Abstract
Saliva has become an attractive body fluid for on-site, remote, and real-time monitoring of oral and systemic health. At the same time, the scientific community needs a saliva-centered information platform that keeps pace with the rapid accumulation of new data and knowledge by annotating, refining, and updating the salivary proteome catalog. We developed the Human Salivary Proteome (HSP) Wiki as a public data platform for researching and retrieving custom-curated data and knowledge on the saliva proteome. The HSP Wiki is dynamically compiled and updated based on published saliva proteome studies and up-to-date protein reference records. It integrates a wide range of available information by funneling in data from established external protein, genome, transcriptome, and glycome databases. In addition, the HSP Wiki incorporates data from human disease–related studies. Users can explore the proteome of saliva simply by browsing the database, querying the available data, performing comparisons of data sets, and annotating existing protein entries using a simple, intuitive interface. The annotation process includes both user feedback and curator committee review to ensure the quality and validity of each entry. Here, we present the first overview of features and functions the HSP Wiki offers. As a saliva proteome–centric, publicly accessible database, the HSP Wiki will advance the knowledge of saliva composition and function in health and disease for users across a wide range of disciplines. As a community-based data- and knowledgebase, the HSP Wiki will serve as a worldwide platform to exchange salivary proteome information, inspire novel research ideas, and foster cross-discipline collaborations. The HSP Wiki will pave the way for harnessing the full potential of the salivary proteome for diagnosis, risk prediction, therapy of oral and systemic diseases, and preparedness for emerging infectious diseases.
Database URL: https://salivaryproteome.nidcr.nih.gov/
Keywords: saliva, biofluid, proteins, database, omics, diagnostics
Introduction
Saliva is a complex exocrine fluid that is mainly composed of salivary gland secretions but with additional contributions from various extrinsic sources including blood, gingival crevicular fluid, oral mucosa and immune cells, nasopharyngeal secretions, and microbes (Dawes et al. 2015). Salivary composition is highly dynamic because external and internal factors influence to varying and unpredictable degrees the relative contributions of these sources that make up whole-mouth saliva. Saliva facilitates mechanical preprocessing and enzymatic predigestion of food, including wetting of dry food, dissolving of taste components, forming a food bolus for chewing, and lubricating it for easier swallowing. Saliva also moisturizes the oral integuments and protects the enamel layer of teeth from erosion, abrasion, and attrition (Lendenmann et al. 2000; Ruhl 2012). In addition, saliva serves as a modulator of oral microbiome homeostasis by tolerating resident microbes in the mouth and shielding the host from intruding pathogens, thus becoming a vehicle of host defense (Cross and Ruhl 2018).
Saliva is desirable for point-of-care diagnostics because of its noninvasive and cost-effective sample collection (Bonne and Wong 2012). Its potential for monitoring oral and systemic diseases has recently gained increased attention during the current COVID-19 pandemic (Huang et al. 2020). Saliva has been effectively employed to detect the virus (Teo et al. 2021) and track the immune response to SARS-CoV-2 (Isho et al. 2020; Huang et al. 2021). Compared with blood, saliva is readily available even by laypersons, which makes it attractive for on-site, remote, and real-time monitoring of health and disease (Tabak 2007). Many blood plasma components are present in saliva, providing a potential noninvasive method for the diagnosis of systemic diseases (Pfaffe et al. 2011). One of the major obstacles, however, is the high biovariability of salivary proteins. Salivary flow, epithelial leakage, gingival and periodontal inflammation, and enzymatic degradation are among the main confounding parameters influencing salivary composition (Helmerhorst et al. 2018). Nevertheless, while diagnostic applications based on quantitative analysis of salivary proteins remain a challenge, several Food and Drug Administration–approved saliva-based tests have been developed, including ones for detection of HIV infection (OraQuick), drug use (Oratect), and SARS-CoV2 infection (SalivaDirect).
In 2008, a large-scale multicenter study identified more than 1,000 proteins in salivary gland secretions and defined the first comprehensive catalog of the human salivary proteome (Denny et al. 2008). Many additional proteome data sets of whole saliva and minor salivary gland secretions have been published since then (Siqueira et al. 2008; Bandhakavi et al. 2009; Jehmlich et al. 2013). With the availability of more advanced proteomics technology, newer data sets provide substantially richer information that allows for quantitative comparisons. As proteomics continues evolving, the scientific community needs a saliva-centered information platform to effectively deposit, annotate, analyze, and exchange the rapidly accumulating data and knowledge to further advance the field.
Various strategies have been leveraged to annotate large biological data sets (Stein 2001; Reeves et al. 2009). One strategy is to rely on dedicated curators for database maintenance, as exemplified by the Human Oral Microbiome Database, which catalogs more than 700 bacterial species in the human mouth and continues to expand (Escapa et al. 2018). An alternative strategy is community-wide open annotation or crowdsourcing (Giles 2007; Sparks et al. 2016; Griffith et al. 2017), with the wiki being one of the most popular frameworks to catalyze collaborative content creation. WikiPathways (Slenter et al. 2018) is an example of earlier, successful wiki-based efforts to promote community annotation of biological pathways using transcriptomics, proteomics, and metabolomics data. Similar wiki databases have been established for general gene and protein annotations (Mons et al. 2008; Good et al. 2012), specific model organisms (Stover et al. 2012; Michna et al. 2016), and biomedical applications (Cariaso and Lennon 2012; Ives et al. 2017).
