Abstract
Phosphatidylglycerol is a crucial phospholipid found ubiquitously in biological membranes of prokaryotic and eukaryotic cells. The phosphatidylglycerol phosphate (PGP) synthase (PgsA), a membrane-embedded enzyme, catalyzes the primary reaction of phosphatidylglycerol biosynthesis. Mutations in pgsA frequently correlate with daptomycin resistance in Staphylococcus aureus and other prevalent infectious pathogens. Here we report the crystal structures of S. aureus PgsA (SaPgsA) captured at two distinct states of the catalytic process, with lipid substrate (cytidine diphosphate-diacylglycerol, CDP-DAG) or product (PGP) bound to the active site within a trifurcated amphipathic cavity. The hydrophilic head groups of CDP-DAG and PGP occupy two different pockets in the cavity, inducing local conformational changes. An elongated membrane-exposed surface groove accommodates the fatty acyl chains of CDP-DAG/PGP and opens a lateral portal for lipid entry/release. Remarkably, the daptomycin resistance-related mutations mostly cluster around the active site, causing reduction of enzymatic activity. Our results provide detailed mechanistic insights into the dynamic catalytic process of PgsA and structural frameworks beneficial for development of antimicrobial agents targeting PgsA from pathogenic bacteria.
Keywords: Phosphatidylglycerol, Synthase, Staphylococcus aureus, Daptomycin resistance, Membrane protein, Crystal structure
Abbreviations: PG, phosphatidylglycerol; PGP, phosphatidylglycerol phosphate; PgsA, phosphatidylglycerol phosphate synthase A; CDP-DAG, cytidine diphosphate-diacylglycerol; CL, cardiolipin; G3P, glycerol 3-phosphate; MRSA, methicillin-resistant Staphylococcus aureus; LCP, lipidic cubic phase
Graphical abstract
Highlights
-
•
PgsA uses a trifurcated amphipathic cavity for binding of substrates or products.
-
•
Conversion of CDP-DAG to PGP induces local conformational changes in PgsA.
-
•
Daptomycin-resistant mutations of PgsA mostly lead to reduced catalytic activity.
-
•
A structure-based five-state model is proposed for the synthesis of PGP by PgsA.
1. Introduction
Lipids are a group of hydrophobic or amphipathic organic compounds and have diverse biological functions by serving as membrane components, fuel, vitamins, hormones, lung surfactants, etc (Masoro, 1977). Lipid biosynthesis and formation of lipid membranes are crucial for the emergence and evolution of various cellular organisms from three domains of life, namely Archaea, Bacteria and Eukarya (Lombard et al., 2012). Phospholipids are an important class of lipids with a phosphate-containing hydrophilic head group and two hydrophobic fatty acyl tails, serving as the major components of biological membrane in addition to membrane proteins (Raetz, 1978). In prokaryotic and eukaryotic cells, phospholipids fulfill indispensable roles by contributing to formation of biological membranes and participating in various fundamental biochemical processes (Dowhan, 1997; Chauhan et al., 2016). In biological membranes, phospholipids assemble into a lipid bilayer structure with low permeability to ions, hydrophilic metabolites and biological macromolecules such as proteins and nucleic acids. Such feature is essential for the establishment of asymmetric environments across the membrane, distinguishing the intracellular domain from the extracellular space. Moreover, phospholipids also give biological membrane the adaptability to tolerate fluctuation of temperature and osmotic pressure, enable membrane to bend and deform, provide a variety of signal molecules (such as phosphoinositide and phosphatidylserine), and contribute specific lipid cofactors and compatible environments to support the proper function and assembly of membrane proteins (Chauhan et al., 2016).
Glycerophospholipids are the dominant type of phospholipids with multitudinous structures and functions (Bell et al., 1981). Among various glycerophospholipids with different head groups, phosphatidylglycerol (PG) is a pivotal anionic phospholipid present in bacteria, plants and animals (Furse, 2017). PG and its derivatives (aminoacyl PG and cardiolipin/CL) are the most abundant phospholipids in Staphylococcus aureus, a Gram-positive bacterium (Roy, 2009). Meanwhile, PG and CL are the major anionic phospholipids in Escherichia coli (a Gram-negative bacterium) membrane besides phosphatidylethanolamine/PE, the most abundant zwitterionic phospholipid in E. coli (Raetz and Dowhan, 1990). In plants, PG is the only major phospholipid in the photosynthetic membranes of chloroplasts and has important functions for the electron transport process during photosynthesis, chloroplast development and chilling tolerance (Wada and Murata, 2007). In mammals, PG is crucial for lung function as it is one of the major phospholipids found in lung surfactants and modulates the surface activity of lung, and it may also regulate innate immunity and inhibit infections caused by respiratory viruses (Veldhuizen et al., 1998; Numata et al., 2010, 2012). Besides, PG is also involved in activation of RNA synthesis and nuclear protein kinase C as well as inhibition of platelet activating factor and phosphatidylcholine (PC) transfer in mammalian cells (Furse, 2017).
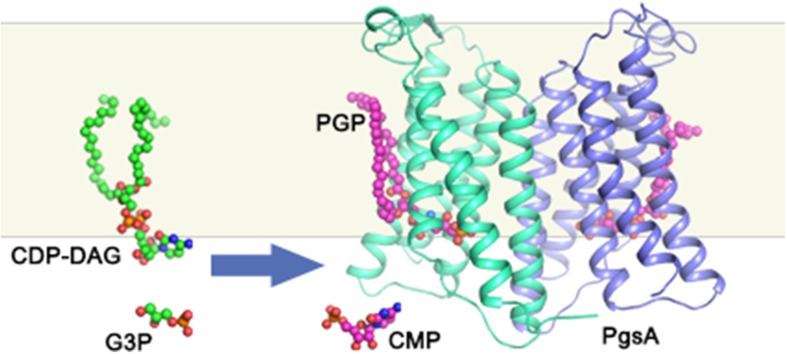
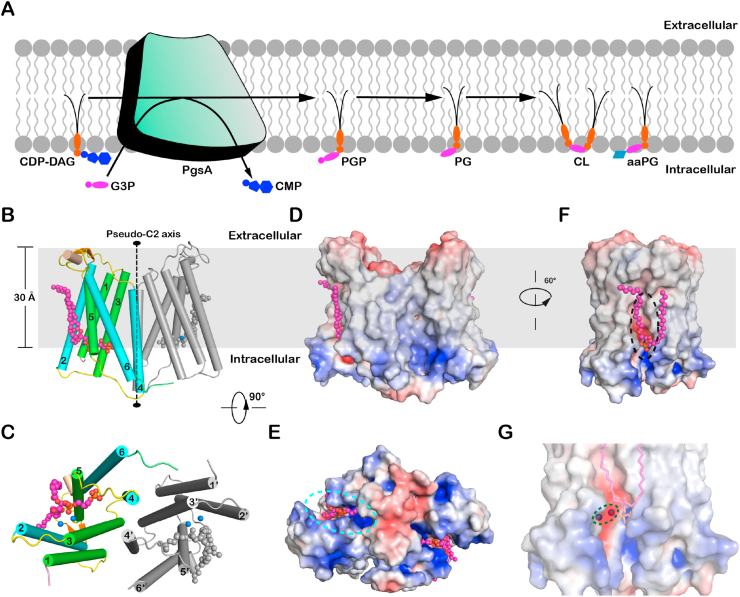
As the PG precursor, phosphatidylglycerol phosphate (PGP) is synthesized through the cytidine diphosphate-diacylglycerol (CDP-DAG)−dependent pathway, a fundamental phospholipid biosynthesis process present in both prokaryotic and eukaryotic cells (Blunsom and Cockcroft, 2020; Jennings and Epand, 2020). The bacterial PGP synthase PgsA is a membrane-embedded enzyme that converts CDP-DAG and glycerol 3-phosphate (G3P) into PGP and cytidine monophosphate (CMP) at the interface between membrane and cytosol (Zhang and Rock, 2008) (Fig. 1A). To further generate PG, PGP is dephosphorylated by phosphatases to remove the terminal phosphate group (Lu et al., 2011). Subsequently, PG can be utilized by the cardiolipin synthase as the substrate to produce CL (Raetz and Dowhan, 1990). Alternatively, it can be used by the aminoacyl phosphatidylglycerol synthases to produce aminoacyl PG (aaPG), an important class of PG derivatives crucial for bacteria to adapt to environmental changes and resist cationic antimicrobial peptides or antibiotics (Roy, 2009; Ernst and Peschel, 2011). Moreover, PG can be converted into palmitoyl-PG by a phospholipase/palmitoyltransferase named PagP (PhoPQ-activated gene P) in the outer membrane of Gram-negative bacteria (Dalebroux et al., 2014).
Fig. 1.
Biological function and overall structure of SaPgsA.
