Abstract
This study aimed to explore the prognostic value of SATB1, SMAD3, and TLR2 expression in non–small-cell lung carcinoma patients with clinical stages I-II. To investigate, we evaluated immunohistochemical staining to each of these markers using tissue sections from 69 patients from our cohort and gene expression data for The Cancer Genome Atlas (TCGA) cohort. We found that, in our cohort, high expression levels of nuclear SATB1n and SMAD3 were independent prognostic markers for better overall survival (OS) in NSCLC patients. Interestingly, expression of cytoplasmic SATB1c exhibited a significant but inverse association with survival rate, and it was an independent predictor of unfavorable prognosis. Likewise, TLR2 was a negative outcome biomarker for NSCLC even when adjusting for covariates. Importantly, stratification of NSCLCs with respect to combined expression of the three biomarkers allowed us to identify subgroups of patients with the greatest difference in duration of survival. Specifically, expression profile of SATB1n-high/SMAD3high/TLR2low was associated with the best OS, and it was superior to each single protein alone in predicting patient prognosis. Furthermore, based on the TCGA dataset, we found that overexpression of SATB1 mRNA was significantly associated with better OS, whereas high mRNA levels of SMAD3 and TLR2 with poor OS. In conclusion, the present study identified a set of proteins that may play a significant role in predicting prognosis of NSCLC patients with clinical stages I-II.
Keywords: non-small-cell lung cancer, prognostic factor, SATB1, SMAD3, TLR2
Introduction
According to the World Health Organization (WHO), lung cancer is the second most frequent type of cancer and the leading cause of cancer mortality worldwide, being responsible for approximately 11.6% of the total number of new cancer cases and 1.8 million deaths in 2018. 1 This high mortality rate is often attributed to disease recurrence, resistance to chemotherapy, and advanced-stage diagnosis. Despite significant developments in oncological management in recent years, the 5-year relative survival rate for people with all types of lung cancer is just 19% (men 16% and women 22%). 2 Therapeutic recommendations depend primarily on tumor stage, histology, size, and position of the cancer, together with patient-specific factors (e.g., age, comorbidity, and pulmonary function). 3 Cigarette smoking is the most important risk factor for lung cancer. Globally, cigarette smoking is linked to approximately 80% of lung cancer deaths. Additional factors contribute to lung cancer development such as secondhand smoking, air pollution, exposure to radon, asbestos, and other carcinogens, poor diet, and indoor emission of fuel burning. 4
There are 2 main types of lung cancer: small-cell lung carcinoma (SCLC, approx. 15% cases) and non–small-cell lung carcinoma (NSCLC, approx. 85% cases).5,6 NSCLC is further classified into 3 main subtypes: squamous cell carcinoma (SCC) (25–30%), adenocarcinoma (ADC) (40%), and large cell carcinoma (LCC) (10–15%), and several other less common subtypes, such as adenosquamous carcinoma and sarcomatoid carcinoma. 7 Patients with early-stage NSCLC usually undergo surgical resection, which remains the best therapeutic option for long-term survival. Unfortunately, around two thirds of cases undergo late-stage diagnoses, when the cancer has already metastasized. While the 5-year relative survival rate for operable early-stage disease is up to 70–90%, it drops dramatically to around 1% for stage IV NSCLC. The median overall survival (OS) of these patients is around 10 months depending on treatment, histology type, and other factors.8–10
Given that NSCLC represents a major health problem accounting for large numbers of deaths, the identification of prognostic biomarkers for these patients is crucial to enhance survival. A number of studies have shown that several nuclear matrix proteins (NMPs) are dramatically deregulated in various cancers.11–24 Undoubtedly, a greater understanding of the relationship between this group of proteins and cancer can be very useful from a clinical point of view.
Special AT-rich binding protein 1 (SATB1) is a higher-order chromatin organizer and global transcriptional regulator. 11 This 763-amino acid protein identified in thymocytes is encoded by the SATB1 human gene located on chromosome 3p23. SATB1 is a known factor that binds to AT-rich sequences known as base unpairing regions (BURs) in the matrix attachment regions (MARs) of DNA. SATB1 anchored to BURs provides a “docking site” on the nuclear matrix for chromatin remodeling/modifying enzymes and transcription factors. Strikingly, SATB1 regulates a number of genes, even those located on distant chromosomes, and therefore, it is referred to as a “genome organizer.” 12 Under physiological conditions, SATB1 is involved in T-cell development, cellular homeostasis, early erythroid differentiation, and responses to various stimuli. 13 Besides physiological processes, this protein is thought to be an important factor in numerous malignancies. Abnormal expression of SATB1 has been reported in various types of cancers, including breast, 14 gastric, 15 lung,16,17 laryngeal squamous cell carcinoma, 18 colorectal, 19 endometrial, 20 prostate, 21 liver 22 ovarian, 23 and bladder cancers. 24 Despite being differentially expressed, a comprehensive understanding of the role of SATB1 protein in NSCLC is hampered by the limited number of studies on the topic, a large heterogeneity between and within histological subtypes, as well as research discrepancies between cell lines and clinical NSCLC tumors.16,25-27 It is, therefore, urgent to study the significance of SATB1 in additional NSCLC cohorts and to uncover potential culprits of NSCLC that may overlap with SATB1 to affect patient prognosis.
SMADs are a family of intracellular proteins that transmit signals from the transforming growth factor-β (TGFβ) superfamily of receptors. 28 There are 3 distinct subgroups of SMADs based on their different roles in TGFβ family signal transduction: R-SMADs (receptor-regulated), which include SMADs 1, 2, 3, 5, and 8, Co-SMADs (common partner), that is, SMAD4, and I-SMADs (inhibitory), which comprises SMAD6 and SMAD7.29,30 As numerous reports suggest, SMAD signaling seems to be relevant to the pathogenesis of several cancers.31–33 However, the role of SMAD3 in tumorigenesis is not clear, as it has been shown to function as both a tumor suppressor and prometastatic factor. 34
Toll-like receptors (TLRs) are a family of immune receptors expressed by antigen presenting cells, fibroblasts, epithelial, and cancer cells. 35 In both normal and tumor cells, they play important roles in the regulation of inflammatory responses, cell proliferation, and apoptosis.36,37 TLRs can recognize a variety of pathogen-associated molecular patterns (PAMPs) to induce various immune responses. According to cellular localization, TLRs are divided into 2 major subtypes: extracellular (TLR1, TLR2, TLR5, TLR6, and TLR10) and intracellular (TLR3, TLR7, TLR8, and TLR9).38,39 In the cell line studies, TLR2 was suggested to be a potential therapeutic target in lung adenocarcinoma, 40 as well as a specific mediator between lung cancer cells and mesenchymal stem cells present within the tumor microenvironment, facilitating cross-talk leading to the promotion of tumor-supportive phenotypic changes of mesenchymal cells. 41 However, TLR2 represents a double-edge sword and may also act as tumor suppressor. 42 Uncovering the prognostic significance of TLR2 in NSCLC is therefore expected to provide better understanding of its role in the biology of this tumor.
For the purpose of the present study, we selected solely SMAD3 and TLR2 since the convergence of these proteins with one another, 43 as well as with SATB1 signaling,44,45 has been previously reported; however, there are no studies on their joint evaluation in NSCLC samples. According to the report by Mikami et al., TGFβ receptor-SMAD3/4 signaling pathway is positively involved in TLR2 induction via a dual mechanism involving functional cooperation with the NF-κB pathway and MAPK phosphatase 1 (MKP-1)-dependent inhibition of p38 MAPK, a negative regulator for TLR2 induction. It showed that TβR-Smad3/4 signaling acts as a positive regulator for host defense and immune response by increasing the expression of TLR2 during respiratory bacterial infections. 43 Lung cancer patients often present with pulmonary bacterial infections, and this coincides with a poor prognosis.46–48 Although the underlying mechanisms for pulmonary infection-triggered lung cancer development are still not fully understood, TLR signaling seems to play an important role. For instance, Ye et al. revealed that NSCLC cells were competent and active in sensing Gram-negative bacteria through TLRs, which fueled their aberrant metabolic features to promote tumor outgrowth and metastasis.49,50 Besides, SATB1, SMAD3, and TLR2 may be also related by their cellular and tissue functions, as each of these proteins is known to regulate the dynamic equilibrium of apoptosis, invasion, migration, proliferation, immune modulation, and inflammation,34,42,51 the disturbance of which is strongly implicated in lung carcinogenesis. Moreover, as far as we are aware, at the protein level, TLR2 has not been previously evaluated as a biomarker candidate for the prediction of clinical outcome in NSCLC. Likewise, SMAD3 is also underexplored in this group of cancer patients. All these make SATB1, SMAD3, and TLR2 interesting candidates to be explored as the individual, and especially combined biomarkers for prognostication of NSCLC patients.
