Main text
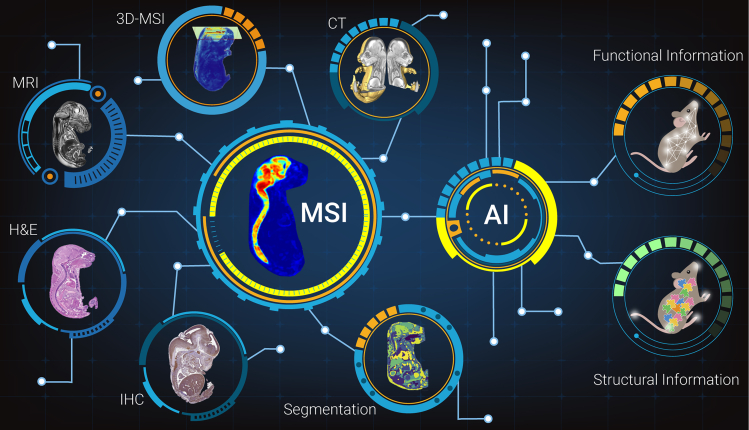
Using advanced molecular imaging techniques, we can acquire the primary disturbance of spatial and temporal information based on histological variations, which has the potential to differentiate the intra-/extraheterogeneity and to predict disease progression or metastasis status in biological systems. Most of the biological and chemical imaging techniques depend on “inherent properties” to visualize the target compounds and their interactions from different forms of samples, such as elemental bond information, the mass-to-charge ratio (m/z), molecular fragment, ion mobility, and functional groups. However, no single technique to date is capable of capturing the overall chemical and biological information simultaneously from complex biological processes. Multi-modal strategy, which intends to overcome the limitation of individual techniques and to acquire more “hidden” information, provides a fresh and valuable perspective on investigating the biological processes with higher spatial or spectral resolutions. Therefore, comprehensive methodologies with two or more analytical techniques display a great attraction. By integrating mass spectrometry imaging (MSI) and other multiplex analytical techniques, the MSI-based multi-modal technique can help us clarify the occurrence, diagnosis, personalized treatment, and prognosis of diseases, as well as embryonic development (Figure 1).
Figure 1.
Take a typical mouse fetus imaging for example to clarify the characteristics of MSI-based multi-modal technique
The molecular variations of mouse fetus are revealed by combing the MSI results with other analytical techniques, such as histological features, image segmentation results, 3D-MSI reconstruction, and in vivo imaging information from MRI and CT. Figures are reprinted from Anal. Chem. Guo, L., et al. 93, 4788–4793, Copyright (2021), with permission from American Chemical Society; NeuroImage. Cleary, J.O., et al. Magnetic resonance virtual histology for embryos: 3D atlases for automated high-throughput phenotyping, 54, 769–778, Copyright (2021), with permission from Elsevier; Sci. Bull. Zhao, C., et al. Airborne fine particulate matter induces cognitive and emotional disorders in offspring mice exposed during pregnancy, 66, 578–591, Copyright (2021), with permission from Elsevier; Development. Wong, M.D., et al. A novel 3D mouse embryo atlas based on micro-CT, 139, 3248–3256, Copyright (2021), with permission from the Company of Biologists Ltd.; Front. Endocrinol. Danilova, T., et al. Mesencephalic astrocyte-derived neurotrophic factor (MANF) is highly expressed in mouse tissues with metabolic function, 10, 765, Copyright (2021), with permission from Frontiers Research Foundation.
MSI can take advantage of the molecular detectability of mass spectrometry and spatial resolution of imaging techniques, and achieve the information of the qualitative, quantitative, and spatial distribution of molecules without labeling. To date, MSI can provide 4D-based spatial omics simultaneously that is, including m/z, ion mobility, abundance, and spatial distribution. By combining MSI and molecular biotechnology, researchers investigated the molecular mechanisms in environmental pollutant (such as bisphenol S [BPS])-induced proliferation and deterioration of breast tumors. They found that intratumor heterogeneities in BPS exposure individuals are closely related to the spatial distribution of lipid and protein markers, and the dysregulation of protein function and rearrangement of metabolites could be used to identify the tumor growth.1
With the advancement of imaging technology and data integration methods, the MSI-based multi-modal technique has shown its great potential for biochemical analysis in the past years, such as metabolism and spatial distribution of a drug, tumor heterogeneity-related molecular phenotype identification, target screening of organs exposed to environmental pollutants, and biomarker screening associated with disease progression.1 The emphases of MSI-based multi-modal techniques is placed on MSI, including matrix-assisted laser desorption/ionization (MALDI)-TOF, desorption electrospray ionization, secondary ion mass spectrometry (SIMS), and MALDI-Fourier transform (FT)-ion cyclotron resonance (ICR), as well as well-established imaging techniques, such as in vivo magnetic resonance imaging (MRI), positron emission tomography (PET), or FT infrared (FTIR) microscopy imaging (Figure 1). These imaging techniques have high complementarity and flexibility in the analytical outcomes of multi-modal imaging in 2D and 3D space by combining high sensitivity, specificity, and spatial resolution. Because of the incomparable advantage of information acquisition, the MSI-based multi-modal technique has the potential to become a future generation of standard strategies for biochemical analysis.
