Significance
ATP-sensitive potassium channel (KATP) modulates membrane excitability according to the intracellular metabolic state: a high ATP/ADP ratio increases excitability by closing the K+ channel pore, while a low ATP/ADP ratio reduces excitability by opening it. Such intricate regulation is achieved by allowing ATP and ADP to act upon two different subunits in KATP that exert opposing effects on the channel’s gate. Previous structural studies have focused on conformations of KATP with a closed pore. This paper presents a structure of KATP with an open pore and points to a specific mechanism of allosteric regulation.
Keywords: ion channel, KATP, ATP-sensitive potassium channel
Abstract
KATP channels are metabolic sensors that translate intracellular ATP/ADP balance into membrane excitability. The molecular composition of KATP includes an inward-rectifier potassium channel (Kir) and an ABC transporter–like sulfonylurea receptor (SUR). Although structures of KATP have been determined in many conformations, in all cases, the pore in Kir is closed. Here, we describe human pancreatic KATP (hKATP) structures with an open pore at 3.1- to 4.0-Å resolution using single-particle cryo-electron microscopy (cryo-EM). Pore opening is associated with coordinated structural changes within the ATP-binding site and the channel gate in Kir. Conformational changes in SUR are also observed, resulting in an area reduction of contact surfaces between SUR and Kir. We also observe that pancreatic hKATP exhibits the unique (among inward-rectifier channels) property of PIP2-independent opening, which appears to be correlated with a docked cytoplasmic domain in the absence of PIP2.
K ATP, a K+ channel that is gated by intracellular ATP and ADP (1–7), functions in many different cells including pancreatic β-cells (6), heart (8), skeletal muscle (9), smooth muscle (10), and neurons (11). By regulating K+ permeability as a function of cytoplasmic ATP and ADP concentrations, KATP links membrane electrical excitability to a cell’s energy budget (1). In pancreatic β-cells, this KATP-mediated link couples insulin secretion to serum glucose concentration (5, 6, 12–16). KATP is thus a pharmacological target for the treatment of type II diabetes (17–19).
KATP consists of an inward-rectifier potassium channel (Kir) surrounded by four sulphonylurea receptors (SUR) that belong to the ABC transporter family (20–23) (Fig. 1A). Kir, a tetramer with four identical subunits, contains an ATP-binding site on the cytoplasmic domain (CTD) of each subunit (24–26). This site binds ATP with higher affinity than ADP (27). When ATP binds, pore closure is favored and thus the ATP site on Kir is referred to as inhibitory (1, 24). Each SUR subunit contains two adenosine nucleotide binding sites nestled in between two nucleotide binding domains (NBDs) (28, 29). These sites are formed when the NBDs engage each other (a process called dimerization) (28, 30, 31). One site, termed the degenerate site because it is incapable of mediating ATP hydrolysis, binds both ATP and ADP. The other, termed the consensus site, mediates ATP hydrolysis and favors ADP binding. Notably, and in contrast to most ABC transporters, Mg2+-ADP alone is sufficient to dimerize the NBDs (32)—and when dimerization occurs, pore opening is favored (27). The opposing influence of ATP and ADP is central to the regulation of KATP gating in cells (1).
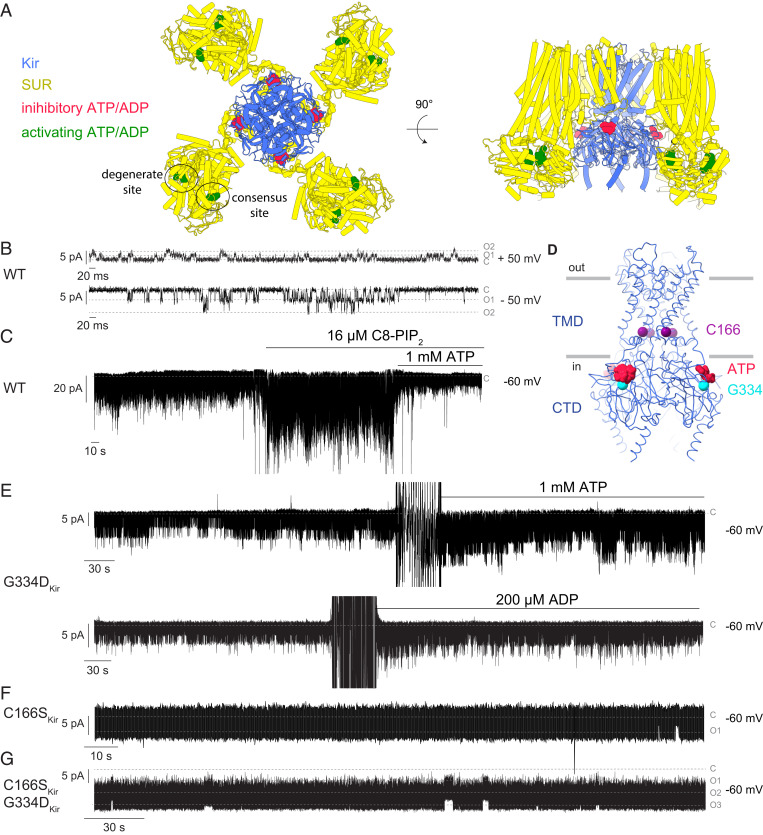
Fig. 1.
Functional validation of purified human KATP (hKir6.2-hSUR1). (A) Locations of inhibitory (red) and activating (green) ATP and ADP in KATP. Kir subunit is colored in blue, and SUR subunit is colored in yellow. In all recordings, the membranes do not contain PIP2. A total of 2 mM MgCl2 was included in recording buffers. Currents are plotted according to physiological conventions such that inward current is negative. (B) Representative single-channel recording of reconstituted WT hKATP at two membrane voltages. Current levels for closed and one and two simultaneously opened channels are labeled as C, O1, and O2. (C) WT hKATP was activated by C8-PIP2 and inhibited by ATP. Although not shown in the figure, ATP inhibition also occurs in the absence of PIP2. (D) Locations of C166 (purple spheres) and G334 (cyan spheres) in the structural model of WT hKir6.2. Inhibitory ATP is colored in red. (E) hKATP (G334DKir) was activated by both ATP and ADP. (F) Representative single-channel recording of hKATP (C166SKir). Current levels for closed and one opened channel are labeled as C and O1. (G) Representative single-channel recording of hKATP (C166SKir, G334DKir). Current levels for closed and one to three simultaneously opened channels are labeled as C, O1, O2, and O3.
Even though many molecular structures of KATP have been determined (32–39), we still cannot explain how ATP and ADP regulate the gate. All of the structures show the same closed conformation (32–39), so we cannot correlate conformational changes near the binding sites with those near the gate. In this paper, we describe a method for expressing and isolating a human pancreatic KATP (hKATP) complex composed of independent polypeptides. We show that hKATP channels in a reconstituted system exhibit physiological and pharmacological properties similar to those in cells. Then, through mutagenic alteration of the inhibitory ATP-binding site and a gate residue, we produce hKATP that exhibits high open probability and no ATP inhibition. Using single-particle cryo-electron miscroscopy (cryo-EM), we characterize hKATP with an open pore. From this structure, we correlate protein conformational changes that connect the ATP and ADP regulatory sites to the gate.
Results
Purification, Reconstitution, and Structure Determination of Human KATP.
As a first step, we developed a protocol to purify wild-type (WT) hKATP from HEK293S GnTl− cells in which both hSUR1 and hKir6.2 were coexpressed as independent polypeptides. By optimizing the amount of hSUR1, hKir6.2, and expression host cells, we obtained WT hKATP that exhibited the same gel-filtration retention volume as the fusion construct in which the C terminus of hSUR1 was covalently fused to the N terminus of hKir6.2 through a peptide linker (23, 33) (SI Appendix, Fig. S1 A and B). This suggested that the majority of purified WT hKATP molecules preserved full stoichiometry (i.e., every hKir6.2 tetramer is associated with four hSUR1 molecules).
