Abstract
We sought to elucidate the mechanisms underlying the aerobic dechlorination of the persistent organic pollutants hexachlorobenzene (HCB) and pentachlorophenol (PCP). We performed genomic and heterologous expression analyses of dehalogenase genes in Nocardioides sp. PD653, the first bacterium found to be capable of mineralizing HCB via PCP under aerobic conditions. The hcbA1A2A3 and hcbB1B2B3 genes, which were involved in catalysing the aerobic dechlorination of HCB and PCP, respectively, were identified and characterized; they were classified as members of the two-component flavin-diffusible monooxygenase family. This was subsequently verified by biochemical analysis; aerobic dechlorination activity was successfully reconstituted in vitro in the presence of flavin, NADH, the flavin reductase HcbA3, and the HCB monooxygenase HcbA1. These findings will contribute to the implementation of in situ bioremediation of HCB- or PCP-contaminated sites, as well as to a better understanding of bacterial evolution apropos their ability to degrade heavily chlorinated anthropogenic compounds under aerobic conditions.
Keywords: HCB, PCP, TC-FDM, dehalogenase, Nocardioides
Introduction
Hexachlorobenzene (C6Cl6; HCB) is an organochlorine fungicide that has been used worldwide since the 1940s.1) Although effective as a disinfectant for the seeds of onions, sorghum, and cereal crops such as wheat, barley, oats, and rye, the use of HCB was discontinued in many countries in the 1970s, owing to its toxicity and environmental persistence, with an estimated half-life in soil of 3–6 years4); in 2001, it was listed as a persistent organic pollutant (POP) at the Stockholm Convention. Nevertheless, HCB remains a widely distributed environmental contaminant. In Japan, although HCB is not registered as a pesticide, it has been found as an impurity in technical pentachlorophenol (C6HCl5O; PCP), which is used mainly in rice paddies, thereby causing co-contamination. PCP has been widely used as a biocide, wood preservative, and disinfectant, as well as a component in anti-fouling paint.2) Although its half-life is generally less than 10 weeks in soil, PCP may persist for many years at heavily contaminated sites where levels exceed the toxicity threshold of soil microorganisms or in cold northern climates.3) It is toxic to a diverse range of organisms, both plants and animals, with this toxicity being attributed to an interference with oxidative phosphorylation, and is particularly acute and chronic in fish. In 2015, PCP (and its salts and esters) was added to Annex A of the Stockholm Convention as a POP.2)
Although highly chlorinated organic compounds such as POPs tend to be recalcitrant to biodegradation, dechlorination can contribute to mitigating their toxicity and persistence in the environment, and is consequently considered an important chemical reaction. Notably, certain bacteria have evolved an ability to dechlorinate and metabolize such anthropogenic chemicals5–7); accordingly, the underlying evolutionary processes and practical applications for in situ bioremediation have recently attracted considerable interest. Dehalococcoides mccartyi strain CBDB1, the first bacterium found to be capable of HCB dechlorination, is the most extensively studied anaerobic bacterium with respect to organohalide respiration, for which it uses polyhalogenated benzenes as growth-supporting electron acceptors,8,9) and generates the end products 1,3,5-trichlorobenzene, 1,3-dichlorobenzene, and 1,4-dichlorobenzene. From the cell extracts of this strain, CbrA, a member of the family of corrinoid/iron-sulfur cluster-containing reductive dehalogenases, has been identified for its dehalogenase activity on 1,2,3,4-tetrachlorobenezene, 1,2,3-trichlorobenzene, and pentachlorobenzene.10) The complete genome sequence of D. mccartyi revealed the presence of 32 reductive dehalogenase homologs, and it is accordingly anticipated that a further novel HCB reductive dehalogenase gene will be identified in strain CBDB1.11)
Compared with the anaerobic degradation of chlorinated organic compounds, relative few studies have provided insights regarding the aerobic biodegradation of HCB. A genetically engineered F87W-Y96F-V247L mutant of CYP101 (P450cam) from Pseudomonas putida has been found to aerobically dechlorinate HCB to yield PCP.12) By creating space for HCB chlorine atoms in the catalytic site of CYP101, the introduction of a further L244A mutation was found to give rise to enhanced HCB oxidative-dehalogenation activity,13) and a gene cassette encoding this mutant CYP101 (F87W-Y96F-L244A-V247L) has been used to transform the complete PCP-degrader Sphingobium chlorophenolicum ATC C 39723, such that this strain can degrade HCB without accumulating toxic intermediates.14)
