Summary
Elongated hypocotyl5 (HY5) is a key transcription factor that promotes photomorphogenesis. Constitutive photomorphogenic1 (COP1)–Suppressor of phytochrome A-105 (SPA) E3 ubiquitin ligase complex promotes ubiquitination and degradation of HY5 to repress photomorphogenesis in darkness. HY5 is also regulated by phosphorylation at serine 36 residue. However, the kinase responsible for phosphorylation of HY5 remains unknown.
Here, using extensive in vitro and in vivo biochemical, genetic, and photobiological techniques, we have identified a new kinase that phosphorylates HY5 and demonstrated the significance of phosphorylation of HY5 in Arabidopsis thaliana.
We show that SPA proteins are the missing kinases necessary for HY5 phosphorylation. SPAs can directly phosphorylate HY5 in vitro, and the phosphorylated HY5 is absent in the spaQ background in vivo. We also demonstrate that the unphosphorylated HY5 interacts strongly with both COP1 and SPA1 and is the preferred substrate for degradation, whereas the phosphorylated HY5 is more stable in the dark. In addition, the unphosphorylated HY5 actively binds to the target promoters and is the physiologically more active form. Consistently, the transgenic plants expressing the unphosphorylated form of HY5 display enhanced photomorphogenesis.
Collectively, our study revealed the missing kinase responsible for direct phosphorylation of HY5 that fine-tunes its stability and activity to regulate photomorphogenesis.
Keywords: Arabidopsis, HY5, phosphorylation, photomorphogenesis, suppressor of phyA-105 (SPA)
Introduction
Light is a key environmental factor that influences diverse developmental processes throughout the entire plant life cycle (Whitelam & Halliday, 2007). Plants have evolved four classes of photoreceptors to monitor the surrounding light conditions: red/far-red light-sensing phytochromes, blue/ultraviolet (UV)A light-sensing cryptochromes and phototropins, and UVB light-sensing UVR8 (Chen et al., 2004; Paik & Huq, 2019). Interestingly, all the light signals perceived by different photoreceptors converge to a downstream transcription factor elongated hypocotyl5 (HY5) to control diverse growth programs (Gangappa & Botto, 2016). For example, dark-grown Arabidopsis seedlings undergo skotomorphogenesis, displaying closed, yellowish cotyledons, and long hypocotyls. Upon light irradiation, seedlings undergo photomorphogenesis, which includes open, wide and green cotyledons, and short hypocotyls (Gommers & Monte, 2018). The dark-to-light transition mainly causes the accumulation of HY5 proteins and then triggers cascades of downstream gene expressions. Indeed, HY5 binds to nearly one-third of the Arabidopsis genes (Lee et al., 2007; Gangappa & Botto, 2016; Burko et al., 2020) and regulates a wide range of plant developmental programs, including flowering time, Chl and anthocyanin biosynthesis, primary and lateral root development, and shade and high-temperature responses (Oyama et al., 1997; Ang et al., 1998; Holm et al., 2002; Andronis et al., 2008; Nozue et al., 2015; Gangappa & Botto, 2016).
The level of HY5 protein is regulated by constitutive photomorphogenic1 (COP1)–suppressor of phytochrome A-105 (SPA) E3 ubiquitin ligase complex (Hoecker, 2017). Both COP1 and SPA are crucial repressors of photomorphogenesis. COP1 protein is enriched in nucleus in the dark and depleted from nucleus in the light (Subramanian et al., 2004; Pacín et al., 2014; Balcerowicz et al., 2017). Thus, in darkness, COP1–SPA complex induces ubiquitination and degradation of HY5 and possibly other positive transcription factors in the nucleus (Hoecker, 2017; Han et al., 2020). However, upon light irradiation, the activity of the COP1–SPA E3 ubiquitin ligase complex is inhibited by photoreceptors (Ordoñez-Herrera et al., 2015; Sheerin et al., 2015; Xu et al., 2015). The reduction of COP1 in the nucleus and the light-induced inhibition of COP1 activity contribute to the accumulation of HY5 and other positive regulators to promote photomorphogenesis.
The function of COP1 as a RING-type E3 ubiquitin ligase is evolutionarily conserved in higher eukaryotes. It consists of an N-terminal zinc finger, a central coiled-coil (CC), and a C-terminal WD40 repeats domain, which is essential for proper COP1 function (Deng et al., 1992; Han et al., 2020). The SPA family of genes are only found in the green lineages. Arabidopsis has four SPA genes (SPA1–SPA4) (Laubinger et al., 2004; Hoecker, 2017). The SPA family of proteins also contain the central CC and the C-terminal WD40 repeats domain, which function similar to the respective domains of COP1 (Hoecker et al., 1999). In addition, SPA proteins contain an N-terminal serine/threonine (Ser/Thr) kinase domain which was recently found to have kinase activity on phytochrome interacting factors (PIF1 and PIF4), key negative regulators of photomorphogenesis (Paik et al., 2019; Lee et al., 2020). SPA1 and COP1 can interact with each other through their CC domains. SPA proteins are important for COP1 function: the presence of SPA proteins can enhance the activity of COP1 in vitro and in vivo (Saijo et al., 2003; Seo et al., 2003). Both cop1 and spaQ (spa1spa2spa3spa4 quadruple) mutants exhibit constitutive photomorphogenesis in the darkness. In plants, COP1 forms multiple complexes with SPA proteins in a tissue and developmental-stage-specific manner. Previous studies showed that the WD40 repeat domain of both COP1 and SPA1 interacts with the N-terminal domain of HY5, and the COP1–SPA complex promotes ubiquitination and subsequent degradation of HY5 and many other transcription factors in darkness (Torii et al., 1998; Hardtke et al., 2000; Saijo et al., 2003; Hoecker, 2017; Han et al., 2020; Kathare et al., 2020).
In addition to COP1–SPA-mediated degradation in darkness, HY5 is also regulated by phosphorylation at serine 36 residue (Hardtke et al., 2000). The unphosphorylated form of HY5 interacts more strongly with COP1 and is preferentially degraded in the dark, but it becomes more abundant in light-grown seedlings (Hardtke et al., 2000). Because serine 36 is located within a conserved Casein Kinase II (CKII) phosphorylation site, these authors also hypothesized that a light-regulated CKII might be responsible for phosphorylation of HY5. However, the identity of the kinase remained elusive. Given the known physical interactions and the recently described SPA kinase activity, it prompted us to examine whether HY5 is a new substrate of SPA kinase. Here, we provide in vitro and in vivo evidence supporting the conclusion that SPA proteins are the missing kinase for HY5 phosphorylation.
Materials and Methods
Plant materials and growth conditions
Wild-type (WT) Col-0, various mutants, and transgenic plants in the Col-0 background of A. thaliana were used in this study, unless indicated otherwise. Plants were grown in soil under 24 h light at 22 ± 0.5°C. Tandem affinity purification-SPA1, luciferase (LUC)-SPA1/spaQ, and LUC-mSPA1/spaQ transgenic plants were reported previously (Paik et al., 2019). To generate HY5, HY5-S36A, and HY5-S36D transgenic lines, the HY5 36th serine (AGC) was changed into alanine (GCC) and aspartic acid (GAC), respectively, using a Quickchange II site-directed mutagenesis kit (200523; Agilent, Cedar Creek, TX, USA) and cloned into pB7FWG vectors, then transformed into hy5-2l5, spaQ, and WT backgrounds. The transformants were selected in the presence of Basta. Multiple transgenic lines with the same overexpressed HY5 proteins were used for analyses.
Vector constructions and protein purification
Maltose-binding protein (MBP)-COP1 and MBP-SPA1 were prepared as described previously (Xu et al., 2014; Paik et al., 2019). For purification of glutathione S-transferase (GST)-HY5 and its mutant proteins, HY5, HY5-S36A, and HY5-S36D were cloned into pGEX4T-1. Each plasmid was transformed into BL21(DE3) cells. Protein expression was induced under 16°C for overnight with 0.1 mM isopropyl β-d-1-thiogalactopyranoside. Collected cells were sonicated in binding buffer (100 mM Tris-Cl, pH 7.5, 150 mM sodium chloride (NaCl), 0.2% Tergitol NP-40, 1 × Protease inhibitor cocktail, and 1 mM phenylmethylsulfonyl fluoride (PMSF)) and purified using GST agarose beads (20211; Pierce Biotechnology Inc., Waltham, MA, USA). Proteins were eluted with the elution buffer (Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM, 10 mM glutathione, 10% Tergitol NP-40, 10% glycerol, 1 mM PMSF, 1× protease inhibitor cocktail) into separate fractions. The eluted proteins were analyzed on sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) gel and used for kinase assays and pull-down assay.
For cloning of the pYES2-(m)SPA1-green fluorescent protein (GFP) expression vectors, the full-length SPA1 and mSPA1 sequences were PCR amplified using the SPA1_fwd and SPA1_rev primers (Supporting Information Table S1). The GFP sequence was separately PCR amplified using GFP_fwd 1 and GFP_rev primers (Table S1). The expression vector pYES2 was initially digested with Eco53kI and gel purified. From each DNA (SPA1, GFP, and digested pYES2) 40 fmol was combined with a Gibson assembly master mix (E2611S; NEB, Ipswich, MA, USA) and ligated according to the manufacturer’s instruction. Cloned pYES2-(m)SPA1-GFP was transformed and purified in Saccharomyces cerevisiae as previously described for the pYES2-PHYB-GFP purification method (Paik et al., 2019).
For cloning of pYES2-SPA2(3/4)-GFP expression vectors, the full-length SPA2(3/4) sequences were PCR amplified using SPA2 (3/4)_fwd and SPA2(3/4)_rev primers (Table S1). The expression vector pYES2 with GFP tag was PCR amplified from pYES2-SPA1-GFP vector using GFP_fwd 2 and pYES2_rev primers (Table S1). SPA2(3/4) and pYES2-GFP were then combined with a Gibson assembly master mix. Cloned pYES2-SPA2 (3/4)-GFP was transformed and purified from S. cerevisiae as already described herein.
