitment is found after DNA damage, and a similar defect is found in USP7-depleted cells (51). In chronic lymphocytic leukemia (CLL), inhibition of USP7 suppresses HRR and induces cell death in a manner independent of ATM-p53 (1). Additionally, E3 ligase RNF168 deubiquitinates the histones that function as a signal in the recruitment of DNA damage factors (21), and controls the access of proteins related to DDR to chromatin (11, 12). USP7 has been shown to regulate cell cycle checkpoint activation, in response to DNA damage, through the deubiquitination and stabilization of RNF168, RNF169, and the histone demethylases, plant homeodomain finger‐containing protein 8 (PHF8) (4, 8, 11, 12).
Protein XPC plays a vital part in recognizing damage, functions as a lesion sensing factor (10), and initiates NER by interacting with USP7 following DNA damage induced by UV (1, 10). ANXA1 (Annexin‐1), one of the substrates of USP7, acts as a stress protein or protective protein during the UV‐induced DDR (53). Additionally, UVSSA (UV-stimulated scaffold protein A) functions as an initiating factor of transcription‐coupled nucleotide excision repair (TC-NER) (44). During the UV-induced TC-NER process, USP7 interacts with RNA polymerase II and UVSSA to stabilize CSB (cockyane syndrome B) that also plays a vital role in the regulation of TC-NER, in response to DNA damage (1). Studies have shown that USP7 depletion causes TC-NER deficiency, such as reduced UV survival and decreased recovery in RNA synthesis following UV (44, 45).
USP7 regulates the stability of trans-lesion synthesis (TLS) DNA polymerase, polη, via a USP7‐HLTF‐PCNA molecular network, in response to DNA damage (54, 55). Following genotoxic stress, USP7 is involved in the regulation of error-free bypass replication through the interaction with helicase-like transcription factor (HLTF), and helps elongate nascent daughter strand DNA through Rad18 (46) (68). The defect in elongation of nascent DNA strands is found in cells depleted of USP7 after UV exposure, which may be due to the regulation of USP7 on PCNA sliding clamp ubiquitination (46, 55, 69, 70). In addition, USP7 also plays a role in DNA damage tolerance by regulating Rad18 stability (8). Lower levels of HLTF and Rad18, suppressed PCNA ubiquitination, and suppressed Polη foci formation are all found in cells depleted of USP7 (35).
2.3 USP7 and Cancers
Accumulated evidence has shown that abnormal expression and activity of USP7 are associated with a variety of cancers. Here is a summary of the mechanisms of USP7 for effecting tumor initiation and progression through interaction with its downstream proteins, as detailed below.
2.3.1 Cell Proliferation
USP7 regulates MDM2/MDMX-p53 circuitry and controls the stabilities of related proteins, thereby influencing cell proliferation, cancer initiation, and progression (31). In prostate cancer, inhibition of USP7 expression is found to have an anti-proliferative effect on cancer cells (71). USP7 also influences the cell cycle by stabilizing the Ki-67 antigen (49), which has been shown to help maintain mitosis and heterochromatin (72). In addition, USP7 regulates the target of the cell cycle, cyclin A2, through the deubiquitination and stabilization of PHF8 demethylase, which acts as an up-regulator of cyclin A2 (48, 52). Moreover, upregulation of USP7, PHF8, and cyclin A2 has also been found in breast cancers, colon and rectum cancers (48).
2.3.2 Cell Migration
In colorectal cancer (CRC) (50) and medulloblastoma (73), USP7 overexpression contributes to the increased rate of cell proliferation and migration through its association with histone LSD1 and DNMT1 (4). In prostate cancer, emerging evidence has proved that USP7 contributes to tumor migration and invasion through stabilizing EZH2 (74). The decreased rate of cell migration and invasion is found in PC3 and DU145 depleted of USP7 (74). Combined treatment with USP7 and EZH2 inhibitors has been shown to reduce the migration and invasion of cancer cells (74). In osteosarcoma (OS), overexpression of USP7 significantly improves the ability of cell migration and invasion, while USP7 depletion has the opposite effects, indicating the key role of USP7 in regulating the migratory and invasive ability of osteosarcoma cells (75).
2.3.3 Cell Metastasis
USP7 is involved in tumor metastasis through deubiquitination and localization of PTEN, which results in the inactivation of PTEN (57, 58). It has been shown that USP7 promotes epithelial-mesenchymal transition (EMT) of OS cells through Wnt/β-catenin signaling (75), which is one of the key parts in promoting cell metastasis (76). In non-small cell lung cancers (NSCLC), USP7 dysregulation is also related to EMT and cell metastasis, leading to poor prognosis (77). Moreover, in both epithelial ovarian cancer (78) and prostate cancer (31), the overexpression of USP7 is found to promote cell invasion, the increased expression level of which is also related to poor survival of ovarian cancer patients (78).
2.3.4 Cell Immunosuppression
During an innate immune response, USP7 has been found to diminish NFκB through its interaction with ICP0 (22, 23). ICP0-USP7 complex participates in the deubiquitination and inactivation of both IKK-γ and TRAF6 (22), leading to the inhibition of innate immunity. Besides, IKK-γ polyubiquitination and IκB degradation can be suppressed when USP7 interacts with HSCARG (20, 21), which subsequently inhibits NF-κB signaling (20). It has also been shown that the recruitment of USP7 by HSV-1 results in HSV-1 evasion of innate immune response (79). Moreover, recent studies have identified the role of USP7 as a target of VP24 from the Ebola virus, which is a protein that plays a pivotal part in innate immune evasion (32).
In the stage of the adaptive immune response, USP7 promotes tumor immune escape through deubiquitination and stabilization of both Foxp3 and Tip60 (31), which play key roles in inhibiting Treg and Teff (59, 63). Conversely, USP7 knockdown results in the instability of Foxp3, which subsequently impairs the immunosuppressive function of Treg (62, 80), and promotes tumor suppression mediated by the immune system (63). In addition, induced lethal systemic autoimmunity has been found in mouse model with USP7 depletion (63). Taken together, all of these data indicate the essential role of USP7 in modulating the immune response.
2.3.5 Cell Apoptosis
Studies have demonstrated the duality of USP7 functions in cancer progression. In other words, USP7 displays tumor-promoting and/or -suppressive functions under different conditions ( Figure 4 ).
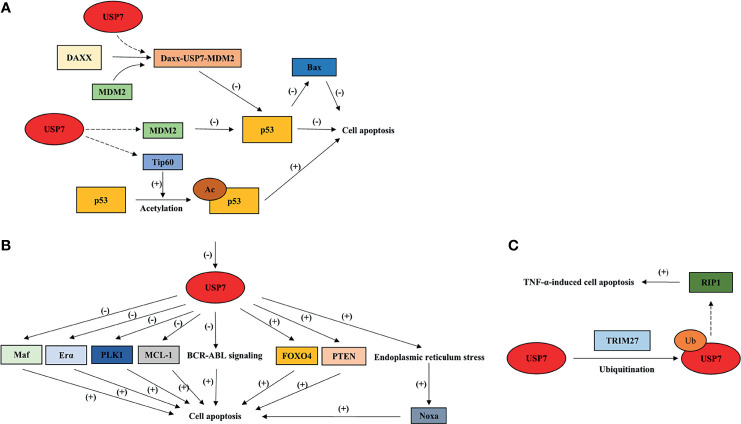
Figure 4.
