Significance
The regulatory roles of cytokinin (CK) signaling on metabolic plasticity of plant response to salt stress remain widely unknown. A comprehensive metabolome and transcriptome analysis of CK-signaling–defective Arabidopsis thaliana histidine-containing phosphotransfer protein ahp2,3,5 and type-B Arabidopsis response regulator arr1,10,12 triple mutants under nonsaline and saline conditions revealed that CK signaling induces a reprogramming of gene-to-metabolite networks involved in Arabidopsis response to salinity. CK signaling modulates prestress and poststress accumulations of sugars, amino acids, and anthocyanins as well as membrane lipid reprogramming as an emerging mechanism of salinity adaptation in Arabidopsis. Our results provide insights into CK-signaling–mediated regulation of gene-to-metabolite networks in response to salt stress, enabling the efficient application of CK biology in stress tolerance-oriented plant biotechnology.
Keywords: cytokinin signaling, comparative metabolomics, comparative transcriptomics, regulatory network, salt stress
Abstract
Cytokinin (CK) in plants regulates both developmental processes and adaptation to environmental stresses. Arabidopsis histidine phosphotransfer ahp2,3,5 and type-B Arabidopsis response regulator arr1,10,12 triple mutants are almost completely defective in CK signaling, and the ahp2,3,5 mutant was reported to be salt tolerant. Here, we demonstrate that the arr1,10,12 mutant is also more tolerant to salt stress than wild-type (WT) plants. A comprehensive metabolite profiling coupled with transcriptome analysis of the ahp2,3,5 and arr1,10,12 mutants was conducted to elucidate the salt tolerance mechanisms mediated by CK signaling. Numerous primary (e.g., sugars, amino acids, and lipids) and secondary (e.g., flavonoids and sterols) metabolites accumulated in these mutants under nonsaline and saline conditions, suggesting that both prestress and poststress accumulations of stress-related metabolites contribute to improved salt tolerance in CK-signaling mutants. Specifically, the levels of sugars (e.g., trehalose and galactinol), amino acids (e.g., branched-chain amino acids and γ-aminobutyric acid), anthocyanins, sterols, and unsaturated triacylglycerols were higher in the mutant plants than in WT plants. Notably, the reprograming of flavonoid and lipid pools was highly coordinated and concomitant with the changes in transcriptional levels, indicating that these metabolic pathways are transcriptionally regulated by CK signaling. The discovery of the regulatory role of CK signaling on membrane lipid reprogramming provides a greater understanding of CK-mediated salt tolerance in plants. This knowledge will contribute to the development of salt-tolerant crops with the ability to withstand salinity as a key driver to ensure global food security in the era of climate crisis.
Cellular responses in plants to salinity require a new state of cellular homeostasis, which can be achieved by reprogramming metabolic pathways in various subcellular compartments, including calcium influx, activation of phospholipase D (PLD) and phosphatidic acid (PA) production, activation of the antioxidant system, changes in cell wall and plasma membrane composition, and increased endocytosis of aquaporins to regulate water transport (1–6). The regulation and integration of these cellular processes, however, are still poorly understood.
Metabolomics is a powerful tool that enables one to explore biological and physiological alterations in plants induced by environmental or genetic perturbations (7, 8). Primary metabolites, such as sugars, amino acids, and lipids, are required for proper plant development, while secondary metabolites like alkaloids, flavonoids, polyphenols, and terpenoids have more targeted functions (7, 8). Both types of metabolites play roles in plant response to a specific stress (8, 9). For example, various sugars (e.g., galactinol, raffinose, and trehalose) and stress-inducible amino acids (e.g., proline, branched-chain amino acids [BCAAs; leucine, isoleucine, and valine], and γ-aminobutyric acid [GABA]) are induced by different types of abiotic stresses in different plant species (10, 11). Metabolism of lipids in plants, which are implicated in maintaining the structure and function of cell membranes, is also reprogrammed in responses to abiotic stresses (12, 13). Additionally, anthocyanins, which are types of flavonoids, are considered to act as antioxidants that detoxify stress-induced reactive oxygen species (ROS) (14, 15). Accumulation of these metabolites under abiotic stress conditions enables plants to maintain ROS-homeostasis and undergo osmotic adjustments that help stabilize proteins and membranes (15, 16). Thus, profiling of primary and secondary metabolites can broaden our knowledge of the metabolic networks regulating plant growth and stress responses.
Several reports have demonstrated the involvement of cytokinins (CKs) and CK-signaling components in regulating stress adaptation in plants (17–21). CK homeostasis in Arabidopsis thaliana is tightly regulated by ISOPENTENYLTRANSFERASES (IPTs) and CK OXIDASES/DEHYDROGENASES (CKXs), key CK biosynthetic and catabolic enzymes, respectively (18). CK signaling in Arabidopsis is mediated by a canonical, multistep His-Asp phosphorelay, which includes the CK sensors ARABIDOPSIS HISTIDINE KINASES (AHKs), HISTIDINE-CONTAINING PHOSPHOTRANSFER PROTEINS (AHPs), and RESPONSE REGULATORS (ARRs) that are further classified as type-A and type-B ARRs. Phosphorylation of type-B ARRs activates downstream target genes, including type-A ARRs, leading to various developmental, morphological, biochemical, and physiological changes that enable plants to adapt to various environmental stresses (21–24). Among the 5 authentic AHPs and 11 type-B ARRs, triple loss-of-function of AHP2, AHP3, and AHP5, or ARR1, ARR10, and ARR12 results in almost completely defective CK signaling (25, 26). AHP and ARR genes have redundant functions in CK-mediated processes, as evidenced by the assessment of various CK-mediated shoot-related and root-related traits in various single, double, and triple ahp and arr mutants (25, 26). Reduced CK content in CK-deficient Arabidopsis plants also results in enhanced salt tolerance (18), suggesting that CKs negatively regulate salt stress signaling in Arabidopsis. CK-deficient Arabidopsis ipt1,3,5,7 mutant plants grown under salt stress exhibit up-regulation of many stress-responsive genes, including HIGH-AFFINITY K+ TRANSPORTER 1 (HKT1;1) (19), which may help maintain a proper K+/Na+ ratio. Ectopic expression of the MsCKX gene of alfalfa (Medicago sativa) in Arabidopsis using the CaMV35S promoter also enhanced salt tolerance of transgenic plants by maintaining high K+/Na+ ratio, enhancing the activities of antioxidant enzymes and increasing the expression levels of several stress-related genes, like DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A and DELTA-1-PYRROLINE-5-CARBOXYLATE SYNTHASE 1, as well as ion transport-related and H+ pump activity-related genes SALT OVERLY SENSITIVE 1 and HKT1 (27).
Consistent with these results, Arabidopsis IPT8-overexpressing transgenic plants with increased levels of endogenous CKs exhibited inhibited primary root growth, decreased activities of ROS-detoxifying enzymes, and reduced salt tolerance (28). In support of these findings, Arabidopsis CK-signaling ahp2,3,5 and arr1,12 mutant plants displayed salt-tolerant phenotypes with higher survival rates under saline conditions than WT plants (17, 29). It is important to note that positive regulatory roles of CKs in plant adaptation to abiotic stresses, like drought, heat, and salinity have also been reported (30–35). For example, creeping bentgrass (Agrostis stolonifera) transgenic lines harboring the Agrobacterium tumefaciens ipt gene driven by the SENESCENCE-ASSOCIATED GENE 12 promoter (SAG12-ipt) showed increased endogenous CK levels, as well as a more extensive root system and less cellular damage under drought stress due to an increase in the activities of CK-induced ROS-scavenging antioxidant enzymes, when compared with wild-type (WT) plants (31). Furthermore, the SAG12-ipt creeping bentgrass plants exhibited delayed leaf senescence and stimulated tiller formation and root production, in comparison with WT, in response to heat stress (32). Additionally, cotton (Gossypium hirsutum) transgenic lines ectopically expressing the A. tumefaciens ipt gene under the control of the cotton CYSTEINE PROTEINASE promoter displayed higher chlorophyll and CK contents, delayed leaf senescence, and improved growth performance, as compared with WT plants, in response to salt stress (34). These examples indicate that the roles of CKs in plant adaptation to abiotic stresses are quite complex, with both positive and negative regulatory functions.
Despite the increasing body of evidence on the relationship between CK metabolism and CK signaling with plant adaptation to various abiotic stresses, including salinity, the downstream metabolic changes involved in this adaptive process remain to be elucidated. In the present study, we examined changes in primary and secondary metabolism in Arabidopsis WT and two ahp2,3,5 and arr1,10,12 triple mutant lines to better understand CK-mediated metabolic regulation in response to salinity. We also integrated metabolite profiles with transcriptome data to obtain a deeper insight into the role of CK signaling in controlling gene-to-metabolite networks underlying plant adaptation to high salinity.
Results
ahp2,3,5 and arr1,10,12 Mutants Exhibit Enhanced Salt Tolerance.
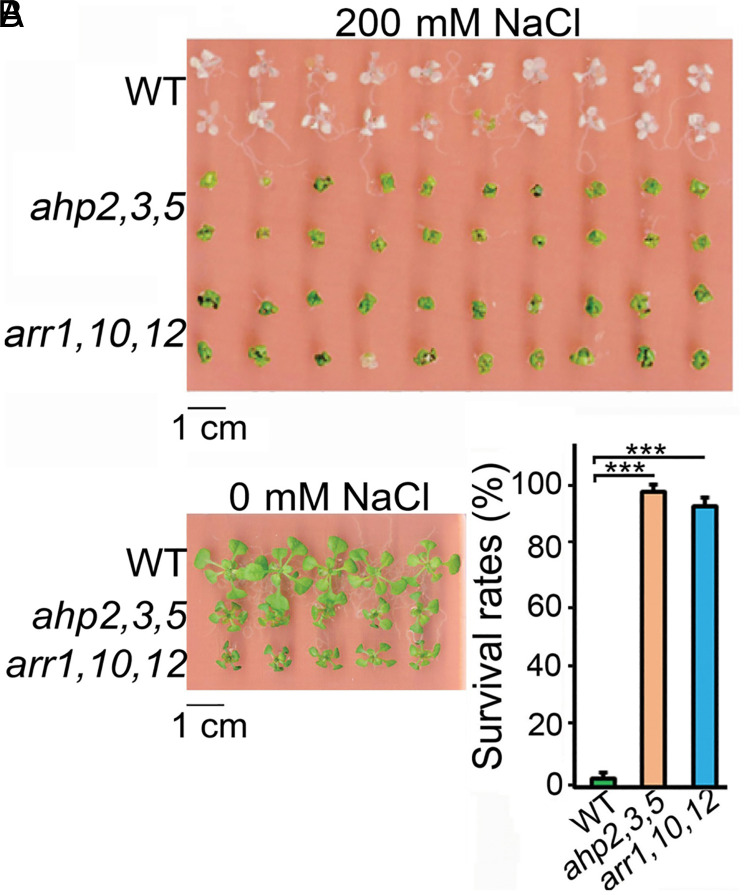
Ten-day-old seedlings of WT and CK-signaling ahp2,3,5 and arr1,10,12 mutants grown on full-strength Murashige and Skoog (MS)-agar plates were transferred to 0.5× MS-agar plates supplemented with 200 mM NaCl and maintained for 6 d to verify their salt tolerance. This “same-plate” method ensured that the genotypes were exposed to the identical salt concentration (36). Similar to the results of a previous report (17), ahp2,3,5 plants displayed over 95% survival to salinity, compared with less than 5% survival of WT plants (Fig. 1). The arr1,10,12 mutant also exhibited higher salt tolerance (>90% survival) than WT plants (Fig. 1). These data indicate that loss-of-function of redundant CK-signaling AHP or ARR genes, which results in almost complete CK insensitivity (25, 26), alleviates the inhibitory effect of the CK-signaling pathway on Arabidopsis tolerance to salt stress.
