Significance
The ribosome has the central role in the translation of RNA. The elongation function of most eukaryotic ribosomes is arrested by the drug cycloheximide (CHX). The site of CHX binding is known. In this work, we determine the structure of a translating ribosome of a filamentous fungus arrested by CHX. By doing so, the specific state of the ribosome arrested in translation is revealed. This work is significant because it provides a structural basis to establish CHX’s mechanism of action. It provides a model for a fungal ribosome and insights into the structure and function of eukaryotic ribosomes through comparative analyses of its features with those from other species.
Keywords: translation, protein synthesis, cryo-electron microscopy
Abstract
Ribosomes translate RNA into proteins. The protein synthesis inhibitor cycloheximide (CHX) is widely used to inhibit eukaryotic ribosomes engaged in translation elongation. However, the lack of structural data for actively translating polyribosomes stalled by CHX leaves unanswered the question of which elongation step is inhibited. We elucidated CHX’s mechanism of action based on the cryo-electron microscopy structure of actively translating Neurospora crassa ribosomes bound with CHX at 2.7-Å resolution. The ribosome structure from this filamentous fungus contains clearly resolved ribosomal protein eL28, like higher eukaryotes but unlike budding yeast, which lacks eL28. Despite some differences in overall structures, the ribosomes from Neurospora, yeast, and humans all contain a highly conserved CHX binding site. We also sequenced classic Neurospora CHX-resistant alleles. These mutations, including one at a residue not previously observed to affect CHX resistance in eukaryotes, were in the large subunit proteins uL15 and eL42 that are part of the CHX-binding pocket. In addition to A-site transfer RNA (tRNA), P-site tRNA, messenger RNA, and CHX that are associated with the translating N. crassa ribosome, spermidine is present near the CHX binding site close to the E site on the large subunit. The tRNAs in the peptidyl transferase center are in the A/A site and the P/P site. The nascent peptide is attached to the A-site tRNA and not to the P-site tRNA. The structural and functional data obtained show that CHX arrests the ribosome in the classical PRE translocation state and does not interfere with A-site reactivity.
Cycloheximide (CHX) is the most widely used laboratory inhibitor of eukaryotic protein synthesis (1). Alma J. Whiffen, a mycologist working at Upjohn, initially identified it in 1946 as a product of Streptomyces griseus fermentation that she named actidione (diketone produced by an actinomycete); actidione inhibited the growth of fungi but not bacteria (2–4). In 1963, CHX’s function to inhibit eukaryotic protein synthesis was demonstrated using a cell-free translation system (CFTS) from Saccharomyces pastorianus (5). The first induced mutations conferring CHX resistance were isolated and genetically mapped in the model fungus Neurospora crassa (6). Around 2 y later, the direct action of CHX to inhibit translation in a mammalian system was demonstrated using active translation lysates prepared from anucleate rabbit reticulocytes (7). The early literature on CHX and related molecules has been thoroughly and contemporaneously reviewed (8).
Structures of the Saccharomyces cerevisiae ribosome (9) and the human ribosome (10), containing CHX tightly bound at the ribosome E site, have given significant insight into its mechanism of action. Based on the position of the CHX binding site, CHX is proposed to interfere with the translocation of P-site transfer RNA (tRNA) to the E site. However, both of these CHX-containing structures were obtained using vacant ribosomes lacking tRNA and therefore did not establish the specific step(s) in the translation cycle at which CHX inhibits elongation. If E-site translocation is blocked as predicted, then this structure is consistent with ribosomes in the pretranslocation (PRE) state with either the nascent peptide on the P-site tRNA and the incoming amino acid on A-site tRNA or in the PRE state with the nascent peptide transferred to A-site tRNA. Both models appear in the literature, with single-molecule biophysical studies supporting the latter (1, 11). Furthermore, the ribosome subunits can have different, relative rotational orientations even while containing A/A and P/P tRNA: A classical PRE state can be discriminated from a rotated PRE* state (12). Thus, there are a variety of states in which CHX could potentially arrest the ribosome even prior to tRNA translocation.
Here, we determined the cryo-electron microscopy (cryo-EM) structure of translating N. crassa ribosomes with CHX at 2.7-Å resolution. To accomplish this, we isolated polysomes from actively growing cells to which CHX was added and maintained CHX at a high concentration throughout all subsequent procedures until the vitrification step of cryo-EM sample preparation. We built a model for the N. crassa ribosome that differs from S. cerevisiae ribosomes and more closely resembles higher-eukaryotic ribosomes in that it contains ribosomal protein eL28. Comparisons of structures with CHX bound to ribosomes showed that the CHX-binding position is highly conserved in N. crassa, S. cerevisiae, and human ribosomes. However, unlike the canonical CHX-bound structures of vacant yeast and human ribosomes (9, 10), which contained an Mg2+ ion adjacent to the CHX-binding pocket, the N. crassa ribosome contained spermidine (SPD) in place of the Mg2+ ion. We sequenced previously identified N. crassa mutations that confer CHX resistance. All amino acid changes in these mutants, including an amino acid change at a previously unidentified position among eukaryotic mutations that confer CHX resistance, map to conserved residues in the CHX-binding pocket. In addition, P/P- and A/A-site tRNAs were present in the N. crassa structure, and the nascent peptide was resolved on the A-site tRNA. Finally, CHX did not appear to interfere with termination as it does with elongation in an N. crassa cell-free translation extract, consistent with CHX interfering with the translocation of tRNA to the E site but not peptidyl transfer events at the A site. The observation that terminating ribosomes are not arrested by CHX could be significant for analyses of the levels of ribosomes mapping to termination codons when CHX is included in ribosome-profiling analyses.
This work provides a structural basis for a mechanistic understanding of CHX action. It also provides a high-resolution model for a fungal ribosome that differs from the S. cerevisiae ribosome.
Results
Overall N. crassa Ribosome Structure.
The original structures of eukaryotic ribosomes containing CHX were obtained using X-ray crystallography or cryo-EM by adding CHX to nontranslating vacant ribosomes (9, 10). Here, we obtained structures of actively translating ribosomes inhibited by CHX from the filamentous fungus N. crassa using cryo-EM and the workflow outlined in SI Appendix, Fig. S1. CHX was added to growing N. crassa cells, and clarified extracts were prepared, layered, and fractionated on sucrose gradients. Ribosomes were collected from the polysome-containing region of the gradients (SI Appendix, Fig. S1A) and prepared for cryo-EM imaging. CHX reversibly binds to ribosomes (13); therefore, we sought to maximize CHX association with ribosomes throughout the isolation and imaging procedures. To accomplish this, we added a high concentration of CHX (2 mg/mL) to the growth medium to arrest translation in vivo and maintained this concentration of CHX at all subsequent steps to obtain samples on grids for imaging.
