Abstract
Objectives
To develop and validate a clinical risk score that can accurately quantify the probability of SARS-CoV-2 infection in patients presenting to an emergency department without the need for laboratory testing.
Design
Cohort study of participants in the Canadian COVID-19 Emergency Department Rapid Response Network (CCEDRRN) registry. Regression models were fitted to predict a positive SARS-CoV-2 test result using clinical and demographic predictors, as well as an indicator of local SARS-CoV-2 incidence.
Setting
32 emergency departments in eight Canadian provinces.
Participants
27 665 consecutively enrolled patients who were tested for SARS-CoV-2 in participating emergency departments between 1 March and 30 October 2020.
Main outcome measures
Positive SARS-CoV-2 nucleic acid test result within 14 days of an index emergency department encounter for suspected COVID-19 disease.
Results
We derived a 10-item CCEDRRN COVID-19 Infection Score using data from 21 743 patients. This score included variables from history and physical examination and an indicator of local disease incidence. The score had a c-statistic of 0.838 with excellent calibration. We externally validated the rule in 5295 patients. The score maintained excellent discrimination and calibration and had superior performance compared with another previously published risk score. Score cut-offs were identified that can rule-in or rule-out SARS-CoV-2 infection without the need for nucleic acid testing with 97.4% sensitivity (95% CI 96.4 to 98.3) and 95.9% specificity (95% CI 95.5 to 96.0).
Conclusions
The CCEDRRN COVID-19 Infection Score uses clinical characteristics and publicly available indicators of disease incidence to quantify a patient’s probability of SARS-CoV-2 infection. The score can identify patients at sufficiently high risk of SARS-CoV-2 infection to warrant isolation and empirical therapy prior to test confirmation while also identifying patients at sufficiently low risk of infection that they may not need testing.
Trial registration number
Keywords: COVID-19, accident & emergency medicine, epidemiology
Strengths and limitations of this study.
Patients were enrolled in a large, geographically distributed network of Canadian urban, regional and rural emergency departments, with strict data quality and cleaning protocols to ensure reliability of collected data.
In addition to clinical variables, we also included the average daily incidence of SARS-CoV-2 infections in a patient’s health region, which is an essential predictor of the probability of a patient’s risk of COVID-19 infection.
Some missing data required either multiple imputation or classification of missing categorical variables as being absent, but the overall missingness of data in this registry is very low.
Although the data collection for the Canadian COVID-19 Emergency Department Rapid Response Network registry relies on abstraction from health records, this approach has been shown to be reliable in our study sites when compared with prospective data collection.
This risk score was developed using data from patients enrolled in the first 9 months of the pandemic when rates of influenza were low, so the score may need to be revalidated and refined in the future to reflect the influence of influenza, the emergence of variant strains of SARS-CoV-2 and widespread population immunisation on patients’ risk of infection.
Introduction
To date, the WHO has reported 190 million diagnosed cases of COVID-19 with 4.2 million fatalities.1 Despite the availability of vaccines to prevent COVID-19, incomplete population-level immunisation and the emergence of variants of concern means that hospitals around the world need to continue to identify and isolate patients with suspected COVID-19 from the time they arrive in the emergency department until their SARS-CoV-2 test results are available. In acutely ill patients, clinicians may need to initiate empirical therapy immediately. A quantitative risk score that can accurately predict the probability of a positive SARS-CoV-2 test result would guide initial isolation and empirical therapy prior to nucleic acid amplification test (NAAT) test result availability while identifying patients with sufficiently low probability of COVID-19 who may not require testing or isolation.
Many risk prediction tools have been developed to predict the probability of SARS-CoV-2 infection.2–14 A living systematic review of these models concluded that most were generated using poor methodological approaches and none were ready for widespread use.2 Most published risk prediction tools, including one identified as promising by the living systematic review, included early laboratory or imaging findings, thus precluding their utility to guide immediate isolation and clinical decisions at the time of first clinical contact. Other risk prediction tools using machine learning included laboratory and imaging results and can only be implemented in hospitals using electronic health records with integrated decision support. None of these models accounted for the prevalence of COVID-19 disease in the local population, which is an important risk predictor, and most only included patients from the early stages of the pandemic.2
The objective of this study is to develop a clinical risk score to predict the probability of a positive SARS-CoV-2 nucleic acid test in a large, generalisable population of patients presenting to emergency departments using only clinical characteristics and indicators of local SARS-CoV-2 incidence. This risk score is intended to guide SARS-CoV-2 testing, isolation and empirical therapy decisions without relying on other laboratory testing or diagnostic imaging. This score could be invaluable in settings that may not have access to adequate resources for timely SARS-CoV-2 testing.
Methods
This analysis uses data from the Canadian COVID-19 Emergency Department Rapid Response Network (CCEDRRN; pronounced ‘SED-rin’). CCEDRRN is an ongoing multicentre, pan-Canadian registry that has been enrolling consecutive patients presenting to emergency departments with suspected COVID-19 disease in hospitals in 8 of 10 Canadian provinces since 1 March 2020.15 Information on the network, including detailed methods and participating sites, is available elsewhere.15 Sites and enrolment periods are shown in the, online supplemental appendix, table 1. Additional information on network sites is available in the online supplemental network appendix. This study follows the methodological and reporting recommendations outlined in the transparency in reporting of a multivariable prediction model for individual diagnosis and prognosis criteria.16 The CCEDRRN data collection form includes prespecified demographic and social variables, vital signs, symptoms and comorbid conditions (derived from the International Severe Acute Respiratory and Emerging Infection Consortium reporting form),17 18 exposure risk variables, hospital laboratory and diagnostic imaging test results, SARS-CoV-2 NAAT results and patient outcomes. Data were abstracted at each site using electronic medical record extraction where available as well as manual review of either electronic or paper charts (depending on site-specific documentation practices) by trained research assistants who were blinded to the potential predictor variables at the time of data collection. Reliability of health record data abstraction was evaluated by comparison with prospective data collection in a sample of patients and found to be reliable.15
Table 1.