In vitro kinase assay
For SPA1 kinase assay, about 500 ng of Pichia pastoris purified SPA1-strep and 1 μg of Escherichia coli purified GST-HY5 or GST-HY5-S36A fusion proteins were used. For mSPA1 kinase assay, SPA1-GFP and mSPA1-GFP were purified from S. cerevisiae as described earlier herein. For SPA2(3/4) kinase assay, SPA2(3/4)-GFP were purified from S. cerevisiae as described earlier herein. All kinase assays were performed in a kinase buffer (50 mM Tris, pH 7.5, 4 mM β-mercaptoethanol, 1 mM EDTA, 10 mM magnesium chloride). Phosphorus-32 radiolabeled gamma-ATP (BLU502A; Perkin Elmer, Waltham, MA, USA) was added to the reaction and incubated at 28°C for 1 h, unless indicated otherwise. SDS sample buffer (6×) was added to stop the reaction, and the boiled proteins were separated on SDS-PAGE gel. The gels were dried and exposed to a phosphor screen and then scanned with Typhoon FLA 9500 (GE Healthcare, Chicago, IL, USA).
HY5 mobility shift assay
To observe GFP-HY5 mobility shift in Col-0 and spaQ, total protein was separated in a 10 cm x 10.5 cm 7% SDS-PAGE or 4–15% gradient SDS-PAGE (456–1084; Bio-Rad) or 8% SDS-PAGE gel containing 15–20 μM Phos-tag acrylamide (AAL-107; Wako Pure Chemical Industries, Osaka, Japan). For regular SDS-PAGE gel, total proteins were extracted from 4-d-old light-grown or dark-grown seedlings with extraction buffer (100 mM Tris·HCl (pH 6.8), 20% glycerol, 5% SDS, 20 mM dithiothreitol, 1 mM PMSF, 1× protease inhibitor, and 100 μM bortezomib). The extracts were cleared by centrifugation and then incubated with or without 400 U ml−1 alkaline phosphatase, calf intestinal (CIP, M0290; NEB) at 37°C for 30 min. The reaction mixtures were terminated by adding 6× SDS loading buffer and boiling at 99°C for 10 min. Immunoblotting analyses were performed with anti-GFP and anti-Tubulin antibodies. For Phos-tag gel, immunoprecipitated proteins were treated with or without CIP, then analyzed by immunoblotting. The in vivo co-immunoprecipitation (co-IP) assay is described later.
Protein extraction and Western blot analyses
To analyze HY5 abundance in dark-to-light and light-to-dark transition, seeds were surface sterilized and grown in the dark or continuous white light (100 μmol m−2 s−1) for 4 d or followed the respective conditions (continues dark or light treatment, different hours of dark or light treatment, 100 μmol m−2 s−1) as described in the figure legends. To analyze HY5 abundance in mutants and transgenic lines, seedlings were grown in continuous dark for 4 d. To analyze (m)SPA1 abundance in LUC-SPA1/spaQ and LUC-mSPA1/spaQ transgenic lines, seedlings were grown in continuous dark for 4 d. For total protein extraction, whole seedlings were collected and ground in 100 μl denaturing extraction buffer (100 mM Tris pH 7.5, 1 mM EDTA, 8 M urea, 1× protease inhibitor cocktail (59; Sigma-Aldrich), 2 mM PMSF) and cleared by centrifugation at 20 200 g for 10 min at 4°C. Samples were boiled for 10 min with 6× SDS sample buffer and separated on a 10% SDS-PAGE gel, blotted onto polyvinylidene difluoride membranes, and probed with corresponding antibodies. Antibodies used in these studies are anti-HY5 (R1245-2; Abiocode Inc., Agoura Hills, CA, USA), anti-GFP (ab6556 for immunoprecipitation; Abcam, Cambridge, MA, USA), anti-LUC (A11120; Thermofisher, Waltham, MA, USA), anti-Tubulin antibodies (BML-PW8770-0025; Enzo Life Sciences, Farmingdale, NY, USA). Secondary horseradish peroxidase (HRP)-bound antibodies were visualized with Super Signal West Pico Chemiluminescent substrate (Pierce Biotechnology Inc.) and developed with an X-ray film or GBox-F3 Syngene Imager. The intensity of the HY5 and Tubulin bands from three independent blots was quantified using ImageJ software (US National Institutes of Health, Bethesda, MD, USA), and the HY5 values were divided by the Tubulin values to generate a ratio for each sample. The HY5 level in the light (L) or at 12 h light was set to 1 from these ratios, respectively, and the relative values of the other time points were then calculated. These relative values are shown as line graphs in each figure in addition to the blots. Student’s t-test was used to analyze the significant difference.
In vitro pull-down assay
For in vitro pull-down assays, MBP-COP1, MBP-SPA1, GST-HY5, GST-HY5-S36A and GST-HY5-S36D fusion proteins were prepared as described previously. A 1 μg protein sample was used for each of them. All protein combinations were incubated with 20 ml of amylose resin in the binding buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.6% Tween 20, and 1 mM DTT) for 3 h. The beads were collected and washed six times with 5 min of rotation each time in binding buffer. The bound HY5 was detected by anti-GST-HRP conjugate (RPN1236; GE Healthcare Bio-Sciences). Membranes were developed and visualized as described earlier. The intensity of the GST-HY5 band from three independent blots was quantified using ImageJ software and normalized to added COP1 and SPA1 proteins. Further, the ratio of the first clear band was set to 1 for each blot.
In vivo co-immunoprecipitation assay
For Co-IP experiments, homozygous HY5-GFP, HY5-S36A and HY5-S36D transgenic seedlings were grown in dark for 4 d and then treated with 40 μM bortezomib (LC Laboratories, Woburn, MA, USA) for at least 4 h. Total proteins were extracted from 1 g tissue with 1 ml protein extraction buffer. After 15 min centrifugation at 16 000 g at 4°C in darkness, 100 μl supernatant of each sample was reserved as total, and the remainder was incubated with Dynabeads Protein A (10002D; Life Technologies Co., Carlsbad, CA, USA) bound with anti-GFP antibody (ab9110; Abcam). Twenty microliter Dynabeads with 1 μg antibody were used for individual sample. After 2 h incubation in the dark at 4°C, beads were washed three times with 1 ml protein extraction buffer with 0.2% NP40. Immuno-precipitated proteins were analyzed by immunoblotting.
RNA extraction and quantitative RT-PCR
The quantitative reverse transcription (RT)-PCR (qRT-PCR) analysis was performed as described with minor variations (Shor et al., 2017). Total RNA was isolated from 4-d-old dark-grown seedlings followed by 3 h light treatment or from 4-d-old light-grown seedlings using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). Total RNA (1 μg) was treated with DNase I to eliminate genomic DNA and then reverse transcribed using SuperScript III (Life Technologies) as per the manufacturer’s protocol. Real-time PCR was performed using the Power SYBR Green RT-PCR Reagents Kit (Applied Biosystems, Foster City, CA, USA) in a 7900HT Fast Real-Time PCR machine (Applied Biosystems). PP2A was used as a control to normalize the expression data. The resulting cycle threshold values were used to calculate the levels of expression of different genes relative to PP2A, as suggested by the manufacturer (Applied Biosystems). The primer sequences used for qRT-PCR are listed in Table S1.
Chromatin immunoprecipitation–quantitative PCR assay
Three biological replicates of HY5 and HY5-S36A and HY5-S36D seedlings grown in the dark for 4 d and moved to simulated white light for 3 h were used for chromatin immunoprecipitation (ChIP)–quantitative PCR (qPCR) analysis. ChIP experiments were performed as previously described (Shor et al., 2017). Anti-GFP (ab6556 for immunoprecipitation; Abcam) antibody was used for immunoprecipitation. After elution, reversing crosslinks, and DNA purification, the amount of each precipitated DNA fragment was detected by real-time qPCR using the specific primers listed in Table S1. Three biological replicates were performed, and three technical repeats were carried out for each biological replicate. Student’s t-test was used to analyze the significant difference.
Measurement of hypocotyl lengths
For the measurement of hypocotyl length under dark, red light (Rc), and far-red light (FRc) with different intensities, images of 150 seedlings (30 seedlings for each line with three independent biological replicates) at each light intensity condition were taken and then measured using the publicly available ImageJ software (http://rsb.info.nih.gov/ij/). Seeds were plated on Murashige & Skoog (MS) medium without sugar and kept in the dark for 3 d at 4°C. Seeds were then exposed to 3 h of white light (100 μmol m−2 s−1) to induce germination and then kept in the dark for 21 h. The dark-grown seedlings were exposed to far-red light (34 μmol mol−2 s−1) for an additional 10 min before being put in darkness. All the other plates were then put in conditions for 3 d as described in the figures. Light fluence rates were measured using a spectroradiometer (model EPP2000; StellarNet, Tampa, FL, USA) as described previously (Shen et al., 2005).
Light treatments
For Western blot, co-IP, ChIP–qPCR and RT-PCR samples, seeds were surface sterilized and plated on MS growth medium without sucrose on filter paper and kept in the dark for 3 d at 4°C. Seeds were then exposed to 3 h of white light (100 μmol mol−2 s−1) to induce germination and then put in the respective conditions (continuous dark or light treatment, different hours of dark or light treatment) as described in the figure legends. White light (100 μmol m−2 s−1) was used in all light treatment.