To facilitate the translation of research findings into saliva-based diagnostic tests and therapeutics, we developed the Human Salivary Proteome Wiki (HSP Wiki) as a community resource for depositing, annotating, and investigating salivary proteomic data. The HSP Wiki platform integrates an array of information and analytic tools to catalyze knowledge dissemination and discovery. The platform is specifically designed for researchers and clinicians from multiple backgrounds interested in salivary gland biology, salivary diagnostics, proteomics, and bioinformatics.
Building the HSP Wiki
Data Conversion and Normalization
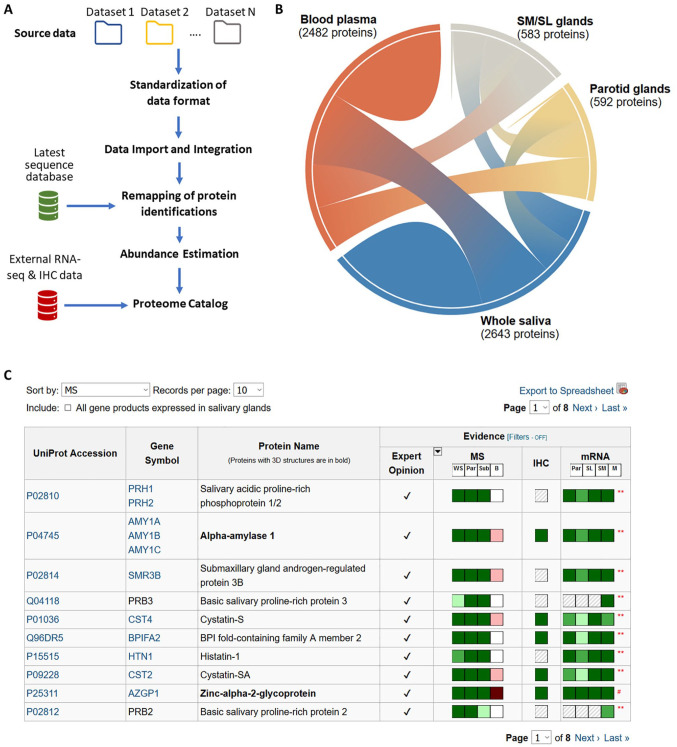
We developed a workflow to integrate and standardize mass spectrometry (MS) data generated by different research groups using different techniques and data formats (Fig. 1A). The first step was to prepare and convert data sets into a standard format that captures peptide and protein identifications and metadata necessary for unambiguous interpretation of its content (Deutsch et al. 2015). A standardized input system allows querying and comparison across different studies. Output results were remapped to a common set of identifiers. Earlier studies used the International Protein Index (IPI) database, which was decommissioned in 2011. Recent studies use protein sequences and identifiers from the UniProt Knowledgebase (UniProt Consortium 2019). Thus, the original IPI identifiers had to be converted to equivalent UniProt counterparts. In addition, periodic updates of sequence databases lead to revision of sequence identities and protein identifiers. For instance, some identifications originally mapped to hypothetical proteins and are no longer valid in the latest database. Therefore, in addition to preserving the original protein accessions and their links to UniProt identifiers, we also incorporated the capability to update the identification results by remapping them using the most up-to-date version of the reviewed protein sequences in UniProt/Swiss-Prot. For protein quantitation, we used spectral counts as a proxy measure for abundance levels of proteins in saliva, as many of the studies in the database lack direct quantitative information. Abundance levels were normalized by counting the total number of spectra produced in each experiment and the number of potential peptide sequences in a given protein sequence.
Figure 1.
Development of the human salivary proteome catalog. (A) Overview of the workflow developed to standardize and integrate data sets submitted in variable data formats and contents and to update identification results using the most up-to-date protein sequences. (B) Chord diagram to visualize the contribution of ductal secretions of the major salivary glands and blood plasma to the complex mixture of proteins in whole saliva. (C) Protein catalog table, which uses color indicators to depict different types of experimental and editorial evidence available in support of including corresponding proteins in the catalog. The color indicators include 3 levels of shades from dark to light, indicating the levels of protein abundance as estimated by mass spectrometry (MS) spectral counts, by immunohistochemistry (IHC), or by mRNA abundance (green color indicators). Striped patterns indicate that information is not available. White indicates an absence of protein or mRNA. For the display in this figure, the filter function has been applied to show only proteins that are upregulated in saliva based on glandular mRNA expression. B, blood plasma (red color indicators); M, minor salivary glands; MS, protein abundances in secretions; mRNA, RNA levels in tissues; Par, parotid gland; Sub, submandibular/sublingual glands; WS, whole saliva.