Next, we examined whether WT hKATP channels reconstituted into synthetic lipid bilayers (40) replicate functional properties documented in cells (12, 41–43). Single-channel recordings showed a conductance of ∼78 pS at −50 mV and ∼28 pS at 50 mV (Fig. 1B), in agreement with previously reported values showing that KATP is a weak inward rectifier (44). Channel activity increased upon addition of 16 µM C8-PIP2 and decreased upon addition of 1 mM ATP (Fig. 1C), consistent with previous patch-clamp recordings showing that WT KATP is activated by PIP2 and inhibited by millimolar concentrations of ATP (41, 42). We note that, in contrast to the prevailing assumption (26), PIP2 is not absolutely required for KATP opening (Fig. 1 B and C). In this respect, KATP (Kir6.2) is distinct from classical inward rectifiers (Kir2) and G protein–gated inward rectifiers (Kir3), which exhibit barely detectable activity in the absence of PIP2 in lipid bilayers (40, 45).
Because a main aim of this study is to investigate an open conformation, we introduced two point mutations into hKir6.2 (Fig. 1D) that are known to favor the open conformation (27, 46). One of these mutations, G334D (G334DKir), is located in the CTD of Kir and abolishes ATP binding at the inhibitory site (27, 47, 48) (Fig. 1D). The second mutation, C166S (C166SKir), located at the inner helix of Kir (Fig. 1D), is known to increase the open probability of KATP in cells (46, 49). hKATP constructs with either single mutation (G334D or C166S) or both mutations (G334D and C166S) exhibited gel-filtration profiles similar to WT.
Addition of either ATP or ADP increased the open probability of the G334DKir mutant as described previously for this mutant expressed in cells (Fig. 1E) (27). This outcome is consistent with abolition of the inhibitory ATP-binding site on Kir so that activation through the SUR sites is uncovered. Because PIP2 was not added in the experiment shown (Fig. 1E) , again we observe that PIP2 is not essential for KATP opening or SUR regulation. Both the single C166SKir mutant (Fig. 1F) and the double mutant, G334DKir and C166SKir (Fig. 1G), renders KATP locked open, independent of whether KATP activators are present.
To examine the structure of this locked-open channel, we collected cryo-EM micrographs of the double mutant in the presence of 10 mM Mg2+-ATP and 0.1 mM Mg2+-ADP (SI Appendix, Fig. S1C). We used these high concentrations to promote the dimerized forms of hSUR1, which we expect are associated with activated conformations of KATP. We chose the double mutant (C166SKir, G334DKir) because the single mutant C166SKir, despite its high basal open probability (Fig. 1F), is still sensitive to ATP inhibition (46). Micrographs showed substantial heterogeneity of hKATP (C166SKir, G334DKir) particles (SI Appendix, Fig. S1C), noticeable even in 2D classes as blurred SUR components relative to Kir (SI Appendix, Fig. S1D). We first resolved the Kir component by symmetry expansion and focused refinement on hKir6.2 (C166S, G334D) (SI Appendix, Fig. S2), which yielded a 3.1-Å–resolution map (Fig. 2 A and B and SI Appendix, Figs. S2 and S3A). 3D classification indicated that nearly all particles belonged to a single class, consistent with the observation that functionally the hKir6.2 (C166S, G334D) double mutant channel is locked in an open conformation (Fig. 1G).
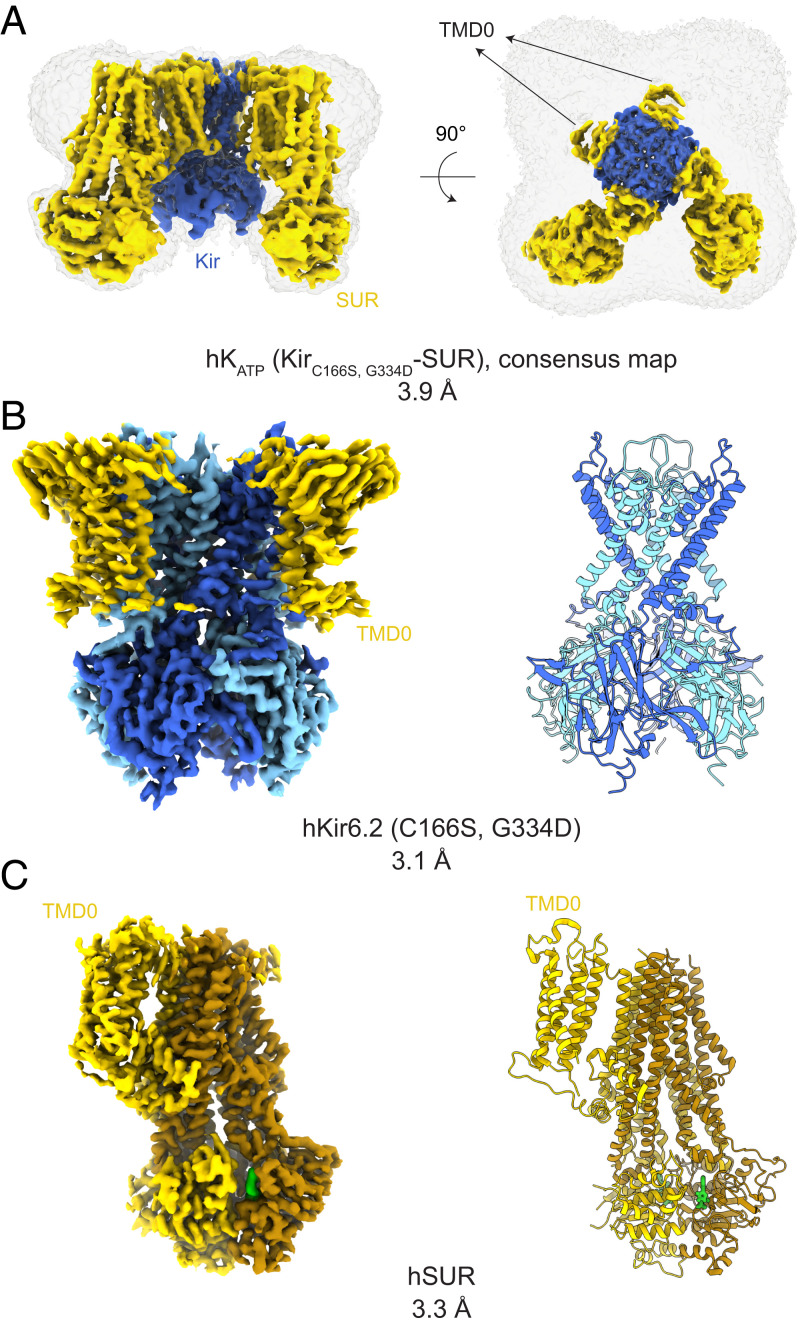
Fig. 2.