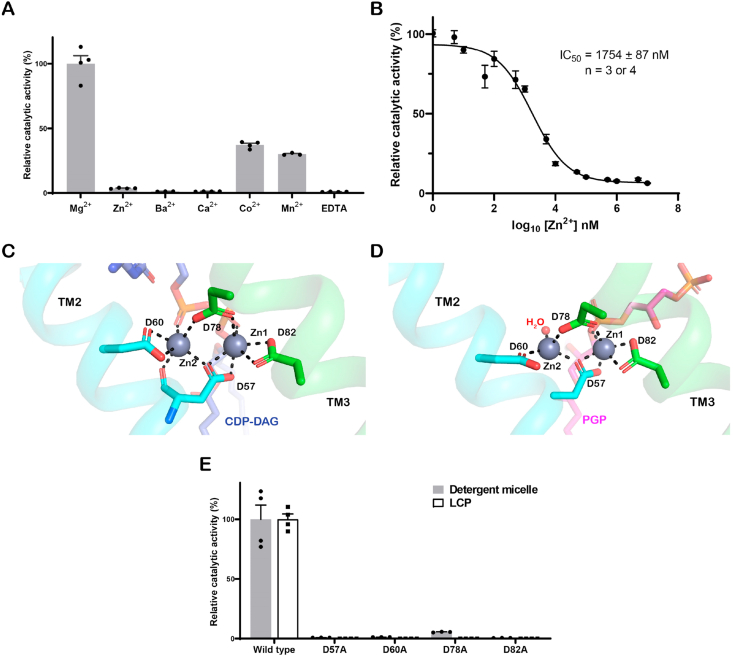
(A) A cartoon diagram showing the functional role of SaPgsA in biosynthesis of PG and its derivatives in the membrane. (B and C) The overall structure of SaPgsA dimer shown in cartoon model viewed along membrane plane (B) or membrane normal from cytoplasmic side (C). Color codes: green, TM1, TM3 and TM5; cyan, TM2, TM4 and TM6; violet, the N-terminal region; lime, the C-terminal region; yellow, loop regions between adjacent TMs; orange, the β-hairpin in the TM1-TM2 loop region; wheat, the short α-helix between TM5 and TM6. The lipid molecules (PGP) are shown as magenta or grey sphere models, whereas the Zinc ions are presented as marine blue spheres. For clarity, one protomer is shown in colored mode, whereas the adjacent protomer is in light grey. (D–F) The electrostatic potential surface representation of SaPgsA. The views are along the membrane plane from two different angles (D, F) or along membrane normal from intracellular side (E). Color codes: blue, electropositive; white, neutral; red, electronegative. The grey box in the background of B, D and F indicates the approximate position of the hydrophobic region of membrane according to the result of prediction through the Positioning of Proteins in Membrane (PPM) server. The cyan dashed elliptical ring in E indicates the position of active-site cavity within the SaPgsA monomer, while the black dashed elliptical ring in F labels the approximate location of a lateral portal connecting the active site with lipid bilayer. (G) A close-up view of the active site center with a bimetal binding site. The green dashed elliptical ring indicates the location of two metal ions in the active site of SaPgsA. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Dysfunctional mutation of pgsA leads to drastic reduction of PG, CL and PG derivatives in E. coli membrane which are essential for cell growth and viability (Miyazaki et al., 1985). Moreover, pgsA is frequently associated with daptomycin resistance in pathogenic bacteria posing threats to human health (Peleg et al., 2012; Hines et al., 2017; Tran et al., 2019). Daptomycin is a cyclic lipopeptide antibiotic used for treating serious infections caused by methicillin-resistant S. aureus (MRSA), vancomycin-resistant Enterococci (VRE) and other Gram-positive pathogens (Kirkpatrick et al., 2003). Accumulating evidences show that mutations in pgsA correlate with daptomycin resistance in several Gram-positive bacteria such as S. aureus, Bacillus subtilis, Corynebacterium striatum, Staphylococcus capitis and Streptococcus oralis (Peleg et al., 2012; Hines et al., 2017; Tran et al., 2019; Goldner et al., 2018; Hachmann et al., 2009; Hagiya et al., 2018; Casanova et al., 2017). Furthermore, SaPgsA has recently been identified as a potential antibacterial target to eradicate methicillin-resistant S. aureus (MRSA) persisters (Lu et al., 2020).
Cyanobacteria with dysfunctional PgsA exhibit impaired assembly, oligomerization and function of both photosystem I (PSI) and photosystem II (PSII) (Apostolova et al., 2008). Besides, PGP synthase gene expression is regulated by factors that affect mitochondrial development, making the enzyme an excellent indicator for mitochondrial membrane biogenesis (Gaynor et al., 1991). A previous study found that the activity of mitochondrial PGP synthase was increased by 21% and 98% in 15- and 22-month-old rats respectively, in comparison to the 2-month-old animals suffering from spontaneously hypertensive heart failure (Saini-Chohan et al., 2009). Moreover, a recent study proposed that mitochondrial cardiolipin synthases might evolve from prokaryotic PgsA through the neofunctionalization of the bacterial ancestor (Luevano-Martinez and Duncan, 2020).
Despite that PgsA has important biological functions and is closely associated with antibiotic resistance of pathogenic bacteria, little is known about its structure, catalytic mechanism and functional link between PgsA mutations and daptomycin resistance. To shed light into the catalytic mechanism of PgsA and assist the future development of antibacterial agents, we have solved the crystal structures of SaPgsA with PGP and CDP-DAG bound to the active site at resolutions of 2.5 Å and 3.0 Å, respectively. Moreover, the daptomycin resistance-related mutations are mapped onto the structure and enzymatic activity assays have been carried out to characterize the functional effect of the mutations. Structure-based mutagenesis and enzymatic assay results allow us to propose a working model for PGP biosynthesis. Meanwhile, our high-resolution structures of SaPgsA may also serve as a framework for rational design of antimicrobial drugs.
2. Results
2.1. Structure determination and overall structural features of SaPgsA
Purified SaPgsA was reconstituted into lipid cubic phase (LCP), a lipid-based matrix providing membrane-like environment for crystallization of membrane proteins (Caffrey and Cherezov, 2009). The initial phases of SaPgsA−PGP complex structure were solved through the ab initio phasing and chain tracing method (Supplementary Figs. 1A and B) (Caballero et al., 2018). SaPgsA molecules were packed in two-dimensional lattices, which further stacked orderly in the third dimension through interlayer contacts to form the type I membrane-protein crystal (Supplementary Fig. 1C). The structures of SaPgsA were refined to 2.5 Å (PGP−bound state) and 3.0 Å (CDP-DAG−bound state) resolution, respectively (Supplementary Table 1). For clarity, the higher resolution SaPgsA−PGP complex structure is used for overall fold analysis.
As shown in Fig. 1B and C, SaPgsA forms a homodimer with two protomers related by a central pseudo two-fold symmetry axis and each protein monomer forms a compact fold with six transmembrane α-helices (TMs 1-6). The SaPgsA dimer is not only observed in the crystal grown in LCP (Caffrey, 2015), but is also present in detergent solution (Supplementary Fig. 1D). The dimerization interface of SaPgsA is stabilized mostly through the extensive hydrophobic interactions between TM3 and TM4’ (the prime symbol indicates TMs from the adjacent protomer) and between TM4 and TM4’ in the membrane-embedded region (Fig. 1B and C).
While SaPgsA belongs to the CDP-alcohol phosphatidyltransferase (CDP-OH_P_transf) superfamily (http://pfam.xfam.org/family/CDP-OH_P_transf), its overall structure differs from the other members of CDP-OH_P_transf superfamily with known structures as it shares fairly low sequence identity (∼20%) with them (Supplementary Figs. 2A and C). CDP-OH_P_transf enzymes are all integral membrane proteins catalyzing the scission of a phosphoric anhydride bond to release CMP from a CDP-alcohol and concomitant formation of a new phosphodiester bond in the presence of divalent cation (Chauhan et al., 2016). They contain a strictly conserved motif (D1xxD2G1xxAR … G2xxxD3xxxD4) that is crucial for catalysis (Supplementary Figs. 2A and B) (Nogly et al., 2014; Sciara et al., 2014; Clarke et al., 2015; Grave et al., 2019; Belcher Dufrisne et al., 2020). As proposed previously (Clarke et al., 2015), CDP-OH_P_transf superfamily can be further classified into three families (A, B and C) basing on membrane topology and evolutionary relationship (Supplementary Fig. 3). Most of prokaryotic PgsAs belong to family A which has not been structurally characterized. The mechanism of product formation, binding and release steps in the catalytic process mediated by CDP-OH_P_transf enzyme remains unclear.
Analysis through the Positioning of Proteins in Membrane (PPM) server (Lomize et al., 2012) indicates that most of the SaPgsA is embedded in the membrane (Fig. 1B and D). The amino-terminal region of SaPgsA is fairly short with only two residues. In comparison, the structures of phosphatidylglycerol phosphate synthase (PIPS) from Renibacterium salmoninarum and Mycobacterium tuberculosis possess much longer N-terminal regions forming a juxta-membrane helix (Clarke et al., 2015; Grave et al., 2019). On the extracellular side, all six TMs of SaPgsA are positioned underneath the membrane surface (Fig. 1B), creating a concave surface which may induce local membrane deformation (Fig. 1D). Two extracellular regions, namely the long TM1-2 loop and the TM5-6 loop with a short four-residue α-helix, are positioned near the membrane surface, whereas a much shorter TM3-4 loop is located below the membrane surface (Fig. 1B and C). On the intracellular side, all TMs of SaPgsA including the shortest one (TM5) extend out of the membrane surface and protrude into cytoplasm (Fig. 1B). For the structures of other CDP-OH_P_transf members, such as AF2299 and PIPS from Mycobacterium kansasii (Sciara et al., 2014; Belcher Dufrisne et al., 2020), TM5s are all completely buried in the membrane instead (Supplementary Fig. 2C).
Curiously, a large open cavity is harbored in the membrane-embedded core of SaPgsA (Fig. 1E). This cavity, surrounded by TM2, TM3, TM4 and TM5, has an amphipathic surface (Fig. 1D and F). While TM4 and TM5 are straight α-helices, TM2 and TM3 each contains a conserved kink vital for enzymatic activity (Nogly et al., 2014). The cavity has a hydrophilic portal connected with the cytoplasm and a lateral hydrophobic portal opening toward the membrane (Fig. 1E and F). Such a feature differs from the one in the di-myo-inositol-1,3′-phosphate-1'phosphate synthase from Archaeoglobus fulgidus (AfDIPPS) (Nogly et al., 2014), as AfDIPPS contains two small pockets opening to the cytoplasm to acquire and accommodate two hydrophilic substrates. The intracellular entrance of SaPgsA is mainly shaped by the TM2-3 and TM4-5 loops located on the intracellular surface, while the lateral membrane-exposed portal is located at the groove between TM2 and TM5. The lateral portal may serve as the gateway for hydrophobic substrate to enter the cavity from the membrane or for the lipid product to return to the membrane. Similarly, the hydrophilic substrate (G3P) or product (CMP) might use the intracellular entrance to access or exit from the active site. In support of this hypothesis, the surface around the intracellular entrance is overall electropositive and may thus facilitate the entry of substrate and release of product via electrostatic attraction.
2.2. An amphipathic cavity with an electropositive pocket for PGP binding
To understand the mechanism of product formation, binding and release process, we have solved the structure of SaPgsA in complex with its product PGP (Supplementary Table 1, SaPgsA−PGP). The 2Fo-Fc and the omit electron density maps fit well with the structural model of PGP (Supplementary Fig. 4A). Consistently, mass spectrometry analysis on the lipid sample extracted from purified SaPgsA protein revealed characteristic peaks of intact PGP and its fragments (Supplementary Fig. 4B). It is noteworthy that PGP was co-purified along with SaPgsA protein and preserved during protein purification and crystallization process.
