Abstract
In mammals, nuclear localization of U-snRNP particles requires the snRNA hypermethylated cap structure and the Sm core complex. The nature of the signal located within the Sm core proteins is still unknown, both in humans and yeast. Close examination of the sequences of the yeast SmB, SmD1, and SmD3 carboxyl-terminal domains reveals the presence of basic regions that are reminiscent of nuclear localization signals (NLSs). Fluorescence microscopy studies using green fluorescent protein (GFP)-fusion proteins indicate that both yeast SmB and SmD1 basic amino acid stretches exhibit nuclear localization properties. Accordingly, deletions or mutations in the NLS-like motifs of SmB and SmD1 dramatically reduce nuclear fluorescence of the GFP-Sm mutant fusion alleles. Phenotypic analyses indicate that the NLS-like motifs of SmB and SmD1 are functionally redundant: each NLS-like motif can be deleted without affecting yeast viability whereas a simultaneous deletion of both NLS-like motifs is lethal. Taken together, these findings suggest that, in the doughnut-like structure formed by the Sm core complex, the carboxyl-terminal extensions of Sm proteins may form an evolutionarily conserved basic amino acid-rich protuberance that functions as a nuclear localization determinant.
Splicing of nuclear pre-mRNAs in yeast and mammals occurs in the multicomponent complex called the spliceosome whose formation involves the ordered assembly of the U1, U2, and U4/U6.U5 snRNPs, together with a multitude of non-snRNP-associated protein factors on the pre-mRNA substrate (27, 67). In both systems, except for the U6 snRNP, each particle is composed of a U snRNA, a set of specific proteins associated with one particular snRNP, and a set of common (or core) proteins shared by all snRNPs. This last group is composed of the Sm proteins B, B′ (in mammals), D1, D2, D3, E, F, and G, which assemble around the Sm site of the snRNA (34). In addition to the common Sm core proteins, the yeast and metazoan U6, U4/U6, and U4/U6.U5 snRNPs also contain seven distinct proteins called Lsm (like Sm) exhibiting clear homology to the Sm proteins (1, 7, 19, 21, 39, 55, 62, 68).
The Sm and Lsm proteins are highly conserved in all eukaryotic organisms. They contain the Sm domain, which consists of two conserved regions separated by a linker of variable length. Seven residues are highly conserved, and at many positions, the physicochemical property of the amino acid is maintained (7, 21, 62). The Sm motif is important for Sm protein function. Indeed, mutations at various positions in this motif, and mostly the hydrophobic conserved residues, abolish protein-protein interactions between Sm partners and hinder Sm core RNP complex formation (5, 21). Based on the crystal structures of two human Sm protein complexes (SmB/SmD3 and SmD1/SmD2), a model in which the seven Sm proteins form a doughnut-like structure has been proposed (26).
Assembly of the eukaryotic U snRNPs is a multistep process following an ordered pathway (36, 47). After transcription by RNA polymerase II, the snRNAs are exported to the cytoplasm. The Sm proteins, which are stored in the cytoplasm (35, 57), then assemble onto the snRNA Sm site. This binding allows hypermethylation of the snRNA 7-methyl cap by a methylase to form a methyl-2,2,7-guanosine cap structure (35, 46). This binding also generates a bipartite nuclear localization signal (NLS) composed of the Sm core complex and the snRNA cap structure (11, 13, 14, 20). This signal will permit the import of the newly made snRNPs to the nucleus. Addition of snRNP-specific proteins to the core snRNP, in the cytoplasm and/or the nucleus, completes the assembly of functional snRNPs (42). Competition experiments indicate that snRNPs are imported by specific receptors not shared by other classes of nuclear proteins (12, 24, 40). It has also been shown that U snRNP import is mediated by importin β, which functions in this process without the NLS-specific importin α receptor (44). Recently, the import receptor for the m3G cap structure has been identified in mammals (23). This protein, called snurportin1, enhances the m3G-cap-dependent nuclear import of U snRNPs. Snurportin1, which is an importin α-like adapter, recognizes only the m3G cap but not the Sm core NLS, indicating that at least two distinct import receptors interact with the snRNP bipartite NLS (23).
Immunofluorescence staining experiments with mammalian cells allowed the detection of a complex subcellular localization of snRNPs. In the nucleus, in addition to a diffuse nucleoplasmic snRNP staining, the snRNP particles are localized in speckles, coiled bodies, and nucleoli (6, 30, 65, 66). The snRNP protein complexes are also localized into the cytoplasmic compartment as discrete punctuate structures which are uniformly distributed and which may represent storage particles of the snRNP core protein complexes or staging centers for snRNP core particle assembly (72).
Interest in snRNP biogenesis has increased since it has been shown that defects in this process are correlated with a human motor neuron degenerative genetic disease, spinal muscular atrophy (15, 33, 37, 45). Most of the studies of snRNP biogenesis have been performed in metazoan systems. In yeast, recent analyses indicate a conservation of the Sm protein-protein interaction sites, suggesting that snRNP structure would be conserved and that snRNP assembly could follow a pathway similar to that in mammals (5). However, it is not yet clear if, in yeast, snRNAs transit through the cytoplasm during snRNP formation. In order to get more information on the mechanisms governing snRNP biogenesis, the yeast system was further exploited. The goal of this study was to define more precisely the molecular basis of the NLS carried by the Sm core complex. In this regard, yeast SmB, SmD1, and SmD3 proteins possess, in their carboxyl-terminal regions, stretches of basic amino acids that are reminiscent of nuclear import signals found in nuclear proteins as well as in ribosomal proteins. Although similar motifs are not found in the human counterparts, it is noteworthy that the human SmB, SmD1, and SmD3 proteins also possess C-terminal extensions rich in basic amino acids (see Discussion). In this report, using fluorescence microscopy studies combined with mutational and functional analyses, I show that the yeast SmB and SmD1 NLS-like motifs exhibit nuclear localization properties. The results suggest that a basic amino acid-rich protuberance formed by the C-terminal extensions of Sm proteins may represent the nuclear localization determinant of the Sm core complex, in both yeast and human cells.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The wild-type YDL401 strain (MATa ura3-52 trp1 his3-Δ200 leu2-Δ1 gal2 galΔ108) (29) was obtained from B. Séraphin and was used for subcellular localization studies of green fluorescent protein (GFP)-Sm fusion alleles and for whole-cell extract preparation. The YAMB strain (YPH background) (64) (MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-Δ/ade2-Δ ade3-Δ/ade3-Δ trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1) was obtained from A. Camasses and was used for SMB and SMD1 gene disruptions. Derived haploid strains were used for phenotypic analyses. Yeast strains were grown using standard procedures and media as previously described (10). Yeast transformations were performed by the lithium acetate method (22).
Gene disruptions.
DNA fragments carrying precise disruptions of SMB and SMD1 genes were generated by PCR using the KAN-MX4 cassette (70) as template with the following oligonucleotides: for SMB, S1BK (5′-AGAACCATCCGTATAACAGGTTTCCAGTAAAGTAAATAACTCGTACGCTGCAGGTCGAC) and S2BK (5′-TGCGTACACAAAAAAAGTATACGGAAACTATATTAGACTACATCGATGAATTCGAGCTCG), and for SMD1, S1D1K (5′-AGATGACTCCAAGTATCGTTTATAAATCGTCGAGAAAAAGATCGTACGCTGCAGGTCGAC) and S2D1K (5′-ACCTCTTCTTGGCCTTTTATTCGCTGGTGCACCAAAATCGCATCGATGAATTCGAGCTCG).
To generate chromosomal disruptions of the SMB and SMD1 genes, the PCR-generated fragments were transformed in the diploid YAMB strain. Kan+ transformants were isolated on selective medium, and deletions of the SMB and SMD1 genes were verified by PCR. After sporulation and dissection of the resulting tetrads, only two spores were always recovered and all viable spores were Kan−, confirming that both SMB and SMD1 genes are essential (16, 53, 55). For each disruption, the lethal phenotypes were complemented by the corresponding wild-type gene placed under the GAL1 promoter.
A second yeast smd1::LEU2 deletion strain was constructed with the YAMB strain by using an smd1Δ::LEU2 XbaI-PstI DNA fragment in which SMD1 sequences were replaced by the LEU2 gene (51) (kindly provided by B. Rymond). After transformation of a YAMB diploid strain with this DNA fragment, diploids growing on −Leu plates were selected. Hemizygotes were sporulated, and the resulting tetrads were dissected. Tetrad progeny showed a 2:2 segregation of the LEU2 marker. An smd1::LEU2 spore carrying the pGFP-SmD1 plasmid was obtained after sporulation and dissection of an smd1::LEU2/SMD1 diploid carrying the pGFP-SmD1 plasmid (URA3 marker).
