Significance
Cancer chemotherapeutic drugs that induce mitotic errors may cause tumor regression in part through the induction of interferon signaling. To test this idea, we measured the ability of antimitotic drugs with different mechanisms to activate the cGAS–STING–interferon pathway. Only microtubule stabilizers and MPS1 inhibitors activated cGAS, and this correlated with their ability to generate cGAS-coated chromatin bridges. We propose that chromatin bridges activate cGAS through a tension-dependent mechanism that depends on cytokinesis. Our results may explain the clinical failure of antimitotic drugs and help to design improved drugs.
Keywords: mitosis, cancer, cGAS, interferon, chemotherapy
Abstract
Mitotic errors can activate cyclic GMP–AMP synthase (cGAS) and induce type I interferon (IFN) signaling. Current models propose that chromosome segregation errors generate micronuclei whose rupture activates cGAS. We used a panel of antimitotic drugs to perturb mitosis in human fibroblasts and measured abnormal nuclear morphologies, cGAS localization, and IFN signaling in the subsequent interphase. Micronuclei consistently recruited cGAS without activating it. Instead, IFN signaling correlated with formation of cGAS-coated chromatin bridges that were selectively generated by microtubule stabilizers and MPS1 inhibitors. cGAS activation by chromatin bridges was suppressed by drugs that prevented cytokinesis. We confirmed cGAS activation by chromatin bridges in cancer lines that are unable to secrete IFN by measuring paracrine transfer of 2′3′-cGAMP to fibroblasts, and in mouse cells. We propose that cGAS is selectively activated by self-chromatin when it is stretched in chromatin bridges. Immunosurveillance of cells that fail mitosis, and antitumor actions of taxanes and MPS1 inhibitors, may depend on this effect.
Mitotic errors contribute to birth defects, aging, carcinogenesis, and cancer therapy. They occur at a low frequency in normal cells, a higher frequency in cancer cells, and a much higher frequency if mitosis occurs in the presence of chemotherapeutics (1, 2). The genetic consequences of mitotic errors include structural rearrangements such as chromothripsis (3) and numerical aberrations termed aneuploidy. A singular instance of structural or numerical defect can cause sustained genetic instability (4, 5). The cytological consequences of mitotic errors include micronuclei and chromatin bridges. Micronuclei exhibit abnormal nuclear transport and a high frequency of DNA damage in the subsequent cell cycle (6, 7). They are also prone to rupture, which exposes their chromatin to the cytoplasm (8). Chromatin bridges are caused by dicentric chromosomes, merotelic attachments, and catenations (9–11). They are typically resolved during anaphase (11) but can remain intact into the subsequent interphase when they become highly stretched due to tension from cell migration (10). Stretched chromatin bridges exhibit compromised nuclear envelopes, DNA damage, and ultimately break through actin-mediated traction forces or endonuclease activity (4, 10). Broken chromatin bridges retract into the primary nucleus or become encapsulated into micronuclei (12).
In addition to genetic and cytological consequences, mitotic errors induce inflammation and immunosurveillance through the activation of the viral DNA sensor cyclic GMP–AMP synthase (cGAS) (13, 14). Upon binding to DNA, cGAS synthesizes 2′3'-cyclic GMP–AMP (cGAMP) which in turn activates STING followed by TBK1 and IRF3, ultimately leading to induction of type I interferon (IFN) expression and secretion (15). cGAS was originally proposed to discriminate viral from self-DNA by cytoplasmic localization of viral DNA (16). Additional regulatory mechanisms have now been identified which include cGAS inhibition by nucleosomes, competition with BANF1, and posttranslational modifications (17–20). Species differences between mouse and human cGAS exist and include different sensitivities to DNA substrate length and propensity to form condensates with DNA (21, 22). cGAMP can move between cells to activate STING in a paracrine manner in cell culture (23) and in tumors (24). This may allow efficient signal propagation from cancer cells that have evolved blocks to IFN secretion. IFN activates adaptive immune responses, which makes cGAS activation an attractive therapeutic strategy to sensitize tumors to immune checkpoint inhibitors (25, 26).
Antimitotic drugs (A-Ms) are a class of cancer chemotherapeutics that perturb mitosis and greatly increase the frequency of mitotic errors (27). They are ideal tool compounds to investigate cGAS activation after mitotic failure because they induce chromosome missegregation in distinct manners that are independent of DNA damage. Taxanes are an important class of clinical A-Ms which stabilize microtubules (MTs) and induce solid-tumor regression. At saturating concentrations in cell culture, taxanes induce a prolonged mitotic arrest leading to cell death (28). Whether this mechanism is responsible for their tumor regression activity remains controversial (29, 30). Potent and specific A-Ms that target Aurora A kinase, Aurora B kinase, Polo-like kinase 1, and KIF11 were tested in cancer patients but found to lack tumor-regression activity for reasons that remain unclear (31, 32). We proposed that the special therapeutic activity of taxanes may depend on cGAS activation (33). Here, we test this idea by comparing the ability of different A-Ms to activate cGAS and correlating this with cytological defects. Unexpectedly, we found a key role for chromatin bridges in cGAS activation which could explain the higher clinical efficacy of taxanes.
Results
Coculture Assay to Measure IFN Secretion.