Results
SPA1 can directly phosphorylate Serine-36 on HY5 proteins in vitro
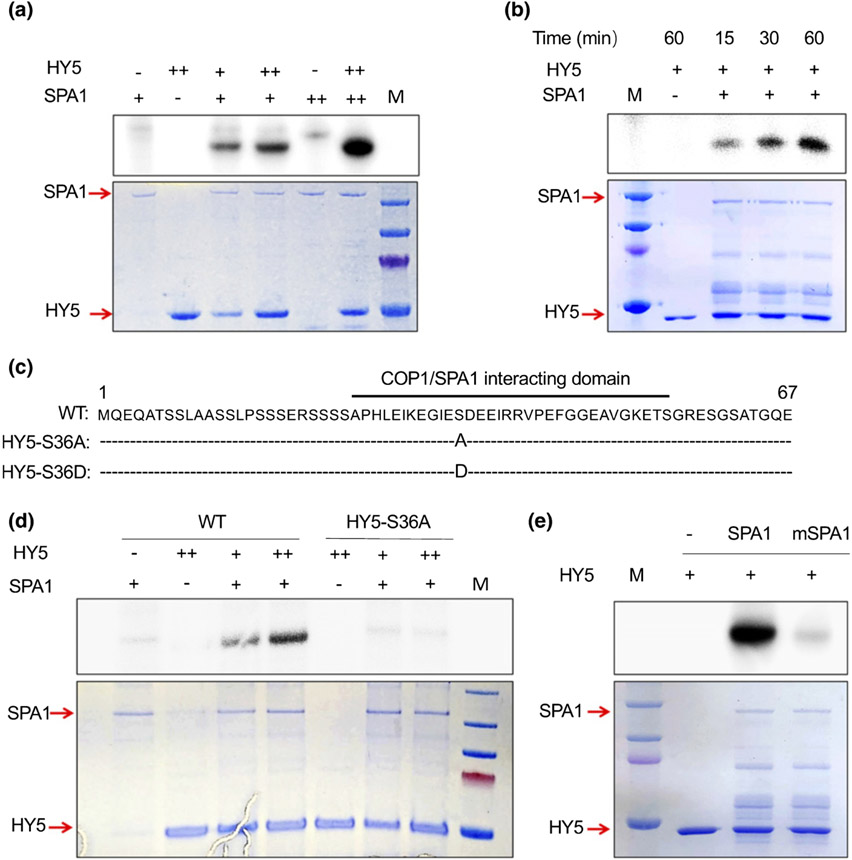
Our recent studies show that SPA1 acts as an Ser/Thr kinase for both PIF1 and PIF4 (Paik et al., 2019; Lee et al., 2020). Since COP1–SPA E3 ubiquitin ligase complex interacts with HY5 proteins in the dark to promote its degradation, we hypothesize that HY5 might be a new substrate of SPA kinase. To test whether SPA can phosphorylate HY5 protein, we first purified strep-tagged full-length SPA1 protein from a eukaryotic expression host (P. pastoris) and GST-tagged HY5 from bacteria (E. coli), and performed an in vitro kinase assay. We found that SPA1 directly phosphorylates HY5 in vitro in a concentration-dependent manner (Fig. 1a,d). To check if this phosphorylation activity is changed by the incubation time, we conducted the kinase assay over time (Fig. 1b). With increasing HY5 protein and incubation time, we observed stronger phosphorylation signals. These results suggest that SPA1 phosphorylates HY5 in a concentration and time-dependent manner (Fig. 1a,b).
Fig. 1.
Suppressor of phytochrome A-105 (SPA)1 acts as a serine/threonine protein kinase and directly phosphorylates elongated hypocotyl5 (HY5) on the 36th serine (Ser-36) in vitro. (a) Full-length SPA1 protein purified from Pichia pastoris phosphorylates HY5 protein in vitro in a concentration-dependent manner (autoradiogram on top panel) with increasing protein levels of HY5 or SPA1. The bottom panel shows the protein levels in a Coomassie-stained gel. −, no protein added; +, protein added; ++, doubled amount of protein added; M, a protein marker. (b) Time-dependent kinase assays of full-length SPA1 on HY5 (autoradiogram in the upper panel, reaction time for 15, 30, or 60 min). The lower panel shows the protein levels in a Coomassie-stained gel. −, no protein added; +, protein added; M, a protein marker. (c) Single phosphorylation site of HY5 is located at Ser-36 in its constitutive photomorphogenic1 and SPA1 interacting domain. Ser-36 was then replaced with alanine (S36A) and aspartic acid (S36D) to generate a nonphosphorylation form and a phospho-mimicking form of HY5, respectively. (d) SPA1 protein purified from P. pastoris phosphorylates wild type (WT) HY5 but not the nonphosphorylation mutant of HY5 in vitro (autoradiogram in upper panel). The lower panel shows the protein levels in a Coomassie-stained gel. −, no protein added; +, protein added; ++, doubled amount of protein added; M, a protein marker. (e) A conserved amino acid mutation on the SPA1 kinase domain (mSPA1) reduces the phosphorylation activity of SPA1 on HY5 (autoradiogram in upper panel). An in vitro kinase assay was performed using purified SPA1-green fluorescent protein (GFP) and mSPA1-GFP proteins purified from Saccharomyces cerevisiae and glutathione S-transferase-HY5 protein purified from Escherichia coli. The lower panel shows the protein level in a Coomassie-stained gel. −, no protein added; +, protein added; M, a protein marker.
To map the phosphorylation sites in HY5, we used GST-HY5 to conduct in vitro phosphorylation assays using SPA1 as a kinase and performed mass-spectrometry analyses. These data revealed a single phosphorylation site (Ser-36) under these conditions, which is located at the 36th serine at the N-terminus of HY5 (Dataset S1). A previous study showed that a deletion of the first 40 amino acids of HY5 completely abolished the interaction with COP1 (Hardtke et al., 2000). A 36 amino acid stretch between the 25th and 60th residues of HY5 proteins was then defined as a COP1 interacting domain. In addition, HY5 interacts with SPA1 through its N-terminal domain (Saijo et al., 2003). Thus, the mapped phosphorylation site of HY5 is within its interaction domain for both COP1 and SPA (Fig. 1c). To address the significance of the phosphorylation site, we replaced the Ser-36 with alanine (S36A) or aspartic acid (S36D) to generate nonphosphorylation mutant and phospho-mimicking mutant, respectively. The in vitro kinase assay showed that mutant recombinant protein (HY5-S36A) cannot be phosphorylated by SPA1 (Fig. 1d), supporting that Ser-36 residue might be the single phosphorylation site of HY5 under these conditions.
SPA1 kinase domain is conserved in SPA1 sequences from multiple plants (Fig. S1A) (Paik et al., 2019). The R517 residue in SPA1 is part of a conserved glutamic acid–arginine (Arg) salt bridge that defines eukaryotic protein kinases (Yang et al., 2012) and has recently been shown to be critical for its biological function (Holtkotte et al., 2016; Paik et al., 2019; Lee et al., 2020). To investigate the importance of SPA1-Ser/Thr kinase activity for HY5 phosphorylation, we used the point mutant version of SPA1 (mSPA1) that has the R517E mutation in the kinase domain. The in vitro kinase assay confirmed a significantly reduced phosphorylation activity of mSPA1 on HY5 (Fig. 1e). These data collectively suggest that SPA1 is a bona fide kinase for HY5.
Furthermore, all the SPA proteins contain an N-terminal Ser/Thr kinase domain (Fig. S1A). To investigate whether other SPAs could phosphorylate HY5, we performed in vitro kinase assays using purified GFP-tagged full-length SPA2 (3/4) protein from a eukaryotic expression host (S. cerevisiae) and GST-tagged HY5 from bacteria (E. coli). The kinase assay results show that SPA2, SPA3, and SPA4 can also directly phosphorylate HY5 (Fig. S1B), suggesting that all four SPAs have redundant kinase activity.
SPAs are necessary for phosphorylation of HY5 in vivo
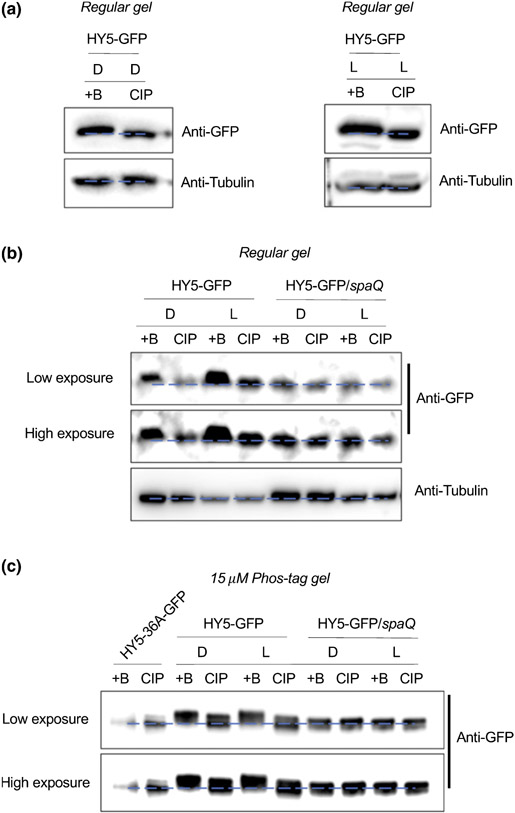
To investigate the phosphorylation status of HY5 in vivo, we generated HY5-overexpressing transgenic plants (HY5-GFP) in WT background and purified HY5-GFP proteins from transgenic seedlings grown in the dark or in the light. In immunoblot analysis, we observed band mobility shift on a regular SDS-PAGE gel after treatment with the native CIP in both dark and light-grown seedlings (Figs 2a, S2A,B), indicating that HY5 is phosphorylated under both dark and light conditions.
Fig. 2.