To construct a yeast strain deleted simultaneously of SMB and SMD1 genes, an smd1::LEU2 spore (a mating type) carrying the pGFP-SmD1 plasmid was mated to an smb::KAN strain (α mating type) carrying a pGAL1-SmB plasmid (TRP1 marker). After mating, diploids were selected on −Ura −Trp selective plates. Diploids were sporulated, and the resulting tetrads were dissected. An smd1::LEU2 smb::KAN spore carrying the pGFP-SmD1 and pGAL1-SmB plasmids was selected. This yeast strain was called YRB110. YRB110 was transformed with the pGFP-H-SmD1ΔNLS plasmid carrying the HIS3 selective gene. Transformants were cured of plasmid pGFP-SmD1 (URA3 marker) by growth on plates containing galactose and 5-fluoroorotic acid. The resulting strain was named YRB120 (MATa smd1::LEU2 smb::KAN pGFP-H-SmD1ΔNLS pGAL1-SmB) and was used for phenotypic studies.
Plasmid constructions.
Plasmid were constructed by standard methods (56). To construct the pGFP-SmB, pGFP-SmD1, pGFP-SmD2, pGFP-SmD3, and pGFP-SmG fusion alleles, the corresponding Sm coding sequences were purified as an NcoI-XhoI fragment (the NcoI being blunt ended with Klenow enzyme) after digestion of pBS1280, pBS1282, pBS1284, pBS1286, and pBS1105 (5), respectively. These fragments were transferred into pGFP-Nfus (URA3 CEN) vector (43) previously cut with XmaI, blunt ended with Klenow enzyme, and cut with XhoI. The pGFP-SmF fusion allele was constructed by subcloning a BamHI-XhoI DNA fragment (the BamHI site being blunt ended with Klenow enzyme) obtained from the pKS(−)SmF plasmid (5) into pGFP-Nfus cut with XmaI, blunt ended with Klenow enzyme, and cut with XhoI. The pGFP-SmE fusion allele was constructed by subcloning a BamHI-XhoI DNA fragment (the BamHI site being blunt ended with Klenow enzyme) obtained from the pACTII-SmE plasmid (5) into pGFP-Nfus cut with XmaI, blunt ended with Klenow enzyme, and cut with XhoI. In all those plasmids, the GFP-Sm fusion alleles are placed under the inducible MET25 promoter (41, 43). The sequences of cloning junctions were verified by dideoxynucleotide sequencing.
To place the SMB gene under the GAL1 promoter, an NcoI-EcoRI (the NcoI site being blunt ended with Klenow enzyme) fragment obtained from pBS1280 (5) was purified and cloned into pGAL1 (TRP1 CEN) vector (2) digested with BamHI, blunt ended with Klenow enzyme, and cut with EcoRI. This plasmid was named pGAL1-SmB. To place the SMD1 gene under the GAL1 promoter, a PCR product was amplified from pGFP-SmD1 plasmid using oligonucleotides GFP (5′-ACGAAAAGAGAGATCACATGATC) and RBD1End (5′-GTGACAGTGAATTCGACAACGACA). The PCR product was digested with BamHI-EcoRI and cloned into the corresponding sites of the pUGAL1 (URA3 CEN) vector. This plasmid was called pUGAL1-SmD1.
Regions encompassing the NLS-like portions of the SmB (residues 104 to 147), SmD1 (residues 127 to 146), and SmD3 (residues 83 to 101) coding sequences were amplified by PCR with the following specific primer pairs that generate BamHI and EcoRI sites at the 5′ and 3′ ends, respectively: for SmB-NLS, RBspeA (5′-AAGCCGCTAGGATCCAAGAAGG) and RBspeB (5′-CCCTAGAATTCCTTTAAGTATGCTTCG), for SmD1-NLS, RB-D1Bam (5′-ATTGCAAATGGATCCAGCAAAAAG) and RB-D1End (5′-GTGACAGTGAATTCGACAACGACA), and for SmD3-NLS, RB-D3Bam (5′-TTAAAGAATGGATCCTTATTCAAA) and RB-D3Eco (5′-TACAATGATGAATTCGTTTCTGCC). The PCR products were digested with BamHI and EcoRI and ligated into the BamHI and EcoRI sites of a pGFP-GFP vector (URA3 CEN) carrying two in-frame GFPs. This vector was constructed as follows: pGFP-Nfus plasmid (43) was digested with EcoRI, blunt ended with mung bean nuclease, and cut with BamHI. The GFP gene-containing fragment was purified and cloned into pUG36 vector previously cut with SpeI, blunt ended with Klenow enzyme, and digested with BamHI. The pUG36 (URA3 CEN) vector is identical to the pGFP-Nfus vector except that it has unique BamHI and EcoRI sites. All PCR-generated insertions were sequenced to insure that mutations were not introduced into the sequences.
Construction of GFP-Sm mutant alleles.
To construct GFP-SmB mutant alleles, the following specific pairs of primers were used: for pGFP-SmBΔNLSΔC (residues 1 to 103 of SmB), RBspeC (5′-ACTAGTGGATCCCCCGGCATGGTA) and RBspeD (5′-GTCTTTCCGAATTCTATAGTAGCGGCT), for pGFP-SmB-Cter (residues 149 to 196 of SmB), RBspeE (5′-GCGAAGCATACTGGATCCAATTCTAGG) and RBspeF (5′-CTATATTAGAATTCACTACATCAA), and for pGFP-SmBΔC (residues 1 to 147 of SmB), RBspeC (5′-ACTAGTGGATCCCCCGGCATGGTA) and RBspeB (5′-CCCTAGAATTCCTTTAAGTATGCTTCG). After PCR amplification using the pGFP-SmB plasmid as template, the products were cloned as BamHI-EcoRI fragments into BamHI-EcoRI-digested vector puG36 (URA3 CEN). To construct pGFP-SmD1ΔNLS (residues 1 to 115 of SmD1) (URA3 CEN plasmid) and pGFP-H-SmD1ΔNLS (HIS3 CEN plasmid), a PCR product was amplified from pGFP-SmD1 plasmid using oligonucleotides GFP (5′-ACGAAAAGAGAGATCACATGATC) and RBD1Eco (5′-GACCCGATCGAATTCAGGAATTAAGT). The PCR product was cut with BamHI-EcoRI and cloned into the corresponding sites of pUG36 (URA3 CEN) and pUG34 (HIS3 CEN) vectors (43). The wild-type SMD1 gene was also cloned into pUG34 (HIS3 CEN) by using the same strategy with oligonucleotides GFP (5′-ACGAAAAGAGAGATCACATGATC) and RBD1End (5′-GTGACAGTGAATTCGACAACGACA). The plasmid was called pH-GFP-SmD1.
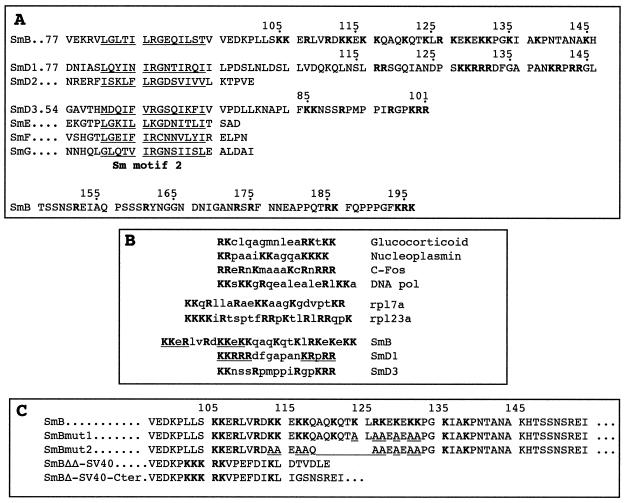
Oligonucleotide-directed mutagenesis of the SMB gene was performed by using the megaprimer strategy (63) with appropriate primers and pGFP-SmB as template. The following mutagenic oligonucleotides were synthesized: SmBmut1 (5′-TCCCGGCGCTGCCTCTGCCTCTGCTGCTGCTAGTGCCGTCTGC) and SmBmut2 (5′-TCCCGGCGCTGCCTCTGCCTCTGCCGCTTGTGCCGCTTCCGCTGCATCTCTCACTAG). The sequences of the mutants were verified by DNA sequencing, and the structures of the generated alleles are shown in Fig. 1C.
FIG. 1.
Amino acid sequence comparison of the C-terminal extensions of yeast Sm proteins. (A) The C-terminal domains of the yeast SmB, SmD1, and SmD3 proteins contain regions rich in basic amino acids (in bold). The evolutionarily conserved Sm motif 2 is underlined. The numbers at the left correspond to the position of the first amino acid shown in the sequence. (B) Comparison of the NLS-like motifs found in the yeast Sm proteins with other nuclear localization motifs. The positions of the basic amino acid-rich regions in the yeast Sm proteins are as follows: SmB, residues 105 to 132; SmD1, residues 128 to 144; and SmD3, residues 85 to 101. The sequences of SmB and SmD1 showing similarities with classical monopartite SV40-type NLSs are underlined. (C) Mutations generated in the SmB NLS-like motif. Basic amino acids are shown in bold. The mutated and deleted positions are underlined in the sequences of the SmBmut1 and SmBmut2 alleles.