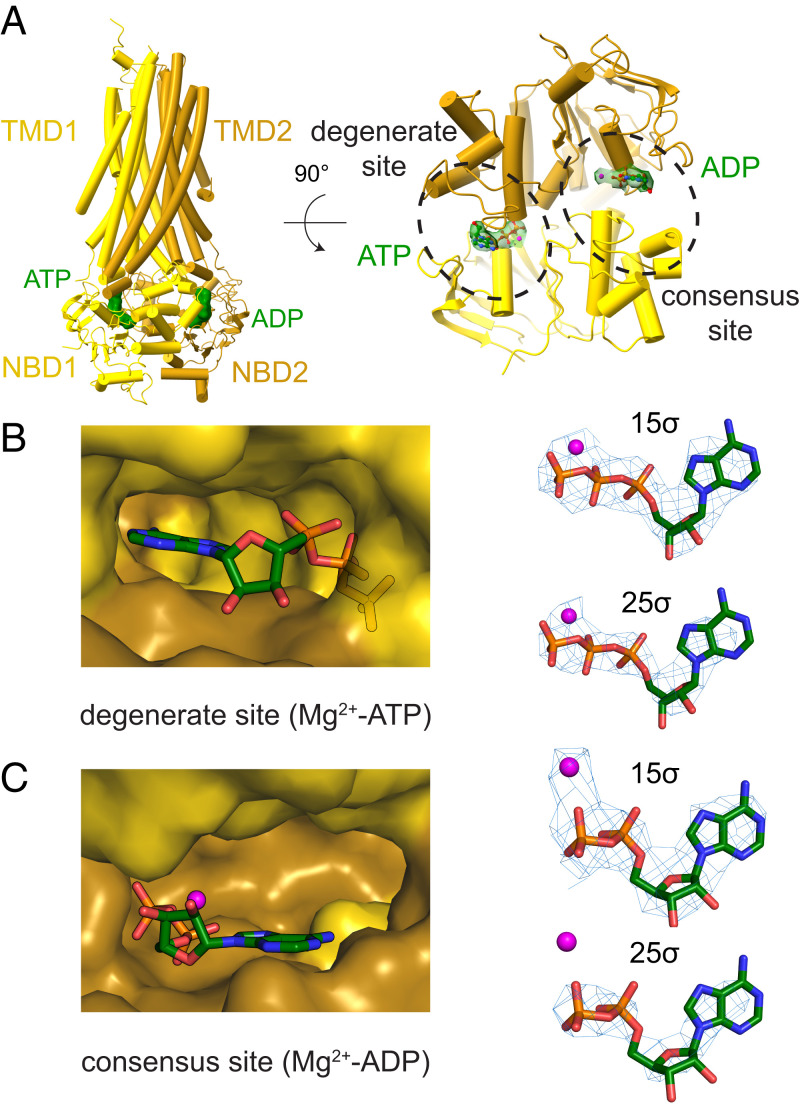
Cryo-EM map and structural models for hKATP (Kir6.2C166S, G334D-SUR1). (A) A 3.9-Å consensus cryo-EM map of KATP before symmetry expansion shows hKir6.2 (C166S, G334D) (blue) and hSUR1 (yellow). A map at low contour level covering the complete molecule is shown in gray. TMD0s from all four SURs show clear density. (B) A 3.1-Å cryo-EM map and corresponding structural model of hKir6.2 (C166S, G334D) obtained by focused refinement. Four channel subunits are colored in different shades of blue. TMD0 from SUR are colored in yellow. (C) A 3.3-Å cryo-EM map and corresponding structural model of hSUR1 obtained by focused refinement. TMD0, TMD1, and NBD1 are colored in yellow, while TMD2 and NBD2 are colored in dark brown. Mg2+-ATP and Mg2+-ADP are colored in green.
Allosteric Communication between the Inhibitory ATP Site on Kir and the Gate.
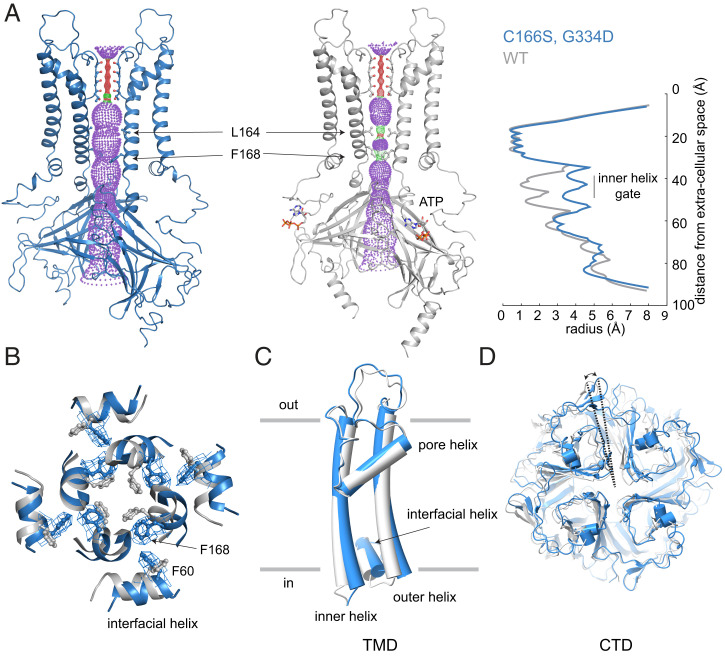
The structure of hKir6.2 (C166S, G334D) has an open pore (Fig. 3A). The radius of the ion pathway at the inner helix gate is ∼3.3 Å compared to ∼1.0 Å in hKir6.2 (WT) (Fig. 3A) (33). Expansion of the ion pathway is associated with rotation of the L164 and F168 side chains (Fig. 3 A and B) and modest conformational changes within the intracellular half of the inner and outer helices (Fig. 3C). Rotation of the CTD (Fig. 3D) is also observed, reminiscent of the “T” and “R” states of Kir6.2 described in the ATP-bound WT KATP structures with a closed pore (32, 38).
Fig. 3.
Global and local conformational changes of hKir6.2 associated with pore opening. hKir6.2 (C166S, G334D) in an ATP-free state is colored blue and hKir6.2 WT in an ATP-bound state is colored gray. (A) Comparison of the ion conduction pathway in the two structures. Kir subunits in the front and back are excluded for better visualization. Pore radii for ion conduction pathways were calculated using the program HOLE, shown as a stippled surface in structural models and plotted against distance from the extracellular surface. For analyses in (B–D), structural models were superposed according to the selectivity filter. (B) Comparison of the inner helix gate (viewed from the extracellular side). (C) Comparison of TMD. (D) Comparison of CTD (viewed from the extracellular side).
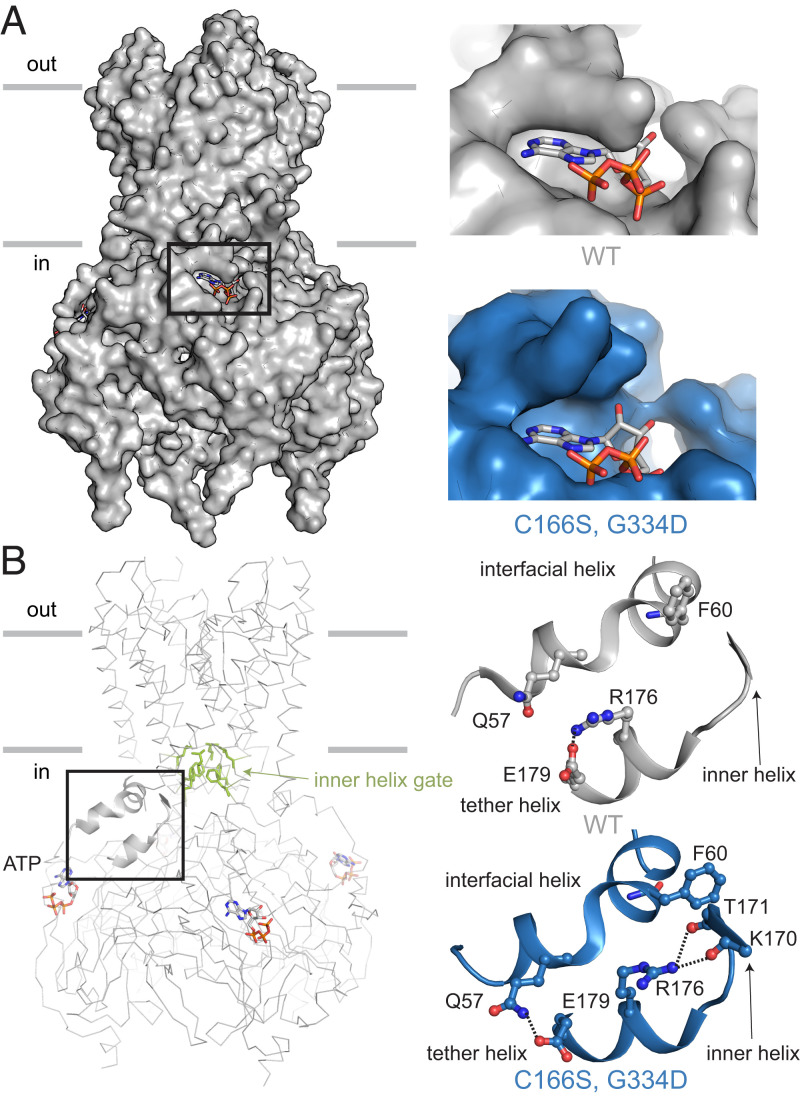
In addition to pore opening, the hKir6.2 (C166S, G334D) mutant channel exhibits structural rearrangements that involve the inhibitory ATP-binding site. Density for ATP at this site is absent (SI Appendix, Fig. S4), and the ATP-binding pocket is wider than in the ATP-bound, WT channel (Fig. 4A). Conformational rearrangements are also observed within the interfacial and tether helices that are located in between the ATP-binding site and the inner helix gate (Fig. 4B). Several conformation-dependent side chain interactions are highlighted in Fig. 4B. One of these in particular, F60 on the interfacial helix, appears well-positioned to stabilize the side chain of F168 when it rotates away from the pore axis to widen the gate (Fig. 3B). These structural differences between the WT, ATP-bound channel and the mutant without ATP bound to the inhibitory site imply that the interfacial and tether helices communicate the occupancy state of the inhibitory site to regulate the channel’s gate.
