Abstract
In all of the transposition reactions that have been characterized thus far, synapsis of two transposon ends is required before any catalytic steps (strand nicking or strand transfer) occur. In V(D)J recombination, there have been inconclusive data concerning the role of synapsis in nicking. Synapsis between two 12-substrates or between two 23-substrates has not been ruled out in any studies thus far. Here we provide the first direct tests of this issue. We find that immobilization of signals does not affect their nicking, even though hairpinning is affected in a manner reflecting its known synaptic requirement. We also find that nicking is kinetically a unireactant enzyme-catalyzed reaction. Time courses are no different between nicking seen for a 12-substrate alone and a reaction involving both a 12- and a 23-substrate. Hence, synapsis is neither a requirement nor an effector of the rate of nicking. These results establish V(D)J recombination as the first example of a DNA transposition-type reaction in which catalytic steps begin prior to synapsis, and the results have direct implications for the order of the steps in V(D)J recombination, for the contribution of V(D)J recombination nicks to genomic instability, and for the diversification of the immune repertoire.
V(D)J recombination assembles the exons that encode the antigen binding domain of immunoglobulin and T-cell receptor genes during the generation of B and T cells (12, 27). In the genome, each of the elements to be recombined, V (variable), D (diversity), and J (joining), is located adjacent to a specific DNA sequence called a recombination signal sequence (RSS), which directs the recombination. The RSS contains a conserved heptamer (5′-CACAGTG-3′) and nonamer (5′-ACAAAAACC-3′) separated by a nonconserved spacer of either 12 or 23 bp, termed 12RSS and 23RSS, respectively (18). The nucleotides of the V, D, or J segment that are immediately adjacent to the heptamer are referred to as the coding end nucleotides. During V(D)J recombination, DNA double-strand breaks are introduced at the junction between the RSS and the adjacent coding V, D, or J segment; the corresponding broken ends are called either the signal ends or the coding ends, accordingly. V(D)J recombination typically involves two DNA double-strand breaks: one at an element with a 12-RSS and the other at an element with a 23-RSS. This feature is known as the 12/23 rule (45). Completion of V(D)J recombination depends on the proper joining of the two coding ends to each other and of the two signal ends to one another through a general DNA double-strand break repair pathway known as nonhomologous end joining (26).
The genes encoding the recombination enzymes that cleave the DNA and initiate V(D)J recombination are called recombination activation genes (RAGs). The RAG protein complex consists of two gene products: RAG1 and RAG2 (4, 20, 34, 40, 44). RAG-mediated cleavage occurs in two steps (30). First, a nick is introduced at the heptamer-coding sequence junction 5′ to the heptamer. This results in a 3′ hydroxyl group at the end of the coding sequence and a 5′ phosphate group attached to the heptamer end. Second, the 3′ hydroxyl group of the coding sequence attacks the antiparallel strand in a direct transesterification reaction (12), which cleaves the DNA into a hairpin-structured coding end and a blunt signal end. This general type of DNA cleavage is also observed in other DNA transposition systems, such as Tn10 transposition (24). The organization of the RAG1 and RAG2 genes in the genome is consistent with an origin from a DNA transposon (35). Indeed, it was found recently that recombinant RAG proteins have transposase activity, in that they can insert DNA fragments with signal ends into other DNA molecules (1, 21, 35, 37), though such activity has not been documented in vivo. It is believed that V(D)J recombination evolved from an ancient DNA transposition event (35, 37).
In all of the transposition reactions characterized thus far, synapsis of two transposon ends is required before any strand nicking or strand transfer (double-strand breakage) occurs (2, 15, 31, 33, 38, 41, 48). It is important to note that two like ends (e.g., two right ends) can equivalently supplant a left and a right end for all steps in some transposition systems (3).
For in vitro V(D)J recombination, it has been clear that nicking does not require the presence of both a 12- and a 23-signal in solution (47). Based on this, some have inferred that nicking is entirely independent of synapsis (12). This conjecture overlooks the possibility that two identical ends (e.g., two 12-signals) could provide synapsis equivalent to a 12/23 synapsis. In fact, direct and indirect (competition) assays of synapsis have documented that 12/12 or 23/23 synapsis occurs either at only moderately (10-fold) reduced levels (19) or at levels similar to those for 12/23 synapsis (51).
In contrast with the assumption that nicking is independent of synapsis, data from others suggests that synapsis can actually stimulate nicking in V(D)J recombination (9). However, the inference of stimulation is based on a control which assumes that RAG binding at one signal is independent of RAG binding at a second signal located 89 bp away on the same DNA fragment. There is no data assuring that such close signals do not interfere with one another (8, 9, 42). In addition, the interpretation of this type of data is complicated by the fact that binding and nicking at one signal may result in rapid collisional rates with the second tethered signal (zero order kinetics), eliminating the ability to determine the kinetic dependence of nicking on synapsis (see Discussion).