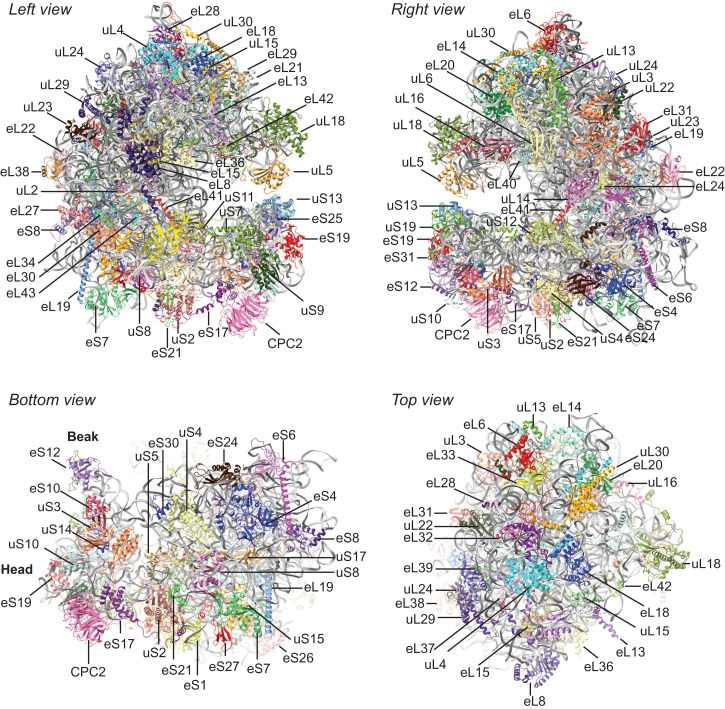
The cryo-EM map of the 80S ribosome at an overall resolution of 2.7 Å (SI Appendix, Fig. S1 C and D) was determined using the approach in SI Appendix, Fig. S1B. We modeled N. crassa ribosomal RNAs (rRNAs) and ribosomal proteins and fitted them into the cryo-EM map (Fig. 1 and SI Appendix, Fig. S1E and Tables S1 and S2). This model of the N. crassa 80S ribosome included 93.6% of 26S, 100% of 5S, 100% of 5.8S, 98.5% of 18S rRNA nucleotides, 42 ribosomal proteins in the large subunit (LSU), and 32 ribosomal proteins in the small subunit (SSU). Heterogeneity of 5S rRNAs has been shown in N. crassa (14); the 5S rRNA model in our structure is based on the major α 5S rRNA sequence. As is seen with other 80S eukaryotic ribosome structures, N. crassa CPC-2 (15), the homolog of RACK1/ASC1, a nonribosomal protein, is stably associated and similarly positioned on the 40S subunit (Fig. 1). The N. crassa ribosome structure obtained also contains A-site and P-site tRNAs, CHX, SPD, and nascent peptide, as discussed below. Additionally, cryo-EM densities for 260 of the 269 Mg2+ ions identified in the S. cerevisiae ribosome structure 6T4Q (16) were confirmed in the N. crassa data. Finally, in this CHX-stalled conformation, regions of the ribosome corresponding to the P1 stalk and L1 stalk were not resolved, and therefore, uL10, uL11 (P1 stalk), and uL1 (L1 stalk) are not in the modeled structure.
Fig. 1.
Positions of N. crassa proteins associated with the 80S N. crassa ribosome. N. crassa proteins were fitted to the N. crassa cryo-EM map as described in the text. Left and right views are rotated 180° as are top and bottom views. Proteins are individually colored; rRNAs are all gray. Models for tRNAs, mRNA, CHX, SPD, Mg2+, and nascent peptide are not shown.
We next compared the overall structure of the N. crassa ribosome to S. cerevisiae ribosomes (SI Appendix, Fig. S2A) and human ribosomes (SI Appendix, Fig. S2B). N. crassa and S. cerevisiae ribosomes are overall similar except for a striking difference at the top of the LSU (SI Appendix, Fig. S2A), comprising rRNA expansion segments ES7L, ES15L, and ribosomal protein eL6 (17, 18). Here, the N. crassa ribosome, like the human ribosome, contains ribosomal protein eL28, while the S. cerevisiae ribosome does not (SI Appendix, Fig. S2A and Movie S1). N. crassa eL28 is encoded by NCU06210, which is annotated as a hypothetical protein in the National Center for Biotechnology Information Database (NCBI Gene ID: 3879074). The interaction of N. crassa ES7L with eL28 is similar to that in the human ribosome (SI Appendix, Fig. S3 A and B). We looked at this difference between the fungal ribosomes more closely. Most Saccharomycotina, including S. cerevisiae and Kluyveromyces lactis, lack an eL28 gene, and there is no such protein in these two yeasts’ ribosome structures (SI Appendix, Fig. S3 C and D). The structured region of the rRNA near N. crassa and human eL28 is not structured in purified S. cerevisiae and K. lactis ribosomes (SI Appendix, Fig. S3 A–D). However, the rRNAs from these yeasts contain the corresponding sequences. Interestingly, while S. cerevisiae ES7 is unstructured in this region in the absence of additional factors (SI Appendix, Fig. S3C), when the ribosome is associated with NatA (19), a protein complex that binds at the ribosome exit pore (SI Appendix, Fig. S3E), ES7 adapts a different structure which is closer to the N. crassa ES7 structure (SI Appendix, Fig. S3 F and G). An additional difference between the N. crassa and S. cerevisiae structures is that the S. cerevisiae ribosome contains density that corresponds to a structured L1 stalk and eIF5A (the large, orange-shaded area in the S. cerevisiae ribosome left view, SI Appendix, Fig. S2A), while N. crassa does not. Finally, the most striking differences between the N. crassa and human ribosomes (SI Appendix, Fig. S2B) are that human ribosomal rRNAs contain longer expansion segments than fungal rRNAs that can contribute to the structure (17).
Localization of CHX in the N. crassa Ribosome.
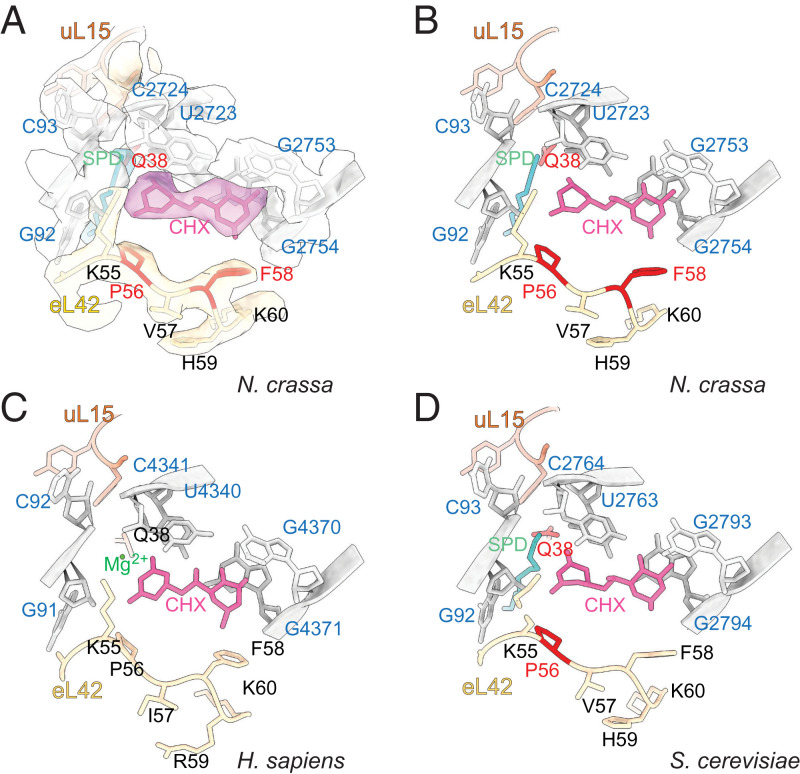
We observed well-resolved, extra density for CHX near the E site of the LSU (Fig. 2 A and B and SI Appendix, Fig. S4A). CHX is located in a region demarcated by eL42, conserved 26S rRNA residues, and uL15. Comparison of the position of CHX in the N. crassa ribosome with its positions in the S. cerevisiae and human ribosomes reveals strong conservation of its orientation in the ribosome and the rRNA and ribosomal proteins surrounding it (Compare Fig. 2 B–D).
Fig. 2.
