Significance
Microscale interactions between marine phytoplankton and their bacterial microbiomes can influence ecosystem functioning and global biogeochemical cycling through complex exchanges of metabolites and sophisticated ecological processes. Previous investigation of the phytoplankton microbiome has not focused on the role of a host’s underlying genetic background. Through examination of a single phytoplankton species’ microbiome across the global ocean, we found that host genotype strongly influenced microbiome community composition, with associations that potentially persist across generations and ocean basins but assemble rapidly (within days). The long-term association of microbiomes with host genetic background could explain the evolution and maintenance of intricate phytoplankton–bacteria interactions.
Keywords: phytoplankton–bacteria interactions, microbiome, phycosphere, population genetics, host–microbe interactions
Abstract
Phytoplankton support complex bacterial microbiomes that rely on phytoplankton-derived extracellular compounds and perform functions necessary for algal growth. Recent work has revealed sophisticated interactions and exchanges of molecules between specific phytoplankton–bacteria pairs, but the role of host genotype in regulating those interactions is unknown. Here, we show how phytoplankton microbiomes are shaped by intraspecific genetic variation in the host using global environmental isolates of the model phytoplankton host Thalassiosira rotula and a laboratory common garden experiment. A set of 81 environmental T. rotula genotypes from three ocean basins and eight genetically distinct populations did not reveal a core microbiome. While no single bacterial phylotype was shared across all genotypes, we found strong genotypic influence of T. rotula, with microbiomes associating more strongly with host genetic population than with environmental factors. The microbiome association with host genetic population persisted across different ocean basins, suggesting that microbiomes may be associated with host populations for decades. To isolate the impact of host genotype on microbiomes, a common garden experiment using eight genotypes from three distinct host populations again found that host genotype influenced microbial community composition, suggesting that a process we describe as genotypic filtering, analogous to environmental filtering, shapes phytoplankton microbiomes. In both the environmental and laboratory studies, microbiome variation between genotypes suggests that other factors influenced microbiome composition but did not swamp the dominant signal of host genetic background. The long-term association of microbiomes with specific host genotypes reveals a possible mechanism explaining the evolution and maintenance of complex phytoplankton–bacteria chemical exchanges.
Interactions between marine phytoplankton and bacteria can exert a profound influence on ecosystem function and biogeochemical cycling, impacting rates of primary production, phytoplankton aggregation, organic carbon export, and nutrient cycling (1–3). As an aquatic analog of the plant rhizosphere, the most intimate relationships between phytoplankton and bacteria exist in the phycosphere, the region immediately surrounding a phytoplankton cell, where molecules can be exchanged despite the effects of turbulence and diffusion (4). Relationships between phytoplankton and bacteria in the phycosphere range from cooperative to competitive (5). For example, during exponential growth, phytoplankton actively secrete amino acids that are taken up by bacteria, despite potentially significant energy costs (6). In turn, some bacteria synthesize essential vitamins and growth hormones that stimulate phytoplankton productivity (7, 8). During phytoplankton senescence, formerly “friendly” bacteria can become pathogenic, producing algicides that lyse phytoplankton cells and release organic carbon to the environment (9). These complex ecological interactions have been investigated in the laboratory for specific phytoplankton–bacteria pairs (3). However, the persistence and phylogenetic breadth of these relationships for both host and microbiome remain open questions (10–12).
In terrestrial habitats, clear linkages exist between the genetic background (i.e., host genotype and population genetic structure) of foundational plant species and the organisms that rely on them. For example, genetic variation within tree species can influence the structure of associated epiphytic, mycorrhizal, and invertebrate communities (13–15) through mutualism, parasitism, commensalism, facilitation, and competition (reviewed in ref. 16). These associations extend to plant-associated bacteria, whose abundance, composition, and diversity reflect intraspecific trait variation among host genotypes (17). Because microbes regulate processes such as decomposition, nutrient dynamics, and energy flow, the influence of intraspecific genetic variation in plants on their associated bacteria extends the effects of community genetics to ecosystem processes (18). In contrast to terrestrial habitats, seawater allows both bacteria and their phytoplankton hosts to drift with nearly unlimited dispersal across the global ocean. It is unknown whether planktonic communities mirror those in terrestrial habitats, where host genetics can shape bacterial community composition and influence ecosystem processes (16), or whether dispersal in the dilute marine environment overwhelms the formation of close and specific relationships between phytoplankton and their associated microbiota. Although phytoplankton drift freely across the global ocean with nearly unlimited dispersal and divide primarily asexually, they still possess clear genetic structure. Phytoplankton species are organized into genetically distinct populations that possess phenotypic trait variation, are associated with specific environmental conditions, and undergo large increases in abundance, known as blooms (19–23). Furthermore, genetically distinct phytoplankton populations persist on time scales of decades to centuries (23, 24), providing ample opportunity for populations to develop specific relationships with other microbes. Few studies have evaluated the microbiomes of multiple strains within a phytoplankton species. Some studies found that individual phytoplankton species may possess a core microbiome, with a consistent set of bacterial phylotypes and metabolic potentials (25, 26), while others found that phytoplankton strains supported distinct microbiomes (10) and differed in their growth responses to the same bacterial strain (7, 27, 28). These intriguing findings have not been rigorously examined in light of the genetic background of the host phytoplankton species. Given that plant genotype often (29–31) but not always (32) drives host microbiomes and given the differences between terrestrial and planktonic habitats, understanding to what extent the genetic background of phytoplankton species influences microbiome composition is critical to understanding the nature of their interactions and parsing the roles of both partners in global biogeochemical cycles.