The pPS815 vector (URA3 2μm) carries the NLS of simian virus 40 (SV40) large T antigen (Tag) at the N terminus of a GFP–β-galactosidase (GFP–β-Gal) reporter protein (31). This fusion is placed behind the ADH1 promoter and was used to generate different SV40 NLS-GFP-SmB fusion alleles as follows. The pGFP-SmB and pGFP-SmB mutant plasmids (pUG36 vector background) were cut with PmlI and XhoI and blunt ended by Klenow treatment. The PmlI-XhoI fragments were purified and cloned into pPS815 vector digested with PmlI and XbaI and blunt ended with Klenow enzyme. The resulting plasmids were named pSV40/NLS-GFP-SmB, pSV40/NLS-GFP-SmBΔNLSΔC, pSV40/NLS-GFP-SmBmut1, and pSV40/NLS-GFP-SmBmut2. Correct cloning of the fragment was verified by digestion with PmlI and sequencing.
To construct plasmid pGFP-SmBΔΔ-SV40 carrying the NLS of SV40 Tag, a PCR product was amplified from pGFP-SmB plasmid using oligonucleotides GFP (see above) and BSV40 (5′-AAGTTAGATATCGAATTCCGGGACCTTTCTCTTCTTTTTTGGCTTATCCTCCACCACTGTGGATAAGATCTGTTCTCCTCTTAG). The PCR product was cut with BamHI-EcoRI and cloned into the corresponding sites of pUG36 (URA3 CEN) vector. The structure of the generated allele is shown in Fig. 1C. Plasmid pGFP-SmBΔ-SV40-Cter was constructed as follows. A PCR fragment carrying the C-terminal domain of SmB was PCR amplified as described above using oligonucleotides RBspeE and RBspeF. The PCR fragment was digested with BamHI-XhoI (the BamHI site being blunt ended with Klenow enzyme) and cloned into plasmid pGFP-SmBΔΔ-SV40 previously digested with ClaI and XhoI, the ClaI site being blunt ended with Klenow enzyme. The structure of the generated alleles is shown in Fig. 1C.
Preparation of yeast extracts, Western blot analysis, and immunoprecipitations.
To analyze the stability of GFP-Sm fusion proteins, yeast whole-cell extracts were prepared as previously described (5, 61). For Western analysis, 5 optical density (OD) equivalents of extracts were mixed with dye, boiled 10 min, and loaded on sodium dodecyl sulfate (SDS)-denaturing polyacrylamide gels (28) or Tricine-SDS-denaturing polyacrylamide gels (58). After transfer to nitrocellulose membrane by electroblotting, the immunoblots were probed with anti-GFP antibodies (Molecular Probes) and then incubated with secondary antibodies conjugated to peroxidase (Promega Corp., Madison, Wis.). The blots were visualized by enhanced chemiluminescence (ECL; Amersham) according to the manufacturer's instructions.
For the immunoprecipitations, 10 μg of anti-GFP antibodies (Boehringer) was coupled to 40 μl of protein A-Sepharose (0.1 g/ml; Pharmacia) in 300 μl of buffer B (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, and 0.1% sodium azide) for 2 h at 4°C. The beads were washed four times with 1 ml of the same buffer, added to 5 OD equivalents of extracts brought to 300 μl of buffer B, and rolled for 2 h at 4°C. The beads were washed four times with 1 ml of buffer B. The RNA was extracted from the pellets by adding 400 μl of proteinase K buffer (100 mM Tris-Cl [pH 7.4], 125 mM EDTA, 150 mM NaCl, 1% SDS) and subjected to Northern analysis as described previously (3).
Light microscopy.
The localization of GFP-Sm fusion proteins was examined in living yeast cells. Cells transformed with pGFP-Sm constructs were grown in either liquid or solid synthetic selective media to early log phase. Cells were washed with phosphate-buffered saline, mounted on slides (10), and examined by fluorescence microscopy. Samples were observed using a Nikon fluorescence microscope. Images were acquired with an PhotonicScience camera.
RESULTS
The C-terminal extensions of yeast SmB, SmD1, and SmD3 contain NLS-like motifs.
All Sm proteins contain the Sm domain, which consists of two blocks of conserved amino acids (Sm motif 1 and Sm motif 2) separated by a nonconserved region (7, 21, 62). In addition, the SmB, SmD1, and SmD3 proteins from all species have C-terminal extensions beyond the Sm motif as compared to the other Sm proteins. A close inspection of the C-terminal protein sequences of the yeast Sm proteins reveals the presence of stretches rich in lysine and arginine residues (Fig. 1A). The region of yeast SmB between positions 105 and 135 contains 16 basic amino acids. In yeast SmD1, of the last 20 amino acids, 9 are arginine or lysine, and in SmD3, seven residues in the C-terminal domain are basic. These clusters of basic residues present similarities with a classical monopartite SV40 Tag-type NLS (8) as well as a bipartite NLS which is defined as two basic clusters separated by a spacer region of any 10 amino acids (8, 48). They also show homologies to nuclear import signals for human ribosomal proteins, such as rpS6 (60), rpL7a (52), or rpL23a (25). A common feature of those last signals is their very basic nature and a greater complexity as compared with the classical NLS. The similarities of the basic amino acid-rich stretches found in SmB, SmD1, and SmD3 to classical, bipartite, and ribosomal proteins nuclear import signals are shown in Fig. 1B. The basic domains found in the yeast Sm proteins were termed NLS-like motifs to distinguish them from classical NLSs.
The NLS-like motifs of SmB and SmD1 exhibit nuclear localization properties.
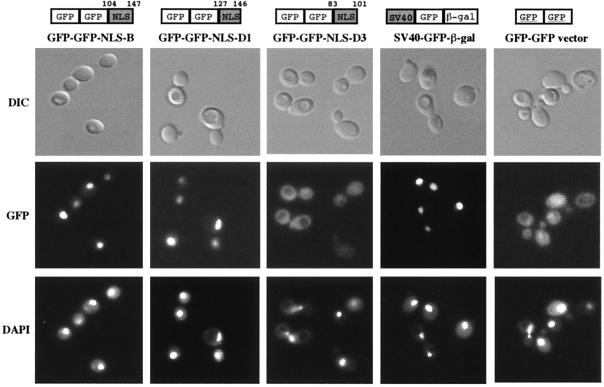
To test whether the NLS-like motifs found in SmB, SmD1, and SmD3 may function as a nuclear targeting signal, they were fused to the C terminus of a dimeric GFP-GFP reporter protein (see Materials and Methods; Fig. 2). Immunoblotting experiments using GFP-specific antibodies demonstrated that expression of these constructs in wild-type yeast cells results in the synthesis of fusion proteins of predicted lengths (data not shown). The intracellular localization of the fusion proteins was visualized in living cells. As shown in Fig. 2, as is the case for the classical NLS of SV40 Tag, the basic motif of either SmB or SmD1 targets the reporter protein to the nucleus, whereas the dimeric GFP-GFP protein alone gives rise to a diffuse cytoplasmic staining. This result demonstrates that the NLS-like motifs of SmB and SmD1 function as nuclear targeting elements in vivo. In contrast, the dimeric GFP-GFP protein fused to the basic motif of SmD3 does not accumulate in the nuclear compartment, indicating that this motif does not possess nuclear localization properties.
FIG. 2.
Localization properties of the putative NLS-like motifs identified in yeast SmB, SmD1, and SmD3 proteins. The NLS-like motif from each indicated Sm protein was fused to the C-terminal domain of a dimeric GFP-GFP reporter protein. The portion of protein sequence used is indicated above each construct. These fusion proteins as well as a fusion carrying the NLS of SV40 Tag at the C terminus of a GFP–β-Gal reporter protein (SV40-GFP-β-Gal) were expressed in wild-type cells. Cells were observed by using differential interference contrast (DIC), and GFP was detected by fluorescence microscopy (GFP). The position of the nuclei was visualized with DAPI (4′,6′-diamidino-2-phenylindole).
Cellular localization of yeast Sm proteins.