Characteristics and selected outcomes of enrolled patients
| Derivation (n=21 743) | Validation (n=5922) | |
| Age in years, median (IQR) | 57 (38–73) | 56 (37–73) |
| Female (%) | 10 992 (50.5) | 3085 (52.1) |
| Arrival from, n (%) | ||
| Home | 19 879 (91.4) | 5429 (91.7) |
| Long-term care/rehabilitation facility/corrections facility | 1000 (4.6) | 262 (4.4) |
| No fixed address/shelter/single room occupancy | 574 (2.6) | 201 (3.4) |
| Interhospital transfer | 290 (1.3) | 30 (0.5) |
| Risk for infection, n (%) | ||
| Healthcare worker | 505 (2.3) | 567 (9.6) |
| Household/caregiver contact | 566 (2.6) | 161 (2.7) |
| Institutional exposure (eg, LTC, prison) | 1354 (6.2) | 213 (3.6) |
| Microbiology lab | 4 (0.0) | 8 (0.1) |
| Travel | 924 (4.2) | 344 (5.8) |
| Other | 1320 (6.1) | 449 (7.6) |
| Unknown | 5415 (24.9) | 1856 (31.3) |
| No documented risk for infection | 10 028 (46.1) | 1075 (18.1) |
| Arrival vital signs, median (IQR) | ||
| Body temperature | 36.7 (36.3–37.1) | 36.8 (36.5–37.1) |
| Heart rate | 91 (79–107) | 90 (78–105) |
| Oxygen saturation | 97 (95–98) | 97 (95–99) |
| Respiratory rate | 18 (18–20) | 18 (16–20) |
| Systolic blood pressure | 133 (118–150) | 136 (120–149) |
| Common comorbid conditions, n (%) | ||
| Active malignant neoplasm (cancer) | 1678 (7.7) | 333 (5.6) |
| Asthma | 1699 (7.8) | 468 (7.9) |
| Atrial fibrillation | 1598 (7.3) | 402 (6.8) |
| Chronic kidney disease | 1214 (5.6) | 321 (5.4) |
| Chronic lung disease (not asthma/pulmonary fibrosis) | 1729 (8) | 583 (9.8) |
| Chronic neurological disorder (not dementia; eg, stroke/TIA, seizure disorder) | 1310 (6) | 400 (6.8) |
| Congestive heart failure | 1450 (6.7) | 368 (6.2) |
| Coronary artery disease | 1591 (7.3) | 449 (7.6) |
| Dementia | 734 (3.4) | 188 (3.2) |
| Diabetes | 2583 (11.9) | 916 (15.5) |
| Dialysis | 198 (0.9) | 28 (0.5) |
| Dyslipidaemia | 2375 (10.9) | 543 (9.2) |
| Hypertension | 6320 (29.1) | 1697 (28.6) |
| Hypothyroidism | 1344 (6.2) | 397 (6.7) |
| Mild liver disease | 280 (1.3) | 90 (1.5) |
| Moderate/severe liver disease | 245 (1.1) | 88 (1.5) |
| Obesity (clinical impression) | 284 (1.3) | 108 (1.8) |
| Organ transplant | 128 (0.6) | 19 (0.3) |
| Rheumatological disorder | 1122 (5.2) | 258 (4.4) |
| Other | 10 075 (46.3) | 2174 (36.7) |
| Past malignant neoplasm (cancer) | 936 (4.3) | 256 (4.3) |
| Psychiatric condition/mental health diagnosis | 2967 (13.6) | 831 (14) |
| Pulmonary fibrosis | 80 (0.4) | 26 (0.4) |
| Symptoms reported, n (%) | ||
| Abdominal pain | 2725 (12.5) | 540 (9.1) |
| Altered consciousness/confusion | 1456 (6.7) | 322 (5.4) |
| Bleeding (haemorrhage) | 330 (1.5) | 22 (0.4) |
| Chest pain (includes discomfort or tightness) | 4242 (19.5) | 974 (16.4) |
| Chills | 2045 (9.4) | 594 (10) |
| Conjunctivitis | 49 (0.2) | 26 (0.4) |
| Cough | 7724 (35.5) | 2663 (44.9) |
| Diarrhoea | 2140 (9.8) | 526 (8.9) |
| Dizziness/vertigo | 1521 (7) | 300 (5.1) |
| Dysgeusia/anosmia | 140 (0.6) | 33 (0.6) |
| Ear pain | 144 (0.7) | 30 (0.5) |
| Fatigue/malaise | 3361 (15.5) | 924 (15.6) |
| Fever | 5055 (23.2) | 1580 (26.7) |
| Headache | 2144 (9.9) | 624 (10.5) |
| Hemoptysis (bloody sputum) | 298 (1.4) | 66 (1.1) |
| Joint pain (arthralgia) | 296 (1.4) | 82 (1.4) |
| Lower chest wall indrawing | 10 (0) | 7 (0.1) |
| Lymphadenopathy | 67 (0.3) | 21 (0.4) |
| Muscle aches (myalgia) | 1575 (7.2) | 517 (8.7) |
| Nausea/vomiting | 4219 (19.4) | 935 (15.8) |
| No recorded symptoms | 2113 (9.7) | 431 (7.3) |
| Runny nose (rhinorrhoea) | 1061 (4.9) | 501 (8.5) |
| Seizures | 205 (0.9) | 42 (0.7) |
| Shortness of breath (dyspnoea) | 8537 (39.3) | 2383 (40.2) |
| Skin rash | 241 (1.1) | 38 (0.6) |
| Skin ulcers | 27 (0.1) | <5 |
| Sore throat | 3024 (13.9) | 985 (16.6) |
| Sputum production | 1507 (6.9) | 401 (6.8) |
| Wheezing | 582 (2.7) | 130 (2.2) |
| Tobacco use, n (%) | 1852 (8.5) | 616 (10.4) |
| Illicit substance use, n (%) | 1219 (5.6) | 353 (6.0) |
| Oxygen required in ED, n (%) | 1919 (8.8) | 627 (10.6) |
| Hospital admission, n (%) | 9913 (45.6) | 2446 (41.3) |
| In-hospital death, n (%) | 753 (3.5) | 213 (3.6) |
| 7-day average incident COVID-19 cases, median (IQR) | 1.3 (0.73.2) | 0.96 (0.5–1.3) |
| SARS-CoV-2 positive, n (%) | 940 (4.3) | 227 (3.8) |
ED, emergency department; LTC, long-term care; TIA, transient ischaemic attack.
