Abstract
Neuroblastoma (NB) is an rare type of tumor that almost affects children age 5 or younger due to its rapid proliferation ability. The overall survival rate of patients with advanced NB is not satisfactory. Ribosomal proteins (RPs) play a critical role in the development and progress of cancer. However, the contribution of RPL35 in NB has not been proven. In this study, we reveal that RPL35 is upregulated in NB tissues and the upregulation of RPL35 promotes proliferation and migration of NB while RPL35 knockdown significantly restrained the proliferation of NB cells. In terms of mechanism, glycolysis was decreased and the mitochondrial respiration was increased with knockdown of RPL35 in NB cells, indicating that RPL35 function as a positive regulator in aerobic glycolysis. Importantly, our data indicated that RPL35 deficiency decreased HIF1α expression both in mRNA and protein levels. Western blot analysis showed that RPL35 knockdown has a negative regulatory effect on the ERK pathway, and RPL35 modulated aerobic glycolysis in part through its regulation of the RPL35/ERK/HIF1α axis. Overall, RPL35 functions as a positive regulator of aerobic glycolysis, and the RPL35/ERK/HIF1α axis could be a potential therapeutic target for the therapy of NB.
Keywords: Neuroblastoma, RPL35, aerobic glycolysis, HIF1α, ERK
Introduction
Neuroblastoma (NB), which almost only occurs in early childhood, is a malignant tumor of the nervous system [1,2]. NB can be classified into high-, medium-, low-, and very low-risk groups. Patients in the low-risk group can resolve themselves without any treatment, while even with high-intensity comprehensive treatments like surgery, radiotherapy, chemotherapy, and immunotherapy, the survival rate of high-risk NB is still very low [3,4]. Neuroblastoma is characterized by significant clinical and biological heterogeneity, and its pathogenetic targets are poorly defined [5,6].
Under normoxia, mammalian cells absorb glucose and catalyze glucose with a series of glucose catalytic enzymes to produce pyruvate, which enters the mitochondria and generates a lot of energy, called oxidative phosphorylation. Tumor cells under normoxia can also absorb a large amount of glucose, while the produced pyruvate does not undergo oxidative phosphorylation, but form a large amount of lactic acid, called aerobic glycolysis [7,8]. Aerobic glycolysis links rapid glucose fermentation with the rapid proliferation and progression of cancer cells [9]. Under hypoxic conditions, aerobic glycolysis provides a carbon skeleton, and NADPH and ATP serve as the basis for the synthesis of proliferating cancer cells [10]. The signaling pathway mediated by hypoxia-inducible factor (HIF)-1α plays a major role in aerobic glycolysis. It can change glucose metabolism to glycolysis phenotype, enabling tumor cells to live under hypoxic stress conditions [11,12]. The rapid proliferation of cancer cells requires higher energy consumption and a faster rate of biosynthesis, which is mainly achieved by increasing the rate of ribosome production. Ribosomal proteins (RPs), as the main components of ribosomes, play a key role in strictly regulated protein synthesis [13,14]. Except for protein synthesis regulation, RPs also have important ribosomal functions in many cellular physiological processes, including regulating cell proliferation and differentiation, cell death, cell migration, invasion, and death [15,16]. RPL35 is a component of the 60S ribosomal subunit that plays key roles in mRNA translation and protein synthesis [17-19]. Recently, it was discovered that RPL35 interacts with eukaryotic translation elongation factor 2, thereby controlling the synthesis of CSN2 protein [20]. It was recently reported that RPL35 expression related to poor patient outcome with NB [21]. However, it is unknown whether RPL35 expression is causally related to NB development and, if so, what the underlying mechanism is.
In the present study, we report that RPL35 is aberrantly, highly expressed in NB tissues and the overexpression of RPL35 promotes proliferation and migration of NB while RPL35 knockdown significantly inhibited the proliferation of NB cells. Mechanistically, knockdown of RPL35 inhibited aerobic glycolysis and decreased HK2 and LDHB expression levels of NB cells. RPL35 regulates HIF1α transcriptional activity through ERK activation in NB. Therefore, RPL35/ERK/HIF1α axis is expected to be a potential target for NB treatment.
Materials and methods
Patients and tissue specimens
15 NB samples and 15 normal samples were collected from the First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital. The research ethics committee of the First Affiliated Hospital of Shandong University of Medical Sciences & chifoshan Hospital of Shandong Province (2019KJTYL009) approved the study in accordance with the declaration of Helsinki.
Cell culture and reagents
The cell line DG and NB cell lines SH-SY5Y, NB-1643, NB-1691, SK-N-BE(2), BE(2)-C, SK-N-AS, and IMR32 were purchased from the National Infrastructure of Cell Line Resource (Beijing, China). All cell lines were cultured in DMEM (GIBCO, USA) medium with 10% FBS. All cell lines were maintained in a humidified incubator at 37°C and 5% CO2 in the air.
Lentivirus vector construction and transfection
The lentivirus carrying knockdown and overexpression sequences for RPL35 was prepared by OBiO (Shanghai, China). The shRNAs to RPL35 (shRPL35) and negative control (shNC) were produced by OBiO (Shanghai, China). The shRNA sequence against RPL35 is 5’-AAAAGGGGCGCATTGTCAATAAATTGGATCCAATTTATTGACAATGCGCCCC-3’ and 5’-AAAAGGCGCATTGTCAATAAAGCATTGGATCCAATGCTTTATTGACAATGCGCC-3’. Lipofectamine 3000 Reagent (Thermo, USA) was used for plasmids transfection into NB cells.
Western blotting
Using RIPA lysis buffer (with protease and phosphatase inhibitor), the NB tissue and normal tissue were minced, homogenized, and digested. NB cells were collected from the culture plate and lysed with precooled RIPA lysis buffer. The supernatant of protein suspensions was collected after centrifuged at 17,000 g for 20 minutes at 4°C. Prepare protein samples for PAGE gel electrophoresis. The protein was then transferred to the 0.2 μm PVDF membrane (BioRad) and immunoblotted with related antibodies. The listed antibodies were used: RPL35 (ab121244), HIF1α (NB100-105), LDHB (sc-133123), HK2 (ab209807), p-Erk (ab229912), Erk (ab184699), β-actin (ab8226).