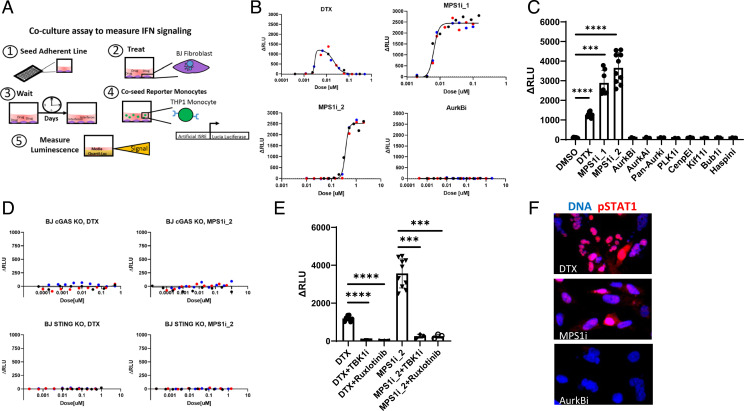
We developed a two-cell coculture assay to measure type I IFN secretion caused by drug-induced mitotic failure in multiwell plates. hTERT-BJ-5Ta (BJ) immortalized fibroblasts are known to have an intact cGAS–STING pathway (34). To detect IFN produced by BJ cells, we coseeded THP1 STING−/− reporter monocytes that express secreted Lucia luciferase under the control of an IFN response element (Fig. 1A). Multiple tests were used to validate this assay and show that it requires an intact cGAS–STING pathway in BJ cells (SI Appendix, Supplementary Information Text and Fig. S1).
Fig. 1.
MT stabilizers and MPS1 inhibitors induce IFN secretion through the cGAS–STING axis. (A) Schematic of the luciferase coculture assay for measuring secreted IFN. The assay is as follows: 1) The adherent line is seeded in a 96-well plate, 2) BJ cells are exposed to drug, 3) BJ cells are cultured in drug for multiple days, 4) STING−/− reporter monocytes which express L. luciferase protein under an IFN response element are coseeded with the BJ cells for 18 h, and 5) luciferase is assayed with a plate reader. Luminescence signal reports on paracrine IFN signaling which originates in the BJ cells. (B) Dose–response for IFN signaling measured with the coculture luciferase assay. DTX produces a bell-shaped curve centered around 10 nM. MPS1 inhibitors (MPS1i_1 and MPS1i_2) produce a hyperbolic response. AurkB inhibition did not induce detectable IFN. ΔRLU is the measured signal with the background luminescence detected in vehicle controls subtracted. Different-color markers represent independent experiments. (C) IFN response induced in BJ cells by a panel of A-Ms. The doses for each drug are reported in Table 1. Only MT stabilizers and MPS1 inhibitors induce IFN signaling. Markers represent independent experiments. (D) BJ cells that lack either cGAS or STING do not produce IFN in response to MT stabilizers and MPS1 inhibitors. Different-color markers represent independent experiments. (E) TBK1 inhibition (MRT67307; 500 nM) and JAK 1/2 inhibition (ruxlotinib; 500 nM) suppress IFN induction by MT stabilizers and MPS1 inhibitors. Markers represent independent experiments. (F) BJ cells were treated with A-M for 3 d and stained for phospho-STAT1 (pSTAT1), a marker of IFN signaling. pSTAT1 showed increased expression and nuclear localization after treatment of BJ cells with DTX (20 nM) or MPS1i_1 (20 nM) but not after AurkB inhibition (100 nM). All error bars denote SD. ***P ≤ 0.001, ****P ≤ 0.0001.
MT Stabilizers and MPS1 Inhibitors Activate the cGAS–IFN Pathway.
We assembled a panel of A-Ms (Table 1) with diverse mechanisms and clinical efficacy (35–37). Only two drug classes generated robust and reproducible IFN responses after 2 to 3 d of exposure: the MT stabilizer docetaxel (DTX) and two MPS1 inhibitors, BAY-1217389 [MPS1_1 (38)] and CFI-402257 [MPS1i_2 (39)] (Fig. 1B). In dose–response assays, DTX produced a maximal IFN signal at ∼10 nM which is in line with estimates for intratumoral concentrations (40). DTX did not induce signal from reporter monocytes in the absence of fibroblasts (SI Appendix, Fig. S2A). Higher doses of DTX decreased IFN signal which we interpret to be due to cytotoxicity from prolonged mitotic arrest (SI Appendix, Supplementary Information Text). Induction of IFN by MPS1 inhibitors followed a hyperbolic curve (Fig. 1B). Evidence for on-target activity of MT stabilizers and MPS1 inhibitors in these assays is described in SI Appendix, Supplementary Information Text and Fig. S2.
Table 1.
Overview of the A-Ms studied
| Target | Drug | Abbreviation | Clinical status | % clinical response | Primary dose studied, nM | IFN detected? |
| MT dynamics | Docetaxel | DTX | First-line therapy | Ovarian: 25 to 41; NSCLC: 19 to 25; head and neck: 32 to 42 |
10 | Yes |
| MPS1 | BAY-1217389 CFI-402257 |
MPS1i_1 MPS1i_2 |
In clinical trials | NA | 20 750 |
Yes |
| Aurora B kinase | Barasertib | AurkBi | Failed trials | 0 | 250 | No |
| Aurora A kinase | Alisertib | AurkAi | Failed trials | 0, 1 | 150 | No |
| Pan-Aurora kinase | Tozasertib | Pan-Aurki | Failed trials | 0 | 250 | No |
| Polo-like kinase 1 | Volasertib | PLK1i | Failed trials | 4.6 | 100 | No |
| Centromere protein E | GSK-923295 | CenpEi | Failed trials | 3 | 250 | No |
| Kif11 | Ispinesib | Kif11i | Failed trials | 0, 0 | 2.5 | No |
| Bub1 | BAY-1816032 | Bub1i | Preclinical | N/A | 1,000 | No |
| Haspin | CHR-6494 | Haspini | Preclinical | N/A | 250 | No |
The table describes the drug targets, drug names, drug abbreviations, clinical status, select reported clinical response rates, the typical experimental dose studied in this article, and our own observations of IFN induction. Percent clinical response refers to objective response (combined complete and partial responses as defined by RECIST scoring) as a monotherapy in solid tumors (35–37). The primary doses studied refer to the concentrations used in our experiments when dose–responses are not shown and unless otherwise noted. These doses are based on literature values and our own observations. IFN detected refers to whether we observed IFN signal in the coculture assay with BJ WT cells. N/A: not applicable; NSCLC: non-small cell lung cancer.
