Significance
α-oxoacid dehydrogenase are large, evolutionary-conserved multienzymatic complexes that carry out key oxidative reactions in central metabolism. For decades, these complexes were thought to share a similar organization in all aerobic organisms, structured around a hollow core resulting from the high oligomeric arrangement of the acyltransferase component. Here, we show that Actinobacteria, one of the largest eubacterial phyla, break this “dogma” owing to a distinct trimeric acyltransferase core. We provide a structural explanation to this different oligomeric architecture and show how this feature, related to a fusion of two genes coding for 2-oxoglutarate dehydrogenase, constitutes a trait of Actinobacteria providing a different perspective on the evolution of these metabolic complexes.
Keywords: Actinobacteria, central metabolism, macromolecular complexes, pyruvate dehydrogenase, integrative structural biology
Abstract
α-oxoacid dehydrogenase complexes are large, tripartite enzymatic machineries carrying out key reactions in central metabolism. Extremely conserved across the tree of life, they have been, so far, all considered to be structured around a high–molecular weight hollow core, consisting of up to 60 subunits of the acyltransferase component. We provide here evidence that Actinobacteria break the rule by possessing an acetyltranferase component reduced to its minimally active, trimeric unit, characterized by a unique C-terminal helix bearing an actinobacterial specific insertion that precludes larger protein oligomerization. This particular feature, together with the presence of an odhA gene coding for both the decarboxylase and the acyltransferase domains on the same polypetide, is spread over Actinobacteria and reflects the association of PDH and ODH into a single physical complex. Considering the central role of the pyruvate and 2-oxoglutarate nodes in central metabolism, our findings pave the way to both therapeutic and metabolic engineering applications.
The α-oxoacid dehydrogenase complexes constitute a family of three-partite, ubiquitous metabolic complexes devoted to the oxidative decarboxylation of α-oxoacids and the concomitant production of reducing equivalents in form of NADH (1). Three such complexes are known: the pyruvate dehydrogenase (PDH), that feeds the tricarboxylic acid (TCA) cycle with carbon units in form of acetyl-CoA; the 2-oxoglutarate dehydrogenase (ODH), part of the oxidative branch of the TCA cycle; and the branched chain α-ketoacid dehydrogenase (BCKDH), involved in the catabolism of aliphatic amino acids. These large tripartite complexes share a common molecular architecture organized around a core made by the E2 component, a flexible, multidomain protein which bears the acyltransferase activity required to transfer the acyl group from the decarboxylated substrate to the CoA-SH acceptor; the number of E2 subunits and the symmetry of the core depend on the complex and the species (1–5). First shown by the crystal structure of Azotobacter vinelandii E2p, the E2 C-terminal catalytic core assumes an obligate homotrimeric state much similar to chloramphenicol acetyltransferase (6), with which it also shares the catalytic mechanism (7). The observed, higher-order oligomerization states are made possible by intermolecular trimer–trimer interactions (TTIs) mediated by a well-conserved, C-terminal 310 hydrophobic helix which makes intermolecular symmetric interactions (5), then confirmed on other E2 enzymes and sometimes described as “knobs and sockets” (3). These interactions make symmetric, highly oligomeric states which adopt, in most cases, either an octahedral 432 symmetry, eight E2 homotrimers being positioned at the vertexes of a cube, or an icosahedral 532 symmetry, with 20 trimers assembled as a dodecahedron; the number of subunits depends on the complex and the species (1). More recently, the presence of an irregularly shaped, 42-mer E2 assembly was described in the archaeon Thermoplasma acidophilum (8), although this peculiar oligomeric state is still based on the same kind of interactions between the C-terminal helices. Thus, the oligomeric state of the core, responsible for the large size of the complex, was observed in all analyzed complexes and is a trend commonly accepted to be universally conserved in Eubacteria, Archaea, and Eukarya. While the reasons for the presence (and evolutionary conservation) of such huge macromolecular scaffolds remain unclear (9), active site coupling (transfer of acyl groups between lipoyl domains within the core) has often been proposed as the major advantage (1, 10). Also, despite the tripartite organization of these complexes as separate E1/E2/E3 enzymes had always been considered as universal, Corynebacterium glutamicum was shown to be deprived of an E2o component (specific to the ODH complex) and to rather possess an E2o succinyl transferase domain fused to E1o, in a protein called OdhA (11, 12). The same situation has then been confirmed for the model Mycobacterium smegmatis (13). As the lipoyl binding domain of E2o is absent from the E2o-E1o fusion, the ODH activity depends on functional lipoyl groups provided in trans and proven to be supplied by E2p from the PDH complex (14), therefore suggesting the presence of a mixed PDH/ODH supercomplex. By using an integrative structural biology approach, we describe here how C. glutamicum E2p, that was expected to serve as the core of the mixed complex, breaks the rule about the oligomeric state of acyltransferase E2 enzymes, reducing its size to the minimal, catalytically active trimeric unit. We also provide evidence supporting these features as a common trait of Actinobacteria.
Results
M. tuberculosis and C. glutamicum E2p Are Both Trimeric in Solution.
To get insight into the oligomeric state of E2p in Corynebacteriales, full-length E2p from both M. tuberculosis (MtE2p_FL) and C. glutamicum (CgE2p_FL) were successfully expressed in Escherichia coli and purified to homogeneity. Analytical ultracentrifugation (AUC) was used to assess the oligomeric assembly of the proteins (Table 1 and SI Appendix, Fig. S1). Strikingly, the ratio between the estimated molecular weight in solution and the one predicted from the sequence indicated a trimeric state in solution for both samples, with no experimental evidence of higher oligomeric states. The same result was observed for the CgE2p catalytic domain alone (CgE2p_CD, comprising residues Leu437 to Leu675). No larger assembly, suggestive of a 24- or 60-mer state, was observed for any of these proteins in the conditions tested (Table 1). To check for proper functionality of the recombinant proteins, we determined the specific transacylase activity of full-length proteins according to the spectrophotometric assay by Hall and Weitzman (15) (SI Appendix, Fig. S2). Both proteins were indeed able to catalyze the transfer of acetyl units from acetyl-CoA to free dihydrolipoamide, in turn generated by commercial dihydrolipoamide dehydrogenase with the concomitant oxidation of NADH (SI Appendix, Fig. S2), resulting in a specific activity of 17.3 ± 0.9 nmol ⋅ min−1 ⋅ mg−1 for CgE2p_FL versus 2.1 ± 0.1 nmol ⋅ min−1 ⋅ mg−1 for MtE2p_FL.
Table 1.
Constructs, predicted molecular masses, and estimated properties from AUC and SAXS
| Sample | Uniprot accession No. | Construct boundaries (residue number) | Predicted molecular mass (kDa) | AUC molecular mass estimation (kDa) | Extrapolated frictional ratio | Estimated number of protomers (AUC) | SAXS molecular mass estimation (Bayesian) (kDa) | Estimated number of protomers (SAXS) |
| CgE2p_FL | Q8NNJ2 | 1–635 | 70.9 | 221 | 2.85 | 3.1 | 243 | 3.4 |
| CgE2p_CD | Q8NNJ2 | 437–635 | 25.4 | 78 | 1.40 | 3.1 | 77 | 3.0 |
| MtE2p_FL | P9WIS7 | 1–553 | 57.1 | 141 | 1.71 | 2.5 | 318 | 5.6 |
| MtE2b_FL | O06159 | 1–393 | 41.2 | 731 | 1.63 | 17.7 | 1,179 | 28.6 |
| MtE2b_CD | O06159 | 165–393 | 24.6 | 600 | 1.48 | 24.3 | 479 | 19.4 |
The absence of the expected higher oligomeric structure was further confirmed by size-exclusion chromatography coupled to SAXS (small angle X-ray scattering) analysis, where the molecular mass was estimated using specific tools available in the ATSAS suite (16, 17) (Table 1 and SI Appendix, Table S1). For both CgE2p_FL and CgE2p_CD, the trimeric form was predominant in solution, whereas for MtE2p_FL the analysis showed a molecular weight estimation that could correspond to a dimer of trimers (Table 1). However, no evidence of a cubic, dodecameric, or any other higher oligomeric state was observed for any of the three proteins. For CgE2p_CD, constant radius of gyration (Rg) through the intensity peak indicated the presence of a unique oligomeric species (SI Appendix, Fig. S3); Rg and Dmax experimental values were 27.7 and 84.7 Å, respectively (SI Appendix, Table S1 and Fig. S3). The dimensionless Kratky plot showed a Gaussian bell shape that decreases to zero, suggestive of a folded, globular, and compact structure (SI Appendix, Fig. S3) and consistently with the AUC results that indicated a trimeric CgE2p_CD protein with low-friction coefficient. For the two full-length proteins, MtE2p_FL and CgE2p_FL, the dimensionless Kratky plot showed a non-Gaussian bell shape followed by a shoulder reaching a plateau above zero, consistent with a properly folded, but not globular, multidomain protein with flexible linkers (SI Appendix, Fig. S3). This result, together with the high-friction coefficient observed by AUC, was suggestive of well-folded, highly elongated proteins in both cases.
