Abstract
Design and fabrication of effective biomimetic vasculatures constitutes a relevant and yet unsolved challenge, lying at the heart of tissue repair and regeneration strategies. Even if cell growth is achieved in 3D tissue scaffolds or advanced implants, tissue viability inevitably requires vascularization, as diffusion can only transport nutrients and eliminate debris within a few hundred microns. This engineered vasculature may need to mimic the intricate branching geometry of native microvasculature, referred to herein as vascular complexity, to efficiently deliver blood and recreate critical interactions between the vascular and perivascular cells as well as parenchymal tissues. This review first describes the importance of vascular complexity in labs- and organs-on-chips, the biomechanical and biochemical signals needed to create and maintain a complex vasculature, and the limitations of current 2D, 2.5D, and 3D culture systems in recreating vascular complexity. We then critically review available strategies for design and biofabrication of complex vasculatures in cell culture platforms, labs- and organs-on-chips, and tissue engineering scaffolds, highlighting their advantages and disadvantages. Finally, challenges and future directions are outlined with the hope of inspiring researchers to create the reliable, efficient and sustainable tools needed for design and biofabrication of complex vasculatures.
Keywords: biofabrication, vasculature, labs-on-chips, organs-on-chips, tissue engineering, additive manufacturing, computer-aided design and engineering
1. Introduction
1.1. The importance of vascular complexity in vascular–parenchymal interactions
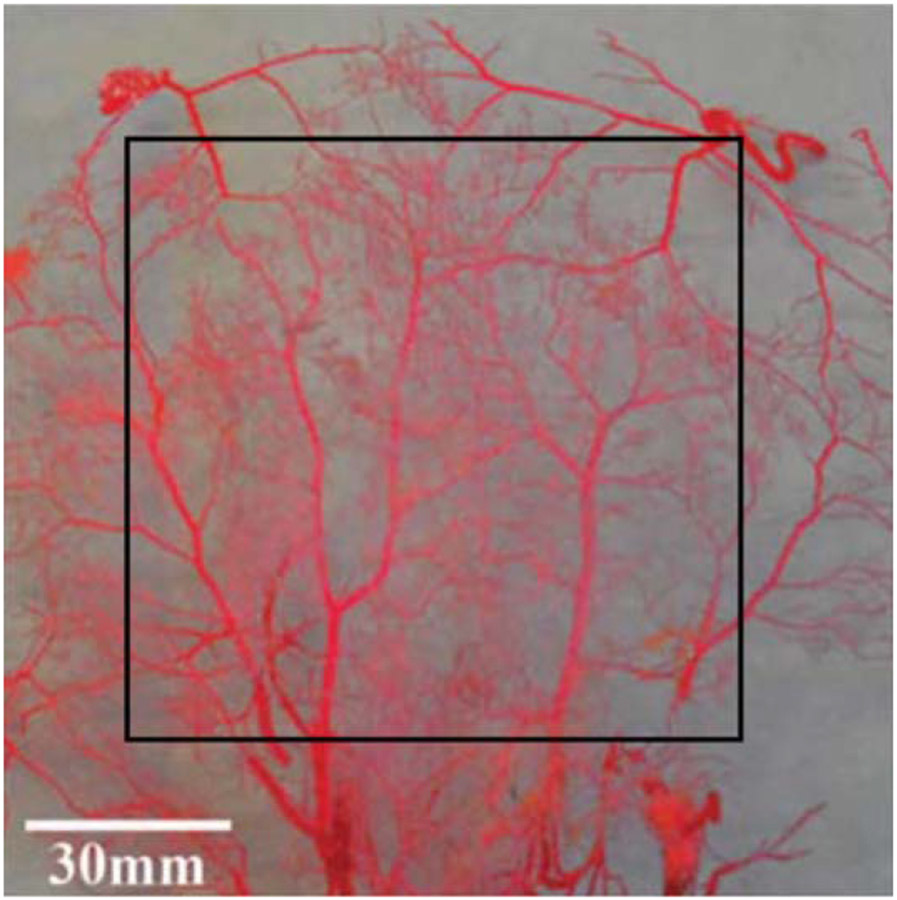
The vasculature distributes blood from the heart to the rest of the body and back again. Blood distribution is critical to oxygen and nutrient delivery, to waste removal from tissues, and to the distribution of hormones, blood cells, fluids, and heat throughout the body. Blood leaves the heart through a single blood vessel, the aorta, which then branches repeatedly through a tightly genetically controlled mechanism into the smaller arteries of the macrovasculature. Further down the vascular tree, the microvasculature forms a dynamic hierarchical branched structure unique to each individual as vessel size decreases from the arterioles to capillaries. Microvascular development is likely prescribed in the genome by a specific set of recursive patterning rules that are repeatedly applied to maintain all cells within about 100 μm of a blood vessel [1] as well as to deliver blood with minimal work required by the heart [2]. In this review, we use the term microvascular complexity to define this hierarchical branched geometry, which is perhaps best described visually (figure 1).
Figure 1.
Rabbit dermal vascular cast [3]. Reproduced from [3]. © IOP Publishing Ltd. All rights reserved.
Recapitulating the complex microvascular structure is essential to any tissue engineered organ, whether for eventual implantation or for in vitro study of disease, i.e. by means of labs- and organs-on-chips or microfluidic systems. While whole organ perfusion can be determined by dividing blood flow through the supplying artery by organ mass, local perfusion within the organ is highly variable and correlated with vascular structure [4, 5]. Microvascular structure and the resulting perfusion heterogeneity are important in modeling vasculature-parenchymal tissue interactions because they are coupled to many flow physical parameters that determine microvascular function, including: blood flow rate, average flow, velocity and effective vascular permeability [6], which are essential for oxygen transport and exchange [7-10]; microvascular pressure and wall shear stress [11], which determine endothelial cell function; thermal conductivity, which is important for body temperature regulation with changing metabolism and environmental conditions [12, 13]; blood cell deformation [14, 15], which impacts thrombosis and inflammation; and microvascular remodeling [16-18], which is critical to development, growth, healing and disease. In fact, studies have shown that neglecting microvascular structure and subsequent blood flow heterogeneity can result in 20%–50% underestimation of important blood flow characteristics such as capillary permeability [19].
In addition, microvascular structure is highly dynamic and variable in different organ systems in health and disease [20]. Corrosion casting, which is used to visualize microvascular complexity by perfusing blood vessels with resin that is then cured prior to dissolving the surrounding tissue, has been used to observe microvascular structure and quantify important differences in morphogenesis and in normal developed organs [21, 22]. In some cases, for example pancreatic islet distribution, microvascular structure can even determine the parenchymal tissue structure [23]. Significant disease-related changes in microvascular structure have been measured in the coronary circulation in cardiac hypertrophy and heart failure [24-28], pulmonary arterial hypertension [29], diabetic retinopathy and glaucoma [10, 30], as well as in the liver, lung, and heart with aging [31-33]. Microvascular structure also has predictive diagnostic value, including distinguishing ischemic from non-ischemic tissue [34], differentiating healthy from cancerous tissue [35-40] or benign from malignant lesions [41], and in monitoring tumor treatment response [42-44]. Retinal vasculature fractal analysis was even shown to predict future coronary heart disease mortality [45]. Since microvascular geometric complexity differs in individuals in both health and disease, recreating microvascular complexity in in vitro systems is critical to enabling personalized medicine.
Although vascular complexity is critical to tissue engineered organs and labs- and organs-on-chips that model both health and disease, the design and fabrication of 3D microvascular structures is challenging. Despite significant advances in both imaging and biofabrication, we do not yet have a comprehensive tool set to engineer a complex vasculature within a tissue engineered structure or lab- or organ-on chip. In this review, we outline the background, state-of-the-art, and future of the design and biofabrication of complex microvasculatures. Specifically, we describe biomechanical and biochemical signals that can be used to promote complex vascular network formation, discuss limitations of traditional 2D, 2.5D and 3D vascular models, and evaluate the advantages and disadvantages of current design and biofabrication strategies for creating in vitro systems that model vascular complexity and vascular–parenchymal interactions. Finally, we outline challenges and future directions in creating in vitro tissue models that include vascular complexity. We focus on a design for (bio-) manufacturing approach, integrating engineering design methodologies with biofabrication processes to promote ease of manufacture, while achieving the required biophysical and biochemical complexity for cell and tissue development. We hope that this review will inspire new multi-scale, multi-material, multi-phase and multi-dimensional strategies that are critical to progress in bioinspired and biomimetic design and manufacturing of complex 3D and 4D microvascular structures for tissue engineered and organ-on-chip systems. Only when we can build fully vascularized tissues, with an arteriole that connects to both an external blood supply (e.g. tissue recipient vasculature) on one end and a complex microvasculature on the other end, will we be able to understand multiscale biotransport and its impact on physiological systems in health and disease [46].
2. Biomechanical and biochemical signals to promote complex vascular network formation
Both biomechanical and biochemical signals are critical to stimulating endothelial cells to create a complex vasculature within another engineered tissue. Both types of stimuli should be taken into account when designing innovative labs- and organs-on-chips, in which precise interactions among the vasculature and parenchymal tissue are sought. Furthermore, the behavior and fate of stem cells, which may be needed to create a device that changes with time in a physiologically biomimetic way, are regulated by biomechanical and biochemical signals which come from their microenvironment, called the stem cell niche [47, 48]. The spatial-temporal configuration and deployment of biomechanical and biochemical signals can further be changed by the cells themselves. Thus the design, implementation, and operation of biomedical microsystems that include a complex vasculature becomes even more difficult due to interactions among the various stimuli and cells in the biodevice.
A wide variety of biomechanical stimuli are well known to affect vascular endothelial cells and blood vessel formation. Perhaps the most established is the effect of fluid shear stress from the flowing blood. Endothelial cells are highly sensitive to both low and high shear stress levels, and shear stress that moves outside of the normal range appears to stimulate new vasculature formation perhaps through paracrine chemical signaling [49-51]. The vasculature also experiences both tensile and compressive stresses due to blood pulsation and forces from the parenchymal tissue. Stretch in particular has been shown to induce angiogenesis and alter extracellular matrix fiber patterning, which in turn changes vascular network geometry [52-54]. More recently, the stiffness of the vascular wall and the parenchymal tissue have proven critical to endothelial cell function. Extracellular matrix stiffness impacts endothelial cell migration, Matrix metalloproteinase (MMP) and growth factor expression, and even growth factor response [55-57]. Extracellular matrix, porosity, viscoelasticity, roughness and surface topography can further regulate endothelial and stem cell function [58]. Additional mechanical stimuli can also be applied to cell culture platforms and microsystems, including: mechanical vibrations, typically by means of piezoelectric resonators [59]; pulsed fluid excitations, using peristaltic and diaphragm (micro)-pumps, which work using principles similar to those pumping mechanisms in human organism [60]; and artificially produced pressure losses, narrowings, blockages and leaks in microfluidic models, so as to mimic the effect of different physiological interactions and even disease processes [61].
Biochemical stimuli are also key for controlling microvascular network formation, evolution, and parenchymal tissue interactions in organ-on-chip models. Growth factors, in particular vascular endothelial growth factor (VEGF) and fibroblast growth factor-2 (FGF2), stimulate angiogenesis and vasculogenesis when included in the right spatio-temporal configuration [62]. MMPs are critical to breaking down the extracellular matrix to enable vascular network formation and modification. Growth factors and MMPs may be deposited in carefully designed chemical gradients within the biodevices by taking advantage of material properties, or they may be produced by the vascular or parenchymal cells in response to a primary stimulus [63]. Since biomechanical and biochemical stimuli generally occur concurrently and often interact, particularly interesting possibilities arise in microsystems capable of combining biomechanical and biochemical stimuli for controlled modulation of cellular responses [51, 57, 64]. Some of the smallest and more functional biomimetic models of physiological structures include both types of stimuli at protein and cell size scales [65, 66].
3. Limitations of conventional 2D, 2.5D, and 3D vascular models
3.1. 2D culture models
The majority of cell culture continues to rely on 2D mono- and co-culture of vascular cells in polystyrene tissue culture dishes. Traditional tissue culture is useful for isolating cell phenotype in response to a specific biochemical stimulus and, in some specific cases, for examining cell–cell interactions using simple side-byside or layer-by-layer co-cultures. However, 2D cell culture lacks many important biomechanical and biochemical aspects of the vascular microenvironment, including both apical and basolateral adhesions with the surrounding extracellular matrix; adhesive and paracrine interactions with a diversity of vascular and parenchymal cells, including smooth muscle cells, pericytes, and stem cells; signaling activation from soluble factors and their spatial gradients, such as nutrients, oxygen, and growth factors; and mechanobiological cues from the substrate topography and stiffness [67]. For example, endothelial cell invasion of a 3D collagen matrix requires a signaling complex formed of adhesion molecules and a membrane-anchored metalloproteinase, among others, whereas endothelial migration in 2D culture does not [68]. In 3D culture, cancer cells form structures that resemble their in vivo architecture, and cells within these 3D structures alter critical intracellular signaling pathways to become more resistant to chemotherapy than cells in 2D culture [69-71]. Furthermore, the ability of cancer cells to recruit new blood vessels through angiogenesis is regulated through integrin engagement in 3D but not 2D culture [72]. Thus conventional 2D cell culture cannot emulate the complex biochemical and biomechanical interactions within the vasculature itself, much less the interactions between the vasculature and parenchymal tissue.