Real-time PCR
All RNA of tissues and cells were extracted by Trizol reagent (Invitrogen, USA), purified RNA was used to reverse-synthesize cDNA template using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher). The real-time qPCR primers sequence are listed following: RPL35 (F: 5’-AGCTCTCTAAGATCCGAGTCG-3’, R: 5’-GAACACGGGCAATGGATTTCC-3’), GLUT1 (F: 5’-ATTGGCTCCGGTATCGTCAAC-3’, R: 5’-GCTCAGATAGGACATCCAGGGTA-3’), HK2 (F: 5’-ATGGGGCTCCAACGAGTTAC-3’, R: 5’-TTTCCTGCCATACACCCACAA-3’), LDHA (F: 5’-TTGACCTACGTGGCTTGGAAG-3’, R: 5’-GGTAACGGAATCGGGCTGAAT-3’), LDHB (F: 5’-TGGTATGGCGTGTGCTATCAG-3’, R: 5’-TTGGCGGTCACAGAATAATCTTT-3’), and PDK1 (F: 5’-CTGTGATACGGATCAGAAACCG-3’, R: 5’-TCCACCAAACAATAAAGAGTGCT-3’) were normalized to GAPDH (F: 5’-GGAGCGAGATCCCTCCAAAAT-3’, R: 5’-GGCTGTTGTCATACTTCTCATGG-3’). Each PCR experiment was performed at least 3 times independently, and the relative expression value was expressed by 2-ΔΔCt.
In vivo tumor growth model and lung metastases model
100 μL with 1×107 NB cells that were transfected with shRPL35 or shNC were subcutaneously injected into 28-day-old female BALB/c nude mice (n=5 per group) in saline. We examined the volume of NB every 5 days, sacrificed the mice, and isolated the tumor for weight after 30 days. For the experimental metastasis studies, NB cells were injected into the tail vein of 28-day-old female BALB/c nude mice (2×106 per mouse, n=5 per group). After 28 days, all mice were sacrificed and then subjected to immunohistochemical analysis and hematoxylin/eosin (HE) staining. All animal surgery were approved by the Institutional Animal Care and Use Committee of Fujian Medical University Union Hospital (No. FJMU IACUC20190061).
Immunohistochemical (IHC) staining
The tumor tissues were embedded with OCT and quickly frozen in the -80°C after weighting. 5-μm-thick sections were made in a Cryostat (Leica, Germany). Slides were blocked with 3% BSA at RT for 2 hours at 37°C. The first antibody against RPL35, Ki67 and CD34 was incubated at RT for 2 hours at 37°C. The second antibody was incubated for 1 hour at RT, and then uses diaminobenzidine (DAB) as the chromogen.
Wound-healing assay
The NB cells were planted into 35-mm dishes at a density of 5×105/mL. Scrape the NB cells when the confluence of cells reaches ~90% using pipette tips and culture in an FBS-free medium. Images were taken at 0 and 24 hours after the scraping.
Transwell assay and colony-formation assay
Polycarbonate membrane filter coated with gelatin and a 24-well perforated chamber (Corning) of Matrigel was used for transwell assay and colony-formation assay. The NB cells (5×104/mL) were suspended in DMEM. Add 600 μL of DMEM (HyClone, FBS) containing 20% FBS to the 100 μL lower chamber. NB cells were dyed with 0.1% hexamethylpararosaniline (Sigma-Aldrich) for 20 minutes and washed 2 times with PBS after 24 hours. To observe the colony-formation ability of NB cells, NB cells were seeded into 12-well plates (1×103/mL), and after 2 weeks of culture, the colonies were stained with 0.1% hexamethylpararosaniline, and then washed with PBS twice. To compare the migration ability of NB cells, the visible violet colonies were counted and analyzed with Image J.
Cell viability assay (CCK8)
The cell proliferation of the NB cells was detected by Cell Counting Kit-8 (CCK-8, Bestbio) assay. The NB cells were plated in 96-well plates with 5×103 cells per well overnight. The supernatant was completely removed, and then the CCK-8 solution was added to the wells and incubated at 37°C in the dark for 4 hours. Finally, a microplate reader was used to measure the absorbance of all the wells at 450 nm. The measurement was performed on 3 independent experiments.
Measurement of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) assay
The measurement of ECAR and OCR were measured and evaluated were performed as previously described [24], seahorse Bioscience XF96 Extracellular Flux Analyzer was used to measure the mitochondrial function and glycolytic capacity of NB cells according to the manufacturer’s instructions of the Seahorse XF Cell Mito Stress Test Kit and Glycolysis Stress Test Kit (Seahorse Bioscience, Billerica, MA, USA).
Promoter activity with the dual luciferase assay
Dual-Luciferase Reporter Assay System (Promega) was used to evaluate the luciferase activity, HIF1α promoter region covering from -2500 to +200 was amplified and cloned into a pGL3-Basic vector to generate the pGL3-HIF1α-firefly luciferase construct. Renilla luciferase-expressing vector pGL4.73 was purchased from Promega (Madison, WI, USA).
Statistical analysis
Data are shown as mean ± SEM. GraphPad Prism software (version 8.0) was used for statistical analyses. Statistical significance was analyzed by using unpaired Student’s t-test or one-way analysis of variance (ANOVA) followed with Bonferroni’s multiple as appropriate. A P-value <0.05 was considered statistically significant.
Results
RPL35 is upregulated in human neuroblastoma
Immunohistochemical staining revealed that RPL35 was highly expressed in NB patients, compared to normal dorsal root ganglia from interrupted pregnancies (Figure 1A). Kaplan-Meier curves of 28 patients showed that high RPL35 levels were correlated with poor overall survival in NB patients (P=0.028, Figure 1B). We then compare the expression of RPL35 in NB tissues, 5 NB tissues from NB patients and 5 normal dorsal root ganglia from interrupted pregnancies were analyzed by Western blot. Compared with normal tissues, the protein level of RPL35 is more highly in NB tissue (Figure 1C). Next, RPL35 expression level in NB tissue and normal tissue from different clinical samples was measured by Q-PCR, and we found RPL35 is increased significantly in NB samples (Figure 1D). Interestingly, inside the NB tissue samples, the mRNA level of RPL35 is significantly higher in the MYCN positive group. As an oncogene related to poor survival, MYCN is essential for aerobic glycolysis in NB [22]. Then, we measured the expression of RPL35 in different NB cell lines (DG, SH-SY5Y, NB-1643, NB-1691, SK-N-BE(2), BE(2)-C, SK-N-AS, and IMR32) at the protein level. RPL35 was highly expressed in NB cell lines, especially in SH-SY5Y and BE(2)-C (Figure 1E, 1F). These results suggest that RPL35 is upregulated in NB.
Figure 1.