CHX and SPD are both present in the N. crassa ribosome E site in actively translating ribosomes arrested by CHX. (A) Densities corresponding to CHX, SPD, and nearby N. crassa ribosomal components are indicated as surfaces and models as sticks and ribbons. (B–D) Comparisons of models of CHX and SPD or Mg2+ in the translating N. crassa ribosome (B); translating S. cerevisiae ribosome (C) (PDB ID: 6TNU); vacant human ribosome (D) (PDB ID: 5LKS). CHX is fuchsia; SPD is cyan; rRNA is gray; and eL42 and uL15 are yellow and orange, respectively, with residues at which mutations in the corresponding organism are known to result in CHX-resistance shaded red.
Importantly, additional density was present close to the N. crassa CHX binding site. We identified it as SPD (Fig. 2 A and B and SI Appendix, Fig. S5A). SPD was also recently modeled in a similar site in the submitted structure of the S. cerevisiae ribosome containing bound CHX and eIF5A (Protein Data Bank [PDB] ID: 6TNU, ref. 20) (SI Appendix, Fig. S5B). In cryo-EM maps of human ribosomes containing CHX, modeled Mg2+ or unmodeled SPD appear to be present (SI Appendix, Fig. S5 C and D) (10, 21). In other structures of human and S. cerevisiae ribosomes containing E/E-site or P/E-site tRNA (16, 21), unmodeled SPD is also present in a similar position (SI Appendix, Fig. S5 E and F). Thus, SPD binding to this region of the eukaryotic ribosome near the CHX binding site appears to be a common feature. Its structural and functional role(s), which remain to be determined, may well be of interest given its close vicinity to such an important functional site of the ribosome.
The comparisons of ribosomes containing CHX and containing E-site tRNA (SI Appendix, Fig. S5) support the proposed functional role of CHX to prevent the binding of deacylated tRNA (9, 10). CHX would clash with tRNA as the tRNA entered the E site upon transition from the P/P into the hybrid P/E position after peptidyl transfer.
N. crassa Mutations that Cause CHX Resistance Map to the CHX Binding Site.
CHX-resistant alleles of two identified genes that confer CHX resistance were obtained from the Fungal Genetics Stock Center. Three alleles of cyh-1 (NCU00706) and two alleles of cyh-2 (NCU03806) were sequenced (SI Appendix, Table S3). The cyh-1 mutations, which affect eL42, were P56L (two independent alleles) and F58L. The two alleles of cyh-2 are two different mutations of the same residue in uL15: Q38K and Q38L. Like ribosomal mutations in S. cerevisiae and other organisms that result in CHX resistance, these mutations are close to the CHX binding site; the affected residues are indicated in red in Fig. 2. In addition, one of these residues identified by mutations in N. crassa and S. cerevisiae, uL15 Q38, and another residue identified in S. cerevisiae, H39 (22), are also in close proximity to SPD (Fig. 2 and Movie S2). All of the N. crassa mutants were more resistant to CHX than the wild-type in vivo (SI Appendix, Fig. S6) and in CFTSs derived from these strains (SI Appendix, Fig. S7A). In contrast, mutant and wild-type CFTSs were similarly sensitive to hygromycin B (HYG) (SI Appendix, Fig. S7B), an aminoglycoside translation inhibitor that binds near the ribosome A site (23), consistent with CHX and HYG targeting different regions of the LSU. These data, including the identification of a new mutation at a conserved residue that affects CHX resistance (F58L) and that maps to the CHX binding site, highlight the importance of the specific amino residues in this region for modulating the sensitivity of these ribosomes to inhibition by CHX.
The Positions of tRNAs, Nascent Peptide, Messenger RNA, and Ribosome Components Show that CHX-Arrested Ribosomes Are in the Classical PRE Translocation State.
The structure of the CHX binding site is highly conserved in eukaryotic ribosomes. Ribosomes go through many different conformations during the various steps in the elongation cycle (12). At what step(s) does CHX inhibit elongation? We could answer this question by analyses of the peptidyltransferase center in the N. crassa ribosome structure. The CHX–ribosome structure determined here differs from previously published CHX-containing structures obtained from vacant ribosomes to which CHX was added (9, 10) and from subsets of translating ribosomes that contained CHX but were affinity-purified based on some other distinguishing feature (20, 21). The structure of the N. crassa ribosome containing CHX shows A/A- and P/P-site tRNAs (Fig. 3A). We wanted to know whether any subclass(es) of these ribosomes had tRNA in hybrid or other states, since mammalian ribosomes purified with CHX were observed to be in three major classes: classical–PRE, hybrid–PRE, and POST (posttranslocation) (21). Therefore, we did a three-dimensional (3D) classification of these particles into 10 subclasses. Every subclass had A/A- and P/P-site tRNA, and none had P/E- or A/P-site hybrid tRNA (SI Appendix, Fig. S8). These data indicated that the purification procedure we used maintained ribosomes in a CHX-arrested, classical PRE state.
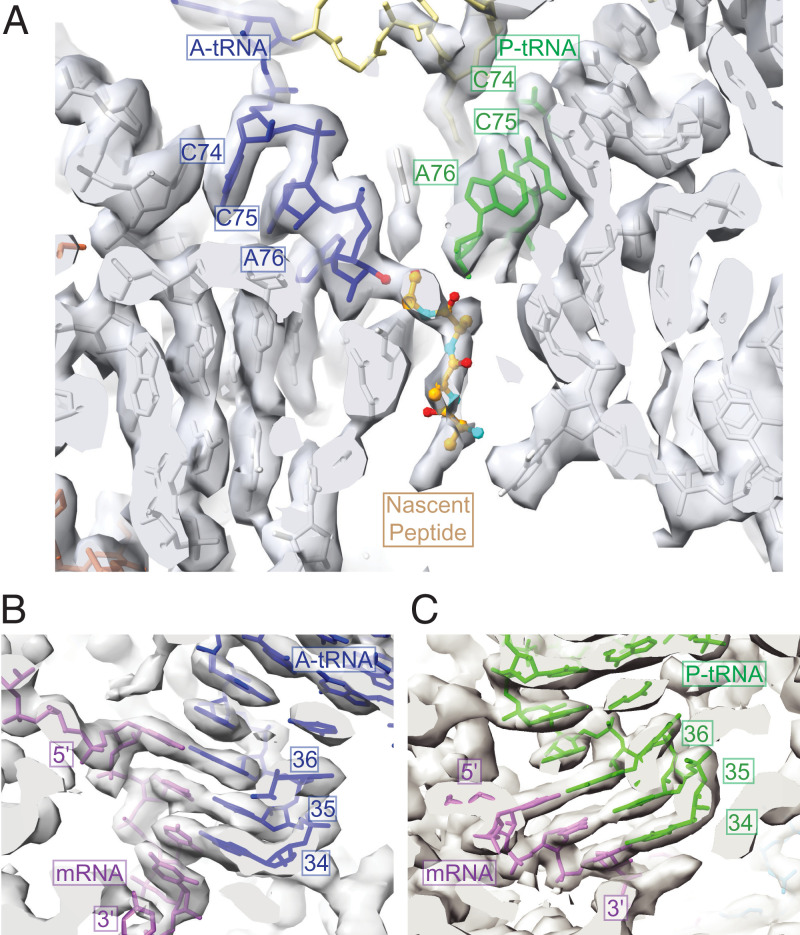
Fig. 3.
CHX arrests ribosomes in the PRE translocation state with the nascent peptide attached to the A-site tRNA. (A) A- and P-site tRNAs at the peptidyltransferase center, with the nascent peptide attached to the A-site tRNA CCA end as shown by the EM density. (B) A-site tRNA anticodon interaction with the mRNA A-site codon. (C) P-site tRNA anticodon interaction with the mRNA P-site codon. A-site tRNA, blue; P-site tRNA, green; mRNA, purple; nascent peptide, various colors; rRNA, gray; uL16, yellow; and uL3, orange.