The experimental design of MSI-based multi-modal techniques focuses on the characteristics of MSI and complements them with another imaging technique by recurring operation of data registration and integration, acquiring a new dataset more informative than any individual technique. An appropriate experiment should be designed according to experimental objectives, throughput, etc. Generally, MSI, as a basic imaging modality, is applied to guide the other modalities. Data integration of MSI and microscopy with a histological performance at a cellular resolution, is used widely for prediction of molecular distribution at a higher chemical specificity and spatial resolution in this “guided acquisition approach.”2 In addition, it could also be an improved guiding relationship, such as in infrared spectroscopic imaging to guide MALDI-FT-ICR-MSI, enabling the multi-modal and multiplex imaging of the same tissue section.
Image registration and data integration have a decisive effect on successful multi-modal imaging. Image registration, which is spatial mapping of pixels from MSI and other imaging datasets, should be considered as image alignment according to specific biological significance. The inaccurate image registration between two imaging modalities is tightly linked to the poor properties of feature or decision level fusion on the consequent process of data integration. It also requires previous biological knowledge and algorithms to improve the fusion accuracy. Reasonable image registration and data integration require advanced data processing, including the consideration of tissue deformation, signal enhancement, the extraction of qualitative, quantitative, and spatial location information, complementary information identification, optimization of fusion strategies, etc. The visualization of multi-modal imaging also plays an important role in mining and feedback of results, ensuring the accuracy of image registration and data integration, as well as explaining the biological significance of multi-modal results. Some methods have been proposed for medical image fusion in recent years, such as M2aia, Elastix, and DeepReg. These methods also work with MSI-based multi-modal techniques.
The main challenge of MSI-based multi-modal imaging is to ensure biological relevance and export accurate results. The precise combination of the imaging modalities, the extraction, and the fusion of complementary information that targets a specific biological problem is not a trivial task. This integration of high complementary imaging techniques is still in its early stage, and there are more values to be revealed that will influence the complex biological processes. Overall, two important technical issues need further to be resolved.
In vivo imaging
Most MSI techniques can only use to ex vivo tissue sections. Construction of an MSI-based multi-modal technique for specific in vivo study remains a challenging issue because of the great differences between MSI and in vivo imaging technique in sample preparation, experimental design, and data acquisition mode. To realize MSI-based in vivo imaging in biological analysis, advanced noninvasive imaging techniques, such as MRI or PET, need to be introduced to clarify the molecular features of biological specimens in vivo firstly, and then simultaneously reveal metabolic variation by combining with MSI and classical histological approaches. With the improvement of consistency and repeatability between MSI and in vivo imaging, the combination of ex vivo MSI and in vivo noninvasive PET/MRI is expected to become an efficient technique for demonstrating the molecular mechanisms of cancer and other diseases during generation, diagnosis, development, and prognosis.3,4
Algorithm and benchmark
Data analysis, which extracts the complementary information of biological significance from different imaging modalities to achieve a remarkable outcome, is critical for multi-modal imaging. Even though several algorithms have been proposed for data analysis on multi-modal imaging, the development of an MSI-based multi-modal technique is still challenging because of the unique properties of MSI, such as high dimensionality, low signal-to-noise ratio, extensive missing values, time-consuming qualitative analysis, and highly correlated ion images. Developing efficient algorithms for data imputation, compound identification, and dimensionality reduction would help to improve the quality of MSI-based multi-modal imaging. Furthermore, the lack of benchmark datasets acts as a drag on algorithm development. Benchmark datasets provide yardsticks for performance evaluation and parameter optimization in algorithm development, and play an important role in MSI-based multi-modal data analysis. Meanwhile, it is urgent for researchers to develop more disease-specific benchmarks for MSI-based multi-modal techniques. It is noteworthy that, in recent years, deep learning approaches have been applied successfully for various fields, including multi-modal imaging. Deep learning can achieve end-to-end learning in an unsupervised or supervised way, which could remove the burden of feature engineering and the reliance on biological domain experts. As we can expect, deep learning would be the main trend of data analysis in MSI-based multi-modal imaging in the future, especially with the help of suitable benchmarks.
In summary, numerous forms of imaging techniques have been developed to combine with MSI. The trends in MSI-based multi-modal technique are from the early 2D to current 3D or 4D imaging, from in vitro to in vivo multi-modal imaging, from metabolism to expression patterns of genes or proteins. In the future, with the advancement of imaging techniques and algorithms, MSI-based multi-modal techniques will go through dynamic imaging instead of static imaging during the developmental processes of normal or disease states.
Acknowledgments
Declaration of interests
The authors declare no competing interests.
Published Online: August 12, 2021
Contributor Information
Jiyang Dong, Email: jydong@xmu.edu.cn.
Zongwei Cai, Email: zwcai@hkbu.edu.hk.
References
- 1.Zhao C., Yong T., Zhang Y., et al. Breast cancer proliferation and deterioration-associated metabolic heterogeneity changes induced by exposure of bisphenol S, a widespread replacement of bisphenol A. J. Hazard Mater. 2021;414:125391. doi: 10.1016/j.jhazmat.2021.125391. [DOI] [PubMed] [Google Scholar]
- 2.Plas R.V., Yang J., Spraggins J., et al. Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods. 2015;12:366–372. doi: 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha T.K., Khatib-Shahidi S., Yankeelov T.E., et al. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat. Methods. 2008;5:57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wehrl H.F., Schwab J., Hasenbach K., et al. Multimodal elucidation of choline metabolism in a murine glioma model using magnetic resonance spectroscopy and 11C-choline positron emission tomography. Cancer Res. 2013;73:1470–1480. doi: 10.1158/0008-5472.CAN-12-2532. [DOI] [PubMed] [Google Scholar]