Here, we assessed host–microbe interactions using the model marine phytoplankton Thalassiosira rotula, a cosmopolitan species characterized by high genotypic and phenotypic diversity, which is subdivided into genetically distinct populations (24, 33). We examined whether the T. rotula microbiome was influenced by host genetic background, either at the genotype or population level. We combined a study of the microbiomes of 81 environmental genotypes (representing eight genetically distinct populations) sampled from around the world (Fig. 1 and Table 1) with a common garden experiment explicitly designed to evaluate the influence of host genetic background on microbiome assembly in the absence of other environmental changes. The union of these approaches allowed us to examine the role of host genetics in setting bacterial assemblage composition and to determine whether a core microbiome might exist in a globally distributed, pelagic phytoplankton.
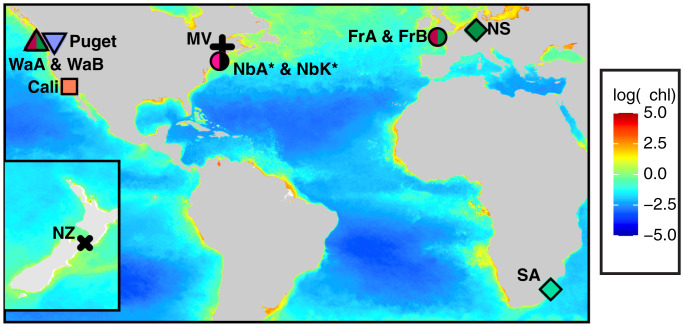
Fig. 1.
Global sampling locations of the phytoplankton host T. rotula populations [symbol colors denote populations identified in Whittaker and Rynearson (24), and black indicates populations identified in this study] and their associated microbiomes (Table 1 and SI Appendix, Table S1). Three sites (Wa, Nb, and Fr) were resampled (symbols with two colors), and whole seawater was collected twice from Narragansett Bay (Nb) for microbiome comparisons with the in situ whole seawater bacterial community (asterisks). The base map is a composite of log annual average chlorophyll a concentrations (milligrams/meter−3, 2010) (https://oceandata.sci.gsfc.nasa.gov/directaccess/MODIS-Aqua/Mapped/Annual/9km/chlor_a/).
Table 1.
Global sampling site information for T. rotula and associated bacteria
| Ocean basin | Sample date | Sample site | Sample name | Host population | No. of host microbiomes sequenced |
| Atlantic | 1/26/10 | Narragansett Bay Plankton Time Series, USA | NbA* | 2 | 5 |
| 10/15/10 | Narragansett Bay Plankton Time Series, USA | NbK*,† | 8 | 4 | |
| 10/14/10 | Martha's Vineyard Coastal Observatory, USA | MV† | 7 | 8 | |
| 3/18/10 | North Sea, Helgoland Roads Time Series, Germany | NS | 4 | 8 | |
| 3/9/10 | SOMLIT-Astan Time Series, France | FrA | 1 | 8 | |
| 3/24/10 | SOMLIT-Astan Time Series, France | FrB | 4 | 7 | |
| Pacific | 2/16/10 | Olympic Peninsula, USA | WaA | 1 | 8 |
| 3/29/10 | Olympic Peninsula, USA | WaB | 4 | 7 | |
| 4/14/10 | Puget Sound, USA | Puget | 6 | 7 | |
| 3/22/10 | Newport Beach Pier, USA | Cali | 3 | 7 | |
| 10/15/10 | Greta Point, New Zealand | NZ | NA | 7 | |
| Indian | 11/19/10 | Durban, South Africa | SA | 5 | 5 |
Additional sample metadata are in SI Appendix, Table S1.
*Whole seawater was collected from Narragansett Bay on two occasions to determine bacterioplankton community composition.
†Host populations genotyped in this study.
Results and Discussion
Characterizing the T. rotula Microbiome.
Marine phytoplankton and bacteria have well-established biogeochemical linkages through bulk pools of nutrients and organic carbon, although the specificity of exchanges and phylogenetic breadth of the partnerships between these organisms remain areas of active investigation (3). Here, we use a rich dataset of 81 single-cell environmental isolates of the phytoplankton species T. rotula and their co-isolated bacterial microbiomes paired with controlled laboratory experiments to examine the relationship between phytoplankton host genetic background and bacterial microbiome. Because environmental T. rotula microbiomes were obtained from washed single cells or chains, the microbiomes we recovered were largely from bacterial cells physically associated with the phytoplankton host. In order to increase the likelihood that bacterial sequences examined were derived from the in situ algal microbiome, we instituted a number of controls, which likely eliminated some real microbial diversity but reduced false relationships and removed potential contaminants. These included washing phytoplankton cells prior to culturing to reduce free-living bacteria to an estimated <1 cell per freshly-isolated isolated phytoplankton, using a low–organic matter medium to support algal-derived bacterial growth and employing strict quality controls for sequence analyses (SI Appendix, SI Results and Discussion).
