Abstract
GPR56/ADGRG1 is a versatile adhesion G protein-coupled receptor important in the physiological functions of the central and peripheral nervous systems, reproductive system, muscle hypertrophy, immune regulation, and hematopoietic stem cell generation. By contrast, aberrant expression or deregulated functions of GPR56 have been implicated in diverse pathological processes, including bilateral frontoparietal polymicrogyria, depression, and tumorigenesis. In this review article, we summarize and discuss the current understandings of the role of GPR56 in health and disease.
Keywords: Adhesion GPCR, G protein, Ligand, Signaling
1. Introduction
Representing the largest transmembrane receptor family in the human proteome, the G protein-coupled receptors (GPCRs) are hallmarked by the characteristic seven transmembrane (7TM) helices and normally couple to heterotrimeric G proteins upon receptor activation [1,2]. There are approximately 800 human GPCRs involved in a plethora of physiological processes such as vision, olfaction, and taste [1,2]. The ligands/binding partners of GPCRs range from subatomic particles (photons) to ions, small organic molecules (lipids, neurotransmitters), peptides, hormones, and proteins [1,3,4]. Functional disruption of GPCRs leads to diverse pathologies, and hence they are the major targets of medicinal drugs in use today. Overall, GPCRs are proteins of functional and medicinal importance [5,6].
Recently, the GPCR superfamily was further divided into five different subfamilies based on their phylogenetic similarities, namely the Glutamate, Rhodopsin, Adhesion, Frizzled, and Secretin (GRAFS classification system) [2]. The human adhesion GPCRs (aGPCRs) form the second-largest GPCR subfamily with a total of 33 members [[7], [8], [9], [10], [11]]. Evolutionarily, aGPCRs are very ancient and are found in all vertebrates, primitive animals, and even ancient metazoans [7,8,10,12]. Phylogenetic analyses have predicted that aGPCRs probably evolved ∼1275 million years ago from the common ancestor of eukaryotes, and it is thought that the Secretin GPCR subfamily is likely derived from the ancestral aGPCRs [9]. Distinguished from other GPCRs, aGPCRs are signified by their unusually large extracellular region (ECR), which contains the GPCR auto-proteolysis inducing (GAIN) domain that mediate auto-proteolytic cleavage of these receptors [13,14]. This novel auto-proteolytic modification leads to the generation of two non-covalently associated receptor subunits, an N-terminal fragment (NTF) (α subunit) and a C-terminal fragment (CTF) (β subunit) [[13], [14], [15], [16]]. To date, aGPCRs have been implicated in diverse physiological and pathological processes such as in embryonic development, nervous and reproductive systems, immune regulation, and stem cell maintenance [10]. In this review article, we will discuss the functional role of aGPCR GPR56/ADGRG1 in health and disease.
1.1. Overview of GPR56/ADGRG1 protein
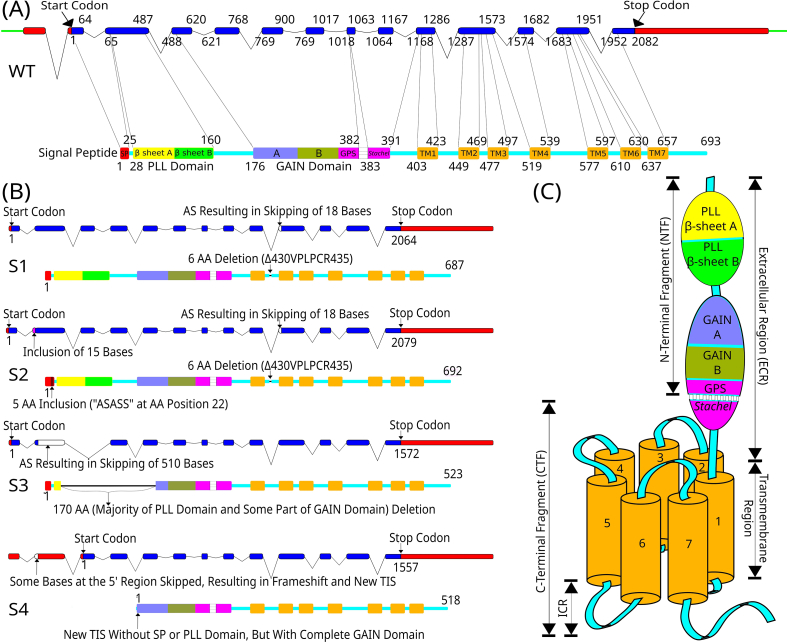
The ADGRG1 gene spans ∼45 kb of DNA on human chromosome 16q21 and comprises of 14 exons with a coding region of 2082 base pairs (bp) from the exon 2 to 14 [Fig. 1A] [17,18]. Located next to it on the same locus are two different aGPCR genes, namely GPR97/ADGRG3 and GPR114/ADGRG5, suggesting a strong evolutionary relationship [19]. GPR56/ADGRG1 is expressed in the brain, heart, thyroid, kidney, testis, pancreas, and skeletal muscle [17,20]. It has at least four protein variants due to alternative RNA splicing and 17 alternative translation initiation sites in the first non-coding exon, all of which have distinct expression profiles in human [Fig. 1B] [16,[21], [22], [23]]. GPR56 contains two different protein domains in its ECR, namely the PLL (Pentraxin/Laminin/neurexin sex-hormone-binding globulin-Like) and GAIN domains [Fig. 1]. The PLL domain is located at the most N-terminus and is unique to GPR56 [16]. Structural analysis has shown that the GAIN domain of GPR56 is unusually small among aGPCRs; its GAIN-A sub-domain contains only three α-helices whereas those of Latrophilin-1 and BAI-3 have six [13,14,16]. The post-translational auto-proteolytic cleavage of GPR56 occurs between Leu-382 and Thr-383 at the GPCR proteolytic site (GPS) within the GAIN-B sub-domain, with His-381, Leu-382, and Thr-383 acting as the catalytic triad [16].
Fig. 1.
Organization of the GPR56 gene and receptor protein variants. (A) Schematic diagrams of the GPR56 gene (upper panel) and the encoding receptor protein (lower panel). The GPR56 gene consists of 14 exons, of which the exons 2–14 code for the receptor protein. The figure depicts the full-length GPR56 gene with a length of 2082 base pairs (Intron: line; Exon: solid bar; coding region: blue; non-coding region: red). The corresponding wild-type (WT) protein isoform is composed of 693 amino acids (Signal Peptide: red; PLL domain-β sheet A: yellow; PLL domain-β sheet B: green; GAIN-subdomain A: purple; GAIN-subdomain B: dark green; GPS region and Stachel peptide: pink; TM regions: orange; Intracellular and extracellular loops: cyan) [16,22]. (B) Schematic diagrams depicting the four GPR56 alternatively spliced isoforms, S1–S4. The corresponding protein structures are depicted below the gene structure [16,22,24]. (C) The schematic cartoon of the structural organization of GPR56 receptor protein. ICR: Intracellular Region.
The unique 133-amino acid long PLL domain is a 12-stranded β-sandwich domain, forming two β-sheets (β-sheet A and B) and sharing only a weak homology with either pentraxin or laminin/neurexin/sex-hormone-binding globulin domain [16]. The PLL domain attenuates the receptor's basal activity and is necessary for GPR56's role in supporting oligodendrocyte development [16]. Interestingly, the GPR56 splice variant 4 lacks the entire PLL domain, but is specifically involved in microglia-mediated synaptic pruning [16,24]. A total of seven N-glycosylation sites are identified in the ECR of GPR56 [25]. N-glycosylation affects GPR56 receptor protein trafficking, cell surface expression, and even ligand-binding affinity. As such, it has been reported that N-glycosylation decreased the binding affinity of GPR56 to tissue transglutaminase 2 (TG2), but did not affect its binding to another ligand, collagen III [[25], [26], [27]]. Apart from collagen III and TG2, many other endogenous protein ligands and binding partners have been identified for GPR56, making it no longer an orphan receptor [28]. These include tetraspanins CD9/CD81, progastrin, phosphatidylserine (PS), and laminin, which function either as a scaffold protein, co-receptor, or cognate ligand [24,[29], [30], [31]]. In addition, a small-molecule agonist, 3-α-acetoxydihydrodeoxygedunin (3-α-DOG), and an antagonist, dihydromunduletone (DHM), as well as a distinct glycosaminoglycan (GAG) binding partner, heparin, were also identified recently for GPR56 [[32], [33], [34], [35]].
1.2. Activation and signaling mechanisms of GPR56
GPR56 receptor activation can be induced by diverse mechanisms, including the tethered ligand (Stachel)-dependent/independent and autoproteolysis-independent modes [33,[36], [37], [38], [39]]. There is a wealth of experimental evidence suggesting that GPR56-NTF functions as a repressor of basal GPR56 signaling. As such, the NTF-deleted GPR56 mutants are constitutively active, characterized by increased SRF and NFAT activity, TGF-α shedding, β-arrestin 2 binding, and ubiquitination of the receptor [36,40]. In line with the well-established tethered-agonist activation mechanism of aGPCRs [37,41], the binding of cognate ligands or receptor-specific activating antibodies (Abs) or other interacting-molecule complexes trigger GPR56 activation and downstream Gα12/13-RhoA signaling cascade by inducing the irreversible NTF shedding and subsequent exposure of the tethered Stachel agonist peptide [Fig. 2, Fig. 3] [33,[36], [37], [38],42]. This Stachel peptide is sometimes also referred as the stalk region, or the β-strand 13 of the GAIN-B sub-domain. More precisely, the agonistic property of this peptide is imparted by the TYFAVLM sequence, which is sufficient to activate GPR56 receptor and increase the SRF activity when added ectopically [37]. The increased SRF and NFAT activity is due to the RhoA signaling by Gα12/13 and calcium-ion channel activation by Gβγ, respectively [36,38]. Interestingly, the homophilic N terminus-N terminus trans-activation of GPR56 is also observed, with amino acids between 228 and 342 being essential [40]. Recently, it has also been shown that the deletion of the entire Stachel sequence of GPR56 still leaves the receptor constitutively active, however it only leads to NFAT activity and TGF-α shedding, but not the SRF activity, hence establishing the Stachel-independent mechanism of receptor activation [36,38].