Fig. 4.
Conformational coupling between the inhibitory ATP-binding pocket and inner helix gate in Kir. hKir6.2 (C166S, G334D) in the ATP-free state is colored blue, and rKir6.2 WT in the ATP-bound state is colored gray. Structural models were superposed according to the CTD. (A) Comparison of the inhibitory ATP-binding pocket in the two structures. In hKir6.2 (C166S, G334D), no cryo-EM density for an ATP molecule was observed. (B) Comparison of the hydrogen bond network involving the interfacial helix, tether helix, and inner helix. Residues that form the inner helix gate (L164 and F168) are shown as green sticks. The inner and tether helices are from the same subunit, while the interfacial helix is from a neighboring subunit.
Molecular Architecture of hSUR1 and Its Interaction with Kir.
The structure of hSUR1 was determined using symmetry expansion with focused classification and refinement, which yielded a 3.3-Å–resolution map (Fig. 2 A and C and SI Appendix, Figs. S2 and S3B). All SUR subunits had dimerized NBDs (Fig. 5A), as in earlier reports (32, 33). Mg2+-ATP was present at the degenerate site (Fig. 5 A and B) and Mg2+-ADP at the consensus site (Fig. 5 A and C). The presence of Mg2+-ADP at the consensus site could either reflect an equilibrium preference for Mg2+-ADP or kinetic trapping after hydrolysis of ATP, as is observed in the multidrug transporter MRP1 (50). However, because the consensus site in hKATP is relatively open to solution compared to the hKATP degenerate site (Fig. 5 A–C) or the MRP1 consensus site (50), the equilibrium preference hypothesis (i.e., selectivity for Mg2+-ADP over Mg2+-ATP) seems to be a more likely explanation.
Fig. 5.
Identities of adenosine nucleotides in hSUR. (A) Overall structure of hSUR. TMD1 and NBD1 are colored yellow; TMD2 and NBD2 brown. (B) Cryo-EM densities and structural models of the degenerate site and (C) consensus site. A magnesium ion is colored in magenta; ATP or ADP molecules are shown as sticks.
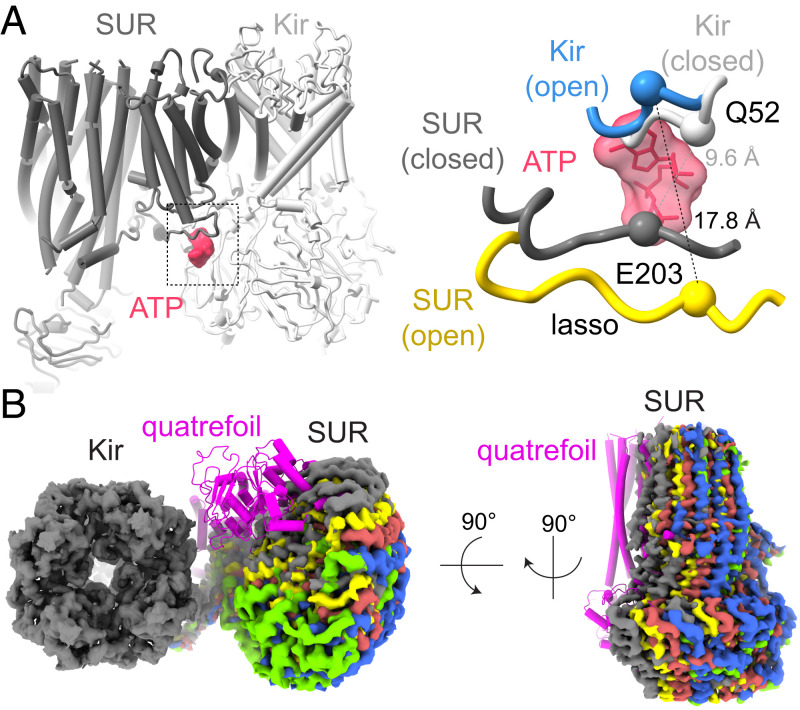
To evaluate the relationship between Kir and SUR, instead of focusing on Kir or SUR alone, we focused on the Kir tetramer and a single SUR as a whole to calculate cryo-EM maps in the 3.7- to 4.0-Å resolution range (SI Appendix, Fig. S2). Compared to previous structures in which the pore of KATP is closed (32–36, 38, 39), several differences are noted. The “lasso motif” of SUR, which contacts Kir near the intracellular membrane surface, is displaced away from Kir: the alpha carbon distance between Q52 on Kir and E203 on SUR is increased from 9.6 Å in the closed channel to 17.8 Å in the open channel (Fig. 6A). This structural difference is compatible with the finding that cross-linking this same pair of residues locks KATP closed (51).
Fig. 6.
Conformational changes in hSUR when coupled to an open pore. (A) Changes in the SUR lasso motif. Inhibitory ATP is colored red; KATP with a closed pore, gray. In KATP with an open pore, Kir is colored blue; SUR yellow. Backbone Cα atoms for Q52 (Kir) and E203 (SUR) are shown as spheres. (B) Cryo-EM density for SUR at five different positions (shown in different colors) relative to Kir. The location of SUR in the quatrefoil configuration (33) is shown in magenta.
Because the inhibitory ATP-binding site is buttressed by the lasso motif (Fig. 6A), the absence of bound ATP likely contributes to the high degree of flexibility between Kir and SUR (Fig. 6B). Moreover, it is possible that the inhibitory ATP-binding site modulates SUR-mediated regulation—and vice versa—through its ability to strengthen or weaken the interface between Kir and SUR.
Discussion
This paper presents a KATP structure with an open inner helix gate, characterized by a widening to ∼3.3-Å radius at the narrowest constriction (Fig. 3A). Although this degree of pore opening is less than that observed in open voltage-gated potassium channels (>4-Å radius) (52–54), the radius approaches that of a hydrated K+ ion (∼4-Å radius). It was assumed, prior to ever seeing the first structure of an ion channel, that an ion channel’s gate must open at least as wide as a hydrated ion. But now that we have observed many ion channel structures under conditions in which they are activated, and in many cases their pores are not wider than a hydrated ion (55–59), it seems that the original assumption might have been incorrect. A slightly narrow tunnel lined with dynamic side chains apparently can provide a good passage for ions.
We also observe that when the pore’s gate is open, the inhibitory ATP-binding site also opens—too wide for ATP to fit snugly. On structural grounds, it would thus appear that ATP should bind with higher affinity to the closed conformation of the channel. In other words, the structure is consistent with the observation that ATP binding to the inhibitory site favors the closed pore. Conformational coupling between the inhibitory site and the gate is mediated by changes in the tether and interfacial helices, which form a bridge for allosteric communication between the ATP-binding site and the gate.