Therefore, the current study was designed to evaluate the immunohistochemical (IHC) expression of SATB1, SMAD3, and TLR2 in 69 formalin-fixed paraffin-embedded tissue samples (FFPE) from NSCLC patients with clinical stages I to II. The research included the reference of obtained results to OS of patients, clinicopathological data, and also the analysis of the correlation between the chosen proteins. Importantly, the combined prognostic value of these 3 proteins was also evaluated. Finally, we examined mRNA expression of these markers in the context of patient survival by utilizing the TCGA dataset.
Materials and methods
The study was conducted on archival FFPE tissue samples collected between 2010 to 2014 from 69 patients diagnosed with NSCLC in Franciszek Łukaszczyk Oncology Center of Bydgoszcz. Histopathological evaluation of each tumor sample was performed by 2 independent pathologists for the purpose of selecting a representative study group at the Department of Clinical Pathomorphology, Collegium Medicum in Bydgoszcz, Nicolaus Copernicus University in Toruń. All tumors were reclassified according the standardized TNM eighth edition classification of The American Joint Committee on Cancer (AJCC) criteria. To avoid excessive study complexity, cohort included ADC, SCC, and LCC, while all other histological types were excluded from the series. The inclusion and exclusion criteria is summarized in Supplementary Figure 1. The study protocol has been approved by The Ethics Committee of Nicolaus Copernicus University in Toruń, Ludwik Rydygier Collegium Medicum in Bydgoszcz (approval number KB 336/2018). All methods used were performed in accordance with applicable principles of good laboratory practice.
FFPE tissue blocks with representative tumor areas were cut using a manual rotary microtome (Accu-Cut, Sakura, Torrance, CA, USA) to 4.0 μm thick. Sections were placed on high-adhesive glass slides (SuperFrost Plus; Menzel-Glaser, Braunschweig, Germany) and dried at 60°C for 1 h. IHC staining was performed using DakoAutostainer Link 48 (Dako, Agilent Technologies, USA) or BenchMark® Ultra automated slide processing system (Ventana Medical Systems, Tucson, AZ, USA). Standardization and optimization of the IHC method were performed using instructions provided by the antibody manufacturers, and data are available in the Human Protein Atlas (http://www.proteinatlas.org). 52
IHC staining of SATB1 and SMAD3 was performed using Dako Autostainer Link 48 automated slide staining platform (Dako) and the FLEX + visualization system. Tissue sections were deparaffinized and rehydrated prior to antigens retrieval using a high-pH buffer (Dako, Agilent Technologies, USA) for 20 min in PT Link pre-treatment module (Dako) at 95–98°C. Next, slides were treated with 3% H2O2 for 10 min at room temperature (RT) to inhibit endogenous peroxidase activity. Then, the preparations were incubated with 3% bovine serum albumin (BSA) solution for 15 min at RT to block non-specific antibody binding sites. The incubation with anti-SATB1 antibody (1:200; cat. no: ab109122, Abcam, Cambridge, MA, USA) and anti-SMAD3 antibody (1:100; cat. no: ab28379) was performed for 30 min at RT. Next, the sections were incubated with the secondary horseradish peroxidase (HRP, Dako) labeled antibody for 20 min at RT. Subsequently, 3,3-diaminobenzidine (DAB) was used to enable localization of the antigen–antibody complex. The tissue sections were counterstained in hematoxylin and washed with PBS buffer. Then, slides were dehydrated in increasing ethanol concentrations (80, 90, 96, and 99.8%), and finally, tissue sections were cleared in xylenes (I–IV), mounted using mounting medium, and examined.
IHC staining of TLR2 was performed using BenchMark® Ultra automated slide processing system (Ventana Medical Systems, Tucson, AZ, USA). Slides were deparaffinized and rehydrated in EZ Prep solution (Ventana Medical Systems) for 8 min at 72°C. Antigen retrieval was achieved in a high-pH Cell Conditioning (CC1) solution for 64 min. Next, incubation with the primary anti-TLR2 antibody (1:200; cat. no: ab24192, Abcam) was performed for 32 min. Antibody detection was performed using VentanaUltraView DAB Detection Kit (Ventana Medical Systems). The tissue sections were counterstained with Hematoxylin for 12 min and one drop of Bluing Reagent for 4 min. Finally, tissue sections were washed in tap water followed by dehydration in increasing ethanol concentrations (80, 90, 96, and 99.8%). Xylene was used to clear the sections, followed by mounting medium and coverslips prior to observation.
Protein expression was analyzed using an ECLIPSE E400 microscope (Nikon Instruments Europe, Amsterdam, Netherlands) at 20× and 40× magnification. All sections were reviewed separately by 2 independent pathologists without knowledge of the patient’s clinical data. The scoring system for SATB1 (SATB1n) and SMAD3 nuclear, as well as TLR2 cytoplasmic immunoreactivity, was determined by adding the multiplication of the fraction of stained cells (FSC) and the percentage of cells at each staining intensity level with the staining intensity ordinal value (scored from 0 for “no staining” to 3+ for “strong staining”), according to a modified H-score with the formula: [1 × (FSC × % cells 1+) + 2 × (FSC × % cells 2+) + 3 × (FSC × % cells 3+)], whereby FSC was calculated based on the number of stained cells per 1000 cells of the same type. The final staining score, ranging from 0 to 300, was then segregated into positive (high) and negative (low) expression on the basis of a specific discriminatory threshold established by the Evaluate Cutpoints software. 53 The cut-off values for positive and negative SATB1n, SMAD3, and TLR2 were as follows: <1; ≥1, <230; ≥230, and <1; ≥1, respectively. For evaluation of cytoplasmic SATB1 (SATB1c), slides were scored as either positive or negative based on the presence (+) or absence (−) of cytoplasmic tumor cell staining.
In our analyses, we also examined the prognostic significance of SATB1, SMAD3, TLR2 mRNA levels in The Cancer Genome Atlas (TCGA) cohort. The survival and gene expression data for the cohort of 630 NSCLC patients were obtained from www.cBioPortal.org and UCSC Xena Browser (http://xena.ucsc.edu/). The RNA-sequencing (RNA-seq) datasets were normalized using the DESeq2 method. The data was split into low-level and high-level expression groups according to cut-off points established in the Evaluate Cutpoints software. 53 Our analyses only included stage I and II cases.
Statistical analyses were performed using GraphPad Prism v 7.01 (GraphPad Software, La Jolla, CA, USA) and SPSS version 26.0 software (IBM Corporation, Armonk, NY, USA). A two-tailed Chi-squared test or Fisher’s exact test was used to assess the significance among the clinical factors and the H-scores evaluated by pathologists. Spearman’s correlation coefficient was used to assess the correlations between the expression of SATB1, SMAD3, and TLR2. Survival curves were plotted using the Kaplan–Meier method, and the differences were evaluated using a log-rank test, counting OS time from the date of operation to the date of death of any cause or the date of last follow-up. The proportionality assumption was verified by graphical examination and by testing for significant interactions when each variable was entered as a time-based covariate. Univariate and multivariate survival analyses were performed with Cox proportional hazard regression for variables that satisfied proportional hazards assumption. The hazard ratios (HRs) and 95% confidence intervals (95% CIs) were also calculated. Multivariate Cox proportional hazards models were built for each tumor marker after data were adjusted for covariates, including gender (male vs female), age (≤62 years vs >62 years), and AJCC pathological stage (stage I vs stage II). A P-value of ≤.05 was considered statistically significant.
Results
A total of 69 patients (47 male; 68.1% and 22 female; 31.9%) diagnosed with NSCLC were included in this study with a mean age at diagnosis of 63 (range 46–82 years). The most common histological type was SCC (n=37; 53.6%), followed by ADC (n=27; 39.1%) and LCC (n=5; 7.2%). According to histological differentiation, tumors were divided into G2: moderately differentiated (intermediate grade) and G3: poorly differentiated (high grade). There were 17 pT3 (24.6%) cases, 27 pT2 (39.1%) cases, and 25 pT1 (36.2%) cases. Most tumors were diagnosed at stage I (56.5%) while 30 cases (43.5%) were diagnosed at stage II. Clinical stages IA, IB, IIA, and IIB were found, respectively, in 22 (31.9%), 15 (21.7%), 9 (13%), and 23 (33.3%) patients. Postsurgical survival data was available for all patients. The median follow-up time was 1990 days and 48 (69.6%) patients died during follow-up. Clinicopathological characteristics of patients of this cohort is summarized in Table 1.