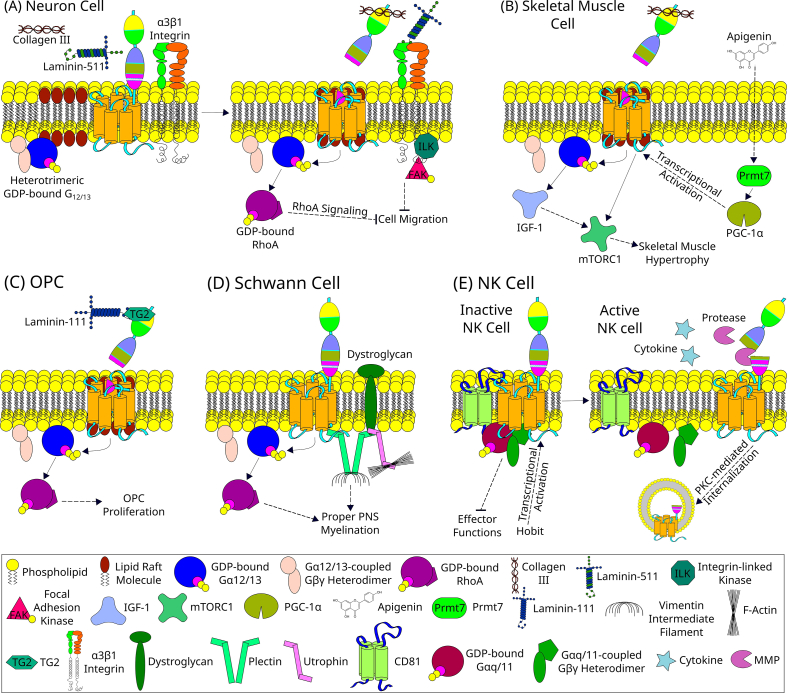
Fig. 2.
GPR56 signaling in physiological processes. (A) In a neuron cell, collagen III acts as a ligand of GPR56 [28]. During the inactive state, GPR56 is found in detergent-soluble non-lipid raft regions of the plasma membrane [102]. However, in the presence of collagen III, the NTF of GPR56 binds to the collagen III which subsequently leads to removal of NTF from the CTF [103]. The shedding of NTF exposes the tethered Stachel agonistic peptide within the CTF [33]. The removal of NTF is also followed by the translocation of CTF to lipid rafts [103]. The tethered Stachel sequence then binds to and induces the conformational changes in CTF to activate the Gα12/13 protein [28]. Upon the GDP-GTP exchange, this heterotrimeric G-protein complex dissociates into Gα12/13 and Gβγ subunits. The Gα12/13 subunit then activates RhoA, which activates various downstream signaling molecules to finally result in the inhibition of migration of the neuron [28]. Some observations suggest a possible interaction of GPR56 with integrin α3β1 [43]. Laminin-511 is known to bind to the integrin α3β1. However, the details of this interaction of GPR56 with integrin remains elusive. (B) In the skeletal muscle cells, resistance exercises and apigenin, a natural flavone found in many edible plants, induces muscle hypertrophy [48,49]. Apigenin enters the cell and activates Prmt7, which then directly activates PGC-1α [49]. The PGC-1α is a transcriptional coactivator which enhances GPR56 and collagen III expression, thereby finally inducing muscle hypertrophy via the IGF-1-mTORC1 pathway [48,49]. (C) In an oligodendrocyte progenitor cell (OPC), the tripartite signaling complex formed of TG2 released by microglia, laminin-111 from ECM, and GPR56 on its cell surface induce the myelin formation and repair in CNS by promoting OPC proliferation and inhibiting its premature differentiation to oligodendrocytes via the Gα12/13-RhoA signaling pathway [47,96]. (D) In a Schwann cell, GPR56 in association with other transmembrane and cytoskeletal linker proteins, like dystroglycan and plectin, induces proper PNS myelination by cytoskeletal remodeling via the RhoA pathway and physical interaction with plectin [29]. (E) Hobit is the primary driver of GPR56 expression in human cytotoxic NK cells [54]. GPR56 in association with CD81 inhibits the effector functions of cytotoxic NK cells during inactive state [19,54]. However, NK cell activation leads to the cleavage of a portion of GPR56 ECR and induces PKC-mediated internalization of the GPR56 receptor, thereby removing its inhibitory effector functions [54]. The solid lines represent direct interaction, whereas the dotted lines indicate indirect pathways with potential additional intermediate(s).
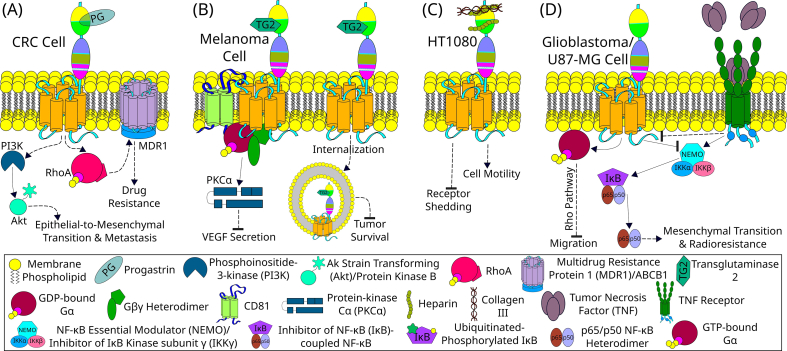
Fig. 3.
GPR56 signaling in pathological processes. (A) In a colorectal cancer (CRC) cell, progastrin (PG) binds to GPR56 and promotes the epithelial-to-mesenchymal transition and metastasis by the PI3K/Akt pathway. Moreover, GPR56 also promotes drug resistance by promoting MDR1 expression via the RhoA pathway [[83], [84], [85]]. (B) GPR56 inhibits the metastasis and tumor survival in a melanoma cell by internalization and degradation of TG2-bound GPR56 thereby controlling the concentration of TG2 in ECM which is a major promoter of tumor survival [46,104]. An alternative mechanism observed in melanoma cells is the interaction of GPR56 with tetraspanins such as CD81, which recruit Gαq/11(Gβγ) heterotrimer and finally inhibits VEGF secretion, and hence the angiogenesis, via the PKCα pathway [72,104]. (C) In the HT1080 cell line, heparin has been demonstrated to bind to the NTF of GPR56 [32,104]. One of the major heparin-binding sites of GPR56 overlaps with the collagen III-binding site, hence making heparin capable of modulating the binding and signaling by collagen III [32]. It is observed that the binding of heparin reduces NTF shedding and promotes cell motility and adhesion [32]. (D) In U87-MG cells, GPR56 inhibits cell migration via the Gαq and Rho pathway [81]. Moreover, GPR56 also inhibits mesenchymal transition and radioresistance by inhibiting the NF-κB signaling pathway upstream of NEMO [82]. TNF activates the TNF receptor which promotes mesenchymal transition and radioresistance by activating NF-κB signaling pathway and downregulating GPR56 indirectly [82]. The solid lines represent direct interaction, whereas the dotted lines indicate indirect pathways with potential additional intermediate(s).
The small-molecule agonist 3-α-DOG can activate the GPS autoproteolysis-deficient GPR56-F385A mutant [33,35]. Likewise, a panel of human GPR56-specific monobodies was shown to be able to activate the autoproteolysis-deficient F385A mutant [39]. These observations led to the proposal of the proteolysis-independent receptor activation mechanism, wherein the small agonist binds to the receptor and cause its conformational change, thereby activating the receptor without requiring its autoproteolysis and the exposure of the tethered Stachel agonist peptide [33,39]. In short, GPR56 mediates diverse signaling mechanisms in concert with its interaction with tissue-/cell type-specific ligands to achieve various physiological and pathological functions [Fig. 2, Fig. 3].
2. Physiological roles of GPR56/ADGRG1
GPR56/ADGRG1 is involved in a wide variety of physiological functions [Fig. 2]. The most studied ones are in the central and peripheral nervous systems (CNS/PNS), the immune system, differentiation of hematopoietic stem and progenitor cells (HSPCs), the reproductive system, and muscular hypertrophy [Table 1].
Table 1.