Database Structure and Integration
The HSP Wiki comprises 2 interconnected subdatabases. The first one stores experimental data at the local BioMart server, a data warehouse optimized for querying large data sets. Currently, the HSP Wiki encompasses more than 800 individual MS data acquisition files from 6 studies, covering whole saliva and the ductal secretions of parotid, submandibular, and sublingual glands both from healthy individuals and subjects suffering from oral cancer or Sjögren’s syndrome (Appendix Table 1). These files form the backbone of the salivary proteome catalog. The second subdatabase stores knowledge facts of salivary proteins and uses a data structure and interface similar to Wikipedia. Building on top of protein metadata from UniProt, we incorporated additional structural and expression data tailored to the specific interests of the salivary research community. These include RNA-Seq and immunohistochemistry-based tissue expression data from the Human Protein Atlas (Uhlén et al. 2015), MS-based abundance data of blood plasma proteins from Peptide Atlas (Schwenk et al. 2017), and glycan structures from UniCarb-DB (Campbell et al. 2014). Recently, transcriptomics data from the 3 major salivary glands have also been incorporated into the Wiki (Saitou et al. 2020). This diverse set of data empowers the community to interrogate salivary multimodal data sets, including gene expression at different levels, and to explore additional protein annotations (Fig. 1B).
Data Display
A comprehensive protein table forms the centerpiece of the HSP Wiki knowledgebase, which concisely and intuitively summarizes the complex information residing in the experimental subdatabase (Fig. 1C). The listing is compiled dynamically based on the proteomics data stored in the database, ensuring that the catalog remains as accurate as evidence permits. The table uses visual keys to indicate gene product abundances in salivary gland ductal secretions and whole saliva as estimated by RNA-seq, MS, and immunostaining. The goal is to provide rich, unbiased evidence to help users explore the dynamic and complex nature of saliva. For example, users can choose proteins with a high abundance in whole saliva but that lack expression in blood plasma.
Features of the HSP Wiki
Here, we highlight selected user-relevant features of the Wiki. For a comprehensive list of features and user guide, please visit our website at https://salivaryproteome.nidcr.nih.gov/public/index.php/Help:Contents.
User Interface and Tools
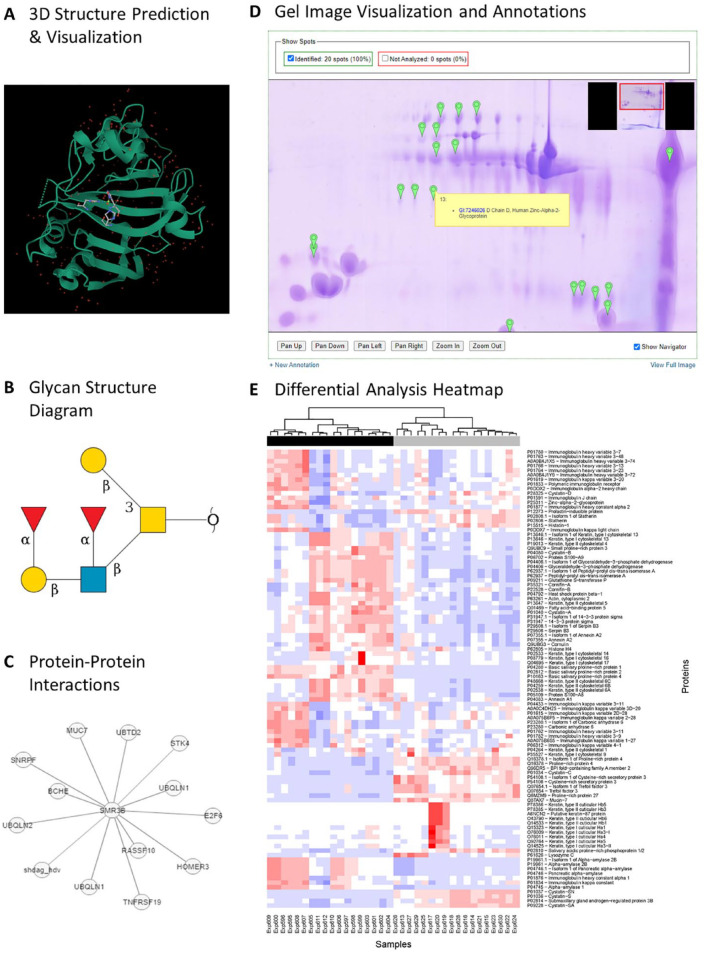
The HSP Wiki incorporates a wide range of tools for users to search and analyze different contents. Users can query the wiki contents using simple free text or an advanced search targeting specific fields; they can also search for experimental data by tissue types, disease, and other associated metadata. Many of the tools available in the wiki also leverage publicly accessible programming interfaces or open-source software developed by the National Center for Biotechnology Information, European Bioinformatics Institute, and other bioinformatics institutes. Common sequence analysis tools, such as BLAST and InterProScan, enable users to search for proteins with similar features or functional domains. Tools are also available to visualize and predict (by sequence homology) 3-dimensional (3D) structures (Fig. 2A). Glycan structure diagrams are shown for some proteins, such as Mucin 7 (Fig. 2B). In addition, the Wiki provides a search interface to IntAct (a curated database of experimentally determined protein and chemical interactions) to explore protein interactions and biological pathways (Fig. 2C).