In single-channel recordings, one observes that KATP exhibits bursting behavior, meaning periods of rapid opening and closing on the millisecond timescale (bursts) are separated by periods of inactivity (interbursts): the transitions between burst and interburst states reflect a slower kinetic process (Fig. 1B). Addition of ATP influences this slower kinetic process by increasing the mean interburst time and reducing the mean burst time (60). Furthermore, the C166S mutation within the inhibitory ATP-binding site reduces the mean interburst time (46). These kinetic properties, together with the structural findings in this study, lead us to conclude that the relatively slow process of transition between burst and interburst intervals in single-channel records reflects closing and opening of the inner helix gate. That ATP lengthens the interburst interval implies that ATP stabilizes the channel conformation in which ATP fits snugly into its site (i.e., the closed conformation). That ATP shortens the burst interval implies that ATP can bind in the more open site (albeit presumably with lower affinity), where it either destabilizes the open conformation or lowers a kinetic barrier in the transition to the closed conformation.
The large conformational changes involving the inner helices, the tether and interfacial helices, and the ATP-binding site are compatible with the relatively slow rates of transition between the burst and interburst states in channel records. The relatively faster transitions within a burst, which are not sensitive to ATP (60, 61), likely reflect smaller conformational changes that occur elsewhere along the ion conduction pathway (62).
The structure of SUR with bound ATP and ADP in the degenerate and consensus sites, respectively, is similar to structures determined previously (32, 33). However, the relationship between SUR and Kir is altered owing to disruption of the interfaces that connect these subunits. This is why SUR exhibited such a high degree of static disorder (Figs. 2A and 6B), necessitating focused refinement to visualize a specific orientation. The interface formed by the lasso motif near the cytoplasmic surface buttresses the inhibitory ATP site, which may be the reason why ATP influences the status of the interface. This observation leads us to speculate that occupation of the inhibitory ATP site might serve the dual function of stabilizing the closed conformation of the pore and permitting SUR to engage Kir. But how this would tie into the ability of SUR to regulate gating is unclear to us. In our view, the mechanism of SUR regulation remains as enigmatic as ever.
This study also demonstrates that Kir6.2 in pancreatic KATP is an unusual inward-rectifier potassium channel with respect to its activation by PIP2. In a reconstituted system with synthetic lipid membranes, we find that hKATP maintains some activity in the absence of PIP2 (Fig. 1 B and C), in contrast to other inward-rectifier channels such as GIRK (Kir3) and Kir2 studied in the same reconstitution system (40, 45). This functional difference between hKATP and GIRK or Kir2 has a structural correlate. In the absence of PIP2, the CTD of hKATP remains docked (i.e., engaged with the pore) (32–36, 38, 39), while the CTD in GIRK as well as in Kir2 are extended (63, 64); PIP2 binding is required to produce the docked conformation in the latter two channels (58, 64) (SI Appendix, Fig. S5). Possibly related, in pancreatic KATP, lipid densities are observed in the presumed PIP2 binding pocket in both the WT closed channel (34, 35, 38, 39) and the double mutant open channel (SI Appendix, Fig. S6). Thus, lipids other than PIP2 might support the CTD-docked conformation. We note that the recently determined Kir6.1 (in KATP from smooth muscle) adopts a CTD-undocked conformation in the PIP2-free state, similar to GIRK and Kir2 (65). Thus, the PIP2-independent basal activity might be a unique trait of the pancreatic KATP containing Kir6.2.
Materials and Methods
Cell Lines.
Sf9 cells were cultured in Sf-900 II SFM medium (GIBCO) supplemented with 100 U/mL penicillin and 100 U/mL streptomycin at 27 °C. HEK293S GnTl− cells were cultured in Freestyle 293 medium (GIBCO) supplemented with 2% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin at 37 °C.
Construct Design.
Synthetic complementary DNAs (cDNAs) encoding human SUR1 (hSUR1) and human Kir6.2 (hKir6.2) were cloned into modified pEG BacMam vectors containing a C-terminal PreScission protease cleavage site followed by enhanced green fluorescent protein (eGFP) (66). A stop codon was inserted at the C-terminal end of hSUR1 so that expressed hSUR1 had no eGFP tag. C166S and G334D point mutations were introduced using Q5 site-directed mutagenesis kit (NEB).
Protein Expression and Purification.
hKATP composed of hSUR1 and hKir6.2 was expressed in HEK293S GnTl− cells using the BacMam method. Briefly, bacmids carrying either hSUR1 or hKir6.2 were generated by transforming DH10Bac Escherichia coli cells with the corresponding pEG BacMam constructs. Baculoviruses were produced by transfecting Sf9 cells with the bacmids using Cellfectin II. Baculoviruses after two (hKir6.2) or three (hSUR1) rounds of amplification were used for cell transduction. HEK293S GnTl− cells were grown in suspension at 37 °C until they reached a density of ∼1.5 × 106 cells/mL, at which point 6% (vol/vol) hKir6.2 and 24% (vol/vol) hSUR1 baculoviruses were added. The cells were allowed to grow at 37 °C for another 12 h, before 10 mM sodium butyrate was added and the growth temperature was shifted to 30 °C. The cells were kept at 30 °C for another 24 h before harvest.
Protein purification was carried out at 4 °C and was finished within 16 h. Cell pellet from 8-L culture was resuspended in 80 mL buffer containing 20 mM Tris·HCl pH 8.5, 500 mM KCl, 0.5 mM CaCl2, 3 mM MgCl2, 2 mM ATP, 0.2 mM ADP, 2 mM DTT supplemented with 10 µg/mL leupeptin, 10 µg/mL pepstatin A, 5 µg/mL E-64, 500 µg/mL AEBSF, and ∼100 µg/mL DNase. The cell pellet was stirred until homogeneous and then gently disrupted using a Dounce homogenizer. The resulting lysate was clarified by centrifugation at 39,000 × g for 1 h. The crude membrane pellet was resuspended with a Dounce homogenizer in 120 mL buffer containing 25 mM Tris·HCl pH 8.5, 625 mM KCl, 0.625 mM CaCl2, 3.75 mM MgCl2, 2.5 mM ATP, 0.25 mM ADP, 2.5 mM DTT supplemented with 12.5 µg/mL leupeptin, 12.5 µg/mL pepstatin A, 6.25 µg/mL E-64, 625 µg/mL AEBSF, and ∼125 µg/mL DNase. The membrane suspension was then mixed with 60 mL 6% glyco-diosgenin (GDN) (m/v, dissolved in H2O) to bring the final concentration of GDN to 1.2% (crude membrane pellet was about 20 mL). The mixture was stirred for 2 h and then the solubilized membranes were clarified by centrifugation at 39,000 × g for 30 min. The resulting supernatant was incubated with GFP nanobody-coupled Sepharose resin (prepared in-house) (67) for 2 h. The resin was then washed with 10 column volumes of wash buffer containing 20 mM Tris·HCl pH 8.5, 300 mM KCl, 3 mM MgCl2, 2 mM ATP, 0.2 mM ADP, 2 mM DTT, and 0.006% GDN (m/v). The resin was resuspended with one column volume of wash buffer and PreScission protease (prepared in-house) was added to the resin to a final concentration of ∼0.08 mg/mL. After 1.5 h, eluted protein was collected from the flow-through and combined with subsequent washes with four column volumes of wash buffer. The protein was then concentrated to ∼800 µL using a 15-mL Amicon concentrator with 100-kDa molecular weight cutoff. Concentrated protein was then clarified by centrifugation at 25,000 × g for 2 min before being further purified by size-exclusion chromatography using Superose 6 Increase column (10/300 GL). Because samples for reconstitution and for cryo-EM grid preparation were purified with different size-exclusion buffers, the buffer compositions will be described in individual sections.
Reconstitution and Bilayer Recording.