Table 1.
Immunoreactivity results for SATB1, SMAD3, TLR2 in association with clinicopathological characteristics of patients with NSCLC.
| Cases (n = 69) | SATB1n | SATB1c | SMAD3 | TLR2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | - | P-value | + | - | P-value | + | - | P-value | + | - | P-value | ||
| Histological type | |||||||||||||
| ADC | 27 | 9 | 18 | .068 | 7 | 20 | .6344 | 7 | 20 | .0629 | 7 | 20 | .0054 |
| SCC | 37 | 23 | 14 | 6 | 31 | 15 | 22 | 1 | 36 | ||||
| LCC | 5 | 2 | 3 | 1 | 4 | 4 | 1 | 0 | 5 | ||||
| Gender | |||||||||||||
| Female | 22 | 9 | 13 | .4403 | 8 | 14 | .0502 | 9 | 13 | .792 | 3 | 19 | .7034 |
| Male | 47 | 25 | 22 | 6 | 41 | 17 | 30 | 5 | 42 | ||||
| Age | |||||||||||||
| <62 | 37 | 14 | 23 | .0547 | 8 | 29 | >.9999 | 18 | 19 | .0506 | 4 | 33 | >.9999 |
| >63 | 32 | 20 | 12 | 6 | 26 | 8 | 24 | 4 | 28 | ||||
| Histologic grade | |||||||||||||
| G2 | 19 | 10 | 9 | .7918 | 5 | 14 | .5083 | 3 | 16 | .0265 | 4 | 15 | .2025 |
| G3 | 50 | 24 | 26 | 9 | 41 | 23 | 27 | 4 | 46 | ||||
| pT status | |||||||||||||
| T1 | 25 | 12 | 13 | .9411 | 3 | 22 | .368 | 10 | 15 | .0269 | 1 | 24 | .1514 |
| T2 | 27 | 14 | 13 | 6 | 21 | 14 | 13 | 3 | 24 | ||||
| T3 | 17 | 8 | 9 | 5 | 12 | 2 | 15 | 4 | 13 | ||||
| pN status | |||||||||||||
| N0 | 63 | 33 | 30 | .1981 | 14 | 49 | .3348 | 25 | 38 | .3978 | 8 | 55 | >.9999 |
| N1 | 6 | 1 | 5 | 0 | 6 | 1 | 5 | 0 | 6 | ||||
| Stage | |||||||||||||
| I | 39 | 19 | 20 | >.9999 | 7 | 32 | .7637 | 19 | 20 | .0450 | 4 | 35 | .7204 |
| II | 30 | 15 | 15 | 7 | 23 | 7 | 23 | 4 | 26 | ||||
SATB1n = nuclear immunoreactivity of SATB1; SATB1c = cytoplasmic immunoreactivity of SATB1; HR = hazard ratio; CI = confidence intervals.
Significant P-values (P < .05) are indicated in bold.
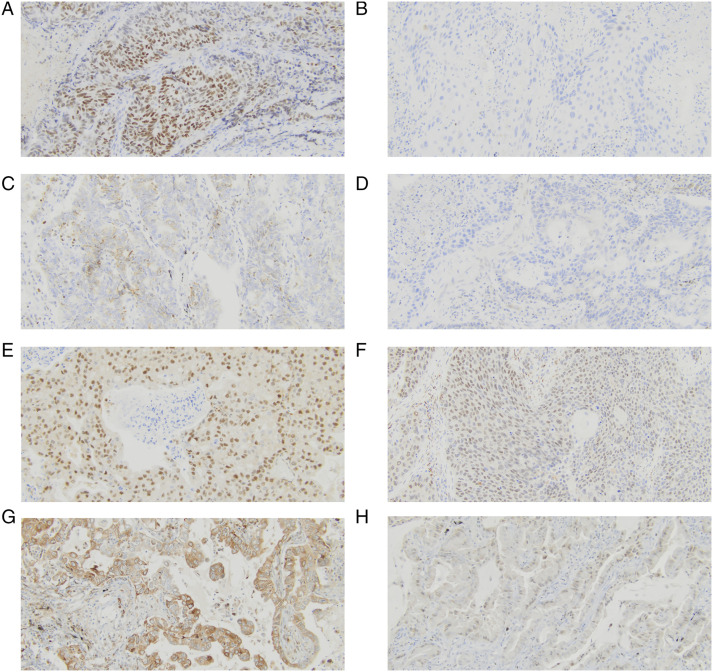
IHC staining of SATB1 was detected in the nuclear and cytoplasmic compartments of NSCLC cells (Figure 1A–1D). Positive nuclear immunoreactivity of SATB1 was found in 34 (49.28%) NSCLC cases, whereas the remaining 35 (50.72%) were negative. Cytoplasmic staining of SATB1 was present in 14 (20.29%) cases. The positive expression of SATB1n trending towards an association with histological type (P = .068). Positive expression of SATB1n was more common in SCC (n=23; 62.16%) than in LCC (n=2; 40%) and ADC (n=9; 33.33%). Moreover, positive expression of SATB1n was more frequently detected in older (62.5%) than younger people (37.83%), although this was not a significant association (P = .0547). In turn, expression of SATB1n was not associated with gender, histological grade, stage, pT, and pN status (P > .05). The association of SATB1c expression with gender was of borderline significance (P = .0502). Positive expression of SATB1c was more common in female (36.36%) than male (12.77%). Expression of SATB1c was not correlated with histological type, histological grade, age, stage, pT, and pN status. The relationship between SATB1 expression and NSCLC clinicopathological features is summarized in Table 1.
Figure 1.
Immunohistochemical analysis of SATB1n, SATB1c, SMAD3, and TLR2 expression in NSCLC tissues (primary magnification ×20). (A) Strong positive (+3) nuclear staining for SATB1, (B) Negative expression of nuclear staining for SATB1, (C) Positive cytoplasmic staining for SATB1, (D) Negative expression of cytoplasmic SATB1, (E) Strong positive (+3) staining for SMAD3, (F) Negative expression of SMAD3, (G) Strong positive (+3) staining for TLR2, (H) Negative expression of TLR2.
IHC staining of SMAD3 was detected in the nuclear compartments of NSCLC cells of 26 (37.68%) NSCLC cases (Figure 1E–1F). The relationship between SMAD3 expression and clinicopathological features was analyzed and demonstrated the association with histological grade (P = .0265), pT status (P = .0269), and tumor stage (P = .045). SMAD3 overexpression was more frequently detected in poorly (n=23; 46.00%) differentiated tumors than in moderately differentiated ones (n=3; 15.79%) (P = .00265). The ratio of SMAD3 overexpression was also significantly higher in patients with pT2 (n=14; 51.85%) NSCLCs than in those with pT1 (n=10; 40.00%) and pT3 (n=2; 11.76%) tumors (P = .0269). Moreover, the overexpression of SMAD3 was more frequently detected in stage I (n=19; 48.72%) tumors than in those with stage II (23.33%) (P = .0450). The prevalence of SMAD3 overexpression was higher in LCC (80%) than in SCC (40.54%) and ADC (25.93%) although this was not a significant association (P = .0629). In addition, high SMAD3 levels were trending towards a correlation with age (P = .0506), being more frequently detected in younger (48.65%) than older people (25%). The expression status of SMAD3 was not associated with gender and pN status. The relationship between SMAD3 expression and NSCLC clinicopathological features is summarized in Table 1.
IHC staining of TLR2 was detected in the cytoplasmic compartments of 8 (11.6%) NSCLC cells (Figure 1G–1H). The ratio of TLR2 overexpression was more common in ADC (n = 7; 25.93%) than in SCC (n = 1; 2.7%) and LCC (n = 0; 0%) (P = .0054). The expression status of TLR2 was not correlated with gender, age, histological grade, stage, pT, and pN status. Representative images of IHC staining for all histological types are demonstrated in Supplementary Figure 2.