Each environmental T. rotula isolate represented a unique multilocus genotype (Dataset S1) and possessed a diverse microbiome; after strict quality controls, we identified 278 bacterial amplicon sequence variants (ASVs) that clustered at the 99.9% identity level, with a range of eight to 51 bacterial ASVs and an average of 24.4 ± 9.6 ASVs per T. rotula environmental genotype (Dataset S2). Our results are consistent with previous reports of bacterial richness ranging from 1 to 125 operational taxonomic units (OTUs; roughly equivalent to ASVs) per phytoplankton isolate (11, 12, 34), including eight to 80 OTUs per T. rotula isolate (10, 35). Furthermore, the T. rotula-associated bacterial communities were significantly different from the in situ bacterioplankton communities (SI Appendix, Fig. S1 and Dataset S2; Analysis of Similarities (ANOSIM) R = 0.80, P = 0.001), highlighting that microbiome composition does not simply reflect water column bacterioplankton composition (3).
The T. rotula-associated bacterial communities from the environmental genotypes were dominated by Proteobacteria (83.5% of all sequences), including γ-proteobacteria (61.3%) and α-proteobacteria (21.3%) (Dataset S2). However, no single ASV was present in all microbiomes. The most frequent ASVs in the T. rotula microbiomes were two α-proteobacteria Loktanella (ASVs 743 and 1,938) and Actinobacteria Rhodococcus (ASV 24), present in 59%, 44%, and 56% of all environmental genotypes, respectively (SI Appendix, Fig. S2). The relative abundance of individual ASVs per host genotype ranged widely, from 0 to >90% in a microbiome (SI Appendix, Fig. S3). For example, when present, the ASV most likely to occur in a microbiome, Loktanella 743, had a relative median abundance of 4.41% but a range of 0 to >80% in any given microbiome. The absence of a common ASV across microbiomes and high degree of variability in microbiome composition indicates that the cosmopolitan phytoplankton T. rotula sampled from across the global ocean lacks a “core” microbiome. Instead, T. rotula cells may associate with closely related lineages of bacteria that serve similar ecological roles but are adapted to distinct environmental conditions. With few exceptions (25), the lack of a core microbiome stands in contrast to most surveys of microbiome communities from multicellular eukaryotes (36) and marine algae (12).
Although no single bacterial ASV was present in all microbiomes, similarities existed at the taxonomic order level, with the order Alteromonadales observed in 80 of the 81 environmental isolates. The Alteromonadales are known to associate with a number of phytoplankton lineages (12, 37). Here, we supplied all required substrates for algal growth in the media, including vitamins and iron; thus, T. rotula may have benefited from the bacterial reduction of both oxidative stress and waste compounds (5). Alternately, under these replete conditions, T. rotula may have derived no benefit from the bacteria, with the relationship being driven by bacterial metabolism of phytoplankton-derived organic material. Overall, these data suggest a strong relationship of the Alteromonadales with T. rotula.
The Role of Host Genetic Background in Structuring T. rotula Microbiomes.
In order to better understand the factors that shape T. rotula-associated microbiomes, we examined the roles of stochasticity, environmental selection, and host genetic background. To test how stochastic effects, such as dispersal, ecological drift, and random death and birth events (38) influenced the microbiomes of environmental isolates, we used the normalized stochastic ratio of the Jaccard matrix (NSTjac), which quantifies the data structure ranging from deterministic (0%) to completely stochastic (100%). The NSTjac averaged 36.2 ± 18.4% across all diatom microbiomes, indicating while some stochasticity was evident in the data structure, deterministic effects strongly impacted the microbiomes of environmental isolates.
To determine whether host genotype or environmental factors explained the composition of host microbiomes, we first examined microbiome composition in light of the T. rotula genetic background. Even at the bacterial class level, there appeared to be stark differences in the microbiomes of different host genotypes (SI Appendix, Fig. S4). Variation among microbiomes was not evenly spread across the global dataset. Instead, we found that phytoplankton isolates from the same water sample consistently coisolated with a more similar bacterial community than isolates from different water samples (ANOSIM R statistic = 0.83, P = 0.001), with strong separation between cocultured and in situ bacterial communities (SI Appendix, Fig. S1). Of the microbiomes we analyzed, the host was subdivided into eight genetically distinct populations from three ocean basins (Fig. 2A and SI Appendix, Tables S1 and S2). By grouping T. rotula genotypes into populations, we observed the same T. rotula population in different locations (e.g., North Sea [NS] and Olympic Peninsula B [WaB]) (24) as well as single locations from which different populations were sampled (e.g., Narragansett Bay Plankton Time Series A and K [NbA and Nbk]). We then performed multiple tests to determine how host population and environmental factors shaped the composition of the T. rotula microbiome. First, microbiome composition was most strongly and equally correlated with host population membership and sea surface temperature (SST) (bioenv correlations both 0.51). Interestingly, the genetic composition of the host population was also strongly influenced by temperature (24), suggesting possible collinearity in the dataset. To further examine the relationship between host microbiome, host population, and SST, variance partitioning was conducted using two tests with different assumptions. We found that regardless of the variance-partitioning test used, host population explained by far the largest portion of variance in microbiome composition (adonis R2 = 0.47 P = 0.001; varpart R2 = 0.35), followed distantly by SST (adonis R2 = 0.057 P = 0.001; varpart R2=0.04), and the interaction between SST and phytoplankton population (adonis R2 = 0.038 P = 0.001; varpart R2 = 0.01). We found no correlation between microbiome composition and chlorophyll a concentration, salinity, and T. rotula concentration. We cannot rule out other unmeasured factors (e.g., nutrients) that could affect both the microbiome and algal population structure (20, 26). Although temperature is known to shape microbiome composition and physiology (39), genetic background of the algal host has not previously been shown to play a role in microbiome community composition.