Figure 2.
Selected features available for Human Salivary Proteome (HSP) Wiki users. (A) A tool for 3D protein structure visualization. (B) Glycan structures linked to mucin MUC7 and other salivary proteins have been imported from UniCarb-DB. (C) A network diagram displaying protein-protein interactions from proteins imported from the IntAct database. (D) A tool developed to visualize and annotate 1D and 2D gel images within the Wiki. (E) A heat map generated from a tool that performs differential abundance analysis of proteomic data sets stored in the Wiki that users can choose for comparisons.
We also developed custom tools for analyzing the experimental data stored in the Wiki. One tool allows for gel image analysis and annotation of both 1D and 2D images (Walz et al. 2006). Once gel files are uploaded, the processing pipeline will automatically detect lanes in 1D gel images, and any existing annotations will be imported (Fig. 2D). Users can also dynamically annotate the image with additional protein identification information at specific positions. Another custom tool is a differential expression analysis function that allows users to select experiments from the BioMart database and compare protein abundance levels between 2 groups of samples (Fig. 2E). More detailed usage examples are presented below in the form of a comparison between parotid and submandibular/sublingual gland proteins and of oral cancer biomarker discovery (see the section “Usage Examples of Analytic Tools”).
Data Upload, Knowledgebase Annotation
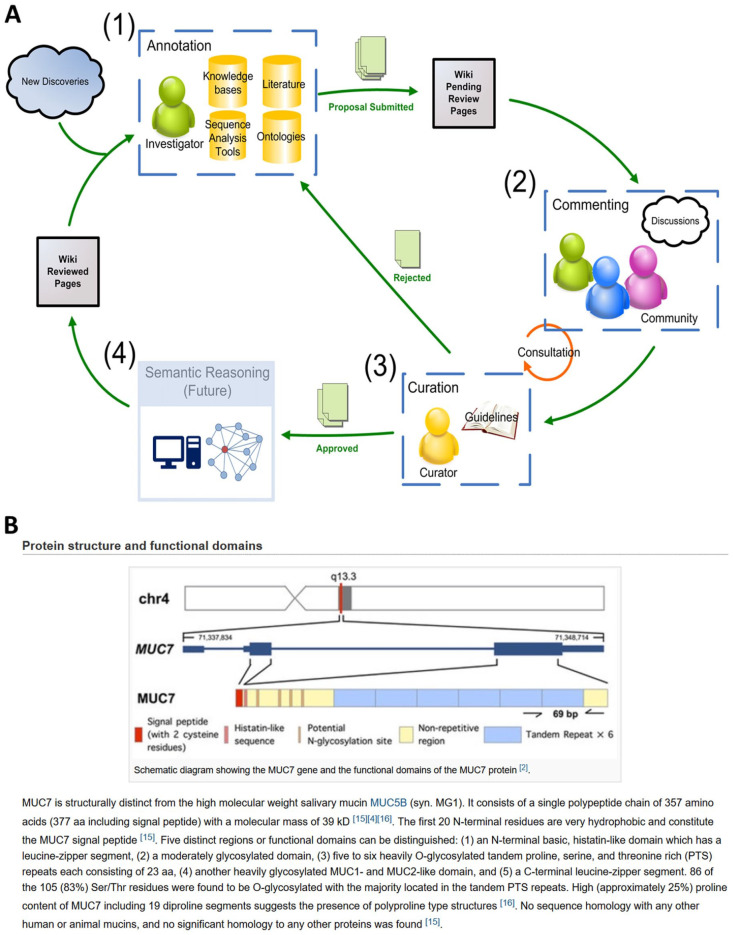
The HSP Wiki serves as an annotation and functional discovery platform for the salivary proteome. The entire HSP Wiki framework is built to support these functions. Figure 3A presents a schematic representation of the annotation workflow. The protein pages are seeded with UniProt annotations. Following the UniProt convention, annotations are divided into 2 sections, Comments and Features, which describe the biological properties and sequence position-specific information for a given protein, respectively. Any registered user can submit proposals to add, remove, or modify annotations and become an annotator. The Wiki provides many tools for annotators to gather evidence to support their annotations (see “User Interface and Tools” section above). Appendix Table 2 describes the individual fields that make up a protein annotation. The HSP Wiki follows the UniProt framework to be compatible with the prepopulated UniProt annotations. For some annotation types, the Description field can be populated with terms from designated ontologies. The annotator can look up the appropriate term by either searching or browsing the selected ontology from a pop-up menu. Currently, the Wiki has 13 ontologies and controlled vocabularies. The Annotation Source field tracks the source of each annotation. This information helps users to determine the origin of annotations, which could be derived from an automatic script or a specific author. This field may serve as an incentive for researchers to annotate the proteins they study, as others may use these credits to infer expertise or seek collaborations.
Figure 3.