Overview of USP7 effects on cell apoptosis by interacting with its substrates. This model summarizes how USP7 effects p53-dependent cell apoptosis (A) and p53-independent cell apoptosis (B, C). Dotted arrow: deubiquitination; Brackets with a “+”: promotional effect; Brackets with a “-”: inhibitory effect. USP7, ubiquitin-specific peptidase 7; FOXO4, forkhead box O4; TRIM27, tripartite motif protein 27; TNF-α, tumor necrosis factor-alpha.
USP7 effects p53-dependent apoptosis through its interaction with p53 and negative regulator of p53, MDM2. In most cases, USP7 shows a higher affinity to MDM2, leading to p53 ubiquitination, which indicates the tumor-promoting effect of USP7. Daxx plays an anti-apoptotic function in cancer cells through the formation of ternary complex USP7-MDM2-Daxx, which stabilizes MDM2, and later promotes degradation of p53 (5). In NSCLC, defect of USP7 leads to p53 upregulation, subsequently causing p53-induced apoptosis through p53 downstream target, Bax (81). In colorectal cancer cell lines, it has been revealed that FAM188B may be the mediator that controls the combination between USP7 and p53/MDM2 (82). The interaction between USP7 and FAM188B effects p53 stability, which may be one of the reasons why p53-dependent apoptosis is induced by FAM188B downregulation (82). Notably, USP7 also induces p53-dependent apoptosis by interacting with Tip60, which is involved in K120 acetylation that is located in the DNA-binding domain of p53 (35).
Accumulated studies have also shown the apoptotic function of USP7 inhibition in a p53-independent manner. In chronic myelogenous leukemia (CML), USP7 is required for stabilization of BCR-ABL and activation of BCR-ABL signaling (83).Inhibiting USP7 was shown to cause BCR-ABL destabilization and to trigger apoptotic signaling pathways (83). In CLL cells, USP7 inhibition induces cell apoptosis by restoring nuclear localization of PTEN (84). Additionally, FOXO4 transcriptional activity is promoted through USP7 inhibition, which induces apoptosis in an HDM2-independent manner (79). In multiple myeloma cell (MM) cells, Maf transcriptional activity is promoted by USP7 through deubiquitination and stabilization of Maf (85). Inhibiting USP7 leads to Maf downregulation, thereby inducing apoptosis (85). USP7 also promotes apoptotic escape of breast cancer cells by deubiquitinating and stabilizing Erα (86). Silencing or inhibiting USP7 induces apoptosis and inhibits cell growth in breast cancer (86). Moreover, USP7 inhibition may cause aberrant mitosis and apoptosis through downregulation and degradation of PLK1 (87). Studies have also shown that USP7 participates in the stabilization of anti-apoptotic protein MCL-1, leading to the upregulation of MCL-1 levels in tumor cells (88). In addition, inhibiting USP7 induces endoplasmic reticulum stress triggered by ubiquitinated proteins accumulation, which promotes NOXA expression levels and leads to NOXA-induced apoptosis (89). Interestingly, USP7 also promotes TNF-induced apoptosis through its combination with TRIM27 and receptor-interacting protein 1 (RIP1) (90), indicating the tumor-suppressive role of USP7.
Collectively, USP7 plays a complex role in regulating apoptosis. Specific mechanisms that could explain how USP7 interacts with its substrates in apoptosis are needed to explore the different roles of USP7 in various cancers.
3 Regulation of USP7 Activity and Expression Levels
With multiple downstream target proteins, USP7 is also influenced by a plethora of regulators. Here is a review of the mechanisms of USP regulation, including the regulation of its expression levels and deubiquitinase activity.
3.1 Transcription Regulation
Several transcription factors are involved in the regulation of USP7 ( Table 2 ). USP7 ubiquitination is found to be catalyzed by ICP0, a protein derived from a virus that functions as an E3 (96), leading to USP7 degradation and decreased levels (95). Downregulation of USP7 mediated by signal transducer and activator of transcription 3 (STAT3) has been found in colon cancer (94). Forkhead box O6 (FOXO6) upregulates USP7 transcription by binding to its promoter (91). In lung carcinoma, cell proliferation is found to be inhibited through the promoted expression of USP7 induced by FOXO6 (91). In acute lymphoblastic leukemia, NOTCH1 contributes to the upregulation of USP7 levels, and USP7, in turn, influences the stability of NOTCH1, which possibly forms a positive-feedback interaction between the two proteins (92). And the similar feedback loop has been demonstrated between USP7 and PHF8 (48, 93). Some regulators of USP7 can indirectly influence its downstream substrates, and thus regulate related processes. For instance, PPM1G reverses USP7’s functions on MDM2 and p53 following the down-regulation of USP7 caused by PPM1G (64). Moreover, LKB1 activation induces HuR to translocate from the nucleus to the cytoplasm, leading to stabilization of USP7 mRNAs and phosphorylation of p53, thereby influencing apoptosis (97). Remarkably, the expression of USP7 is also found to be enhanced by the HIV-1 virus, because USP7 plays an essential part in regulating the stability of the trans-activator of transcription (TAT) (21), which is involved in regulating the levels of HIV genes (98).
Table 2.
Regulators of USP7 levels.
| Regulators | References | |
|---|---|---|
| Up-regulators | FOXO6 a | (91) |
| NOTCH1 | (92) | |
| PHF8 b | (48, 93) | |
| Down-regulators | STAT3 c | (94) |
| PPM1G d | (64) | |
| ICP0 | (95) | |
Forkhead box O6.
Plant homeodomain finger‐containing protein 8.
Signal transducer and activator of transcription 3.
Protein phosphatase 1G.
3.2 Posttranslational Modifications
USP7 activity is regulated through PTMs, mainly including ubiquitination and phosphorylation, as detailed below ( Table 3 ).
Table 3.
Regulators of USP7 activity.
| Regulators | Effects | References |
|---|---|---|
| ICP0 | Ubiquitination | (95) |
| Trip12 | (99) | |
| TRIM27 | (90) | |
| CK2 a | Phosphorylation | (64) |
| BCR-ABL | (100) | |
| PPM1G | Dephosphorylation | (64) |
Casein kinase 2.
3.2.1 Ubiquitination
The region where USP7 is ubiquitinated is located near the site where it binds to the E3 ubiquitin ligase ICP0 (96). ICP0 functions as a catalyzer in USP7 ubiquitination, leading to its degradation and decreased levels (95). The feedback interaction is found between USP7 and ICP0, as USP7 could in turn participate in the deubiquitination of ICP0 (95). Interestingly, the ubiquitination of USP7 is also found in cells that do not express ICP0 (5), indicating that USP7 could be ubiquitinated in an ICP0-independent way. However, the specific DUB and E3 ligases that participate in the ubiquitination of USP7 have not been identified (5). Recently, another E3 ligase Trip12 has also been shown to ubiquitinate USP7 (99). In addition, polyubiquitinated USP7 promotes TNF-α induced apoptosis by interacting with E3 ligase TRIM27 on Lys869 and RIP1 (90). However, USP7 protease activity does not always require ubiquitination, without which USP7 is still able to deubiquitinate MDM2 and p53 (5).