CgE2p_CD Possesses a Unique C-Terminal Region Characterized by an Insertion that Blocks the Protein in Its Trimeric Form.
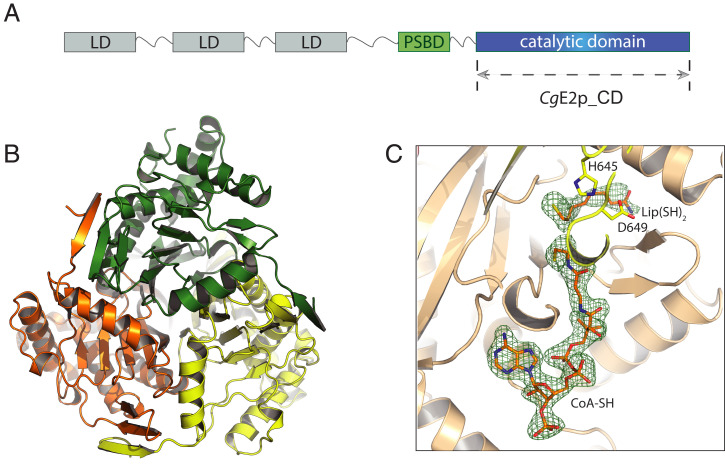
To understand the reasons for CgE2p behaving as a trimeric protein and not assuming a higher-oligomerization state, we characterized it structurally, starting by extensive crystallization trials of its catatlyic core (CgE2p_CD; Fig. 1A). Crystals were obtained from the protein in the absence of ligands, in the presence of the acceptor substrate CoA and in a ternary complex with CoA and dihydrolipoamide (SI Appendix, Table S2). X-ray diffraction showed that the samples belonged to three different crystallographic space groups, with resolutions ranging from 2.10 to 1.35 Å. In all the crystal forms observed, CgE2p_CD showed the expected, chloramphenicol acetyltransferase-like fold, with the same, obligate homotrimeric assembly (Fig. 1B) first observed for the A. vinelandii ortholog (AvE2p_CD) (6). The active site environment at the interface between subunits in the functional trimer was also conserved (SI Appendix, Fig. S4A), with His645 and Asp649 as the equivalent residues to His610 and Asn614 in AvE2p_CD. However, the crystal structure provided no evidence of any higher-order quaternary assembly. To investigate whether the trimeric assembly of CgE2p_CD was still competent to substrate binding, as indicated by its detectable transacylase activity (3.9 ± 0.1 nmol ⋅ min−1 ⋅ mg−1 with acetyl-CoA as the donor substrate), we proceeded to the cocrystallization trials with CoA, dihydrolipoamide [Lip(SH)2], dithiothreitol (DTT) (as an antioxidant), or their possible combinations. Cocrystallization with CoA yielded high-resolution diffracting crystals (SI Appendix, Table S2) which show a full-occupancy complex, with no significant conformational change with respect to the apo structure. Overall, the structure was similar to the equivalent complex from A. vinelandii [Protein Data Bank (PDB): 1EAD; (7)], with the adenosine 3′-phosphate moiety occupying basically the same position in both enzymes, CoA assuming an “IN” conformation with the panthetheine chain entering the active site tunnel, and the guanidinium group of Arg480 (Arg450 in AvE2p_CD) making a salt bridge with the CoA 3′-phosphate (SI Appendix, Fig. S4B). Likewise, electron density maps indicated the oxidation of the CoA terminal thiol group, as in AvE2p_CD (SI Appendix, Fig. S4B).
Fig. 1.
Crystal structure of CgE2p_CD showing a trimeric protein with no trimer–trimer interactions. (A) Schematic representation of the domain architecture of CgE2p. LD: lipoyl domain; PSBD: peripheral subunit-binding domain. (B) Ribbon representation of the apo form of CgE2p_CD showing its homotrimeric structure, with protomers highlighted in orange, green, and yellow, respectively. (C) Zoomed view of the active site region of ternary complex formed by CgE2p_CD with its substrates CoA-SH and dihydrolipoamide [Lip(SH)2]. The polder omit electron density map for both ligands, contoured at the 4 σ level, is shown as a green mesh.
In addition, a ternary complex was obtained by cocrystallization with CoA and free dihydrolipoamide, similarly to AvE2p_CD (7), although it belonged to a different crystalline form with respect to the CoA complex (SI Appendix, Table S2). This structure showed the same, IN conformation for the CoA pantetheine chain and the dihydrolipoamide molecule in an orientation close, albeit not identical to the one observed for the corresponding complex in AvE2p_CD (PDB: 1EAB; Fig. 1C and SI Appendix, Fig. S5).
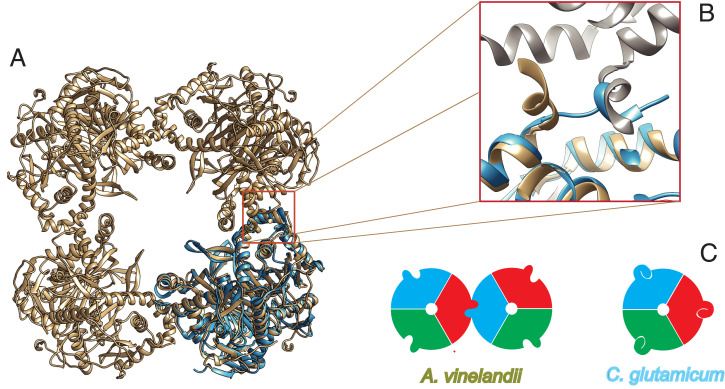
Overall, CgE2p_CD was very similar to the available structures of homologous acyltransferase domains from other E2 enzymes, with rmsds of 1.1 to 1.3 Å over 220 to 225 aligned residues, like E2p from Bacillus stearothermophilus (5) (PDB: 1B5S, with which the rms difference is 1.10 Å over 222 residues), SucB (E2o) from E. coli (18) (PDB: 1SCZ, rmsd 1.29 Å over 211 residues), and E2b from Bos taurus (3) (PDB: 2II4, average rmsd 1.3 Å over 221 to 223 aligned residues), despite the low-sequence identity (lower than 25% in all cases; SI Appendix, Fig. S6). However, all previously known enzymes shared a higher-order oligomeric assembly displaying 432 or 532 symmetry, with the functional homotrimers acting as the “building blocks” that interact with each other through their C-terminal, 310 helices (1, 5). Interestingly, this helix showed the most striking differences when CgE2p_CD was superimposed to AvE2p_CD (Fig. 2 A and B). While in AvE2p_CD this trimer–trimer-interacting helix (TTI helix) was roughly perpendicular to the preceding helix and protruded outward, positioning its hydrophobic residues into a pocket on the facing trimer, in the case of the corynebacterial enzyme, the helix laid on the same monomer surface, with the C-terminal hydrophobic residues occupying the same pocket in an intramolecular interaction (Fig. 2B). In other words, the position of the C-terminal helix for CgE2p_CD coincided with the position of the same, incoming helix from the facing trimer in the A. vinelandii cubic structure (Fig. 2B). As a consequence, the knobs and sockets interaction between trimers is replaced by an intramolecular interaction involving the same features, hindering any trimer–trimer intermolecular interaction and therefore forcing the protein in an homotrimeric state (Fig. 2C).