No specific tRNAs are represented in the peptidyltransferase centers of polysomal ribosomes. Therefore, we adapted the tRNA model for a eukaryotic ICG–anticodon tRNA that decodes Arg codons (PDB ID: 6T4Q) (16) to model the tRNA densities of both A- and P-site tRNAs. Fitting the models to the tRNA densities allowed visualization of both A-site and P-site tRNAs in the peptidyltransferase center (Fig. 3A) and visualization of A-site and P-site messenger RNA (mRNA)–tRNA codon–anticodon interactions (Fig. 3 B and C). Importantly, we observed extra density corresponding to the nascent peptide attached to the 3′-hydroxyl group of the A-site tRNA but not the P-site tRNA (Fig. 3A). The map’s local resolution at the position of this peptide–tRNA bond was ∼2.6 Å (SI Appendix, Fig. S4B). The density corresponding to the nascent peptide was not fully modeled because of the presence of many different nascent peptide species and nascent peptide conformations in polysomes. In contrast to the limited resolution of nascent peptide density, the density corresponding to the ribosome exit tunnel, including the constriction site important for nascent peptide interactions with the ribosome, could be confidently modeled (SI Appendix, Fig. S9).
The positions of the A/A- and P/P-site tRNAs and the nascent peptide’s attachment to the A-site tRNA indicated that the N. crassa ribosome arrested by CHX is in the PRE state. Several conformations of the PRE state in human ribosomes can be discriminated by the relative orientations of the SSU and LSU and the positions of tRNAs (12). The conformation of the N. crassa ribosome with CHX (Fig. 4A), assessed by comparing the relative SSU–LSU configurations, better resembled the human classical-1 PRE state determined in the absence of CHX (Fig. 4B) than the rotated PRE* state (Fig. 4C).
Fig. 4.
The N. crassa CHX-bound ribosome is in the classical PRE state conformation. (A–C) Bottom view of the 80S ribosomes from N. crassa and H. sapiens low pass filtered to 10 Å for clarity. The 60S subunits are all aligned to the N. crassa 60S subunit. (A) The 40S subunit of N. crassa ribosome (blue). N. crassa mRNA is purple. (B) The 40S subunits of N. crassa ribosome (blue) and H. sapiens classical-1 PRE state ribosome (pink, EMD-2909) align well. (C) The 40S subunit of H. sapiens PRE* state ribosome (pink, EMD-2906) is rotated slightly counterclockwise compared to the 40S subunit of N. crassa ribosome (blue). (D–F) N. crassa uS19 (tan) and human uS19 (fuchsia) have similar contacts and conformations in the CHX-bound PRE state ribosome. The view given is similar to that in Bhaskar et al. (21) for comparison of the position of the uS19 C-terminal tail. The density of mRNA is low pass filtered to 10 Å for clarity, and the surface is displayed in purple. The C-terminal tails of both uS19 proteins interact with mRNA, A-site tRNA (blue), and P-site tRNA (green). (D) N. crassa uS19 position (tan) in the ribosome. (E) Human uS19 position (fuchsia) in the ribosome (EMD-10668, PDB ID: 6Y0G). tRNA densities were extracted from the cryo-EM map using ChimeraX. (F) S. cerevisiae uS19 position (rose) in the ribosome (EMD-10537, PDB ID: 6TNU).
Additional structural analyses support the assignment of the CHX-stalled N. crassa ribosome to the classical PRE state. Recently, the C-terminal tail of human uS19 was shown to change conformation depending on the functional state of the ribosome (21). This uS19 domain is positioned between the A- and P-site tRNAs near the anticodon–codon interaction with mRNA. Consistent with the N. crassa ribosome assignment to the classical PRE state, the position of N. crassa uS19 C-terminal tail in the CHX-stalled conformation is highly similar to the human uS19 in the classical PRE conformation (Fig. 4 D and E). These regions of N. crassa and human uS19 share identical amino acid sequences, while the S. cerevisiae uS19 C-terminal tail differs in sequence (SI Appendix, Fig. S10A). When comparing uS19 structures in N. crassa, human, and S. cerevisiae ribosomes containing CHX, all three uS19 structures are similar, except that the S. cerevisiae model lacks this C-terminal tail (SI Appendix, Fig. S10B). Thus, the A- and P-site tRNAs, uS19, and CHX are similarly positioned in the N. crassa and human structures, based on both models and cryo-EM maps, showing that CHX-bound ribosomes in both species are in the classical PRE state. However, unlike the case for the N. crassa ribosome, we do not see density corresponding to the nascent peptide on the A-site tRNA in the corresponding cryo-EM map for the mammalian, classical PRE ribosome (EMD-10668) (21), possibly because the peptide is disordered.
In conclusion, the structural data indicate that the CHX-arrested, translating N. crassa ribosome is in the classical-1 PRE state with the nascent peptide transferred to the A-site tRNA.
CHX Does Not Inhibit a Translation Termination Event.
CHX does not interfere with A-site reactivity during elongation since the nascent peptide is transferred to the A-site tRNA. Therefore, in the presence of CHX, termination should still occur since release factors enter the A site to cause chain termination, and chain termination does not require tRNA translocation (24). Analyses of the positions of ribosomes in fungal CFTSs by primer–extension inhibition (toeprint) assays demonstrated that adding CHX before adding RNA to the system results in stabilization of ribosomes with the initiation codons of open reading frames (ORFs) in the P site and the second ORF codon in the A site (25). This is consistent with CHX causing elongation arrest at initiation codons. Toeprinting of CFTS translating RNA in the absence of CHX reveals the positions of ribosomes engaged in initiation, elongation, and termination (26). Early evidence that CHX does not interfere with termination was obtained using toeprinting to map the positions of ribosomes on mRNA in an S. cerevisiae CFTS (26). Since we had the structure of the N. crassa ribosome with CHX, we looked at the effect of CHX on termination kinetically using an N. crassa CFTS. We toeprinted ribosomes on a luciferase (LUC) reporter RNA with an upstream ORF (uORF) specifying the arginine attenuator peptide (AAP), which causes well-established, arginine-dependent transient stalling of ribosomes with a termination codon in the A site (27). If CHX did not inhibit termination, then ribosomes should be released from stalling through eventual, successful termination. However, if CHX inhibited termination, then the transient, Arg-dependent termination stall should be stabilized by CHX, as are elongation stalls. The results show that while CHX stabilizes ribosomes engaged in elongation (Fig. 5, indicated by arrowheads and brackets), it does not stabilize terminating ribosomes (Fig. 5, indicated by stars). Specifically, toeprint analyses of CFTS, to which CHX had been added for 0, 2.5, or 5 min, showed progressively decreased signals of ribosomes stalled at termination codons (Fig. 5, compare lanes 2, 3, and 4 and compare lanes 6, 7, and 8), consistent with termination occurring in its presence. Stalling of ribosomes at the AAP termination codon, which is Arg dependent, is most obvious when CFTS were analyzed immediately (0 min) after adding CHX (Fig. 5, compare lanes 2 and 6). In striking contrast, after 5 min in CHX, there was little difference in the signal from ribosomes at the AAP termination codon in low or high Arg (Fig. 5, compare lanes 4 and 8). This indicates that ribosomes at the termination codon are not stabilized by CHX. In contrast, toeprints corresponding to ribosomes engaged in elongation within the AAP-coding region, including those with the initiation codon in the ribosome P site, remained stable or increased during incubation with CHX, consistent with their expected stabilization by CHX.
Fig. 5.
