Significance
Sleep loss disrupts consolidation of hippocampus-dependent memory. To understand the cellular basis for this effect, we quantified RNAs associated with translating ribosomes in cytosol and on cellular membranes of different hippocampal neuron populations. Our analysis suggests that while sleep loss (but not learning) alters numerous ribosomal transcripts in cytosol, learning has dramatic effects on transcript profiles for less–well-characterized membrane-bound ribosomes. We demonstrate that postlearning sleep deprivation occludes already minimal learning-driven changes on cytosolic ribosomes. It simultaneously alters transcripts associated with metabolic and biosynthetic processes in membrane-bound ribosomes in excitatory hippocampal neurons and highly active, putative “engram” neurons, respectively. Together, these findings provide insights into the cellular mechanisms altered by learning and their disruption by subsequent sleep loss.
Keywords: synaptic plasticity, bioinformatics, memory consolidation, ribosomes, translation
Abstract
The hippocampus is essential for consolidating transient experiences into long-lasting memories. Memory consolidation is facilitated by postlearning sleep, although the underlying cellular mechanisms are largely unknown. We took an unbiased approach to this question by using a mouse model of hippocampally mediated, sleep-dependent memory consolidation (contextual fear memory). Because synaptic plasticity is associated with changes to both neuronal cell membranes (e.g., receptors) and cytosol (e.g., cytoskeletal elements), we characterized how these cell compartments are affected by learning and subsequent sleep or sleep deprivation (SD). Translating ribosome affinity purification was used to profile ribosome-associated RNAs in different subcellular compartments (cytosol and membrane) and in different cell populations (whole hippocampus, Camk2a+ neurons, or highly active neurons with phosphorylated ribosomal subunit S6 [pS6+]). We examined how transcript profiles change as a function of sleep versus SD and prior learning (contextual fear conditioning; CFC). While sleep loss altered many cytosolic ribosomal transcripts, CFC altered almost none, and CFC-driven changes were occluded by subsequent SD. In striking contrast, SD altered few transcripts on membrane-bound (MB) ribosomes, while learning altered many more (including long non-coding RNAs [lncRNAs]). The cellular pathways most affected by CFC were involved in structural remodeling. Comparisons of post-CFC MB transcript profiles between sleeping and SD mice implicated changes in cellular metabolism in Camk2a+ neurons and protein synthesis in highly active pS6+ (putative “engram”) neurons as biological processes disrupted by SD. These findings provide insights into how learning affects hippocampal neurons and suggest that the effects of SD on memory consolidation are cell type and subcellular compartment specific.
The role of sleep in promoting synaptic plasticity and memory consolidation is an enduring mystery (1). For the past two decades, transcriptomic (2–6) and proteomic (7–10) profiling of the mammalian brain after sleep versus sleep deprivation (SD) has provided insights regarding general functions of sleep for the brain. Such work provided the initial basis for the synaptic homeostasis (SHY) hypothesis for sleep function (2), which proposes that synapses are broadly “downscaled” during sleep. However, the function of such a process in memory consolidation, and its occurrence during postlearning sleep, remains a matter of debate (1, 11). Observations from in vivo electrophysiology have led to the conclusion that specific patterns of activity present during learning experiences may be replayed during subsequent sleep. Such a mechanism could be instructive with regard to memory storage [i.e., by selectively reactivating “engram neurons” engaged during prior learning (12) and promoting synaptic plasticity in those neurons]. Indeed, recent findings suggest that in structures such as sensory cortex, reactivation of engram neurons during the first few hours of postlearning sleep plays a critical role in consolidating new memories (12). However, while recent work has demonstrated long-lasting transcriptional changes in engram neurons (13), it remains unknown how sleep-associated reactivation of these neurons (and other features of brain physiology associated with sleep) would affect intracellular pathways involved in synaptic plasticity (1, 14).
More recently, transcriptomic and proteomic profiling of synaptic or axonal organelles has been used to better understand the effects of learning (15) or of sleep versus wake (10) on synaptic function. However, to date, there has been no experimental work aimed at characterizing cellular changes during sleep-dependent memory consolidation—that is, those occurring during postlearning sleep. Here, we use a well-established mouse model of sleep-dependent memory consolidation—contextual fear memory (CFM) —to study this process. CFM can be encoded in a single learning trial (contextual fear conditioning [CFC]; placement in a novel environmental chamber followed by a foot shock). Memory for this context-shock pairing is consolidated via hippocampus-dependent mechanisms over the next few hours (16). Critically, CFM consolidation can be disrupted by SD over the first 5 to 6 h following CFC (17–21). Over this same post-CFC interval, disruption of either neuronal activity (16), transcription (22, 23), or translation (24–27) in the dorsal hippocampus can likewise disrupt consolidation. This suggests that an activity- and sleep-dependent mechanism, impinging on biosynthetic pathways in the hippocampus, is essential for consolidation.
To shed light on these mechanisms, we characterized changes to ribosome-associated messenger RNA (mRNAs) from different hippocampal cell populations [including all Camk2a+ excitatory neurons and the subset of highly active hippocampal neurons expressing Ser244/247 phosphorylated S6 (pS6+) (28, 29)] as a function of both sleep versus SD and prior CFC. By quantifying mRNA profiles on ribosomes differentially localized to the cytosol and cellular membranes, we find that while the majority of changes to transcripts on cytosolic ribosomes vary as a function of sleep versus wake, the majority of transcript changes on membrane-bound (MB) ribosomes vary as a function of learning. Our findings reveal subcellular functions for postlearning sleep and suggest cellular mechanisms by which sleep could selectively promote memory storage.
Results
Translating Ribosome Affinity Purification Isolates Cytosolic and MB Ribosomal Transcripts from Different Hippocampal Neuron Subpopulations.
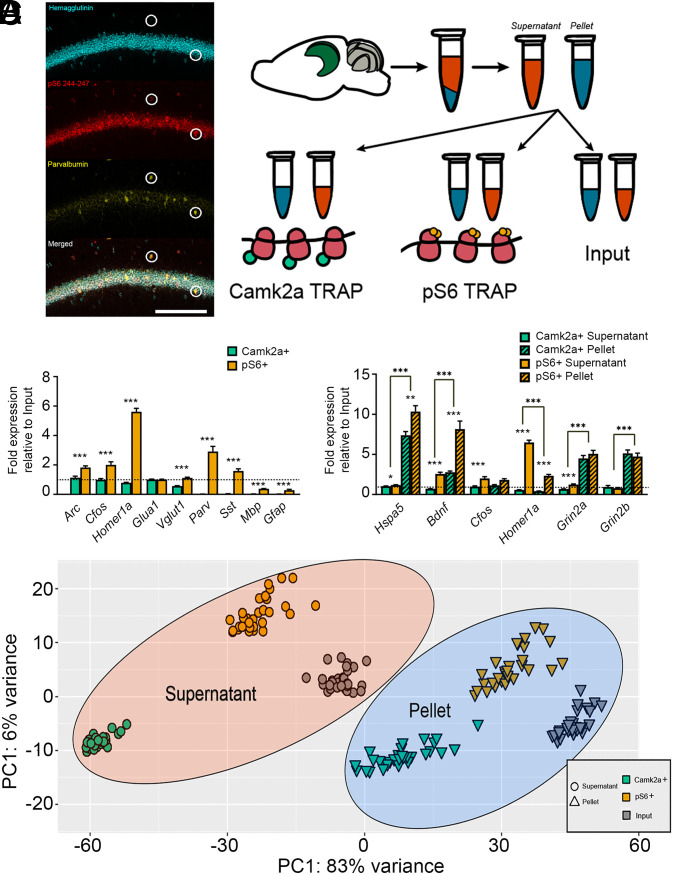
To quantify the effects of sleep and learning on hippocampal mRNA translation, we employed two translating ribosome affinity purification (TRAP) techniques. First, to quantify ribosome-associated mRNAs in excitatory neurons, B6.Cg-Tg(Camk2a-cre)T29-1Stl/J mice were crossed to the B6N.129-Rpl22tm1.1Psam/J mouse line (30, 31). Offspring from this cross express hemagglutinin (HA)-tagged ribosomal protein 22 (HA-Rpl22) in excitatory (Camk2a+) neurons (Fig. 1 A, Left). Second, to quantify mRNAs associated with ribosomes in active hippocampal neurons, we used an antibody targeting the terminal phosphorylation sites (Ser244/247) of ribosomal protein S6 (pS6) (28) (Fig. 1 A, Left). These sites are phosphorylated in neurons as the result of high neuronal activity, by ERK (32), mTOR-dependent kinase S6K1/2 (33), and CK1 (34), leading to translation initiation (34) and possibly translation of select transcripts (33). This strategy allowed us to compare mRNAs expressed in the whole hippocampus (Input) with those associated with ribosomes in either Camk2a+ or highly active (pS6+) neuronal populations from the same hippocampal tissue. To further test how mRNA translation varies as a function of ribosomes’ subcellular localization, we centrifuged our homogenized hippocampal tissue and collected samples from supernatant (presumptive cytosolic) and pellet (presumptive membrane-containing) fractions (35). From both fractions, we compared whole-hippocampus (Input) transcripts with transcripts isolated by TRAP from excitatory neurons (Camk2a+) and more highly active (pS6+) neuron populations (Fig. 1A).
Fig. 1.
TRAP-based profiling of hippocampal cell populations and isolation of subcellular fractions. (A, Left) Confocal images showing expression of HA (Camk2a), phosphorylated S6 (pS6), and parvalbumin in area CA1 of dorsal hippocampus. Highlighted neurons are parvalbumin, pS6, and HA. (Scale bar, 100 μm.) (Right) Schematic of protocol for isolating mRNAs from subcellular fractions and different cell populations using TRAP. (B) Camk2a+ (cyan) and pS6+ (orange) TRAP mRNA enrichment values were calculated (versus Input) for activity-dependent (Arc, Cfos, Homer1a), excitatory neuron (Glua1, Vglut1), inhibitory neuron (Parv, Sst), and glial (Mbp, Gfap) transcripts. *** indicates P < 0.001 for enrichment value differences between Camk2a+ and pS6+ neuronal populations (t test, n = 7/group). (C) Camk2a+ and pS6+ TRAP enrichment in supernatant (solid bars) and pellet (hatched bars) fractions (versus Input) for transcripts encoding secreted (Bdnf), transmembrane (Grin2a, Grin2b), ER (Hspa5), and cytosolic (Cfos, Homer1a) proteins. (t test, n = 9/group, *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively). (D) PCA plot (variance stabilizing transformation [VST], Deseq2) for RNA-seq data from the three cell populations (Input, Camk2a+ neurons, and pS6+ neurons) and two fractions (supernatant and pellet). Data are shown for n = 30 biological replicates (i.e., bilateral hippocampi) from 30 total mice across the four treatment groups.