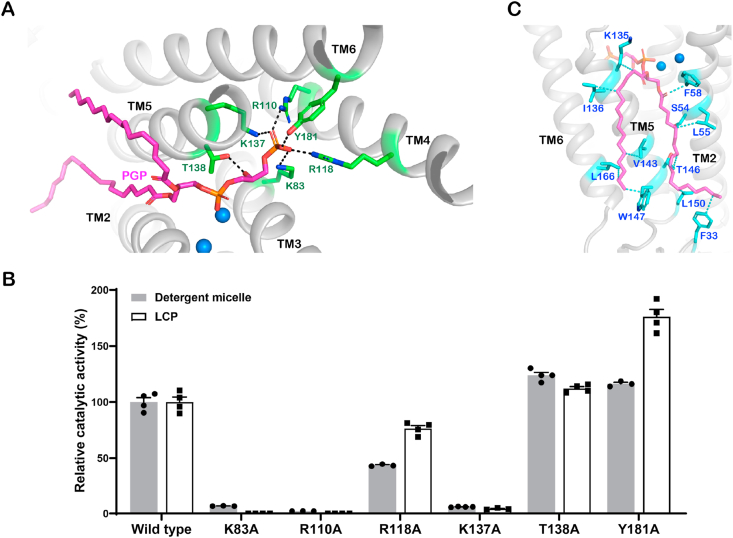
In the SaPgsA−PGP complex structure, each protomer binds one endogenous PGP molecule in the active site (Fig. 2A). The PGP binding site is embedded mostly in the hydrophobic region and harbored inside the large intracellular cavity. The hydrophilic head group of PGP is positioned nearby the bimetal catalytic center within the electropositive region of the cavity exposed to the cytoplasm (Fig. 2A). The 3′-phosphoryl moiety of PGP is sandwiched between TM3 and TM5, and interacts with the evolutionarily conserved Lys83, Arg110, Arg118, Lys137 and Tyr181 (Fig. 2A and Supplementary Fig. 2B). The positively charged Arg118 and hydroxyl-containing Tyr181 are close to the intracellular surface of membrane, and may attract the negatively charged substrate G3P into the active site. A characteristic feature of the PGP binding site lies in the strongly electropositive region made of three positively charged residues (Lys83, Arg110 and Lys137). Such arrangement is favorable for neutralizing the negative charges on the G3P head group of PGP. Consistently, K83A, R110A, R118A and K137A mutants exhibited activity lower than the wild type. In contrast, the Y181A mutation increased the activity, as the mutation may facilitate entry of G3P into the active site by opening the pathway wider (Fig. 2A and B). Besides, the backbone 3-phosphoryl moiety of PGP is located between TM2 and TM5 (Fig. 2A). Thr138 on TM5 forms a hydrogen bond with the 2′-hydroxyl of PGP, and the loss of this interaction may facilitate PGP release from the active center. Therefore, T138A exhibited slightly higher activity than the wild type (Fig. 2B). On the other hand, the 1-acyl chain of PGP extends along the groove between TM2 and TM5, whereas the 2-acyl chain is positioned on the hydrophobic surface of TM5 and TM6 (Fig. 2C). Since CMP is absent in the SaPgsA−PGP complex structure, it may represent an intermediate state of the catalytic process when the product PGP has formed and after CMP has been released.
Fig. 2.
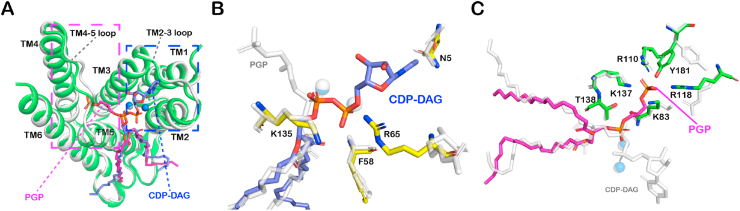
The PGP binding site in SaPgsA.
(A) Interactions between PGP and SaPgsA through salt bridges and hydrogen bonds. Six evolutionarily conserved residues are colored in green. Black dashed lines indicate the polar interactions between PGP head group and adjacent residues with bond lengths at 2.3–3.0 Å. The two blue spheres indicate two metal ions in the active. (B) The relative catalytic activity of six point mutants related to the PGP−binding site. The error bars indicate ± SEM with n = 3 or 4. The solid grey bars represent the activity data measured with WT and mutant enzymes solubilized in detergent, whereas the white bars represent the data measured with enzymes embedded in LCP. (C) Hydrophobic and van der Waals interactions between the acyl chains of PGP and SaPgsA. Relevant residues are colored in cyan. The cyan dashed lines indicate the interactions between PGP acyl chains and adjacent residues at distances of 3.2–3.9 Å. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. The CDP-DAG-binding site partially overlaps with that of PGP
To unravel the substrate binding sites of SaPgsA, we performed crystallization trials of the enzyme in the presence of both CDP-DAG and G3P at a molar ratio of 1:10:40 (SaPgsA: CDP-DAG: G3P). Although the amount of G3P added into the crystallization mixture is in excess relative to the protein and CDP-DAG molecule, no recognizable G3P densities were observed in the active site. Nevertheless, the 2Fo-Fc and omit electron density maps show clear densities for CDP-DAG (Supplementary Fig. 4C), and one CDP-DAG molecule is found in the active site of each monomer.
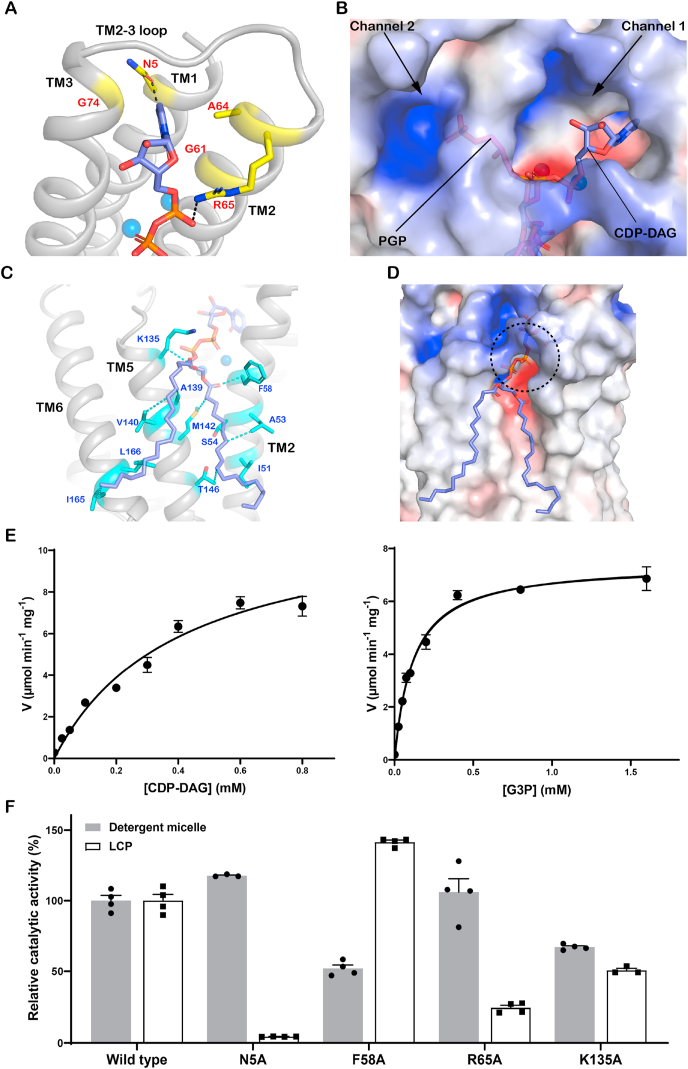
The CDP moiety of CDP-DAG interacts with Asn5 and Arg65 through hydrogen bonds and salt bridges, and the nucleotide ring wedges into a pocket delineated by TM1, TM2, TM3 and TM2-3 loop (Fig. 3A). Among the eight highly conserved amino acid residues (D57xxD60G61xxA64R65 … G74xxxD78xxxD82) in the active site of SaPgsA, Gly61, Ala64, Arg65 and Gly74 outline the pocket for accommodating the nucleotide moiety of CDP-DAG. The two glycine residues may provide local structural flexibility for the release of CMP. A wide opening surrounded by electropositive surface extends from the binding site of the CDP-DAG nucleotide ring to the cytosol, potentially serving as the exit portal for CMP (Fig. 3B, Channel 1). The acyl chains of CDP-DAG are located at the groove between TM2 and TM5, like those of PGP observed in the SaPgsA−PGP complex structure (Fig. 3C and D). The binding site may allow the enzyme to acquire CDP-DAG from membrane through lateral diffusion.
Fig. 3.
The binding site of CDP-DAG in SaPgsA.
(A) Interactions of the CDP moiety of CDP-DAG with SaPgsA. The black dashed lines indicate hydrogen bonds and salt bridges at bond lengths of 2.4 Å or 3.1 Å. (B) Putative CMP release channel (Channel 1) and G3P entrance channel (Channel 2). The PGP molecule from the SaPgsA−PGP complex structure is superposed onto the SaPgsA−CDP-DAG complex structure to indicate the putative G3P location. (C) Interaction of the fatty acyl chains of CDP-DAG with adjacent amino acid residues in SaPgsA. The relevant residues are highlighted in cyan. The cyan dashed lines indicate hydrophobic and van der Waals interactions at distance of 3.1–4.0 Å. (D) The putative entrance for CDP-DAG head group as indicated by the black dashed circle. (E) Substrate-dependent kinetic measurement of SaPgsA activity in detergent micelle. The data points are plotted as mean value ± standard error (SEM as indicated by the error bars. n=3 or 4). For those data points with small errors, the error bars are buried within the symbols. (F) The relative catalytic activity of four alanine mutants related to the CDP-DAG−binding site. The error bars indicate ± SEM with n = 3 or 4. The solid grey bars represent the activity data measured with WT and mutant enzymes solubilized in detergent, whereas the white bars represent the data measured with enzymes embedded in LCP. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The [CDP-DAG] and [G3P]-dependent kinetic assay results indicate SaPgsA has Km values of 0.40 mM for CDP-DAG and 0.12 mM for G3P (Fig. 3E, measured in detergent micelle). The enzyme exhibits little or no substrate-induced cooperativity as the Hill coefficient was close to 1 (0.90 and 1.04 for CDP-DAG and G3P respectively), suggesting the two protomers likely perform catalytic function independently. To be consistent with the crystallization environment, we also reconstituted SaPgsA into LCP and assessed its activity using a direct and continuous spectroscopic assay (Supplementary Fig. 5A) (Li and Caffrey, 2011). SaPgsA was active in the gel-like LCP media, supporting the view that SaPgsA was properly folded in the LCP bilayer and that the structures were of functional relevance (Supplementary Fig. 5). The [G3P]-dependent (Km of 0.37 mM) kinetics in LCP closely resemble those measured in detergent (Supplementary Fig. 5C). Likewise, the PGP-binding site mutants showed similar relative activities (vs wild type) in detergents or LCP (Fig. 2B). Curiously, the [CDP-DAG]-dependent kinetic profile differs from the one measured in detergent (Supplementary Fig. 5C). While the exact reason for this difference is unclear, the results are not unexpected considering the dramatic differences between the two types of local environments around the enzyme. Putatively, the activity difference may reflect slower substrate-diffusion in the viscous LCP or that the enzyme acquires and converts the lipid substrate at different rates in different environments.