bmjopen-2021-055832supp001.pdf (130.3KB, pdf)
bmjopen-2021-055832supp002.pdf (2.2MB, pdf)
Each consecutive, eligible patient enrolled in the registry was assigned a CCEDRRN unique identifier. Trained research assistants entered anonymised participant data into a REDCap database (V.10.9.4; Vanderbilt University, Nashville, Tennessee, USA). Regular data quality checks including verification of extreme or outlying values were performed by each participating site, coordinated by the CCEDRRN coordinating centre.
Participants
We included data from consecutive patients tested for SARS-CoV-2 at 32 CCEDRRN sites. From each site’s start date forward, we included consecutive eligible patients aged 18 years and older who had a biological sample (swab, endotracheal aspirate and bronchoalveolar lavage) specimen collected for NAAT on their index emergency department visit or, if admitted, within 24 hours of emergency department arrival. For patients with multiple emergency department encounters involving COVID-19 testing, we only used the first encounter in this analysis.
We excluded patients who had a positive SARS-CoV-2 NAAT within 14 days prior to their emergency department visit, patients with cardiac arrest prior to emergency department arrival and those with missing outcome data.
Predictors
Candidate predictors were chosen based on clinical consensus and availability within the CCEDRRN registry. Predictors included known risk factors for SARS-CoV-2 infection, including work as a healthcare provider, institutional living (ie, long-term care and prison), close personal or household contacts with SARS-CoV-2 infection and symptoms including cough, anosmia or dysgeusia, fever, myalgias and vital signs on emergency department arrival. The full list of candidate variables and their definitions are available in the (online supplemental appendix table 2).
In addition to these clinical variables, the 7-day average incident COVID-19 case count was calculated for the health region of each participating site using publicly available epidemiological data.19 For each calendar day within each health region represented in the study, we calculated the average daily incident rate of new infections per 100 000 population over the preceding 7 days. This 7-day average incidence was assigned to each patient based on the date of their index emergency department encounter and the health region of the forward sortation area of their postal code of residence. For patients with no fixed address, we allocated them to the health region of the hospital in which they were tested. As publicly available incident COVID-19 case data were not available for the early pandemic, we imputed values for the first 5 weeks of the pandemic by modelling the reported COVID-19 cases that had accumulated in every health region over time using linear interpolation (0.1% missing).
Outcome
The primary outcome of this analysis was the diagnosis of SARS-CoV-2 infection using a criterion standard of a positive NAAT at the time of index emergency department visit or within 14 days after the index encounter.
Sample size and precision
The 46 candidate predictors had 52 degrees of freedom and with an expected SARS-CoV-2 infection rate of 5%, a sample size of 1040 was sufficient for the derivation cohort based on an anticipated event rate of less than 20% and a requirement for 20 outcomes per degree of freedom.20 Over 21 000 patients were available for the derivation cohort at the time of analysis, providing more than sufficient data for reliable prediction modelling.
Model development and validation
We randomly assigned study sites to the derivation and validation cohorts with the goal of assigning 75% of eligible patients and outcome events to the derivation cohort and 25% to the validation cohort. Thus, the derivation and validation cohorts are geographically distinct. Within the derivation cohort, candidate predictors were examined for colinearity and missing or extreme values. In the presence of colinearity, one predictor was dropped from the set of candidate predictors. Five multiple imputations were used for continuous variables with missing data. Patients with values of ‘not recorded’ for categorical variables (eg, smoking and need for supplemental oxygen) were assumed to have the reference value (ie, ‘no’) for that categorical variable. The initial logistic regression model considered all candidate predictors, with continuous predictors fit with restricted cubic splines with three knots. The strengths of associations between predictors and outcome were assessed using an analysis of variance plot to inform the df to allocate to each predictor. The model was fit again with these changes. A fast step-down procedure reduced the model to key predictors based on an Akaike’s information criterion stopping rule with a threshold of 120 to enable a model with a relatively small number of predictors that would be clinically easy to use. Internal bootstrap validation with 1000 bootstrap samples was conducted to provide an optimism-corrected C-statistic. Continuous predictors were categorised based on the relationship between the spline function and outcome.
We then developed the points-based CCEDRRN COVID-19 Infection Score (CCIS) using a nomogram to assign integer point values for each variable included in the derived model. Discrimination of the score was evaluated using the c-statistic. Calibration was evaluated using calibration curves and comparison of observed and expected outcomes. Diagnostic performance was evaluated using sensitivity and specificity, predictive values and likelihood ratios at different point thresholds.
We then evaluated the discrimination, calibration and performance characteristics of the CCIS in an external validation cohort of patients from geographically distinct study sites who were not part of the derivation cohort.
Validation of previously published models
We used our combined (derivation and validation) study cohort to externally validate the COvid Rule out Criteria (CORC) score developed by Kline et al 3 (although we were not able to include race and ethnicity variables as these are not reliably recorded or reported in most Canadian hospitals). We compared measures of discrimination and calibration, along with sensitivity and specificity of risk score values for the CCIS and CORC (with race and ethnicity variables removed). We split each score into categories of low, moderate and high risk for SARS-CoV-2 infection. Low risk was defined as a score having a sensitivity for ruling out infection of 95% or higher. High risk was defined as a score having a specificity for ruling in infection of 95% or higher. We compared the performance of the two scores by calculating net reclassification improvement across low-risk, moderate-risk and high-risk categories.21 22
Role of the funding sources
The funding organisations had no role in the study conduct, data analysis, manuscript preparation or submission.