The precedent in the other transposition systems makes it even more important that the dependence of nicking on synapsis be directly tested. The only suggestion that there might be a difference between V(D)J recombination and the other biochemically characterized transposition reactions concerning synapsis prior to any catalysis is that the hairpin step is much more efficient with both the 12- and 23-signals present in the solution than with reactions having only one type of signal present (19, 47, 51). In contrast, the nicking step appears to be similarly efficient whether one or both types of signals are present (9, 47, 51, 53). It is important to note that this in no way indicates that synapsis is unnecessary for nicking. It simply indicates that if synapsis is required for both nicking and hairpinning, then the 12/23 rule is operating only at the hairpinning step. This could simply be indicative of a more stringent three-dimensional structure of the protein-DNA complex at the later step.
The uncertainty about synapsis at the nicking step affects how one views all of the subsequent steps. If nicking is independent of synapsis, then a synaptic step that has functional consequences must occur between the key catalytic steps of nicking and hairpinning (53). Hence, the ordering of the steps requires a test of the synaptic requirement at the point of nicking.
In the present study, we unambiguously demonstrate that nicking is independent of synapsis in a system where synapsis was prevented by immobilization of the DNA substrate on streptavidin beads. Moreover, the initial rate of nicking is consistent with a unireactant enzyme-catalyzed kinetic model. The kinetic analysis provides additional insights regarding the time required for nicking. These findings distinguish the mechanism of the RAG proteins from other characterized transposases in a key aspect, and they have implications for the development of an immune receptor repertoire.
MATERIALS AND METHODS
DNA substrates.
Nonbiotinylated oligonucleotides were synthesized by Operon Technologies, Inc. (Alameda, Calif.). A DNA substrate containing either a 12RSS or a 23RSS was made by annealing two complementary oligonucleotides. The 12-substrate for the initial rate assay was made by annealing KY28 (5′ GAT CAG CTG ATA GCT ACC ACA GTG CTA CAG ACT GGA ACA AAA ACC CTG CT 3′) and KY29 (5′ TAG CAG GGT TTT TGT TCC AGT CTG TAG CAC TGT GGT AGC TAT CAG CTG AT 3′). The 23-substrate for the initial rate assay was made by annealing KY36 (5′ GAT CAG CTG ACA GTA GCA CAG TGG TAG TAC TCC ACT CTC TGG CTG TAC AAA AAC CCT GCT 3′) and KY37 (5′ TAG CAG GGT TTT TGT ACA GCC AGA GAG TGG AGT ACT ACC ACT GTG CTA CTG TCA GCT GAT 3′).
Biotinylated oligonucleotides were synthesized by MWG Biotech (High Point, N.C.). The biotin group is located at the 3′ end of the bottom strand of each biotinylated DNA substrate (Fig. 1A). The biotinylated 12-substrate was made by annealing KY108 (5′ CCC TTC CTT GAT CAG CTG ATA GCT ACC ACA GTG CTA CAG ACT GGA ACA AAA ACC CTG CT 3′) and KY109 (5′ AGC AGG GTT TTT GTT CCA GTC TGT AGC ACT GTG GTA GCT ATC AGC TGA TCA AGG AAG GGX 3′) (X = biotin). The biotinylated 23-substrate was made by annealing KY110 (5′ CTG ACA GTA GCA CAG TGG TAG TAC TCC ACT CTC TGG CTG TAC AAA AAC CCT GCT 3′) and KY112 (5′ AGC AGG GTT TT T GTA CAG CCA GAG AGT GGA GTA CTA CCA CTG TGC TAC TGT CAG 3′). The nonbiotinylated 12-substrate used in the immobilized cleavage assay was made by annealing KY108 and KY124 (which differs from KY109 by not having a 3′ biotin group). The nonbiotinylated 23-substrate used in the immobilized cleavage assay was made by annealing KY110 and KY111 (which differs from KY112 by not having a 3′-biotin group). The DNA substrates have the biotin group located at the 3′ end of the bottom strand (Fig. 1A).
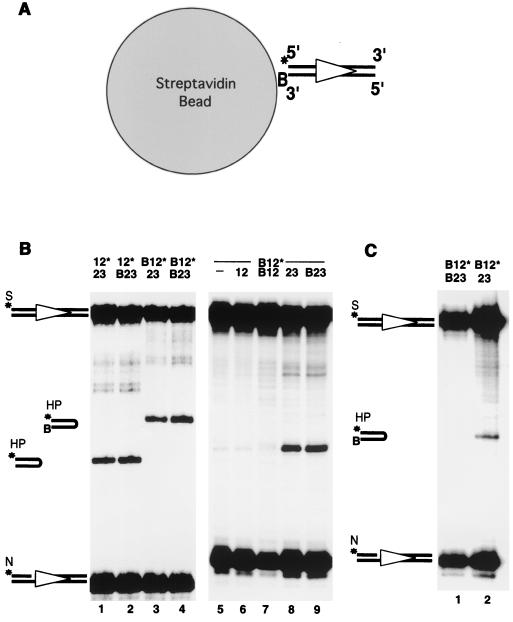
FIG. 1.