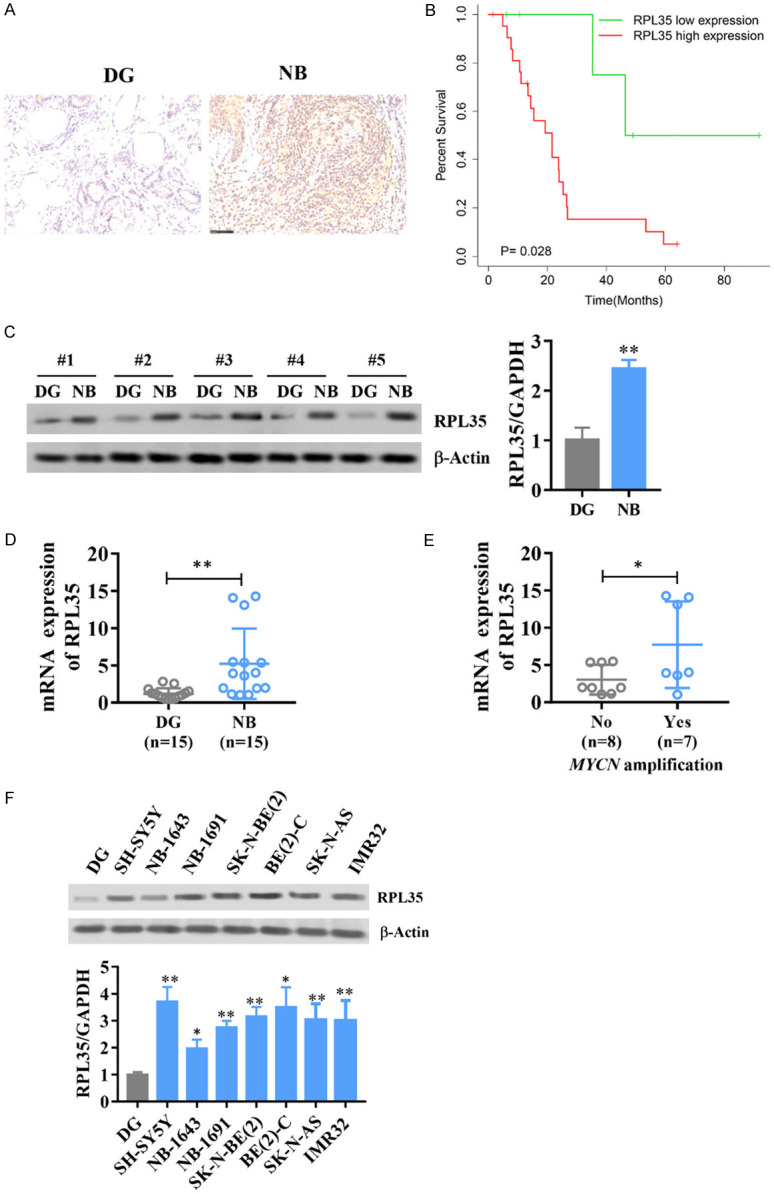
RPL35 is upregulated in human neuroblastoma. A. Immunohistochemistry showing RPL35 expression in examples of NB tumor patient samples and normal tissue from different clinical samples. B. Kaplan-Meier curve showing overall survival of 28 NB patients with high or low RPL35 expression. C. Western blot analysis of RPL35 in NB tissue and normal tissue from different clinical samples. D, E. The mRNA level of RPL35 in NB tissue and normal tissue from different clinical samples. F. The protein level of RPL35 in different NB cell lines (DG, SH-SY5Y, NB-1643, NB-1691, SK-N-BE(2), BE(2)-C, SK-N-AS and IMR32). *P<0.05, **P<0.01.
RPL35 accelarates the proliferation and migration of NB
In order to observe the function of RPL35 in NB, we transfected NB cells with RPL35 or control sequence carried by lentivirus. We measured the cell viability at different times (0 h, 24 h, 48 h, 72 h, and 96 h) after infection and we found that SH-SY5Y and BE(2)-C cells infected with RPL35 shows better cell viability compared with the vector control (Figure 2A). Next, we used colony formation assay on SH-SY5Y and BE(2)-C cells that transfected with RPL35 or vector and found that RPL35 increased the colony formation of NB cells (Figure 2B). By measuring the migration and invasion ability of NB cells, we found that the up-regulation of RPL35 increased the ability of cell migration and invasion of NB (Figure 2C-E).
Figure 2.
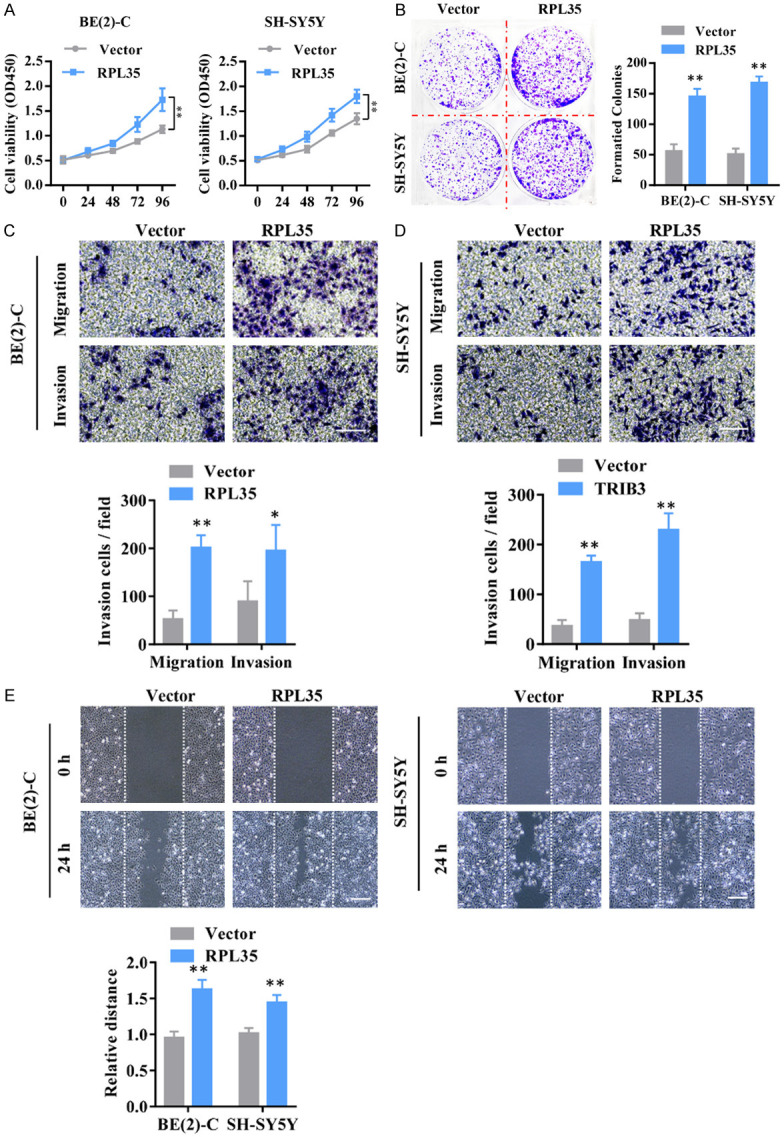
RPL35 promotes the proliferation and migration of NB. A. The cell viability of SH-SY5Y and BE(2)-C at different times (0 h, 24 h, 48 h, 72 h, and 96 h) after infection with Lenti-RPL35 or Lenti-vector were evaluated by CCK8. B. The cloning formation of SH-SY5Y and BE(2)-C cells after infected with Lenti-RPL35 or Lenti-vector. C, D. The cell invasive ability measured by transwell assay. E. The migration potential evaluated by the wound healing assay. Scale bar =100 μm. *P<0.05, **P<0.01.