Patient and public involvement
The CCEDRRN governance structure includes patient representatives on the Executive Committee, Scientific Steering Committee, Protocol Review and Publications Committee, Data Access and Monitoring Committee and Knowledge Translation Committee. The network also has a Patient Engagement Committee composed of patient partners from across Canada. Patient partners provided input into study design and selection of outcomes for all CCEDRRN analyses and provide advice on knowledge sharing and translation strategies.
Results
This analysis is based on 27 665 patients consecutively enrolled from 32 participating emergency departments between March and October 2020 (figure 1, online supplemental appendix table 1). Sites and enrolment periods contributing patient data are shown in the online supplemental appendix table 1. Of the included patients, 1167 (4.2%) had a positive SARS-CoV-2 NAAT result, including 1133 who had a positive initial test and 34 who tested positive after a negative (27) or indeterminate (7) initial NAAT.
Figure 1.
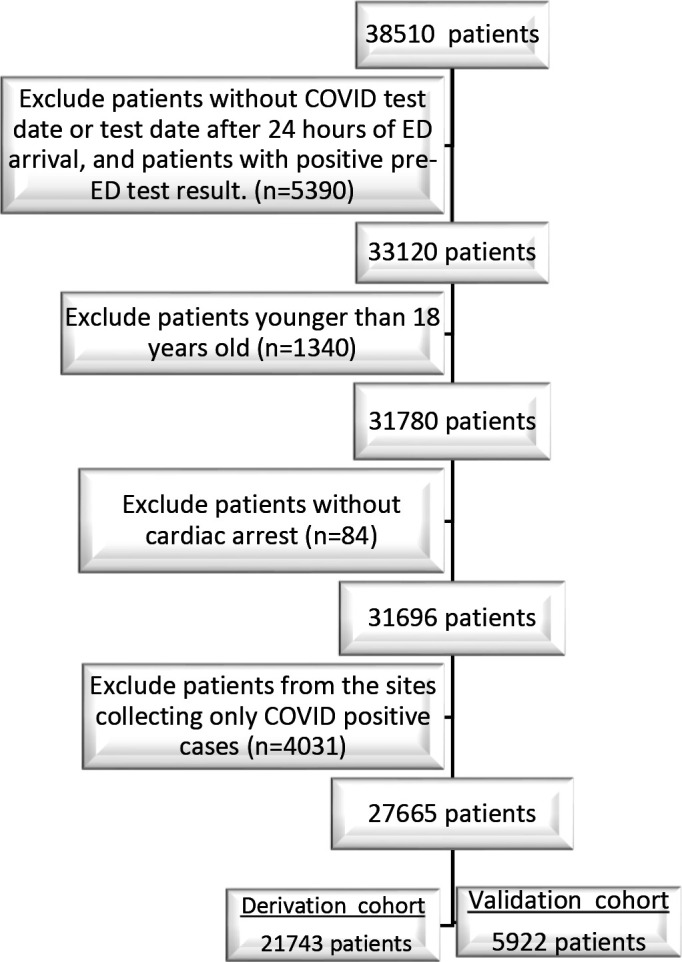
Flow diagram of patients through the .
ED, emergency department.
The study cohort was subdivided into a derivation cohort (21 743 patients from 16 sites, 940 (4.3%) SARS-CoV-2 positive) and a separate external validation cohort (5922 patients from 16 different sites, 227 (3.8%) SARS-CoV-2 positive). Demographic and clinical characteristics of the derivation and validation cohorts are shown in table 1. No continuous variable requiring multiple imputation had more than 3.4% missingness (online supplemental appendix table 2).
In the derivation cohort, we derived a 10-variable model to predict the probability of a patient having a positive SARS-CoV-2 NAAT. The regression coefficients and ORs for each variable in the model are shown in table 2. The c-statistic for the derived model was 0.851 with excellent calibration.
Table 2.
Adjusted associations between model predictor variables and SARS-CoV-2 nucleic acid test results
| Variable/score component | Regression coefficient (SE) | Adjusted OR (95% CI) | Score value |
| 7-day average incident COVID-19 cases | |||
| 0–2 daily cases per 100 000 population | – | – | 0 |
| 2–7.99 daily cases per 100 000 population | 1.22 (0.09) | 3.38 (2.85 to 4.00) | 1 |
| ≥8 daily cases per 100 000 population | 2.21 (0.10) | 9.09 (7.53 to 10.97) | 2 |
| Institutional exposure (eg, LTC, prison) or travel from country with known cases within 14 days | 0.88 (0.09) | 2.40 (2.01 to 2.87) | 1 |
| Healthcare worker/microbiology lab | 1.10 (0.16) | 3.02 (2.22 to 4.10) | 1 |
| Household/caregiver contact | 1.83 (0.12) | 6.25 (4.92 to 7.93) | 2 |
| Temperature | |||
| <36 and no self-reported fever | −0.75 (0.3) | 0.47 (0.28 to 0.80) | -1 |
| 36–37.4 and no self-reported fever | – | – | 0 |
| ≥37.5 or self-reported fever | 1.21 (0.08) | 3.36 (2.88 to 3.91) | 1 |
| Supplemental oxygen delivered in the ED | 0.98 (0.1) | 2.66 (2.18 to 3.24) | 1 |
| Cough | 0.85 (0.08) | 2.33 (2.01 to 2.71) | 1 |
| Dysgeusia/anosmia | 2.03 (0.24) | 7.60 (4.76 to 12.15) | 2 |
| Muscle aches (myalgia) | 0.7 (0.11) | 2.02 (1.64 to 2.48) | 1 |
| Current tobacco user | −1.13 (0.21) | 0.32 (0.21 to 0.49) | -1 |
ED, emergency department; LTC, long-term care.
