Abstract
The unprecedented rate of extinction calls for efficient use of genetics to help conserve biodiversity. Several recent genomic and simulation-based studies have argued that the field of conservation biology has placed too much focus on conserving genome-wide genetic variation, and that the field should instead focus on managing the subset of functional genetic variation that is thought to affect fitness. Here, we critically evaluate the feasibility and likely benefits of this approach in conservation. We find that population genetics theory and empirical results show that conserving genome-wide genetic variation is generally the best approach to prevent inbreeding depression and loss of adaptive potential from driving populations toward extinction. Focusing conservation efforts on presumably functional genetic variation will only be feasible occasionally, often misleading, and counterproductive when prioritized over genome-wide genetic variation. Given the increasing rate of habitat loss and other environmental changes, failure to recognize the detrimental effects of lost genome-wide genetic variation on long-term population viability will only worsen the biodiversity crisis.
Keywords: genomics, extinction, population dynamics
Decades of theoretical (1) and empirical (2, 3) research suggest that conserving genome-wide genetic variation improves population viability. Maintaining genetic variation and adaptive potential (the ability of a population to evolve adaptively in response to selection; usually measured as narrow sense heritability [the proportion of phenotypic variance attributed to additive genetic effects]) and avoiding inbreeding depression (reduced fitness of individuals whose parents are related) are central motivations for maintaining large, connected natural populations. Principles of genetics and evolution have therefore played a large role in conservation biology since its inception (4, 5). The genomics revolution has inspired biologists to leverage genome analysis to advance conservation beyond what was possible with traditional genetics. Numerous studies have sequenced genomes of nonmodel organisms of conservation concern to understand population history, inbreeding depression, and the genetic basis of adaptation. A particularly exciting area of research has been to determine when and how functional genetic information can advance conservation.
Several recent studies suggest that too much emphasis has been placed on genome-wide genetic variation in conservation biology. For example, persistence of small populations for long periods of time despite low genetic variation, and the collapse of the Isle Royale wolf population after the infusion of genetic variation via immigration, have been interpreted as a challenge to the idea that genetic variation generally increases population viability (6–12). Additionally, a weak relationship between conservation status and genetic variation has been used to argue that genome-wide (presumably neutral) genetic variation is of little importance to conservation (11). Several authors have thus advocated for an approach that focuses on functional genetic variation that is thought to directly affect fitness (including minimizing deleterious genetic variation) in place of the traditional emphasis on conserving genome-wide genetic variation (6–8, 11).
Here, we evaluate the theoretical and empirical basis of this challenge to the importance of genome-wide genetic variation and show that its premise is inconsistent with population genetic theory and empirical findings. While it is clear that functional genetic information can advance conservation, deemphasizing the maintenance of genome-wide genetic variation would increase the extinction risk of threatened populations.
Is Genetic Variation Predictive of Inbreeding and Inbreeding Depression?
Inbreeding depression is thought to be driven mainly by homozygous and identical by descent, deleterious, partially recessive alleles (13), with lethal and small-effect deleterious alleles contributing substantially (14). Two segments of DNA are identical by descent when they both descend from a single haploid genome in a recent ancestor. The constant input of new deleterious mutations (15–19) makes inbreeding depression a ubiquitous phenomenon that can push populations toward extinction (2, 20–23). One of the foundational predictions of theoretical population genetics is that the rate of loss of mean heterozygosity (, the heterozygous fraction of an individual’s genome) per generation (Δ = 1/2Ne) is identical to the rate of increase in mean individual inbreeding (, the individual inbreeding coefficient: the identical-by-descent fraction of an individual’s genome), which is Δ = 1/2Ne (24). is therefore expected to be entirely predictive of (24–29).
A more difficult but crucial question is whether genome-wide genetic variation (π, nucleotide diversity: expected proportion of nucleotide differences between randomly chosen pairs of haploid genomes in a population) is predictive of inbreeding depression. Deleterious alleles are lost in small populations due to selection and genetic drift (30, 31), but they are also more often expressed in homozygotes in smaller populations due to inbreeding. Selective purging (selective elimination of deleterious, partially recessive alleles that are exposed to purifying selection via inbreeding) of large-effect deleterious alleles following inbreeding combined with genetic drift may therefore result in low inbreeding load (a measure of the potential for inbreeding to reduce fitness, measured by the number of lethal equivalents, which is a set of alleles that would on average cause death when homozygous) and little inbreeding depression in the most highly inbred populations with the lowest π. However, the presence of purging does not imply that high fitness is maintained in small populations with low π.
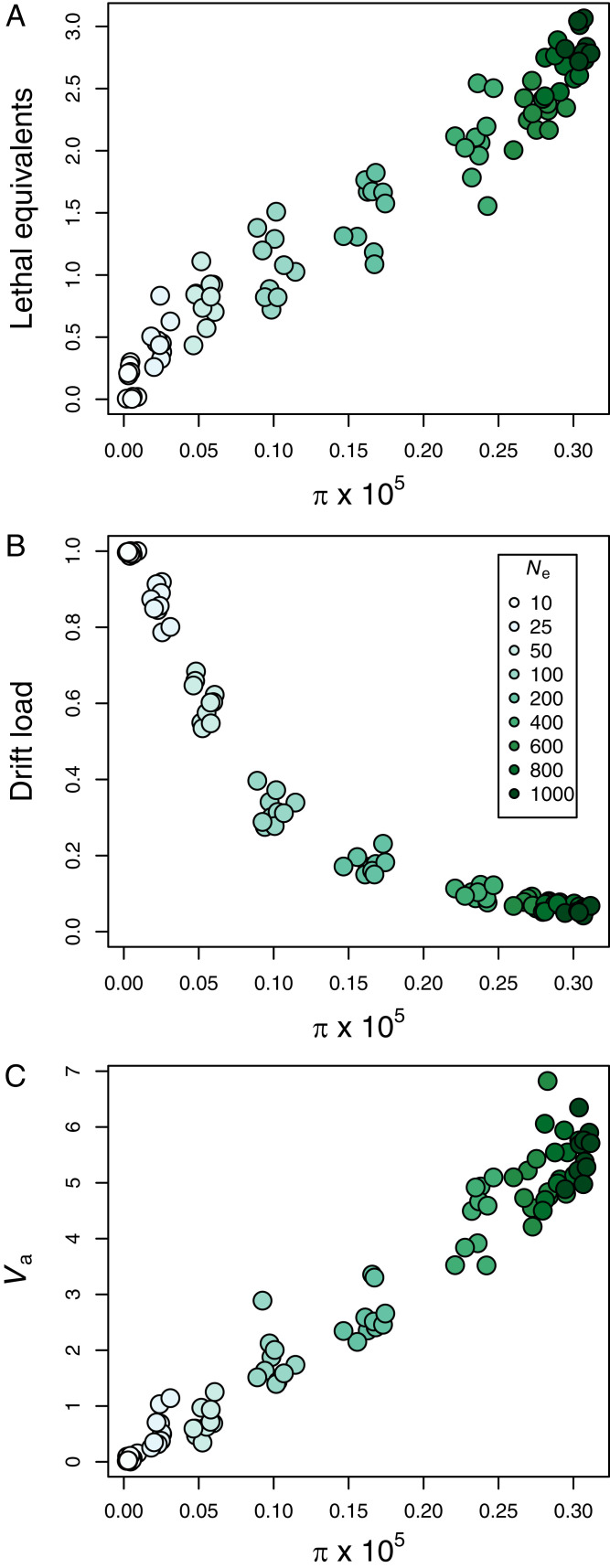
Population genetics theory predicts that larger populations will have higher neutral (24) and deleterious genetic variation (32, 33). This is illustrated in Fig. 1, where simulated large populations have higher π (24) and higher inbreeding load (32–34) arising from segregating partially recessive deleterious alleles. These simulations assume empirically supported models of fitness and dominance (h) effects (SI Appendix). h is the dominance coefficient: a derived allele is recessive when h = 0 (heterozygous genotypes have the same mean fitness as homozygous wildtypes), dominant when h = 1 (heterozygous genotypes have the same mean fitness as homozygous derived allele genotype), and additive when h = 0.5 (heterozygous genotypes have fitness midway between the alternative homozygous genotypes). Smaller populations have lower π due to genetic drift, and fewer lethal equivalents due to genetic drift and purging. However, despite having fewer lethal equivalents, chronically smaller populations have lower mean fitness due to partially recessive deleterious alleles being expressed following inbreeding, and some reaching high frequency or fixation (i.e., high drift load [the reduction in mean fitness of a population, due to homozygosity for deleterious allele]). Therefore, a negative relationship is expected between π and drift load for populations at mutation–drift–selection equilibrium.