Fig. 1.
Phenotype and survival rates of Arabidopsis CK-signaling ahp2,3,5 and arr1,10,12 mutants, relative to WT plants, grown for 6 d on 0.5× Murashige and Skoog-agar plates supplemented with 200 mM NaCl. (A) Phenotypes of the investigated genotypes at 0 mM and 200 mM NaCl. (B) Survival rates (%) of the investigated genotypes at 200 mM NaCl. Values represent means and SEs calculated from three independent replicates (n = 3, 20 plants per genotype per replicate). Asterisks indicate significantly higher survival rates of ahp2,3,5 and arr1,10,12 compared with WT plants (***P < 0.001; Student’s t test).
Comprehensive Metabolite Profiling of WT, ahp2,3,5, and arr1,10,12 Plants under Saline and Nonsaline Conditions.
The metabolomes of 10-d-old WT, ahp2,3,5, and arr1,10,12 plants transferred to media supplemented with 0 mM (nonsaline) or 200 mM NaCl (saline) for 24 h were analyzed to determine the metabolic reprogramming that occurred in ahp2,3,5 and arr1,10,12 mutant plants to enable these mutants to survive under salt stress. Whole seedlings from each genotype were collected for comprehensive primary and secondary metabolite analyses using both gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) and liquid chromatography-quadruple TOF-MS (LC-Q-TOF-MS). A total of 79 primary and secondary metabolites, including 16 sugars, 24 amino acids, 4 polyamines, 9 tricarboxylic acid cycle-related metabolites, 6 phenolics, 2 phytosterols, and 18 other general metabolites, were successfully assigned using GC-TOF-MS (Dataset S1). On the other hand, 127 primary and secondary metabolites, including 15 flavonoids, 13 glucosinolates, and 99 lipid-related metabolites, were successfully assigned using LC-Q-TOF-MS (Datasets S2 and S3).
These 206 metabolites were all evaluated by unsupervised multivariate principal component analysis (PCA) and dendrogram clustering to determine the interrelated effects of high salinity on the metabolic profiles of the investigated genotypes (SI Appendix, Fig. S1). Most of the variance (51.4%) in the metabolite abundances was captured by PC1, while a lower proportion of variance (28.6%) was captured by PC2 (SI Appendix, Fig. S1A). Lipid-related metabolites, including polyunsaturated triacylglycerols (e.g., TAG_52:4, TAG_52:5, and TAG_52:6) and linoleic acid, contributed more to PC1, while other lipid-related (e.g., polyunsaturated monogalactosyldiacylglycerol [MGDG_34:3], glucosylceramide [GlcCer_d18:1/h16:0], saturated digalactosyldiacylglycerol [DGDG_34:0], and polyunsaturated phosphatidylcholine [PC_34:4]), and glucosinolate-related (e.g., 5-methylthiopentyl glucosinolate [5-MTP-GLS] and 3-methylsulfinylpropyl glucosinolate [3-MSOP-GLS]) metabolites contributed more highly to PC2 (SI Appendix, Fig. S1B). The PCA results collectively indicate that lipid-related and glucosinolate-related metabolites are the key metabolites involved in the responses of ahp2,3,5 and arr1,10,12 mutant plants to salinity. PC1 also captured genotype-related differences, while PC2 displayed separation based on the nonsaline and saline conditions (SI Appendix, Fig. S1C). Based on K-mean clusters of the 206 metabolites, the investigated genotypes exhibited three distinct groups of WT, ahp2,3,5, and arr1,10,12 plants under nonsaline (WT-C, ahp2,3,5-C, and arr1,10,12-C, respectively) and saline (WT-S, ahp2,3,5-S, and arr1,10,12-S, respectively) conditions (SI Appendix, Fig. S1C). Specifically, ahp2,3,5-C and arr1,10,12-C clustered in group 1, ahp2,3,5-S and arr1,10,12-S in group 2, and WT-C and WT-S plants in group 3 (SI Appendix, Fig. S1C). Similar to the PCA, dendrogram clustering also separated the investigated genotypes into three clusters containing the same members in each cluster (SI Appendix, Fig. S1D). The collective results of the PCA and dendrogram clustering indicated that the metabolic changes in ahp2,3,5 and arr1,10,12 mutants were similar under nonsaline and saline conditions but different from those observed in WT plants.
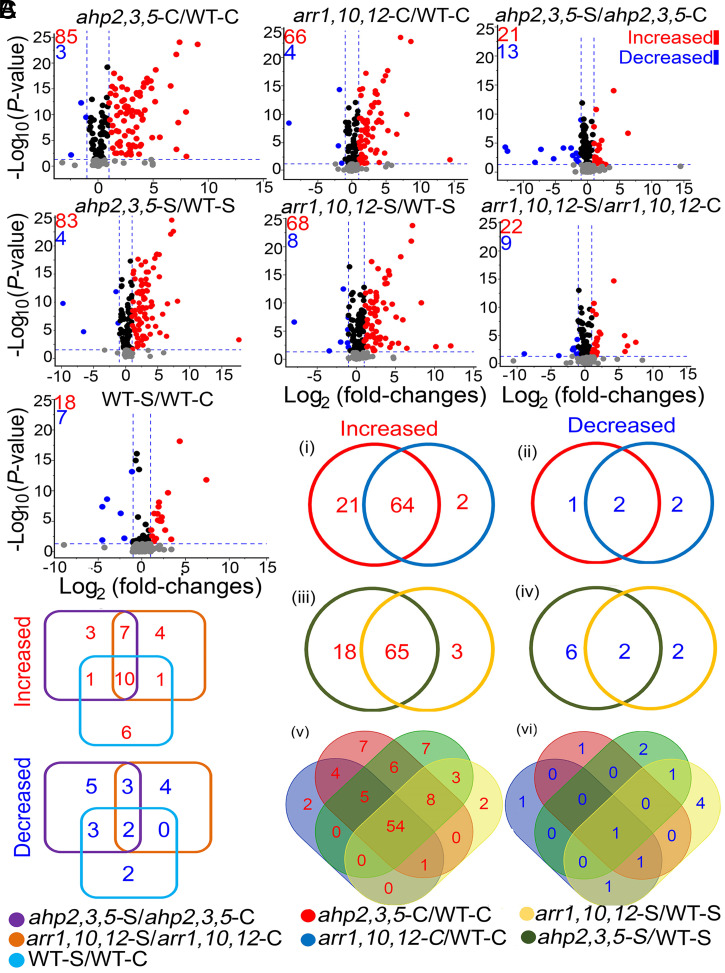
The false-discovery rate (FDR) approach was then implemented to identify the differentially produced metabolites (DPMs) in the CK-signaling mutant and WT plants under nonsaline and saline conditions. Among the 206 identified metabolites, 21, 22, and 18 metabolites increased [log2 (fold-changes) ≥ 1; q-values ≤ 0.05], and 13, 9, and 7 metabolites decreased [log2 (fold-changes) ≤ −1; q-values ≤ 0.05] in the ahp2,3,5-S/ahp2,3,5-C, arr1,10,12-S/arr1,10,12-C, and WT-S/WT-C comparisons, respectively (Fig. 2A and Dataset S4). A comparison of the mutants with WT plants under nonsaline or saline conditions revealed an increase in 85 and 66 metabolites and a decrease in 3 and 4 metabolites in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons, respectively, while 83 and 68 metabolites increased and 4 and 8 metabolites decreased in the ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S comparisons, respectively (Fig. 2A and Dataset S4). These results collectively indicate that in terms of the number of the altered metabolites, a greater metabolic change occurred, under nonsaline and saline conditions, when ahp2,3,5 and arr1,10,12 mutants were compared with WT plants, while a smaller metabolic change occurred in each individual genotype in response to salinity.
Fig. 2.
Volcano plots and Venn diagrams of DPMs in ahp2,3,5 and arr1,10,12 mutants, relative to WT plants, grown under nonsaline (ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C) and saline (ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S) conditions, as well as for each genotype, under saline versus nonsaline conditions (ahp2,3,5-S/ahp2,3,5-C, arr1,10,12-S/arr1,10,12-C, and WT-S/WT-C comparisons). (A) Volcano plots of significantly increased [log2 (fold-changes) ≥ 1; q-values ≤ 0.05], and decreased [log2 (fold-changes) ≤ −1; q-values ≤ 0.05] metabolites in the investigated comparisons. Blue-dashed lines represent the q-value and fold-change threshold. Red and blue points highlight the increased and decreased metabolites, respectively, in the investigated comparisons. (B) Venn diagrams of overlapping metabolites in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C (i and ii); ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S (iii and iv); and ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (v and vi). (C) Venn diagrams of overlapping metabolites in the ahp2,3,5-S/ahp2,3,5-C, arr1,10,12-S/arr1,10,12-C, and WT-S/WT-C comparisons. Three independent biological replicates (n = 3) from WT and CK-signaling mutant genotypes were collected for the metabolome (primary and secondary metabolites, except lipids) analysis, while five biological replicates (n = 5) were collected for lipidome analysis.
Metabolite Correlation and Network Analysis of WT, ahp2,3,5, and arr1,10,12 Plants in Response to Salinity.