We first validated cell type–specific transcript enrichment from Camk2a+ and pS6+ cell populations, using qPCR. Consistent with recent data (36), relative to Input mRNA, Camk2a+ TRAP produced similar mRNA levels of Arc, Cfos, Homer1a, Glua1, and Vglut1 transcripts and reduced expression of interneuron and glial cell markers. In light of hemizygosity for HA-Rpl22, this is consistent with roughly twofold enrichment of excitatory markers relative to Input. Highly active pS6+ neurons’ mRNA profiles displayed greater enrichment of activity-driven transcripts (Arc, Cfos, and Homer1a) and interneuron-specific transcripts (Pvalb and Sst) than Camk2a+ neurons and comparable levels of excitatory neuron-specific mRNAs (Glua1 and Vglut1) (Fig. 1 A and B). These data support the interpretation that pS6 is present in a subset of highly active Camk2a+ neurons in the hippocampus, along with select populations of interneurons (20).
We next used qPCR to initially characterize subcellular enrichment of mRNAs differentially expressed in supernatant and pellet fractions. Previous reports have found that isolating pellet ribosomes selectively enriches for endoplasmic reticulum (ER)- and dendrite-localized transcripts (35). To test whether transcripts encoding ER and dendritic proteins are differentially present in supernatant versus pellet, we first analyzed Hspa5 (encoding the resident ER chaperone BIP). Hspa5 was significantly more enriched in the pellet fractions than the supernatant fractions of both Camk2a+ and pS6+ neurons (Fig. 1C; Camk2a+: supernatant, 1.03 × Input; pellet, 7.38 × Input; P < 0.001, t test; pS6+: supernatant, 1.17 × Input; pellet, 10.33 × Input; P < 0.001). Similarly, Bdnf, Grin2a, and Grin2b mRNAs (encoding the secreted growth factor and glutamatergic receptor subunits) were more enriched on ribosomes isolated from the pellet fraction compared to the supernatant fraction. In contrast, Homer1a [encoding a truncated form of synaptic scaffolding protein Homer1, whose transcript is initially translated in the soma (37, 38)] was more abundant on supernatant ribosomes in both neuron populations, and the immediate early gene transcript Cfos was equally abundant in both fractions. These results support the idea that ribosome-associated transcripts observed in pellet and supernatant fractions encode proteins with predicted enrichment on cell membranes and in cytosol, respectively. To further validate this interpretation, we further characterized transcript profiles from different cell populations (Camk2a+, pS6+, Input) and subcellular fractions (supernatant, pellet) using a nonbiased approach - RNA sequencing (RNA-seq). The biological replicates used for this analysis consisted of biological hippocampi from individual mice (i.e., one mouse/sample; total n = 30 mice across 4 treatment groups described below in the following Results subsection). Principal component analysis (PCA) analysis revealed six discrete clusters of RNA-seq–based mRNA expression profiles, with PC1 accounting for 83% of the total expression variance (PC2 contributed only 6%). These clusters segregated based on the sample’s origin (Fig. 1D), with supernatant and pellet fractions each forming a similar grouping of three distinct clusters. Critically, three clusters were present in both the supernatant and pellet fractions’ data; these two sets of three clusters showed the same relationship in PC space relative to one another and segregated based on cell type (Camk2a+, pS6+, or Input). No obvious subclusters (i.e., representing the four treatment groups of the mice; see the following subsection of Results, below) were present within this PC space.
To characterize transcripts differentially localized to supernatant and pellet fractions, we next calculated the relative mRNA abundance in the two fractions from Camk2a+ neurons, pS6+ neurons, and Input (i.e., whole hippocampus) (SI Appendix, Figs. S1 A and B and S2A) using Deseq2 (39). Again, for this analysis, each mouse’s hippocampi (n = 30 samples, from 30 mice across the 4 behavioral conditions) constituted a biological replicate. The 2,000 transcripts that were most differentially expressed between the fractions (based on adjusted P value) were then characterized using the database for annotation, visualization and integrated discovery (DAVID)’s cellular component annotation (40). Transcripts more abundant in the supernatant fraction from both neuron populations (and Input) encoded proteins with functions localized to the cytoplasm and nucleus. Transcripts more abundant in the pellet fraction encoded proteins with functions localized to the plasma membrane, ER, Golgi apparatus, and synapses (SI Appendix, Figs. S1 C and D and S2B). Thus, for subsequent analyses, we refer to supernatant and pellet ribosomal fractions as cytosolic and MB ribosomes, respectively. Signaling and metabolic pathways with components highly represented among mRNAs more abundant on cytosolic ribosomes were assessed using Ingenuity Pathway Analysis (IPA) canonical pathways (SI Appendix, Fig. S1 E and F). These included cytosol-localized cellular function pathways such as ubiquitination, nucleotide excision repair, hypoxia signaling, and sumoylation pathways. In contrast, transcripts more abundant on MB ribosomes enriched for components of signaling pathways involved in synaptic (GABAergic receptor, glutamatergic receptor, and endocannabinoid signaling) and ER (e.g., unfolded protein response) functions.
To further investigate subcellular localization of transcripts representing cellular pathways critically involved in hippocampal function, we examined signaling pathways represented by transcripts selectively expressed in both cytosolic and MB fractions. Because both learning and sleep affect synaptic structure and function (10, 41–43), we first focused on the synaptogenesis signaling pathway. In Camk2a+ neurons, transcripts more abundant on MB ribosomes encoded secreted proteins (e.g., Bdnf), transmembrane proteins including AMPA, NMDA, and ephrin receptors (e.g., Gria1, Gria2, Gria3, Grin2a, Grin2b, Grin2c, Epha1, and Epha2), and membrane-associated enzymes (Plcγ) (SI Appendix, Fig. S1G). mRNAs more abundant on cytosolic ribosomes encoded intracellular complexes including adaptor proteins (Crk, Shc) and kinases (Cdk5, Lmk1, Gsk3b, Mapk1, Mapk2, and P70S6K) in the synaptogenesis pathway. Components of the CREB signaling pathway (another pathway regulated by both learning and sleep) (18, 44–47) were also highly represented among transcripts, which differentially localized to both cytosolic versus MB ribosomes, in both Camk2a+ and pS6+ neuronal populations (SI Appendix, Fig. S1 E and F). mRNAs encoding enzymes which regulate CREB transcriptional activity selectively localized to ribosomes in either compartment (e.g., Polr2c, encoding the RNA polymerase subunit, in the cytosolic fraction; Adcy1, encoding adenylate cyclase, in the MB fraction). mRNAs encoding G protein coupled receptors and ion channels (metabotropic glutamate receptors, ER IP3 receptors, calcium channel subunits, AMPA and NMDA receptor subunits) localized exclusively to the MB fraction, while those encoding transcription factors and kinases localized primarily to the cytosolic fraction (SI Appendix, Fig. S1H). In contrast to Camk2a+ and pS6+ neuron populations, the CREB signaling pathway was not represented among mRNAs differentially localized to subcellular fractions of Input (i.e., whole hippocampus; SI Appendix, Fig. S2), suggesting that differential localization of mRNAs encoding CREB signaling components are more pronounced in neurons than other hippocampal cell types.
Functional categories represented in the two subcellular fractions in Input RNA followed a pattern largely similar to that seen in Camk2a+ and pS6+ neuronal populations. However, in contrast to profiles from neuronal populations, the signaling pathway category that was most represented by differentially localized mRNAs between the two Input fractions was the protein ubiquitination pathway (SI Appendix, Fig. S2). This finding suggests more dramatic subcellular segregation of mRNAs encoding ubiquitin pathway components in nonneuronal hippocampal cell types (i.e., glial cells).
Learning and Sleep Loss Have Divergent Effects on Cytosolic and MB Ribosome-Associated mRNA Profiles.
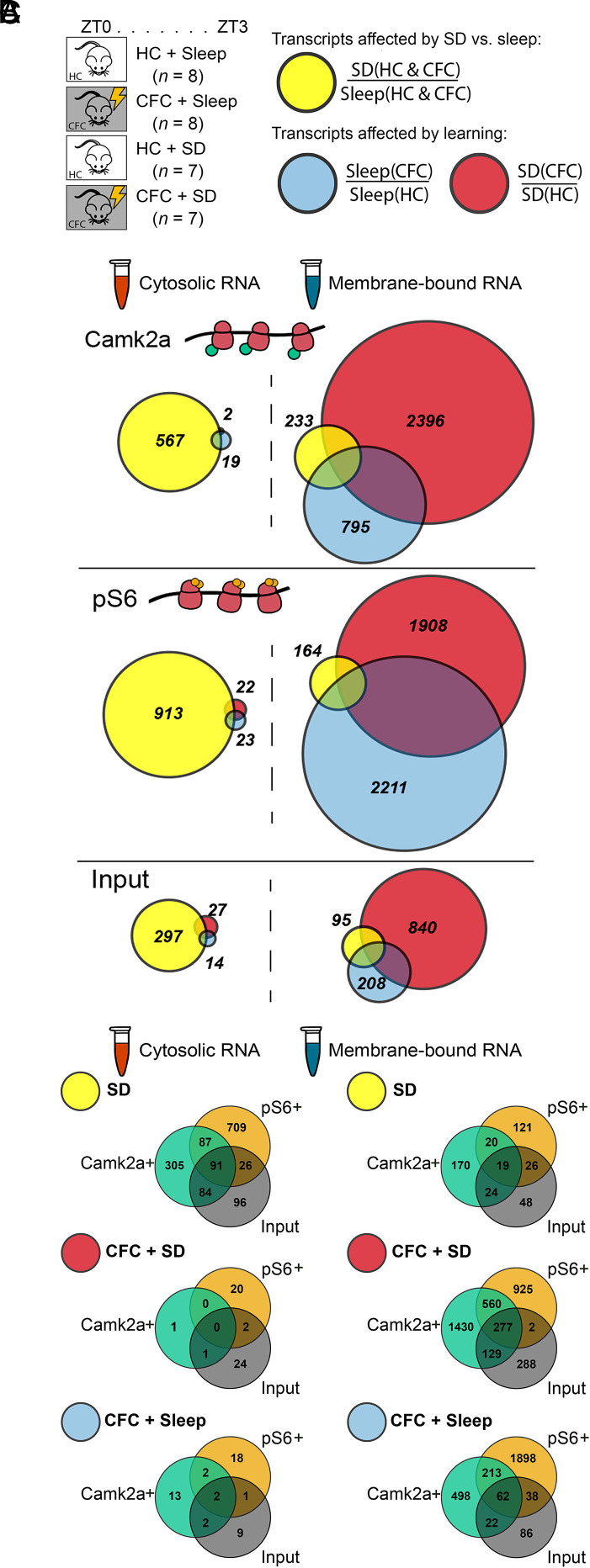
Because both learning and sleep alter hippocampal activity, intracellular signaling, and function (18, 19, 48, 49), we next tested how cytosolic and MB ribosome-associated transcripts in different neuron types were affected by prior training on a hippocampus-dependent memory task. Consistent with previous findings from our laboratory and others’ (17–21), post-CFC SD disrupted CFM consolidation in the transgenic mice used in this study (SI Appendix, Fig. S3). We also tested how these transcript profiles were affected by a brief (3-h) period of subsequent sleep or SD. This 3-h time post-CFC window was selected to correspond to peak post-CFC learning-induced changes in hippocampal network activity (20, 49, 50), and the time course over protein synthesis is required for CFM consolidation (26, 27). At lights on (Zeitgeber time [ZT] 0; i.e., the beginning of the rest phase), mice were either left undisturbed in their home cage (HC) or underwent single-trial CFC. Over the next 3 h, mice in CFC and HC control groups were either permitted ad libitum sleep (Sleep) or underwent SD in their HC by gentle handling (Fig. 2A). Freely sleeping HC + Sleep and CFC + Sleep mice spent 76 ± 2% and 73 ± 4%, respectively, of the 3-h time window prior to euthanasia in a sleeping state. This is consistent with our prior data showing that ∼60 to 75% of the first 6 h following CFC are spent in nonrapid eye movement (non-REM) sleep, and roughly 3 to 7% in REM sleep (19, 49, 50). These manipulations were followed by RNA isolation and sequencing as described above in the preceding Results subsection. Effects of learning and sleep loss on mRNA abundance were quantified for each cell population (Camk2a+, pS6+, or Input) and subcellular fraction (cytosolic or MB; e.g., pS6+ MB) to preserve gene-level inferences made by the Deseq2 model.