Fig. 1.
Relationship of nucleotide diversity () with the inbreeding load (lethal equivalents) (A), drift load (B), and additive genetic variance in a quantitative trait (Va) (C). The data are from the 1,000th generation of 10 simulated populations with nine different constant effective population sizes (Ne).
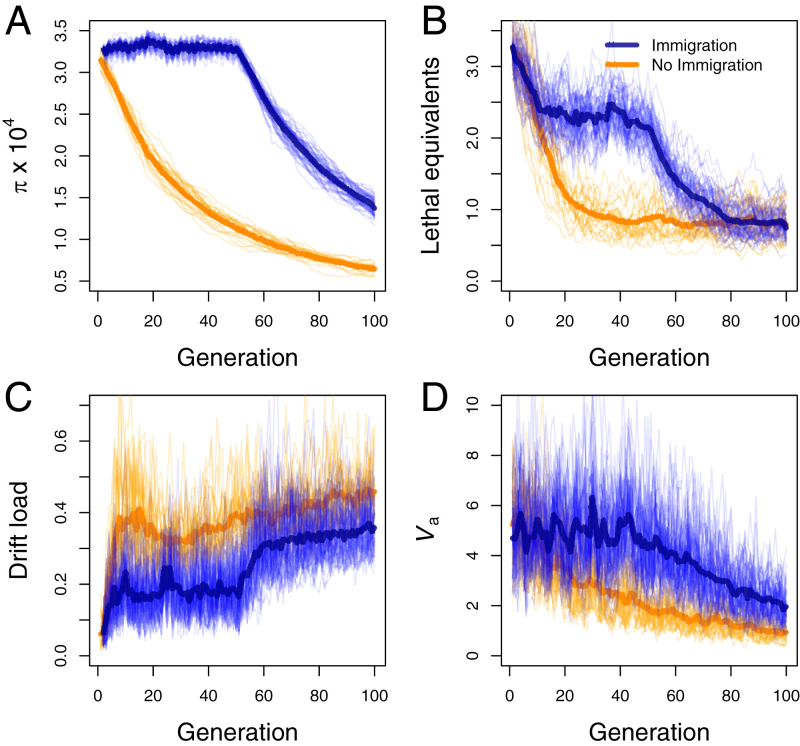
Equilibrium levels of π and drift load are not expected in populations with fluctuating population size or immigration rate. A common scenario with high conservation relevance is isolated populations that have experienced recent bottlenecks. The simulated data in Fig. 2 show that genome-wide π declines over time following a bottleneck, as expected from classical theory (24) (Fig. 2A). This pattern is paralleled by lethal equivalents (Fig. 2B) owing to the loss of deleterious alleles via genetic drift and purging of deleterious alleles expressed in homozygotes due to inbreeding (30, 31). However, the deleterious alleles remaining after a bottleneck often go to high frequency or fixation. This results in individuals being homozygous for increasingly more deleterious alleles (higher drift load; Fig. 2C) as π declines inexorably during a sustained bottleneck, the same pattern expected for small populations at equilibrium (Fig. 1). It is notable, however, that π, inbreeding load, and drift load can change at substantially different rates following a bottleneck. For example, drift load can become quite high before π declines substantially following a bottleneck (Fig. 2 A and C). However, small populations that already have low π are also expected to have low mean fitness, due to ever-increasing drift load, which demonstrates that π is a good indicator of drift load and mean fitness. Occasional immigration can be sufficient to maintain high π and low drift load in small populations (Fig. 2). This is one reason why maintaining connectivity is a priority in conservation biology, and why genetic rescue (increase in population growth or reduction in genetic load arising from the immigration of individuals with new alleles) is an effective tool for managing small, isolated populations (30, 35, 36).
Fig. 2.
Genetic effects of bottlenecks with and without immigration. Nucleotide diversity () (A), number of lethal equivalents (B), drift load (C), and the additive genetic variance in a quantitative trait (Va) (D) are shown for 100 generations after a simulated bottleneck in isolated populations (orange) and with five immigrants every two generations up to generation 50 (blue). Population size was held constant at Ne = 1,000 for 1,000 generations before the bottleneck and then at Ne = 25 starting at generation 0. The thin lines show the results from 25 replicates. The thick lines represent the mean across 25 replicates. Immigrants during the first 50 generations are from a population with Ne = 500 that split from the receiving population the generation of the bottleneck. Details of the simulation model and parameters are provided in SI Appendix.
Empirical data show that purging does not eliminate the extinction threat posed by inbreeding. Pedigree-based studies have yielded mixed results with regard to purging, with typically only a small portion of inbreeding depression being removed after sustained inbreeding in small populations (37–39). Analyses of 60 genomes from seven ibex species found that species which went through the most severe bottlenecks had more deleterious alleles (40). Alpine ibex, which were once reduced to 100 individuals, had fewer highly deleterious alleles but more mildly deleterious alleles compared to Iberian ibex (bottleneck size 1,000 individuals). Empirical genetic data suggest small populations have higher drift load (40–42) which has resulted in lower population growth in populations with lower genetic variation (2, 3). In agreement with theoretical expectations outlined above, these data suggest that purging is insufficient to maintain high fitness in the face of strong genetic drift and inbreeding. Thus, the presence of genomic signatures of purging should not be taken as evidence for the absence of inbreeding depression, or for demographic stability of small populations.
The relationship between π and fitness is obviously complicated, particularly immediately after a bottleneck (Fig. 2). Populations with the lowest π and highest inbreeding will also have the lowest inbreeding load, on average, due to reduced deleterious genetic variation via genetic drift and purging. However, these same genetically depauperate populations will typically have lower fitness than larger, genetically diverse populations, on average, due to ever-increasing drift load (Figs. 1 and 2). The bottom line is that reduced fitness is generally expected in small, isolated, genetically depauperate populations, due to inbreeding depression and the accumulation of drift load, and that maintaining genetic variation and population connectivity will increase long-term viability.
Is Genome-Wide Genetic Variation Predictive of Adaptive Potential?