Crowdsourcing opportunities available in the Human Salivary Proteome (HSP) Wiki. (A) Workflow for the annotation and curation process in the HSP Wiki that leverages the collective expertise of the research community. (1) A researcher has just finished a study and adds his findings to the Wiki. (2) After the proposal is submitted, the community will have the opportunity to register their feedback. (3) With expert knowledge on the subject matter, a curator reviews the proposal, taking into consideration the feedback gathered from the community. The curator may approve or reject all or parts of the proposal. If rejected, the researcher can resubmit the rejected changes after revisions. If approved, the new content will be incorporated into the Wiki. An additional step is possible, in which (4) the annotations in the Wiki can be fed to machine learning tools for discovering implicit relationships among entities. A curator will need to examine the output from these tools to ensure that the machine-generated knowledge is accurate before incorporating it into the Wiki. (B) As demonstrated in this screenshot of a sample expert commentary page, users with recognized expertise on certain proteins are invited to provide descriptions including unique properties of these proteins in saliva.
An annotator can put 1 or more annotations in a proposal basket and submit them as a single proposal for examination. When a proposal is submitted, affected pages will be locked with a message on top to indicate changes. Any user can view the proposed changes on the Proposed protein page, which is a copy of the original page with the proposed annotations incorporated and highlighted. Users can comment or debate about the proposed annotations using discussion forums within the Wiki.
Digital Curation
User-submitted contents, except invited commentaries (Fig. 3B), will go through a curation process. The curation workflow ensures that community annotations in the Wiki are accurate and supported by sound scientific evidence. Experts from different backgrounds form the curation committee. This team evolves dynamically based on the expertise and ability to contribute and improve the database. Members of the curation committee can view the status of all outstanding submissions and claim any proposals waiting for review. Once a proposal is claimed, the curator will inspect the proposal and communicate with the annotator. The status message on the affected pages will display the responsible curator’s name. The transparency of this process empowers collaborations and leverages the collective knowledge of the community. The curator will accept or reject the proposal in whole or in part and notify the annotator by email. Accepted changes are incorporated into the corresponding pages, and users can track revisions using the built-in page history feature.
Usage Examples of Analytic Tools
Differential Expression Analysis to Compare Glandular Salivary Proteomes
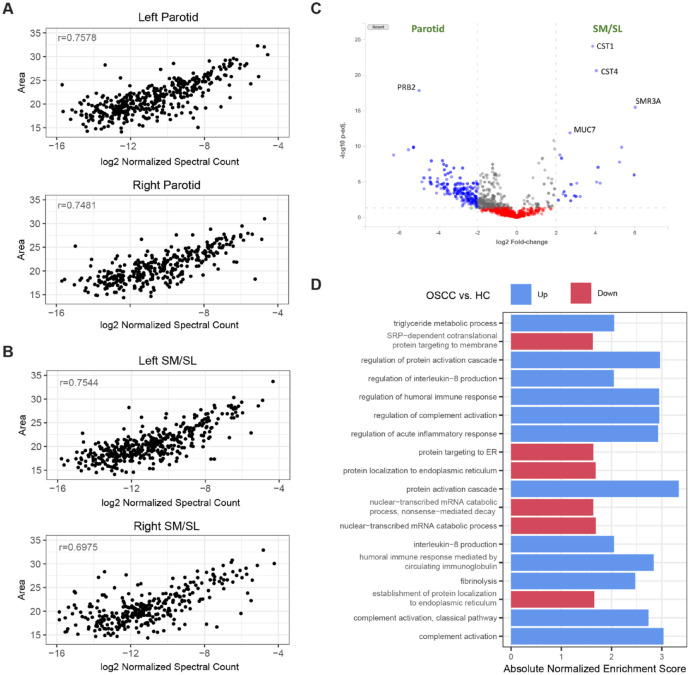
Users can employ the HSP Wiki differential expression analysis tool to facilitate exploration and secondary analysis of the MS data sets within the database, for instance, to compare protein levels in selected data sets. We demonstrate here how to use this tool to perform differential expression analysis of a recently published comparison of the parotid and submandibular/sublingual glands using liquid chromatography tandem MS (Schulte et al. 2019). Spectral count–based quantitation in the Wiki correlated well with quantitation based on peak area used in the original study (Fig. 4A, B). The HSP Wiki differential expression analysis tool replicated the main findings that cystatins-S, -SN, and mucin 7 are core submandibular/sublingual proteins, whereas PRB2 is highly associated with parotid secretions. The heat map and volcano plot produced by the tool highlight proteins that are significantly differentially expressed (Figs. 2D, 4C).
Figure 4.
Differential analyses comparing glandular ductal secretions and saliva samples from a biomarker discovery study demonstrate the capabilities of the Human Salivary Proteome (HSP) Wiki. (A, B) The correlation of protein abundance levels estimated by the peak area (Top3 approach) and spectral counting methods in parotid and submandibular/sublingual (SM/SL) glandular ductal saliva, respectively. Pearson’s correlation coefficients for representative samples are shown. (C) Volcano plot showing differential protein abundances between the parotid and SM/SL glands. The plot was generated using the DE analysis tool from the Wiki. Proteins with statistically significant fold changes ≥log2 are shown as blue dots. Gene symbols for the most gland specifically expressed proteins are shown. (D) Enriched Gene Ontology (GO) biological processes from gene set enrichment analysis (GSEA) of the differential abundance results between salivary proteomes of oral squamous cell carcinoma (OSCC) subjects and healthy controls. Only pathways with adjusted P values ≤0.05 and ≥66% leading gene representation (i.e., enrichment signals coming from at least 66% of proteins in the pathway) are shown.