GPR56 in physiology.
| Section | Physiological function | The role of GPR56/ADGRG1 | References |
|---|---|---|---|
| 1.1 | Cerebral Cortex Development and Patterning | Proper cerebral cortex development and designing by inhibiting the migration of neurons hence preventing neuronal over migration and breaching of pial basement membrane | [17,23,28] |
| 1.2 | Rostral Cerebellum Development | Adhesion of rostral granule cells to extracellular matrix proteins such as laminin and fibronectin | [45] |
| 1.3 | CNS Myelination: Oligodendrocyte Progenitor Cell (OPC) Proliferation and Oligodendrocyte (OL) Development | Regulation of myelination by positively regulating OPC proliferation and negatively regulating pro-OL differentiation to OL | [31,47,96] |
| 1.4 | PNS Myelination by Schwann Cells (SCs) | Proper radial axonal sorting in SCs by cytoskeletal remodeling | [29] |
| 1.5 | Neural Stem and Progenitor Cells Maintenance | Maintenance of stemness or inhibition of differentiation | [69] |
| 1.6 | Synaptic Remodeling in CNS | Microglia-mediated synaptic pruning via detection of phosphatidylserine on apoptotic synapses | [24] |
| 1.7 | Stress and Depression | Anti-depressant-like activities | [70,97] |
| 2.1 | Hematopoietic Stem and Progenitor Cells | Endothelial-to-Hematopoietic Transition (EHT) | [57,59] |
| 2.2 | Hemostatic Shear Force Sensor | Platelet collagen-responsder and shear force sensor during hemostasis | [55] |
| 2.3 | Cytotoxic Lymphocytes Steady State | Inhibition of cytotoxicity and migration of cytotoxic lymphocytes during steady or inactivated state | [19] |
| 3 | Muscle Hypertrophy | Overload-induced or Apigenin-induced muscle hypertrophy | [48,49] |
| 4 | Pancreas/islet β-cell | Protection of β-cells from cytokine-induced apoptosis; increasing calcium-ion levels; glucose-induced insulin secretion | [60] |
| 5 | Adipose Tissue | Adipogenesis | [63] |
| 6 | Reproduction System Development | Asymmetric testis development and male fertility in mammals | [64] |
2.1. Nervous system (CNS/PNS)
GPR56 is expressed in Tuj1+ migrating neurons, Cajal-Retzius cells, radial glial cells, and multiple cell types from the preplate and the marginal zones, including the ventricular and sub-ventricular zones of the developing brains [28,43]. In the developing cerebrum, GPR56 couples to Gα12/13 and activates the RhoA pathway upon binding to collagen-type III alpha-1 (COL3A1) expressed by meningeal fibroblasts at the pia surface/marginal zone, thereby inhibiting neuronal (over)migration and hence properly laminating the cerebral cortex [28]. As such, the COL3A1 homozygous null mutation phenocopies the over migration of neurons, breaching of the pial basement membrane (BM), and cobblestone-like cortical malformation, all of which were similarly observed in the cerebrum of GPR56 homozygous null mutants [28].
In addition, it was reported that the loss of integrin-α3 (Itga3) leads to enhanced phenotypic severity of GPR56 deletion in a dose-dependent manner with Itga3−/−Gpr56−/− being most severely affected [44]. These enhanced phenotypic severities in Itga3 and Gpr56 double-knockout mice include the earlier manifestation of abnormalities, larger neuronal ectopias in number and size, and reduced collagen III-mediated neuronal migration inhibition in vitro compared to controls [44]. GPR56 and α3 integrin co-localize in radial glial cells and rostral preplate neurons, and α3β1 integrin does not bind to collagen III, but it does bind to laminin-511 [44]. It is hence possible that the α3β1 integrin coordinates with GPR56 via an undefined mediator to regulate cortical development [Fig. 2A]. Granule cells of the rostral cerebellum express GPR56 as well, and these cells in Gpr56-null mutants have a reduced binding affinity to laminin-111 and fibronectin compared to those of wild-type (WT) mice. These phenotypes are likely due to the defective TG2-binding of GPR56-null granule cells since TG2 can interact with GPR56, laminin, and fibronectin [31,45,46].
GPR56 is also involved in the myelin formation and repair in CNS neurons via a tripartite signaling complex of microglial TG2, laminin in ECM, and GPR56 on oligodendrocyte progenitor cells (OPCs) via the Gα12/13-Rho signaling pathway [Fig. 2C] [30,46,47]. GPR56 acts as a positive regulator of OPC proliferation, but a negative regulator of OPC differentiation to oligodendrocytes (OLs) [47]. The myelin sheath thickness of myelinated axons, however, does not differ significantly between Gpr56 null-mutant and WT [47]. GPR56 coupled to Gα12/13 is also involved in the myelination of PNS neurons by cytoskeletal remodeling in Schwann cells (SCs) via the RhoA pathway and by physical interaction with plectin, a large cytoskeletal linker protein, upon receptor activation [Fig. 2D] [29].
In addition to the cortical lamination, and repair as well as formation of myelin, GPR56 was recently found to be involved in the microglia-mediated synaptic remodelling by serving as a receptor for phosphatidylserine (PS) exposed on the membrane of apoptotic synapses, thus facilitating their engulfment and removal by microglial cells. Interestingly, this synaptic pruning function of GPR56 is specifically mediated by its splice variant 4 isoform [Fig. 1B] expressed by microglial cells [24].
In short, GPR56 regulates brain cortical patterning and proper cerebral cortex development, rostral cerebellar development, nerve axon myelination and myelin repair in both CNS and PNS, OPC proliferation, inhibition of OPC differentiation to OLs in the CNS, and proper radial axonal sorting by SCs in the PNS, including synaptic remodelling.
2.2. Muscle hypertrophy
Apigenin (4′,5,7-trihydroxyflavone), a natural flavone abundant in various edible plants, and resistance exercises induce muscle hypertrophy by Prmt7-PGC-1α-ADGRG1 that activates Gα12/13 and downstream signaling through the IGF1-Akt-mTOR pathway [Fig. 2B] [48,49]. Expression of the PGC-1α isoform 4 (PGC-1α4) transcription coactivator is sufficient for enhancing GPR56 and collagen III expressions, thereby inducing muscle hypertrophy [48]. Interestingly, GPR56 may act at an early stage of myotube fusion and regulate myoblast commitment to differentiation, possibly through SRF to promote the transcription of myogenic regulators. However, GPR56 overall is dispensable for muscle development since the absence of GPR56 only produces a mild phenotype due to unknown compensatory mechanisms [50].
PCBP2 (Poly(C)-binding protein 2) negatively regulates angiotensin II (Ang II)-induced hypertrophy of cardiomyocytes by regulating the GPR56 mRNA stability and its degradation at the transcriptional or post-transcriptional level [51]. PCBP2 mRNA and protein levels are low in human failing or hypertrophic hearts, overload-mediated hypertrophic hearts in mice, and neonatal rat cardiomyocytes treated with Ang II, isoproterenol, or phenylephrine to induce hypertrophy [51]. This decreased PCBP2 levels significantly increase the amounts of Ang II, GPR56, and fetal genes such as Nppa, Nppb, and Myh7, hence inducing an Ang II-mediated increase in cardiomyocyte size and protein synthesis (hypertrophy) [51].
In a recent report, all-trans-retinoic-acid-induced or β-carotene-stimulated myotubes and the soleus muscles of β-carotene treated mice were shown to enhance Tg2 mRNA expression and secretion of TG2 into the ECM [52]. Extracellular TG2 promotes phosphorylation of Akt, mTOR, and ribosomal p70S6K through GPR56 to induce protein synthesis and hypertrophy in myotubes without requiring its cross-linking abilities [52]. These results suggest that although GPR56 is not necessary for overall muscle development due to possible compensatory mechanisms, it is required for resistance exercise-induced muscle hypertrophy, Ang II or overload-induced cardiomyocyte hypertrophy, and myotube function in general.
2.3. Immune system and HSPC
In the immune system, GPR56 acts as a specific cell surface marker of pan-cytotoxic leukocytes. Its expression is restricted to NK cells and cytotoxic T lymphocyte subsets, including CD8+, CD4+, and γδ T cells [19,53]. GPR56 expression in human cytotoxic lymphocytes is primarily driven by Hobit and overlaps with that of cytolytic enzymes such as perforin, granzyme A and granzyme B [19,54]. GPR56 inhibits the cytotoxic activity and migration ability of NK cells by coupling to CD81 [19,54]. GPR56 downregulation in cytotoxic lymphocytes upon cell activation is protein kinase C-mediated, which is also known to associate with the intracellular region of CD81 [Fig. 2E] [54]. Platelets also express GPR56, which functions as a collagen responder and shear force sensor during hemostasis [55]. It is important to note that the expression profiles of GPR56 and Hobit in human and murine immune cells differ significantly. As such, while GPR56 and Hobit are co-expressed in human peripheral NK and T cells, both molecules are absent in murine peripheral NK and T cells [54].
Recently, a novel KLR/GPR56-based classification system (KLRB1, KLRG1, GPR56/ADGRG1, and KLRF1) was proposed for CD4+ memory T cells that better define the functional states of these cells than the classical CD45RA/CCR7-based classification [56]. Therefore, based on the production potential of selective cytokines (TNF and INF-γ), CD4+ memory T cells are divided into high, medium, low, and exhausted subgroups that can be defined by the unique expression patterns of distinct surface KLRs and GPR56 [56].
GPR56 is expressed abundantly by HSPCs during definitive hematopoiesis in the embryo and adult bone marrow, but its levels are reduced significantly upon lineage restriction and differentiation [57]. The highly enriched CD34−c-Kit+Sca-1+Lin+ hematopoietic stem cell (HSC) fraction expresses GPR56, and the GPR56+ fraction contains almost all of the long-term repopulating (LTR) cells, hence implicating GPR56 as a potential surface marker for LTR HSCs [58]. GPR56 is a transcriptional target of heptad transcription factors (TFs) and is involved in endothelial-to-hematopoietic transition (EHT) [59]. Moreover, heptad TFs and GPR56 are essential for HSC generation during EHT in healthy as well as leukemic human hematopoietic cells, and this role of GPR56 is conserved even in zebrafish [59]. GPR56 is also necessary to overcome repeated proliferative stress induced by serial bone marrow transplantations [57]. However, despite all these, GPR56 is mostly dispensable in context to HSPCs since GPR56 deficiency does not lead to any significant impairment in HSPC maintenance, migration, or function during steady-state or myeloablative stress-induced hematopoiesis [57].