3.2. 2.5D culture models
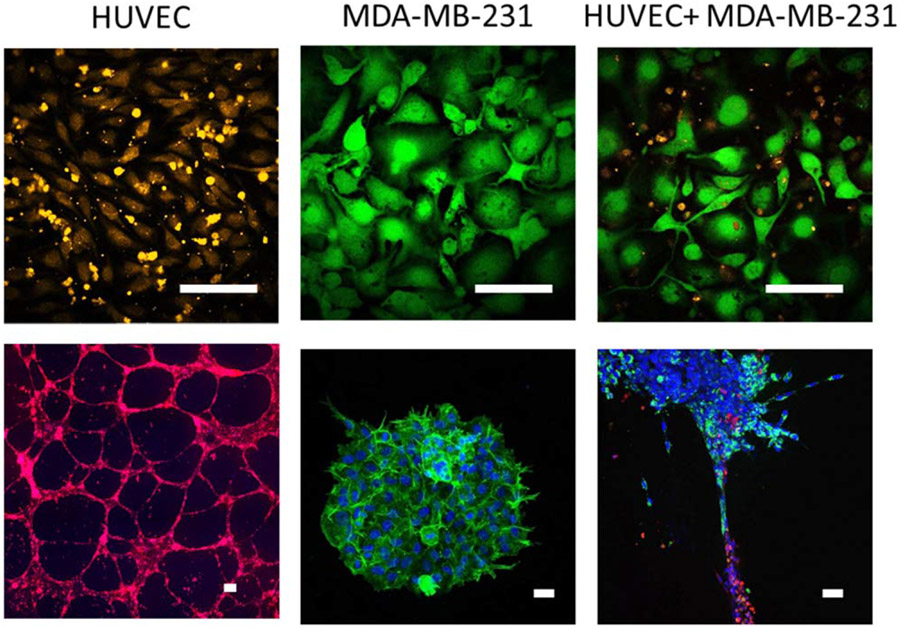
2.5D culture, in which cells are cultured on top of a thin extracellular matrix layer, provides improvements over strictly 2D cell culture systems by providing a substrate that is more physiologically relevant in terms of both biochemical composition and biomechanics. A variety of matrix-based 2.5D assays were developed to study new blood vessel formation from a pre-existing vascular network (angiogenesis). A simple and widely used assay for creating capillary-like structures relies on plating endothelial cells at low density on Matrigel, a laminin and growth factor-rich extracellular matrix isolated from Engelbreth-Holm-Swarm tumors [73]. Since this assay is 2.5D, it is simple to set up, image and analyze. In our recent work, we demonstrated that this system can be used to study endothelial-parenchymal interactions that do not occur in traditional 2D culture. When human umbilical vein endothelial cells (HUVEC) were co-cultured with the breast cancer cell line MDA-MB-231 in 2D culture for 24 h, the breast cells grew on top of the endothelial cells, resulting in endothelial cell death. We then used 2.5D Matrigel culture to separately form HUVEC into capillary-like structures and MDA-MB-231 cells into spheroids. When the MDA-MB-231 spheroids were pipetted out of their Matrigel culture and onto the endothelial capillary-like tubes, both cell types remained viable for up to 96 h and breast cancer cells were observed to migrate out of the spheroid and along the endothelial tubes within 24 h (figure 2). This example illustrates how heterogeneous cell interactions differ in 2D culture as compared to 2.5D or 3D culture. However, even in this 2.5D system, the endothelial network is still formed primarily of cell cords or processes, rather than tubes with lumens, which means that cancer cell intravasation cannot be observed nor can the effects of blood flow be studied.
Figure 2.
Conventional 2D cell culture does not enable the study of vascular–parenchymal interactions. (top) Human umbilical vein endothelial cells (HUVEC) (labeled with orange CellTracker) were cultured for 24 h with the breast cancer cell line MDA-MB-231 (labeled with green Calcein) in 2D culture. Breast cancer cells grew on top of HUVEC, resulting in endothelial cell death. (bottom) In contrast, when HUVEC (labeled with red CellTracker) were cultured in 3D tubes in Matrigel, and MDA-MB-231 3D spheroids (labeled for nuclei with Hoescht (blue) and integrin α6 (green), also formed in Matrigel) were pipetted onto the endothelial tubes, both cell types remained viable for up to 96 h and breast cancer cells migrated out of the spheroid and along the endothelial tubes within 48 h. Scale bar = 100 μm.
Transwell® or Boyden chambers enable co-culture of different cell types in a 2.5D configuration. These systems are comprised of a well with a permeable, micro-porous membrane assembled into a tissue culture dish. One type of cells is typically grown within the well, and a chemoattractant can be placed on the opposing side or a second type of cells can be grown on the bottom surface of the well or on the bottom of the tissue culture dish. Boyden chambers have been extensively used to investigate cell migration, invasion, and cell–cell interactions [74]. For example, endothelial cell migration through a fibronectin-coated Boyden chamber was shown to increase in response to VEGF, and smooth muscle cell migration was inhibited by an intact endothelial monolayer [75, 76]. Boyden chambers have also been used to study cross-talk between tumor cells and the microenvironment, in particular the effect of cancer-asociated fibroblasts on cancer cell migration and invasion [77]. Tumor-endothelium interactions, including transendothelial cancer cell migration in metastasis, have also been extensively studied using Boyden chambers [78-80]. However, Boyden chambers impose an artificial membrane between the two cell types, which limits physical interactions between cells (e.g. myoendothelial gap junctions) and extracellular matrix; present a stiffer, polymeric surface between the cells; maintain each cell type in a 2D configuration; and enable only transient chemotactic gradients to form. Thus Boyden chambers are not effective in modeling vascular complexity, either with or without parenchymal interactions.
More recently, Boyden chamber-type culture has been combined with microfluidics in the development of vascularized organs-on-chips, in which endothelial cells are cultured on the opposite side of an intermediate porous membrane from parenchymal cells on a single microdevice to recreate an important blood-tissue interface [81]. Organ-on-chip development has been facilitated by novel manufacturing techniques to create thin, porous membranes [82]. In alternative designs, gates or openings have been micro-manufactured across PDMS membranes to connect parallel adjacent channels and let co-cultured vascular and parenchymal cells interact [83, 84]. For example, human blood–brain barrier-on-a-chip devices, which recapitulate physiologically relevant flow rates and tightly controlled endothelial barrier function, were created by culturing brain microvascular endothelial cells and brain cells (e.g. astrocytes, neurons, microglia) on opposite sides of a porous membrane or on the inside and outside of a hydrogel channel [85-87]. A commercially available device manufactured by Flocel provides an in vitro model of the blood–brain barrier through micro-porous tubes immersed in a cylindrical chamber. Inside the tubes, endothelial cells are cultured under the appropriate fluid flow conditions, while outside the tubes, astrocytes and glial cells are cultured with stimuli from growth factors. The neural cells can then interact with the endothelial cells through the tube pores in a biomimetic way. In a human lung-on-a-chip, microvascular endothelial cells were cultured on one side of a porous matrix-coated membrane, and alveolar epithelial cells were cultured on the other side [88]. By flowing medium over the endothelial cells and air over the epithelial cells coupled with stretching the membrane, the device mimicked flow from blood and air as well as strain from breathing. Hepatocytes and endothelial cells were co-patterned using dielectrophoresis, or hepatocytes were cultured on the opposite side of a nanoporous membrane from an endothelial cell lined microfluidic channel to create liver-on-a-chip models [89-91].
Organs-on-chips have potential as personalized, reasonable and sustainable alternatives for physiological and pathophysiological research, as compared to conventional animal models. These systems allow real-time imaging and analysis coupled with the tight control typical of in vitro system. Unfortunately, their relatively simple geometry causes them to suffer from many of the same challenges as Boyden chamber, and the 2.5D configuration does not permit the full recapitulation of microvascular complexity. However, by recreating multicellular culture and mechanical forces applied by vascular perfusion, these biomimetic microsystems can partially emulate cell and biochemical microvascular interactions and some of the significant functionalities of complex organs, which may enhance our understanding of vascular–parenchymal interactions and their role in disease.
3.3. 3D culture models
Microfluidic systems allow the 3D recreation of microvascular structures, either molded within the polymeric system or within an extracellular matrix gel. Microfluidics can reproduce both microvascular hemodynamics and biochemical signals at a physiologically relevant scale [92-94]. Microfluidic-based strategies enable researchers to expose vascular cells to precisely controlled flow due to the low Reynolds number in small channels; mimic the microvascular geometry; and make high throughput experimentation possible through reduced reagent usage, multiplexed configurations, ease of manufacture, and dynamic imaging capability [93], for which the use of transparent materials in their manufacture is beneficial. Flow can be applied in microfluidic systems through hydrostatic pressure, syringe or peristaltic pumps, pneumatic valves or electrokinetics, among other options, to achieve steady, pulsatile, or oscillating flow [95, 96]. In addition, some devices can synergistically apply flow on samples that are cyclically stretched or have different substrate stiffness, so that physiological mechanical forces can be applied to both the apical and basal sides of the endothelial monolayer [97, 98].
Microfluidic microvascular systems can be templated in polydimethylsiloxane (PDMS) using photolithography, which enables the cheap and rapid manufacture of complex structures at high resolution. However, PDMS is relatively impermeable, stiffer than most tissues, and does not promote cell attachment unless it is functionalized. Therefore, many vascular microfluidic models have shifted to incorporate 3D hydrogel microenvironments to mimic the extracellular matrix. These hydrogels can enable time-responsive, 4D systems, depending on the properties of the incorporated hydrogels. The challenge is to create a hydrogel that is strong enough to withstand the flow pressure and that sends adequate signals for the cells and tissues under development. Endothelialized channels have been created using both synthetic and natural hydrogels, including polyethylene glycol diacrylate (PEGDA), alginate, collagen, fibrin, silk, and agarose among others [99-103]. These channels can be created by patterning gels and then bonding them to flat layers [99], by molding the hydrogel around a cylindrical structure such as a needle [104], by using micromolded dissolvable templates (e.g. gelatin, sugar, salt, Pluronic®) to create a microfluidic network [105-107], or by additive or subtractive photopatterning [108-110]. When coupled with secondary channels to create soluble factor gradients, these microfluidic chambers become powerful tools to study angiogenesis in response to both biochemical and biomechanical cues [111-114].
More recently, vascularized cancer-on-a-chip devices have been created with a microvasculature in a hydrogel that can be laden with tumor cells. In these systems, endothelial cells embedded within Matrigel, collagen, or another extracellular matrix protein self-assemble to form a 3D, branched structure model [115-117]. In one case, spheroids composed of endothelial and tumor cells were embedded in fibrin matrix containing fibroblasts. The endothelial cells sprouted out of the spheroid and into the matrix, enabling spheroid vascularization including the visualization of tumor cell intravasation [118]. In a more complex model, endothelial cells were mixed into the extracellular matrix, which was then injected into the center of a microfluidic system with two perfused side channels. The endothelial cells self-assembled into complex, perfusable vascular networks, which were then co-cultured with cancer cells to determine drug efficacy [119]. These systems have enabled in vitro studies of vasculature-tumor cell interactions critical to metastasis and drug delivery [120-123]; however, these sprouting networks are difficult to control in terms of their morphology and no studies have yet shown that these systems recapitulate a similar microvascular complexity to that observed in healthy states or in cancer.
While significant advances have been made in creating in vitro microvascular models, there remains an unfortunate tradeoff between engineering a specific vascular geometry, which is largely done in PDMS, and replicating the vascular biochemical and biomechanical microenvironment, which is better achieved within hydrogels. In fact, both are needed to recreate and study microvascular complexity, since channels with non-physiological geometry and/or non-physiologic mechanical and biochemical properties induce endothelial dysfunction [98, 124, 125]. The challenge becomes even greater when vascular complexity is to be incorporated in tissue engineered organs, in which the microvasculature must be fabricated within a variety of macro- and microenvironments (e.g. mechanical properties, surface topography, porosity and pore distribution) in varied engineered tissues [126]. The temporal dimension (4D) must also be considered as these multi-material, multi-scale, multi-phase and multi-dimensional engineered tissues develop towards mature organ formation. In the future, more complex devices will be needed to assess, model and understand how a physiologically relevant microvascular structure impacts vascular cell dynamics and parenchymal tissue interactions in health and disease. In the remainder of this review, we will describe recent advances in imaging, computer-aided engineering (CAE), materials science and technology, micro- and nano-manufacturing, and surface functionalization that can be used to recreate the complex microvasculature through precise control of materials geometry and surface properties [67].
4. Biofabrication of in vitro systems that model vascular complexity
4.1. Design and analysis of vascular complexity
The design of vascular complexity into labs- and organs-on-chips or engineered tissues begins with quantifying the properties of microvascular geometry in vivo. Traditional techniques to describe vascular patterns rely on mean vessel diameter, mean vessel segment length, branch angle, vessel area density or vessel length density, among others. When applied to the design of microvascularized in vitro devices, these parameters fail to fully recapitulate microvascular properties because they do not take into account the complex microvascular geometry [127]. Alternative quantitative methods to describe complex structures each have advantages and disadvantages. Hierarchical branching can be used to define the branching order using simple measures; however, this technique works best in parent-child branch architectures and is less suitable for vascular networks that lack hierarchical structure but instead are more homogenously distributed (e.g. microcirculation). Imaging techniques, such as micro-CT, enable detailed 3D microvascular mapping, but since in most cases they lack the temporal dimension, vessel order and flow cannot be evaluated and therefore cannot be designed into an engineered replicate [128]. Contrast-enhanced ultrasound using micro-bubble destruction can enhance imaging techniques to visualize both architecture and flow direction, and thereby enable a person’s specific vasculature to be replicated; however, these techniques are time-consuming and expensive and therefore impractical for widespread implementation [34]. Fractals perhaps best describe vascular complexity. Fractal analysis, as well as variants including multi-fractals, lacunarity, and the Lindenmeyer system, has been used to analyze microvascular geometry in tumor, pulmonary, myocardial, renal, skeletal muscle, and cerebral perfusion [20, 32, 129-134]. For example, perfusion heterogeneity, which is independent of the measured volume size, can be effectively described as a self-similar or fractal quantity. Unfortunately, as we describe later in this review, fractal analysis are not as easily converted into a design for biomanufacturing due to current limitations in computational tools.
Once vascular complexity has been imaged and quantified, it then must be converted into an engineered design. Design is most quickly implemented when supported by software that directly connects to manufacturing tools. CAE refers to the use of software to aid in these engineering tasks, and in its broadest sense includes computer-aided design (CAD) and manufacturing (CAM). Here we focus first on CAD for designing vascular complexity and then on CAE as the simulation tool for verifying CAD model geometries, materials, and the effects of loads and boundary conditions.