Fig. 2.
Cytosolic ribosomal transcripts are altered primarily by SD, while MB ribosomal transcripts are altered primarily by learning. (A, Left) Experimental paradigm for RNA-seq experiments, showing the four treatment groups. n values indicate the number of mice per group; one mouse’s bilateral hippocampi constituted one biological replicate. At lights on, mice were either left in their HC or underwent single-trial CFC. All mice were then either permitted ad libitum sleep or were SD over the following 3 h. (Right) Transcript comparisons for quantifying effects of SD (yellow) included both HC and CFC animals. To quantify effects of CFC, CFC + Sleep (blue), and CFC + SD (red), mice were analyzed separately. Following behavioral manipulations, cytosolic and MB fractions for different cell populations were isolated as described in Fig. 1. (B) Proportional Venn diagrams reflect the number of significantly altered transcripts in each cell populations and subcellular fractions (i.e., Camk2a+/MB), based on comparisons shown in A. Complete transcript lists for each comparison are available in Datasets S4, S5, and S6. (C) Transcripts altered by SD and CFC in different subcellular fractions in Camk2a+, pS6+, and Input populations were used to construct Venn diagrams. Full transcript lists are included in Dataset S7.
We first assessed the specific effects of SD alone (comparing SD and Sleep conditions) by combining data sets from naive (HC) and recently trained (CFC) mice (Fig. 2A, yellow). Learning and novel sensory experiences during SD are often a caveat for identification of brain biochemical which changes due to wake per se (11). We thus pooled samples from the HC and CFC groups for this specific portion of the Deseq2 analysis, with the aim of more reliably identifying the most robust transcript changes driven by SD, while disregarding changes that might occur primarily as a function of learning (11). We then quantified the effects of learning (comparing CFC and HC conditions) separately in sleeping and SD mice (Fig. 2A, red [SD], blue [Sleep]). Comparing the two latter sets of transcript changes is useful for identifying those occurring during successful CFM consolidation (in sleep) and those occurring in a condition where consolidation fails (SD). Venn diagrams (shown in Fig. 2B) illustrate the relative number of significant transcript changes resulting from these comparisons. SD had a relatively large effect on cytosolic ribosomal mRNAs (Camk2a+: 567 transcripts; pS6+: 913 transcripts; Input: 297 transcripts) compared to the effect of learning, which had extremely modest effects on cytosolic ribosomal transcripts. Conversely, MB ribosomal mRNAs were dramatically altered by learning in both SD (CFC + SD: Camk2a+, 2,396 transcripts; pS6+, 1,908 transcripts; Input, 840 transcripts) and Sleep groups (CFC + Sleep: Camk2a+, 795 transcripts; pS6+, 2,211 transcripts; Input, 208 transcripts). In contrast, relatively few MB ribosomal mRNAs were altered by SD (Camk2a+: 233 transcripts; pS6+: 164 transcripts; Input: 95 transcripts). These results suggest that SD and learning differentially affect ribosomal mRNA profiles based on their subcellular localization. SD (versus Sleep) appears to have more pronounced effects (i.e., significantly affecting a larger number of transcripts) in the cytosol, and learning appears to have more pronounced effects on MB ribosomes. Moreover, relatively few transcripts significantly affected by sleep or learning overlapped between the different cell populations, as shown in Fig. 2C. Together these data suggest that effects of learning and sleep on ribosome-associated transcripts are highly cell type and subcellular compartment specific.
SD Primarily Affects Cytosolic Ribosomal mRNAs Involved in Transcription Regulation.
Far more cytosolic ribosome–associated mRNAs were significantly altered by SD (compared with relatively few changes driven by CFC) in Camk2a+ neurons (SD: 567 transcripts, CFC: 20 transcripts), pS6+ neurons (SD: 913 transcripts, CFC: 43 transcripts), and to a lesser extent, Input (whole hippocampus) mRNA (SD: 297 transcripts, CFC: 37 transcripts) (Fig. 3 and SI Appendix, Fig. S4). We characterized the molecular and cellular pathways altered by SD-induced changes to cytosolic ribosome transcripts. Molecular functions most affected by SD alone (based on adjusted P values <0.1 for transcripts using IPA annotation) overwhelmingly favored transcriptional regulation and RNA processing in both Camk2a+ and pS6+ neurons, as well as in Input (Fig. 3 B, Top). Previous transcriptome analysis has shown that mRNAs encoding transcription regulators are more abundant following brief SD in the hippocampus (Fos, Elk1, Nr4a1, Creb, and Crem1) (4–6, 36) and neocortex (Per2, Egr1, and Nr4a1) (2, 36). Consistent with those findings, SD increased the abundance of multiple mRNAs encoding transcription factors and upstream regulators in all cell populations, including E2f6, Elk1, Erf, Fosl1, Fosl2, Fos, Fosb, Lmo4, Taf12, Xbp1, Atf7, Artnl2, Atoh8, Bhlhe40(Dec1), Crebl2, Crem, Egr2, Nfil3, and Ubp1. Transcripts significantly affected in Camk2a+ and pS6+ neurons overlapped partially with those reported in previous SD experiments, including components of pathways for AMPK, PDGF, ERK/MAPK, IGF1, and ER stress signaling (Fig. 3 B, Bottom) (4–6, 24, 51). However, only a small fraction of mRNAs recently reported to be altered by SD in whole hippocampus (5), hippocampal Camk2a+ neurons’ ribosome-associated profiles (6), or whole forebrain (10) overlapped with SD-affected mRNAs in cytosolic fractions of any cell population (SI Appendix, Fig. S5). In pS6+ neurons only, mRNAs encoding components of the PI3K/AKT and TGF-B signaling pathways were down-regulated in the cytosolic fraction after SD (Fig. 3 B, Bottom), suggesting that activity in these pathways may be higher during sleep.
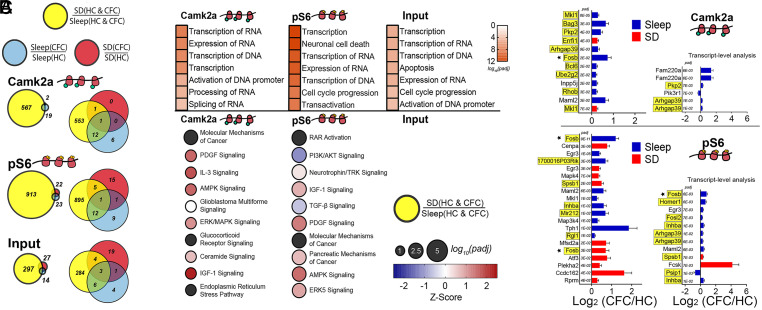
Fig. 3.
mRNAs altered by SD on cytosolic ribosomes encode transcriptional regulators. (A) Proportional and overlapping Venn diagrams of transcripts significantly altered by SD, CFC + Sleep, and CFC + SD in cytosolic fractions from Camk2a+ neurons, pS6+ neurons, and Input. (B, Top) The seven most-enriched molecular and cellular function categories (ranked by Padj value) for transcripts altered by SD alone in Camk2a+ neurons, pS6+ neurons, and Input. (Bottom) The 10 most-enriched canonical pathways of SD-affected transcripts are listed in order Padj value (indicated by circle diameter), with z-scores reflecting direction of pathway regulation (indicated by hue). There were no significant canonical pathways present in the Input fraction. (C, Left) The 10 transcripts most significantly affected in CFC + Sleep (blue) and CFC + SD (red) conditions for Camk2a+ (Top) and pS6+ (Bottom) neurons, ranked by Padj value. Transcripts that were also significantly altered as a function of SD alone are highlighted in yellow. (Right) Results of transcript-level analysis (56) show transcripts for transcript isoforms altered in Camk2a+ (Top) and pS6+ (Bottom) neurons following CFC. Transcript isoforms that were significantly altered as a function of SD are highlighted in yellow. Functional category analysis available in Dataset S8.
We next performed upstream regulator analysis to characterize transcript changes, which might be due to SD-associated transcriptional regulation. Results of this analysis provide both a P value for the significance of mRNAs’ regulation by a specific common upstream regulator and a z-score indicating the direction of the regulated mRNAs’ fold change (i.e., transcriptional activation or suppression). Taken together, these values predict the activation state of specific gene regulator complexes during SD (52). In line with prior meta-analysis of SD-induced transcripts (53), Creb1 was identified as the transcriptional regulator whose downstream target’s were most consistently affected across all cytosolic transcripts (SI Appendix, Fig. S6A). Creb1 transcript itself was not increased following SD, although Crebl2 and Crem mRNAs were both significantly increased, and multiple Creb1 transcriptional targets (e.g., Fos, Arc, Fosb, Egr2, Nfil3, Nr4a2, Bag3, and Irs2) were up-regulated in the cytosolic fraction of both Camk2a+ and pS6+ neuronal populations (SI Appendix, Figs. S6B and S7) following SD.
