Significance
Motility is one of the most striking bacterial behaviors common among many species. Motile bacteria can expand rapidly into previously unoccupied habitats, thus increasing fitness. Fast expansion is fostered by the sensing of gradients which the bacteria generate themselves and is further boosted by cell growth. Here we analyze the integrated dynamics of these processes mathematically. The relations obtained provide an analytical description of the expansion process and its dependence on core bacterial and environmental characteristics, like the growth rate, the abundance of chemical attractants in the surroundings, and the detection limit of the attractants.
Keywords: bacterial chemotaxis, range expansion, Keller–Segel model, Fisher wave, front propagation
Abstract
Bacterial cells navigate their environment by directing their movement along chemical gradients. This process, known as chemotaxis, can promote the rapid expansion of bacterial populations into previously unoccupied territories. However, despite numerous experimental and theoretical studies on this classical topic, chemotaxis-driven population expansion is not understood in quantitative terms. Building on recent experimental progress, we here present a detailed analytical study that provides a quantitative understanding of how chemotaxis and cell growth lead to rapid and stable expansion of bacterial populations. We provide analytical relations that accurately describe the dependence of the expansion speed and density profile of the expanding population on important molecular, cellular, and environmental parameters. In particular, expansion speeds can be boosted by orders of magnitude when the environmental availability of chemicals relative to the cellular limits of chemical sensing is high. Analytical understanding of such complex spatiotemporal dynamic processes is rare. Our analytical results and the methods employed to attain them provide a mathematical framework for investigations of the roles of taxis in diverse ecological contexts across broad parameter regimes.
As a fundamental part of their life cycle, bacteria spread by dispersing into and colonizing new habitats. Many species of bacteria navigate in these new habitats by sensing gradients of certain chemicals and biasing their flagellum-based swimming to move themselves along these gradients (1, 2). This process, known as chemotaxis, is among the most extensively investigated topics in molecular biology (1, 3–7) and was observed in diverse microbial habitats such as the gut (8); the soil (9); leaves (10, 11); and marine environments such as the phycosphere, sinking marine particles, and coral reefs (2, 12–14). Further, chemotaxis is employed by many eukaryotic cells such as the free-living Dictyostelium (15) and is an important element of many tissue-forming processes involved in embryogenesis (16), neuronal patterning (17), wound healing (18), and tumor metastasis (19).
Beyond promoting the movements by individual cells, chemotaxis also drives the collective movement of cells leading to emergent patterns and behaviors at the population level (20, 21). Such collective dynamics have been best studied with bacteria in culture plates and microfluidic devices. For example, when Escherichia coli cells are inoculated at the center of a soft agar plate replete with nutrients, consumption of preferred chemicals (referred to as attractants) results in collective cell movement up self-generated attractant gradients (22), leading to the emergence of striking migrating bands that propagate radially outward from the inoculation site (23–25). These migrating bands typically comprise one or two peaks in population density, which stand in contrast to the predictions of canonical models of front propagation and population expansion (26–28); they also expand at much faster speeds than predicted by canonical models. These population-level changes can strongly shape fitness and ecological interactions as recent laboratory studies have shown (29–32).
The first attempt to understand these migrating bands mathematically was made by Keller and Segel, who recovered a traveling-wave solution using a pair of reaction–diffusion–convection equations to describe the density of bacterial populations and the concentration of the attractant they consume (33). While being highly influential, the Keller–Segel (KS) model neglected cell growth, a substantial factor in the expansion process. It further required unrealistic assumptions on attractant sensing without which the migrating bands lose stability (34). Subsequent modeling efforts including cell growth managed to recover the stability of the bands, but their predictions did not match major experimental observations such as the sharply peaked density profiles and their rapid migration speeds (31, 35–38).
Recent work by Cremer et al. (39) demonstrated that the major features of the migrating bands for E. coli in soft agar can be accurately captured using a model in which bacterial growth is independent of the attractant. Numerical solutions to their growth-expansion (GE) model quantitatively described not only the boosted speed of the migrating band but also the signature spatial profile of the bands and their dependence on molecular parameters (39). Their results established the role of attractants as an environmental cue exploited by bacteria independent of possible nutritional values to promote rapid expansion.
The success of the GE model in describing E. coli in soft agar raises the possibility that the phenotype of rapid expansion and distinct density bands might also occur for chemotactic systems in the wild, in situations where growth, diffusion, and chemotaxis dominate. However, from the numerical work of Cremer et al. (39), it is not clear what aspects of their results are generalizable given that both bacterial and environmental characteristics can be vastly different in the wild. For example, bacteria living near sulphidic sediments move more than 30 times faster than E. coli (39, 40), while bacterial motility is significantly reduced by high viscosity in the gut (41, 42). Addressing the generalizability of the GE model requires a detailed mathematical analysis of the interplay of 1) growth, 2) diffusion, and 3) chemotaxis, preferably with analytical solutions. While growth and diffusion have been studied together in the canonical models of front propagation (26–28), as have diffusion and chemotaxis in the KS model (33, 36, 43–45), a sufficient understanding of the interplay of all three is still lacking.
Toward obtaining such an understanding, we describe here a detailed analytical study of the GE model. Through a heuristic analysis, we derive analytic relations that describe the dependence of the expansion speed and density profile on important molecular, cellular, and environmental parameters, including the rate of cell growth, the diffusivity and availability of the attractants, the motility and sensitivity of the bacteria, carrying capacity, and the limit of attractant sensing. Our analysis reveals the key condition for the population to attain rapid expansion speed and suggests a very broad parameter regime for which rapid expansion can be expected.
GE Model
In the GE model the evolution of the bacterial density, ρ, in space and time (t) is given by
| [1] |
The growth of the population is given by the logistic equation where r is the growth rate and ρc is the carrying capacity of the system. The nondirected run-and-tumble movement of bacteria is described by a diffusion-like term with the motility coefficient , while directed movement along the gradient of the attractant concentration a is described by a convection term with the drift velocity , where
| [2] |
χ0 is the chemotactic coefficient which describes how cells translate the sensing of the local attractant gradients into directed movement. The value of χ0 depends on the strain, the internal cell state, the medium cells move through (e.g., liquid medium or soft agar), and the type of attractant being used (46). am describes the finite sensitivity of the attractant-sensing receptors (47, 48) and ensures that for . Finally, the dynamics of the attractant are determined by its diffusion and consumption by the bacteria:
| [3] |
where Da is the molecular diffusion coefficient of the attractant, μ is the rate of uptake of the attractant by the bacteria, and ak is the Michaelis–Menten constant describing attractant uptake. We note that the GE model defined by Eqs. 1–3 is a slight simplification of the one studied numerically in (39). However, the simplifications do not significantly impact our results, even at the quantitative level (see SI Appendix, Fig. S2, for comparison with the generalized GE model used in ref. 39).
Without growth (r = 0), the GE model resembles the original KS model which additionally also assumed negligible attractant diffusion, i.e., Da = 0, and infinitesimal sensitivity in sensing, i.e., am = 0. The latter assumption of the KS model is necessary for stable traveling waves (33, 34) as otherwise the portion of the band with is not able to migrate as fast as the rest of the band and falls behind, leading to a gradually diminishing and slowing band. Many models have attempted to replenish the bands by including cell growth (35, 49–53), and while they are able to recover stability, they fail to reproduce experimental observations such as fast expansion speeds and a trailing region of increasing bacterial density behind the migrating band.
In the absence of chemotaxis (), the GE model reduces to the Fisher–Kolmogorov–Petrovsky–Piskunov (F-KPP) equation which describes expansion by growth and nondirected motion alone (26, 27, 54). The F-KPP equation has been extensively studied in the theory of front propagation (28) and has been used canonically to describe range expansion into unoccupied habitats (55–57), including the expansion dynamics of nonmotile bacteria in colony growth and long-range dispersal (58–62). While growth and nondirected motion movement result in a traveling-wave solution with a stable expansion speed given by (known as the Fisher speed) (26–28, 63), it is not sufficient to explain the high expansion speeds of the bacterial front observed in populations of chemotactic bacteria (39). Indeed, as we will see in Parameter Dependences of the Expansion Speed, the expansion speed for the GE model can lead to expansion speeds orders of magnitude higher than the Fisher speed.
Remarkably, while the two different reaction–diffusion models (KS and F-KPP) fail to even qualitatively describe the experimental observation of fast-moving stable migrating bands by themselves, when combined together, they are able to quantitatively explain the prominent features of bacterial chemotaxis for a broad range of physiological and environmental conditions (39).