Suppressor of phytochrome A-105s (SPAs) are necessary for the phosphorylation of elongated hypocotyl5 (HY5) in vivo. (a) Immunoblots showing phosphorylation of HY5-green fluorescent protein (GFP) under both dark and light conditions. Seedlings were grown in either dark (D) or continuous white light (L) for 4 d before sampling, and extracted proteins were then separated on 7% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels. The slow-migrating bands are phosphorylated forms of HY5-GFP, as indicated by the phosphatase treatment. CIP, Calf intestinal alkaline phosphatase; +B, inactivated boiled CIP. Tubulin proteins were used as loading control. (b) Immunoblots showing a defect of HY5 phosphorylation in spaQ mutant background compared with wild-type. Seedlings were grown in either dark (D) or continuous white light (L) for 4 d before sampling, and extracted proteins were then separated on 7% SDS-PAGE gels. The slow-migrating bands are phosphorylated forms of HY5-GFP, as indicated by the phosphatase treatment. +B, inactivated boiled CIP. Tubulin proteins were used as loading control. (c) Immunoblots showing a defect in HY5 phosphorylation in spaQ mutant background compared with wild-type in gels containing 15 μM Phos-tag. HY5-GFP proteins were immunoprecipitated from either 4-d-old dark (D) or continuous white light (L)-grown seedlings and then separated on 8% SDS-PAGE gels containing 15 μM Phos-tag. The slow-migrating bands are phosphorylated forms of HY5-GFP. The unphosphorylated form of HY5-36A-GFP protein extracted from 4-d-old light-grown seedlings was used as a control. +B, inactivated boiled CIP. Note that higher level of HY5-GFP was loaded for dark samples in all panels to better assess the difference in migration.
To examine the in vivo effect of SPAs on the HY5 phosphorylation, we then generated transgenic plants overexpressing HY5-GFP in spaQ background (HY5-GFP/spaQ). Strikingly, the phosphorylation and band shift of HY5 by CIP treatment observed in WT was completely abolished in the spaQ mutants both grown in the dark and light (Fig. 2b). The HY5 phosphorylation status was further examined by utilizing Phos-tag-containing SDS-PAGE gels (Figs 2c, S2C). In WT plants, HY5-GFP showed clear mobility shift after CIP treatment in the presence of 20 μM Phos-tag (Fig. S2C). However, in spaQ mutant, no mobility shift was observed under these conditions and HY5-GFP showed a faster migrating band than that in WT (Fig. 2c), suggesting a complete absence of phosphorylation of HY5 in vivo. Taken together, these results indicate that SPAs are responsible for HY5 phosphorylation in vivo under both light and dark conditions.
SPA1 kinase domain is essential for its biological function
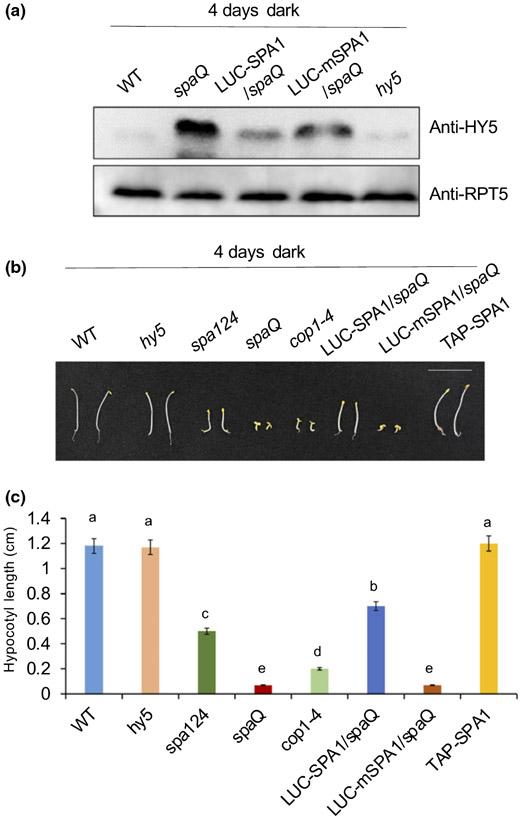
To confirm the role of SPAs in the regulation of HY5, we measured the level of endogenous HY5 protein in different mutants and transgenic lines grown in the dark for 4 d (Fig. S3). As expected, we hardly detected HY5 protein signals in WT seedlings grown in the dark, whereas HY5 proteins were significantly accumulated in both cop1 and spaQ mutants (Fig. S3A). In spa triple mutants (spa123 and spa124), weaker bands of HY5 protein were detected, indicating that four SPA members have redundant functions in regulating HY5 levels (Fig. S3B). To clarify the biological role of SPA1 kinase activity, we then used the transgenic lines overexpressing similar amounts of SPA1 (LUC-SPA1) and mSPA1 (LUC-mSPA1) in spa quadruple mutants (spaQ) background (Fig. S4). The HY5 level was more reduced by introduction of LUC-SPA1 compared with LUC-mSPA1 in the spaQ background (Fig. 3a), suggesting that the reduction in kinase activity of mSPA1 is deficient in degrading HY5 in the dark. This result is also consistent with the LUC-mSPA1 transgenic seedlings phenotype in the dark, which failed to rescue the constitutive photomorphogenesis, whereas LUC-SPA1 can largely rescue the constitutive photomorphogenic phenotype of spaQ (Fig. 3b,c).
Fig. 3.
Suppressor of phytochrome A-105 (SPA)1 kinase domain is necessary for its kinase activity and biological function. (a) Immunoblots showing elongated hypocotyl5 (HY5) accumulation in the different genotypes. Seedlings were grown in the dark for 4 d before sampling for protein extraction; endogenous anti-HY5 antibody and anti-RPT5 antibody were used. RPT5 protein was used as loading control. (b) Photograph showing the seedling phenotypes of different genotypes of Arabidopsis thaliana grown in darkness for 4 d. Bar, 10 mm. (c) A bar graph shows the hypocotyl length of corresponding genotypes grown under dark for 4 d. Error bars indicate SD (n > 30). The letters ‘a’ to ‘e’ indicate statistically significant differences among means of hypocotyl lengths of the genotypes shown based on the one way ANOVA followed by Tukey's honestly significant difference (P < 0.05).
HY5 phosphorylation affects its interaction with COP1 and SPA1
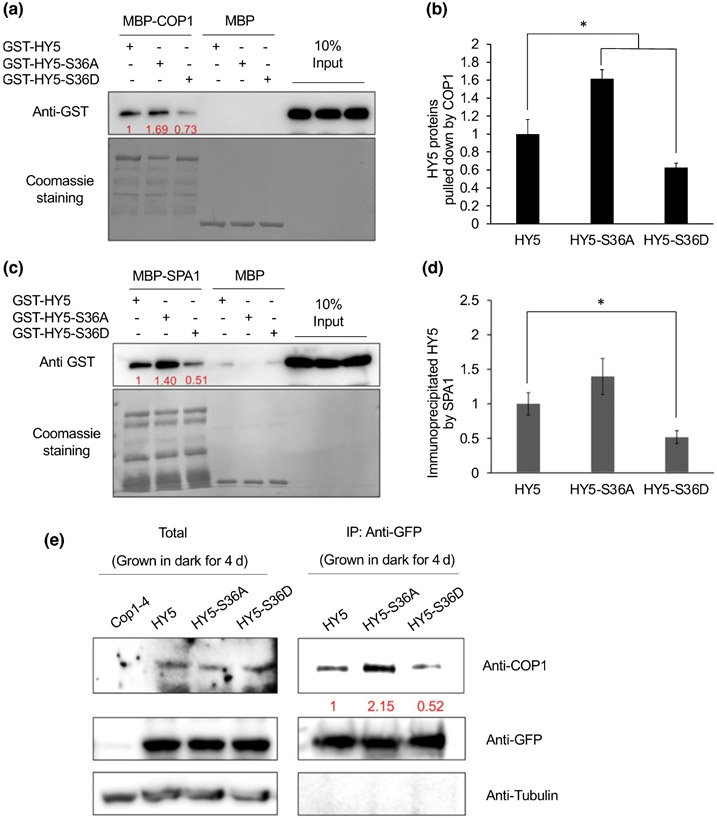
Degradation of HY5 largely depends on its interaction with COP1–SPA complex in the nucleus (Hardtke et al., 2000; Osterlund et al., 2000; Saijo et al., 2003). HY5 interacts with both COP1 and SPA1 through its N-terminal domain (Hardtke et al., 2000; Saijo et al., 2003), and the phosphorylation site resides within the interacting domain (Fig. 1c). Using plant extracts as a source of kinase and commercially available lambda phosphatase, it was shown that the unphosphorylated form of HY5 interacts with COP1 more strongly than the phosphorylated form does (Hardtke et al., 2000). Since we have identified the kinase necessary for phosphorylation of HY5 and also created the phospho-null and phospho-mimic forms of HY5, we therefore examined whether the phosphorylation of HY5 affects its interactions with COP1 and SPA1. To address this, we performed in vitro pull-down assays with purified fusion proteins, MBP-COP1 and MBP-SPA1. Each of the recombinant GST-fused WT HY5, HY5-S36A, and HY5-S36D proteins was precipitated by MBP-COP1 or MBP-SPA1. Interestingly, the results show that the nonphosphorylated form of HY5 (HY5-S36A) protein has a higher affinity to both COP1 and SPA1, whereas the phospho-mimicking form of HY5 (HY5-S36D) protein has a significantly lower affinity to both COP1 and SPA1 than with WT HY5 (Fig. 4a-d). We also conducted an in vivo co-IP assay with transgenic lines overexpressing similar amounts of HY5-GFP, HY5-S36A-GFP and HY5-S36D-GFP in hy5 mutant background (Fig. 4e). When immunoprecipitated using GFP antibody, four times more COP1 protein was co-immunoprecipitated in the HY5-S36A-GFP line than that in the HY5-S36D-GFP line (Fig. 4e). Taken together, both in vitro and in vivo data suggest that phosphorylation alters the affinity of HY5 to COP1–SPA complex and may also affect its accumulation and biological functions, as previously shown (Hardtke et al., 2000).
Fig. 4.