CFC-Induced Changes in Cytosolic Ribosome–Associated mRNAs Are Occluded by Subsequent SD.
Hippocampal fear memory consolidation in the hours following CFC relies on both sleep (17–19) and CREB-mediated transcription (13, 54). With Creb1 activity high during SD, we were curious what effect SD would have on the abundance of ribosome-associated transcripts involved in memory consolidation. As discussed in the preceding Results subsection above, few cytosolic ribosome–associated mRNAs were altered by CFC, regardless of subsequent sleep (Figs. 2 and 3). In Camk2a+ neurons, 19 cytosolic ribosome–associated transcripts were altered (compared to HC + Sleep controls) in CFC + Sleep mice, whereas only 2 transcripts were significantly altered (compared to HC + SD controls) in CFC + SD mice. Of those, most (13/19 from CFC + Sleep, 2/2 from CFC + SD) were also significantly increased by SD alone (Fig. 3A). Ribosome-associated mRNAs whose abundance was significantly affected by both SD and CFC in Camk2a+ neurons included activity-dependent transcripts such as Fosb, Arhgap39(Vilse), and Errfi1 (Fig. 3C, yellow). For comparison, in Input (i.e., whole hippocampus), slightly more cytosolic ribosome–associated mRNAs were significantly altered after CFC + SD (27 transcripts) versus CFC + Sleep (14 transcripts). Of these, 6/27 and 9/14, respectively, were similarly significantly affected by SD alone (Fig. 3A).
These initial data suggested that changes in cytosolic ribosome–associated transcripts following SD alone could occlude some of the transcript changes triggered by CFC, that is, making effects of CFC itself on transcript abundance undetectable. To better characterize how this might affect the neurons that are most activated by CFC (which could represent CFC “engram neurons”), we compared cytosolic transcripts affected by CFC and SD in pS6+ neurons. While similar numbers of transcripts were altered by CFC followed by sleep or SD (23 versus 22), only two transcripts (Fosb and Egr3) were similarly affected in both CFC + Sleep and CFC + SD mice (Fig. 3 A and C). Of the transcripts altered on cytosolic ribosomes after CFC in pS6+ neurons, several (12/23 of those affected in freely sleeping mice, 6/22 of those affected in SD mice) were also regulated by SD alone (Fig. 3A), including Fosb, Egr3, Arghap39(Vilse), Gpr3, Ssh2, Inhba, Rnf19a, and Cdkn1a (Fig. 3C, yellow). Because SD alters a significant number of transcripts involved in RNA splicing/processing (Fig. 3B), and splice isoforms play critical roles in synaptic plasticity and memory storage (55), we next assessed effects of SD and CFC on differentially spliced mRNA isoforms using transcript-level analysis (56) (Fig. 3C). On cytosolic ribosomes from pS6+ neurons, both the activity-dependent splice isoform of Homer1 scaffolding protein (Homer1a) and the activity-dependent splice isoform of Fosb, which encodes a highly stable protein (ΔFosb), were significantly increased after CFC followed by ad libitum sleep but were not significantly affected by CFC in SD mice (Fig. 3C). These isoforms were also increased as a function of SD alone, suggesting another mechanism by which SD could occlude changes to pS6+ neurons initiated by learning (Fig. 3C, yellow).
To validate and extend these findings, and to better characterize the persistence of CFC-induced cellular changes, we harvested hippocampi from CFC and HC mice following 5 h of SD or ad libitum sleep (i.e., a later time point with respect to learning). Using qPCR, we first quantified mRNA levels for splice isoforms of Fosb and Homer1 in the cytosolic fraction of whole hippocampus (Input), Camk2a+ neurons, and pS6+ neurons. Similar to what was observed after 3 h of SD, 5 h of SD increased expression of both Fosb and its long-lasting splice isoform ΔFosb, regardless of prior CFC (SI Appendix, Fig. S8 A, Left). Compared with Input, Fosb and ΔFosb transcripts were relatively de-enriched on cytosolic ribosomes from Camk2a+ neurons, but both were highly enriched on cytosolic ribosomes pS6+ neurons (SI Appendix, Fig. S8 A, Right). This finding is consistent with neural activity regulating both S6 phosphorylation (28) and Fosb and ΔFosb transcript abundance (57, 58). To measure CFC-driven changes in these transcripts, we normalized Fosb and ΔFosb transcripts in CFC + Sleep or CFC + SD mice to that of the corresponding HC control group. In both pS6+ neurons and Input, ΔFosb increased following CFC, relative to HC controls, regardless of subsequent sleep or SD. In other words, CFC had an apparent effect on ΔFosb abundance, even after the effects of SD alone on ΔFosb expression were accounted for. However, on ribosomes taken from all Camk2a+ neurons, both Fosb and ΔFosb transcripts increased in CFC + Sleep mice, but this increase was occluded by increases in the transcripts caused by SD alone (36) in CFC + SD mice (SI Appendix, Fig. S8 A, Bottom).
We also used qPCR to quantify the relative expression of Homer1 and its splice variant Homer1a in cytosolic fractions after CFC and 5 h subsequent sleep or SD. Homer1 itself was modestly affected by 5 h of SD, while its splice variant Homer1a was dramatically elevated, consistent with earlier findings (36, 59, 60) (SI Appendix, Fig. S8 B, Left). Homer1 was de-enriched in both Camk2a+ and pS6+ neurons relative to Input, while Homer1a was enriched only in pS6+ neurons (consistent with regulation by neuronal activity) (SI Appendix, Fig. S8 B, Right). Similar to ΔFosb, CFC increased Homer1a across all cell populations in mice allowed ad libitum sleep, but this increase was occluded in Camk2a+ neurons by SD (SI Appendix, Fig. S8 B, Bottom).
We quantified additional cytosol-localized transcripts for proteins with known functions in hippocampal plasticity and memory to test the effects of CFC and 5 h subsequent sleep or SD (SI Appendix, Fig. S9). These included transcripts increased in our Desq2 analysis following SD alone and either unaffected by CFC (Cfos and Arc), altered only in CFC + SD mice (Atf3), or altered only in CFC + Sleep mice (1700016P03Rik). We also visualized Egr3, which was unaffected by SD but increased by CFC in both CFC + SD and CFC + Sleep groups. All transcripts except Egr3 were altered by 5 h SD in Camk2a+ and pS6+ neuron populations. In pS6+ neurons, Cfos and the long noncoding RNA (lncRNA) transcript 1700016P03Rik (61, 62) remained significantly elevated as a function of learning 5 h following CFC; SD either fully or partially occluded these learning-associated changes (SI Appendix, Fig. S9). No significant CFC-induced changes in these transcripts were detectable on cytosolic ribosomes from Camk2a+ neurons at 5 h post-CFC.
Taken together, these data support the hypothesis that CFC-associated changes in activity-regulated transcripts at cytosolic ribosomes are likely occluded by subsequent SD (Discussion). This effect, which is most pronounced for highly active (putative engram) hippocampal neurons, constitutes a plausible mechanism for memory consolidation disruption by SD.
SD Affects MB Ribosomal Transcripts Involved in Receptor-Mediated Signaling, ER Function, and Protein Synthesis.
Fewer mRNAs were altered as a function of SD alone on MB ribosomes compared with cytosolic ribosomes—where most observed changes were driven by SD rather than CFC (Figs. 2 and 4A and SI Appendix, Fig. S10). Changes in MB ribosomal transcripts due to SD were also dwarfed by more numerous changes to MB ribosome–associated transcripts following CFC. These changes differed substantially between Camk2a+ neurons, pS6+ neurons, and Input; thus, canonical pathways represented by transcripts altered in the three populations by SD also differed. Critically, no canonical pathways were significantly enriched by SD-induced transcript changes in either Camk2a+ neurons or Input. On MB ribosomes from both Input and Camk2a+ cell populations, SD-altered transcripts included components of cellular pathways that were significantly affected in SD-regulated transcripts on cytosolic ribosomes (Fig. 3B) and with transcripts affected by SD in prior whole-hippocampus transcriptome studies (4–6, 24, 51). These included components of the AMPK (Chrm5, Irs2, Pfkfb3, Ppm1f, Prkab2, Prkag2, and Smarcd2), IGF1 (Elk1 and Rasd1), IL-3 (Crkl and Foxo1), relaxin (Gnaz, Pde4b, Smpdl3a), and neuregulin (Errfi1) signaling pathways in Camk2+ neurons and components of glucocorticoid receptor signaling (Elk1, Gtf2e2, Prkab2, Prkag2, Rasd1, Smarcd2, Taf12, Fos, Krt77, Ptgs2, and Tsc22d3), unfolded protein response (Hspa5 and Pdia6), and ER stress (Calr and Xbp1) pathways in both Camk2a+ neurons and Input. Critically, however, only 30 (6%) of the 511 mRNAs previously reported to be altered by SD in whole hippocampus (4) overlapped with SD-affected mRNAs in the MB fraction of whole hippocampus (i.e., Input; SI Appendix, Fig. S4). This suggests that even within these commonly identified cellular pathways, individual transcripts altered by SD on MB ribosomes may differ substantially from those reported previously.
In contrast to SD-driven changes in Camk2a+ neurons and Input, SD-altered transcripts from pS6+ neurons’ MB ribosomes significantly enriched for several canonical pathways. These included the neurotrophin/TRK (Atf4, Bdnf, Fos, Ngf, Plcg1, and Spry2), corticotropin releasing hormone (Crh and Vegfa), ERK5 (Il6st and Rasd1), and EIF2 (Eif2b3, Hspa5, Ptbp1, and Rpl37a) signaling pathways and the human embryonic stem cell pluripotency pathway (Bmp2, Inhba, and Wnt2). Thus, the major pathways affected by SD among MB ribosome–associated transcripts comprise receptor signaling pathways, protein synthesis regulation, and ER function.
Learning-Related Changes in MB Ribosomal Transcripts Diverge Based on Subsequent Sleep or SD.