Fig. 2.
Divergence of T. rotula populations and their associated microbiomes. (A) PCoA showing genetic divergence (FST) among phytoplankton populations identified using microsatellites. Each point indicates a genetically distinct T. rotula population (P < 0.05, Bonferroni-corrected). Colors and site locations are as in Fig. 1 and Table 1. MV and NbK are identified genetic populations of T. rotula and are plotted with previously identified populations (24). No genetic population was determined for the New Zealand sample (NZ) due to insufficient sample size. Image credit: Data reprinted with permission from ref. 24. (B) Non-metric multidimensional scaling (NMDS) of a log-normalized Bray–Curtis dissimilarity matrix of T. rotula microbiome 99.9% identity 16S ASV composition. Each point represents the microbiome of one T. rotula environmental isolate. Host isolates were sampled from different locations (shapes) and different populations (colors). Ellipses represent the 80% CI around the mean for microbiomes from each genetically distinct host phytoplankton population (stress = 0.19).
We observed a significant relationship between host population and the taxonomic composition of its microbiome (Fig. 2B, ANOSIM R Statistic = 0.66, P = 0.001). Environmental genotypes from the same host population had microbiomes with compositional similarities even when those genotypes originated from different ocean basins (e.g., populations 1 and 4, Fig. 2B). For example, the microbiomes of T. rotula population 1 sampled from the north Pacific and north Atlantic (SOMLIT-Astan Time Series, France [FrA] and Olympic Peninsula A [WaA]) shared 20 ASVs (43 and 30% of the ASVs in FrA and WaA microbiomes, respectively). Two ASVs, α-proteobacteria Loktanella (ASV 743) and Actinobacteria Rhodococcus (ASV 24), were present in all population 1 genotypes from both ocean basins. The microbiome similarities among environmental genotypes from different ocean basins are notable, because rates of surface water circulation in the global ocean can translate into decades-long separation of individuals from the same host population (40).
Overall, compositional divergence between microbiomes was significantly correlated with genetic divergence between host phytoplankton populations (FST) (Fig. 3A) (mantel r = 0.45, P = 0.01; linear regression R2 = 0.17, F = 6.63, P = 0.02). In contrast, geographic distance had no significant impact on the phytoplankton microbiome (Fig. 3B) (mantel r = 0.18, P = 0.12; linear regression R2 = 0.02, F = 1.82, P = 0.18). Positive geographic distance–decay relationships are signatures of neutral processes (41), evidenced by the biogeographic patterns of the global bacterioplankton community (42), and have been shown to occur in some plankton microbiomes (43). In contrast, our findings suggest that phytoplankton microbiomes sampled at different times or places (10) may reflect the genetic background of the host phytoplankton and are not simply the result of colonization by the bacteria responding most rapidly to phytoplankton exudates, as models have predicted (44).
Fig. 3.
Distance decay of T. rotula microbiomes. (A) Host normalized genetic distance ((FST)/(1-FST)) versus microbiome dissimilarity (Bray–Curtis). Mantel test for matrix correlation: r = 0.45 and P = 0.01; Regression analysis: R2 = 0.17, F = 6.63, and P = 0.02. (B) Pairwise comparison of log geographic distance between sampling sites and log T. rotula microbiome dissimilarity (Bray–Curtis). Mantel test for matrix correlation: r = 0.17 and P = 0.11; Regression analysis: R2 = 0.01, F = 1.60, and P = 0.22.
To explicitly test whether distinct bacterial communities could assemble in response to T. rotula with different genetic backgrounds in the absence of potentially confounding environmental differences, we performed a common garden experiment under controlled conditions using host cells freshly isolated from the field (Table 2 and Dataset S3) (35, 45, 46). Eight host genotypes were made axenic, inoculated with a <1-µm natural seawater bacterial community, incubated for 5 d, and then the entire contents of the flasks were harvested by filtering. The final concentrations of host T. rotula were high (22,575 ± 10,129 cells/mL −1), similar to levels observed during phytoplankton blooms (47). Subsequent sequencing of treatment flasks revealed a total of 419 ASVs, with an average of 330.1 ± 35.4 ASVs per genotype versus 350.3 ± 3.0 per bacterial control at the time of harvest (Fig. 4A and Dataset S4). The higher diversity of bacteria observed in this experiment compared to the survey of environmental genotypes (24.4 ± 9.6 bacterial ASVs per T. rotula isolate) can be attributed to factors including retention of nongrowing cells and inclusion of both algal-attached and free-living bacteria. Despite capturing both the free-living and attached bacterial community, there was similarity between the microbiome composition of the common garden T. rotula and the environmental isolates. In the environmental isolates, the two most frequently occurring bacterial classes (Gammaproteobacteria [62.05% of isolates] and Alphaproteobacteria [21.57%]), bacterial orders (Alteromonadales [98.77%] and Rhodobacterales [92.60%]), and bacterial families (Rhodobacteraceae [92.59%] and Colwelliaceae [75.31%]) were present in 100% of the common garden T. rotula microbiomes. Furthermore, the two most frequently occurring bacterial genera in the environmental isolates (Loktanella [70.37% of isolates] and Colwellia [69.14%]) were present in 66.67% and 100% of the common garden isolates, respectively. Loktanella was not present in the common garden microbiomes of two distinct PopC genotypes (NbQ-B4 and NbQ-B6), representing potential genotypic specific preferences for certain bacteria.