Phosphorylation of elongated hypocotyl5 (HY5) affects its interaction with constitutive photomorphogenic1 (COP1) and suppressor of phytochrome A-105 (SPA)1. (a–d) Recombinant maltose-binding protein (MBP)-COP1, MBP-SPA1, and glutathione S-transferase (GST)-HY5 (and mutant forms of HY5) proteins were purified from Escherichia coli. (a) In vitro pull-down assay shows that nonphosphorylated forms of HY5 (GST-HY5-S36A) have higher affinity to MBP-COP1. GST-HY5 and all other phosphorylation mutant proteins were pulled down by MBP-COP1 using maltose agarose beads. The pellet fraction was eluted and analyzed by immunoblotting using anti-GST and anti-MBP antibodies or stained by Coomassie. (b) A bar graph showing the interaction between COP1 and wild-type HY5 and mutant HY5 proteins. Error bars indicate SD (n = 3). The asterisk indicates a significant difference between wild-type HY5 and mutant HY5 based on Student’s t-test (P < 0.05). (c) In vitro pull-down assay shows that nonphosphorylated forms of HY5 (GST-HY5-S36A) have higher affinity to MBP-SPA1. GST-HY5 and all other phosphorylation mutant proteins were pulled down by MBP-SPA1 using maltose agarose beads. The pellet fraction was eluted and analyzed by immunoblotting using anti-GST and anti-MBP antibodies or stained by Coomassie. (d) A bar graph showing the interaction between SPA1 and wild-type HY5 and mutant HY5 proteins. Error bars indicate SD (n = 3). The asterisk indicates a significant difference between wild-type HY5 and mutant HY5 based on Student’s t-test (P < 0.05). The numbers below anti-GST blots (in a and c) indicate the relative band intensities of immunoprecipitated HY5 normalized to added COP1 and SPA1 proteins, respectively. The ratio of the first clear band was set to 1 for each blot. (e) In vivo co-immunoprecipitation assay shows that the nonphosphorylated form of HY5 protein strongly interacts with COP1. IP, immunoprecipitation. Homozygous HY5, HY5-S36A and HY5-S36D-green fluorescent protein (GFP) seedlings were grown at 22°C in continuous dark for 4 d and then treated with 40 μM bortezomib for at least 4 h. The total proteins were extracted and incubated with protein A beads bound with anti-GFP antibody (rabbit). The total and precipitated proteins were examined by immunoblotting using antibodies against COP1, GFP, and Tubulin (mouse). cop1-4 mutant was used as the negative control. The numbers below anti-COP1 blots indicate the relative band intensities of co precipitated COP1 normalized to those of precipitated HY5-GFP, HY5-S36A and HY5-S36D. The ratio of the first clear band was set to 1 for each blot.
Unphosphorylated HY5 degrades and accumulates faster than phosphorylated HY5
As a key positive regulator in seedlings photomorphogenesis, HY5 accumulates in response to the light and degrades in the dark (Osterlund et al., 2000). Previous studies have shown that the interaction between HY5 and COP1 or SPA1 is required for the degradation of HY5 in a polyubiquitination-dependent manner (Osterlund et al., 2000; Saijo et al., 2003). As we observed that phosphorylation alters HY5-interaction affinity to COP1 and SPA1, we hypothesized that different interaction affinity may further affect HY5 stability. To verify our hypothesis, we examined the ubiquitination status of HY5 (S36A/S-36D)-GFP in vivo. WT HY5, HY5-S36A and HY5-S36D proteins were immunoprecipitated from dark-grown transgenic seedlings, pretreated with proteasome inhibitor (bortezomib), and then probed with anti-GFP and anti-Ub antibodies. Strikingly, the HY5-S36A has significantly higher ubiquitination level, whereas HY5-S36D has significantly reduced ubiquitination level compared with WT HY5 (Fig. 5).
Fig. 5.
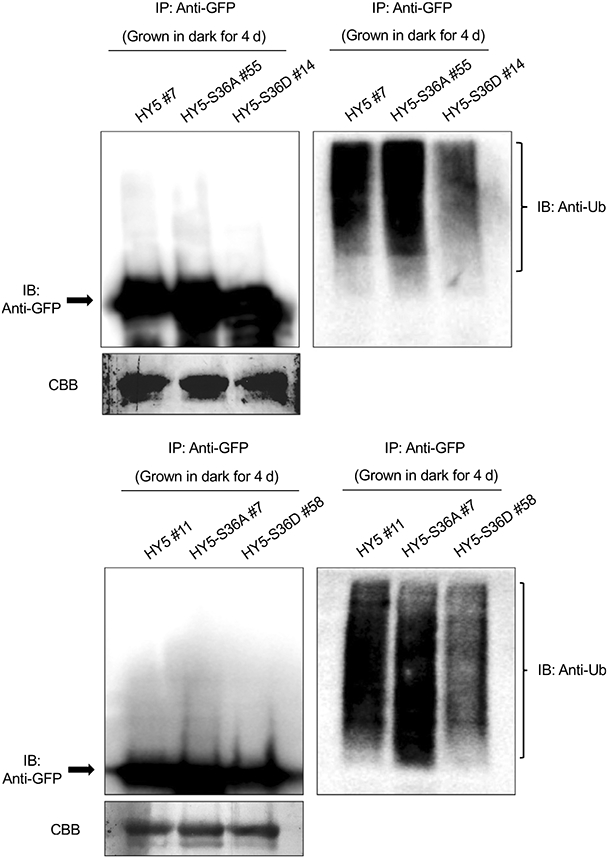
Phosphorylation alters elongated hypocotyl5 (HY5) ubiquitination level in vivo. Immunoblots (IB) showing the relative HY5 (S36A/S36D)-green fluorescent protein (GFP) protein level (left) and their ubiquitination status in response to dark (right). IP, immunoprecipitation. Total protein was extracted from 4-d-old dark-grown seedlings pretreated with the proteasome inhibitor (40 μM bortezomib) for 4 h before protein extraction. HY5 (S36A/S36D)-GFP was immunoprecipitated using anti-GFP antibody (rabbit) from protein extracts. The immunoprecipitated samples were then separated on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and probed with anti-GFP (left, Mouse) or anti-Ub (right, Mouse) antibodies. The upper smear bands are polyubiquitinated HY5 (S36A/S36D). Upper: line HY5 #7, HY5-S36A #55 and HY5-S36D #14 were used; lower: line HY5 #11, HY5-S36A #7 and HY5-S36D #58 were used. Arrows indicate the HY5 (S36A/S36D)-GFP protein. The lower panel (CBB) shows the protein level in a Coomassie-stained membrane.
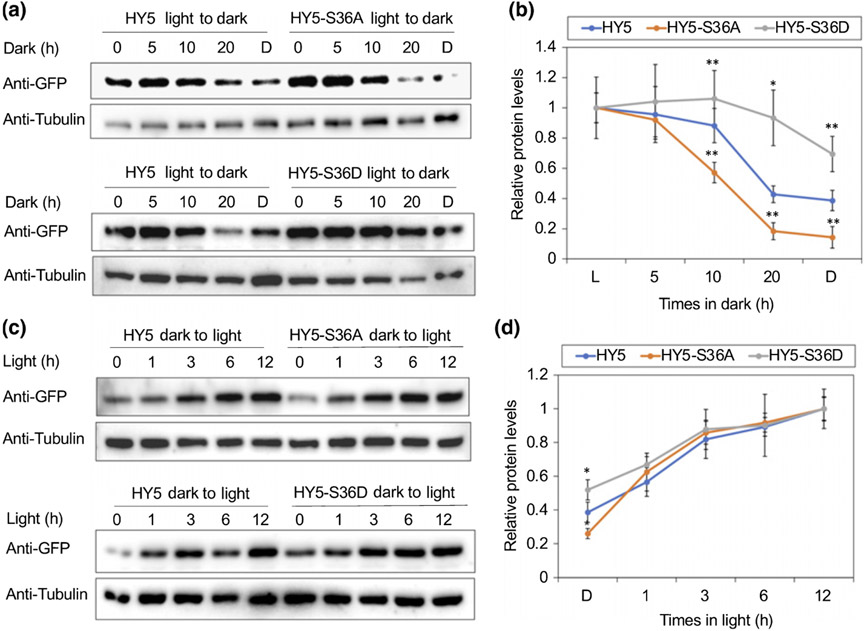
We also tested the protein levels of HY5, HY5-S36A and HY5-S36D transgenic seedlings that were grown in continuous light for 4 d and then transferred into dark for several hours (5, 10, 20 h) or grown in continuous dark for 4 d. The result shows that nonphosphorylated HY5 (HY5-S36A) is degraded faster during the light-to-dark transition, whereas the phospho-mimic form of HY5 (HY5-S36D) is more stable, even in the dark (Fig. 6a,b). Specifically, the degradation rate of nonphosphorylated HY5 was significantly increased after 5 h of dark transition (Fig. 6b). On the contrary, the degradation of phospho-mimicking HY5 was not obvious in the first 10 h of dark transition and its degradation was observed only after longer exposure of darkness (Fig. 6b). Our results suggest that the nonphosphorylated form of HY5 is the preferred substrate for degradation and the phospho-mimic form of HY5 is not prone to be degraded. This is consistent with our observation as well as a previously published report that the nonphosphorylated form of HY5 has higher binding affinity to COP1 and SPA1 and higher ubiquitination level, whereas the phospho-mimic form of HY5 has much lower affinity to both proteins and a lower ubiquitination level (Hardtke et al., 2000). These results indicate that the phosphorylation of HY5 results in weak interaction with COP1–SPA complex and subsequent ubiquitination status, making the HY5 proteins more stable in the dark.
Fig. 6.