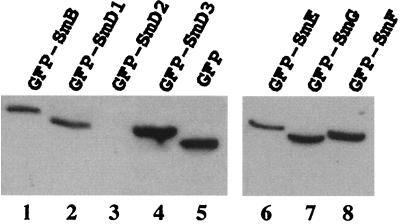
If carboxyl-terminal domains of SmB and SmD1 alone confer nuclear import of the GFP-GFP reporter protein, full-length SmB and SmD1 proteins should also show nuclear localization when fused to GFP. To test this, the subcellular localization of the yeast SmB and SmD1 proteins was analyzed along with the other yeast Sm proteins. Translational fusions were constructed by placing the whole sequence of the different Sm proteins at the C terminus of GFP. The constructs were transformed into wild-type yeast cells which were grown in −Ura medium to maintain the GFP-Sm fusion containing plasmid. Growth assays were performed under repressing conditions (presence of methionine in the medium), since the repression of the MET25 promoter results in a rest activity of 10 to 20% (41). To characterize the expression of the fusion proteins, whole-cell lysates were prepared from yeast cells carrying the different constructs, and proteins were analyzed by Western blotting using anti-GFP antibodies. With the exception of GFP-SmD2 (Fig. 3, lane 3), expression of the GFP-Sm fusion constructs in wild-type cells results in the synthesis of fusion proteins of predicted lengths and at approximately similar levels (Fig. 3). The cause of the instability of the GFP-SmD2 fusion protein is unknown and has not been further investigated.
FIG. 3.
Western analysis of GFP-Sm fusion proteins. Equivalent amounts of cell extracts prepared from wild-type strains carrying the indicated GFP-Sm fusion proteins were fractionated by SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-GFP antibodies. Both panels represent two independent migrations. The GFP-SmD2 fusion protein (lane 3) was not stably expressed for unknown reasons. Control extract was made from a wild-type strain carrying the GFP vector alone (lane 5). The predicted molecular masses of the proteins are as follows: GFP-SmB, 55 kDa; GFP-SmD1, 45 kDa; GFP-SmD2, 42 kDa; GFP-SmD3, 37 kDa; GFP-SmE, 39 kDa; GFP-SmF, 38 kDa; GFP-SmG, 36 kDa; GFP, 27 kDa.
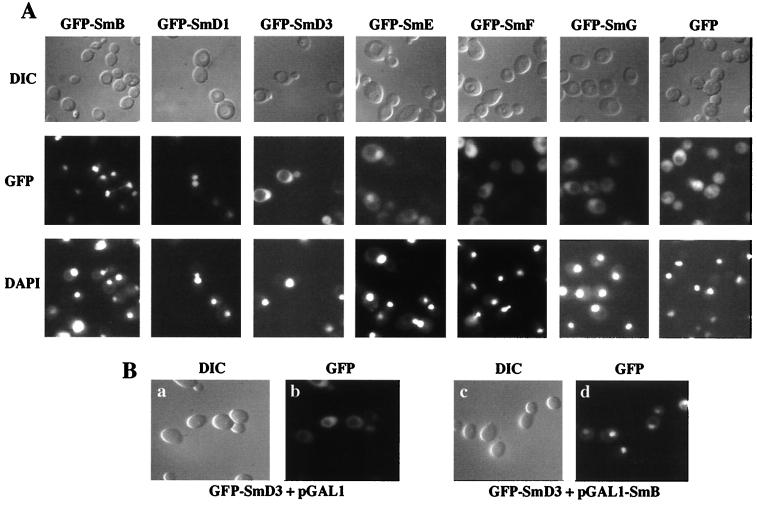
Microscopic GFP fluorescence examination of the different tagged Sm proteins indicate that GFP-SmB and GFP-SmD1 fusion proteins are localized in the nucleus (Fig. 4A). In contrast, a diffuse cytoplasmic staining is observed for the GFP-SmD3 fusion protein, although a nuclear signal is also observed for this construct. This nuclear signal may be due to the association of the GFP-SmD3 fusion protein with the SmB protein. Indeed, the existence of yeast SmB-SmD3 complex has been reported both in mammals and yeast (5, 16, 17, 21, 32, 47). The effect of an overproduction of SmB on the subcellular localization of the GFP-SmD3 protein was therefore tested. A yeast strain carrying the GFP-SmD3 fusion was transformed with a plasmid carrying the SMB gene under the GAL1 promoter (pGAL1-SmB) or with an empty pGAL1 vector. As shown in Fig. 4B, panel d, the GFP-SmD3 fusion protein locates predominantly in the nuclear compartment under galactose-inducing conditions of the SMB gene. In contrast, under the same inducing conditions, the subcellular localization of the GFP-SmD3 protein is not modified in a strain carrying the empty pGAL1 vector (Fig. 4B, panel b). These experiments demonstrate that overexpression of SmB produces a nuclear localization of the GFP-SmD3 fusion protein. Concerning the other yeast GFP-Sm protein fusions, the corresponding panels in Fig. 4A show that GFP-SmE and GFP-SmG tagged proteins are found both in nuclear and cytoplasmic compartments, whereas no specific nuclear accumulation of the GFP-SmF fusion is observed, as is the case for the GFP protein.
FIG. 4.
Cellular localizations of yeast GFP-Sm fusion proteins. (A) The indicated GFP-Sm fusion proteins were expressed in a wild-type strain, and living cells were observed using differential interference contrast (DIC) and by fluorescence microscopy (GFP). The position of the nuclei was visualized with DAPI (see text for details). (B) GFP-SmD3 localizes to the nucleus upon SmB overexpression. The YAMB haploid wild-type strain was transformed with the GFP-SmD3 fusion protein and with either the empty pGAL1 vector (a and b) or the pGAL1-SmB plasmid (c and d). Transformants were grown in galactose-containing medium, and the cells were observed using differential interference contrast (DIC) and fluorescence microscopy (GFP).
Incorporation of GFP-Sm fusion proteins into snRNP particles.
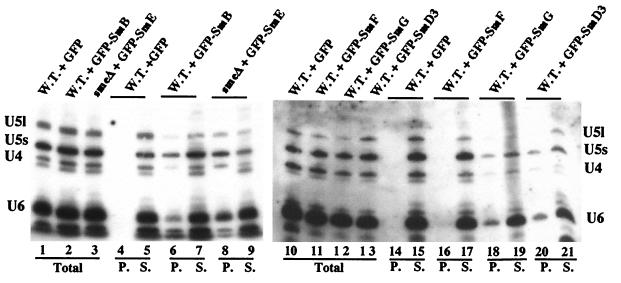
To determine if the GFP-Sm fusion proteins behave like wild-type Sm proteins, the incorporation of the tagged proteins into snRNP particles was analyzed by immunoprecipitation of snRNPs from yeast cells using anti-GFP antibodies (Fig. 5). Whole-cell extracts were prepared from wild-type strains carrying the indicated GFP-Sm fusion (Fig. 5, lanes 6, 7, and 16 to 21) and, as control, from a wild-type strain expressing only GFP protein (Fig. 5, lanes 4, 5, 14, and 15). Since the GFP-SmE allele can complement a chromosomal deletion of the SME1 gene (data not shown), an extract was also prepared from an smeΔ::HIS3 strain (4) carrying the GFP-SmE fusion as sole source of SmE protein (Fig. 5, lanes 8 and 9). Equal amounts of extracts were incubated with anti-GFP antibodies bound to protein A-Sepharose beads (see Materials and Methods) and RNA, which was recovered from the total extract (Fig. 5, Total), the supernatant (Fig. 5, lanes S), and the immunoprecipitates (Fig. 5, lanes P), and were analyzed by Northern hybridization. As shown in Fig. 5, the snRNAs were found in the immunoprecipitates from the strains expressing GFP-SmB (lane 6), GFP-SmE (lane 8), GFP-SmG (lane 18), and GFP-SmD3 (lane 20) but not from the strain expressing GFP-SmF (lane 16) and not from the control strain expressing GFP (lanes 4 and 14). The U6 snRNA was also precipitated from strains expressing GFP-SmB (lane 6), GFP-SmE (lane 8), GFP-SmG (lane 18), and GFP-SmD3 (lane 20) due to its association with the U4 snRNA in the U4/U6 and U4/U6.U5 snRNPs (34).
FIG. 5.
Incorporation of the GFP-Sm fusion proteins into snRNPs. Strains containing the indicated GFP-Sm fusion protein were grown under conditions maintaining the plasmid. Whole-cell extracts were prepared, and snRNPs were immunoprecipitated with anti-GFP antibodies. RNA was extracted from the supernatants (S), the pellets (P), and equivalent aliquots of the total lysates (Total), separated on denaturing polyacrylamide gels, and subjected to Northern analysis. Hybridization was with probes specific for the yeast U4, U5, and U6 snRNAs. W.T., wild-type strain; smeΔ, strain carrying a chromosomal deletion of the SmE gene (4).
These experiments demonstrate that the tested GFP-Sm fusion proteins, with the exception of GFP-SmF, behave like wild-type Sm proteins. This is further supported by the fact that the GFP-SmB and the GFP-SmD1 fusions (see below) as well as the GFP-SmE protein complement a chromosomal deletion of their respective genes. The ability of these fusion proteins to replace the endogenous corresponding proteins demonstrates that they are assembled into functional snRNPs and are capable of promoting subsequent spliceosome assembly steps.