Toeprinting shows that terminating ribosomes release from RNA in the presence of CHX but elongating ribosomes do not. In vitro synthesized, capped, and polyadenylated LUC reporter mRNA containing the wild-type AAP uORF was translated in the N. crassa CFTS in the presence of 10 µM (−) Arg or 2 mM (+) Arg (low or high Arg). CHX (final concentration 0.5 µg/µl) was added to the CFTS reactions either before the addition of mRNA (time T0) or after translation was underway for 10 min (steady-state translation, time T10). Steady-state reactions to which CHX was added were incubated in the presence of CHX for an additional 0, 2.5, or 5 min as indicated. The sequence of the mRNA can be directly deduced from the sequencing lanes reading from top to bottom. The positions of the uORF start and stop codons, and the LUC start codon, are boxed in the nucleotide sequence. The primer extension (toeprint) products corresponding to elongating ribosomes that have the uORF and LUC initiation codons in the P site are indicated by arrowheads; terminating ribosomes with the uORF termination codon in the A site are indicated with a star; and elongating ribosomes stalled within the uORF coding region are marked by a bracket. Control samples of RNA in reaction mixtures without extract and reaction mixtures containing extract but not mRNA are as indicated.
Discussion
CHX is commonly used to inhibit eukaryotic translation and is an important reagent for ribosome-profiling analyses. Thus, it is crucial to understand its action during translation. Despite extensive biochemical work, however, CHX has only been studied from a structural perspective in the context of vacant 80S ribosomes (9, 10). Here, we focused on its action on translating ribosomes. We purified N. crassa ribosomes from polysome fractions isolated from cells in which translation was arrested by CHX and obtained their structure through single-particle cryo-EM. These data provide direct structural evidence that CHX’s mechanism of action is to arrest ribosomes in the classical PRE state and do not support alternative models for its mechanism of action.
In N. crassa ribosomes containing CHX, the tRNAs are in the A/A and P/P sites, and there is no E-site tRNA. CHX has arrested these ribosomes in the classical PRE state with the nascent peptide attached to the A-site tRNA. The bases for assigning the CHX-arrested structure to the classical PRE state are as follows. First, the respective positions of the SSU and the LSU are most similar to the respective positions of human ribosomes in the classical-1 PRE state. Second, the uS19 C-terminal tail is resolved in the N. crassa structure and is in a conformation nearly identical to that of uS19 in human ribosomes in the classical PRE state. The classical PRE state is so far the only ribosome conformation for which this uS19 conformation has been observed (21). Furthermore, the N. crassa ribosome, like the human ribosome in the classical PRE state, does not contain eIF5A and has an unstructured L1 stalk. Also, consistent with arrest in the classical PRE state, the CHX-arrested N. crassa ribosome does not contain eEF2, which is associated with ribosomes in the process of translocation (12), or eEF1A, which is associated with POST-state ribosomes (28). This structural determination of the CHX-arrested state of ribosomes in polysomes as classical PRE with the peptide on the A-site tRNA is consistent with single-molecule biophysical data (11).
The use of high concentrations of CHX throughout the structural analysis procedure used here resulted in a population of ribosome particles that lacked any significant ribosome subpopulations containing tRNA in the P/E site. In contrast, in a study in which structural analyses followed purification of ribosomes with lower concentrations of CHX, which also included affinity purification steps, significant subpopulations of ribosomes lacking CHX and ribosomes with tRNAs in hybrid translocation states are also observed (21). Recent measurements of the dissociation constant of CHX from ribosomes yielded values of 1 ± 0.4 μM for S. cerevisiae ribosomes and 4 ± 1 μM for human ribosomes (29). A 1-μM dissociation constant corresponds to a half-life of 0.7 s for a first-order reaction (30). Because CHX binding is not irreversible, when it leaves, there is an increased kinetic chance of moving to the hybrid state occurring before CHX rebinding. The high concentrations of CHX we used could minimize these occurrences. The dissociation of CHX resulting in procession through the hybrid state could cause an additional round of elongation, which could be followed by subsequent CHX rebinding and arrest. Such considerations could be the basis for an alternative way to account for the proposed differences in CHX and lactimidomycin mechanisms of action that are distinct from mechanistic differences arising because of differences in the structures of these molecules (1).
The structure of CHX binding in translating N. crassa ribosomes is similar to the corresponding structures in vacant yeast and human ribosomes (9, 10). Thus, CHX is not altered by the presence of A/A- and P/P-site tRNA. Molecular analyses of the classical N. crassa CHX-resistant mutations (all of the alleles of cyh-1 and cyh-2 available from the Fungal Genetics Stock Center) showed that they affected residues at the binding site. Mutations at residues previously identified as important for CHX resistance were directly affected by some alleles: uL15 residue Q38 and eL42 residue P56. A mutation at an additional eL42 residue, F58L, was also discovered.
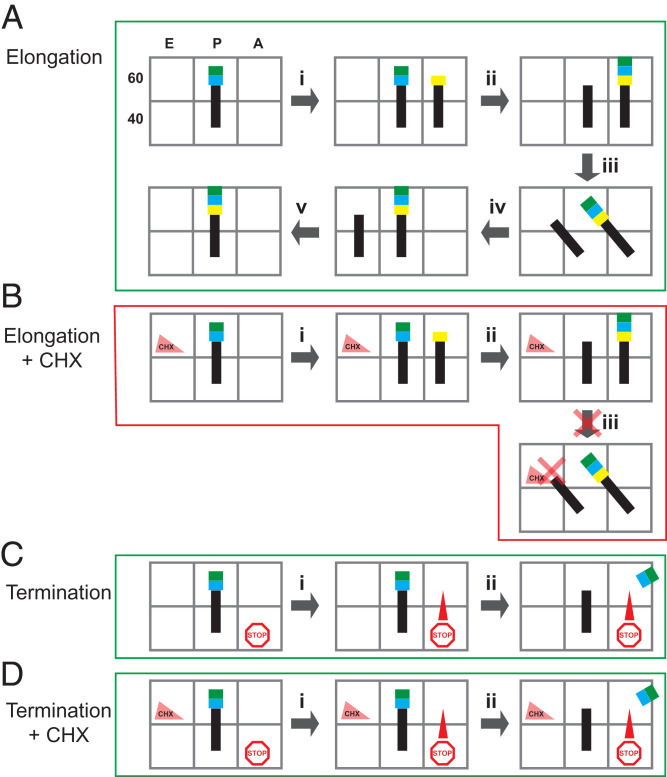
These data can be integrated into the model initially proposed for CHX action, based on structural determination of its location in nontranslating (vacant) ribosomes in which CHX acts by clashing with occupancy of the E-site tRNA (9), but the structure of the translating ribosome’s peptidyltransferase center was not known. Fig. 6 diagrams this integrated model for CHX action. Normally, in elongation (Fig. 6A), aminoacyl tRNA enters the A site (Fig. 6 A, i), and peptidyltransferase activity transfers the peptide to it from the P-site tRNA (Fig. 6 A, ii). Translocation of the tRNAs occurs (Fig. 6 A, iii and iv), and deacylated tRNA leaves the E site (Fig. 6 A, v). When CHX is in the E site (Fig. 6B), aminoacyl tRNA entry into the A site and transfer of the peptide onto the A-site aminoacyl tRNA occur (Fig. 6 B, i and ii), but translocation cannot happen. Therefore, the ribosome is arrested in the classical PRE state with the peptide on the A-site tRNA. Consistent with this model, our data indicate that CHX does not interfere with termination, because termination does not require translocation of tRNA (Fig. 6 C and D).
Fig. 6.