The ability of populations to adapt to changing environmental conditions (adaptive potential) is fundamental for persisting through environmental change (43, 44). A core insight from theoretical genetics is that adaptation requires additive genetic variance (Va) for the selected trait(s) (45). A lack of Va can limit a population’s response to selection and eventually lead to extinction (43, 44, 46). As with other types of genetic variation, Va is affected by mutation at loci affecting the trait, selection, migration, and genetic drift (47). We therefore expect, from first principles, that larger populations will have higher π and higher Va than small populations on average (Fig. 1) and thus that π should be correlated with Va. Despite strong theoretical support, determining the strength and importance of this relationship in real populations, especially those of conservation concern, has generated longstanding controversy (48).
Basic population genetic theory shows that population size and connectivity play major roles in determining Va and thus adaptive potential. Isolated populations below a certain size should lose Va due to genetic drift more rapidly than it is replenished via mutation (47). Additionally, recently bottlenecked populations that have lost π will eventually also lose Va and evolutionary potential in the absence of immigration (Fig. 2). However, while the eventual reduction in Va in small populations is inevitable, the initial effects of a bottleneck on Va can be complex. Recently bottlenecked populations may show decreases, stability, or even short-term increases in Va due to the conversion of dominant or epistatic variance into Va as allele frequencies change due to genetic drift (49–51). This potential conversion of nonadditive to additive variation in bottlenecked populations is highly stochastic across traits and populations, and is one of the processes that can cloud the relationship between molecular and quantitative trait variation (52). Nonetheless, the two important takeaways are 1) although bottlenecks can complicate the prediction of declining Va for any given trait in small populations, Va will be reduced on average, especially for traits with primarily additive inheritance; and 2) eventually, the inexorable decline in π in very small populations means that all small populations will eventually lose Va and their ability to adapt to environmental change. Adaptive potential in such populations will be severely limited unless Va is replenished by new mutations or migration from differentiated populations (35) (Fig. 2).
The hypothesis that small populations harbor less Va has been tested empirically in both laboratory and field settings. Most experimental studies show declines in Va and weaker responses to selection in small populations or following bottlenecks (53–55). On the other hand, field studies often find a weak association between Va and genome-wide genetic variation when comparing across populations (48, 56); this weak relationship is likely due to a combination of factors, none of which refute the two takeaways described above.
As discussed above, empirical results suggest that Va may initially increase after a bottleneck due to the conversion of epistatic and dominance variance to Va (50, 57), and then decline after substantial inbreeding accumulates. Further, Va is expected to vary among traits and populations depending on genetic architecture, mutation rate, and the mode and history of selection. In practice, most studies are unable to account for these factors and are generally only able to assess a few traits per species/population. Estimates of Va for each trait are also typically based on a modest number of families. Although the number of traits, populations, and species studied has increased, determining the total Va for fitness in a given population of conservation concern is not an attainable goal. Additionally, the vast majority of the best-characterized species with respect to Va in the wild (i.e., most of the species included in refs. 48 and 56 metaanalyses) are common. The species and populations in which the relationship between Va and genetic variation is expected to be strongest, namely, declining species of conservation concern, tend to be the most difficult to characterize.
Arguably the most important point is that the loss of genetic variation in small and/or bottlenecked populations is inevitable and will eventually lead to reduced Va and reduced adaptive potential, regardless of short-term and stochastic outcomes. Isolated populations that remain small are unlikely to recover substantial Va due to the slow rate of mutation and the counteracting loss of variation to genetic drift, and the lack of adaptive potential is problematic for long-term viability (43, 44, 47).
What Is the Relationship between Genome-Wide Genetic Variation and Population Viability?
The central question regarding the role of genetic variation in conservation is whether populations with lower are less likely to persist. Genetic effects on the persistence of a particular population are difficult to predict with certainty because there are many factors involved that are difficult to evaluate, including mating system and demographic history (32, 33), current and future environmental conditions (58), and the extent to which soft selection (where an individual’s fitness depends on its phenotype or genotype relative to others in the same population) versus hard selection (where an individual’s absolute fitness depends only on its phenotype or genotype and is independent of the phenotypes or genotypes of other individuals in the population) predominate (59, 60). Additionally, the highly stochastic demography of small populations, which is exacerbated by inbreeding depression (61), means that widely divergent outcomes can be expected across populations with the same environmental, demographic, and genetic starting conditions. However, theoretical empirical studies have yielded broadly applicable insights into the effects of genetic variation and inbreeding on population viability.
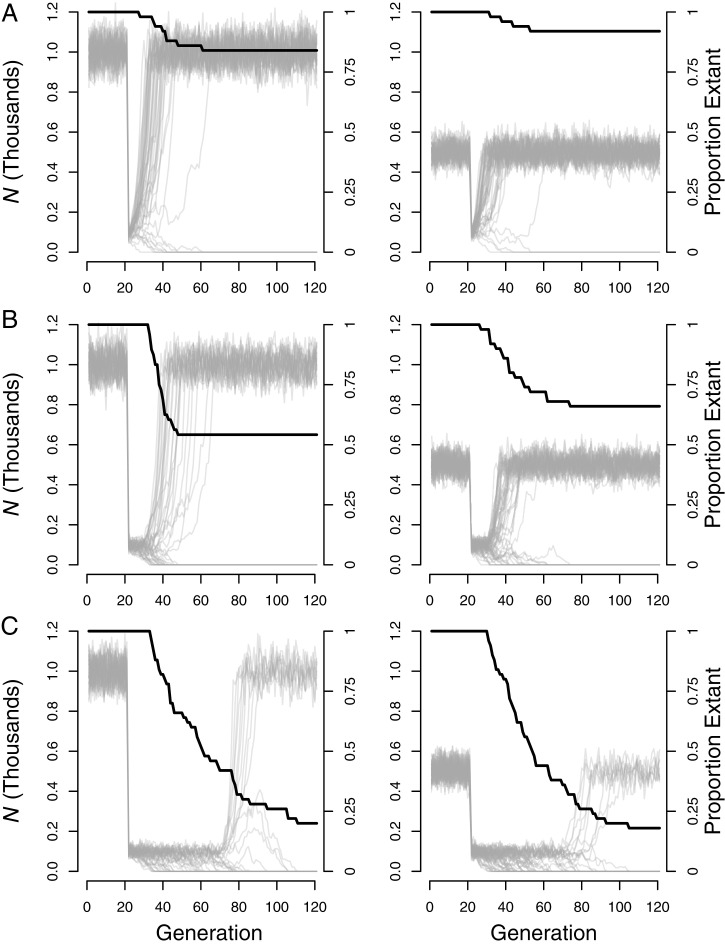
Population genetics theory predicts that small, isolated populations with low genetic variation are more likely to go extinct due to genetic effects than larger, more genetically diverse populations under empirically supported mutational assumptions (19, 22, 23, 62). De novo mutations following a bottleneck are expected to cause eventual extinction of very small, genetically depauperate populations via mutational meltdown (extinction of a population due to the synergistic interactions of population decline, genetic drift, and the accumulation of deleterious alleles) (SI Appendix, Fig. S1) (19). The average time to extinction is shorter under the more realistic scenario where bottlenecked populations carry deleterious mutations at the outset (Fig. 3). However, the extinction rate depends strongly on bottleneck duration, with longer restrictions conferring increased extinction due to both demographic stochasticity and the constant increase in drift load. Short-lived bottlenecks are one scenario where viability may sometimes be higher for historically smaller, less genetically diverse populations that have fewer deleterious alleles at the outset of the bottleneck due to historical genetic drift and purging (Figs. 1 and 3 A and B). However, this assumes inbreeding depression is the only genetic challenge operating, and simultaneous selection caused by environmental change may reverse this relationship. Longer bottlenecks in isolated populations are expected to result in very high extinction rates due to mutational meltdown, regardless of the abundance of deleterious alleles at the outset (19) (Fig. 3C).