Table 2.
Common garden experiment population names, collection dates, number of T. rotula host isolates genotyped, and number of host isolates (and their names) used for the common garden experiment
| Population name | Site name | Collection date | No. of host isolates genotyped | No. host isolates in the common garden experiment | Isolate names |
| PopA | NbO | 3/20/17 | 18 | 3 | NbO-A4, NbO-A5, NbO-D4 |
| PopB | NbP | 5/23/17 | 38 | 2 | NbP-C5, NbP-YE5 |
| PopC | NbQ | 11/13/17 | 23 | 3 | NbQ-B4, NbQ-B6, NbQ-B7 |
All T. rotula isolates were collected from the Narragansett Bay Plankton Time Series (Table 1).
Fig. 4.
Bacterial community composition associated with T. rotula genotypes in a common garden experiment. (A) Percentage of relative abundance of bacterial taxonomic orders, obtained using 16S rRNA gene sequencing, after a 5-d common garden experiment. Eight T. rotula genotypes were grown in triplicate (lower x-axis labels), selected from three genetically distinct populations (upper x-axis), and inoculated (Inoc) with a <1 μm natural seawater bacterial community (Bact_T0) taken from a whole seawater sample (WSW_T0). Bacterial controls (Bacteria, upper x-axis) were analyzed on day 5 and were run in quadruplicate. (B) NMDS of a normalized Bray–Curtis dissimilarity matrix of 99.9% identity 16S rRNA gene ASV composition of bacterial communities from different T. rotula genotypes and populations (shapes, colors) and from bacterial controls (Bacteria) at the time of harvest. Ellipses represent 80% CI for the microbiomes from each genetically distinct phytoplankton population (stress = 0.08).
By evaluating replicates for each genotype as well as multiple genotypes within T. rotula populations, the common garden experiment allowed us to examine the relative importance of stochastic effects as well as the influence of two levels of host genetic background on bacterial composition: the underlying genotype of the host and host membership in a particular genetic population. Deterministic processes appeared to play a larger role than stochasticity in shaping host microbiomes in the common garden experiment (NSTjac 23.0% [PopA], 39.0% [PopB], and 37.8% [PopC]). Consistent with a key role for selection, we found that replicate flasks from the same T. rotula common garden genotype yielded highly reproducible bacterial communities (Fig. 4B; ANOSIM, R = 0.93, significance = 0.001; adonis R2 = 0.76, P = 0.001). Next, we examined the role of population. The T. rotula used in the common garden experiment originated from three genetically distinct populations (SI Appendix, Tables S3 and S4). We found that genotypes from the same population had more similar bacterial communities than those from other populations (Fig. 4B and Dataset S4; ANOSIM, R = 0.52, significance = 0.001; adonis R2 = 0.38, P = 0.001). Remarkably, the population-specific effects were evident rapidly, within the 5-d incubation period, suggesting that even stronger differences might have emerged over longer time frames but with concurrent increases in “bottle effects” (48, 49). While the inoculum communities were dominated by SAR11, the microbiome cocultured with T. rotula was enriched in fast-growing organisms (e.g., Vibrio, Marinomonas) similar to those in Fu (46). Marine bacterioplankton are known to respond rapidly to primary producer–derived microenvironments including nutrient plumes (50), lysed phytoplankton (44), and both real and synthetic phytoplankton (35, 51). Here, we showed that marine bacteria rapidly differentiate their composition in the presence of phytoplankton with different genetic backgrounds.
Despite similarities among the microbiomes of phytoplankton isolates from the same genetic population, T. rotula-associated bacterial communities exhibited variability both among genotypes from a single population and among replicates of a single genotype, suggesting that other mechanisms, such as stochastic and priority effects (52–54), also likely played a role in community assembly. As we showed, stochastic effects, while apparent, were not the dominant drivers shaping T. rotula microbiome composition. Priority effects could influence the microbiome through historical contingencies shaping the pool of available microbes as well as through colonization of phytoplankton surfaces (55). The common garden experiment could have been especially susceptible to priority effects, since we examined community assembly using axenic genotypes, a scenario in which abundant and fast-growing phylotypes may dominate over phycosphere-adapted taxa.
Microbiome variation among genotypes within the same host population may well be explained by a phenomenon that we describe here as genotypic filtering, similar to the process of environmental filtering (56) or host filtering (57). Genotypic variation is associated with high levels of phenotypic variation in phytoplankton, including T. rotula (33, 58). Prior observations of extensive phenotypic variation of T. rotula together with recent evidence that exometabolites from different phytoplankton species predictably influence microbiome composition (51), suggest that different T. rotula genotypes may release distinct organic compounds that select for or attract different bacterial associates. Phytoplankton like T. rotula have been shown to promote the growth and attachment of beneficial bacteria and inhibit colonization of opportunistic bacteria through secondary metabolites (59). Phenotypic variation among genotypes may thus lead to genotypic filtering, a mechanism not previously attributed to phytoplankton-associated bacterial communities. The taxonomic extent of genotypic filtering in the phytoplankton is unknown. Intriguingly, there is evidence of phytoplankton-intraspecific variation in exudate profiles, which are well-known to influence bacterial composition and activities (60) and evidence of genotype-specific responses to bacteria (7), both of which suggest that genotypic filtering may be a widespread phenomenon in the phytoplankton.