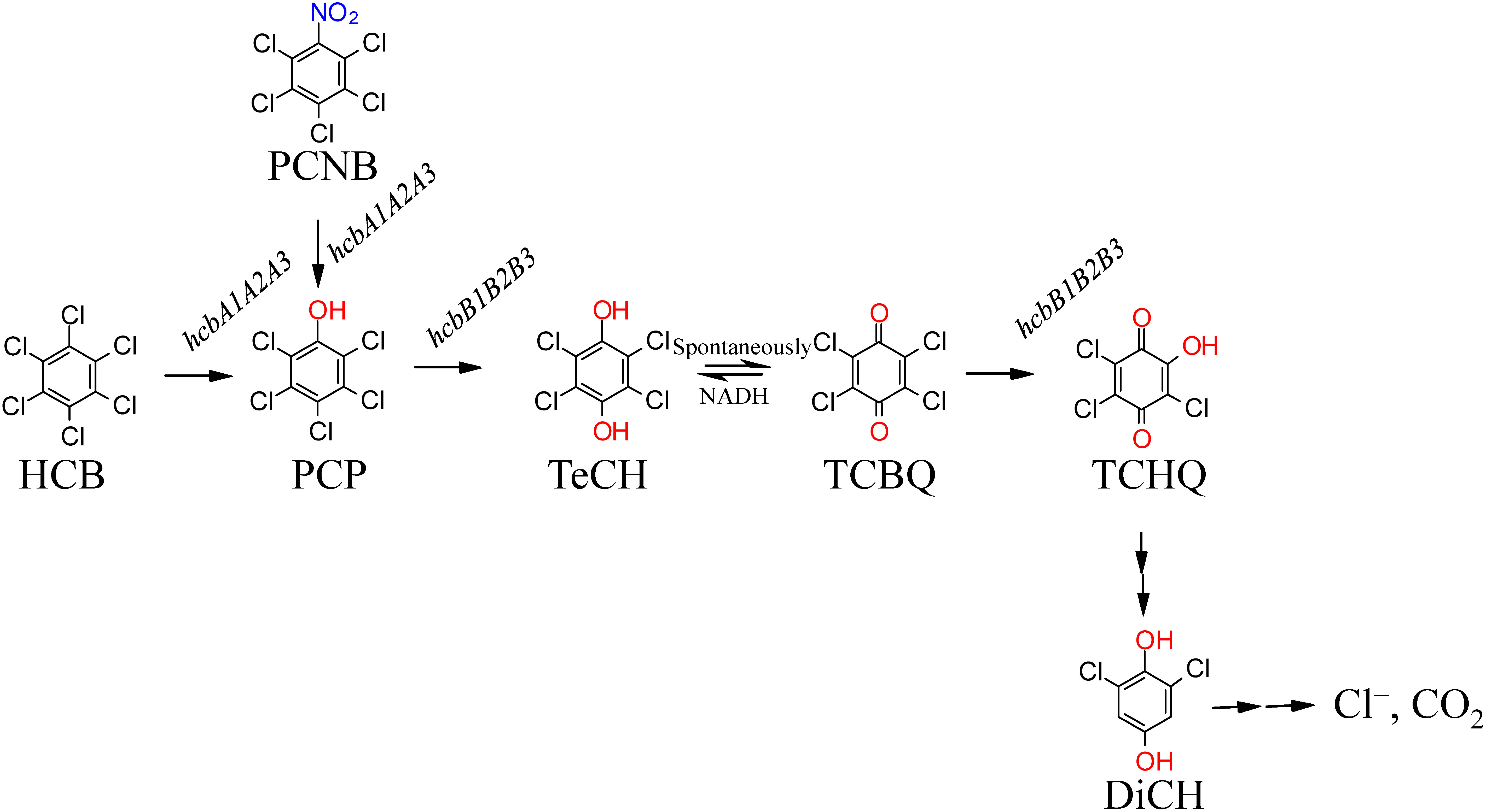
Among naturally occurring bacteria, Liu et al. 2009 found that species in the genera Azospirillum and Alcaligenes were dominant members of the HCB-metabolizing community isolated from contaminated soil.15) Nocardioides sp. PD653 was identified as the first bacterium capable of mineralizing HCB under aerobic conditions.16) This strain was originally isolated as a pentachloronitrobenzene (PCNB)-degrader from the soil of a Chinese cabbage field, in which PCNB-resistant bacteria had emerged, and was subsequently found to be capable of aerobically degrading HCB via the intermediates PCP, 2,3,5,6-tetrachloro-p-hydroquinone (TeCH), and 2,6-dichloro-p-hydroquinone (DiCH) (Fig. 1). Given that to date more than 77 PCP-degrading bacteria have been isolated under both aerobic and anaerobic conditions,17) it can be speculated that the bottleneck limiting HCB metabolism lies in the initial step involving the dechlorination of HCB to form PCP, the underlying mechanism of which is incompletely understood. Furthermore, although mechanisms associated with the pcp gene-dependent degradation of PCP in S. chlorophenolicum strain ATC C 39723 have been investigated in detail,18,19) these genes have only been detected in closely related gram-negative strains,20,21) and not in gram-positive strains, including PD653. Consequently, it can be speculated that gram-positive strains have evolved novel degradative mechanisms.
Fig. 1. Proposed pathway for the aerobic degradation of HCB and PCNB by PD653. HCB; hexachlorobenzene, PCNB; pentachloronitrobenzene, PCP; pentachlorophenol, TeCH; 2,3,5,6-tetrachloro-p-hydroquinone, TCBQ; 2,3,5,6-tetrachloro-p-benzoquinone, TCHQ; 2,5,6-trichloro-p-hydroquinone, DiCH; 2,6-dichloro-p-hydroquinone. (Reprinted from Ref. 58).
Elucidating the mechanisms underlying the initial step of HCB metabolism in PD653 is expected to contribute not only to enhancing the efficiency of in situ bioremediation but also to gaining a better understanding of the evolutionary processes associated with the bacterial metabolism of highly chlorinated anthropogenic compounds. To achieve these goals, we sought in the present study to identify the genes involved in the dechlorination of HCB and PCP. Furthermore, the aerobic dechlorination of HCB was successfully reconstituted by HcbA1A3 in vitro, and the inducibility of the hcbB3 gene encoding the novel PCP-dehalogenase was analysed.
1. Identification of hcbA genes involved in catalysing aerobic HCB dechlorination22)
Prior to our examination of the genes contributing to the dechlorination of HCB, we initially isolated Nocardioides sp. PD653-B2 (GenBank accession number: LC196157) during the subculturing of strain PD653. This derivative strain can degrade PCP, although not HCB. Neither is it able to denitrate PCNB to PCP, although it has been found to transform this to pentachloroaniline, a major metabolite detected in PCNB-contaminated soil.23) These observations thus tended to indicate that genes associated with the oxidative dehalogenation of HCB and denitration of PCNB had either been deleted or mutated in this strain. Consequently, we compared the draft genomes of PD653 and PD653-B2; having identified the candidate genes conserved only in PD653, we performed the respective heterologous expression analyses.
1.1. Comparative genomic analysis of PD653 and its derivative strain PD653-B2
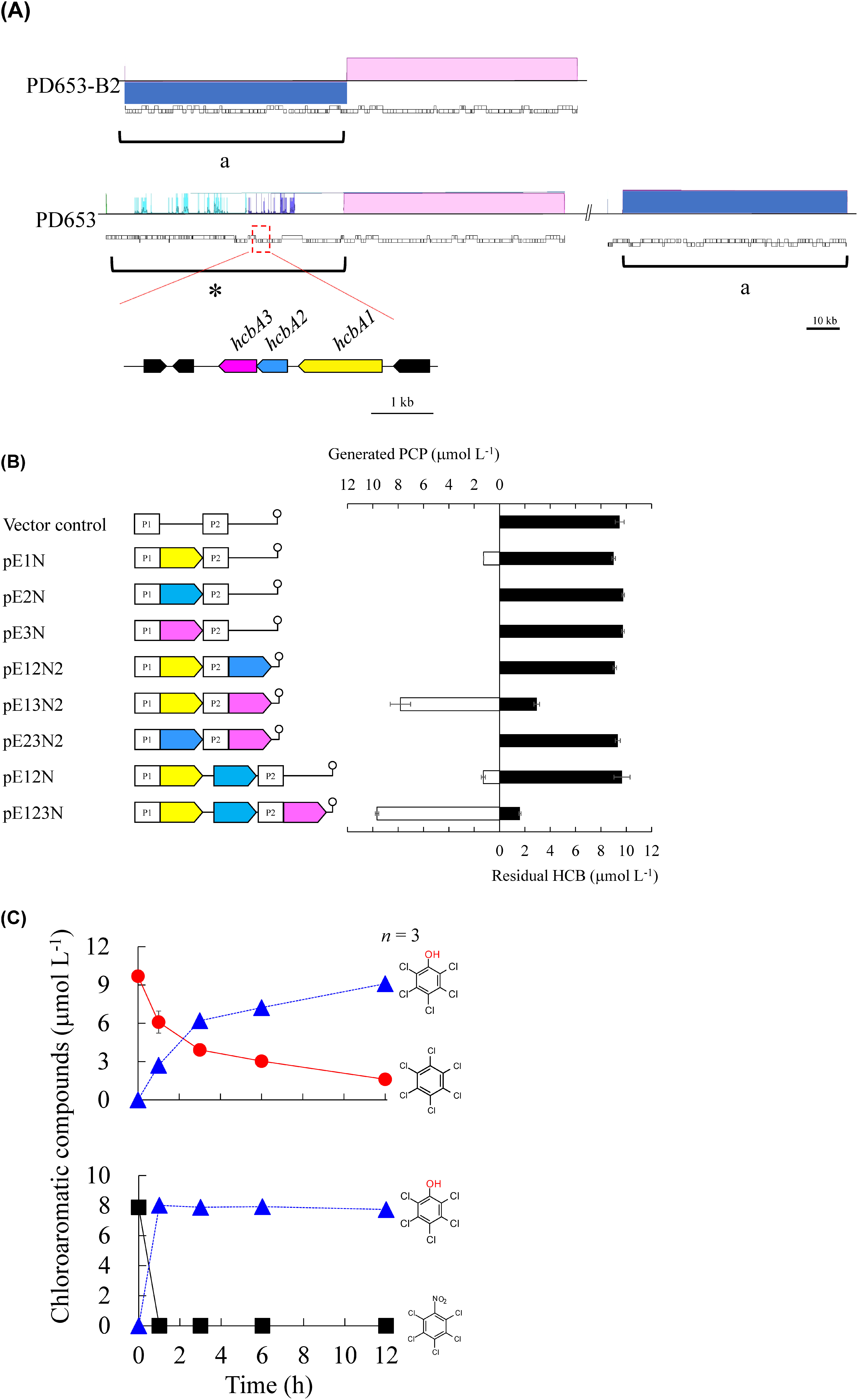
The estimated genome sizes of PD653 and PD653-B2 are 5.08 Mb (87 contigs; DDBJ/EMBL/GenBank numbers DBJG01000001 to DBJG01000087) and 4.99 Mb (81 contigs; DDBJ/EMBL/GenBank numbers DBJE01000001 to DBJE01000081), respectively. Comparative genomic analysis using the Mauve tool24) revealed a region of interest from positions 1 to 71,874 of contig 22 (GenBank accession no. BDJG01000022), in which a deletion of 96 coding sequences was detected in the PD653-B2 sequence (indicated by an asterisk in Fig. 2A). Within this region, an approximately 60-kb segment of the PD653-B2 genome, corresponding to contig 64 in PD653 (GenBank accession no. GDJG01000064), was found to be translocated and inverted, thereby indicating that a genome rearrangement had occurred during the derivation of PD653-B2 from PD653 (Fig. 2A). Three coding sequences, namely, PD653_2189 (hcbA1), PD653_2188 (hcbA2), and PD653_2187 (hcbA3), were selected as candidate genes, based on their annotation, and then subjected to heterologous expression (Fig. 2A). The deduced amino acid sequence of HcbA1 was similar to that of Ese, which is classified as a member of the two-component flavin-diffusible monooxygenase (TC-FDM) family involved in the endosulfan and endosulfan sulfate metabolism of Arthrobacter sp. KW (432 bits; 49% identity).25) The deduced amino acid sequence of HcbA2 was similar to that of EmoB, an NADH:FMN oxidoreductase in the TC-FDM system for the degradation of EDTA by Mesorhizobium sp. BNC1 (77 bits; 37% identity).26) The deduced amino acid sequence of HcbA3 contained a conserved domain (smart00903) typical of the flavin-reductase components classified in the TC-FDM family, including TftC,27) PheA2,28) and HpaC.29)
Fig. 2. A) Comparative analysis of the Nocardioides sp. PD653 and PD653-B2. The region present only in PD653 is indicated with an asterisk (*). The translocated segments are marked “a”. (Reprinted and partially modified form Ref. 22.) B) The candidate genes ORF1 (hcbA1), ORF2 (hcbA2), and ORF3 (hcbA3) are marked with yellow, azure, and magenta arrows, respectively. C) Comparison of dechlorination activity for E. coli cells transformed using expression vectors. Solid and open bars indicate residual HCB and PCP in the culture, respectively. Each error bar indicates the standard error for triplicate samples. ORF1 (hcbA1), ORF2 (hcbA2), and ORF3 (hcbA3) are marked with yellow, azure, and magenta arrows, respectively. The stem-loop symbol indicates the position of the terminator. P1 and P2 denote T7 promoters 1 and 2, respectively. (Reprinted by permission from American Society of Microbiology: Appl. Environ. Microbiol. Ref. 22.) D) Time courses for HCB degradation (●), PCNB (■), and PCP generation (▲) by E. coli/pE123N. Each concentration shown is the mean (n=3) with the standard deviation.
1.2. Dechlorination of HCB and PCNB by recombinant E. coli cells
We assayed the HCB oxidative dehalogenase activity of recombinant Escherichia coli BL21 (DE3) cells harbouring the pETDuet-1 plasmid (Novagen, Madison, WI, USA), which has two multiple cloning sites (MCS). We accordingly established that co-expression of hcbA1 (inserted at MCS1) and hcbA3 (inserted at MCS2) was an essential prerequisite for HCB degradation activity in recombinant E. coli cells, whereas no apparent activity was detected when hcbA1, hcbA2, and hcbA3 were independently inserted into E. coli (Fig. 2B). Interestingly, we found that co-expression of hcbA1 and hcbA3 led to not only the dechlorination of HCB but also the denitration of PCNB to form PCP (Fig. 2C), and that insertion of hcbA2 downstream of hcbA1 at MCS1 contributed to a moderate enhancement of the HCB degradation activity from 68.7 to 83.0%, using an initial concentration of 10 µmol L−1 HCB, thus indicating that these genes are involved in the initial HCB dechlorination step. Notably, under oxygen-limiting conditions [(O2)<0.5 mg L−1], the HCB dechlorination activity was completely lost, whereas the re-introduction of O2 restored activity, providing convincing evidence that O2 is essential for HcbA1 activity. This result was consistent with the deduced amino acid sequence analysis, suggesting that HcbA1 can be classified as a monooxygenase. Furthermore, given the deduced amino acid sequence of the encoded protein, we assume that hcbA3 encodes a putative flavin reductase, which along with hcbA1 comprises the TC-FDM system.
Although bacteria utilize TC-FDM family enzymes for a diverse range of reactions, including bioluminescence,30) oxidation of aromatic compounds,31) degradation of chelating agents,32) desulphurization of sulfonated compounds,33) and biosynthesis of antibiotics,34) these enzymes are all characterized by a common feature: TC-FDM systems comprise a flavin-dependent monooxygenase and partner reductase components. In these systems, the reductase enzyme generates reduced flavins via the reduction of oxidized flavin, with the reducing equivalents being provided by a pyridine nucleotide. Subsequently, the reduced flavins are transferred to oxygenase enzymes for monooxygenation reactions, accompanied by the formation of a hydroperoxyflavin or peroxyflavin intermediate. Furthermore, the genes that encode the reductase and monooxygenase enzymes are often located in the same operon. This feature was also found to be common to the hcbA genes, all three genes of which were transcribed polycistronically.