Fig. 3.
Population viability during bottlenecks from carrying capacity K = 1,000 (Left) and K = 500 (Right) to K = 100. The bottlenecks were 2 (A), 10 (B), and 50 (C) generations in length. The black line shows the proportion of extant populations. Gray lines show population size for each of 50 replicate simulations in each scenario.
Empirical studies of population dynamics arguably provide the strongest evidence for the broad benefits of increased genetic variation for population viability. Numerous studies have almost universally found that populations with higher genetic variation have increased population growth and viability (63). For example, lower genetic variation was associated with reduced population growth in Alpine ibex (3) and increased local extinction in Glanville fritillary butterflies (2). Inbred laboratory lines of animals, which quickly lose genetic variation, often become extinct substantially more rapidly than control lines (64, 65). Additionally, the infusion of genetic variation via natural (66) and facilitated immigration (genetic rescue) nearly always increases population growth (35, 36, 67, 68) either by masking of deleterious recessive alleles or by infusing adaptive genetic variation.
The collapse of the Isle Royale wolf population after a mainland male immigrated to the small population has been interpreted as a counterexample to the efficacy of genetic rescue (8). However, detailed documentation indicates that results from this unusual system are unsuitable as a general example of the likely demographic outcome of genetic rescue attempts (67, 69, 70). The immigration of only a single male into Isle Royale makes it unusual in the context of managed genetic rescue attempts, which typically involve translocation of multiple individuals into a small population (e.g., refs. 71–73). The single migrant male wolf dominated and increased reproduction, resulting in genetic rescue. However, his extremely high reproduction resulted in very high inbreeding within two generations and the subsequent dramatic population decline (67, 69, 70). This male was likely just an opportunistic, successful migrant from the nearest population. It is unclear whether he carried an exceptional number of deleterious alleles that drove the subsequent decline, or whether inbreeding following exceptionally high reproduction of any individual would have led to a similar demographic outcome.
Recovery of some populations from severe bottlenecks, and persistence of some populations despite small Ne and low genetic variation, are often cited as a challenge to the idea that low genetic variation and inbreeding reduce population viability (6, 8, 9, 11, 74–77). Soulé (ref. 5, p. 178) pointed out the fundamental flaw of this argument, which he referred to as the “fallacy of the accident” nearly 35 y ago: The only observable populations that have experienced bottlenecks are those that survived. The potentially numerous populations that went extinct under similar conditions are unobservable. Counting extant, genetically depauperate populations is therefore an unreliable metric of the extinction risk posed by lost genetic variation and inbreeding. Theoretical population genetics and population ecology both predict that some populations will survive bottlenecks, and some lucky ones will persist for long periods at small population size. However, such cases are likely the rare exception, the lottery winners, so to speak (5, 67).
The most immediate threats to small, genetically depauperate populations are demographic stochasticity and inbreeding depression. However, long-term population persistence will, in most cases, require populations to adapt to environmental change (climate change, novel diseases, invasive species, etc.) (44, 78). Rapid adaptation to new conditions is possible but requires sufficient genetic variation and relatively large population size (53, 79). All of the material above highlights the fundamental importance of maintaining large, connected, genetically diverse populations. Long-term population viability requires having both manageable genetic load (the reduction in fitness due to effects of both segregating and fixed deleterious alleles) and adaptive potential associated with genome-wide genetic variation.
Simulation-Based Inferences of the Effects of Genetic Variation and Inbreeding on Population Viability
Simulation-based studies showed, long ago, that inbreeding depression can substantially increase extinction risk (23, 80). However, our increasing understanding of deleterious mutation parameters (e.g., deleterious mutation rates, and the distribution of fitness effects [DFE]) combined with the availability of sophisticated, user-friendly simulation software (81) will likely advance our understanding of inbreeding depression and purging within the field of conservation.
While there is much to learn about deleterious mutation parameters, a lot is known about the most important elements. First, deleterious mutations arise frequently (15, 16, 82–84), and large-effect deleterious alleles appear to be a major driver of inbreeding depression (14, 85–87). For example, lethal alleles arose via mutation at a rate of ∼3% per diploid genome in Drosophila (14). Inbreeding depression appeared to be largely due to highly deleterious alleles originating in a subset of pedigree founders in sheep and mice (86, 87). Lethal and other large-effect deleterious alleles are frequently observed in small natural populations, humans, and model organisms (14, 83, 85, 88–90). The majority of humans and Drosophila likely carry one or more recessive lethal alleles (85, 89, 90). Deleterious mutations appeared at a rate of U = 1.2 per diploid genome per generation in Drosophila (15) and U = 1.6 in hominids (16). Mutation accumulation studies show that the DFE for deleterious mutations is strongly bimodal, with most mutations having small to moderate effects (e.g., |s| < 0.25) and a minority being lethal or semilethal (82).
Second, the degree of dominance (h) is strongly related to mutation effect size. Direct observation of dominance effects in yeast and Drosophila suggest that nearly neutral deleterious mutations are slightly recessive on average (h slightly less than 0.5), and highly deleterious mutations (e.g., |s| > 0.25) are nearly fully recessive (h very near zero), with h declining exponentially as s increases in size (14, 91, 92). There is still much uncertainty regarding deleterious mutation parameters (see discussion below). However, the best available information suggests that reasonable values of U are >1, the DFE is strongly bimodal, and dominance declines substantially with increasing size of s. These findings guide the simulations presented above (details in SI Appendix).
Recently, results from genetically explicit simulations were used to argue that genome-wide genetic variation is of little importance to population viability, and that purging is likely to prevent extinction (8, 11, 74). However, these studies excluded large-effect deleterious mutations (SI Appendix, Fig. S2) and assumed values of U that were between 2.6 and 92.3 times lower than the best estimate of U in Drosophila (Table 1). As a result, these models (8, 11, 74) produce substantially weaker inbreeding depression (<0.05 to ∼1 lethal equivalent) than observed in real populations, where the median number of lethal equivalents for juvenile survival in captive mammals was 3.1 (93), and 12 for total fitness in wild mammals (23) (SI Appendix, Fig. S3). There is substantial uncertainty in deleterious mutation rates, and the DFE, particularly for nonmodel organisms. However, the discrepancy between the assumed mutation parameters and the resulting inbreeding depression in the aforementioned studies (8, 11, 74) and the best available empirical estimates (Table 1 and SI Appendix, Fig. S3) yield results that underestimate the importance of genetic variation in conservation, and the efficacy of genetic rescue as a tool in conservation.
Table 1.
Deleterious mutation rates used in previous simulation-based analyses of inbreeding depression and genetic rescue
| Study | Mutation target size | Mutation rate | Proportion deleterious | U * | UDrosophila/U† |
| Teixeira and Huber (11) | 1,000 exons | 1 × 10−5/exon/generation | 0.66 | 0.013 | 92.3 |
| Robinson et al. (74) | 2,000 genes × 1,000 bp | 1 × 10−8/bp/generation | 0.7 | 0.028 | 42.9 |
| Kyriazis et al. (8) | 20,000 genes × 1,500 bp | 1 × 10−8/bp/generation | 0.77 | 0.462 | 2.6 |
*U is calculated as 2 × mutation target size × mutation rate × proportion of mutations that are deleterious.