In short, GPR56/ADGRG1 modulates human cytotoxic NK cell effector functions in part by negatively regulating cell cytotoxicity and migration, involves in the platelet–collagen interaction and hemostatic response, but is mostly dispensable in context to HSPCs. Further explorations will be required to better understand the role of GPR56 in human HSPCs and immune cells.
2.4. Pancreas/islet β-cell
GPR56 is the most abundantly expressed GPCR in human and mouse pancreatic islets, and its expression is strongly correlated with many essential genes for β-cell function and type-2 diabetes risk [60]. Extracellular collagen III found at the peri-islet basement membrane and islet capillaries are the putative ligand of GPR56 in pancreatic β-cells [60]. It protects β-cells from cytokine-induced apoptosis, increases calcium-ion levels, and potentiates glucose-induced insulin secretion [60]. Despite these roles, GPR56 is mostly dispensable for in vitro glucose-induced insulin secretion, glucose tolerance in adult mice, or normal islet vascularization or innervation [61]. Interestingly, reduced GPR56 expression was identified while studying the effects of GPR142 knockdown in rat INS-1832/13 insulinoma cell line [62]. Hence, there might be some regulatory cross-talk between GPR142 and GPR56. These results suggest that modulation of GPR56 signaling could possibly improve islet transplantation outcomes and may even be a potential target for treating type-2 diabetes.
2.5. Adipogenesis
Adipogenesis is the formation of adipocytes from stem cells, and in vitro studies have shown that GPR56 is necessary for adipogenesis, although in vivo validations are yet to be done. Compared to lean control rats, adipose tissues of genetically obese Zucker rats have low GPR56-transcript levels [63]. Furthermore, low endogenous GPR56 protein levels are found in 3T3-L1 preadipocytes, and GPR56 knock-out by genome editing abolishes adipogenesis in 3T3-L1 cells [63]. Interestingly, only mitotic cells present cell surface GPR56 staining, suggesting that its localization may be regulated during cell division and might contribute to the modulation of adipogenesis [63].
2.6. Reproduction
The absence of GPR56 causes asymmetric partial disruption of seminiferous tubules in the embryonic gonads just after the initial establishment of testis cords in mouse [64]. Therefore, it seems to act as a spatial or temporal inducer of asymmetry in cord remodeling during male gonad development. Sertoli cells express GPR56, and the absence of its expression leads to improper formation of seminiferous tubules during embryogenesis, leading to reduced male fertility [64]. Interestingly, only the mesonephric side of the testis cords is disrupted. In contrast, the coelomic side remains predominantly intact, thereby proposing the first-ever genetic evidence for the asymmetric formation of testis cords in mouse gonads [64]. Spermatogenesis still occurs in the normal parts of the Gpr56-knockout adult mice testes, leading to partial fertility in these mice [64].
GPR56 is a highly female-biased gene on the embryonic day 6 of chicken embryos. It is strongly expressed in female gonads, primarily in the ovary's cortex, compared to male gonads in which GPR56 expression is weak [65]. During experimental sex reversal, GPR56 expression is down-regulated in embryonic female gonads treated with fadrozole, an aromatase inhibitor [65]. A more recent study showed that GPR56 is involved in Mullerian duct development in the chicken embryo [66]. GPR56 mRNA is present in the inner Mullerian duct epithelium during the elongation phase of the duct formation [66]. Interestingly, collagen III expression is also observed in the Mullerian duct during the same developmental stages, suggesting a possible role of collagen III as a ligand for GPR56 in these tissues [66]. Knockdown of GPR56 expression using in ovo electroporation results in variably truncated ducts and loss of expression of both epithelial and mesenchymal markers of duct development, whereas in vitro overexpression leads to enhanced cell proliferation and cell migration [66]. Hence, GPR56 plays an essential role in avian Mullerian duct development through duct elongation regulation. It is also essential for asymmetric testis cord remodeling in male mammals.
3. Pathological roles of GPR56/ADGRG1
Since GPR56/ADGRG1 is involved in diverse physiological functions, it is not surprising that many pathological disorders were manifested due to abnormal functioning of this receptor, mainly in the nervous system and cancer development [Fig. 3] [Table 2].
Table 2.
GPR56 in pathology.
| Section | Pathological function | GPR56 expression level | The role of GPR56/ADGRG1 | Special Remarks | References |
|---|---|---|---|---|---|
| 1 | Bilateral Fronto-Parietal Polymicrogyria (BFPP) | Mutation(s) in coding region of GPR56 gene | Neuronal migration inhibition and hence in the proper development of the cerebral cortex and rostral cerebellum. | Mutations in GPS (GPCR Proteolytic Site) results in a phenotypically more severe case of BFPP | [17,45] |
| 2.1.1 | EVI1high Acute Myeloid Leukemia (AML) | Upregulated | Probable role in cellular growth and apoptosis | Highly expressed in CD34+CD38−EVI1high Leukemic Stem Cell (LSC) fraction | [74] |
| 2.1.2 | Leukemic Stem Cells (LSCs) of AML | Upregulated | Chemosensitivity | Identification of LSCs beyond CD34+ fractions, and high expression significantly correlating with OS and chemotherapy resistance | [75,77,[98], [99], [100]] |
| 2.1.3 | Cytarabine-resistant AML | Upregulated | Chemosensitivity to cytarabine | GPR56 SNP rs1376041 G > A minor allele is highly associated with high GPR56 mRNA expression and cytarabine resistance in leukemic cells | [76] |
| 2.2 | Metastatic Melanoma | Downregulated | Tumor cell migration, angiogenesis and ECM remodeling | Downregulation correlates with higher metastatic potential melanoma | [71] |
| 2.3.1 | Glioma/Glioblastoma | Upregulated | Glioblastoma cell adhesion | GPR56 expression is correlated with cellular transformation phenotypes | [80] |
| 2.3.2 | Radioresistant glioblastoma | Downregulated | Inhibitor of mesenchymal differentiation | GPR56low presents with a poor prognosis in glioblastoma patients largely due to increased differentiation to radioresistant mesenchymal glioblastomas | [82] |
| 2.4 | Esophageal Squamous Cell Carcinoma (ESCC) | Upregulated | Cell migration | GPR56 nuclear expression significantly correlates with nodal invasion/metastasis in ESCC | [88,89] |
| 2.5 | Acute Lymphoblastic Leukemia (ALL) | Upregulated | – | – | [79] |
| 2.6 | Non-small-cell Lung Carcinoma | Upregulated | Cell proliferation and invasion capacity | GPR56 expression significantly correlates with the TNM stage of NSCLC and OS | [90] |
| 2.7 | Multidrug-Resistant Lung Adenocarcinoma | Higher promoter methylation of GPR56 isoform 3 | Multidrug resistance | GPR56, MT1G and RASSF1 can be used as a potential methylation marker associated with acquired MDR lung adenocarcinoma | [91] |
| 2.8 | Osteosarcoma | Upregulated | Cell proliferation and invasive capacity | GPR56 highly correlates with TNM stage and OS in osteosarcoma patients | [94] |
| 2.9 | Ovarian Cancer | Upregulated | Cellular proliferation | GPR56 expression significantly correlates with advanced FIGO stage, lymph node metastasis and OS; GPR56 can act as an independent prognostic factor | [86] |
| 2.10 | Colorectal cancer (CRC) | Upregulated | CRC cell proliferation, metastasis and invasion via epithelial-to-mesenchymal transition | GPR56 expression significantly correlates to malignant progression of the primary tumor and poorer prognosis | [84] |
| 2.11 | Gastric Cancer | Downregulated | Probable role in cellular growth, migration, and invasion | GPR56 is under the direct control of VEZT, which is significantly correlated with TNM stages | [92] |
| 3 | Rheumatoid Arthritis (RA) | High levels of serum soluble GPR56 | – | GPR56 positively correlates with rheumatoid factor and elevated tumor necrosis factor in RA patients | [101] |
| 4 | Depression | Downregulated | – | Treatment of anti-depressants, duloxetine or fluoxetine, leads to upregulation of GPR56 expression levels | [70] |
3.1. BFPP
Bilateral frontoparietal polymicrogyria (BFPP), a novel brain cortical malformation, is an autosomal recessive genetic disorder caused by homozygous mutations in the coding region of GPR56 gene [17]. BFPP patients usually display five common clinical features and three characteristic MRI signatures [Table 3] [17,18]. In addition, BFPP is also associated with West syndrome and Lennox-Gastaut syndrome in multiple cases [67]. The histopathology of BFPP is cobblestone-like lissencephaly characterized by neuronal ectopias breaching the pial BM of the cerebral cortex caused by the over migration of post-mitotic neurons from deep as well as superficial cortical layers into the marginal zone [68]. This breaching of pial BM is associated with discontinuity of ECM constituents of pial BM, such as laminin, Col-III, Col-IV, and nidogen [68]. Neuronal progenitor/precursor cells (NPCs) and migrating neurons in their early phases of differentiation (GPR56+Nestin+ cells) express GPR56 as well [68,69]. However, the fully differentiated GFAP-positive-astrocytes, βIII-tubulin-positive neurons, or migrating neurons in their later phases of differentiation (GPR56−Nestin+ cells) do not express GPR56 [69]. GPR56 expression is required for proper positioning, adhesion, and inhibition of neuronal migration upon activation by COL3A1 presented on meningeal fibroblasts in the cerebral cortex during neurogenesis and cerebral cortical development [28,69].