Subcellular localizations of GFP-Sm mutant fusion alleles.
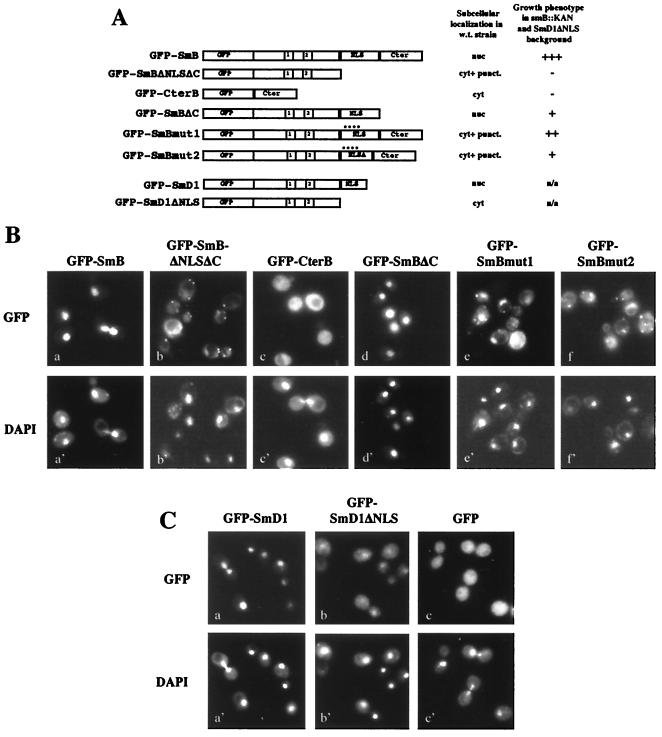
Given that GFP-SmB and GFP-SmD1 fusion proteins can substitute for the wild-type corresponding protein in vivo (see below), mutant alleles carrying mutations in the NLS-like motifs of SmB and SmD1 were constructed to test the functional importance of these sequences (Fig. 6A). These constructs were transformed into haploid wild-type cells, and the intracellular localizations of the Sm mutant fusion proteins were observed by fluorescence microscopy (Fig. 6B). Whereas the GFP-CterB fusion distributes uniformly in the cytoplasm (Fig. 6B, panel c) as does the empty GFP vector (Fig. 6C, panel c), the GFP-SmBΔC fusion protein localizes to the nucleus (Fig. 6B, panel d) like the full-length GFP-SmB fusion protein (Fig. 6B, panel a). In contrast, although a light nuclear signal can be observed in some cells, the GFP-SmBΔNLSΔC fusion allele gives rise mainly to a cytoplasmic staining visualized as heterogeneously distributed fluorescent sites (Fig. 6B, panel b).
FIG. 6.
Intracellular localizations of GFP-SmB and GFP-SmD1 mutant fusion alleles in wild-type cells. (A) Schematic representation of the GFP-Sm mutant alleles. The structures of the mutant fusion constructs are schematically shown: GFP represents the reporter protein, 1 and 2 represent the conserved Sm1 and Sm2 motifs, NLS represents the NLS-like motifs of SmB and SmD1, and Cter represents the SmB C-terminal region (residues 148 to 196). The mutations generated in the GFP-SmBmut1 and GFP-SmBmut2 alleles are detailed in Fig. 1C. A summary of the subcellular localizations in wild-type cells and the growth phenotypes of the mutants in an smb::KAN SmD1ΔNLS context (see Fig. 7B) is shown at the right. nuc, nucleus; cyt. + punct., cytoplasmic staining with fluorescent sites; +++, viable; −, lethal; ++ and +, viable with growth defect; n/a, not applicable. (B) Subcellular localizations of the GFP-SmB mutant alleles. The GFP-SmB fusion alleles were expressed in wild-type cells, and GFP was detected in living cells by fluorescence microscopy. The position of the nuclei was visualized with DAPI. (C) Subcellular localizations of the GFP-SmD1 fusion alleles. The GFP-SmD1 fusion alleles were expressed in wild-type cells, and GFP and DAPI were detected in living cells by fluorescence microscopy.
To test whether mutations in the basic clusters of the SmB NLS-like motif affect nuclear localization, two additional mutants were constructed. GFP-SmBmut1 carries mutations in six positively charged residues which are changed to alanine in the second basic cluster of the NLS-like motif, whereas GFP-SmBmut2 carries a deletion of seven amino acids (residues 119 to 125) as well as nine mutations of basic residues to alanine (Fig. 1C). Examination of subcellular localization of those mutants in wild-type cells shows that they are both found in cytoplasmic dots (Fig. 6B, panels e and f), as is the GFP-SmBΔNLSΔC mutant (Fig. 6B, panel b). As seen for this last mutant, light nuclear fluorescence is also observed for the GFP-SmBmut1 and GFP-SmBmut2 mutant alleles. The GFP-SmD1ΔNLS fusion protein locates predominantly in the cytoplasm (Fig. 6C, panel b) compared to the wild-type GFP-SmD1 fusion, which locates in the nucleus (Fig. 6C, panel a). For the GFP-SmD1ΔNLS mutant allele, fluorescent sites are also present in the cytoplasm, although they are less visible than for the GFP-SmBΔNLSΔC protein.
Taken together, these results (summarized in Fig. 6A) demonstrate that the NLS-like motifs of SmB and SmD1 are important for optimal nuclear localizations of their respective reporter proteins.
Growth phenotypes of GFP-Sm mutant alleles.
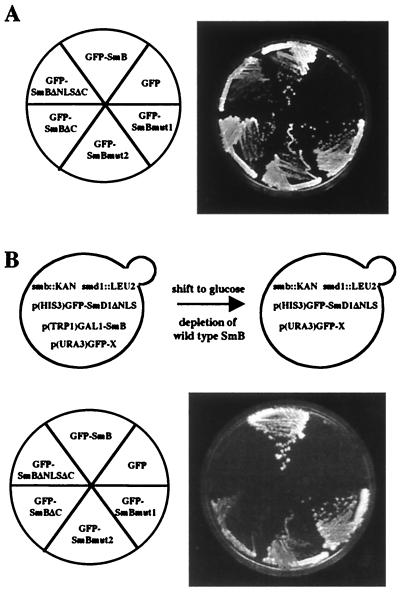
If the NLSs from SmB and SmD1 are essential for Sm protein nuclear import and function, one would expect that deletion of these motifs will hinder the import process and therefore give rise to yeast growth defects. This was tested by constructing chromosomal SMB and SMD1 deletion strains (see Materials and Methods). The lethality of each smb::KAN and smd1::KAN strain can be rescued by complementation with plasmids carrying their respective genes under the GAL1 promoter (data not shown). To determine the effect of the GFP-Sm mutant alleles in vivo, the plasmids carrying the mutant fusions were transformed into smb::KAN and smd1::KAN strains carrying the pGAL1-SmB and pGAL1-SmD1 gene constructs, respectively. Transformed cells did not show any growth defect on galactose-containing media, indicating that the mutants had no dominant phenotype. When placed on glucose-containing media, which repress the production of wild-type SmB protein, all GFP-SmB mutant fusion proteins, except the GFP-CterB allele (data not shown), sustain yeast cell growth like the GFP-SmB protein, whereas strain smb::KAN carrying the empty GFP vector is lethal (Fig. 7A). The strain containing the GFP-SmBΔNLSΔC allele grows more slowly than the strains containing other GFP-SmB mutant alleles, giving rise to smaller colonies after 4 days at 30°C (Fig. 7A). The GFP-SmD1ΔNLS mutant protein is also capable of replacing the GFP-SmD1 fusion protein without any obvious growth defect (data not shown).
FIG. 7.
Growth phenotypes of GFP-SmB mutant fusion alleles. (A) An smb::KAN strain carrying a pGAL1-SmB gene was transformed with the indicated GFP-SmB fusion alleles. The different strains, which grow on galactose-based media, were streaked on glucose-based media, which repress the production of the wild-type SmB gene. The phenotypes were observed after 4 days at 30°C. (B) Simultaneous deletion of the SmB- and SmD1-NLS-like motifs gives rise to yeast cell lethality. Above, strain YRB120 carries chromosomal disruptions of the SMB and SMD1 genes. This strain is able to grow on galactose-based media, since it carries the SmB gene under the GAL1 promoter [p(TRP1)GAL1-SmB plasmid] and the GFP-SmD1ΔNLS mutant allele [p(HIS3)GFP-SmD1ΔNLS plasmid]. This strain was transformed with different constructs of GFP-SmB mutant alleles carried by a URA3 plasmid [p(URA3)GFP-X plasmid]. When placed on glucose-based media, the wild-type SmB gene is repressed, allowing the determination of the growth phenotype of the GFP-SmB mutant alleles. Below, growth assay. The growth phenotypes of the indicated GFP-SmB mutants were determined after 4 days at 30°C.