†UDrosophila is the deleterious mutation rate per diploid genome, UDrosophila =1.2 (15).
Is the Relationship between Genetic Variation and Conservation Status Informative of the Importance of Genetic Variation for Population Viability?
It has been suggested that a weak relationship between genetic variation and conservation status (e.g., International Union for Conservation of Nature [IUCN] Red List) means that genome-wide genetic variation is uninformative of extinction risk (11). However, this relationship is not universally expected, even though extinction risk is strongly affected by genome-wide genetic variation.
First, a lag is expected between reduced population size and the loss of genetic variation. Most threatened populations initially decline due to nongenetic factors (e.g., habitat loss, disease, climate change). Thus, multiple generations are required for a substantial reduction in genetic variation, even after severe bottlenecks (Fig. 2A). Threatened populations that became small due to nongenetic factors may still have high genetic variation due to this lag. Second, failing to control for other factors that influence genetic variation [e.g., Ne, dispersal, generation time, and mutation rate (11)] can obscure the relationship between genetic variation and conservation status. In contrast, a study controlling for phylogeny (a proxy for the aforementioned confounding factors) showed a significant relationship between genetic variation and conservation status (94).
Differences among studies in the measures of genetic variation can further obscure true relationships between genetic variation and conservation status. Estimates of genetic variation for different species used in comparative studies vary widely in the number of sampled individuals and populations, and in the regions of the genome analyzed. Some studies estimate species-wide genetic diversity from a single individual (11, 95, 96) and compare different genetic data types across species (6, 96). Using single genomes to estimate species-wide genetic diversity is problematic because the individuals chosen may not be representative of the species as a whole [e.g., captive individuals (95)]. Rather, multiple individuals and populations are necessary to accurately reflect a species’ distribution of genetic variation (97, 98). Additionally, estimates of genetic diversity are affected by reference genome quality (99), mapping bias (100, 101), the methods used to measure genetic variation (e.g., whole genome sequencing, reduced representation sequencing, RNA sequencing), and bioinformatics approaches (98, 99). Thus, sampling, genetic markers, and analyses should be standardized when measuring the relationship between genetic variation and conservation status.
Lastly, IUCN Red List status is an imperfect index of extinction risk, because it is a subjective measure of population viability. The IUCN Red List is important for monitoring biodiversity, but the guidelines used to categorize threat levels within the Red List are subject to user interpretation, which can lead to inconsistent assessments (102–106). The imperfect relationship between IUCN Red List status and extinction risk means that Red List status is an inappropriate surrogate for extinction risk in assessing the relationship between genome-wide diversity and extinction risk. Together, these issues suggest that the weak relationship between genetic variation and conservation status has little bearing on the importance of genome-wide genetic variation for extinction risk.
What Is the Role of Functional Genetic Variation in Conservation?
The widespread availability of genomic data for nonmodel organisms has rapidly advanced our understanding of the genetic basis and evolution of fitness-related traits in natural populations (e.g. refs. 107–111). This revolution has raised the question of how to effectively integrate functional genetic information into conservation practice (112–115). It has repeatedly been suggested that genetic assessment and management of threatened populations should be focused on variation at particular loci that affect particular fitness traits (11, 116–118). However, such gene-targeted conservation approaches are always difficult, and are prone to failure for several reasons.
First, understanding the genetic basis of fitness remains extremely complicated and challenging (112, 114). While some important traits in natural populations are affected by loci with very large effects, most traits are determined by many small-effect loci (119–121). A comprehensive understanding of the genetic basis of such traits is out of reach for nonmodel organisms (122). To accurately understand the locus-specific effects on a trait and fitness requires information on dominance and pleiotropy, epistasis, genotype-by-environment interactions, and the amount of linkage disequilibrium with other loci influencing the trait or other fitness components (112). These factors are expected to vary among traits and to differ for the same trait among species and potentially among populations within a species (e.g., ref. 107). Therefore, substantial effort is necessary to understand the conservation relevance of a particular genetic variant and predict whether the benefits of gene-targeted conservation actions outweigh potential detrimental effects (112, 114).
A classic example of the potential for undesirable outcomes of gene-targeted conservation management is the suggestion that genetic management of captive and wild populations should be designed around maintaining genetic variation at the major histocompatibility complex (MHC) (11, 116, 117, 123). The MHC has been studied in great detail in humans because of its importance in immunity, organ transplantation, and autoimmune disease, but its organization is poorly understood in most other vertebrates. Although there is strong evidence for its adaptive importance, some variants have detrimental effects, and the adaptive effects of other variants appear to be environmentally dependent (124). Detailed examination of the fitness effects of MHC alleles and haplotypes is necessary to determine how much maintaining MHC variation enhances fitness.
Additionally, as highlighted multiple times over the last 35 y (112, 125–129), basing conservation management on a small subset of loci risks increasing the loss of genetic variation elsewhere in the genome. Such efforts would be counterproductive unless the gain in mean fitness associated with gene-targeted management is greater than the loss in fitness associated with lost genome-wide genetic variation (112). This highlights the challenges and pitfalls of gene-targeted conservation. When recommendations for maintaining genome-wide genetic variation versus particular adaptive variants are in conflict, a cost–benefit analysis of the two approaches should be performed and a composite solution identified (112). Recent cases where genomic analyses have revealed that large-effect loci play a key role in traits of conservation importance (e.g., refs. 107, 108, 110, and 130) will be the first to empirically test the efficacy of gene-targeted conservation approaches.
Discussion
Genomic data should be used to challenge findings from population genetics theory and previous empirical data that form the basis for genetic management of small populations. Recent genomic studies provide useful fodder to determine how to effectively use genomic data to improve conservation in ways that were previously impossible. Examples are emerging of how understanding functional genetic variation could improve recommendations to conserve imperiled populations (107, 108, 110, 130), making genomic data more useful for conservation than ever before. However, genomic data have not discredited the decades’ worth of evidence that inbreeding depression, mutational meltdown, and loss of adaptive potential are major threats to conservation.
Identifying genetic variants that affect fitness traits undoubtedly advances understanding of the genetic basis of adaptation, and that is important in itself (131). However, placing conservation priority on a small, apparently adaptive portion of the genome ignores what may be the vast majority of variation elsewhere in the genome that will fuel adaptation to unpredictable future conditions (112, 114, 125, 126). This approach is reminiscent of the “adaptationist programme” that Gould and Lewontin (132) criticized >40 y ago for being overly enamored with adaptive explanations for interesting traits (“spandrels”) without considering that they might have arisen by accident, and that they are but one part of the whole, complex organism (114). Now, as then, we should avoid the temptation to place undue priority on putatively adaptive loci [“molecular spandrels” (133)] without first considering the rest of the genome. Our inability to predict future changes in genotype-by-environment interactions should lead us to recognize the importance of genome-wide genetic variation (including presently neutral variation), and, more importantly, the factors that make it possible—large livable habitats and natural patterns of connectivity among them. Conserving genetic variation across the whole genome is almost certainly the most reliable approach to conserve the genetic variation that matters.
We know of no convincing evidence that supports abandoning the focus on genome-wide genetic variation in exchange for a focus on functional variation. The recent simulation studies that have been used to discount the importance of genome-wide genetic variation in conservation (8, 11, 74) are based on assumptions that are inconsistent with the preponderance of empirical data on the genetics of inbreeding depression and its effect on population viability (see above). Some small populations may not suffer strong inbreeding depression, and some may not rebound following the introduction of genetic variation. However, as pointed out in the formative years of conservation biology, we must resist the temptation to dismiss the extinction risks associated with lost genetic variation in small populations (5).