Microbiome stability among genotypes separated by global-scale geographic distances may be maintained in part by the phytoplankton life cycle. Prior work has posited that phytoplankton microbiomes are short lived with a duration of just 1 to 2 d, equivalent to the lifespan of phytoplankton cells (51). While it is possible that microbiomes reassemble on daily timescales, our results suggest that additional mechanisms may act to promote the long-term similarity of the phytoplankton microbiome, such as vertical transfer of bacteria during reproduction (61). The host T. rotula divides primarily asexually, yielding two daughter cells that each receive from the mother cell one-half of a hard, silicified cell-wall covering known as a frustule (62), which presumably contains a near-complete microbiome able to colonize the newly-formed half frustule. Sexual reproduction in phytoplankton like T. rotula occurs on time scales of once per year to once every 40 y, although the details are not well known (62). As sexually generated offspring cast off both halves of the parent frustule and a new frustule is formed, microbiome colonization would occur from bacteria in the surrounding water column. It is interesting to note that in colony-forming phytoplankton like T. rotula, it has been observed that only a few cells in the monoclonal colony tend to undergo sexual reproduction (62), providing a route for microbiome colonization from neighboring cells and long-term stability of microbiome community composition.
The process of T. rotula microbiome community assembly can be explained through an ecological framework of selection, drift, dispersal, and diversification (53, 63). For selection and drift, the NSTjac (38) of both the environmental isolates (36.2 ± 18.4%) and common-garden experiment (33.3 ± 8.9%) indicates that the genotype selection effect is dominated by deterministic rather than stochastic factors, which provides a foundation for exploring assembly mechanisms in future studies. With regards to dispersal, we find that the same genetic populations of T. rotula sampled from different locations carry common ASVs, suggesting reassociation of microbiomes and maintenance of certain microbiome partners despite geographic separation. Finally, we found evidence of genotypic filtering in T. rotula microbiomes, which could lead to the diversification and maintenance of different populations of T. rotula.
Here, across both global environmental samples and a common-garden experiment, we observed that host genotype shapes the microbiomes of a single-celled phytoplankton. The association of bacteria with host genetic background observed here has the potential to impact host ecology and biogeochemical cycling. For example, microbiomes are known to alter growth and survival rates of the host (64, 65) and expand host metabolic potential (26). The evolutionary fingerprint of the phytoplankton host on bacterial microbiomes opens routes of inquiry into factors that initiate and maintain microbiome composition and function. In particular, the long-term association (or frequent reassociation) of bacteria to a genetically-restricted subset of a species sets up the potential for significant eco-evolutionary dynamics to occur in both partners and provides a mechanism for the evolution and maintenance of recently identified sophisticated molecular exchanges between phytoplankton and bacteria (3).
Materials and Methods
Global Environmental Sampling.
Surface water containing T. rotula cells was collected 12 times from nine locations in the Atlantic, Pacific, and Indian Oceans (Fig. 1, Table 1, and SI Appendix, Table S1) (24) and single cells or chains of T. rotula were isolated and genotyped (SI Appendix, SI Materials and Methods). We amplified the V3-V4 region of the 16S ribosomal ribonucleic acid (rRNA) gene (SI Appendix, SI Materials and Methods) and paired-end sequencing (2 × 250 bp) of the amplicons was performed on a MiSeq (Illumina) using version 2 chemistry at the Duke Genome Sequencing and Analysis Core Facility. Quality control, ASV clustering at 99.9% identity, and statistical analysis are described in SI Appendix, SI Materials and Methods.
Common Garden Experiment.
Single-cell isolates of T. rotula were collected from the Narragansett Bay Long-Term Plankton Time Series site three times in 2017 (March 20 [NbO/PopA], May 23 [NbP/PopB], and November 13 [NbQ/PopC]) (Table 2), genotyped using microsatellite markers, and maintained in axenic culture (SI Appendix, SI Materials and Methods). Direct comparisons were not made between the common garden and global environmental T. rotula genotypes, because we modified PCR and fragment analysis protocols compared to Whittaker and Rynearson (24) and used different 16S rRNA gene primers between the two datasets (SI Appendix, SI Materials and Methods). Eight axenic genotypes were chosen to undergo the common garden experiment described in SI Appendix, SI Materials and Methods. After the 5-d experiment, we amplified the V4-V5 region of the 16S rRNA gene of the triplicate common garden genotypes, quadruplicate bacterial controls, and starting inoculum, sequenced the amplicons on a MiSeq. 2 × 250 bp run (Illumina), and performed bioinformatic and statistical analysis (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
This research was supported by Gordon and Betty Moore Foundation Award GBMF3768 (to D.E.H. and T.A.R.) and NSF Award OCE-1638834 (to T.A.R.). Part of the research was conducted with a graduate stipend (to O.A.) and using instrumentation supported by NSF Established Program to Stimulate Competitive Research (EPSCoR) Awards OIA-1004057 and OIA-1655221. We also acknowledge Zhao Wang for assistance with sequencing preparation.