The GE model describes a system of nonlinear coupled partial differential equations (Eqs. 1–3) that has a degree of 4 and is accompanied by appropriate initial values and boundary conditions. For our system, we specify the initial values to be a localized profile for ρ (any localized profile converges to the same steady state solution) and a uniform attractant concentration denoted by a0. In one dimension and with x denoting the spatial coordinate, we look for a stable traveling-wave solution of the form
where c > 0 is the expansion speed. This converts the system of coupled partial differential equations to two one-dimensional ordinary differential equations as follows:
| [4] |
| [5] |
In Eqs. 4 and 5, we have taken an additional simplifying assumption that am = ak. For the well-characterized model organism E. coli, both the uptake and sensitivity of the major attractant aspartate are (39, 64–67). Relaxing this assumption affects the results only weakly, as will be discussed below.
Eqs. 4 and 5 are supplemented by boundary conditions that describe limiting values for the bacterial density and attractant concentration far from the front:
| [6] |
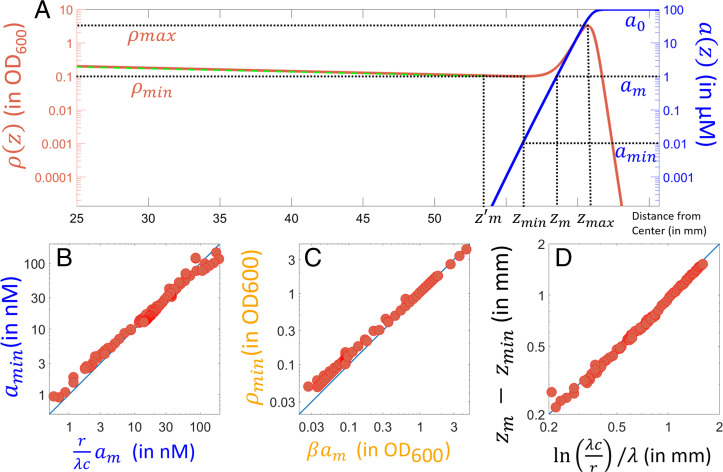
Fig. 1 shows the numerically obtained steady state profiles emerging from Eqs. 4 and 5 using experimentally established model parameters (39). All numerical solutions were obtained using finite element simulations (68, 69) (Materials and Methods). The density profile (solid red line) has a distinct peak at the front which defines the appearance of the migrating band observed in experiments (23, 24, 39) and can be divided into three distinct regimes: the growth regime (left of the density trough), the chemotaxis regime (the rising part of the density profile), and the diffusion regime (right of the density peak), as indicated in Fig. 2B. Such a division reflects the fact that as we will show in the sections dedicated to each regime, in each of these regimes, either the growth, chemotaxis, or diffusion term dominates in Eq. 4.
Fig. 1.
Profiles of bacterial density (solid red line, in optical density measured at a wavelength of 600 nm []), drift velocity (dashed green line, in mm/h), and attractant concentration (dotted blue line, in mM) for a steadily expanding population 14.5 h after the inoculation. Arrows indicate the different regimes used in the analytical consideration. Model parameters used are adapted from those determined in ref. 39 and are provided in SI Appendix, Table S1 (this simulation used the low-motility parameters).
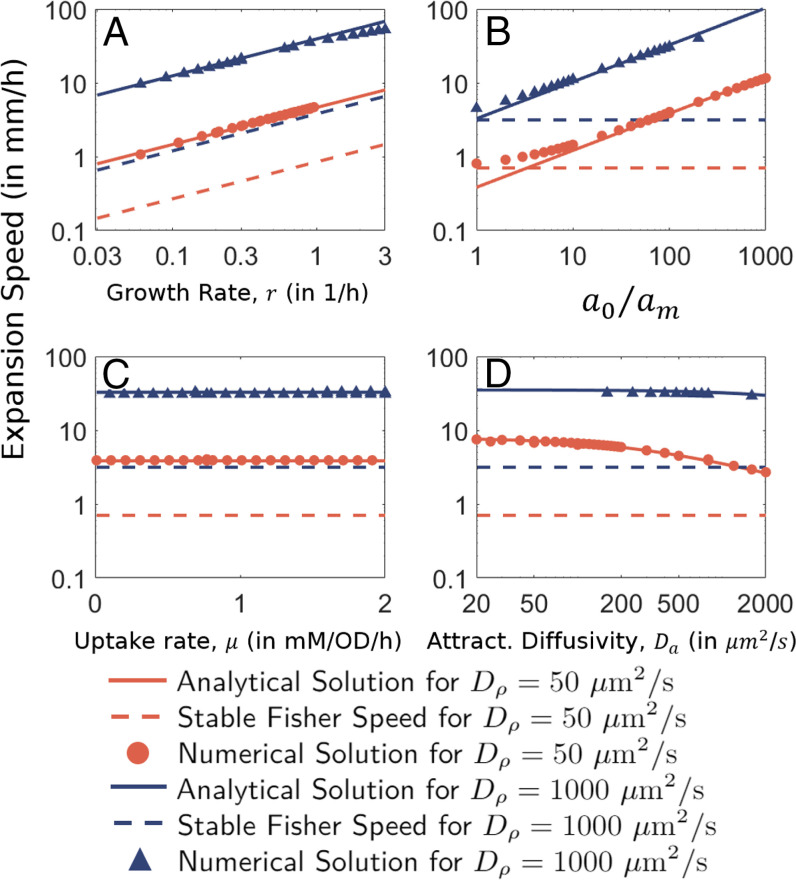
Fig. 2.
Dependence on (A) growth rate r, (B) uptake rate μ, (C) relative attractant levels , and (D) attractant diffusion Da. Analytical relation for the expansion speed (Eq. 23) is shown by solid lines ( in red and blue, respectively). The corresponding Fisher speeds, , are denoted by corresponding dashed lines. Numerical solutions of the GE model (Eqs. 4 and 5) are shown by corresponding symbols. Unless specified, all parameter values are the default values given in SI Appendix, Table S1.
Chemotaxis Regime
Heuristic Derivation of the Expansion Speed
We first analyze the most striking feature of the traveling wave, the density bulge. Initially, we consider Eqs. 4 and 5 in the limit of (this assumption will be relaxed later). We start with the following ansatz:
| [7] |
with β being a proportionality constant. This reduces Eq. 5 to a homogeneous linear differential equation in a(z) with constant coefficients. The solution to such an equation is an exponential function, , with λ satisfying
| [8] |
The ansatz Eq. 7 also simplifies Eq. 4 considerably, with the penultimate term on the right-hand side (RHS) of Eq. 4 now proportional to . Another consequence of the ansatz is that , a relation that will be used often in our calculations. With the ansatz, Eq. 4 simplifies to
| [9] |
To proceed further, we consider the case that growth is much smaller than chemotactic drift so that the term proportional to r on the RHS of Eq. 9 may be neglected. This requires that both of the following conditions be true: The first is a condition on the parameters such that
| [10] |
which is equivalent to assuming that the timescale of growth is much larger than the timescale of chemotactic drift, and thus, the two timescales may be separated. As we will show later, this assumption corresponds to a broad, biologically relevant parameter regime and is independent of the growth rate itself (because λc turns out to be proportional to r). The second is a condition on the values of attractant concentration a(z),
| [11] |
As we will show in Growth Regime and the Density Trough, the quantity on the RHS of condition [11] is approximately the value of the attractant concentration at the trough of the density profile (i.e., the left boundary of the chemotaxis regime). Thus, for growth that is small compared to chemotactic drift (i.e., condition [10]), Eq. 9 becomes independent of a(z) in the chemotaxis regime. This means Eq. 4 is a linear equation involving , a(z), and their derivatives, and it (self-consistently) admits the ansatz Eq. 7 as a solution. With the last term in Eq. 9 neglected, the solution to λ is readily obtained, i.e.,
| [12] |
where the solution λ = 0 is rejected as it does not solve Eq. 8. In this regime, the solution to the attractant concentration can be explicitly written as
| [13] |
where zm is defined by .