Fig. 2.
A unique, C-terminal TTI helix orientation is responsible for the lack of higher-order oligomerization in CgE2p. (A) Ribbon representation of the trimeric CgE2p_CD (blue) superimposed to AvE2p_CD (brown), which forms a 24-mer cubic assembly (PDB: 1EAB). (B) Zoomed view on the superimposed TTI region, showing how the TTI helix in CgE2p occupies the equivalent position of the incoming TTI helix in AvE2p. (C) Schematic representation of the TTIs in AvE2p exemplifying canonical E2 enzymes (Left), with different colors representing protomers and the protruding extension representing the C-terminal TTI helix; the situation observed in CgE2p (Right), where the TTI helix is involved in intramolecular interactions that hinder trimer–trimer associations.
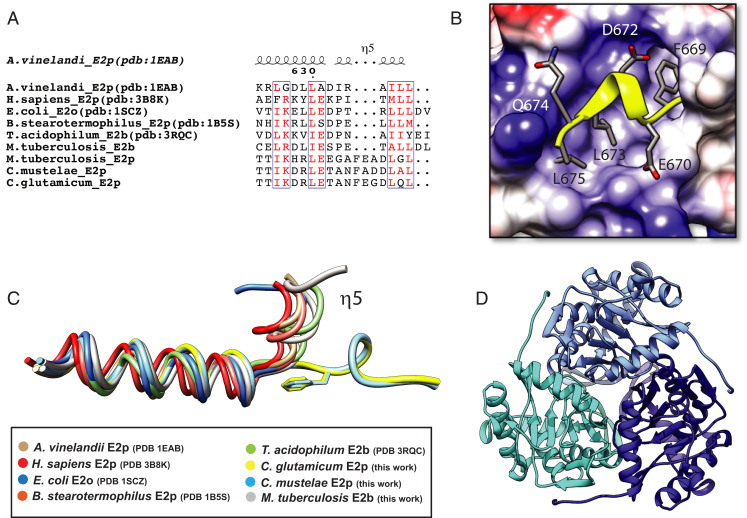
The above differences were also reflected by the distribution of the hydrophobic residues (Fig. 3A), which in oligomeric AvE2p are clustered at the helix C terminus (SI Appendix, Fig. S6), whereas in CgE2p they are arranged at one side of the helix, thus preserving the mostly hydrophobic nature of the helix–pocket interaction (Fig. 3B). This property is exemplified by the replacement of Leu637 in AvE2p for a polar residue (Gln674) in CgE2p, which reflects the new amphipathic nature of the helix. Most importantly, the different orientation of the terminal helix in CgE2p_CD is due to the presence of a three-residue insertion (Phe669-Glu670-Gly671) in CgE2p (Fig. 3A). This insertion allows the C-terminal helix to change its orientation (Fig. 3C) and lay on the surface to reach the accommodating pocket in cis (Fig. 3B), thus replacing an intermolecular interaction by an intramolecular one (Fig. 2C). To confirm that this structural arrangement was not due to the truncated CgE2p_CD protein, a new CgE2p construct, including the peripheral subunit binding domain (PSBD), was also overexpressed in E. coli, purified, and crystallized, yielding a new, 2.23-Å resolution structure (SI Appendix, Table S2) showing the catalytic domain in addition to an N-terminal, 13-residue extension that represent part of the flexible linker connecting to the PSBD (SI Appendix, Fig. S7). Noteworthy, no difference in the oligomeric state, nor in the position of the C-terminal, 310 helix was observed compared to the previous structures (SI Appendix, Fig. S7). In addition, the E2p ortholog from M. tuberculosis, MtE2p, also possesses a three-residue insertion (Phe547-Glu548-Ala549) in an equivalent position just preceding the predicted 310 terminal helix (Fig. 3A), suggesting that the phenylalanine-containing insertion (PCI) could be responsible for the lack of higher-order oligomerization states. To check this hypothesis, we generated a series of seven synthetic CgE2p_CD constructs coding mutant versions of the catalytic domain, in which the C-terminal end bearing the PCI and following helix were replaced by the corresponding ones from E. coli SucB (EcE2o), the closest E2 enzyme in terms of sequence homology having a known structure (44% sequence identity for the catalytic domain; SI Appendix, Fig. S6). The designed mutants were distinguished by a few additional amino acid substitutions to adapt the binding pocket to an intermolecular interaction, incrementing the number of substitutions (SI Appendix, Fig. S8). The construct expressing mutant M3 was selected for the best solubility profile upon overexpression in E. coli; this mutein showed a polydisperse size-exclusion chromatography profile that indicates the presence of a continuum of higher–molecular weight species (SI Appendix, Fig. S9). However, its specific transacetylase activity (3.2 ± 0.6 nmol ⋅ min−1 ⋅ mg−1) was not significantly affected when compared to the wild-type construct expressing the catalytic core (3.9 ± 0.1 nmol ⋅ min−1 ⋅ mg−1).
Fig. 3.
A three-residue insertion (PCI) is responsible for the unique TTI helix orientation that hinders intertrimer interactions. (A) Sequence alignment, limited to the C-terminal region, between several E2 enzymes of known structure and the actinobacterial proteins studied in this work (i.e., MtE2b, MtE2p, CmE2p, and CgE2p). (B) Cartoon representation of the CgE2p TTI helix (yellow) over its binding pocket, colored according to its surface electrostatic charges. (C) Structural superimposition of the same C-terminal region shown in A, showing the distinct orientation of the TTI helix (η5) in CgE2p_CD (yellow) and CmE2p_CD (cyan), described in this work. Please note that the same helix assumes the canonical orientation in MtE2b (gray). (D) Ribbon representation of the homotrimeric structure of CmE2p_CD, with protomers indicated by distinct colors.
The PCI Is a Common Feature in Actinobacteria.
To analyze the distribution of the PCI, we performed sequence analysis of the E2-coding genes in representative genomes distributed within the phylum Actinobacteria (SI Appendix, Fig. S10). Our results show that the PCI was not only present in Corynebacteriales, including M. tuberculosis and C. glutamicum, but more generally in sequences from two classes, namely Actinobacteria (also called Actinomycetales that represent the biggest class inside the phylum Actinobacteria) and Acidimicrobiia (SI Appendix, Fig. S10). The high conservation of the Phe residue present in the insertion emphasizes the importance of its interaction within the context of the hydrophobic pocket. Other residues involved in H-bond interactions with the pocket (mainly Glu670, Leu675, and Leu677) were also conserved (SI Appendix, Fig. S10). Interestingly, although the PCI is conserved, the TTI helix region has lost some of the C-terminal residues and even the full C-terminal helix is predicted to be absent in some of the organisms, in agreement with the absence of the intertrimer interactions in which this helix is involved.
PCI and E2 Oligomerization.
To confirm the relationship between the presence of the three-residue, actinobacterial-specific, C-terminal insertion (PCI) and the oligomerization order of E2 proteins from the Actinobacteria class, we followed two parallel paths. First, we structurally analyzed the orthologous protein from Corynebacterium mustelae, a recently described species from the same genus (19), as an example of ortholog bearing the same insertion. The catalytic domain of the predicted E2p enzyme from this species (CmE2p_CD, residues 428 to 666) was crystallized in complex with CoA and its structure solved by molecular replacement to 2.50-Å resolution (SI Appendix, Table S2). The crystal structure shows a homotrimeric arrangement of CmE2p_CD (Fig. 3D) with no significant structural changes with respect to the C. glutamicum counterpart (rmsd of 0.79 Å over the whole trimer) and without evidence of any relevant, higher-order oligomeric state. In particular, the C-terminal 310 helix had the same relative orientation as in CgE2p_CD (Fig. 3C) and showed the same intramolecular interaction over the trimer surface, with the three-residue insertion (including the well-conserved Phe) occupying an equivalent position in both structures (Fig. 3 A and C). The active site environment, including bound CoA, was also superimposable to CgE2p_CD (SI Appendix, Fig. S11).