Cleavage with immobilized DNA substrate indicates that nicking is independent of synapsis. (A) Immobilization of DNA substrates on streptavidin agarose beads. The 12-substrate is depicted by an open triangle. B designates the biotin group. A star indicates the position of the radioisotope label. (B) Biotinylation does not interfere with 12/23 regulated in vitro nicking (N) and hairpin (HP) formation. S, substrate. All panel B reactions were performed in the absence of streptavidin agarose beads. The combinations of 12- and 23-substrates are indicated on the top of the gels. A dash indicates no DNA substrate. Other symbols are the same as described above. (C) Nicking and hairpin formation on an immobilized DNA substrate. Lane 1, both 12- and 23-substrates were biotinylated and immobilized on the streptavidin agarose beads. Lane 2, biotinylated 12-substrate was immobilized on the beads. Freely diffusible nonbiotinylated 23-substrate was added separately after the immobilization of the 12-substrate.
Oligonucleotides were labeled at the 5′ end with [γ-32P]ATP (3,000 Ci/mmol) (New England Nuclear Research Products, Boston, Mass.) and T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) according to the manufacturer's instructions. Unincorporated radioisotope was removed by using G-25 Sephadex (Amersham/Pharmacia, Piscataway, N.J.) spin column chromatography. To make the double-stranded DNA substrate, labeled oligonucleotides were mixed with an equal amount of unlabeled complementary oligonucleotides in a buffer containing 10 mM Tris-hydrochloride, pH 8.0, and 100 mM NaCl. The mixture was heated at 95°C for 5 min and allowed to cool down to room temperature for 1 h.
Protein expression and purification.
Fusion proteins of maltose-binding protein (MBP) and core regions of RAG1 (amino acids 384 to 1008) and RAG2 (amino acids 1 to 388) were expressed and purified from baculovirus-infected insect cells as previously described (30, 47, 51). Glutathione S-transferase (GST)-fused truncated mouse RAG1 (amino acids 330 to 1040) and RAG2 (amino acids 1 to 383) were coexpressed in the human epithelial cell line 293T and purified as previously described (5, 39, 49). C-terminally truncated mouse high-mobility-group 1 protien (HMG1) was expressed in bacteria as a six-histidine-tagged protein and purified over a Ni-nitrilotriacetic acid column (Qiagen Inc., Valencia, Calif.) (51). Protein concentrations were determined using the Bradford method (Bio-Rad, Hercules, Calif.). The molar concentration of the RAGs was calculated assuming two RAG1 and two RAG2 polypeptides (tetramer). (In this regard, it is noteworthy that recent experiments indicate a trans cleavage mechanism by a complex with two RAG1s and one or two RAG2s [P. Swanson, personal communication], suggesting a trimer or tetramer stoichiometry.)
Nicking and hairpinning of DNA substrates bound on streptavidin beads.
Exactly 0.2 pmoles of biotinylated DNA substrates in 100 μl of buffer (25 mM morpholinepropanesulfonic acid [pH 7.0], 5 mM MgCl2, 30 mM KCl, 30 mM potassium glutamate) was mixed with 1 μl of streptavidin-agarose suspended in 100 μl of the same buffer and incubated at 37°C for 30 min. The beads were collected by low-speed centrifugation and washed twice with 100 μl of cleavage buffer. The cleavage reaction was initiated by the addition of 100 nM HMG1 and 200 ng of the RAG proteins and incubated for 30 min at 37°C. The mixture was then extracted with phenol and chloroform followed by ethanol precipitation. The DNA pellet was redissolved in 10 μl of formamide, heated for 5 min at 100°C, and then immediately quenched with ice water.
Initial rate of nicking.
A 10-μl reaction mixture containing 25 mM morpholinepropanesulfonic acid (pH 7.0), 5 mM MgCl2, 30 mM KCl, 30 mM potassium glutamate, 100 nM HMG1, 10 nM RAG proteins, 2 nM 32P-labeled 12-substrate, and various amounts of unlabeled identical 12-substrate was incubated for periods of up to 5 min at 37°C. The reaction was stopped by the addition of 0.1% sodium dodecyl sulfate and 20 mM EDTA. The mixture was then extracted with phenol and chloroform followed by ethanol precipitation. The DNA pellet was redissolved in 10 μl of formamide, heated for 5 min at 100°C, and then immediately quenched with ice water.
Constants in the kinetic equations were determined by curve fitting using DeltaGraph v4.5 (SPSS Inc., Chicago, Ill.).
Denaturing polyacrylamide gel electrophoresis.
Reaction products were separated on 15% polyacrylamide gels containing 7 M urea in 1× Tris-borate-EDTA buffer. Gels were visualized by autoradiography by using a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager 445SI and quantified with ImageQuaNT software (version 1.0).
RESULTS
Nicking does not require synapsis.
As a first test of a synaptic requirement for nicking, we immobilized the biotinylated 12-substrate on streptavidin agarose beads. The amount of biotinylated 12-substrate loaded onto the streptavidin beads was less than 1% of the binding capacity, ensuring that each DNA molecule tethered on the beads was separated maximally from other DNA molecules (Fig. 1A). This allowed us to perform the nicking and hairpinning assay under conditions such that synapsis was prevented.
In free solution, the biotinylated DNA substrates undergo nicking and hairpin formation with an efficiency comparable to that of their nonbiotinylated counterparts (Fig. 1B, lanes 1 to 4). When biotinylated substrates are immobilized on the streptavidin beads in the presence of free partner signals, they still form hairpins (Fig. 1C, lane 2). Biotinylation of the DNA substrates does not interfere with the 12/23 rule of this reaction (Fig. 1B, compare lanes 5, 6, and 7 to lanes 8 and 9). In addition, a bound biotinylated 12-substrate mixed with a free 12-substrate does not result in hairpin formation (data not shown), assuring that the immobilization does not permit the 12/23 rule to be violated.
