Abstract
A major pathway of eukaryotic mRNA turnover occurs by deadenylation-dependent decapping that exposes the transcript to 5′→3′ exonucleolytic degradation. A critical step in this pathway is decapping, since removal of the cap structure permits 5′→3′ exonucleolytic digestion. Based on alterations in mRNA decay rate from strains deficient in translation initiation, it has been proposed that the decapping rate is modulated by a competition between the cytoplasmic cap binding complex, which promotes translation initiation, and the decapping enzyme, Dcp1p. In order to test this model directly, we examined the functional interaction of Dcp1p and the cap binding protein, eukaryotic translation initiation factor 4E (eIF4E), in vitro. These experiments indicated that eIF4E is an inhibitor of Dcp1p in vitro due to its ability to bind the 5′ cap structure. In addition, we demonstrate that in vivo a temperature-sensitive allele of eIF4E (cdc33-42) suppressed the decapping defect of a partial loss-of-function allele of DCP1. These results argue that dissociation of eIF4E from the cap structure is required before decapping. Interestingly, the temperature-sensitive allele of eIF4E does not suppress the decapping defect seen in strains lacking the decapping activators, Lsm1p and Pat1p. This indicates that these activators of decapping affect a step in mRNA turnover distinct from the competition between Dcp1 and eIF4E.
The turnover of mRNAs is a significant aspect of the differential gene expression in the eukaryotic cell (for reviews, see references 5, 11, 23, and 36). It has become clear, at least in yeast, that mRNAs with different decay rates are largely degraded by a single general pathway of mRNA turnover. In this pathway, mRNAs are first deadenylated, which allows the transcript to become a substrate for a decapping reaction catalyzed by the decapping enzyme encoded by the DCP1 gene (6, 14, 22, 29–31). Once decapped, the mRNAs are then susceptible to 5′→3′ exonucleolytic degradation by the Xrn1p exoribonuclease (22, 29, 30). Several observations suggest a similar pathway of degradation is likely to exist in other eukaryotic cells. For example, deadenylation can be the first step in mammalian mRNA turnover (42, 47). Moreover, deadenylated decapped intermediates in turnover can be detected in mammals and in Chlamydomonas (12, 18). In addition, several proteins functioning in mRNA decapping in yeast (Dcp1p, Dcp2p, Xrn1p, Pat1p [also known as Mrt1p], and Lsm1p) have homologs throughout the eukaryotic kingdom (4, 15, 37, 38, 46).
In yeast, multiple lines of evidence suggest that the decapping of mRNAs is an important control point in the regulation of mRNA half-life. In the deadenylation-dependent decapping pathway of mRNA turnover, the basis for the differential decay rates of individual yeast mRNAs is that transcripts differ in their rates of deadenylation and decapping. Short-lived mRNAs decap rapidly, while longer-lived mRNAs decap more slowly (29, 30). In addition, in the deadenylation-independent pathway of mRNA turnover, transcripts bypass the need for deadenylation and very rapidly decap (32). Given these observations, in order to understand differential mRNA stability, it will be critical to determine the mechanisms that modulate the rates of decapping.
One hypothesis for the control of decapping is that the rate of decapping is controlled by a steric competition for the cap structure between the decapping enzyme and the translation initiation machinery (13, 23, 40). This proposal was originally based on the fact that the cap structure is crucial for the ability of an mRNA to initiate translation efficiently due to its ability to assemble the cytoplasmic cap binding complex, which ultimately directs ribosome loading to the 5′ end (3, 34). A competition between the decapping enzyme and eukaryotic translation initiation factor 4E (eIF4E) (the cap binding protein) is supported by two observations. First, conditional alleles of eIF4E (e.g., cdc33-42) lead to faster decapping at the restrictive temperature (40). Second, inhibition of translation initiation due to the insertion of strong secondary structures in the 5′ untranslated region (UTR) leads to faster decapping (30). However, several observations suggest that the interrelationship between translation initiation and decapping may be more complex. For example, mutations in the eIF3 translation initiation complex, which acts downstream of the cytoplasmic cap binding complex in the translation initiation process, also promote faster mRNA degradation (40). In addition, decreasing the translation initiation rate by creating a poor AUG context also leads to faster rates of mRNA decapping (25).
These observations suggest two possible relationships between the translation initiation complex and the decapping enzyme. First, the decapping rate of a transcript could be a function of its overall translation rate per se, and not a reflection of a physical competition between the cap binding complex and the decapping enzyme. Alternatively, the decapping rate of the transcript could be due to a competition between the cap binding complex and the Dcp1p, with the stability of the cap binding complex on the mRNA being influenced by the overall success of the translation initiation process. The critical difference between these two views is that in the latter model, the cap binding complex must dissociate before decapping can occur. However, these models cannot be currently distinguished by the available in vivo evidence, since any change to the cap binding complex also changes the translation rate.
In order to determine whether the cap binding complex directly competes with the decapping enzyme for the cap structure, we examined the effects of the cap binding protein eIF4E on decapping in vitro by using purified Dcp1p and eIF4Ep. The advantage of this system is that it is fully defined, and any indirect effects of eIF4E on decapping due to translation per se have been prevented, since there are no other translation factors in the reaction. These experiments have shown that eIF4Ep binding to the cap structure was sufficient to inhibit Dcp1p decapping activity. We have also recapitulated this observation in vivo by showing that a mutation in eIF4E can suppress the decapping defect caused by the partial-loss-of-function allele, dcp1-1. These results argue that eIF4E bound to the cap structure protects the mRNA from decapping and indicate that dissociation of eIF4E from the cap structure will be a critical step in mRNA turnover.
MATERIALS AND METHODS
Yeast strains.
The genotypes of all of the strains (Saccharomyces cerevisiae) used in this study are listed in Table 1, and the strains were grown in standard media. All strains have GAL1 upstream activating sequence-regulated PGK1pG and MFA2pG genes, as well as the LEU2 gene, collectively termed LEU2pm, integrated at the CUP1 locus (21).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| yRP841 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 21 |
| yRP1063 | MATα dcp1-1 leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 21 |
| yRP1315 | MATaleu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [CDC33/URA3] | 40 |
| yRP1321 | MATaleu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | 39 |
| yRP1463 | MATα leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [GST-eIF4E/URA3] | This study |
| yRP1464 | MATaleu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG [GST-ts-4E/URA3] | This study |
| yRP1465 | MATadcp1-1 leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1466 | MATα dcp1-1 leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | This study |
| yRP1467 | MATaleu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG his3::URA3 | This study |
| yRP1468 | MATaleu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG his3::URA3 [cdc33-42/TRP1] | This study |
| yRP1469 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG his3::URA3 lsm1::HIS3 | This study |
| yRP1470 | MATaleu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG his3::URA3 lsm1::HIS3 [cdc33-42/TRP1] | This study |
| yRP1471 | MATaleu2 lys2 prt1-63 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1472 | MATadcp1-1 leu2 lys2 prt1-63 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1372 | MATahis4-539 leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG pat1::LEU2 | 45 |
| yRP1473 | MATaleu2 lys2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG his3::URA3 pat1::HIS3 [cdc33-42/TRP1] | This study |
Vector construction.