3.2.2 Phosphorylation
Casein kinase 2 (CK2) improves the affinity of USP7 or USP7 isoform (USP7S) to HDM2 or MDM2 by phosphorylating USP7 or USP7S at Ser18, while dephosphorylating USP7 or USP7S enhances their affinity for p53 (64). In addition, the effect of USP7/USP7S on HDM2 and p53 could be reversed by the ATM-dependent phosphatase PPM1G (64). Activated ATM signaling promotes USP7/USP7S dephosphorylation mediated by PPM1G, inactivates and downregulates USP7/USP7S, and leads to decreased levels of MDM2 and p53 upregulation (64). Moreover, in chronic leukemia, it has been shown that BCR-ABL could aberrantly phosphorylate USP7 at Tyr243, contributing to the function of USP7 as a deubiquitinase for PTEN (8, 100). And dysregulated PTEN caused by phosphorylated USP7 is associated with the pathogenesis of leukemia (100).
4 Conclusion
This review summarizes the physiological and pathological functions of USP7 in immune signaling, DNA damage response, and cancers, as well as the regulatory mechanisms of its expression and activity. However, in respect of the various functions of USP7 in cell processes, the association between USP7 and cancers, and USP7 targeted cancer therapy, there are still many fields that need to be further explored.
In order to illuminate the real roles of USP7 in carcinogenesis, it is requisite to further investigate the dual effects of USP7 in tumor regulation. Additionally, considering the critical part that tumor suppressor p53 plays in various cancers, targeting the USP7-MDM2-p53 axis will still be one of the popular topics in the future. Further research is needed to explore whether there are other regulatory factors of USP7 on p53 and MDM2, and USP7-specific transcription inhibitors and factors, the discovery of which could have future implications for the study of cancer with an intact p53 regulatory axis.
Accumulated studies have shown p53 mutation and p53-independent role of USP7 in quite a few cancer cases. For example, in melanoma cells, USP7 expression levels are found to increase in a p53-independent manner (101), and inhibiting USP7 results in endoplasmic reticulum stress and DNA damage, which is in a way independent of p53 (102, 103). Most USP7 inhibitors are developed based on wild type (WT) p53, however, a recent study has shown that inhibiting USP7 plays a suppressive role in tumor progression with both WT p53 and p53 mutation (104). All of these indicate that USP7 may effect carcinogenesis by interacting with other substrates, which needs to be further explored.
Given the association between USP7 and a few signaling pathways in tumor progression [such as NF-κB pathway (105), Wnt/β-Catenin pathway (106), Hippo pathway (107), and NOTCH pathway (108)], more related mechanistic investigations on USP7 regulation are needed to develop new targeting strategies in cancer therapy.
In addition to the nuclear deubiquitinase role, USP7 also participates in the recycling of endosomal proteins (109). Exploring additional functions of USP7 is required for improving our understanding of its roles in biological and pathological processes.
In the process of developing USP7 inhibitors, there are still some issues that remain to be solved. Theoretically, for instance, the full-length structure of USP7 has not been fully studied, and co-crystal structures of USP7 in complex with small molecules also remain unclear. How USP7 is activated, how does USP7 recognize its various substrates, and how USP7 inhibitors work? A complete and deeper understanding of these fundamentals of molecular theory and mechanisms are essential to the development of USP7 inhibitors. In general, many reported USP7 inhibitors so far have been shown to have poor selectivity, low specificity, and low efficacy (low micromolar potency). Unsatisfactory pharmacokinetic characteristics of USP7 inhibitors also limit their evaluation in vivo. In order to improve their selectivity and specificity, manipulation of USP7’s affinity to specific target substrates may be feasible, which requires support on the theoretical basis of USP7 recognition mechanisms. Further studies should also focus on the development of better screening methods, which will undoubtedly improve the accuracy and efficiency of inhibitor evaluation. Additionally, it appears that quite a few USP7 inhibitors could also target other related DUBs (such as USP47, which has high homology with USP7) with similar potency. As a result, this kind of cross effect adds difficulty in determining the contribution of USP7 inhibition to the effect of the tested compound in cancer progression.
According to the discussion above, the development of USP7 inhibitors is still in a primary and long-term stage. However, notably, there have also been some breakthroughs in recent years. The inhibition mechanism of P22077 and P50429 on USP7 has been revealed by Pozhidaeva et al. (110). Gavory et al. have reported the first reversible USP7 inhibitor with high efficacy (nanomolar potency) and high selectivity, which has been shown to have no interaction with USP47 when targeting USP7 (111). Schauer et al. reported the first irreversible USP7 inhibitor with high efficacy (sub-nanomolar potency) and high selectivity (112). Moreover, P5091 has been shown to have efficacy and very low toxicity in tumor treatment in vivo (113–115). In sum, all of the issues are not insurmountable for the development of more ideal inhibitors.
Excitingly, the combination of USP7 inhibitors and several current major cancer treatments has been shown to possess promising therapeutic prospects, mainly including chemotherapy, radiotherapy, and immunotherapy (such as PD-L1 treatment), which will be one of the study directions in the future. For example, the resistance to bortezomib appears to be overcome through the combination of USP7 inhibitors P22077 and cytotoxic drugs used for chemotherapy (4, 116). USP7 inhibition may also improve the effectiveness of Olaparib treatment by increasing drug sensitivity through CCDC6 downregulation (117, 118). In addition, given the essential roles of PHF8 and CHK1 that act as USP7 substrates in DNA damage response, combination of USP7 inhibitors and chemotherapy/radiotherapy may reduce breast cancer cell resistance (42, 63), which is expected to accelerate the development of breast cancer therapy. Besides, targeting USP7 inhibits cell apoptosis in response to chemo-/radiotherapy (5), indicating the promising role of USP7 inhibitors in reducing adverse reactions in cancer treatment. Moreover, during anti-tumor immunotherapy, it has been shown that USP7 inhibition causes PD-L1 increased level (119) and IL-10 downregulation by attenuating Treg function (120), which may contribute to enhancing the therapeutic effect of PD-L1 treatment. Meanwhile, given the suppressive function of USP7 inhibitors in regulating Treg cells (31), a non-negligible concern about the safety of inhibitors is raised: Whether the host immune response will be impaired due to functional inhibition of Treg? Therefore, in order to ensure the availability and effectiveness of USP7 inhibitors in cancer therapy, evaluation of drug safety is very necessary, which requires more data from in vivo experiments.
In sum, clinical application of targeting USP7 possesses a broad prospect, and research on USP7 is necessary, which still has a long way to go.