Although population genetics theory has done a remarkably good job of predicting patterns now observable in genomic data, many questions remain unanswered that will improve the utility of genomic data in conservation. For example, how prevalent is soft selection? The presence of soft selection could help explain some of the instances where populations persist for long periods despite inbreeding (59, 60). How much do U and the DFEs for deleterious mutations vary among taxa? U may be rather consistent within some taxonomic groups (e.g., mammals) where the number of genes is strongly conserved (134). Nevertheless, variation among taxa in gene number, mutation rate, and the amount of intergenic DNA that is subject to deleterious mutation is an important consideration for assessing the fitness effects of inbreeding. Lastly, while it is clear that the distribution of mutation fitness effects is bimodal (82), understanding the specific shape of this distribution, and how much this varies among taxa, is important for our understanding of the extinction risks associated with small population size and inbreeding.
Genomic data will undoubtedly continue to be used to revisit and refine insights gained since genetics was first applied to conservation and to understand the extinction process (4, 5, 46, 135). So far, genomic data have reinforced earlier findings showing that genome-wide genetic variation is key to population viability. Given the increasing rate of habitat loss and fragmentation, failing to recognize and mitigate the effects of lost genome-wide genetic variation would only exacerbate the biodiversity crisis.
Supplementary Material
Acknowledgments
We thank P. Hohenlohe, R. Waples, A. García-Dorado, and three anonymous reviewers for comments on previous drafts. S.W.F. was supported by NSF Grant DEB 2016569. D.A.T. was supported by National Institute of General Medical Sciences of the NIH under Award RL5GM118990. W.C.F. was supported by NSF Grants DEB 1413925, DEB 1754821, and DEB 1838282. S.H. was supported by University of Wisconsin–Milwaukee College of Letters and Science.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104642118/-/DCSupplemental.
Data Availability
All data for this study are included in the article and/or in the SI Appendix. Simulation computer codes are available in Zenodo at https://doi.org/10.5281/zenodo.5513957.
References
- 1.Lande R., Shannon S., The role of genetic variation in adaptation and population persistence in a changing environment. Evolution 50, 434–437 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Saccheri I., et al. , Inbreeding and extinction in a butterfly metapopulation. Nature 392, 491–494 (1998). [Google Scholar]
- 3.Bozzuto C., Biebach I., Muff S., Ives A. R., Keller L. F., Inbreeding reduces long-term growth of Alpine ibex populations. Nat. Ecol. Evol. 3, 1359–1364 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Frankel O., Soulé M. E., Conservation and Evolution (CUP Archive, 1981). [Google Scholar]
- 5.Soulé M. E., Viable Populations for Conservation (Cambridge University Press, 1987). [Google Scholar]
- 6.Robinson J. A., et al. , Genomic flatlining in the endangered island fox. Curr. Biol. 26, 1183–1189 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Robinson J. A., et al. , Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci. Adv. 5, eaau0757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyriazis C. C., Wayne R. K., Lohmueller K. E., Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett. 5, 33–47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin P. A., et al. , Reference genome and demographic history of the most endangered marine mammal, the vaquita. Mol. Ecol. Resour. 21, 1108–1020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westbury M. V., et al. , Hyena paleogenomes reveal a complex evolutionary history of cross-continental gene flow between spotted and cave hyena. Sci. Adv. 6, eaay0456 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teixeira J. C., Huber C. D., The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. U.S.A. 118, e2015096118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue Y., et al. , Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348, 242–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlesworth D., Willis J. H., The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Simmons M. J., Crow J. F., Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11, 49–78 (1977). [DOI] [PubMed] [Google Scholar]
- 15.Haag-Liautard C., et al. , Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445, 82–85 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Eyre-Walker A., Keightley P. D., High genomic deleterious mutation rates in hominids. Nature 397, 344–347 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Lynch M., Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. U.S.A. 107, 961–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldane J., The effect of variation of fitness. Am. Nat. 71, 337–349 (1937). [Google Scholar]
- 19.Lynch M., Conery J., Burger R., Mutation accumulation and the extinction of small populations. Am. Nat. 146, 489–518 (1995). [Google Scholar]
- 20.Keller L. F., Waller D. M., Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (2002). [Google Scholar]
- 21.Frankham R., Genetics and extinction. Biol. Conserv. 126, 131–140 (2005). [Google Scholar]
- 22.Mills L. S., Smouse P. E., Demographic consequences of inbreeding in remnant populations. Am. Nat. 144, 412–431 (1994). [Google Scholar]
- 23.O’Grady J. J., et al. , Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol. Conserv. 133, 42–51 (2006). [Google Scholar]
- 24.Wright S., Evolution in Mendelian populations. Genetics 16, 97–159 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell J. E., Visscher P. M., Goddard M. E., Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 11, 800–805 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Speed D., Balding D. J., Relatedness in the post-genomic era: Is it still useful? Nat. Rev. Genet. 16, 33–44 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Jacquard A., Inbreeding: One word, several meanings. Theor. Popul. Biol. 7, 338–363 (1975). [DOI] [PubMed] [Google Scholar]
- 28.Malécot G., The Mathematics of Heredity (W. H. Freeman, 1970). [Google Scholar]
- 29.Kardos M., et al. , Genomic consequences of intensive inbreeding in an isolated wolf population. Nat. Ecol. Evol. 2, 124–131 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Hedrick P. W., Garcia-Dorado A., Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol. Evol. 31, 940–952 (2016). [DOI] [PubMed] [Google Scholar]
- 31.García-Dorado A., Understanding and predicting the fitness decline of shrunk populations: Inbreeding, purging, mutation, and standard selection. Genetics 190, 1461–1476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitlock M. C., Selection, load and inbreeding depression in a large metapopulation. Genetics 160, 1191–1202 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlesworth D., Charlesworth B., Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18, 237–268 (1987). [Google Scholar]
- 34.Morton N. E., Crow J. F., Muller H. J., An estimate of the mutational damage in man from data on consanguineous marriages. Proc. Natl. Acad. Sci. U.S.A. 42, 855–863 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteley A. R., Fitzpatrick S. W., Funk W. C., Tallmon D. A., Genetic rescue to the rescue. Trends Ecol. Evol. 30, 42–49 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Frankham R., Genetic rescue of small inbred populations: Meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 24, 2610–2618 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Frankham R., Gilligan D. M., Morris D., Briscoe D. A., Inbreeding and extinction: Effects of purging. Conserv. Genet. 2, 279–284 (2001). [Google Scholar]