Phosphorylation alters elongated hypocotyl5 (HY5) degradation and accumulation rate. (a) Immunoblots showing the degradation pattern of HY5, HY5-S36A and HY5-S36D proteins in response to dark. Transgenic seedlings were grown in continuous light for 4 d and then transferred to dark (D) for 5, 10 or 20 h, or grown in continuous dark for 4 d. Anti-green fluorescent protein (GFP) antibody and anti-tubulin antibody were used for this assay. Tubulin proteins were used as loading control. Multiple lines were tested, and lines HY5 #7, HY5-S36A #55 and HY5-S36D #14 are shown as representative lines. (b) A line graph shows the relative rate of degradation of HY5 in response to the dark treatment. The band intensities of HY5 and Tubulin were measured using the ImageJ tool. For each line, the HY5 level in the light (L) was set to 1 and the relative HY5 levels in response to dark were then calculated. Error bars indicate SD (n = 3). The asterisks indicate statistically significant differences between wild-type HY5 and two mutant lines based on Student’s t-test (*, P < 0.05; **, P < 0.005). (c) Immunoblots showing the accumulation pattern of HY5, HY5-S36A and HY5-S36D proteins in response to light. Transgenic seedlings were grown in continuous dark for 4 d and then transferred to light for 1, 3, 6 or 12 h. Anti-GFP antibody and anti-tubulin antibody were used for this assay. Tubulin proteins were used as loading control. Multiple lines were tested, and lines HY5 #7, HY5-S36A #55 and HY5-S36D #14 are shown as representative lines. (d) A line graph shows the relative accumulation rate of HY5 in response to the light treatment. The band intensities of HY5 and Tubulin were measured using the ImageJ tool. For each line, the HY5 level in the 12 h light was set to 1 and the relative HY5 levels in response to light were then calculated. Error bars indicate SD (n = 3). The asterisks indicate statistically significant differences between wild-type HY5 and two mutant lines based on Student’s t-test (*, P < 0.05).
In addition, we compared the accumulation rates of each mutant forms of HY5 during dark-to-light transition (Fig. 6c,d). The accumulation of HY5 in response to light is very fast, and the accumulation occurs within 1 h of light exposure. It reaches to the peak in 3 h (Fig. 6d). The nonphosphorylated form of HY5 appears to be more sensitive to the light irradiation, as it shows relatively rapid and higher HY5 protein accumulation within the first 1 h after light irradiation, even though it is not statistically significant (Fig. 6d). Taken together, our results suggest that phosphorylation of HY5 results in an altered response rate to dark and light. As a negative regulator of HY5, SPA1 phosphorylates HY5 and the phosphorylated HY5 remains stable in the dark, which suggests a COP1–SPA–HY5 negative feedback loop may exist.
HY5 phosphorylation affects its molecular and physiological activity
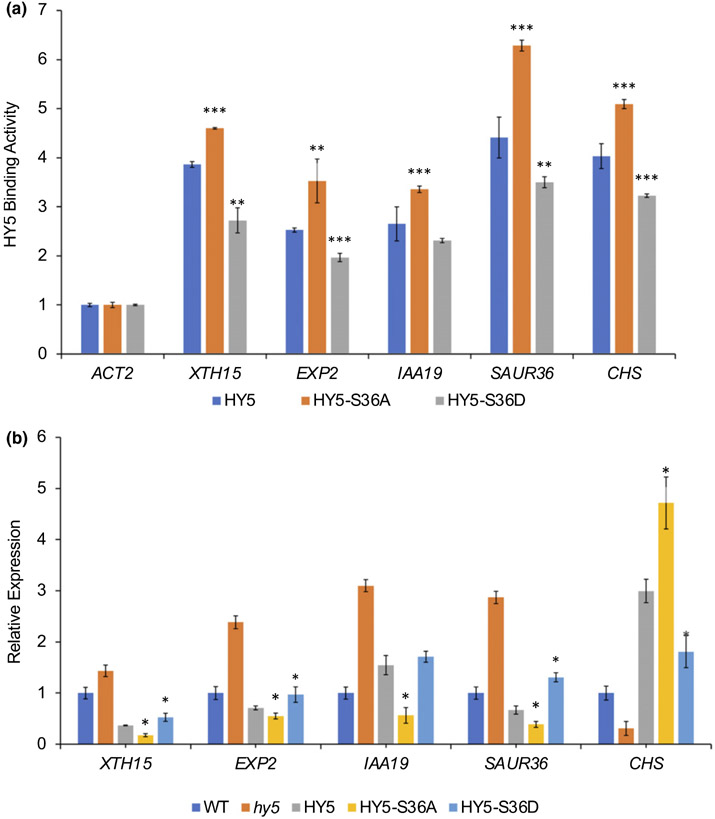
Previous studies have reported that phosphorylation can affect the binding affinity of HY5 to its target DNAs in vitro (Hardtke et al., 2000). To investigate whether phosphorylation alters the in vivo DNA bindings of HY5, we performed ChIP–qPCR assays. We selected several well-known HY5 target genes (XTH15, EXP2, IAA19, SAUR36 and CHS). Because HY5 protein accumulates to the peak after 3 h of light activation during dark-to-light transition (Fig. 6d), we performed ChIP–qPCR with transgenic seedlings grown in the dark for 4 d and then transferred to the light for an additional 3 h. Surprisingly, nonphosphorylated forms of HY5 enriched to the target loci significantly higher than the WT HY5. On the other hand, the phospho-mimic form of HY5 enriched to target loci slightly lower than the WT HY5 (Figs 7a, S5A), indicating that unphosphorylated HY5 has higher affinity to its target DNA, which is largely consistent with a previous report (Hardtke et al., 2000).
Fig. 7.
Unphosphorylated elongated hypocotyl5 (HY5) is more active in binding targets and regulating gene expression. (A) Chromatin immunoprecipitation–quantitative PCR (qPCR) results show the targeting of HY5 and two phosphorylation mutant proteins. For this assay, each transgenic seedling was grown in dark for 4 d and then transferred into light for an additional 3 h. Three biological replicates were performed, and three technical repeats were carried out for each biological replicate; error bars indicate SD (n = 3). Lines HY5 #7, HY5-S36A #55 and HY5-S36D #14 were used. The asterisks indicate statistically significant differences between wild-type HY5 and mutant HY5 based on Student’s t-test (**, P < 0.005; ***, P < 0.0005). (b) Reverse transcription qPCR results show transcript levels of selected HY5 target genes in HY5 and two phosphorylation mutant lines (HY5 #7, HY5-S36A #55 and HY5-S36D #14) along with wild-type and hy5 as controls. Three biological replicates were performed, and three technical repeats were carried out for each biological replicate. For this assay, each transgenic seedling was grown in dark for 4 d and then transferred into light for an additional 3 h. Error bars indicate SD (n = 3). The asterisks indicate statistically significant differences between wild-type HY5 and two mutant lines based on Student’s t-test (*, P < 0.05).
To examine whether the altered association of HY5 with its target loci affects their levels of transcript abundance, we then performed real-time qRT-PCR with the three transgenic lines grown under the same condition along with WT and hy5 as controls. Consistent with higher association, the expression of three auxin signaling pathway genes (SAUR36, IAA19 and EXP2) and a growth gene (XTH15), which are known HY5 repressed genes, was strongly repressed in the HY5-S36A line (Figs 7b, S5B). However, HY5-S36D displayed higher expression of some of these target genes compared with the WT HY5. Conversely, an HY5-activated gene (CHS) displayed the opposite pattern of expression (Figs 7b, S5B). Our data suggest that the phosphorylation of HY5 affects the transcriptional activity of HY5 by changing its binding ability to target loci in vivo. The levels of HY5 protein and me3ssenger RNA were similar among HY5, HY5-S36A, and HY5-S36D lines after 3 h light irradiation or grown in continuous light for 4 d (Figs S6, S7), and all the proteins were similarly stabilized under light (Fig. S7). Therefore, the differences in the association of phosphorylated and unphosphorylated HY5 and transcription of target loci are due to altered HY5 activity, and not due to different levels of expression of transgenes.
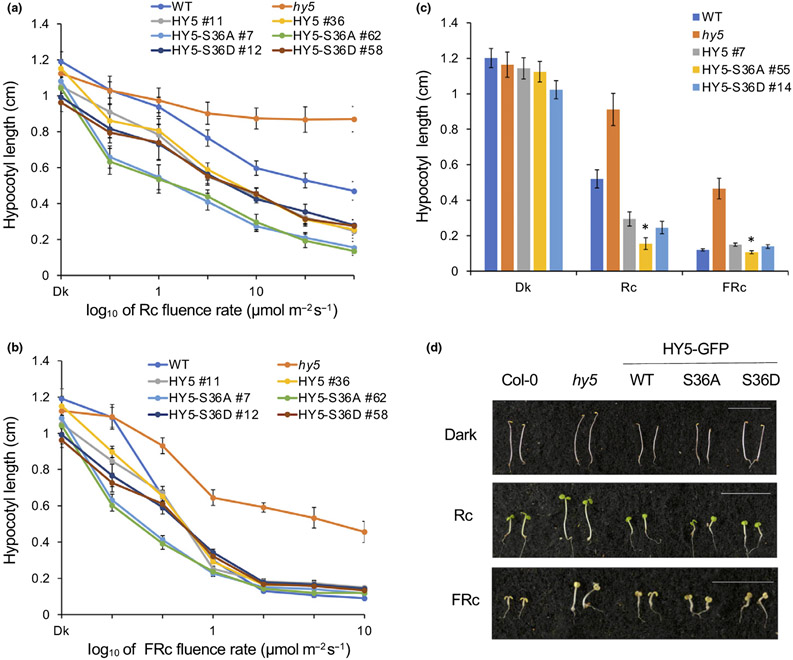
Furthermore, to check whether the phosphorylation mutants of HY5 result in any phenotypic changes, we measured the hypocotyl length of seedlings of the phosphorylation mutant lines grown under dark for 4 d or continuous red (Rc) or far-red (FRc) light for 4 d with increasing light intensities (Fig. 8a,b). In both red light and far-red light conditions, hy5 mutant showed relatively longer hypocotyl, which was rescued by overexpressing HY5 protein (Fig. 8c,d). Notably, the HY5-S36A line showed shorter hypocotyls than others under most of Rc conditions and low intensity of FRc conditions (Fig. 8a,b), even though all the lines had similar hypocotyl lengths in the dark. HY5-S36A hypocotyl length was strongly reduced once exposed to light. Taken together, our results clearly showed that higher binding affinity of unphosphorylated HY5 is physiologically more active than phosphorylated HY5 in regulating photomorphogenesis, especially during rapid light response.