The glutathione S-transferase (GST)-eIF4E and GST-ts-4E genes were created by PCR of the eIF4E gene from pMDA-101 carrying either a wild-type eIF4E gene or the eIF4E/cdc33-42 allele (2) with the oligonucleotides GGGAATTCCATATGTCCGTTGAAGAAGTTAG (oRP969) and GGTACTAGTCTAGACATGATGACTTTATACGTG (oRP970). These fragments were digested with NdeI and XbaI and ligated into the yeast GST expression vector pRP966 between the NdeI and XbaI sites to yield plasmids pRP967 and pRP968. This CEN URA3 plasmid expresses either GST-eIF4Ep or GST-ts-4Ep from the inducible GAL1 promoter.
The His-eIF4E and His-ts-4E genes were created by PCR of the eIF4E gene from pMDA-101 carrying either a wild-type eIF4E gene or the eIF4E-42 allele with the oligonucleotides CCGCCGCATATGTCCGTTGAAGAAGTTAGCAAG (oRP971) and CGGAAAAGGATCCTTACAAGGTGATTGATGGTTGAG (oRP972). These fragments were digested with NdeI and BamHI and ligated into the Escherichia coli expression vector pET-16b (Novagen) between the NdeI and BamHI sites to yield pRP969 and pRP970. These plasmids express either His-eIF4E or His-ts-4E from the inducible T7 promoter.
All plasmids were fully sequenced to ensure that all mutations, junctions, and tags were properly generated.
Protein purifications.
His-Dcp1p utilized in these assays was purified as previously described (6).
GST-eIF4E protein used in the experiments shown in Fig. 2 and 4 was purified by a protocol adapted from reference 44. Briefly, 1 liter of strain yRP1463 containing pRP967 was grown in selective media containing 2% galactose to late log phase and harvested by centrifugation. Cells were washed once with double-distilled H2O and then resuspended in 12 ml of buffer A (100 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 7 mM 2-mercaptoethanol, 30 mM HEPES [pH 7.5]) containing Complete (Boehringer-Mannheim) protease inhibitors. Acid-washed beads (24 g) were added to the cell suspension, and cells were lysed by five cycles of vortexing for 30 s and 1 min of cooling on ice. The lysate was clarified by two 5-min spins at 15,500 rpm in an SS34 rotor. The resulting supernatant was added to 500 μl of m7GDP-agarose resin prepared as described in reference 17 and preequilibrated in buffer A. The slurry was allowed to rock on a platform shaker at 4°C for 2 h and then was chromatographed at 4°C by gravity flow. The flowthrough was collected and reapplied to the column. The column was washed three times with 10 ml of buffer B (100 mM KCl, 0.2 mM EDTA, 0.01% Triton X-100, 0.5 mM PMSF, 7 mM 2-mercaptoethanol, and 20 mM HEPES [pH 7.4]) containing Complete protease inhibitors and washed three times with 10 ml of buffer B plus 0.1 mM GDP. The eIF4E protein was eluted in five 250-μl washes with buffer B plus 0.1 mM m7GTP. The GST-eIF4E protein was dialyzed into buffer containing 50 mM KCl, 20 mM HEPES (pH 7.4), 7 mM 2-mercaptoethanol, 0.2 mM EDTA, and 0.01% Triton X-100. The protein was aliquoted and frozen at −80°C.
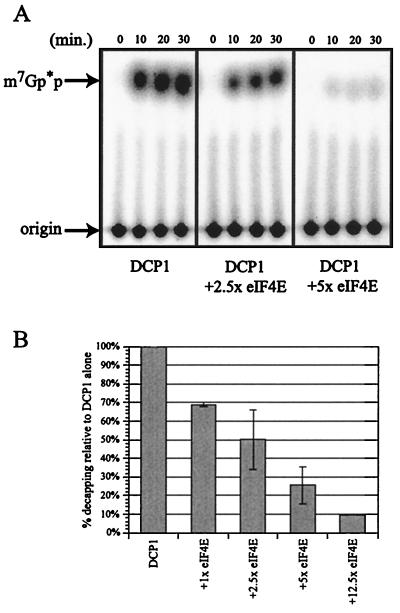
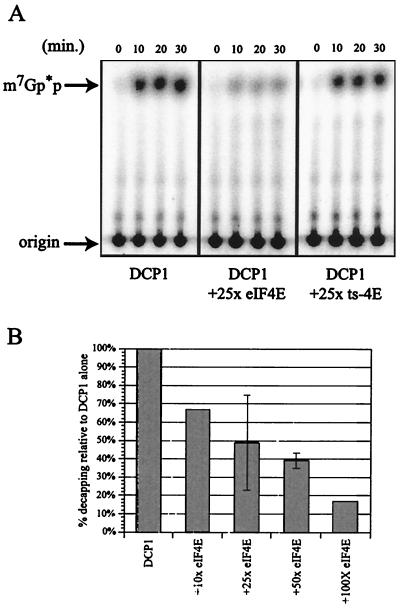
FIG. 2.
Purified eIF4E inhibits decapping. (A) Analysis of decapping activity by DCP1 alone (left panel) and increasing amounts of GST-eIF4E (purified with a cap affinity column): either 8.3 pmol of GST-eIF4Ep (middle panel [2.5×]) or 16.7 pmol of GST-eIF4Ep (right panel [5×]). Decapping by His-DCP1p was assayed over a 30-min time course with aliquots removed at the designated time points. The products of the reaction were resolved by thin-layer chromatography. (B) Decapping is inhibited upon addition of eIF4E. The histogram shows the amount of decapping activity obtained with increasing amounts of eIF4E. Relative activity was determined by assaying decapping ± eIF4E in multiple experiments and is expressed as the amount of decapping relative to that with DCP1 alone (set at 100%). The 30-min time point is shown on the histogram.