CAD is a primary part of computer-aided tissue engineering (CATE), [135, 136] which can be connected with additive manufacturing technologies to fabricate advanced biostructures made from novel biomaterials [137]. CAD can be used to design 2D UV-lithographic masks for vascular-like channel creation, 2.5D chips with integrated planar vasculatures, and 3D complex vasculatures within tissue constructs. CAD software uses Euclidean geometry operations, such as extrusions, grooves, holes, sweeps along guides or surfaces through curves, together with Boolean operations, pattern-based design processes and matrix-based procedures, to create complex geometries. More complex structures can be designed using geometries based on constructal law [138], fractal descriptions [139] and topological optimization procedures [126, 140]. These may involve mechanical, thermal, fluidic or even mass-transport phenomena to achieve complex and in many cases biomimetic geometries. Computational models can be created to minimize or maximize a given function under a system of constraints in vascular network design. Examples include minimizing perfusion work [141-143], minimizing vascular volume fraction while maintaining oxygen concentration[144], maximizing extravascular transport efficiency [144, 145], and maximizing transmural pressure to maintain vascular stability [146]. These innovative CAD approaches are currently being used to create biodevices with improved biomimicry and design-controlled knowledge-based vascular structures.
The CAD model then serves as input for CAE, which uses the finite element method (FEM) to produce integrated analysis of the complex biodevice design. FEM software solves complex engineering problems through mesh discretization of a continuous domain into a set of discrete elements (connected by nodes) and by transforming initial partial differential and integral equations into an approximate system of ordinary differential equations (forced to be valid in the nodes) for final numerical integration. This method is especially well-suited to solving partial differential equations over a complicated domain or geometry when the domain changes during the whole simulation, when the desired precision varies over the system under study or when the solution lacks smoothness. These characteristics are particularly useful for mechanical, static and dynamic, structural, thermal, fluidic and electromagnetic models of multiscale biomaterials and biodevices, including complex engineered vasculatures in labs- and organs-on-chips and tissue engineered constructs. For instance, fluid dynamics simulations in scaffolds with inner dendrite fractal-like structures were used assess their potential benefits for nutrient delivery [147]. Vascular complexity has also been considered in combined studies of the effects of scaffold physical and degradation properties on cell and tissue growth with respect to nutrient transport, drug distribution, and debris elimination [148-150]. CAE has potential to combine models working at different scales, from molecular to cellular to organ level, to promote multi-scale modeling approaches and enhance our toolset for creating biodevices with vascular complexity.
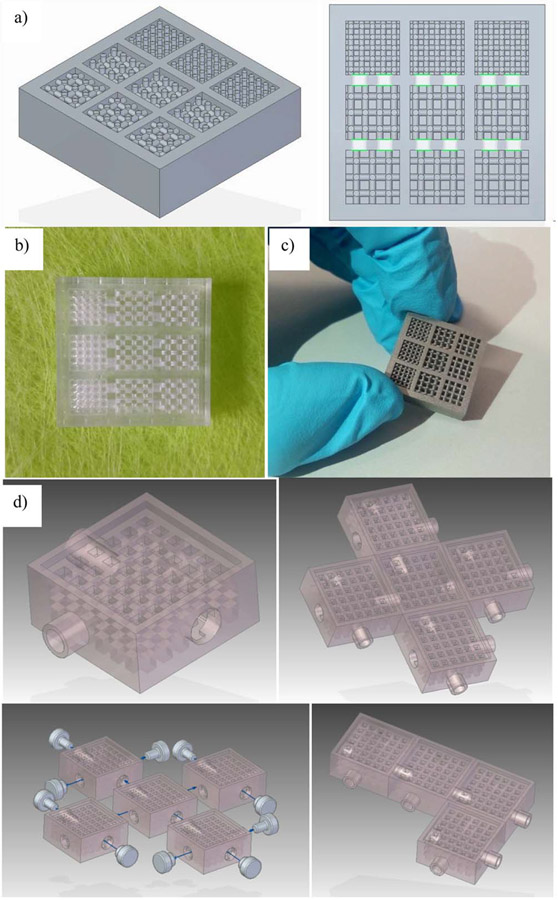
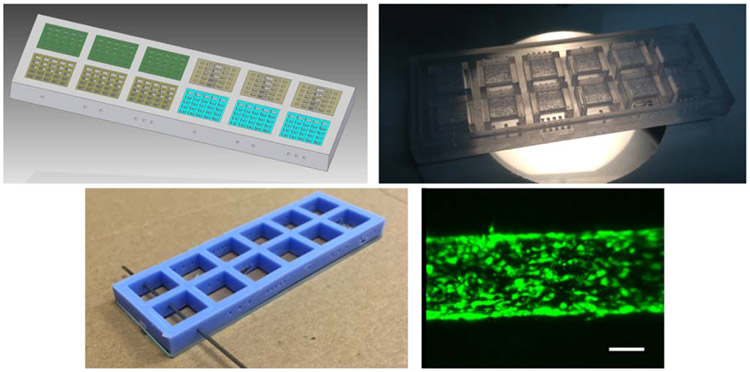
To highlight the versatility and potential of connecting design and computational modeling with advanced micro-manufacturing, some case studies are presented. Figure 3 presents a multi-chamber organ-on-chip, in which chambers arranged in series incorporate different cell types and their optimal 3D matrices. The chambers are connected through micro-perforated walls to promote vascularization between the chambers and thus enable fluid flow and paracrine interactions between the different cell types. This multi-chamber system can be used for example to study how the microvasculature affects stem cell differentiation or cell phenotypic expression in a multicellular milieu while maintaining each cell type in its preferred matrix. After the CAD model is realized (figure 3(a)), the multi-chamber organ-on-chip can be rapid prototyped first by laser stereolithography for conceptual manufacturability tests (figure 3(b)) and later by additive selective laser sintering of titanium powder for cell studies (figure 3(c)). This multi-chamber system stands out for including three parallel sets of three interconnected chambers integrating scaffolds with different porosities, hence enabling multiplexed experiments for analyzing, in a single device, the impact of porosity, addition of growth factors and flow rate, on cell behavior and fate. A related system based on scaffolds integrated within fluidic chambers with interconnectable inlets and outlets follows a ‘plug-and-play’ approach. Different functional tissues can be cultured in the chambers and connected in varied configurations to rapidly establish interconnected multi-organ-on-chip systems (figure 3(d)). In this case, the fluid flow rates can be directly derived from the CAD files, which helps in pump selection tasks and performance optimization.
Figure 3.
Combining computer-aided design and rapid prototyping resources for the straightforward development of in vitro systems that model vascular complexity and interaction among cells and tissues. (a) CAD of inter-connected multi-scaffold chambers. (b) Conceptual prototype for geometrical validation by laser stereolithography and (c) selective laser sintered device in alloy appropriate for culture. (d) Modular plug and play alternative design shows how these chambers can be connected to create larger structures. Support with CAD modeling: Beatriz Lopez and Jesús Puertas.
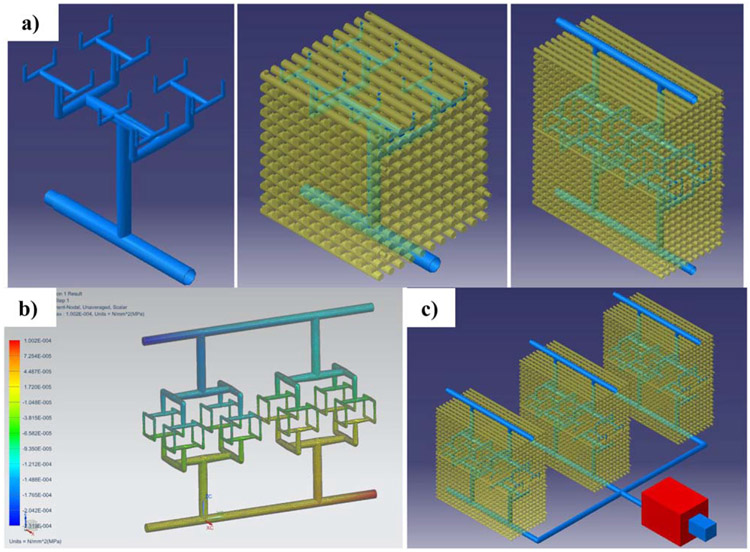
Figure 4 shows the CAD model of modular tissue engineering constructs with a 3D branching microvasculature designed using the constructal approach (figure 4(a)). The vascular network and the 3D scaffold structure for mechanical support can be manufactured by additive processes, as detailed in the next section. This construct is also modular and could be connected to other building blocks for increased complexity. Once the CAD vascular network is obtained, CAE resources (e.g. FEM simulations) can be used to analyze flow velocities, their associated shear stresses, and pressure drops through the system at different volume flow rates. For example, the system in figure 4(b) shows a pressure loss of 33 kPa across the construct. NX (Siemens PLM Solutions) and Catia v.5 (Dassault Systèmes) are used as CAD and engineering resources for the conceptual designs and simulations of figures 3 and 4. Laser stereolithography (SLA-3500 by 3D Systems) with epoxy resin (Accura® 60 by 3D Systems) and selective laser sintering of titanium powder (Materialise NV) are used for analyzing manufacturability of the different concepts.
Figure 4.
Combining computer-aided design and simulations for engineering biomimetic constructs. (a) Three-dimensional vasculature following the constructal approach and modular tissue constructs. (b) Fluid flow simulation for selecting the adequate pumping system for a (c) high-throughput system and completing the design of modular plug and play systems with scaffold chambers and vascular channels.
4.2. Manufacturing vascular complexity
Since several review papers have described broad strategies for creating vasculatures in labs- and organs-on-chips and in tissue engineered constructs [151-153], we instead focus in depth on biofabrication techniques that can be used to manufacture a 3D complex vasculature. While microvascular self-assembly can be induced after construct fabrication through growth factors, this is costly and slow and therefore impractical for widespread use. Spatially defined endothelial patterns can also accelerate engineered tissue vascularization after implantation [154]. It is therefore essential to be able to design and manufacture a complex microvasculature at the same time as the initial construct. Each technique has advantages and disadvantages, depending on the desired geometrical complexity, the device type and final application (i.e. in vitro device or final implant), and on the types of cells and tissues being created. We present the most interesting available technologies, analyze their advantages and disadvantages, and present some challenges and strategies for achieving complex vascular geometry. For clarity we grouped the technologies into several categories, including subtractive methods, additive methods and biochemical patterning techniques.
4.2.1. Subtractive methods
Conventional subtractive manufacturing, in which material is removed from the starting shape to produce the final product, has been miniaturized to create micromachining methods that enable the creation of features smaller than 100 μm. Computer numerical control (CNC) micromilling, which is primarily done in plastics and metals, uses cutting tools as small as 25 μm to create molds for further fabrication or to directly create the final part [155]. Micromilling enables rapid transfer of a 3D CAD file into the final part, often in less than an hour, and therefore is useful for rapid prototyping. However, micromilling has significant limitations in precision, which is determined by cutting tool dimensions and micromill accuracy; surface roughness, which can affect cell function as well as optical transparency for microscopy; and the ability to create internal features, which means that micromilling is primarily useful for 2.5D applications such as microfluidics. Laser micromachining has the advantages of being able to accurately place small features with controlled cutting depth in a wide variety of materials. Nearly 20 years ago, the Vacanti group micromachined silicon to create a template for microvascular patterning. The microvascular networks were then lifted off the template and folded into 3D tissues [156]. While this technique similarly has the advantage of allowing designed geometry to be directly transferred into the part, laser micromachining also has the disadvantage of limited ability to create inner geometries as well as slower manufacturing speeds. In addition, the molding and transfer steps are labor-intensive and may require new molds to be produced for each engineered tissue.
Current subtractive methods move far beyond CNC machining. For example, phase-separation procedures can be considered subtractive, as the liquid or vapor phases are eliminated to leave a porous structure. 3D porous structures for tissue regeneration and repair have also been created using sol–gel and foaming techniques [157]. In one example, sol–gel glass foams of silicon dioxide and calcium oxide were shown to induce macrophages to differentiate into osteoclasts, osteoblasts to deposit mineralized bone, and endothelial cells to form tube-like structures between cell clusters, suggesting that these cells could possibly form a functional microvasculature [158]. These techniques have the advantage of creating a highly porous structure through which endothelial cells can be seeded to create a complex microvasculature, as well as the ability to select materials that enable growth factor controlled release for microvascular self-assembly. The primary disadvantage of phase-separation is the challenge in controlling the biomaterial micro-structure and hence the microvascular geometry and personalized vasculature design. However, the use of micro-fluidic devices for foam generation may improve homogeneity and control over the biomaterial and vascularized tissue final properties [159, 160].
For improved design of complex microvascular geometry, sacrificial molds or inserts provide a promising alternative. In the simplest case, needles are inserted into the device prior to hydrogel addition and then removed once the matrix has gelled. The technique forms hollow tubes that can be as small as 100 μm in diameter after the needles are removed [104, 105]. While the tubes are straight and homogeneous, a more complex vasculature can be formed by fabricating multiple tubes and promoting microvascular formation between the molded tubes [161]. Straight channels can also be created through viscous fingering, which occurs when a less viscous fluid is used to displace a more viscous fluid, was used to pattern a cylindrical channel within a hydrogel [162]. A PDMS microfluidic channel was filled with collagen, which was then briefly incubated at 37 °C to initiate polymerization and increase solution viscosity. Cell culture medium was then passively pumped through the microfluidic channel to create a single continuous lumen along the channel length. This procedure has since been used to create models of angiogenesis as well as the blood–brain barrier [113, 163]. Alternatively, sacrificial molds or inserts with more complex geometry can be created using design-controlled additive manufacturing (commonly 3D printing). For example, fugitive inks or water-soluble materials were rapid cast on 3D printed complex microvascular structures, assembled into a hydrogel microdevice, and then dissolved after gel polymerization occurred [78, 120, 121]. Alternatively, the sacrificial material can be directly printed within the hydrogel. Examples include: omnidirectional printing of fugitive inks within photocrosslinkable hydrogels [122], printed alginate templates within fibrin gels [123], 3D printing of Pluronic® [125], which dissolves when cooled (while other bioinks typically gel when cooled), among others. Hence, multi-material 3D printing can be employed to generate both the structure and the sacrificial element to be dissolved, extracted or eliminated to enable vascularization. While sacrificial molds have the advantage of creating controlled complex vascular geometries in a variety of materials, they remain limited to relatively large microvascular diameters and fairly simple structures. Thus these sacrificial molds do not yet enable engineering of physiologic microvascular complexity.