Reconstitution of hKATP was accomplished by two methods. For both methods, a lipid mixture composed of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) at 2:1:1 (w:w:w) was used. Briefly, different lipid stocks were mixed in chloroform, washed by pentane twice, dried under an argon stream, and stored at −20 °C. Before use, dried lipid was warmed up under vacuum overnight and was then hydrated to 25 mg/mL by gently rotating with H2O at room temperature. After the lipid was fully dispersed in H2O, it was sonicated using a water bath sonicator until opalescent.
In the first reconstitution method, hKATP was purified with a size-exclusion buffer containing 20 mM Tris·HCl pH 8.5, 300 mM KCl, 2 mM DTT, and 0.00105% GDN (m/v). GDN was present at only 0.5 critical micelle concentration (CMC) to facilitate its future removal. Subsequently, the peak fractions were pooled and were concentrated to 1 to 2 mg/mL using a 2-mL Amicon concentrator with 100-kDa molecular weight cutoff. After being concentrated in 0.5 CMC GDN, the majority of protein sample was still monodispersed as confirmed by fluorescence-detection size-exclusion chromatography using tryptophan fluorescence. For this reconstitution method, sonicated lipid vesicles were destabilized with C12E10 by incubating them at room temperature for 1 h using 20 mg/mL lipid (vesicles), 10 mM K-HEPES pH 7.5, 300 mM KCl, and 1% C12E10. C12E10 was chosen at a concentration that was not able to fully solubilize lipid vesicles. C12E10-destabilized vesicles (20 mg/mL) were then mixed with hKATP (1-2 mg/mL in 0.00105% GDN) at 1:1 volume ratio and then incubated at 4 °C for 1 h. The protein–lipid mixture (normally 100 to 500 µL) was then dialyzed (dialysis tubing with 50-kDa molecular weight cutoff) against 2 L buffer containing 10 mM K-HEPES pH 7.5, 300 mM KCl, and 2 mM DTT, with bio-beads (Bio-Rad) in the buffer at about 0.5 g bio-beads per liter of dialysis buffer. The dialysis was performed at 4 °C. The dialysis buffer was exchanged every 6 h twice, after which (∼18 h postpurification) the protein–lipid mixture was directly mixed with bio-beads at a ratio of 70 mg bio-beads (wet weight) per 100 µL protein–lipid sample. The protein–lipid sample was incubated with bio-beads at 4 °C for 16 h and then aliquoted, flash frozen under liquid nitrogen, and stored at −80 °C.
In the second reconstitution method, hKATP was purified with a size-exclusion buffer containing 20 mM Tris·HCl pH 8.5, 300 mM KCl, 2 mM DTT, and 0.006% GDN (m/v), after which the peak fractions were pooled and concentrated to 1 to 2 mg/mL using a 2-mL Amicon concentrator with 100-kDa molecular weight cutoff. Sonicated lipid vesicles were destabilized with GDN by incubating them at room temperature for 2 h in 20 mg/mL lipid (vesicles), 10 mM K-HEPES pH 7.5, 300 mM KCl, and 2% GDN. GDN was used at a high concentration in order to fully solubilize the lipids into mixed micelles. Then, GDN-solubilized lipids (20 mg/mL) were mixed with hKATP (1 to 2 mg/mL in 0.006% GDN) at 1:1 volume ratio and then incubated at 4 °C overnight (∼8 to 12 h) to allow thorough mixing of protein micelles and lipid-GDN mixed micelles. Then, GDN was removed by adding shots of methyl-β-cyclodextrin (MBCD) solution every 6 to 8 h for four times. Every shot of MBCD solution contained 18 mM MBCD (in 10 mM K-HEPES pH 7.5 and 300 mM KCl) with the same volume as the initial volume of GDN-solubilized lipid used to mix with protein such that the molar ratio between every shot of MBCD and GDN was 1.2. Therefore, in the end, the absolute amount of MBCD in molar units was 4.8 times that of GDN, and the total volume was diluted threefold. Then, the vesicles were pelleted by ultracentrifugation at 48,000 rpm for 10 min and resuspended in a buffer containing 10 mM K-HEPES pH 7.5, 300 mM KCl, and 2 mM DTT. Finally, vesicles were aliquoted, flash frozen under liquid nitrogen, and stored at −80 °C.
Both reconstitution methods produced functional KATP current in synthetic lipid membranes. However, as it is not clear how much MBCD was carried over in the second method, we normally used the first method for KATP reconstitution. All recordings reported in this work were acquired using protoliposomes produced with the first reconstitution method.
Bilayer recordings were performed with a recording chamber as described previously (40). Briefly, 20 mg/mL lipid solution in decane composed of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) at 2:1:1 (w:w:w) was painted over a ∼100-µm hole on a piece of polyethylene terephthalate transparency film that separated two sides of the recording chamber made of polyoxymethylene. The recording buffer, composed of 10 mM K-HEPES pH 7.5, 150 mM KCl, and 2 mM MgCl2, was added to both sides of the recording chamber. Before bilayer recordings, a frozen protoliposome aliquot was thawed on ice and then KCl solution was added to a final concentration of 1 M. The protoliposomes were sonicated in a water bath sonicator at room temperature briefly before being added to the lipid bilayer. Voltage across the lipid bilayer was controlled with an Axopatch 200B amplifier in whole-cell mode. The analog current signal was lowpass filtered at 1 kHz (Bessel) and digitized at 10 kHz with a Digidata 1440A digitizer. Digitized data were recorded with software pClamp (Molecular Devices). The orientations of the channels were determined by rectification (difference in unitary conductance for inward and outward currents). Bilayer membranes were useful if the CTDs of channels faced the accessible side of the recording chamber. To investigate how channel current responded to different ligands, ligand stock solution was mixed with the recording buffer (600 µL). The stock concentrations for ligand solutions were as follows: C8-PIP2 (10 mg/mL in H2O), ATP (500 mM in H2O, pH adjusted to 7.5 with KOH), and ADP (12 mM in H2O, pH adjusted to 7.5 with KOH).
Cryo-EM Grid Preparation and Data Acquisition.
For cryo-EM grid preparation, hKATP (C166SKir, G334DKir) was purified with a size-exclusion buffer containing 20 mM Tris·HCl pH 8.5, 150 mM KCl, 11 mM MgCl2, 1 mM ATP, 0.2 mM ADP, 2 mM DTT, and 0.006% GDN (m/v). Only the fractions corresponding to the left half of the peak were pooled (12 to 13 mL in SI Appendix, Fig. S1 A and B) and concentrated to 6.83 mg/mL using a 0.5-mL Amicon concentrator with 100-kDa molecular weight cutoff. Then, ATP stock solution (500 mM, dissolved in H2O, pH adjusted to 7.5 with KOH) was added to a concentrated hKATP (C166SKir, G334DKir) sample directly to achieve a final concentration of 10 mM. The sample was incubated on ice for 30 min before grid preparation.
To prepare cryo-EM grids, 3.5 µL sample was pipetted onto glow-discharged Quantifoil R1.2/1.3 400 mesh holey carbon Au grids. After 20 s at 16 °C with a humidity of 100%, the grids were blotted for 1.5 s with a blotting force of 0 and flash frozen in liquid ethane using an FEI Vitrobot Mark IV (FEI).
For cryo-EM data acquisition, grids were loaded onto a 300-keV Titan Krios transmission electron microscope with a Gatan K2 Summit detector. A total of 1,624 micrographs were recorded in superresolution mode with a 100-µm objective aperture using SerialEM (68). The micrographs had a physical pixel size of 1.3 Å (superresolution pixel size of 0.65 Å) and a nominal defocus range of 1.0 to 2.0 µm. The micrographs were recorded with 12-s exposure (0.2 s per frame) with a dose rate of 8 e−/pixel/s, which gave a total cumulative dose of ∼57 e−/Å2 (0.95 e−/Å2/frame).
Cryo-EM Image Processing.