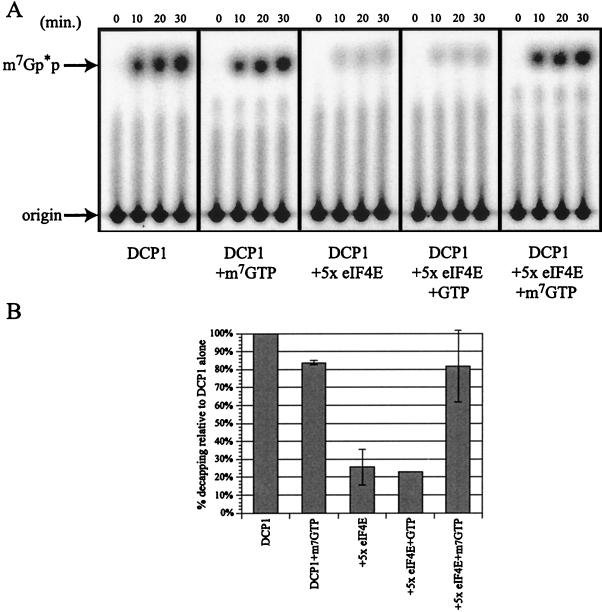
FIG. 4.
Inhibition of decapping by eIF4E is relieved upon m7GTP addition. (A) Analysis of decapping activity by DCP1 ± m7GTP (left two panels) and in the presence of eIF4E ± GTP and m7GTP (right three panels). Decapping by His-DCP1 was assayed over a 30-min time course, with aliquots removed at the designated time points. The products of the reaction were resolved by thin-layer chromatography. (B) Relative decapping activity in the presence of eIF4E and m7GTP. Relative activity was determined by assaying decapping ± eIF4E and m7GTP in multiple experiments and is expressed as the amount of decapping relative to DCP1 alone (set at 100%). The 30-min time point is shown on the histogram.
The GST-eIF4E (strain yRP1463 containing plasmid pRP967) and GST-ts-4E (strain yRP1464 containing plasmid pRP968) proteins used in Fig. 3 were purified as described above with the following modifications. Twelve milliliters of the postlysis supernatants was added to 500 μl of glutathione-Sepharose (Pharmacia), prepared as recommended by the manufacturer, and preequilibrated in buffer C (50 mM Tris [pH 7.5], 0.1% NP-40, 2 mM EDTA, 100 mM NaCl) containing Complete protease inhibitors. The slurry was allowed to rock on a platform shaker at 4°C for 2 h and then chromatographed at 4°C by gravity flow. The flowthrough was collected and reapplied to the column. The column was washed three times with 10 ml of buffer C. The eIF4E proteins were eluted in three 250-μl washes with 25 mM reduced glutathione. The GST-eIF4E and GST-ts-4E proteins were dialyzed into buffer containing 50 mM NaCl, 30 mM Tris (pH 8.0), 0.2 mM EDTA, 7 mM 2-mercaptoethanol, and 0.01% Triton X-100. The proteins were aliquoted and frozen at −80°C.
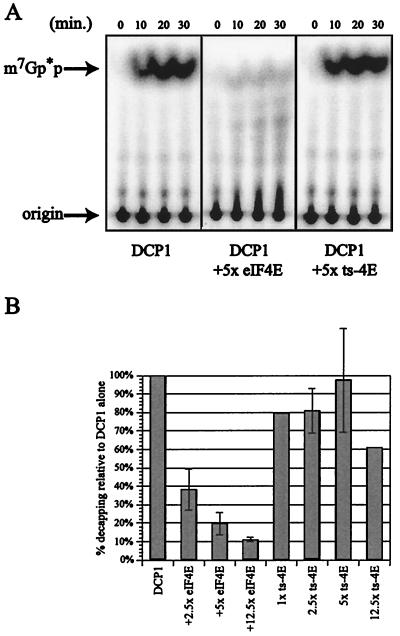
FIG. 3.
Decapping is not inhibited by a nonfunctional allele of eIF4E. (A) Analysis of decapping activity by DCP1 alone (left panel) and in the presence of either eIF4E (middle panel; 16.7 pmol of GST-eIF4E) or a nonfunctional allele of eIF4E (right panel, labeled ts-4E; 16.7 pmol of GST-ts-4E) (both purified with a glutathione affinity column). Decapping by His-DCP1 was assayed over a 30-min time course, with aliquots removed at the designated time points. The products of the reaction were resolved by thin-layer chromatography. (B) Relative decapping activity in the presence of increasing amounts of either GST-eIF4E or GST-ts-4E. Relative activity was determined by assaying decapping ± eIF4E in multiple experiments and is expressed as the amount of decapping relative to DCP1 alone (set at 100%). The 30-min time point is shown on the histogram.
His-eIF4E and His-ts-4E were purified as recommended by the manufacturer (Novagen). Briefly, 100-ml cultures of BL-21 cells containing either a His-eIF4E (pRP969) or a His-ts-4E (pRP970) plasmid were grown in Luria broth medium at 37°C to an optical density of 0.6. The cultures were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h and then harvested by centrifugation. After elution of the proteins from a nickel-nitrilotriacetic acid column (Qiagen) with 1× elution buffer, the proteins were dialyzed first into a mixture of 50 mM KCl, 20 mM HEPES (pH 7.5), and 5 mM EDTA to separate the proteins from the nickel and then into a mixture of 50 mM KCl, 20 mM HEPES (pH 7.5), 7 mM 2-mercaptoethanol, 0.2 mM EDTA, and 0.01% Triton X-100. The proteins were aliquoted and frozen at −80°C.
All purified protein preparations were analyzed by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) methods on 12% polyacrylamide gels (24). Protein size markers were purchased from Gibco-BRL.
Substrate preparation.
Uncapped MFA2 mRNAs lacking poly(A) tails were synthesized in vitro by using the Riboprobe in vitro transcription system (Promega). The DNA template used was as previously described (26). T7 transcription was done as recommended by the manufacturer with 1 to 2 μg of template DNA at 37°C for 2 h. The DNA template was digested with 1 μl of RQ1 DNase. The resulting uncapped transcript was purified by phenol-chloroform extraction and Sephadex G-50 chromatography. The samples were precipitated and resuspended in 20 μl of diethyl pyrocarbonate-H2O.
The mRNAs were capped as previously described (26).
Decapping assays.