To obtain a relation for the expansion speed c and its dependence on the model parameters, we note that Eqs. 8 and 12 are by themselves insufficient since there are three unknown quantities: c, λ, and β. To obtain a defined solution, we thus invoke the boundary conditions at z = + ∞ well outside the chemotaxis regime (Eq. 6). This is done by integrating Eqs. 4 and 5 from a position in the chemotaxis regime to z = + ∞. For Eq. 4 with , we obtain
| [14] |
where is the total bacterial population to the right of . Note that Eq. 14 is exact and independent of our ansatz. For located in the chemotaxis regime, we plug in our ansatz Eq. 7 and Eqs. 13 to 14, yielding
| [15] |
Note that while the term with growth rate r was negligible in Eq. 9, it cannot be neglected in the integral form as it involves contributions by outside of the chemotaxis regime. Using Eq. 9, Eq. 15 simplifies to
| [16] |
Now, while Eq. 16 provides us another equation for and λ, we have a new unknown, . However, another relation for is obtained by integrating both sides of Eq. 5 from to , yielding
| [17] |
where . We show in SI Appendix, section S5, that for . Neglecting in Eq. 17 and using Eq. 8, we obtain
| [18] |
Eqs. 16 and 18 allow us to eliminate and explicitly obtain the proportionality constant of the ansatz Eq. 7,
| [19] |
The explicit value of β now allows us to solve for λ and c using Eqs. 8 and 12:
| [20] |
| [21] |
From Eqs. 20 and 21, we find that the condition amounts to the following condition on the parameters:
| [22] |
Thus, the requirement for our ansatz to hold translates to an equivalent condition on the chemotactic model parameters that is independent of the growth rate r. As detailed in Parameter Dependences of the Expansion Speed, this parameter regime is typical for the study of migrating bands, with a few fold below Da for bacteria in soft agar, and comparable to Da for motile cells in liquid medium, while am is several orders of magnitude smaller than a0.
Parameter Dependences of the Expansion Speed
To validate our heuristic approach we compared the derived relation for the expansion speed, Eq. 21, with numerical simulations, obtaining an excellent match for a broad range of model parameters. We show the dependences on growth rate, uptake rate, background attractant concentration, and the attractant diffusion coefficient in Fig. 2.
First, there is a square root dependence on the growth rate r, as validated by numerical results in Fig. 2A. This demonstrates that the well-known square root dependence of cF, the Fisher speed, on growth rate is preserved in the GE model. The expansion speed is further increased by the square root of the relative background attractant concentration, (Fig. 2B). However, the expansion speed c does not depend on the specific rate of attractant uptake μ (Fig. 2C) nor the inoculum population size (as the steady state bulge size is an emergent property, independent of the initial population size). The independence on μ is particularly counterintuitive since it is the uptake of attractant that establishes the attractant gradient which, in turn, drives the chemotactic movement. The independence on μ is in contrast to the KS model, which predicts that (where NKS is the inoculum population size), but is in agreement with experimental results (23, 39). We will show in The Growth–Leakage Balance that our solution for c can be similarly expressed in terms of μ and N0, the size of the density bulge. However, unlike the KS solution, N0 is here an emergent quantity that turns out to be inversely proportional to μ. Thus, the dependence on μ cancels out, making the expansion speed independent of μ.
The most nontrivial aspect of Eq. 21 is perhaps the predicted dependence of the expansion speed c on the attractant diffusion coefficient Da (Fig. 2D), which was not considered in most previous models (33, 34, 70, 71). Although this dependence itself is not so strong, it significantly affects the dependence of c on the cellular motility characteristics as we discuss next.
To see how the expansion speed depends on the cellular motility parameters and χ0 we first note that and χ0 result from the run-and-tumble dynamics and are thus both proportional to , where v0 is the run velocity, and τ is the average duration of runs. The ratio results from the properties of the flagella motor, the ligand/chemotactic receptor interaction, and the chemotactic signaling network (46). To better describe the differences, we here define the chemotactic sensitivity, , a dimensionless parameter such that a large value of represents a strong chemotactic response to a ligand. Notably, can vary across a broad range depending on the environment, with for E. coli swimming in soft agar (39) and to in liquid media (39, 72, 73). In contrast, is not expected to be affected by environmental obstacles but by the chemotactic properties of the cell and the type of attractant and is found to vary from 2 to 8 (72, 74). We can thus keep and as independent parameters and write the expansion speed, Eq. 21, as
| [23] |
The predicted comparison with numerical solutions confirms the dependence on cellular parameters: Notably, for high cellular motility, , Eq. 23 gives , as seen in Fig. 3 A and B (solid dark blue lines show the analytical prediction for ). The thick cyan lines show a square root fit. Meanwhile, in the range and thus has a linear dependence on the motility parameter and chemotactic sensitivity (thick yellow lines).
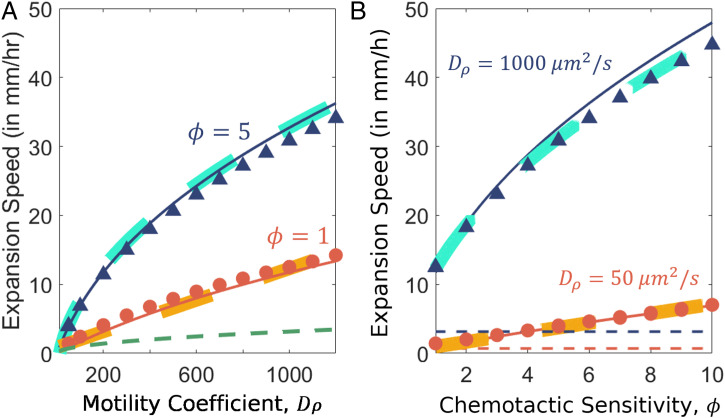
Fig. 3.
Dependence of expansion speed on motility parameters. (A) Dependence on cellular motility . Numerical solutions for and are shown by red circles and dark blue triangles, respectively. Analytical solutions following Eq. 23 are shown by corresponding solid red and blue lines. The green dashed line represents the stable Fisher speed, , the minimum expansion speed of our system. (B) Dependence on the chemotactic sensitivity, . Numerical solutions for and are shown by red and dark blue circles, respectively. Analytic solutions following Eq. 23 are shown by the corresponding solid lines. Thick yellow and cyan dashed lines are best fits for the respective values of and to demonstrate that for and that if is large compared to Da. Unless specified, all parameter values are the default values given in SI Appendix, Table S1.
The dependence of the expansion speed on the value of Da (Fig. 2D) and its relation to (Fig. 3) reveals a crucial role of the molecular diffusion of the attractant, which has historically been assumed to be of a much smaller scale than the motility-induced bacterial diffusion and chemotaxis (33, 34, 36, 44, 70, 71, 75). Large Da can be understood to result in a smoothening of the attractant gradient, thereby slowing down chemotaxis. In fact, for extremely large values of Da, we note that the bacterial population is unable to establish a gradient in the attractant concentration, and our analysis fails to hold as seen in the self-consistency condition [22]. Quantitatively, the molecular diffusivity () well exceeds the chemotactic coefficient and the effective cell diffusivity of E. coli in soft agar () (39). Hence, condition [22] is satisfied for for aspartate, thus explaining the deviation seen at small for (see red circles in Fig. 2B).
We also verified the dependence of the expansion speed on itself for (Fig. 3B). For , the numerical values do not match the analytical values, as they are beyond the regime of self-consistency discussed above. Outside of this regime, the traveling-wave solution transitions to the pulled wave dynamics of the F-KPP equation, with a lower bound on the expansion speed given by the Fisher speed () (SI Appendix, Fig. S3).
Effect of Carrying Capacity
Next, we consider the effect of a finite carrying capacity ρc and the corresponding effect on expansion. To do so, we follow a similar approach as in Heuristic Derivation of the Expansion Speed; see SI Appendix, section S6, for details of the calculations performed. Incorporating the effect of ρc leads us to the following form for the expansion speed:
| [24] |
where is the expansion speed for infinitely large carrying capacities, as given by Eq. 21, and γ is a dimensionless function determined by the shape of the density bulge. While we are unable to determine the exact functional form of γ, we find an excellent agreement between the numerical results and analytical solution for the best-fit value of γ (found to be for and for as seen in Fig. 4A.
Fig. 4.
Effect of carrying capacity. (A) Dependence of expansion speed on the ambient attractant concentration when the carrying capacity is finite (). Markers (red circles and blue triangles) indicate numerical values, solid lines indicate analytical predictions as per Eq. 24, and dashed lines indicate analytical predictions with . All results in red are for , and all results in blue are for . (B) The ambient attractant concentration resulting in maximum expansion speed is shown depending on the dimensionless parameter . The analytical solution, Eq. 24, is shown as corresponding solid lines. Dashed lines show the solutions () without a limiting carrying capacity (, as shown in Fig. 3). Different symbols in B denote which model parameter was varied from its default value (square if μ, circle if ρc, triangle if r, and diamond if am) for (red) and (blue). For details, refer to SI Appendix, Supplemental Methods, and to SI Appendix, Table S2, for range of values used for each parameter. Parameters have the default values from SI Appendix, Table S1, unless specified.