Likewise, we analyzed MtE2b, the predicted E2 component of the BCKDH complex in M. tuberculosis (20). The rationale for the choice of this protein came from two observations. First, the analysis of its primary sequence did not show any insertion at the C terminus, in contrast with CgE2p, CmE2p, and MtE2p from the same species (Fig. 3A), suggesting that it may assume a higher oligomeric state. However, an unpublished model of its catalytic domain issued from a structural genomics initiative (PDB: 3L60) showed a trimeric arrangement, apparently challenging our hypothesis. We therefore designed a similar construct to overexpress MtE2b_CD (residues 165 to 393) in E. coli with an N-terminal, cleavable His6-tag and purified the recombinant protein. Crystallization experiments carried out after cleavage of the N-terminal His6-tag yielded bipyramid shaped crystals belonging to the cubic space group F432 and diffracting X-rays up to 1.5-Å resolution (SI Appendix, Table S2). The crystal structure, solved by molecular replacement using the coordinates available from the PDB, showed, unlike the 3L60 model, the same 24-mer cubic arrangement first observed for AvE2p_CD or bovine E2b (3) (SI Appendix, Fig. S12). The terminal 310 helix assumed the same canonical conformation, being involved in the knobs and sockets TTI. To verify the oligomerization state in solution of this construct as well as the entire MtE2b protein, we carried out solution studies on both MtE2b_FL and MtE2b_CD. Both AUC data and ab initio models obtained from SAXS data were consistent with a large assembly for both constructs (Table 1), indicative of the presence of the canonical, 24-mer cubic assembly (SI Appendix, Figs. S1 and S13). We also found MtE2b_FL able to catalyze transacylase activity using isobutyryl-CoA as the donor substrate, with specific activity values (2.5 ± 0.2 nmol ⋅ min−1 ⋅ mg−1) comparable to those measured for MtE2p_FL, which, in contrast, is trimeric (2.1 ± 0.1 nmol ⋅ min−1 ⋅ mg−1 with acetyl-CoA as the donor substrate).
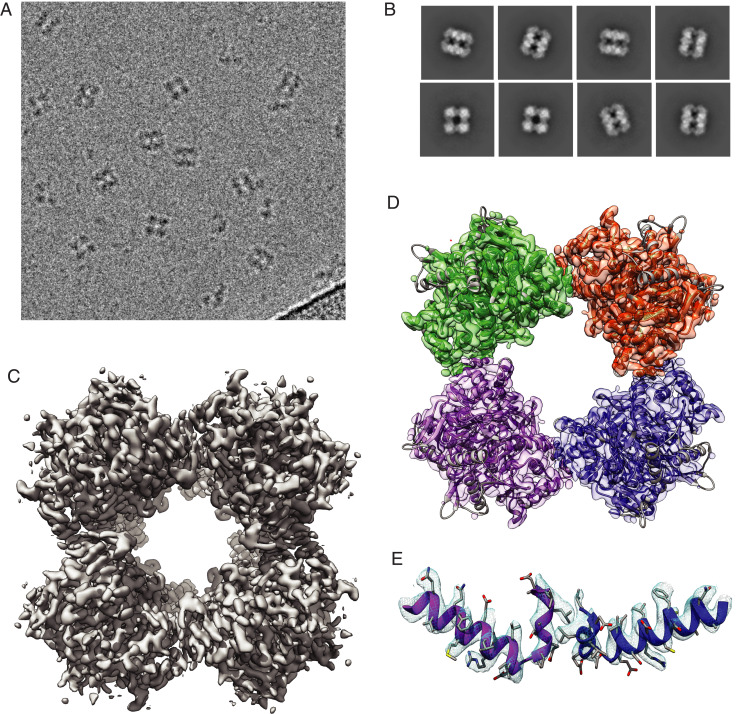
We then took advantage of the oligomerization state of MtE2b to analyze recombinant proteins from both constructs by single-particle cryo-electron microscopy (cryo-EM). Isolated particles indeed showed a cubic shape structure for both MtE2b_FL and MtE2b_CD proteins, with a particle side of ∼13 nm (Fig. 4A and SI Appendix, Fig. S14), consistently with the crystallographic data. Several rounds of two-dimensional (2D) classification were performed to select a subset of good particles (SI Appendix, Fig. S15), in which the class averages reflected the expected cubic shape and high-resolution features, such as defined α-helices (Fig. 4B), whereas particles showing apparent partial dissociation of the cubic complex were discarded (SI Appendix, Fig. S15). Several ab initio models were calculated and, after three-dimensional (3D) classification, homogeneous refinement, local CTF refinement, and density modification, an EM map at 3.82-Å resolution was obtained, showing the expected cubic assembly (Fig. 4C and SI Appendix, Fig. S15). Real space refinement was then performed to fit the 24-mer, cubic crystallographic assembly into the EM map, showing a very good fitting overall (Fig. 4D), including the TTI helix and its canonical orientation including the knobs and sockets interactions (Fig. 4E). The ensemble of our structural and biophysical data therefore supports a strong link between the three-residue, C-terminal insertion (PCI), the orientation of the terminal 310 helix (TTI), and the E2 oligomeric state.
Fig. 4.
Cryo-EM structure of MtE2b_CD. (A) Micrograph showing individual cubic particles. (B) Representative 2D classes showing different particle orientations. (C) Electron microscopy map of MtE2b_CD, showing a cubic, 24-mer assembly with octahedral symmetry. (D) Cubic assembly of the same MtE2b_CD derived from its 1.5-Å resolution crystal structure (PDB: 6ZZN, this work), reconstituted from crystallographic symmetry and fitted into the electron microscopy map. (E) Zoomed view on the TTI region, showing the TTI helix adopting the canonical orientation observed in other E2 enzymes.
The Presence of the PCI in Actinobacterial E2p Is Related to the Presence of OdhA.
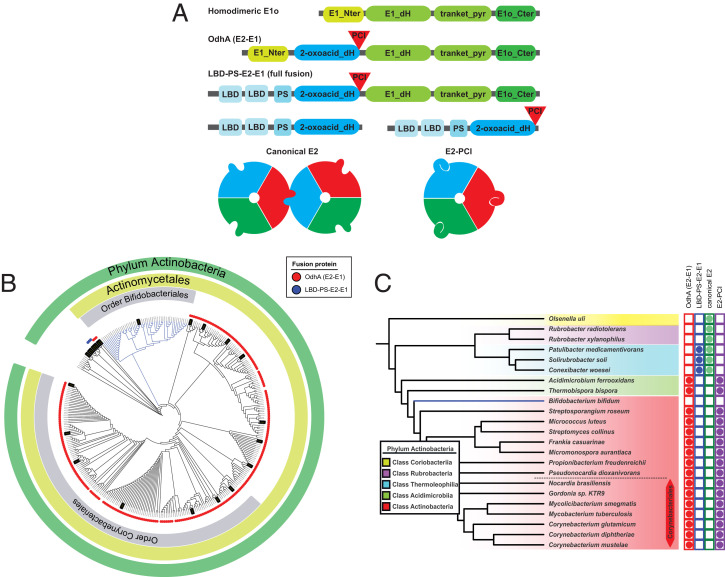
Unlike other canonical E1 proteins, C. glutamicum OdhA has a unique domain architecture characterized by the fusion of an E2 domain to E1o (12, 14), also found in mycobacterial KGD (13) (Fig. 5A). Taking advantage of this unique E2-E1 fusion feature, we retrieved the sequences of OdhA homologs from an extended list of available genomes (SI Appendix, Table S3), seeking for the same association of Pfam domains that identify the E2 and E1o domains, respectively (Fig. 5A). This analysis revealed that, except for a few isolated cases, OdhA homologs are a unique feature of Actinobacteria (Fig. 5B). Inside this phylum, OdhA homologs are widely present in the Actinomycetales class, except for members of the order Bifidobacteriales and the Acidimicrobiia class but not in the classes Coriobacteriia or Rubrobacteria (Fig. 5C). The three known members from the class Thermoleophilia show a unique architecture, in which the first domain is replaced by two lipoyl domains and an E3 binding domain (Fig. 5A), thus bearing a full fusion of E2 and E1o enzymes. Unlike OdhA, such an enzyme, which is so far uncharacterized, could perform both the oxidative decarboxylation and the transacylation steps without the need for lipoyl domains provided in trans. Although speculative at this stage, this hypothesis is coherent with the predicted absence, in this sequence, of the N-terminal, α-helical segment, which is found in OdhA and in other E1o enzymes and predicted to be required for the interaction with E2 (Fig. 5A).