When both 12- and 23-substrates were immobilized on the beads, although nicking of the 12-substrate was evident, no hairpin formation could be detected (Fig. 1C, lane 1). The lack of hairpinning demonstrated that immobilization of the DNA on the streptavidin beads was able to block synapsis. When 23-substrate was nonbiotinylated and, therefore, free to synapse with the tethered 12-substrate, we detected not only the nicked product but also the hairpinned product (Fig. 1C, lane 2). This indicated that the tethered 12-substrate was able to undergo hairpinning when synapsis was possible. In contrast to hairpin formation, nicking of the 12-substrate was not affected when synapsis was blocked (Fig. 1C, lane 1), indicating that nicking does not require synapsis. In corresponding experiments, normal levels of nicking could be detected on bead-immobilized 23-substrates in the absence of 23/23 synapsis (data not shown).
From these results, we conclude that nicking does not require and is not stimulated by synapsis of any two sites in V(D)J recombination. Efficient hairpin formation, on the other hand, does require synapsis between a 12- and a 23-RSS. This lack of dependence of nicking on synapsis is unique to V(D)J recombination, because other transposition systems require synapsis for any catalysis to initiate (15, 31, 33, 38, 41, 48).
Initial rate kinetics for the nicking step.
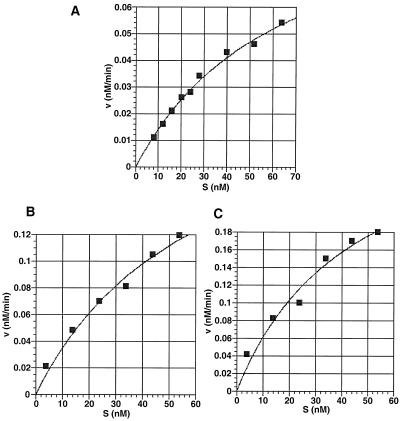
If nicking is independent of synapsis, kinetically it should be a unireactant enzymatic reaction (in which the enzyme binds one substrate at a time and acts on it catalytically), and it should fit the rate equation v = kN[E0][S]/(Km+[S]). In this equation, kN is the catalytic constant for nicking, Km is the Michaelis-Menten constant for the binding of the 12- or 23-substrate, [E0] is the concentration of the total active RAGs, and [S] is the concentration of the free 12- or 23-substrate. To determine if the unireactant kinetic model fits the nicking reaction, we measured the initial rate of the nicking reaction at different substrate concentrations. The substrates used in this assay contain consensus RSSs (18) and optimal coding end sequences for maximal nicking and hairpinning (53). When plotting the initial velocity against the substrate concentration, we found that the data fit the kinetic model of a unireactant enzyme-catalyzed reaction quite well (Fig. 2). The constants for the kinetic equation were determined by curve fitting (Table 1). The initial rate measurements were performed with both GST-fused RAG proteins (Fig. 2A) and MBP-fused RAG proteins (Fig. 2B and C). Importantly, we found that the type of fusion protein does not alter the unireactant nature of the enzyme-catalyzed reaction.
FIG. 2.
Initial rates of nicking as a function of substrate concentration. The plot of the initial rate (v0) against the initial 12-substrate or 23-substrate concentration (S0). (A) Nicking of the 12-substrate by the GST-RAGs. (B) Nicking of the 12-substrate by the MBP-RAGs. (C) Nicking of the 23-substrate by the MBP-RAGs. Curve fitting was done with DeltaGraph 4.5, and the kinetic constants of best fit are listed in Table 1.
TABLE 1.
| DNA substrate | Coding end sequence | K12 or K23 (nM) | kN (min−1) |
|---|---|---|---|
| KY28/29 | 5′-TAGCTAC-12RSS-3′ | 60 | 0.6 |
| KY36/37 | 5′-ACAGTAG-23RSS-3′ | 67 | 0.5 |
Coding end sequences in the second column represent the top strand sequence (Fig. 1). K12 and K23 are the Michaelis-Menten constants (Km) for the 12- and 23-substrate, respectively. kN is the catalytic constant (turnover number) for nicking. For MBP-RAGs, K12 = 59 nM and kN12 = 0.07 min−1; K23 = 45 nM and kN23 = 0.13 min−1.
For a 23-substrate, nicking is also a kinetically unireactant reaction (Fig. 2C). The Km (and kcat) is very similar to that of the 12-substrate (Table 1). Therefore, the difference between a 12- and a 23-substrate does not significantly alter the nicking constants. We have done time course studies of nicking in which a 12-substrate is alone or 12- and 23-substrates are both present (53). The nicking efficiencies were indistinguishable from each other. Hence, whether the reaction mixture includes only one or both types of substrates, the nicking kinetics show the same unireactant behavior.
Burst kinetics and functional stoichiometry.