Subtractive manufacturing can also be used to improve physiologically relevant cell–cell interactions in labs- and organs-on-chips after cells have been introduced by removing the artificial membranes that separate endothelial cells from parenchymal cells. For example, chitosan membranes (<100 μm thick) were created at the interface of an acidic chitosan solution and a basic buffer solution within a microfluidic channel [164]. After cells were seeded on either side of the chitosan membrane, the membrane was then removed using an acidic solution. This technique was then used to create a blood–brain barrier device in which endothelial cells and astrocytes were able to directly interact [165]. Human astrocytes in Matrigel were seed on one side of a chitosan membrane. The chitosan membrane was then removed through brief exposure to acetic acid, after which brain microvascular endothelial cells were seeded into the empty side of the channel to grow on the Matrigel surface. This same technique could be used with other sacrificial hydrogels like alginate to enable direct cell–cell interactions in an organ-on-a-chip system. While this technique has the advantage of enabling direct endothelial-parenchymal interactions, it still is limited by the lack of vascular geometric complexity.
4.2.2. Additive methods
Additive manufacturing has revolutionized biofabrication (and also the traditional manufacturing sectors) [166]. In additive manufacturing, materials are added usually layer-by-layer to create the final part. Additive manufacturing can now be achieved with a wide variety of materials, from metals to ceramics to extracellular matrix proteins and even cells themselves. While each technique has its own specifications, the layer-by-layer approach likely has the highest potential for controlling geometric complexity to achieve biomimetic and even personalized vascularized constructs. Since there are now many additive manufacturing techniques, we split them into two groups: non-biological materials and biological materials.
4.2.2.1. Additive manufacturing of non-biological materials
Electrospinning uses electric force to draw a charged polymer jet out of a spinneret and deposit it onto a grounded collector. A spinning collector can be used to create structures with aligned fibers, and the spinneret design can be varied to create small fibers (down to hundreds of nanometers) and core–shell fibers made of two immiscible materials. In some cases, the inner material can be removed to produce hollow fibers. Due to the manufacturing conditions, it remains difficult to electrospin biological materials and therefore proteins and cells are usually added after the fabrication is complete. However in one case, fibrinogen and polylactic acid (PLA) were electrospun into either randomly distributed or aligned nanofibers. Endothelial cells on the aligned nanofibers showed an elongated shape and increased motility, suggesting that this material could be used for guided neovascularization [167]. While electrospinning is one of the only manufacturing techniques to truly create nanofibrous materials that guide endothelial cell growth, this technique does not allow complex geometries to be fabricated according to a specific design and since the polymeric mats are usually dense, cell infiltration is often poor resulting in primarily 2.5D devices.
Fused deposition modeling is the most widely used 3D printing method for conventional additive manufacturing. Thermoplastic materials such as acrylonitrile butadiene styrene (ABS) and PLA are melted and extruded through a nozzle in a computer-controlled layer-by-layer process that eventually builds a 3D part. By using a support removable material, complex controlled geometries can be fabricated directly from CAD files. However, traditional 3D printing has several significant disadvantages, including the inability to print biological materials, the limited precision (with features generally on the scale of hundreds of microns), and the relatively slow fabrication speed. Therefore, fused deposition modeling has primarily been used to create molds or to print sacrificial materials which are later removed to form channels [168, 169]. In one example, a polyvinyl alcohol (PVA) network was printed within a PLA supporting material. The PLA was then dissolved and replaced with a HepG2 cells embedded within crosslinked gelatin. Finally, the PVA was dissolved with water to create the microvascular network within the HepG2-gelatin biomaterial [170]. In another example, electrospun fibers were molded around a PVA sacrificial template to form microvascular channels within the structure [171]. Unfortunately the large size and limited geometrical complexity of these sacrificial templates means that traditional 3D printing has limited applications in creating a complex vasculature; however, this technique has inspired many other additive manufacturing techniques that start to overcome these limitations.
Selective laser sintering expands the types of materials supported by additive manufacturing. In this technique, powdered polymers, ceramics, or metals are fused together by scanning a high power laser over the powder bed in a pattern determined from a CAD file. As each cross-sectional layer is added, the remaining powder supports the part features, which enables complex geometries to be created without supporting materials. Perfusable channels were created within porous 3D tissue engineering scaffolds by selective laser sintering polycaprolactone with sodium chloride as the porogen [172]. Selective laser sintering has the advantages of rapidly creating biocompatible parts with high strength and stiffness and complex open internal structures. However, precision limits the technique to feature sizes greater than 50 μm, biological materials cannot be used due to the laser intensity, interior features must all be open so that extra powder can be removed, and the cost of powder materials is high. Thus this technique does not allow the design of a true microvasculature, nor can microvascular self-assembly easily be induced within the material through growth factors.
Microstereolithography also uses a laser to transfer a CAD shape into a 3D part. In this layer-by-layer process, a UV laser draws the design cross section into a photopolymer resin, eventually creating a complex geometry with feature sizes down to tens of microns. In one example, laser stereolithography was used as a mold for PDMS casting of a multichamber system for reliably creating linear vascularized channels using a needle or other sacrificial material (figure 5). This simple system was then modified to enhance vascular complexity by creating a vascular mold with a bifurcation and varied vessel size, as well as parenchymal tissue wells in the space around the vasculature. By varying the branching pattern (hierarchy, branch size and number, branch angles, etc) and the parenchymal tissue well distance from the vasculature, we can now use this device to determine how varied aspects of vascular geometric complexity affect tissue properties such as biotransport (figure 6). Advances in multiphoton lithography (also called direct laser writing) enable fabrication of features with 100 nm resolution, albeit with a cost in fabrication time [173, 174]. In one case, confocal microscopy was used to image retinal microvasculature. The pattern was then recreated using a combination of simple shapes, which were used as input for the scanning confocal laser to recreate the microvascular pattern within a photocrosslinkable polyethylene glycol (PEG)-RGDS hydrogel. Endothelial cells encapsulated within the hydrogel organized into capillary-like tubule structures, forming a retinal model that was personalized to a given patient [109]. While the need for photopolymers limits the materials that can be used, ceramic particles can be added to the photopolymer suspension to create dense ceramic components. Stereolithography and its derivative techniques rapidly create strong parts with complex geometries, and the use of bio-photopolymers enables the use of biological materials (detailed in the next section). However, supports are still needed to create certain structures, and the systems remain expensive especially for high precision applications.
Figure 5.
A multi-chamber system with inserted functionally graded scaffolds and different number of vascular channels to promote performing several experiments with a single device. (top) Device CAD design and laser stereolithography prototype of rapid mold for PDMS casting of the outer device frame. (bottom) Final PDMS frame, for needle placement, scaffold insertion or hydrogel casting and vasculature formation by needle removal, obtained by vacuum casting and placed upon microscope slide and fluorescently labeled endothelial cells (labeled with Calcein, green) lining the inside of a collagen channel in the device. Scale bar = 100 μm.
Figure 6.
A hydrogel-based complex vasculature, complete with bifurcation, varied vessel size, and wells for co-culture of a parenchymal cell type at varying distances from the vasculature to study biotransport effects. (left) PDMS mold for the device and (right) hydrogel device seeded with endothelial cells (green) in the vascular channels and breast cancer cells (red) in the adjacent wells. Scale bar = 3 mm.
Additive manufacturing processes to create complex metal geometries for later vascularization are limited. However, recent efforts to add electrodeposition to the additive manufacturing toolbox have expanded the ability to create metal parts and objects with nanometer level vertical resolution. In electrochemical fabrication (EFAB) and its second generation technology (MICA), a metal (often nickel) is electrodeposited in a specific pattern using a micromask, after which a sacrificial metal (often copper) is deposited. The surface is then flattened prior to depositing the next layer. After all layers are deposited, the sacrificial metal is removed to create the final part [175]. While this technique enables creation of complex geometries with 5–20 μm features in biocompatible metals and alloys, the process is not true additive manufacturing due to the need for the mask, part size is limited, and biological components can only be added after the fabrication process is complete. EFAB has also been used to deposit and align adhesive extracellular matrix proteins into tissue engineered structures or on biological electrodes [176, 177]. Thus there is potential that EFAB could enable vasculature fabrication into multicellular structures, potentially with electrical signaling or sensing.
4.2.2.2. Additive manufacturing of biological materials
In many cases, adding biological components such as proteins as cells after manufacture is challenging due to slow or limited penetration inside the non-biological material. Therefore several of the aforementioned techniques have been adapted to include proteins and cells directly in the fabrication process. The inclusion of biological components necessarily limits the use of high temperatures and voltages, toxic materials and solvents, and extended fabrication times. However, the advantage of directly creating the 3D culture system or tissue often outweighs these limitations.
3D bioprinting uses the layer-by-layer approach to deposit bioinks composed of cells and biocompatible hydrogels into pre-designed tissue-like structures. Multiple cell types, including endothelial cells, can be bioprinted in different hydrogels and into specific locations by using multiple nozzles. Endothelial cells have been successfully bioprinted in alginate, gelatin, fibrin, and Matrigel® among many others [178-181]. Furthermore, scaffolds can be printed with intrinsic channels or a gridded structure to enable flow through the bioconstruct [161, 182]. Ink jet printers, which use drop-on-demand technology to precisely place picoliters of bioink, can be used to achieve smaller features including bifurcations down to around 100 μm [183, 184]. Despite the significant advantages of being able to print biomaterials and cells together into a pre-determined 3D design, 3D bioprinting generally produces feature sizes on the order of hundreds of microns and therefore cannot be used to create a complex microvasculature except by inducing vascular self-assembly through growth factors.
Stereolithography has also been modified to use photopolymerizable biopolymers with cells of different types encapsulated within the material. In live cell stereolithography, UV lasers can directly pattern multiple biopolymers, depending on the way in which the pre-polymer solution is added, with excellent cell viability over time [185]. Since the line-by-line writing approach in stereolithography is time-consuming and may impact material integrity at the line interfaces, alternative photopolymerization techniques have been developed to overcome these limitations. Digital light processing (DLP) based biofabrication, in which a digital micromirror array rather than a physical mask controls photopolymer illumination, improves upon the speed, resolution, and therefore scalability of live cell stereolithography. In one case, endothelial cells were directly printed into the designed vascular channels without using a sacrificial material and formed lumen-like structures [186]. These new techniques enable production of larger parts with good cell viability, while maintaining the advantages of sub-micron resolution 3D fabrication of biopolymers and cells into complex geometries. Thus light-based biofabrication strategies may hold the highest potential for building complex microvasculatures into tissue engineered constructs and labs- or organs-on-chips.
To achieve even greater resolution in single cell placement, laser assisted cell printing techniques such as laser-induced forward transfer (LIFT) and its variations allow cell level resolution. In these processes, cells are attached to a laser transparent print ribbon using a biopolymer, usually Matrigel®. The ribbon is moved over a receiving substrate, which is often also a biopolymer such as Matrigel®. Individual cells are then propelled off the ribbon and onto the receiving substrate using a pulsed laser beam. Endothelial cells have been placed in precise patterns with other cell types via direct laser writing, for example to create hepatic sinusoid-like structures [187, 188]. Other laser based single cell patterning techniques such as optical tweezers enable single endothelial cells to be placed into a main channel and then guided down branch channels [189]. While direct cell writing is unparalleled in terms of the cell patterning precision, there are significant disadvantages including the limited ability to pattern and build in the z dimension, difficulty in producing the ribbon, and the potential damage to the cell by the laser and deposition process.
Biostructures of several cubic millimeters in size, with micrometric features, have been created using a combination of biomimetic designs and novel additive manufacturing processes combining 3D printing and laser-based polymerization of photo cross-linkable resins, hydrogels and polymers [190, 191]. Growth factor incorporation into the bioprinted materials or bioinks can be used to further improve vascularization after fabrication. These additive 3D prototyping approaches can form biological structures with [192] and without [193] scaffold support. On a smaller scale, micro-fluidic bio-printing combines printed bio-inks and encapsulated living cells to generate heterogeneous 3D tissue constructs with highly defined biomimetic structures and vascularization potential [194]. The advent of high-performance low-cost systems (i.e. Inkredible 3D printer system by CellInk and Biobot1 from Biobots) and of the fabber movement and related open-source hardware and software approaches (i.e. RepRap), as alternative to the initial and much more expensive bioprinting systems (i.e. Bioplotter by EnvisionTec), is promoting the exponential growth of bioprinting. In addition, conventional 3D prototyping machines can be converted into ‘bio-plotters’ and ‘cell-printers’ [195] to manufacture biosubstrates with incorporated living cells and nutrients, thus enhancing vascular network formation within a complex 3D tissue [196].
Microfluidic systems can also be combined with additive manufacturing techniques to develop models of vascular–parenchymal interactions. In the commercially available OrganoPlate® by Mimetas, adjacent lanes of cell-laden Matrigel and liquids (usually flowing cell culture medium) are patterned using phaseguides [197]. Phaseguides are lines of material of either a different surface wettability than the rest of the chamber or a geometrical change that make it energetically advantageous for the hydrogel to advance along the phaseguide before crossing it [198]. Cells suspended in liquid Matrigel are taken up into the phaseguide defined channels by capillary action, after which the Matrigel is gelled. The remaining channels are then filled with either cell culture medium or other cell types suspended in Matrigel. Since the phaseguides are significantly smaller than the chamber height, gradients and cell–cell communications are possible among the adjacent channels. This combination of additive manufacturing in a microfluidic channel has since been used to create a variety of organ-on-a-chip systems, including lung, liver, and breast cancer [199-201].