To further explore the function of RPL35 in NB progression, we infected SH-SY5Y and BE(2)-C cells with lentivirus carrying short-hairpin RNA (shRNA) targeting RPL35 (sh-RPL35-1 and sh-RPL35-2). Colony formation assay, transwell assay, and wound healing assay were performed to see the effects of RPL35 knockdown on the proliferation and migration of NB cells and found that RPL35 down-regulation suppressed the proliferation and migration of NB cells (Figure 3A-D). These results demonstrated that RPL35 participate in the process of the proliferation and migration of NB cells.
Figure 3.
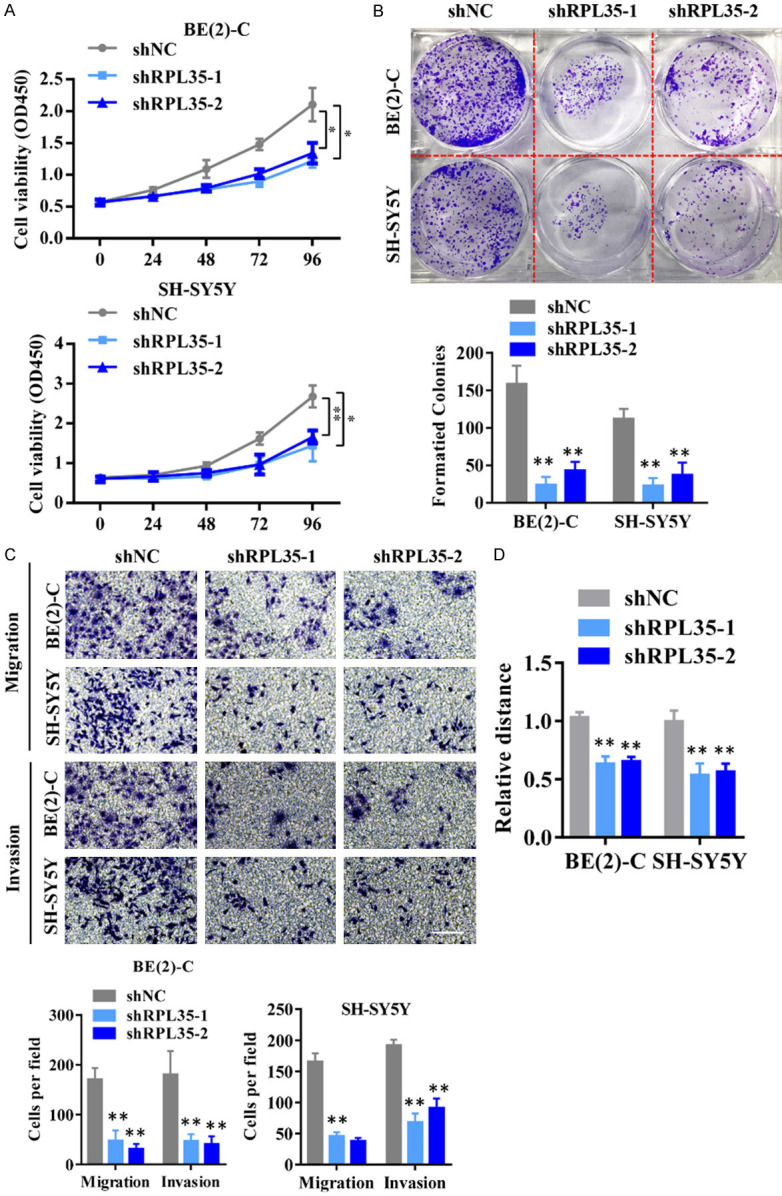
Knockdown of RPL35 impairs proliferation and migration of NB cells. A. The effect of RPL35 knockdown on SH-SY5Y and BE(2)-C cells viability was detected by CCK-8 assay at the indicated time. B. The effect of RPL35 knockdown on the cloning formation of SH-SY5Y and BE(2)-C cells. C. The effect of RPL35 knockdown on migration ability and invasion ability was determined after shRNA transfection in SH-SY5Y and BE(2)-C cells using a migration assay and a transwell assay. D. The migration potential of SH-SY5Y and BE(2)-C cells treated with shNC and shRPL35 were evaluated by the wound healing test. Scale bar =100 μm. *P<0.05, **P<0.01.
Knockdown of RPL35 leads to suppression of neuroblastoma progression
We next used a xenograft mouse model to explore the effect of RPL35 knockdown on cell growth of NB. SH-SY5Y cells stably infected with shRPL35 carried by lentivirus (lenti-shRPL35) were subcutaneous injected into BALB/c nude mice. We found that shRPL35 decreased the tumor volume and tumor weight significantly after 4 weeks (Figure 4A, 4B). Besides, IHC analysis showed that Ki-67 positive cells were decreased; meanwhile, CD34 positive blood vessels were also reduced in all shRPL35 groups than the control group (Figure 4C). These results from a mouse xenograft model indicate that RPL35 knockdown suppresses the growth of NB cell-derived tumors in vivo. To evaluate whether RPL35 influences the metastasis of NB cells in vivo, RPL35 knockdown SH-SY5Y cells were injected into the tail vein of BABL/c nude mice. The HE staining for the lungs of different groups shows that RPL35 knockdown decreased the occupation of NB cells in the lungs (Figure 4D). Besides, we found that shRPL35 effectively increased the survival rate of nude mice injected with SH-SY5Y cells compared to those in the control group (Figure 4E).
Figure 4.