An intriguing prediction of Eq. 24 is a peak in the relation between c and a0 whose existence is numerically confirmed (Fig. 4A). Thus, too much attractant actually reduces the expansion speed, i.e., the expansion speed of the population cannot be arbitrarily increased merely by increasing the ambient attractant concentration, but is limited ultimately by the physiological and molecular parameters. To understand this nonmonotonic dependence, we note that in Eq. 24, the effect of ρc is insignificant for , i.e., if ρc is large compared to the highest density expected from the ansatz Eq. 7 when . For sufficiently large a0 such that , the quantity (which describes the amount of attractant taken up by bacteria at the peak density, where , in one doubling time) becomes small, and the population is unable to take up the attractant fast enough to generate a substantial gradient in a(z). The lack of a substantial gradient, in turn, leads to mitigated expansion speeds. As a result, for small ρc, Eq. 24 simplifies to . Thus, for small carrying capacity, c increases with μ and decreases with a0, qualitatively similar to the relation found by Keller and Segel (). In both solutions, the decrease in expansion speed with a0 results from a bound on the total population of the density bulge that limits its ability to consume the attractant.
We note that the existence of a peak in expansion speed for varying background attractant concentrations was observed experimentally and reported already over 30 y ago (39, 76) but was believed to be due to receptor saturation. Our analytical solution in Eq. 24, validated by simulations (Fig. 4A), provides an excellent quantitative explanation of this phenomenon even in the absence of receptor saturation.
The attractant concentration corresponding to the maximum expansion speed is found to be
| [25] |
and is validated numerically in Fig. 4B. The resulting maximum expansion speed is , and the corresponding carrying capacity is proportional to . Thus, for the population to maximize its expansion speed at high attractant concentrations, a very high carrying capacity is required. As the carrying capacity is typically no more than a few OD for aerobically grown cells, the attractant concentration for the maximum expansion speed, , is not expected to be above (Eq. 25 and Fig. 4A).
Case of
If we relax the assumption that ak = am and take as our ansatz , we note an additional term in Eq. 9 that is of the order
| [26] |
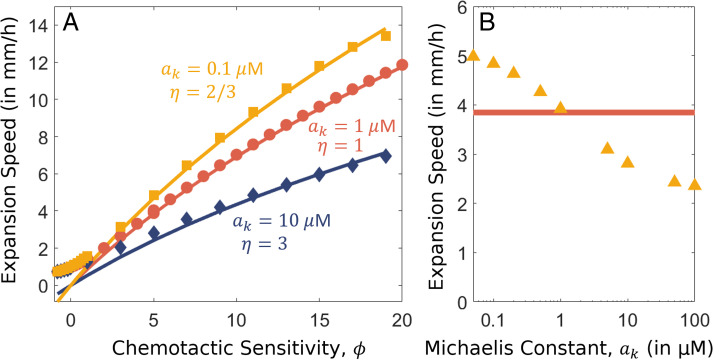
relative to the dominant chemotactic drift term. It is due to this term that our ansatz Eq. 7 fails to hold if . A similar term is found in Eq. 17. While trivially negligible if ak = am, the terms are also negligible for and as . Thus, we expect our analysis of the chemotaxis regime (and the growth regime, which we perform in Growth Regime and the Density Trough) to also be applicable for the case that as long as . However, when , our ansatz will not hold, and the value of a(z) where switches from being relatively constant as in the growth regime to rising exponentially as in the chemotaxis regime is undetermined by our current analysis. We expect the transition to be at , between ak and am, as both of these values are crucial in determining the transition in . The coupled nature of and a(z) make it difficult to determine η exactly. Such an assumption leads to a similar expression for expansion speed but where replaces am in the final form. We find an excellent agreement with numerical results for for just one fitting parameter, η, which we find to be ∼2/3 for and for . The range of exponential speeds for different values of ak while keeping am fixed at 1 is shown in Fig. 5 A and B. Notably, c is seen to decrease only twofold for a 2,000-fold increase in ak, from 50 nM to 100 M for standard parameters (Fig. 5B), while if both ak and am increase 2,000-fold, c would decrease 45-fold (Fig. 2B).
Fig. 5.
Effect of varying Michaelis constant, ak. (A) Dependence of expansion speed on the chemotactic sensitivity, , for different values of ak and . Solid lines indicate analytical solutions for corresponding best fit values of η, and markers denote the numerical solutions. Results for , and are shown in yellow, red, and blue, respectively. (B) Dependence of the expansion speed on model parameter ak. The numerical solutions obtained for are represented by yellow triangles, and the analytic solution found in Eq. 21 for is shown by the red line. Parameters have the default values from SI Appendix, Table S1, unless specified.
Diffusion Regime and the Density Peak
Next, we describe the dynamics of the propagating density profile at its asymptotic front. This is the diffusion regime which lies to the right of the density peak (Fig. 1), where the exponential increase of the concentration of the attractant observed in the chemotaxis regime is curtailed by the right boundary condition, i.e., . Here the drift velocity becomes and thus negligible as . The equation for is no longer affected by the attractant, and the dynamics are thus described by the F-KPP equation. The solution is
| [27] |
where ρ0 is a proportionality constant (see Eq. 33) and cD is the speed of propagation of the asymptotic front.
For the front to be a part of the stationary solution that propagates at the same speed as the chemotaxis regime, c (Eq. 21), we must have cD = c, which well exceeds the F-KPP speed, . It is well known for the F-KPP equation that if the dynamical system admits a uniformly translating front solution with , then the front solution corresponding to the traveling speed c is the stable solution (28). For the case that the front is asymptotic, the initial conditions are compact, and the right boundary condition is the unstable state, , the steeper front solution is selected for (28) (see SI Appendix, section S7A, for a brief description). Thus, our dynamical system selects for a solution with the leading asymptotic behavior given by
| [28] |
for the diffusion regime.
We then turn to the form of a(z) in the diffusion regime. As in this regime, Eq. 5 becomes
| [29] |
This is a nonhomogeneous linear differential equation in a(z) with the solution
| [30] |
where a1 is an undetermined constant of integration. The leading behavior is determined by whichever exponential term decays more slowly as :
| [31] |
As to the peak itself, it can be shown (see SI Appendix, section S9, for details) that the attractant concentration at the peak, , is given by the bound
| [32] |
With some further approximations (that and that the ansatz Eq. 7 extends throughout the rising region of the density bulge), we obtain that
| [33] |
where is the maximal population density at the front (i.e., the density peak) and is the minimal population density left of the peak (i.e., the density dip at the transition between the chemotaxis and growth regimes). A numerical verification of Eqs. 32 and 33 for a broad parameter range is shown in SI Appendix, Fig. S4.
Growth Regime and the Density Trough
Next, we turn to the growth regime, which is the region with exponential density profile trailing the density bulge (Fig. 1B). In this regime, the increase in as drives the attractant concentration to zero according to Eq. 5; i.e., as . Consequently, and
| [34] |
in the growth regime, sufficiently to the left of the density trough. Later in this section, we will quantitatively define the condition where the v term is negligible compared to c. Here we briefly describe characteristics of the solution when this condition holds.
Eliminating the term associated with chemotactic drift removes the dependence of on a(z) in Eq. 4, with the only remaining processes determining being growth and diffusion. Thus, we recover the F-KPP equation, with the solution , where
| [35] |
cG being the traveling velocity of the growth regime. As in the diffusion regime, here cG must be the same as c, the speed of the chemotaxis regime, in order for Eq. 4 to admit a stationary solution. Since , the two solutions for are and . It is well established for the F-KPP equation that for a solution to move stably at a speed exceeding cF, its front must be shallower than λF; see ref. 77 and SI Appendix, section S7B. Hence, is selected. Thus, the form of density sufficiently to the left in the growth regime must be given by
| [36] |
ρ1 being a proportionality constant that sets the z scale as will be specified in this section.
To understand how the front of the growth regime is set, we focus on the transition region between the growth and chemotaxis regimes (located close to the density trough). A magnified view of this transition region is shown in Fig. 6A, with denoting the location of the density minimum.
Fig. 6.
Transition from the chemotaxis to the growth regime. (A) Steady expansion profiles of (solid red line) and a(z) (solid blue line) for the standard parameters (SI Appendix, Table S1; ). The profile of as predicted by the ansatz Eq. 7 is shown using the dashed green line. Dashed horizontal lines indicate distinct values of a and ρ as indicated. (B–D) Numerically obtained values of , , and for a broad variation of parameters. Seven model parameters in Eqs. 4 and 5 (other than ρc, which was >1, 000 for all results here) were varied across many decades (see SI Appendix, Supplemental Methods, for details of what was done and SI Appendix, Table S3, for the range of values investigated). Blue lines show y = x to demonstrate agreement with the predicted values of , and .