We then identified overlapping DPMs in ahp2,3,5 and arr1,10,12 mutants relative to WT plants under nonsaline and saline conditions based on the construction of a Venn diagram (Fig. 2 B and C and Dataset S5). A total of 64 and 2 metabolites increased and decreased, respectively, under nonsaline conditions in ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons (Fig. 2 B, i and ii and Dataset S5), whereas 65 and 2 metabolites increased and decreased, respectively, under saline conditions in ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S (Fig. 2 B, iii and iv and Dataset S5). Notably, 54 metabolites exhibited increased levels in both mutants, relative to WT plants, under both saline and nonsaline conditions, suggesting that the lack of CK signaling in these mutants causes constitutive accumulation of these metabolites independent of salinity (Fig. 2 B, v and Dataset S5). Only one metabolite, 3-MSOP-GLS, decreased in both mutants regardless of saline conditions (Fig. 2 B, vi and Dataset S5). In addition, 10 metabolites increased and 2 metabolites (D-arabinose and fumaric acid) decreased in both of the mutants and the WT in response to salinity (Fig. 2C and Dataset S5). The overlapping 55 DPMs in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons indicate that the alterations in metabolites related to flavonoid and lipid metabolism are more specific to the mutant genotypes under saline and nonsaline conditions (Fig. 2 B, v and vi and Dataset S5).
On the other hand, the overlapping 12 DPMs in the ahp2,3,5-S/ahp2,3,5-C, arr1,10,12-S/arr1,10,12-C, and WT-S/WT-C comparisons indicate that changes in metabolites related to amino acid and sugar metabolism appear to be common metabolic responses to salinity in both of the mutants and the WT (Fig. 2C and Dataset S5). A total of 83 (80 increased and 3 decreased) overlapping DPMs were identified in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (Dataset S5). These 83 DPMs were classified into six main classes: 1) sugars, 2) amino acids and polyamines, 3) lipids and sterols, 4) flavonoids and phenolics, 5) glucosinolates, and 6) other general metabolites (SI Appendix, Fig. S2 A–E). Higher metabolite levels were generally observed in the mutants than in the WT under saline and nonsaline conditions, as illustrated in a heatmap hierarchical clustering (SI Appendix, Fig. S2 A–E and Dataset S5). The heatmap analysis revealed a segregation pattern similar to the segregation of genotypes obtained by PCA and dendrogram clustering (SI Appendix, Fig. S1), further confirming the metabolic differences between the two mutants and WT (SI Appendix, Fig. S2 A–E). It is plausible that the 83 identified DPMs may be the key metabolites playing an integral role in the salt tolerance exhibited by the ahp2,3,5 and arr1,10,12 mutant genotypes.
Subsequently, we conducted a "genotype–genotype" correlation analysis based on Pearson correlation coefficients (Pcc) generated for the 83 overlapping DPMs to identify the relationships among the examined genotypes under nonsaline and saline conditions (SI Appendix, Fig. S2F). Results indicated strong positive correlations between ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C (r = 0.88) comparisons under nonsaline conditions, ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S (r = 0.86) under saline conditions, and between ahp2,3,5-S/WT-S and ahp2,3,5-C/WT-C (r = 0.88) in saline versus nonsaline conditions (SI Appendix, Fig. S2F). Slightly weaker positive correlations were identified between arr1,10,12-S/WT-S and arr1,10,12-C/WT-C (r = 0.80), arr1,10,12-S/WT-S and ahp2,3,5-C/WT-C (r = 0.80), and between ahp2,3,5-S/WT-S and arr1,10,12-C/WT-C (r = 0.72) (SI Appendix, Fig. S2F). Additionally, a heatmap of "metabolite–metabolite" correlation based on Pcc demonstrated that 80 of the 83 DPMs showed high positive correlations, while the remaining 3 metabolites—3-MSOP-GLS, MGDG_34:1, and ribose—exhibited negative correlations among ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (SI Appendix, Fig. S3 and Dataset S6). The genotype–genotype and metabolite–metabolite correlation analyses provided further evidence that ahp2,3,5 and arr1,10,12 mutants exhibit similar metabolic changes, when compared with WT plants, under nonsaline and saline conditions.
Next, metabolite pathway analysis was conducted using the MetaboAnalyst to identify pathways associated with the 83 overlapping DPMs. Results indicated that most of the DPMs were enriched in the pathways associated with “cyanoamino acid metabolism,” “glycerolipid metabolism,” “alanine, aspartate and glutamate metabolism,” “phenylalanine metabolism,” and “flavone and flavonol biosynthesis” (SI Appendix, Fig. S4). All of the 83 DPMs were converted into Kyoto Encyclopedia of Genes and Genomes compound IDs (KEGG IDs), which were mapped into specific metabolic pathways using the KEGG mapper for Arabidopsis along with their metabolite levels identified in the examined genotypes under nonsaline and saline conditions (SI Appendix, Fig. S5). Results revealed that the reprogramming of lipid metabolism-related and flavonoid metabolism-related pathways was more profound in the CK-signaling–defective ahp2,3,5 and arr1,10,12 mutants compared with WT plants under nonsaline and saline conditions (SI Appendix, Fig. S5).
Comprehensive Transcriptome Analysis of WT, ahp2,3,5, and arr1,10,12 Plants in Response to Salt Stress.
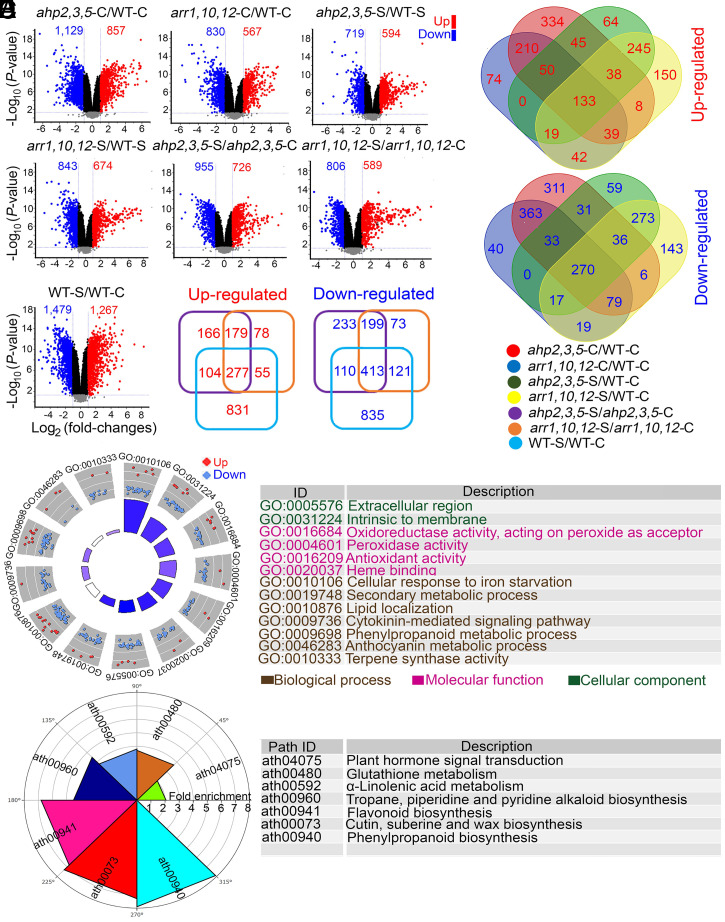
Ten-day-old WT, ahp2,3,5, and arr1,10,12 plants were also collected at 24 h posttreatment with salt stress for transcriptome analysis using microarray technology to assess the metabolic changes in response to salinity at molecular level. Results of the transcriptome analysis are provided in Dataset S7. Log2 (fold-changes) ≥ 1 (up-regulated) and ≤ −1 (down-regulated) with q-values ≤ 0.05 were used as the minimum cutoffs to identify differentially expressed genes (DEGs) that are robustly regulated by CK signaling in the examined genotypes under nonsaline and saline conditions. The analysis revealed that 857 and 567 genes were up-regulated, and 1,129 and 830 genes were down-regulated in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons, respectively (Fig. 3A and Dataset S7). A total of 594 and 674 genes were up-regulated, while 719 and 843 genes were down-regulated in ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S comparisons, respectively (Fig. 3A and Dataset S7). Additionally, 726, 589, and 1,267 genes were up-regulated, while 955, 806, and 1,479 genes were down-regulated in ahp2,3,5, arr1,10,12, and WT plants, respectively, in response to salinity (Fig. 3A and Dataset S7). The constructed Venn diagram illustrated that there were 403 overlapping DEGs (133 up-regulated and 270 down-regulated genes) in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (Fig. 3B and Dataset S8), and 690 overlapping DEGs (277 up-regulated and 413 down-regulated genes) in the ahp2,3,5-S/ahp2,3,5-C, arr1,10,12-S/arr1,10,12-C, and WT-S/WT-C comparisons (Fig. 3C and Dataset S8). The 403 overlapping DEGs represent the core changes in the transcriptome that may contribute to salt tolerance in the mutant genotypes and that are modulated by the repression of CK signaling.
Fig. 3.
Heatmap hierarchical clustering and genotype–genotype correlations of 83 metabolites differentially produced in ahp2,3,5 and arr1,10,12 mutants, relative to WT plants, grown under nonsaline (ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C) and saline (ahp2,3,5-S/WT-S and arr1,10,12-S/WT comparisons) conditions. (A–E) Heatmap hierarchical clusters of sugars (A), amino acids and polyamines (B), lipids and sterols (C), flavonoids, phenolics, and glucosinolates (D), and other general metabolites (E) in the investigated comparisons. (F) Genotype–genotype correlations based on the Pcc of DPMs in the investigated comparisons. The metabolite production levels in the heatmaps are a z-score–normalized data matrix. Red and blue colors indicate increased and decreased levels of metabolites, respectively, as indicated by the colored scales.
Gene Ontology and KEGG Enrichment Analysis, and Regulatory Network Analysis of WT, ahp2,3,5, and arr1,10,12 Plants in Response to Salt Stress.