2. Biochemical characterization of the NADH : FMN oxidoreductase HcbA335)
We postulated that the mechanism underlying the initial HCB-dechlorination step in PD653 is closely associated with the TC-FDM system; purified HcbA1 was indeed found to dechlorinate HCB in the presence of an E. coli flavin reductase (Fre),36) FMN, and NADH, resulting in the formation of flavin-N5-oxide,37) thereby supporting our hypothesis and implying the presence of the partner reductase component of HcbA1. Given our observation that co-expression of hcbA1 and hcbA3 led to HCB dehalogenase activity in E. coli cells, we believe that HcbA3 plays a role in supplying reduced flavin in vivo. However, as we have yet to conduct the requisite biochemical analyses, it remains unclear whether oxidative HCB dehalogenation is actually catalysed by a TC-FDM system.
2.1. Flavin-binding studies of purified C-terminally histidine-tagged HcbA3
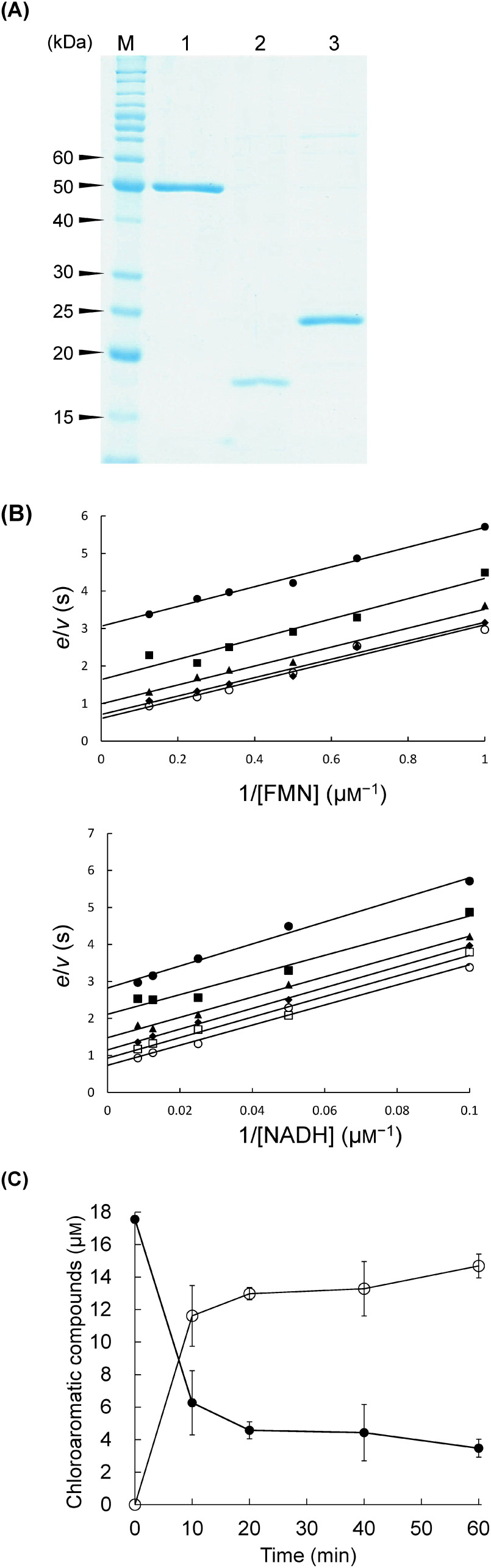
Recombinant C-terminally histidine-tagged HcbA1, HcbA2, and HcbA3 were overexpressed in Rhodococcus erythropolis L88 and subsequently purified (Fig. 3A). Notably, each of the purified protein preparations was colourless and lacked the typical absorption spectra of flavin-containing flavoproteins, thereby indicating that the purified proteins were deficient of any bound flavin cofactors. The binding affinity was determined spectrofluorometrically, yielding an average Kd value of 0.75±0.17 µM for the FMN–HcbA3C-His complex. Moreover, we detected no fluorescence quenching when riboflavin and FAD were titrated with HcbA3C-His, nor when flavins were titrated with HcbA1C-His or HcbA2C-His, thus revealing that whereas HcbA3C-His was able to bind to FMN, HcbA1C-His and HcbA2C-His were not.
Fig. 3. (Reprinted from Ref. 35.) A) Purified C-terminally histidine-tagged HcbA1, HcbA2, and HcbA3 as shown by SDS-PAGE. Lane M, marker proteins; lane 1, oxidative HCB dehalogenase HcbA1C-His; lane 2, putative flavin reductase HcbA2C-His; lane 3, NADH:FMN oxidoreductase HcbA3 C-His. B) Double reciprocal plots of HcbA3C-His steady-state kinetics. Top: assays were performed using 25 nM HcbA3C-His, 1–8 µM FMN, and various fixed concentrations of NADH, including 10 µM (●), 20 µM (■), 40 µM (▲), 80 µM (◆), and 120 µM (○). Bottom: assays were performed using 25 nM HcbA3C-His, 10–120 µM NADH, and various fixed concentrations of FMN, including 1 µM (●), 1.5 µM (■), 2 µM (▲), 3 µM (◆), 4 µM (○), and 8 µM (□). C) Conversion of HCB (●) to PCP (○) via oxidative dehalogenation of HcbA1C-His-HcbA3C-His. Each error bar indicates the standard error for triplicate samples.
2.2. Flavin binding studies
Steady-state kinetic measurements of HcbA3C-His were performed by monitoring the oxidation of NADH to NAD+ (ε340=6.22 mM−1 cm−1) using a UV-Vis spectrophotometer. When NADH was used as a pyridinic substrate for HcbA3C-His in combination with FMN, we detected distinct activity; in contrast, no activity was observed when using NADPH as a substrate. Compared with FAD and riboflavin, we identified FMN as the preferred flavin substrate of HcbA3C-His, and therefore we measured the steady-state kinetic parameters of HcbA3C-His in the presence of both FMN and NADH. Initial rates of NADH oxidation were observed to correspond to typical Michaelis–Menten kinetics, and Lineweaver–Burk plots showed parallel patterns for a ping-pong mechanism (Fig. 3B). Consequently, the steady-state kinetic parameters were determined from a fit of the data to Eq. (1):
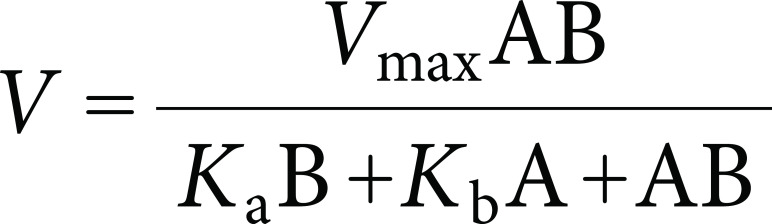 |
(1) |
where A and B are the substrate concentrations, and Ka and Kb represent the Michaelis constants for substrates A and B, respectively. According to this equation, the actual values of Vmax and kcat were the same for both substrates (NADH and FMN); the average Km values for NADH and FMN were 51.66±11.58 µM and 4.43±0.69 µM, respectively; the Vmax and kcat values for both substrates were 2.21±0.86 µM and 66.74±5.91 s−1, respectively. On the basis of these observations, we thus concluded that HcbA3 is an NADH:FMN oxidoreductase. The distinct specificity of HcbA3 indicates that FMN may be a major co-substrate for the HCB oxidative dehalogenation reaction catalysed by HcbA1A3 in strain PD653.