In contrast to the sparsely expressed CFC-driven transcript changes observed on cytosolic ribosomes, the majority of transcript changes on MB ribosomes were driven by CFC (Figs. 2 and 4A). Critically, learning-induced changes in MB fraction transcripts diverged in all cell populations, depending on whether CFC was followed by 3 h of ad libitum sleep or 3 h of SD. In contrast to the high degree of overlap between SD-driven and CFC-driven transcript changes in the cytosol, on MB ribosomes, mRNAs affected by SD showed significantly less proportional overlap (2 to 10%) with CFC-induced changes (Fig. 4A). These data suggest that translational profiles of MB ribosomes are most selectively affected by prior learning but that the specific mRNAs associated with MB ribosomes also vary dramatically as a function of postlearning sleep or SD.
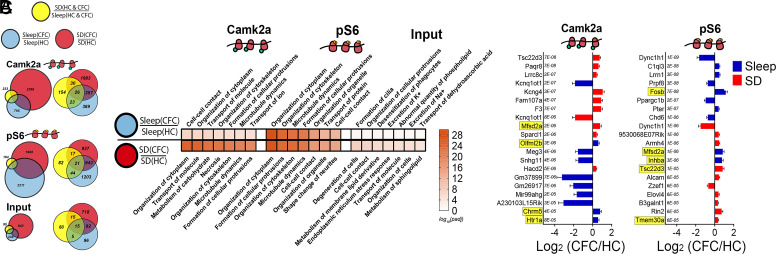
Fig. 4.
Transcripts altered by CFC on MB ribosomes encode regulators of neuronal morphology, intracellular trafficking, and lncRNAs. (A) Proportional and overlapping Venn diagrams of transcripts significantly altered by SD, CFC + Sleep, and CFC + SD in MB fractions from Camk2a+ neurons, pS6+ neurons, and Input. (B) The seven most-significant molecular functions (ranked by Padj value) for transcripts altered by CFC + Sleep (Top) and CFC + SD (Bottom) in Camk2a+ neurons, pS6+ neurons, and Input. (C) The 10 transcripts most significantly affected in CFC + Sleep (blue) and CFC + SD (red) conditions for Camk2a+ and pS6+ neurons, ranked by Padj value. Transcripts that were also significantly altered as a function of SD alone are highlighted in yellow. Functional category analysis available in Dataset S10.
We first characterized the cellular and molecular functions of MB ribosomal mRNAs altered as a function of CFC and subsequent sleep or SD. For Camk2a+ and pS6+ neurons, the most enriched functional categories largely overlapped and represented similar molecular categories in CFC + Sleep and CFC + SD mice—including organization of cytoplasm, organization of cytoskeleton, microtubule dynamics, cell–cell contact, and formation of protrusions (Fig. 4B). In contrast, few of these categories were enriched in Input MB fractions. There, the most enriched functional categories for transcripts altered by CFC + Sleep included excretion of sodium and potassium, formation of cilia, organization of cellular protrusions, desensitization of phagocytes, and abnormal quantity of phospholipid. Alterations in Input mRNAs following CFC + SD also enriched for functional categories not represented in neuronal MB fractions, including cell degeneration, metabolism of membrane lipid derivative, metabolism of sphingolipid, and ER stress response. Together, these data suggest that CFC may alter similar membrane-associated cellular functions in Camk2a+ and pS6+ neuronal populations, regardless of subsequent sleep or SD. In contrast, CFC may have distinct and pronounced effects on membrane-associated functions in other hippocampal cell types (e.g., glia), and these effects may diverge based on the animal’s sleep state.
To further characterize changes in MB-associated ribosomal transcripts following learning, we first compared significantly altered mRNAs (based on adjusted P value) in Camk2a+ and pS6+ neurons following CFC in sleeping and SD conditions (Fig. 4C). At Camk2a+ neurons’ MB ribosomes, CFC + Sleep led to increased abundance for mRNAs encoding transmembrane receptors (Chrm5 and Htr1a) and dramatically decreased abundance for multiple lncRNAs including Kcnq1ot1, Meg3, Mir99ahg, and unannotated transcripts (e.g., Gm37899 and Gm26917) (Fig. 4C). Many other lncRNAs showed reduced abundance on MB ribosomes following CFC (including Neat1, Malat1, Mirg, and Ftx). With the exception of Mirg and Ftx, these lncRNAs were also significantly reduced following CFC + SD. CFC + SD led to the most significant transcript increases on MB ribosomes for Lrrc8c (encoding an acid sensing, volume-regulated anion channel) and antiadhesive extracellular molecules (Sparcl1/Hevin), adhesion molecules (F3/Contactin1), transmembrane receptors (Paqr8), potassium modifiers (Kcng4), ER-tethered lipid synthesis molecules (Hacd2), and actin regulators (Fam107a).
In highly active (pS6+) neurons, Dync1h1 was the most significantly altered transcript following CFC and was dramatically reduced in both freely sleeping and SD mice (Fig. 4C). Dync1h1 encodes the main retrograde motor protein in eukaryotic cells, supporting retrograde transport in axons and dendrites (63). To a lesser extent, its abundance was also significantly reduced on MB ribosomes from Camk2a+ neurons. Dync1h1 was not reduced as a function of SD itself in either neuron population, suggesting that decreases in Dync1h1 are specific to the postlearning condition.
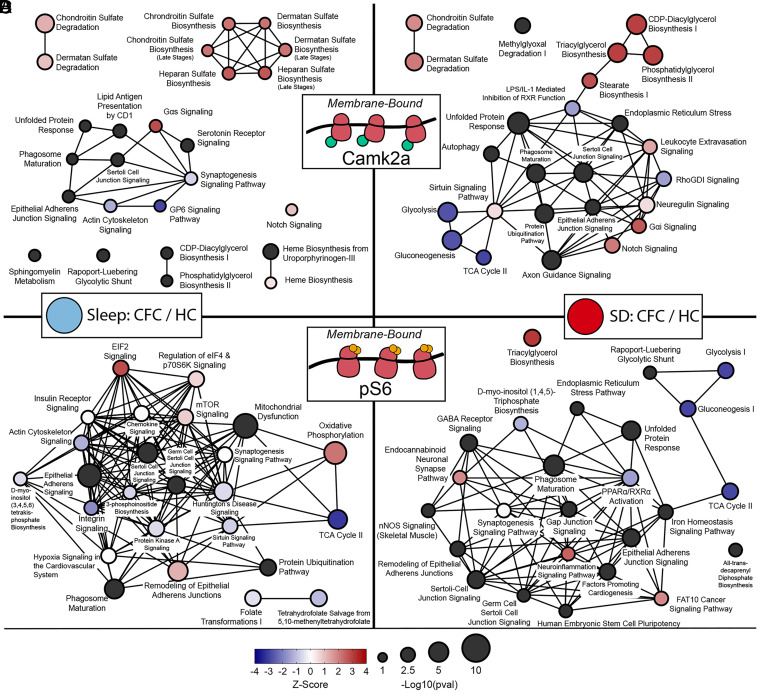
We next constructed canonical pathway networks affected by CFC + Sleep or CFC + SD to visualize the signaling and metabolic pathways differently altered in the two conditions. Canonical pathways are represented as hubs and connected through common transcript components. Here, hub sizes were weighted by their corresponding P value and shaded to indicate their z-score (blue indicating a decrease in the pathway following CFC, whereas red indicates an increase) (Fig. 5). Network comparisons of MB ribosome–associated transcript changes in Camk2a+ neurons revealed overlapping hubs significant in both CFC + Sleep (Fig. 5A) and CFC + SD conditions (Fig. 5D). These hubs represented chondroitin sulfate degradation, unfolded protein response, notch signaling, phagosome maturation, sertoli cell junction signaling, and epithelial adherens signaling canonical pathways. However, the significance values for these pathway hubs—similar to the number of transcripts altered in Camk2a+ neurons after CFC—were markedly higher in SD mice relative to sleeping mice. For example, the overlap and centrality of sertoli cell junction signaling, phagosome maturation, and epithelial adherens junction signaling pathways suggest common transcripts altered in both freely sleeping and SD mice after CFC. Upon closer inspection, while some tubulin transcripts were decreased following CFC in both Sleep and SD groups (Tuba1a and Tuba1b), a large number of tubulin-encoding mRNAs were decreased only after SD (Tuba4a, Tubb2a, Tubb3, Tubb4a, Tubb4b, and Tubg1) (Dataset S10). Similarly, while unfolded protein response–related transcripts were moderately elevated following CFC + Sleep (Calr, Mbtps1, P4hb, Sel1l, and Syvn1), substantially more mRNAs associated with the unfolded protein response were increased after CFC + SD (Amfr, Calr, Canx, Cd82, Cebpz, Dnajc3, Edem1, Eif2ak3, Hsp90b1, Hspa5, Mapk8, Mbtps1, Nfe2l2, Os9, P4hb, Sel1l, Syvn1, Ubxn4, and Xbp1).
Fig. 5.
MB ribosomal transcript networks affected by CFC vary as a function of subsequent sleep or SD. Canonical pathway network analysis of transcripts altered on MB ribosomes from Camk2a+ (A and B) or pS6+ (C and D) neurons following CFC + Sleep (A, C) or CFC + SD (B, D). Hub size and color denote Padj value and z-score, respectively, in each condition, while connecting lines indicate commonly expressed genes between hubs. Canonical pathways are available in Dataset S10.
In Camk2a+ neurons, CFC + SD also altered the expression of MB ribosome–associated mRNAs linked to metabolic pathways that were unaffected in the CFC + Sleep group (Fig. 5 A and B). For example, CFC + SD increased abundance of transcripts related to lipid (triacylglycerol, phosphatidylglycerol, cdp-diacylglycerol) biosynthesis, including mRNAs encoding 1-acylglycerol-3-phosphate O-acyltransferases (Agpat2, Agpat3, and Agpat4), ELOVL fatty acid elongases (Elvol1, Elovl2, and Elovl6), and phospholipid phosphatases (Plpp3). CFC + SD also decreased the abundance of transcripts related to glucose metabolic pathways (glycolysis, gluconeogenesis, and TCA Cycle), including Aldoa, Adloc, Eno1, Eno2, Gapdh, Gpi1, Pfkl, and Pkm (Dataset S10). Taken together with results shown in Fig. 4, these data indicate that Sleep and SD lead to divergent changes in the bioenergetic genes present at MB Camk2a+ ribosomes following learning.