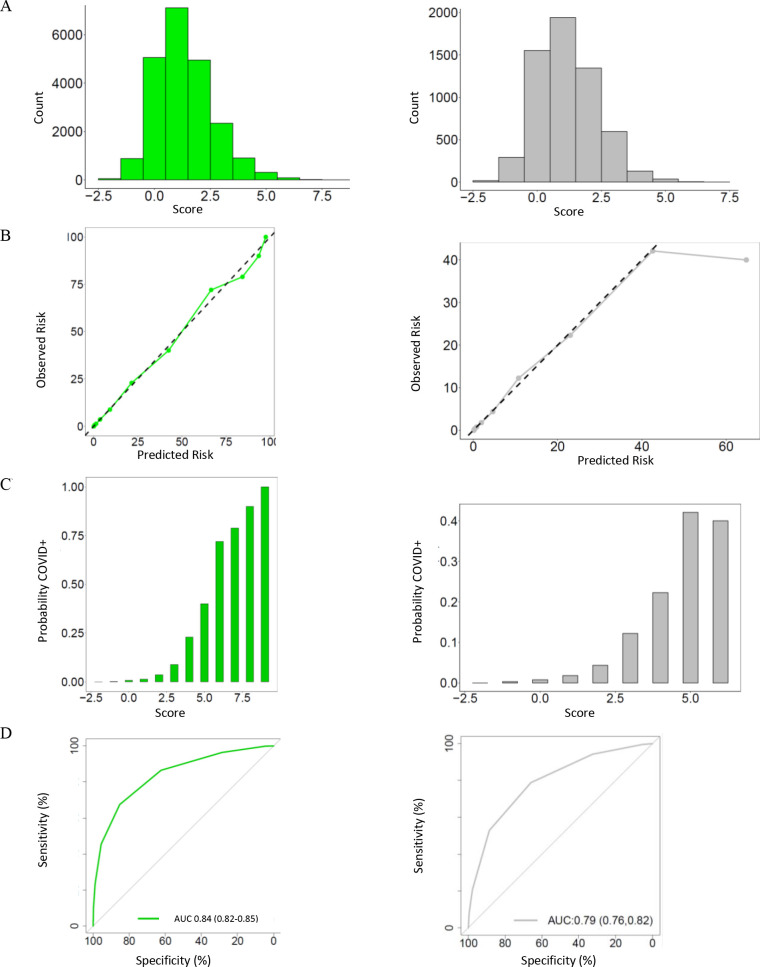
We created a points-based CCIS using rounded regression coefficients with a range of −2 to 9 points (table 2). The c-statistic of the CCIS in the derivation cohort was 0.838 (0.824–0.852) with excellent calibration (figure 2). A score of zero or less ruled out a positive SARS-CoV-2 test result in 5996/21 743 patients (27.6%) with a sensitivity of 96.6% (95% CI 95.2 to 97.7). A score of 4 or more was observed in 1338/21 743 patients (6.2%) and had a specificity of 95.6 (95% CI 95.3 to 95.8) indicating a low frequency of false positives (online supplemental appendix table 3).
Figure 2.
Distribution and performance of the CCEDRRN COVID-19 infection score in the derivation cohort (left panel) and validation cohorts (right panel): (A) distribution of the score, (B) observed in-hospital mortality across the range of the score, (C) predicted versus observed probability of in-hospital mortality and (D) receiver operating characteristic curve with area under the curve and associated 95% CI. CCEDRRN, Canadian COVID-19 Emergency Department Rapid Response Network.
We then quantified the performance of the CCIS in our external validation cohort. In this cohort, the c-statistic for the points-based risk score was 0.792 (figure 2). A score of zero or less ruled out a positive SARS-CoV-2 test result in 1863/5925 patients (31.4%) with a sensitivity of 94.3% (95% CI 90.4 to 96.9). A score of 4 or more was observed in 174/5925 patients (2.9%) and had a specificity of 97.8 (95% CI 97.4 to 98.1) indicating a low frequency of false positives (table 3).
Table 3.
Performance metrics for the CCEDRRN COVID-19 Infection Score for ruling in or ruling out SARS-CoV-2 infection at different score cut-off values in the validation cohort
| Score cut-off | n (%) |
Sensitivity
(%, 95% CI) |
Specificity
(%, 95% CI) |
LR+ | LR− |
COVID+
n (%) |
| Rule out: | ||||||
| ≤−2 | 17 (0.3) | 100 (98.4 to 100) | 0.3 (0.2 to 0.5) | 1 | NA | 0 (0) |
| ≤−1 | 310 (5.2) | 99.6 (97.6 to 100) | 5.4 (4.9 to 6.1) | 1.1 | 0.1 | 1 (0.3) |
| ≤0 | 1863 (31.5) | 94.3 (90.4 to 96.9) | 32.5 (31.3 to 33.7) | 1.4 | 0.2 | 13 (0.7) |
| ≤1 | 3806 (64.3) | 78.9 (73.0 to 84.0) | 66.0 (64.7 to 67.2) | 2.3 | 0.3 | 48 (1.3) |
| ≤2 | 5152 (87.0) | 52.9 (46.2 to 60.0) | 88.6 (87.7 to 89.4) | 4.6 | 0.5 | 107 (2.1) |
| ≤3 | 5748 (97.1) | 20.7 (15.6 to 26.6) | 97.8 (97.4 to 98.1) | 9.3 | 0.8 | 180 (3.1) |
| Rule in: | ||||||
| ≥3 | 770 (13.0) | 52.9 (46.2 to 59.5) | 88.6 (87.7 to 89.4) | 4.6 | 0.5 | 120 (15.6) |
| ≥4 | 174 (2.9) | 20.7 (15.6 to 26.6) | 97.8 (97.4 to 98.1) | 9.3 | 0.8 | 47 (27.0) |
| ≥5 | 44 (0.7) | 7.9 (4.8 to 12.2) | 99.5 (99.3 to 99.7) | 17.4 | 0.9 | 18 (40.9) |
| ≥6 | 6 (0.1) | 0.9 (0.1 to 3.2) | 99.9 (99.8 to 100) | 12.5 | 1 | 2 (33.3) |
| ≥7 | 1 (<0.1) | 0 (0 to 1.6) | 100.0 (99.9 to 100) | NA | 1 | 0 (0) |
CCEDRRN, Canadian COVID-19 Emergency Department Rapid Response Network; LR+, positive likelihood ratio; LR−, negative likelihood ratio.