These experiments demonstrate that integrity of the NLS-like motifs of SmB and SmD1 is not required for viability. The fact that the SmB and SmD1 protein functions are not impeded by mutations in their NLS-like motifs could be explained by a functional redundancy of both sequences.
Concomitant deletions of the SmB and SmD1 NLS-like motifs produce yeast cell lethality.
If the SmB- and SmD1-NLS-like motifs are functionally redundant, deletions of both sequences should impair their functions and produce a yeast cell growth defect. This was tested, as described in the legend to Fig. 7B, in strain YRB120 carrying simultaneously smb::KAN and smd1::LEU2 disrupted alleles. Due to the production of wild-type SmB protein, the strains containing the different constructs grow on galactose-containing media (data not shown). On glucose, cells carrying the full-length GFP-SmB fusion protein grow and cells carrying the empty GFP vector die (Fig. 7B, lower panel). Under the same conditions, the GFP-SmBΔNLSΔC mutant fusion protein is unable to support growth in a GFP-SmD1ΔNLS background, whereas the GFP-SmBΔC mutant fusion strain is viable but presents a growth defect (Fig. 7B). Growth curve determination shows that the generation time of this mutant is threefold longer than that of a strain expressing the GFP-SmB fusion protein. The GFP-SmBmut1 and GFP-SmBmut2 mutant strains are also viable although they show growth defects, since both mutants give rise to smaller colonies than the GFP-SmB fusion (Fig. 7B). The growth defect is stronger for the GFP-SmBmut2 allele which is more severely mutated in the NLS-like motif than the GFP-SmBmut1 allele (Fig. 1C). Comparable growth phenotypes are observed at 25°C, the growth defect of the GFP-SmBΔC, GFP-SmBmut1, and GFP-SmBmut2 mutant alleles being exacerbated at this temperature. Growth phenotypes have not been analyzed at 37°C, since the YRB120 strain carrying the GFP-SmB fusion protein grows very poorly at these conditions.
These experiments show that simultaneous deletions of the SmB- and SmD1-NLS-like motifs give rise to yeast lethality, demonstrating the functional redundancy of both sequences. Furthermore, they indicate a correlation between the number of basic amino acids present in the NLS-like motif of SmB and yeast growth defect. Finally, these results show that the C-terminal region of SmB (residues 148 to 196) is also important for optimal growth of yeast cells.
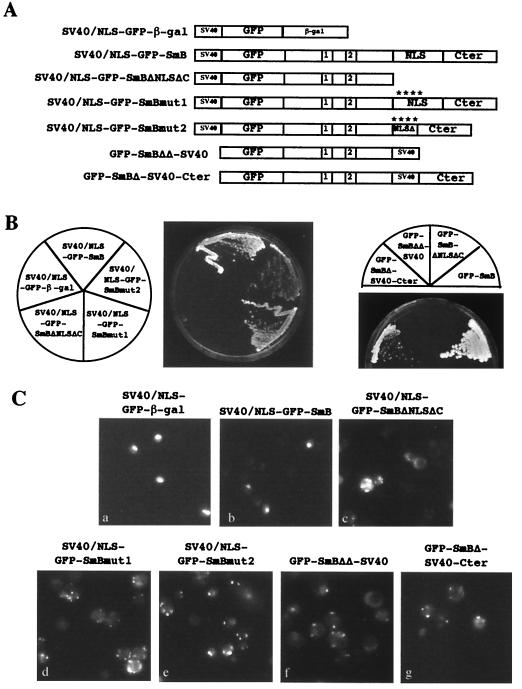
The SV40 Tag NLS cannot efficiently replace the NLS-like motif of SmB.
The results described above show that mutations in the NLS-like motif of SmB produce mislocalization of GFP-SmB mutant alleles and induce a growth defect in an smb::KAN SmD1ΔNLS background strain. To test whether the NLS from SV40 Tag can functionally substitute for the NLS-like motif of SmB, different SmB mutants were fused in frame to a GFP reporter protein carrying, at its N terminus, the NLS of SV40 (Fig. 8A). The NLS of SV40 was also introduced at the 3′ end of the GFP-SmBΔNLSΔC construct (pGFP-SmBΔΔ-SV40 plasmid). Finally, the NLS of SV40 was used to replace the NLS-like region (residues 101 to 148) of SmB (pGFP-SmBΔ-SV40-Cter plasmid). The structure of the generated alleles is shown in Fig. 1C. The different constructs were transformed into the smb::KAN strain carrying the pGAL1-SmB plasmids, and their growth phenotypes were determined as described in Fig. 7A. All the strains with GFP-SmB mutant alleles carrying the NLS of SV40 Tag were viable when tested on galactose-containing and glucose-containing media (data not shown), showing that the fusions are able to complement an smb::KAN-deleted strain in an SmD1 wild-type background. To test whether the SV40 NLS containing GFP-SmB alleles can complement a simultaneous deletion of the NLS-like motifs of SmB and SmD1, these mutants were transformed into the YRB120 strain, and their growth phenotypes were determined as described in Fig. 7B. On galactose-containing media, all mutant fusions grew, indicating that no mutant had any obvious dominant phenotype (data not shown). When placed on glucose-containing media, the strain containing the SV40 NLS-GFP-SmB fusion grew, whereas the strain carrying the SV40 NLS–GFP–β-Gal vector was lethal (Fig. 8B). The strain carrying the GFP-SmBΔΔ-SV40 fusion was also unable to grow (Fig. 8B, right panel). In contrast, a strain carrying the GFP-SmBΔ-SV40-Cter allele was not lethal although it showed a severe growth defect when compared to the GFP-SmB fusion (Fig. 8B, right panel). The growth phenotypes of the SV40 NLS-GFP-SmBΔNLSΔC, SV40 NLS-GFP-SmBmut1, and SV40 NLS-GFP-SmBmut2 fusion alleles are identical to those of the GFP-SmBΔNLSΔC, GFP-SmBmut1, and GFP-SmBmut2 mutants, respectively (compare Fig. 8B and 7B).
FIG. 8.
Growth phenotypes and intracellular localizations of SV40 NLS containing GFP-SmB mutant alleles. (A) Schematic representation of the GFP-SmB mutant alleles carrying the NLS motif of SV40 Tag. The structures of the mutant fusion constructs are as described in the legend to Fig. 6A. (B) Growth assay. The constructs were transformed in the YRB120 strain, and their phenotypes were determined as described in the legend to Fig. 7B. (C) Subcellular localizations of the SV40 NLS containing GFP-SmB fusion alleles in wild-type cells as detected by fluorescence microscopy.
The fact that no improvement of growth is observed for the N terminus-fused SV40 NLS containing GFP-SmB fusion alleles may be due to a mislocalization of the fusion proteins, as observed for the corresponding GFP-SmB mutant alleles in wild-type cells (Fig. 6B). To test this, the subcellular localizations of the SV40 NLS containing GFP-Sm fusion proteins were examined in the same strain. As shown in Fig. 8C, the SV40 NLS-GFP-SmB (panel b) is located in the nuclear compartment, as is the case for the SV40 NLS–GFP–β-Gal fusion (panel a). In contrast, the SV40 NLS-GFP-SmBΔNLSΔC and GFP-SmBΔΔ-SV40 fusion proteins are located in cytoplasmic dots or patches (Fig. 8C, panels c and f, respectively). This is also the case for the SV40 NLS-GFP-SmBmut1, SV40 NLS-GFP-SmBmut2, and GFP-SmBΔ-SV40-Cter fusion proteins (Fig. 8C, panels d, e, and g, respectively). For these three mutants, a light nuclear staining can also be observed, showing that they retain low nuclear localization properties.
These results demonstrate that addition of an SV40 NLS to the N terminus of the GFP-SmB mutant alleles does not correct their nuclear localization defects and their growth phenotypes. However, substitution of the NLS-like region of SmB with the NLS of SV40 allows yeast to sustain growth, albeit inefficiently.
DISCUSSION
In this study, using GFP-tagged wild-type Sm proteins and Sm mutant alleles, I show that the NLS-like motifs of yeast SmB and SmD1 proteins exhibit nuclear localization properties. Both motifs are important for nuclear localization of their respective proteins. However, deletion of either motif did not impair yeast cell growth while simultaneous deletion of both SmB- and SmD1-NLS-like sequences produces yeast cell lethality, showing that these motifs have redundant essential functions. Taken together, the results suggest that, in the heptameric ring model formed by the Sm core complex, a basic amino acid-rich protuberance composed of the positively charged residues found in the C-terminal regions of Sm proteins, may represent a nuclear localization determinant, both in yeast and human cells.