We performed a similar canonical pathway network analysis on transcripts altered on MB ribosomes from pS6+ neurons following CFC (Fig. 5 C and D). Many of the same pathways altered by CFC in Camk2a+ neurons (in both Sleep and SD conditions) were also observed in pS6+ neurons—including sertoli cell junction signaling, epithelial adherens signaling, and phagosome maturation—suggesting some overlap. Pathways affected in the CFC + SD condition in Camk2a+ neurons included lipid and carbohydrate pathways affected in pS6+ neurons. Interestingly, in the CFC + Sleep condition (where CFM is being consolidated), there was an increased abundance of MB ribosome–associated transcripts representing protein translation regulatory pathways (eIF2, regulation of eIF4 and p70S6K, and mTOR signaling pathways). This change, critically, was not present in CFC + SD mice. In both sleeping and SD mice, MB ribosomal transcripts, which decreased in abundance after CFC, included eukaryotic initiation factors (Eif3a, Eif3c, Eif3l, Eif4a1, Eif4g1, Eif4g3), mTOR, and Tsc1. However, in mice allowed post-CFC sleep, transcripts related to the small ribosomal subunit were elevated in pS6+ neurons, including Rps12, Rps14, Rps17, Rps19, Rps20, Rps21, Rps23, Rps24, Rps26, Rps28, Rps29, and Rps6. This suggests that following CFC, sleep may promote an increase in overall protein synthesis capacity, which occurs in the most active hippocampal neurons. The fact that these changes occur selectively on MB ribosomes suggest that this increased synthetic capacity may be cell compartment specific.
Discussion
Our present results demonstrate not only that ribosome-associated transcripts are altered in the hippocampus as a function of 1) learning and/or 2) sleep versus sleep loss but also as a function of 3) the cell population being profiled and 4) the subcellular location of the ribosomes. We find that the latter aspect (i.e., location of ribosomes within the cell) is a major contributor to the observed effects of learning and subsequent sleep or SD on hippocampal ribosome transcript profiles. Neuronal ribosomes have long been known to segregate by cell compartment, present either as “free-floating” (i.e., cytosolic) or MB complexes, which are easily separated by centrifugation (64). These populations are known to engage in compartmentalized translation of mRNAs. The advent of TRAP has yielded insights into the specialized functions of ribosomes in different cellular compartments. Cytosolic ribosomes are known to process mRNAs encoding proteins with functions in the cytosolic compartment, including transcription factors and kinases. MB ribosomes typically associate with the rough ER and translate mRNAs encoding secreted or integral membrane proteins. Available data, from nonneural cell types, suggest that the two translational environments are biochemically distinct and can be differentially regulated—for example, by cellular stress (65). Where ribosomes have been isolated from subcellular compartments in neurons (e.g., in Purkinje neurons) (35), MB ribosome fractions have been shown to enrich for ER-associated ribosomes and for ribosomes in the dendritic compartment engaged in local translation. Our present findings reflect this, demonstrating that the transcript profiles of MB and cytosolic ribosomes among hippocampal neurons are highly distinctive (Fig. 1 and SI Appendix, Figs. S1 and S2).
Many forms of hippocampus-dependent memory are disrupted (in human subjects and animal models) by either pre- or postlearning sleep loss (1, 66–68). Indeed, sleep loss seems to disrupt plasticity mechanisms within the hippocampus more dramatically than in other brain areas (41, 69). The underlying mechanisms by which sleep loss leads to these changes, and disrupts memory mechanisms, have remained elusive. Transcriptome profiling of the effects of sleep loss alone on the hippocampus has indicated that SD increases expression of genes involved in transcriptional activation and down-regulates expression of genes involved in transcriptional repression, ubiquitination, and translation (4–6). While we found that the same cellular pathways are affected in the cytosolic fraction as in prior studies (Fig. 3B), specific transcripts affected by SD in neither the cytosolic nor the MB ribosomal transcripts showed a large degree of overlap with transcripts affected by SD in those studies (SI Appendix, Fig. S5). Our present data add to this by demonstrating that in the whole hippocampus, Camk2a+ neurons, or pS6+ neurons, the majority of purely SD-driven changes to transcripts are present in the cytosolic fraction and on cytosolic ribosomes (Figs. 2B and 3). While SD-driven mRNA changes also occur on MB ribosomes, these changes are relatively few in number (Figs. 2B and 4). Pathways affected by SD—across neuronal populations and across both subcellular compartments—were those linked to regulation of transcription, consistent with prior findings (4–6, 36) and the AMPK, IL-3, IGF1, and PDGF signaling pathways. Critically, AMPK (70) and IGF1 signaling (71) have been implicated in homeostatic sleep responses—that is, changes in sleep architecture of brain oscillations—following SD. Thus, it is tempting to speculate that sleep loss could lead to subsequent changes in sleep brain dynamics through changes in intracellular signaling in neurons.
However, two unanswered questions are 1) which SD-associated changes in specific transcripts’ synthesis or translation provide a plausible mechanism to disrupt hippocampal memory consolidation, and 2) what cell types within the hippocampus are critically affected by SD following learning. This study aimed to address this in the context of a form of hippocampus-dependent memory consolidation (CFM), which is critically dependent on postlearning sleep. Work from our laboratory and others has shown that disruption of sleep within the first few hours following CFC is sufficient to disrupt CFM consolidation (17, 19, 20). While some of systems-level mechanisms occurring during post-CFC sleep have been implicated in the consolidation process (19, 20, 49, 50, 72, 73), almost nothing is known about the cellular mechanisms mediating sleep (or SD) effects on CFM consolidation.
We were surprised that very few transcript changes were induced on cytosolic ribosomes by CFC, in comparison with the large number of cytosolic ribosomal mRNAs affected by SD alone. However, of those cytosolic transcripts altered by CFC, almost all 1) were similarly affected in either CFC + Sleep or CFC + SD conditions, and 2) were similarly altered by SD itself (Fig. 3C). This makes sense in light of the fact that many mRNAs altered on cytosolic ribosomes after CFC are known to be transcribed or translated in an activity-dependent manner. For example, Creb1-regulated cytosolic transcripts were up-regulated by SD; this effect was evident across hippocampal Input, the Camk2a+ neuronal population, and the highly active pS6+ neuron population. Importantly, post-CFC SD fully occluded learning-induced changes in activity-regulated transcripts present on cytosolic ribosomes in Camk2a+ neurons and partially occluded analogous changes in Input and pS6+ neurons. In other words, changes in these transcripts attributable to CFC (in comparison with HC controls) were no longer detectable if mice subsequently underwent SD, due to similar changes being evoked by SD alone. The transcript changes which show SD-mediated occlusion include increases in Fosb (and its isoform Δfosb, which encodes a highly stable protein) and Homer1 (and its short isoform Homer1a) (SI Appendix, Figs. S8 and S9). These transcript isoforms encode proteins that are critically linked to synaptic plasticity and memory, including CFM (74, 75). Because here we examined transcript levels across neuron populations, rather than in single neurons, there are at least two plausible explanations for the SD-driven occlusion we observe. First, SD and CFC may evoke similar intracellular events within individual neurons. This scenario is analogous to the way that cellular changes driven by LTP and some forms of learning occlude one another in the hippocampus (76). Such a mechanism could cause occlusion like we observe here due to saturation of an intracellular mechanism (e.g., reaching the limit of a transcriptional response) by the combination of learning and SD. Alternatively, the type of occlusion we see after SD could also be consistent with a neuron population-based effect—that is, while CFC induces transcript changes in a subset of learning-activated neurons, additional (nonengram) neurons undergo these same changes during SD. This seems plausible, in light of the fact that overall levels of these transcripts increase more as a function of SD alone versus learning alone. Regardless of the precise mechanism, the timing with which SD occludes changes in these transcripts’ abundance (3 to 5 h following CFC) coincides a critical window for post-CFC sleep, essential for CFM consolidation (17, 19). Thus, this change to cytosolic ribosomes’ transcripts could represent a plausible mechanism for memory disruption by sleep loss.
This occlusion-based interpretation of how sleep loss disrupts information storage is consistent with the central idea behind the SHY hypothesis of sleep function —namely, that sleep is essential for reducing signal-to-noise in brain circuits (11, 77, 78). SHY proposes that this reduction is effected, ultimately, through brain-wide reductions in synaptic strength. The hypothesis is supported by both biochemical and physiological data but is not without controversy (1, 11). Critically, the transcriptomic data, which provided support for the hypothesis—comprising transcripts that would correspond to the cytosolic ribosomal fraction—are consistent with our present analysis of SD’s impact on transcripts on cytosolic ribosomes. While it is unclear whether synaptic strength changes are being effected by SD, it is clear that activity-driven transcriptional machinery does change during SD, in a manner that could interfere with newly encoded information in the hippocampus. However, we find that SD-driven changes to cytosolic ribosomal transcripts represent only a small fraction of the neuronal biology that is dynamically changing as a function of learning and subsequent sleep.
In contrast to the relative paucity of transcripts altered on cytosolic ribosomes by prior learning, CFC affected a surprisingly large number of mRNAs on MB ribosomes (Figs. 2B and 4A). In general, CFC induced changes in MB ribosome–associated transcripts encoding proteins associated with neuronal structural remodeling—from cellular pathways involved in cytoskeletal remodeling, intracellular transport, and cell–cell interactions (Fig. 4B). Some changes were also highly surprising and unexpected—for example, the significant reduction in ribosome-associated lncRNAs on MB ribosomes in the Camk2a+ neuron population after CFC (Fig. 4C). Critically, the precise transcripts and—in some cases—the cellular pathways altered after CFC diverged dramatically based on whether learning was followed by sleep or SD (Figs. 4 and 5). These differences provide a wealth of information regarding potential mechanisms for SD-related disruption of CFM. For example, our present findings suggest that in nonneuronal cell types in the hippocampus, CFC induces a unique set of transcript changes—which are present in the MB fraction of Input but are absent from MB fractions of neuronal ribosomes. Increased abundance of transcripts related to energy metabolism, particularly those encoding mitochondrial proteins, glucose transporters, and proteins related to glycogen metabolism, are commonly observed following SD (2–4). Unlike previous reports, our data suggest that in Camk2a+ neurons, cellular metabolic/energetic pathways may be selectively disrupted when CFC is followed by SD but not when CFC is followed by sleep (Figs. 4 and 5). Thus, it may be that SD disrupts CFM consolidation by increasing the metabolic demands on the hippocampus.