In a combined cohort of patients (derivation and validation combined), we compared the discrimination and diagnostic performance of the CCIS to the CORC score. The CCIS had a c-statistic of 0.837 compared with 0.750 for the CORC score (with race/ethnicity variables removed) (online supplemental appendix figure 1). A CCIS of zero or less ruled out SARS-CoV-2 infection in 28.4% of patients with a sensitivity of 96.1% (online supplemental appendix table 4), whereas a CORC score of negative one or less ruled out SARS-CoV 2 infection in 9.9% of patients with 97.4% (online supplemental appendix table 5) sensitivity. Compared with the CORC score (with race/ethnicity variables removed), the CCIS showed substantial net reclassification improvement net reclassification improvement (NRI=0.310,)(online supplemental appendix table 6).
Discussion
We have derived and validated a simple clinical risk score, the CCIS, to predict the probability of a positive SARS-CoV-2 NAAT in patients presenting to emergency departments. It uses only clinical variables available at the patient’s bedside, along with a common publicly available measure of community COVID-19 incidence. In this study population, the score ruled out SARS-CoV-2 infection with 96.1% sensitivity in almost one-third of patients. It also identified patients at high risk of infection with over 95% specificity.
In addition to clinical variables, we also included the average daily incidence of SARS-CoV-2 infections in a patient’s health region, which is an essential predictor of the probability of a patient’s risk of COVID-19 infection. Although access to timely incidence data may be challenging in under-resourced health systems, this information is publicly reported in many health jurisdictions. In practice, the local incidence would likely need to be shared within an emergency department on a daily basis. We developed data-driven cut-offs for categorisation of low, moderate and high incidence for calculation the CCIS. Thus, the clinician would only need to know whether local incidence is high, moderate or low to use this score, and the incidence category changes slowly over time. Patients who live and work in separate health regions could be assigned the higher incidence value at hospital presentation for a conservative risk estimate. Patients in areas with high disease burden will automatically score 2 points, meaning that few patients in these settings will be classified as low risk. Therefore, symptomatic patients would all warrant testing. This underscores the need for liberal isolation and testing practices in settings with high rates of community SARS-CoV-2 transmission.
The CCIS has several important clinical applications. The ability to differentiate patients with high or low probability of COVID-19 disease could guide safe and effective patient isolation or cohorting from the time of hospital arrival, prior to the availability of SARS-CoV-2 test results. Identification of patients with extremely low risk of SARS-CoV-2 infection may even allow safe omission of testing, which will minimise testing resource utilisation in settings with limited testing capacity. Identifying patients with a high probability of SARS-CoV-2 infection can help prioritise use of rapid antigen testing and initiation of effective empiric therapy in critically ill patients prior to availability of NAAT results. By presenting risk estimates and sensitivity for all risk score values, we allow end-users to choose cut-offs for ruling-in and ruling-out SARS-CoV-2 infection that make sense for their setting and application.
Several other risk prediction instruments have been developed to predict positive COVID-19 test results in undifferentiated patients. These tools were developed in studies with substantial methodological limitations and incorporate variables not immediately available at the time of a patient’s hospital arrival, so are not useful to guide early isolation, testing and treatment decisions.2 None of these risk prediction tools considered the prevalence of disease in the population. Prevalence can substantially change the approach to testing and cohorting, and this will become increasingly important as prevalence rates drop and selective rather than liberal testing may be more appropriate.
US-based investigators recently reported the development3 and validation25 of the CORC score using only clinical variables. The CORC score contains several similar variables to the CCIS. The CORC score included race and ethnicity as predictor variables, which may limit the generalisability of the CORC score beyond the urban American population in which it was developed, as it does not reflect the international diversity of ethnic backgrounds. Moreover, it is unlikely race or ethnicity represents a biological risk. The association between race and ethnicity and SARS-CoV-2 infection in the CORC score likely reflects other sociodemographic and geographic predictors of the risk of COVID-19 infection in the American population.25 The CCIS uses the 7-day average local incidence as an estimate of population risk. We believe this approach is more generalisable across populations and better reflects individual patients’ pretest probability of SARS-CoV-2 infection.26
Strengths and limitations
The cohorts used to derive and validate the rule included comprehensive data on consecutive eligible patients from a large, geographically distributed network of Canadian urban, regional and rural emergency departments. Strict data quality protocols and data cleaning protocols ensured the reliability of collected data. This score may be employed at the time of a patient’s arrival to hospital, does not require the use of additional laboratory testing or imaging nor the use of electronic calculators or electronic medical records for implementation.
Some missing data required either multiple imputation or classification of missing categorical variables as being absent. The overall missingness of data in this registry is very low.15 Although the data collection for the CCEDRRN registry relies on abstraction from health records, this approach has been shown to be reliable in our study sites when compared with prospective data collection. 15
The clinical variables in the model are not likely to be sensitive to changes in geographical changes in SARS-CoV-2 epidemiology. The variable of travel from a country with high incidence may become less informative as the pandemic has spread globally and ‘hot spots’ change. However, high-prevalence areas may change over time, meaning that the risk factor of travel from a region with a high prevalence is likely to still be informative.
This risk score was developed using data from patients enrolled in the first nine months of the pandemic when rates of influenza were low. As such, the score may need to be revalidated and refined in the future to reflect the influence of influenza, the emergence of variant strains of SARS-CoV-2 and widespread population immunisation on patients’ risk of infection.
Conclusion
We derived and successfully validated the CCIS to accurately predict the probability of SARS-CoV-2 nucleic acid test results in patients presenting to emergency departments. The CCIS uses clinical variables, accounts for the incidence of SARS-CoV-2 in the community and is ready for immediate clinical use. This score has potential utility to guide early decisions around SARS-CoV-2 test utilisation, patient isolation and empirical therapy for patients solely based on clinical assessment.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Ms Amber Cragg in the preparation of this manuscript. We would like to thank the University of British Columbia clinical coordinating centre staff, legal, ethics, privacy and contract staff and the research staff at each of the participating institutions in the network outlined in the attached supplement. The network would not exist today without the dedication of these professionals. Thank you to all our patient partners who shared their lived experiences and perspectives to ensure that the knowledge we cocreate addresses the concerns of patients and the public. Creating the largest network of collaboration across Canadian Emergency Departments would not have been feasible without the tireless efforts of emergency department chiefs, and research coordinators and research assistants at participating sites. Finally, our most humble and sincere gratitude to all our colleagues in medicine, nursing and the allied health professions who have been on the frontlines of this pandemic from day 1 staffing our ambulances, emergency departments, intensive care units and hospitals bravely facing the risks of COVID-19 to look after our fellow citizens and after one another. We dedicate this network to you (online supplemental appendix, table 1).