A gene ontology (GO) enrichment analysis (37) was conducted to provide further insight into biological attributes of the 403 overlapping DEGs. The enrichment analysis (Fisher’s extract test and Bonferroni corrections; q-values ≤ 0.05) classified the 403 overlapping DEGs into three main GO categories: 1) biological process (BP), 2) molecular function (MF), and 3) cellular component (CC) (Fig. 3D). The overlapping DEGs in the BP category had highly enriched GO terms related to “cellular response to iron ion starvation” (GO:0010106), “secondary metabolic process” (GO:0019748), “lipid localization” (GO:0010876), “cytokinin-mediated signaling pathway” (GO:0009736), and “phenylpropanoid metabolic process” (GO:0009698) (Fig. 3D). The top enriched GO terms for the overlapping DEGs in the MF category were “oxidoreductase activity, acting on peroxide as acceptor” (GO:0016684), “peroxidase activity” (GO:0004601), “antioxidant activity” (GO:0016209), and “heme binding” (GO:0020037) (Fig. 3D). Finally, “extracellular region” (GO:0005576) and “intrinsic to membrane” (GO:0031224) were among the top enriched GO terms of the overlapping DEGs in the CC category (Fig. 3D).
Next, the 403 overlapping DEGs were mapped into the KEGG database to identify the key metabolic pathways in which these DEGs were significantly enriched (Fig. 3E). Results revealed that the overlapping DEGs were significantly (q-values ≤ 0.05) enriched in seven metabolic pathways, namely “phenylpropanoid biosynthesis,” “cutin, suberin, and wax biosynthesis,” “flavonoid biosynthesis,” “tropane, piperidine, and pyridine alkaloid biosynthesis,” “α-linolenic acid metabolism,” “glutathione metabolism,” and “plant hormone signal transduction” (Fig. 3E). In support of the transcriptome data and the results of GO and KEGG analyses, reverse transcription-quantitative PCR (RT-qPCR) analysis also revealed the up-regulation of several anthocyanin/flavonoid biosynthesis-related genes, including LEUCOANTHOCYANIDIN DIOXYGENASE/ANTHOCYANIDIN SYNTHASE (LDOX/ANS), DIHYDROFLAVONOL 4-REDUCTASE (DFR/TT3), and UDP-GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE (UF3GT) (7), in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (SI Appendix, Fig. S6 A–D and Dataset S7). Similarly, the transcript levels of HYDROXYSTEROID DEHYDROGENASE 6 (HSD6) and OLEOSIN (e.g., OLEO1 and OLEO2) genes, which are involved in lipid homeostasis and signaling (38–40), were also up-regulated in ahp2,3,5 and arr1,10,12 mutants, relative to WT plants, under saline and nonsaline conditions, as shown by the transcriptome and RT-qPCR data (SI Appendix, Fig. S6 E and F and Dataset S7).
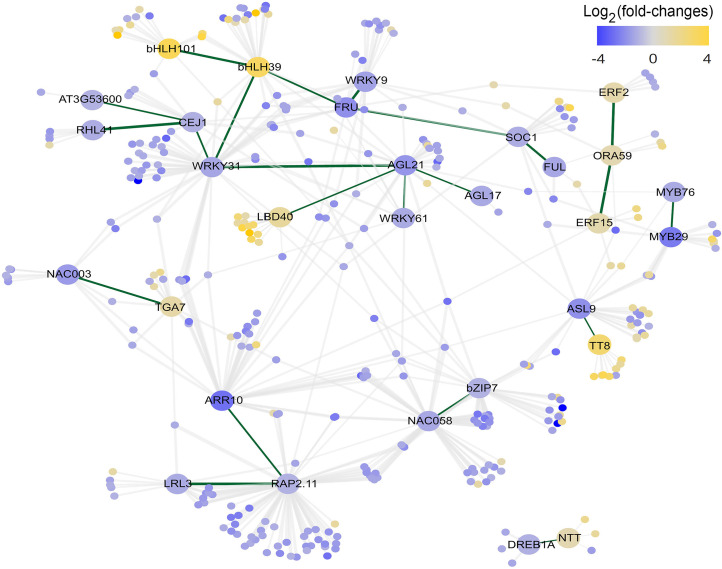
Next, a total of 1,215 overlapping DEGs were identified in the comparison between the transcriptome datasets of ahp2,3,5 and arr1,10,12 mutants with the transcriptome dataset of WT plants obtained under nonsaline and saline conditions. These DEGs were subjected to a regulatory network analysis (Dataset S9), and 89 genes encoding transcription factors (TFs) potentially associated with enhanced salt tolerance in the ahp2,3,5 and arr1,10,12 mutants were identified (Dataset S9). Next, an ARACNe-inferred coexpression subnetwork of the 1,215 DEGs was created to identify the TF modules associated with the salt tolerance of the mutants. The TFs were further filtered to identify TFs participating in TF–TF interactions to establish TF modules in the subnetwork (Fig. 4). Eight modules were found to be present in the final subnetwork of 30 TFs, 7 of which involved 2 or 3 TFs (Dataset S9). Notably, the eighth module contained 14 TFs (Dataset S9), of which 11 TFs were down-regulated, while 3 TF-encoding genes, LATERAL ORGAN BOUNDARIES DOMAIN-CONTAINING PROTEIN 40 (LBD40), BASIC HELIX-LOOP-HELIX 39 (bHLH39), and bHLH101 were up-regulated in the mutant genotypes under nonsaline and/or saline conditions (Fig. 4).
Fig. 4.
KEGG pathways associated with the 83 metabolites differently produced in ahp2,3,5 and arr1,10,12 mutants, relative to WT plants, grown under nonsaline (ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C) and saline (ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S comparisons) conditions. The metabolite production levels in the heatmaps are a z-score–normalized data matrix. KEGG IDs of the DPMs were submitted to KEGG mapper to identify specific pathways. Red and blue colors indicate increased and decreased levels of metabolites, respectively, as indicated by the colored scales. 4-amino-butyrate, GABA; cyanidin 3-O-[2-O-(β-d-xylopyranosyl)-6-O-(E-p-coumaroyl)-β-d-glucopyranoside]-5-O-[6-O-(malonyl)-β-d-glucopyranoside], A5; cyanidin 3-O-[2-O-(2-O-(E-sinapoyl)-β-d-xylopyranosyl)-6-O-(4-O-E-p-coumaroyl)-β-d-glucopyranoside]-5-O-[6-O-(malonyl)-β-d-glucopyranoside], A9; cyanidin 3-O-[2-O-(2-O-(E-sinapoyl)-β-d-xylopyranosyl)-6-O-(4-O-(β-d-glucopyranosyl)-E-p-coumaroyl)-β-d-glucopyranoside]-5-O-[β-d-glucopyranoside], A10; cyanidin 3-O-[2-O-(2-O-(E-sinapoyl-β-d-xylopyranosyl)-6-O-(4-O-(β-d-glucopyranosyl)-(E-p-coumaroyl)-β-d-glucopyranoside)-5-O-[6-O-(malonyl)-β-d-glucopyranoside], A11; kaempferol-diHex-Rha, KHR; kaempferol-3-Rha-7-Glu, KRG; quercetin-3-O-α-L-rhamnopyranosyl(1, 2)-β-D-glucopyranoside-7-O-α-L-rhamnopyranoside, F4; rhamnoside, Rha.
Results of the GO, KEGG, and regulatory network analyses collectively revealed the enrichment of genes involved in secondary metabolism (e.g., anthocyanin, flavonoid, terpene, and lipid biosynthetic pathways), nutrient assimilation (e.g., iron, sulfate, and sugar transports), and oxidoreductase activities, which is consistent with the accumulations of several metabolites related to flavonoids, lipids, and sterols in ahp2,3,5 and arr1,10,12 plants, relative to WT plants, under nonsaline and/or saline conditions.
Discussion
CKs and various members of their signaling networks—such as AHK2, AHK3, AHK4, AHP2, AHP3, AHP5, ARR1, ARR10, and ARR12—have been reported to function as negative regulators of plant responses to various abiotic stresses, including drought and salinity (17, 18, 29, 41, 42) (Fig. 1). For example, the Arabidopsis arr1,12 double mutant showed a higher seed germination rate than WT did at 150 mM NaCl (29). Additionally, at young seedling stage, both arr1 and arr12 single mutants, as well as the arr1,12 double mutant, exhibited higher survival rates than WT at 200 mM NaCl, and arr1,12 mutant was the least affected by salt stress (29). Likewise, the double mutants ahp2,3, ahp2,5, and ahp3,5, and double mutants for any mutation combinations of ARR1, ARR10, and ARR12 exhibited enhanced drought tolerance relative to WT; however, their level of drought tolerance was less than that of the respective triple mutant (17, 21). These results indicate functional redundancy of these CK-signaling members in regulating Arabidopsis responses to stresses, as observed in shoot-related and root-related traits (25, 26). Although the loss-of-function analysis of CK signaling partially revealed the physiological and molecular mechanisms responsible for the adaptation of Arabidopsis plants to salinity, the downstream metabolic changes have not been clarified. Thus, in this study, integrated metabolome and transcriptome analyses of Arabidopsis WT, and the defective CK-response triple mutants ahp2,3,5 and arr1,10,12 were conducted to elucidate the metabolic plasticity regulated by CK signaling in plant response to salinity.
Analysis of the metabolome data in ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons under nonsaline conditions revealed an increase in 64 metabolites involved in sugar, amino acid, flavonoid, and lipid metabolism, while 2 metabolites (ribose and 3-MSOP-GLS) decreased (Figs. 2 B, i and ii, SI Appendix, Figs. S2 A–E and S5 and Dataset S5). These data indicate that the defects in CK signaling placed the metabolism of these metabolites under a stage ready to react to unfavorable conditions. Similarly, salt stress induced the accumulation of 65 metabolites involved in sugar, amino acid, flavonoid, and lipid metabolism in ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S comparisons, while decreasing the levels of 2 metabolites, MGDG_34:1 and 3-MSOP-GLS (Figs. 2 B, iii and iv, SI Appendix, Figs. S2 A–E and S5 and Dataset S5). Notably, under both nonsaline and saline conditions, 54 metabolites increased, while only 3-MSOP-GLS decreased in both ahp2,3,5 and arr1,10,12 compared with WT plants (Figs. 2 B, v and vi, SI Appendix, Figs. S2 A–E and S5 and Dataset S5). These results suggest that the defects in CK signaling result in the metabolic reprogramming of specific metabolic pathways that are preactivated in the two triple CK-signaling mutants, enabling them to rapidly adapt to and survive unfavorable saline conditions.