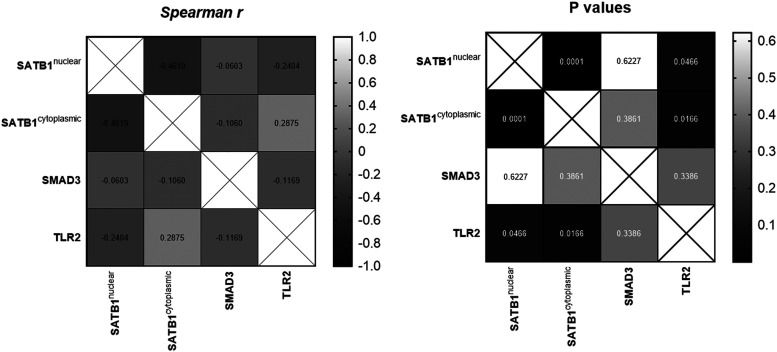
A weak negative and significant association was confirmed between the expression of SATB1n and TLR2 (P = .0466, Spearman coefficient r = −.2404). Furthermore, weak positive and significant association was found between SATB1c and TLR2 expression (P = .0166, Spearman coefficient r = .2875). In addition, a moderately negative association was confirmed between the expression of SATB1n and SATB1c (P = .0001, Spearman coefficient r = −.4619). In the entire cohort, the expression of SMAD3 was not significantly correlated with the expression of SATB1 and TLR2 (Figure 2).
Figure 2.
Correlation between SATB1n, SATB1c, SMAD3, and TLR2 expression in lung cancer tissues. Correlation values are presented in a heat map (Spearman correlation test).
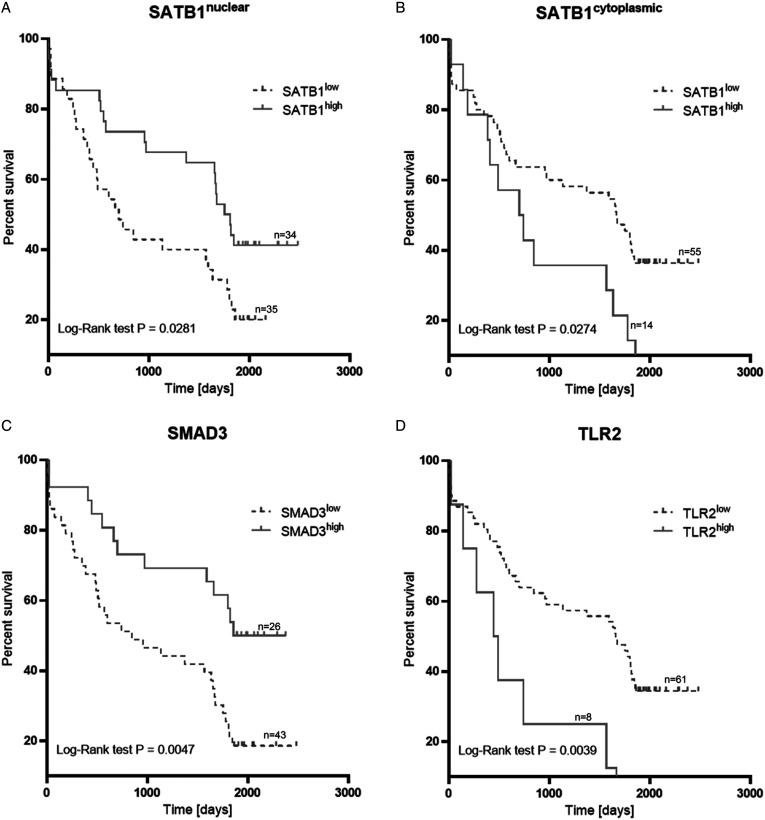
Kaplan–Meier survival analysis indicated that NSCLC patients with a high level of SATB1n expression (median OS 1781 days) had higher OS rates (log-rank test P = .028) than those with SATB1n low expression level (median OS 701 days). NSCLC patients with the presence of SATB1c expression (median OS 722.5 days) had lower OS rates (log-rank test P = .0274) than those with low-level expression (median OS 1668 days). Kaplan–Meier analysis also revealed the significance of SMAD3 and TLR2 expression for NSCLCs. We found that high expression of TLR2 correlated with decreased OS rates (log-rank test P = .0039), and high expression of SMAD3 correlated with increased OS rates (log-rank test P = .0047) (Figure 3). Median OS periods for TLR2 high and TLR2low, as well as SMAD3high and SMAD3low were 467 days/1662 days and 2116 days/846 days, respectively.
Figure 3.
Overall survival analysis according to the expression of SATB1n (A), SATB1c (B), SMAD3 (C), and TLR2 (D).
Univariate analysis demonstrated that positive SATB1n expression was significantly associated with a better survival prognosis (HR .53, 95%CI .30–.94, P = .031), and it persisted as an independent prognostic factor for improved OS in multivariate analysis after adjustment for age, gender, and stage (HR .49, 95%CI .27–.90, P = .022). In the case of SATB1c, the univariate Cox analysis revealed its presence predicted an unfavorable OS (HR 2.04, 95%CI 1.07–3.88, P = .031). When examined in multivariate analysis, SATB1c remained as an independent prognostic factor in terms of OS (adjusted HR 2.11, 95%CI 1.07–4.16, P = .030). Likewise, TLR2 was a significant predictor of poor OS in both univariate (HR 3.02, 95%CI 1.37–6.66, P = .006) and multivariate (adjusted HR 3.00, 95%CI 1.36–6.63, P = .007) analysis. Furthermore, univariate analysis showed a longer OS was significantly correlated with high SMAD3 expression (HR .41, 95%CI .21–.78, P = .006), a result that was maintained during multivariate analysis following adjustment for covariates (HR = .40, 95% CI .20–.78; P = .007). Results for univariate and multivariate analysis are summarized in Table 2 and Table 3, respectively.
Table 2.
Univariate analysis of prognostic factors by Cox proportional hazard model (n = 69).
| Variable | Univariate analysis | |||
|---|---|---|---|---|
| HR | 95% CI | P-value | ||
| SATB1n | .53 | .30 | .94 | .031 |
| SATB1c | 2.04 | 1.07 | 3.88 | .031 |
| SMAD3 | .41 | .21 | .78 | .006 |
| TLR2 | 3.02 | 1.37 | 6.66 | .006 |
| Gender | 1.02 | .56 | 1.87 | .96 |
| Age | 1.03 | .99 | 1.07 | .17 |
| Stage | .84 | .48 | 1.48 | .55 |
| pT status | .68 | .36 | 1.29 | .24 |
| pN status | 1.7 | .67 | 4.32 | .27 |
SATB1n = nuclear immunoreactivity of SATB1; SATB1c = cytoplasmic immunoreactivity of SATB1; HR = hazard ratio; CI = confidence intervals.
Significant P-values (P < .05) are indicated in bold.
Table 3.
Multivariate analysis of prognostic factors by Cox proportional hazard model (n = 69).
| Variable | Multivariate analysis: SATB1n | Variable | Multivariate analysis: SATB1c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95.0% CI | P-value | HR | 95.0% CI | P-value | ||||
| SATB1n | .49 | .27 | .90 | .022 | SATB1c | 2.11 | 1.07 | 4.16 | .030 |
| Gender | .90 | .49 | 1.66 | .74 | Gender | .86 | .46 | 1.61 | .64 |
| Age | 1.23 | .69 | 2.22 | .48 | Age | 1.08 | .61 | 1.90 | .80 |
| Stage | 1.19 | .67 | 2.10 | .55 | Stage | 1.12 | .63 | 1.98 | .70 |
SATB1n = nuclear immunoreactivity of SATB1; SATB1c = cytoplasmic immunoreactivity of SATB1; HR = hazard ratio; CI = confidence intervals.
Likelihood ratio P, adjusted for gender, age and stage. Significant P-values (P < .05) are indicated in bold.
Having established the significance of SATB1, SMAD3, and TLR2 as single prognostic markers in our cohort of NSCLC patients, we also examined the impact of their combined expression on OS. As shown by Kaplan–Meier analysis, the best OS was observed for patients whose NSCLCs simultaneously expressed SATB1n and SMAD3 at high level and TLR2 at low level. In turn, patients whose NSCLCs had opposite expression profile of the 3 proteins had dramatically shorter OS (undefined vs 490 days; P < .0001) (Figure 4A). A univariate analysis of a combined 3-protein panel of SATB1n-high/SMAD3high/TLR2low was associated with better survival prognosis (HR .19, 95%CI .06–.62, P = .006) (Table 4) and was a potent independent prognostic marker for NSCLC patients when examined in a multivariate analysis (adjusted HR .19, 95%CI .06–.63, P =.007) (Table 5). The worst OS was seen for SATB1n-lowTLR2high co-expressing tumors with a particularly short median OS (445 days), while the patients with the opposite expression profile had median OS of 1807 days (P = .0003) (Figure 4B).