The maximal initial rate, Vmax, of a particular enzymatic reaction depends on the enzyme concentration. The catalytic constant, or kcat (named kN for nicking), which reflects the catalytic efficiency, can be deduced based on the equation Vmax = kcat [E]. However, it is difficult to determine the actual RAG protein concentration in a cleavage reaction. First, the stoichiometry of the active RAG complex has not been clear (4, 10, 44); this means that the real molar concentration cannot be directly deduced from the measured protein concentration. Second, the RAG preparation is not homogeneous because it consists of two gene products that are usually coexpressed. It can contain RAG1-RAG2 tetramers, RAG1-RAG2 dimers, RAG1-RAG1 dimers, and RAG2 oligomers (4, 5). Although it is reported that a tetramer of two RAG1 and two RAG2 molecules is the active RAG complex, it is not currently possible to purify the tetramer in the quantities necessary for kinetic studies (T. Bailin and M. Sadofsky, personal communication).
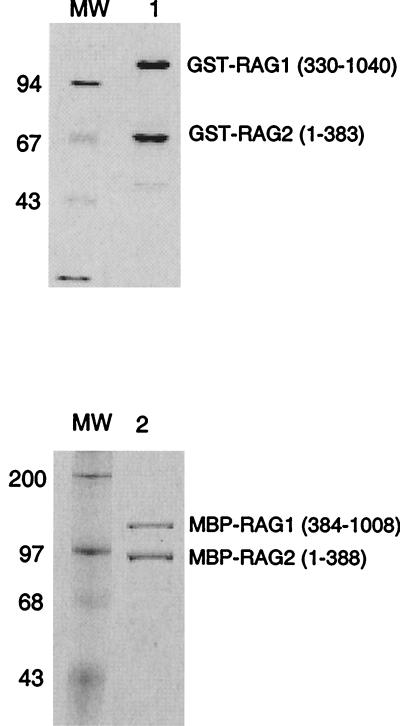
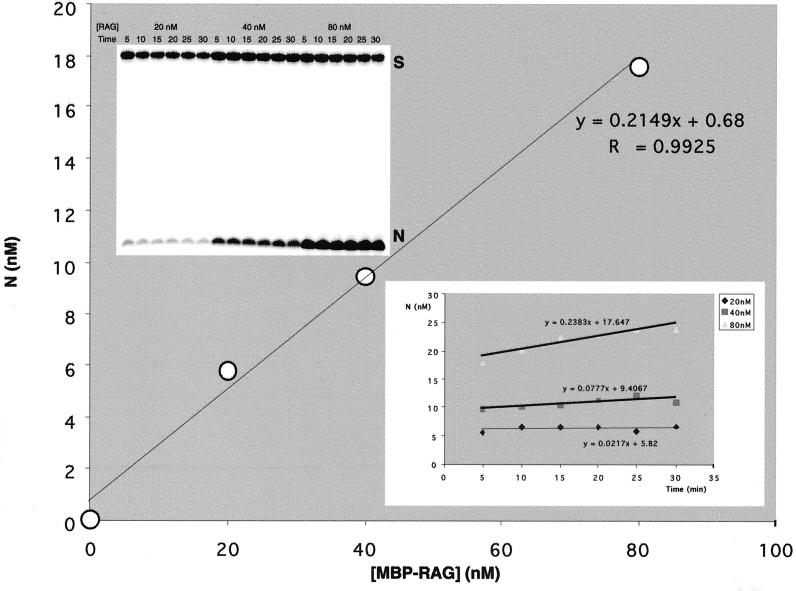
It is important to note that regardless of its absolute stoichiometric composition, the fraction of the RAG protein preparation that is active can be accurately determined. Coomassie staining of the purified RAG proteins used here showed that there was not marked proteolysis or contamination with other proteins (Fig. 3). To determine the concentration of the active RAG proteins, we used the burst kinetic assay described previously (7). Here, the RAG proteins were preincubated with the 12-substrate in Ca2+, which allows protein-DNA binding (20, 44). Then Mg2+ was added to initiate the nicking reaction (Fig. 4, upper inset gel). For each RAG concentration, the initial burst of the reaction was determined by extrapolating the accumulation of the nicked product back to time zero (Fig. 4, lower inset). The product bursts were then plotted as a function of the enzyme concentration; the molarity can be determined from the slope of this secondary plot (Fig. 4). For the MBP-RAG proteins, we found that 21.5% of the RAG protein was active, based on a RAG tetramer stoichiometry (Fig. 4). We also analyzed our GST-fused RAG proteins and found that 3.2% of the GST-RAG proteins were active (data not shown). Therefore, the percentage of active RAG proteins does not dramatically affect its kinetic behavior over the range of 3.2 to 21.5%. Moreover, the changing of the N-terminal fusion domain also does not markedly alter the kinetics of nicking.
FIG. 3.
Coomassie staining of the RAG fusion proteins used in kinetic studies. The GST-RAGs were purified using glutathione agarose (see Materials and Methods). The MBP-RAGs were purified using nickel-nitrilotriacetic acid resin followed by amylose resin (see Materials and Methods). Molecular weight (MW) markers, given in thousands, are shown on the left side of the gel. The position of each RAG protein band is indicated on the right side of the gel.
FIG. 4.