Model for CHX action. E, P, and A sites of the 60S and 40S subunits are cartooned as open boxes; tRNAs are black lines; amino acids are blue, green, and yellow boxes; and CHX is a triangle in the 60S E site. (A) For a single-elongation cycle, in the absence of CHX, the aminoacyl-tRNA enters the empty A site (i), followed by nascent peptide transfer to the A-site tRNA (ii). Translocation occurs (iii and iv), and the deacylated tRNA leaves the E site to complete the cycle (v). (B) When CHX is bound in the E site, the aminoacyl-tRNA enters the empty A site (i), and transfer of the nascent peptide to the A-site tRNA occurs (ii). However, CHX in the E site prevents translocation of the P-site tRNA to the E site, arresting the ribosome in the PRE translocation state with the polypeptide attached to the A-site tRNA and deacylated tRNA in the P site. (C) For termination, a stop codon in the A site recruits release factor (i) resulting in release of the nascent peptide without translocation (ii). (D) When CHX is bound in the E site, termination occurs because translocation is not required.
The loss of ribosomes with a termination codon in their A site from an mRNA in the continued presence of CHX, while ribosomes engaged in elongation on that mRNA are stabilized, has implications for assessing signals from terminating ribosomes in ribosome-profiling (ribo-seq) studies employing CHX to maintain ribosomes association with mRNA. The levels of ribosomes observed at termination codons could depend on differential effects of preparation and analysis conditions that do not have similar consequences for levels of ribosomes arrested during elongation because these latter ribosomes are generally stabilized by CHX.
We observed an additional density close to the N. crassa CHX binding site and unambiguously identified it as SPD (Fig. 2 A and B and SI Appendix, Fig. S5A). SPD is a polycation and is thought to function similarly to Mg2+. Indeed, this SPD site is occupied by Mg2+ in the structure of the vacant human 80S ribosome bound with CHX (SI Appendix, Fig. S5C) and the vacant S. cerevisiae ribosome bound with CHX (9, 10). We sought to determine the generality of this finding of SPD instead of Mg2+. SPD was recently modeled in the submitted structure of the S. cerevisiae ribosome containing bound CHX and eIF5A (20), and it is at the corresponding site (SI Appendix, Fig. S5B). While not modeled, densities for SPD at the corresponding sites are also present in S. cerevisiae ribosome without CHX but with E-site tRNA and human ribosomes with CHX or with E-site tRNA (SI Appendix, Fig. S5 D–F, respectively). These data, therefore, identify a conserved SPD binding site in eukaryotic ribosomes. It is worth noting that when we looked at 269 assigned Mg2+ binding sites in the S. cerevisiae ribosome in the N. crassa ribosome, we saw density corresponding to 260 of these Mg2+ molecules; in no case did we see density corresponding to SPD in these locations. Thus, while Mg2+ and SPD could potentially function interchangeably, it appears that the SPD binding site near the CHX binding site is different from other Mg2+ binding sites. In a published structure of rabbit 80S ribosome stalled on a poly(A) tail, two SPD molecules were provisionally included in the model (PDB ID: 6SGC) near the CCA 3′ end of P-site tRNA (31). However, we do not see corresponding densities in the N. crassa structure nor do we see density corresponding to SPD in the E site in this rabbit ribosome structure. Finally, some binding sites of SPD have been identified in high-resolution, prokaryotic ribosome structures [PDB ID codes: 7NHN (32), 4YBB (33), and 5IT8 (34)]. We compared these structures with the N. crassa ribosome structure and did not see densities for those SPD molecules in the corresponding regions of the N. crassa ribosome.
As is the case for the CHX-bound N. crassa ribosome, S. cerevisiae ribosomes that contain both CHX and eIF5A also have the nascent peptide attached to the A/A-site tRNA (20). However, in the presence of eIF5A, the L1 stalk is structured, and the E-site’s structure, therefore, differs from that of the N. crassa ribosome. Furthermore, the C-terminal tail of uS19 is not resolved in this S. cerevisiae structure (Fig. 4F), consistent with the idea that a structured uS19 C-terminal tail, as observed in the N. crassa and human classical PRE ribosomes, is a distinct feature of that translational step. Thus, while there are similarities between the N. crassa and S. cerevisiae structures, as they each contain CHX, SPD, and nascent peptide on the A/A tRNA, these ribosomes are not in the same state. Therefore, the S. cerevisiae ribosome likely represents an eIF5A-specific state distinct from the classical PRE state seen in the presence of CHX alone.
This ribosome structure from the Ascomycete N. crassa represents a near atomic structure of a cytosolic ribosome from a filamentous fungus. The structure of the N. crassa mitochondrial ribosome was also recently determined (35). The structure of the N. crassa cytosolic ribosome differs from the Ascomycete yeast S. cerevisiae in that it contains eL28 while the yeast ribosome does not. Thus, as with the demonstration of the structure of a human-like translation factor eIF3 in N. crassa that is more complex than S. cerevisiae eIF3 (36) and the existence of mammalian-like, exon junction complex– mediated, nonsense-mediated mRNA decay in N. crassa that S. cerevisiae lacks (37, 38), this structure provides insight into important similarities and differences in the eukaryotic translational machinery. The 80S K. lactis ribosome structure (39) also lacks eL28; the yeast K. lactis, like S. cerevisiae, is a Saccharomycetes member. Interestingly, while eL28 is present in some protozoa, it is also missing in the ribosome structure from the protozoan Trichomonas vaginalis (18). How the loss of eL28 figures in these organisms’ evolutionary history and the consequences of its presence versus absence for ribosome function are questions that remain to be answered.
In summary, we obtained the structure of the translating N. crassa ribosome arrested by CHX. This structure provides direct evidence that CHX arrests ribosomes in the classical PRE state with the nascent peptide on the A-site tRNA. The reactivity of the A site in the presence of CHX is also demonstrated by toeprinting analyses which showed that termination, unlike elongation, is not arrested by CHX. These studies also revealed that the N. crassa ribosome is structurally different from its budding yeast counterpart in containing eL28 and, in this respect, more closely resembles the ribosomes of mammals. Most fungi have eL28, and the N. crassa ribosome structure could be paradigmatic for these species, which include important Ascomycete pathogens such as Aspergillus fumigatus and Basidiomycete pathogens such as Cryptococcus neoformans. This work thus provides a structural basis for understanding CHX action and provides a model for these fungal ribosomes.
Materials and Methods
Strains and Culture Conditions.
The N. crassa wild-type strain FGSC 2489 (74-OR23-1V A) and CHX-resistant N. crassa strains (SI Appendix, Table S3) were obtained from the Fungal Genetics Stock Center (40) and maintained as described (37, 41). Homokaryons of all strains were obtained by microconidiation (42).
Preparation, Analyses, and Purification of N. crassa Polysomes.
N. crassa polysomes were prepared and analyzed, as previously described, with minor changes (43). Suspension of conidia was inoculated onto 50 mL solid Vogel’s sucrose medium in a 250-mL flask. The culture was incubated at room temperature for 14 d, and conidia were harvested through two layers of cheesecloth. The number of conidia was counted, and the concentration was calculated. Conidia were inoculated to 500 mL Vogel’s sucrose medium to make the final concentration of 1 × 107 conidia/mL, and the culture was incubated at 32 °C with orbital shaking (180 rpm). After about 6 h, when 90% of the conidia show germ tubes, CHX (2 mg/mL) was added to the culture 5 min before harvesting, and the germlings were harvested by vacuum filtration onto Whatman 541 filter paper. The mycelial pad was peeled off the filter paper and weighed. Mycelial pads (0.25 g each) were transferred to 2-mL screwcap tubes containing ice-cold 0.75 mL polysome extraction buffer (100 mM KCl, 20 mM Hepes-KOH [pH 7.5], 2 mM magnesium acetate, 15 mM 2-mercaptoethanol, and 2 mg/mL CHX) and 0.5 g zirconia/silica beads (0.5-mm diameter). The tubes were disrupted using a beadbeater at 4 °C for 50 s and centrifuged at 16,100 × g at 4 °C for 5 min. Supernatants (0.45 mL) were transferred to fresh 2-mL screwcap tubes, frozen in liquid nitrogen, and stored at −80 °C.