Superresolution image stacks were gain-normalized, binned by 2 with Fourier cropping, and corrected for beam-induced motion using MotionCor2 (69). Parameters for the contrast transfer function were estimated from motion-corrected summed images without dose-weighting using the program GCTF (70). All subsequent cryo-EM image processing was performed on dose-weighted motion-corrected summed images. A total of 633,136 particles were picked by Laplacian-of-Gaussian auto-picking in RELION 3.0 (71). Particles were then cleaned by four rounds of 2D classification in cryoSPARC2 (72), leaving 212,341 particles, which were then subjected to ab initio reconstruction in C1 requesting two classes (cryoSPARC2). The model for the good class was first manually rotated to align with the C4 symmetry axis in Chimera (73), then particles belonging to the good class (151,967) were refined in C1 against the rotated reference model by nonuniform refinement (74) using cryoSPARC2. Bayesian particle polishing in RELION (75) using the consensus model after nonuniform refinement did not improve the resolution or the cryo-EM map. And because using improved models from subsequent data processing steps for Bayesian particle polishing also had little improvement on the resulting cryo-EM map, no Bayesian particle polishing was performed for the entire data processing.
To calculate a cryo-EM map for hKir6.2 by focused refinement, after obtaining the consensus model, the particles were expanded fourfold by C4 symmetry expansion in RELION (76). The symmetry-expanded particles (607,868) were reimported into cryoSPARC2 for refinement. The particles were first refined locally (using nonuniform refinement option) with a loose mask covering both the protein and micelle density and then further refined locally (using the nonuniform refinement option) with a tight mask covering hKir6.2 and four TMD0 densities in hSUR1.
To calculate a cryo-EM map for hSUR1, local refinement (with nonuniform refinement option) was carried out with a mask covering hSUR1. The resulting density was not well-resolved and had many broken regions, suggesting structural heterogeneity. Therefore, particles were sorted by 3D classification in RELION without alignment using a tight mask covering hSUR1. Particles belonging to the good class (144,706) were further refined in cryoSPARC2 using nonuniform local refinement with the same mask used for 3D classification.
To obtain cryo-EM maps for hKir6.2-hSUR1, the particles belonging to the good hSUR1 class were first refined locally (using the nonuniform refinement option) in cryoSPARC2 while applying a mask covering hKir6.2 and four TMD0 densities in SUR. This operation ensured that hKir6.2 was aligned in all particles. Then, 3D classification without alignment in RELION was carried out using a mask covering hKir6.2, four TMD0 densities in SUR, plus a large volume surrounding one of the SURs, assuming that this region covered all possible SUR locations. This procedure resulted in five good classes having 23,783, 24,689, 18,583, 28,719, and 18,522 particles, which belonged to positions 1 to 5 in Fig. 6B and SI Appendix, Fig. S2. These five classes were first locally refined in cryoSPARC2 (using the nonuniform refinement option) with the same mask used for 3D classification. However, further nonuniform local refinement using masks either covering hKir6.2, four SUR TMD0s and the lasso motif, or covering SUR TMD1-2 and SUR NBDs resulted in maps with higher resolution and better quality. Therefore, we used maps belonging to these two halves of SUR for model building.
For all cryo-EM maps, the mask-corrected Fourier shell correlation (FSC) curves were calculated in cryoSPARC2, and the resolutions were reported based on the 0.143 criterion (77). Local resolutions were calculated using cryoSPRAC2.
Initial models were obtained by docking components from the hKATP structure (Protein Data Bank [PDB] ID: 6C3P) solved with a fusion construct (33) into the cryo-EM maps. The models were then iteratively edited and refined in Coot (78) and PHENIX (79). The quality of the final models was evaluated by MolProbity (80). All figures were prepared using PyMOL (81), Chimera (73), and ChimeraX (82).
Supplementary Material
Acknowledgments
We thank Mark Ebrahim, Johanna Sotiris, and Honkit Ng at the Evelyn Gruss Lipper Cryo-EM Resource Center of Rockefeller University for assistance with cryo-EM data collection, Dr. Chia-Hsueh Lee for advice in cryo-EM sample preparation and data processing, Dr. Weiwei Wang for advice in bilayer recording, Dr. Kenneth Pak Kin Lee and Dr. Xiao Tao for advice in general biochemistry and electrophysiology, Yi Chun Hsiung for assistance with insect and mammalian cell cultures, Dr. Jue Chen for advice on SUR structure and ABC transporter mechanism, and the R.M. laboratory and C.Z. laboratory members for helpful discussions. C.Z. is supported by the Jane Coffin Childs Memorial Fund Fellowship. R.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112267118/-/DCSupplemental.
Data Availability
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB), and the coordinates have been deposited in the PDB under the following accession numbers: focused on Kir (C166S G334D) (EMD-24839, PDB ID 7S5T), focused on SUR (EMD-24840, PDB ID 7S5V), focused on Kir and SUR, position 1 (EMD-24842, PDB ID 7S5X), position 2 (EMD-24843, PDB ID 7S5Y), position 3 (EMD-24844, PDB ID 7S5Z), position 4 (EMD-24845, PDB ID 7S60), and position 5 (EMD-24846, PDB ID 7S61).
References
- 1.Nichols C. G., KATP channels as molecular sensors of cellular metabolism. Nature 440, 470–476 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Tantama M., Martínez-François J. R., Mongeon R., Yellen G., Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 4, 2550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kakei M., Kelly R. P., Ashcroft S. J., Ashcroft F. M., The ATP-sensitivity of K+ channels in rat pancreatic B-cells is modulated by ADP. FEBS Lett. 208, 63–66 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Dunne M. J., Petersen O. H., Intracellular ADP activates K+ channels that are inhibited by ATP in an insulin-secreting cell line. FEBS Lett. 208, 59–62 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Rorsman P., Trube G., Glucose dependent K+-channels in pancreatic beta-cells are regulated by intracellular ATP. Pflugers Arch. 405, 305–309 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Cook D. L., Hales C. N., Intracellular ATP directly blocks K+ channels in pancreatic B-cells. Nature 311, 271–273 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Lederer W. J., Nichols C. G., Nucleotide modulation of the activity of rat heart ATP-sensitive K+ channels in isolated membrane patches. J. Physiol. 419, 193–211 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noma A., ATP-regulated K+ channels in cardiac muscle. Nature 305, 147–148 (1983). [DOI] [PubMed] [Google Scholar]
- 9.Spruce A. E., Standen N. B., Stanfield P. R., Voltage-dependent ATP-sensitive potassium channels of skeletal muscle membrane. Nature 316, 736–738 (1985). [DOI] [PubMed] [Google Scholar]
- 10.Standen N. B., et al. , Hyperpolarizing vasodilators activate ATP-sensitive K+ channels in arterial smooth muscle. Science 245, 177–180 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Karschin C., Ecke C., Ashcroft F. M., Karschin A., Overlapping distribution of K(ATP) channel-forming Kir6.2 subunit and the sulfonylurea receptor SUR1 in rodent brain. FEBS Lett. 401, 59–64 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Ashcroft F. M., Kakei M., Kelly R. P., Sutton R., ATP-sensitive K+ channels in human isolated pancreatic B-cells. FEBS Lett. 215, 9–12 (1987). [DOI] [PubMed] [Google Scholar]
- 13.Tarasov A., Dusonchet J., Ashcroft F., Metabolic regulation of the pancreatic beta-cell ATP-sensitive K+ channel: A pas de deux. Diabetes 53 (suppl. 3), S113–S122 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Ashcroft F. M., ATP-sensitive potassium channelopathies: Focus on insulin secretion. J. Clin. Invest. 115, 2047–2058 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koster J. C., Permutt M. A., Nichols C. G., Diabetes and insulin secretion: The ATP-sensitive K+ channel (K ATP) connection. Diabetes 54, 3065–3072 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Ashcroft F. M., The Walter B. Cannon Physiology in Perspective Lecture, 2007. ATP-sensitive K+ channels and disease: From molecule to malady. Am. J. Physiol. Endocrinol. Metab. 293, E880–E889 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Sturgess N. C., Ashford M. L., Cook D. L., Hales C. N., The sulphonylurea receptor may be an ATP-sensitive potassium channel. Lancet 2, 474–475 (1985). [DOI] [PubMed] [Google Scholar]
- 18.Rubaiy H. N., The therapeutic agents that target ATP-sensitive potassium channels. Acta Pharm. 66, 23–34 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Feldman J. M., Glyburide: A second-generation sulfonylurea hypoglycemic agent. History, chemistry, metabolism, pharmacokinetics, clinical use and adverse effects. Pharmacotherapy 5, 43–62 (1985). [DOI] [PubMed] [Google Scholar]