Fig. 8.
Unphosphorylated elongated hypocotyl5 (HY5) displays enhanced photomorphogenesis. (a, b) Line graphs show hypocotyl lengths of each HY5-overexpressing transgenic line (HY5, HY5-S36A and HY5-S36D) in response to (a) red light (Rc) and (b) far-red light (FRc) with increasing light intensities. Hypocotyl lengths in the dark shown in (a) and (b) were measured from the same plate. Error bars indicates SD (n > 30). (c) Bar graph showing the hypocotyl lengths of wild-type (WT) Col-0, hy5 mutant, HY5, HY5-S36A and HY5-S36D overexpressing lines grown under dark (Dk), red (Rc, 6 μmol m−2 s−1), and far-red light (FRc, 1 μmol m−2 s−1) conditions for 4 d. Error bars indicates SD (n > 30). The asterisk indicates significant difference between WT HY5 and two mutant lines based on Student’s t-test (P < 0.05). (d) Photographs show the hypocotyl lengths of various genotypes of Arabidopsis thaliana in continuous dark, Rc or FRc. Seeds of various genotypes were grown on Murashige & Skoog medium without sucrose for 4 d. Bar, 10 mm.
Discussion
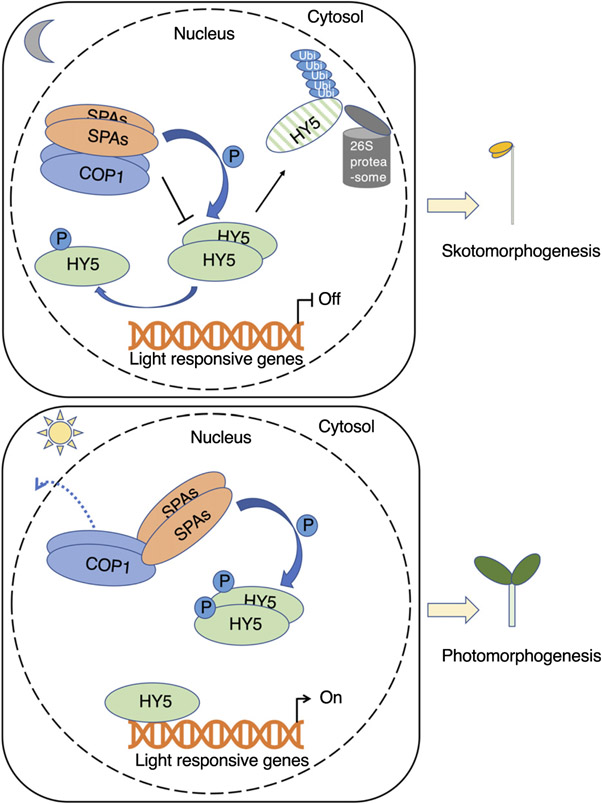
HY5 protein plays a pivotal role in photomorphogenesis. Therefore, it is not surprising that a number of HY5 binding targets are involved in diverse developmental processes. In Arabidopsis, more than 60% of early-induced genes by phyA or phyB are HY5 direct targets, which strongly supports the notion that HY5 is one of the high hierarchical regulators of the transcriptional cascade for photomorphogenesis (Gangappa & Botto, 2016). The regulation of HY5 by light includes posttranslational modifications such as phosphorylation and poly-ubiquitination that are controlled by phytochromes and COP1–SPA E3 ubiquitin ligase complex. Previous studies have hypothesized that a light-regulated CKII-like kinase might phosphorylate HY5 and regulate its abundance and activity (Hardtke et al., 2000; Gangappa & Botto, 2016). Despite the phosphorylation in HY5 was reported > 20 yr ago, the kinase responsible for HY5 phosphorylation remained elusive. Here, we provide a mechanistic view on how HY5 is phosphorylated and ubiquitinated by a cognate kinase-E3 ubiquitin ligase (COP1–SPA) to modulate its regulatory activity in photomorphogenesis (Fig. 9).
Fig. 9.
A model showing suppressor of phytochrome A-105 (SPA)-mediated phosphorylation of elongated hypocotyl5 (HY5) to fine-tune photomorphogenesis. In the dark (upper panel), constitutive photomorphogenic1 (COP1)–SPA protein complexes function as E3 ubiquitination ligases and interact preferentially with the unphosphorylated HY5 for ubiquitination and degradation by the 26S proteasome; thus, light-responsive genes are turned off, and seedlings show skotomorphogenesis. In addition, SPAs phosphorylate HY5, and the phosphorylated HY5 has less affinity for the COP1–SPA complex and is relatively more stable. Thus, seedlings can maintain a small pool of HY5 in the dark. Upon light irradiation (lower panel), the remaining phosphorylated HY5 (or potentially a dephosphorylated form) can rapidly activate light responses. COP1–SPA complex is also reorganized, reducing the E3 ligase activity of the COP1–SPA complex, thus stabilizing HY5 under light. COP1 is also excluded from the nucleus under prolonged light conditions (dashed arrow), thus stabilizing HY5 and other positive factors that promote photomorphogenesis. Unphosphorylated HY5 binds more efficiently to the promoters of downstream genes and promotes photomorphogenesis, whereas SPAs phosphorylate HY5 to avoid overphotomorphogenesis under light.
In this study, we provide evidence that SPAs are the kinases for HY5 and, thus, regulate its stability and activity. We observed strong phosphorylation signals in in vitro kinase assays, and the phosphorylation of HY5 was significantly reduced in spaQ mutant compared with that in WT in vivo (Figs 1, 2). We also found that SPA1 phosphorylates HY5 through only one phosphorylation site, which is Ser-36 (Fig. 1d), which is consistent with a previous report (Hardtke et al., 2000). The mutation of this site abolished the phosphorylation of HY5. Previously, CKII was hypothesized to be a likely candidate kinase of HY5 (Hardtke et al., 2000). CKII was also shown to be a kinase for PIFs and another positively acting basic helix–loop–helix transcription factor HFR1 (Park et al., 2008; Bu et al., 2011). However, the in vitro kinase assay using a kinase-containing fraction of enriched seedling extract in a previous study is not sufficient to conclude that CKII is the kinase (Hardtke et al., 2000), given that other components may also exist in the extract. By contrast, we present strong in vitro and in vivo evidence that SPAs are the key kinases necessary for HY5 phosphorylation. Hardtke et al. (2000) also reported that the kinase activity responsible for HY5 phosphorylation might be regulated by light and that the unphosphorylated HY5 may accumulate more under light for efficient light response. However, we observed phosphorylation of HY5 under both light and dark conditions (Fig. 2a). Previously, SPA1 was shown to be recruited by phyB for light-induced PIF1 phosphorylation (Paik et al., 2019). Further studies are necessary to dissect whether the SPA-mediated phosphorylation of HY5 is light regulated or not.
It has been suggested that the kinase domain of SPA1 may provide structural information critical for SPA1 function (Holtkotte et al., 2016). SPA1R517E mutant shows a defect in PIF1 degradation and seed germination in response to light (Paik et al., 2019). Similarly, SPA1R517E (mSPA1) transgenic lines failed to rescue the seedlings’ de-etiolation phenotype of spaQ (Fig. 3b). Since the Arg 517 in SPA1 is well conserved in many plants (Paik et al., 2019), it is possible that the structural integrity of the kinase domain is important for its proper function, whereas the biochemical basis remains unknown. Our study shows that the kinase domain of SPA1 may act as a molecular scaffold for potential protein–protein interaction. We observed considerable HY5 accumulation in the SPA1R517E line (Fig. 3a), which explained the de-etiolation phenotype of SPA1R517E mutant, showing photomorphogenesis in the dark. Moreover, the accumulation of HY5 in SPA1R517E mutant in the dark resulting from the failure of degradation by COP1–SPA complex confirmed our hypothesis that the kinase domain of SPA1 might act as a molecular scaffold for protein–protein interaction.
Phosphorylation of transcription factors is a common modification that can influence their biological properties, such as multimerization or nucleocytoplasmic partitioning in both plants and animals. In plant photomorphogenesis, phosphorylation has been observed for many transcription factors (Pham et al., 2018). Other than PIF1, light can also induce phosphorylation of PIF3 at multiple sites, and phosphorylated PIF3 is subject to the degradation by light-responsive BTB protein (LRB) and EIN3-binding F box protein (EBF) E3 ligases (Ni et al., 2013, 2014; Dong et al., 2017). Phosphorylation can also regulate the transcriptional activity of PIF4, which affects diurnal hypocotyl elongation and also influences translocation of PIF7, which regulates shade-induced stem elongation (Bernardo-García et al., 2014; Huang et al., 2018). In this study, we demonstrated that the phosphorylation of HY5 results in lower binding affinity to COP1 and SPA1, which reduces HY5 ubiquitination and, in turn, stabilizes HY5 proteins in the dark (Figs 4-6). Therefore, HY5 abundance is regulated apparently by two parallel pathways. One is the oscillation of the availability of COP1 in the nucleus, and the other is the specific phosphorylation of HY5, which modulates HY5’s ability to interact with COP1–SPA complex. Considering that HY5 is a key positive regulator of photomorphogenesis at the early seedling stage, this mechanism may help maintain a small pool of less active HY5 in the dark so that seedlings can give a rapid initial response during dark-to-light transition. Thus, SPAs are acting both negatively and positively to regulate HY5 level and activity, forming a negative feedback loop between HY5 and COP1–SPA (Fig. 9). This is consistent with a recent study showing that accumulation of HY5 in the dark leads to an increase in the COP1–SPA complex, and thus its own degradation (Burko et al., 2020).