Author Contributions
Conceptualization, TO. Writing—original draft preparation, LZ and TO. Writing— review and editing, LZ, TO, ML, and TH. Visualization, ML, TH, AMHS, and WM. Supervision, AMHS, WM, and NZ. All authors contributed to the article and approved the submitted version.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81760447(TO); grant no. 81960247(NZ), Project of Science and Technology Department of Jiangxi Province (grant no. S2019QNJJB1056) (NZ), grant no. 20202BABL206099 (WM) and Jiangxi Provincial Education Department Project [grant no.GJJ180054 (TO); grant no. GJJ180116(NZ)].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Yeasmin Khusbu F, Chen FZ, Chen HC. Targeting Ubiquitin Specific Protease 7 in Cancer: A Deubiquitinase With Great Prospects. Cell Biochem Funct (2018) 36(5):244–54. doi: 10.1002/cbf.3336 [DOI] [PubMed] [Google Scholar]
- 2. Chen YJ, Wu H, Shen XZ. The Ubiquitin-Proteasome System and its Potential Application in Hepatocellular Carcinoma Therapy. Cancer Lett (2016) 379(2):245–52. doi: 10.1016/j.canlet.2015.06.023 [DOI] [PubMed] [Google Scholar]
- 3. Hershko A, Ciechanover A. The Ubiquitin System. Annu Rev Biochem (1998) 67:425–79. doi: 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 4. Nininahazwe L, Liu B, He C, Zhang H, Chen ZS. The Emerging Nature of Ubiquitin-Specific Protease 7 (USP7): A New Target in Cancer Therapy. Drug Discov Today (2021) 26(2):490–502. doi: 10.1016/j.drudis.2020.10.028 [DOI] [PubMed] [Google Scholar]
- 5. Zhou J, Wang J, Chen C, Yuan H, Wen X, Sun H. USP7: Target Validation and Drug Discovery for Cancer Therapy. Med Chem (2018) 14(1):3–18. doi: 10.2174/1573406413666171020115539 [DOI] [PubMed] [Google Scholar]
- 6. Komander D, Rape M. The Ubiquitin Code. Annu Rev Biochem (2012) 81:203–29. doi: 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 7. Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex With Ubiquitin Aldehyde. Cell (2002) 111(7):1041–54. doi: 10.1016/s0092-8674(02)01199-6 [DOI] [PubMed] [Google Scholar]
- 8. Pozhidaeva A, Bezsonova I. USP7: Structure, Substrate Specificity, and Inhibition. DNA Repair (Amst) (2019) 76:30–9. doi: 10.1016/j.dnarep.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarkari F, La Delfa A, Arrowsmith CH, Frappier L, Sheng Y, Saridakis V. Further Insight Into Substrate Recognition by USP7: Structural and Biochemical Analysis of the HdmX and Hdm2 Interactions With USP7. J Mol Biol (2010) 402(5):825–37. doi: 10.1016/j.jmb.2010.08.017 [DOI] [PubMed] [Google Scholar]
- 10. He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, et al. Ubiquitin-Specific Protease 7 Regulates Nucleotide Excision Repair Through Deubiquitinating XPC Protein and Preventing XPC Protein From Undergoing Ultraviolet Light-Induced and VCP/p97 Protein-Regulated Proteolysis. J Biol Chem (2014) 289(39):27278–89. doi: 10.1074/jbc.M114.589812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu Q, Sharma N, He J, Wani G, Wani AA. USP7 Deubiquitinase Promotes Ubiquitin-Dependent DNA Damage Signaling by Stabilizing RNF168. Cell Cycle (2015) 14(9):1413–25. doi: 10.1080/15384101.2015.1007785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. An L, Jiang Y, Ng HH, Man EP, Chen J, Khoo US, et al. Dual-Utility NLS Drives RNF169-Dependent DNA Damage Responses. Proc Natl Acad Sci USA (2017) 114(14):E2872–81. doi: 10.1073/pnas.1616602114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pfoh R, Lacdao IK, Georges AA, Capar A, Zheng H, Frappier L, et al. Crystal Structure of USP7 Ubiquitin-Like Domains With an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target Usp7. PloS Pathog (2015) 11(6):e1004950. doi: 10.1371/journal.ppat.1004950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, et al. Molecular Mechanism for USP7-Mediated DNMT1 Stabilization by Acetylation. Nat Commun (2015) 6:7023. doi: 10.1038/ncomms8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma J, Martin JD, Xue Y, Lor LA, Kennedy-Wilson KM, Sinnamon RH, et al. C-Terminal Region of USP7/HAUSP Is Critical for Deubiquitination Activity and Contains a Second Mdm2/P53 Binding Site. Arch Biochem Biophys (2010) 503(2):207–12. doi: 10.1016/j.abb.2010.08.020 [DOI] [PubMed] [Google Scholar]
- 16. van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 Transcriptional Activity Is Regulated by Monoubiquitination and USP7/HAUSP. Nat Cell Biol (2006) 8(10):1064–73. doi: 10.1038/ncb1469 [DOI] [PubMed] [Google Scholar]
- 17. Faesen AC, Luna-Vargas MP, Sixma TK. The Role of UBL Domains in Ubiquitin-Specific Proteases. Biochem Soc Trans (2012) 40(3):539–45. doi: 10.1042/BST20120004 [DOI] [PubMed] [Google Scholar]
- 18. Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP Activation by its C-Terminal Ubiquitin-Like Domain and Allosteric Regulation by GMP-Synthetase. Mol Cell (2011) 44(1):147–59. doi: 10.1016/j.molcel.2011.06.034 [DOI] [PubMed] [Google Scholar]
- 19. Colleran A, Collins PE, O'Carroll C, Ahmed A, Mao X, McManus B, et al. Deubiquitination of NF-κb by Ubiquitin-Specific Protease-7 Promotes Transcription. Proc Natl Acad Sci USA (2013) 110(2):618–23. doi: 10.1073/pnas.1208446110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li T, Guan J, Li S, Zhang X, Zheng X. HSCARG Downregulates NF-κb Signaling by Interacting With USP7 and Inhibiting NEMO Ubiquitination. Cell Death Dis (2014) 5(5):e1229. doi: 10.1038/cddis.2014.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bojagora A, Saridakis V. USP7 Manipulation by Viral Proteins. Virus Res (2020) 286:198076. doi: 10.1016/j.virusres.2020.198076 [DOI] [PubMed] [Google Scholar]