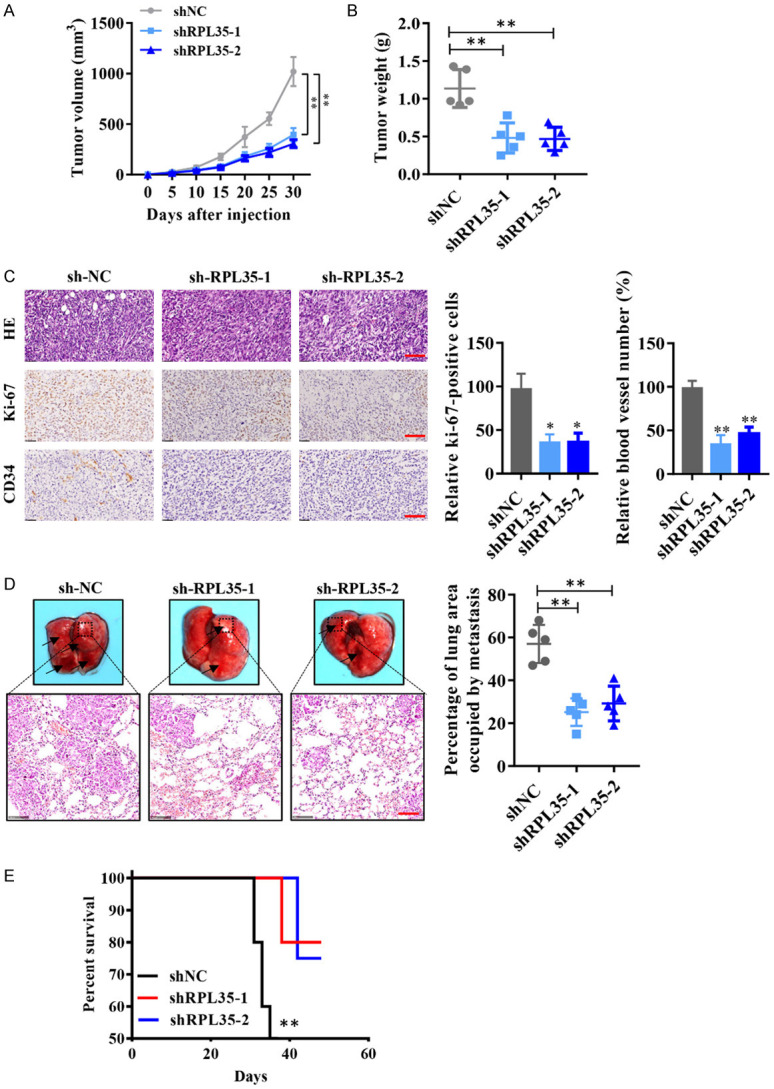
Knockdown of RPL35 leads to suppression of NB progression in vivo. A. Quantitative analysis of the NB tumor volume at the indicated time. B. Quantitative analysis of the NB tumor weight. C. HE staining, Ki-67, and CD34 immunohistological staining of tumors from shNC and shRPL35 cells. Scale bar =50 μm. D. HE staining for lung occupancy of shRPL35 or shNC expressing NB cells. Scale bar =100 μm. E. The survival rate of nude-mice after NB cell implantation. *P<0.05, **P<0.01.
Downregulation of RPL35 suppresses glycolysis of NB cells in vitro
The enhanced aerobic glycolysis can promote the over proliferation of cancer cells [23-25]; We therefore examined whether shRPL35 negatively regulates aerobic glycolysis. During aerobic glycolysis, cancer cells use glucose to produce lactic acid rather than the mitochondrial respiratory pathway. We used the Seahorse extracellular flux analyzer to measure glycolysis and mitochondrial respiration, where ECAR reflects aerobic glycolysis and OCR indicates mitochondrial respiration. ECAR measurement shows that shRPL35 reduced glycolysis and glycolytic capacity (Figure 5A, 5B). During aerobic glycolysis, mitochondrial respiration is impaired, resulting in a decrease in oxygen consumption, which can be reflected by OCR measurement. We observed that shRPL35 enhanced OCR, indicating that it has a positive effect on mitochondrial respiration (Figure 5C, 5D). By the use of the glycolytic inhibitor 2-DG (2 mM), we detected the proliferation of RPL35 overexpressed and knockdown NB cells. We found that RPL35 knockdown decreased the proliferation of NB cells, while the extra addition of 2-DG abolished the effect of RPL35 on the proliferation of NB (Figure 5E). Similarly, upregulation of RPL35 increased the proliferation of NB cells, and 2-DG abolished the effect of RPL35 on the proliferation of NB (Figure 5F). These results indicated that the effect of RPL35 on NB proliferation was mainly through the glycolytic pathway, and RPL35 is a positive regulator of aerobic glycolysis.
Figure 5.
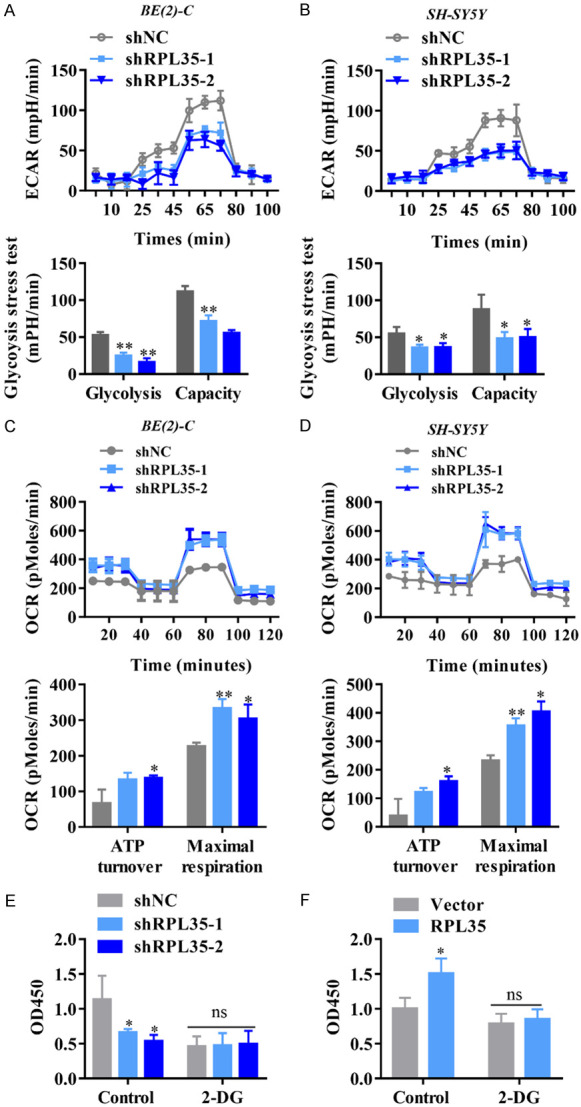
Downregulation of RPL35 suppresses glycolysis of NB cells in vitro. (A, B) Diagram and quantitative of ECAR results obtained by Seahorse extracellular flux analyzer to determine the impact of shRPL35 on aerobic glycolysis in SH-SY5Y and BE(2)-C cells. (C, D) Diagram and quantitative of OCR measurement with Seahorse analyzer to confirm the role of shRPL35 in mitochondrial respiration. (E, F) The proliferation of NB cells that stably knockdown (E) or overexpressed (F) of RPL35 by the administration of the glycolytic inhibitor 2-DG (2 mM) measured by CCK8. *P<0.05, **P<0.01.