Decapping reactions were assayed at 30°C over a 30-min time course. All reactions depicted on histograms are an average of multiple experiments. The reaction mixtures generally contained ∼2.3 fmol of m7G[32P]pppMFA2 mRNA, 3.1 pmol of DCP1p, 50 mM Tris (pH 7.6), 5 mM MgCl2, 50 mM NH4Cl, 1 mM dithiothreitol (DTT), and 1 μl of RNasin in a volume of 15 μl. eIF4E proteins were added at the concentrations noted in the figure legends. Bovine serum albumin (BSA) was added to all reaction mixtures to maintain constant total protein concentrations. GTP and m7GTP were added at a concentration of 1 mM. Time courses experiments were carried out by withdrawing an aliquot of the reaction mixture at various time points. Decapping in each aliquot was stopped by adding 1 μl of 0.5 M EDTA to the aliquot and placing it on ice. The products of the reaction were separated by polyethyleneimine-cellulose thin-layer chromatography developed in 0.45 M (NH4)2SO4 and detected with a Molecular Dynamics PhosphorImager.
mRNA analysis.
Transcriptional pulse-chase experiments used to track a synchronous pool of mRNAs were done as previously described (40). Briefly, cells were grown to mid-log phase in medium containing raffinose, shifted to 38°C for 1 h, harvested, and resuspended in medium containing galactose to induce the transcripts; finally, the cells were again harvested, and dextrose was added to shut off transcription.
mRNA was isolated as described previously (10). RNase H and polyacrylamide Northern (RNA) assays were done with 40 μg of RNA as previously described (31).
RESULTS
Purification of eIF4E.
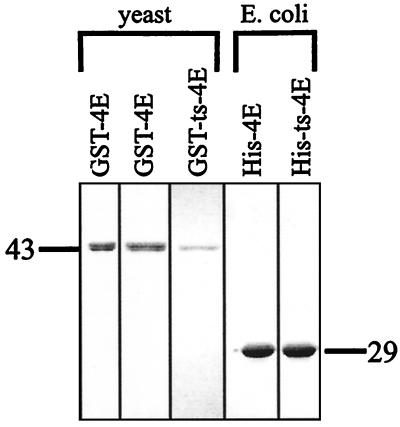
In order to examine the effects of eIF4E on decapping by Dcp1p in vitro, we first purified the two proteins. Dcp1p was obtained from a previous purification by using a His-Dcp1p fusion expressed in yeast as previously described and is highly purified based on silver staining (26). eIF4E was initially purified from yeast as a GST-eIF4E fusion protein. This GST-eIF4E fusion protein was functional, since it complemented an eIF4EΔ (cdc33Δ) strain and was also able to bind to a cap column (Fig. 1) (data not shown). Although eIF4E can be directly purified out of yeast by a cap affinity column, we utilized a GST-eIF4E fusion to allow the parallel purification of a mutant version of eIF4E (cdc33-42), which is unable to bind the cap column. In addition, both wild-type and mutant eIF4E were purified from E. coli by using His-tagged versions of the proteins. We utilized the His tag to purify eIF4E from E. coli, since we have observed that in our hands, purification of GST, or any GST-tagged protein, from E. coli leads to the copurification of nucleolytic activity (data not shown). In all cases, the preparations were highly purified and free of additional polypeptides as assayed by Coomassie stain (Fig. 1).
FIG. 1.
Purification of various eIF4E proteins. Analysis of GST-eIF4E and His-eIF4E purified in various manners by Coomassie-stained SDS-PAGE. The leftmost column shows GST-eIF4E purified from a yeast extract by m7GDP chromatography. The second and third columns show GST-eIF4E and GST-ts-4E purified from yeast extracts by glutathione chromatography. The two right columns show His-eIF4E and His-ts-4E purified from E. coli extracts by nickel column chromatography. The relevant protein size marker (in kilodaltons) is shown next to each panel. The size differences between the GST-tagged and His-tagged versions of eIF4E come about through the size of the particular epitope added to the eIF4E protein.
eIF4E blocks decapping activity in vitro.
The effects of these purified eIF4E proteins on decapping were then examined by using an in vitro decapping assay, which consists of adding purified Dcp1p to in vitro-transcribed, cap-labeled mRNA and assaying for the release of the 32P-labeled cap structure by thin-layer chromatography (6, 26, 48). To allow comparison across experiments, the amount of cap released by Dcp1p alone was set at 100% for each experiment. An important result was that following the addition of increasing amounts of GST-eIF4E purified from yeast over a cap affinity column to the decapping reaction, there was a decreasing amount of cap released over time. At a 12.5 M excess of eIF4E protein, relative to Dcp1p, decapping activity was decreased almost 8.5-fold (Fig. 2). BSA was added to all reaction mixtures to maintain a constant protein concentration, but BSA alone had no inhibitory effect on the activity of Dcp1p (data not shown). Addition of eIF4E by itself had no effect on the integrity of the substrate (data not shown).
To test whether the ability of eIF4E to inhibit Dcp1p activity was due to eIF4E binding to the cap structure, two experiments were performed. First, we examined the effect on decapping of a purified mutant, GST-eIF4E, which is unable to bind the cap structure in vitro (2). Since the temperature-sensitive allele of eIF4E is unable to bind a cap column, this protein was purified over a glutathione column. The wild-type eIF4E protein was also purified in this manner to control for the purification procedure. Addition of wild-type eIF4E protein again leads to inhibition of DCP1, indicating that the method of protein purification has no effect on the activity of eIF4E in vitro (Fig. 3). However, addition of up to a fivefold molar excess of the mutant eIF4E protein had no effect on the decapping activity of the Dcp1 protein (Fig. 3). This result suggests that the eIF4E protein blocks decapping due to its interaction with the cap structure.
A second test of whether the ability of eIF4E to inhibit Dcp1p was due to eIF4E binding to the cap structure was to take advantage of the different effects of the cap analog m7GTP on eIF4E and Dcp1p. In this case, the decapping activity of Dcp1p was not affected by m7GTP (26), whereas eIF4E is known to be dissociated from the cap structure by m7GTP, which serves as an alternative binding site (43). As seen previously, addition of m7GTP to a decapping assay had minimal effect on Dcp1p activity (Fig. 4). However, addition of m7GTP to a reaction mixture containing Dcp1p and eIF4E restored decapping. GTP, which is not bound effectively by eIF4E, had no effect on eIF4E's ability to block Dcp1p. We interpret these observations to indicate that the inhibition of Dcp1p by eIF4E requires binding of eIF4E to the cap structure.