- 22. Daubeuf S, Singh D, Tan Y, Liu H, Federoff HJ, Bowers WJ, et al. HSV ICP0 Recruits USP7 to Modulate TLR-Mediated Innate Response. Blood (2009) 113(14):3264–75. doi: 10.1182/blood-2008-07-168203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pozhidaeva AK, Mohni KN, Dhe-Paganon S, Arrowsmith CH, Weller SK, Korzhnev DM, et al. Structural Characterization of Interaction Between Human Ubiquitin-Specific Protease 7 and Immediate-Early Protein ICP0 of Herpes Simplex Virus-1. J Biol Chem (2015) 290(38):22907–18. doi: 10.1074/jbc.M115.664805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jäger W, Santag S, Weidner-Glunde M, Gellermann E, Kati S, Pietrek M, et al. The Ubiquitin-Specific Protease USP7 Modulates the Replication of Kaposi's Sarcoma-Associated Herpesvirus Latent Episomal DNA. J Virol (2012) 86(12):6745–57. doi: 10.1128/jvi.06840-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chavoshi S, Egorova O, Lacdao IK, Farhadi S, Sheng Y, Saridakis V. Identification of Kaposi Sarcoma Herpesvirus (KSHV) Virf1 Protein as a Novel Interaction Partner of Human Deubiquitinase Usp7. J Biol Chem (2016) 291(12):6281–91. doi: 10.1074/jbc.M115.710632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee HR, Choi WC, Lee S, Hwang J, Hwang E, Guchhait K, et al. Bilateral Inhibition of HAUSP Deubiquitinase by a Viral Interferon Regulatory Factor Protein. Nat Struct Mol Biol (2011) 18(12):1336–44. doi: 10.1038/nsmb.2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Y, Hayward GS. The Ubiquitin E3 Ligase RAUL Negatively Regulates Type I Interferon Through Ubiquitination of the Transcription Factors IRF7 and IRF3. Immunity (2010) 33(6):863–77. doi: 10.1016/j.immuni.2010.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ching W, Koyuncu E, Singh S, Arbelo-Roman C, Hartl B, Kremmer E, et al. A Ubiquitin-Specific Protease Possesses a Decisive Role for Adenovirus Replication and Oncogene-Mediated Transformation. PloS Pathog (2013) 9(3):e1003273. doi: 10.1371/journal.ppat.1003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, et al. Proteomic Profiling of the Human Cytomegalovirus UL35 Gene Products Reveals a Role for UL35 in the DNA Repair Response. J Virol (2012) 86(2):806–20. doi: 10.1128/JVI.05442-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, et al. Protein Profiling With Epstein-Barr Nuclear Antigen-1 Reveals an Interaction With the Herpesvirus-Associated Ubiquitin-Specific Protease HAUSP/Usp7. J Biol Chem (2003) 278(32):29987–94. doi: 10.1074/jbc.M303977200 [DOI] [PubMed] [Google Scholar]
- 31. Qi SM, Cheng G, Cheng XD, Xu Z, Xu B, Zhang WD, et al. Targeting USP7-Mediated Deubiquitination of MDM2/MDMX-P53 Pathway for Cancer Therapy: Are We There Yet? Front Cell Dev Biol (2020) 8:233. doi: 10.3389/fcell.2020.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vidal S, El Motiam A, Seoane R, Preitakaite V, Bouzaher YH, Gomez-Medina S, et al. Regulation of the Ebola Virus VP24 Protein by SUMO. J Virol (2019) 94(1):e01687-19. doi: 10.1128/JVI.01687-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A Role for the Tip60 Histone Acetyltransferase in the Acetylation and Activation of ATM. Proc Natl Acad Sci USA (2005) 102(37):13182–7. doi: 10.1073/pnas.0504211102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dar A, Shibata E, Dutta A. Deubiquitination of Tip60 by USP7 Determines the Activity of the P53-Dependent Apoptotic Pathway. Mol Cell Biol (2013) 33(16):3309–20. doi: 10.1128/MCB.00358-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Valles GJ, Bezsonova I, Woodgate R, Ashton NW. USP7 Is a Master Regulator of Genome Stability. Front Cell Dev Biol (2020) 8:717. doi: 10.3389/fcell.2020.00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rufini A, Tucci P, Celardo I, Melino G. Senescence and Aging: The Critical Roles of P53. Oncogene (2013) 32(43):5129–43. doi: 10.1038/onc.2012.640 [DOI] [PubMed] [Google Scholar]
- 37. Reed SM, Quelle DE. P53 Acetylation: Regulation and Consequences. Cancers (Basel) (2014) 7(1):30–69. doi: 10.3390/cancers7010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour Suppression: Disruption of HAUSP Gene Stabilizes P53. Nature (2004) 428(6982):1 p following 486. doi: 10.1038/nature02501 [DOI] [PubMed] [Google Scholar]
- 39. Sacco JJ, Coulson JM, Clague MJ, Urbe S. Emerging Roles of Deubiquitinases in Cancer-Associated Pathways. IUBMB Life (2010) 62(2):140–57. doi: 10.1002/iub.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becker K, Marchenko ND, Palacios G. Moll UM. A Role of HAUSP in Tumor Suppression in a Human Colon Carcinoma Xenograft Model. Cell Cycle (2008) 7(9):1205–13. doi: 10.4161/cc.7.9.5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alonso-de Vega I, Martin Y, Smits VA. USP7 Controls Chk1 Protein Stability by Direct Deubiquitination. Cell Cycle (2014) 13(24):3921–6. doi: 10.4161/15384101.2014.973324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-Mediated Stabilization of ZEB1 Promotes DNA Damage Response and Radioresistance Through CHK1. Nat Cell Biol (2014) 16(9):864–75. doi: 10.1038/ncb3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 Counteracts SCFbetaTrCP- But Not APCCdh1-Mediated Proteolysis of Claspin. J Cell Biol (2009) 184(1):13–9. doi: 10.1083/jcb.200807137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, et al. UV-Sensitive Syndrome Protein UVSSA Recruits USP7 to Regulate Transcription-Coupled Repair. Nat Genet (2012) 44(5):598–602. doi: 10.1038/ng.2230 [DOI] [PubMed] [Google Scholar]
- 45. Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, et al. Mutations in UVSSA Cause UV-Sensitive Syndrome and Destabilize ERCC6 in Transcription-Coupled DNA Repair. Nat Genet (2012) 44(5):593–7. doi: 10.1038/ng.2228 [DOI] [PubMed] [Google Scholar]
- 46. Zlatanou A, Sabbioneda S, Miller ES, Greenwalt A, Aggathanggelou A, Maurice MM, et al. USP7 Is Essential for Maintaining Rad18 Stability and DNA Damage Tolerance. Oncogene (2016) 35(8):965–76. doi: 10.1038/onc.2015.149 [DOI] [PubMed] [Google Scholar]
- 47. Oh YM, Yoo SJ, Seol JH. Deubiquitination of Chfr, a Checkpoint Protein, by USP7/HAUSP Regulates Its Stability and Activity. Biochem Biophys Res Commun (2007) 357(3):615–9. doi: 10.1016/j.bbrc.2007.03.193 [DOI] [PubMed] [Google Scholar]
- 48. Wang Q, Ma S, Song N, Li X, Liu L, Yang S, et al. Stabilization of Histone Demethylase PHF8 by USP7 Promotes Breast Carcinogenesis. J Clin Invest (2016) 126(6):2205–20. doi: 10.1172/JCI85747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang C, Lu J, Zhang QW, Zhao W, Guo JH, Liu SL, et al. USP7 Promotes Cell Proliferation Through the Stabilization of Ki-67 Protein in non-Small Cell Lung Cancer Cells. Int J Biochem Cell Biol (2016) 79:209–21. doi: 10.1016/j.biocel.2016.08.025 [DOI] [PubMed] [Google Scholar]