Figure 4.
Overall survival analysis according to the combination of the protein panel: SATB1nSMAD3TLR2 (A), SATB1nTLR2 (B).
Table 4.
Univariate analysis of prognostic factors by Cox proportional hazard model for combined expression of proteins (n = 69).
| Variable | Univariate analysis: SATB1/SMAD3/TLR2 | |||
| HR | 95% CI | P-value | ||
| Others | Ref | |||
| SATB1n-high/SMAD3high/TLR2low | .19 | .06 | .62 | .006 |
| SATB1n-low/SMAD3low/TLR2high | 2.34 | .90 | 6.11 | .082 |
SATB1n = nuclear immunoreactivity of SATB1; SATB1c = cytoplasmic immunoreactivity of SATB1; HR = hazard ratio; CI = confidence intervals.
Significant P-values (P < .05) are indicated in bold. Cases designated ‘others’ grouped the remaining combinations of expression patterns.
Table 5.
Multivariate analysis of prognostic factors by Cox proportional hazard model for combined expression of proteins (n = 69).
| Variable | Multivariate analysis: SATB1/SMAD3/TLR2 | |||
|---|---|---|---|---|
| HR | 95% CI | P-value | ||
| Others | Ref | |||
| SATB1n-high/SMAD3high/TLR2low | .19 | .06 | .63 | .007 |
| SATB1n-low/SMAD3low/TLR2high | 1.87 | .68 | 5.17 | .225 |
| Gender | 1.00 | .54 | 1.85 | .989 |
| Age | 1.03 | .99 | 1.07 | .156 |
| Stage | .62 | .34 | 1.13 | .116 |
SATB1n = nuclear immunoreactivity of SATB1; SATB1c = cytoplasmic immunoreactivity of SATB1; HR = hazard ratio; CI = confidence intervals.
Likelihood ratio P, adjusted for gender, age and stage. Significant P-values (P < .05) are indicated in bold. Cases designated ‘others’ grouped the remaining combinations of expression patterns.
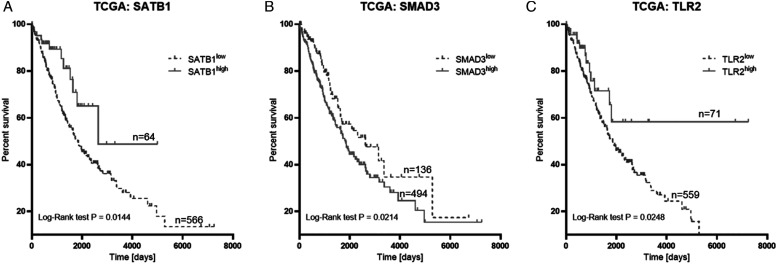
The analysis of prognostic significance of SATB1 mRNA levels in the TCGA cohort revealed that SATB1 overexpression was significantly associated with better OS of NSCLC patients (P = .0144). The median OS times were 1830 and 2639 days for low and high expression groups, respectively. Furthermore, the TCGA dataset showed that SMAD3 overexpression was associated with significantly shorter OS (2620 days vs 1830 days; P = .0214). Finally, Kaplan–Meier survival analysis revealed that high TLR2 expression was significantly associated with better survival time (1841 days) of NSCLC patients in comparison to those with its low expression level (634 days; P = .0248) (Figure 5).
Figure 5.
Overall survival analysis of SATB1 (A), SMAD3 (B), TLR2 (C) mRNA levels in TCGA cohort.
Discussion
NSCLC is among the most frequently diagnosed malignancies and the leading cause of cancer-related death worldwide. Early diagnosis and treatment of NSCLC is a prerequisite for increasing survival. Those patients with early-stage NSCLC but identified as low risk require less aggressive therapy, while those classified as high risk might be good candidates for adjuvant therapy. Therefore, it is essential to establish diagnostic and prognostic markers that can identify early-stage NSCLC patients who require more aggressive therapy.
In the past few years, research has been conducted to assess the expression level and role of SATB1 in many kinds of human tumors, including lung cancer. However, there are contradictory results about SATB1 expression levels, especially when examining prognostic and clinicopathological features. Several studies have shown discrepancies regarding SATB1 expression in lung tumors compared to normal lung tissues.16,25,27 Due to the lack of data on SATB1 expression in normal bronchial tissues in ourcohort, it was not possible to verify these findings.17,54–56 Several attempts have also been made to investigate the correlation between SATB1 expression and clinicopathological features of NSCLC patients. In our investigation, we observed cytoplasmic and nuclear staining of the SATB1 protein in cancer cells. Nuclear SATB1 positivity was 49.28% and higher than the rates reported by Glatzel-Plucinska et al. and Selinger et al.16,25 We presume that the discrepancy between the positivity rates may be due to differences in the experimental design, for example, evaluation of IHC reactions, selected cut-offs, antibody clones, and the subjectivity of the pathologists interpretation. Several studies from other authors have reported that SCCs show markedly higher SATB1 expression level compared to ADC.16,25,27 A similar trend was also reported in our study for SATB1n, but the data were not significant (P = .068). In our cohort, the SATB1n positivity rate occurred more frequently in SCC (62.16%) than ADC (33.33%). SCCs and ADCs differ in gene expression profile, cellular origin, and also targeting mutations, and this may be responsible for the differences in the frequency of SATB1 positive cases. Furthermore, a congruous relationship has also been demonstrated by Glatzel-Plucinska et al. in SCC and ADC cell lines. Moreover, the previous research has shown that SATB1 expression was associated with the degree of tumor differentiation in clinical ADC and SCC samples, although this could not be confirmed in our study, possibly due to the smaller cohort size. In addition, our study included only moderately and poorly differentiated tumors. Selinger et al. noticed that a high SATB1 level was associated with an early disease stage. The patients in our cohort had stage I or II disease; therefore, we could not make comparisons with more advanced clinical stages. Several studies have found a significant associations between SATB1 expression and OS of NSCLC patients. Our cohort study showed that patients with high SATB1n expression had better median OS than did patients with SATB1n underexpression (1781 vs 701 days). Moreover, and based on the TCGA dataset, we found that SATB1 overexpression was also significantly associated with better OS of NSCLC patients. Furthermore, we revealed that an elevated SATB1 expression was an independent prognostic factor that predicted superior survival in NSCLC patients, which was also confirmed by Glatzel-Plucinska et al. 16 Additionally, Selinger et al. 25 have demonstrated that a loss of SATB1 expression was a negative prognostic factor for patients with SCC. However, our examination of cytoplasmic SATB1 showed these patients had lower OS rates than those with SATB1c underexpression. Furthermore, we demonstrated that high SATB1c expression was an independent prognostic factor which predicted poor OS. To the best of our knowledge, this is the first study to show that SATB1c was significantly associated with OS of NSCLC patients. The findings may imply that depending on subcellular localization, SATB1 has an opposite prognostic significance in terms of OS in early-stage NSCLC patients. Interestingly, Selinger et al. also observed cytoplasmic staining of SATB1, but this staining pattern was not associated with survival or any other clinicopathological parameters. Other cited authors omitted the estimation of the cytoplasmic fraction of SATB1 or combined it with the nuclear fraction, which seems unjustified in view of our results. Undoubtedly, our results suggested an association of both nuclear and cytoplasmic SATB1 expression with patient OS, highlighting the role of SATB1 in tumor progression.