Fig. 5.
Distribution of OdhA orthologs and PCI-bearing E2 enzymes in the Actinobacteria phylum. (A) Schematic representation of the different domain associations. Green blocks indicate Pfam domains associated to homodimeric E1o enzymes, while blue blocks refer to domains composing E2 enzymes. The respective Pfam accession numbers are indicated in the Material and Methods section. LBD: lipoyl binding domain; PS: PSBD. (B) Distribution of OdhA orthologs (red dots) or full E2o-E1o fusion enzymes (LBD-PS-E2-E1; blue dots) in the phylum Actinobacteria. Branches highlighted in black correspond to genomes selected to be analyzed for the PCI presence in E2. (C) Representative sequences from the actinobacterial tree, showing the correlation between the presence of OdhA-like enzymes or full E2o-E1o fusion enzymes, and PCI-bearing enzymes versus canonical E2 enzymes.
Noteworthy, the correlation between the PCI in E2 and the presence of an OdhA homolog (devoid of a lipoyl binding domain) could be observed for all the representative genomes (Fig. 5C). Both features are present in classes Actinobacteria and Acidimicrobiia and absent in the classes Thermoleophilia, Rubrobacteria, or Coriobacteria. Since OdhA requires E2p to perform its activity in the context of the PDH/ODH supercomplex, it is tempting to speculate that loss of E2 higher oligomerization could be a feature evolved to optimize the assembly of such a mixed complex, in which an E2o-E1o fusion enzyme has to compete with E1p for the same lipoyl groups provided by a shared E2p. Along the same line, this might explain why this “minimal size” E2p was only detected in the Actinobacteria phylum.
Discussion
The ensemble of enzymatic reactions that make up central metabolism and concur to the production of reducing equivalents in heterotrophic organisms is commonly considered to be extremely conserved. The same has longtime been believed for the respective enzymes and their assemblies, for which most of our current knowledge comes from studies made decades ago on model organisms, which, for prokaryotes, were in most cases represented by E. coli or B. subtilis. 2-oxoacid dehydrogenase complexes, which are key players in central metabolism, were also objects of intensive study during this “golden age” of enzymology, with most of our knowledge coming from seminal work carried out, over several decades, by the groups led by renowned biochemists like Richard N. Perham (1), Lester Reed (21), and John Guest (22). Their extensive biochemical work was then supported by structural evidence that started being published during the ‘90s, initiated by the work performed in Wim Hol’s laboratory on the diazotroph, gram-negative model A. vinelandii, and the first determination of a crystal structure of AvE2p_CD (6), which in turn confirmed observations dating back to the ‘60s on E. coli E2p (23) and E2o (24). The following structural studies have mostly focused on the PDH complexes, especially the ones from model species like E. coli, B. stearothermophilus, or their counterparts from human or yeast (1, 25, 26), while ODH and BCKDH complexes, despite a number of biochemical studies (27), were largely assumed to share PDH-like molecular structures. In all cases, 2-oxoacid dehydrogenase complexes have all been considered to conform to the “rule” of a hollow, large E2 core showing either cubic (432) or icosahedral (532) symmetry, assembled according to an ensemble of equivalent, and quasi-equivalent interactions involving the C-terminal end of each E2 monomer (5). The more unusual 42-mer E2 assembly described in T. acidophilum (8), albeit deviating from the standard symmetry, still possesses a hollow core whose assembly is based on the same kind of interactions between C-terminal helices and thus does not represent a real exception to the rule. Although active site coupling has generally been believed to be the main benefit arising from the presence of large multimeric assemblies (1), questions were already raised in the past, when the same team that described the T. acidophilum E2 structure showed that precluding the formation of higher-E2 oligomers did not lead to loss of catalytic activity (9), in contrast with previous reports. Consistently with our results, recent work showed that PDH and ODH activities in C. glutamicum are associated to molecular assemblies that are smaller than in E. coli, proposing a hexameric E2p core for the PDH/ODH supercomplex from size-exclusion chromatography experiments (28). Our work here thus shows how Corynebacteriales, and most Actinomycetales, break the “dogma” regarding the 3D architecture of 2-oxoacid dehydrogenase complexes, providing a simple structural explanation to the lack of larger E2 molecular assemblies and at the same time raises new questions about the origin and evolution of these enzymatic machineries.
Actinobacteria represent one of the largest and ubiquitous prokaryotic phyla, with more than 50 families. Its members include the Streptomyces genus, one of the largest bacterial genera and a major source of antibiotics, as well as relevant human pathogens, such as M. tuberculosis and cell factories as C. glutamicum. Despite the wide applications of C. glutamicum in the biotech industry, which range from the production of amino acids to that of biofuels (29), relatively little is known about how Corynebacteriales regulate their central metabolism. The evidence reported here supports a strong correlation between the presence of E2o and E1o activities on a single OdhA-like polypeptide and the loss of highly oligomeric structure of the PDH core, two properties that are most likely related to the dependence of both the PDH and ODH activities on the lipoyl domains provided by a single E2p protein and, therefore, their coexistence in a mixed multienzyme machinery. The presence of such kind of supercomplex, first hypothesized in 2006 for C. glutamicum (11), is in turn suggestive of the presence of regulation mechanisms that might well be unique to Actinobacteria. In this respect, the discovery that the FHA protein OdhI is a phospho-dependent regulator of OdhA in C. glutamicum (11, 30) paved the way to show that ODH activity in Corynebacteriales—including M. tuberculosis and M. smegmatis—is regulated by a signal transduction pathway triggered by the Ser/Thr kinase PknG in response to nutrient availability (31–34). So far, the presence of an FHA protein as a metabolic regulator, which in mycobacteria also acts on glutamate dehydrogenase and glutamate synthase (34, 35), has only been described in Corynebacteriales and might be related to the unusual architecture of the PDH/ODH supercomplex. An additional ODH allosteric regulator is acetyl-CoA, identified as an allosteric activator of M. smegmatis KGD (13) but seemingly common in Corynebacteriales, as it was earlier reported as an activator of corynebacterial ODH (36). As acetyl-CoA is the product of the PDH reaction, positive feedback mechanisms by product channeling from PDH to ODH in a common complex are possible. On the other hand, little is known about the regulation of the PDH reaction in Actinobacteria, apart from the known involvement of the RamB transcriptional regulator in the expression of the aceE (E1p) gene in C. glutamicum (37).
Recent developments in metabolomics have started to unravel the mechanisms of metabolic regulation in M. tuberculosis (38, 39), a particularly important facet as metabolic plasticity is a key capability of the pathogen to switch from active replication to dormant states. The strategy of targeting central metabolism for new drugs has been suggested by several groups, mainly because of the presence of unique M. tuberculosis pathways that are crucial for pathogen survival and latent state persistence within the host (38, 40, 41). The peculiar architecture of the PDH/ODH supercomplex might therefore be exploited for drug design purposes, especially considering that the chemical inhibition of DlaT (MtE2p) has been shown to kill nonreplicating mycobacteria (40). On the other hand, considering that both pyruvate and 2-oxoglutarate are essential metabolites and precursors of a number of compounds of biotechnological interest, many of which industrially produced via C. glutamicum (42), our findings also pave the way to metabolic engineering applications.
Materials and Methods
Plasmid Construction.
C. glutamicum open reading frames were amplified from genomic DNA by PCR and cloned in pET-28a vector by restriction-free cloning (43), while expression constructs for the other proteins (CmE2p, MtE2p, and MtE2b) and for all the CgE2p_CD mutants were provided by Genscript (Leiden). In all cases, a sequence coding for a His6-tag followed by the TEV (tobacco etch virus) protease cleavage site (ENLYFQG) were fused to the N terminus of the protein of interest. Constructs were made to express the following: for CgE2p (Uniprot accession No. Q8NNJ2), the full-length protein (CgE2p_FL, residues 1 to 675), the catalytic domain (CgE2p_CD, corresponding to residues 437 to 675), and the catalytic core and the PSBD (CgE2p-ΔLBDs, corresponding to residues 372 to 675). For CmE2p (Uniprot accession No. A0A0G3H170), the catalytic core was expressed (CmE2p_CD, residues 428 to 666), while for MtE2p (Uniprot accession No. P9WIS7), the full-length protein was expressed (MtE2p_FL, residues 1 to 553).