Knockdown of RPL35 downregulates glycolytic proteins HK2 and LDHB
Aerobic glycolysis promotes malignant cell transformation and tumor progression [25,26], while glycolysis inhibition impairs the growth and metastasis of many tumor cells [27,28], indicating an effective treatment for tumors. To understand the mechanisms by which RPL35 knockdown inhibits glucose metabolism, the expression of several key glycolytic genes include glucose transporter 1 (GLUT1), hexokinase 2 (HK2), lactate dehydrogenase A (LDHA), LDHB, and phosphoinositide-dependent kinase-1 (PDK1) in RPL35-knockdown and control SH-SY5Y and BE(2)-C cells were measured. Downregulation of RPL35 decreased the mRNA of HK2 and LDHB in NB cells (Figure 6A, 6B). We also observed the increased protein and mRNA levels of HK2, LDHB in NB tissues (Figure 6C, 6D). Among them, HK2 is a key kinase that phosphorylates glucose to produce glucose-6-phosphate, a rate-limiting and irreversible step of glycolysis. LDHB is a key glycolytic enzyme that catalyzes the interconversion of pyruvate and lactate. These results indicate that RPL35 enhances glucose metabolism by increasing the expression of glycolytic genes.
Figure 6.
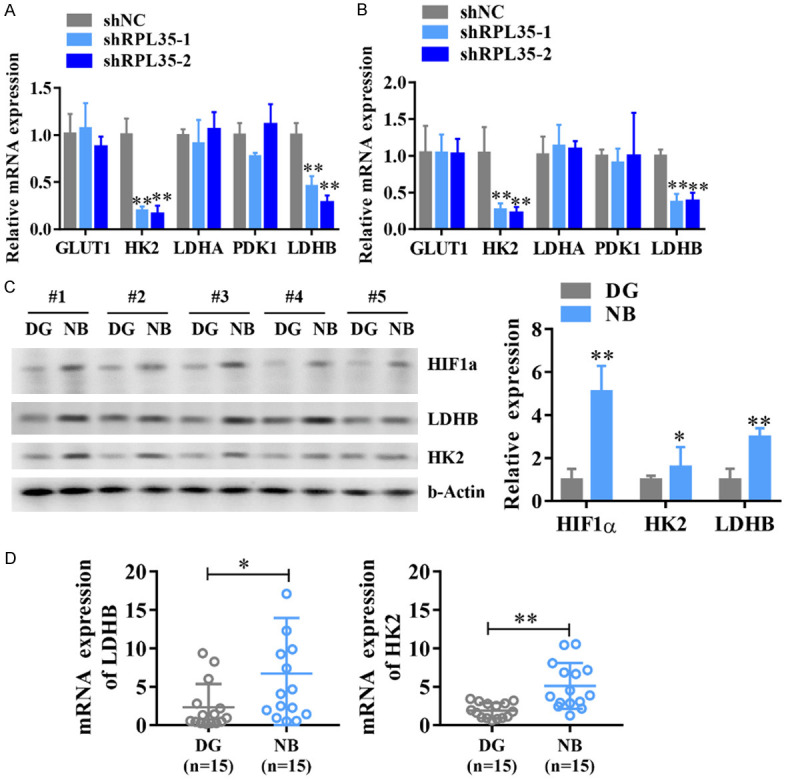
shRPL35 downregulates glycolytic proteins HK2 and LDHB. A, B. The Q-PCR analysis of GLUT1, HK2, LDHA, LDHB, and PDK1 mRNA level in shRPL35 and control SH-SY5Y and BE(2)-C cells. C. The protein levels of HK2, LDHB, and HIF1α in different NB tissues. D. The mRNA levels of HK2, LDHB in different NB tissues. *P<0.05, **P<0.01.
RPL35 regulate HIF1α transcriptional activity through Erk activation in NB
Many glycolytic genes including PGK1, GLUT1, HK2, PGM, LDHA, and MCT are downstream targets of HIF1α [29]. Interestingly, we also found protein level of HIF1α is increased in NB tissues (Figure 6C). Therefore, we hypothesized that RPL35 might regulate HIF1α transcriptional activity to enhance the expression of glycolytic genes. To test this hypothesis, we transfected RPL35 expression plasmid, together with a HIF1α promoter-reporter, into HEK293T cells. We found that RPL35 positively regulated HIF1α transcriptional activity in a dose-dependent manner, reflecting the positive role of RPL35 in the regulation process of HIF1α (Figure 7A). The ERK pathway promotes the nuclear translocation of HIF1α [30] or further enhances the transcriptional activity of HIF-1α induced by hypoxia [31]. Therefore, we hypothesized that RPL35 might regulate HIF1α transcriptional activity through activation of the ERK signaling pathway. We observed the decreased level of phosphorylation of ERK as well as the downregulation of HIF1α in RPL35 knockdown NB cells which indicates that ERK activation participates in the RPL35 regulating HIF1α expression in NB cells (Figure 7B). To test the role of ERK in regulating HIF1α transcriptional activity in NB cells, we transfected shRPL35 with and without ERK2-mutant expression plasmid, together with a HIF1α promoter-reporter, into HEK293T cells. We observed that shRPL35 negatively regulated HIF1α transcriptional activity while ERK2-mutant mostly abolished this inhibition effect in NB cells (Figure 7C). To assess the role of the RPL35/ERK axis in regulating glycolysis in NB cells, we performed ECAR with Seahorse Extracellular Flux Analyzer. As observed, ERK2-mutant overexpression abolished the inhibition of shRPL35 on ECAR in NB cells, indicating the positive role of the RPL35/ERK/HIF1α axis in aerobic glycolysis regulation (Figure 7D).
Figure 7.