A comparative analysis of the metabolome data derived from ahp2,3,5-S/ahp2,3,5-C, arr1,10,12-S/arr1,10,12-C, and WT-S/WT-C comparisons was also carried out to identify the common and specific metabolic responses of mutant and WT plants to salinity (Fig. 2C and Dataset S5). Results revealed that 10 metabolites related to sugar and amino acid metabolism increased, while 2 metabolites—arabinose and fumaric acid—decreased in both of the mutants and WT plants under salinity (Fig. 2C and Dataset S5). The two mutants and WT exhibited similar fold-changes in sugars and amino acids under saline conditions; thus, the changes in the two metabolic pathways in the mutants and WT plants appeared to be a common metabolic response to salinity (Fig. 2C and Dataset S5). In contrast, the analyses of 54 increased and 1 decreased metabolites identified in the mutants, relative to the WT under nonsaline and saline conditions, indicate that the accumulations of metabolites related to flavonoid and lipid metabolism are primarily detected in the mutant genotypes (Figs. 2 B, v and vi, SI Appendix, Figs. S2 A–E and S5 and Dataset S5). These data suggest that these pathways are negatively regulated by CK signaling.
Integration of the Metabolome and Transcriptome Analyses of Primary Metabolites in ahp2,3,5, arr1,10,12, and WT Plants under Nonsaline and Saline Conditions.
Sucrose and myo-inositol increased in WT and mutant plants under salinity, thus representing a common metabolic response to salt stress (SI Appendix, Fig. S2A and Dataset S5), while trehalose, isomaltose, raffinose, and galactinol were more abundant in the mutant genotypes, compared with WT plants, under nonsaline and saline conditions (SI Appendix, Fig. S2A and Dataset S5). Additionally, the transcriptome and RT-qPCR data revealed that the transcript levels of GALACTINOL SYNTHASE 4 (GOLS4), encoding an enzyme involved in the first reaction in the biosynthesis of galactinol and raffinose (43), were lower in the CK-signaling mutant genotypes, relative to the WT, under both nonsaline and saline conditions (SI Appendix, Fig. S6 G and H and Dataset S7). Similarly, STACHYOSE SYNTHASE (STS), encoding an enzyme that catalyzed the conversion of galactinol and raffinose into stachyose (43), was down-regulated in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons (SI Appendix, Fig. S5 G and H and Dataset S7). These results indicate that the defects in CK signaling may suppress the conversion of galactinol and raffinose into stachyose, rather than induce their biosynthesis in the mutant genotypes. Sugars not only serve as a building block and energy source in support of plant growth, but also function as signaling molecules and osmoprotectants that stabilize cellular membranes and maintain turgor, a prerequisite for plant survival under stress conditions (44). For example, exogenous application of trehalose to rice (Oryza sativa) helped maintain root integrity and shoot growth under saline conditions (45, 46). The protective role of trehalose was attributed to the preservation of active ion pumps, which selectively excludes Na+ ion accumulation in chloroplasts, induction of antioxidant enzyme activity and osmolyte production, and reduction of lipid peroxidation under salt stress (45). Furthermore, raffinose not only can function as a compatible solute, but also as a ROS scavenger and liposome protectant due to its high oligomeric length (47). The protective functions of sugars may contribute to the cellular adjustment of inner pressures and the maintenance of cell membrane integrity in CK-signaling mutant plants under stress conditions (17, 24, 48).
An increase in the levels of lysine, glutamate, ornithine, threonine, alanine, histidine, proline, and serine was also a common metabolic response to salinity in both WT and CK-signaling mutants (SI Appendix, Fig. S2B and Dataset S5). Interestingly, enhanced drought tolerance of creeping bentgrass SAG12-ipt transgenic lines was also attributed to the higher accumulation of some amino acids (49). Specifically, in response to drought, proline more highly increased in SAG12-ipt than WT plants, whereas alanine, valine, and GABA increased in SAG12-ipt and decreased in WT plants (49). In contrast, the levels of serine and threonine decreased in SAG12-ipt compared with WT plants under drought (49). It is worth noting that Arabidopsis transgenic lines ectopically expressing barley (Hordeum vulgare) HvCKX2 using the CaMV35S promoter showed an increase in arginine and ornithine contents, whereas Arabidopsis transgenic plants harboring an A. tumefaciens ipt gene driven by the CaMV35S promoter exhibited a considerable decrease in arginine and ornithine contents, suggesting that CKs positively regulate the conversion of arginine to polyamines (50). Collectively, these findings indicate that changes in CK homeostasis may alter the metabolism of certain amino acids, which might be helpful in plant stress adaptation.
In this study, BCAAs, typical stress-induced amino acids, were also more highly produced in the mutants than in WT plants under saline and nonsaline conditions (SI Appendix, Figs. S2B and S5 and Dataset S5). A defect in BCAA biosynthesis in Arabidopsis plants resulted in a phenotype with increased sensitivity to salt stress (51, 52), suggesting that BCAAs contribute to the adaptation of A. thaliana to salt stress. GABA was also observed to accumulate in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons, but decreased in the ahp2,3,5-S/WT-S and arr1,10,12-S/WT-S comparisons, while glutamate, a GABA precursor, exhibited the opposite trend in the same comparisons (SI Appendix, Figs. S2B and S5 and Dataset S5). The GABA homeostasis observed in the CK-signaling mutants was correlated with the up-regulation of GLUTAMATE DECARBOXYLASE 4 (GAD4), encoding an enzyme involved in the conversion of glutamate to GABA (53, 54), under nonsaline conditions, and its down-regulation under saline conditions (SI Appendix, Fig. S6 G and H and Dataset S7). In addition, the GABA shunt, which comprises glutamate-GABA-succinate route, also plays a major role in the maintenance of the carbon/nitrogen balance and subsequent energy production, particularly under abiotic stress conditions (53, 54).
Our results imply that CK signaling may control the glutamate/GABA ratio to maintain energy flux, or glutamate is simply induced by salt stress, as glutamate is not only a precursor for the biosynthesis of GABA, but also for the synthesis of proline, polyamine, and ornithine (55). In addition, the plant-specific type I membrane protein-encoding GLUTAMINE DUMPER 6 (GDU6) gene, encoding a protein involved in cell efflux of a wide spectrum of amino acids (56), was also up-regulated in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (SI Appendix, Fig. S6G and Dataset S7). It is possible that this feature may contribute to the higher levels of amino acids in the CK-signaling mutants under nonsaline and saline conditions.
Integration of the Metabolome and Transcriptome Analyses of Flavonoids in ahp2,3,5, arr1,10,12, and WT Plants under Nonsaline and Saline Conditions.
The increases of flavonoid-related metabolites in the CK-signaling mutants under nonsaline and saline conditions were highly correlated with the results obtained from the GO and KEGG enrichment analyses using the transcriptome data (Fig. 3, SI Appendix, Figs. S2D, S5, and S6 A–D, SI Appendix, and Dataset S7). This result indicates that the defect in CK signaling constitutively reprogrammed the regulation of the flavonoid pathway, thereby priming the salt-stress-adaptive responses in the mutant genotypes. Three flavonols (two kaempferol and one quercetin glucosides), and four anthocyanin glucosides (A5, A9, A10, and A11) were highly accumulated in ahp2,3,5 and arr1,10,12 mutants, relative to the WT, under saline and nonsaline conditions (SI Appendix, Figs. S2D and S5 and Dataset S5). Collectively, the results indicate that most of the metabolic changes observed in the two CK-signaling mutants were well correlated with the changes in transcript levels of genes involved in the identified metabolic pathways. It appears that the defect in CK signaling enhances the expression of numerous genes, inducing the accumulation of metabolites that are associated with increased salt tolerance in the mutant plants.
The MYB-bHLH-WD40-repeat (MBW) ternary protein complex is an important regulator of the latter steps of flavonoid biosynthesis (7). Transcriptome and RT-qPCR data revealed the up-regulation of several MYB genes involved in flavonol biosynthesis (57), including MYB11 in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons, and MYB111 in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, and arr1,10,12-S/WT-S comparisons (SI Appendix, Fig. S6 C and D and Dataset S7). MYB111 overexpression induces flavonoid accumulation and enhances salt tolerance in Arabidopsis, while its mutation decreases Arabidopsis salt tolerance (58). Likewise, PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1/MYB75), PAP2/MYB90, TRANSPARENT TESTA GLABRA 2 (TTG2), and TT8 genes, which encode TFs that function as master regulators of anthocyanin biosynthesis in Arabidopsis (59), were also up-regulated in the ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (SI Appendix, Fig. S6 C and D and Dataset S7).
These results were further supported by a gene–metabolite correlation analysis of the 403 overlapping DEGs and 83 overlapping DPMs identified in ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S (SI Appendix, Fig. S7 and Dataset S10). Correlation analysis revealed two clustering groups for genes (G1-cluster and G2-cluster) and two clusters for metabolites (M1-cluster and M2-cluster) (SI Appendix, Fig. S7). Metabolites grouped in the M1-cluster showed positive correlation with genes grouped in G1-cluster (SI Appendix, Fig. S7 and Dataset S10). Specifically, anthocyanin-related metabolites (M1 to M4) exhibited positive correlations (r = 0.61 to 0.99) with anthocyanin biosynthesis-related regulatory genes, such as PAP1, PAP2, and TT8 (SI Appendix, Fig. S7 and Dataset S10), indicating that flavonoid biosynthesis is transcriptionally regulated by CK signaling in Arabidopsis under nonsaline and saline conditions.
A previous study also suggested that the arr1,10,12 mutant plants exhibited sugar-induced anthocyanin accumulation as a result of the activation of the flavonoid MBW transcriptional complex and UF3GT gene (60). An increase in the total anthocyanin content was consistently observed in the arr1,10,12 triple mutant plants, which contributed to its enhanced drought-tolerant phenotype, compared with WT (21). Similarly, sucrose-induced expression of PAP1 in sucrose-treated Arabidopsis PAP1-Dominant (pap1-D) mutant plants results in increased anthocyanin accumulation, relative to sucrose-treated WT plants, and a higher survival rate of the pap1-D mutant plants under saline conditions (61). Taken together, these results indicate that the defect in CK signaling induced the expression of several regulatory genes like MYB11, MYB111, PAP1, PAP2, TTG2, and TT8, thereby modulating the pre- and poststress accumulations of flavonols and anthocyanins. This modulation appears to represent an important mechanism for salt stress adaptation in the CK-signaling ahp2,3,5 and arr1,10,12 mutants.
Integration of Metabolome and Transcriptome Analyses of Lipids and Phytosterols in ahp2,3,5, arr1,10,12, and WT Plants under Nonsaline and Saline Conditions.