Table 3.
| Feature | Description | |
|---|---|---|
| 1 | Mental Retardation | Moderate to severe mental retardation in all patients and limited verbal language in most cases. |
| 2 | Motor Developmental Delay | Developmental milestones are achieved at much later ages. |
| 3 | Seizures | Reported in most BFPP patients. They mostly have symptomatic generalized epilepsy. Types of seizures varying among the patients include tonic, atonic, atypical absence, and myoclonic. Some BFPP cases are also associated with Lennox-Gastaut syndrome, characterized by a severe form of generalized seizure of multiple types. Specific electroencephalogram (EEG) abnormality called slow spike-and-wave pattern is also observed in BFPP patients while awake and generalized fast rhythms while asleep. |
| 4 | Cerebellar Signs | Cerebellar signs such as truncal ataxia, finger dysmetria, and rest tremor are observable in most BFPP patients. |
| 5 | Dysconjugate Gaze | Many BFPP cases report the patient's gaze as esotropia, nystagmus, exotropia, strabismus, or a history of squint eye. |
| 6 | Bilateral Polymicrogyria | Abnormally thickened cortex with many small ridges or folds in a bilaterally symmetrical manner with decreasing anterior-to-posterior gradient of severity. |
| 7 | White Matter Defect | Random white matter signal changes are observed in all BFPP patients bilaterally. |
| 8 | Brainstem and Cerebellar Hypoplasia | BFPP patients present with small brain stem and vermis in many cases. |
The developing granule cells of the rostral cerebellum also express GPR56. Analyses of Gpr56-knockout mice and BFPP-diagnosed human fetus have revealed highly severe malformation of rostral cerebellum characterized by the fusion of adjacent lobules I-III, disrupted layering of neurons and glia, and fragmentation of the pial BM [45]. As discussed in the section Nervous system (CNS/PNS), it is probably due to the loss of the ability of these Gpr56−/− granule cells to bind to TG2. No other defects in cell proliferation, migration, or neurite growth are known in Gpr56-knockout rostral cerebellum granule cells. Another typical feature observed in BFPP patients is the presence of white matter abnormalities indicative of myelination defect. Indeed, GPR56/ADGRG1 plays a crucial role in myelination in CNS, as described in the section Nervous system (CNS/PNS).
3.2. Depression and other nervous system-related pathologies
GPR56 is also associated with nervous system-related pathologies other than BFPP. A 15-bp deletion in a regulatory element <150 bp upstream of the transcriptional start site of the non-coding exon 1 m (e1m) of the GPR56 gene leads to severe bilateral cortical disruptions around the Sylvian region, predominantly in the Broca's region, of the primary language area [23]. Additionally, GPR56 was found recently to be a reliable treatment-response biomarker of depression patients treated with anti-depressants [70]. Specifically, treatment of anti-depressants, duloxetine or citalopram, leads to upregulated GPR56 expression only in the responder cohorts [70]. By contrast, chronic stress in mice leads to a significant reduction of GPR56 expression in the blood, prefrontal cortex and dorsal hippocampal areas, but not in the nucleus accumbens or ventral hippocampal region [70]. Such down-regulated GPR56 expression in mice results in depression-like behavior, which is reversed following the treatment of antidepressants [70]. The gene sets associated with AKT-, GSK3-, and EIF4-pathways were found to be the most upregulated in GPR56 agonist-treated cells compared to controls in the functional characterization of the anti-depressant role of GPR56 [70]. As these pathways have previously been implicated highly in depression and anti-depressant action of several drugs, including selective serotonin reuptake inhibitors and ketamine, GPR56 most probably uses these pathways for agonist-mediated anti-depressant activities [70].
3.3. Cancers
Invasion and migration of tumor cells are highly dependent on their interaction with ECM and a delicate balance of cell adhesion and detachment. GPR56 promotes or inhibits cancer development in a cell type- and/or tumorigenic stage-specific manner in different cancers [Fig. 3] [Table 2].
3.3.1. Melanoma
In 1999, A. J. Zendman et al. discovered an inverse correlation of the TM7XN1 (later found to be GPR56) expression level with the metastatic potential of human melanoma cell lines [71]. Upon further exploration, it was found that multiple mechanisms are involved in the role of GPR56 in melanoma tumorigenesis. One is due to the inhibition of vascular endothelial growth factor (VEGF) secretion by GPR56, thereby reducing the number of blood vessels at the site of metastatic melanomas [Fig. 3B] [22,72]. TG2 was identified as the ECM ligand of GPR56 in melanoma cells, and GPR56-mediated TG2 internalization and degradation is considered a possible anti-tumorigenic mechanism [Fig. 3B] [46]. As TG2 is a major cross-linking enzyme of ECM that promotes fibronectin and focal adhesion kinase (FAK) deposition, its internalization followed by degradation likely provides a control over tumor tissue ECM environment [46]. These ECM modifications help in impeding the expansion of micro-metastases to macro-metastases of melanoma [73]. On the other hand, activation of GPR56 was shown by us recently to promote melanoma cell migration by coupling to Gα12/13 and IL-6 up-regulation in a RhoA-dependent manner [42]. The roles of GPR56 in melanoma development are likely dependent on the stage of cancer cells and the activation mechanisms involved.
3.3.2. Acute myeloid and lymphoblastic leukemia (AML/ALL)
GPR56 is an excellent cell surface marker for AML. It identifies CD34+ AML cells as well as CD34- leukemia stem cell (LSC) fraction and is correlated with various prognostic factors such as low overall survival (OS) and high chemo-resistance [[74], [75], [76], [77]]. The presence of GPR56 SNP rs1376041 G > A minor allele A is associated with high GPR56 mRNA expression, which in turn is positively associated with cytarabine resistance in leukemic cells of AML patients [76]. GPR56 silencing in AML cells leads to a significant increase in apoptosis and cytarabine sensitivity [76]. These results suggest that GPR56 might also be a potential therapeutic target in AML, especially in the chemo-resistant AML.
GPR56 expression is high in EVI1high (Ecotropic Viral Integration site-1) AML cells [74]. It is associated with high cell-adhesion and anti-apoptotic ability [74]. EVI1 directly binds to the promoter region of GPR56 and regulates its expression in these EVI1high AML cells [74]. Furthermore, therapeutically useful and novel pyrrole-imidazole polyamides (PIPs) have been developed that can specifically bind to the GPR56 promoter and inhibit the binding of EVI1, thereby inhibiting its expression and thus suppressing the cell growth and promoting apoptosis [77,78]. The expression levels of GPR56 and white blood cell count at diagnosis are significantly associated in ALL [79]. In addition, GPR56 is upregulated in T-ALL compared to precursor B-ALL [78].
3.3.3. Glioblastoma
Glioblastomas have upregulated expression levels of GPR56, which co-localize with α-actinin on the leading edges of the extending cell membranes, thereby suggesting a role in cellular migration and adhesion [80]. Receptor activation in U87-MG human glioma cells inhibits cell migration via Gαq and Rho pathway [Fig. 3D] [81]. The mesenchymal transition of glioblastoma occurs naturally and in response to therapy. Clinically, a correlation of higher radioresistance and shorter survival is associated with glioblastoma of the mesenchymal subtype [82]. Interestingly, upregulated GPR56 expression in proneural and classical glioblastomas cells is lost during their mesenchymal transition [82]. Conversely, silencing or downregulation of GPR56 promotes mesenchymal differentiation and radioresistance of proneural glioma initiating cells [82]. It is thought that GPR56, by inhibiting NF-κB signaling, might act as an inhibitor of mesenchymal differentiation of glioblastoma and beyond [82]. Altogether, these results suggest that GPR56low expression might correlate with a poor prognosis and that GPR56 can act as a diagnostic marker of glioma/glioblastomas.
3.3.4. Colorectal cancer (CRC)
GPR56 levels are significantly upregulated in CRC cells of CRC patients. It is reported to associate with progastrin and is expressed by epithelial cells found near the base of the colonic crypts and marks the long-lived colonic progenitor cells [83]. Progastrin specifically binds to GPR56 expressing cells, and overexpression of progastrin leads to an increase in GPR56 expressing cells, while treating azoxymethane (AOM), a carcinogen, leads to an increase in GPR56 mRNA in progastrin overexpressing mouse colonic mucosa compared to WT mice [83]. GPR56 is required for progastrin-dependent inhibition of apoptosis of colonic epithelial and CRC cells [Fig. 3A] [83,84]. The high expression of GPR56 correlates well with malignant progression of primary tumors, poorer prognosis, and drug-resistance, compared to low GPR56 expression [84]. GPR56 increases cell migration of CRC cells by stimulating epithelial-to-mesenchymal transition via PI3K/AKT signaling activation [84]. LGR5 (Leucine-rich repeat-containing G-protein-coupled Receptor 5) is a cancer stem cell marker in CRC and is reported to interconvert with more drug-resistant LGR5- cancer cells [85]. Interestingly, a decrease in levels of LGR5 is associated with an increase in GPR56 levels and enhanced resistance to drugs such as irinotecan and 5-fluorouracil [85]. Knockdown of GPR56, however, restores the sensitivity of these cells to the drugs [85]. Enhanced expression of ABC (ATP-Binding Cassette) transporters, MDR1 (ABCB1) in particular, is reported to cause this GPR56-mediated enhancement of drug-resistance via the RhoA-mediated signaling pathway [Fig. 3A] [85].
3.3.5. Ovarian cancer
High GPR56 expression is observed in epithelial ovarian cancer cells compared to normal ovarian tissues and adjacent ovarian tissues [86]. It is significantly correlated with advanced FIGO (International Federation of Gynecology and Obstetrics) stage, lymph node metastasis, and OS, and can also act as an independent prognostic factor [86]. GPR56 silencing leads to inhibitory effects on cell proliferation, migration ability, and invasion of ovarian cancer cells [86]. GPR56 knockdown leads to the downregulation of RhoA-GTP and upregulation of E-cadherin [86]. DMU-214 (3′-Hydroxy-3,4,5,4′-Tetramethoxy-stilbene), a metabolite of cytotoxic resveratrol analogue DMU-212, exerts significantly higher biological activity than the parent compound [87]. The anti-migratory and anti-proliferative activity of DMU-214 on ovarian cancer cells is probably by downregulating the mRNA and protein levels of GPR56, SRF, and RGCC (Regulator of Cell Cycle) by 60–75% [87]. Overall, GPR56 has the potential for diagnostic and therapeutic roles in ovarian cancer.