- 50. Felle M, Joppien S, Németh A, Diermeier S, Thalhammer V, Dobner T, et al. The USP7/Dnmt1 Complex Stimulates the DNA Methylation Activity of Dnmt1 and Regulates the Stability of UHRF1. Nucleic Acids Res (2011) 39(19):8355–65. doi: 10.1093/nar/gkr528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Su D, Ma S, Shan L, Wang Y, Wang Y, Cao C, et al. Ubiquitin-Specific Protease 7 Sustains DNA Damage Response and Promotes Cervical Carcinogenesis. J Clin Invest (2018) 128(10):4280–96. doi: 10.1172/JCI120518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, et al. PHF8 Mediates Histone H4 Lysine 20 Demethylation Events Involved in Cell Cycle Progression. Nature (2010) 466(7305):508–12. doi: 10.1038/nature09272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park JJ, Lim KH, Baek KH. Annexin-1 Regulated by HAUSP is Essential for UV-Induced Damage Response. Cell Death Dis (2015) 6(2):e1654. doi: 10.1038/cddis.2015.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zlatanou A, Stewart GS. Damaged Replication Forks Tolerate USP7 to Maintain Genome Stability. Mol Cell Oncol (2016) 3(1):e1063571. doi: 10.1080/23723556.2015.1063571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qian J, Pentz K, Zhu Q, Wang Q, He J, Srivastava AK, et al. USP7 Modulates UV-Induced PCNA Monoubiquitination by Regulating DNA Polymerase Eta Stability. Oncogene (2015) 34(36):4791–6. doi: 10.1038/onc.2014.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 Control Ubiquitination and Stability of the Maintenance DNA Methyltransferase Dnmt1. J Cell Biochem (2011) 112(2):439–44. doi: 10.1002/jcb.22998 [DOI] [PubMed] [Google Scholar]
- 57. Chen CY, Chen J, He L, Stiles BL. PTEN: Tumor Suppressor and Metabolic Regulator. Front Endocrinol (Lausanne) (2018) 9:338. doi: 10.3389/fendo.2018.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Álvarez-Garcia V, Tawil Y, Wise HM, Leslie NR. Mechanisms of PTEN Loss in Cancer: It's All About Diversity. Semin Cancer Biol (2019) 59:66–79. doi: 10.1016/j.semcancer.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 59. Wang F, Wang L, Wu J, Sokirniy I, Nguyen P, Bregnard T, et al. Active Site-Targeted Covalent Irreversible Inhibitors of USP7 Impair the Functions of Foxp3+ T-Regulatory Cells by Promoting Ubiquitination of Tip60. PloS One (2017) 12(12):e0189744. doi: 10.1371/journal.pone.0189744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiao Y, Huang Q, Wu Z, Chen W. Roles of Protein Ubiquitination in Inflammatory Bowel Disease. Immunobiology (2020) 225(6):152026. doi: 10.1016/j.imbio.2020.152026 [DOI] [PubMed] [Google Scholar]
- 61. Lu Y, Kim NM, Jiang YW, Zhang H, Zheng D, Zhu FX, et al. Cambogin Suppresses Dextran Sulphate Sodium-Induced Colitis by Enhancing Treg Cell Stability and Function. Br J Pharmacol (2018) 175(7):1085–99. doi: 10.1111/bph.14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu J, Kumar S, Wang F, Wang H, Chen L, Arsenault P, et al. Chemical Approaches to Intervening in Ubiquitin Specific Protease 7 (USP7) Function for Oncology and Immune Oncology Therapies. J Med Chem (2018) 61(2):422–43. doi: 10.1021/acs.jmedchem.7b00498 [DOI] [PubMed] [Google Scholar]
- 63. Wang L, Kumar S, Dahiya S, Wang F, Wu J, Newick K, et al. Ubiquitin-Specific Protease-7 Inhibition Impairs Tip60-Dependent Foxp3+ T-Regulatory Cell Function and Promotes Antitumor Immunity. EBioMedicine (2016) 13:99–112. doi: 10.1016/j.ebiom.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-Dependent Downregulation of USP7/HAUSP by PPM1G Activates P53 Response to DNA Damage. Mol Cell (2012) 45(6):801–13. doi: 10.1016/j.molcel.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Malapelle U, Morra F, Ilardi G, Visconti R, Merolla F, Cerrato A, et al. USP7 Inhibitors, Downregulating CCDC6, Sensitize Lung Neuroendocrine Cancer Cells to PARP-Inhibitor Drugs. Lung Cancer (2017) 107:41–9. doi: 10.1016/j.lungcan.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 66. Kumagai A, Dunphy WG. Claspin, a Novel Protein Required for the Activation of Chk1 During a DNA Replication Checkpoint Response in Xenopus Egg Extracts. Mol Cell (2000) 6(4):839–49. doi: 10.1016/s1097-2765(05)00092-4 [DOI] [PubMed] [Google Scholar]
- 67. Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 Pathways in DNA Damage Signaling and Cancer. Adv Cancer Res (2010) 108:73–112. doi: 10.1016/b978-0-12-380888-2.00003-0 [DOI] [PubMed] [Google Scholar]
- 68. Qing P, Han L, Bin L, Yan L, Ping WX. USP7 Regulates the Stability and Function of HLTF Through Deubiquitination. J Cell Biochem (2011) 112(12):3856–62. doi: 10.1002/jcb.23317 [DOI] [PubMed] [Google Scholar]
- 69. Kashiwaba S, Kanao R, Masuda Y, Kusumoto-Matsuo R, Hanaoka F, Masutani C. USP7 Is a Suppressor of PCNA Ubiquitination and Oxidative-Stress-Induced Mutagenesis in Human Cells. Cell Rep (2015) 13(10):2072–80. doi: 10.1016/j.celrep.2015.11.014 [DOI] [PubMed] [Google Scholar]
- 70. Masuda Y, Kanao R, Kawai H, Kukimoto I, Masutani C. Preferential Digestion of PCNA-Ubiquitin and P53-Ubiquitin Linkages by USP7 to Remove Polyubiquitin Chains From Substrates. J Biol Chem (2019) 294(11):4177–87. doi: 10.1074/jbc.RA118.005167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen ST, Okada M, Nakato R, Izumi K, Bando M, Shirahige K. The Deubiquitinating Enzyme USP7 Regulates Androgen Receptor Activity by Modulating Its Binding to Chromatin. J Biol Chem (2015) 290(35):21713–23. doi: 10.1074/jbc.M114.628255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sun X, Kaufman PD. Ki-67: More Than a Proliferation Marker. Chromosoma (2018) 127(2):175–86. doi: 10.1007/s00412-018-0659-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Callegari K, Maegawa S, Bravo-Alegria J, Gopalakrishnan V. Pharmacological Inhibition of LSD1 Activity Blocks REST-Dependent Medulloblastoma Cell Migration. Cell Commun Signal (2018) 16(1):60. doi: 10.1186/s12964-018-0275-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee JE, Park CM, Kim JH. USP7 Deubiquitinates and Stabilizes EZH2 in Prostate Cancer Cells. Genet Mol Biol (2020) 43(2):e20190338. doi: 10.1590/1678-4685-GMB-2019-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zeng Q, Li Z, Zhao X, Guo L, Yu C, Qin J, et al. Ubiquitinspecific Protease 7 Promotes Osteosarcoma Cell Metastasis by Inducing Epithelialmesenchymal Transition. Oncol Rep (2019) 41(1):543–51. doi: 10.3892/or.2018.6835 [DOI] [PubMed] [Google Scholar]