Previously, we have shown that for (defined by ; Fig. 6A) in the chemotaxis regime, cell density is given by the ansatz Eq. 7, with the attractant concentration a(z) given by Eq. 13. We showed that the validity of this ansatz required , i.e., Eq. 9. However, even with , this condition will eventually break down for , for , including possibly the vicinity of (Fig. 6A). Thus, in order to address the density profile in the transition region, we cannot rely on the ansatz Eq. 7 anymore.
Here we extend our ansatz to a modified form which we will show to be valid for both the chemotaxis and growth regimes, including all of the transition region:
| [37] |
Clearly, for , Eq. 37 recovers the form of density established for the growth regime, i.e., Eq. 36, with . For where a(z) is given by Eq. 13 in the chemotaxis regime, Eq. 37 becomes
where the last approximation results from for our parameter regime . Furthermore, we can verify that the modified ansatz Eq. 37 satisfies Eq. 4 for intermediate range of a(z), leaving behind a linear equation for a(z) that is the same as that obtained in the chemotaxis regime, with the same solution (Eq. 13) (SI Appendix, section S7B). Our modified ansatz thus leads to the following form for the cell density:
| [38] |
which we claim to be valid for the entire regime (for ), including the vicinity of the density trough located at .
We can now use the expression given by Eq. 38 to work out characteristics of the solution in the transition region. By setting , we obtain (for ):
| [39] |
| [40] |
| [41] |
These results are validated numerically for a range of parameters (Fig. 6 B–D).
We can determine the left boundary of the transition region, , by finding the range of where Eq. 38 is described by the simple exponential form in Eq. 36 (dashed green line, Fig. 6A). This can be estimated by setting the asymptotic form
| [42] |
to . Using Eq. 40 for , we find
| [43] |
In other words, Eq. 42 can be written as . Note that because according to Eq. 43 for , for ; i.e., the density function on the left side of is constant with relative variation of the order of . [We can verify the self-consistency of the ansatz Eq. 38 by using it to compute the drift velocity and hence evaluate the spatial domain where the condition [34] is satisfied. We find that [34] is satisfied for or from Eq. 43.]
To summarize, the transition region between the chemotaxis and growth regimes ranges from where the distances from to zm and are given by Eqs. 39 and 43, respectively. The total width of the transition zone is
| [44] |
Note that the time it takes for the wave front to migrate across the transition region is . Thus, the key condition for our results, , corresponds simply to , i.e., a separation of timescale between expansion and population growth. This is a condition which we expect to hold for most expanding populations.
The Growth–Leakage Balance
We can finally use the explicit solution for to connect the dynamics in the chemotaxis and growth regimes. We consider the total bacterial population to the right of a position , which is comoving with the population: . The change in over time is given formally by
| [45] |
where
obtained from taking time derivative of using Eq. 1 is the leakage flux which includes the loss of cells across the position in the laboratory frame due to chemotaxis and diffusion, and the last term in Eq. 45 describes the growth of the cells in the region .
In the absence of growth r = 0, Novick-Cohen and Segel (34) showed that incorporating the lower Weber cutoff to the KS model led to the loss of cells from the front and subsequently the slowdown of the migrating wave front. We see from Eq. 45 that the incorporation of growth, even at very low rates, allows the migrating wave front to replenish itself and thereby maintain stability.
In the stationary state (), quantities in the moving frame have no time dependence, i.e., . Thus,
| [46] |
which is just Eq. 14 with v(z) given by a(z) that solves the stationary equations, Eqs. 4 and 5. Earlier, we solved Eq. 14 using the ansatz Eq. 7 that holds only in the chemotaxis regime with . We can repeat the calculation using Eqs. 38 and 13 derived from our ansatz Eq. 37. We find the leakage flux to be very weakly z-dependent in the vicinity of the density trough, i.e.,
| [47] |
where
| [48] |
Since according to Eqs. 39 and 43, we conclude that up to an order of . Consequently, N(z) is also nearly z-independent, reflecting the sharply peaked structure of the density front. For convenience, we define as the size of the population in the density bulge. The above results then lead to an important biological relation,
| [49] |
with the bulge size given by
| [50] |
Eq. 49 describes a balance of the growth of the cells in the front and their leakage behind the front, as depicted in Fig. 7. At a given instance (time t0), the wave front is shown as the dashed red line in the laboratory frame. The front region, composed of N0 cells, grows at a rate rN0. This growth is balanced by cells leaving the front (i.e., across the black dashed line indicating ), with flux . At some time δt later, the front has traversed a distance . The total amount of cells leaving the front during this time is . The corresponding density of the cells left behind the propagating front is (shown as the purple region in Fig. 7A). The cells left behind will grow at the rate r. For δt much smaller than the doubling time, the density behind the front will not have grown much and will thus remain at (Fig. 7A). We have shown that this is the case for the time it takes for the front to traverse the width of the trough region (Eq. 44). After a time that is large compared to the doubling time, the population size at the back will become (Fig. 7B). Given that , we have
| [51] |
Fig. 7.
Schematic of the dynamics of the transition between chemotaxis and growth regimes. (A) In a short time , the density bulge shown near x0 (dotted red line) moves forward to be near (solid red line). In that time, the density bulge grows by an amount and is diminished by leakage given by an amount . During steady expansion, these values match the expressions given by our ansatz (Eqs. 7 and 37). The leaked cells are deposited behind the density bulge where the bacterial density is roughly constant for a distance δx. Thus, , and the total deposition over time δt, given by , is also equal to . (B) After a long time , the density bulge moves to be near a position (dashed red line). Cells behind the density bulge grow at a rate r, and the density thus accumulates as .
Thus, the trailing exponential density profile in Eq. 51, while looking like a moving front, is merely a result of the exponential growth of a stationary population, which is seeded by the traveling wave front at density and speed c.
Finally, we note that the picture depicted in Fig. 7A can be used directly to predict the value or without going through detailed calculation: since the bacteria are concentrated in the density bulge, the removal of the attractant is almost entirely due to uptake by cells in the density bulge. This gives us the mass conservation condition∗
| [52] |
The growth–leakage balance rN0 = J0 then gives . The consideration described in Fig. 7A then immediately gives the result that the density left behind the front bulge, which would be , is given by . Thus, we obtain a surprisingly simple result,
| [53] |
independent of the other details of the system.
We can also use the expression for thus obtained to calculate the consumption of attractant around the density trough. Using and a(z) from Eq. 13, Eq. 5 becomes
| [54] |
This relation together with the proportionality between λ and c, Eq. 12, immediately gives the central result on the expansion speed, Eq. 21. This simple line of consideration reveals the underlying origin of the dependence of the expansion speed on : the growth–leakage balance relates the ambient concentration a0 to the trough density (Eq. 53), and the balance between attractant uptake and drift/diffusion at the trough relates c and λ to and am.
Discussion
To reveal the underlying dynamics governing chemotaxis-driven population expansion, we analyzed the experimentally verified GE model mathematically (39). Following an extensive traveling-wave analysis we were able to describe the density and attractant profiles throughout the chemotaxis and growth regimes (Fig. 6 and Eqs. 13 and 37). We determined the expansion speed (Eq. 21) and the slope λ which specifies the width of the migrating band (Eq. 20).
Our results, which are in excellent agreement with numerical simulations for a broad range of model parameters tested (Figs. 2–6), recover many key experimentally observed relations of the expansion speed to biological and environmental parameters (39) that previous models based on the KS model had failed to capture (36, 37). Notably, while our model agrees with the KS model near the density bulge, with the same relation between expansion speed and the size of the peak (; Eq. 52), the size of the peak itself is not a constant as in the KS model but an emergent quantity. Consequently, the expansion speed depends on many of the model parameters.
First, the expansion speed depends on the ratio of the initial attractant concentration to the lower limit of attractant sensitivity (i.e., ) for large carrying capacity. For finite carrying capacity our analysis predicts the nonmonotonic dependence of expansion speed with initial attractant concentration, providing an explanation for this long-known experimental observation (76): For lower attractant concentrations, increasing concentration increases the size of the bulge and hence promotes faster expansion. However, for higher concentrations, the carrying capacity limits the size of the bulge, and expansion speed decreases with increasing attractant concentration as it takes longer for the bulge to consume the attractant and establish a gradient (Eq. 24 and Fig. 4A).