3.3.6. Other cancers
GPR56, TG2, and NF-κB are concomitantly expressed in esophageal squamous cell carcinoma (ESCC), and their expression levels are significantly correlated with ESCC nodal invasion and metastasis [88,89]. GPR56 positively regulates tumor growth and invasion capacity of non-small-cell lung carcinoma (NSCLC) [90]. Methylation of the GPR56 gene is linked to multidrug resistance in lung adenocarcinomas [91]. In the tissues and blood of gastric cancer patients, Vezatin (VEZT), a putative tumor suppressor adherens junctions transmembrane protein that is highly correlated with the TNM (Tumor, Nodes, Metastases) stage, depth of cancer invasion, and lymphatic metastasis, is inactivated by hypermethylation of its promoter region compared to healthy controls [92]. Upon restoring the expression of VEZT in gastric cancer cell lines, tumor growth, adhesion, cell cycle, and migration or invasion is inhibited, probably by the downregulation of its direct target genes, TCF19 and CDC42, and by the upregulation of another direct target gene, GPR56 [91,92]. GPR56 expression levels are significantly increased in TCF-4J (T Cell Factor 4 isoform J) compared with TCF-4K (T Cell Factor 4 isoform K) expressing hepatocellular carcinoma cells [93]. GPR56 positively regulates tumor proliferation and invasive capacity of osteosarcoma, and is highly correlated with the TNM stage and OS of osteosarcoma patients, suggesting GPR56 to be a potential independent unfavorable prognostic factor [94].
4. Conclusion and future perspectives
Since its discovery in 1999 by two independent groups via two entirely different approaches, GPR56 is one of the most-studied members of the aGPCR family [10,21,71]. Numerous studies discussed herein have also established it as a critical receptor in health and disease. Its functional role in various physiological and pathological processes described above makes it a considerable target for developing novel diagnostic tools and therapeutics for the relevant diseases/disorders such as cancer progression and immune-system dysfunction. Indeed, with the increasing understanding of its activation and signaling mechanisms as well as the functional link of GPR56 to the migration, metastasis, and radio-/chemo-resistance of diverse cancers that are the major causes of cancer treatment failure [73,85], GPR56 might be an useful diagnostic and/or prognostic marker for certain cancers, as well as potential cancer-targeted therapeutics [95].
Conflict of interest
The authors declare that there was no commercial or financial relationship during this study that could be construed as a potential conflict of interest.
Acknowledgments
HHL is supported by grants from the Chang Gung Memorial Hospital (CMRPD1K0132, CMRPD1K0212, CMRPD1K0221 and CMRPD1K0301-2) and the Ministry of Science and Technology, Taiwan (MOST-107-2320-B-182-006).
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Kobilka B.K. G protein coupled receptor structure and activation. Biochim Biophys Acta Biomembrs Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fredriksson R., Lagerström M.C., Lundin L.G., Schiöth H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 3.Kroeze W.K., Sheffler D.J., Roth B.L. G-protein-coupled receptors at a glance. J Cell Sci. 2003;116:4867–4869. doi: 10.1242/jcs.00902. [DOI] [PubMed] [Google Scholar]
- 4.Ji T.H., Grossmann M., Ji I. G protein-coupled receptors I. Diversity of receptor-ligand interactions. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 5.Hauser A.S., Attwood M.M., Rask-Andersen M., Schiöth H.B., Gloriam D.E. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H.H. G-protein-coupled receptors and their (Bio) Chemical significance win 2012 nobel prize in chemistry. Biomed J. 2013;36:118–124. doi: 10.4103/2319-4170.113233. [DOI] [PubMed] [Google Scholar]
- 7.Fredriksson R., Schiöth H.B. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 8.Kamesh N., Aradhyam G.K., Manoj N. The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis. BMC Evol Biol. 2008;8:129. doi: 10.1186/1471-2148-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan A., Almén M.S., Fredriksson R., Schiöth H.B. The origin of GPCRs: identification of mammalian like Rhodopsin, adhesion, glutamate and frizzled GPCRs in fungi. PloS One. 2012;7 doi: 10.1371/journal.pone.0029817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamann J., Aust G., Araç D., Engel F.B., Formstone C., Fredriksson R., et al. International union of basic and clinical pharmacology. XCIV. adhesion G protein-coupled receptors. Pharmacol Rev. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordström K.J., Fredriksson R., Schiöth H.B. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol. 2008;8:9. doi: 10.1186/1471-2148-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putnam N.H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 13.Araç D., Boucard A.A., Bolliger M.F., Nguyen J., Soltis S.M., Südhof T.C., et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prömel S., Langenhan T., Araç D. Matching structure with function: the GAIN domain of Adhesion-GPCR and PKD1-like proteins. Trends Pharmacol Sci. 2013;34:470–478. doi: 10.1016/j.tips.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Lin H.H., Chang G.W., Davies J.Q., Stacey M., Harris J., Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 16.Salzman G.S., Ackerman S.D., Ding C., Koide A., Leon K., Luo R., et al. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron. 2016;91:1292–1304. doi: 10.1016/j.neuron.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piao X., Hill R.S., Bodell A., Chang B.S., Basel-Vanagaite L., Straussberg R., et al. Protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 18.Jin Z., Luo R., Piao X. GPR56 and its related diseases. Prog Mol Biol Transl Sci. 2009;89:1–13. doi: 10.1016/S1877-1173(09)89001-7. [DOI] [PubMed] [Google Scholar]
- 19.Peng Y.M., van de Garde M.D., Cheng K.F., Baars P.A., Remmerswaal E.B., van Lier R.A., et al. Specific expression of GPR56 by human cytotoxic lymphocytes. J Leukoc Biol. 2011;90:735–740. doi: 10.1189/jlb.0211092. [DOI] [PubMed] [Google Scholar]
- 20.Liu M., Parker R.M., Darby K., Eyre H.J., Copeland N.G., Crawford J., et al. GPR56, a novel secretin-like human G-protein-coupled receptor gene. Genomics. 1999;55:296–305. doi: 10.1006/geno.1998.5644. [DOI] [PubMed] [Google Scholar]
- 21.Bjarnadóttir T.K., Geirardsdóttir K., Ingemansson M., Mirza M.A., Fredriksson R., Schiöth H.B. Identification of novel splice variants of Adhesion G protein-coupled receptors. Gene. 2007;387:38–48. doi: 10.1016/j.gene.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.E., Han J.M., Park C.R., Shin K.J., Ahn C., Seong J.Y., et al. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J Canc Res Clin Oncol. 2010;136:47–53. doi: 10.1007/s00432-009-0635-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae B.I., Tietjen I., Atabay K.D., Evrony G.D., Johnson M.B., Asare E., et al. Evolutionarily dynamic alternative splicing of GPR56 Regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T., Chiou B., Gilman C.K., Luo R., Koshi T., Yu D., et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020;39 doi: 10.15252/embj.2019104136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z., Tietjen I., Bu L., Liu-Yesucevitz L., Gaur S.K., Walsh C.A., et al. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16:1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- 26.Luo R., Jin Z., Deng Y., Strokes N., Piao X. Disease-associated mutations prevent GPR56-collagen III interaction. PloS One. 2012;7 doi: 10.1371/journal.pone.0029818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzman G.S., Zhang S., Fernandez C.G., Araç D., Koide S. Specific and direct modulation of the interaction between adhesion GPCR GPR56/ADGRG1 and tissue transglutaminase 2 using synthetic ligands. Sci Rep. 2020;10:16912. doi: 10.1038/s41598-020-74044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo R., Jeong S.J., Jin Z., Strokes N., Li S., Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ackerman S.D., Luo R., Poitelon Y., Mogha A., Harty B.L., D'Rozario M., et al. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J Exp Med. 2018;215:941–961. doi: 10.1084/jem.20161714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little K.D., Hemler M.E., Stipp C.S. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-gαq/11 association. Mol Biol Cell. 2004;15:2375–2387. doi: 10.1091/mbc.E03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giera S., Luo R., Ying Y., Ackerman S.D., Jeong S.J., Stoveken H.M., et al. Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. ELife. 2018;7 doi: 10.7554/eLife.33385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang N.Y., Chang G.W., Huang Y.S., Peng Y.M., Hsiao C.C., Kuo M.L., et al. Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J Cell Sci. 2016;129:2156–2169. doi: 10.1242/jcs.174458. [DOI] [PubMed] [Google Scholar]