Differential Expression Analysis to Compare Healthy and Disease-Associated Salivary Proteomes
Additional downstream analyses can also be performed using external analysis tools. Raw MS data from a study comparing saliva proteomes between patients with oral squamous cell carcinoma (OSCC) and healthy controls (Lin et al. 2019) were deposited into the Wiki database. Of 2,246 proteins identified from MaxQuant analyses, 103 proteins were significantly differentially expressed between the control and OSCC groups. Of 39 proteins with predictive power in discriminating the 2 groups, 29 were also significantly different (false discovery rate ≤ 0.05) based on the spectral counting quantification method used by the Wiki. After exporting the results to the R software, the entire set of proteins was ranked based on test statistics computed by the Wiki, and gene-set enrichment analysis against Gene Ontology (GO) pathways demonstrated consistent findings with the original report and conclusions (Lin et al. 2019), including blood coagulation, acute inflammatory response, humoral immune response, and protein activation cascade as top enriched pathways (Fig. 4D). In addition, the Wiki analysis showed that triglyceride and interleukin-8 regulation were also enriched in the saliva of OSCC patients.
Future Development of the HSP Wiki
In the future, the HSP Wiki will interact through automated data workflows with additional relevant databases hosting sequence, structure, posttranslational modification information, and oral microbiome data. The Human Oral Microbiome Database (HOMD, http://www.homd.org/) and the glycosylation database GlyGen (https://www.glygen.org/) are anticipated extensions. Since the interactions of oral bacteria with salivary proteins are primarily mediated by glycans expressed either on the proteins or the bacterial surface (Cross and Ruhl 2018), a glycome-centric database could serve as the interface between proteome and microbiome in the oral cavity. These future integrative pipelines will provide novel insights into functional salivary biology. Another possibility is to add salivary proteome information from other organisms (e.g., primates [Thamadilok et al. 2020] and rodents [Stopka et al. 2016]) for interspecies comparisons to gain novel insights into the functions of saliva (Stearns 2020).
It will be paramount for the HSP Wiki to integrate the ability to compare salivary proteome composition between health and disease states, including Sjögren syndrome, salivary gland malignancies, oral cancer, dental caries, periodontal diseases, and systemic diseases. For risk prediction, it will be important to obtain access to other databases providing global population-wide information on genetic variants and disease correlations.
Discussion
We established the HSP Wiki as a public data platform for researching and retrieving custom-curated data and knowledge information on the saliva proteome. The HSP Wiki is useful across a wide range of disciplines in advancing the knowledge of saliva composition and function in health and disease, and serves as a worldwide research platform to retrieve and exchange salivary proteome information, inspire novel ideas, and foster cross-discipline collaborations. This community-based data and knowledgebase will help to harness the full potential of the salivary proteome for diagnosis, risk prediction, and therapy of oral and systemic diseases and to increase preparedness for future emerging diseases and pandemics. Already, the HSP Wiki experiences considerable user traffic and has been used as a valuable resource for scientific investigation (Saitou et al. 2020). Similar to other databases in the Big Data era, integrating multimodal data from multiple sources into the HSP Wiki is not trivial. Thus far, the main challenge in building the HSP Wiki has been integrating UniProt data and the proteomics data generated by different research groups. Currently, only identification data are stored in the Wiki, and data upload is performed manually by system administrators, but the platform was designed for add-on improvements and automated systems. Quantitative data on disease states will facilitate biomarker discovery and functional explorations. Since a wide range of instruments and software tools are employed by the proteomics community, it is important to adhere to applicable data standards, such as mzML (Martens et al. 2011), to effectively manage data and maintain consistency across input files.
One inherent challenge of an open-platform, community-based database will be how to sieve through the user input and process the sheer amount of data. In addition, with constantly improving proteomics analytical methods will come along improved data input formats. The challenge will be to keep the HSP Wiki database flexible enough to adapt to such new formats, yet make sure that newly deposited data sets remain comparable with earlier data sets. Will the community-based open platform be sufficient to maintain quality data and up-to-date evidence-based knowledge information? How much will the curation committee have to control and channelize the community-based input? Lastly, how will we ensure sustainability? Many resources failed to attract enough usage not because of design issues but because of community disinterest (Waldrop 2008). Effective incentives consist of citations to user-derived scientific publications on appropriate Wiki pages, acknowledgments for data input, and other possibilities of claiming authorship for knowledge input. A wide-ranging and diverse community engagement will be critical for keeping this unique salivary proteome database up to date. To accomplish this goal, we are planning a series of activities including symposia to bring contributors and users together.
The HSP Wiki is a database specifically tailored to the needs of a particular scientific community interested in studying the human salivary proteome and its interactions. The database allows the large-scale manual or semi-automated reconstruction of functional networks derived from saliva. The Wiki aims to serve as a common denominator for system biology investigations of human proteins. Success and user engagement of the HSP Wiki will be monitored through website activities, citations, scientific discovery, and future integrative networking with other Omics initiatives including the microbiome and glycome. We believe that the HSP Wiki will improve salivary sciences, saliva-based diagnostics, precision medicine, and dentistry, and ultimately facilitate personalized treatment for both oral and systemic diseases.