For MtE2b (Uniprot accession No. O06159), two constructs were designed: the first one corresponding to the full-length protein (MtE2b_FL, residues 1 to 393) and the second one corresponding to the catalytic core region (MtE2b_CD, defined as residue 165 to 393). All plasmids were verified by DNA sequencing.
Protein Purification.
Each plasmid was transformed into E. coli BL21DE3 using standard protocols. In the case of CgE2p_FL and CgE2p_CD, transformed cells were growth in Lysogeny Broth (LB) medium at 37 °C until optical density measure at 600 nm reach ∼0.6. Afterward, temperature was dropped to either 18 °C or 30 °C (SI Appendix, Table S4) and incubated for 18 h after adding 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). For MtE2p_FL, MtE2b_FL, MtE2b_CD, CgE2p-ΔLBDs and constructs expressing mutants generated on CgE2p_CD, autoinduction medium adapted from Studier (44) was used, with slight modifications. In this case, bacterial cells were grown at 37 °C for 4 h, following an overnight incubation at either 30 or 18 °C (SI Appendix, Table S4). Cells were recovered by centrifugation and washed twice with the corresponding lysis buffer (SI Appendix, Table S4). Cell lysis was performed using a CF2 cell disruptor (Constant Systems Ltd.). The soluble fraction was separated from debris by centrifugation at 26,800 g for 1 h. The recombinant proteins were purified by Ni2+-IMAC on HisTrap columns (GE Healthcare). Fractions, as confirmed by SDS-PAGE, containing the protein of interest were dialyzed against gel filtration buffer (SI Appendix, Table S4), supplemented with 1 mM DTT, for 12 h at 18 °C after adding His-tagged TEV protease (45) at a 1:30 ratio (w/w) to remove the N-terminal His6-tag from the recombinant protein. The cleaved, recombinant proteins were then separated from the His-tagged TEV protease (and the cut N-terminal portion) by gravity–flow separation on Ni-NTA agarose (Qiagen). Finally, the recovered recombinant proteins were further purified by size-exclusion chromatography using a Sephacryl S-400 16/60 column (GE Healthcare). Fractions containing the protein were pulled, concentrated, and flash frozen in liquid nitrogen.
Transacylase Assay.
Transacylase activity of CgE2p, MtE2p, and MtE2b was determined by the spectrophotometric method first described by Hall and Weitzman (15), with slight modifications, following transacylation in the reverse direction from an acyl-CoA donor to free dihydrolipoamide, generated by the dihydrolipoamide dehydrogenase reaction (SI Appendix, Fig. S2). The assay mixture contained 0.1 M Tris⋅HCl pH 7.6, 10 mM MgCl2, 1 mM EDTA, 0.4 mM NADH, 0.2 mM lipoamide, 0.25 U of lipoamide dehydrogenase from bovine intestinal mucosa (Enzyme Commission number 1.8.1.4, Sigma-Aldrich L-6777), and an E2 protein: 22.5 μg CgE2p_FL (1.6 μM), 300 to 600 μg MtE2p_FL (26.2 to 52.4 μM), or 120 to 240 μg MtE2b_FL (14.6 to 29.2 μM). For catalytic domain constructs, quantities added were 110 μg for CgE2p_CD (21.1 μM) or 69 μg for the M3 mutant (12.1 μM). The approach to equilibrium in lipoamide dehydrogenase reaction and the background rate of NADH oxidation was followed for 30 min; then, transacylation was started by adding 0.15 mM acetyl-CoA (SI Appendix, Fig. S2) or 1 mM isobutyryl-CoA to the assay mixture for both E2p proteins or MtE2b_FL, respectively, to a final reaction volume of 200 μL. Subtraction of endogenous rate of NADH oxidation from the final rate after acyl-CoA addition gave a measure for the dihydrolipoamide transacylase activity. The linear region of the progress curves, generally limited to 1 min (SI Appendix, Fig. S2), was used to calculate the steady-state velocities. Extinction coefficient of NADH used for calculations is 6.22 mM−1 ⋅ cm−1.
Lipoamide dehydrogenase, as well as acyl-CoA substrates were not limiting components, as increasing their concentrations did not affect the final transacylase activity. The measurements were performed at least as triplicates and resulting activities presented in nmol ⋅ min−1 ⋅ mg−1 of protein.
SEC-SAXS and Ab Initio Modeling.
SAXS experiments were performed either at the SOLEIL synchrotron (Saint-Aubin) on the SWING beamline, or at the ESRF (European Synchrotron Radiation Facility, Grenoble) on BM29 (CgE2p_CD sample). Experimental settings are provided in SI Appendix, Table S1. The protein samples were injected, through an Agilent HPLC system, into a size-exclusion chromatography column (Superose 6 Increase 5/150 GL, GE Healthcare Bio-Sciences) previously equilibrated with the corresponding buffer (SI Appendix, Table S4) and eluted directly into the SAXS flow-through capillary cell at a flow rate of 0.2 mL/min. Frames were collected continuously during the whole-protein elution time.
Frames were analyzed using either the FOXTROT software developed at Synchrotron Soleil (for data collected on Swing), or the RAW software (46). Frames corresponding to the buffer were averaged and subtracted from sample frames. The forward scattering I(0) and Rg for each sample subtracted frame were derived from Guinier approximation; the I(0) versus frame curve was used to select the region corresponding to the protein’s elution peak (SI Appendix, Fig. S3). Frames corresponding to the elution peak and with constant Rg were averaged. All subsequent data processing was performed using software part of the ATSAS suite (16), starting from PRIMUS, which was used to recalculate I(0), and Rg, while GNOM was used to calculate the pair-distance distribution function P(r) and Dmax (17). The POROD volume and other tools available in ATSAS were used to estimate the molecular weights of the proteins (47). The dimensionless Kratky plot was calculated according to previous work (48, 49) using the value determined by Guinier analysis.
Ab initio modeling was also performed using software from the ATSAS suite. For MtE2b_CD, 10 independent models were generated using DAMMIF, with no imposed symmetry. In both cases, the best models were obtained assuming oblate anisometry. The models were then compared and align with DAMSEL and DAMSUP, and output files were generated using DAMAVER, DAMFILT, and DAMSTART, all within ATSAS. Finally, the DAMSTART output was provided to DAMMIN to obtain a dummy atom model fitting the data.
AUC.
Sedimentation velocity (SV) AUC assays were performed using a Beckman Coulter ProteomeLab XL-I AUC equipped with UV-Vis absorbance and Raleigh interference detection systems, using the 8-hole Beckman An-50Ti rotor at 20 °C. Experiments were performed at 10,000 rpm for MtE2b_CD and MtE2b_FL and 36,000 rpm for MtE2p_FL, CgE2p_FL and CgE2p_CD. Three concentrations of each protein were prepared for this experiment in their corresponding buffer and loaded into AUC cells. During the run, SV was followed using by measuring absorbance at 290 nm. SEDFIT 15.01 [available at https://sedfitsedphat.github.io/ (50)] was used to calculate the sedimentation coefficient distribution C(s), then corrected to standard conditions to get the final standard values. These coefficients were plotted as a function of the different concentrations and an extrapolation to zero concentration was made to obtain the standard value for the main oligomer. From these values, molecular mass and friction ratio were obtained.
Crystallization.