On the basis of its secondary structure, the putative flavin reductase HcbA3 is predicted to be similar to TftC,27) HpaC,29) PheA2,28) and CobR.38) Among these reductases, TftC and PheA2 catalyse the reduction of flavin via a sequential and ping-pong bi-bi mechanism, respectively.28,39) In the TC-FDM family of enzymes, in addition to PheA2, the NADPH-preferring flavin reductase FRP also follows this type of mechanism in single enzyme assays.40) Both PheA2 and FRP contain flavins as prosthetic groups, which mediate proton transfer from pyridinic substrates to exogenously added flavin substrates; however, HcbA3C-His lacks such flavin cofactors, indicating that a mechanism independent of flavin cofactor involvement plays a role in HcbA3 oxidoreductase activity.
2.3. Reconstitution of the oxidative HCB dehalogenase activity of HcbA1A3 in vitro
Although we demonstrated that HcbA3C-His generated reduced flavin in vitro, it was important to investigate whether this activity led the dehalogenation of HCB by HcbA1C-His. To this end, we initially determined the optimal molar ratio of HcbA1 and HcbA3 to be 7 : 1. A curve showing the degradation of HCB by these enzymes at this ratio is presented in Fig. 3C. Briefly, 0.129 µg HCB was dehalogenated and 0.124 µg PCP was generated by 1 µg HcbA1C-His in 10 min, clearly demonstrating that oxidative HCB dehalogenation can be mediated by a TC-FDM system in bacteria. This finding will enable us to investigate the mechanism involved in reduced flavin transfer, thereby indicating whether this is a diffusion process or a direct transfer flavin involving protein–protein interactions (PPIs), given that reduced flavins generated in HcbA3-catalysed reactions tend to be unstable, on account of their susceptibility to oxidation by molecular oxygen. Several examples involving such PPIs between monooxygenase and flavin reductase for the transfer of reduced flavin have been reported in the literature, including FRP–luciferase in Vibrio harveyi,41) SsuE–SsuD in E. coli,42) EmoA–EmoB in Mesorhizobium sp. BNC1,26) and PrnF–PrnD in Pseudomonas fluorescens Pf-5.43) However, despite adopting several different approaches, including a pull-down assay and fluorometric titration, we failed to obtain any conclusive evidence for the presence of specific interactions between HcbA1 and HcbA3. Accordingly, we provisionally hypothesise that no transient interaction occurs with respect to flavin transfer.
3. Identification of novel hcbB genes catalysing the dechlorination of PCP44)
Numerous PCP-degrading microorganisms have been reported to date, and strategies have been established for evaluating the PCP-degrading microbial community in situ based on molecular biological techniques.45,46) In this regard, S. chlorophenolicum ATC C 39723 is one of the most extensively studied model bacteria capable of mineralizing PCP.18) During the process of PCP metabolism, strain ATC C 39723 recruits PcpB, a single flavin-containing protein and PCP-4 monooxygenase (AAF15368; encoded by pcpB).47) Notably, pcpB or its allele have been detected in sphingomonads isolated from geographically discrete contaminated sites, thereby indicating the widespread distribution of this gene via horizontal gene transfer.20,48) In contrast, in other bacterial genera, including PCP-degrading gram-positive strains such as PD653, almost nothing is known regarding the genes and enzymes involved in PCP metabolism. To the best of our knowledge, this has only been demonstrated in Mycobacterium chlorophenolicum PCP-I, in which membrane-associated cytochrome P-450 is involved in hydroxylation.49) For example, on the basis of Southern blot analysis, Orser et al. have shown that pcpB probes fail to hybridize with the digested genomic DNA of this strain,50) which thus tends to imply the involvement of another catabolic gene in the initial dechlorination of PCP. Consequently, in the present study, a primary objective was to identify the putative genes involved in the degradation of PCP by PD653.
3.1. Prediction of PCP catabolic genes based on RNA-seq analysis
RNA-seq analysis revealed that 47 of the 5087 assessed genes showed a greater than four-fold difference in mRNA levels between the PD653 cells exposed to HCB and those of the non-exposed control (p>0.05). Among these, we selected three coding sequences, PD653_1112 (hcbB1), PD653_1113 (hcbB2), and PD653_1114 (hcbB3), for annotation and amino acid sequence analysis. We accordingly found that the amino acid sequence deduced for PD653_1112 (hcbB1) was similar to the protein Rv1155 (PDB number 1W9A_B) in Mycobacterium tuberculosis (140 bit, 52% identity).51) The closest related protein to ORF1 was identified as a putative pyridoxine/pyridoxamine 5′-phosphate oxidase (accession number KMO83753) from Mycobacterium chlorophenolicum (187 bit, 65% identity). The protein showing the closest relationship to PD653_1113 (hcbB2) is the xylose isomerase-like TIM barrel (KMO83752) from M. chlorophenolicum (248 bit, identity 50%), whereas the deduced amino acid sequence of PD653_1114 (hcbB3) was similar to TftD (577 bit, 54% identity), a FADH2-dependent monooxygenase that catalyses the sequential hydroxylation of 2,4,5-trichlorophenol to 2,5-dichlorohydroquinone, followed by 5-chlorohydroxyquinol (UniProtKB/Swiss-Prot: O87009.2), in Burkholderia cepacia AC1100.27,52,53) Given that TC-FDM has previously been characterized as a key catabolic enzyme of chloroaromatic compounds, we hypothesized that either hcbB1 or hcbB2 encodes a reductase component that supplies reduced flavins that facilitates degradation of PCP by the HcbB3.