SMAD is a critical intracellular mediator of TGFβ signaling from the cell surface to the nucleus, and the subject of this investigation, SMAD3, is involved in regulating gene activity, cell proliferation, differentiation, and cell death. A number of previous studies demonstrated that SMAD3-mediated TGFβ signaling is implicated in tumor angiogenesis, tissue invasion, and metastasis. Scientists agree that the overexpression of SMAD3 is involved in the regulation of various physiological, as well as pathological processes, including carcinogenesis. However, its role in tumorigenesis is not clear.57,58 The results of the present study indicated that SMAD3 was overexpressed in 37.68% of NSCLC cases. Our study further demonstrated that SMAD3 expression was significantly correlated with histopathological grade, pT status, and stage, but not with gender and pN status. Moreover, the overexpression of SMAD3 was trending towards a correlation with histological type and age. In our cohort, it was more likely to have SMAD3 positivity in poorly differentiated cancer cells than in those that were moderately differentiated, which could indicate that SMAD3 is involved in the progression of NSCLC. On the other hand, our results revealed an association of SMAD3 with favorable prognostic variables, such as smaller tumor size and more frequent occurrence in stage I tumors. In recent years, several studies have been carried out to examine the relationship between SMAD3 and the prognosis of cancer patients.33,59,60 In contrast to the cited results, we found that patients with high SMAD3 expression had better median OS than did patients with SMAD3 underexpression. Cox regression analyses showed that overexpression of SMAD3 was a favorable independent prognostic factor for OS. This discordance between studies may be due to different study population or ethnicity-related differences in NSCLC biology. Moreover, Niu et al. used tissue microarrays (TMAs), but the heterogeneity of whole tumor samples could confound microarray analysis, so in our study we stained whole-tissue sections. Notably, in our report, we included only patients in stage I to II of NSCLC, whereas Marwitz et al. did not provide patient stage information from their cohort, and Niu et al. presented stage data for 18 (15.13%) patients only, all of whom had stage III and IV lung cancer. Therefore, it is most probable that in NSCLC, SMAD3 may act as a negative regulator of carcinogenesis and improve patient survival in early disease stages, while it functions in the opposite manner in advanced cancers, resulting in diminished survival in patients. It is not unexpected since, based on experimental model systems, a similar duality of function under different tumor stages has been reported for TGFβ, also in the case of lung cancer. In the experimental studies on other tumor types, this paradoxical effect of TGFβ has been shown to be mediated through , for example, SMAD3. 61 On the other hand, our analyses based on TCGA have shown that the expression of SMAD3 mRNA was significantly negatively correlated with patient survival, which is in agreement with Niu et al.59,60 Pan et al. 62 revealed that SMAD3 was a tumor suppressor for post-progression survival but an oncogene for progression-free survival. In turn, Zeng et al. 63 showed that increased SMAD3 mRNA levels were not related to OS in NSCLC patients from the CBioPortal cohort. These results suggest an inverse role for SMAD3 protein and mRNA in the clinical behavior of NSCLC. A mismatch between mRNA and protein levels has been widely reported in the literature.64–66 The difference between mRNA and protein expression with respect to prognostic significance confirms the importance of comprehensive tumor analyses. However, it cannot be assumed that the amount of mRNA is directly correlated with protein expression. Noteworthily, posttranscriptional and posttranslational mechanisms may influence protein levels, and increased mRNA levels may produce only small amounts of detectable proteins. 67 Based on our results, we can conclude that SMAD3 protein is perhaps functionally associated with better prognosis in early-stage NSCLC, whereas the contrary is true for SMAD3 mRNA. However, our conclusions are constrained by the fact that presented results come from disparate research population, that is, for proteins from our own cohort and mRNAs from the TCGA cohort. Definitely, to validate this concept, it is necessary to estimate mRNA and protein expression in one cohort of patients. 68
TLR2 is a member of the TLR family, mainly expressed by immune cells, but as they are also expressed on tumor cells. Thus, in recent years, a lot of scientific interest has focused on TLR expression and their functions in cancer cells, and a growing body of evidence underscores the correlation between TLR expression and cancer prognosis. In recent years, numerous studies have indicated that TLR2 is expressed on neoplastic cells from several solid tumors, such as brest, 69 gastrin, 70 colon, 71 oral, 72 and pancreatic cancer. 73 In addition, it has been revealed that activation of TLR2 promotes cancer progression and metastasis through different cell-intrinsic mechanisms, 74 and its expression closely associates with patient prognosis.69,70,75 To our knowledge, the present study is the first to investigate the correlation between TLR2 protein expression and clinicopathological features in patients with clinical stages I to II NSCLC. In our cohort, TLR2 was overexpressed in 11.6% of NSCLC cases and TLR2 positivity rate more frequently occurred in ADC (25.9%) than SCC (2.7%) and LCC (0%). These results suggest that ADC tumors express higher levels of TLR2 than SCC and LCC. A similar relationship was observed by Gergen et al., but in this case, it was found in ADC and non-adenocarcinoma cell lines. 54 Furthermore, several authors have investigated the correlation between TLR2 expression and patient survival.69,70,73,75,76 Bauer et al. provided evidence for a significant association between high mRNA expression of TLR2 in TCGA cohort of NSCLC patients. According to their report, expression of TLR2 is associated with improved survival outcomes in NSCLC, and our analysis of TCGA data supports this finding. However, and contrary to mRNA expression, we found using IHC that high TLR2 protein levels were significantly associated with worse OS of NSCLC patients. Moreover, we demonstrated that TLR2 was a significant predictor of poor OS in both univariate and multivariate analyses. These results suggest opposite role for TLR2 protein and mRNA in the clinical behavior of stage I to II NSCLCs. To confirm this supposition, it is necessary to estimate mRNA and protein expression in the same cohort of patients. These results suggest an opposite role for TLR2 protein and mRNA in the clinical behavior of stage I to II NSCLCs. Notably, TLR2 is not only expressed in tumor cells but also in immune cells, endothelial and epithelial cells, serving as internal staining controls. IHC staining enables for quantitative evaluation of proteins in a morphological and subcellular context, an advantage of this method over gene expression analyses. To confirm this supposition, similar to our findings with SMAD3, it is necessary to estimate mRNA and protein expression in the same cohort of patients following laser tissue microdissection.
Lastly, given the functional relationship between SMAD3 and SATB1, 77 as well as a potential link between the latter and TLR2,44 we attempted to determine the effect of the combined expression of these proteins on OS of NSCLC patients. Kaplan–Meier survival analysis demonstrated that the subset of patients with tumors that co-expressed high levels of SATB1 and SMAD3, and simultaneously low levels of TLR2 had the best OS, and the combined expression of the 3 markers better predicted patient survival than looking at each marker individually. Furthermore, a combined 3-protein panel of SATB1n-high/SMAD3high/TLR2low emerged as a powerful independent prognostic factor associated with better outcome. Therefore, our analyses showed that utilizing IHC to examine the combined expression of SATB1n, SMAD3, and TLR2 could be more helpful for predicting the prognosis of NSCLC patients than single markers. Given that our study group is limited in number, it is necessary to confirm this finding in large-scale studies.
In summary, our cohort study revealed that nuclear SATB1 expression was associated with favorable patient survival, whereas cytoplasmic SATB1 had a significant correlation with poor outcome. Interestingly, nuclear and cytoplasmic SATB1 appeared to be independent markers either for better or worse prognosis, respectively. These data suggest that a differential subcellular expression of SATB1 plays an important role in the pathology and/or progression of NSCLC, and clinically, it has an inverse prognostic significance. Furthermore, herein we found that high expression levels of SMAD3 and TLR2 were independent prognostic markers associated with favorable and poor survival, respectively. Importantly, stratification of NSCLCs with respect to combined expression of the 3 biomarkers allowed us to identify subgroups of patients with the greatest difference in survival. Specifically, the expression profile of SATB1n-high/SMAD3high/TLR2low was associated with the best OS, and it was superior to each single protein alone in predicting patient prognosis. Based on the TCGA dataset, we revealed that overexpression of SATB1 mRNA was significantly associated with better OS, while high mRNA levels of SMAD3 and TLR2 were negatively correlated with patient survival. Overall, each of these proteins may have a potential clinical utility for determining a patient’s prognosis in stage I and II NSCLC patients by themselves, although when a combined 3 protein panel is used; this provides a stronger indication of OS than each marker alone. However, our results should be interpreted cautiously due to existence of some limitations. This study needs validation with a larger patient cohort, and in prospective and multicenter studies.
Supplemental Material
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748211056697 for Prognostic Significance of TLR2, SMAD3 and Localization-dependent SATB1 in Stage I and II Non–Small-Cell Lung Cancer Patients by Justyna Durślewicz, Anna Klimaszewska-Wiśniewska, Jakub Jóźwicki, Paulina Antosik, Marta Smolińska-Świtała, Maciej Gagat, Adam Kowalewski and Dariusz Grzanka in Cancer Control
Supplemental Material, sj-pdf-2-ccx-10.1177_10732748211056697 for Prognostic Significance of TLR2, SMAD3 and Localization-dependent SATB1 in Stage I and II Non–Small-Cell Lung Cancer Patients by Justyna Durślewicz, Anna Klimaszewska-Wiśniewska, Jakub Jóźwicki, Paulina Antosik, Marta Smolińska-Świtała, Maciej Gagat, Adam Kowalewski and Dariusz Grzanka in Cancer Control
Notations
- ADC
- adenocarcinoma
- AJCC
- The American Joint Committee on Cancer
- BUR
- base unpairing region
- FFPE
- paraffin-embedded tissue samples
- IHC
- immunohistochemical
- LCC
- large cell carcinoma
- MAR
- matrix attachment region
- NMP
- nuclear matrix protein
- NSCLC
- non–small-cell lung carcinoma
- OS
- overall survival
- PAMP
- pathogen-associated molecular pattern
- SATB1
- special AT-rich binding protein 1
- SCC
- squamous cell carcinoma
- SCLC
- small-cell lung carcinoma
- TGFβ
- transforming growth factor-β
- TLR
- toll-like receptor
- WHO
- World Health Organization
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported by research task MN-SDL-11/WL/2019 within the frame-work of basal research activity (Nicolaus Copernicus University in Toruń, Faculty of Medicine, Collegium Medicum in Bydgoszcz).