- 38.Lacy R. C., Ballou J. D., Effectiveness of selection in reducing the genetic load in populations of Peromyscus polionotus during generations of inbreeding. Evolution 52, 900–909 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Crnokrak P., Barrett S. C., Perspective: Purging the genetic load: A review of the experimental evidence. Evolution 56, 2347–2358 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Grossen C., Guillaume F., Keller L. F., Croll D., Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nat. Commun. 11, 1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathur S., DeWoody J. A., Genetic load has potential in large populations but is realized in small inbred populations. Evol. Appl. 14, 1540–1557 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan A., et al. , Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2023018118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch M., Lande R., “Evolution and extinction in response to environmental change” inBiotic Interactions and Global Change, Kareiva P. M., Kingsolver J. G., Huey R. B., Eds. (Sinauer, Sunderland, MA, 1993), pp. 234–250. [Google Scholar]
- 44.Kardos M., Luikart G., The genomic architecture of fitness is a major driver of population viability during rapid environmental change. Am. Nat. 197, 511–525 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Falconer D. S., Mackay T. F. C., Introduction to Quantitative Genetics (Pearson, ed. 4, 1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bürger R., Lynch M., Evolution and extinction in a changing environment: A quantitative‐genetic analysis. Evolution 49, 151–163 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Lande R., Barrowclough G., “Effective population size, genetic variation, and their use in population management” in Viable Populations for Conservation, Soulé M., Ed. (Cambridge University Press, Cambridge, Unitd Kingdom, 1987), chap. 6, pp. 87–123. [Google Scholar]
- 48.Reed D. H., Frankham R., How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55, 1095–1103 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Goodnight C. J., Epistasis and the effect of founder events on the additive genetic variance. Evolution 42, 441–454 (1988). [DOI] [PubMed] [Google Scholar]
- 50.Bryant E. H., McCommas S. A., Combs L. M., The effect of an experimental bottleneck upon quantitative genetic variation in the housefly. Genetics 114, 1191–1211 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodnight C. J., On the effect of founder events on epistatic genetic variance. Evolution 41, 80–91 (1987). [DOI] [PubMed] [Google Scholar]
- 52.Barton N. H., Turelli M., Effects of genetic drift on variance components under a general model of epistasis. Evolution 58, 2111–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Frankham R., et al. , Do population size bottlenecks reduce evolutionary potential? Anim. Conserv. 2, 255–260 (1999). [Google Scholar]
- 54.Willi Y., Van Buskirk J., Hoffmann A. A., Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433–458 (2006). [Google Scholar]
- 55.de Villemereuil P., et al. , Little adaptive potential in a threatened passerine bird. Curr. Biol. 29, 889–894.e883 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Mittell E. A., Nakagawa S., Hadfield J. D., Are molecular markers useful predictors of adaptive potential? Ecol. Lett. 18, 772–778 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Lacy R. C., Malo A. F., Alaks G., Maintenance of genetic variation in quantitative traits of a woodland rodent during generations of captive breeding. Conserv. Genet. 19, 789–802 (2018). [Google Scholar]
- 58.Meagher S., Penn D. J., Potts W. K., Male-male competition magnifies inbreeding depression in wild house mice. Proc. Natl. Acad. Sci. U.S.A. 97, 3324–3329 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agrawal A. F., Ecological determinants of mutation load and inbreeding depression in subdivided populations. Am. Nat. 176, 111–122 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Bell D. A., Kovach R. P., Robinson Z. L., Whiteley A. R., Reed T. E., The ecological causes and consequences of hard and soft selection. Ecol. Lett. 24, 1505–1521 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Goodman D., “The demography of chance extinction” in Viable Populations for Conservation, Soulé M. E., Ed. (Cambridge University Press, Cambridge, UK, 1987), pp. 11–34. [Google Scholar]
- 62.Lande R., Risk of population extinction from fixation of new deleterious mutations. Evolution 48, 1460–1469 (1994). [DOI] [PubMed] [Google Scholar]
- 63.Lacy R. C., Importance of genetic variation to the viability of mammalian populations. J. Mammal. 78, 320–335 (1997). [Google Scholar]
- 64.Bowman J., Falconer D., Inbreeding depression and heterosis of litter size in mice. Genet. Res. 1, 262–274 (1960). [Google Scholar]
- 65.Wright L. I., Tregenza T., Hosken D. J., Inbreeding, inbreeding depression and extinction. Conserv. Genet. 9, 833 (2008). [Google Scholar]
- 66.Åkesson M., et al. , Genetic rescue in a severely inbred wolf population. Mol. Ecol. 25, 4745–4756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ralls K., Sunnucks P., Lacy R. C., Frankham R., Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 251, 108784 (2020). [Google Scholar]
- 68.Fitzpatrick S. W., et al. , Genomic and fitness consequences of genetic rescue in wild populations. Curr. Biol. 30, 517–522.e515 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Hedrick P., Robinson J., Peterson R. O., Vucetich J. A., Genetics and extinction and the example of Isle Royale wolves. Anim. Conserv. 22, 302–309 (2019). [Google Scholar]
- 70.Hedrick P. W., Peterson R. O., Vucetich L. M., Adams J. R., Vucetich J. A., Genetic rescue in Isle Royale wolves: Genetic analysis and the collapse of the population. Conserv. Genet. 15, 1111–1121 (2014). [Google Scholar]
- 71.Hogg J. T., Forbes S. H., Steele B. M., Luikart G., Genetic rescue of an insular population of large mammals. Proc. Biol. Sci. 273, 1491–1499 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson W. E., et al. , Genetic restoration of the Florida panther. Science 329, 1641–1645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westemeier R. L., et al. , Tracking the long-term decline and recovery of an isolated population. Science 282, 1695–1698 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Robinson J. A., Brown C., Kim B. Y., Lohmueller K. E., Wayne R. K., Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr. Biol. 28, 3487–3494.e3484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caro T. M., Laurenson M. K., Ecological and genetic factors in conservation: A cautionary tale. Science 263, 485–486 (1994). [DOI] [PubMed] [Google Scholar]
- 76.Simberloff D., The contribution of population and community biology to conservation science. Annu. Rev. Ecol. Syst. 19, 473–511 (1988). [Google Scholar]
- 77.Harcourt A., Population viability estimates: Theory and practice for a wild gorilla population. Conserv. Biol. 9, 134–142 (1995). [Google Scholar]
- 78.Frankel O. H., Genetic conservation: Our evolutionary responsibility. Genetics 78, 53–65 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ørsted M., Hoffmann A. A., Sverrisdóttir E., Nielsen K. L., Kristensen T. N., Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLoS Genet. 15, e1008205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lacy R. C., VORTEX: A computer simulation model for population viability analysis. Wildl. Res. 20, 45–65 (1993). [Google Scholar]
- 81.Haller B. C., Messer P. W., SLiM 2: Flexible, interactive forward genetic simulations. Mol. Biol. Evol. 34, 230–240 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Eyre-Walker A., Keightley P. D., The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8, 610–618 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Trask A. E., et al. , Evidence of the phenotypic expression of a lethal recessive allele under inbreeding in a wild population of conservation concern. J. Anim. Ecol. 85, 879–891 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Ralls K., Ballou J. D., Rideout B. A., Frankham R., Genetic management of chondrodystrophy in California condors. Anim. Conserv. 3, 145–153 (2000). [Google Scholar]
- 85.Ballinger M. A., Noor M. A. F., Are lethal alleles too abundant in humans? Trends Genet. 34, 87–89 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Lacy R. C., Alaks G., Walsh A., Hierarchical analysis of inbreeding depression in Peromyscus polionotus. Evolution 50, 2187–2200 (1996). [DOI] [PubMed] [Google Scholar]
- 87.Casellas J., Piedrafita J., Caja G., Varona L., Analysis of founder-specific inbreeding depression on birth weight in Ripollesa lambs. J. Anim. Sci. 87, 72–79 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Ralls K., Ballou J. D., Genetic status and management of California condors. Condor 106, 215–228 (2004). [Google Scholar]