In the most active (pS6+) neurons, CFC + Sleep leads to regulation of numerous pathways linked to protein synthesis regulation, including a widespread increase in MB ribosomal mRNAs encoding the translational machinery itself. This change is not seen when sleep is followed by SD. The pS6+ neuron population is likely mixed—containing a subset of the most highly active Camk2a+ neurons (i.e., those activated by experience) and some interneurons as well (20, 79). Indeed, S6 phosphorylation may even occur selectively in specific neuronal synapses activated by learning (29). Nonetheless, the fact that learning- and sleep-dependent changes in transcripts differs between the pS6+ population and either the whole hippocampus (Input) or the whole Camk2a+ population suggests that ribosomal function differs markedly as a function of state- and experience-dependent neuronal (or synaptic) activation. Thus, it is tempting to speculate that in the neurons and synapses most active following memory encoding (putative “engram neurons”), long-lasting changes to protein synthesis on cell membranes may play a critical role in subsequent memory consolidation. Neuropharmacological and biochemical studies have suggested that disruption of both hippocampal cAMP signaling and protein synthesis by SD may prevent memory consolidation (18, 24, 26, 27, 45). Our present data suggest that CFC may initiate changes to these pathways in specific subpopulations of hippocampal neurons and that these changes are subsequently facilitated by post-CFC sleep.
Recently, TRAP has been used to characterize compartment-specific ribosomal transcripts of amygdala-projecting cortical axons during cued fear memory consolidation (15). However, the methods used in this study (as is true for most transcriptome and TRAP studies) likely primarily report transcript changes associated with cytosolic rather than MB ribosomes. Here, we show that the vast majority of changes due to learning itself are expressed at MB ribosomes (Figs. 2B and 4)—with surprisingly few CFC-induced changes to cytosolic ribosome–associated mRNAs (Fig. 3). Recent comparisons of hippocampal ribosome–associated and total mRNA abundance suggests that cytosolic and MB ribosome–associated mRNAs are distinctly regulated with regard to translation efficiency (80). Thus, understanding the effects of both learning and subsequent sleep on structures like the hippocampus will require further investigation into their effects on translation happening at the membrane.
An important caveat in considering our present findings is that they represent a specific 3-h post-CFC time point. This time point is likely to be highly informative for understanding CFM consolidation mechanisms, as it represents the time of peak post-CFC learning-induced changes in hippocampal network activity (20, 49, 50) and a window during which protein synthesis is required for CFM consolidation (26, 27). However, it is plausible that the time course of ribosome-associated transcript changes in the cytosolic and membrane compartments differs following CFC. If this were the case, it is possible that learning-induced changes to ribosome-associated transcripts are present in the cytosolic compartment at either earlier or later time points. It is also plausible that sleep loss may alter transcripts on membrane-associated ribosomes at earlier or later time points. Thus, significant future study will be needed to determine the relative timing of CFC- and sleep-induced transcript changes on ribosomes in these two cellular compartments.
How universal are the sleep-dependent mechanisms for memory consolidation that our present data suggest? For example, are the same neural mechanisms present in other brain regions in the context of sleep-dependent information storage? It seems likely that very basic neurobiological mechanisms—such as activity-driven transcript regulation and segregation of specific transcripts between cytosolic and MB ribosomes—would be similar throughout the brain. Moreover, recent data from our laboratory have shown that certain features of postlearning sleep, such as coordinated network oscillations (14, 81), are required for new information storage in both hippocampus (19, 50) and neocortex (12, 82). However, our data have also shown that many cell type–specific effects of SD differ between hippocampus and neocortex (20, 36, 69). Future studies are needed to determine how consistent these differential subcellular responses to learning and subsequent sleep are, across different cell types and brain regions. However, consolidation of various types of memory, across species, share common cellular substrates (1, 14, 83), with postlearning mRNA translation being a vital element. Changes in the activity patterns of neurons and the activation of particular intracellular pathways during postlearning sleep share common features, across brain structures and species (1, 14). Our present findings have illustrated several sleep-dependent postlearning cellular processes, which affect pathways vital for learning and memory. Future studies will determine whether these processes underlie sleep-dependent memory consolidation events in the other brain circuits, following diverse forms of learning.
Materials and Methods
All animal husbandry and experimental procedures were approved by the University of Michigan Institutional Animal Care and Use Committee (Public Health Service Animal Welfare Assurance no. D16-00072 [A3114-01]). All mice were maintained on a 12:12 h light/dark cycle (lights on at 8 AM) with food and water provided ad libitum. B6.Cg-Tg(Camk2a-cre)T29-1Stl/J mice (Jackson) were crossed to B6N.129-Rpl22tm1.1Psam/J mice (Jackson) to express HA-tagged Rpl22 protein in Camk2a+ neurons. Double-transgenic mice were individually housed with beneficial enrichment for 1 wk prior to experiments and were habituated to daily handling (5 min/day) for 5 d prior to experiments. For RNA-seq experiments, mice were randomly assigned to one of four groups: HC + Sleep (n = 8), HC + SD (n = 7), CFC + Sleep (n = 8), or CFC + SD (n = 7). Each mouse’s bilateral hippocampi comprised one biological replicate for sequencing, and no samples were pooled between mice. Beginning at lights on (8 AM), half of the mice underwent single-trial CFC as described previously (19, 49, 50). Briefly, at lights on (ZT 0) mice were placed in a novel conditioning chamber (Med Associates) and were allowed 2.5 min of free exploration time prior to delivery of a 2-s, 0.75 mA foot shock through the chamber’s grid floor. After 3 min total in the chamber, mice were returned to their original HC. As a control for the effects of learning, HC controls remained in their HC during this time. HC + SD or CFC + SD mice were then kept awake continuously by gentle handling (SD; consisting of cage tapping, nest disturbance, and if necessary, stroking with a cotton-tipped applicator) over the next 3 h for all RNA-seq studies or 5 h for all qPCR experiments. HC + Sleep and CFC + Sleep mice were permitted ad libitum sleep in their HC for the same time interval.
RiboTag TRAP was performed as previously described (31) by indirect conjugation (84), separating MB and free-floating ribosomes (35). Following homogenization and centrifugation, the resulting supernatant (cytosolic fraction) was transferred to a new tube while the pellet (MB fraction) was resuspended in homogenization buffer. Both MB and cytosolic fractions were separated into Input, Camk2a+, and pS6+ fractions. For isolating ribosomes from Camk2a+ populations, fractions were incubated with 1:40 anti-HA antibody (Abcam, ab9110) (85). To isolate ribosomes from highly active (pS6+) neurons, fractions were incubated with 1:25 anti-pS6 244-247 (ThermoFisher 44-923G) (28). Homogenate-antibody solution was added directly to Protein G Dynabeads (ThermoFisher, 10009D) for incubation. After washing conjugated beads and removing the supernatant, RNA was eluted by vortexing the beads vigorously in 350 μL RLT (Qiagen, 79216). Eluted RNA was purified using RNeasy Micro kit (Qiagen).
RNA-seq was carried out at the University of Michigan’s DNA Sequencing Core. Amplified complementary DNA libraries were prepared using Takara’s SMART-seq v4 Ultra Low Input RNA Kit (Takara 634888) and sequenced on Illumina’s NovaSEq. 6000 platform. Reads mapped to unique transcripts were counted with featureCounts (86). Differential expression analyses were run with Deseq2 on all 30 hippocampal samples, with bilateral hippocampi from each mouse constituting a biological replicate (39). To characterize the differences between the effects of SD and CFC, significantly altered transcripts were analyzed using IPA. Gene ontology (GO) analyses were performed in IPA and DAVID’s Functional Annotation tool. For subcellular fraction comparisons, 2,000 of the top cytosolic (Log2 Fold Change [Log2FC] > 0) and MB (Log2FC < 0) differentially expressed transcripts (ranked by adjusted P values) were run through IPA's Canonical Pathways analysis. To characterize differences in common metabolic pathways between cytosolic and MB fractions, hierarchical clustering was used to visualize the most differentially expressed transcripts. Since signaling pathways were less overlapping between the MB and cytosolic fraction, they were ranked by enrichment P values. Those transcripts were then run through DAVID’s Functional Annotation tool, selecting for cellular composition to describe the cellular compartment the corresponding protein relates to. Data were plotted in Fragments Per Million and their correlation value (R) calculated in the ViDger R package (87).
Complete materials and methods are in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to members of the S.J.A. laboratory and to Drs. Natalie Tronson and Ryan Mills for helpful feedback on this manuscript. This work was supported by research grants from the NIH (DP2 MH 104119 and RO1 NS118440), the Human Frontiers Science Program (N023241-00_RG105) to S.J.A., and the Bioinformatics Core of the University of Michigan Biomedical Research Core Facilities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108534118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
Change History
January 31, 2022: The SI Appendix has been updated.