Surprisingly, all other A-Ms tested failed to produce measurable signal in our coculture assay (at concentrations reported in Table 1). In dose–response assays, the Aurora B kinase inhibitor (AurkBi) barasertib failed to produce detectable IFN signal over 3 logs of concentration centered on the published half maximal inhibitory concentration (IC50) [Fig. 1 B and C (41)].
We next verified that the IFN secreted after DTX and MPS1i exposure was induced by the cGAS–STING pathway. Fibroblasts where cGAS or STING was deleted using CRISPR engineering failed to produce IFN signal following treatment with MT stabilizers or MPS1 inhibitors (Fig. 1D and SI Appendix, Fig. S2K). The luciferase signal was also suppressed by concurrent treatment with TBK1 or JAK1/2 inhibitors (Fig. 1E). These results confirm that IFN induction by A-Ms is induced by cGAS–STING activation.
To confirm IFN induction with an alternative assay, we stained BJ cells for phospho-STAT1 after a 3-d drug exposure and observed nuclear localization in response to DTX and MPS1i but not AurkBi (Fig. 1F). This confirms the coculture assay result.
Overall, our results suggest that IFN secretion through the cGAS–STING pathway is not a general consequence of mitotic failure but rather selectively induced by MT stabilizers and MPS1 inhibitors. To explore possible therapeutic relevance of IFN induction by MT stabilizers, we performed drug-washout experiments that model clinical exposure. Washout converted the bell-shaped concentration–response curves to hyperbolic, consistent with a role for IFN induction in tumor regression (SI Appendix, Suplementary Information Text and Fig. S3).
Chromatin Bridges Correlate with cGAS Activation after A-M Treatment.
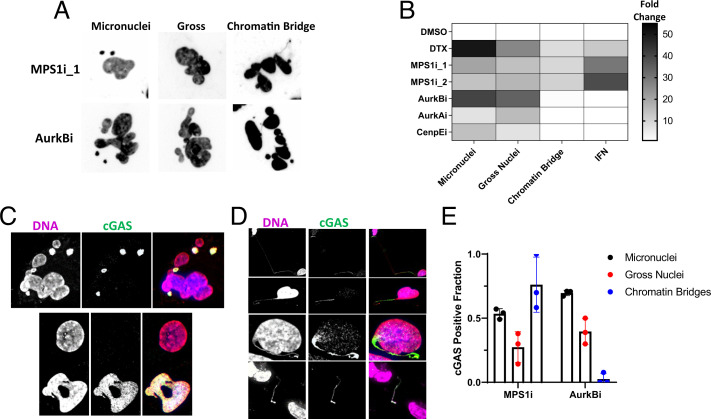
To understand why MT stabilizers and MPS1 inhibitors activate cGAS and other A-Ms do not, we quantified defects in nuclear morphology after A-M exposure using three classifications: micronuclei, gross nuclei, and chromatin bridges (Fig. 2A and SI Appendix, Supplementary Information Text and Fig. S4). Only chromatin bridges which spanned two daughter nuclei were scored. In drugs that prevented cytokinesis, thin threads of DNA were sometimes observed connecting chromatin masses within the same polyploid cell. These were difficult to resolve microscopically and were not scored.
Fig. 2.
Quantification of aberrant chromatin structures and cGAS localization produced by A-Ms. (A) Example nuclear morphologies produced by MPS1 inhibitors or Aurora B kinase inhibitors and visualized with a DNA dye. Aberrant structures are classified as micronuclei, gross nuclei, or chromatin bridges. Chromatin bridges appear as the small thin strands connecting independent nuclear objects. (B) A heatmap that summarizes the observed results for abnormal nuclear structures and IFN signal across different A-Ms. Chromatin bridges, not micronuclei, correlate with IFN signaling. Data were normalized to the corresponding vehicle control value. (C) Images of cGAS-positive micronuclei and gross nuclei produced after mitotic failure. cGAS was visualized with immunofluorescence. (D) Examples of cGAS-positive chromatin bridges. Observed chromatin bridges were either intact (first row), broken on one end (second and third rows), or broken on both ends (fourth row). cGAS was visualized with immunofluorescence. (E) Quantification of cGAS positivity on different types of aberrant chromatin structures produced after either MPS1 inhibition (750 nM; MPS1i_2) or AurkB inhibition (250 nM; barasertib). cGAS-positive micronuclei and gross nuclei showed similar frequencies after treatment with either drug. cGAS-positive chromatin bridges distinguish the two types of A-Ms. Each marker represents an independent experiment in which three to five FOVs were scored. All errors bars denote SD.
cGAS Localizes to All Aberrant Chromatin after Mitotic Errors.
We next localized cGAS after mitotic errors by immunofluorescence (SI Appendix, Fig. S4E). A fraction of interphase nuclei stained positive for cGAS even under control conditions. Micronuclei and gross nuclei induced by all drugs were cGAS-positive (Fig. 2C). Staining signal was lost in cGAS−/− BJ cells which validated the antibody (SI Appendix, Fig. S4F). Chromatin bridges, which were only observed after treatment with MT stabilizers or MPS1 inhibitors, stained strongly for cGAS (Fig. 2D). cGAS-positive chromatin bridges were only induced by drugs that induced IFN, while cGAS-positive micronuclei were strongly induced by drugs that did not (Fig. 2E). cGAS:DNA intensity ratios were difficult to compare between aberrant chromatin structures due to their widely varying size. This ratio appeared much higher in bridges, where the DNA density was low, compared with micronuclei, suggesting cGAS must be recruited to DNA at high local density to activate.