For the four residues (Asn5, Phe58, Arg65 and Lys135) interacting directly with CDP-DAG, the corresponding alanine mutants behave very differently in detergents and LCP (Fig. 3F). In detergents, F58A and K135A exhibited reduced activity, while N5A and R65A showed activity close to the wild type. Considering that CDP-DAG can diffuse easily in the detergent-based environments, N5A and R65A may have no significant effect on the process of CDP-DAG access to the active-center. By contrast, N5A and R65A showed much lower activity than the wild type in LCP. In LCP, diffusion of CDP-DAG is restricted in two dimensions, presumably at a slower rate than in detergents. The enzyme may rely on Asn5 and Arg65 to attract and position CDP-DAG head group in the active site. In addition, the charge interaction between Lys135 and the α-phosphoryl moiety of CDP-DAG was also important, probably by steering the substrate to a proper position for catalysis. Phe58 is proximal to the fatty acyl chains of CDP-DAG and at the lateral portal for CDP-DAG entry (Fig. 3C). The F58A mutation may change the local structure, potentially influencing the access of CDP-DAG. As Phe58 is exposed on a hydrophobic surface, the activity of F58A mutant appears strongly dependent on the local environments, showing higher relative activity (vs the wild type) in LCP than in detergent (Fig. 3F).
2.4. A bimetal binding site is located at the active-site center
Divalent cations are essential for the activity of CDP-OH_P_transf enzymes (Nogly et al., 2014; Hirabayashi et al., 1976; Chang and Kennedy, 1967). Previous studies indicated PgsA activity is highly dependent on Mg2+ (Li and Caffrey, 2011; Hirabayashi et al., 1976; Carman and Wieczorek, 1980). As shown in Fig. 4A and Supplementary Fig. 5B, the activity of SaPgsA is highest in the condition with Mg2+, whereas it becomes less active in the presence of Zn2+, Cd2+, Co2+ or Mn2+ and inactive with Ba2+, Ca2+ or EDTA. Besides, SaPgsA activity is inhibited by increasing concentration of Zn2+ with the half maximal inhibitory concentration (IC50) at ∼1.8 μM (Fig. 4B). Notably, the SaPgsA crystals were grown in high concentrations (0.2 or 0.3 M) of Zn2+. The structures of SaPgsA contain two metal ions as suggested by the 2Fo-Fc electron density map, they were identified as zinc ions according to the anomalous difference Fourier density map (Supplementary Fig. 6). The two Zn2+-binding sites in SaPgsA structures are validated by the metal-ligand geometry and valence analysis result (Supplementary Fig. 7) (Zheng et al., 2017). A previous theoretical study indicated that Zn2+ can substitute Mg2+ directly in its octahedral binding site or readjust into a tetrahedral geometry during the exchange (Dudev and Lim, 2001). Moreover, Zn2+ and Mg2+ share similar electronegativity and ionic radius (Zn2+, 0.88 Å; Mg2+, 0.86 Å). Therefore, the binding mode of Mg2+ should closely resemble those of Zn2+ ions observed in the SaPgsA structures.
Fig. 4.
The bimetal center in SaPgsA.
(A) The dependence of PGP synthase activity of SaPgsA on different divalent metal ions. (B) Analysis on the concentration-dependent inhibitory effect of Zn2+ on SaPgsA enzymatic activity in detergent micelle. The error bars indicate ± SEM with n = 3 or 4. For those data points with small errors, the error bars are buried within the symbols. (C and D) Detailed view of the bimetal binding sites in the SaPgsA−CDP-DAG complex structure (C) and the SaPgsA−PGP complex structure (D). Asp57 and Asp60 on TM2 are depicted as cyan sticks, while Asp78 and Asp82 on TM3 are colored in green. CDP-DAG is shown as light blue stick, whereas PGP is shown as magenta stick. Black dashed lines indicate the coordination bonds at lengths of 2.0–2.3 Å. (E) The relative catalytic activity of metal-binding site mutants compared to the wild type. In (A) and (E), the error bars indicate ± SEM with n = 3 or 4. The solid grey bars represent the activity data measured with WT and mutant enzymes solubilized in detergent, whereas the white bars represent the data measured with enzymes embedded in LCP. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The SaPgsA structures contain two zinc ions (Zn1 and Zn2) in the active site center. Zn1 and Zn2 are located at the bottom surface of the membrane-embedded cavity, and close to the lateral portal (Fig. 1G). They are coordinated by Asp57, Asp60, Asp78 and Asp82 from a conserved sequence motif (D57xxD60G61xxA64R65 … G74xxxD78xxxD82) at bond lengths of 2.0–2.3 Å (Supplementary Fig. 6). Meanwhile, Zn1 and Zn2 are bridged by Asp57 with bidentate coordination constraining the inter-zinc distance to 3.5 Å or 3.7 Å. Such a bimetal center structure tends to increase the pKa of surrounding residues, thereby creating an acidic anion microenvironment to facilitate bond formation (Nogly et al., 2014). In detail, Zn1 is coordinated by Asp57, Asp78, Asp82 and the β-phosphoryl moiety of CDP-DAG, while Zn2 is coordinated by Asp57, Asp60, Asp78 and the α-phosphoryl moiety of CDP-DAG (Fig. 4C). Similarly, in the SaPgsA−PGP complex structure, a water molecule and the backbone 3-phosphoryl moiety of PGP replace the α-phosphoryl and β-phosphoryl moieties of CDP-DAG respectively (Fig. 4D). Evidently, Zn1 coordinates the β-phosphoryl moiety of CDP-DAG in a proper position for the nucleophilic attack launched by the 1-OH of G3P. On the other hand, Zn2 may be involved in the scission of the α-β phosphoric anhydride bond of CDP-DAG, an important step to secure the irreversibility of catalysis (Luevano-Martinez and Duncan, 2020). The four acidic residues (Asp57, Asp60, Asp78 and Asp82), well positioned for coordination of Zn2+ and Zn2+-mediated CDP-DAG binding, are indispensable for SaPgsA activity, as removal of the metal-coordinating carboxyl group of the acidic residues by alanine mutation abolished enzyme activity (Fig. 4E).
2.5. SaPgsA exhibits distinct local conformations at the two different states
Strikingly, the structure of SaPgsA−PGP complex differs from that of SaPgsA−CDP-DAG complex in terms of local conformations. While the common phosphatidyl moieties align relatively well, the CMP head group of CDP-DAG and the G3P head group of PGP point to nearly opposite directions (Fig. 5A). As a result, the side chain of Asn5 on TM1 in the CDP-DAG−bound state moves outward by ∼1 Å in respect to the corresponding residue in the PGP−bound state to avoid steric hindrance with the cytosine group of CDP-DAG. Meanwhile, the side chain of Arg65 moves toward the α-phosphoryl moiety of CDP-DAG due to electrostatic attraction (Fig. 5B). Likewise, the residues involved in binding the G3P head group of PGP adopt different rotameric conformations compared to those in the CDP-DAG−bound state (Fig. 5C). Specifically, Lys83, Arg110, Arg118, Lys137 and Tyr181 from one protomer move toward the 3′-phosphoryl moiety of PGP due to electrostatic attraction, while Arg110′ and Tyr181′ in the adjacent protomer move slightly away from it to avoid steric hindrance. The TM4-5 loop shifts toward TM6 by 1–3 Å so that the substrate-binding cavity opens wider in comparison with the CDP-DAG−bound state (Fig. 5A). On the intracellular surface of SaPgsA−CDP-DAG complex structure, two channels (Channel 1 and Channel 2) connect the active site with cytoplasm and are separated by Lys75 from TM3 and Ala130 from TM4-5 loop (Fig. 3B). While Channel 1 may serve as the portal for CMP release, Channel 2 likely guides the entry of G3P to the active site. In the PGP−bound state, the TM4-5 loop moves toward TM6, expanding Channel 1 at the cost of diminishing Channel 2. On the other hand, binding of CDP-DAG to the active site may induce conformational changes leading to the formation of Channel 2 to embrace G3P. Such reversible seesaw-like movement may facilitate the substrate entry and product release cycle during catalysis.
Fig. 5.
Structural dynamics of SaPgsA in response to CDP-DAG or PGP binding.
(A) Local conformational differences between the structures of SaPgsA−CDP-DAG complex (white) and the SaPgsA−PGP complex (green). The blue dashed box indicates conformational changes induced by CDP-DAG binding. The magenta dashed box indicates the conformational changes induced by the binding of PGP head group. PGP and CDP-DAG are presented as stick models in magenta and blue, respectively. (B) A zoom-in view of the conformation changes of amino acid residue side chains induced by CDP-DAG binding. The amino acid residues involved in binding the head group of CDP-DAG are presented as stick models in yellow (carbon), blue (nitrogen) and red (oxygen). For comparison, the reference structure (SaPgsA−PGP complex) superposed on the one in complex with CDP-DAG is shown in silver. (C) An expanded view of the side chain conformation changes induced by PGP binding. The amino acid residues involved in binding the head group of PGP are presented as stick models in green (carbon), blue (nitrogen) and red (oxygen). As a reference, the structure of SaPgsA−CDP-DAG superposed on the one in complex with PGP is shown in silver. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.6. A possible trade-off mechanism for PgsA-related daptomycin resistance
Due to misuse of antibiotics, development of antibiotic-resistance in S. aureus poses a global threat to human health (Lowy, 1998). Unfortunately, daptomycin treatment of MRSA infections can fail in more than 20% of total clinical cases due to the occurrence of daptomycin resistance (Stefani et al., 2015). Previous studies reported frequent occurrence of pgsA mutations in clinical and laboratory derived daptomycin-resistant S. aureus (Peleg et al., 2012; Hines et al., 2017; Casanova et al., 2017), Streptococcus oralis (Grave et al., 2019) and Staphylococcus capitis (Hagiya et al., 2018) strains. To further explore the mechanism of PgsA's involvement in daptomycin resistance of Gram-positive pathogen, we constructed eight SaPgsA single mutants which are directly from the daptomycin-resistant S. aureus or from Streptococcus oralis and Staphylococcus capitis strains (Tran et al., 2019; Hagiya et al., 2018).