Lipidome profiles revealed a prominent increase in lysophosphatidylcholines (LysoPCs having both saturated 16:0 and unsaturated 18:2 lipids), unsaturated TAGs (e.g., 50:1, 50:2, 52:4, 52:5, 52:6, 52:7, 54:9, and 56:6) and lipid precursors, including glycerol, glycerol-3-phosphate hexadecanoate, sterols (e.g., campesterol and sitosterol), linoleic acid, and α-linolenic acid, in the ahp2,3,5-C/WT-C, arr,1,10,12-C/WT-C, ahp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (SI Appendix, Figs. S2C and S5 and Dataset S5). For example, sulfoquinovosyl diacylglycerols (SQDGs) and DGDG increased in the ahp2,3,5-C/WT-C and ahpp2,3,5-S/WT-S comparisons, while glucosinolate 3-MSOP-GLS decreased in all four ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, ahpp2,3,5-S/WT-S, and arr1,10,12-S/WT-S comparisons (SI Appendix, Figs. S2C and S5 and Dataset S5). The data on lipid reprogramming were consistent with the transcript profiles of CK-signaling mutants versus WT plants under nonsaline and saline conditions (Dataset S7). For example, the expression of G3P TRANSPORTER/PERMERASE (G3PT/G3PP), which is involved in G3P transport between the cytosol and the plastids as the initial step in lipid biosynthesis (62), was up-regulated in the ahp2,3,5-C/WT-C and arr1,10,12-C/WT-C comparisons (SI Appendix, Fig. S6E and Dataset S7). Similarly, the transcript levels of UDP-SULFOQUINOVOSYL SYNTHASE (SQD1) and GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2 (GPT2), which are involved in SQDG biosynthesis and the transport of glucose-6-phosphate into plastids for lipid accumulation, respectively (38, 39), were also up-regulated in ahp2,3,5-C/WT-C, arr1,10,12-C/WT-C, and ahp2,3,5-S/WT-S comparisons (SI Appendix, Fig. S6E and Dataset S7). In agreement with these results, gene–metabolite correlation analysis also revealed positive correlations; for example, in the case of SQD1 gene expression with SQDG_36:4 and SQDG_36:5 contents (r = 0.82 and 0.94, respectively), and of FATTY ACID SYNTHASE (FAD) gene expression with TAG_50:1, TAG_50:2, TAG_52:4 and TAG_52:7 contents (r = 0.75 to 0.98) (Dataset S10).
Lipids affect membrane integrity, fluidity, and permeability, and play important roles in signal transduction to regulate plant growth and responses to environmental stresses, including salinity (40, 63). For example, phosphatidic acid produced from phospholipids by PLD acts as a key signaling molecule in plant responses to saline conditions, and Arabidopsis PLD-deficient pldα1 mutant plants exhibit a drastic decrease in salt tolerance relative to WT plants (64). A higher sterol/phospholipid ratio is also reported to be a characteristic feature in salt-tolerant canola (Brassica napus) cultivars (65), and thought to be involved in the regulation of ion homeostasis under salt stress (63). Furthermore, salinity induces the accumulation of linoleic and α-linolenic acids in Arabidopsis, which contributes to the activation of plasma membrane H+-ATPase activity to control K+/Na+ homeostasis, thus serving as a crucial factor in the survival of plants under salt stress (66). Results of these studies strongly support our findings, which also revealed higher accumulations of α-linolenic acid, linoleic acid, glycerol, G3P, hexadecanoate, SQDGs, LysoPCs, TAGs, and sterols, but decreased levels of MGDG_34:1 and 3-MSOP-GLS in the CK-signaling mutants, relative to WT plants, under nonsaline and saline conditions (SI Appendix, Figs. S2C and S5 and Dataset S5). Salinity-induced lipid reprogramming was also evident in the GO and KEGG analyses of CK-signaling mutants (Fig. 3 D and E). Collectively, our data suggest that the defect in CK signaling induces changes in the lipid profiles in mutant plants, which help maintain membrane function, ion transport and K+/Na+ homeostasis in these plants subjected to salt stress.
Among the identified lipids, TAGs highly accumulated in the CK-signaling mutants, relative to WT plants, especially under saline conditions (SI Appendix, Figs. S2C and S5 and Dataset S5). The accumulation of TAGs is frequently observed in senescing leaves in responses to various abiotic stresses and exogenous phytohormone applications (67, 68). For example, increased synthesis of TAGs in Arabidopsis and wheat (Triticum aestivum) represents an essential adaptive response to high temperature stress (69, 70), while TAG turnover in leaves of sweet potato (Ipomoea batatas) seedlings is activated to maintain energy flux for the ATP-dependent defensive responses against salinity (71). Furthermore, a lower content of MGDG in bundle sheath cell chloroplasts than mesophyll cell chloroplasts was shown to be associated with improved salt tolerance in maize (Zea mays) (72). In responses to general osmotic and nutrient-deficient stresses, a decrease in MGDG contents and increase in TAG contents in Arabidopsis and Craterostigma plantagineum plants resulted from the conversion of MGDG, contributed to the removal of excess MGDG to reduce membrane lipid peroxidation, and the accumulation of toxic products of thylakoid membrane degradation, thereby stabilizing cell membranes (73, 74).
Several genes involved in oleosin and streoleosin lipid droplet (LD) formation—such as OLEO1, OLEO2, and HSD6, respectively—were up-regulated in the two CK-signaling mutants under nonsaline and saline conditions (SI Appendix, Fig. S6 E and F and Dataset S7). Similarly, ABA INSENSETIVE 3 (ABI3), ABI4, and ABI5, which encode key TFs involved in TAG formation (73, 75, 76), were also up-regulated in the CK-signaling mutants, relative to the WT, especially under saline conditions (SI Appendix, Fig. S6 E and F). Oleosins embedded on the surface of TAGs contribute to LD stabilization and the prevention of oil body fusions, and have been linked to improved salt tolerance in Arabidopsis (76–78), perhaps by increasing TAG biosynthesis (79). In Arabidopsis, ABI3, ABI4, and ABI5 TFs positively modulate the expression of DIACYLGLYCEROL ACYLTRANSFERASE 1 that encodes a rate-limiting enzyme involved in TAG biosynthesis, thereby contributing to TAG accumulation and consequently adaptation to abiotic stresses (73, 75, 76). Recently, ABI5 has been shown to act together with MYB49 in modulating cuticle deposition and antioxidant capacity in leaves of Arabidopsis, thereby improving salt tolerance (80). The up-regulation of ABI3, ABI4, ABI5, OLEO1, OLEO2, and OLEO3 in the CK-signaling mutants (SI Appendix, Fig. S6 E and F) suggests that the ABA-dependent TAG biosynthetic pathway is a downstream target of CK signaling.
The TF-Regulatory Network Associated with Salt Tolerance in ahp2,3,5 and arr1,10,12 Mutants.
A total of 30 TFs, of 89 TFs derived from 1,215 overlapping DEGs in ahp2,3,5 and arr1,10,12 mutants, relative to WT plants and under nonsaline and saline conditions, exhibited TF–TF interactions and were categorized into 8 modules in a final subnetwork (Fig. 4 and Dataset S9). Among these eight modules, the AP2/ERF module consisted of three up-regulated genes—ERF2, ORA59, and ERF15—that serve as activators of stress-responsive genes (Fig. 4). A recent study (81) reported ORA59 to be an important regulatory gene in ethylene/jasmonic acid signaling. ORA59 up-regulates NITRATE TRASNPORTER 1.8 expression in Arabidopsis, and hence salinity-induced nitrate reallocation to roots as a trade-off between plant growth and stress adaptation. Similarly, ectopic expression of GmERF135, a soybean (Glycine max) ortholog of Arabidopsis ERF2, in Arabidopsis enhanced the growth rate of transgenic plants under saline and drought conditions, relative to WT and erf2 mutant plants (82). Results of our regulatory network analysis, combined with previous reports, suggest that ERF family members play a critical role in adaptation of the CK-signaling ahp2,3,5 and arr1,10,12 mutants to salt stress. In addition, bHLH101 and bHLH39, which were present in the largest module along with other TF-encoding genes, were also up-regulated (Fig. 4). These genes are highly expressed in response to iron deficiency in several plant species, including Arabidopsis, tomato (Solanum lycopersicum), rice, maize, and soybean, as part of an adaptive mechanism to improve iron uptake and maintain iron homeostasis (83). In addition to the important roles of iron in all major functions of plant metabolism, from chlorophyll biosynthesis to energy transfer, iron also participates in alleviation of salt stress by activating antioxidant enzymes (83). We hypothesize that iron homeostasis is maintained through the regulatory role of basic helix–loop–helix (bHLH) TFs, which may represent one of the mechanisms underlying the salt tolerance exhibited by CK-signaling mutants. MYB29 and MYB76 are also positive regulators of aliphatic glucosinolate (e.g., 4-methylthiobutyl [4MT]-GSL) biosynthesis in Arabidopsis (84), and were down-regulated in the CK-signaling mutants (Fig. 4). This finding is consistent with the observed decrease in 3-MSOP-GLS contents in the CK-signaling mutants (SI Appendix, Fig. S2D and Dataset S5). Additionally, glucosinolate levels are negatively regulated by ABA signaling (84). Thus, the decrease in 3-MSOP-GLS is in agreement with the previous reports of increased ABA responsiveness in ahp2,3,5 and arr1,10,12 mutants (17, 21). Increased ABA responsiveness can result in the up-regulation of many ABA-responsive genes, thereby contributing to salt tolerance in the ahp2,3,5 and arr1,10,12 mutants. The collective data indicate that CK signaling regulates a complex regulatory network in responses to saline conditions, comprising various TFs and their associated genes.
In conclusion, results of our integrated metabolome and transcriptome analyses demonstrated the pre- and poststress accumulations of numerous primary and secondary metabolites that could be involved in the improved salt tolerance exhibited by CK-signaling mutants, relative to WT plants. The mutant genotypes and WT plants both exhibited similar metabolic changes in the contents of amino acid-related and sugar-related metabolites in response to salt stress, which may represent a common adaptive response. Changes in the contents of flavonoid-related and lipid-related metabolites, however, were specific to the mutants under nonsaline and saline conditions. Our results also suggest that WT plants may need a longer time to undergo the metabolomic adaptive responses than the two CK-signaling mutant plants. In addition, our findings revealed strong correlations between the metabolism of flavonoids, lipids, and sterols with the expression of genes related to their biosynthetic pathway. Our study collectively provides a strategy for manipulating CK signaling to modulate lipid metabolism during plant response to salt stress. These efforts should ultimately lead to the development of improved salt-tolerant crop plants in the era of climate change.
Materials and Methods
Plant Material and Stress Treatments.