Stacking of 2D and 2.5D materials or microsystems can also be used either as an alternative or as a complement to additive manufacturing techniques. As example of this approach, 3D stratified tissues created by stacking cell sheets in co-culture with endothelial cells led to pre-vascular network formation in vitro and promoted neovascularization after implantation in vivo [202]. More recently, researchers engineered a prevascularized cell sheet for tissue regeneration by culturing human bone marrow-derived mesenchymal stem cells (hMSCs) to form a thick cell sheet, and then seeding human umbilical vein endothelial cells (HUVEC) on the hMSC sheet. In vitro, the hMSC sheets promoted HUVEC migration to form horizontal and vertical networks. In vivo, many blood vessels grew into the hMSC/HUVEC sheets after implantation. These prevascularized hMSC/HUVEC sheets were then folded to form a 3D construct using a modified cell sheet engineering technique [203]. Similar stacking procedures were used to develop sandwiched micro-environments, which were used to manipulate cell phenotype and differentiation [204-206]. Once vascularized, these stacked cell sheets can be used in tissue engineering, for example of cardiac tissue [207]. Unfortunately the stacking procedure is labor intensive, and therefore difficult to scale up to mass manufacturing or to use to create personalized in vitro models.
4.2.3. Biochemical patterning (adhesive ligands, growth factors and others)
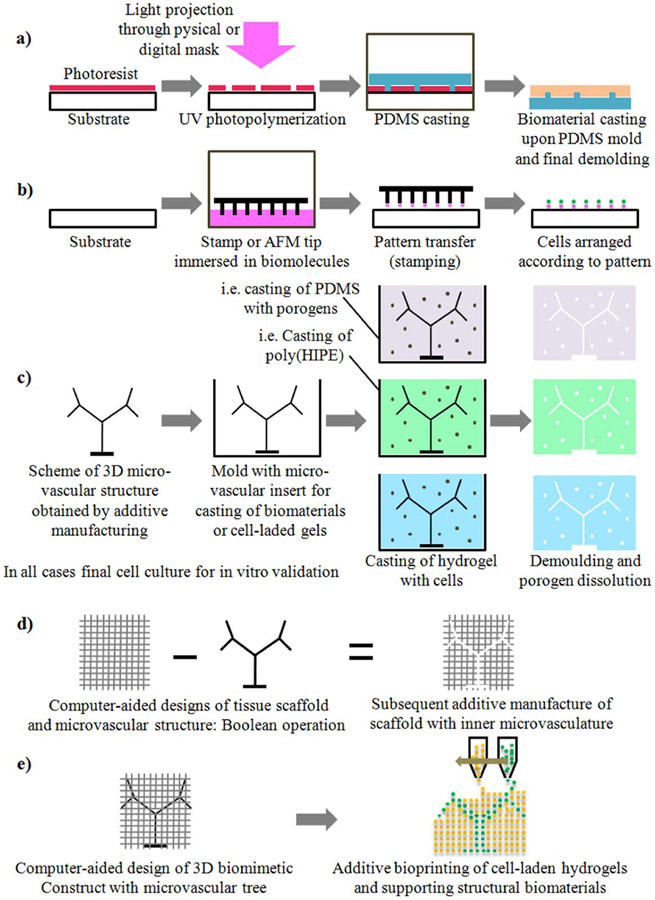
Biochemical patterning started with soft lithography, in which soft stamps are used to pattern planar surfaces. Cells can then attach in desired locations, forming complex patterns and cellular circuits which promote the first stages of vascularization [208]. The soft stamps are obtained by casting PDMS into micro-manufactured molds, which can be micro-machined by laser ablation or high-precision CNC machining, or additively manufactured by laser stereolithography. Typical stamp features are limited to 75–100 μm, which may not be small enough if single cell interactions are desired. A higher degree of precision can be achieved by patterning surfaces with biomolecules using atomic force microscope tips, a procedure called dip-pen nanolithography, which acts at the molecular scale to create biomimetic single cell patterns [209, 210]. In spite of the advantages of low cost and micron-level precision, these 2D and 2.5D technologies are limited in terms of geometrical complexity and the protein patterns may not be stable in the long term. Thus these techniques are more suited to labs and organs-on-chips than to 3D tissue constructs.
To achieve biochemical patterning in 3D structures, light-based procedures, using either physical or digital masks, provide interesting alternatives in terms of attainable size and precision. For instance, VEGF and RGD photopatterned in micron-scale regions within a poly(ethylene glycol) hydrogel using laser scanning lithography enabled endothelial cells to form tubules with lumens [211]. To create more biomimetic structures, an image-guided micropatterning method was used to directly transfer 3D vascular patterns derived from labeled tissues into hydrogel scaffolds via two-photon laser scanning photolithography. This process used both structural and biochemical cues to guide endothelial cells into recapitulating complex vascular structures into a 3D hydrogel [109]. E-beam lithography enables protein patterning at even smaller size features, but the method is costly and time consuming. These lithographic techniques can be used on many different materials, is precise, and has resolution down to hundreds of nanometers. However, 3D applications remain difficult and thus these techniques are also more applicable to 2 or 2.5D cultures and systems.
Summarizing, figure 7 schematically presents strategies for the development of lab- and organ-on-chip systems and tissue engineering constructs with incorporated complex vasculature. Table 1 presents a comparative overview of technologies, detailing materials, attainable precision, limiting aspects and key applications in the field of study. Also included are processes specifically designed for mass production, including micro-injection molding and hot-embossing. The desired multi-scale, multi-material, multi-phase and multi-dimensional systems rely on synergistic combinations among them, as further analyzed in the section on current challenges and future research directions. We believe that the combined use of present approaches, together with technological advances to come in the next five years, will enable versatile, multi-scale, multi-material, multi-phase, time-responsive complex vasculatures within biohybrid devices and tissues.
Figure 7.
Schematic representation of alternative technologies for the development of (lab and organ)-on-chip systems and tissue engineering constructs with incorporated vascular structures: (a) UV photolithography combined with soft lithography for 2.5D microvascular sheets or fluidic layers. (b) Biochemical patterning by stamping with AFM tips or with arranged micropillars. (c) Soluble mold inserts and casting with porogens towards final 3D porous and vascularized structures. (d) Lattice or porous tissue scaffolds with inner microvasculature controlled from design. (e) Bioprinting with gels laden with different cell types for design controlled vascular structures.
Table 1.
Comparative summary of technologies and materials for biodevices with vasculatures.
| Technology | Details | Materials | Advantages | Limitations | Applications | References |
|---|---|---|---|---|---|---|
| Subtractive methods | ||||||
| CNC micro-machining | 15–75 μm | Metals and polymers | Control of geometry from design stage | Precision, inner details are difficult to obtain. | 2.5D micro-systems, 3D textured implants | [155, 212, 213] |
| Laser micro-machining | 5–50 μm | Organic and inorganic materials | Control of geometry from design stage | Manufacture speed, geometrical limitations | Textured micro-systems and implants | [212, 213] |
| Phase separation, emulsion, foaming, insert-based techniques | 500 nm–50 μm | polymers, poly(HIPEs) | 3D porous, fractal-like and biomimetic geometries | The geometry cannot be defined from the design stage | Mainly prototypes of 3D cell culture matrices | [157-160] |
| Additive manufacturing methods | ||||||
| Conventional 3D printers | 300–400 μm | Thermo-plastics, ABS, PLA | Complex and controlled geometries | Precision, presence of supports | Mainly conceptual prototypes | [168-171] |
| Selective laser sintering/melting | 50–250 μm | Ceramics, metals and alloys | Complex and controlled geometries | Precision, cost of materials | Complex 3D implants for hard tissues | [172] |
| MICA process | 5–20 μm | Metals and medical alloys | Complex and controlled geometries | Cost of materials, reduced overall part size | Complex micro-systems, 3D scaffolds and implants | [175] |
| Laser (micro-) stereolithography/Digital light processing | 10–100 μm | Mainly epoxy resins and bio-photo-polymers | Complex and controlled geometries | Materials, presence of supports | Complex micro-systems, 3D scaffolds and implants | [185, 186] |
| Two-photon polymerization/Direct laser writing | 250–750 nm | Photo-polymers, hydrogels, Ormocer® | Complex and controlled geometries, high-precision | Manufacture speed and low final part size | Complex micro-systems and niches, 3D scaffolds | [186] |
| Lithography-based ceramic manufacture | 50–250 μm | Ceramics and bio-ceramics | Complex and controlled geometries | Materials, presence of supports | Complex micro-systems, 3D scaffolds, implants | [214] |
| Bioplotters and bioprinters (using bioinks and cell-laden materials) | 300–400 μm | Biomaterials, bio-inks, bio-composites | Complex and controlled geometries, biomaterials | Precision, presence of supports, final biodevice stability | Mainly prototypes of cell culture matrices | [178-181] |
| 3D live cell lithography | 500 nm–1 μm, cell level | Bio-photo-polymers with cells | Complex and controlled geometries | Limited part size for cell viability and vascularization | Complex micro-systems and 3D scaffolds with cells | [215] |
| Laser assisted cell printing (i.e. LIFT) | Cell level | Bio-inks and cells | Precision of cell patterning | Limited z dimension | 2.5D micro-systems | [216] |
| Biochemical patterning methods, surface modification and mass-replication techniques | ||||||
| UV lithography | 100–500 nm | Silicon, polymers, glass | Precision, successful industrial process | 3D applications, i.e. implants or scaffolds, are complex to achieve | 2.5D microsystems, labs- and organs-on-chips, cell culture | [212, 213] |
| Soft lithography | 1–10 μm | PDMS replicas, gels | Low-cost, easy lab process for prototypes | 3D applications, i.e. implants or scaffolds, are complex to achieve | 2.5D microsystems, labs- and organs-on-chips, cell culture | [208] |
| Stamping and AFM patterning of biomolecules | 500 nm–500 μm | Several substrates | Precision of cell patterning | Some patterns are not stable in the long term | 2.5D microsystems, labs- and organs-on-chips, cell culture | [209, 210] |
| X-ray/e-beam lithography | 500 nm | Polymers, i.e. PMMA, SU8 | Precision, aspect ratios from 10 to 50 | 3D complex geometries cannot be obtained, time consuming. | 2.5D microsystems, labs- and organs-on-chips, cell culture | [217] |
| Micro-injection molding and hot-embossing | 500 nm–250 μm | Polymer replicas | Processes for mass-production | 3D complex geometries cannot be obtained | 2.5D microsystems, labs- and organs-on-chips, cell culture | [217] |
5. Challenges and future directions
5.1. Integration of design and manufacturing procedures
A primary challenge in fabricating complex biomimetic vascular structures is the limitations inherent in current imaging and design software. CAD programs use Boolean, patterning and matrix-based operations to create micrometric details and microstructures. These programming algorithms increase file sizes into the Gbyte scale, resulting in files that cannot be adequately handled by the related automated manufacturing resources. The ‘universal’ .stl, .igs, .dxf…, formats are not optimal, especially for fractal-based designs that better describe natural system complexity [139]. Such fractal features, as well as other mathematical descriptions of porous structures and vasculatures, can be described and programmed in just one line of code in other software, while their conventional CAD geometrical description unnecessarily increases file size. The shift to algorithmic rather than descriptive geometry in CAD programs is key to promoting advanced design and manufacturing of bioinspired and biomimetic complex vasculatures within labs- and organs-on-chips, as well as within tissue engineered constructs. Continuous advances in the software used to convert medical images into 3D and 4D inputs for CAD tasks (most medical imaging software used in hospitals already includes .stl file generation) will further enable the development of personalized biomimetic vascularized microsystems for studying physiological phenomena [218].
5.2. Manufacturing technologies with increased precision and operative scale range
Additive manufacturing continues to have important challenges in reliably and repeatably creating complex vasculatures within engineered biodevices. Most additive manufacturing technologies do not provide the required precision for constructing micron-scale details (table 1). At the same time, those technologies with higher precision are not capable of manufacturing objects larger than a few cubic millimeters, which proves ineffective for most medical needs. Consequently, technologies with both increased precision and operative scale range are needed. In one recent development, hierarchical metallic metamaterials with 3D features that ranged from the nanometer to centimeter scale and overall part sizes of several centimeters were created using a high-resolution, large-area additive manufacturing technique [219]. Similar techniques must be developed using biomaterials, bioinks, extracellular matrix components and even living cells as printing materials.
Other current research in processes that produce both increased precision and operative scale range focuses on combinations of synergistic technologies. For example, 3D and 4D printing techniques can be used to create the larger vessels, while smaller vessels can be obtained with biomaterials that stimulate micro-vasculature formation [128]. Combinations of additive and subtractive laser-based procedures have also been used to manufacture the components of vascularized organs-on-chips [83]. A master model was obtained using additive laser stereolithography, while laser ablation was applied to generate the smaller details. Final metallization for mold insert creation connected this process with mass-production techniques, for example micro-injection molding of thermoplastic materials. Alternatively, multi-scale approaches can be developed in materials acceptable for cell culture by combining lithographic and high-precision additive manufacturing technologies [220]. Such combinatorial approaches can be further exploited down to the molecular size scale if additional combinations, for instance with nano-pen lithography, are developed [209, 210]. Systematically combining and analyzing potential synergies among the technologies described in table 1 can lead to truly multi-scale, multi-material and time-responsive systems that better emulate the interactions between the vasculature and the parenchymal tissues.
5.3. Collaboration and education
For complex 3D and 4D biofabrication of complex vascular structures to become a reality, it is essential to encourage collaboration among researchers from multiple fields including biology, medicine, pharmacy, physics, chemistry and engineering. Collaborative design methodologies and online resources and platforms to promote such collaboration play an important role. For instance, the ‘Chips and Tips’ website designed by the Royal Chemical Society provides ideas and solutions through interactions with colleagues regarding practical issues frequently encountered in the laboratory. Similar collaborative platforms for the integrated design and manufacture of complex vasculatures may also help to enhance progress, especially if linked with open-access software similar to recent advances in open-source medical devices.