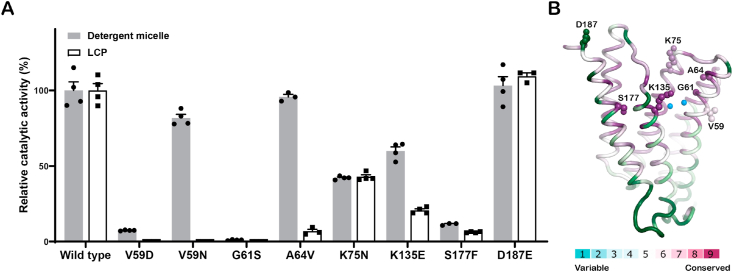
Among the eight SaPgsA mutants (V59D, V59N, G61S, A64V, K75N, K135E, S177F and D187E) tested, all except A64V and D187E exhibited reduced activity when compared to the wild type (Fig. 6A). As Asp187 is distant from the active site or the putative substrate entrance channels, the D187E mutation has little effect on the activity. In contrast, Val59, Gly61 and Ala64 are all located on TM2 nearby the characteristic and conserved helix kink essential for catalysis. In addition, Val59, Gly61 and Ala64 in the conserved “D57xxD60G61xxA64R65” motif are proximal to the bimetal catalytic center (Fig. 6B). Therefore, the V59D and G61S mutations result in dramatic or complete loss of activity. As for V59N and A64V, they exhibit activity close to the wild type in detergent micelle, but show much lower activity in LCP. The different amphipathic environments may have dramatic influence on the binding site of CDP-DAG. Lys75 is located at TM3 near the strictly conserved residues Gly74 and Asp78 as well as the bimetal catalytic center. K75N mutation resulted in ∼50% loss of activity (Fig. 6A). Presumably, the Asn75 residue introduced by mutation might form hydrogen bond with the hydroxyl groups of the ribose and distort/dislocate the substrate. Such mutation may also affect the release of PGP as it is located nearby the 3-phosphoryl moiety of PGP. Besides, Ser177 on TM6 is near the 3′-phosphoryl moiety of PGP. The S177F mutant also displayed dramatic reduction of activity. The mutation introduced a bulky side chain potentially causing severe clashes with PGP or G3P.
Fig. 6.
The daptomycin-resistant point mutants of SaPgsA.
(A) The relative enzymatic activity of the daptomycin-resistant point mutants. The error bars indicate ± SEM with n = 3 or 4. The solid grey bars represent the activity data measured with WT and mutant enzymes solubilized in detergent, whereas the white bars represent the data measured with enzymes embedded in LCP. (B) Structural mapping of the daptomycin-resistant mutations in SaPgsA. The residues are colored on the basis of the conservation score of each residue. The amino acid residues subject to mutation for activity assay are shown as sphere models. For clarity, only one protomer of the SaPgsA dimer is shown.
Collectively, the dysfunctional mutations of SaPgsA might result in a significant decrease of PG content in S. aureus membrane. A recent study indicates that PG is involved in forming a tripartite complex with Ca2+-daptomycin and undecaprenyl-coupled cell envelope precursors, so as to support the action of daptomycin on bacterial cells by targeting and interrupting cell wall biosynthesis (Grein et al., 2020). In case that PG content is lowered in the mutant strains, daptomycin might not be able to target cell wall biosynthesis, as it could not recruit enough PG to form the tripartite complex. As a result, the S. aureus strains carrying the pgsA mutations with null or low activity develop increased resistance (or reduced susceptibility) to daptomycin. Reduction of SaPgsA activity might effectively attenuate the bactericidal activity of daptomycin even though it would cause slower cell growth. Thus, S. aureus may trade-off PG biosynthesis with drug resistance to survive the antibiotic treatment while keeping anionic phospholipid content at minimal level.
3. Discussion
PG, CL and their derivatives have pivotal functions in various biological processes within prokaryotic and eukaryotic cells (Furse, 2017; Roy, 2009; Paradies et al., 2014), in addition to their fundamental roles as the building blocks of biological membranes. Back in 1960s, the activity of PgsA was discovered and the enzyme was partially purified from E. coli (Chang and Kennedy, 1967), and the bacterial pgsA gene was cloned in 1980s (Ohta et al., 1981). Further enzymatic kinetic study on E. coli PgsA suggested the enzyme adopts an ordered sequential Bi-Bi mechanism and CDP-DAG at high concentration may form a dead-end complex with the enzyme so as to inhibit the catalytic process (Hirabayashi et al., 1976). Notably, SaPgsA has recently been identified as a promising antibacterial target because of its vital role in the formation of bacterial cell membrane and cell wall (Lu et al., 2020). Based on the preliminary molecular docking result, we propose that the cajaninstilbene-acid analogue may specifically occupy the CDP-DAG binding site in SaPgsA and lead to inhibition of its activity (Supplementary Fig. 8) (Lu et al., 2020). Since CDP-DAG was previously found to inhibit PgsA activity by forming the so-called dead-end complex (Hirabayashi et al., 1976), the structures of SaPgsA with CDP-DAG bound may provide useful insights to inspire the design and development of PgsA inhibitors as antibiotic drugs. Moreover, as the products of enzymes often serve to inhibit the activity (Frieden and Walter, 1963), the structure of SaPgsA−PGP complex may offer a different angle for the design of inhibitors.
For a water-insoluble membrane-bound enzymes involved in lipid metabolism, a kinetic model, termed “surface dilution kinetics”, was developed to describe the action of lipid-dependent enzymes (Carman et al., 1995). The model considers both three-dimensional bulk interactions and two-dimensional surface interactions occurring between the enzymes and lipid substrates in the catalytic processes, so that both the bulk concentration and surface concentration of lipids are included in defining the kinetic parameters of lipid-dependent enzymes. As SaPgsA is also a lipid-dependent enzyme, its catalytic reaction should also follow the “surface dilution kinetics”, especially for the assay in detergent micelle. For the case in LCP, the three-dimensional bulk interactions between SaPgsA and lipid substrate should be minimal, as both the enzyme and lipid substrate are mostly confined in the lipidic phase so that they may not be able to diffuse freely in the aqueous solution. Therefore, two-dimensional surface interactions between enzyme and lipid substrate may dominate in the catalytic process of SaPgsA reconstituted in LCP. As the phospholipid substrate/product may change the physical properties of LCP such as bilayer thickness and curvature (Cherezov et al., 2002), the effect of such changes on SaPgsA activity need to be considered for more accurate analysis on the enzymatic kinetics in LCP. For future studies, detailed comparative analysis on the kinetics behaviors of SaPgsA in detergent micelle versus in LCP, by varying the bulk concentration of CDP-DAG with a constant surface concentration of CDP-DAG (mol fraction of CDP-DAG relative to CDP-DAG + Triton X-100/monoolein), will be necessary for assessing the influence of different media on the kinetic behaviors of SaPgsA.
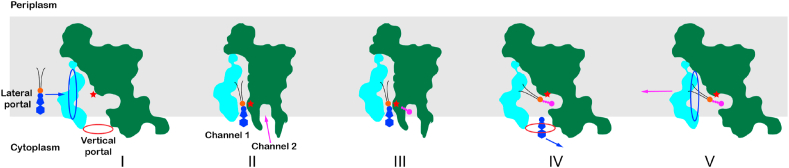
Based on the structural observation and functional analysis results, we propose a five-state working model and a putative nucleophilic substitution mechanism to account for the dynamic catalytic process mediated by SaPgsA (Fig. 7 and Supplementary Fig. 9). Firstly, CDP-DAG from the membrane enters the active site through a lateral portal. Similar lateral portal/entrance is also observed in many other membrane-bound enzymes or transporters utilizing lipids as substrates to catalyze reactions at the membrane-water interface (Dufrisne et al., 2017), such as CDP-DAG synthetase (Liu et al., 2014), Phospho-MurNAc-pentapeptide translocase MraY (Chung et al., 2013), Phosphatidylinositol-phosphate synthase (Clarke et al., 2015), G3P acyltransferase PlsY (Li et al., 2017), phosphatidylglycerophosphate phosphatase B (Fan et al., 2014), etc. Upon CDP-DAG binding, the SaPgsA protein may go through a conformational change around the membrane-perpendicular portal near the intracellular surface. The portal rearranges into two channels (Channel 1 and Channel 2) and the entry of G3P is guided by Channel 2, while Channel 1 is occupied by the head group of CDP-DAG. Secondly, the β-phosphorus of CDP-DAG (as an electrophile) and the 1-OH of G3P (as a nucleophile) are activated by the bimetal center and the overall acidic microenvironment of the active-site center. Zn1 serves to pull electrons from the β-phosphorus group towards the coordinating oxygen, making the β-phosphorus more electrophilic. When G3P is steered into the active site with its 1-OH in line with the di-phosphoryl of CDP-DAG, the nucleophilic attack launched by the 1-OH of G3P initiates a series of electron rearrangements and leads to the formation of new P-O bond between G3P and the β-phosphoryl of CDP-DAG (Supplementary Fig. 9). Thirdly, as the reaction proceeds, the products (PGP and CMP) form and CMP is released to cytosol through the expanded Channel 1. To further free up the active site, PGP diffuses outward to the bulk membrane through the same lateral portal used for CDP-DAG entry (Fig. 7).
Fig. 7.
A multi-state cartoon model accounting for the catalytic process mediated by SaPgsA.
The grey shading indicates the estimated membrane region. The red and blue elliptical rings indicate the vertical and lateral portals for the entry of G3P and CDP-DAG (or for the release of CMP and PGP) respectively. Among the five putative states (I-V), State II represents the one observed in the SaPgsA−CDP-DAG complex structure, whereas State V corresponds to the state of the SaPgsA−PGP complex structure. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Eukaryotic cardiolipin synthase catalyzes the conversion of CDP-DAG and PG to cardiolipin and CMP in mitochondria (Supplementary Fig. 10A) (Schlame, 2008). It is closely related to PgsA and they belong to the same family in the CDP-OH_P_transf superfamily (Supplementary Fig. 3A). Abnormalities in mitochondrial CL content are associated with several human diseases, such as Barth syndrome, myocardial ischemia/reperfusion injury, diabetes and Parkinson's disease (Paradies et al., 2019). Besides, knockdown of cardiolipin synthase induces mitochondrial elongation in human cells (Matsumura et al., 2018). While human cardiolipin synthase shares 29% sequence identity with SaPgsA, the amino acid residues around the CDP-DAG and PGP binding sites in SaPgsA are mostly conserved in human cardiolipin synthase (Supplementary Figs. 10B and C). As the structures of eukaryotic cardiolipin synthases are still unknown, the SaPgsA structure can potentially serve as a homologous model for future researches on mammalian cardiolipin synthases.