Footnotes
Twitter: @GameYadav
Correction notice: This article has been corrected since it was first published. ADM and CMH are joint first authors.
Collaborators: Canadian COVID-19 Emergency Department Rapid Response Network (CCEDRRN) investigators for the Network of Canadian Emergency Researchers and the Canadian Critical Care Trials Group: Patrick Fok, Hana Wiemer, Samuel Campbell, Kory Arsenault, Tara Dahn, Kavish Chandra, Patrick Archambault, Joel Turner, Éric Mercier, Greg Clark, Sébastien Robert, Raoul Daoust, Laurie Morrison, Steven Brooks, Ivy Cheng, Krishan Yadav, Michelle Welsford, Rob Ohle, Justin Yan, Rohit Mohindra, Megan Landes, Tomislav Jelic, Ankit Kapur, Phil Davis, Andrew McRae, Brian Rowe, Katie Lin, Stephanie VandenBerg, Jake Hayward, Jaspreet Khangura, Corinne Hohl, Daniel Ting, Maja Stachura, Frank Scheuermeyer, Balijeet Braar, Craig Murray, John Taylor, Ian Martin, Sean Wormsbecker, Matt Bouchard, Lee Graham.
Contributors: CMH, ADM, LJM, RR and JP conceived the study, with input on the design and selection of variables from the other contributors. CMH, LJM, PMA, SCB, PD and EL obtained funding on behalf of the Canadian COVID-19 Emergency Department Rapid Response Network (CCEDRRN) investigators. CMH, ADM, PMA, SCB, IC, PD, JH, BR, RO, MW and KY facilitated data collection along with other members of the CCEDRRN and can verify the underlying data. RR and JP developed the analytic plan. SV performed the analysis, with assistance from GG and RR, including accessing and verification of underlying data. All contributors provided input on interpretation of findings. ADM, CMH and RR drafted the manuscript with additional input from all contributors. ADM and CH are the guarantors of this work.
Funding: The Canadian Institutes of Health Research (447679), Ontario Ministry of Colleges and Universities (C-655–2129), Saskatchewan Health Research Foundation (5357), Genome BC (COV024 and VAC007) Fondation du CHU de Québec (Octroi No. 4007). Sero-Surveillance and Research (COVID-19 Immunity Task Force Initiative) provided peer-reviewed funding. The BC Academic Health Science Network and BioTalent Canada provided non-peer reviewed funding.
Disclaimer: These organisations are not-for-profit and had no role in study conduct, analysis or manuscript preparation.
Competing interests: ADM, CMH, RR, PMA, SCB, IC, PD, JH, EL, RO, BR, MW, KY, LJM and JP are coinvestigators on the funding sources listed in the funding statement and have no additional competing interests. GG and SV have no competing interests.
Ethics Approval: The CCEDRRN network protocol was approved with a waiver of informed consent by the University of British Columbia Research Ethics Board (UBC REB H20-01015) and subsequently by the research ethics boards of all participating institutions.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
Canadian COVID-19 Emergency Department Rapid Response Network (CCEDRRN) investigators for the Network of Canadian Emergency Researchers and the Canadian Critical Care Trials Group:
Patrick Fok, Hana Wiemer, Samuel Campbell, Kory Arsenault, Tara Dahn, Kavish Chandra, Patrick Archambault, Joel Turner, Éric Mercier, Greg Clark, Sébastien Robert, Raoul Daoust, Laurie Morrison, Steven Brooks, Ivy Cheng, Krishan Yadav, Michelle Welsford, Rob Ohle, Justin Yan, Rohit Mohindra, Megan Landes, Tomislav Jelic, Ankit Kapur, Phil Davis, Andrew McRae, Brian Rowe, Katie Lin, Stephanie VandenBerg, Jake Hayward, Jaspreet Khangura, Corinne Hohl, Daniel Ting, Maja Stachura, Frank Scheuermeyer, Balijeet Braar, Craig Murray, John Taylor, Ian Martin, Sean Wormsbecker, Matt Bouchard, and Lee Graham
Data availability statement
Data are available on reasonable request. For investigators who wish to access Canadian COVID-19 Emergency Rapid Response Network data, proposals may be submitted to the network for review and approval by the network’s peer-review publication committee, the data access and management committee and the executive committee, as per the network’s governance. Information regarding submitting proposals and accessing data may be found at https://www.ccedrrn.com
Data availability statement
For investigators who wish to access Canadian COVID-19 Emergency Rapid Response Network data, proposals may be submitted to the network for review and approval by the network’s peer-review publication committee, the data access and management committee and the executive committee, as per the network’s governance. Information regarding submitting proposals and accessing data may be found at https://canadiancovid19ednetworkorg/. For investigators who wish to access Canadian COVID-19 Emergency Rapid Response Network data, proposals may be submitted to the network for review and approval by the network’s peer-review publication committee, the data access and management committee and the executive committee, as per the network’s governance. Information regarding submitting proposals and accessing data may be found at https://www.ccedrrn.com
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. World Health Organization . WHO Coronavirus Dashboard, 2021. Available: https://covid19.who.int/ [Accessed 19 Nov 2021].