Another key issue for advancing biofabrication technologies is to create teaching-learning activities to educate future researchers, designers, manufacturers and even marketers of these technologies. A simple and reproducible infrastructure to inspire the next generation to innovate in biofabrication might include hands-on workshops using modular kits that illustrate the basic biofabrication concepts. For example, researchers at MIT developed low-cost kits for diagnostic-oriented microfluidic systems, which are even compatible with LEGO, so that K-12 students can easily construct a lab-on-a-chip. The kits have been so successful that they led to spin-off MEDIkits [221]. The extension of this approach to biofabrication will inspire the next generation of bioengineers who will advance our current technologies.
6. Conclusions
Recent progress aimed at recreating complex vasculatures and their interactions with parenchymal tissues in vitro have laid the foundations for biomimetic labs- and organs-on-chips. These devices create advanced cell culture niches that enable new ways of effectively and sustainably modeling and studying disease. Unique combinations of computational design and modeling resources, and a wide set of synergistic micro- and nano-manufacturing techniques, help to improve the operational and dimensional ranges covered by these biodevices (from single cells to interacting cell colonies and tissues) and to achieve realistic representations of relevant physiological processes. Our personal view is that there will be remarkable advances in the upcoming five years, which will provide researchers with a wide range of reliable, efficient and sustainable tools to create in vitro models of disease and advance medical professionals towards a future of personalized medicine.
References
- [1].Metzger RJ and Krasnow MA 1999. Genetic control of branching morphogenesis Science 284 1635–9 [DOI] [PubMed] [Google Scholar]
- [2].Murray CD 1926. The physiological principle of minimum work: I. The vascular system and the cost of blood volume Proc. Natl Acad. Sci. USA 12 207–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mondy WL. et al. Computer-aided design of microvasculature systems for use in vascular scaffold production. Biofabrication. 2009;1:035002. doi: 10.1088/1758-5082/1/3/035002. [DOI] [PubMed] [Google Scholar]
- [4].Bassingthwaighte JB 1970. Blood flow and diffusion through mammalian organs Science 167 1347–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bassingthwaighte JB, King RB and Roger SA 1989. Fractal nature of regional myocardial blood flow heterogeneity Circ. Res 65 578–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang SF et al. 2015. A permeability model for power-law fluids in fractal porous media composed of arbitrary cross-section capillaries Physica A 437 12–20 [Google Scholar]
- [7].Beard DA and Bassingthwaighte JB 2001. Modeling advection and diffusion of oxygen in complex vascular networks Ann. Biomed. Eng 29 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Beard DA 2001. Computational framework for generating transport models from databases of microvascular anatomy Ann. Biomed. Eng 29 837–43 [DOI] [PubMed] [Google Scholar]
- [9].Pittman RN 2005. Oxygen transport and exchange in the microcirculation Microcirculation 12 59–70 [DOI] [PubMed] [Google Scholar]
- [10].Gandica Y. et al. Hypoxia inv ascular networks: a complex system approach to unravel the diabetic paradox. Plos One. 2014;9:e113165. doi: 10.1371/journal.pone.0113165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pries AR and Secomb TW 2005. Control of blood vessel structure: insights from theoretical models Am. J. Physiol. Heart. Circ. Physiol 288 H1010–5 [DOI] [PubMed] [Google Scholar]
- [12].Li L et al. 2014. A comprehensive study of the effective thermal conductivity of living biological tissue with randomly distributed vascular trees Int. J. Heat Mass Transfer 72 616–21 [Google Scholar]
- [13].Li L and Yu BM 2013. Fractal analysis of the effective thermal conductivity of biological media embedded with randomly distributed vascular trees Int. J. Heat Mass Transfer 67 74–80 [Google Scholar]
- [14].Secomb TW, Hsu R and Pries AR 2002. Blood flow and red blood cell deformation in nonuniform capillaries: effects of the endothelial surface layer Microcirculation 9 189–96 [DOI] [PubMed] [Google Scholar]
- [15].Davis JM 2014. On the linear stability of blood flow through model capillary networks Bull. Math. Biol 76 2985–3015 [DOI] [PubMed] [Google Scholar]
- [16].Pries AR, Reglin B and Secomb TW 2001. Structural adaptation of microvascular networks: functional roles of adaptive responses Am. J. Physiol. Heart Circ. Physiol 281 H1015–25 [DOI] [PubMed] [Google Scholar]
- [17].Pries AR, Reglin B and Secomb TW 2003. Structural response of microcirculatory networks to changes in demand: information transfer by shear stress Am. J. Physiol. Heart Circ. Physiol 284 H2204–12 [DOI] [PubMed] [Google Scholar]
- [18].le Noble F et al. 2005. Control of arterial branching morphogenesis in embryogenesis: go with the flow Cardiovasc. Res 65 619–28 [DOI] [PubMed] [Google Scholar]
- [19].Bassingthwaighte JB and Levin M 1981. Analysis of coronary outflow dilution curves for the estimation of cellular uptake rates in the presence of heterogeneous regional flows Basic Res. Cardiol 76 404–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gould DJ et al. 2011. Multifractal and lacunarity analysis of microvascular morphology and remodeling Microcirculation 18 136–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lorthois S and Cassot F 2010. Fractal analysis of vascular networks: insights from morphogenesis J. Theor. Biol 262 614–33 [DOI] [PubMed] [Google Scholar]
- [22].Cassot F et al. 2006. A novel three-dimensional computer-assisted method for a quantitative study of microvascular networks of the human cerebral cortex Microcirculation 13 1–18 [DOI] [PubMed] [Google Scholar]
- [23].Jo J. et al. The fractal spatial distribution of pancreatic islets in three dimensions: a self-avoiding growth model. Phys. Biol. 2013;10:036009. doi: 10.1088/1478-3975/10/3/036009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kassab GS et al. 1993. Morphometry of pig coronary arterial trees Am. J. Physiol 265 H350–65 [DOI] [PubMed] [Google Scholar]
- [25].Kassab GS, Lin DH and Fung YC 1994. Morphometry of pig coronary venous system Am. J. Physiol 267 H2100–13 [DOI] [PubMed] [Google Scholar]
- [26].Kassab GS 2000. The coronary vasculature and its reconstruction Ann. Biomed. Eng 28 903–15 [DOI] [PubMed] [Google Scholar]
- [27].Huo YL and Kassab GS 2016. Scaling laws of coronary circulation in health and disease J. Biomech 49 2531–9 [DOI] [PubMed] [Google Scholar]
- [28].Gong YJ et al. 2016. Intraspecific scaling laws are preserved in ventricular hypertrophy but not in heart failure Am. J. Physiol. Heart Circ. Physiol 311 H1108–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Helmberger M. et al. Quantification of tortuosity and fractal dimension of the lung vessels in pulmonary hypertension patients. Plos One. 2014;9:e87515. doi: 10.1371/journal.pone.0087515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ciancaglini M et al. 2015. Fractal dimension as a new tool to analyze optic nerve head vasculature in primary open angle glaucoma In Vivo 29 273–9 [PubMed] [Google Scholar]
- [31].Warren A et al. 2008. Effects of old age on vascular complexity and dispersion of the hepatic sinusoidal network Microcirculation 15 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Levin DL et al. 2007. Effects of age on pulmonary perfusion heterogeneity measured by magnetic resonance imaging J. Appl. Physiol 102 2064–70 [DOI] [PubMed] [Google Scholar]
- [33].Chen X. et al. Growth, ageing and scaling laws of coronary arterial trees. J. R. Soc. Interface. 2015;12:20150830. doi: 10.1098/rsif.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Charalampidis D et al. 2006. Anatomy and flow in normal and ischemic microvasculature based on a novel temporal fractal dimension analysis algorithm using contrast enhanced ultrasound IEEE Trans. Med. Imaging 25 1079–86 [DOI] [PubMed] [Google Scholar]
- [35].Wardlaw G, Wong R and Noseworthy MD 2008. Identification of intratumour low frequency microvascular components via BOLD signal fractal dimension mapping Phys. Med 24 87–91 [DOI] [PubMed] [Google Scholar]
- [36].Goh V et al. 2009. Assessment of the spatial pattern of colorectal tumour perfusion estimated at perfusion CT using two-dimensional fractal analysis Eur. Radiol 19 1358–65 [DOI] [PubMed] [Google Scholar]
- [37].Di Ieva A et al. 2008. Euclidean and fractal geometry of microvascular networks in normal and neoplastic pituitary tissue Neurosurgical Rev. 31 271–80 [DOI] [PubMed] [Google Scholar]
- [38].Sanghera B et al. 2012. Reproducibility of 2D and 3D fractal analysis techniques for the assessment of spatial heterogeneity of regional blood flow in rectal cancer Radiology 263 865–73 [DOI] [PubMed] [Google Scholar]
- [39].Dimitrakopoulou-Strauss A, Strauss LG and Burger C 2001. Quantitative PET studies in pretreated melanoma patients: a comparison of 6-[F-18] fluoro-L-dopa with F-18-FDG and O-15-water using compartment and noncompartment analysis J. Nucl. Med 42 248–56 [PubMed] [Google Scholar]
- [40].Grizzi F. et al. Quantitative evaluation and modeling of two-dimensional neovascular network complexity: the surface fractal dimension. BMC Cancer. 2005;5:14. doi: 10.1186/1471-2407-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Acharya UR et al. 2012. Non-invasive automated 3D thyroid lesion classification in ultrasound: a class of thyroscan systems Ultrasonics 52 508–20 [DOI] [PubMed] [Google Scholar]
- [42].Rose CJ et al. 2007. Quantifying heterogeneity in dynamic contrast-enhanced MRI parameter maps Med. Image Comput. Comput. Assist. Interv 10 376–84 [DOI] [PubMed] [Google Scholar]
- [43].Alic L et al. 2011. Heterogeneity in DCE-MRI parametric maps: a biomarker for treatment response? Phys. Med. Biol 56 1601–16 [DOI] [PubMed] [Google Scholar]
- [44].O’Connor JP et al. 2011. DCE-MRI biomarkers of tumour heterogeneity predict CRC liver metastasis shrinkage following bevacizumab and FOLFOX-6 Br. J. Cancer 105 139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liew G et al. 2011. Fractal analysis of retinal microvasculature and coronary heart disease mortality Eur. Heart J 32 422–9 [DOI] [PubMed] [Google Scholar]
- [46].Zakrzewicz A, Secomb TW and Pries AR 2002. Angioadaptation: keeping the vascular system in shape News Physiol. Sci 17 197–201 [DOI] [PubMed] [Google Scholar]
- [47].Caplan AI 1991. Mesenchymal stem cells J. Orthop. Res 9 641–50 [DOI] [PubMed] [Google Scholar]
- [48].Caplan AI 2008. All MSCs are pericytes? Cell Stem Cell 3 229–30 [DOI] [PubMed] [Google Scholar]
- [49].Baum O et al. 2004. Endothelial NOS is main mediator for shear stress-dependent angiogenesis in skeletal muscle after prazosin administration Am. J. Physiol. Heart Circ. Physiol 287 H2300–8 [DOI] [PubMed] [Google Scholar]
- [50].Mathew JG, Basehore S and Clyne AM 2017. Fluid shear stress and fibroblast growth factor-2 increase endothelial cell-associated vitronectin Appl. Bionics Biomech 2017 9040161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Reisig K and Clyne AM 2010. Fibroblast growth factor-2 binding to the endothelial basement membrane peaks at a physiologically relevant shear stress Matrix Biol. 29 586–93 [DOI] [PubMed] [Google Scholar]
- [52].Figueroa DS, Kemeny SF and Clyne AM 2014. Glycated collagen decreased endothelial cell fibronectin alignment in response to cyclic stretch via interruption of actin alignment J. Biomech. Eng 136 101010. [DOI] [PubMed] [Google Scholar]
- [53].Rivilis I et al. 2002. Differential involvement of MMP-2 and VEGF during muscle stretch- versus shear stress-induced angiogenesis Am. J. Physiol. Heart Circ. Physiol 283 H1430–8 [DOI] [PubMed] [Google Scholar]
- [54].Belle J et al. 2014. Stretch-induced in tussuceptive and sprouting angiogenesis in the chick chorioallantoic membrane Microvasc. Res 95 60–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhao D et al. 2018. Substrate stiffness regulated migration and angiogenesis potential of A549 cells and HUVECs J. Cell Physiol 233 3407–17 [DOI] [PubMed] [Google Scholar]
- [56].Canver AC et al. 2016. Endothelial directed collective migration depends on substrate stiffness via localized myosin contractility and cell-matrix interactions J. Biomech 49 1369–80 [DOI] [PubMed] [Google Scholar]
- [57].Sack KD, Teran M and Nugent MA 2016. Extracellular matrix stiffness controls VEGF signaling and processing in endothelial cells J. Cell Physiol 231 2026–39 [DOI] [PubMed] [Google Scholar]
- [58].Samadikuchaksaraei A et al. 2014. Stem cells as building blocks A2—Lanza, Robert Principles of Tissue Engineering ed Langer R and Vacanti J 4th edn (Boston: Academic; ) ch 4 pp 41–55 [Google Scholar]
- [59].Apicella A and Aversa R 2012. A biomimetic and biomechanical approach for tissue engineering: hybrid nanomaterials and a piezoelectric tunable bending apparatus for mechanically stimulated osteoblast cells growth Int. Conf. on Biomedical Electronics and Devices [Google Scholar]
- [60].Voyvodic PL, Min D and Baker AB 2012. A multichannel dampened flow system for studies on shear stress-mediated mechanotransduction Lab Chip 12 3322–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wong KHK et al. 2012. Microfluidic models of vascular functions Annu Rev Biomed Eng. 14 205–30 [DOI] [PubMed] [Google Scholar]
- [62].Martino MM. et al. Extracellular matrix and growth factor engineering for controlled angiogenesis in regenerative medicine. Front. Bioeng. Biotechnol. 2015;3:45. doi: 10.3389/fbioe.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kant RJ and Coulombe KLK 2018. Integrated approaches to spatiotemporally directing angiogenesis in host and engineered tissues Acta Biomater. 69 42–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Patel NS, Reisig KV and Clyne AM 2013. A computational model of fibroblast growth factor-2 binding to endothelial cells under fluid flow Ann. Biomed. Eng 41 154–71 [DOI] [PubMed] [Google Scholar]