We also measured DNA damage following mitotic errors by staining for γH2A.X, a sensitive marker of DNA damage (42). We did not observe a correlation between DNA damage and cGAS activation (SI Appendix, Supplementary Information Text and Fig. S5). Lack of correlation of IFN secretion with micronuclei or DNA damage suggests that stretched chromatin bridges are unique in their ability to activate cGAS following mitotic errors.
cGAS Activation Requires Cytokinesis and Stretching Forces on Chromatin.
To test whether blocking chromatin-bridge formation would suppress IFN signal after failed mitosis, we treated cells with a constant concentration of MPS1i sufficient to produce chromatin bridges and titrated drugs that block cytokinesis by different mechanisms. A CenpE inhibitor which does not block cytokinesis was used as a negative control (43). Mechanisms of cytokinesis interference by various drugs, and further discussion on the design of this assay, are described in SI Appendix, Supplementary Information Text.
All drugs that block cytokinesis suppressed IFN induction by MPS1i (Fig. 3 A and B). IC50 values for the reduction of IFN signal were consistent with on-target activity in all cases. The results for AurkB and AurkA inhibition were confirmed with structurally dissimilar drugs. AurkB inhibition did not block IFN signaling induced by an ATM inhibitor which activates cGAS in a mitosis-independent manner [Fig. 3C and SI Appendix, Fig. S1C (44)].
Fig. 3.
cGAS activation requires cytokinesis and stretching forces on chromatin. (A) A-M combinations antagonize IFN induction. BJ cells were simultaneously treated with an inflammatory dose of MPS1 inhibitor (MPS1_2; 750 nM) and titrations of other A-Ms which interfere with cytokinesis and chromatin-bridge formation. The majority of tested A-Ms show dose-dependent suppression of the IFN signal produced by MPS1 inhibition. CenpE inhibition is not known to interfere with cytokinesis and is considered a negative control. The IFN fraction represents the measured signal relative to MPS1 inhibition alone. Different-color markers represent independent experiments. (B) CytoD, an actin inhibitor, suppressed IFN produced by MPS1 inhibition. Different-color markers represent independent experiments. (C) Cytokinesis interference does not suppress IFN signaling generally. BJ cells were simultaneously treated with an inflammatory dose of ATM inhibitor (KU-55933; 40 μM) and a titration of AurkBi. DNA damage caused by ATM inhibition activated the cGAS–STING–IFN axis irrespective of AurkBi dose. Different-color markers represent independent experiments. (D) Example images of cGAS-positive micronuclei observed after A-M combinations with MPS1i_2 (750 nM). Notably, only CenpE inhibition permitted cGAS-positive chromatin-bridge formation when combined with MPS1 inhibition. This combination also retained IFN signaling. (E) Frequencies of aberrant chromatin structures produced by A-M combinations. A-M combinations which suppressed IFN signal in A also suppressed chromatin-bridge formation. Doses for drug combinations are described in Table 1. Markers represent independent experiments. (F) Model for how A-Ms affect mitotic failure and IFN signal. Only low-dose MT stabilizers and MPS1 inhibitors produce stretched chromatin bridges because they cause chromosome missegregation events without preventing spindle formation or cytokinesis. All A-Ms tested produce cGAS-positive micronuclei. Chromatin-bridge formation is directly perturbed by many A-Ms through targeting spindle formation (AurkAi and KIF11) or cytokinesis (CytoD and AurkBi) or both (CA4). All error bars denote SD. ns, P > 0.05; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Cells treated with drug combinations contained high levels of cGAS-positive micronuclei (Fig. 3D), but only combinations in which cGAS-positive chromatin bridges were still present activated cGAS (Fig. 3E).
Based on the data in Figs. 1, 2, and 3, we propose the model in Fig. 3F.
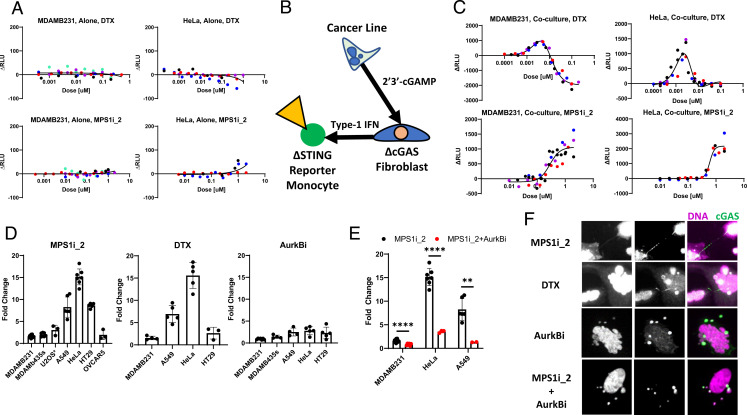
Paracrine cGAMP Assay to Measure cGAS Activation in Cancer Cells and Mouse Cells.
To generalize our results, and test their relevance for drug action in tumors, we sought to measure cGAS activation by mitotic errors in human cancer and mouse cells. Many cancer lines are unable to secrete IFN in response to aberrant DNA or cGAMP [SI Appendix, Fig. S1 E and F (34, 45, 46)]. Consistent with these reports, two cancer lines, MDAMB231 and HeLa, failed to produce detectable IFN secretion after treatment with either DTX or MPS1i (Fig. 4A). We therefore developed a cGAS activation assay that measures paracrine spread of cGAMP from cancer cells to cGAS−/− BJ fibroblasts, which then secrete IFN that is detected by reporter STING−/− THP1 monocytes (Fig. 4B). This triculture assay revealed strong basal cGAMP production in some lines (SI Appendix, Fig. S7 A and B). MDAMB231 cells, which are known to have chronically activated cGAS (45), exhibited the largest basal signal. Importantly, cocultures of cancer lines with STING−/− BJ cells, or in the presence of a TBK1 inhibitor, failed to produce detectable IFN which validates the assay principle (SI Appendix, Fig. S7C). The triculture assay provides a sensitive and convenient technique to detect cGAS activity in cancer lines which lack IFN secretion and may model the tumor microenvironment. It also allowed analysis of mouse cells despite species specificity of the IFN–interferon receptor interaction (SI Appendix, Supplementary Information Text and Fig. S8).