eIF4E purified from yeast is known to copurify various other translation initiation factors, most notably eIF4Gp and Caf20p (27). Although these proteins could only be present at low levels in our preparations, based on silver staining assays, it is a formal possibility that the inhibition of decapping we observed was actually due to a contaminating protein that copurified with wild-type eIF4E. To rule out this possibility, we purified histidine-tagged versions of both the wild-type and temperature-sensitive allele of eIF4E from E. coli, which contains no endogenous cap binding proteins. Consistent with our earlier results, the wild-type eIF4E from E. coli inhibited the decapping activity of Dcp1p, while the temperature-sensitive allele of eIF4E had no effect on decapping (Fig. 5). However, significantly larger amounts of protein were needed to achieve the same degree of inhibition, as seen with protein purified from yeast. The likely explanation for why the E. coli-produced protein was less active at inhibiting decapping comes from the fact that a large portion of the eIF4E made in E. coli exists in an inactive form (16). Since we purified the eIF4E based on an epitope tag, to allow purification of the mutant eIF4E, it is highly likely that only a portion of the purified protein is functional. Second, the possibility exists that during purification of eIF4E from yeast, small amounts of eIF4G are copurified. Since addition of eIF4G to eIF4E increases the affinity of eIF4E for the cap structure, this may allow the eIF4E purified from yeast to show a greater degree of inhibition in a decapping reaction. Nevertheless, the observation that eIF4E purified from E. coli does inhibit decapping indicates that eIF4Ep, when bound to the cap structure of a mRNA, is sufficient to inhibit Dcp1p's ability to decap the mRNA.
FIG. 5.
eIF4E protein purified from E. coli has similar activity to eIF4E purified from yeast. (A) Analysis of decapping activity by DCP1 alone (left panel) and in the presence of either eIF4E (middle panel; 71.8 pmol of His-eIF4E) or a nonfunctional allele of eIF4E (right panel, labeled ts-4E; 71.8 pmol of His-ts-4E) purified from E. coli. Decapping by His-DCP1 was assayed over a 30-min time course, with aliquots removed at the designated time points. (B) Relative decapping activity in the presence of increasing amounts of His-eIF4E. Relative activity was determined by assaying decapping ± eIF4E in multiple experiments and is expressed as the amount of decapping relative to DCP1 alone (set at 100%). The 30-min time point is shown on the histogram.
Mutations in eIF4E can suppress dcp1 mutants in vivo.
The observation that eIF4E inhibits decapping in vitro suggests a simple relationship between eIF4E and Dcp1p, wherein a competition for the cap structure leads to either translation initiation or decapping, depending on which protein binds to the cap. This model predicts that mutations in Dcp1p should be suppressible by loss-of-function alleles of eIF4E in vivo. In order to test this prediction, we created a strain carrying the partially functional dcp1-1 allele in combination with the temperature-sensitive allele of eIF4E. This allele of dcp1 gives reduced levels of decapping at 24°C and is essentially null for decapping at 37°C in this strain background.
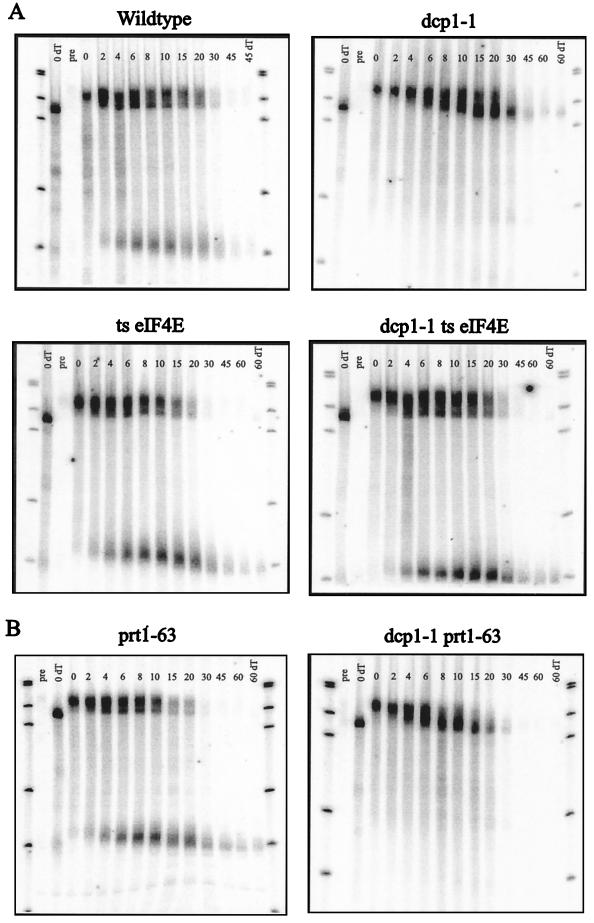
To examine decapping in this experiment, we utilized a transcriptional pulse-chase experiment. In this procedure, we utilized the MFA2pG gene under control of the GAL1 upstream activation sequence (14). This gene also carries an insertion of a poly(G) tract in its 3′ UTR that inhibits the 5′→3′ exonuclease and thereby traps an mRNA decay intermediate (14). In these specific experiments, strains were grown in raffinose-containing medium at 24°C and then shifted to 38°C for 1 h to inactivate the temperature-sensitive eIF4E protein. Galactose was added to induce transcription of the MFA2 gene for 6 min, followed by dextrose addition to repress the MFA2 gene. This burst of transcription created a population of mRNAs that were all full length with long poly(A) tails. Time points were examined after transcriptional repression to allow observation of the decay rate of the full-length mRNA, the deadenylation kinetics, and the appearance of the poly(G) fragment, which also gives an estimate for the rate of decapping.
In this assay, the MFA2pG transcript from a wild-type strain deadenylated rapidly [based on differences in the full-length mRNA relative to a sample with the poly(A) tail removed], and upon deadenylation to an oligo(A) length, the mRNA rapidly decapped, leading to the production of the poly(G) fragment (Fig. 6). In contrast, the full-length oligoadenylated MFA2pG mRNA from the dcp1-1 strain was stabilized, and essentially no fragment was produced (Fig. 6). An important observation is that the temperature-sensitive eIF4E mutation now suppressed the decapping defect seen in the dcp1-1 strain and restored both degradation of the full-length mRNA and the appearance of mRNA decay fragment. This suppression must be due to increased function of the Dcp1-1p, since temperature-sensitive eIF4E dcp1Δ strains show no decapping at high temperature (40). This observation is consistent with the in vitro data presented above and argues that there is a competition between Dcp1p and eIF4Ep for the cap substrate in vivo. Moreover, this observation suggests that the Dcp1-1p, which retains approximately 15% of wild-type decapping activity (21), has a defect in decapping due to an inability to compete with eIF4E (see Discussion).
FIG. 6.