4. Materials and methods
4.1. Cloning, expression and purification
The gene encoding SaPgsA was synthesized (GenScript, China) with optimized codon usage for protein expression in E. coli. The target sequence was ligated into pET-15b vector between the NdeI and BamHI restriction enzyme sites. All point mutants of SaPgsA were introduced through QuickChange site-directed mutagenesis and verified by sequencing. Sequences of all the primers used in this study are listed in Supplementary Table 2. Plasmids were transformed into E. coli C41(DE3) or C43(DE3) competent cells, then overexpressed in Terrific Broth medium at 37 °C. Once the cell density reached to OD600 of 1.0, the inducer isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added with a final concentration of 0.5 mM. The induction of wild type and point mutant was continued for 2–3 h at 37 °C and 12–15 h at 16 °C, respectively. Cells were harvested by centrifugation at 9,000×g for 15 min at 4 °C and stored at −80 °C refrigerator.
Protein purification procedures were performed at 4 °C. 50 g frozen cell paste was resuspended in 400 ml lysis buffer (50 mM Tris-HCl, 200 mM NaCl, 20 mM imidazole, pH 8.0) and then homogenized by using the T10 basic homogenizer (IKA, Germany). Cells were broken by passed through a French press (ATS Engineering, Canada) at pressure of 900–1000 bar for 4-5 times. The cell lysate was centrifuged at 12,000×g for 20 min to remove cell debris. The supernatant was centrifuged further at 160,000×g for 1 h to yield the membrane pellet. The membrane fraction was resuspended and homogenized in 100 ml lysis buffer, then solubilized with 1.5% (w/v) of n-dodecyl-β-D-maltopyranoside (β-DDM, Anatrace, USA). After solubilization for 2–3 h, insoluble materials were removed by centrifugation at 41,000×g for 30 min. The supernatant was then incubated with 3 ml pre-equilibrated Ni-NTA resin (GE Healthcare, USA). After 2–3 h gentle agitation, the resin was packed to a gravity column and washed sequentially with 15 ml buffer A (25 mM Tris-HCl, 300 mM NaCl, 10% glycerin, 20 mM imidazole, 0.1% β-DDM, pH 7.5) and 15 ml buffer B (25 mM Tris-HCl, 300 mM NaCl, 10% glycerin, 70 mM imidazole, 0.05% β-DDM, pH 7.5). The protein was eluted with 12 ml buffer C (25 mM Tris-HCl, 300 mM NaCl, 10% glycerin, 300 mM imidazole, 0.05% β-DDM, pH 7.5), and immediately diluted by 2-folds volumes of buffer D (25 mM Tris-HCl, 300 mM NaCl, 10% glycerin, 0.05% β-DDM, pH 7.5). The protein sample was concentrated to 10–15 mg ml−1 (approximated by A280 absorbance) using a 50 kDa cut-off concentrator (Millipore, USA) and further purified by size exclusion chromatography (SEC) on a Superdex 200 increase 10/300 GL column (GE Healthcare, USA) in buffer E (10 mM Tris-HCl, 300 mM NaCl, 0.05% β-DDM, pH 7.5). Sharp monodisperse peak protein fractions of SEC were pooled and concentrated to 40–45 mg ml−1. And then, the protein samples were flash frozen in liquid nitrogen and stored at −80 °C refrigerator.
4.2. Crystallization
Crystals of SaPgsA were grown in lipidic cubic phase (LCP) at 4 °C or 20 °C (Caffrey and Cherezov, 2009). Initially, the protein solution was homogenized with 1.5-folds volumes molten 9.9 MAG (Molecular Dimensions, USA) using a gas-tight coupled syringe device (Hampton Research, USA) at room temperature (20–22 °C). Crystallization trials were set up by covering 40 nl of LCP bolus with 800 nl of precipitant solution onto a 96-well glass sandwich plate (FAstal BioTech, China) using a Gryphon robot (Art Robbins Instruments, USA). The glass sandwich plate was immediately sealed with a glass coverslip, then stored and imaged in Rock Imager 1000 (Formulatrix, USA) at 20 °C. Extensive crystallization trials yielded needle-like crystals, which were grown in 10–40% (v/v) 1,4-butanediol, 0.2 M or 0.3 M zinc acetate, and 0.1 M imidazole/HCl (pH 5.0–8.5). Plate-like crystals mostly appeared in 20–25 days and matured to 100 × 100 × 20 μm about 40–60 days at 4 °C. Meanwhile, we also obtained substrate-bound crystals in the crystallization condition containing CDP-DAG (Avanti, USA) and G3P (Sigma, USA). As for CDP-DAG, the amount of CDP-DAG was calculated on the basis of 10-folds of the protein mole number. The 9.9 MAG doped with CDP-DAG was prepared before it was used, and involved dissolving CDP-DAG in chloroform, adding it to molten 9.9 MAG in the appropriate amount, vortexing, and evaporating the chloroform with nitrogen gas for 3–4 min, then dried by a vacuum desiccator for 6–8 h. On the other hand, the protein solution was incubated with about 40-folds (molar ratio) of G3P at 4 °C for 1–2 h. The substrate-bound crystals appeared after 10 days and reached full size in 30–60 days at 20 °C. Desired wells were opened with a glass cutter (Hampton Research, USA) and the crystals were harvested using 50–100 μm Dual Thickness Microloops LD (MiTeGen, USA) at 4 °C or 20 °C, respectively. The crystals were immediately flash frozen in liquid nitrogen without additional cryoprotection.
4.3. Data collection and processing
X-ray diffraction data was collected at beamline BL18U1 of Shanghai Synchrotron Radiation Facility (SSRF, China) in the National Center for Protein Sciences Shanghai (NCPSS, China), using a PILATUS3 S 6M Detector (X-ray wavelength 0.9785 Å or 0.9793 Å). The near-Zn absorption K-edge anomalous data was collected at SPring-8 (Hyogo, Japan) beamline BL41XU, using an EIGER X 16M detector (X-ray wavelength 1.2820 Å). Diffraction images were indexed and integrated using XDS(Kabsch, 2010), scaled and merged with AIMLESS in the CCP4 software (Evans, 2006, 2011). The crystals are in the C2 space group, and diffracted X-ray to 2.5 Å (PGP−bound) and 3.0 Å (CDP-DAG−bound) resolution at synchrotron radiation source, respectively.
4.4. Structure determination and refinement
Initial phases were obtained through ab initio phasing and chain tracing method using ARCIMBOLDO-LITE program with 10 copies of a helix containing 18 residues as an initial estimated models (Caballero et al., 2018; Sammito et al., 2015). Structure refinement was carried out through alternating between cycles of refinement in PHENIX (Refine) and manual building in Coot (Afonine et al., 2012; Emsley et al., 2010). Lipid, solvent and water molecules were added manually in the model for refinement at later stages when their electron densities were well identified. The SaPgsA−PGP complex structure was refined to 2.5 Å resolution with Rwork/Rfree values of 0.213/0.250, respectively. The SaPgsA−CDP-DAG complex structure was solved by molecular replacement using the SaPgsA−PGP complex structure as search model, and refined to 3.0 Å with Rwork/Rfree values of 0.251/0.299. Data collection and refinement statistics are listed in Table S1. All structure figures were generated with PyMOL (Schrodinger, 2015). The orientation and position of SaPgsA protein in the membrane was calculated by using the PPM web server (Lomize et al., 2012). The conservation score of each residue in SaPgsA presented in Fig. 6B was calculated by the ConSurf web server (Landau et al., 2005). The omit maps for PGP (Supplementary Fig. 4A) and CDP-DAG (Supplementary Fig. 4C) were calculated using PHENIX (Polder) (Liebschner et al., 2017).
4.5. Oligomeric state analysis through SEC-MALS
To analyze the oligomeric state of SaPgsA in detergent micelle, the size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) method was applied. 50 μl SaPgsA sample was injected into the WTC-030S5 SEC column (Wyatt, USA) and eluted at 0.5 ml min−1 in buffer E. Data collection and analysis were performed with Astra 5 software (Wyatt, USA). The specific refractive index (dn dc−1) value of protein at 0.185 ml g−1 and that of β-DDM at 0.144 ml g−1 were used for data processing. The total molecular masses and individual masses of the proteins and the detergent were determined with Astra 5 software using protein conjugate analysis. The peak overlap and peak broadening corrections were carried out with Astra 5 software. Experiments were performed use protein sample with concentration at 1.13 mg ml−1, 4.54 mg ml−1 and 5.12 mg ml−1, respectively. All results showed that SaPgsA was at dimeric state in detergent micelle. For clarity, only 4.54 mg ml−1 sample was presented in Supplementary Fig. 1D.
4.6. Identification of PGP by mass spectrometry
To extract lipids, purified SaPgsA (5.82 mg ml−1, 200 μl) was mixed with 40 mg SM2 Biobeads (Bio-rad, USA) and gently shaking of the mixture was carried out overnight at 4 °C to remove detergent. The sample was centrifuged to remove detergent and Biobeads, then dried by a vacuum desiccator. A mixture of 60 μl chloroform, 120 μl methanol and 38 μl ultrapure water was added to the sample and then the tube was vortexed once every 2 min for 10 times. Then, a mixture of 60 μl chloroform and 60 μl of 2 M KCl was added and vortexed once every 2 min for 7 times. The mixture was centrifuged for 5 min at 18,800×g and the lower phase was separate out in a new tube. The selected lower phase sample was dried by a vacuum desiccator. Finally, add 25 μl chloroform and 25 μl methanol to the above tube, and the sample was ready for the following lipid mass spectrometry. The lipid mass spectrometry was performed according to the protocol described before (Cai et al., 2015). Since there is no PGP mass spectrometry data in the existing database and the standard PGP sample is not commercially available, we manually searched for putative PGP molecule signal from total lipidomic data by identifying characteristic fragment peaks corresponding to those of previous published data (Lu et al., 2011).