Fig. 4.
A-Ms activate cGAS in cancer lines through chromatin-bridge formation. (A) MDAMB231 and HeLa cells were exposed to either DTX or MPS1_2 for 3 d. IFN signaling was not detected for either cell line in the coculture assay. ΔRLU is the measured signal with the background luminescence detected in vehicle controls subtracted. Different-color markers represent independent experiments. (B) Schematic of the modified coculture luciferase assay, referred to as the triculture assay, which was designed to detect cGAS activation in cancer cells through paracrine cGAMP signaling. (C) MPS1 inhibitors and MT stabilizers activate cGAS in cancer lines. Both MDAMB231 and HeLa cells showed dose-dependent responses to DTX and MPS1 inhibition when measured with the triculture assay. The response curves mirror the shape of IFN induction in the BJ cells. The negative ΔRLU in the DTX MDAMB231 response represents cytotoxicity that reduced signal below the high basal signal. Different-color markers represent independent experiments. (D) MPS1 inhibitors and MT stabilizers activate cGAS across a panel of cancer cells. Cancer lines were treated with either 1 nM DTX, 750 nM MPS1i_2, or 250 nM AurkBi for 3 d. They showed variable levels of IFN response to A-M treatment. Of the cancer lines that showed evidence of cGAS activation in the triculture assay, MPS1 inhibition and DTX treatment induced larger responses than AurkB inhibition. Markers represent independent experiments. (E) AurkB inhibitors reduce cGAS activation in MPS1i-treated cancer cells. Cancer cells were treated with MPS1i_2 (750 nM) in combination with AurkBi (250 nM) for 3 d. Markers represent independent experiments. (F) HeLa cells treated with different A-Ms. cGAS-positive chromatin bridges are produced after treatment with MPS1i and DTX but not by AurkBi or the combination of AurkBi and MPS1i_2. Notably, conditions which exhibited minimal cGAS activation contained abundant cGAS-positive micronuclei. All error bars denote SD. **P ≤ 0.01, ****P ≤ 0.0001.
Chromatin Bridges, Not Micronuclei, Activate cGAS in Cancer Cells.
In two cancer lines, DTX and MPS1 inhibition caused cGAS activation with dose–response curves similar to those measured in BJ cells (Fig. 4C; compare with Fig. 2B). Negative ΔRLU values represent cytotoxicity from drugs that reduced luminescence below the basal signal. We then treated a collection of different cancer cells with A-Ms at on-target doses (Fig. 4D). Cancer lines displayed variable levels of signal induction, reported as fold change relative to their basal signal, in response to DTX and MPS1 inhibition. Notably, lines which were highly responsive to MPS1i and DTX, such as HeLa cells, only showed minimal signal after AurkB inhibition. To further assay cGAS activity in HeLa cells, we treated with camptothecin, a known inflammatory DNA-damaging agent [SI Appendix, Fig. S7D (47)] or with a small panel of A-Ms (SI Appendix, Fig. S7E). Camptothecin strongly induced cGAS activation in HeLa cells. In contrast, the A-Ms induced minimal signal and suggest that errors in mitosis do not typically activate cGAS. Inhibition of AurkB blocked the additional cGAS activation induced by MPS1i in multiple lines, confirming a requirement for cytokinesis in cGAS activation after mitotic failure (Fig. 4). cGAS localization data confirmed that cGAS localized to chromatin bridges but not to micronuclei, correlating with activation in cancer cells (Fig. 4F and SI Appendix, Fig. S7G) as well as mouse cells (SI Appendix, Fig. S8B).
Discussion
We used a panel of A-Ms to cause mitotic errors through distinct mechanisms and measured resulting chromatin abnormalities and IFN secretion in immortalized fibroblasts, mouse fibroblasts, and cancer cell lines. Unexpectedly, we observed that chromatin bridges, not micronuclei, activate cGAS after a failed mitosis in human and mouse cells despite both structures recruiting cGAS. Lack of IFN secretion by cells with abundant cGAS-positive micronuclei, for example following AurkBi exposure, shows that localization is not a reliable proxy for cGAS activation.
Our conclusion that abundant cGAS-positive micronuclei generated by A-Ms do not activate cGAS in human cell lines appears to contradict previous reports (13, 14). We ruled out human–mouse differences as the cause of the discrepancy (SI Appendix, Supplementary Information Text and Figs. S6 and S8). Thin chromatin bridges are difficult to image and may have been missed or underestimated in previous studies. It is also possible that micronuclei formed in other ways can activate cGAS, for example following irradiation. cGAS activation after irradiation is difficult to ascribe solely to mitotic errors because it simultaneously inflicts interphase DNA damage, oxidative stress, and up-regulation of genes such as TREX1 which combine to modulate cGAS activation in complex ways (48–50). IFN signaling detected 5 to 7 d postirradiation could be an indirect effect of chromatin bridges breaking and becoming micronuclei or micronuclei incurring massive DNA damage in subsequent cell cycles (7). Our work used mitosis-specific perturbations and a fibroblast line in which mitotic errors cause G1 arrest, which prevents the complex outcomes seen in cells from undergoing multiple cell cycles with damaged chromatin. Our data argue against the idea that acute DNA damage in micronuclei is sufficient to drive cGAS activation in either mouse or human cells. We observed distinct patterns of DNA damage and cGAS localization on chromatin bridges, but further studies are required to clarify this relationship.