Mutations in translation initiation factors have different effects on the decapping defect of a partially functional DCP1 allele. (A) A mutation in the eIF4E protein can suppress the decapping defect of a partially functional DCP1 allele. (B) A mutation in the PRT1 protein cannot suppress the decapping defect of a partially functional DCP1 allele. Transcriptional pulse-chase analysis of the MFA2pG mRNA was used to examine the levels of fragment RNA produced through decapping-dependent degradation of the body of the message. The time points used after a 6-min transcriptional induction and subsequent repression are shown above each lane. The bottom band is the decay intermediate stabilized by the poly(G) insertion in the 3′ UTR. The 0 dT lane is the 0-min time point in which the poly(A) tail has been completely removed by cleavage with RNase H and oligo(dT). This allows for comparison of poly(A) tail lengths over the time course. The blots were probed with oligonucleotide oRP140 (5′-ATATTGATTAGATCAGGAATTCC-3′). ts, temperature sensitive; pre, preinduction time point.
Mutations in eIF4E do not suppress the decapping defects seen in lsm1Δ, pat1Δ, or dcp2Δ strains.
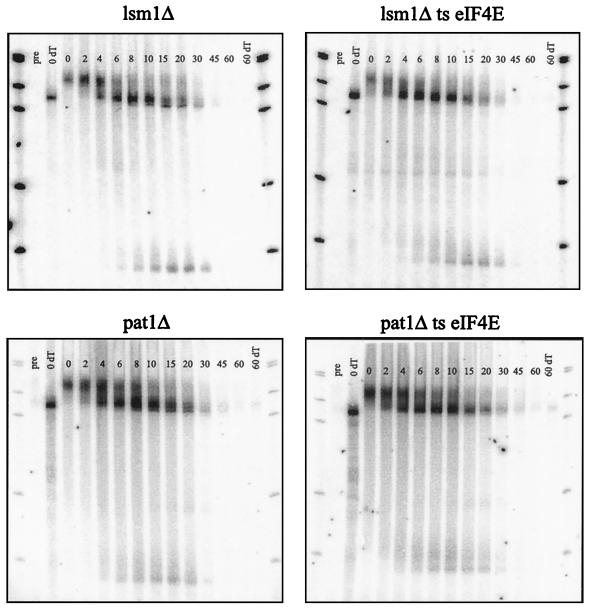
The observations presented above argue that one step in mRNA decapping is a competition between the cap binding protein and the decapping enzyme. This suggests that one manner in which decapping will be modulated is by proteins that influence this competition. One prediction of this model is that mutations in proteins that regulate the decapping reaction, either by affecting eIF4E binding to the cap, or by activating Dcp1p function, will be suppressed by the temperature-sensitive allele of eIF4E in a manner analogous to that of the dcp1-1 lesion. Given this logic, we asked if defects in other proteins that lead to an inhibition of decapping can be suppressed by the temperature-sensitive eIF4E allele. Mutations in the dcp2, lsm1, and pat1 genes were used in this analysis. Dcp2p is a protein that interacts with Dcp1p and is necessary for decapping (15). Several observations indicate that Dcp2p is required for the production of active Dcp1 enzyme, but is not required for the Dcp1p to function once it has been made. For example, Dcp1p purified from a dcp2Δ strain was completely inactive (15). In contrast, Dcp1p produced in a DCP2 cell, but separated from Dcp2p by a high-salt wash, was fully functional (T. Dunckley, M. Tucker, and R. Parker, submitted for publication). Lsm1p is a member of a seven-protein complex that binds to mRNA and is necessary for efficient decapping (7–9, 45). Pat1p interacts with the Lsm complex and is also required for efficient decapping (46). Deletions of each of these three genes were combined with the temperature-sensitive allele of eIF4E, and the levels of decapping were analyzed from each strain by using the same transcriptional pulse-chase analysis described earlier.
An interesting observation is that the temperature-sensitive eIF4E/cdc33-42 mutation did not suppress the decapping defect seen in the lsm1Δ, pat1Δ, and dcp2Δ mutants (Fig. 7) (data not shown). Quantification of the decay rate of the full-length MFA2 mRNA or the levels of poly(G) fragment produced in each strain shows that the eIF4E mutation in combination with either a lsm1Δ or pat1Δ mutation gives identical mRNA decay, as seen in the lsm1Δ or pat1Δ strains alone (data not shown). The failure of the temperature-sensitive eIF4E allele to suppress the dcp2Δ mutation may not be that surprising, since this allele abolishes all decapping and fails to produce active decapping enzyme (15). However, the failure of the temperature-sensitive eIF4E allele to suppress the decapping defect in the lsm1Δ or the pat1Δ strains is likely to be highly significant, because the decapping defect seen in these strains is less severe than the defect seen in the dcp1-1 allele (compare Fig. 6 to Fig. 7). This indicates that the ability of a temperature-sensitive eIF4E lesion to suppress a decapping defect is not a function of the strength of the decapping defect per se, but instead depends on the exact change in the decapping reaction. This observation strongly implies that Lsm1p and the associated Lsm-Pat1p complex (45) affect a step in mRNA decapping distinct from the competition between Dcp1p and eIF4Ep (see Discussion).
FIG. 7.
A mutation in the eIF4E protein cannot suppress the decapping defect of a lsm1Δ or a pat1/mrt1Δ mutant. Transcriptional pulse-chase analysis of the MFA2pG mRNA was used to examine the levels of fragment RNA produced through decapping-dependent degradation of the body of the message. The time points examined after a 6-min transcriptional induction and subsequent repression are shown above each lane. The bottom band is the decay intermediate stabilized by the poly(G) insertion in the 3′ UTR. The blots were probed with oligonucleotide oRP140. ts, temperature sensitive; pre, preinduction time point.
A second interesting observation from these experiments is that there was an additional shortening of the mRNA from the 3′ end in the lsm1Δ and pat1Δ strains similar to what has been previously described for the lsm1 mutant strains (7). This phenomenon was seen in the continued shortening of the full-length mRNA after deadenylation was complete leading to the production of an mRNA lacking ∼10 to 15 additional nucleotides at the 3′ end. This difference can be more clearly seen in the low levels of the mRNA decay fragment produced in lsm1Δ and pat1Δ strains, which was shorter than the corresponding fragment seen in wild-type cells by approximately 15 nucleotides (Fig. 7). However, this difference in 3′ trimming into the mRNA body was not simply due to an inhibition of decapping, since dcp1Δ or dcp2Δ strains, which are completely blocked for decapping, did not show 3′ trimming into the body of the mRNA (compare Fig. 6 and 7) (6, 15). One interpretation of these results is that the Lsm-Pat1p complex normally binds to the 3′ end of the mRNA, and following deadenylation, this complex inhibits additional 3′ trimming into the mRNA body.