Author Contributions
W.W. Lau, M. Hardt, M. Freire, S. Ruhl, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; Y.H. Zhang, contributed to conception, design, and data interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211014432 for The Human Salivary Proteome Wiki: A Community-Driven Research Platform by W.W. Lau, M. Hardt, Y.H. Zhang, M. Freire and S. Ruhl in Journal of Dental Research
Acknowledgments
We are grateful for valuable scientific discussions and advice provided over the years by many HSP Wiki contributors, most prominently Drs. Susan Fisher, Sven Gorr, Frank Oppenheim, Walter Siqueira, and David Wong. We also thank Calvin Johnson, Beecher Greenman, Robyn Wyrick, Zana Coulibaly, and Anthony Fletcher from CIT for computational and technical support. The HSP Wiki has been supported by the National Institute of Dental and Craniofacial Research (NIDCR), and its development was actively driven forward by NIDCR officials Drs. Preethi Chander, Penny Burgoon, Amanda Melillo, Lillian Shum, Eleni Kouslevari, Douglas Sheeley, and Larry Tabak. Our special thanks go to Drs. Supaporn Thamadilok, Yanbao Yu, and Steven Hall for data input and technical help, and to Raja Mazumder and William York (GlyGen database) for advice on how to add glycosylation data to the HSP Wiki protein catalog.
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institute of Dental and Craniofacial Research (NIDCR) grants 3R01DE019807-04S1 to S.R. and JCVI Innovation fund to M.F. and by the Intramural Research Program of Center for Information Technology at the National Institutes of Health.
ORCID iDs: M. Hardt  https://orcid.org/0000-0002-2300-216X
https://orcid.org/0000-0002-2300-216X
Y.H. Zhang  https://orcid.org/0000-0001-8277-8008
https://orcid.org/0000-0001-8277-8008
M. Freire  https://orcid.org/0000-0003-4906-7698
https://orcid.org/0000-0003-4906-7698
References
- Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. 2009. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 8(12):5590–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne NJ, Wong DT. 2012. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 4(10):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Nguyen-Khuong T, Hayes CA, Flowers SA, Alagesan K, Kolarich D, Packer NH, Karlsson NG. 2014. Validation of the curation pipeline of UniCarb-DB: building a global glycan reference MS/MS repository. Biochim Biophys Acta. 1844:108–116. [DOI] [PubMed] [Google Scholar]
- Cariaso M, Lennon G. 2012. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 40:D1308–D1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BW, Ruhl S. 2018. Glycan recognition at the saliva-oral microbiome interface. Cell Immunol. 333:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, Pedersen AML, Villa A, Ekström J, Proctor GB, Vissink A, Aframian D, McGowan R, Aliko A, Narayana N, et al. 2015. The functions of human saliva: a review sponsored by the World Workshop on Oral Medicine VI. Arch Oral Biol. 60(6):863–874. [DOI] [PubMed] [Google Scholar]
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, et al. 2008. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 7(5):1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Albar JP, Binz P-A, Eisenacher M, Jones AR, Mayer G, Omenn GS, Orchard S, Vizcaíno JA, Hermjakob H. 2015. Development of data representation standards by the human proteome organization proteomics standards initiative. J Am Med Inform Assoc. 22(3):495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems. 3(6):e00187–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles J. 2007. Key biology databases go wiki. Nature. 445(7129):691–691. [DOI] [PubMed] [Google Scholar]
- Good BM, Clarke EL, de Alfaro L, Su AI. 2012. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 40:D1255–D1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, Ainscough BJ, Ramirez CA, Rieke DT, Kujan L, et al. 2017. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 49(2):170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst EJ, Dawes C, Oppenheim FG. 2018. The complexity of oral physiology and its impact on salivary diagnostics. Oral Dis. 24(3):363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Pérez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Conde CD, Gasmi B, Stein S, Beach M, et al. 2021. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med [epub ahead of print 25 Mar 2021]. doi: 10.1038/s41591-021-01296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Pérez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Domínguez Conde C, Gasmi B, Stein S, Beach M, et al. 2020. Integrated single-cell atlases reveal an oral SARS-CoV-2 infection and transmission axis. medRxiv. doi: 10.1101/2020.10.26.20219089 [DOI] [Google Scholar]
- Isho B, Abe KT, Zuo M, Jamal AJ, Rathod B, Wang JH, Li Z, Chao G, Rojas OL, Bang YM, et al. 2020. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Science Immunol. 5(52):eabe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives C, Campia I, Wang R-L, Wittwehr C, Edwards S. 2017. Creating a structured AOP knowledgebase via ontology-based annotations. Appl In Vitro Toxicol. 3(4):298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehmlich N, Dinh KHD, Gesell-Salazar M, Hammer E, Steil L, Dhople VM, Schurmann C, Holtfreter B, Kocher T, Völker U. 2013. Quantitative analysis of the intra- and inter-subject variability of the whole salivary proteome.J Periodontal Res. 48(3):392–403. [DOI] [PubMed] [Google Scholar]