- [65].Griep LM et al. 2013. BBB ON CHIP: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function Biomed. Microdevices 15 145–50 [DOI] [PubMed] [Google Scholar]
- [66].Chen LJ and Kaji H 2017. Modeling angiogenesis with micro- and nanotechnology Lab Chip 17 4186–219 [DOI] [PubMed] [Google Scholar]
- [67].Yao X, Peng R and Ding J 2013. Cell-material interactions revealed via material techniques of surface patterning Adv. Mater 25 5257–86 [DOI] [PubMed] [Google Scholar]
- [68].Sacharidou A et al. 2010. Endothelial lumen signaling complexes control 3D matrix-specific tubulogenesis through interdependent Cdc42- and MT1-MMP-mediated events Blood 115 5259–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Petersen OW et al. 1992. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells Proc. Natl Acad. Sci. USA 89 9064–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Riedl A et al. 2017. Comparison of cancer cells in 2D versus 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses J. Cell Sci 130 203–18 [DOI] [PubMed] [Google Scholar]
- [71].Wu Z. et al. Bioprinting three-dimensional cell-laden tissue constructs with controllable degradation. Sci. Rep. 2016;6:24474. doi: 10.1038/srep24474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fischbach C et al. 2009. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement Proc. Natl Acad. Sci. USA 106 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kubota Y et al. 1988. Role of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary-like structures J. Cell Biol 107 1589–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Boyden S 1962. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes J. Exp. Med 115 453–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mastyugin V et al. 2004. A quantitative high-throughput endothelial cell migration assay J. Biomol. Screen 9 712–8 [DOI] [PubMed] [Google Scholar]
- [76].Gorog P and Kovacs IB 1998. Inhibition of vascular smooth muscle cell migration by intact endothelium is nitric oxide-mediated: interference by oxidised low density lipoproteins J. Vasc. Res 35 165–9 [DOI] [PubMed] [Google Scholar]
- [77].Santi A et al. 2015. Cancer associated fibroblasts transfer lipids and proteins to cancer cells through cargo vesicles supporting tumor growth Biochim. Biophys. Acta 1853 3211–23 [DOI] [PubMed] [Google Scholar]
- [78].Flores-Perez A et al. 2018. Angiogenesis analysis by in vitro coculture assays in transwell chambers in ovarian cancer Methods Mol. Biol 1699 179–86 [DOI] [PubMed] [Google Scholar]
- [79].Simoneau B, Houle F and Huot J 2012. Regulation of endothelial permeability and transendothelial migration of cancer cells by tropomyosin-1 phosphorylation Vasc. Cell 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Li YH and Zhu C 1999. A modified Boyden chamber assay for tumor cell transendothelial migration in vitro Clin Exp Metastasis 17 423–9 [DOI] [PubMed] [Google Scholar]
- [81].Bhatia SN and Ingber DE 2014. Microfluidic organs-on-chips Nat. Biotechnol 32 760–72 [DOI] [PubMed] [Google Scholar]
- [82].Le-The H et al. 2018. Large-scale fabrication of free-standing and sub-mum PDMS through-hole membranes Nanoscale 10 7711–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Díaz Lantada A 2016. Handbook on microsystems for enhanced control of cell behavior Fundamentals, Design and Manufacturing Strategies, Applications and Challenges (Berlin: Springer; ) [Google Scholar]
- [84].Gori M. et al. investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. Plos One. 2016;11:e0159729. doi: 10.1371/journal.pone.0159729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Booth R and Kim H 2012. Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB) Lab Chip 12 1784–92 [DOI] [PubMed] [Google Scholar]
- [86].Prabhakarpandian B et al. 2013. SyM-BBB: a microfluidic blood brain barrier model Lab Chip 13 1093–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Partyka PP et al. 2017. Mechanical stress regulates transport in a compliant 3D model of the blood-brain barrier Biomaterials 115 30–9 [DOI] [PubMed] [Google Scholar]
- [88].Huh D et al. 2010. Reconstituting organ-level lung functions on a chip Science 328 1662–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ho CT et al. 2006. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap Lab Chip 6 724–34 [DOI] [PubMed] [Google Scholar]
- [90].Carraro A et al. 2008. In vitro analysis of a hepatic device with intrinsic microvascular-based channels Biomed. Microdevices 10 795–805 [DOI] [PubMed] [Google Scholar]
- [91].Lee PJ, Hung PJ and Lee LP 2007. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture Biotechnol. Bioeng 97 1340–6 [DOI] [PubMed] [Google Scholar]
- [92].Whitesides GM 2006. The origins and the future of microfluidics Nature 442 368–73 [DOI] [PubMed] [Google Scholar]
- [93].Wong KH et al. 2012. Microfluidic models of vascular functions Annu. Rev. Biomed. Eng 14 205–30 [DOI] [PubMed] [Google Scholar]
- [94].Young EW 2013. Advances in microfluidic cell culture systems for studying angiogenesis J. Lab Autom 18 427–36 [DOI] [PubMed] [Google Scholar]
- [95].Shao J et al. 2009. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress Lab Chip 9 3118–25 [DOI] [PubMed] [Google Scholar]
- [96].Chin LK et al. 2011. Production of reactive oxygen species in endothelial cells under different pulsatile shear stresses and glucose concentrations Lab Chip 11 1856–63 [DOI] [PubMed] [Google Scholar]
- [97].Zheng W et al. 2012. A microfluidic flow-stretch chip for investigating blood vessel biomechanics Lab Chip 12 3441–50 [DOI] [PubMed] [Google Scholar]
- [98].Galie PA et al. 2015. Application of multiple levels of fluid shear stress to endothelial cells plated on polyacrylamide gels Lab Chip 15 1205–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cabodi M et al. 2005. A microfluidic biomaterial J. Am. Chem. Soc 127 13788–9 [DOI] [PubMed] [Google Scholar]
- [100].Bettinger CJ et al. 2007. Silk fibroin microfluidic devices Adv. Mater 19 2847–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ling Y et al. 2007. A cell-laden microfluidic hydrogel Lab Chip 7 756–62 [DOI] [PubMed] [Google Scholar]
- [102].Du Y et al. 2011. Sequential assembly of cell-laden hydrogel constructs to engineer vascular-like microchannels Biotechnol. Bioeng 108 1693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zheng Y et al. 2012. In vitro microvessels for the study of angiogenesis and thrombosis Proc. Natl Acad. Sci. USA 109 9342–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Chrobak KM, Potter DR and Tien J 2006. Formation of perfused, functional microvascular tubes in vitro Microvasc. Res 71 185–96 [DOI] [PubMed] [Google Scholar]
- [105].Golden AP and Tien J 2007. Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element Lab Chip. 7 720–5 [DOI] [PubMed] [Google Scholar]
- [106].Miller JS et al. 2012. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues Nat. Mater 11 768–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wu W, DeConinck A and Lewis JA 2011. Omnidirectional printing of 3D microvascular networks Adv. Mater 23 H178–83 [DOI] [PubMed] [Google Scholar]
- [108].Hahn MS, Miller JS and West JL 2006. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior Adv. Mater 18 2679 [Google Scholar]
- [109].Culver JC et al. 2012. Three-dimensional biomimetic patterning in hydrogels to guide cellular organization Adv. Mater 24 2344–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kloxin AM et al. 2009. Photodegradable hydrogels for dynamic tuning of physical and chemical properties Science 324 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Sudo R et al. 2009. Transport-mediated angiogenesis in 3D epithelial coculture FASEB J. 23 2155–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Jeong GS et al. 2011. Microfluidic assay of endothelial cell migration in 3D interpenetrating polymer semi-network HA-Collagen hydrogel Biomed. Microdevices 13 717–23 [DOI] [PubMed] [Google Scholar]
- [113].Bischel LL et al. 2013. Tubeless microfluidic angiogenesis assay with three-dimensional endothelial-lined microvessels Biomaterials 34 1471–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kim C et al. 2015. A quantitative microfluidic angiogenesis screen for studying anti-angiogenic therapeutic drugs Lab Chip 15 301–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Davis GE and Camarillo CW 1996. An alpha 2 beta 1 integrin-dependent pinocytic mechanism involving intracellular vacuole formation and coalescence regulates capillary lumen and tube formation in three-dimensional collagen matrix Exp. Cell Res 224 39–51 [DOI] [PubMed] [Google Scholar]
- [116].Stitt AW et al. 2005. Impaired retinal angiogenesis in diabetes: role of advanced glycation end products and galectin-3 Diabetes 54 785–94 [DOI] [PubMed] [Google Scholar]
- [117].Montesano R, Orci L and Vassalli P 1983. In vitro rapid organization of endothelial cells into capillary-like networks is promoted by collagen matrices J. Cell Biol 97 1648–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ehsan SM et al. 2014. A three-dimensional in vitro model of tumor cell intravasation Integr. Biol 6 603–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Phan DTT et al. 2017. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications Lab Chip 17 511–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Song JW. et al. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PLoS One. 2009;4:e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang S. et al. Study on invadopodia formation for lung carcinoma invasion with a microfluidic 3D culture device. PLoS One. 2013;8:e56448. doi: 10.1371/journal.pone.0056448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jeon JS. et al. In vitro model of tumor cell extravasation. PLoS One. 2013;8:e56910. doi: 10.1371/journal.pone.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zervantonakis IK et al. 2012. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function Proc. Natl Acad. Sci. USA 109 13515–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yang X et al. 2011. Traffic of leukocytes in microfluidic channels with rectangular and rounded cross-sections Lab Chip 11 3231–40 [DOI] [PubMed] [Google Scholar]
- [125].Orr AW et al. 2005. The subendothelial extracellular matrix modulates NF-kappaB activation by flow: a potential role in atherosclerosis J. Cell Biol 169 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Place ES, Evans ND and Stevens MM 2009. Complexity in biomaterials for tissue engineering Nat. Mater 8 457–70 [DOI] [PubMed] [Google Scholar]
- [127].Hlatky L, Hahnfeldt P and Folkman J 2002. Clinical application of antiangiogenic therapy: microvessel density, what it does and doesn’t tell us J. Natl Cancer Inst 94 883–93 [DOI] [PubMed] [Google Scholar]
- [128].Wilson SH et al. 2002. Simvastatin preserves the structure of coronary adventitial vasa vasorum in experimental hypercholesterolemia independent of lipid lowering Circulation 105 415–8 [DOI] [PubMed] [Google Scholar]
- [129].Michallek F and Dewey M 2014. Fractal analysis in radiological and nuclear medicine perfusion imaging: a systematic review Euro. Radiol 24 60–9 [DOI] [PubMed] [Google Scholar]
- [130].Bauer WR et al. 2001. Fast high-resolution magnetic resonance imaging demonstrates fractality of myocardial perfusion in microscopic dimensions Circ. Res 88 340–6 [DOI] [PubMed] [Google Scholar]
- [131].Grant PE and Lumsden CJ 1994. Fractal analysis of renal cortical perfusion Investigative Radiology 29 16–23 [DOI] [PubMed] [Google Scholar]
- [132].Kalliokoski KK et al. 2001. Perfusion heterogeneity in human skeletal muscle: fractal analysis of PET data Eur. J. Nucl. Med 28 450–6 [DOI] [PubMed] [Google Scholar]
- [133].Warsi MA, Molloy W and Noseworthy MD 2012. Correlating brain blood oxygenation level dependent (BOLD) fractal dimension mapping with magnetic resonance spectroscopy (MRS) in Alzheimer’s disease Magn. Reson. Mater. Phys. Biol. Med 25 335–44 [DOI] [PubMed] [Google Scholar]
- [134].Yasar O, Lan SF and Starly B 2009. A Lindenmayer system-based approach for the design of nutrient delivery networks in tissue constructs Biofabrication 1 045004. [DOI] [PubMed] [Google Scholar]
- [135].Sun W et al. 2004. Computer-aided tissue engineering: overview, scope and challenges Biotechnol. Appl. Biochem 39 29–47 [DOI] [PubMed] [Google Scholar]
- [136].Sun W and Lal P 2002. Recent development on computer aided tissue engineering—a review Comput. Methods Programs Biomed 67 85–103 [DOI] [PubMed] [Google Scholar]
- [137].Giannitelli SM et al. 2014. Current trends in the design of scaffolds for computer-aided tissue engineering Acta Biomater. 10 580–94 [DOI] [PubMed] [Google Scholar]
- [138].Bejan A and Lorente S 2010. The constructal law of design and evolution in nature Phil. Trans. R. Soc. B 365 1335–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lantada AD et al. 2013. Fractals in tissue engineering: toward biomimetic cell-culture matrices, microsystems and microstructured implants Expert Rev. Med. Devices 10 629–48 [DOI] [PubMed] [Google Scholar]
- [140].Almeida HD and Bartolo PJD 2010. Virtual topological optimisation of scaffolds for rapid prototyping Med. Eng. Phys 32 775–82 [DOI] [PubMed] [Google Scholar]
- [141].Hoganson DM et al. 2010. Principles of biomimetic vascular network design applied to a tissue-engineered liver scaffold Tissue Eng. A 16 1469–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Emerson DR et al. 2006. Biomimetic design of microfluidic manifolds based on a generalised Murray’s law Lab Chip 6 447–54 [DOI] [PubMed] [Google Scholar]
- [143].Janakiraman V, Mathur K and Baskaran H 2007. Optimal planar flow network designs for tissue engineered constructs with built-in vasculature Ann. Biomed. Eng 35 337–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Truslow JG and Tien J 2011. Perfusion systems that minimize vascular volume fraction in engineered tissues Biomicrofluidics 5 22201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Radisic M et al. 2005. Mathematical model of oxygen distribution in engineered cardiac tissue with parallel channel array perfused with culture medium containing oxygen carriers Am. J. Physiol. Heart Circ. Physiol 288 H1278–89 [DOI] [PubMed] [Google Scholar]