Mutations in PRT1 do not suppress dcp1 mutants in vivo.
Previous work has shown that mutations in several different translation initiation factors can cause an increase in the rate of decapping (40). The mutation which causes the greatest change in mRNA half-life is the prt1-63 allele, which is a component of the eIF3 complex involved in delivering the 40S ribosomal subunit to the cap binding complex (20, 33). A mutation in the Prt1 protein is thought to prevent this delivery from occurring, which may destabilize the cap binding complex, leading to faster rates of decapping. For this reason, we determined whether the prt1-63 mutation, which leads to faster decapping, could suppress the decapping defect of the dcp1-1 allele similarly to the temperature-sensitive eIF4E mutation.
As described previously with the temperature-sensitive eIF4E allele, prt1-63 was combined with the dcp1-1 allele, and transcriptional pulse-chase analysis was performed to examine the levels of decapping from the double mutant strain. As shown in Fig. 6B, the prt1-63 mutation looks similar to the wild-type strain for the MFA2pG mRNA, but does have shorter half-lives and increased levels of fragment being produced. In contrast, the strain containing both the prt1-63 and the dcp1-1 mutations looks identical to the strain containing the dcp1-1 mutation alone. This result shows that a mutation that inactivates the eIF3 complex is not able to suppress the decapping defect of the dcp1-1 allele. This implies that the function of the Prt1 protein is distinct from the competition between eIF4Ep and Dcp1p. When eIF3 is not fully functional, eIF4E and the cap binding complex are still an effective block to the partially functional Dcp1-1 protein (see Discussion).
DISCUSSION
The cap binding protein eIF4E is an inhibitor of decapping.
Several lines of evidence indicate that when the cap binding protein, eIF4E, is bound to the cap structure, mRNA decapping is blocked. First, eIF4E protein purified from yeast or E. coli is sufficient to inhibit Dcp1p in vitro (Fig. 2 and 5). This inhibition is due to eIF4E binding the cap structure, since m7GTP can abolish the inhibition and restore decapping (Fig. 4). Moreover, a mutant version of eIF4E defective in cap binding fails to inhibit decapping (Fig. 3). Second, defects in eIF4E increase the rate of decapping in yeast cells (40). Third, the temperature-sensitive allele of eIF4E can suppress the decapping defect of a dcp1-1 mutant at the restrictive temperature (Fig. 6). Since the dcp1-1 allele is known to reduce decapping activity to approximately 15% of wild-type levels (21), the simplest interpretation is that the competition between eIF4E and Dcp1p is at the level of Dcp1p interaction with the cap structure (Fig. 8). The combination of these in vivo and in vitro observations strongly argues that eIF4E is an inhibitor of decapping and imply that dissociation of eIF4E from the cap structure is required before mRNA decapping and degradation can occur.
FIG. 8.
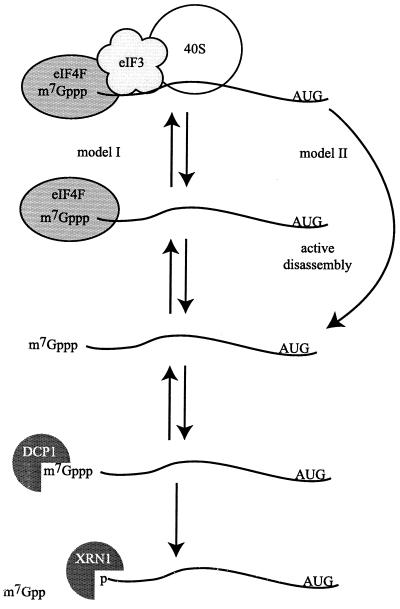
A model representing how translation initiation and decapping may compete for the mRNA cap structure. The left-hand side (model I) shows translation initiation in various states of assembly. When eIF4F is bound to the cap structure, the Dcp1p may compete for cap binding; once recognition of the cap occurs by the Dcp1p, decapping immediately targets the mRNA for degradation. The right-hand side (model II) diagrams an active mode of disassembly for the translation initiation machinery, leading to an unprotected mRNA.
We have demonstrated that eIF4E can inhibit Dcp1p in vitro; however, excess amounts of eIF4E are required to achieve this effect. Consistent with this observation, several lines of evidence suggest a similar situation may exist in vivo. First, eIF4E binds capped mRNA weakly by itself (estimated Kd of 1 to 10 μM) (35) and is thought to normally interact with mRNAs in conjunction with other RNA binding proteins, most notably eIF4G (19, 27). In contrast, the Km for Dcp1p interacting with capped mRNA has been estimated at 10 nM (26), suggesting that much larger amounts of purified eIF4E would be needed in an in vitro reaction to block Dcp1p activity. This is potentially an important difference, since it implies that the critical changes that lead to weakened eIF4E binding and subsequent decapping may be the loss of protein-protein interactions between eIF4E and proteins, such as eIF4G and, indirectly, Pab1p, that stabilize its interaction with the mRNA in vivo.
Distinct functions of the Lsm-Pat1p complex and Dcp1p in the decapping of mRNAs.
Several observations suggest that the functions of the Lsm-Pat1p complex and the Dcp1p in the decapping of mRNAs are distinct. For example, although the temperature-sensitive eIF4E allele suppresses the decapping defect of the dcp1-1 lesion in vivo, the temperature-sensitive eIF4E allele had no affect on the decapping defect in lsm1Δ or pat1Δ strains (Fig. 6 and 7). These results indicate that decapping is a multistep process and suggest that the Lsm and Pat1 proteins act at a stage distinct from the competition between Dcp1p and eIF4E.
There are several possible manners in which the Lsm and Pat proteins could promote decapping and subsequent degradation. First, this protein complex could act downstream of the Dcp1p and eIF4E competition, perhaps to promote catalysis by Dcp1p once it has bound the cap structure. This possibility would also provide a rationale for the association of the Lsm-Pat1p complex with the Xrn1p (8), the ribonuclease responsible for 5′→3′ digestion of the mRNA body following decapping (22). The interaction with Xrn1p also raises the formal possibility that these proteins also affect the 5′→3′ exonucleolytic digestion following decapping. However, three observations suggest that these proteins are not normally required for 5′→3′ exonucleolytic degradation. First, examination of the mRNA species that accumulate in lsm or pat1 mutants indicates that they retain the cap structure (7, 21, 41, 45). Second, the finding that some mRNA fragment arising by 5′→3′ digestion is produced in these mutants suggests that whenever an mRNA is decapped, 5′→3′ digestion proceeds normally. Third, the decapping and 5′→3′ exonucleolytic degradation of mRNAs containing early nonsense codons are normal in lsm and pat1 strains (7, 9, 21). Thus, although unlikely for the reasons discussed above, we cannot rule out the formal possibility that the failure of the temperature-sensitive allele of eIF4E to suppress the decapping defect in lsm1Δ or pat1Δ strains is due to an additional block to 5′→3′ decay that only occurs when the decapping enzyme is defective. Nevertheless, it is clear that the function of the Lsm complex and Pat1p must include effects at a step distinct from the Dcp1p and eIF4E competition.