4.7. Activity assay with radioisotope labeled substrate
SaPgsA enzymatic activity was measured by the incorporation of [14C]-G3P into the chloroform-soluble PGP (Chang and Kennedy, 1967; Ohta et al., 1981). A 50 μl reaction mixture contains 1.6 mM [14C]-G3P (specific activity at 129.3 mCi mmol−1, PerkinElmer, USA), 0.8 mM CDP-DAG (Avanti, USA), 100 mM Tris-HCl (pH 8.0), 100 mM MgCl2, 20 mM Triton X-100 (Sigma, USA) and 100 nM enzyme. The reaction system was incubated at 27 °C for 15 min. To terminate the reaction, 180 μl chloroform/methanol/concentrated HCl (1:2:0.02, v/v/v) was added to the system, followed by addition of 60 μl of chloroform and 60 μl of 2 M KCl per tube. The tube was vortexed for 1 min and centrifuged for 15 min at 2,400×g to separate the organic phase from the lower aqueous phase. Then 100 μl lower organic phase was pipetted into a new tube and centrifuged at 2,400×g for 15 min. For the measurement of chloroform-soluble PGP radioactivity, 40 μl lower organic phase was mixed with 1 ml OptiPhase Supermix Cocktail scintillation liquid (PerkinElmer, USA) and vortexed for 2 min. The sample was subjected to liquid scintillation counting in the Wallac 1450 MicroBeta trilux liquid scintillation and luminescence counter (PerkinElmer, USA). Three or four parallel repeat experiments were performed for the measurement of each data point.
In order to analyze the influence of divalent metal ions on the activity of SaPgsA, 100 mM MgCl2 in the reaction buffer was replaced with 100 mM ZnCl2, BaCl2, CaCl2, CoCl2, MnCl2 or 10 mM EDTA, respectively. For the CDP-DAG- or G3P-dependent kinetic assays, 50 nM wild type SaPgsA enzyme was used and the concentration of CDP-DAG/G3P was held constant at 0.8 mM/1.6 mM, whereas the concentration of the other substrate was varied from 0.025 to 1.6 or 0.025–0.8 mM as shown. For all the activity assays in detergent, G3P (≥95%, Cat. 94124, Sigma, USA) of high purity was used. The data were analyzed and plotted using GraphPad Prism 8.0.
4.8. Activity assay in LCP
LCP was prepared using 9.9 MAG doped with CDP-DAG in different proportions as described above. Based on the previous protocol (Li and Caffrey, 2011), 2 μl sticky LCP was dispensed onto the sidewall of a UV-transparent 96-well plate (Cat. 8404, Thermo Scientific, USA). 200 μl pre-warmed buffer F (50 mM Tris-HCl, 0.1 mM EDTA, G3P and divalent metal ion at various concentration, pH 8.0) was loaded to each well. The reaction was immediately monitored in a plate reader with 271 nm absorbance at 30 °C. Using CMP standards prepared as stock solutions in 0.1 N HCl (Ploeser and Loring, 1949), the molar extinction coefficient of CMP in the assay buffer was calibrated as 9,175 M−1 cm−1. For CDP-DAG Km determination: 50 μg ml−1 of SaPgsA, 0–3.5 mol% CDP-DAG, 1 mM G3P (Cat. 94124, Sigma, USA) and 0.1 M MgCl2 were used. For G3P Km determination: 50 μg ml−1 of SaPgsA, 2 mol% CDP-DAG, 0–10 mM G3P and 0.1 M MgCl2 were used. For the activity assays of single point mutants: 100 μg ml−1 of enzyme, 0.5 mol% CDP-DAG, 15 mM glycerol phosphate isomeric mixture (Cat. G6501, Sigma, USA) and 0.1 M MgCl2 were used. For effect of divalent metal ions on SaPgsA activity: 50 μg ml−1 SaPgsA, 0.5 mol% CDP-DAG, 15 mM glycerol phosphate isomeric mixture and various metals used as follows (MgCl2, 0.1 M; ZnCl2, 0.63 mM; MnCl2, 0.4 mM; CoCl2, 12.5 mM; CdCl2, 2.5 mM). The glycerol phosphate isomeric mixture contains the β-isomer (G2P) and the racemic α-isomers (G1P and G3P). While High purity G3P (≥95%) was used for the assays of [CDP-DAG] and [G3P] kinetics in LCP, the glycerol phosphate isomeric mixture at high concentrations (15 mM) was used for other activity assay in LCP to reduce cost, as we found that G1P and G2P did not inhibit SaPgsA activity even at 10-fold molar excess of G3P. For measuring Km values, three independent experiments were carried out, each with three such technical replicates. Other assays were performed with three or four technical replicates where the same batch of LCP was dispensed into three or four wells. The data were analyzed and plotted using GraphPad Prism 8.0.
The concentration for the soluble substrate G3P is reported as molarity, whereas that for CDP-DAG is reported as mole fractions (in proportion to the LCP host lipid) owing to the uncommon assay setup. Specifically, CDP-DAG is, macroscopically speaking, confined in the gel-like LCP media despite that there is a vast excess of soaking solution in the assay. The volume of the entire assay system used for calculating molar concentrations is irrelevant to the effective concentration of CDP-DAG in the LCP bilayer. Therefore, the CDP-DAG concentration for the activity assay in LCP was presented as mole fractions.
Accession numbers
The atomic models and structure factors have been deposited with the Protein Data Bank under the accession number 7DRJ and 7DRK. Enzymatic activity assay data presented in Figs. 2B, 3E, 3F, 4A, 4B, 4E, 6A, S5B and S5C are accessible through https://doi.org/10.17632/3d6w948vf5.1 webpage.
Author contributions
B.Y. cloned, expressed and purified the wild type and mutants of SaPgsA, crystallized SaPgsA, collected and processed X-ray diffraction data, solved and refined the structures, and carried out activity assay with radioisotope-labeled substrate. H.Y. performed SaPgsA activity assay in LCP under D.L.'s supervision. B.Y., D.L and Z.L. analyzed the structures and catalytic mechanism. B.Y. and Z.L. wrote the initial draft and all authors participated in revising the manuscript. Z.L conceived and supervised the project.
CRediT authorship contribution statement
Bowei Yang: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization. Hebang Yao: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing. Dianfan Li: Methodology, Validation, Writing – review & editing, Supervision, Funding acquisition. Zhenfeng Liu: Conceptualization, Methodology, Validation, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Isabel Usón at the Institute of Molecular Biology of Barcelona for the guidance in using the ARCIMBOLDO-LITE program and helpful discussion; the staff members at BL18U1 of Shanghai Synchrotron Radiation Facility (SSRF) and BL41XU of SPring-8 for their assistance during X-ray data collection; Y. Wang, H. J. Zhang, X. X. Yu and Z. S. Xie at the Protein Science Research Facility of the Institute of Biophysics (IBP) for their help in LCP crystallization, activity assay, static light-scattering experiments and mass spectrometry analysis respectively; Y. Yin for her efforts in protein expression and purification; X. B. Liang for technical assistance on biochemistry, crystal handling and X-ray diffraction data collection. The project is financially supported by the National Natural Science Foundation of China (31925024 and 31670749, Z.L.; 31870726, D.L.), the Strategic Priority Research Program of CAS (XDB37020101, Z.L.; XDB37020204, D.L.), the Basic Frontier Science Research Program of CAS (ZDBS-LY-SM003) and CAS Facility-based Open Research Program.
Handling editor: Natalie Strynadka.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.11.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova E.L., Domonkos I., Dobrikova A.G., Sallai A., Bogos B., Wada H., et al. Effect of phosphatidylglycerol depletion on the surface electric properties and the fluorescence emission of thylakoid membranes. J. Photochem. Photobiol., B. 2008;91:51–57. doi: 10.1016/j.jphotobiol.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Belcher Dufrisne M., Jorge C.D., Timoteo C.G., Petrou V.I., Ashraf K.U., Banerjee S., et al. Structural and functional characterization of phosphatidylinositol-phosphate biosynthesis in mycobacteria. J. Mol. Biol. 2020;432:5137–5151. doi: 10.1016/j.jmb.2020.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R.M., Ballas L.M., Coleman R.A. Lipid topogenesis. J. Lipid Res. 1981;22:391–403. [PubMed] [Google Scholar]
- Blunsom N.J., Cockcroft S. CDP-diacylglycerol synthases (CDS): gateway to phosphatidylinositol and cardiolipin synthesis. Front Cell Dev Biol. 2020;8:63. doi: 10.3389/fcell.2020.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero I., Sammito M., Millan C., Lebedev A., Soler N., Uson I. ARCIMBOLDO on coiled coils. Acta Crystallogr D Struct Biol. 2018;74:194–204. doi: 10.1107/S2059798317017582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr F Struct Biol Commun. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M., Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T., Shu Q., Hou J., Liu P., Niu L., Guo X., et al. Profiling and relative quantitation of phosphoinositides by multiple precursor ion scanning based on phosphate methylation and isotopic labeling. Anal. Chem. 2015;87:513–521. doi: 10.1021/ac503224j. [DOI] [PubMed] [Google Scholar]
- Carman G.M., Wieczorek D.S. Phosphatidylglycerophosphate synthese and phosphatidylserine synthase activites in Clostridium perfringens. J. Bacteriol. 1980;142:262–267. doi: 10.1128/jb.142.1.262-267.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G.M., Deems R.A., Dennis E.A. Lipid signaling enzymes and surface dilution kinetics (∗) J. Biol. Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- Casanova N.G., Ruiz M.S., Bellido J.L.M. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev. Española Quimioter. 2017;30:391–396. [PubMed] [Google Scholar]
- Chang Y.Y., Kennedy E.P. Biosynthesis of phosphatidyl glycerophosphate in Escherichia coli. J. Lipid Res. 1967;8:447–455. [PubMed] [Google Scholar]
- Chauhan N., Farine L., Pandey K., Menon A.K., Butikofer P. Lipid topogenesis--35years on. Biochim. Biophys. Acta. 2016;1861:757–766. doi: 10.1016/j.bbalip.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V., Clogston J., Misquitta Y., Abdel-Gawad W., Caffrey M. Membrane protein crystallization in meso: lipid type-tailoring of the cubic phase. Biophys. J. 2002;83:3393–3407. doi: 10.1016/S0006-3495(02)75339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.C., Zhao J., Gillespie R.A., Kwon D.-Y., Guan Z., Hong J., et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke O.B., Tomasek D., Jorge C.D., Dufrisne M.B., Kim M., Banerjee S., et al. Structural basis for phosphatidylinositol-phosphate biosynthesis. Nat. Commun. 2015;6:8505. doi: 10.1038/ncomms9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z.D., Matamouros S., Whittington D., Bishop R.E., Miller S.I. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Dudev T., Lim C. Metal selectivity in metalloproteins: Zn2+ vs Mg2+ J. Phys. Chem. B. 2001;105:4446–4452. [Google Scholar]