- 2. Update to living systematic review on prediction models for diagnosis and prognosis of covid-19. BMJ 2021;372:n236. 10.1136/bmj.n236 [DOI] [PubMed] [Google Scholar]
- 3. Kline JA, Camargo CA, Courtney DM, et al. Clinical prediction rule for SARS-CoV-2 infection from 116 U.S. emergency departments 2-22-2021. PLoS One 2021;16:e0248438. 10.1371/journal.pone.0248438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McDonald SA, Medford RJ, Basit MA, et al. Derivation with internal validation of a multivariable predictive model to predict COVID-19 test results in emergency department patients. Acad Emerg Med 2021;28:206–14. 10.1111/acem.14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kurstjens S, van der Horst A, Herpers R, et al. Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. Clin Chem Lab Med 2020;58:1587–93. 10.1515/cclm-2020-0593 [DOI] [PubMed] [Google Scholar]
- 6. Lippi G, Henry BM, Hoehn J, et al. Validation of the Corona-Score for rapid identification of SARS-CoV-2 infections in patients seeking emergency department care in the United States. Clin Chem Lab Med 2020;58:e311–3. 10.1515/cclm-2020-1121 [DOI] [PubMed] [Google Scholar]
- 7. Sung J, Choudry N, Bachour R. Development and validation of a simple risk score for diagnosing COVID-19 in the emergency room. Epidemiol Infect 2020;148:e273. 10.1017/S0950268820002769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwab P, DuMont Schütte A, Dietz B, et al. Clinical predictive models for COVID-19: systematic study. J Med Internet Res 2020;22:e21439. 10.2196/21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plante TB, Blau AM, Berg AN, et al. Development and external validation of a machine learning tool to rule out COVID-19 among adults in the emergency department using routine blood tests: a large, multicenter, real-world study. J Med Internet Res 2020;22:e24048. 10.2196/24048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soltan AAS, Kouchaki S, Zhu T, et al. Rapid triage for COVID-19 using routine clinical data for patients attending Hospital: development and prospective validation of an artificial intelligence screening test. Lancet Digit Health 2021;3:e78–87. 10.1016/S2589-7500(20)30274-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cabitza F, Campagner A, Ferrari D, et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin Chem Lab Med 2020;59:421–31. 10.1515/cclm-2020-1294 [DOI] [PubMed] [Google Scholar]
- 12. Jehi L, Ji X, Milinovich A, et al. Individualizing risk prediction for positive coronavirus disease 2019 testing: results from 11,672 patients. Chest 2020;158:1364–75. 10.1016/j.chest.2020.05.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joshi RP, Pejaver V, Hammarlund NE, et al. A predictive tool for identification of SARS-CoV-2 PCR-negative emergency department patients using routine test results. J Clin Virol 2020;129:104502. 10.1016/j.jcv.2020.104502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Formica V, Minieri M, Bernardini S, et al. Complete blood count might help to identify subjects with high probability of testing positive to SARS-CoV-2. Clin Med 2020;20:e114–9. 10.7861/clinmed.2020-0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hohl CM, Rosychuk RJ, McRae AD, et al. Development of the Canadian COVID-19 emergency department rapid response network population-based registry: a methodology study. CMAJ Open 2021;9:E261–70. 10.9778/cmajo.20200290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moons KGM, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1–73. 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 17. WHO . Clinical characterisation protocol (CCP), version 3.1/3.2. Oxford University, 2020. [Google Scholar]
- 18. ISARIC/WHO Clinical Characterisation Protocol for Severe Emerging Infections [COVID-19] [UPH]. Health Res. Available: /planning-and-improving-research/application-summaries/research-summaries/isaricwho-clinical-characterisation-protocol-for-severe-emerging-infections/ [Accessed 4 Jun 2021].
- 19. Health regional Archive (public view), 2019. Available: https://resources-covid19canada.hub.arcgis.com/datasets/3aa9f7b1428642998fa399c57dad8045/data?layer=1 [Accessed 4 Jun 2021].
- 20. Visit 6 manual 17: ARIC neurocognitive exam. Available: https://www2.cscc.unc.edu/aric/
- 21. Leening MJG, Vedder MM, Witteman JCM, et al. Net reclassification improvement: computation, interpretation, and controversies: a literature review and clinician's guide. Ann Intern Med 2014;160:122–31. 10.7326/M13-1522 [DOI] [PubMed] [Google Scholar]
- 22. Kerr KF, Wang Z, Janes H, et al. Net reclassification indices for evaluating risk prediction instruments: a critical review. Epidemiology 2014;25:114–21. 10.1097/EDE.0000000000000018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing, 2020. [Google Scholar]
- 24. Harrell . rms: regression modeling Strategies., version 6.2-0, 2021. Available: https://cran.r-project.org/web/packages/rms/index.html
- 25. Nevel AE, Kline JA. Inter-Rater reliability and prospective validation of a clinical prediction rule for SARS-CoV-2 infection. Acad Emerg Med 2021;28:Epub ahead of print:761–7. 10.1111/acem.14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vyas DA, Eisenstein LG, Jones DS. Hidden in Plain Sight - Reconsidering the Use of Race Correction in Clinical Algorithms. N Engl J Med 2020;383:874–82. 10.1056/NEJMms2004740 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2021-055832supp001.pdf (130.3KB, pdf)
bmjopen-2021-055832supp002.pdf (2.2MB, pdf)
Data Availability Statement
Data are available on reasonable request. For investigators who wish to access Canadian COVID-19 Emergency Rapid Response Network data, proposals may be submitted to the network for review and approval by the network’s peer-review publication committee, the data access and management committee and the executive committee, as per the network’s governance. Information regarding submitting proposals and accessing data may be found at https://www.ccedrrn.com
For investigators who wish to access Canadian COVID-19 Emergency Rapid Response Network data, proposals may be submitted to the network for review and approval by the network’s peer-review publication committee, the data access and management committee and the executive committee, as per the network’s governance. Information regarding submitting proposals and accessing data may be found at https://canadiancovid19ednetworkorg/. For investigators who wish to access Canadian COVID-19 Emergency Rapid Response Network data, proposals may be submitted to the network for review and approval by the network’s peer-review publication committee, the data access and management committee and the executive committee, as per the network’s governance. Information regarding submitting proposals and accessing data may be found at https://www.ccedrrn.com