The two Arabidopsis CK-signaling mutants, ahp2,3,5 (ahp2-1, ahp3, and ahp5-2, CS860161) and arr1,10,12 (arr1-3, arr10-5, and arr12-1, CS39992) used in this study are in Col-0 background, and were obtained from the Arabidopsis Biological Resource Center and ref. 25, respectively. The salt tolerance assay was performed using the plate method, as previously described (18). For metabolome and transcriptome analyses, 10-d-old WT and CK-signaling mutant plants grown on Murashige and Skoog-agar plates containing 0.8% (wt/vol) agar and 3% (wt/vol) sucrose were transferred onto 0.5× MS-agar media (without sucrose) supplemented with 0 mM (nonsaline) or 200 mM NaCl (saline), and maintained for 24 h as described in Nishiyama et al. (19). Whole-plant samples from each genotype were collected at 24 h posttreatment, and frozen immediately in liquid nitrogen for further processing. Three independent biological replicates (n = 3; 20 plants per replicate) from each genotype were collected for the metabolome (primary and secondary metabolites, except lipids) and transcriptome analyses using GC-TOF-MS and LC-Q-TOF-MS for the former, and a microarray for the latter. One of the replicated salt stress-treated samples from the ahp2,3,5 genotype was identified as an outlier and not included in the transcriptome analysis. Five biological replicates (n = 5; 20 plants per replicate) were collected for lipidome analysis using LC-Q-TOF-MS.
Metabolome Analysis.
Previously established methods were used to extract the plant materials and perform the GC-TOF-MS analysis of primary metabolites (except lipids) and some secondary metabolites (85, 86), and the LC-Q-TOF-MS analysis of flavonols, anthocyanins, glucosinolates, and lipids (87). All of the raw GC-TOF-MS data were preprocessed using MATLAB 7.0.4 (The Math Works, https://www.mathworks.com). The resultant data matrix was normalized using cross-contribution compensating multiple-standard normalization (88). Metabolites obtained by GC-TOF-MS were identified and annotated as previously described (85). MASSBANK (89), KNApSAcK, and MS2T (90) databases were used for peak annotation and prediction of secondary metabolites obtained by LC-Q-TOF-MS (87). DPMs were identified using the LIMMA package (91). Peak selection in the lipidome data were conducted using Waters MarkerLynx XS v4.1, and peak annotation was performed using MS-DIAL software (92) and commercially available standards.
All 206 of the identified metabolites were subjected to multivariate PCA and dendrogram clustering using the “factoextra” and “FactoMine” packages in R v3.5.1 (7). The Benjamini–Hochberg procedure (FDR, i.e., q ≤ 0.05), and log2 (fold-changes) ≥ 1 and log2 (fold-changes) ≤ −1 as minimum cutoffs were used to identify increased and decreased metabolites in all comparisons. The filtered metabolites were then subjected to Volcano plot and heatmap clustering using R v3.5.1. A Venn diagram was constructed by using the default parameters of Bioinformatics and Evolutionary Genomics web-based tool (https://bioinformatics.psb.ugent.be/webtools/Venn/) to identify the number of overlapping DPMs in various comparison. A genotype–genotype correlation analysis based on Pcc was carried out using the “ggplot2” package in R v3.5.1. A metabolite–metabolite correlation heatmap was constructed using the “ggcorrplot” package in R v3.5.1, and a KEGG pathway analysis of the identified metabolites was conducted using the MetaboAnalysis v4.0 database (https://www.metaboanalyst.ca) and KEGG mapper (KEGG Mapper-Search Pathway, https://www.genome.jp).
Transcriptome Analysis.
Hybridization to the Arabidopsis Oligo 44K microarray (v4.0, Agilent Technologiess) was performed as previously described (19). All arrays were scanned with a microarray scanner (G2505B, Agilent Technologies) and analyzed using the LIMMA package (91). The q-values ≤ 0.05, and log2 (fold-changes) ≥ 1 and (log2 fold-changes) ≤ −1 were used as minimum cutoffs for the identification of up-regulated and down-regulated genes, respectively, in all comparisons. The up-regulated and down-regulated genes were visualized using Volcano plot with the aid of the “EnhancedVolcano” package in R v3.5.1. GO enrichment analysis of the DEGs was carried out using PANTHER14.1 (http://go.pantherdb.org/webservices/go/overrep.jsp) and AmiGO (37). The Arabidopsis Genome Initiative (AGI) of the DEGs were mapped into the Arabidopsis “ath” KEGG reference database using the “clusterProfiler” and “pathview” packages in R v3.5.1 (93) to identify the key metabolic pathways associated with the identified DEGs. An ARACNe-inferred seedling network was obtained using previously described methodology for A. thaliana microarray experiments using the ATH1-121501 platform downloaded from ArrayExpress (94, 95). The ArrayQualityMetrics (96) R package was run for each experiment, and low-quality samples were discarded. The remaining high-quality, CEL files of Col-0 samples were converted to text format using the apt-cel-convert utility from the Affymetrix Power Tools suite, and the microarray name in the file was modified to a custom string matching the version 22 TAIRG ATH1121501 CustomCDF (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp) package name files. Two pairs of cdf/probe_tab files were used, the CustomCDF (full dataset) and TF-only cdf files (TF-only dataset). The TF-only cdf file was obtained by masking the CustomCDF file using the cdf_masker.pl Perl script (https://github.com/ricardo-aaron/cdf_masker), and the list of A. thaliana TFs from the PlantTFDB database (97). The TF-only probe_tab file was obtained by filtering the CustomCDF probe_tab file using the PlantTFDB gene identifiers (http://planttfdb.gao-lab.org/). Expression data were then normalized using gcrma, and used as input for the ARACNe algorithm (98) . TF-to-non-TF interactions from the full dataset and TF–TF interactions from the TF-only dataset were merged in order to recreate a complete network. A subnetwork of DEGs in a [(ahp2,3,5-C + arr1,10,12-C + ahp2,3,5-S + arr1,10,12-S)/4 − (WT-C + WT-S)/2] comparison was then obtained using igraph (https://igraph.org) and plotted using Cytoscape (99).
RT-qPCR.
Total RNA extraction, cDNA synthesis and RT-qPCR were conducted as previously described (18, 19). The POLYUBIQUITIN10 (UBQ10) gene was used as a reference gene. Gene-specific primers used in the RT-qPCR analyses are listed in Dataset S11. Three independent biological replicates (n = 3; 20 plants per replicate) were used in the analysis.
Supplementary Material
Acknowledgments
The authors thank Dr. J. J. Kieber and the Arabidopsis Biological Resource Center for providing the ahp2,3,5 mutant; Dr. G. E. Schaller for the arr1,10,12 mutant; and Mr. M. Kobayashi, Mr. M. Suzuki, Mr. K. Takano, and Dr. T. Mori for their technical assistance. This work was supported by the grants from the Basic Science program from CONACytT-Mexico (Grant 00126261) and the Governor University Research Initiative program (05-2018) from the State of Texas (L.H.-E.).
Footnotes
Reviewers: B.H., The State University of New Jersey; and M.J., Jawaharlal Nehru University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105021118/-/DCSupplemental.
Data Availability
All data are included and freely available in this article. The raw microarray data and the detailed protocol were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE94885). The raw metabolite data were submitted to MetaboLights (accession no. MTBLS2532). All other study data are included in the main text and/or supporting information.
References
- 1.Munns R., Gilliham M., Salinity tolerance of crops—What is the cost? New Phytol. 208, 668–673 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Abiala M. K., Abdelrahman M., Burritt D. J., Tran L. P., Salt stress tolerance mechanisms and potential applications of legumes for sustainable reclamation of salt-degraded soils. Land Degrad. Dev. 29, 3812–3822 (2018). [Google Scholar]
- 3.Fu Y., Yang Y., Chen S., Ning N., Hu H., Arabidopsis IAR4 modulates primary root growth under salt stress through ROS-mediated modulation of auxin distribution. Front. Plant Sci. 10, 522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szymańska K. P., Polkowska-Kowalczyk L., Lichocka M., Maszkowska J., Dobrowolska G., SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int. J. Mol. Sci. 20, 143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shabala L., et al. , Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of barley (Hordeum vulgare) to salinity stress. Plant Physiol. 172, 2445–2458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park H. J., Kim W. Y., Yun D. J., A new insight of salt stress signaling in plant. Mol. Cells 39, 447–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelrahman M., et al. , Widely targeted metabolome and transcriptome landscapes of Allium fistulosum-A. cepa chromosome addition lines revealed a flavonoid hot spot on chromosome 5A. Sci. Rep. 9, 3541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelrahman M., et al. , Comparative metabolome and transcriptome analyses of susceptible Asparagus officinalis and resistant wild A. kiusianus reveal insights into stem blight disease resistance. Plant Cell Physiol. 61, 1464–1476 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Mundim F. M., Pringle E. G., Whole-plant metabolic allocation under water stress. Front. Plant Sci. 9, 852 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joshi V., Joung J. G., Fei Z., Jander G., Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 39, 933–947 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Fàbregas N., Fernie A. R., The metabolic response to drought. J. Exp. Bot. 70, 1077–1085 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Sui N., et al. , Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front. Plant Sci. 9, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinta G., et al. , Dynamics of metabolic responses to periods of combined heat and drought in Arabidopsis thaliana under ambient and elevated atmospheric CO2. J. Exp. Bot. 69, 2159–2170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe S., et al. , Arabidopsis molybdenum cofactor sulfurase ABA3 contributes to anthocyanin accumulation and oxidative stress tolerance in ABA-dependent and independent ways. Sci. Rep. 8, 16592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boestfleisch C., Papenbrock J., Changes in secondary metabolites in the halophytic putative crop species Crithmum maritimum L., Triglochin maritima L. and Halimione portulacoides (L.) Aellen as reaction to mild salinity. PLoS One 12, e0176303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo R., et al. , Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 10, ply016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishiyama R., et al. , Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. U.S.A. 110, 4840–4845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama R., et al. , Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishiyama R., et al. , Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One 7, e32124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golan Y., Shirron N., Avni A., Shmoish M., Gepstein S., Cytokinins induce transcriptional reprograming and improve Arabidopsis plant performance under drought and salt stress conditions. Front. Environ. Sci. 4, 63 (2016). [Google Scholar]
- 21.Nguyen K. H., et al. , Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. U.S.A. 113, 3090–3095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veerabagu M., et al. , The interaction of the Arabidopsis response regulator ARR18 with bZIP63 mediates the regulation of PROLINE DEHYDROGENASE expression. Mol. Plant 7, 1560–1577 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Lomin S. N., et al. , Studies of cytokinin receptor-phosphotransmitter interaction provide evidences for the initiation of cytokinin signalling in the endoplasmic reticulum. Funct. Plant Biol. 45, 192–202 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Cortleven A., et al. , Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 42, 998–1018 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Argyros R. D., et al. , Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchison C. E., et al. , The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18, 3073–3087 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S., et al. , Overexpression of the cytokinin oxidase/dehydrogenase (CKX) from Medicago sativa enhanced salt stress tolerance of Arabidopsis. J. Plant Biol. 62, 374–386 (2019). [Google Scholar]