Phosphorylation of transcription factors to regulate their stability and DNA binding capacity is common in eukaryotic cells (Hunter, 2007). Previous ChIP-chip analysis showed that HY5 binds directly to the promoters of genes related to auxin signaling, ethylene signaling, and GA signaling. Moreover, it was shown that HY5 is necessary for the rapid transcription of those genes during the dark-to-light transition, which eventually allows the accumulation of Chl and anthocyanin for photosynthesis (Lee et al., 2007; Burko et al., 2020). We show here that unphosphorylated HY5 has a stronger binding affinity to its target promoters, such as G-box and ACE-box in XTH15, EXP2, IAA19, CHS and SAUR36, than phosphorylated HY5 does (Fig. 7a). This is consistent with the gene expression (Fig. 7b) and phenotype of the HY5-S36A transgenic line, which shows shorter hypocotyl length than WT HY5 does (Fig. 8). HY5 activity is also regulated by interaction with other transcription factors, including the BBX factors (Song et al., 2020). In fact, HY5 lacks any transcriptional activation domain. A recent study showed that interaction with BBX20/21/22 proteins is necessary for activation of gene expression by HY5 (Bursch et al., 2020). Whether phosphorylation of HY5 has any influence on interaction with other transcription factors awaits further studies.
In summary, the data presented here describe SPA proteins as the missing kinases for phosphorylation of HY5 (Fig. 9). Thus, the COP1–SPA complex is acting both positively and negatively to regulate HY5 abundance and activity to fine-tune photomorphogenesis. Our data also show that, by modulating phosphorylation of HY5 and altering its protein stability and activity, plants can rapidly respond to light irradiation and also avoid overphotomorphogenesis, which would be of great advantage for seedlings in the constantly changing natural environment.
Supplementary Material
Fig. S1 SPAs have conserved kinase domain and function as kinases of HY5.
Fig. S2 Phosphorylation of HY5 occurs in both dark and light conditions.
Fig. S3 Accumulation of HY5 protein in different genotypes.
Fig. S4 Immunoblots showing LUC-(m)SPA1 protein levels in the spaQ mutant background.
Fig. S5 Unphosphorylated HY5 is more active in binding targets and regulating gene expression.
Fig. S6 Transcript of HY5 in dark-to-light transition.
Fig. S7 Protein and transcript levels of HY5 in HY5 and two phosphorylation mutant lines.
Table S1 Primer sequences used in experiments described in the text.
Mass-spectrometry analyses for identifying the phosphorylation site in HY5.
Acknowledgements
We thank Dr Xing Wang Deng for sharing the cop1 mutants and Dr Ute Hoecker for sharing the spaQ mutant. We thank the Huq and Sung lab members for critical reading of the manuscript. This work was supported by the grants from the National Science Foundation (MCB-2014408 to EH and IOS-1656764 to SS) and National Institute of Health (NIH) (GM-114297 to EH and GM-100108 to SS) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no competing financial interests.
Footnotes
Supporting Information
Additional Supporting Information may be found online in the Supporting Information section at the end of the article.
References
- Andronis C, Barak S, Knowles SM, Sugano S, Tobin EM. 2008. The Clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Molecular Plant 1: 58–67. [DOI] [PubMed] [Google Scholar]
- Ang L-H, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng X-W. 1998. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Molecular Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Balcerowicz M, Kerner K, Schenkel C, Hoecker U. 2017. SPA proteins affect the subcellular localization of COP1 in the COP1/SPA ubiquitin ligase complex during photomorphogenesis. Plant Physiology 174: 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Daviere J-M, Prat S. 2014. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes & Development 28: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Zhu L, Yu L, Dennis M, Lu X, Person M, Tobin E, Browning K, Huq E. 2011. Phosphorylation by CK2 enhances the rapid light-induced degradation of PIF1. Journal of Biological Chemistry 286: 12066–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y, Seluzicki A, Zander M, Pedmale UV, Ecker JR, Chory J. 2020. Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32: 967–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch K, Toledo-Ortiz G, Pireyre M, Lohr M, Braatz C, Johansson H. 2020. Identification of BBX proteins as rate-limiting cofactors of HY5. Nature Plants 6: 921–928. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. 2004. Light signal transduction in higher plants. Annual Review of Genetics 38: 87–117. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, Feldman KA, Quail PH. 1992. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Dong J, Ni W, Yu R, Deng XW, Chen H, Wei N. 2017. Light-dependent degradation of PIF3 by SCF-EBF1/2 promotes a photomorphogenic response in Arabidopsis. Current Biology 27: 2420–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF. 2016. The multifaceted roles of HY5 in plant growth and development. Molecular Plant 9: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Gommers CMM, Monte E. 2018. Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiology 176: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Huang X, Deng XW 2020. The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Communications 1: e100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. 2000. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO Journal 19: 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. 2017. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Current Opinion in Plant Biology 37: 63–69. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. 1999. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499. [DOI] [PubMed] [Google Scholar]
- Holm M, Ma L, Qu LJ, Deng XW. 2002. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes & Development 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkotte X, Dieterle S, Kokkelink L, Artz O, Leson L, Fittinghoff K, Hayama R, Ahmad M, Hoecker U. 2016. Mutations in the N-terminal kinase-like domain ofthe repressor ofphotomorphogenesis SPA1 severely impair SPA1 function but not light responsiveness in Arabidopsis. The Plant Journal 88: 205–218. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhang Q, Jiang Y, Yang C, Wang Q, Li L. 2018. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7: e31636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2007. The age ofcrosstalk: phosphorylation, ubiquitination, and beyond. Molecular Cell 28: 730–738. [DOI] [PubMed] [Google Scholar]
- Kathare PK, Xu X, Nguyen A, Huq E. 2020. A COP1-PIF-HEC regulatory module fine-tunes photomorphogenesis in Arabidopsis. The Plant Journal 104: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U. 2004. The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Paik I, Huq E. 2020. SPAs promote thermomorphogenesis by regulating the phyB-PIF4 module in Arabidopsis. Development 147: dev189233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Chalkley RJ, Pham TND, Guan S, Maltby DA, Burlingame AL, Wang Z-Y, Quail PH. 2013. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang Z-Y, Quail PH. 2014. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Tat AV, Kumar Devisetty U, Robinson M, Mumbach MR, Ichihashi Y, Lekkala S, Maloof JN. 2015. Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genetics 11: e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordoñez-Herrera N, Fackendahl P, Yu X, Schaefer S, Koncz C, Hoecker U. 2015. A cop1 spa mutant deficient in COP1 and SPA proteins reveals partial co-action of COP1 and SPA during Arabidopsis post-embryonic development and photomorphogenesis. Molecular Plant 8: 479–481. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. 2000. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. 1997. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Development 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ. 2014. Rapid decline in nuclear COSTITUTIVE PHOTOMORPHOGENESIS1 abundance anticipates the stabilization ofits target ELONGATED HYPOCOTYL5 in the light. Plant Physiology 164: 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Chen F, Ngoc Pham V, Zhu L, Kim J-I, Huq E. 2019. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nature Communications 10: e4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Huq E. 2019. Plant photoreceptors: multi-functional sensory proteins and their signaling networks. Seminars in Cell & Developmental Biology 92: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-J, Ding L, Dai M, Lin R, Wang H. 2008. Multisite phosphorylation of Arabidopsis HFR1 by casein kinase II and a plausible role in regulating its degradation rate. Journal of Biological Chemistry 283: 23264–23273. [DOI] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. 2018. Phytochromes and phytochrome interacting factors. Plant Physiology 176: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. 2003. The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes & Development 17: 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. 2003. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y-D, Huq E, Hiltbrunner A 2015. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. 2005. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize seedling photomorphogenesis in Arabidopsis. The Plant Journal 44: 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shor E, Paik I, Kangisser S, Green R, Huq E. 2017. PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytologist 215: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Bian Y, Liu J, Sun Y, Xu D. 2020. B-box proteins: pivotal players in light-mediated development in plants. Journal of Integrative Plant Biology 62: 1293–1309. [DOI] [PubMed] [Google Scholar]
- Subramanian C, Kim BH, Lyssenko NN, Xu X, Johnson CH, von Arnim AG. 2004. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proceedings of the National Academy of Sciences, USA 101:6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, McNellis TW, Deng X-W. 1998. Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO Journal 17: 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G, Halliday K. 2007. Light and plant development. Oxford, UK: Blackwell Publishing. [Google Scholar]
- Xu X, Paik I, Zhu L, Bu Q, Huang X, Deng XW, Huq E. 2014. PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26: 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E. 2015. Illuminating progress in phytochrome-mediated light signaling pathways. Trends in Plant Science 20: 641–650. [DOI] [PubMed] [Google Scholar]
- Yang J, Wu J, Steichen JM, Kornev AP, Deal MS, Li S, Sankaran B, Woods VL, Taylor SS. 2012. A conserved Glu–Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold. Journal of Molecular Biology 415: 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 SPAs have conserved kinase domain and function as kinases of HY5.
Fig. S2 Phosphorylation of HY5 occurs in both dark and light conditions.
Fig. S3 Accumulation of HY5 protein in different genotypes.
Fig. S4 Immunoblots showing LUC-(m)SPA1 protein levels in the spaQ mutant background.
Fig. S5 Unphosphorylated HY5 is more active in binding targets and regulating gene expression.
Fig. S6 Transcript of HY5 in dark-to-light transition.
Fig. S7 Protein and transcript levels of HY5 in HY5 and two phosphorylation mutant lines.
Table S1 Primer sequences used in experiments described in the text.
Mass-spectrometry analyses for identifying the phosphorylation site in HY5.