- 33.Zhu B., Luo R., Jin P., Li T., Oak H.C., Giera S., et al. GAIN domain-mediated cleavage is required for activation of G protein- coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J Biol Chem. 2019;294:19246–19254. doi: 10.1074/jbc.RA119.008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoveken H.M., Bahr L.L., Anders M.W., Wojtovich A.P., Smrcka A.V., Tall G.G. Dihydromunduletone is a small-molecule selective adhesion G protein-coupled receptor antagonist. Mol Pharmacol. 2016;90:214–224. doi: 10.1124/mol.116.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoveken H.M., Larsen S.D., Smrcka A.V., Tall G.G. Gedunin- and khivorin-derivatives are small-molecule partial agonists for adhesion G protein-coupled receptors GPR56/ADGRG1 and GPR114/ADGRG5. Mol Pharmacol. 2018;93:477–488. doi: 10.1124/mol.117.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kishore A., Hall R.A. Disease-associated extracellular loop mutations in the adhesion G protein-coupled receptor G1 (ADGRG1; GPR56) differentially regulate downstream signaling. J Biol Chem. 2017;292:9711–9720. doi: 10.1074/jbc.M117.780551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoveken H.M., Hajduczok A.G., Xu L., Tall G.G. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc Natl Acad Sci U S A. 2015;112:6194–6199. doi: 10.1073/pnas.1421785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kishore A., Purcell R.H., Nassiri-Toosi Z., Hall R.A. Stalk-dependent and stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and Bai1 (ADGRB1) J Biol Chem. 2016;291:3385–3394. doi: 10.1074/jbc.M115.689349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salzman G.S., Zhang S., Gupta A., Koide A., Koide S., Araç D. Stachel-independent modulation of GPR56/ADGRG1 signaling by synthetic ligands directed to its extracellular region. Proc Natl Acad Sci U S A. 2017;114:10095–10100. doi: 10.1073/pnas.1708810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paavola K.J., Stephenson J.R., Ritter S.L., Alter S.P., Hall R.A. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem. 2011;286:28914–28921. doi: 10.1074/jbc.M111.247973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liebscher I., Schön J., Petersen S.C., Fischer L., Auerbach N., Demberg L.M. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014;9:2018–2026. doi: 10.1016/j.celrep.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang N.Y., Peng Y.M., Juang H.H., Chen T.C., Pan H.L., Chang G.W., et al. GPR56/ADGRG1 activation promotes melanoma cell migration via NTF dissociation and CTF-mediated Gα12/13/RhoA signaling. J Invest Dermatol. 2017;137:727–736. doi: 10.1016/j.jid.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 43.Jeong S.J., Luo R., Li S., Strokes N., Piao X. Characterization of G protein-coupled receptor 56 protein expression in the mouse developing neocortex. J Comp Neurol. 2012;520:2930–2940. doi: 10.1002/cne.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong S.J., Luo R., Singer K., Giera S., Kreidberg J., Kiyozumi D., et al. GPR56 functions together with α3β1 integrin in regulating cerebral cortical development. PloS One. 2013;8 doi: 10.1371/journal.pone.0068781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koirala S., Jin Z., Piao X., Corfas G. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci. 2009;29:7439–7449. doi: 10.1523/JNEUROSCI.1182-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L., Friedland S., Corson N., Xu L. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res. 2014;74:1022–1031. doi: 10.1158/0008-5472.CAN-13-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackerman S.D., Garcia C., Piao X., Gutmann D.H., Monk K.R. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Gα12/13 and RhoA. Nat Commun. 2015;6:6122. doi: 10.1038/ncomms7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White J.P., Wrann C.D., Rao R.R., Nair S.K., Jedrychowski M.P., You J.S., et al. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci U S A. 2014;111:15756–15761. doi: 10.1073/pnas.1417898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang Y.J., Son H.J., Choi Y.M., Ahn J., Jung C.H., Ha T.Y. Apigenin enhances skeletal muscle hypertrophy and myoblast differentiation by regulating Prmt7. Oncotarget. 2017;8:78300–78311. doi: 10.18632/oncotarget.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu M.P., Doyle J.R., Barry B., Beauvais A., Rozkalne A., Piao X., et al. G-protein coupled receptor 56 promotes myoblast fusion through serum response factor- and nuclear factor of activated T-cell-mediated signalling but is not essential for muscle development in vivo. FEBS J. 2013;280:6097–6113. doi: 10.1111/febs.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Si Y., Ma N., Mei J. The RNA-binding protein PCBP2 inhibits Ang II-induced hypertrophy of cardiomyocytes though promoting GPR56 mRNA degeneration. Biochem Biophys Res Commun. 2015;464:679–684. doi: 10.1016/j.bbrc.2015.06.139. [DOI] [PubMed] [Google Scholar]
- 52.Kitakaze T., Yoshikawa M., Kobayashi Y., Kimura N., Goshima N., Ishikawa T., et al. Extracellular transglutaminase 2 induces myotube hypertrophy through G protein-coupled receptor 56. Biochim Biophys Acta Mol Cell Res. 2020;1867:118563. doi: 10.1016/j.bbamcr.2019.118563. [DOI] [PubMed] [Google Scholar]
- 53.Della Chiesa M., Falco M., Parolini S., Bellora F., Petretto A., Romeo E., et al. GPR56 as a novel marker identifying the CD56dull CD16+ NK cell subset both in blood stream and in inflamed peripheral tissues. Int Immunol. 2010;22:91–100. doi: 10.1093/intimm/dxp116. [DOI] [PubMed] [Google Scholar]
- 54.Chang G.W., Hsiao C.C., Peng Y.M., Vieira Braga F.A., Kragten N.A., Remmerswaal E.B., et al. The adhesion G protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep. 2016;15:1757–1770. doi: 10.1016/j.celrep.2016.04.053. [DOI] [PubMed] [Google Scholar]
- 55.Yeung J., Adili R., Stringham E.N., Luo R., Vizurraga A., Rosselli-Murai L.K., et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc Natl Acad Sci U S A. 2020;117:28275–28286. doi: 10.1073/pnas.2008921117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Truong K.L., Schlickeiser S., Vogt K., Boës D., Stanko K., Appelt C., et al. Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4+ memory T cells. Nat Commun. 2019;10:2263. doi: 10.1038/s41467-019-10018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao T.N., Marks-Bluth J., Sullivan J., Gupta M.K., Chandrakanthan V., Fitch S.R., et al. High-level Gpr56 expression is dispensable for the maintenance and function of hematopoietic stem and progenitor cells in mice. Stem Cell Res. 2015;14:307–322. doi: 10.1016/j.scr.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tokoro Y., Yamada Y., Takayanagi S.I., Hagiwara T. 57R2A, a newly established monoclonal antibody against mouse GPR56, marks long-term repopulating hematopoietic stem cells. Exp Hematol. 2018;59:51–59. doi: 10.1016/j.exphem.2017.12.001. e1. [DOI] [PubMed] [Google Scholar]
- 59.Kartalaei P.S., Yamada-Inagawa T., Vink C.S., de Pater E., van der Linden R., Marks-Bluth J., et al. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J Exp Med. 2015;212:93–106. doi: 10.1084/jem.20140767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunér P., Al-Amily I.M., Soni A., Asplund O., Safi F., Storm P., et al. Adhesion G protein-coupled receptor G1 (ADGRG1/GPR56) and pancreatic β-cell function. J Clin Endocrinol Metab. 2016;101:4637–4645. doi: 10.1210/jc.2016-1884. [DOI] [PubMed] [Google Scholar]
- 61.Olaniru O.E., Pingitore A., Giera S., Piao X., Castañera González R., Jones P.M., et al. The adhesion receptor GPR56 is activated by extracellular matrix collagen III to improve β-cell function. Cell Mol Life Sci CMLS. 2018;75:4007–4019. doi: 10.1007/s00018-018-2846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Al-Amily I.M., Dunér P., Groop L., Salehi A. The functional impact of G protein-coupled receptor 142 (Gpr142) on pancreatic β-cell in rodent. Pflugers Arch. 2019;471:633–645. doi: 10.1007/s00424-019-02262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al Hasan M., Roy P., Dolan S., Martin P.E., Patterson S., Bartholomew C. Adhesion G-protein coupled receptor 56 is required for 3T3-L1 adipogenesis. J Cell Physiol. 2020;235:1601–1614. doi: 10.1002/jcp.29079. [DOI] [PubMed] [Google Scholar]
- 64.Chen G., Yang L., Begum S., Xu L. GPR56 is essential for testis development and male fertility in mice. Dev Dyn. 2010;239:3358–3367. doi: 10.1002/dvdy.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayers K.L., Lambeth L.S., Davidson N.M., Sinclair A.H., Oshlack A., Smith C.A. Identification of candidate gonadal sex differentiation genes in the chicken embryo using RNA-seq. BMC Genom. 2015;16:704. doi: 10.1186/s12864-015-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roly Z.Y., Major A.T., Fulcher A., Estermann M.A., Hirst C.E., Smith C.A. Adhesion G-protein-coupled receptor, GPR56, is required for Müllerian duct development in the chick. J Endocrinol. 2020;244:395–413. doi: 10.1530/JOE-19-0419. [DOI] [PubMed] [Google Scholar]
- 67.Parrini E., Ferrari A.R., Dorn T., Walsh C.A., Guerrini R. Bilateral frontoparietal polymicrogyria, Lennox-Gastaut syndrome, and GPR56 gene mutations. Epilepsia. 2009;50:1344–1353. doi: 10.1111/j.1528-1167.2008.01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li S., Jin Z., Koirala S., Bu L., Xu L., Hynes R.O., et al. GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28:5817–5826. doi: 10.1523/JNEUROSCI.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bai Y., Du L., Shen L., Zhang Y., Zhang L. GPR56 is highly expressed in neural stem cells but downregulated during differentiation. Neuroreport. 2009;20:918–922. doi: 10.1097/WNR.0b013e32832c92d7. [DOI] [PubMed] [Google Scholar]