An alternative possibility is that the Lsm-Pat1p complex acts upstream of the Dcp1p-eIF4E competition either to recruit Dcp1p to the mRNA or to affect the process of translation initiation in a manner that ultimately affects the stability of the interaction between the cap binding protein and the cap. This hypothesis is suggested by the observations that two-hybrid interactions have been detected between the Lsm complex and three proteins involved in the process of AUG recognition (17): eIF2, which delivers the tRNA to the ribosome; eIF2b, which is involved in GTP-GDP exchange of eIF2 (for review, see reference 28); and RPS28, which is a ribosomal protein localized to the site of mRNA decoding (1). Future experiments investigating the proteins involved in mRNA decay should provide insight into these important issues.
Possible mechanisms for eIF4E release from the cap structure.
Our results argue that dissociation of eIF4E, or the larger eIF4F complex, from the cap structure is a key step in facilitating decapping. Given this, a critical issue is what triggers disassembly of the cap binding complex from mRNA. In principle, there are two, nonexclusive, mechanisms that are consistent with current data. In the first model, the stability of eIF4F on the cap complex is a function of a series of reversible assembly and disassembly steps, and any change that drives one of these reactions toward the disassembled state would favor decapping (Fig. 8, model I). Thus, defects in eIF4E would promote decapping, because they would increase the disassembly of eIF4F from the mRNA. Similarly, mutations in eIF3 could promote decapping, because they allow the eIF4F-mRNA complex an increased chance to disassemble. However, the nature of the effect would be different in each case. Thus, for prt1-63, the lesion would presumably limit the interaction of eIF3 and 40S subunits to the cap binding complex, still allowing the normal competition between eIF4E and Dcp1p. This would explain why a prt1-63 mutation fails to suppress the decapping defect of dcp1-1. In contrast, the temperature-sensitive eIF4E allele would directly affect the eIF4E-Dcp1p competition, thereby enhancing the decapping rate in wild-type cells and suppressing the decapping defect in dcp1-1 strains.
A related, but fundamentally similar, model is that disassembly of the translation initiation complex may in some cases be an active process that promotes removal or degradation of the translation initiation complex (Fig. 8, model II), perhaps in response to the failure to complete a translation initiation event. The latter model might also explain why any defect in translation initiation up to AUG recognition promotes mRNA decapping, whereas inhibition of translation elongation stabilizes mRNAs (reviewed in reference 39). It will be important in future work to determine the mechanisms by which the translation initiation complex is disassembled.
ACKNOWLEDGMENTS
This work was supported by the Howard Hughes Medical Institute and by a grant to R.P. from the National Institutes of Health (GM45443).
We thank Elizabeth Little for the GST expression vector used in this work and Rhett Michelson for technical comments on the manuscript. In addition, we thank the members of the Parker laboratory, in particular John Jacobs Anderson, for their support and contributions to the preparation of the manuscript.
REFERENCES
- 1.Alksne L E, Anthony R A, Liebman S W, Warner J R. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci USA. 1993;90:9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmann M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A K. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashkirov V I, Scherthan H, Solinger J A, Buerstedde J M, Heyer W D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 6.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 7.Boeck R, Lapeyre B, Brown C E, Sachs A B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caponigro G, Muhlrad D, Parker R. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker C J, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 14.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 15.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edery I, Altmann M, Sonenberg N. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene. 1988;74:517–525. doi: 10.1016/0378-1119(88)90184-9. [DOI] [PubMed] [Google Scholar]
- 17.Fromont-Racine M, Mayes A E, Brunet-Simon A, Rain J C, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs J D, Legrain P. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gera J F, Baker E J. Deadenylation-dependent and -independent decay pathways for α1-tubulin mRNA in Chlamydomonas reinhardtii. Mol Cell Biol. 1998;18:1498–1505. doi: 10.1128/mcb.18.3.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 20.Hannig E M. Protein synthesis in eukaryotic organisms: new insights into the function of translation initiation factor eIF-3. Bioessays. 1995;17:915–919. doi: 10.1002/bies.950171103. [DOI] [PubMed] [Google Scholar]
- 21.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.LaGrandeur T, Parker R. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA. 1999;5:420–433. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaGrandeur T E, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanker S, Muller P P, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 28.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 30.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 32.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 33.Naranda T, MacMillan S E, Hershey J W. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- 34.Rhoads R E. Cap recognition and the entry of mRNA into the protein synthesis initiation cycle. Trends Biochem Sci. 1988;13:52–56. doi: 10.1016/0968-0004(88)90028-x. [DOI] [PubMed] [Google Scholar]
- 35.Rhoads R E, Joshi-Barve S, Rinker-Schaeffer C. Mechanism of action and regulation of protein synthesis initiation factor 4E: effects on mRNA discrimination, cellular growth rate, and oncogenesis. Prog Nucleic Acid Res Mol Biol. 1993;46:183–219. doi: 10.1016/s0079-6603(08)61022-3. [DOI] [PubMed] [Google Scholar]
- 36.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rother R P, Frank M B, Thomas P S. Purification, primary structure, bacterial expression and subcellular distribution of an oocyte-specific protein in Xenopus. Eur J Biochem. 1992;206:673–683. doi: 10.1111/j.1432-1033.1992.tb16973.x. [DOI] [PubMed] [Google Scholar]
- 38.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz D C, Parker R. The interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae: translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 40.Schwartz D C, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 43.Sonenberg N, Morgan M A, Merrick W C, Shatkin A J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci USA. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 45.Tharun S, He W, Mayes A E, Lennertz P, Beggs J D, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 46.Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 48.Zhang S, Williams C J, Hagan K, Peltz S W. Mutations in VPS16 and MRT1 stabilize mRNAs by activating an inhibitor of the decapping enzyme. Mol Cell Biol. 1999;19:7568–7576. doi: 10.1128/mcb.19.11.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]