The crystallization experiments were performed at either 18 or 4 °C by the sitting drop vapor diffusion technique in 96-well plates, according to established protocols at the Crystallography Core Facility of the Institut Pasteur (51). Crystallization conditions were the following: for CgE2p_CD in apo form, 0.1 M Hepes-Na pH 7.5, 5% PEG4000, 30% 2-methyl-2,4-penthane diol, at 4 °C; for CgE2p_CD in complex with CoA, 0.1 M Hepes-Na pH 7.5, 1.56 M trisodium citrate, at 4 °C; for CgE2p_CD in ternary complex with CoA and dihydrolipoamide, 0.1 M Tris⋅HCl pH 7.3, 1.62 M trisodium citrate, at 18 °C; for CgE2p_ΔLBDs, 30% PEG1500 at 18 °C; for CmE2p_CD in complex with CoA, 0.1 M Hepes-Na pH 7.5, 150 mM NaCl, 30% PEG4000, at 18 °C; and for MtE2b_CD, 0.1M Imidazole pH 6.5, 4 M NH4+ acetate at 18 °C.
Data Collection and Structure Solution.
Diffraction data were acquired from crystals maintained at 100 K, either at the SOLEIL Synchrotron on the beamline Proxima-1 or Proxima-2A or on the automated beamline MASSIF-1 at the ESRF (52). The data were processed with XDS (53), run through either XDSME (available at https://github.com/legrandp/xdsme) or autoPROC (54), and scaled with Aimless, available from the CCP4 suite (55), or STARANISO, as provided within autoPROC. The structures were solved by molecular replacement through the program PHASER (56). Coordinates of the E. coli E2p catalytic domain [PDB: 4N72; (57)] were used as the search model to first solve the structure of CgE2p_CD in apo form, which, in turn, served as the molecular replacement search model for the following datasets, including CmE2p_CD, but not MtE2b_CD whose structure was solved by using the previously released coordinates of the same protein (PDB: 3L60). All rebuilding and adjustments of the models were performed with COOT (58). The refinement was carried out with BUSTER, applying local structure similarity restraints for noncrystallography symmetry (59) and a Translation–Libration–Screw model. “Polder” maps for the identification of ligand density were calculated with the specific tool within the PHENIX suite (60), while chemical dictionaries for ligands were generated with the Grade server (http://grade.globalphasing.org). Validation of models was performed with MOLPROBITY (61) and the validation tools in PHENIX (62). Data collection, refinement, model statistics, and PDB accession codes for coordinates and structure factors are indicated in SI Appendix, Table S2. Graphical representations were rendered with Pymol (63).
CryoEM.
For grid preparation, 3.5 µL of 0,2 mg/mL MtE2b_CD were applied into a Lacey Carbon Film on 200 Mesh Copper Grids. Grid vitrification was performed in a Vitrobot apparatus (Thermo Fisher Scientific) with 95% humidity, ashless filter paper (Standard Vitrobot Filter Paper, Ø55/20 mm, Grade 595; Electron Microscopy Sciences), and using a blot time of 3.5 s while blotting force was set to zero. Grids were stored in liquid nitrogen and collected in a 200 keV Thermo Fisher Scientific Glacios Cryo-Transmission Electron Microscope. Images were recorded using a Falcon III direct electron detector camera in linear mode. The images were collected using EPU software 2.2.0.65REL (Thermo Fisher Scientific), with a defocus range from −0.8 to −2.6 μm. The pixel size was set to 0.96 Å. A total of 37 frames per movie were collected, with an overall dose of 42.5 e−/Å2.
Movies frames were aligned using Motiocor2 (64, 65), and contrast transfer function (CTF) estimation was performed using Gctf (66). Movies were imported and analyzed in Cryosparc v2.15 (67). Using the curate exposure feature, 1,235 out of 1,476 movies, followed by 2,483 out of 3,533 movies from a second data collection, were selected for further analysis. A first round of manual picking was performed, and after two rounds of 2D classification, the three most populated classes were selected for template-based particle picking against a dataset containing the selected micrographs. The selected particles were cleaned using the “inspect pick” function of Cryosparc and three rounds of 2D classification, and a final dataset of 40,805 and 168,705 good particles, corresponding to collection session 1 and 2, respectively, was selected. A round of ab initio and 3D classification into four classes was then performed to select a subset of 111,891 particles that could be refined to 5.8 Å. Following local CTF refinement and imposing octahedral O3 symmetry, the dataset could be refined to 3.92 Å. Finally, density modification was applied using the tool resolve_cryo_em from the PHENIX suite (68), reaching a final map at 3.82-Å resolution. Model fitting into cryo-EM maps was performed using the programs UCSF Chimera (69) and phenix.real_space_refine (70). The final, sharpened map from Cryosparc as well as the density-modified map from resolve_cryo_em were deposited to the Electron Microscopy Data Bank (EMDB) under the accession code 11600. Figures were generated and rendered with UCSF Chimera.
Bioinformatic Analysis.
Multiple sequence alignments were performed using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) (71). Further alignment details were carried out in AliView version 1.26 (72) . Sequences from MtE2b, MtE2p, CgE2p, CmE2p, and E2 proteins with known structure were compared to the representative sequences to account for the PCI presence; the software ESPript 3.0 (https://espript.ibcp.fr) (73) was then used to generate alignment pictures providing secondary structure elements from a reference structure. The presence of the PCI was analyzed in the region corresponding to the C-terminal α-helix (residues 630 to 637 in AvE2p).
To identify odhA-like genes, the gene search tool from IMG/M was used (Integrated Microbial Genomes and Microbiomes system version 5.0) (74); to simplify the analysis, only genomes annotated as finished were selected. For each genome, odhA-like genes were identified by their conserved Pfam domain associations, including the following: PF16078 (“2-oxogl_dehyd_N”), PF00198 (“2-oxoacid dH”), PF00676 (“E1-dh”), PF02279 (“Transket_pyr”), and PF16870 (“OxoGdeHyase_C”). All the finished genomes corresponding to the phylum Actinobacteria and two nonfinished genomes corresponding to the class Thermoleophila were selected from IMG/M available genomes (only one finished genome was available for Thermoleophila). After elimination of duplicated genomes, odhA-like genes and E2-E1–like genes were identified using their corresponding Pfam architecture. For the selected genomes, a tree was created using the JGI distance tree tool, using the alignment of 16S genes based on the SILVA database and the “dnadist” and “neighbor” tools from the PHYLIP package (available at https://evolution.genetics.washington.edu/phylip/). Figures were built using the Interactive Tree of Life version 4 online tool (75).
To analyze whether any correlation could be detected between odhA-like genes and the presence of the PCI in E2p genes, a representative pool of genomes from the larger orders inside Actinobacteria class (namely Corynebacteriales, Streptomycetales, Propionibacteriales, Micromonosporales, Microccocales, and Bifidobacteriales), as well as representatives from the other classes inside the phylum Actinobacteria were selected. For each genome, predicted E2 proteins were identified using the signature pfam domains (listed in the paragraph above) and aligned.
Supplementary Material
Acknowledgments
This work was funded by the Agence Nationale de la Recherche (ANR) through the projects SUPERCPLX (ANR-13-JSV8-0003) and METACTINO (ANR-18-CE92-0003), both granted to M.B., and by institutional grants from the Institut Pasteur and the CNRS. We are grateful to the core facilities at Institut Pasteur C2RT (Centre for Technological Resources and Research), in particular to A. Haouz, P. Weber, and C. Pissis (Crystallography); S. Brûlé and B. Baron (Molecular Biophysics); M. Matondo, T. Chaze, and T. Douché (Proteomics); and J.-M. Winter, S. Tachon, and M. Vos (NanoImaging). We also gratefully acknowledge F. Gubellini (Structural Microbiology Unit) for her help in EM sample preparation and J.J. Pierella Karlusich (Ecole Normale Superieure, Paris), A. Thureau (Synchrotron Soleil), and V. Bunik (Lomonosov University, Moscow) for many helpful discussions. The NanoImaging Core at Institut Pasteur was created with the help of a grant from the French Government’s “Investissements d’Avenir” program (EQUIPEX CACSICE—“Centre d’analyse de systèmes complexes dans les environnements complexes,” ANR-11-EQPX-0008) and is acknowledged for support with cryo-EM sample preparation, image acquisition, and analysis. We also acknowledge the synchrotron sources Soleil (Saint-Aubin, France) and ESRF (Grenoble, France) for granting access to their facilities and their staff for helpful assistance on the respective beamlines. L.Y. and A.B. are both part of the Pasteur—Paris University (PPU) International PhD program; L.Y. was funded by the Wuhan Institute of Biological Products Co. Ltd. (Wuhan, People’s Republic of China), subsidiary company of China National Biotec Group Company Limited, and by a doctoral fellowship from the China Scholarship Council.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.P.S.d.C. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112107118/-/DCSupplemental.