- 70.Belzeaux R., Gorgievski V., Fiori L.M., Lopez J.P., Grenier J., Lin R., et al. GPR56/ADGRG1 is associated with response to antidepressant treatment. Nat Commun. 2020;11:1635. doi: 10.1038/s41467-020-15423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zendman A.J., Cornelissen I.M., Weidle U.H., Ruiter D.J., van Muijen G.N. TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential 1. FEBS Lett. 1999;446:292–298. doi: 10.1016/s0014-5793(99)00230-6. [DOI] [PubMed] [Google Scholar]
- 72.Yang L., Chen G., Mohanty S., Scott G., Fazal F., Rahman A., et al. GPR56 regulates VEGF production and angiogenesis during melanoma progression. Canc Res. 2011;71:5558–5568. doi: 10.1158/0008-5472.CAN-10-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Millar M.W., Corson N., Xu L. The adhesion G-protein-coupled receptor, GPR56/ADGRG1, inhibits cell-extracellular matrix signaling to prevent metastatic melanoma growth. Front Oncol. 2018;8:8. doi: 10.3389/fonc.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saito Y., Kaneda K., Suekane A., Ichihara E., Nakahata S., Yamakawa N., et al. Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia. 2013;27:1637–1649. doi: 10.1038/leu.2013.75. [DOI] [PubMed] [Google Scholar]
- 75.Daria D., Kirsten N., Muranyi A., Mulaw M., Ihme S., Kechter A., et al. GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia. 2016;30:1734–1741. doi: 10.1038/leu.2016.76. [DOI] [PubMed] [Google Scholar]
- 76.Bargal S.A., Rafiee R., Crews K.R., Wu H., Cao X., Rubnitz J.E., et al. Genome-wide association analysis identifies SNPs predictive of in vitro leukemic cell sensitivity to cytarabine in pediatric AML. Oncotarget. 2018;9:34859–34875. doi: 10.18632/oncotarget.26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daga S., Rosenberger A., Quehenberger F., Krisper N., Prietl B., Reinisch A., et al. High GPR56 surface expression correlates with a leukemic stem cell gene signature in CD34-positive AML. Canc Med. 2019;8:1771–1778. doi: 10.1002/cam4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saha H.R., Kaneda-Nakashima K., Shimosaki S., Suekane A., Sarkar B., Saito Y., et al. Suppression of GPR56 expression by pyrrole-imidazole polyamide represents a novel therapeutic drug for AML with high EVI1 expression. Sci Rep. 2018;8:13741. doi: 10.1038/s41598-018-32205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silveira V.S., Scrideli C.A., Moreno D.A., Yunes J.A., Queiroz R.G., Toledo S.C., et al. Gene expression pattern contributing to prognostic factors in childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2013;54:310–314. doi: 10.3109/10428194.2012.710330. [DOI] [PubMed] [Google Scholar]
- 80.Shashidhar S., Lorente G., Nagavarapu U., Nelson A., Kuo J., Cummins J., et al. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24:1673–1682. doi: 10.1038/sj.onc.1208395. [DOI] [PubMed] [Google Scholar]
- 81.Ohta S., Sakaguchi S., Kobayashi Y., Mizuno N., Tago K., Itoh H. Agonistic antibodies reveal the function of GPR56 in human glioma U87-MG cells. Biol Pharm Bull. 2015;38:594–600. doi: 10.1248/bpb.b14-00752. [DOI] [PubMed] [Google Scholar]
- 82.Moreno M., Pedrosa L., Paré L., Pineda E., Bejarano L., Martínez J., et al. GPR56/ADGRG1 inhibits mesenchymal differentiation and radioresistance in glioblastoma. Cell Rep. 2017;21:2183–2197. doi: 10.1016/j.celrep.2017.10.083. [DOI] [PubMed] [Google Scholar]
- 83.Jin G., Sakitani K., Wang H., Jin Y., Dubeykovskiy A., Worthley D.L., et al. The G-protein coupled receptor 56, expressed in colonic stem and cancer cells, binds progastrin to promote proliferation and carcinogenesis. Oncotarget. 2017;8:40606–40619. doi: 10.18632/oncotarget.16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji B., Feng Y., Sun Y., Ji D., Qian W., Zhang Z., et al. GPR56 promotes proliferation of colorectal cancer cells and enhances metastasis via epithelial-mesenchymal transition through PI3K/AKT signaling activation. Oncol Rep. 2018;40:1885–1896. doi: 10.3892/or.2018.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang S., Chatterjee T., Godoy C., Wu L., Liu Q.J., Carmon K.S. GPR56 drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-mediated mechanism. Mol Canc Res. 2019;17:2196–2207. doi: 10.1158/1541-7786.MCR-19-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Z., Huang Z., Yang W., Li Z., Xing S., Li H., et al. Expression of orphan GPR56 correlates with tumor progression in human epithelial ovarian cancer. Neoplasma. 2017;64:32–39. doi: 10.4149/neo_2017_104. [DOI] [PubMed] [Google Scholar]
- 87.Nowicki A., Skupin-Mrugalska P., Jozkowiak M., Wierzchowski M., Rucinski M., Ramlau P., et al. The effect of 3′-hydroxy-3,4,5,4′-tetramethoxy-stilbene, the metabolite of the resveratrol analogue DMU-212, on the motility and proliferation of ovarian cancer cells. Int J Mol Sci. 2020;21:1100. doi: 10.3390/ijms21031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sud N., Sharma R., Ray R., Chattopadhyay T.K., Ralhan R. Differential expression of G-protein coupled receptor 56 in human esophageal squamous cell carcinoma. Canc Lett. 2006;233:265–270. doi: 10.1016/j.canlet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 89.Kausar T., Sharma R., Hasan M.R., Tripathi S.C., Saraya A., Chattopadhyay T.K., et al. Clinical significance of GPR56, transglutaminase 2, and NF-κB in esophageal squamous cell carcinoma. Cancer Invest. 2011;29:42–48. doi: 10.3109/07357907.2010.512597. [DOI] [PubMed] [Google Scholar]
- 90.Song Y., Li A., Zhang L., Duan L. Expression of G protein-coupled receptor 56 is associated with tumor progression in non-small-cell lung carcinoma patients. OncoTargets Ther. 2016;9:4105–4112. doi: 10.2147/OTT.S106907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo R., Wu G., Li H., Qian P., Han J., Pan F., et al. Promoter methylation profiles between human lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP) cells and its progenitor A549 cells. Biol Pharm Bull. 2013;36:1310–1316. doi: 10.1248/bpb.b13-00153. [DOI] [PubMed] [Google Scholar]
- 92.Miao R., Guo X., Zhi Q., Shi Y., Li L., Mao X., et al. VEZT, a novel putative tumor suppressor, suppresses the growth and tumorigenicity of gastric cancer. PloS One. 2013;8 doi: 10.1371/journal.pone.0074409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tomimaru Y., Koga H., Yano H., de la Monte S., Wands J.R., Kim M. Upregulation of T-cell factor-4 isoform-responsive target genes in hepatocellular carcinoma. Liver Int Off J Int Assoc Study Liver. 2013;33:1100–1112. doi: 10.1111/liv.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Z., Gao P., Li Z. Expression of G Protein-coupled receptor 56 is an unfavorable prognostic factor in osteosarcoma patients. Tohoku J Exp Med. 2016;239:203–211. doi: 10.1620/tjem.239.203. [DOI] [PubMed] [Google Scholar]
- 95.Bassilana F., Nash M., Ludwig M.G. Adhesion G protein-coupled receptors: opportunities for drug discovery. Nat Rev Drug Discov. 2019;18:869–884. doi: 10.1038/s41573-019-0039-y. [DOI] [PubMed] [Google Scholar]
- 96.Giera S., Deng Y., Luo R., Ackerman S.D., Mogha A., Monk K.R., et al. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat Commun. 2015;6:6121. doi: 10.1038/ncomms7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suzuki G., Kanda Y., Nibuya M., Hiramoto T., Tanaka T., Shimizu K., et al. Stress and electroconvulsive seizure differentially alter GPR56 expression in the adult rat brain. Brain Res. 2007;1183:21–31. doi: 10.1016/j.brainres.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 98.Pabst C., Bergeron A., Lavallée V.P., Yeh J., Gendron P., Norddahl G.L., et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood. 2016;127:2018–2027. doi: 10.1182/blood-2015-11-683649. [DOI] [PubMed] [Google Scholar]
- 99.Maiga A., Lemieux S., Pabst C., Lavallée V.P., Bouvier M., Sauvageau G., et al. Transcriptome analysis of G protein-coupled receptors in distinct genetic subgroups of acute myeloid leukemia: identification of potential disease-specific targets. Blood Canc J. 2016;6 doi: 10.1038/bcj.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jentzsch M., Bill M., Grimm J., Schulz J., Schuhmann L., Brauer D., et al. High expression of the stem cell marker GPR56 at diagnosis identifies acute myeloid leukemia patients at higher relapse risk after allogeneic stem cell transplantation in context with the CD34+/CD38- population. Haematologica. 2020;105 doi: 10.3324/haematol.2019.229260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tseng W.Y., Jan Wu Y.J., Yang T.Y., Chiang N.Y., Tsai W.P., Gordon S., et al. High levels of soluble GPR56/ADGRG1 are associated with positive rheumatoid factor and elevated tumor necrosis factor in patients with rheumatoid arthritis. J Microbiol Immunol Infect. 2018;51:485–491. doi: 10.1016/j.jmii.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 102.Chiang N.Y., Hsiao C.C., Huang Y.S., Chen H.Y., Hsieh I.J., Chang G.W., et al. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J Biol Chem. 2011;286:14215–14225. doi: 10.1074/jbc.M110.183830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luo R., Jeong S.J., Yang A., Wen M., Saslowsky D.E., Lencer W.I., et al. Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PloS One. 2014;9 doi: 10.1371/journal.pone.0100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang K.Y., Lin H.H. The activation and signaling mechanisms of GPR56/ADGRG1 in melanoma cell. Front Oncol. 2018;8:304. doi: 10.3389/fonc.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]