3.2. Biotransformation of PCP by recombinant E. coli cells
Our examination of the PCP dissipation activity of recombinant E. coli cell cultures obtained for the different enzyme constructs generated in this study revealed that co-expression of hcbB1 and hcbB3 or hcbB2 and hcbB3 led to significant PCP dissipation activity (Fig. 4A). On the basis of the predicted HCB metabolic pathway in PD653 (Fig. 1), we presumed that an intermediate compound generated by PCP metabolism is TeCH, although it was unclear whether this compound would accumulate in the culture or be further metabolism in vivo. Hence, to establish the fate of this putative intermediate we used the cleared lysate of induced E. coli cells harbouring hcbB1 and hcbB3. This enabled us to detect trace amounts of TeCH, based on GC-MS analysis, which thereby tends to indicate that these hcbB genes encode an enzyme that catalyses the conversion of PCP to TeCH. Moreover, analysis of the downstream pathway by recombinant E. coli cells co-expressing hcbB2–hcbB3 revealed that TeCH was spontaneously converted to TCBQ (2,3,5,6-tetrachloro-p-benzoquinone), followed by dechlorination to 2,5,6-trichloro-p-hydroquinone (TCHQ). Collectively, these observations enabled us to establish that the identified hcbB genes can contribute to a two-step dechlorination of PCP to yield TCHQ via the intermediate conversion to TeCH (Fig. 1), which is similar to the sequential hydroxylation of 2,4,5-trichlorophenol to 5-chlorohydroxyquinol via 2,5-dichlorohydroquinone catalysed by TftD. Thus, HcbB3 and TftD would appear to have comparable biochemical functions. Both hcbB1 and hcbB2 appear to encode a flavin-reductase component for HcbB3, although they do not have close homology to TftC,27) a flavin reductase component of TftD,
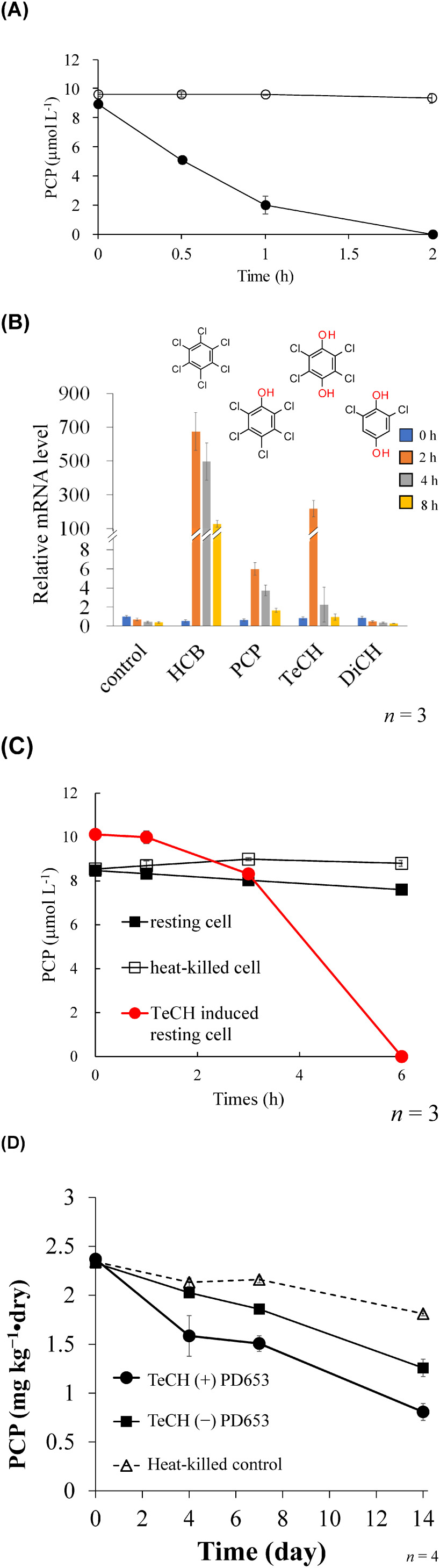
Fig. 4. (Reprinted from Ref. 44.) A) Time courses for PCP degradation (●) by E. coli BL21 (DE3) carrying hcbB2-hcbB3 and PCP concentrations (○) in the vector control cultures. Each concentration shown is the mean (n=3) with the standard deviation. B) Expression of the hcbB3 gene in PD653 by hexachlorobenzene (HCB) and its degradation intermediates, pentachlorophenol (PCP), 2,3,5,6-tetrachloro-p-hydroquinone (TeCH), and 2,6-dichloro-p-hydroquinone (DiCH). Gene expression was calculated relative to the rpoB gene using the cycle threshold (2−ΔΔCt) method. C) Induction of hcbB3 gene in PD653 resting cells by 2,3,5,6-tetrachloro-p-hydroquinone (TeCH) resulted in an advanced PCP-degradation activity. D) Dissipation of PCP by PD653 in artificially contaminated sand.
3.3. Transcriptional analysis of a novel PCP dehalogenase gene hcbB3
To confirm the RNA-seq data and investigate the effect of HCB and of the compounds formed during its transformation, we monitored the expression of hcbB3 via RT-qPCR. The transcriptional properties determined based on RNA-seq were consistent with the RT-qPCR results, showing that hcbB3 was significantly upregulated by HCB (Fig. 4B). Indeed, after 2, 4, and 8 hr of exposure to HCB, the expression of hcbB3 was found to be 675-, 497-, and 127-fold higher, respectively, than that in the control, which had been not exposed to HCB. In addition, it was observed that within 2 hr of exposure to TeCH, the expression of hcbB3 increased 217-fold compared with that of the control. We thus established that the expression of hcbB3 is upregulated not only by HCB, but also by TeCH. Given that PD653 metabolizes PCP via TeCH and that TeCH induces the expression of hcbB3 (Fig. 4B), we speculate that metabolism by PD653 might promote an upregulation of hcbB3 in the presence of PCP.
As shown in Fig. 4C, the resting PD653 cells exposed to 10 mg L−1 TeCH clearly showed higher PCP-degrading activity compared with cells in the absence of exogenous TeCH, with the PCP degradation rate at 0.19 µmol L−1 hr−1 being increased to 1.68 µmol L−1 hr−1 by TeCH treatment. In this regard, it is often the case that the substrate inducing catabolic gene expression is not the initial substrate but a degradative intermediate.54–56) However, it is interesting that hcbB3 was induced by the initial substrate HCB. We optimized the conditions for the induction of hcbB3 in PD653 cells by applying TeCH, and thereafter inoculating the induced PD653 cells into sand (pH 6.19, total carbon=0.98%, total nitrogen=0.08%, C/N ratio=12.25) artificially contaminated with 2.5 mg kg−1 dry PCP. Under these conditions, after a 14-day incubation in the presence of the induced cells, we obtained a degradation rate of 56.9% compared with that of 33.0% observed using non-induced cells (Fig. 4C).
Concluding remarks
In the present study, in which we undertook a genomic analysis of Nocardioides sp. PD653, an actinobacterium capable of mineralizing HCB under aerobic conditions, we identified a group of hcbA genes encoding enzymes that catalyse the oxidative dehalogenation of HCB, and hcbB genes associated with a two-step dechlorination of PCP. The enzymes encoded by hcbA1 and hcbB3 can be classified as members of the TC-FDM family that have been reported to be involved in diverse reactions, including the degradation of pesticide molecules comprising an aromatic ring.57) Biochemical analysis revealed that HcbA3 can be classified as an NADH:FMN oxidoreductase, which we demonstrated could function as a partner reductase component for the HCB monooxygenase HcbA1 in vitro, although we postulate that there is no transient interaction between these two components. Furthermore, co-expression of hcbB genes in E. coli cells revealed that newly discovered PCP-dehalogenase HcbB3, which was similar to chlorophenol monooxygenase TftD, can exhibit the PCP-degrading activity with HcbB1 or HcbB2. It was demonstrated that induction of hcbB3 using intermediate metabolite TeCH significantly improved the PCP-degradation rate by PD653.