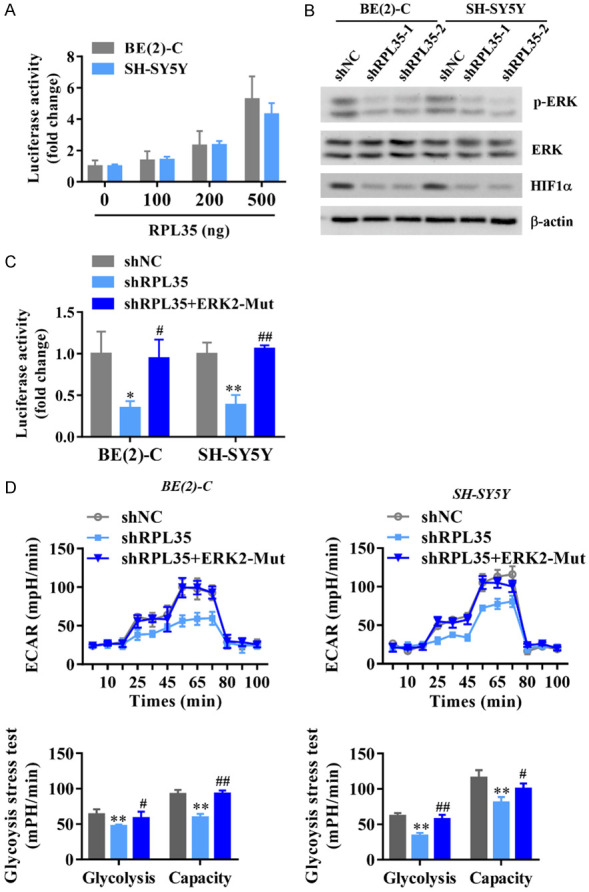
RPL35 regulates HIF1α transcriptional activity through Erk activation in NB. A. HIF1α transcriptional activity in NB cells overexpressed with different doses of RPL35 measured with the dual-luciferase assay. B. Western blot analysis of ERK phosphorylation as well as the HIF1α expression in RPL35 knockdown NB cells. C. ERK2-mutant mostly abolished the inhibition effect of shRPL35 on HIF1α transcriptional activity in NB cells. D. Diagram and quantitative of ECAR results obtained by Seahorse extracellular flux analyzer to determine the impact of ERK-mutant to shRPL35’s effect on aerobic glycolysis in SH-SY5Y and BE(2)-C cells. *P<0.05, **P<0.01 vs. shNC. #P<0.05, ##P<0.01 vs. shRPL35.
Discussion
In this study, we reported that RPL35 is abnormally expressed in NB tissues, and the overexpression of RPL32 promotes the proliferation and migration of NB, while RPL35 knockdown significantly inhibits the proliferation of NB cells. Besides, we first uncovered the previously unrecognized role of RPL35 which promotes aerobic glycolysis through upregulation of HIF1α via RPL35/ERK/HIF1α axis in NB cells.
RPs are the major component of the ribosome and play important roles in the process of protein synthesis. Surprisingly, besides their basic protein synthesis function, RPs also possess key functions in many other cellular processes. RPL3 was reported to facilitate the apoptosis of p53-mutated lung cancer cells and colon cancer cells with fluorouracil (5-FU) treatment [32,33]. RPS6 was reported significantly increased and phosphorylated in nonsmall cell lung cancer (NSCLC) and that knockdown of RPS6 inhibits NSCLC cell growth by inducing cell-cycle arrest [34]. It has been found that RPL35 is a component of the 60S ribosomal subunit and interacts with eukaryotic translation elongation factor 2 thereby control CSN2 protein synthesis [20]. However, as a member of RPs, the extra ribosomal functions of RPL35 have never been found. Here, we demonstrated RPL35 promotes aerobic glycolysis through upregulation of HIF1α via RPL35/ERK/HIF1α axis in NB cells, which is a previously unrecognized extra ribosomal function of RPL35.
In order to maintain tumorigenesis and aggressiveness, even if there is enough oxygen, tumor cells still have a unique metabolic preference to convert glucose into lactic acid, called aerobic glycolysis [35,36]. Activation of oncogenes or inactivation of tumor suppressors contributes to the expression of glycolytic genes in tumor cells [37,38]. Small molecular organic compounds (such as 3-bromopyruvate or 2-deoxyglucose (2-DG)) inhibit aerobic glycolysis and have potential therapeutic value for tumors [39,40]. Therefore, it is important to study modulators of aerobic glycolysis to improve the therapeutic efficiencies of NB. Previous studies have shown that hypoxia activation can induce the activation of the ERK signaling pathway, and ERK activity regulated the protein stability of HIF1α. Therefore, we checked the activation state of ERK1/2 in RPL35 down-regulated NB cells and observed a decreased activation of ERK1/2. Subsequent measurements showed that RPL35 regulated HIF1α and glycolysis in an ERK-dependent manner.
Taken together, this study reported that RPL35 is upregulated in NB tissues, and the RPL35 promotes proliferation and migration of NB cells in vitro and in vivo, and discovered a novel function of RPL35 in the modulation of glycolysis and provided the possible molecular mechanism. These results identified that the RPL35/ERK/HIF1α axis as a promising target for the diagnosis and treatment of NB.
Acknowledgements
The present study is supported by Joint Funds for the innovation of science and Technology-Fujian province (2018Y9017).