- Lendenmann U, Grogan J, Oppenheim FG. 2000. Saliva and dental pellicle—a review. Adv Dent Res. 14:22–28. [DOI] [PubMed] [Google Scholar]
- Lin Y-H, Eguez RV, Torralba MG, Singh H, Golusinski P, Golusinski W, Masternak M, Nelson KE, Freire M, Yu Y. 2019. Self-assembled STrap for global proteomics and salivary biomarker discovery. J Proteome Res. 18(4):1907–1915. [DOI] [PubMed] [Google Scholar]
- Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, et al. 2011. mzML—a community standard for mass spectrometry data. Mol Cell Proteomics. 10(1):R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michna RH, Zhu B, Mäder U, Stülke J. 2016. SubtiWiki 2.0—an integrated database for the model organism Bacillus subtilis. Nucleic Acids Res. 44(D1):D654–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mons B, Ashburner M, Chichester C, van Mulligen E, Weeber M, den Dunnen J, van Ommen G-J, Musen M, Cockerill M, Hermjakob H, et al. 2008. Calling on a million minds for community annotation in WikiProteins. Genome Biol. 9(5):R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. 2011. Diagnostic potential of saliva: current state and future applications. Clin Chem. 57(5):675–687. [DOI] [PubMed] [Google Scholar]
- Reeves GA, Talavera D, Thornton JM. 2009. Genome and proteome annotation: organization, interpretation and integration. J R Soc Interface. 6(31):129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl S. 2012. The scientific exploration of saliva in the post-proteomic era: from database back to basic function. Expert Rev Proteomics. 9(1):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Gaylord EA, Xu E, May AJ, Neznanova L, Nathan S, Grawe A, Chang J, Ryan W, Ruhl S, et al. 2020. Functional specialization of human salivary glands and origins of proteins intrinsic to human saliva. Cell Rep. 33(7):108402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte F, Hasturk H, Hardt M. 2019. Mapping relative differences in human salivary gland secretions by dried saliva spot sampling and nanoLC-MS/MS. Proteomics. 19(20):e1900023. [DOI] [PubMed] [Google Scholar]
- Schwenk JM, Omenn GS, Sun Z, Campbell DS, Baker MS, Overall CM, Aebersold R, Moritz RL, Deutsch EW. 2017. The human plasma proteome draft of 2017: building on the human plasma peptideatlas from mass spectrometry and complementary assays. J Proteome Res. 16(12):4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira WL, Salih E, Wan DL, Helmerhorst EJ, Oppenheim FG. 2008. Proteome of human minor salivary gland secretion. J Dent Res. 87(5):445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, Mélius J, Cirillo E, Coort SL, Digles D, et al. 2018. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 46(D1):D661–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks R, Lau WW, Tsang JS. 2016. Expanding the immunology toolbox: embracing public-data reuse and crowdsourcing. Immunity. 45(6):1191–1204. [DOI] [PubMed] [Google Scholar]
- Stearns SC. 2020. Frontiers in molecular evolutionary medicine. J Mol Evol. 88(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. 2001. Genome annotation: from sequence to biology. Nat Rev Genet. 2(7):493–503. [DOI] [PubMed] [Google Scholar]
- Stopka P, Kuntová B, Klempt P, Havrdová L, Černá M, Stopková R. 2016. On the saliva proteome of the Eastern European house mouse (Mus musculus musculus) focusing on sexual signalling and immunity. Sci Rep. 6:32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover NA, Punia RS, Bowen MS, Dolins SB, Clark TG. 2012. Tetrahymena genome database Wiki: a community-maintained model organism database. Database. 2012:bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak LA. 2007. Point-of-care diagnostics enter the mouth. Ann N Y Acad Sci. 1098:7–14. [DOI] [PubMed] [Google Scholar]
- Teo AKJ, Choudhury Y, Tan IB, Cher CY, Chew SH, Wan ZY, Cheng LTE, Oon LLE, Tan MH, Chan KS, et al. 2021. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci Rep. 11(1):3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamadilok S, Choi K-S, Ruhl L, Schulte F, Kazim AL, Hardt M, Gokcumen O, Ruhl S. 2020. Human and nonhuman primate lineage-specific footprints in the salivary proteome. Mol Biol Evol. 37(2):395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Proteomics: tissue-based map of the human proteome. Science. 347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop M. 2008. Big data: Wikiomics. Nature. 455(7209):22–25. [DOI] [PubMed] [Google Scholar]
- Walz A, Stühler K, Wattenberg A, Hawranke E, Meyer HE, Schmalz G, Blüggel M, Ruhl S. 2006. Proteome analysis of glandular parotid and submandibular-sublingual saliva in comparison to whole human saliva by two-dimensional gel electrophoresis. Proteomics. 6(5):1631–1639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211014432 for The Human Salivary Proteome Wiki: A Community-Driven Research Platform by W.W. Lau, M. Hardt, Y.H. Zhang, M. Freire and S. Ruhl in Journal of Dental Research