Determining the active RAG concentration by burst kinetics. The top inset shows the nicking of a 12-substrate with 20, 40, and 80 nM total concentrations of RAGs for a 30-min time course. These results were graphed as the nicked product versus time (lower inset); the y intercepts from this plot are the burst at each enzyme concentration. The major figure shows the burst (y intercept from the lower inset plot) as a function of the RAG concentration; the slope of this plot gives the fraction of active MBP-RAGs in the protein preparation. The slope is 0.215, indicating that 21.5% of the RAGs are catalytically active. CaCl2 was the source of the 1 mM Ca2+, and MgCl2 was the source of the 5 mM Mg2+.
In addition to the burst kinetics, we used an independent functional stoichiometry assay (36) to determine the active fraction of our GST-RAG proteins. In this method, we plotted the initial rate as a function of the RAG concentration. The accumulation of nicked product plateaus at between 60 and 85% of the total input substrate concentration (data not shown). When we plotted the initial rate as a function of the RAG concentration, the maximal initial rate (Vmax) could be achieved. For a fixed substrate concentration (0.4 nM), the initial rate plateaus at a RAG concentration of approximately 20 nM (data not shown). This indicates that the active fraction of the RAGs is about 2.0%. This is consistent with the results obtained with the burst kinetic study (3.2%) for the GST-RAG proteins.
DISCUSSION
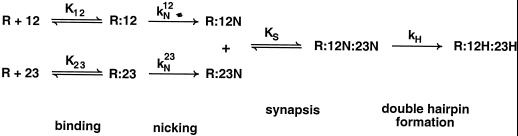
We have shown that immobilization of DNA substrates on streptavidin beads, which blocks synapsis, still permits their nicking but not their hairpinning. These are the first data to demonstrate that synapsis of two signals is not required for the nicking step, making it unique in this respect among transposition-type reactions. Our results demonstrate that the nicking step of V(D)J recombination occurs as a one-substrate enzymatic reaction. The nicking of the 12-substrate alone is a unireactant enzyme-catalyzed reaction, and the same is true for the nicking of the 23-substrate alone. This indicates that even if a RAG complex binds to a 12- and a 23-RSS simultaneously, it would still nick them independently. Based on the results here, we can infer a kinetic scheme for the RAG-mediated nicking and hairpinning during the initiation of V(D)J recombination (Fig. 5).
FIG. 5.
Kinetic scheme for the binding, nicking, synapsis, and hairpin formation steps mediated by the RAG complex (RAG1, RAG2, and HMG1). 12 represents a 12-substrate (12RSS and adjacent DNA). Equilibrium constants are depicted with K, and rate constants are depicted with k. R represents the active RAG complexes. R:12 is the RAG complex bound to the substrate. 12N is the nicked form of the 12-substrate. 12H is the hairpinned form of the 12-substrate. R:12N:23N is the synaptic complex of nicked 12- and 23-substrates.
Comparison to other studies on nicking in V(D)J recombination.
It has been shown that in a solution containing only 12-substrates, those 12-substrates could undergo nicking (8, 9, 14, 46, 47). However, in all of those previous studies, it was unclear whether two 12-substrates might synapse to permit the nicking step or the 12-substrates were being nicked individually (and prior to any synapsis). The same applies to reactions involving only 23-substrates. Our immobilization and kinetics studies clearly indicate that the nicking step occurs as a one-substrate reaction, even when both 12- and 23-substrates are present.
It is interesting to compare our results with a previous study that examined time courses of nicking in an effort to understand the relationship between nicking and synapsis (9). In that study, DNA targets containing two signals on each DNA molecule were used. Most nicking was asynchronous, because nicking occurs at one signal, even though the other signal on the same DNA molecule was unnicked. The total nicking of a 12/12 DNA target occurred at a rate similar (within twofold) to that for a 12/23 target. All of these results are consistent with our findings that the nicking step is a unireactant enzyme-catalyzed process.
In that previous study (9), a minor fraction of the total nicking was termed synchronous because both signals were nicked. It was argued that synapsis stimulates nicking, because a 13-fold decrease in synchronous nicking was observed when the intersignal space was reduced from 279 to 89 bp. We assume that synapsis is inhibited at the shorter intersignal length. However, inferences based on these two-signal substrates have the following complications. First, the apparent synchronized nicking may not actually be a coordinated event upon synapsis. That is, it may not be synchronous when examined at a higher temporal resolution. The majority of the nicking is asynchronous and shows little difference among the 12/12, 23/23, and 12/23 substrates. Second, the changing of the intersignal distance does not rule out the possibility of steric interference when two RAG complexes bind to two closely located RSSs. Such interference could readily explain the lower rate of double nicking of the 89-bp intersignal substrate. It is important to note that the previous study does not directly address the issue of whether synapsis is required for nicking, because two different DNA molecules might be synapsing prior to nicking under those experimental conditions.
Physiologic significance of a unireactant nicking step.
Chromatin structure regulates the local accessibility of RAG recombinases (6, 16, 32, 43). The immunoglobulin intronic enhancers have been found to be essential for opening up the chromatin (6, 11, 16, 22, 23, 32, 43). The fact that DJ recombination typically precedes the V to DJ recombination at the immunoglobulin H locus may reflect a gradual chromosomal structural change initiated from the enhancer near the JH region and extending into the distal V cluster. Our determination that nicking can occur without synapsis means that it is quite possible that the JH segments may exist in the nicked form before any VH and D segments become accessible to the RAGs.