- 89.Gao Z., Waggoner D., Stephens M., Ober C., Przeworski M., An estimate of the average number of recessive lethal mutations carried by humans. Genetics 199, 1243–1254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Narasimhan V. M., et al. , Health and population effects of rare gene knockouts in adult humans with related parents. Science 352, 474–477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agrawal A. F., Whitlock M. C., Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187, 553–566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deng H.-W., Lynch M., Estimation of deleterious-mutation parameters in natural populations. Genetics 144, 349–360 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ralls K., Ballou J. D., Templeton A., Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv. Biol. 2, 185–193 (1988). [Google Scholar]
- 94.Spielman D., Brook B. W., Frankham R., Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U.S.A. 101, 15261–15264 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Consortium Z.; Zoonomia Consortium, A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corbett-Detig R. B., Hartl D. L., Sackton T. B., Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13, e1002112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pečnerová P., et al. , High genetic diversity and low differentiation reflect the ecological versatility of the African leopard. Curr. Biol. 31, 1862–1871.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Armstrong E. E., et al. , Long live the king: Chromosome-level assembly of the lion (Panthera leo) using linked-read, Hi-C, and long-read data. BMC Biol. 18, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.A. Prasad, E. D. Lorenzen, M. V. Westbury, Evaluating the role of reference-genome phylogenetic distance on evolutionary inference. Mol. Ecol. Res., 10.1111/1755-0998.13457 (2021). [DOI] [PubMed]
- 100.Brandt D. Y., et al. , Mapping bias overestimates reference allele frequencies at the HLA genes in the 1000 genomes project phase I data. G3: Genes, Genomes. G3 (Bethesda) 5, 931–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gopalakrishnan S., et al. , The wolf reference genome sequence (Canis lupus lupus) and its implications for Canis spp. population genomics. BMC Genomics 18, 495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clark J. A., May R. M., Taxonomic bias in conservation research. Science 297, 191–192 (2002). [DOI] [PubMed] [Google Scholar]
- 103.Cardoso P., Borges P. A., Triantis K. A., Ferrández M. A., Martín J. L., Adapting the IUCN Red List criteria for invertebrates. Biol. Conserv. 144, 2432–2440 (2011). [Google Scholar]
- 104.Akçakaya H. R., Butchart S. H., Mace G. M., Stuart S. N., Hilton‐Taylor C., Use and misuse of the IUCN Red List Criteria in projecting climate change impacts on biodiversity. Glob. Change Biol. 12, 2037–2043 (2006). [Google Scholar]
- 105.Trull N., Böhm M., Carr J., Patterns and biases of climate change threats in the IUCN Red List. Conserv. Biol. 32, 135–147 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Hayward M. W., et al. , Ambiguity in guideline definitions introduces assessor bias and influences consistency in IUCN Red List status assessments. Front. Ecol. Evol. 3, 10.3389/fevo.2015.00087 (2015). [Google Scholar]
- 107.Thompson N. F., et al. , A complex phenotype in salmon controlled by a simple change in migratory timing. Science 370, 609–613 (2020). [DOI] [PubMed] [Google Scholar]
- 108.Barson N. J., et al. , Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 528, 405–408 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Pearse D. E., et al. , Sex-dependent dominance maintains migration supergene in rainbow trout. Nat. Ecol. Evol. 3, 1731–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Epstein B., et al. , Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat. Commun. 7, 12684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Küpper C., et al. , A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48, 79–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kardos M., Shafer A. B. A., The peril of gene-targeted conservation. Trends Ecol. Evol. 33, 827–839 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Shafer A. B., et al. , Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 30, 78–87 (2015). [DOI] [PubMed] [Google Scholar]
- 114.Pearse D. E., Saving the spandrels? Adaptive genomic variation in conservation and fisheries management. J. Fish Biol. 89, 2697–2716 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Funk W., Forester B. R., Converse S. J., Darst C., Morey S., Improving conservation policy with genomics: A guide to integrating adaptive potential into US Endangered Species Act decisions for conservation practitioners and geneticists. Conserv. Genet. 20, 115–134 (2018). [Google Scholar]
- 116.Hughes A. L., MHC polymorphism and the design of captive breeding programs. Conserv. Biol. 5, 249–251 (1991). [Google Scholar]
- 117.Manlik O., et al. , Is MHC diversity a better marker for conservation than neutral genetic diversity? A case study of two contrasting dolphin populations. Ecol. Evol. 9, 6986–6998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laikre L., Hereditary defects and conservation genetic management of captive populations. Zoo Biol. 18, 81–99 (1999). [Google Scholar]
- 119.Boyle E. A., Li Y. I., Pritchard J. K., An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Manolio T. A., et al. , Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang J., et al. , Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kardos M., Husby A., McFarlane S. E., Qvarnström A., Ellegren H., Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Mol. Ecol. Resour. 16, 727–741 (2016). [DOI] [PubMed] [Google Scholar]
- 123.Oliver M. K., Piertney S. B., Selection maintains MHC diversity through a natural population bottleneck. Mol. Biol. Evol. 29, 1713–1720 (2012). [DOI] [PubMed] [Google Scholar]
- 124.Garrigan D., Hedrick P. W., Perspective: Detecting adaptive molecular polymorphism: Lessons from the MHC. Evolution 57, 1707–1722 (2003). [DOI] [PubMed] [Google Scholar]
- 125.Miller P. S., Hedrick P. W., MHC polymorphism and the design of captive breeding programs: Simple solutions are not the answer. Conserv. Biol. 5, 556–558 (1991). [Google Scholar]
- 126.Vrijenhoek R. C., Leberg P. L., Let’s not throw the baby out with the bathwater: A comment on management for MHC diversity in captive populations. Conserv. Biol. 5, 252–254 (1991). [Google Scholar]
- 127.Haig S. M., Ballou J. D., Derrickson S. R., Management options for preserving genetic diversity: Reintroduction of Guam rails to the wild. Conserv. Biol. 4, 290–300 (1990). [Google Scholar]
- 128.Hedrick P. W., Brussard P. F., Allendorf F. W., Beardmore J. A., Orzack S., Protein variation, fitness, and captive propagation. Zoo Biol. 5, 91–99 (1986). [Google Scholar]
- 129.Lacy R. C., Should we select genetic alleles in our conservation breeding programs? Zoo Biol. 19, 279–282 (2000). [Google Scholar]
- 130.Jones M. R., et al. , Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360, 1355–1358 (2018). [DOI] [PubMed] [Google Scholar]
- 131.Ellegren H., Sheldon B. C., Genetic basis of fitness differences in natural populations. Nature 452, 169–175 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Gould S. J., Lewontin R. C., The spandrels of San Marco and the Panglossian paradigm: A critique of the adaptationist programme. Proc. R. Soc. Lond. B Biol. Sci. 205, 581–598 (1979). [DOI] [PubMed] [Google Scholar]
- 133.Barrett R. D., Hoekstra H. E., Molecular spandrels: Tests of adaptation at the genetic level. Nat. Rev. Genet. 12, 767–780 (2011). [DOI] [PubMed] [Google Scholar]
- 134.Demuth J. P., De Bie T., Stajich J. E., Cristianini N., Hahn M. W., The evolution of mammalian gene families. PLoS One 1, e85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allendorf F. W., Genetics and the conservation of natural populations: Allozymes to genomes. Mol. Ecol. 26, 420–430 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for this study are included in the article and/or in the SI Appendix. Simulation computer codes are available in Zenodo at https://doi.org/10.5281/zenodo.5513957.