- Dufrisne M.B., Petrou V.I., Clarke O.B., Mancia F. Structural basis for catalysis at the membrane-water interface. Biochim. Biophys. Acta. 2017;1862:1368–1385. doi: 10.1016/j.bbalip.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C.M., Peschel A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 2011;80:290–299. doi: 10.1111/j.1365-2958.2011.07576.x. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Jiang D., Zhao Y., Liu J., Zhang X.C. Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7636–7640. doi: 10.1073/pnas.1403097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden E., Walter C. Prevalence and significance of the product inhibition of enzymes. Nature. 1963;198:834–837. doi: 10.1038/198834a0. [DOI] [PubMed] [Google Scholar]
- Furse S. Is phosphatidylglycerol essential for terrestrial life? J. Chem. Biol. 2017;10:1–9. doi: 10.1007/s12154-016-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor P.M., Hubbell S., Schmidt A.J., Lina R.A., Minskoff S.A., Greenberg M.L. Regulation of phosphatidylglycerolphosphate synthase in Saccharomyces cerevisiae by factors affecting mitochondrial development. J. Bacteriol. 1991;173:6124–6131. doi: 10.1128/jb.173.19.6124-6131.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldner N.K., Bulow C., Cho K., Wallace M., Hsu F.F., Patti G.J., et al. Mechanism of high-level daptomycin resistance in Corynebacterium striatum. mSphere. 2018;3 doi: 10.1128/mSphereDirect.00371-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grave K., Bennett M.D., Hogbom M. Structure of Mycobacterium tuberculosis phosphatidylinositol phosphate synthase reveals mechanism of substrate binding and metal catalysis. Commun. Biol. 2019;2:175. doi: 10.1038/s42003-019-0427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein F., Muller A., Scherer K.M., Liu X., Ludwig K.C., Klockner A., et al. Ca(2+)-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 2020;11:1455. doi: 10.1038/s41467-020-15257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann A.B., Angert E.R., Helmann J.D. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 2009;53:1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiya H., Sugawara Y., Kimura K., Hamaguchi S., Nishi I., Hayashi M., et al. Emergence of daptomycin non-susceptible coagulase-negative Staphylococci in patients with cardiovascular device infections: two cases report investigated by whole genome analysis. Medicine (Baltim.) 2018;97 doi: 10.1097/MD.0000000000013487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines K.M., Waalkes A., Penewit K., Holmes E.A., Salipante S.J., Werth B.J., et al. Characterization of the mechanisms of daptomycin resistance among gram-positive bacterial pathogens by multidimensional lipidomics. mSphere. 2017;2 doi: 10.1128/mSphere.00492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi T., Larson T.J., Dowhan W. Membrane-associated phosphatidylglycerophosphate synthetase from Escherichia coli: purification by substrate affinity chromatography on cytidine 5'-diphospho-1,2-diacyl-sn-glycerol sepharose. Biochemistry. 1976;15:5205–5211. doi: 10.1021/bi00669a002. [DOI] [PubMed] [Google Scholar]
- Jennings W., Epand R.M. CDP-diacylglycerol, a critical intermediate in lipid metabolism. Chem. Phys. Lipids. 2020;230:104914. doi: 10.1016/j.chemphyslip.2020.104914. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick P., Raja A., LaBonte J., Lebbos J. Daptomycin. Nat Rev Drug Discov. 2003;2:943–944. doi: 10.1038/nrd1258. [DOI] [PubMed] [Google Scholar]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Caffrey M. Lipid cubic phase as a membrane mimetic for integral membrane protein enzymes. P Natl. Acad Sci. USA. 2011;108:8639–8644. doi: 10.1073/pnas.1101815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tang Y., Wu Y., Zhao S., Bao J., Luo Y., et al. Structural insights into the committed step of bacterial phospholipid biosynthesis. Nat. Commun. 2017;8:1691. doi: 10.1038/s41467-017-01821-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr D Struct Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yin Y., Wu J., Liu Z. Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis. Nat. Commun. 2014;5:4244. doi: 10.1038/ncomms5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard J., López-García P., Moreira D. The early evolution of lipid membranes and the three domains of life. Nat. Rev. Microbiol. 2012;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- Lomize M.A., Pogozheva I.D., Joo H., Mosberg H.I., Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Lu Y.H., Guan Z., Zhao J., Raetz C.R. Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 2011;286:5506–5518. doi: 10.1074/jbc.M110.199265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Hou W., Xu X.F., Chen Q., Li Z., Lin J., et al. Biological evaluation and chemoproteomics reveal potential antibacterial targets of a cajaninstilbene-acid analogue. Eur. J. Med. Chem. 2020;188:112026. doi: 10.1016/j.ejmech.2019.112026. [DOI] [PubMed] [Google Scholar]
- Luevano-Martinez L.A., Duncan A.L. Origin and diversification of the cardiolipin biosynthetic pathway in the Eukarya domain. Biochem. Soc. Trans. 2020;48:1035–1046. doi: 10.1042/BST20190967. [DOI] [PubMed] [Google Scholar]
- Masoro E.J. Lipids and lipid metabolism. Annu. Rev. Physiol. 1977;39:301–321. doi: 10.1146/annurev.ph.39.030177.001505. [DOI] [PubMed] [Google Scholar]
- Matsumura A., Higuchi J., Watanabe Y., Kato M., Aoki K., Akabane S., et al. Inactivation of cardiolipin synthase triggers changes in mitochondrial morphology. FEBS Lett. 2018;592:209–218. doi: 10.1002/1873-3468.12948. [DOI] [PubMed] [Google Scholar]
- Miyazaki C., Kuroda M., Ohta A., Shibuya I. Genetic manipulation of membrane phospholipid composition in Escherichia coli: pgsA mutants defective in phosphatidylglycerol synthesis. P Natl. Acad Sci. USA. 1985;82:7530–7534. doi: 10.1073/pnas.82.22.7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogly P., Gushchin I., Remeeva A., Esteves A.M., Borges N., Ma P., et al. X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism. Nat. Commun. 2014;5:4169. doi: 10.1038/ncomms5169. [DOI] [PubMed] [Google Scholar]
- Numata M., Chu H.W., Dakhama A., Voelker D.R. Pulmonary surfactant phosphatidylglycerol inhibits respiratory syncytial virus–induced inflammation and infection. Proc. Natl. Acad. Sci. U.S.A. 2010;107:320–325. doi: 10.1073/pnas.0909361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata M., Kandasamy P., Nagashima Y., Posey J., Hartshorn K., Woodland D., et al. Phosphatidylglycerol suppresses influenza A virus infection. Am. J. Respir. Cell Mol. Biol. 2012;46:479–487. doi: 10.1165/rcmb.2011-0194OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta A., Waggoner K., Radominska-Pyrek A., Dowhan W. Cloning of genes involved in membrane lipid synthesis: effects of amplification of phosphatidylglycerophosphate synthase in Escherichia coli. J. Bacteriol. 1981;147:552–562. doi: 10.1128/jb.147.2.552-562.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies G., Paradies V., De Benedictis V., Ruggiero F.M., Petrosillo G. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta. 2014;1837:408–417. doi: 10.1016/j.bbabio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Paradies G., Paradies V., Ruggiero F.M., Petrosillo G. Role of cardiolipin in mitochondrial function and dynamics in health and disease: molecular and pharmacological aspects. Cells. 2019;8 doi: 10.3390/cells8070728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A.Y., Miyakis S., Ward D.V., Earl A.M., Rubio A., Cameron D.R., et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploeser J.M., Loring H.S. The ultraviolet absorption spectra of the pyrimidine ribonucleosides and ribonucleotides. J. Biol. Chem. 1949;178:431–437. [PubMed] [Google Scholar]
- Raetz C.R. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol. Rev. 1978;42:614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C.R., Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 1990;265:1235–1238. [PubMed] [Google Scholar]
- Roy H. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life. 2009;61:940–953. doi: 10.1002/iub.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini-Chohan H.K., Holmes M.G., Chicco A.J., Taylor W.A., Moore R.L., McCune S.A., et al. Cardiolipin biosynthesis and remodeling enzymes are altered during development of heart failure. J. Lipid Res. 2009;50:1600–1608. doi: 10.1194/jlr.M800561-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammito M., Millan C., Frieske D., Rodriguez-Freire E., Borges R.J., Uson I. ARCIMBOLDO_LITE: single-workstation implementation and use. Acta Crystallogr D Biol Crystallogr. 2015;71:1921–1930. doi: 10.1107/S1399004715010846. [DOI] [PubMed] [Google Scholar]
- Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L.L.C. 2015. The PyMOL Molecular Graphics System. Version 1.8. [Google Scholar]
- Sciara G., Clarke O.B., Tomasek D., Kloss B., Tabuso S., Byfield R., et al. Structural basis for catalysis in a CDP-alcohol phosphotransferase. Nat. Commun. 2014;5:4068. doi: 10.1038/ncomms5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani S., Campanile F., Santagati M., Mezzatesta M.L., Cafiso V., Pacini G. Insights and clinical perspectives of daptomycin resistance in Staphylococcus aureus: a review of the available evidence. Int. J. Antimicrob. Agents. 2015;46:278–289. doi: 10.1016/j.ijantimicag.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Tran T.T., Mishra N.N., Seepersaud R., Diaz L., Rios R., Dinh A.Q., et al. Mutations in cdsA and pgsA Correlate with Daptomycin Resistance in Streptococcus mitis and S. oralis. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.01531-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuizen R., Nag K., Orgeig S., Possmayer F. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 1998;1408:90–108. doi: 10.1016/s0925-4439(98)00061-1. [DOI] [PubMed] [Google Scholar]
- Wada H., Murata N. The essential role of phosphatidylglycerol in photosynthesis. Photosynth. Res. 2007;92:205–215. doi: 10.1007/s11120-007-9203-z. [DOI] [PubMed] [Google Scholar]
- Zhang Y.M., Rock C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- Zheng H., Cooper D.R., Porebski P.J., Shabalin I.G., Handing K.B., Minor W. CheckMyMetal: a macromolecular metal-binding validation tool. Acta Crystallogr D Struct Biol. 2017;73:223–233. doi: 10.1107/S2059798317001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.