Disclosure of conflict of interest
None.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Hematol Oncol Clin North Am. 2010;24:65–86. doi: 10.1016/j.hoc.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Verissimo CS, Molenaar JJ, Fitzsimons CP, Vreugdenhil E. Neuroblastoma therapy: what is in the pipeline? Endocr Relat Cancer. 2011;18:R213–231. doi: 10.1530/ERC-11-0251. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawara A, Li Y, Izumi H, Muramori K, Inada H, Nishi M. Neuroblastoma. Jpn J Clin Oncol. 2018;48:214–241. doi: 10.1093/jjco/hyx176. [DOI] [PubMed] [Google Scholar]
- 5.Aygun N. Biological and genetic features of neuroblastoma and their clinical importance. Curr Pediatr Rev. 2018;14:73–90. doi: 10.2174/1573396314666180129101627. [DOI] [PubMed] [Google Scholar]
- 6.Swift CC, Eklund MJ, Kraveka JM, Alazraki AL. Updates in diagnosis, management, and treatment of neuroblastoma. Radiographics. 2018;38:566–580. doi: 10.1148/rg.2018170132. [DOI] [PubMed] [Google Scholar]
- 7.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 8.Ganapathy-Kanniappan S. Molecular intricacies of aerobic glycolysis in cancer: current insights into the classic metabolic phenotype. Crit Rev Biochem Mol Biol. 2018;53:667–682. doi: 10.1080/10409238.2018.1556578. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Ren B, Yang G, Wang H, Chen G, You L, Zhang T, Zhao Y. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol Life Sci. 2020;77:305–321. doi: 10.1007/s00018-019-03278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 11.Weng ML, Chen WK, Chen XY, Lu H, Sun ZR, Yu Q, Sun PF, Xu YJ, Zhu MM, Jiang N, Zhang J, Zhang JP, Song YL, Ma D, Zhang XP, Miao CH. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1alpha pathway suppression. Nat Commun. 2020;11:1869. doi: 10.1038/s41467-020-15795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu D, Linders A, Yamak A, Correia C, Kijlstra JD, Garakani A, Xiao L, Milan DJ, van der Meer P, Serra M, Alves PM, Domian IJ. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ Res. 2018;123:1066–1079. doi: 10.1161/CIRCRESAHA.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graifer D, Karpova G. Roles of ribosomal proteins in the functioning of translational machinery of eukaryotes. Biochimie. 2015;109:1–17. doi: 10.1016/j.biochi.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Draper DE, Reynaldo LP. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 1999;27:381–388. doi: 10.1093/nar/27.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Hum Genomics. 2010;4:327–344. doi: 10.1186/1479-7364-4-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 18.Preiss T, W Hentze M. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 19.Pestova TV, Hellen CU. The structure and function of initiation factors in eukaryotic protein synthesis. Cell Mol Life Sci. 2000;57:651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang N, Hu L, Liu C, Gao X, Zheng S. 60S ribosomal protein L35 regulates β-casein translational elongation and secretion in bovine mammary epithelial cells. Arch Biochem Biophys. 2015;583:130–139. doi: 10.1016/j.abb.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Liu PY, Tee AE, Milazzo G, Hannan KM, Maag J, Mondal S, Atmadibrata B, Bartonicek N, Peng H, Ho N, Mayoh C, Ciaccio R, Sun Y, Henderson MJ, Gao J, Everaert C, Hulme AJ, Wong M, Lan Q, Cheung BB, Shi L, Wang JY, Simon T, Fischer M, Zhang XD, Marshall GM, Norris MD, Haber M, Vandesompele J, Li J, Mestdagh P, Hannan RD, Dinger ME, Perini G, Liu T. The long noncoding RNA lncNB1 promotes tumorigenesis by interacting with ribosomal protein RPL35. Nat Commun. 2019;10:5026. doi: 10.1038/s41467-019-12971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliynyk G, Ruiz-Perez MV, Sainero-Alcolado L, Dzieran J, Zirath H, Gallart-Ayala H, Wheelock CE, Johansson HJ, Nilsson R, Lehtio J, Arsenian-Henriksson M. MYCN-enhanced oxidative and glycolytic metabolism reveals vulnerabilities for targeting neuroblastoma. iScience. 2019;21:188–204. doi: 10.1016/j.isci.2019.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin Y, Hu Q, Ji S, Xu J, Dai W, Liu W, Xu W, Sun Q, Zhang Z, Ni Q, Yu X, Zhang B, Xu X. Homeodomain-interacting protein kinase 2 suppresses proliferation and aerobic glycolysis via ERK/cMyc axis in pancreatic cancer. Cell Prolif. 2019;52:e12603. doi: 10.1111/cpr.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Q, Qin Y, Ji S, Xu W, Liu W, Sun Q, Zhang Z, Liu M, Ni Q, Yu X, Xu X. UHRF1 promotes aerobic glycolysis and proliferation via suppression of SIRT4 in pancreatic cancer. Cancer Lett. 2019;452:226–236. doi: 10.1016/j.canlet.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li HM, Yang JG, Liu ZJ, Wang WM, Yu ZL, Ren JG, Chen G, Zhang W, Jia J. Blockage of glycolysis by targeting PFKFB3 suppresses tumor growth and metastasis in head and neck squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:7. doi: 10.1186/s13046-016-0481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren JG, Seth P, Ye H, Guo K, Hanai JI, Husain Z, Sukhatme VP. Citrate suppresses tumor growth in multiple models through inhibition of glycolysis, the tricarboxylic acid cycle and the IGF-1R pathway. Sci Rep. 2017;7:4537. doi: 10.1038/s41598-017-04626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan W, Peng K, Li M, Qin L, Tong Z, Yan J, Shen B, Yu C. Histone demethylase JMJD1A promotes urinary bladder cancer progression by enhancing glycolysis through coactivation of hypoxia inducible factor 1alpha. Oncogene. 2017;36:3868–3877. doi: 10.1038/onc.2017.13. [DOI] [PubMed] [Google Scholar]
- 30.Liu XH, Kirschenbaum A, Lu M, Yao S, Dosoretz A, Holland JF, Levine AC. Prostaglandin E2 induces hypoxia-inducible factor-1alpha stabilization and nuclear localization in a human prostate cancer cell line. J Biol Chem. 2002;277:50081–50086. doi: 10.1074/jbc.M201095200. [DOI] [PubMed] [Google Scholar]
- 31.Hur E, Chang KY, Lee E, Lee SK, Park H. Mitogen-activated protein kinase kinase inhibitor PD98059 blocks the trans-activation but not the stabilization or DNA binding ability of hypoxia-inducible factor-1alpha. Mol Pharmacol. 2001;59:1216–1224. doi: 10.1124/mol.59.5.1216. [DOI] [PubMed] [Google Scholar]
- 32.Russo A, Saide A, Cagliani R, Cantile M, Botti G, Russo G. rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFkappaB upon 5-FU treatment. Sci Rep. 2016;6:38369. doi: 10.1038/srep38369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo A, Maiolino S, Pagliara V, Ungaro F, Tatangelo F, Leone A, Scalia G, Budillon A, Quaglia F, Russo G. Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles. Oncotarget. 2016;7:79670–79687. doi: 10.18632/oncotarget.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B, Zhang W, Gao J, Chen H, Jiang L, Liu D, Cao Y, Zhao S, Qiu Z, Zeng J, Zhang S, Li W. Downregulation of ribosomal protein S6 inhibits the growth of non-small cell lung cancer by inducing cell cycle arrest, rather than apoptosis. Cancer Lett. 2014;354:378–389. doi: 10.1016/j.canlet.2014.08.045. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015;356:156–164. doi: 10.1016/j.canlet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson H, Lindgren D, Mandahl Forsberg A, Mulder H, Axelson H, Johansson ME. Primary clear cell renal carcinoma cells display minimal mitochondrial respiratory capacity resulting in pronounced sensitivity to glycolytic inhibition by 3-Bromopyruvate. Cell Death Dis. 2015;6:e1585. doi: 10.1038/cddis.2014.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M, De Cola A, Scavo E, Carollo R, D’Agostino D, Forli F, D’Aguanno S, Todaro M, Stassi G, Di Ilio C, De Laurenzi V, Urbani A. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014;5:e1336. doi: 10.1038/cddis.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]