For polysome analyses, 400 μL (about 18 A260 units) homogenate was layered on 12-mL linear sucrose gradients, which contain 10 to 50% (weight/weight) sucrose, 10 mM Hepes-KOH (pH 7.5), 70 mM ammonium acetate, 4 mM magnesium acetate, and 2 mg/mL CHX. Gradients were centrifuged using a Beckman SW41 rotor at 41,000 rpm at 4 °C for 2 h. BioComp Gradient Station Model 153 and Gilson Fraction Collector FC203B were used to generate the sucrose gradients and to analyze and fractionate the gradients, respectively. Polysome profiles were generated, and 26 fractions were collected from each gradient, flash frozen in liquid nitrogen, and stored at −80 °C.
Fractions corresponding to the polysomes (disomes and greater) from multiple gradients were pooled, washed, and concentrated using Corning Spin-X UF 6 mL Centrifugal Concentrators (100,000 molecular weight cut-off). The washing buffer was the buffer A that has been used to prepare N. crassa cell-free extract for in vitro translation [30 mM Hepes-KOH pH 7.6, 100 mM KOAc pH 7.0, 3 mM Mg(OAc)2 pH 7.0, 2 mM DTT] supplemented with 2 mg/mL CHX. For each centrifugation, about 15 mL washing buffer was used, and the volume was concentrated to about 1 mL. The washing/concentrating was repeated six times to remove sucrose. Finally, the ribosomes were concentrated to about A260 = 10.
Cryo-EM Sample Preparation.
Cryogrids were prepared as previously described (44). A total of 3 μL purified N. crassa polysomes was applied onto glow-discharged C-Flat 2/1 holey grids. The grids, equilibrated at 16 °C and 100% relative humidity, were plunged into liquid ethane to freeze them using a Vitrobot Mark III (FEI company).
Cryo-EM Single-Particle Data Acquisition.
The frozen grids were loaded in a Titan Krios cryoelectron microscope (Thermo Fisher Scientific) operated at 300 kV, condenser lens aperture 50 μm, spot size 7, and parallel beam with an illuminated area of 1.08 μm in diameter. The nominal magnification was at 130,000×, corresponding to a calibrated sampling of 1.06 Å per physical pixel. Movie stacks were collected automatically using EPU software on a K2 direct electron camera equipped with a Bioquantum energy filter with an energy slit of 20 eV (Gatan), operating in counting mode at a recording rate of 5 raw frames/s and a total exposure time of 5 s, yielding 25 frames/stack, and a total dose of 32 e−/Å2. A total of 1,836 movie stacks were collected.
Cryo-EM Image Processing.
All micrographs were imported into RELION-3.1 (45) for image processing. The motion correction was performed using MotionCor2 (46). The defocus and astigmatism were determined using CTFFIND4 (47). All particles were autopicked using the convolutional neural network–based script e2boxer.py in EMAN2 (48) and further checked manually to remove bad particles. After 2D classification in RELION-3.1, a total of 206,276 particles were subjected to 3D classification in RELION-3.1. The 3D classification of LSU and SSU was performed in RELION-3.1 to further remove bad particles. Then, a total of 196,154 particles were subjected to Bayesian polishing in RELION-3.1, followed by 3D reconstruction using cryo-SPARC (49). A sharpening B-factor of −65.4 Å2 was applied to the resulting cryo-EM map to yield the final sharpened map at 2.7-Å global resolution estimated by the 0.143 criterion of the Fourier Shell Correlation (FSC) curve. Local resolution maps were determined in RELION-3.1.
Molecular Modeling.
The sequences of N. crassa 5S, 5.8S, 18S, and 26S rRNA used to build the model were obtained through NCBI (GenBank IDs K02469.1, M10692.1, FJ360521.1, and FJ360521.1, respectively). Template-based comparative modeling was performed using modeRNA (50). The S. cerevisiae 80S structures (PDB ID codes: 4U3U and 6T4Q) (9, 16) were chosen as the templates. The sequences were aligned using the SILVA ACT website (https://www.arb-silva.de/aligner/) (51). The structures of the rRNA expansion segments were manually built using Rosetta (52) and Coot (53) with the aid of the secondary structures that were predicted by RNAfold WebServer (rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (54). The rRNA structures were refined using Rosetta and PHENIX (55).
The sequences of N. crassa ribosomal proteins were obtained from the UniProt database. For each ribosomal protein, the initial homology model was generated using MODELER (56) and then refined into the cryo-EM map using Rosetta (52). The best model was then selected based on both the geometry and the fitting scores.
The tRNA model used for A- and P-site tRNAs was the eukaryotic ICG-anticodon tRNA that decodes Arg codons (PDB ID: 6T4Q) (16). The mRNA model and nascent peptide model were adapted from the mRNA model and nascent peptide model in a human classical PRE ribosome structure (PDB ID: 6Y0G) (21).
CHX and SPD modeling was done with the electronic Ligand Builder and Optimization Workbench (57). The models of Mg2+ ions were adapted from the S. cerevisiae ribosome structure [PDB ID: 6T4Q (16)].
The models of all rRNA, ribosomal proteins, tRNA, mRNA, nascent peptide, CHX, SPD, and Mg2+ were then merged into one model, refined using Rosetta and PHENIX and inspected and adjusted using Coot.
The statistics obtained by using PHENIX and MolProbity (58) of the refined 80S ribosome model are listed in SI Appendix, Table S4.
Structural Figures and Movies Preparation.
All figures and movies were made using UCSF Chimera (59) and UCSF ChimeraX (60).
Sequencing of N. crassa cyh-1 and cyh-2 Alleles.
Genomic DNA was isolated as described (43). For each strain, the DNA sequences of both cyh-1 (NCU00706) and cyh-2 (NCU03806) genes were determined by PCR amplification of genomic DNA, followed by sequencing of the PCR product. For cyh-1, oligo pair OLS041 (GACCTCACACATCAACGA)/OLS042 (TTCGCAACCTCGCTACCA) were used for amplification and sequencing and an additional oligo, OLS057 (TCATGTCGCGTCGAGCTCTGT), was also used for sequencing. For cyh-2, oligo pairs CYH2F1 (CGAGACCCGTGAAGCGTCT)/CYH2R1 (TTGAGGATGGGGGCCCAC) and CYH2F2 (GACGATGACGGATAGACC)/CYH2R2 (GGCTTCTGGACGAATGTT) were used for both amplification and sequencing. The cyh-1 sequencing reads were aligned to NCBI Reference Sequence, XM_959447.3, and the cyh-2 sequencing reads were aligned to GenBank, AL513466.1 (which includes cyh-2), to identify the mutations.
Determination of CHX Resistance Levels In Vivo.
For testing CHX resistance, conidia (104 in 50 μL sterile water) from wild-type and mutant strains were inoculated into 16 × 125-mm tubes containing 3 mL Vogel’s minimal medium/2% sucrose/2% agar supplemented with CHX after autoclaving. Cultures were grown for 8 d at room temperature. Tubes were scored for growth and photographed.
Preparation of Cell-Free Translation Systems, Determination of CHX Resistance Levels In Vitro, and Toeprinting.
The procedures for preparation of CFTS, preparation of in vitro–synthesized capped and polyadenylated LUC RNA for translation and for toeprinting without and with CHX were as previously described (26, 41). For testing CHX sensitivity and Hyg sensitivity, drugs at the indicated concentrations were added at T0.