Most yeast GFP-Sm fusion proteins assemble into functional snRNPs.
That the majority of yeast GFP-Sm fusion proteins behave like wild-type Sm proteins is supported by the immunoprecipitation studies and by the functional in vivo approaches. These results imply that the fusion proteins are assembled into snRNPs following a complete assembly pathway. Failure of the GFP-SmF fusion protein to assemble into snRNPs may reflect its inability to associate with the SmE partner. Consistent with this explanation, a glutathione S-transferase-SmF fusion protein (GST-SmF) is unable to interact with in vitro-translated 35S-SmE in an in vitro binding assay (R. Bordonné, unpublished results), whereas a GST-SmE fusion protein binds efficiently to in vitro-translated 35S-SmF (5). Therefore, the presence of a Tag at the N terminus of the SmF protein may induce a misfolding of the fusion protein which impairs its function.
Fluorescence microscopy analyses revealed that the yeast GFP-Sm mutant alleles deleted or mutated in the NLS-like motif generate cytoplasmic punctuated structures in a wild-type strain (Fig. 6B). It is interesting to compare such yeast sites of staining with the described discrete punctuate sites observed recently in mammalian fibroblasts in immunofluorescence microscopic studies using five anti-Sm monoclonal antibodies (72). In these studies, dots, distributed extensively and evenly in the cytoplasm, are visible in addition to the expected intense nuclear staining representing the speckled distribution of snRNP proteins in the nucleus. These cytoplasmic punctuated sites may reflect the cytoplasmic pools of snRNP core proteins and represent storage particles of the snRNP core protein complexes or staging centers for snRNP core particle assembly (72). It is tempting to suggest that the fluorescent sites observed for the yeast GFP-SmB alleles mutated in the NLS-like motif may also represent storage and/or assembly structures for yeast Sm core particles. Alternatively, rather than a specialized compartment for Sm core protein assembly, the cytoplasmic punctuate distribution observed for some yeast GFP-SmB mutant alleles could also reflect an aggregation and/or precipitation of the mutated proteins due to their cytoplasmic retention. Further studies are needed to distinguish between those possibilities.
Nuclear import signal of the Sm core complex.
Examination of subcellular localization of the GFP-SmB and GFP-SmD1 alleles mutated in the NLS-like motifs suggest that these motifs represent nuclear import determinants (Fig. 6). Although these results cannot totally rule out the possibility that the NLS-like motifs of SmB and SmD1 might be required for efficient assembly or stabilization of the Sm core complex, this seems unlikely for the following reasons. First, unique deletion of each motif does not affect the viability of yeast GFP-SmB (Fig. 7A) and GFP-SmD1 mutants, demonstrating that the different mutant alleles are correctly assembled into snRNPs and are able to perform all subsequent steps of spliceosome formation. Second, coimmunoprecipitation studies with C-terminal truncation mutants of the human Sm proteins B′ and D3 show that the amino-terminal 93 amino acids of SmB′, containing the two Sm motifs, are sufficient for efficient and stable complex formation in vitro with full-length SmD3 (21). Likewise, this study shows that deletions in the C terminus of SmD3 do not affect the capability of SmD3 mutants to interact with SmB′ protein. Third, fragments of human SmB (residues 1 to 91) and SmD3 (residues 1 to 75) proteins lacking the C-terminal extensions form a stable complex even in high salt (26). Finally, the heptameric ring proposed for the human Sm core protein complex has been modeled using SmB-SmD3 and SmD1-SmD2 crystals containing approximately 70 residues of each protein chain (26). Taken together, these observations strongly suggest that mutations in (or deletions of) the NLS-like motifs located in the C-terminal extensions of the Sm proteins do not affect assembly or stabilization of the Sm core complex.
Although the yeast SmB, SmD1, and SmD3 proteins contain portions of sequence exhibiting homologies to nuclear import signals, only the NLS-like motifs of SmB and SmD1 can direct the GFP reporter protein to the nucleus. This could be explained by the fact that only the NLS regions of SmB and SmD1 contain classical monopartite, SV40 Tag-type NLSs (Fig. 1B). In this regard, the viability of the SmBmut2 construct in an smb::KAN SmD1ΔNLS background could be due to the retention, in this mutant, of a region (residues 105 to 108) encompassing a monopartite classical NLS (Fig. 1C). In addition, the SmBΔ-SV40-Cter construct, in which the NLS region of SmB is replaced by the SV40 Tag NLS, is also able to grow, albeit inefficiently (Fig. 8B). These observations suggest that the classical monopartite NLSs found in SmB and SmD1 might indeed be functional.
The NLS-like region of SmD3 may also be part of the determinant specifying nuclear localization in the context of the yeast Sm core complex. Indeed, the nuclear targeting capability of the three NLS-like sequences may be cumulative. In support to this proposal is the fact that the SmB, SmD1, and SmD3 proteins are adjacent in the heptameric ring formed by the human Sm core protein complex (26). Since the proposed heptamer model accounts for only 58% of the total mass of the core proteins (26), in the doughnut-like shape formed by the Sm proteins, the C-terminal extensions of the three Sm proteins could form a basic amino acid-rich protuberance representing the nuclear localization determinant of the Sm core complex. Such a basic amino acid-rich protuberance could mediate protein-protein interactions with other factors like cytoplasmic transporters and/or nuclear import receptors. The existence of a basic protuberance in the complex of the Sm core proteins is coherent with the functional redundancy of the NLS-like motifs of SmB and SmD1, and the proposed model accommodates also the observed correlation between the number of basic residues found in the NLS-like region of GFP-SmB mutant alleles and their growth phenotypes. This suggests that, in the context of the Sm core complex, the number of basic residues present in the protuberance, rather than a linear sequence of amino acids, is an important element for nuclear import. This view is compatible with the inability of the SV40-NLS to substitute for the SmB NLS-like motif when fused to the N terminus of GFP (Fig. 8), since in such a position, the NLS motif of SV40 Tag is not contiguous with the other basic domains of the Sm complex protuberance.
In the context of the Sm core complex, the basic amino acid-rich protuberance formed by the NLS-like motifs of the C-terminal domains of Sm proteins is reminiscent to import signals of a number of human ribosomal proteins, such as rpS6, rpL7a, or rpL23a (25, 52, 60). These import signals, represented by an accumulation of basic amino acids, are presumed to be distinct from the simple basic or bipartite NLSs and are recognized by receptors other than the importin α, which recognizes the classical NLSs (38). Indeed, import studies in a mammalian system have demonstrated that at least four importin β-like transport receptors, namely importin β itself, transportin, RanBP5, and RanBP7, directly bind and mediate import of ribosomal proteins into nuclei (25). Moreover, two importin β-like transport receptors (Yrb4p and Pse1p) have been implicated in yeast rpL25 import (50, 59). Based on these observations, it is noteworthy that U snRNP nuclear import in mammals requires importin β but not the NLS-specific importin α (44). Whether the homologies between ribosomal protein nuclear import domains and the basic amino acid-rich protuberance of the Sm core protein complex have functional significances remains to be demonstrated, and further studies aimed at the identification of the nuclear import receptors recognizing the basic protuberance of the Sm complex are clearly needed to clarify this question.
It is very likely that the nature of the NLS proposed for the yeast Sm core complex will also apply to the human Sm complex. While only the corresponding yeast polypeptides contain portions of sequence exhibiting resemblance to known nuclear import signals, the C-terminal extensions of the mammalian SmD1 and SmD3 proteins contain numerous GR dipeptides as well as multiple lysine and arginine residues and, in addition for SmB, several clusters of proline-rich stretches (18, 32, 49, 54, 69). Given the evolutionary conservation of the lengths of the C-terminal extensions of these proteins, it is possible that the NLS determinant formed by the human Sm core complex and required for human snRNP import is also composed of a protuberance formed by the C-terminal domains of human Sm proteins.
The results described here also suggest a role for the C-terminal extensions of yeast Sm proteins in a step of spliceosome assembly located further than nuclear import of the Sm complex. A role for these regions can be inferred from recent work showing that eight proteins cross-link to pre-mRNA in the yeast commitment complex (71). Among these are the SmB, SmD1, and SmD3 proteins. It is thought that residues in the C-terminal tails of these three proteins might engage direct contact with the pre-mRNA substrate, thereby stabilizing the U1 snRNP–pre-mRNA interaction. This is compatible with the growth phenotype of the GFP-SmBΔC alleles, indicating that residues in the C-terminal domain (residues 148 to 196) of SmB are required for optimal yeast cell growth. Such a function of the C-terminal region of yeast SmB could also be redundant with the C-terminal region of SmD1, since both regions are dispensable for growth in an SmD1 and an SmB wild-type background, respectively.
U snRNP import in yeast.