Second, our results reveal a dependence of the expansion speed on the diffusion of the attractant (Da; Fig. 2D). The effect of multiple diffusion coefficient-like parameters (, and Da) is one of the reasons the GE model is difficult to analyze. In Cremer et al. (39), a scaling theory was developed to describe the dependence of the expansion speed on the chemotaxis coefficient χ0. Assuming that χ0 was the main relevant factor, the scaling theory predicted that . Our analysis here reveal that holds for large Da but for small Da (Fig. 3).
The analytical understanding attained in this work quantitatively supports the role of chemotaxis in range expansion found by Cremer et al. (39). Particularly, bacterial chemotaxis does not necessarily occur to fulfill an immediate nutritional need, nor does it necessarily reflect an attempt to avoid starvation. For example, cells move chemotactically toward attractants they cannot metabolize and also swim in nutrient-replete conditions (23, 25, 39). Instead, chemotaxis could be hard-wired to promote the expansion of bacterial populations into unoccupied territories well before nutrients run out in the existing environment; low levels of attractants thus act as aroma-like cues that establish the direction of expansion and enhance the speed of population movement (39). Subsequently, cells left behind by the migrating band fully occupy the region behind the front by growing at rates determined by nutrient availability. This allows the population to expand rapidly into unoccupied territories while colonizing the traversed territories, without one compromising the other.
Our results also expand upon the general theory of front propagation into unstable states and reveal an alternative mechanism for speed selection. While many studies of front propagation involve modification of the nonlinear growth/reaction term in the original F-KPP equation (28, 78, 79), our model considers a drift term which is a functional of an environmental variable, the attractant concentration. Though the canonical results pertaining to the F-KPP equation are not expected to hold in such a two-variable system, the dynamics in the growth and diffusion regimes in our system are effectively described by the F-KPP equation. While the expansion of an F-KPP wave front pushed by the bulk (as in the diffusion regime) at rates higher than the stable Fisher speed has long been known (28, 54), our results demonstrate how F-KPP wave fronts can also be seeded by a transition regime at the front (as in the growth regime) to attain very large expansion speeds. Alternatively put, by following the chemotacting cells in the leading density bulge, the trailing region can propagate faster than predicted by F-KPP equation based on growth and diffusion alone.
Finally, we note that our analysis assumes a separation of timescales between growth (slow) and chemotactic migration (fast), i.e., , indicating that cell growth is negligible over the timescale at which the population migrates across the width of the density bulge given by . This condition is fulfilled for a broad parameter regime where the background attractant concentration is large () or when the chemotactic coefficient much exceeds the diffusion of the attractant () (condition [22]). For the narrow parameter regime where condition [22] breaks down, e.g., when , the expansion speed approaches the stable Fisher speed cF as it is the expected speed for a pulled wave solution determined by the asymptotic front (28) in the absence of a push from the bulk. This is supported by numerical data (SI Appendix, Fig. S3). A solution to the GE model that includes the small-χ0 regime would provide an analytical connection to the F-KPP equation and thereby provide insight on the transition from the pushed and seeded dynamics observed when to the well-established pulled wave dynamics (28, 58, 80, 81).
Outlook
Beyond an analytical understanding of the interplay between chemotaxis, growth, and diffusion in the case of migrating bands of E. coli in soft agar [for which Cremer et al. (39) had previously established the experimental validity of the GE model], the mathematical relations obtained here offer testable predictions for settings that involve chemotaxis, growth, and diffusion independent of the specifics of the strain or the environment. First, we identify the parameter regime in which chemotaxis leads to a boost in the expansion speed (condition [22]). Outside of this regime, expansion of the population is governed by the F-KPP equation. Second, the formation of a distinct density bulge conforms to the same parameter regime (SI Appendix, section S9) and is thus also confirmed as an identifiable signature of boosted expansion for experiments and observations of bacteria in natural settings.
Quantitatively, we derive a relation between the expansion speed and the population profile in the density peak (Eq. 12) which only involves cellular motility parameters. Thus, the expansion speed can be inferred from just a snapshot of the density profile without requiring multiple measurements over an extended period of time (which can be difficult to do for bacteria outside the laboratory). Conversely, the cellular motility parameters can be inferred if measurements of the expansion speed and density profile are possible. Similarly, we derive a relation between the expansion speed and the trailing region (Eq. 51) which only involves the growth rate. We also connect the expansion speed to the chemoattractant flux generated by a source (Eq. 21). We thus provide a relation between population-level features and molecular/cellular level characteristics for chemotactic bacteria, analogous to what the solution of the F-KPP equation does for undirected motion (26–28). This relation provides an analytical framework for further studies of the effects of evolution (30) and phenotypic variability (5, 82–84) on the spread of the population.
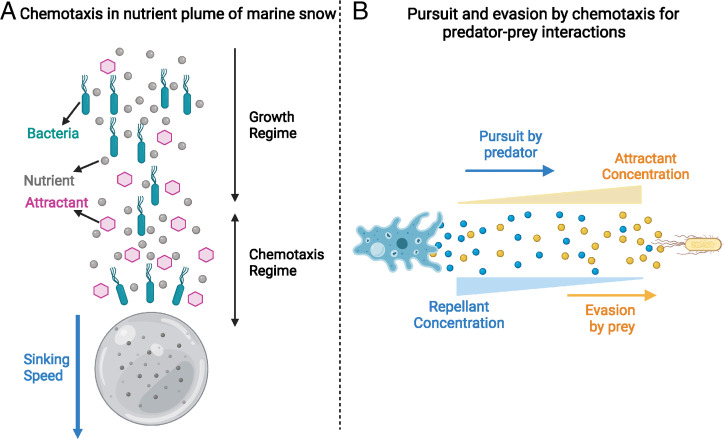
Our results may also be extended beyond expanding bacterial populations in culture plates. Fig. 8A shows an illustration of particulate organic matter, also known as marine snow, which sinks steadily in the ocean (85, 86). Nutrients and attractants generated from metabolic processes on such particles could sustain the growth of bacterial population chasing the sinking particle (87–89). To understand the resultant population distribution, the framework of the present study needs to be further extended to include nutrient and attractant generation, uptake, and diffusion on and near the particles (88, 90).
Fig. 8.
Illustrations of possible extensions of our analysis. (A) Chemotaxis in marine bacteria in the plume of sinking marine particles. The gray bead is the sinking particulate organic matter. A flux of nutrients (gray circles) and attractants (pink hexagons) diffuse into the wake of the particle. The cyan motile bacteria are able to direct their motion up the attractant gradient and thus move toward the sinking particle and keep up with it. (B) Chemotaxis in pursuit–evasion dynamics in a predator–prey system. A predator (shown as a blue amoeba) may pursue gradients of chemicals (attractant; yellow beads) left by a motile prey (a yellow bacterial cell). The motile prey may, in turn, evade the predator by performing chemotaxis and moving away from chemicals (repellent; blue beads) secreted by the predator.
More generally, the biological processes underlying chemotaxis-driven population expansion, sensing, directed movement, and the modification of the environment (as accomplished by uptake of attractant by chemotactic bacteria) are not limited to bacteria and have been reported for eukaryotic cells and higher-order organisms (15, 43). Directed movement may result from sensory cues as shown in nematodes (91) or as a consequence of area-restricted search as shown in beetles preying on aphids (92). The framework developed here may thus be employed to understand the GE dynamics of different organisms in diverse biological scenarios such as the spread of invasive species into a novel habitat (55–57, 93), the movement of cells and cell collectives during development in fruit flies and chickens (16, 94), or cancer metastasis in zebrafish and mammalian cells (19). Using this framework, we may understand the spatiotemporal dynamics of diverse ecological interactions between different species, such as predator–prey interactions where each species can direct their motion toward or away from each other, with the other species (or secretions by each species) playing the role of attractant or repellent (95) (Fig. 8B). The analysis of directed motion at the population level has largely been neglected in favor of the F-KPP dynamics (96) due to the immense body of knowledge developed for undirected motion (26–28). The mathematical understanding developed here presents a convenient framework to study directed population dynamics which has previously lacked analytical tools.
Materials and Methods
To generate all of the numerical results, finite element simulations of the system of equations were performed using FEniCS, a computing platform for solving partial differential equations (68, 69). A one-dimensional mesh of resolution 15 to 50 m was used to simulate a moving window of 30 mm (or 120 mm for very fast fronts). Finite elements of type were used.
The initial bacterial density was specified with in order to initiate a sufficiently localized initial population with a differentiable functional form. The initial attractant concentration was specified to be constant everywhere. Neumann boundary conditions of zero flux were specified on both ends of the simulation domain. A difference equation was then solved to approximate the differential equation in time using a small time step (typically between 2 and 25 s). The resulting solutions were recorded and used for the subsequent iteration of the difference equation.