To reliably quantify cGAS activation, we developed two assays that are affordable and amenable to high-throughput screening (Figs. 1A and 4A). Our triculture assay confirmed robust paracrine spread of cGAMP from cancer cells to fibroblasts (23) and strong basal activation of cGAS in some cancer lines (45, 51). We did not investigate the mechanism of cell-to-cell spread, which was reported to occur via gap junctions (23) and transporter-mediated secretion reuptake (52). Paracrine spread of cGAMP from cancer cells to stroma and leukocytes may allow the whole tumor to mount an IFN response to drug-induced mitotic errors even if the cancer cells have lost their ability to secrete IFN, which is often observed in cancer cell lines.
It is not safe to extrapolate directly from cell-culture data to clinical activity, but if we provisionally accept the hypothesis that IFN secretion contributes to tumor regression, then our data have interesting implications. They suggest taxanes promote tumor regression in part by inducing IFN after failed mitosis, and the clinical failure of high-quality inhibitors targeting Kif11, AurkA, AurkB, and Plk1 may be due to lack of IFN induction. A major caveat to this hypothesis is that most cancer cell lines are unable to secrete IFN in response to MT stabilizers or MPS1 inhibitors. It is possible that taxane-responsive solid tumors retain IFN secretion or that cGAMP spreads from tumor cells to neighboring stroma cells and leukocytes to activate STING. An alternative hypothesis, not mutually exclusive, is that taxanes promote an IFN response in proliferating stromal cells. These ideas could be tested by measuring IFN induction and other inflammatory cytokines in taxane-treated tumors at single-cell resolution. An exciting implication of our data is that MPS1 inhibitors hold clinical promise because they efficiently activate cGAS. However, they are significantly less cytotoxic than taxanes in our short-term assays, which makes clinical efficacy hard to predict.
Lastly, our results have implications for immunosurveillance of failed mitosis in normal tissues. Mitotic errors occur spontaneously and can promote tumors. cGAS activation by stretched chromatin bridges may be an evolved mechanism to alert the immune system to potentially transformed cells. Conversely, the same mechanism could promote tumorigenesis by generating inflammatory niches. Further work is needed to understand immunosurveillance of failed mitosis in vivo, and the roles of cGAS and cGAMP in tumor evolution.
Materials and Methods
Cell Culture.
Solid tumor lines were purchased from ATCC, with the exception of MDAMB231, which was kindly provided by the Sorger laboratory, Harvard Medical School. The U2OS line used expressed fluorescently labeled cGAS and BAF and was kindly shared by Tae Yeon Yoo, Harvard Medical School. hTERT-BJ-5Ta and hTERT-BJ-5Ta cGAS−/− were kindly provided by the Silver laboratory, Harvard Medical School. Engineered THP1 reporter cells were purchased from Invivogen. All the cell lines used in this study were grown under 37 °C and 5% CO2 in a humidified incubator. Cell lines were grown in appropriate media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (volume:volume). Specifically, THP1, MDAMB231, OVCAR5, and HT29 cells were grown in RPMI, and BJ-5Ta, U2OS, HeLa, MDAMB435s, and A549 cells were grown in Dulbecco’s modified Eagle’s medium. Cells were kept below passage 25 and regularly screened for mycoplasma.
Drugs.
MT stabilizers (DTX, MedChemExpress; epothilone-B, MedChemExpress), MT depolymerizers (combetastatin-A4, provided by Sergine Brutus, Harvard Medical School), Aurora kinase A inhibitors (alisertib, MedChemExpress; MK-8745, MedChemExpress), Aurora kinase B inhibitors (barasertib, MedChemExpress; ZM-447439, MedChemExpress), pan-Aurora kinase inhibitors (tozasertib; Haoyuan Chemexpress), MPS1 inhibitors (CFI-402257, MedChemExpress; BAY-1217389, MedChemExpress), haspin inhibitor (CHR-6494, Cayman Chemicals), Bub1 inhibitor (BAY-1816032, MedChemExpress), Eg5 inhibitor (ispinesib, Cayman Chemicals), centromere protein E inhibitor (GSK-923295, Cayman Chemicals), actin depolymerizer (cytochalasin D [CytoD], provided by Christine Field, Harvard Medical School), ATM inhibitor (KU-55933, Selleckchem), TBK1 inhibitor (MRT67307, Cayman Chemicals), and JAK inhibitor (ruxlotinib, MedChemExpress). All drugs were dissolved in dimethyl sulfoxide (DMSO).
Antibodies and DNA Stains.
Anti-cGAS 1:400 (D1D3G; CST 15102), anti–phospho-STAT1 1:400 (Tyr701, 58D6; CST 9167), anti–phospho-γH2A.X 1:400 (Ser139; Millipore 05-636), anti-STING 1:1,000 (D2P2F; CST 13647). Secondary antibodies were purchased from Invitrogen and conjugated with different Alexa Fluors (1:500). Hoechst 33528 (1 μg/mL) or Syto DeepRed (1:1,000; Thermo Fisher) was used for counterstaining DNA.
Coculture Reporter Assay.
The assay is based on the reporter cell protocol outlined by Invivogen but modified to incorporate two cell lines, one IFN producer (i.e., BJ fibroblasts) and one IFN reporter (i.e., THP1). Producer lines were resuspended in fresh media and seeded with a 100-μL volume in a 96-well plate with between 8 and 10,000 cells per well. Cells were treated with compound 6 to 24 h after initial seeding using a D300 digital dispenser and all wells were normalized to the same total volume of solvent, typically DMSO, below 0.1% total volume. Reporter cells were resuspended in fresh media and added to the drugged cells 72 h after drug addition. Reporter cells were seeded with 100,000 cells per well in a volume of 50 μL and left overnight. To assay for luciferase secretion, 10 μL of conditioned media from the coculture wells was mixed with 50 μL of commercial Quanti-Luc reagent (Invivogen) in a black, transparent 96-well plate (Corning) and brought to a plate reader (Victor) to measure luminescence. Experimental conditions were typically performed with technical replicates in triplicate.