Ethics Statement: The study protocol has been approved by The Ethics Committee of Nicolaus Copernicus University in Toruń, LudwikRydygier Collegium Medicum in Bydgoszcz (approval number KB 336/2018).
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Justyna Durślewicz https://orcid.org/0000-0001-7301-5324
Anna Klimaszewska-Wiśniewska https://orcid.org/0000-0002-8493-3806
Jakub Jóźwicki https://orcid.org/0000-0002-2771-413X
References
- 1.World Health Organisation . Latest Global Cancer Data. Cancer Burden Rises to 18.1 Million New Cases and 9.6 Million Cancer Deaths in 2018. Lyon, France: International Agency for Research on Cancer; 2018. [Google Scholar]
- 2.American Cancer Society . Facts & Figures. Atlanta, GA: American Cancer Society; 2019. https://www.cancer.org/. Accessed May, 1 2021. [Google Scholar]
- 3.Lemjabbar-Alaoui H, Hassan OU, Yang Y-W, Buchanan P. Lung cancer: biology and treatment options. Biochim Biophys Acta Rev Cancer. 2015;1856:189-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shankar A, Dubey A, Saini D, et al. Environmental and occupational determinants of lung cancer. Transl Lung Cancer Res. 2019;8:S31-S49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. 2008;83:355-367. [DOI] [PubMed] [Google Scholar]
- 6.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putora PM, Ess S, Panje C, et al. Prognostic significance of histology after resection of brain metastases and whole brain radiotherapy in non-small cell lung cancer (NSCLC). Clin Exp Metastasis. 2015;32:143-149. [DOI] [PubMed] [Google Scholar]
- 9.Jackman DM, Zhang Y, Dalby C, et al. Cost and survival analysis before and after implementation of Dana-Farber clinical pathways for patients with stage IV non-small-cell lung cancer. J Oncol Pract. 2017;13:e346-e352. [DOI] [PubMed] [Google Scholar]
- 10.Rasco DW, Yan J, Xie Y, Dowell JE, Gerber DE. Looking beyond surveillance, epidemiology, and end results: patterns of chemotherapy administration for advanced non-small cell lung cancer in a contemporary, diverse population. J Thorac Oncol. 2010;5:1529-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavan Kumar P, Purbey PK, Sinha CK, et al. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231-243. [DOI] [PubMed] [Google Scholar]
- 12.Kohwi-Shigematsu T, Poterlowicz K, Ordinario E, Han H-J, Botchkarev VA, Kohwi Y. Genome organizing function of SATB1 in tumor progression. Semin Cancer Biol. 2013;23:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521-535. [PMC free article] [PubMed] [Google Scholar]
- 14.Han H-J, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187-193. [DOI] [PubMed] [Google Scholar]
- 15.Lu X, Cheng C, Zhu S, et al. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol Rep. 2010;24:981-987. [DOI] [PubMed] [Google Scholar]
- 16.Glatzel-Plucinska N, Piotrowska A, Grzegrzolka J, et al. SATB1 level correlates with Ki-67 expression and is a positive prognostic factor in non-small cell lung carcinoma. Anticancer Res. 2018;38:723-736. [DOI] [PubMed] [Google Scholar]
- 17.Zhou LY, Liu F, Tong J, Chen QQ, Zhang FW. [Expression of special AT-rich sequence-binding protein mRNA and its clinicopathological significance in non-small cell lung cancer]. J South Med Univ. 2009;29:534-537. [PubMed] [Google Scholar]
- 18.Zhao X-D, Ji W-y, Zhang W, et al. Overexpression of SATB1 in laryngeal squamous cell carcinoma. ORL (Basel). 2010;72:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Frömberg A, Rabe M, Aigner A. Multiple effects of the special at-rich binding protein 1 (SATB1) in colon carcinoma. Int J Cancer. 2014;135:2537-2546. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Wang X, Wang Q. Expression of SATB1 and E-cad in tissues of patients with endometrial carcinoma and the relationship with clinicopathological features. Exp Therap Med. 2018;15:4339-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao L, Yang C, Wang J, et al. SATB1 is overexpressed in metastatic prostate cancer and promotes prostate cancer cell growth and invasion. J Transl Med. 2013;11:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu W, Luo M, Wang Z, et al. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int. 2012;32:1064-1078. [DOI] [PubMed] [Google Scholar]
- 23.Nodin B, Hedner C, Uhlén M, Jirström K. Expression of the global regulator SATB1 is an independent factor of poor prognosis in high grade epithelial ovarian cancer. J Ovarian Res. 2012;5:24-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han B, Luan L, Xu Z, Wu B. Expression and biological roles of SATB1 in human bladder cancer. Tumor Biol. 2013;34:2943-2949. [DOI] [PubMed] [Google Scholar]
- 25.Selinger CI, Cooper WA, Al-Sohaily S, et al. Loss of special at-rich binding protein 1 expression is a marker of poor survival in lung cancer. J Thorac Oncol. 2011;6:1179-1189. [DOI] [PubMed] [Google Scholar]
- 26.Qi H, Fu X, Li Y, et al. SATB1 promotes epithelial-mesenchymal transition and metastasis in prostate cancer. Oncol Letter. 2017;13:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang B, Zhou H, Wang S, Lang XP, Wang X. Effect of silencing SATB1 on proliferation, invasion and apoptosis of A549 human lung adenocarcinoma cells. Oncol Letter. 2016;12:3818-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmierer B, Hill CS. TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970-982. [DOI] [PubMed] [Google Scholar]
- 29.Feng X-H, Derynck R. Specificity and versatility in TGF-β signaling through SMADS. Annu Rev Cell Dev Biol. 2005;21:659-693. [DOI] [PubMed] [Google Scholar]
- 30.Afrakhte M, Morén A, Jossan S, et al. Induction of inhibitory SMAD6 and SMAD7 mRNA by TGF-β family members. Biochem Biophys Res Commun. 1998;249:505-511. [DOI] [PubMed] [Google Scholar]
- 31.Chen C-L, Tsukamoto H, Liu J-C, et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest. 2013;123:2832-2849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Zhang J, Zhang L, Cui H, et al. High expression levels of SMAD3 and SMAD7 at diagnosis predict poor prognosis in acute myeloid leukemia patients undergoing chemotherapy. Cancer Gene Ther. 2019;26:119-127. [DOI] [PubMed] [Google Scholar]
- 33.Kim S-H, Kim K-H, Ahn S, Hyeon J, Park C-K. SMAD3 and SMAD3 phosphoisoforms are prognostic markers of gastric carcinoma. Dig Dis Sci. 2013;58:989-997. [DOI] [PubMed] [Google Scholar]
- 34.Millet C, Zhang YE. Roles of SMAD3 in TGF-β signaling during carcinogenesis. Crit Rev Eukaryot Gene Expr. 2007;17:281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Nardo D. Toll-like receptors: activation, signalling and transcriptional modulation. Cytokine. 2015;74:181-189. [DOI] [PubMed] [Google Scholar]
- 36.Chung YH, Kim D. Enhanced TLR4 expression on colon cancer cells after chemotherapy promotes cell survival and epithelial-mesenchymal transition through phosphorylation of GSK3β. Anticancer Res. 2016;36:3383-3394. [PubMed] [Google Scholar]
- 37.Gambara G, Desideri M, Stoppacciaro A, et al. TLR 3 engagement induces IRF ‐3‐dependent apoptosis in androgen‐sensitive prostate cancer cells and inhibits tumour growth in vivo. J Cell Mol Med. 2015;19:327-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christmas P. Toll-like receptors: sensors that detect infection. Nat Educ. 2010;3:85. [Google Scholar]
- 39.Kawai T, Akira S. Pathogen recognition with toll-like receptors. Curr Opin Immunol. 2005;17:338-344. [DOI] [PubMed] [Google Scholar]