Supplementary Material
Acknowledgments
We are grateful for cryo-EM support from Dr. Wah Chiu. Cryo-EM data were collected at the Stanford Linear Accelerator Center and Stanford under NIH grants (S10OD021600 and U24GM116787). This work was supported by grants NIH R35 GM126966 to D.B.-P., NIH regional cryo-EM data collection consortium U24GM1167 and Welch Foundation A1863 to J.Z., and NIH R21 AI138158 to M.S.S.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111862118/-/DCSupplemental.
Data Availability
The Coulomb potential map and atomic coordinates have been deposited in the Electron Microscopy Databank (EMDB code: EMD-24307) (61) and PDB (PDB ID code: 7R81) (62). All other study data are included in the article and/or supporting information.
References
- 1.Schneider-Poetsch T., et al. , Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whiffen A. J., Bohonos N., Emerson R. L., The production of an antifungal antibiotic by Streptomyces griseus. J. Bacteriol. 52, 610–611 (1946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leach B. E., Ford J. H., Whiffen A. J., Actidione, an antibiotic from Streptomyces griseus. J. Am. Chem. Soc. 69, 474 (1947). [DOI] [PubMed] [Google Scholar]
- 4.Whiffen A. J., The production, assay, and antibiotic activity of actidione, an antibiotic from Streptomyces griseus. J. Bacteriol. 56, 283–291 (1948). [DOI] [PubMed] [Google Scholar]
- 5.Siegel M. R., Sisler H. D., Inhibition of protein synthesis in vitro by cycloheximide. Nature 200, 675–676 (1963). [DOI] [PubMed] [Google Scholar]
- 6.Hsu K. S., The genetic basis of actidione resistance in Neurospora. J. Gen. Microbiol. 32, 341–347 (1963). [DOI] [PubMed] [Google Scholar]
- 7.Colombo B., Felicetti L., Baglioni C., Inhibition of protein synthesis by cycloheximide in rabbit reticulocytes. Biochem. Biophys. Res. Commun. 18, 389–395 (1965). [DOI] [PubMed] [Google Scholar]
- 8.Sisler H. D., Siegel M. R., “Cycloheximide and other glutarimide antibiotics” in Antibiotics 1: Mechanism of Action, Gottlieb D., Shaw P. D., Eds. (Springer-Verlag, Berlin, Heidelberg, 1967), pp. 283–307. [Google Scholar]
- 9.Garreau de Loubresse N., et al. , Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Myasnikov A. G., et al. , Structure-function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun. 7, 12856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budkevich T., et al. , Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 44, 214–224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behrmann E., et al. , Structural snapshots of actively translating human ribosomes. Cell 161, 845–857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wettstein F. O., Noll H., Penman S., Effect of cycloheximide on ribosomal aggregates engaged in protein synthesis in vitro. Biochim. Biophys. Acta 87, 525–528 (1964). [DOI] [PubMed] [Google Scholar]
- 14.Selker E. U., Stevens J. N., Metzenberg R. L., Heterogeneity of 5S RNA in fungal ribosomes. Science 227, 1340–1343 (1985). [DOI] [PubMed] [Google Scholar]
- 15.Müller F., et al. , The cpc-2 gene of Neurospora crassa encodes a protein entirely composed of WD-repeat segments that is involved in general amino acid control and female fertility. Mol. Gen. Genet. 248, 162–173 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Tesina P., et al. , Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J. 39, e103365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armache J.-P., et al. , Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. USA 107, 19748–19753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., et al. , Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Res. 27, 1275–1288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knorr A. G., et al. , Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat. Struct. Mol. Biol. 26, 35–39 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Buschauer R., et al. , The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368, eaay6912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaskar V., et al. , Dynamics of uS19 C-terminal tail during the translation elongation cycle in human ribosomes. Cell Rep. 31, 107473 (2020). [DOI] [PubMed] [Google Scholar]
- 22.A. M. McGeachy, Z. A. Meacham, N. T. Ingolia, An accessible continuous-culture turbidostat for pooled analysis of complex libraries. ACS Synth. Biol. 8, 844–856 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Borovinskaya M. A., Shoji S., Fredrick K., Cate J. H. D., Structural basis for hygromycin B inhibition of protein biosynthesis. RNA 14, 1590–1599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellen C. U. T., Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A. G., Sachs M. S., Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 20, 6453–6463 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C., Amrani N., Jacobson A., Sachs M. S., “The use of fungal in vitro systems for studying translational regulation” in Translation Initiation: Extract Systems and Molecular Genetics, Lorsch J., Ed. (Elsevier, San Diego, 2007), vol. 429, pp. 203–225. [DOI] [PubMed] [Google Scholar]
- 27.Dever T. E., Ivanov I. P., Sachs M. S., Conserved upstream open reading frame nascent peptides that control translation. Annu. Rev. Genet. 54, 237–264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flis J., et al. , tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis. Cell Rep. 25, 2676–2688.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellegrino S., et al. , Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome. Nucleic Acids Res. 47, 3223–3232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard T. D., A guide to simple and informative binding assays. Mol. Biol. Cell 21, 4061–4067 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandrasekaran V., et al. , Mechanism of ribosome stalling during translation of a poly(A) tail. Nat. Struct. Mol. Biol. 26, 1132–1140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowe-McAuliffe C., et al. , Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens. Nat. Commun. 12, 3577 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noeske J., et al. , High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocozaki A. I., et al. , Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors. Proc. Natl. Acad. Sci. U.S.A. 113, 8188–8193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh Y., Naschberger A., Mortezaei N., Herrmann J. M., Amunts A., Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi. Nat. Commun. 11, 5187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith M. D., et al. , Human-like eukaryotic translation initiation factor 3 from Neurospora crassa. PLoS One 8, e78715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Sachs M. S., Control of mRNA stability in fungi by NMD, EJC and CBC factors through 3′UTR introns. Genetics 200, 1133–1148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurilla A., Szőke A., Auber A., Káldi K., Silhavy D., Expression of the translation termination factor eRF1 is autoregulated by translational readthrough and 3'UTR intron-mediated NMD in Neurospora crassa. FEBS Lett. 594, 3504–3517 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Huang B. Y., Fernández I. S., Long-range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation. Proc. Natl. Acad. Sci. U.S.A. 117, 1429–1437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCluskey K., Wiest A., Plamann M., The Fungal Genetics Stock Center: A repository for 50 years of fungal genetics research. J. Biosci. 35, 119–126 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Wu C., Dasgupta A., Shen L., Bell-Pedersen D., Sachs M. S., The cell free protein synthesis system from the model filamentous fungus Neurospora crassa. Methods 137, 11–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebbole D., Sachs M. S., A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37, 17–18 (1990). [Google Scholar]
- 43.Luo Z., Freitag M., Sachs M. S., Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol. 15, 5235–5245 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang K., et al. , Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis. Nucleic Acids Res. 45, 10884–10894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Rother M., Rother K., Puton T., Bujnicki J. M., ModeRNA: A tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 39, 4007–4022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruesse E., Peplies J., Glöckner F. O., SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiMaio F., et al. , Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Gruber A. R., Lorenz R., Bernhart S. H., Neuböck R., Hofacker I. L., The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eswar N., et al. , Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci.10.1002/0471140864.ps0209s50 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen L., et al. , Structure of the translating Neurospora crassa ribosome arrested by cycloheximide. Electron Microscopy Data Bank. https://www.emdataresource.org/EMD-24307. Deposited 25 June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen L., et al. , Structure of the translating Neurospora crassa ribosome arrested by cycloheximide. Protein Data Bank. https://www.rcsb.org/structure/7R81. Deposited 25 June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Coulomb potential map and atomic coordinates have been deposited in the Electron Microscopy Databank (EMDB code: EMD-24307) (61) and PDB (PDB ID code: 7R81) (62). All other study data are included in the article and/or supporting information.