The results presented in this paper suggest that a basic amino acid-rich protuberance composed of the NLS-like motifs found in the C-terminal extensions of Sm proteins represents the nuclear import signal formed by the complex of Sm core proteins, both in yeast and mammals. However, in yeast, it is yet unknown if the snRNAs transit through the cytoplasm or remain in the nucleus during snRNP biogenesis. Therefore, it is possible that yeast snRNP biogenesis might not require nuclear snRNA export and that the complex of Sm core proteins might be nuclear imported as an snRNA-free preassembled complex. In this regard, it is interesting to note that no homologue to snurportin1, the receptor recognizing the snRNA cap structure in mammals, has been found in the yeast genome database although snurportin1 is evolutionarily conserved in Caenorhabditis elegans, Drosophila, mice, and humans (23). Moreover, a recent study provided evidence for the presence, in mammalian fibroblasts, of an snRNA-free spliceosomal Sm protein complex (9), showing that such a heteromer exists in vivo. Further studies are needed to define whether or not yeast snRNAs transit through the cytoplasm and in which compartment of the yeast cell the Sm core is assembled. The tools generated in this work will certainly be helpful in resolving these issues.
ACKNOWLEDGMENTS
I thank Alain Camasses and Brian Rymond for strains and plasmids and Eric Allemand, Edouard Bertrand, Bruno Lapeyre, John Mouaikel, Bertrand Séraphin, Johann Soret, and Jamal Tazi for helpful suggestions and/or critical reading of the manuscript.
This work was supported by the Association Française contre les Myopathies (AFM) and the Centre National de la Recherche Scientifique (CNRS).
REFERENCES
- 1.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum S, Mueller M, Schmid S R, Linder P, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4A-dependent cell-free system. Proc Natl Acad Sci USA. 1989;86:6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordonné R, Banroques J, Abelson J, Guthrie C. Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP-snRNP interactions, and pre-mRNA splicing in vivo. Genes Dev. 1990;4:1185–1196. doi: 10.1101/gad.4.7.1185. [DOI] [PubMed] [Google Scholar]
- 4.Bordonné R, Tarassov I. The yeast SME1 gene encodes the homologue of the human E core protein. Gene. 1996;176:111–117. doi: 10.1016/0378-1119(96)00230-2. [DOI] [PubMed] [Google Scholar]
- 5.Camasses A, Bragado-Nilsson E, Martin R, Séraphin B, Bordonné R. Interactions within the yeast Sm core complex: from proteins to amino acids. Mol Cell Biol. 1998;18:1956–1966. doi: 10.1128/mcb.18.4.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano M T, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper M, Johnston L H, Beggs J D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 9.Filali M, Qiu J, Awasthi S, Fischer U, Monos D, Kamoun M. Monoclonal antibody specific to a subclass of polyproline-Arg motif provides evidence for the presence of an snRNA-free spliceosomal Sm protein complex in vivo: implications for molecular interactions involving proline-rich sequences of Sm B/B′ proteins. J Cell Biochem. 1999;74:168–180. [PubMed] [Google Scholar]
- 10.Fink G R, Guthrie C. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:3–21. [PubMed] [Google Scholar]
- 11.Fischer U, Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U, Darzynkiewicz E, Tahara S M, Dathan N A, Lührmann R, Mattaj I W. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J Cell Biol. 1991;113:705–714. doi: 10.1083/jcb.113.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer U, Sumpter V, Sekine M, Satoh T, Lührmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U, Heinrich J, van Zee K, Fanning E, Lührmann R. Nuclear transport of U1 snRNP in somatic cells: differences in signal requirement compared with Xenopus laevis oocytes. J Cell Biol. 1994;125:971–980. doi: 10.1083/jcb.125.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 16.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 17.Fury M G, Zhang W, Christodoulopoulos I, Zieve G W. Multiple protein:protein interactions between the snRNP common core proteins. Exp Cell Res. 1997;237:63–69. doi: 10.1006/excr.1997.3750. [DOI] [PubMed] [Google Scholar]
- 18.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot H V, Mann M, Séraphin B, Rosbash M, Lührmann R, Fabrizio P. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 19.Gottschalk A, Neubauer G, Banroques J, Mann M, Lührmann R, Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamm J, Darzynkiewicz E, Tahara S M, Mattaj I W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 21.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill J, Donald K A, Griffiths D E, Donald G D. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jäkel S, Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kambach C, Walke S, Young R, Avis J M, de la Fortelle E, Raker V A, Lührmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 27.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamond A I, Carmo-Fonseca M. Localisation of splicing snRNPs in mammalian cells. Mol Biol Rep. 1993;18:127–133. doi: 10.1007/BF00986767. [DOI] [PubMed] [Google Scholar]
- 31.Lee M S, Henry M, Silver P A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 32.Lehmeier T, Raker V, Hermann H, Lührmann R. cDNA cloning of the Sm proteins D2 and D3 from human small nuclear ribonucleoproteins: evidence for a direct D1-D2 interaction. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 34.Lührmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990;1087:256–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 35.Mattaj I W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 36.Mattaj I W. U snRNP assembly and transport. In: Birnstiel M L, editor. Structure and function of the major and minor small nuclear ribonucleoprotein particles. Berlin, Germany: Springer-Verlag; 1988. pp. 303–357. [Google Scholar]
- 37.Mattaj I W. Ribonucleoprotein assembly: clues from spinal muscular atrophy. Curr Biol. 1998;8:R93–R95. doi: 10.1016/s0960-9822(98)70055-7. [DOI] [PubMed] [Google Scholar]
- 38.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 39.Mayes A E, Verdone L, Legrain P, Beggs J D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaud N, Goldfarb D. Microinjected U snRNAs are imported to oocyte nuclei via the nuclear pore complex by three distinguishable targeting pathways. J Cell Biol. 1992;116:851–861. doi: 10.1083/jcb.116.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelissen R L, Will C L, van Venrooij W J, Lührmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 44.Palacios I, Hetzer M, Adam S A, Mattaj I W. Nuclear import of U snRNPs requires importin β. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plessel G, Fischer U, Lührmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raker V A, Plessel G, Lührmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 48.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 49.Rokeach L A, Haselby J A, Hoch S O. Molecular cloning of a cDNA encoding the human Sm-D autoantigen. Proc Natl Acad Sci USA. 1988;85:4832–4836. doi: 10.1073/pnas.85.13.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 51.Roy J, Zheng B, Rymond B C, Woolford J L., Jr Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo G, Ricciardelli G, Pietropaolo C. Different domains cooperate to target the human ribosomal L7a protein to the nucleus and to the nucleoli. J Biol Chem. 1997;272:5229–5235. doi: 10.1074/jbc.272.8.5229. [DOI] [PubMed] [Google Scholar]
- 53.Rymond B C. Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1993;90:848–852. doi: 10.1073/pnas.90.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rymond B C, Rokeach L A, Hoch S O. Human snRNP polypeptide D1 promotes pre-mRNA splicing in yeast and defines nonessential yeast Smd1p sequences. Nucleic Acids Res. 1993;21:3501–3505. doi: 10.1093/nar/21.15.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Sauterer R A, Goyal A, Zieve G W. Cytoplasmic assembly of small nuclear ribonucleoprotein particles from 6 S and 20 S RNA-free intermediates in L929 mouse fibroblasts. J Biol Chem. 1990;265:1048–1058. [PubMed] [Google Scholar]
- 58.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 59.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff F R. Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt C, Lipsius E, Kruppa J. Nuclear and nucleolar targeting of human ribosomal protein S6. Mol Biol Cell. 1995;6:1875–1885. doi: 10.1091/mbc.6.12.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 62.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Séraphin B, Kandels-Lewis S. An efficient PCR mutagenesis strategy without gel purification [correction of purification] step that is amenable to automation. Nucleic Acids Res. 1996;24:3276–3277. doi: 10.1093/nar/24.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sleeman J, Lyon C E, Platani M, Kreivi J P, Lamond A I. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- 66.Spector D L. Nuclear organization of pre-mRNA processing. Curr Opin Cell Biol. 1993;5:442–447. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- 67.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 68.Stevens S W, Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc Natl Acad Sci USA. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Dam A, Winkel I, Zijlstra-Baalbergen J, Smeenk R, Cuypers H T. Cloned human snRNP proteins B and B′ differ only in their carboxy-terminal part. EMBO J. 1989;8:3853–3860. doi: 10.1002/j.1460-2075.1989.tb08563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 71.Zhang D, Rosbash M. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 1999;13:581–592. doi: 10.1101/gad.13.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zieve G W. The cytoplasmic sites of the snRNP protein complexes are punctate structures that are responsive to changes in metabolism and intracellular architecture. Exp Cell Res. 1999;247:249–256. doi: 10.1006/excr.1998.4354. [DOI] [PubMed] [Google Scholar]