- 76. Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of Disease: Epithelial-Mesenchymal Transition–Does Cellular Plasticity Fuel Neoplastic Progression? Nat Clin Pract Oncol (2008) 5(5):280–90. doi: 10.1038/ncponc1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhao GY, Lin ZW, Lu CL, Gu J, Yuan YF, Xu FK, et al. USP7 Overexpression Predicts a Poor Prognosis in Lung Squamous Cell Carcinoma and Large Cell Carcinoma. Tumour Biol (2015) 36(3):1721–9. doi: 10.1007/s13277-014-2773-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang L, Wang H, Tian L, Li H. Expression of USP7 and MARCH7 Is Correlated With Poor Prognosis in Epithelial Ovarian Cancer. Tohoku J Exp Med (2016) 239(3):165–75. doi: 10.1620/tjem.239.165 [DOI] [PubMed] [Google Scholar]
- 79. Nicholson B, Suresh Kumar KG. The Multifaceted Roles of USP7: New Therapeutic Opportunities. Cell Biochem Biophys (2011) 60(1-2):61–8. doi: 10.1007/s12013-011-9185-5 [DOI] [PubMed] [Google Scholar]
- 80. van Loosdregt J, Fleskens V, Fu J, Brenkman AB, Bekker CP, Pals CE, et al. Stabilization of the Transcription Factor Foxp3 by the Deubiquitinase USP7 Increases Treg-Cell-Suppressive Capacity. Immunity (2013) 39(2):259–71. doi: 10.1016/j.immuni.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shin SB, Kim CH, Jang HR, Yim H. Combination of Inhibitors of USP7 and PLK1 has a Strong Synergism Against Paclitaxel Resistance. Int J Mol Sci (2020) 21(22):8629. doi: 10.3390/ijms21228629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Choi ES, Lee H, Sung JY, Lee CH, Jang H, Kim KT, et al. FAM188B Enhances Cell Survival via Interaction With USP7. Cell Death Dis (2018) 9(6):633. doi: 10.1038/s41419-018-0650-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jiang S, Wang X, He Y, Huang H, Cao B, Zhang Z, et al. Suppression of USP7 Induces BCR-ABL Degradation and Chronic Myelogenous Leukemia Cell Apoptosis. Cell Death Dis (2021) 12(5):456. doi: 10.1038/s41419-021-03732-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Carrà G, Panuzzo C, Torti D, Parvis G, Crivellaro S, Familiari U, et al. Therapeutic Inhibition of USP7-PTEN Network in Chronic Lymphocytic Leukemia: A Strategy to Overcome TP53 Mutated/Deleted Clones. Oncotarget (2017) 8(22):35508–22. doi: 10.18632/oncotarget.16348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. He Y, Wang S, Tong J, Jiang S, Yang Y, Zhang Z, et al. The Deubiquitinase USP7 Stabilizes Maf Proteins to Promote Myeloma Cell Survival. J Biol Chem (2020) 295(7):2084–96. doi: 10.1074/jbc.RA119.010724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xia X, Liao Y, Huang C, Liu Y, He J, Shao Z, et al. Deubiquitination and Stabilization of Estrogen Receptor Alpha by Ubiquitin-Specific Protease 7 Promotes Breast Tumorigenesis. Cancer Lett (2019) 465:118–28. doi: 10.1016/j.canlet.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 87. Peng Y, Liu Y, Gao Y, Yuan B, Qi X, Fu Y, et al. USP7 is a Novel Deubiquitinase Sustaining PLK1 Protein Stability and Regulating Chromosome Alignment in Mitosis. J Exp Clin Cancer Res (2019) 38(1):468. doi: 10.1186/s13046-019-1457-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yang P, Xie J, Li Y, Lin HP, Fenske W, Clementino M, et al. Deubiquitinase USP7-Mediated MCL-1 Up-Regulation Enhances Arsenic and Benzo(a)Pyrene Co-Exposure-Induced Cancer Stem Cell-Like Property and Tumorigenesis. Theranostics (2020) 10(20):9050–65. doi: 10.7150/thno.47897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hu T, Zhang J, Sha B, Li M, Wang L, Zhang Y, et al. Targeting the Overexpressed USP7 Inhibits Esophageal Squamous Cell Carcinoma Cell Growth by Inducing NOXA-Mediated Apoptosis. Mol Carcinog (2019) 58(1):42–54. doi: 10.1002/mc.22905 [DOI] [PubMed] [Google Scholar]
- 90. Zaman MM, Nomura T, Takagi T, Okamura T, Jin W, Shinagawa T, et al. Ubiquitination-Deubiquitination by the TRIM27-USP7 Complex Regulates Tumor Necrosis Factor Alpha-Induced Apoptosis. Mol Cell Biol (2013) 33(24):4971–84. doi: 10.1128/MCB.00465-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hu HJ, Zhang LG, Wang ZH, Guo XX. FoxO6 Inhibits Cell Proliferation in Lung Carcinoma Through Up-Regulation of USP7. Mol Med Rep (2015) 12(1):575–80. doi: 10.3892/mmr.2015.3362 [DOI] [PubMed] [Google Scholar]
- 92. Jin Q, Martinez CA, Arcipowski KM, Zhu Y, Gutierrez-Diaz BT, Wang KK, et al. USP7 Cooperates With NOTCH1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clin Cancer Res (2019) 25(1):222–39. doi: 10.1158/1078-0432.CCR-18-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tavana O, Gu W. Modulation of the P53/MDM2 Interplay by HAUSP Inhibitors. J Mol Cell Biol (2017) 9(1):45–52. doi: 10.1093/jmcb/mjw049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yang Z, Huo S, Shan Y, Liu H, Xu Y, Yao K, et al. STAT3 Repressed USP7 Expression is Crucial for Colon Cancer Development. FEBS Lett (2012) 586(19):3013–7. doi: 10.1016/j.febslet.2012.06.025 [DOI] [PubMed] [Google Scholar]
- 95. Boutell C, Canning M, Orr A, Everett RD. Reciprocal Activities Between Herpes Simplex Virus Type 1 Regulatory Protein ICP0, a Ubiquitin E3 Ligase, and Ubiquitin-Specific Protease USP7. J Virol (2005) 79(19):12342–54. doi: 10.1128/jvi.79.19.12342-12354.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein Interaction Domains of the Ubiquitin-Specific Protease, USP7/HAUSP. J Biol Chem (2003) 278(48):47753–61. doi: 10.1074/jbc.M307200200 [DOI] [PubMed] [Google Scholar]
- 97. Frau M, Feo F, Pascale RM. Pleiotropic Effects of Methionine Adenosyltransferases Deregulation as Determinants of Liver Cancer Progression and Prognosis. J Hepatol (2013) 59(4):830–41. doi: 10.1016/j.jhep.2013.04.031 [DOI] [PubMed] [Google Scholar]