Footnotes
Author contributions: O.M.A., D.E.H., and T.A.R. conceived the study and designed experiments; O.M.A, K.A.W., and T.A.R. collected field isolates and determined host population genetic structure; O.M.A. performed the common garden experiment; T.C.W. constructed all 16S rRNA gene libraries; O.M.A., D.E.H., and T.A.R. analyzed the data; and O.M.A. and T.A.R. wrote the initial manuscript and all authors participated in drafting the final version.
The authors declare no competing interest.
This article is a PNAS Direct Submission.
1Present address: Corning School of Ocean Studies, Maine Maritime Academy, Castine, ME 04420.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105207118/-/DCSupplemental.
Data Availability
All 16S rRNA gene sequences from global environmental genotype microbiomes are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject PRJNA494816) under accession nos. SRR8046252 through SRR8046343. All 16S rRNA gene sequences from the common garden genotype microbiomes are available in the NCBI Sequence Read Archive (BioProject PRJNA705708) under accession nos. SRX10227248 through SRX10227277. All environmental data are deposited in the Biological and Chemical Oceanography Data Management Office Database for the environmental isolates (66) (https://www.bco-dmo.org/dataset/860347) and the common garden experiment (67) (https://www.bco-dmo.org/dataset/860381). All remaining study data are included in the article and/or supporting information.
References
- 1.Cole J. J., Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13, 291–314 (1982). [Google Scholar]
- 2.Azam F., Malfatti F., Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Seymour J. R., Amin S. A., Raina J.-B., Stocker R., Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Bell W., Mitchell R., Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277 (1972). [Google Scholar]
- 5.Amin S. A., Parker M. S., Armbrust E. V., Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrer-González F. X., et al. , Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 15, 762–773 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amin S. A., et al. , Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G., Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J., The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sapp M., et al. , Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53, 683–699 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Sison-Mangus M. P., Jiang S., Tran K. N., Kudela R. M., Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J. 8, 63–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behringer G., et al. , Bacterial communities of diatoms display strong conservation across strains and time. Front. Microbiol. 9, 659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbour R. C., Baker S. C., O’Reilly-Wapstra J. M., Harvest T. M., Potts B. M., A footprint of tree-genetics on the biota of the forest floor. Oikos 118, 1917–1923 (2009). [Google Scholar]
- 14.Zytynska S. E., Fay M. F., Penney D., Preziosi R. F., Genetic variation in a tropical tree species influences the associated epiphytic plant and invertebrate communities in a complex forest ecosystem. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 1329–1336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korkama T., Pakkanen A., Pennanen T., Ectomycorrhizal community structure varies among Norway spruce (Picea abies) clones. New Phytol. 171, 815–824 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Whitham T. G., et al. , Community specificity: Life and afterlife effects of genes. Trends Plant Sci. 17, 271–281 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Johnson D., Martin F., Cairney J. W. G., Anderson I. C., The importance of individuals: intraspecific diversity of mycorrhizal plants and fungi in ecosystems. New Phytol. 194, 614–628 (2012). [DOI] [PubMed] [Google Scholar]
- 18.LeRoy C. J., Whitham T. G., Wooley S. C., Marks J. C., Within-species variation in foliar chemistry influences leaf-litter decomposition in a Utah river. J. N. Am. Benthol. Soc. 26, 426–438 (2007). [Google Scholar]
- 19.Rynearson T. A., Armbrust E. V., DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol. Oceanogr. 45, 1329–1340 (2000). [Google Scholar]
- 20.Rynearson T. A., Newton J. A., Armbrust E. V., Spring bloom development, genetic variation, and population succession in the planktonic diatom Ditylum brightwellii. Limnol. Oceanogr. 51, 1249–1261 (2006). [Google Scholar]
- 21.Godhe A., et al. , Physical barriers and environmental gradients cause spatial and temporal genetic differentiation of an extensive algal bloom. J. Biogeogr. 43, 1130–1142 (2016). [Google Scholar]
- 22.Gao Y., et al. , Spatiotemporal genetic structure of regional-scale Alexandrium catenella dinoflagellate blooms explained by extensive dispersal and environmental selection. Harmful Algae 86, 46–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Härnström K., Ellegaard M., Andersen T. J., Godhe A., Hundred years of genetic structure in a sediment revived diatom population. Proc. Natl. Acad. Sci. U.S.A. 108, 4252–4257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittaker K. A., Rynearson T. A., Evidence for environmental and ecological selection in a microbe with no geographic limits to gene flow. Proc. Natl. Acad. Sci. U.S.A. 114, 2651–2656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ajani P. A., et al. , The microbiome of the cosmopolitan diatom Leptocylindrus reveals significant spatial and temporal variability. Front. Microbiol. 9, 2758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frischkorn K. R., Rouco M., Van Mooy B. A. S., Dyhrman S. T., Epibionts dominate metabolic functional potential of Trichodesmium colonies from the oligotrophic ocean. ISME J. 11, 2090–2101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker J. W., Hogle S. L., Rosendo K., Chisholm S. W., Co-culture and biogeography of Prochlorococcus and SAR11. ISME J. 13, 1506–1519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sher D., Thompson J. W., Kashtan N., Croal L., Chisholm S. W., Response of Prochlorococcus ecotypes to co-culture with diverse marine bacteria. ISME J. 5, 1125–1132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner M. R., et al. , Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 12151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peiffer J. A., et al. , Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen J. L., et al. , Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nat. Commun. 8, 433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veach A. M., et al. , Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome 7, 76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittaker K. A., Rignanese D. R., Olson R. J., Rynearson T. A., Molecular subdivision of the marine diatom Thalassiosira rotula in relation to geographic distribution, genome size, and physiology. BMC Evol. Biol. 12, 209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker L., Kemp P., Exploring bacteria–diatom associations using single-cell whole genome amplification. Aquat. Microb. Ecol. 72, 73–88 (2014). [Google Scholar]