In addition to V, D, and J segments, theoretically there are thousands of cryptic sites that are not defined in the genome. A particular nicking event can result from an isolated nicking, a synaptic nicking with any other 12- or 23-RSS, or a synaptic nicking upon pairing with any of the thousands of cryptic sites. Because of this, our inference concerning nicking without synapsis in the genome is not directly testable.
Recently, we showed that the nicking step is rate limiting when the coding end sequence adjacent to the heptamer is suboptimal (TT-heptamer) (53). This feature can explain why some V, D, or J segments are used less frequently than others (13). Our observation here that nicking is independent of synapsis suggests that the V, D, and J segments that have optimal signals and coding ends (for nicking) will exist in the nicked form longer than those with the suboptimal ones (53). The greater fraction of the time in the nicked state might be expected to result in a greater probability that these nicked forms could then enter into synapsis and coordinated hairpin formation. This greater probability could be an important determinant of which V, D, or J segments contribute to the immune repertoire (53). Nicking in a manner that does not rely on synapsis may increase the overall number of V, D, and J segments that are nicked, thereby allowing a wider array of coding segments to contribute to the immune repertoire.
What happens if a RAG complex makes a nick and then dissociates? DNA ligase I is the most abundant ligase in eukaryotic cells, and it is very effective at ligating nicks (28). Hence, religation may occur quite quickly. At some frequency, such nicks may be predisposed to the types of chromosomal instabilities that often involve antigen receptor loci.
Utility of a kinetic description of the nicking step.
It is useful to compare the fraction of active RAGs from the functional stoichiometry and burst kinetics with electrophoretic mobility shift results using the same RAG preparation. When kN is sufficiently small, the dissociation constant KD is equal to Km. It is not possible to determine KD by a gel mobility shift without knowing the active RAG concentration (i.e., the concentration of RAGs that are able to bind the DNA substrate). If we substitute Km for KD, then from our previous electrophoretic mobility shift assay (see Fig. 5 of reference 53), it can be estimated that maximally about 7.5% of the GST-RAG proteins are able to form a complex with DNA substrates ([E] = KD[ES]/[S] ≈ Km[ES]/[S]). This sets an upper limit for the active GST-RAG fraction, because KD must be smaller than Km. This is consistent with our burst kinetics (3.2%) and functional stoichiometry (2.0%) results on the GST-RAGs, which indicate that only a small fraction of the RAG protein preparation is active. Corresponding estimations can be done for the MBP-RAGs. Mobility shift analysis places an upper limit of 50% on the fraction of RAGs active for substrate binding (data not shown). The burst kinetics analysis determination of 21.5% is in line with this estimate. Binding and catalytic activity by only a fraction of the RAG protein preparation probably explain why efficient nicking and hairpinning typically require more RAG protein than DNA substrate (5, 19, 25, 47, 51).
The GST- and MBP-RAGs gave kinetic values that were reasonably similar to each other (Table 1). The Kms were not significantly different for the two fusion forms. There was a 4-fold and an 8.5-fold reduction in the catalytic constant (kcat) with MBP-RAGs for nicking of the 23- and 12-substrates, respectively. These relatively small differences could be due to the steric interference by the bulky MBP fusion. Nevertheless, the constants are still reasonably close given the complete exchange of a 26-kDa GST fusion domain for a 46-kDa MBP fusion domain.
Interestingly, the catalytic constant of the MBP- and GST-RAGs (0.1 and ∼0.6 min−1, respectively) is within the same range as those for some other endonucleases, including the homing-type endonuclease from Pyrobaculum organotrophum (2 min−1) (29), the restriction enzyme BamHI (0.72 min−1 and 1.86 min−1) (17), and EcoRI (13.2 min−1) (52). It is slower than those for some other restriction enzymes, such as EcoRV (63 min−1) (50).
Comparison of V(D)J recombination to other transposition and site-specific recombination reactions.
All other site-specific and transposition recombination reactions require synapsis prior to initiation of the first catalytic step [nicking in the case of V(D)J recombination]. One might have expected that synapsis would also be essential for V(D)J recombination based on this evolutionary trend. Synapsis ensures that nicking at the two recombination sites is coordinated. However, V(D)J recombination is the first such reaction to break this rule. We can only speculate as to the reasons for this deviation from other similar reactions. First, the effects on the immune repertoire discussed earlier may be important forces that may have favored this evolutionary deviation from the other transposition reactions. Second, V(D)J recombination requires that the recombinase find the signals in a genome which is more complex than any other site-specific or transposition reaction. In many transposition reactions, such as in the case of retroviruses, the transposase is packaged within the nucleocapsid; when double-stranded DNA is generated from the infecting RNA, the transposase must bind to the ends of a genome that is only kilobases in length. In the case of prokaryotic transposases, the bacterial genome is relatively small. Based on nicking without synapsis, a potentially wider number of V, D, and J segments may contribute to the repertoire, as has been mentioned. The multisite nature of this transposition reaction and its use in an immune defense process make it advantageous to open the first step of this process to as many V, D, and J segments as possible. For this reason, a unireactant first step may have been a distinct evolutionary advantage.