Acknowledgments
I would like to thank the Pesticide Science Society of Japan for presenting me with an honorary award. This research was conducted mainly at the Institute of Agro-Environmental Sciences, NARO. (formerly the National Institute for Agro-Environmental Sciences: NIAES) I would like to first thank my supervisor, Dr. Kazuhiro Takagi from NIAES, whose expertise was invaluable in formulating research questions and methodology. I acknowledge the Drs. Ryota Kataoka, Hiromasa Kiyota, Akio Iwasaki, Yu Kanesaki, and Fabrice Martin-Laurent for their valuable suggestions. I also acknowledge the members of the NIAES: Drs. Satoru Ishikawa, Masato Kuramata, and Futa Sakakibara. This work was supported by grants from the MEXT-Supported Program for the Strategic Research Foundation at Private Universities 2013 to 2017 (grant no. S1311017), a JSPS Research Fellowships for Young Scientists (17J00825), and the Cooperative Research Programme, Trade and Agriculture (TAD/PROG) OECD/2010.
References
- 1).J. L. Barber, A. J. Sweetman, D. van Wijk and K. C. Jones: Hexachlorobenzene in the global environment: Emissions, levels, distribution, trends and processes. Sci. Total Environ. 349, 1–44 (2005). [DOI] [PubMed] [Google Scholar]
- 2).http://chm.pops.int/TheConvention/ThePOPs/TheNewPOPs/tabid/2511/Default.aspx (Accessed 30, Mar., 2021)
- 3).Report of the Persistent Organic Pollutants Review Committee on the work of its ninth meeting: Risk profile on pentachlorophenol and its salts and esters. https://www.env.go.jp/council/05hoken/y051-155b/ref07.pdf (Accessed 30, Mar., 2021)
- 4).J. Beck and K. E. Hansen: The degradation of quintozene, pentachlorobenzene, hexachlorobenzene and pentachloroaniline in soil. Pestic. Sci. 5, 41–48 (1974). [Google Scholar]
- 5).S. Chaussonnerie, P. L. Saaidi, E. Ugarte, A. Barbance, A. Fossey, V. Barbe, G. Gyapay, T. Brüls, M. Chevallier, L. Couturat, S. Fouteau, D. Muselet, E. Pateau, G. N. Cohen, N. Fonknechten, J. Weissenbach and D. L. Paslier: Microbial degradation of a recalcitrant pesticide: Chlordecone. Front. Microbiol. 7, 2025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).K. Furukawa and T. Miyazaki: Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J. Bacteriol. 166, 392–398 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).R. Imai, Y. Nagata, M. Fukuda, M. Takagi and K. Yano: Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from gamma-hexachlorocyclohexane. J. Bacteriol. 173, 6811–6819 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).L. Adrian, U. Szewzyk, J. Wecke and H. Görish: Bacterial dehalorespiration with chlorinated benzenes. Nature 408, 580–583 (2000). [DOI] [PubMed] [Google Scholar]
- 9).D. Leys, L. Adrian and H. Smidt: Organohalide respiration: Microbes breathing chlorinated molecules. Philos. Trans. R. Soc. B Biol. Sci. 368, 20120316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).L. Adrian, J. Rhnenführer, J. Gobom and T. Hölscher: Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73, 7717–7724 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).M. Kube, A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt and L. Adrian: Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273 (2005). [DOI] [PubMed] [Google Scholar]
- 12).J. P. Jones, E. J. O’Hare and L. L. Wong: Oxidation of polychlorinated benzenes by genetically engineered CYP101 (cytochrome P450(cam)). Eur. J. Biochem. 268, 1460–1467 (2001). [DOI] [PubMed] [Google Scholar]
- 13).X. Chen, A. Christopher, J. P. Jones, S. G. Bell, Q. Guo, F. Xu, Z. Rao and L. L. Wong: Crystal structure of the F87W/Y96F/V247L mutant of cytochrome P-450cam with 1,3,5-trichlorobenzene bound and further protein engineering for the oxidation of pentachlorobenzene and hexachlorobenzene. J. Biol. Chem. 277, 37519–37526 (2002). [DOI] [PubMed] [Google Scholar]
- 14).D. Z. Yan, H. Liu and N. Y. Zhou: Conversion of Sphingobium chlorophenolicum ATCC 39723 to a hexachlorobenzene degrader by metabolic engineering. Appl. Environ. Microbiol. 72, 2283–2286 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).T. Liu, Z. L. Chen and Y. F. Shen: Aerobic biodegradation of hexachlorobenzene by an acclimated microbial community. Int. J. Environ. Pollut. 37, 235–244 (2009). [Google Scholar]
- 16).K. Takagi, A. Iwasaki, I. Kamei, K. Satsuma, Y. Yoshioka and N. Harada: Aerobic mineralization of hexachlorobenzene by newly isolated pentachloronitrobenzene-degrading Nocardioides sp. strain PD653. Appl. Environ. Microbiol. 75, 4452–4458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).L. Bosso and G. Cristinzio: A comprehensive overview of bacteria and fungi used for pentachlorophenol biodegradation. Rev. Environ. Sci. Biotechnol. 13, 387–427 (2014). [Google Scholar]
- 18).D. L. Saber and R. L. Crawford: Isolation and characterization of Flavobacterium strains that degrade pentachlorophenol. Appl. Environ. Microbiol. 50, 1512–1518 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).M. Cai and L. Xun: Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184, 4672–4680 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).M. A. Tiirola, H. Wang, L. Paulin and K. S. Kulomaa: Evidence for natural horizontal transfer of the pcpB gene in the evolution of polychlorophenol-degrading sphingomonads. Appl. Environ. Microbiol. 68, 4495–4501 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).K. T. Leung, S. Campbell, Y. Gan, D. C. White, H. Lee and J. T. Trevors: The role of the Sphingomonas species UG30 pentachlorophenol-4-monooxygenase in p-nitrophenol degradation. FEMS Microbiol. Lett. 173, 247–253 (1999). [DOI] [PubMed] [Google Scholar]
- 22).K. Ito, K. Takagi, A. Iwasaki, N. Tanaka, Y. Kanesaki, F. Martin-Laurent and S. Igimi: Identification of the hcb gene operon involved in catalyzing aerobic hexachlorobenzene dechlorination in Nocardioides sp. strain PD653. Appl. Environ. Microbiol. 83, e00824-e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).K. Osawa, T. Miyamoto and I. Yamamoto: Residue analysis of pentachloronitrobenzene and its related compound in soils by mass fragmentography. J. Pestic. Sci. 9, 339–344 (1984) (in Japanese). [Google Scholar]
- 24).A. C. Darling, B. Mau, F. R. Blattner and N. T. Perna: Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).K. M. Weir, T. D. Sutherland, I. Horme, R. J. Russell and J. G. Oakeshott: A single monooxygenase, ese, is involved in the metabolism of the organochlorides endosulfan and endosulfate in an Arthrobacter sp. Appl. Environ. Microbiol. 72, 3524–3530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).J. Bohuslavek, J. W. Payne, Y. Liu, H. Bolton Jr. and L. Xun: Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1. Appl. Environ. Microbiol. 67, 688–695 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).L. Xun: Purification and characterization of chlorophenol 4-monooxygenase from Burkholderia cepacia AC1100. J. Bacteriol. 178, 2645–2649 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).F. M. Duffner and R. Mueller: A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: Nucleotide sequence and analysis of the genes. FEMS Microbiol. Lett. 161, 37–45 (1998). [DOI] [PubMed] [Google Scholar]