Triculture Reporter Assay.
The assay was performed in an identical manner to the coculture assay, but producer cells consisted of BJ cGAS knockout (KO)fibroblasts mixed with cancer lines at a 1:1 ratio, with 5,000 cells each. The rest of the assay proceeded as described for the coculture assay.
Immunofluorescence.
Cells were grown on no. 1.5 glass coverslips in multiwell plates (Corning) and exposed to drug. Following multiday drug exposure, cells were washed with 37 °C phosphate-buffered saline (PBS) and fixed with either 37 °C 3.2% paraformaldehyde (diluted in PBS) for 10 min at room temperature for cGAS staining or with ice-cold methanol at −20 °C for 10 min when for pSTAT1 staining. Coverslips were then washed with PBS three times and blocked and permeabilized with PBS, 0.1% Triton X-100, 5% bovine serum albumin (BSA) for 1 h at room temperature. Coverslips were then stained with primary antibodies suspended in PBS, 0.1% Triton X-100, 1% BSA overnight at 4 °C. Coverslips were then brought to room temperature and washed three times with PBS, 0.1% Triton X-100 and stained with secondary antibodies for 1 h. Coverslips were then washed three times with PBS, 0.1% Triton X-100 and stained with DNA dye for 30 min. Coverslips were washed three times and mounted in media composed of 20 mM Tris (pH 8), 0.5% N-propyl gallate, and 90% glycerol. Coverslips were sealed with nail-polish hardener (Sally Henson).
Imaging.
Slides were brought to the microscope for imaging. Images (20×) were collected on a Nikon Eclipse Ti2-E inverted microscope equipped with a Nikon CFI Plan Apo Lambda 20×, numerical aperture (NA) 0.75 objective lens. Images were acquired with a SOLA SE V-nIR light engine and an Andor Zyla 4.2 PLUS sCMOS camera and controlled with NIS Element software. Images (60×) were collected on a confocal Nikon Ti inverted microscope equipped with a Nikon CFI Plan Apo Lambda 60×, NA 1.4 objective lens. Z series were acquired with a Spectral Applied Research LMM-5 laser module with solid-state lasers and collected with a Hamamatsu ORCA-R2 cooled charge-coupled device (CCD) camera. MetaMorph software was used for image acquisition.
Automated Scoring of Nuclei and Micronuclei.
We developed an analysis pipeline for the automated segmentation and analysis of nuclei and micronuclei in images. Images were preprocessed with the ImageJ algorithms unsharp mask and fast Fourier transform band-pass filter to improve the detection of micronuclei. The images were then uploaded to the publicly available nucleAIzer segmentation tool (53). Postprocessing of the nucleAIzer output was performed in MATLAB to distinguish nuclei from micronuclei and to reduce oversegmentation of multilobed nuclei. Example results are shown in SI Appendix, Fig. S3. Micronucleus frequencies were normalized by the number of cells scored in the same field of view (FOV) and reported as the number of micronuclei per 100 cells.
Manual Scoring of Gross Nuclei and Chromatin Bridges.
Images were manually scored for gross nuclei and chromatin bridges. Gross nuclei were classified based on multiple factors including binucleation, multinucleation, deranged morphology, and abnormally large size. Chromatin bridges were identified based on obvious DNA spanning two nuclear objects. However, due to the fragile nature of chromatin bridges, bridge fragments and the remnants of bridge protrusions were also included in our classification. Between three and five FOVs were analyzed per independent experiment and the frequency of abnormality was normalized to the total number of cells.
CRISPR KO Cell Line Generation.
KO cell lines were generated with the Synthego KO Kit. Single clones were expanded and sequenced and KO efficiency was estimated with the online Synthego ICE tool. Western blots were performed to confirm STING−/− (SI Appendix, Fig. S2).
Statistical Tests.
Unpaired t tests were used to compute reported P values. When more than two conditions were compared, a one-way ANOVA was used. Prism software (GraphPad) was used for all calculations. The level of statistical significance is represented as follows: not significant (ns), P > 0.05; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Supplementary Material
Acknowledgments
We thank Yukiye Koide and Emma Spady from the Silver laboratory (Harvard Medical School) for assistance with CRISPR−/− cell line generation, Mirra Chung from the Sorger laboratory (Harvard Medical School) for sharing the MDAMB231 cell line, Adrian Salic from the Salic laboratory (Harvard Medical School) for sharing the 3T3 cell line, Alina Simerzin from the Lahav laboratory (Harvard Medical School) for sharing the EMT6 cell line, the Nikon Imaging Center (Harvard Medical School), Laboratory of Systems Pharmacology (Harvard Medical School), and Dr. Jon Kagan for reviewing the manuscript. P.J.F. was supported by a fellowship from the Lynch Foundation and a Ruth L. Kirschstein Fellowship (F31CA254156). This work was funded by NIH Grant GM131753.
Footnotes
Reviewers: D.C., Dartmouth College; and R.H.M., Antoni van Leeuwenhoek Nederlands Kanker Instituut.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103585118/-/DCSupplemental.
Data Availability
All cell lines used in this study are available upon request. Image analysis code and sample image files are freely available in GitHub at https://github.com/patfly/NuclearAbnormalities. All other study data are included in the article and/or SI Appendix.