Data Availability
Coordinates/structure factors and cryo-EM map data have been deposited in Worldwide PDB (6ZZI–6ZZN, and EMD-11600).
References
- 1.Perham R. N., Swinging arms and swinging domains in multifunctional enzymes: Catalytic machines for multistep reactions. Annu. Rev. Biochem. 69, 961–1004 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Milne J. L. S., et al. , Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J. Biol. Chem. 281, 4364–4370 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato M., et al. , A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex. EMBO J. 25, 5983–5994 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne J. L. S., et al. , Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: A multifunctional catalytic machine. EMBO J. 21, 5587–5598 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izard T., et al. , Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. U.S.A. 96, 1240–1245 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattevi A., et al. , Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science 255, 1544–1550 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Mattevi A., Obmolova G., Kalk K. H., Teplyakov A., Hol W. G., Crystallographic analysis of substrate binding and catalysis in dihydrolipoyl transacetylase (E2p). Biochemistry 32, 3887–3901 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Marrott N. L., et al. , The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly. FEBS J. 279, 713–723 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Marrott N. L., et al. , Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex. Biochem. J. 463, 405–412 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Byron O., Lindsay J. G., “The pyruvate dehydrogenase complex and related assemblies in health and disease” in Macromolecular Protein Complexes, J. R. Harris, J. Marles-Wright, Eds. (Subcellular Biochemistry, Springer International Publishing, 2017), pp. 523–550. [DOI] [PubMed] [Google Scholar]
- 11.Niebisch A., Kabus A., Schultz C., Weil B., Bott M., Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 281, 12300–12307 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Usuda Y., et al. , Molecular cloning of the Corynebacterium glutamicum (‘Brevibacterium lactofermentum’ AJ12036) odhA gene encoding a novel type of 2-oxoglutarate dehydrogenase. Microbiology (Reading) 142, 3347–3354 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Wagner T., Bellinzoni M., Wehenkel A., O’Hare H. M., Alzari P. M., Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism. Chem. Biol. 18, 1011–1020 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Hoffelder M., Raasch K., van Ooyen J., Eggeling L., The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. J. Bacteriol. 192, 5203–5211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall E. R., Weitzman P. D., A continuous spectrophotometric assay for the transacylase (E2) component of pyruvate and alpha-oxoglutarate dehydrogenase enzyme complexes. Anal. Biochem. 62, 286–290 (1974). [DOI] [PubMed] [Google Scholar]
- 16.Franke D., et al. , ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 50, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003). [Google Scholar]
- 18.Knapp J. E., et al. , Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 280, 655–668 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Rückert C., Eimer J., Winkler A., Tauch A., Complete genome sequence of the type strain Corynebacterium mustelae DSM 45274, isolated from various tissues of a male ferret with lethal sepsis. Genome Announc. 3, e01012–e01015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venugopal A., et al. , Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe 9, 21–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed L. J., From lipoic acid to multi-enzyme complexes. Protein Sci. 7, 220–224 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guest J. R., Angier S. J., Russell G. C., Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann. N. Y. Acad. Sci. 573, 76–99 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Reed L. J., Fernandez-Moran H., Koike M., Willms C. R., Electron microscopic and biochemical studies of pyruvate dehydrogenase complex of Escherichia coli. Science 145, 930–932 (1964). [DOI] [PubMed] [Google Scholar]
- 24.Derosier D. J., Oliver R. M., Reed L. J., Crystallization and preliminary structural analysis of dihydrolipoyl transsuccinylase, the core of the 2-oxoglutarate dehydrogenase complex. Proc. Natl. Acad. Sci. U.S.A. 68, 1135–1137 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel M. S., Nemeria N. S., Furey W., Jordan F., The pyruvate dehydrogenase complexes: Structure-based function and regulation. J. Biol. Chem. 289, 16615–16623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z. H., McCarthy D. B., O’Connor C. M., Reed L. J., Stoops J. K., The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. U.S.A. 98, 14802–14807 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunik V., Artiukhov A., Vitamin-Dependent Multienzyme Complexes of 2-Oxo Acid Dehydrogenases: Structure, Function, Regulation and Medical Implications (Nova Science Publishers, 2017). [Google Scholar]
- 28.Kinugawa H., et al. , In vitro reconstitution and characterization of pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase hybrid complex from Corynebacterium glutamicum. MicrobiologyOpen 9, e1113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewin G. R., et al. , Evolution and ecology of actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 70, 235–254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz C., Niebisch A., Gebel L., Bott M., Glutamate production by Corynebacterium glutamicum: Dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl. Microbiol. Biotechnol. 76, 691–700 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya N., et al. , An aspartate-specific solute-binding protein regulates protein kinase G activity to control glutamate metabolism in mycobacteria. MBio 9, e00931-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieck B., et al. , PknG senses amino acid availability to control metabolism and virulence of Mycobacterium tuberculosis. PLoS Pathog. 13, e1006399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ventura M., et al. , GarA is an essential regulator of metabolism in Mycobacterium tuberculosis. Mol. Microbiol. 90, 356–366 (2013). [DOI] [PubMed] [Google Scholar]
- 34.O’Hare H. M., et al. , Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70, 1408–1423 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Nott T. J., et al. , An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis. Sci. Signal. 2, ra12 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Shiio I., Ujigawatakeda K., Presence and regulation of alpha-ketoglutarate dehydrogenase complex in a glutamate-producing bacterium, Brevibacterium flavum. Agr. Biol. Chem. 44, 1897–1904 (1980). [Google Scholar]
- 37.Blombach B., Cramer A., Eikmanns B. J., Schreiner M., RamB is an activator of the pyruvate dehydrogenase complex subunit E1p gene in Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 16, 236–239 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Rhee K. Y., et al. , Central carbon metabolism in Mycobacterium tuberculosis: An unexpected frontier. Trends Microbiol. 19, 307–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Carvalho L. P. S., et al. , Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17, 1122–1131 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Bryk R., et al. , Selective killing of nonreplicating mycobacteria. Cell Host Microbe 3, 137–145 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouzy A., et al. , Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 10, e1003928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieschalka S., Blombach B., Bott M., Eikmanns B. J., Bio-based production of organic acids with Corynebacterium glutamicum. Microb. Biotechnol. 6, 87–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unger T., Jacobovitch Y., Dantes A., Bernheim R., Peleg Y., Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172, 34–44 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 45.van den Berg S., Löfdahl P.-A., Härd T., Berglund H., Improved solubility of TEV protease by directed evolution. J. Biotechnol. 121, 291–298 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Hopkins J. B., Gillilan R. E., Skou S., BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Cryst. 50, 1545–1553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hajizadeh N. R., Franke D., Jeffries C. M., Svergun D. I., Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Sci. Rep. 8, 7204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez J., Vachette P., Russo D., Desmadril M., Durand D., Heat-induced unfolding of neocarzinostatin, a small all-beta protein investigated by small-angle X-ray scattering. J. Mol. Biol. 308, 721–743 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Durand D., et al. , NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber P., et al. , High-throughput crystallization pipeline at the crystallography core facility of the institut pasteur. Molecules 24, 4451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowler M. W., et al. , MASSIF-1: A beamline dedicated to the fully automatic characterization and data collection from crystals of biological macromolecules. J. Synchrotron Radiat. 22, 1540–1547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J., et al. , Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex. J. Biol. Chem. 289, 15215–15230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smart O. S., et al. , Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68, 368–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liebschner D., et al. , Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schrödinger, LLC., The PyMOL Molecular Graphics System, version 2.2.0. https://pymol.org/2/ (Accessed 13 November 2021).
- 64.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X., et al. , Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Ramírez-Aportela E., et al. , Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larsson A., AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen I.-M. A., et al. , IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates/structure factors and cryo-EM map data have been deposited in Worldwide PDB (6ZZI–6ZZN, and EMD-11600).