- 29).M. A. Prieto, A. Perez-Aranda and J. L. Garcia: Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J. Bacteriol. 175, 2162–2167 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).E. A. Meighen: Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55, 123–142 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).U. Arunachalam, V. Massey and C. S. Vaidyanathan: p-Hydroxyphenylacetate-3-hydroxylase. A two-protein component enzyme. J. Biol. Chem. 267, 25848–25855 (1992). [PubMed] [Google Scholar]
- 32).H. R. Knobel, T. Egli and J. R. van der Meer: Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J. Bacteriol. 178, 6123–6132 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).E. Eichhorn, J. R. van der Ploeg and T. Leisinger: Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274, 26639–26646 (1999). [DOI] [PubMed] [Google Scholar]
- 34).S. G. Kendrew, S. E. Harding, D. A. Hopwood and E. N. Marsh: Identification of a flavin:NADH oxidoreductase involved in the biosynthesis of actinorhodin. Purification and characterization of the recombinant enzyme. J. Biol. Chem. 270, 17339–17343 (1995). [DOI] [PubMed] [Google Scholar]
- 35).K. Ito, K. Takagi, R. Kataoka and H. Kiyota: Biochemical characterization of NADH:FMN oxidoreductase HcbA3 from Nocardioides sp. PD653 in catalyzing aerobic HCB dechlorination. J. Pestic. Sci. 45, 125–131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).L. Xun and E. R. Sandvik: Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 66, 481–486 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).S. Adak and T. P. Begley: Hexachlorobenzene catabolism involves a nucleophilic aromatic substitution and flavin-N5-oxide formation. Biochemistry 58, 1181–1183 (2019). [DOI] [PubMed] [Google Scholar]
- 38).A. D. Lawrence, S. L. Taylor, A. Scott, M. L. Rowe, C. M. Johnson, S. E. J. Rigby, M. A. Geeves, R. W. Pickersgill, M. J. Howard and M. J. Warren: FAD binding, cobinamide binding and active site communication in the corrin reductase (CobR). Biosci. Rep. 34, e00120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).B. N. Webb, J. W. Ballinger, E. Kim, S. M. Belchik, K. S. Lam, B. Youn, M. S. Nissen, L. Xun and C. Kang: Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Biol. Chem. 285, 2014–2027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).B. Lei and S. C. Tu: Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry 37, 14623–14629 (1998). [DOI] [PubMed] [Google Scholar]
- 41).J. C. Low and S. C. Tu: Energy transfer evidence for in vitro and in vivo complexes of Vibrio harveyi flavin reductase P and luciferase. Photochem. Photobiol. 77, 446–452 (2003). [DOI] [PubMed] [Google Scholar]
- 42).K. Abdurachim and H. R. Ellis: Detection of protein-protein interactions in the alkanesulfonate monooxygenase system from Escherichia coli. J. Bacteriol. 188, 8153–8159 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).J. K. Lee and H. Zhao: Identification and characterization of the flavin:NADH reductase (PrnF) involved in a novel two-component arylamine oxygenase. J. Bacteriol. 189, 8556–8563 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).K. Ito, K. Takagi, Y. Matsushima, A. Iwasaki, N. Tanaka, Y. Kanesaki, F. Martin-Laurent and S. Igimi: Identification of the novel hcbB operon catalyzing the dechlorination of pentachlorophenol in the Gram-positive bacterium Nocardioides sp. strain PD653. J. Pestic. Sci. 43, 124–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).S. Mahmood, G. I. Paton and J. I. Prosser: Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ. Microbiol. 7, 1349–1360 (2005). [DOI] [PubMed] [Google Scholar]
- 46).M. Lanthier, B. Tartakovsky, R. Villemur, G. DeLuca and S. R. Guiot: Microstructure of anaerobic granules bioaugmented with Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 68, 4035–4043 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).L. Xun, E. Topp and C. S. Orser: Confirmation of oxidative dehalogenation of pentachlorophenol by a Flavobacterium pentachlorophenol hydroxylase. J. Bacteriol. 174, 5745–5747 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).K. T. Leung, S. Campbell, Y. Gan, D. C. White, H. Lee and J. T. Trevors: The role of the Sphingomonas species UG30 pentachlorophenol-4-monooxygenase in p-nitrophenol degradation. FEMS Microbiol. Lett. 173, 247–253 (1999). [DOI] [PubMed] [Google Scholar]
- 49).J. S. Uotila, M. S. Salkinoja-Salonen and J. H. A. Apajalahti: Dechlorination of pentachlorophenol by membrane bound enzymes of Rhodococcus chlorophenolicus PCP-I. Biodegradation 2, 25–31 (1991). [DOI] [PubMed] [Google Scholar]
- 50).C. S. Orser, C. C. Lange, L. Xun, T. C. Zahrt and B. J. Schneider: Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175, 411–416 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).S. Canaan, G. Sulzenbacher, V. Roig-Zamboni, L. Scapuccini-Calvo, F. Frassinetti, D. Maurin, C. Cambillau and Y. Bourne: Crystal structure of the conserved hypothetical protein Rv1155 from Mycobacterium tuberculosis. FEBS Lett. 579, 215–221 (2005). [DOI] [PubMed] [Google Scholar]
- 52).J. S. Karns, J. J. Kilbane, S. Duttagupta and A. M. Charkrabarty: Metabolism of Halophenols by 2,4,5-trichlorophenoxyacetic acid-degrading Pseudomonas cepacia. Appl. Environ. Microbiol. 46, 1176–1181 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).B. N. Webb, J. W. Ballinger, E. Kim, S. M. Belchik, K. S. Lam, B. Youn, M. S. Nissen, L. Xun and C. Kang: Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Biol. Chem. 285, 2014–2027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Y. Ohtsubo, Y. Nagata, K. Kimbara, M. Takagi and A. Ohta: Expression of the bph genes involved in biphenyl/PCB degradation in Pseudomonas sp. KKS102 induced by the biphenyl degradation intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid. Gene 256, 223–228 (2000). [DOI] [PubMed] [Google Scholar]
- 55).J. R. van der Meer, W. M. de Vos, S. Harayama and A. J. Zehnder: Microbiol. Mol. Biol. Rev. 56, 677–694 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).M. A. Schell: Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene 36, 301–309 (1985). [DOI] [PubMed] [Google Scholar]
- 57).C. W. Chu, B. Liu, N. Li, S. G. Yao, D. Cheng, J. D. Zhao, J. G. Qiu, X. Yan, Q. He and J. He: A novel aerobic degradation pathway for thiobencarb is initiated by the TmoAB two-component flavin mononucleotide-dependent monooxygenase system in Acidovorax sp. strain T1. Appl. Environ. Microbiol. 83, e01490-e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).K. Takagi: Study on the biodegradation of persistent organic pollutants (POPs). J. Pestic. Sci. 45, 119–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]