- 20.J. P. Clement, 4th, et al. , Association and stoichiometry of K(ATP) channel subunits. Neuron 18, 827–838 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Inagaki N., Gonoi T., Seino S., Subunit stoichiometry of the pancreatic beta-cell ATP-sensitive K+ channel. FEBS Lett. 409, 232–236 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Shyng S., Nichols C. G., Octameric stoichiometry of the KATP channel complex. J. Gen. Physiol. 110, 655–664 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikhailov M. V., et al. , 3-D structural and functional characterization of the purified KATP channel complex Kir6.2-SUR1. EMBO J. 24, 4166–4175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucker S. J., et al. , Molecular determinants of KATP channel inhibition by ATP. EMBO J. 17, 3290–3296 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antcliff J. F., Haider S., Proks P., Sansom M. S., Ashcroft F. M., Functional analysis of a structural model of the ATP-binding site of the KATP channel Kir6.2 subunit. EMBO J. 24, 229–239 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibino H., et al. , Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol. Rev. 90, 291–366 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Proks P., de Wet H., Ashcroft F. M., Activation of the K(ATP) channel by Mg-nucleotide interaction with SUR1. J. Gen. Physiol. 136, 389–405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aittoniemi J., et al. , Review. SUR1: A unique ATP-binding cassette protein that functions as an ion channel regulator. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 257–267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vedovato N., Ashcroft F. M., Puljung M. C., The nucleotide-binding sites of SUR1: A mechanistic model. Biophys. J. 109, 2452–2460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldham M. L., Davidson A. L., Chen J., Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkens S., Structure and mechanism of ABC transporters. F1000Prime Rep. 7, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J. X., et al. , Ligand binding and conformational changes of SUR1 subunit in pancreatic ATP-sensitive potassium channels. Protein Cell 9, 553–567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K. P. K., Chen J., MacKinnon R., Molecular structure of human KATP in complex with ATP and ADP. eLife 6, e32481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li N., et al. , Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Martin G. M., Kandasamy B., DiMaio F., Yoshioka C., Shyng S. L., Anti-diabetic drug binding site in a mammalian KATP channel revealed by Cryo-EM. eLife 6, e31054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin G. M., et al. , Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. eLife 6, e24149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puljung M. C., Cryo-electron microscopy structures and progress toward a dynamic understanding of KATP channels. J. Gen. Physiol. 150, 653–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding D., Wang M., Wu J. X., Kang Y., Chen L., The structural basis for the binding of repaglinide to the pancreatic KATP channel. Cell Rep. 27, 1848–1857.e1844 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Martin G. M., et al. , Mechanism of pharmacochaperoning in a mammalian KATP channel revealed by cryo-EM. eLife 8, e46417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W., Whorton M. R., MacKinnon R., Quantitative analysis of mammalian GIRK2 channel regulation by G proteins, the signaling lipid PIP2 and Na+ in a reconstituted system. eLife 3, e03671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shyng S. L., Nichols C. G., Membrane phospholipid control of nucleotide sensitivity of KATP channels. Science 282, 1138–1141 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Baukrowitz T., et al. , PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science 282, 1141–1144 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Hilgemann D. W., Ball R., Regulation of cardiac Na+, Ca2+ exchange and KATP potassium channels by PIP2. Science 273, 956–959 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Inagaki N., et al. , Reconstitution of IKATP: An inward rectifier subunit plus the sulfonylurea receptor. Science 270, 1166–1170 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Tao X., MacKinnon R., Correlation between structure and function in phosphatidylinositol lipid-dependent Kir2.2 gating. bioRxiv [Preprint] (16 February 2021). 10.1101/2021.02.15.431350. [DOI] [PMC free article] [PubMed]
- 46.Trapp S., Proks P., Tucker S. J., Ashcroft F. M., Molecular analysis of ATP-sensitive K channel gating and implications for channel inhibition by ATP. J. Gen. Physiol. 112, 333–349 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drain P., Li L., Wang J., KATP channel inhibition by ATP requires distinct functional domains of the cytoplasmic C terminus of the pore-forming subunit. Proc. Natl. Acad. Sci. U.S.A. 95, 13953–13958 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masia R., et al. , An ATP-binding mutation (G334D) in KCNJ11 is associated with a sulfonylurea-insensitive form of developmental delay, epilepsy, and neonatal diabetes. Diabetes 56, 328–336 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Usher S. G., Ashcroft F. M., Puljung M. C., Nucleotide inhibition of the pancreatic ATP-sensitive K+ channel explored with patch-clamp fluorometry. eLife 9, e52775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., et al. , Characterization of the kinetic cycle of an ABC transporter by single-molecule and cryo-EM analyses. eLife 9, e56451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratt E. B., Zhou Q., Gay J. W., Shyng S. L., Engineered interaction between SUR1 and Kir6.2 that enhances ATP sensitivity in KATP channels. J. Gen. Physiol. 140, 175–187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long S. B., Tao X., Campbell E. B., MacKinnon R., Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Long S. B., Campbell E. B., Mackinnon R., Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Long S. B., Campbell E. B., Mackinnon R., Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science 309, 903–908 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Sun J., MacKinnon R., Structural basis of human KCNQ1 modulation and gating. Cell 180, 340–347.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park E., MacKinnon R., Structure of the CLC-1 chloride channel from Homo sapiens. eLife 7, e36629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whorton M. R., MacKinnon R., X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature 498, 190–197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen S. B., Tao X., MacKinnon R., Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee C. H., MacKinnon R., Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures. Science 360, 508–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L., Geng X., Drain P., Open state destabilization by ATP occupancy is mechanism speeding burst exit underlying KATP channel inhibition by ATP. J. Gen. Physiol. 119, 105–116 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Enkvetchakul D., Loussouarn G., Makhina E., Shyng S. L., Nichols C. G., The kinetic and physical basis of K(ATP) channel gating: Toward a unified molecular understanding. Biophys. J. 78, 2334–2348 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Proks P., Capener C. E., Jones P., Ashcroft F. M., Mutations within the P-loop of Kir6.2 modulate the intraburst kinetics of the ATP-sensitive potassium channel. J. Gen. Physiol. 118, 341–353 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao X., Avalos J. L., Chen J., MacKinnon R., Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 326, 1668–1674 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niu Y., Tao X., Touhara K. K., MacKinnon R., Cryo-EM analysis of PIP2 regulation in mammalian GIRK channels. eLife 9, e60552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sung M. W., et al. , Vascular KATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides. Proc. Natl. Acad. Sci. U.S.A. 118, e2109441118 (2021). [DOI] [PMC free article] [PubMed]
- 66.Goehring A., et al. , Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirchhofer A., et al. , Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Zivanov J., Nakane T., Scheres S. H. W., A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheres S. H., Processing of structurally heterogeneous Cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Scheres S. H., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8 (2015).
- 82.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB), and the coordinates have been deposited in the PDB under the following accession numbers: focused on Kir (C166S G334D) (EMD-24839, PDB ID 7S5T), focused on SUR (EMD-24840, PDB ID 7S5V), focused on Kir and SUR, position 1 (EMD-24842, PDB ID 7S5X), position 2 (EMD-24843, PDB ID 7S5Y), position 3 (EMD-24844, PDB ID 7S5Z), position 4 (EMD-24845, PDB ID 7S60), and position 5 (EMD-24846, PDB ID 7S61).