References
- 1.Fenech M., Morley A. A., Measurement of micronuclei in lymphocytes. Mutat. Res. 147, 29–36 (1985). [DOI] [PubMed] [Google Scholar]
- 2.Nelson L., et al. , A living biobank of ovarian cancer ex vivo models reveals profound mitotic heterogeneity. Nat. Commun. 11, 822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crasta K., et al. , DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umbreit N. T., et al. , Mechanisms generating cancer genome complexity from a single cell division error. Science 368, eaba0712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passerini V., et al. , The presence of extra chromosomes leads to genomic instability. Nat. Commun. 7, 10754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terradas M., Martín M., Hernández L., Tusell L., Genescà A., Nuclear envelope defects impede a proper response to micronuclear DNA lesions. Mutat. Res. 729, 35–40 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Ly P., et al. , Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 19, 68–75 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatch E. M., Fischer A. H., Deerinck T. J., Hetzer M. W., Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimini D., et al. , Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153, 517–527 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maciejowski J., Li Y., Bosco N., Campbell P. J., de Lange T., Chromothripsis and kataegis induced by telomere crisis. Cell 163, 1641–1654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan K.-L., North P. S., Hickson I. D., BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26, 3397–3409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffelder D. R., et al. , Resolution of anaphase bridges in cancer cells. Chromosoma 112, 389–397 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Harding S. M., et al. , Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie K. J., et al. , cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q., Sun L., Chen Z. J., Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guey B., et al. , BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 369, 823–828 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Pathare G. R., et al. , Structural mechanism of cGAS inhibition by the nucleosome. Nature 587, 668–672 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Kujirai T., et al. , Structural basis for the inhibition of cGAS by nucleosomes. Science 370, 455–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Li S., Role of post-translational modifications of cGAS in innate immunity. Int. J. Mol. Sci. 21, 7842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W., et al. , Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W., Mohr L., Maciejowski J., Kranzusch P. J., cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol. Cell 81, 739–755.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ablasser A., et al. , Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus A., et al. , Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity 49, 754–763.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borden E. C., Interferons α and β in cancer: Therapeutic opportunities from new insights. Nat. Rev. Drug Discov. 18, 219–234 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Wang H., et al. , cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. U.S.A. 114, 1637–1642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen A., Medema R. H., Mitosis as an anti-cancer target. Oncogene 30, 2799–2809 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Gascoigne K. E., Taylor S. S., Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14, 111–122 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Weaver B. A., How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell 25, 2677–2681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komlodi-Pasztor E., Sackett D., Wilkerson J., Fojo T., Mitosis is not a key target of microtubule agents in patient tumors. Nat. Rev. Clin. Oncol. 8, 244–250 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Mitchison T. J., The proliferation rate paradox in antimitotic chemotherapy. Mol. Biol. Cell 23, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan V. C., et al. , Why great mitotic inhibitors make poor cancer drugs. Trends Cancer 6, 924–941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchison T. J., Pineda J., Shi J., Florian S., Is inflammatory micronucleation the key to a successful anti-mitotic cancer drug? Open Biol. 7, 170182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia T., Konno H., Ahn J., Barber G. N., Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowinsky E. K., The development and clinical utility of the taxane class of antimicrotubule chemotherapy agents. Annu. Rev. Med. 48, 353–374 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Penna L. S., Henriques J. A. P., Bonatto D., Anti-mitotic agents: Are they emerging molecules for cancer treatment? Pharmacol. Ther. 173, 67–82 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Traynor A. M., et al. , Phase I dose escalation study of MK-0457, a novel Aurora kinase inhibitor, in adult patients with advanced solid tumors. Cancer Chemother. Pharmacol. 67, 305–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. M. Wengneret al., . Novel Mps1 kinase inhibitors with potent antitumor activity. Mol. Cancer Ther. 15, 583–592 (2016). [DOI] [PubMed] [Google Scholar]
- 39.J. M. Masonet al., . Functional characterization of CFI-402257, a potent and selective Mps1/TTK kinase inhibitor, for the treatment of cancer. Proc. Natl. Acad. Sci. U.S.A 114, 3127–3132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zasadil L. M., et al. , Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 6, 229ra43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.C. O. de Groot, A cell biologist’s field guide to aurora kinase inhibitors. Front. Oncol. 5, 285 (2015). [DOI] [PMC free article] [PubMed]
- 42.Sedelnikova O. A., Rogakou E. P., Panyutin I. G., Bonner W. M., Quantitative detection of (125)IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat. Res. 158, 486–492 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Tanudji M., et al. , Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell 15, 3771–3781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Härtlova A., et al. , DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Bakhoum S. F., et al. , Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau L., Gray E. E., Brunette R. L., Stetson D. B., DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350, 568–571 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Zhao B., et al. , Topoisomerase 1 cleavage complex enables pattern recognition and inflammation during senescence. Nat. Commun. 11, 908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanpouille-Box C., et al. , DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohr L., et al. , ER-directed TREX1 limits cGAS activation at micronuclei. Mol. Cell 81, 724–738.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gehrke N., et al. , Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39, 482–495 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Carozza J. A., et al. , 2'3'-cGAMP is an immunotransmitter produced by cancer cells and regulated by ENPP1. bioRxiv [Preprint] (2019). 10.1101/539312 (accessed 5 February 2019). [DOI]
- 52.Ritchie C., Cordova A. F., Hess G. T., Bassik M. C., Li L., SLC19A1 is an importer of the immunotransmitter cGAMP. Mol. Cell 75, 372–381.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollandi R., et al. , nucleAIzer: A parameter-free deep learning framework for nucleus segmentation using image style transfer. Cell Syst. 10, 453–458.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All cell lines used in this study are available upon request. Image analysis code and sample image files are freely available in GitHub at https://github.com/patfly/NuclearAbnormalities. All other study data are included in the article and/or SI Appendix.