- 98. Rice AP, Mathews MB. Transcriptional But Not Translational Regulation of HIV-1 by the Tat Gene Product. Nature (1988) 332(6164):551–3. doi: 10.1038/332551a0 [DOI] [PubMed] [Google Scholar]
- 99. Liu X, Yang X, Li Y, Zhao S, Li C, Ma P, et al. Trip12 is an E3 Ubiquitin Ligase for USP7/HAUSP Involved in the DNA Damage Response. FEBS Lett (2016) 590(23):4213–22. doi: 10.1002/1873-3468.12471 [DOI] [PubMed] [Google Scholar]
- 100. Morotti A, Panuzzo C, Crivellaro S, Pergolizzi B, Familiari U, Berger AH, et al. BCR-ABL Disrupts PTEN Nuclear-Cytoplasmic Shuttling Through Phosphorylation-Dependent Activation of HAUSP. Leukemia (2014) 28(6):1326–33. doi: 10.1038/leu.2013.370 [DOI] [PubMed] [Google Scholar]
- 101. Xiang M, Liang L, Kuang X, Xie Z, Liu J, Zhao S, et al. Pharmacological Inhibition of USP7 Suppresses Growth and Metastasis of Melanoma Cells In Vitro and In Vivo . J Cell Mol Med (2021) 25(19):9228–40. doi: 10.1111/jcmm.16834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Pawlak A, Bajzert J, Bugiel K, Hernandez Suarez B, Kutkowska J, Rapak A, et al. Ubiquitin-Specific Protease 7 as a Potential Therapeutic Target in Dogs With Hematopoietic Malignancies. J Vet Intern Med (2021) 35(2):1041–51. doi: 10.1111/jvim.16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lee G, Oh TI, Um KB, Yoon H, Son J, Kim BM, et al. Small-Molecule Inhibitors of USP7 Induce Apoptosis Through Oxidative and Endoplasmic Reticulum Stress in Cancer Cells. Biochem Biophys Res Commun (2016) 470(1):181–6. doi: 10.1016/j.bbrc.2016.01.021 [DOI] [PubMed] [Google Scholar]
- 104. Leger PR, Hu DX, Biannic B, Bui M, Han X, Karbarz E, et al. Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 With In Vivo Antitumor Activity. J Med Chem (2020) 63(10):5398–420. doi: 10.1021/acs.jmedchem.0c00245 [DOI] [PubMed] [Google Scholar]
- 105. Li T, Guan J, Li S, Zhang X, Zheng X. HSCARG Downregulates NF-kappaB Signaling by Interacting With USP7 and Inhibiting NEMO Ubiquitination. Cell Death Dis (2014) 5:e1229. doi: 10.1038/cddis.2014.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Novellasdemunt L, Foglizzo V, Cuadrado L, Antas P, Kucharska A, Encheva V, et al. USP7 Is a Tumor-Specific WNT Activator for APC-Mutated Colorectal Cancer by Mediating Beta-Catenin Deubiquitination. Cell Rep (2017) 21(3):612–27. doi: 10.1016/j.celrep.2017.09.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sun X, Ding Y, Zhan M, Li Y, Gao D, Wang G, et al. Usp7 Regulates Hippo Pathway Through Deubiquitinating the Transcriptional Coactivator Yorkie. Nat Commun (2019) 10(1):411. doi: 10.1038/s41467-019-08334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rawat R, Starczynowski DT, Ntziachristos P. Nuclear Deubiquitination in the Spotlight: The Multifaceted Nature of USP7 Biology in Disease. Curr Opin Cell Biol (2019) 58:85–94. doi: 10.1016/j.ceb.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hao YH, Fountain MD, Jr, Fon Tacer K, Xia F, Bi W, Kang SH, et al. USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Mol Cell (2015) 59(6):956–69. doi: 10.1016/j.molcel.2015.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Pozhidaeva A, Valles G, Wang F, Wu J, Sterner DE, Nguyen P, et al. USP7-Specific Inhibitors Target and Modify the Enzyme's Active Site via Distinct Chemical Mechanisms. Cell Chem Biol (2017) 24(12):1501–12.e5. doi: 10.1016/j.chembiol.2017.09.004 [DOI] [PubMed] [Google Scholar]
- 111. Gavory G, O'Dowd CR, Helm MD, Flasz J, Arkoudis E, Dossang A, et al. Discovery and Characterization of Highly Potent and Selective Allosteric USP7 Inhibitors. Nat Chem Biol (2018) 14(2):118–25. doi: 10.1038/nchembio.2528 [DOI] [PubMed] [Google Scholar]
- 112. Schauer NJ, Liu X, Magin RS, Doherty LM, Chan WC, Ficarro SB, et al. Selective USP7 Inhibition Elicits Cancer Cell Killing Through a P53-Dependent Mechanism. Sci Rep (2020) 10(1):5324. doi: 10.1038/s41598-020-62076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pan T, Li X, Li Y, Tao Z, Yao H, Wu Y, et al. USP7 Inhibition Induces Apoptosis in Glioblastoma by Enhancing Ubiquitination of ARF4. Cancer Cell Int (2021) 21(1):508. doi: 10.1186/s12935-021-02208-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang Y, Zhou L, Rouge L, Phillips AH, Lam C, Liu P, et al. Conformational Stabilization of Ubiquitin Yields Potent and Selective Inhibitors of USP7. Nat Chem Biol (2013) 9(1):51–8. doi: 10.1038/nchembio.1134 [DOI] [PubMed] [Google Scholar]
- 115. Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, et al. Discovery of Specific Inhibitors of Human USP7/HAUSP Deubiquitinating Enzyme. Chem Biol (2012) 19(4):467–77. doi: 10.1016/j.chembiol.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 116. Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, et al. USP7 Inhibitor P22077 Inhibits Neuroblastoma Growth via Inducing P53-Mediated Apoptosis. Cell Death Dis (2013) 4:e867. doi: 10.1038/cddis.2013.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Morra F, Luise C, Merolla F, Poser I, Visconti R, Ilardi G, et al. FBXW7 and USP7 Regulate CCDC6 Turnover During the Cell Cycle and Affect Cancer Drugs Susceptibility in NSCLC. Oncotarget (2015) 6(14):12697–709. doi: 10.18632/oncotarget.3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A Small Molecule Inhibitor of Ubiquitin-Specific Protease-7 Induces Apoptosis in Multiple Myeloma Cells and Overcomes Bortezomib Resistance. Cancer Cell (2012) 22(3):345–58. doi: 10.1016/j.ccr.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dai X, Lu L, Deng S, Meng J, Wan C, Huang J, et al. USP7 Targeting Modulates Anti-Tumor Immune Response by Reprogramming Tumor-Associated Macrophages in Lung Cancer. Theranostics (2020) 10(20):9332–47. doi: 10.7150/thno.47137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Fu C, Zhu X, Xu P, Li Y. Pharmacological Inhibition of USP7 Promotes Antitumor Immunity and Contributes to Colon Cancer Therapy. Onco Targets Ther (2019) 12:609–17. doi: 10.2147/OTT.S182806 [DOI] [PMC free article] [PubMed] [Google Scholar]