References
- 1.Puentes-Mestril C., Aton S. J., Linking network activity to synaptic plasticity during sleep: Hypotheses and recent data. Front. Neural Circuits 11, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirelli C., Gutierrez C. M., Tononi G., Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41, 35–43 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Mackiewicz M., et al. , Macromolecule biosynthesis: A key function of sleep. Physiol. Genomics 31, 441–457 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Vecsey C. G., et al. , Genomic analysis of sleep deprivation reveals translational regulation in the hippocampus. Physiol. Genomics 44, 981–991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaine M. E., et al. , Altered hippocampal transcriptome dynamics following sleep deprivation. Mol. Brain 14, 125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons L. C., Chatterjee S., Vanrobaeys Y., Gaine M. E., Abel T., Translational changes induced by acute sleep deprivation uncovered by TRAP-Seq. Mol. Brain 13, 165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirelli C., Pfister-Genskow M., McCarthy D., Woodbury R., Tononi G., Proteomic profiling of the rat cerebral cortex in sleep and waking. Arch. Ital. Biol. 147, 59–68 (2009). [PMC free article] [PubMed] [Google Scholar]
- 8.Ren J., et al. , Quantitative proteomics of sleep-deprived mouse brains reveals global changes in mitochondrial proteins. PLoS One 11, e0163500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirrier J.-E., et al. , Proteomic changes in rat hippocampus and adrenals following short-term sleep deprivation. Proteome Sci. 6, 14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noya S. B., et al. , The forebrain synaptic transcriptome is organized by clocks but its proteome is driven by sleep. Science 366, eaav2642 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Havekes R., Aton S. J., Impacts of sleep loss versus waking experience on brain plasticity: Parallel or orthogonal? Trends Neurosci. 43, 385–393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clawson B. C., et al. , Causal role for sleep-dependent reactivation of learning-activated sensory ensembles for fear memory consolidation. Nat. Commun. 12, 1200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao-Ruiz P., et al. , Engram-specific transcriptome profiling of contextual memory consolidation. Nat. Commun. 10, 2232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puentes-Mestril C., Roach J., Niethard N., Zochowski M., Aton S. J., How rhythms of the sleeping brain tune memory and synaptic plasticity. Sleep (Basel) 42, zsz095 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostroff L. E., et al. , Axon TRAP reveals learning-associated alterations in cortical axonal mRNAs in the lateral amgydala. eLife 8, e51607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daumas S., Halley H., Francés B., Lassalle J. M., Encoding, consolidation, and retrieval of contextual memory: Differential involvement of dorsal CA3 and CA1 hippocampal subregions. Learn. Mem. 12, 375–382 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graves L. A., Heller E. A., Pack A. I., Abel T., Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learn. Mem. 10, 168–176 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vecsey C. G., et al. , Sleep deprivation impairs cAMP signalling in the hippocampus. Nature 461, 1122–1125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ognjanovski N., Broussard C., Zochowski M., Aton S. J., Hippocampal network oscillations rescue memory consolidation deficits caused by sleep loss. Cereb. Cortex 28, 3711–3723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delorme J., et al. , Sleep loss drives acetylcholine- and somatostatin interneuron-mediated gating of hippocampal activity to inhibit memory consolidation. Proc. Natl. Acad. Sci. U.S.A. 118, e2019318118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qureshi M. F., Jha S. K., Short-term total sleep-deprivation impairs contextual fear memory, and contextual fear-conditioning reduces REM sleep in moderately anxious Swiss mice. Front. Behav. Neurosci. 11, 239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira L. M., et al. , Hippocampus and prefrontal cortex modulation of contextual fear memory is dissociated by inhibiting de novo transcription during late consolidation. Mol. Neurobiol. 56, 5507–5519 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Igaz L. M., Vianna M. R., Medina J. H., Izquierdo I., Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J. Neurosci. 22, 6781–6789 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tudor J. C., et al. , Sleep deprivation impairs memory by attenuating mTORC1-dependent protein synthesis. Sci. Signal. 9, ra41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gafford G. M., Parsons R. G., Helmstetter F. J., Consolidation and reconsolidation of contextual fear memory requires mammalian target of rapamycin-dependent translation in the dorsal hippocampus. Neuroscience 182, 98–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourtchouladze R., et al. , Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn. Mem. 5, 365–374 (1998). [PMC free article] [PubMed] [Google Scholar]
- 27.Bekinschtein P., et al. , Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron 53, 261–277 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Knight Z. A., et al. , Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirbhoy P. S., Farris S., Steward O., Synaptic activation of ribosomal protein S6 phosphorylation occurs locally in activated dendritic domains. Learn. Mem. 23, 255–269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz E., Bean J. C., Carey D. P., Quintana A., McKnight G. S., RiboTag: Ribosomal tagging strategy to analyze cell-type-specific mRNA expression in vivo. Curr. Protoc. Neurosci. 88, e77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanz E., et al. , Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U.S.A. 106, 13939–13944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelleher R. J. III, Govindarajan A., Tonegawa S., Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron 44, 59–73 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Biever A., Valjent E., Puighermanal E., Ribosomal protein S6 phosphorylation in the nervous system: From regulation to function. Front. Mol. Neurosci. 8, 75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutchinson J. A., Shanware N. P., Chang H., Tibbetts R. S., Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J. Biol. Chem. 286, 8688–8696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kratz A., et al. , Digital expression profiling of the compartmentalized translatome of Purkinje neurons. Genome Res. 24, 1396–1410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puentes-Mestril C., et al. , Sleep loss drives brain region- and cell type-specific alterations in ribosome-associated transcripts involved in synaptic plasticity and cellular timekeeping. J. Neurosci. 41, 5386–5398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada D., Ozawa F., Inokuchi K., Input-specific spine entry of soma-derived Vesl-1S protein conforms to synaptic tagging. Science 324, 904–909 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Brakeman P. R., et al. , Homer: A protein that selectively binds metabotropic glutamate receptors. Nature 386, 284–288 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dennis G. Jr., et al. , DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, 3 (2003). [PubMed] [Google Scholar]
- 41.Raven F., Meerlo P., Van der Zee E. A., Abel T., Havekes R., A brief period of sleep deprivation causes spine loss in the dentate gyrus of mice. Neurobiol. Learn. Mem. 160, 83–90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brüning F., et al. , Sleep-wake cycles drive daily dynamics of synaptic phosphorylation. Science 366, eaav3617 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Spano G. M., et al. , Sleep deprivation by exposure to novel objects increases synapse density and axon-spine interface in the hippocampal CA1 region of adolescent mice. J. Neurosci. 39, 6613–6625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vecsey C. G., et al. , Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 27, 6128–6140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abel T., et al. , Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell 88, 615–626 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Kandel E. R., The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 5, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J., Phan T. X., Yang Y., Garelick M. G., Storm D. R., Increases in cAMP, MAPK activity, and CREB phosphorylation during REM sleep: Implications for REM sleep and memory consolidation. J. Neurosci. 33, 6460–6468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Havekes R., et al. , Sleep deprivation causes memory deficits by negatively impacting neuronal connectivity in hippocampal area CA1. eLife 5, e13424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ognjanovski N., Maruyama D., Lashner N., Zochowski M., Aton S. J., CA1 hippocampal network activity changes during sleep-dependent memory consolidation. Front. Syst. Neurosci. 8, 61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ognjanovski N., et al. , Parvalbumin-expressing interneurons coordinate hippocampal network dynamics required for memory consolidation. Nat. Commun. 8, 15039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naidoo N., Giang W., Galante R. J., Pack A. I., Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J. Neurochem. 92, 1150–1157 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Krämer A., Green J., Pollard J. Jr., Tugendreich S., Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Liu Y., Briesemann M., Yan J., Computational analysis of gene regulation in animal sleep deprivation. Physiol. Genomics 42, 427–436 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Katche C., et al. , Delayed wave of c-Fos expression in the dorsal hippocampus involved specifically in persistence of long-term memory storage. Proc. Natl. Acad. Sci. U.S.A. 107, 349–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poplawski S. G., et al. , Contextual fear conditioning induces differential alternative splicing. Neurobiol. Learn. Mem. 134 (Pt B), 221–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi L., Pimentel H., Bray N. L., Pachter L., Gene-level differential analysis at transcript-level resolution. Genome Biol. 19, 53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McClung C. A., et al. , DeltaFosB: A molecular switch for long-term adaptation in the brain. Brain Res. Mol. Brain Res. 132, 146–154 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Corbett B. F., et al. , ΔFosB regulates gene expression and cognitive dysfunction in a mouse model of Alzheimer’s disease. Cell Rep. 20, 344–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackiewicz M., Paigen B., Naidoo N., Pack A. I., Analysis of the QTL for sleep homeostasis in mice: Homer1a is a likely candidate. Physiol. Genomics 33, 91–99 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Maret S., et al. , Homer1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. U.S.A. 104, 20090–20095 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aten S., Hansen K. F., Hoyt K. R., Obrietan K., The miR-132/212 locus: A complex regulator of neuronal plasticity, gene expression and cognition. RNA Dis. 3, e1375 (2016). [PMC free article] [PubMed] [Google Scholar]
- 62.Aten S., et al. , miR-132 couples the circadian clock to daily rhythms of neuronal plasticity and cognition. Learn. Mem. 25, 214–229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiavo G., Greensmith L., Hafezparast M., Fisher E. M. C., Cytoplasmic dynein heavy chain: The servant of many masters. Trends Neurosci. 36, 641–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrews T. M., Tata J. R., Protein synthesis by membrane-bound and free ribosomes of the developing rat cerebral cortex. Biochem. J. 124, 883–889 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reid D. W., Nicchitta C. V., Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 16, 221–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Havekes R., Abel T., The tired hippocampus: The molecular impact of sleep deprivation on hippocampal function. Curr. Opin. Neurobiol. 44, 13–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krause A. J., et al. , The sleep-deprived human brain. Nat. Rev. Neurosci. 18, 404–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasch B., Born J., About sleep’s role in memory. Physiol. Rev. 93, 681–766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delorme J. E., Kodoth V., Aton S. J., Sleep loss disrupts Arc expression in dentate gyrus neurons. Neurobiol. Learn. Mem. 160, 73–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chikahisa S., Fujiki N., Kitaoka K., Shimizu N., Séi H., Central AMPK contributes to sleep homeostasis in mice. Neuropharmacology 57, 369–374 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Chennaoui M., et al. , Effect of acute sleep deprivation and recovery on insulin-like growth factor-I responses and inflammatory gene expression in healthy men. Eur. Cytokine Netw. 25, 52–57 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Boyce R., Glasgow S. D., Williams S., Adamantidis A., Causal evidence for the role of REM sleep theta rhythm in contextual memory consolidation. Science 352, 812–816 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Xia F., et al. , Parvalbumin-positive interneurons mediate neocortical-hippocampal interactions that are necessary for memory consolidation. eLife 6, e27868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eagle A. L., et al. , Experience-dependent induction of hippocampal ΔFosB controls learning. J. Neurosci. 35, 13773–13783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clifton N. E., Trent S., Thomas K. L., Hall J., Regulation and function of activity-dependent Homer in synaptic plasticity. Mol. Neuropsychiatry 5, 147–161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitlock J. R., Heynen A. J., Shuler M. G., Bear M. F., Learning induces long-term potentiation in the hippocampus. Science 313, 1093–1097 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Tononi G., Cirelli C., Sleep and synaptic homeostasis: A hypothesis. Brain Res. Bull. 62, 143–150 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Tononi G., Cirelli C., Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81, 12–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raven F., Aton S. J., The engram’s dark horse: How interneurons regulate state-dependent memory processing and plasticity. Front. Neural Circuits 15, 750541 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho J., et al. , Multiple repressive mechanisms in the hippocampus during memory formation. Science 350, 82–87 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Roach J. P., et al. , Resonance with subthreshold oscillatory drive organizes activity and optimizes learning in neural networks. Proc. Natl. Acad. Sci. U.S.A. 115, E3017–E3025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Durkin J., et al. , Cortically coordinated NREM thalamocortical oscillations play an essential, instructive role in visual system plasticity. Proc. Natl. Acad. Sci. U.S.A. 114, 10485–10490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kandel E. R., Dudai Y., Mayford M. R., The molecular and systems biology of memory. Cell 157, 163–186 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Jiang Y., et al. , Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat. Neurosci. 18, 1446–1454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shigeoka T., Jung J., Holt C. E., Jung H., “Axon-TRAP-ribotag: Affinity purification of translated mRNAs from neuronal axons in mouse in vivo” in RNA Detection, Gaspar I., Ed. (Methods in Molecular Biology, Humana Press, New York, 2018), pp. 85–94. [DOI] [PubMed] [Google Scholar]
- 86.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 87.McDermaid A., Monier B., Zhao J., Liu B., Ma Q., Interpretation of differential gene expression results of RNA-seq data: Review and integration. Brief. Bioinform. 20, 2044–2054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.