- 35.Mönnich J., et al. , Niche-based assembly of bacterial consortia on the diatom Thalassiosira rotula is stable and reproducible. ISME J. 14, 1614–1625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datta M. S., et al. , Inter-individual variability in copepod microbiomes reveals bacterial networks linked to host physiology. ISME J. 12, 2103–2113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris J. J., Johnson Z. I., Szul M. J., Keller M., Zinser E. R., Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6, e16805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ning D., Deng Y., Tiedje J. M., Zhou J., A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. U.S.A. 116, 16892–16898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., et al. , Environmental stability impacts the differential sensitivity of marine microbiomes to increases in temperature and acidity. ISME J. 15, 19–28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jönsson B. F., Watson J. R., The timescales of global surface-ocean connectivity. Nat. Commun. 7, 11239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanson C. A., Fuhrman J. A., Horner-Devine M. C., Martiny J. B. H., Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Sunagawa S., et al. , Tara Oceans Coordinators, Ocean plankton. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Rouco M., Haley S. T., Dyhrman S. T., Microbial diversity within the Trichodesmium holobiont. Environ. Microbiol. 18, 5151–5160 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Smriga S., Fernandez V. I., Mitchell J. G., Stocker R., Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 113, 1576–1581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta M. S., Sliwerska E., Gore J., Polz M. F., Cordero O. X., Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7, 11965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dittami S. M., et al. , Host-microbe interactions as a driver of acclimation to salinity gradients in brown algal cultures. ISME J. 10, 51–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rynearson T. A., Armbrust E. V., Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol. Ecol. 14, 1631–1640 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Eilers H., Pernthaler J., Amann R., Succession of pelagic marine bacteria during enrichment: A close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66, 4634–4640 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinhassi J., Berman T., Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69, 199–211 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stocker R., Seymour J. R., Samadani A., Hunt D. E., Polz M. F., Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. U.S.A. 105, 4209–4214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu H., Uchimiya M., Gore J., Moran M. A., Ecological drivers of bacterial community assembly in synthetic phycospheres. Proc. Natl. Acad. Sci. U.S.A. 117, 3656–3662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrenberg S., et al. , Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 7, 1102–1111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemergut D. R., et al. , Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J., et al. , Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 4, e00584-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukami T., Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1–23 (2015). [Google Scholar]
- 56.Kraft N. J. B., et al. , Community assembly, coexistence and the environmental filtering metaphor. Funct. Ecol. 29, 592–599 (2015). [Google Scholar]
- 57.Vályi K., Mardhiah U., Rillig M. C., Hempel S., Community assembly and coexistence in communities of arbuscular mycorrhizal fungi. ISME J. 10, 2341–2351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godhe A., Rynearson T., The role of intraspecific variation in the ecological and evolutionary success of diatoms in changing environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibl A. A., et al. , Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. U.S.A. 117, 27445–27455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bromke M. A., et al. , Metabolomic profiling of 13 diatom cultures and their adaptation to nitrate-limited growth conditions. PLoS One 10, e0138965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Tol H. M., Amin S. A., Armbrust E. V., Ubiquitous marine bacterium inhibits diatom cell division. ISME J. 11, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Round F., Crawford R., Mann D., The Diatoms (Cambridge University Press, 1990). [Google Scholar]
- 63.Vellend M., Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Paul C., Pohnert G., Interactions of the algicidal bacterium Kordia algicida with diatoms: Regulated protease excretion for specific algal lysis. PLoS One 6, e21032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paul C., Mausz M. A., Pohnert G., A co-culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics 9, 349–359 (2013). [Google Scholar]
- 66. O. M. Ahern, T. A. Rynearson, Thalassiosira rotula microbiome global sample. Biological and Chemical Oceanography Data Management Office. https://www.bco-dmo.org/dataset/8603475. Deposited 17 August 2021.
- 67.Rynearson O. M. Ahern, T. A., Thalassiosira rotula common garden experiment. Biological and Chemical Oceanography Data Management Office. https://www.bco-dmo.org/dataset/860381. Deposited 17 August 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA gene sequences from global environmental genotype microbiomes are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject PRJNA494816) under accession nos. SRR8046252 through SRR8046343. All 16S rRNA gene sequences from the common garden genotype microbiomes are available in the NCBI Sequence Read Archive (BioProject PRJNA705708) under accession nos. SRX10227248 through SRX10227277. All environmental data are deposited in the Biological and Chemical Oceanography Data Management Office Database for the environmental isolates (66) (https://www.bco-dmo.org/dataset/860347) and the common garden experiment (67) (https://www.bco-dmo.org/dataset/860381). All remaining study data are included in the article and/or supporting information.