In order to obtain high spatial and temporal resolutions simultaneously, a moving window technique was utilized. In the moving window technique, only a 30-mm (or 120-mm for very fast fronts) interval was simulated at a time. However, when the front of the wave had gone beyond a certain threshold in the simulation domain, the simulation domain was translated to the right, and the attractant concentrations and bacterial densities were extrapolated for the sections of the new simulation domain for which the values were not previously known. This technique holds very well as long as a threshold sufficiently far from the right end of the domain is chosen (this is also desirable to ignore edge effects) such that the linear extrapolation is correct within numerical resolution.
To analyze the simulations and extract the expansion speeds, the position of the maximum drift velocity was recorded for each time step. A linear fit over time was then employed for the position to obtain the expansion speed. Only fits with a small enough sum of residuals were considered. The fits were also curated manually to ensure that the expansion speed was calculated using a period of steady and constant expansion.
Supplementary Material
Acknowledgments
We thank Massimo Vergassola, Lev Tsimring, Roman Stocker, Johannes Keegstra, and Francesco Carrara for helpful discussions and Ying Tang for advice with numerical simulations. This research was supported by Simons Foundation (Grant 542387) and the NSF (Division of Molecular and Cellular Biosciences 2029574).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105138118/-/DCSupplemental.
∗This relation can also be obtained systematically from our solution by using (from Eq. 40) and the expression for β from Eq. 19 in Eq. 50. Since the result for β was invoked, it involves the approximation made following Eq. 17. This reflects the fact that in arriving at Eq. 52, we assumed that attractant uptake is always saturating.
Data Availability
Data and code are made available in GitHub at https://github.com/avaneeshnarla/chemotaxis-traveling-wave. All other study data are included in the article and/or SI Appendix.
References
- 1.Sourjik V., Wingreen N. S., Responding to chemical gradients: Bacterial chemotaxis. Curr. Opin. Cell Biol. 24, 262–268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stocker R., Marine microbes see a sea of gradients. Science 338, 628–633 (2012). [DOI] [PubMed] [Google Scholar]
- 3. Berg H. C., “Chemotaxis” in Quantitative Biology: From Molecular to Cellular Systems, Wall M. E., Ed. (Taylor and Francis, 2013), pp. 341–364. [Google Scholar]
- 4.Wong-Ng J., Celani A., Vergassola M., Exploring the function of bacterial chemotaxis. Curr. Opin. Microbiol. 45, 16–21 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Waite A. J., Frankel N. W., Emonet T., Behavioral variability and phenotypic diversity in bacterial chemotaxis. Annu. Rev. Biophys. 47, 595–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu Y., Quantitative modeling of bacterial chemotaxis: Signal amplification and accurate adaptation. Annu. Rev. Biophys. 42, 337–359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colin R., Sourjik V., Emergent properties of bacterial chemotaxis pathway. Curr. Opin. Microbiol. 39, 24–33 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Lopes J. G., Sourjik V., Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. ISME J. 12, 2736–2747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharf B. E., Hynes M. F., Alexandre G. M., Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 90, 549–559 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Kroupitski Y., et al., Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 75, 6076–6086 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vorholt J. A., Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Barbara G. M., Mitchell J. G., Bacterial tracking of motile algae. FEMS Microbiol. Ecol. 44, 79–87 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Garren M., et al., A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta M. S., Sliwerska E., Gore J., Polz M. F., Cordero O. X., Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devreotes P. N., Zigmond S. H., Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu. Rev. Cell Biol. 4, 649–686 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Dormann D., Weijer C. J., Chemotactic cell movement during development. Curr. Opin. Genet. Dev. 13, 358–364 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Tessier-Lavigne M., Goodman C. S., The molecular biology of axon guidance. Science 274, 1123–1133 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Baggiolini M., Chemokines and leukocyte traffic. Nature 392, 565–568 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Roussos E. T., Condeelis J. S., Patsialou A., Chemotaxis in cancer. Nat. Rev. Cancer 11, 573–587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenner M. P., Levitov L. S., Budrene E. O., Physical mechanisms for chemotactic pattern formation by bacteria. Biophys. J. 74, 1677–1693 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budrene E. O., Berg H. C., Complex patterns formed by motile cells of Escherichia coli. Nature 349, 630–633 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Erban R., Othmer H. G., From individual to collective behavior in bacterial chemotaxis. SIAM J. Appl. Math. 65, 361–391 (2004). [Google Scholar]
- 23.Adler J., Chemotaxis in bacteria. Science 153, 708–716 (1966). [DOI] [PubMed] [Google Scholar]
- 24.Adler J., Effect of amino acids and oxygen on chemotaxis in Escherichia coli. J. Bacteriol. 92, 121–129 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adler J., Chemoreceptors in bacteria. Science 166, 1588–1597 (1969). [DOI] [PubMed] [Google Scholar]
- 26.Fisher R. A., The wave of advance of advantageous genes. Ann. Eugen. 7, 355–369 (1937). [Google Scholar]
- 27.Kolmogorov A. N., Étude de l’équation de la diffusion avec croissance de la quantité de matière et son application à un problème biologique. Bull. Univ. Moskow, Ser. Internat. Sec. A 1, 1–25 (1937). [Google Scholar]
- 28.Van Saarloos W., Front propagation into unstable states. Phys. Rep. 386, 29–222 (2003). [Google Scholar]
- 29.Gude S., et al., Bacterial coexistence driven by motility and spatial competition. Nature 578, 588–592 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Liu W., Cremer J., Li D., Hwa T., Liu C., An evolutionarily stable strategy to colonize spatially extended habitats. Nature 575, 664–668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraebel D. T., et al., Environment determines evolutionary trajectory in a constrained phenotypic space. eLife 6, e24669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda T., et al., Coordination of gene expression with cell size enables Escherichia coli to efficiently maintain motility across conditions. bioRxiv [Preprint] (2021). (Accessed 12 May 2021). [DOI] [PMC free article] [PubMed]
- 33.Keller E. F., Segel L. A., Traveling bands of chemotactic bacteria: A theoretical analysis. J. Theor. Biol. 30, 235–248 (1971). [DOI] [PubMed] [Google Scholar]
- 34.Novick-Cohen A., Segel L. A., A gradually slowing travelling band of chemotactic bacteria. J. Math. Biol. 19, 125–132 (1984). [DOI] [PubMed] [Google Scholar]
- 35.Lapidus I. R., Schiller R., A model for traveling bands of chemotactic bacteria. Biophys. J. 22, 1–13 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller E. F., “Assessing the Keller-Segel model: How has it fared?” in Biological Growth and Spread. W., Jäger, Rost H., Tautu P., Eds. (Springer, 1980), pp. 379–387. [Google Scholar]
- 37.Tindall M. J., Maini P. K., Porter S. L., Armitage J. P., Overview of mathematical approaches used to model bacterial chemotaxis II: Bacterial populations. Bull. Math. Biol. 70, 1570–1607 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Koster D. A., Mayo A., Bren A., Alon U., Surface growth of a motile bacterial population resembles growth in a chemostat. J. Mol. Biol. 424, 180–191 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Cremer J., et al., Chemotaxis as a navigation strategy to boost range expansion. Nature 575, 658–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fenchel T., Motility and chemosensory behaviour of the sulphur bacterium thiovulum majus. Microbiology 140, 3109–3116 (1994). [Google Scholar]
- 41.McDonald D. E., Pethick D. W., Mullan B. P., Hampson D. J., Increasing viscosity of the intestinal contents alters small intestinal structure and intestinal growth, and stimulates proliferation of enterotoxigenic Escherichia coli in newly-weaned pigs. Br. J. Nutr. 86, 487–498 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Berg H. C., Turner L., Movement of microorganisms in viscous environments. Nature 278, 349–351 (1979). [DOI] [PubMed] [Google Scholar]
- 43.Painter K. J., Mathematical models for chemotaxis and their applications in self-organisation phenomena. J. Theor. Biol. 481, 162–182 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Horstmann D., From 1970 until present: The Keller-Segel model in chemotaxis and its consequences. II. Jahresber. Deutsch. Math. Verein. 106, 51–69 (2004). [Google Scholar]