- [146].Truslow JG, Price GM and Tien J 2009. Computational design of drainage systems for vascularized scaffolds Biomaterials 30 4435–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Satoko T, Chiaki M and Soshu K 2012. Biofluid flow simulation of tissue engineering scaffolds with dendrite structures Advances in Bioceramics and Porous Ceramics V ed Narayan R, Colombo P, Halbig M and Mathur S (Hoboken, NJ: Wiley; ) [Google Scholar]
- [148].Sanz-Herrera JA, Garcia-Aznar JM and Doblare M 2009. A mathematical approach to bone tissue engineering Phil. Trans. R. Soc. A 367 2055–78 [DOI] [PubMed] [Google Scholar]
- [149].Kang H, Lin CY and Hollister SJ 2010. Topology optimization of three dimensional tissue engineering scaffold architectures for prescribed bulk modulus and diffusivity Struct. Multidiscip. Optim 42 633–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Hollister SJ 2005. Porous scaffold design for tissue engineering Nat. Mater 4 518–24 [DOI] [PubMed] [Google Scholar]
- [151].Bae H. et al. Building vascular networks. Sci. Trans. Med. 2012;4:160ps23. doi: 10.1126/scitranslmed.3003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Hasan A et al. 2014. Microfluidic techniques for development of 3D vascularized tissue Biomaterials 35 7308–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Malheiro A et al. 2016. Patterning vasculature: the role of biofabrication to achieve an integrated multicellular ecosystem ACS Biomater. Sci. Eng 2 1694–709 [DOI] [PubMed] [Google Scholar]
- [154].Baranski JD et al. 2013. Geometric control of vascular networks to enhance engineered tissue integration and function Proc. Natl Acad. Sci. USA 110 7586–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Guckenberger DJ et al. 2015. Micromilling: a method for ultra-rapid prototyping of plastic microfluidic devices Lab Chip 15 2364–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Kaihara S et al. 2000. Silicon micromachining to tissue engineer branched vascular channels for liver fabrication Tissue Eng. 6 105–17 [DOI] [PubMed] [Google Scholar]
- [157].Netti PA 2014. Biomedical Foams for Tissue Engineering Applications (Waltham, MA: Woodhead Publishing; ) pp 3–39 [Google Scholar]
- [158].Midha S et al. 2013. Bioactive glass foam scaffolds are remodelled by osteoclasts and support the formation of mineralized matrix and vascular networks in vitro Adv. Healthc. Mater 2 490–9 [DOI] [PubMed] [Google Scholar]
- [159].Costantini M et al. 2014. Highly ordered and tunable polyHIPEs by using microfluidics J. Mater. Chem. B 2 2290–300 [DOI] [PubMed] [Google Scholar]
- [160].Costantini M et al. 2015. Microfluidic foaming: a powerful tool for tailoring the morphological and permeability properties of sponge-like biopolymeric Scaffolds ACS Appl. Mater. Interfaces 7 23660–71 [DOI] [PubMed] [Google Scholar]
- [161].Lee VK et al. 2014et al. Creating perfused functional vascular channels using 3D bio-printing technology Biomaterials 35 8092–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Bischel LL, Lee SH and Beebe DJ 2012. A practical method for patterning lumens through ECM hydrogels via viscous finger patterning J. Lab Autom 17 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Herland A. et al. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLoS One. 2016;11:e0150360. doi: 10.1371/journal.pone.0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Luo X et al. 2010. In situ generation of pH gradients in microfluidic devices for biofabrication of freestanding, semi-permeable chitosan membranes Lab Chip 10 59–65 [DOI] [PubMed] [Google Scholar]
- [165].Tibbe MP. et al. Microfluidic gel patterning method by use of a temporary membrane for organ-on-chip applications. Adv. Mater. Technol. 2018;3:1700200. [Google Scholar]
- [166].Lantada AD and Morgado PL 2012. Rapid prototyping for biomedical engineering: current capabilities and challenges Annu. Rev. Biomed. Eng 14 73–96 [DOI] [PubMed] [Google Scholar]
- [167].Gugutkov D et al. 2017. Electrospun fibrinogen-PLA nanofibres for vascular tissue engineering J. Tissue Eng. Regen. Med 11 2774–84 [DOI] [PubMed] [Google Scholar]
- [168].Farina M. et al. 3D printed vascularized device for subcutaneous transplantation of human islets. Biotechnol. J. 2017;12 doi: 10.1002/biot.201700169. [DOI] [PubMed] [Google Scholar]
- [169].Daly AC et al. 2018. 3D printed microchannel networks to direct vascularisation during endochondral bone repair Biomaterials 162 34–46 [DOI] [PubMed] [Google Scholar]
- [170].Pimentel CR et al. 2018. Three-dimensional fabrication of thick and densely populated soft constructs with complex and actively perfused channel network Acta Biomater. 65 174–84 [DOI] [PubMed] [Google Scholar]
- [171].Jeffries EM et al. 2014. Micropatterning electrospun scaffolds to create intrinsic vascular networks Macromol. Biosci 14 1514–20 [DOI] [PubMed] [Google Scholar]
- [172].Niino T. et al. Laser sintering fabrication of three-dimensional tissue engineering scaffolds with a flow channel network. Biofabrication. 2011;3:034104. doi: 10.1088/1758-5082/3/3/034104. [DOI] [PubMed] [Google Scholar]
- [173].Lee SH, Moon JJ and West JL 2008. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration Biomaterials 29 2962–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Moon JJ et al. 2009. Micropatterning of poly(ethylene glycol) diacrylate hydrogels with biomolecules to regulate and guide endothelial morphogenesis Tissue Eng. A 15 579–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Cohen A. MICA freeform versus selective laser melting 2014 [Google Scholar]
- [176].Jang LK. et al. Facile and controllable electrochemical fabrication of cell-adhesive polypyrrole electrodes using pyrrole-RGD peptides. Biofabrication. 2017;9:045007. doi: 10.1088/1758-5090/aa92a2. [DOI] [PubMed] [Google Scholar]
- [177].Nguyen TU. et al. Electrochemical fabrication of a biomimetic elastin-containing bi-layered scaffold for vascular tissue engineering. Biofabrication. 2018;11:015007. doi: 10.1088/1758-5090/aaeab0. [DOI] [PubMed] [Google Scholar]
- [178].Khalil S and Sun W 2009. Bioprinting endothelial cells with alginate for 3D tissue constructs J. Biomech. Eng 131 111002. [DOI] [PubMed] [Google Scholar]
- [179].Buyukhatipoglu K. et al. The role of printing parameters and scaffold biopolymer properties in the efficacy of a new hybrid nano-bioprinting system. Biofabrication. 2009;1:035003. doi: 10.1088/1758-5082/1/3/035003. [DOI] [PubMed] [Google Scholar]
- [180].Buyukhatipoglu K et al. 2010. Bioprinted nanoparticles for tissue engineering applications Tissue Eng. C 16 631–2 [DOI] [PubMed] [Google Scholar]
- [181].Benning L et al. 2018. Assessment of hydrogels for bioprinting of endothelial cells J. Biomed. Mater. Res. A 106 935–47 [DOI] [PubMed] [Google Scholar]
- [182].Ouyang L. et al. Three-dimensional bioprinting of embryonic stem cells directs highly uniform embryoid body formation. Biofabrication. 2015;7:044101. doi: 10.1088/1758-5090/7/4/044101. [DOI] [PubMed] [Google Scholar]
- [183].Cui XF and Boland T 2009. Human microvasculature fabrication using thermal inkjet printing technology Biomaterials 30 6221–7 [DOI] [PubMed] [Google Scholar]
- [184].Christensen K et al. 2015. Freeform inkjet printing of cellular structures with bifurcations Biotechnol. Bioeng 112 1047–55 [DOI] [PubMed] [Google Scholar]
- [185].Chan V et al. 2010. Three-dimensional photopatterning of hydrogels using stereolithography for long-term cell encapsulation Lab Chip 10 2062–70 [DOI] [PubMed] [Google Scholar]
- [186].Zhu W et al. 2017. Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture Biomaterials 124 106–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Barron JA et al. 2004. Biological laser printing: a novel technique for creating heterogeneous 3-dimensional cell patterns Biomed. Microdevices 6 139–47 [DOI] [PubMed] [Google Scholar]
- [188].Nahmias Y and Odde DJ 2006. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic-endothelial sinusoid-like structures Nat. Protocols 1 2288–96 [DOI] [PubMed] [Google Scholar]
- [189].Hocheng H and Tseng C 2008. Mechanical and optical design for assembly of vascular endothelial cells using laser guidance and tweezers Opt. Commun 281 4435–41 [Google Scholar]
- [190].Borchers K. et al. New cytocompatible materials for additive manufacturing of bio-inspired blood vessels systems. Int. Conf. on Biofabrication 2012 [Google Scholar]
- [191].Hoch E et al. 2012. Stiff gelatin hydrogels can be photo-chemically synthesized from low viscous gelatin solutions using molecularly functionalized gelatin with a high degree of methacrylation J. Mater. Sci., Mater. Med 23 2607–17 [DOI] [PubMed] [Google Scholar]
- [192].Ovsianikov A. et al. Laser printing of cells into 3D scaffolds. Biofabrication. 2010;2:014104. doi: 10.1088/1758-5082/2/1/014104. [DOI] [PubMed] [Google Scholar]
- [193].Norotte C et al. 2009. Scaffold-free vascular tissue engineering using bioprinting Biomaterials 30 5910–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Colosi C et al. 2016. Microfluidic bioprinting of heterogeneous 3D tissue constructs using low-viscosity bioink Adv. Mater 28 677–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Kanani C. Cell Printing: A Novel Method to Seed Cells onto Biological Scaffolds. Worcester Polytechnic Institute; 2012. [Google Scholar]
- [196].Jakab K. et al. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication. 2010;2:022001. doi: 10.1088/1758-5082/2/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Trietsch SJ et al. 2013. Microfluidic titer plate for stratified 3D cell culture Lab Chip 13 3548–54 [DOI] [PubMed] [Google Scholar]
- [198].Vulto P et al. 2010. A microfluidic approach for high efficiency extraction of low molecular weight RNA Lab Chip 10 610–6 [DOI] [PubMed] [Google Scholar]
- [199].Jang M. et al. On-chip three-dimensional cell culture in phaseguides improves hepatocyte functions in vitro. Biomicrofluidics. 2015;9:034113. doi: 10.1063/1.4922863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Stucki JD. et al. Medium throughput breathing human primary cell alveolus-on-chip model. Sci. Rep. 2018;8:14359. doi: 10.1038/s41598-018-32523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Lanz HL. et al. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer. 2017;17:709. doi: 10.1186/s12885-017-3709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Asakawa N et al. 2010. Pre-vascularization of in vitro three-dimensional tissues created by cell sheet engineering Biomaterials 31 3903–9 [DOI] [PubMed] [Google Scholar]
- [203].Ren LL. et al. Preparation of three-dimensional vascularized MSC cell sheet constructs for tissue regeneration. Biomed. Res. Int. 2014;2014:301279. doi: 10.1155/2014/301279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Ballester-Beltran J, Lebourg M and Salmeron-Sanchez M 2013. Dorsal and ventral stimuli in sandwich-like microenvironments. Effect on cell differentiation Biotechnol. Bioeng 110 3048–58 [DOI] [PubMed] [Google Scholar]
- [205].Ballester-Beltran J et al. 2014. Fibronectin-matrix sandwich-like microenvironments to manipulate cell fate Biomater. Sci 2 381–9 [DOI] [PubMed] [Google Scholar]
- [206].Ballester-Beltran J et al. 2015. Cell migration within confined sandwich-like nanoenvironments Nanomedicine 10 815–28 [DOI] [PubMed] [Google Scholar]
- [207].Shimizu T 2014. Cell sheet-based tissue engineering for fabricating 3-dimensional heart tissues Circ. J 78 2594–603 [DOI] [PubMed] [Google Scholar]
- [208].Xia YN and Whitesides GM 1998. Soft lithography Annu. Rev. Mater. Sci 28 153–84 [Google Scholar]
- [209].Sekula S et al. 2008. Multiplexed lipid dip-pen nanolithography on subcellular scales for the templating of functional proteins and cell culture Small 4 1785–93 [DOI] [PubMed] [Google Scholar]
- [210].Hirtz M. et al. Multiplexed biomimetic lipid membranes on graphene by dip-pen nanolithography. Nat. Commun. 2013;4:2591. doi: 10.1038/ncomms3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Leslie-Barbick JE et al. 2011. Micron-scale spatially patterned, covalently immobilized vascular endothelial growth factor on hydrogels accelerates endothelial tubulogenesis and increases cellular angiogenic responses Tissue Eng. A 17 221–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Gad-el-Hak M 2005. The MEMS Handbook (Boca Raton, FL: CRC Press; ) [Google Scholar]
- [213].Madou MJ 2011. Fundamentals of Microfabrication (Boca Raton, FL: CRC Press; ) [Google Scholar]
- [214].Schwentenwein M, Schneider P and Homa J 2014. Lithography based ceramic manufacture: a novel technique for additive manufacturing of high performance ceramics Adv. Sci. Technol 88 60–4 [Google Scholar]
- [215].Linnenberger A et al. 2013. Three dimensional live cell lithography Opt. Express 21 10269–77 [DOI] [PubMed] [Google Scholar]
- [216].Koch L et al. 2013. Laser assisted cell printing Curr. Pharm. Biotechnol 14 91–7 [PubMed] [Google Scholar]
- [217].Mappes T et al. 2008. Submicron polymer structures with x-ray lithography and hot embossing Microsyst. Technol 14 1721–5 [Google Scholar]
- [218].Boerckel JD. et al. Microcomputed tomography: approaches and applications in bioengineering. Stem Cell Res. Ther. 2014;5:144. doi: 10.1186/scrt534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Zheng XY. et al. Multiscale metallic metamaterials. Nat. Mater. 2016;15:1100. doi: 10.1038/nmat4694. [DOI] [PubMed] [Google Scholar]
- [220].Hengsbach S and Lantada AD 2014. Rapid prototyping of multi-scale biomedical microdevices by combining additive manufacturing technologies Biomed. Microdevices 16 617–27 [DOI] [PubMed] [Google Scholar]
- [221].Gomez-Marquez J. Design for hack in medicine: MacGyver nurses and Legos are helping us make MEDIKits for better health care Make. Magazine 2011 [Google Scholar]