- 40.Gergen AK, Kohtz PD, Halpern AL, et al. Activation of toll-like receptor 2 promotes proliferation of human lung adenocarcinoma cells. Anticancer Res. 2020;40:5361-5369. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Wang S, Zhu R, Li H, Han Q, Zhao RC. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol Oncol. 2016;9:42-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Lorenzo A, Bolli E, Tarone L, Cavallo F, Conti L. Toll-like receptor 2 at the crossroad between cancer cells, the immune system, and the microbiota. Int J Mol Sci. 2020;21:1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikami F, Lim JH, Ishinaga H, et al. The transforming growth factor-β-Smad3/4 signaling pathway acts as a positive regulator for TLR2 induction by bacteria via a dual mechanism involving functional cooperation with NF-κB and MAPK phosphatase 1-dependent negative cross-talk with p38 MAPK. J Biol Chem. 2006;281:22397-22408. [DOI] [PubMed] [Google Scholar]
- 44.Karim AF, Reba SM, Li Q, Boom WH, Rojas RE. Toll like receptor 2 engagement on CD4+T cells promotes TH9 differentiation and function. Eur J Immunol. 2017;47:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali A, Zhang P, Liangfang Y, et al. KLF17 empowers TGF-β/Smad signaling by targeting Smad3-dependent pathway to suppress tumor growth and metastasis during cancer progression. Cell Death Dis. 2015;6:e1681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akinosoglou KS, Karkoulias K, Marangos M. Infectious complications in patients with lung cancer. Eur Rev Med Pharmacol Sci. 2013;17:8-18. [PubMed] [Google Scholar]
- 47.Qiao D, Wang Z, Lu Y, Wen X, Li H, Zhao H. A retrospective study of risk and prognostic factors in relation to lower respiratory tract infection in elderly lung cancer patients. Am J Cancer Res. 2015;5:423-432. [PMC free article] [PubMed] [Google Scholar]
- 48.Bremnes RM, Al-Shibli K, Donnem T, et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol. 2011;6:824-833. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Wang C, Shan S, Liu X, Jiang Z, Ren T. TLR4/ROS/miRNA-21 pathway underlies lipopolysaccharide instructed primary tumor outgrowth in lung cancer patients. Oncotarget. 2016;7:42172-42182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye M, Gu X, Han Y, Jin M, Ren T. Gram-negative bacteria facilitate tumor outgrowth and metastasis by promoting lipid synthesis in lung cancer patients. J Thorac Dis. 2016;8:1943-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naik R, Galande S. SATB family chromatin organizers as master regulators of tumor progression. Oncogene. 2019;38:1989-2004. [DOI] [PubMed] [Google Scholar]
- 52.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based human protein Atlas. Nat Biotechnol. 2010;28:1248-1250. [DOI] [PubMed] [Google Scholar]
- 53.Ogłuszka M, Orzechowska M, Jędroszka D, Witas P, Bednarek AK. Evaluate cutpoints: adaptable continuous data distribution system for determining survival in Kaplan-Meier estimator. Comput Methods Progr Biomed. 2019;177:133-139. [DOI] [PubMed] [Google Scholar]
- 54.Beer DG, Kardia SLR, Huang C-C, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816-824. [DOI] [PubMed] [Google Scholar]
- 55.Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot SG, Estilo C, Maghami E, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063-3071. [DOI] [PubMed] [Google Scholar]
- 57.Ku J-L, Park S-H, Yoon K-A, et al. Genetic alterations of the TGF-β signaling pathway in colorectal cancer cell lines: a novel mutation in Smad3 associated with the inactivation of TGF-β-induced transcriptional activation. Cancer Lett. 2007;247:283-292. [DOI] [PubMed] [Google Scholar]
- 58.Wolfraim LA, Fernandez TM, Mamura M, et al. Loss of Smad3 in Acute T-Cell Lymphoblastic Leukemia. N Engl J Med. 2004;351:552-559. [DOI] [PubMed] [Google Scholar]
- 59.Niu H, Huang Y, Yan L, et al. Knockdown of SMAD3 inhibits the growth and enhances the radiosensitivity of lung adenocarcinoma via p21 in vitro and in vivo. Int J Biol Sci. 2020;16:1010-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marwitz S, Ballesteros-Merino C, Jensen SM, et al. Phosphorylation of SMAD3 in immune cells predicts survival of patients with early stage non-small cell lung cancer. J Immuno Therap Cancer. 2021;9:e001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han G, Wang X-J. Roles of TGFβ signaling Smads in squamous cell carcinoma. Cell Biosci. 2011;1:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan S, Zhou G, Hu W, Pei H. SMAD-6, -7 and -9 are potential molecular biomarkers for the prognosis in human lung cancer. Oncol Letter. 2020;20:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng Z, Yang Y, Qing C, et al. Distinct Expression and Prognostic Value of Members of SMAD Family in Non-small Cell Lung Cancer. Baltimore, MD: Medicine; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klimaszewska-Wiśniewska A, Buchholz K, Neska-Długosz I, et al. RRM2 and SPDL1 and their prognostic significance in pancreatic adenocarcinoma. Cancers. 2021;1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolet BP, Wolkers MC. Limited but gene-class specific correlation of mRNA and protein expression in human CD8 + T cells. Cold Spring Harb Perspect Biol. 2020. [Google Scholar]
- 66.Kosti I, Jain N, Aran D, Butte AJ, Sirota M. Cross-tissue analysis of gene and protein expression in normal and cancer tissues. Sci Rep. 2016;6:24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ginestier C, Charafe-Jauffret E, Bertucci F, et al. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am J Pathol. 2002;161:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4117:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Liu S, Zhang Y, Yang J. Dysregulation of TLR2 serves as a prognostic biomarker in breast cancer and predicts resistance to endocrine therapy in the luminal B subtype. Frontiers Oncol. 2020;10:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu YD, Yu L, Ying L, et al. Toll‐like receptor 2 regulates metabolic reprogramming in gastric cancer via superoxide dismutase 2. Int J Cancer. 2019;144:3056-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paarnio K, Tuomisto A, Väyrynen SA, et al. Serum TLR2 and TLR4 levels in colorectal cancer and their association with systemic inflammatory markers, tumor characteristics, and disease outcome. APMIS. 2019;127:561-569. [DOI] [PubMed] [Google Scholar]
- 72.Ng LK, Rich AM, Hussaini HM, et al. Toll-like receptor 2 is present in the microenvironment of oral squamous cell carcinoma. Br J Cancer. 2011;104:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lanki MA, Seppänen HE, Mustonen HK, et al. Toll-like receptor 2 and Toll-like receptor 4 predict favorable prognosis in local pancreatic cancer. Tumor Biol. 2018. [DOI] [PubMed] [Google Scholar]
- 74.Chen X, Zhang L, Jiang Y, et al. Radiotherapy-induced cell death activates paracrine HMGB1-TLR2 signaling and accelerates pancreatic carcinoma metastasis. J Exp Clin Cancer Res. 2018;37:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tye H, Kennedy CL, Najdovska M, et al. STAT3-driven upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer Cell. 2012;22:466-478. [DOI] [PubMed] [Google Scholar]
- 76.Beilmann-Lehtonen I, Böckelman C, Mustonen H, Koskensalo S, Hagström J, Haglund C. The prognostic role of tissue TLR2 and TLR4 in colorectal cancer. Virchows Arch. 2020;477:705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stephen TL, Payne KK, Chaurio RA, et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity. 2017;46:51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-ccx-10.1177_10732748211056697 for Prognostic Significance of TLR2, SMAD3 and Localization-dependent SATB1 in Stage I and II Non–Small-Cell Lung Cancer Patients by Justyna Durślewicz, Anna Klimaszewska-Wiśniewska, Jakub Jóźwicki, Paulina Antosik, Marta Smolińska-Świtała, Maciej Gagat, Adam Kowalewski and Dariusz Grzanka in Cancer Control
Supplemental Material, sj-pdf-2-ccx-10.1177_10732748211056697 for Prognostic Significance of TLR2, SMAD3 and Localization-dependent SATB1 in Stage I and II Non–Small-Cell Lung Cancer Patients by Justyna Durślewicz, Anna Klimaszewska-Wiśniewska, Jakub Jóźwicki, Paulina Antosik, Marta Smolińska-Świtała, Maciej Gagat, Adam Kowalewski and Dariusz Grzanka in Cancer Control