- 45.Arumugam G., Tyagi J., Keller-Segel chemotaxis models: A review. Acta Appl. Math. 171, 1–82 (2021). [Google Scholar]
- 46.Si G., Wu T., Ouyang Q., Tu Y., Pathway-based mean-field model for Escherichia coli chemotaxis. Phys. Rev. Lett. 109, 048101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borkovich K. A., Alex L. A., Simon M. I., Attenuation of sensory receptor signaling by covalent modification. Proc. Natl. Acad. Sci. U.S.A. 89, 6756–6760 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg H. C., Tedesco P. M., Transient response to chemotactic stimuli in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 72, 3235–3239 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauffenburger D., Aris R., Keller K., Effects of cell motility and chemotaxis on microbial population growth. Biophys. J. 40, 209–219 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy C. R., Aris R., Traveling waves in a simple population model involving growth and death. Bull. Math. Biol. 42, 397–429 (1980). [DOI] [PubMed] [Google Scholar]
- 51.Lauffenburger D., Kennedy C. R., Aris R., Traveling bands of chemotactic bacteria in the context of population growth. Bull. Math. Biol. 46, 19–40 (1984). [Google Scholar]
- 52.Pedit J. A., Marx R. B., Miller C. T., Aitken M. D., Quantitative analysis of experiments on bacterial chemotaxis to naphthalene. Biotechnol. Bioeng. 78, 626–634 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Hilpert M., Lattice-Boltzmann model for bacterial chemotaxis. J. Math. Biol. 51, 302–332 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Ablowitz M. J., Zeppetella A., Explicit solutions of Fisher’s equation for a special wave speed. Bull. Math. Biol. 41, 835–840 (1979). [Google Scholar]
- 55.Skellam J. G., Random dispersal in theoretical populations. Biometrika 38, 196–218 (1951). [PubMed] [Google Scholar]
- 56.Lubina J. A., Levin S. A., The spread of a reinvading species: Range expansion in the California sea otter. Am. Nat. 131, 526–543 (1988). [Google Scholar]
- 57.Hastings A., et al., The spatial spread of invasions: New developments in theory and evidence. Ecol. Lett. 8, 91–101 (2005). [Google Scholar]
- 58.Gandhi S. R., Yurtsev E. A., Korolev K. S., Gore J., Range expansions transition from pulled to pushed waves as growth becomes more cooperative in an experimental microbial population. Proc. Natl. Acad. Sci. U.S.A. 113, 6922–6927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warren M. R., et al., Spatiotemporal establishment of dense bacterial colonies growing on hard agar. eLife 8, e41093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallatschek O., Nelson D. R., Life at the front of an expanding population. Evolution 64, 193–206 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Hallatschek O., Hersen P., Ramanathan S., Nelson D. R., Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl. Acad. Sci. U.S.A. 104, 19926–19930 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fusco D., Gralka M., Kayser J., Anderson A., Hallatschek O., Excess of mutational jackpot events in expanding populations revealed by spatial Luria-Delbrück experiments. Nat. Commun. 7, 12760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tikhomirov V., “A study of the diffusion equation with increase in the amount of substance, and its application to a biological problem” in Selected Works of A. N. Kolmogorov, Kolmogorov A. N., Ed. (Springer, 1991), pp. 242–270. [Google Scholar]
- 64.Schellenberg G. D., Furlong C. E., Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J. Biol. Chem. 252, 9055–9064 (1977). [PubMed] [Google Scholar]
- 65.Wong-Ng J., Melbinger A., Celani A., Vergassola M., The role of adaptation in bacterial speed races. PLOS Comput. Biol. 12, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaknin A., Berg H. C., Physical responses of bacterial chemoreceptors. J. Mol. Biol. 366, 1416–1423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neumann S., Grosse K., Sourjik V., Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, 12159–12164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alnæs M. S., UFL: A Finite Element Form Language, Logg A., Mardal K. A., Wells G. N., Eds. (Springer, 2012). [Google Scholar]
- 69.Logg A., Mardal K.-A., Wells G., Automated Solution of Differential Equations by the Finite Element Method (Springer, 2012). [Google Scholar]
- 70.Keller E. F., Mathematical aspects of bacterial chemotaxis. Antibiot. Chemother. (1971) 19, 79–93 (1974). [DOI] [PubMed] [Google Scholar]
- 71.Rosen G., On the propagation theory for bands of chemotactic bacteria. Math. Biosci. 20, 185–189 (1974). [Google Scholar]
- 72.Ford R. M., Phillips B. R., Quinn J. A., Lauffenburger D. A., Measurement of bacterial random motility and chemotaxis coefficients: I. Stopped-flow diffusion chamber assay. Biotechnol. Bioeng. 37, 647–660 (1991). [DOI] [PubMed] [Google Scholar]
- 73.Dufour Y. S., Gillet S., Frankel N. W., Weibel D. B., Emonet T., Direct correlation between motile behavior and protein abundance in single cells. PLOS Comput. Biol. 12, e1005041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ford R. M., Lauffenburger D. A., Analysis of chemotactic bacterial distributions in population migration assays using a mathematical model applicable to steep or shallow attractant gradients. Bull. Math. Biol. 53, 721–749 (1991). [DOI] [PubMed] [Google Scholar]
- 75.Ahmed T., Shimizu T. S., Stocker R., Microfluidics for bacterial chemotaxis. Integr. Biol. 2, 604–629 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Wolfe A. J., Berg H. C., Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. U.S.A. 86, 6973–6977 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ebert U., van Saarloos W., Front propagation into unstable states: Universal algebraic convergence towards uniformly translating pulled fronts. Physica D 146, 1–99 (2000). [Google Scholar]
- 78.Dee G. T., van Saarloos W., Bistable systems with propagating fronts leading to pattern formation. Phys. Rev. Lett. 60, 2641–2644 (1988). [DOI] [PubMed] [Google Scholar]
- 79.Van Saarloos W., Front propagation into unstable states. II. Linear versus nonlinear marginal stability and rate of convergence. Phys. Rev. A Gen. Phys. 39, 6367–6390 (1989). [DOI] [PubMed] [Google Scholar]
- 80.Collet P., Eckmann J. P., The stability of modulated fronts. Helv. Phys. Acta 60, 969–991 (1987). [Google Scholar]
- 81.Erm P., Phillips B. L., Evolution transforms pushed waves into pulled waves. Am. Nat. 195, E87–E99 (2020). [DOI] [PubMed] [Google Scholar]
- 82.Fu X., et al., Spatial self-organization resolves conflicts between individuality and collective migration. Nat. Commun. 9, 2177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salek M. M., Carrara F., Fernandez V., Guasto J. S., Stocker R., Bacterial chemotaxis in a microfluidic T-maze reveals strong phenotypic heterogeneity in chemotactic sensitivity. Nat. Commun. 10, 1877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mattingly H. H., Emonet T., A mechanism for migrating bacterial populations to non-genetically adapt to new environments. bioRxiv [Preprint] (2021). (Accessed 22 September 2021).
- 85.Asper V. L., Measuring the flux and sinking speed of marine snow aggregates. Deep-Sea Res. A, Oceanogr. Res. Pap. 34, 1–17 (1987). [Google Scholar]
- 86.Alldredge A. L., Silver M. W., Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20, 41–82 (1988). [Google Scholar]
- 87.Shanks A. L., Trent J. D., Marine snow: Microscale nutrient patches 1. Limnol. Oceanogr. 24, 850–854 (1979). [Google Scholar]
- 88.Kiørboe T., Jackson G. A., Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46, 1309–1318 (2001). [Google Scholar]
- 89.Stocker R., Seymour J. R., Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiørboe T., Formation and fate of marine snow: Small-scale processes with large-scale implications. Sci. Mar. 65, 57–71 (2001). [Google Scholar]
- 91.Ward S., Chemotaxis by the nematode Caenorhabditis elegans: Identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. U.S.A. 70, 817–821 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kareiva P., Odell G., Swarms of predators exhibit “preytaxis” if individual predators use area-restricted search. Am. Nat. 130, 233–270 (1987). [Google Scholar]
- 93.Andow D. A., Kareiva P. M., Levin S. A., Okubo A., Spread of invading organisms. Landsc. Ecol. 4, 177–188 (1990). [Google Scholar]
- 94.Yang X., Dormann D., Münsterberg A. E., Weijer C. J., Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev. Cell 3, 425–437 (2002). [DOI] [PubMed] [Google Scholar]
- 95.Tsyganov M. A., Brindley J., Holden A. V., Biktashev V. N., Quasisoliton interaction of pursuit-evasion waves in a predator-prey system. Phys. Rev. Lett. 91, 218102 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Armsworth P. R., Roughgarden J. E., The impact of directed versus random movement on population dynamics and biodiversity patterns. Am. Nat. 165, 449–465 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code are made available in GitHub at https://github.com/avaneeshnarla/chemotaxis-traveling-wave. All other study data are included in the article and/or SI Appendix.