ACKNOWLEDGMENTS
We are indebted to Irwin H. Segel (University of California, Davis) and Jay Winkler (Caltech, Pasadena, Calif.) for advice on enzyme kinetics. We thank Chih-Lin Hsieh and Fred Chedin for reviewing the manuscript.
This research was supported by NIH grants to M.R.L. M.R.L. is the Rita and Edward Polusky Basic Cancer Research Professor.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Aldaz H, Schuster E, Baker T. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell. 1996;85:257–269. doi: 10.1016/s0092-8674(00)81102-2. [DOI] [PubMed] [Google Scholar]
- 3.Arciszewska L K, Drake D, Craig N L. Transposon Tn7. cis-acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- 4.Bailin T, Mo X, Sadofsky M J. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by the recombination activating genes RAG1 and RAG2. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 6.Cedar H, Bergman Y. Developmental regulation of immune system gene rearrangement. Curr Opin Immunol. 1999;11:64–69. doi: 10.1016/s0952-7915(99)80012-0. [DOI] [PubMed] [Google Scholar]
- 7.Cornish-Bowden A. Fundamentals of enzyme kinetics. London, England: Butterworths; 1979. [Google Scholar]
- 8.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 9.Eastman Q M, Schatz D G. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eastman Q M, Villey I J, Schatz D G. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrester W C, Genderen C V, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 12.Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 13.Gerstein R M, Lieber M R. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 14.Grawunder U, Lieber M R. A complex of RAG-1 and RAG-2 persists on the DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res. 1997;25:1375–1382. doi: 10.1093/nar/25.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haniford D B, Benjamin H W, Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991;64:171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- 16.Hempel W M, Leduc I, Mathieu N, Tripathi R, Ferrier P. Accessibility control of V(D)J recombination: lessons from gene targeting. Adv Immunol. 1998;69:309–352. doi: 10.1016/s0065-2776(08)60610-0. [DOI] [PubMed] [Google Scholar]
- 17.Hensley P, Nardone G, Chirikjian J, Watsney M E. The time-resolved kinetics of superhelical DNA cleavage by BamHI restriction endonuclease. J Biol Chem. 1990;265:15300–15307. [PubMed] [Google Scholar]
- 18.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1067. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 19.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 20.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 21.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein T, Forrester W, Qiu R-G, Grosschedl R. The immunoglobulin mu enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T, Forrester W, Fernandez-Herrero L, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy A K, Guhathakurta A, Kleckner N, Haniford D B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 25.Kim D R, Oettinger M A. Functional analysis of coordinated cleavage in V(D)J recombination. Mol Cell Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieber M R. Pathologic and physiologic double-strand breaks: roles in cancer, aging, and the immune system. Am J Pathol. 1998;153:1323–1332. doi: 10.1016/s0002-9440(10)65716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieber M R. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl T, Barnes D E. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 29.Lykke-Andersen J, Thi-Ngoc H P, Garrett R A. DNA substrate specificity and cleavage kinetics of an archaeal homing-type endonuclease from Pyrobaculum organotrophum. Nucleic Acids Res. 1994;22:4583–4590. doi: 10.1093/nar/22.22.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBlane J F, Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 31.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 32.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. k chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy J, Goff S. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oettinger M A, Schatz D G, Gorka C, Baltimore D. Rag-1 and Rag-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 35.Plasterk R. Ragtime jumping. Nature. 1998;394:718–719. doi: 10.1038/29389. [DOI] [PubMed] [Google Scholar]
- 36.Roman L J, Kowalczykowski S C. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 1989;28:2863–2873. doi: 10.1021/bi00433a018. [DOI] [PubMed] [Google Scholar]
- 37.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 38.Sakai J, Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawchuk D, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 41.Segall A M. Analysis of higher order intermediates and synapsis in the bent-L pathway of bacteriophage site-specific recombination. J Biol Chem. 1998;273:24258–24265. doi: 10.1074/jbc.273.37.24258. [DOI] [PubMed] [Google Scholar]
- 42.Sheehan K M, Lieber M R. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol Cell Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sleckman B P, Gorman J, Alt F W. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 44.Swanson P C, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 46.van Gent D, Hoim K, Paull T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 48.Wei S Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weis-Garcia F, Besmer E, Sawchuk D J, Yu W, Hu Y, Cassard S, Nussenzweig M, Cortes P. V(D)J recombination: in vitro coding joint formation. Mol Cell Biol. 1997;17:6379–6385. doi: 10.1128/mcb.17.11.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wenz C, Hahn M, Pingoud A. Engineering of variants of the restriction endonuclease EcoRV that depend in their cleavage activity on the flexibility of sequences flanking the recognition site. Biochemistry. 1998;37:2234–2242. doi: 10.1021/bi9719197. [DOI] [PubMed] [Google Scholar]
- 51.West R B, Lieber M R. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol Cell Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright D J, Jack W E, Modrich P. The kinetic mechanism of EcoRI endonuclease. J Biol Chem. 1999;274:31896–31902. doi: 10.1074/jbc.274.45.31896. [DOI] [PubMed] [Google Scholar]
- 53.Yu K, Lieber M R. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol Cell Biol. 1999;19:8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]