- 28.Wang Y., Shen W., Chan Z., Wu Y., Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis thaliana. Front. Plant Sci. 6, 1004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason M. G., et al. , Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 64, 753–763 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Merewitz E. B., Gianfagna T., Huang B., Effects of SAG12-ipt and HSP18. 2-ipt expression on cytokinin production, root growth, and leaf senescence in creeping bentgrass exposed to drought stress. J. Am. Soc. Hortic. Sci. 135, 230–239 (2010). [Google Scholar]
- 31.Xu Y., Burgess P., Zhang X., Huang B., Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 67, 1979–1992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y., Tian J., Gianfagna T., Huang B., Effects of SAG12-ipt expression on cytokinin production, growth and senescence of creeping bentgrass (Agrostis stolonifera L.) under heat stress. Plant Growth Regul. 57, 281 (2009). [Google Scholar]
- 33.Liu X., Huang B., Cytokinin effects on creeping bentgrass response to heat stress: II. leaf senescence and antioxidant metabolism. Crop Sci. 42, 466–472 (2002). [Google Scholar]
- 34.Liu Y. D., et al. , Improved salt tolerance and delayed leaf senescence in transgenic cotton expressing the Agrobacterium IPT gene. Biol. Plant. 56, 237–246 (2012). [Google Scholar]
- 35.Ghanem M. E., et al. , Root-synthesized cytokinins improve shoot growth and fruit yield in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 62, 125–140 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verslues P. E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J. K., Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Carbon S., et al. ; AmiGO Hub; Web Presence Working Group, AmiGO: Online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong Y., Zhang W., Wang X., Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ. 33, 627–635 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Okazaki Y., et al. , A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okazaki Y., Saito K., Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 79, 584–596 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Tran L. S., et al. , Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang N. Y., Cho C., Kim N. Y., Kim J., Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J. Plant. Physiol. 169, 1382–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Vinson C. C., et al. , Characterization of raffinose metabolism genes uncovers a wild Arachis galactinol synthase conferring tolerance to abiotic stresses. Sci. Rep. 10, 15258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keunen E., Peshev D., Vangronsveld J., Van Den Ende W., Cuypers A., Plant sugars are crucial players in the oxidative challenge during abiotic stress: Extending the traditional concept. Plant Cell Environ. 36, 1242–1255 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Garcia A. B., et al. , Effects of osmoprotectants upon NaCl stress in rice. Plant Physiol. 115, 159–169 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdallah M. M. S., Abdelgawad Z. A., El-Bassiouny H. M. S., Alleviation of the adverse effects of salinity stress using trehalose in two rice varieties. S. Afr. J. Bot. 103, 275–282 (2016). [Google Scholar]
- 47.Van den Ende W., Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 4, 247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao W. H., et al. , Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143, 707–719 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merewitz E. B., et al. , Elevated cytokinin content in ipt transgenic creeping bentgrass promotes drought tolerance through regulating metabolite accumulation. J. Exp. Bot. 63, 1315–1328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Černý M., et al. , Proteome and metabolome profiling of cytokinin action in Arabidopsis identifying both distinct and similar responses to cytokinin down- and up-regulation. J. Exp. Bot. 64, 4193–4206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing A., Last R. L., A regulatory hierarchy of the Arabidopsis branched-chain amino acid metabolic network. Plant Cell 29, 1480–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C., et al. , Dihydroxyacid dehydratase is important for gametophyte development and disruption causes increased susceptibility to salinity stress in Arabidopsis. J. Exp. Bot. 66, 879–888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shelp B. J., Bown A. W., McLean M. D., Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4, 446–452 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Snowden C. J., Thomas B., Baxter C. J., Smith J. A. C., Sweetlove L. J., A tonoplast Glu/Asp/GABA exchanger that affects tomato fruit amino acid composition. Plant J. 81, 651–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winter G., Todd C. D., Trovato M., Forlani G., Funck D., Physiological implications of arginine metabolism in plants. Front. Plant Sci. 6, 534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratelli R., Voll L. M., Horst R. J., Frommer W. B., Pilot G., Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol. 152, 762–773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brendolise C., et al. , Multiple copies of a simple MYB-binding site confers trans-regulation by specific flavonoid-related R2R3 MYBs in diverse species. Front. Plant Sci. 8, 1864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B., et al. , The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 166, 103807 (2019). [Google Scholar]
- 59.Li P., et al. , Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210, 905–921 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Das P. K., et al. , Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol. Cells 34, 93–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh J.-E., Kim Y. H., Kim J. H., Kwon Y. R., Lee H., Enhanced level of anthocyanin leads to increased salt tolerance in Arabidopsis PAP1-D plants upon sucrose treatment. J. Korean Soc. Appl. Biol. Chem. 54, 79–88 (2011). [Google Scholar]
- 62.Kawai H., et al. , Arabidopsis glycerol-3-phosphate permease 4 is localized in the plastids and involved in the accumulation of seed oil. Plant Biotechnol. 31, 159–165 (2014). [Google Scholar]
- 63.Mansour M., Salama K., Allam H., Role of the plasma membrane in saline conditions: Lipids and proteins. Bot. Rev. 81, 416–451 (2015). [Google Scholar]
- 64.Yu L., et al. , Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Zamani B., Bybordi A., Khorshidi S., Nezami T., Effects of NaCl salinity levels on lipids and proteins of canola (Brassica napus L.) cultivars. Adv. Environ. Biol. 4, 397–403 (2010). [Google Scholar]
- 66.Han X., et al. , A bioassay-guided fractionation system to identify endogenous small molecules that activate plasma membrane H+-ATPase activity in Arabidopsis. J. Exp. Bot. 68, 2951–2962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H. U., et al. , Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol. J. 13, 1346–1359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arisz S. A., et al. , DIACYLGLYCEROL ACYLTRANSFERASE1 contributes to freezing tolerance. Plant Physiol. 177, 1410–1424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mueller S. P., Krause D. M., Mueller M. J., Fekete A., Accumulation of extra-chloroplastic triacylglycerols in Arabidopsis seedlings during heat acclimation. J. Exp. Bot. 66, 4517–4526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narayanan S., Tamura P. J., Roth M. R., Prasad P. V., Welti R., Wheat leaf lipids during heat stress: I. High day and night temperatures result in major lipid alterations. Plant Cell Environ. 39, 787–803 (2016a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y., et al. , Involvement of phosphatidylserine and triacylglycerol in the response of sweet potato leaves to salt stress. Front. Plant Sci. 10, 1086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Omoto E., Iwasaki Y., Miyake H., Taniguchi M., Salinity induces membrane structure and lipid changes in maize mesophyll and bundle sheath chloroplasts. Physiol. Plant. 157, 13–23 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Yang Y., Yu X., Song L., An C., ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiol. 156, 873–883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gasulla F., et al. , The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: A comparative approach. Plant J. 75, 726–741 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Mukhopadhyay P., Tyagi A. K., OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5, 9998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skubacz A., Daszkowska-Golec A., Szarejko I., The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7, 1884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J., et al. , The E3 ligase AtRDUF1 positively regulates salt stress responses in Arabidopsis thaliana. PLoS One 8, e71078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zou J., et al. , Induction of lipid and oleosin biosynthesis by (+)-abscisic acid and its metabolites in microspore-derived embryos of Brassica napus L. cv Reston (Biological responses in the presence of 8′-hydroxyabscisic acid). Plant Physiol. 108, 563–571 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parthibane V., Rajakumari S., Venkateshwari V., Iyappan R., Rajasekharan R., Oleosin is bifunctional enzyme that has both monoacylglycerol acyltransferase and phospholipase activities. J. Biol. Chem. 287, 1946–1954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang P., et al. , The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 43, 1925–1943 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Zhang G.-B., Yi H.-Y., Gong J.-M., The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26, 3984–3998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao M.-J., et al. , The roles of GmERF135 in improving salt tolerance and decreasing ABA sensitivity in soybean. Front. Plant Sci. 10, 940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurt F., Filiz E., Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100 and bHLH101 genes in Arabidopsis, tomato, rice, soybean and maize: Insights into iron (Fe) homeostasis. Biometals 31, 489–504 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Li Y., et al. , Abscisic acid-mediated induction of FLAVIN-CONTAINING MONOOXYGENASE 2 leads to reduced accumulation of methylthioalkyl glucosinolates in Arabidopsis thaliana. Plant Sci. 303, 110764 (2021). [DOI] [PubMed] [Google Scholar]
- 85.Kusano M., et al. , Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 855, 71–79 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Kusano M., et al. , Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst. Biol. 1, 53 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuda F., et al. , MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 57, 555–577 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Redestig H., et al. , Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal. Chem. 81, 7974–7980 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Horai H., et al. , MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Afendi F. M., et al. , KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53, e1 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Smyth G. K., “limma: Linear models for microarray data” in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Gentleman R., Carey V. J., Huber W., Irizarry R. A., Dudoit S., Eds. (Statistics for Biology and Health, Springer, New York, NY, 2005) pp. 397–420. [Google Scholar]
- 92.Tsugawa H., et al. , MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luo W., Brouwer C., Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chávez Montes R. A., et al. , ARACNe-based inference, using curated microarray data, of Arabidopsis thaliana root transcriptional regulatory networks. BMC Plant Biol. 14, 97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Athar A., et al. , ArrayExpress update—From bulk to single-cell expression data. Nucleic Acids Res. 47, D711–D715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kauffmann A., Gentleman R., Huber W., arrayQualityMetrics—A bioconductor package for quality assessment of microarray data. Bioinformatics 25, 415–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin J., et al. , PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Margolin A. A., et al. , ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 (suppl. 1), S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included and freely available in this article. The raw microarray data and the detailed protocol were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE94885). The raw metabolite data were submitted to MetaboLights (accession no. MTBLS2532). All other study data are included in the main text and/or supporting information.